Abstract
Gastrointestinal stromal tumors (GISTs) are the major gastrointestinal mesenchymal tumors with a variable malignancy ranging from a curable disorder to highly malignant sarcomas. Metastasis and recurrence are the main causes of death in GIST patients. To further explore the mechanism of metastasis and to more accurately estimate the recurrence risk of GISTs after surgery, the clinical significance and functional role of collagen triple helix repeat containing-1 (CTHRC1) in GIST were investigated. We found that CTHRC1 expression was gradually elevated as the risk grade of NIH classification increased, and was closely correlated with disease-free survival and overall survival in 412 GIST patients. In vitro experiments showed that recombinant CTHRC1 protein promoted the migration and invasion capacities of primary GIST cells. A luciferase reporter assay and pull down assay demonstrated that recombinant CTHRC1 protein activated noncanonical Wnt/PCP-Rho signaling but inhibited canonical Wnt signaling. The pro-motility effect of CTHRC1 on GIST cells was reversed by using a Wnt5a neutralizing antibody and inhibitors of Rac1 or ROCK. Taken together, these data indicate that CTHRC1 may serve as a new predictor of recurrence risk and prognosis in post-operative GIST patients and may play an important role in facilitating GIST progression. Furthermore, CTHRC1 promotes GIST cell migration and invasion by activating Wnt/PCP-Rho signaling, suggesting that the CTHRC1-Wnt/PCP-Rho axis may be a new therapeutic target for interventions against GIST invasion and metastasis.
Abbreviations: CTHRC1, collagen triple helix repeat containing 1; DFS, disease-free survival; ECM, extracellular matrix; GIST, gastrointestinal stromal tumors; OS, overall survival; qRT-PCR, quantitative real-time polymerase chain reaction
Introduction
Gastrointestinal stromal tumors (GISTs) are mesenchymal neoplasms that usually arise in the stomach or small intestine and typically cause bleeding, anaemia and pain [1]. It is believed that GISTs originate from interstitial cells of Cajal [2] and may also derive from gastrointestinal smooth muscles or gut stem cells [3]. The pathological features of GISTs range from benign neoplasms to fatal sarcomas [1,4]. Most gastrointestinal stromal tumors stain positively for KIT [5,6], Ki67 [7] and anoctamin 1; exon mutations [6] in KIT or PDGFRA genes in approximately 80% or 10% of GISTs, respectively, have been demonstrated [1]. More than 60% of GIST patients can be cured by surgical resection [8,9]. The use of imatinib mesylate (Gleevec; Novartis) adjuvant treatment [8] is recommended in advanced GIST patients with postoperative recurrence risk, and the survival rate can be improved; however, secondary imatinib resistance is common [10].
Micrometastases and overt metastases are the main causes of death in malignant tumors, and this is the case in GIST patients as well [11,12]. Approximately 40% GISTs patients had metastatic lesions when definitively diagnosed, and more than 10% patients exhibited overt metastases [1]. Therefore, developing new predictors that can be used to estimate the risk of metastasis and postoperative recurrence is urgent.
Extracellular matrix (ECM) proteins play important roles in regulating tumor invasion and metastasis [13-15]. Given the secretary property, ECM proteins are also ideal candidates for tumor serum biomarkers and therapeutic targets.
Collagen triple helix repeat containing-1(CTHRC1) is a 28-kD extracellular matrix glycoprotein containing an NH2-terminal signaling peptide for extracellular secretion, a short collagen triple helix repeat of 36 amino acids, and a COOH-terminal globular domain [16].CTHRC1 was initially found in a screen for differentially expressed genes in balloon-injured versus normal rat arteries [16]. It has been reported that the CTHRC1 protein positively regulates the Wnt-PCP pathway by stabilizing formation of the Wnt ligand and Frizzled receptor complex [17] in developmental morphogenesis [17]. CTHRC1 has recently been shown to be highly expressed in human pancreatic cancer tissues [18], hepatocellular carcinoma [13], gastric cancer [19], and colorectal cancer [20], and it promotes invasion and metastasis in these cancers. Several studies revealed that CTHRC1 regulates cancer cell motility and invasiveness through activating the Wnt-PCP pathway [18]. However, the clinical significance and functional role of CTHRC1 in GIST remain unclear. In this study, we first examined the expression of CTHRC1 and its correlation with the clinicopathological parameters of GIST. Then, we further analyzed the relationship between CTHRC1 expression and the survival of GISTs patients and identified CTHRC1 as a novel prognostic factor of GIST. Finally, we demonstrated that CTHRC1 promoted migration and invasion of primary GIST cells through activated Wnt/PCP-Rho signaling.
Materials and Methods
Ethics Statement
We obtained approval from the Regional Ethical Committees, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China for the use of clinical GIST patients' tissues. All the patients joined this study have signed informed consent. Ethical approval number, 2012031.
Patients
The inclusion criteria for our study were as follows: 1) a distinct pathologic diagnosis of GIST (CD117 positive in immunohistochemistry staining) ; 2) primary GIST cases without history of other solid tumors; 3) accepted radical surgery treatment without tumor residual; 4) without any chemotherapy, radiotherapy or other anti-cancer therapies before surgery; 5) availability of complete clinicopathologic and follow-up data; 6) obtained informed consent of patients and approval of the ethics committee of Renji Hospital for the use of samples. A total of 412 GIST cases, pathologic diagnosed and treated range from September 2004 to September 2013, were retrospectively identified from the hospitalization archives of Department of General Surgery, Renji Hospital, Shanghai, China. The paraffin-embedded tissue samples of these patients were used for tissue microarray construction and immunohistochemical staining. The clinicopathologic parameters include patients' age, gender, pathogenic site, histological type, tumor size (cm), number of mitoses/50 high-power fields (HPF), tumor rupture, mutation type and imatinib adjuvant treatment regimens. The risk of GIST behavior was classified into very low, low, intermediate, and high-risk categories according to the modified National Institute of Health (NIH) consensus [21,22]. In our study, the criterion of imatinib adjuvant therapy is at least twelve months uninterrupted drugs at a dose of 400mg/day. All the patients involved in our research accepted physical examination once a month during the first year after surgery and every six months thereafter. High risk GIST patients were accepted computed tomography (CT) or magnetic resonance imaging (MRI) of abdomen and pelvis at three-months intervals during the first three years after surgery, and subsequently at six-months intervals until five years after surgery. Complete follow-up data for GIST patients in cohort were available. Patients were followed until September 2013. Overall survival (OS) was defined as the time from surgery to death or the last follow-up examination. Disease free survival (DFS) was defined from the date of surgery until the detection of tumor recurrence or last observation.
Tissue Microarray Construction
Tissue microarrays were constructed by Suzhou Xinxin Biotechnology ( Xinxin Biotechnology Co, Suzhou, China). Tissue paraffin blocks of GIST samples were stained with hematoxylin-eosin to confirm the diagnoses and marked at fixed points with most typical histological characteristics under a microscope. Two 1.6 mm cores per donor block were transferred into a recipient block tissue microarrayer, and each dot array contained fewer than one hundred and sixty dots. Three-micron-thick sections were cut from the recipient block and transferred onto glass slides using an adhesive tape transfer system for ultraviolet cross linkage.
Immunohistochemistry Stain
The tissue microarray glass slides were baked at 55°C for one hour, and then de-paraffinized gradually through xylene, 50% xylene, gradient concentrations of ethanol until immersed in tap water. Tissue sections were blocked for peroxidase activity with 0.3% Hydrogen peroxide at 37°C for 30mins. Antigen retrieval was carried out via boiling in 10mmol/L citrate buffer (pH6.0) for fifteen mins. Then the tissues were incubated with CTHRC1 antibody (mouse monoclonal antibody, 1:100 dilution, Huaan Biotechnology, Hangzhou, China) overnight at 4°C. Next day, the tissues were washed with phosphate buffer solution (PBS) for three times and incubated with HRP-labelled anti-mouse secondary antibody (1:200dilution, Dako, Carpinteria , CA, USA) for one hour at room temperature. Immunostaining was carried out using diaminobenzidine substrate chromogen (Dako, Carpinteria, CA, USA) method and chromogenic reaction was controlled under microscope. After immunostaining, tissues were immersed into hematoxylin for nuclear staining. The TMA slides were then dehydrated through gradient concentrations of ethanol, cleared with xylene, and coverslipped with neutral balsam (Shenggong , Shanghai, China). The staining results were judged by two pathologists according to criterion as follows: 0: weak, no staining was observed; 1 +, 25% to 50% of the tumor cells were weak or moderate staining; 2 +, strong, more than 50% tumor cells were moderate or strong staining. 1 + and 2 + scores were identified as positive staining, while 0 score means negative staining. Negative controls for primary and secondary antibodies were shown in Figure W3. Total RNA Extraction and Quantitative Real-time PCR Total RNA was extracted from 29 fresh GIST tissues using Trizol reagent (Takara, Dalian, China) followed the manufacturer instructions. The reverse-transcription reactions were carried out with random primers and M-MLV Reverse Transcriptase (Takara, Dalian, China). The 29 cases of cDNA were used for templates of quantitative real-time PCR reaction in SYBR-Green method. All the qPCR reactions were performed on a StepOneTM real-time PCR System (Applied Biosystems, Foster City, CA,USA). Beta-actin was used as an internal control. The 2-△Ct method was used to quantify the relative CTHRC1 expression levels. The forward and reverse CTHRC1 primer sequences were: 5′-TGGTATTTCACATTCAATGGAGCTG-3′ and 5′-TGGGTA- -ATCTGAACAAGTGCCAAC-3′, respectively.
Western Blotting
Fresh GIST tissues were lysed in tissue protein extraction reagent (Invitrogen). Primary GIST cells were lysed in Western and IP lysis buffer (P0013, Beyotime, Jiangsu, China) supplemented with 1mM PMSF (Adamas beta, Shanghai, China). The lysis buffer includes, 20mM Tris (pH7.5), 150mM NaCl, 1%Triton X-100, sodium pyrophosphate, β-glycerophosphate, EDTA, Na3VO4, leupeptin. Proteins were separated by 10% SDS-PAGE under reducing condition, followed by blocking in phosphate-buffered saline/Tween-20 containing 1%BSA (Bovine Serum Albumin). The NC (Nitrocellulose filter membrane) or PVDF (Polyvinylidenefluoride) membrane was incubated with antibodies for CTHRC1(1:1000, mouse, Huaan, Hangzhou, China) , JNK (1:1000, Rabbit source, Cell Signaling Technology), RhoA (1:1000,Rabbit,CST), Rac1(1:1000,Mouse, Millipore) ,Cdc42 (1:1000,Mouse) and species-specific secondary antibodies. Bound the IRDye 680 anti-mouse (LI-COR, 1:20000) and IRDye 800 anti-rabbit (LI-COR, 1:10000) secondary antibodies were revealed by Odyssey imaging system (LI-COR). Wnt5a neutralizing antibody (R&D), Wnt3a neutralizing antibody (R&D), NSC23766 (Rac1 inhibitor, Merck Millipore, effective dose: 50 μM), Y-27632 (ROCK inhibitor, effective dose: 100 μM).
CTHRC1 Recombinant Protein Expression, Purification and Verification
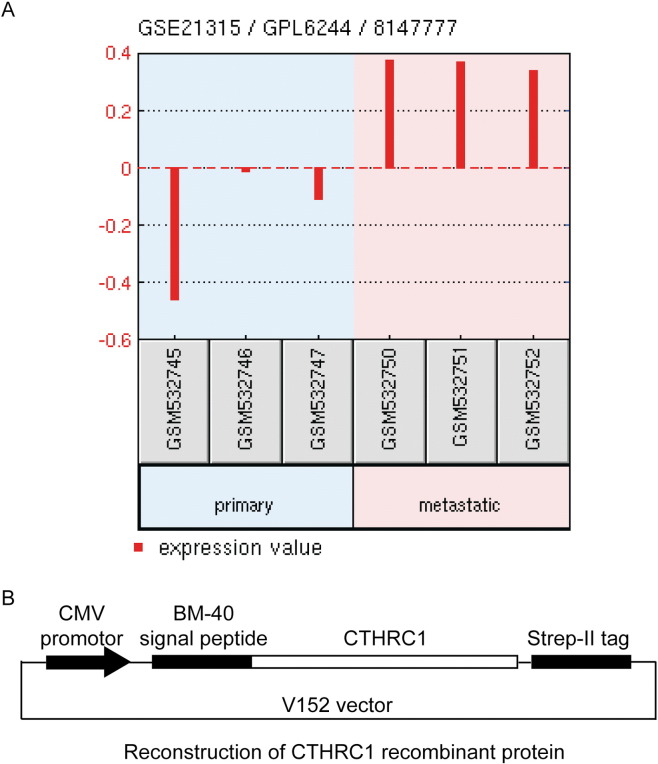
CTHRC1 ORF were cloned into the episomal expression vector V152 (Figure W1B) with pCEP-Pu-Strep II-tag (C-terminal) in-frame and the sequence of the BM-40 (SPARC/osteonectin) signal peptide downstream of the CMV promoter. CTHRC1 was recombinant expressed in EBNA-293 cells after transfecting reconstructed plasmid by using X-tremeGENE 9 DNA Transfecting Reagent (Roche, Mannheim, Germany). Forty eight hours after transfection, the EBNA-293 cells were screening with puromycin (Sigma-Aldrich, St. Louis, MO) at a dose of 2μg/ml in DMEM supplemented with 10% FBS for seven days, then the culture media were collected and applied to the Strep Tactin sepharose column(IBA, Gottingen, Germany). After this, the column was washed with binding buffer and eluted by elution buffer containing 2.5 mM desthiobiotin. The collected fractions were further quantified by Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) and BCA Protein Assay Kit (Pierce. Biotechnology Inc, Rockford, IL) and identified by western blotting assay.
In Vitro Migration and Invasion Assays
For the transwell migration assay, 4×104 primary GIST cells were placed on the top chamber of each insert with the noncoated membrane (Millicell). Cells were trypsinized and resuspended in DMEM and 700-900μL of medium supplemented with 10% fetal bovine serum added rCTHRC1 protein followed gradient doses of 0 nM, 1 nM, 20 nM, 50 nM respectively were injected into the lower chamber. After 24 hours for GIST cells in the migration assays, any cells remaining in the top chambers or on the upper membrane of the inserts were carefully removed. After fixation and staining in a dye solution containing 0.1% crystal violet and 20% methanol, cells adhering to the lower membrane of the inserts were counted and imaged through an IX71 inverted microscope (Olympus Corp. Tokyo, Japan). We carried out invasion assay by adding 100μl matrigel (BD Bioscience, Franklin Lakes, NJ) into top chamber of transwell and placed 8×104 primary GIST cells onto the matrigel. 48 hours later, the transwell for invasion was ceased and staining.
Cell Viability Assay
Cell viability was detected using a standard Cell Counting Kit-8 assay. Primary GIST cells were seeded into 96-well plates (100μl per well) at a density of 3×104 cells per ml. Cells in four divided groups were added rCTHRC1 protein followed gradient doses of 0 nM, 1 nM, 20 nM, 50 nM respectively. We added 10μl of reagent from Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) to each well for detection at day 1, 2, 3, 4, 5. After two hours of incubation at 37°C, the optical density was measured using microplate reader at a wavelength of 450nm.
Cell Isolation and Primary Cell Culture
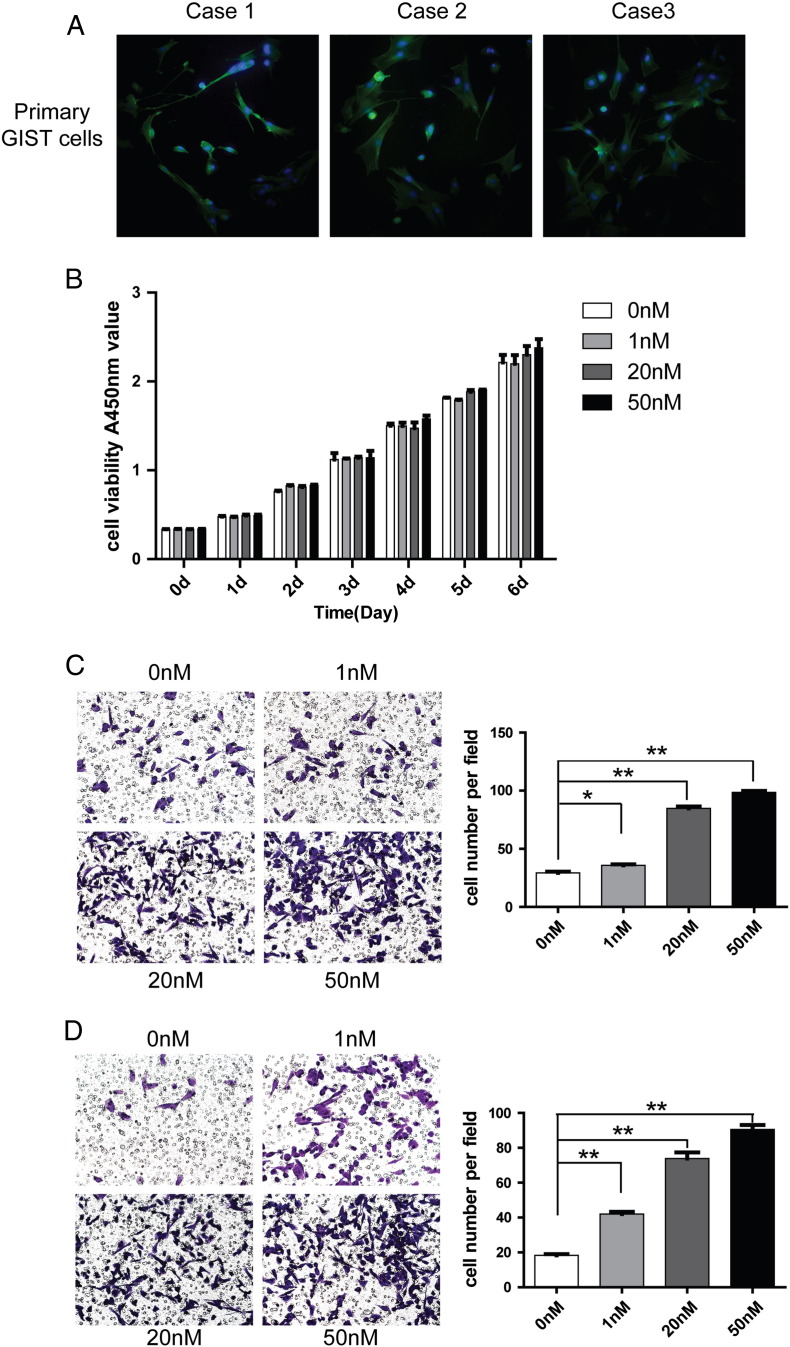
Fresh surgical GIST tissues were gently minced with scissors, washed twice with phosphate-buffered saline (PBS), and then filtered through the steel mesh with 200 μm pore diameter. After washed in cold PBS, cell pellets were resuspended in RPMI-1640 medium supplemented with 20% fetal calf serum (FCS; Gibco, France) and seeded onto culture dishes. The primary GIST cells were cultured in incubator with 5%CO2 and 37 degrees centigrade. The culture medium for primary GIST cells were changed twice every three days . The successfully isolated primary GIST cells were shown in Figure 3A.
Figure 3.
(A) Immunofluorescent staining showed the morphology of primary GIST cells isolated from the GIST tissues of three patients. The green fluorescence represents phalloidin for F-actin staining, whereas the blue fluorescence represents DAPI for nuclear staining. (B) Cell viability of primary GIST cells treated with rCTHRC1 protein at doses of 0 nM, 1 nM, 20 nM, 50 nM were measured using CCK-8 assay for six days. (C) Representative images (left) of GIST cells that migrated to the bottom of transwell filter (8μm, pore diameter) and statistical analysis (right) of the cell migration stimulated with rCTHRC1 protein or vehicle. (D) Representative images (left) of GIST cells that invaded through Matrigel to the bottom of transwell filter (8μm, pore diameter) and statistical analysis (right) of the cell invasiveness stimulated with rCTHRC1 protein or vehicle. The results shown are mean±SD of migration, and invading cells were photographed at 200 × magnification per field. (*, P < 0.05; **, P < 0.01).
Pull Down Assay
Pull down assays were conducted as reported [23] . Primary GSIT cells cultured in 100 mm dishes were serum-starved for 24 hours and treated with rCTHRC1 at a dose of 20 nM or desthiobiotin buffer for 2 hours. The primary antibodies used included the following: mouse primary antibody against Rac1 (Millipore, 1:1000) and rabbit primary antibody against RhoA, Cdc42 (Cell Signaling Technology, 1:1000).
Luciferase Reporter Assay
Primary GIST cells were seeded in 96-well plates and transfected with mixture of 100 ng TCF/catenin reporter plasmid (Wnt/β-catenin signaling), or 100 ng ATF2 reporter plasmid (Wnt/PCP signaling), and 10 ng Renilla following the recommended protocol for the Lipofectamine 2000 transfection system. One group of GIST cells were treated with rCTHRC1 protein at a concentration of 20 nM. After 48 hours of incubation, firefly and Renilla luciferase activities were measured using the dual-luciferase reporter assay system (Promega, Madison, WI) from the cell lysates.
Statistical Analysis
Statistical analyses were conducted using SPSS 16.0 software (Chicago, IL, USA). We performed chi-squared tests in cross tables to assess the relationships between expression levels of CTHRC1 and clinicopathological factors. Overall survival (OS) and Disease-free survival (DFS) were calculated using Kaplan-Meier method. The survival distributions were compared through log-rank test. All statistical tests were two-sided. One-way analysis of variance (ANOVA, Post-hoc testing) was used to compare groups (Table W1). P value less than 0.05 was considered statistically significant.
Results
CTHRC1 Expression Is Gradually Elevated in Accordance with GIST Risk Grades
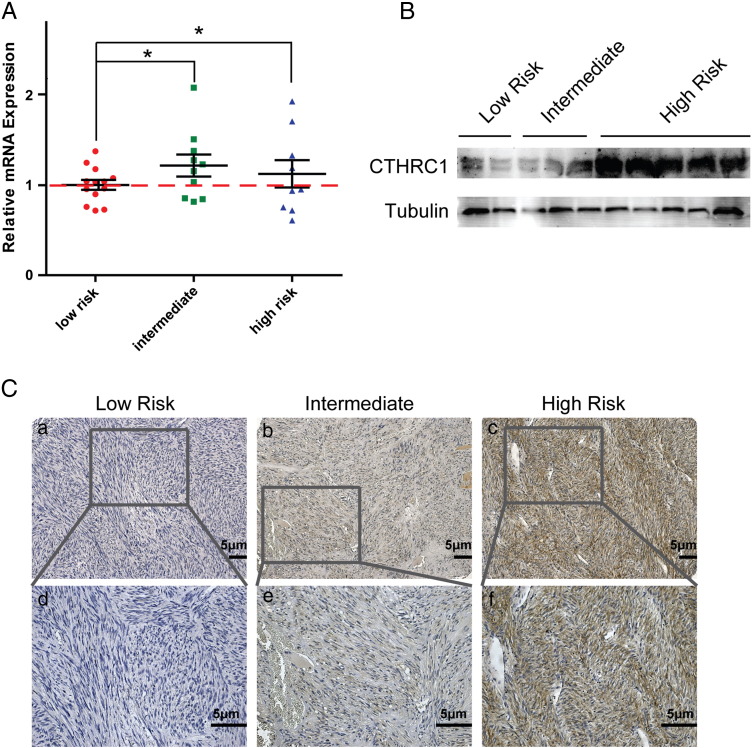
To investigate the CTHRC1 expression level in GIST tumor tissues with different risk grades, we first evaluated the mRNA expression level of CTHRC1 in fresh GIST tissue samples by quantitative PCR (qPCR). The results showed that CTHRC1 mRNA expression levels in GIST tumor tissues of the intermediate- and high-risk groups were higher than those of the low-risk group (Figure 1A). We further compared the protein expression level of CTHRC1 in GIST tissues with different risk grades. The three low-risk, two intermediate-risk and five high-risk samples were analyzed by western blotting. The results showed that the CTHRC1 protein expression level in the high-risk group was significantly higher than that of the intermediate- and low-risk groups (Figure 1B).
Figure 1.
CTHRC1 expression in GIST tissues. (A) Relative mRNA expression of CTHRC1 in low-risk group was significantly lower than those in the intermediate- and high-risk groups. (B) Western blotting analysis showed that CTHRC1 expression in high-risk GIST patients was significantly higher than those of low-risk or intermediate-risk groups. Tubulin was included as a loading control. (C) Representative image of immunohistochemical staining of CTHRC1 in low-risk, intermediate-risk and high-risk GIST tissues. Original magnification: a, b, c, 100 ×; d, e, f, 200 ×. Scale bars, 5 μm (*, P < 0.05).
CTHRC1 Protein Expression Level Is Closely Correlated with Risk Grade of NIH Classification, and Prognosis of GIST
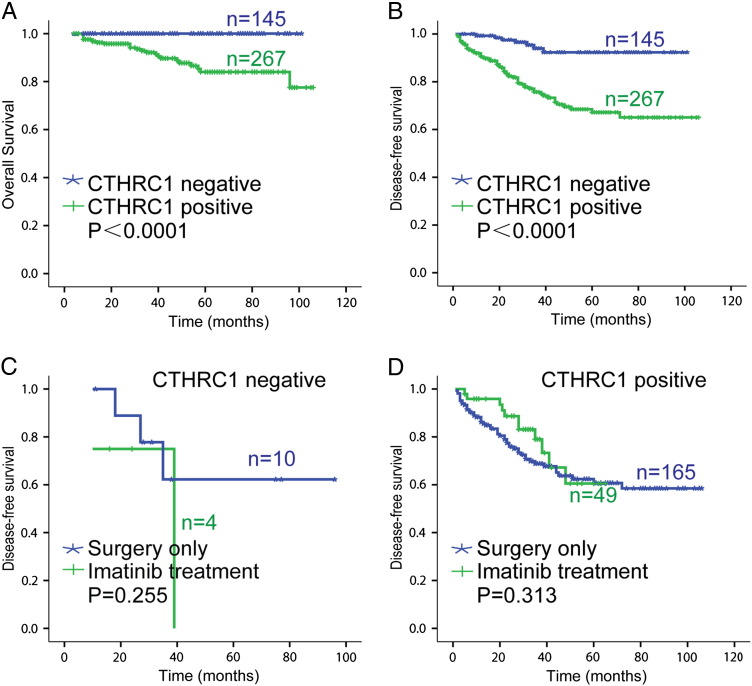
The clinicopathological significance of CTHRC1 was further examined using a tissue microarray which contained 412 GIST tissue samples. The immunohistochemistry staining results showed that 145 (35.2%) cases showed CTHRC1 low expression, 267 (64.8%) cases showed CTHRC1 high expression (Figure 1C). The correlations between CTHRC1 expression and the clinicopathological parameters are shown in Table 1. We found that the expression level of CTHRC1 was higher in the patients with high NIH grade, large tumor size (> 10 cm) or increased mitotic figures than those with low NIH grade, small tumor size (< 10 cm), fewer mitotic figures with statistical significance (P < 0.05). Interestingly, we found that there was a significant difference between male (70.85%) and female (57.67%) GIST patients in frequency of high CTHRC1 levels. The statistical analysis suggested that CTHRC1 expression was not correlated with age, histological type, or tumor rupture. We further investigated the correlation between CTHRC1 expression and overall survival (OS) or disease-free survival (DFS) of GIST patients. The OS in the CTHRC1 negative (low) group (five-year OS rates, 100%, 145/145) was remarkably superior than that in the CTHRC1 positive (high) expression group (five-year OS rates, 90.6%, 242/267) (Figure 2A). The DFS in the CTHRC1 negative (low) group (five-year DFS rates, 95.2%, 138/145) was significantly higher than that in the CTHRC1 positive (high) expression group (five-year DFS rates, 76.8%, 205/267) (Figure 2B). In summary, CTHRC1 expression in GIST tumor tissues was closely correlated with OS and DFS of GIST patients.
Table 1.
Relationship between CTHRC1 expression and clinicopathologic features of GIST patients(*, P < 0.05; **, P < 0.01).
| Variable | CTHRC1(n = 412) |
|||
|---|---|---|---|---|
| Low | High | P | ||
| Age$ | ≤ 59 years | 31 | 49 | 0.458 |
| > 59 years | 114 | 218 | ||
| Gender | Male | 65 | 158 | 0.005 |
| Female | 80 | 109 | ||
| Tumor site | Stomach | 100 | 128 | < 0.001 |
| Small bowel | 28 | 93 | ||
| Colon | 12 | 9 | ||
| Other sites | 5 | 37 | ||
| Tumor size(cm) | ≤ 2 | 30 | 7 | < 0.001 |
| > 2&≤5 | 102 | 61 | ||
| > 5&≤10 | 9 | 127 | ||
| > 10 | 4 | 72 | ||
| Mitoses per 50 HPFs | ≤ 5 | 138 | 170 | < 0.001 |
| > 5&≤10 | 2 | 52 | ||
| > 10 | 5 | 45 | ||
| Modified NIH criteria | Very low risk | 30 | 2 | < 0.001 |
| Low risk | 101 | 51 | ||
| Intermediate risk | 4 | 58 | ||
| High risk | 10 | 156 | ||
| NIH invasion | 1 | 30 | 2 | < 0.001 |
| 2 | 101 | 51 | ||
| 3 | 4 | 58 | ||
| 4 | 5 | 101 | ||
| 5 | 5 | 55 | ||
| Tumor bleeding | No | 131 | 220 | 0.030 |
| Yes | 14 | 47 | ||
Abbreviations: HPF, high power field of the microscope; NIH, National Institutes of Health.
The P value in bold emphasize statistical significance (P<0.001).
Median age of total 412 patients was 59 years.
Figure 2.
(A) Kaplan-Meier analysis of overall survival related to the expression of CTHRC1 in 412 GIST patients. (B) Kaplan-Meier analysis of disease-free survival related to the expression of CTHRC1 in 412 GIST patients. (C) Among CTHRC1-negative intermediate- and high-risk GIST patients, there was no significant difference in DFS between the groups with or without imatinib adjuvant treatment. (D) Among CTHRC1-positive intermediate- and high-risk GIST patients, no significant difference between the imatinib treatment group and the surgery alone group were observed.
Correlation between CTHRC1 Expression and the Efficacy of Imatinib Adjuvant Treatment
According to the NIH classification guideline, intermediate- or high-risk GIST patients require adjuvant treatment with imatinib. In this study, we analyzed the correlation between the efficacy of imatinib adjuvant treatment and CTHRC1 expression. Among 14 CTHRC1- negative intermediate- and high-risk GIST patients, there was no correlation between imatinib treatment and patient prognosis (Figure 2C). The difference in DFS between the imatinib treatment group and the surgery treatment only group was not statistically significant (P = 0.255). Notably, among 214 CTHRC1- positive intermediate- and high-risk GIST patients, the DFS rate in the imatinib treatment group were higher than that in the surgery only group within 3 years of follow-up. However, the differences in 5-year DFS rates in the two groups were not statistically significant (P = 0.313) (Figure 2D). Therefore, the expression of CTHRC1 can not predict the efficacy of imatinib adjuvant treatment in our current study.
Recombinant CTHRC1 Protein Promotes GIST Cell Migration and Invasion in a Dose-dependent Manner
To explore the biological functions of CTHRC1 as a secreted protein, CTHRC1 was recombinantly expressed in EBNA-293 cells, and further purified and verified by western blotting (Figure W2). Then, the purified recombinant CTHRC1 (rCTHRC1) protein was applied to primary GIST cells in a migration and Matrigel invasion assay. Compared to the vehicle group, GIST cell migration and invasion were significantly enhanced by rCTHRC1 protein at doses of 1 nM, 20 nM, and 50 nM (Figure 3, C and D). Moreover, the promotion of cell motility by the rCTHRC1 protein was dose-dependent. However, primary cell viability was not remarkably affected by rCTHRC1 protein based on the cell viability assay (Figure 3B). These results demonstrate that CTHRC1 is a potent pro-invasion factor that facilitates GIST cell invasion in a dose-dependent manner.
CTHRC1 Activates Wnt/PCP-Rho Signaling in Primary GIST Cells
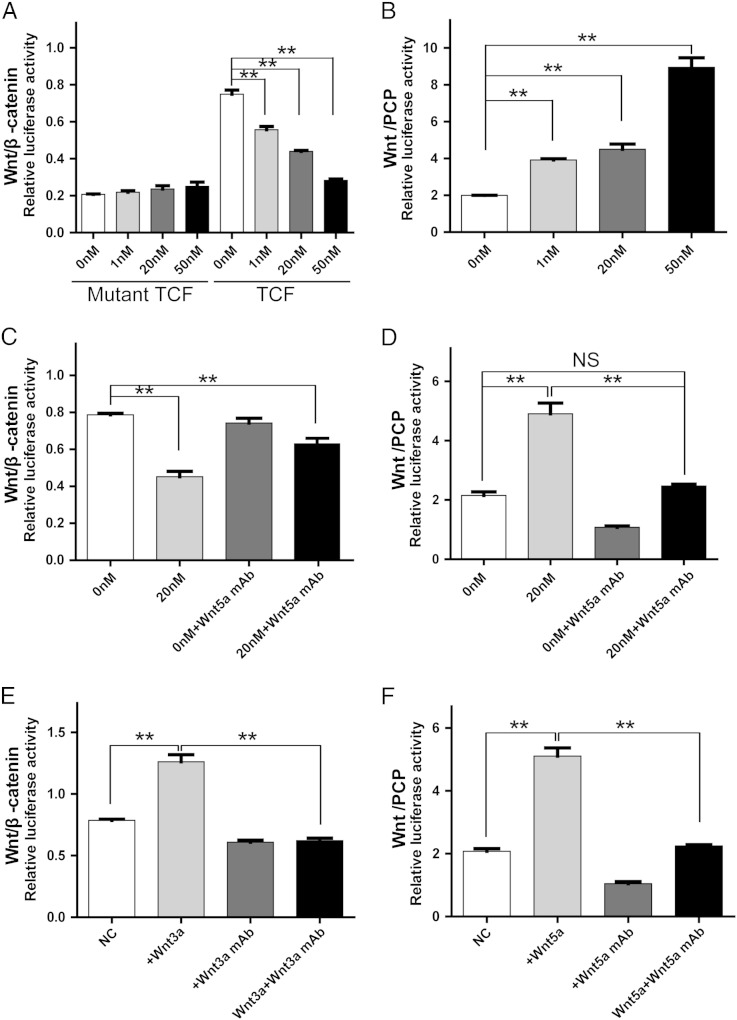
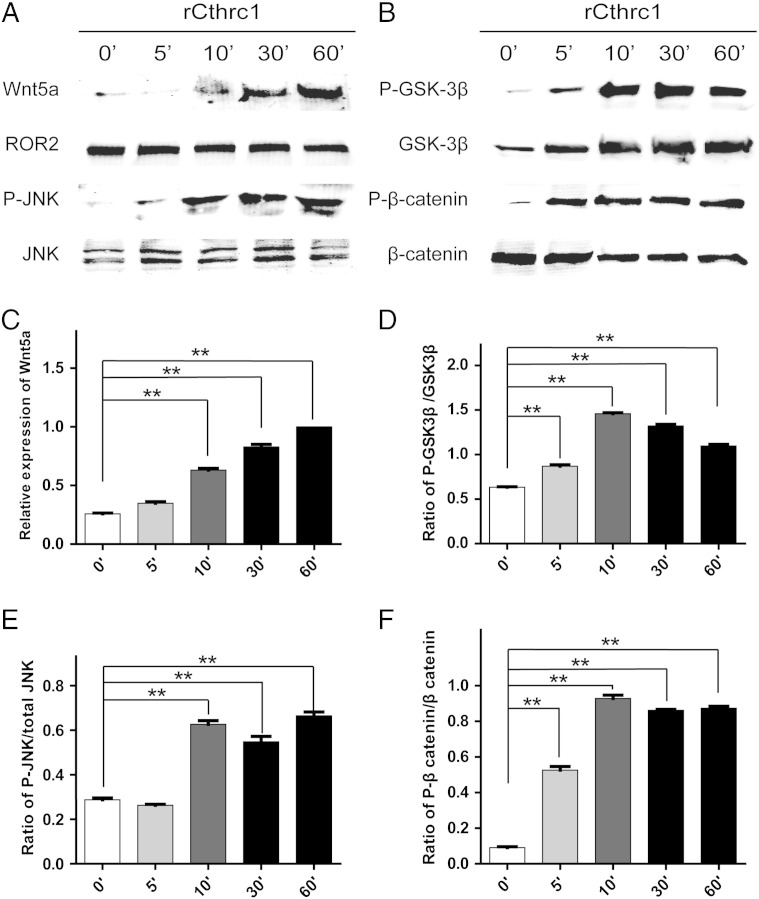
To understand the underlying mechanism by which CTHRC1 promotes GIST cell migration and invasion, we examined the activation of the canonical Wnt pathway and the non-canonical Wnt pathway. GIST cells were transfected with a Wnt//β-catenin reporter plasmid (TCF/catenin plasmid) and negative control counterpart plasmid or non-canonical Wnt/PCP pathway reporter plasmid (ATF2 plasmid). Recombinant CTHRC1 or vehicle control was added 24 hours after transfection, and luciferase activity was determined. The results showed that Wnt/β-catenin signaling was inhibited while the noncanonical Wnt/PCP signaling was activated by rCTHRC1 protein in primary GIST cells (Figure 4, A and B). The effects of rCTHRC1 protein on Wnt signaling was blocked by a Wnt5a neutralizing antibody (Figure 4, C and D). We also verified the block effect of Wnt3a and Wnt5a neutralizing antibodies (Figure 4, E and F). We further confirmed the inhibitory effect of CTHRC1 on Wnt/β-catenin signaling using western blotting assy. The level of phosphorylated β-catenin, which indicates the degradation of β-catenin, was increased in primary GIST cells treated with rCTHRC1 (Figure 6, B and F). In addition, the level of GSK3β, which phosphorylates β-catenin on Ser-33/Ser-37/Thr-41 was increased in rCTHRC1 treated primary GIST cells (Figure 6, B and D). Therefore, we confirmed that CTHRC1 inhibits the canonical Wnt/β-catenin pathway in primary GIST cells.
Figure 4.
(A) Dual-luciferase reporter assay showed that rCthrc protein inhibited Wnt/β-catenin signaling of primary GIST cells in a dose-dependent manner. The results shown are mean±SD of relative firefly/Renilla ratio. (B) Noncanonical Wnt/PCP signaling of GIST cells was activated by rCTHRC1 protein in a dose-dependent manner. (C) The inhibitory effect of rCTHRC1 on Wnt/β-catenin signaling was partially blocked by Wnt5a monoclonal neutralizing antibody. (D) The promoting effect of rCTHRC1 protein on Wnt/PCP signaling was almost blocked by Wnt5a monoclonal neutralizing antibody. (E) The promoting effect of Wnt3a on Wnt/β-catenin signaling was almost blocked by Wnt3a monoclonal neutralizing antibody. (F) The promoting effect of Wnt5a on Wnt/PCP signaling was almost blocked by Wnt5a monoclonal neutralizing antibody. (*, P < 0.05; **, P < 0.01).
Figure 6.
(A) The expression of Wnt5a and the phosphorylation of JNK were examined after treatment with rCTHRC1 by western blotting. (B) The phosphorylation of GSK3β and β-catenin were examined after treatment with rCTHRC1 by western blotting. (C) Quantitative analysis of grey value for Wnt5a using ImageJ software. The relative expression of Wnt5a induced by rCTHRC1 at 0′, 5′, 10′, 30′ was compared with the grey value of Wnt5a induced by rCTHRC1 at 60′. (D) Quantitative analysis of grey value for phospho-GSK3β/total GSK3β ratio using ImageJ software. (E) Quantitative analysis of grey value for phospho-JNK/total JNK ratio using ImageJ software. (F) Quantitative analysis of grey value for phospho-β-catenin/totalβ-catenin ratio using ImageJ software. (*, P < 0.05; **, P < 0.01).
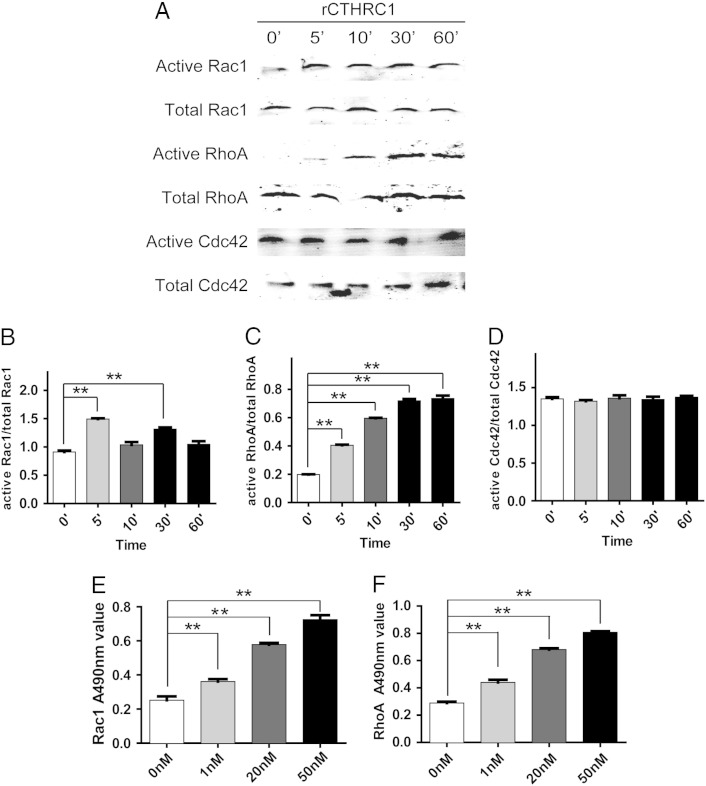
The downstream molecules of the Wnt/PCP pathway mainly include the small GTPase family, such as Rac1, RhoA and Cdc42, which play important roles in cancer cell migration and invasion. Using a Rho GTPases pull-down assay, we found that the rCTHRC1 protein enhanced the activity of RhoA and Rac1 but not Cdc42 (Figure 5, A-D).
Figure 5.
(A) Analysis of the active and total RhoA,Rac1 and Cdc42 in primary GIST cells treated with rCTHRC1 protein by pull-down assay.(B) Quantitative analysis of grey value for active Rac1/ -total Rac1 ratio using ImageJ software. (C) Quantitative analysis of grey value for active RhoA/ -total RhoA ratio using ImageJ software. (D) Quantitative analysis of grey value for active Cdc42/ total Cdc42 ratio using ImageJ software. (E) Rac1 G-LISA assay was used to assess the levels of GTP-bound Rac1 in GIST cells treated with rCTHRC1 protein. (F) RhoA G-LISA assay was used to assess the levels of GTP-bound RhoA in GIST cells treated with rCTHRC1 protein. (*, P < 0.05; **, P < 0.01).
To further confirm the above results, the GLISA assay, another approach to measure the activities of Rho GTPases, was performed. It also demonstrated that the activities of RhoA and Rac1 were significantly enhanced by rCTHRC1 treatment in primary GIST cells, which is consistent with the results of the Rho GTPases pull-down assay (Figure 5, E and F).
Furthermore, the phosphorylation of c-Jun N terminal kinase (JNK), another downstream molecule of the Wnt/PCP pathway, and Wnt5a were also elevated by rCTHRC1 treatment (Figure 6, A, C and E). These results suggested that CTHRC1 may promote GIST cell invasion through the Wnt/PCP-Rho-JNK pathway.
CTHRC1-induced Primary GIST Cell Migration and Invasion Is Wnt5a and Wnt/PCP Signaling-dependent
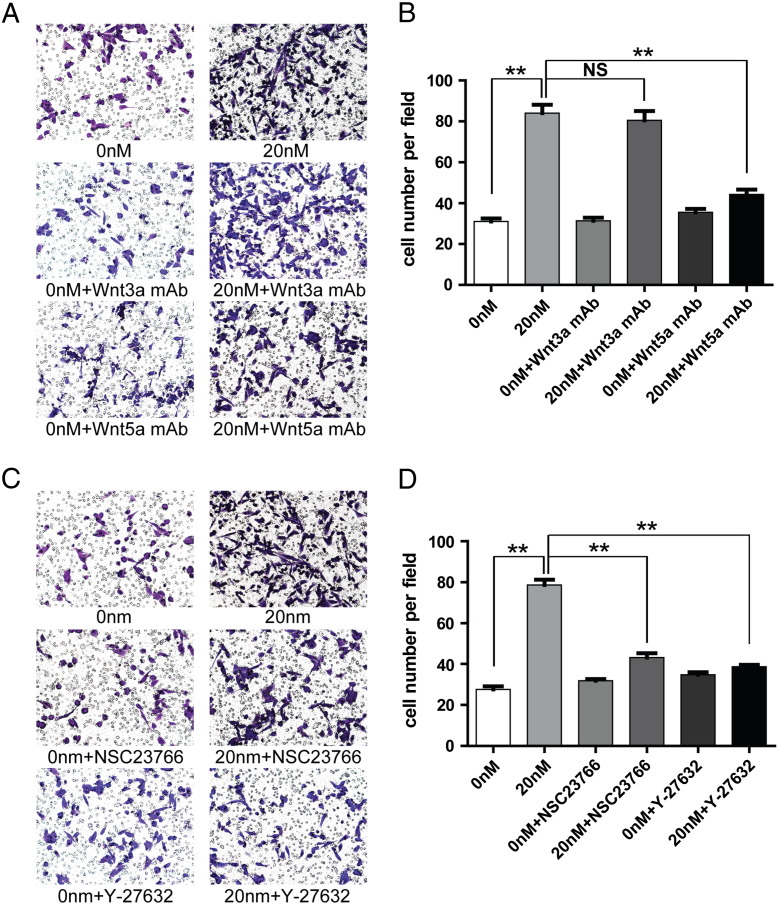
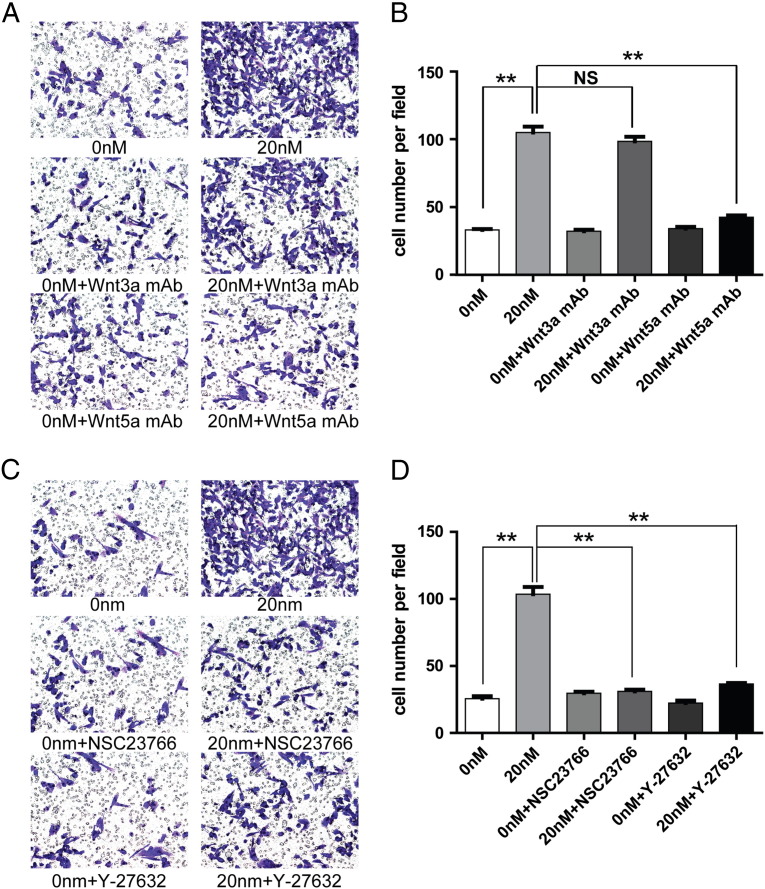
We further investigated whether Wnt3a (a ligand of canonical Wnt/β-catenin pathway) and Wnt5a (a ligand of noncanonical Wnt/PCP pathway) are involved in CTHRC1-induced GIST cell migration and invasion by using neutralizing antibodies of Wnt3a and Wnt5a. The data illustrated that the migration- and invasion-promoting activities of rCTHRC1 at a dose of 20 nM were not affected by the Wnt3a neutralizing antibody (Figure 7, A and B, Figure 8, A and B). However, the promoting effects of rCTHRC1 on GIST cell migration and invasion were almost completely blocked by the Wnt5a neutralizing antibody (Figure 7, A and B, Figure 8, A and B). We further investigated whether the promoting effects of CTHRC1 on GIST cells motility are Wnt/PCP signaling-dependent by using inhibitors of ROCK and Rac1, which are key downstream molecules of the Wnt/PCP pathway. The results showed that both ROCK and Rac1 inhibitors treatment inhibits the promoting effects of rCTHRC1 on GIST cell migration and invasion (Figure 7, C and D, Figure 8, C and D).
Figure 7.
(A) The promotive effect of rCTHRC1 protein was blocked by Wnt5a neutralizing antibody but not blocked by Wnt3a neutralizing antibody shown from cell migration assay in vitro. (B) Quantification analysis of migrated cells were performed for six randomly selected fields (original magnification:200 ×). (C) The promotive effect of rCTHRC1 protein was partially blocked by Rac1 inhibitor (NSC23766) as well as ROCK inhibitor (Y-27632) shown from cell migration assay. (D) Quantification analysis of migrated cells were performed for six randomly selected fields (original magnification:200 ×). (*, P < 0.05; **, P < 0.01).
Figure 8.
(A) The pro-invasion effect of rCTHRC1 protein was blocked by Wnt5a neutralizing antibody but not blocked by Wnt3a neutralizing antibody shown from cell invasion assay in vitro. (B) Quantification analysis of migrated cells were performed for six randomly selected fields (original magnification: 200 ×). (C) The pro-invasion effect of rCTHRC1 protein was partially blocked by Rac1 inhibitor (NSC23766) as well as ROCK inhibitor (Y-27632) shown from cell invasion assay. (D) Quantification analysis of migrated cells were performed for six randomly selected fields (original magnification:200 ×). (*, P < 0.05; **, P < 0.01).
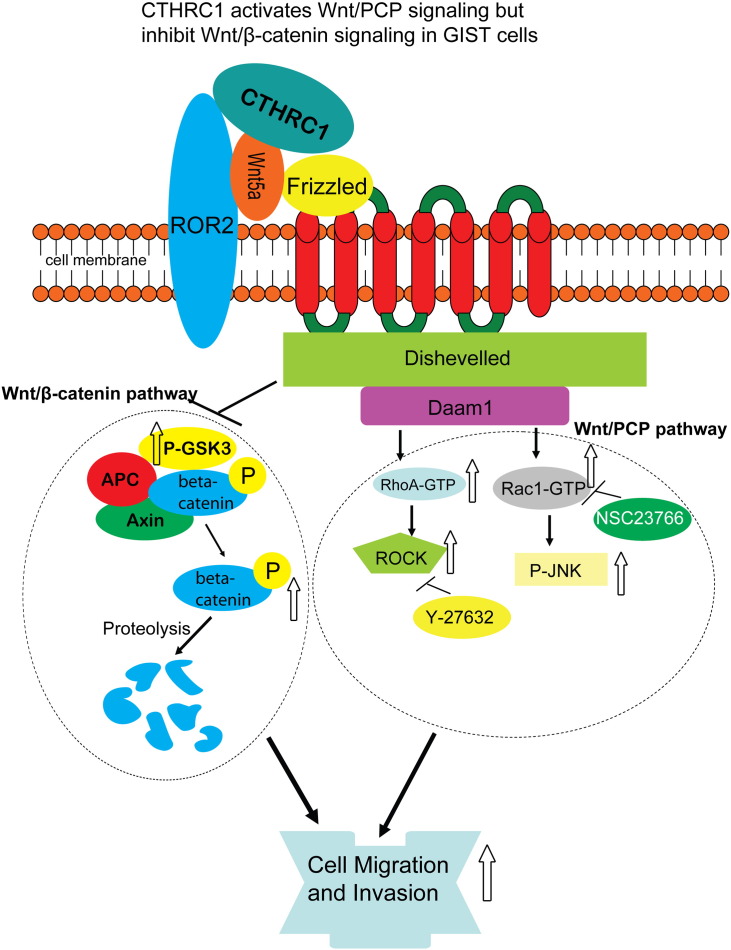
Taken together, these data indicated that the CTHRC1-induced GIST cell migration and invasion is Wnt5a and noncanonical Wnt/PCP signaling dependent (Figure 9).
Figure 9.
CTHRC1 induced cell signaling alteration and its related cell movement.
Discussion
GISTs have a variable malignancy degree ranging from a curable disorder to highly malignant sarcomas [1,8]. The majority of GISTs stain positive for KIT oncoproteins in immunohistochemical assays [3,24]. KIT is a stem cell growth factor receptor that plays pro-proliferative and anti-apoptotic roles in GIST progression [5,6]. GIST patients treated with the KIT targeted inhibitor, − imatinib, showed prolonged median recurrence-free survival of 12 to 24 months [1,21]. Recurrence and metastasis in GIST patients are the major causes of treatment failure or even death [7,25,26]. Thus, new predictive biomarkers for recurrence and an understanding of the mechanisms of GIST metastasis are urgently needed.
By analyzing the GIST microarray dataset (GSE21315) from the GEO database (Figure W1A), we found that CTHRC1 expression in GIST with liver metastasis was remarkably higher than in primary GIST tissues(fold change>3, P < 0.05). This result strongly suggested that CTHRC1 may play important roles in regulating GIST metastasis. The NIH classification published in 2002 was widely accepted as standard for predicting the prognosis of GIST patients [27,28]. According to the NIH classification, the risk assessments are based on tumor size and the number of mitotic figures. We have analyzed the correlation between CTHRC1 expression and GIST clinicopathological parameters and found that the CTHRC1 expression levels were closely related to NIH classification, tumor size and the number of mitotic figures. These analyses suggest that CTHRC1-positive GISTs exhibit a greater likelihood of malignant behavior and more aggressive features. Moreover, there was a significant difference between male and female GIST patients in frequency of high CTHRC1 levels (Table 1). It has been reported that CTHRC1 is associated with attenuated inflammatory arthritis severity in males, but not in females [29,30]. The naive mice assay showed that the expression and inducibility of CTHRC1 were highly dependent on sex [29,30]. Among naive wild-type BALB/c mice, CTHRC1 expression was remarkably higher in males than in females. Moreover, CTHRC1 was one of the major sex-affected differentially expressed genes [29,30]. Therefore, sex disparities may cause the difference between male and female GIST patients in high CTHRC1 expression rates.
The Kaplan-Meier curves analysis revealed that CTHRC1 expression was closely correlated with OS and DFS of GIST patients. GIST patients with CTHRC1-positive tumors had shorter OS and DFS than CTHRC1-negative patients. Therefore, we identified that CTHRC1 is an available predictor of poor prognosis including OS and DFS in GIST patients. In addition, the great clinical value of CTHRC1 in predicting the recurrence risk of postoperative GIST patients may contribute to improving the clinical therapeutic effects.
Tumor microenvironments including components of extracellular matrix protein play crucial roles in promoting tumor invasion and metastasis [31]. CTHRC1, a secreted ECM protein, has been reported to be up-regulated in many solid tumors. In hepatocellular carcinoma, CTHRC1 is up-regulated and promotes tumor invasion and predicts poor prognosis [13]. CTHRC1 plays a promoting role in pancreatic cancer progression and metastasis by enhancing the migration ability of cancer cells [18]. These accumulating data indicate that CTHRC1 is an important regulator of tumor invasion and metastasis in the tumor microenvironment. In the present study, we have found that CTHRC1 expression in GIST tissue is gradually elevated in accordance with risk grading. Based on an in vitro functional assay, CTHRC1 was considered to be an invasion-promoting protein and ultimately contributed to gastrointestinal stromal tumor metastasis and recurrence.
Although the functional roles of CTHRC1 in tumor cell invasion and metastasis have been well established, the underlying mechanisms of how CTHRC1 promotes cancer cell invasion remains unclear. It has been reported that CTHRC1 acts as a Wnt cofactor that selectively activates the PCP pathway in the inner ear developmental process [17]. In the present study, we showed that CTHRC1 promotes GIST cell invasion by activating Wnt/PCP signaling, which is supported by the following evidence. First, the luciferase reporter assay and western blotting showed that recombinant CTHRC1 protein activated the PCP pathway of Wnt signaling of primary GIST cells in a dose-dependent manner. Second, the pro-invasion activity of the rCTHRC1 protein was blocked by the neutralizing antibody of Wnt5a (a ligand for Wnt/PCP pathway [32,33]) and the inhibitors of Rac1 and ROCK (the downstream molecules of Wnt/PCP signaling [34,35]).
CTHRC1 promoted tumor cells migration by activating Rac1 and resulted in metastasis of pancreatic cancer [18]. The overexpression of CTHRC1 promotes tumor invasion by activating RhoA in hepatocellular carcinoma [13]. Accordingly, we demonstrated the promoting effect of CTHRC1 on both RhoA and Rac1 in GIST cells. Moreover, we further verified that the pro-invasion activity of CTHRC1 in GIST cells was dependent on the Wnt5a/PCP-Rho axis by blocking the Wnt5a/PCP-Rho pathway with neutralizing antibodies and specific inhibitors. The noncanonical Wnt/PCP pathway transmits signaling from the cell-surface Frizzled receptor-coupled Wnt5a protein, via the Dvl-RhoA/Rac1-JNK-ATF2/c-Jun cascade [36-38], to the nucleus. Noncanonical Wnt/PCP signaling plays important roles in promoting cell migration [39] and formation of cell protrusions [40,41]. The small Rho GTPases Rac1, RhoA and Cdc42, are key executors of Wnt/PCP related cell migration [42,43]. RhoA controls the assembly of actin to generate contractile forces [44,45], while Rac1 and Cdc42 promote actin polymerization contributing to the formation of protrusive forces [46,47]. Therefore, the efficient cell movement requires synergistic actions of the three Rho GTPases [48-50]. In this study, we have shown that CTHRC1, a secreted protein, transduces outside-in signals through the Wnt/PCP pathway and coordinates the action of the three Rho GTPases to promote GIST cell migration and invasion.
Taken together, we have demonstrated that CTHRC1 expression level is closely correlated with risk grade of NIH classification and prognosis of GIST, indicating that CTHRC1 served as a new predictor of recurrence risk and prognosis in post-operative GIST patients. Furthermore, we have shown that CTHRC1 promotes GIST cell migration and invasion by activating the Wnt/PCP-Rho signaling, suggesting that the CTHRC1- Wnt/PCP-Rho axis may be a new therapeutic target for interventions against GIST invasion and metastasis.
Acknowledgments
The study was supported by the National Science Foundation of China (81071738; 81101600, 81272743 and 81302094), Shanghai Health Bureau Youth Fund (2009Y110). ZGZ and HC were the principal investigator and supervised the implementation of the study. MZM wrote the protocols and, with CZ, XMY, ZZZ and HYY, analyzed the data and interpreted the findings. CZ and MZM were responsible for the collection of clinical data. HM had successfully isolated primary GIST cells from patients' tissues. MZM and CZ carried out the experiments involved in this article. HC, ZGZ, JRG, SLY and WXQ participated in critical revision of the manuscript for important intellectual content. All authors had full access to the primary data and the final analysis and approved the final version of the manuscript.
These authors declare no conflict of interest.
Footnotes
This article refers to supplementary materials, which is designated by Table W1 and Figures W1 to W3 is available online at www.neoplasia.com.
Contributor Information
Ming-Ze Ma, Email: caohuishcn@hotmail.com.
Chun Zhuang, Email: zhuangchun88@163.com.
Xiao-Mei Yang, Email: xmyang@sibs.ac.cn.
Zi-Zhen Zhang, Email: zzzhang16@hotmail.com.
Hong Ma, Email: mahong4899@126.com.
Wen-Ming Zhang, Email: wenming_zhang@163.com.
Haiyan You, Email: bettyyouyou@yeah.net.
Wenxin Qin, Email: wxqin@sjtu.edu.cn.
Jianren Gu, Email: jrgu@shsci.org.
Shengli Yang, Email: slyang@sibs.ac.cn.
Hui Cao, Email: caohuishcn@hotmail.com.
Zhi-Gang Zhang, Email: zzhang@shsci.org.
Appendix A. Supplementary materials
Figure W1.
(A) Analysis of CTHRC1 differential expression in metastatic GIST and primary GIST based on data from GEO Database (GSE21315). (B) Schematic diagram of V152 vector which was used to reconstruct CTHRC1-StrepII recombinant plasmid.
Figure W2.
Verification of affinity purified CTHRC1 protein by Coomassie Brilliant Blue staining and Western blotting.
Figure W3.
Negative controls for primary and secondary antibodies in GIST TMA immunohistochemistry staining assay (original magnification:100 ×).
Table W1.
ANOVA analysis (post-hoc testing) for statistics of figures.
| Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) 1 = low risk,2 = intermediate risk,3 = high risk | (J) 1 = low risk,2 = intermediate risk,3 = high risk | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 1A | ||||||
| 1 | 2 | − 1.53243* | .57014 | .031 | − 2.9405 | -.1244 |
| 3 | − 1.47190* | .58777 | .046 | − 2.9235 | -.0203 | |
| 2 | 1 | 1.53243* | .57014 | .031 | .1244 | 2.9405 |
| 3 | .06053 | .62280 | .995 | − 1.4776 | 1.5986 | |
| 3 | 1 | 1.47190* | .58777 | .046 | .0203 | 2.9235 |
| 2 | -.06053 | .62280 | .995 | − 1.5986 | 1.4776 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) concentration | (J) concentration | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
|
Figure 3B Day0 | ||||||
| 0 | 1 | -.0040000 | .0094810 | .973 | -.034361 | .026361 |
| 20 | -.0160000 | .0094810 | .389 | -.046361 | .014361 | |
| 50 | -.0013333 | .0094810 | .999 | -.031695 | .029028 | |
| 1 | 0 | .0040000 | .0094810 | .973 | -.026361 | .034361 |
| 20 | -.0120000 | .0094810 | .607 | -.042361 | .018361 | |
| 50 | .0026667 | .0094810 | .992 | -.027695 | .033028 | |
| 20 | 0 | .0160000 | .0094810 | .389 | -.014361 | .046361 |
| 1 | .0120000 | .0094810 | .607 | -.018361 | .042361 | |
| 50 | .0146667 | .0094810 | .456 | -.015695 | .045028 | |
| 50 | 0 | .0013333 | .0094810 | .999 | -.029028 | .031695 |
| 1 | -.0026667 | .0094810 | .992 | -.033028 | .027695 | |
| 20 | -.0146667 | .0094810 | .456 | -.045028 | .015695 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) concentration | (J) concentration | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
|
Figure 3B Day1 | ||||||
| 0 | 1 | .0023333 | .0055578 | .973 | -.015465 | .020131 |
| 20 | -.0216667* | .0055578 | .019 | -.039465 | -.003869 | |
| 50 | -.0230000* | .0055578 | .014 | -.040798 | -.005202 | |
| 1 | 0 | -.0023333 | .0055578 | .973 | -.020131 | .015465 |
| 20 | -.0240000* | .0055578 | .011 | -.041798 | -.006202 | |
| 50 | -.0253333* | .0055578 | .008 | -.043131 | -.007535 | |
| 20 | 0 | .0216667* | .0055578 | .019 | .003869 | .039465 |
| 1 | .0240000* | .0055578 | .011 | .006202 | .041798 | |
| 50 | -.0013333 | .0055578 | .995 | -.019131 | .016465 | |
| 50 | 0 | .0230000* | .0055578 | .014 | .005202 | .040798 |
| 1 | .0253333* | .0055578 | .008 | .007535 | .043131 | |
| 20 | .0013333 | .0055578 | .995 | -.016465 | .019131 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) concentration | (J) concentration | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
|
Figure 3B Day2 | ||||||
| 0 | 1 | -.0596667* | .0133083 | .009 | -.102285 | -.017049 |
| 20 | -.0253333 | .0133083 | .299 | -.067951 | .017285 | |
| 50 | -.0746667* | .0133083 | .002 | -.117285 | -.032049 | |
| 1 | 0 | .0596667* | .0133083 | .009 | .017049 | .102285 |
| 20 | .0343333 | .0133083 | .120 | -.008285 | .076951 | |
| 50 | -.0150000 | .0133083 | .684 | -.057618 | .027618 | |
| 20 | 0 | .0253333 | .0133083 | .299 | -.017285 | .067951 |
| 1 | -.0343333 | .0133083 | .120 | -.076951 | .008285 | |
| 50 | -.0493333* | .0133083 | .025 | -.091951 | -.006715 | |
| 50 | 0 | .0746667* | .0133083 | .002 | .032049 | .117285 |
| 1 | .0150000 | .0133083 | .684 | -.027618 | .057618 | |
| 20 | .0493333* | .0133083 | .025 | .006715 | .091951 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) concentration | (J) concentration | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
|
Figure 3B Day3 | ||||||
| 0 | 1 | -.0046667 | .0051962 | .806 | -.021307 | .011973 |
| 20 | -.0223333* | .0051962 | .011 | -.038973 | -.005693 | |
| 50 | -.0283333* | .0051962 | .003 | -.044973 | -.011693 | |
| 1 | 0 | .0046667 | .0051962 | .806 | -.011973 | .021307 |
| 20 | -.0176667* | .0051962 | .038 | -.034307 | -.001027 | |
| 50 | -.0236667* | .0051962 | .008 | -.040307 | -.007027 | |
| 20 | 0 | .0223333* | .0051962 | .011 | .005693 | .038973 |
| 1 | .0176667* | .0051962 | .038 | .001027 | .034307 | |
| 50 | -.0060000 | .0051962 | .669 | -.022640 | .010640 | |
| 50 | 0 | .0283333* | .0051962 | .003 | .011693 | .044973 |
| 1 | .0236667* | .0051962 | .008 | .007027 | .040307 | |
| 20 | .0060000 | .0051962 | .669 | -.010640 | .022640 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) concentration | (J) concentration | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
|
Figure 3B Day4 | ||||||
| 0 | 1 | -.0030000 | .0072111 | .974 | -.026092 | .020092 |
| 20 | .0173333 | .0072111 | .154 | -.005759 | .040426 | |
| 50 | -.0753333* | .0072111 | .000 | -.098426 | -.052241 | |
| 1 | 0 | .0030000 | .0072111 | .974 | -.020092 | .026092 |
| 20 | .0203333 | .0072111 | .086 | -.002759 | .043426 | |
| 50 | -.0723333* | .0072111 | .000 | -.095426 | -.049241 | |
| 20 | 0 | -.0173333 | .0072111 | .154 | -.040426 | .005759 |
| 1 | -.0203333 | .0072111 | .086 | -.043426 | .002759 | |
| 50 | -.0926667* | .0072111 | .000 | -.115759 | -.069574 | |
| 50 | 0 | .0753333* | .0072111 | .000 | .052241 | .098426 |
| 1 | .0723333* | .0072111 | .000 | .049241 | .095426 | |
| 20 | .0926667* | .0072111 | .000 | .069574 | .115759 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) concentration | (J) concentration | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
|
Figure 3B Day5 | ||||||
| 0 | 1 | .0146667 | .0078916 | .316 | -.010605 | .039938 |
| 20 | -.0660000* | .0078916 | .000 | -.091272 | -.040728 | |
| 50 | -.0893333* | .0078916 | .000 | -.114605 | -.064062 | |
| 1 | 0 | -.0146667 | .0078916 | .316 | -.039938 | .010605 |
| 20 | -.0806667* | .0078916 | .000 | -.105938 | -.055395 | |
| 50 | -.1040000* | .0078916 | .000 | -.129272 | -.078728 | |
| 20 | 0 | .0660000* | .0078916 | .000 | .040728 | .091272 |
| 1 | .0806667* | .0078916 | .000 | .055395 | .105938 | |
| 50 | -.0233333 | .0078916 | .071 | -.048605 | .001938 | |
| 50 | 0 | .0893333* | .0078916 | .000 | .064062 | .114605 |
| 1 | .1040000* | .0078916 | .000 | .078728 | .129272 | |
| 20 | .0233333 | .0078916 | .071 | -.001938 | .048605 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) concentration | (J) concentration | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
|
Figure 3B Day6 | ||||||
| 0 | 1 | -.0046667 | .0106797 | .970 | -.038867 | .029533 |
| 20 | -.0750000* | .0106797 | .001 | -.109200 | -.040800 | |
| 50 | -.1590000* | .0106797 | .000 | -.193200 | -.124800 | |
| 1 | 0 | .0046667 | .0106797 | .970 | -.029533 | .038867 |
| 20 | -.0703333* | .0106797 | .001 | -.104533 | -.036133 | |
| 50 | -.1543333* | .0106797 | .000 | -.188533 | -.120133 | |
| 20 | 0 | .0750000* | .0106797 | .001 | .040800 | .109200 |
| 1 | .0703333* | .0106797 | .001 | .036133 | .104533 | |
| 50 | -.0840000* | .0106797 | .000 | -.118200 | -.049800 | |
| 50 | 0 | .1590000* | .0106797 | .000 | .124800 | .193200 |
| 1 | .1543333* | .0106797 | .000 | .120133 | .188533 | |
| 20 | .0840000* | .0106797 | .000 | .049800 | .118200 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) VAR00002 | (J) VAR00002 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 3C | ||||||
| 0 | 1 | − 6.50000* | 1.69558 | .005 | − 11.2458 | − 1.7542 |
| 20 | − 55.50000* | 1.69558 | .000 | − 60.2458 | − 50.7542 | |
| 50 | − 69.16667* | 1.69558 | .000 | − 73.9125 | − 64.4208 | |
| 1 | 0 | 6.50000* | 1.69558 | .005 | 1.7542 | 11.2458 |
| 20 | − 49.00000* | 1.69558 | .000 | − 53.7458 | − 44.2542 | |
| 50 | − 62.66667* | 1.69558 | .000 | − 67.4125 | − 57.9208 | |
| 20 | 0 | 55.50000* | 1.69558 | .000 | 50.7542 | 60.2458 |
| 1 | 49.00000* | 1.69558 | .000 | 44.2542 | 53.7458 | |
| 50 | − 13.66667* | 1.69558 | .000 | − 18.4125 | − 8.9208 | |
| 50 | 0 | 69.16667* | 1.69558 | .000 | 64.4208 | 73.9125 |
| 1 | 62.66667* | 1.69558 | .000 | 57.9208 | 67.4125 | |
| 20 | 13.66667* | 1.69558 | .000 | 8.9208 | 18.4125 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) concentration | (J) concentration | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 3D | ||||||
| 0 | 1 | − 23.66667* | 3.28549 | .000 | − 32.8625 | − 14.4708 |
| 20 | − 55.50000* | 3.28549 | .000 | − 64.6959 | − 46.3041 | |
| 50 | − 72.16667* | 3.28549 | .000 | − 81.3625 | − 62.9708 | |
| 1 | 0 | 23.66667* | 3.28549 | .000 | 14.4708 | 32.8625 |
| 20 | − 31.83333* | 3.28549 | .000 | − 41.0292 | − 22.6375 | |
| 50 | − 48.50000* | 3.28549 | .000 | − 57.6959 | − 39.3041 | |
| 20 | 0 | 55.50000* | 3.28549 | .000 | 46.3041 | 64.6959 |
| 1 | 31.83333* | 3.28549 | .000 | 22.6375 | 41.0292 | |
| 50 | − 16.66667* | 3.28549 | .000 | − 25.8625 | − 7.4708 | |
| 50 | 0 | 72.16667* | 3.28549 | .000 | 62.9708 | 81.3625 |
| 1 | 48.50000* | 3.28549 | .000 | 39.3041 | 57.6959 | |
| 20 | 16.66667* | 3.28549 | .000 | 7.4708 | 25.8625 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) VAR00002 | (J) VAR00002 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 4A (Mutant TCF) | ||||||
| 0 | 1 | -.01200 | .02143 | .941 | -.0806 | .0566 |
| 20 | -.02933 | .02143 | .550 | -.0980 | .0393 | |
| 50 | -.04333 | .02143 | .257 | -.1120 | .0253 | |
| 1 | 0 | .01200 | .02143 | .941 | -.0566 | .0806 |
| 20 | -.01733 | .02143 | .849 | -.0860 | .0513 | |
| 50 | -.03133 | .02143 | .500 | -.1000 | .0373 | |
| 20 | 0 | .02933 | .02143 | .550 | -.0393 | .0980 |
| 1 | .01733 | .02143 | .849 | -.0513 | .0860 | |
| 50 | -.01400 | .02143 | .911 | -.0826 | .0546 | |
| 50 | 0 | .04333 | .02143 | .257 | -.0253 | .1120 |
| 1 | .03133 | .02143 | .500 | -.0373 | .1000 | |
| 20 | .01400 | .02143 | .911 | -.0546 | .0826 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) VAR00002 | (J) VAR00002 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 4A (TCF) | ||||||
| 0 | 1 | .19167* | .02076 | .000 | .1252 | .2582 |
| 20 | .30900* | .02076 | .000 | .2425 | .3755 | |
| 50 | .46600* | .02076 | .000 | .3995 | .5325 | |
| 1 | 0 | -.19167* | .02076 | .000 | -.2582 | -.1252 |
| 20 | .11733* | .02076 | .002 | .0508 | .1838 | |
| 50 | .27433* | .02076 | .000 | .2078 | .3408 | |
| 20 | 0 | -.30900* | .02076 | .000 | -.3755 | -.2425 |
| 1 | -.11733* | .02076 | .002 | -.1838 | -.0508 | |
| 50 | .15700* | .02076 | .000 | .0905 | .2235 | |
| 50 | 0 | -.46600* | .02076 | .000 | -.5325 | -.3995 |
| 1 | -.27433* | .02076 | .000 | -.3408 | -.2078 | |
| 20 | -.15700* | .02076 | .000 | -.2235 | -.0905 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) VAR00002 | (J) VAR00002 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 4B | ||||||
| 0 | 1 | − 1.92467* | .41532 | .007 | − 3.2547 | -.5947 |
| 20 | − 2.50733* | .41532 | .001 | − 3.8373 | − 1.1773 | |
| 50 | − 6.96000* | .41532 | .000 | − 8.2900 | − 5.6300 | |
| 1 | 0 | 1.92467* | .41532 | .007 | .5947 | 3.2547 |
| 20 | -.58267 | .41532 | .531 | − 1.9127 | .7473 | |
| 50 | − 5.03533* | .41532 | .000 | − 6.3653 | − 3.7053 | |
| 20 | 0 | 2.50733* | .41532 | .001 | 1.1773 | 3.8373 |
| 1 | .58267 | .41532 | .531 | -.7473 | 1.9127 | |
| 50 | − 4.45267* | .41532 | .000 | − 5.7827 | − 3.1227 | |
| 50 | 0 | 6.96000* | .41532 | .000 | 5.6300 | 8.2900 |
| 1 | 5.03533* | .41532 | .000 | 3.7053 | 6.3653 | |
| 20 | 4.45267* | .41532 | .000 | 3.1227 | 5.7827 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) 1 = 0nm,2 = 20nm,3 = 0nm+Wnt5a mAb,4 = 20nm+Wnt5a mAb | (J) 1 = 0nm,2 = 20nm,3 = 0nm+Wnt5a mAb,4 = 20nm+Wnt5a mAb | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 4C | ||||||
| 1 | 2 | .33467* | .03334 | .000 | .2279 | .4414 |
| 3 | .04400 | .03334 | .577 | -.0628 | .1508 | |
| 4 | .16100* | .03334 | .006 | .0542 | .2678 | |
| 2 | 1 | -.33467* | .03334 | .000 | -.4414 | -.2279 |
| 3 | -.29067* | .03334 | .000 | -.3974 | -.1839 | |
| 4 | -.17367* | .03334 | .004 | -.2804 | -.0669 | |
| 3 | 1 | -.04400 | .03334 | .577 | -.1508 | .0628 |
| 2 | .29067* | .03334 | .000 | .1839 | .3974 | |
| 4 | .11700* | .03334 | .033 | .0102 | .2238 | |
| 4 | 1 | -.16100* | .03334 | .006 | -.2678 | -.0542 |
| 2 | .17367* | .03334 | .004 | .0669 | .2804 | |
| 3 | -.11700* | .03334 | .033 | -.2238 | -.0102 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) VAR00002 | (J) VAR00002 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 4D | ||||||
| 1 | 2 | − 2.75433* | .27391 | .000 | − 3.6315 | − 1.8772 |
| 3 | 1.07233* | .27391 | .019 | .1952 | 1.9495 | |
| 4 | -.32100 | .27391 | .659 | − 1.1981 | .5561 | |
| 2 | 1 | 2.75433* | .27391 | .000 | 1.8772 | 3.6315 |
| 3 | 3.82667* | .27391 | .000 | 2.9495 | 4.7038 | |
| 4 | 2.43333* | .27391 | .000 | 1.5562 | 3.3105 | |
| 3 | 1 | − 1.07233* | .27391 | .019 | − 1.9495 | -.1952 |
| 2 | − 3.82667* | .27391 | .000 | − 4.7038 | − 2.9495 | |
| 4 | − 1.39333* | .27391 | .004 | − 2.2705 | -.5162 | |
| 4 | 1 | .32100 | .27391 | .659 | -.5561 | 1.1981 |
| 2 | − 2.43333* | .27391 | .000 | − 3.3105 | − 1.5562 | |
| 3 | 1.39333* | .27391 | .004 | .5162 | 2.2705 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) 1 = NC,2 = + Wnt3a,3 = + Wnt3a mAb,4 = Wnt3a+Wnt3a mAb | (J) 1 = NC,2 = + Wnt3a,3 = + Wnt3a mAb,4 = Wnt3a+Wnt3a mAb | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 4E | ||||||
| 1 | 2 | -.47433* | .04523 | .000 | -.6192 | -.3295 |
| 3 | .17733* | .04523 | .019 | .0325 | .3222 | |
| 4 | .16567* | .04523 | .026 | .0208 | .3105 | |
| 2 | 1 | .47433* | .04523 | .000 | .3295 | .6192 |
| 3 | .65167* | .04523 | .000 | .5068 | .7965 | |
| 4 | .64000* | .04523 | .000 | .4951 | .7849 | |
| 3 | 1 | -.17733* | .04523 | .019 | -.3222 | -.0325 |
| 2 | -.65167* | .04523 | .000 | -.7965 | -.5068 | |
| 4 | -.01167 | .04523 | .994 | -.1565 | .1332 | |
| 4 | 1 | -.16567* | .04523 | .026 | -.3105 | -.0208 |
| 2 | -.64000* | .04523 | .000 | -.7849 | -.4951 | |
| 3 | .01167 | .04523 | .994 | -.1332 | .1565 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) 1 = NC,2 = + Wnt5a,3 = + Wnt5a mAb,4 = Wnt5a+Wnt5a mAb | (J) 1 = NC,2 = + Wnt5a,3 = + Wnt5a mAb,4 = Wnt5a+Wnt5a mAb | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 4F | ||||||
| 1 | 2 | − 3.02767* | .20239 | .000 | − 3.6758 | − 2.3795 |
| 3 | 1.02867* | .20239 | .004 | .3805 | 1.6768 | |
| 4 | -.17100 | .20239 | .832 | -.8191 | .4771 | |
| 2 | 1 | 3.02767* | .20239 | .000 | 2.3795 | 3.6758 |
| 3 | 4.05633* | .20239 | .000 | 3.4082 | 4.7045 | |
| 4 | 2.85667* | .20239 | .000 | 2.2085 | 3.5048 | |
| 3 | 1 | − 1.02867* | .20239 | .004 | − 1.6768 | -.3805 |
| 2 | − 4.05633* | .20239 | .000 | − 4.7045 | − 3.4082 | |
| 4 | − 1.19967* | .20239 | .002 | − 1.8478 | -.5515 | |
| 4 | 1 | .17100 | .20239 | .832 | -.4771 | .8191 |
| 2 | − 2.85667* | .20239 | .000 | − 3.5048 | − 2.2085 | |
| 3 | 1.19967* | .20239 | .002 | .5515 | 1.8478 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) VAR00002 | (J) VAR00002 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 5B | ||||||
| 0 | 5 | -.58300* | .05290 | .000 | -.7571 | -.4089 |
| 10 | -.12600 | .05290 | .197 | -.3001 | .0481 | |
| 30 | -.40433* | .05290 | .000 | -.5784 | -.2302 | |
| 60 | -.13600 | .05290 | .150 | -.3101 | .0381 | |
| 5 | 0 | .58300* | .05290 | .000 | .4089 | .7571 |
| 10 | .45700* | .05290 | .000 | .2829 | .6311 | |
| 30 | .17867* | .05290 | .044 | .0046 | .3528 | |
| 60 | .44700* | .05290 | .000 | .2729 | .6211 | |
| 10 | 0 | .12600 | .05290 | .197 | -.0481 | .3001 |
| 5 | -.45700* | .05290 | .000 | -.6311 | -.2829 | |
| 30 | -.27833* | .05290 | .003 | -.4524 | -.1042 | |
| 60 | -.01000 | .05290 | 1.000 | -.1841 | .1641 | |
| 30 | 0 | .40433* | .05290 | .000 | .2302 | .5784 |
| 5 | -.17867* | .05290 | .044 | -.3528 | -.0046 | |
| 10 | .27833* | .05290 | .003 | .1042 | .4524 | |
| 60 | .26833* | .05290 | .003 | .0942 | .4424 | |
| 60 | 0 | .13600 | .05290 | .150 | -.0381 | .3101 |
| 5 | -.44700* | .05290 | .000 | -.6211 | -.2729 | |
| 10 | .01000 | .05290 | 1.000 | -.1641 | .1841 | |
| 30 | -.26833* | .05290 | .003 | -.4424 | -.0942 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) VAR00002 | (J) VAR00002 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 5C | ||||||
| 0 | 5 | -.20667* | .01588 | .000 | -.2589 | -.1544 |
| 10 | -.39767* | .01588 | .000 | -.4499 | -.3454 | |
| 30 | -.52167* | .01588 | .000 | -.5739 | -.4694 | |
| 60 | -.53733* | .01588 | .000 | -.5896 | -.4851 | |
| 5 | 0 | .20667* | .01588 | .000 | .1544 | .2589 |
| 10 | -.19100* | .01588 | .000 | -.2433 | -.1387 | |
| 30 | -.31500* | .01588 | .000 | -.3673 | -.2627 | |
| 60 | -.33067* | .01588 | .000 | -.3829 | -.2784 | |
| 10 | 0 | .39767* | .01588 | .000 | .3454 | .4499 |
| 5 | .19100* | .01588 | .000 | .1387 | .2433 | |
| 30 | -.12400* | .01588 | .000 | -.1763 | -.0717 | |
| 60 | -.13967* | .01588 | .000 | -.1919 | -.0874 | |
| 30 | 0 | .52167* | .01588 | .000 | .4694 | .5739 |
| 5 | .31500* | .01588 | .000 | .2627 | .3673 | |
| 10 | .12400* | .01588 | .000 | .0717 | .1763 | |
| 60 | -.01567 | .01588 | .855 | -.0679 | .0366 | |
| 60 | 0 | .53733* | .01588 | .000 | .4851 | .5896 |
| 5 | .33067* | .01588 | .000 | .2784 | .3829 | |
| 10 | .13967* | .01588 | .000 | .0874 | .1919 | |
| 30 | .01567 | .01588 | .855 | -.0366 | .0679 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) time | (J) time | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 5D | ||||||
| 0 | 5 | .03000 | .03961 | .937 | -.1004 | .1604 |
| 10 | -.01000 | .03961 | .999 | -.1404 | .1204 | |
| 30 | .00667 | .03961 | 1.000 | -.1237 | .1370 | |
| 60 | -.02333 | .03961 | .974 | -.1537 | .1070 | |
| 5 | 0 | -.03000 | .03961 | .937 | -.1604 | .1004 |
| 10 | -.04000 | .03961 | .845 | -.1704 | .0904 | |
| 30 | -.02333 | .03961 | .974 | -.1537 | .1070 | |
| 60 | -.05333 | .03961 | .671 | -.1837 | .0770 | |
| 10 | 0 | .01000 | .03961 | .999 | -.1204 | .1404 |
| 5 | .04000 | .03961 | .845 | -.0904 | .1704 | |
| 30 | .01667 | .03961 | .992 | -.1137 | .1470 | |
| 60 | -.01333 | .03961 | .997 | -.1437 | .1170 | |
| 30 | 0 | -.00667 | .03961 | 1.000 | -.1370 | .1237 |
| 5 | .02333 | .03961 | .974 | -.1070 | .1537 | |
| 10 | -.01667 | .03961 | .992 | -.1470 | .1137 | |
| 60 | -.03000 | .03961 | .937 | -.1604 | .1004 | |
| 60 | 0 | .02333 | .03961 | .974 | -.1070 | .1537 |
| 5 | .05333 | .03961 | .671 | -.0770 | .1837 | |
| 10 | .01333 | .03961 | .997 | -.1170 | .1437 | |
| 30 | .03000 | .03961 | .937 | -.1004 | .1604 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) VAR00002 | (J) VAR00002 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 5E | ||||||
| 0 | 1 | -.11033* | .02696 | .015 | -.1967 | -.0240 |
| 20 | -.32633* | .02696 | .000 | -.4127 | -.2400 | |
| 50 | -.47333* | .02696 | .000 | -.5597 | -.3870 | |
| 1 | 0 | .11033* | .02696 | .015 | .0240 | .1967 |
| 20 | -.21600* | .02696 | .000 | -.3023 | -.1297 | |
| 50 | -.36300* | .02696 | .000 | -.4493 | -.2767 | |
| 20 | 0 | .32633* | .02696 | .000 | .2400 | .4127 |
| 1 | .21600* | .02696 | .000 | .1297 | .3023 | |
| 50 | -.14700* | .02696 | .003 | -.2333 | -.0607 | |
| 50 | 0 | .47333* | .02696 | .000 | .3870 | .5597 |
| 1 | .36300* | .02696 | .000 | .2767 | .4493 | |
| 20 | .14700* | .02696 | .003 | .0607 | .2333 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) VAR00002 | (J) VAR00002 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 5F | ||||||
| 0 | 1 | -.15167* | .01790 | .000 | -.2090 | -.0943 |
| 20 | -.39267* | .01790 | .000 | -.4500 | -.3353 | |
| 50 | -.51900* | .01790 | .000 | -.5763 | -.4617 | |
| 1 | 0 | .15167* | .01790 | .000 | .0943 | .2090 |
| 20 | -.24100* | .01790 | .000 | -.2983 | -.1837 | |
| 50 | -.36733* | .01790 | .000 | -.4247 | -.3100 | |
| 20 | 0 | .39267* | .01790 | .000 | .3353 | .4500 |
| 1 | .24100* | .01790 | .000 | .1837 | .2983 | |
| 50 | -.12633* | .01790 | .000 | -.1837 | -.0690 | |
| 50 | 0 | .51900* | .01790 | .000 | .4617 | .5763 |
| 1 | .36733* | .01790 | .000 | .3100 | .4247 | |
| 20 | .12633* | .01790 | .000 | .0690 | .1837 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) VAR00002 | (J) VAR00002 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 6C | ||||||
| 0 | 5 | -.08967* | .02012 | .008 | -.1559 | -.0234 |
| 10 | -.37167* | .02012 | .000 | -.4379 | -.3054 | |
| 30 | -.56933* | .02012 | .000 | -.6356 | -.5031 | |
| 60 | -.74133* | .02012 | .000 | -.8076 | -.6751 | |
| 5 | 0 | .08967* | .02012 | .008 | .0234 | .1559 |
| 10 | -.28200* | .02012 | .000 | -.3482 | -.2158 | |
| 30 | -.47967* | .02012 | .000 | -.5459 | -.4134 | |
| 60 | -.65167* | .02012 | .000 | -.7179 | -.5854 | |
| 10 | 0 | .37167* | .02012 | .000 | .3054 | .4379 |
| 5 | .28200* | .02012 | .000 | .2158 | .3482 | |
| 30 | -.19767* | .02012 | .000 | -.2639 | -.1314 | |
| 60 | -.36967* | .02012 | .000 | -.4359 | -.3034 | |
| 30 | 0 | .56933* | .02012 | .000 | .5031 | .6356 |
| 5 | .47967* | .02012 | .000 | .4134 | .5459 | |
| 10 | .19767* | .02012 | .000 | .1314 | .2639 | |
| 60 | -.17200* | .02012 | .000 | -.2382 | -.1058 | |
| 60 | 0 | .74133* | .02012 | .000 | .6751 | .8076 |
| 5 | .65167* | .02012 | .000 | .5854 | .7179 | |
| 10 | .36967* | .02012 | .000 | .3034 | .4359 | |
| 30 | .17200* | .02012 | .000 | .1058 | .2382 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) time | (J) time | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 6D | ||||||
| 0 | 5 | -.23467* | .02028 | .000 | -.3014 | -.1679 |
| 10 | -.82533* | .02028 | .000 | -.8921 | -.7586 | |
| 30 | -.68733* | .02028 | .000 | -.7541 | -.6206 | |
| 60 | -.46233* | .02028 | .000 | -.5291 | -.3956 | |
| 5 | 0 | .23467* | .02028 | .000 | .1679 | .3014 |
| 10 | -.59067* | .02028 | .000 | -.6574 | -.5239 | |
| 30 | -.45267* | .02028 | .000 | -.5194 | -.3859 | |
| 60 | -.22767* | .02028 | .000 | -.2944 | -.1609 | |
| 10 | 0 | .82533* | .02028 | .000 | .7586 | .8921 |
| 5 | .59067* | .02028 | .000 | .5239 | .6574 | |
| 30 | .13800* | .02028 | .000 | .0712 | .2048 | |
| 60 | .36300* | .02028 | .000 | .2962 | .4298 | |
| 30 | 0 | .68733* | .02028 | .000 | .6206 | .7541 |
| 5 | .45267* | .02028 | .000 | .3859 | .5194 | |
| 10 | -.13800* | .02028 | .000 | -.2048 | -.0712 | |
| 60 | .22500* | .02028 | .000 | .1582 | .2918 | |
| 60 | 0 | .46233* | .02028 | .000 | .3956 | .5291 |
| 5 | .22767* | .02028 | .000 | .1609 | .2944 | |
| 10 | -.36300* | .02028 | .000 | -.4298 | -.2962 | |
| 30 | -.22500* | .02028 | .000 | -.2918 | -.1582 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) time | (J) time | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 6E | ||||||
| 0 | 5 | .02467 | .02220 | .797 | -.0484 | .0977 |
| 10 | -.33967* | .02220 | .000 | -.4127 | -.2666 | |
| 30 | -.26100* | .02220 | .000 | -.3341 | -.1879 | |
| 60 | -.37767* | .02220 | .000 | -.4507 | -.3046 | |
| 5 | 0 | -.02467 | .02220 | .797 | -.0977 | .0484 |
| 10 | -.36433* | .02220 | .000 | -.4374 | -.2913 | |
| 30 | -.28567* | .02220 | .000 | -.3587 | -.2126 | |
| 60 | -.40233* | .02220 | .000 | -.4754 | -.3293 | |
| 10 | 0 | .33967* | .02220 | .000 | .2666 | .4127 |
| 5 | .36433* | .02220 | .000 | .2913 | .4374 | |
| 30 | .07867* | .02220 | .034 | .0056 | .1517 | |
| 60 | -.03800 | .02220 | .469 | -.1111 | .0351 | |
| 30 | 0 | .26100* | .02220 | .000 | .1879 | .3341 |
| 5 | .28567* | .02220 | .000 | .2126 | .3587 | |
| 10 | -.07867* | .02220 | .034 | -.1517 | -.0056 | |
| 60 | -.11667* | .02220 | .003 | -.1897 | -.0436 | |
| 60 | 0 | .37767* | .02220 | .000 | .3046 | .4507 |
| 5 | .40233* | .02220 | .000 | .3293 | .4754 | |
| 10 | .03800 | .02220 | .469 | -.0351 | .1111 | |
| 30 | .11667* | .02220 | .003 | .0436 | .1897 | |
| data Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) time | (J) time | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 6F | ||||||
| 0 | 5 | -.43533* | .01941 | .000 | -.4992 | -.3715 |
| 10 | -.83733* | .01941 | .000 | -.9012 | -.7735 | |
| 30 | -.77067* | .01941 | .000 | -.8345 | -.7068 | |
| 60 | -.78267* | .01941 | .000 | -.8465 | -.7188 | |
| 5 | 0 | .43533* | .01941 | .000 | .3715 | .4992 |
| 10 | -.40200* | .01941 | .000 | -.4659 | -.3381 | |
| 30 | -.33533* | .01941 | .000 | -.3992 | -.2715 | |
| 60 | -.34733* | .01941 | .000 | -.4112 | -.2835 | |
| 10 | 0 | .83733* | .01941 | .000 | .7735 | .9012 |
| 5 | .40200* | .01941 | .000 | .3381 | .4659 | |
| 30 | .06667* | .01941 | .040 | .0028 | .1305 | |
| 60 | .05467 | .01941 | .104 | -.0092 | .1185 | |
| 30 | 0 | .77067* | .01941 | .000 | .7068 | .8345 |
| 5 | .33533* | .01941 | .000 | .2715 | .3992 | |
| 10 | -.06667* | .01941 | .040 | -.1305 | -.0028 | |
| 60 | -.01200 | .01941 | .969 | -.0759 | .0519 | |
| 60 | 0 | .78267* | .01941 | .000 | .7188 | .8465 |
| 5 | .34733* | .01941 | .000 | .2835 | .4112 | |
| 10 | -.05467 | .01941 | .104 | -.1185 | .0092 | |
| 30 | .01200 | .01941 | .969 | -.0519 | .0759 | |
| data Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) 1 = 0nm,2 = 20nm,3 = 0nm+Wnt3a mAb,4 = 20nm+Wnt3a mAb,5 = 0nm+Wnt5a mAb,6 = 20nm+Wnt5a mAb | (J) 1 = 0nm,2 = 20nm,3 = 0nm+Wnt3a mAb,4 = 20nm+Wnt3a mAb,5 = 0nm+Wnt5a mAb,6 = 20nm+Wnt5a mAb | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 7B | ||||||
| 1 | 2 | − 53.00000* | 4.10916 | .000 | − 65.4984 | − 40.5016 |
| 3 | -.33333 | 4.10916 | 1.000 | − 12.8317 | 12.1651 | |
| 4 | − 49.50000* | 4.10916 | .000 | − 61.9984 | − 37.0016 | |
| 5 | − 4.50000 | 4.10916 | .879 | − 16.9984 | 7.9984 | |
| 6 | − 13.33333* | 4.10916 | .031 | − 25.8317 | -.8349 | |
| 2 | 1 | 53.00000* | 4.10916 | .000 | 40.5016 | 65.4984 |
| 3 | 52.66667* | 4.10916 | .000 | 40.1683 | 65.1651 | |
| 4 | 3.50000 | 4.10916 | .955 | − 8.9984 | 15.9984 | |
| 5 | 48.50000* | 4.10916 | .000 | 36.0016 | 60.9984 | |
| 6 | 39.66667* | 4.10916 | .000 | 27.1683 | 52.1651 | |
| 3 | 1 | .33333 | 4.10916 | 1.000 | − 12.1651 | 12.8317 |
| 2 | − 52.66667* | 4.10916 | .000 | − 65.1651 | − 40.1683 | |
| 4 | − 49.16667* | 4.10916 | .000 | − 61.6651 | − 36.6683 | |
| 5 | − 4.16667 | 4.10916 | .910 | − 16.6651 | 8.3317 | |
| 6 | − 13.00000* | 4.10916 | .038 | − 25.4984 | -.5016 | |
| 4 | 1 | 49.50000* | 4.10916 | .000 | 37.0016 | 61.9984 |
| 2 | − 3.50000 | 4.10916 | .955 | − 15.9984 | 8.9984 | |
| 3 | 49.16667* | 4.10916 | .000 | 36.6683 | 61.6651 | |
| 5 | 45.00000* | 4.10916 | .000 | 32.5016 | 57.4984 | |
| 6 | 36.16667* | 4.10916 | .000 | 23.6683 | 48.6651 | |
| 5 | 1 | 4.50000 | 4.10916 | .879 | − 7.9984 | 16.9984 |
| 2 | − 48.50000* | 4.10916 | .000 | − 60.9984 | − 36.0016 | |
| 3 | 4.16667 | 4.10916 | .910 | − 8.3317 | 16.6651 | |
| 4 | − 45.00000* | 4.10916 | .000 | − 57.4984 | − 32.5016 | |
| 6 | − 8.83333 | 4.10916 | .290 | − 21.3317 | 3.6651 | |
| 6 | 1 | 13.33333* | 4.10916 | .031 | .8349 | 25.8317 |
| 2 | − 39.66667* | 4.10916 | .000 | − 52.1651 | − 27.1683 | |
| 3 | 13.00000* | 4.10916 | .038 | .5016 | 25.4984 | |
| 4 | − 36.16667* | 4.10916 | .000 | − 48.6651 | − 23.6683 | |
| 5 | 8.83333 | 4.10916 | .290 | − 3.6651 | 21.3317 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) 1 = 0nm,2 = 20nm,3 = 0nm+NSC23766,4 = 20nm+NSC23766,5 = 0nm+Y-27632,6 = 20nm+Y-27632 | (J) 1 = 0nm,2 = 20nm,3 = 0nm+NSC23766,4 = 20nm+NSC23766,5 = 0nm+Y-27632,6 = 20nm+Y-27632 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 7D | ||||||
| 1 | 2 | − 51.00000* | 2.31541 | .000 | − 58.0425 | − 43.9575 |
| 3 | − 4.16667 | 2.31541 | .481 | − 11.2092 | 2.8759 | |
| 4 | − 15.50000* | 2.31541 | .000 | − 22.5425 | − 8.4575 | |
| 5 | − 7.16667* | 2.31541 | .044 | − 14.2092 | -.1241 | |
| 6 | − 11.00000* | 2.31541 | .001 | − 18.0425 | − 3.9575 | |
| 2 | 1 | 51.00000* | 2.31541 | .000 | 43.9575 | 58.0425 |
| 3 | 46.83333* | 2.31541 | .000 | 39.7908 | 53.8759 | |
| 4 | 35.50000* | 2.31541 | .000 | 28.4575 | 42.5425 | |
| 5 | 43.83333* | 2.31541 | .000 | 36.7908 | 50.8759 | |
| 6 | 40.00000* | 2.31541 | .000 | 32.9575 | 47.0425 | |
| 3 | 1 | 4.16667 | 2.31541 | .481 | − 2.8759 | 11.2092 |
| 2 | − 46.83333* | 2.31541 | .000 | − 53.8759 | − 39.7908 | |
| 4 | − 11.33333* | 2.31541 | .000 | − 18.3759 | − 4.2908 | |
| 5 | − 3.00000 | 2.31541 | .785 | − 10.0425 | 4.0425 | |
| 6 | − 6.83333 | 2.31541 | .061 | − 13.8759 | .2092 | |
| 4 | 1 | 15.50000* | 2.31541 | .000 | 8.4575 | 22.5425 |
| 2 | − 35.50000* | 2.31541 | .000 | − 42.5425 | − 28.4575 | |
| 3 | 11.33333* | 2.31541 | .000 | 4.2908 | 18.3759 | |
| 5 | 8.33333* | 2.31541 | .013 | 1.2908 | 15.3759 | |
| 6 | 4.50000 | 2.31541 | .397 | − 2.5425 | 11.5425 | |
| 5 | 1 | 7.16667* | 2.31541 | .044 | .1241 | 14.2092 |
| 2 | − 43.83333* | 2.31541 | .000 | − 50.8759 | − 36.7908 | |
| 3 | 3.00000 | 2.31541 | .785 | − 4.0425 | 10.0425 | |
| 4 | − 8.33333* | 2.31541 | .013 | − 15.3759 | − 1.2908 | |
| 6 | − 3.83333 | 2.31541 | .570 | − 10.8759 | 3.2092 | |
| 6 | 1 | 11.00000* | 2.31541 | .001 | 3.9575 | 18.0425 |
| 2 | − 40.00000* | 2.31541 | .000 | − 47.0425 | − 32.9575 | |
| 3 | 6.83333 | 2.31541 | .061 | -.2092 | 13.8759 | |
| 4 | − 4.50000 | 2.31541 | .397 | − 11.5425 | 2.5425 | |
| 5 | 3.83333 | 2.31541 | .570 | − 3.2092 | 10.8759 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) 1 = 0nm,2 = 20nm,3 = 0nm+Wnt3a mAb,4 = 20nm+Wnt3a mAb,5 = 0nm+Wnt5a mAb,6 = 20nm+Wnt5a mAb | (J) 1 = 0nm,2 = 20nm,3 = 0nm+Wnt3a mAb,4 = 20nm+Wnt3a mAb,5 = 0nm+Wnt5a mAb,6 = 20nm+Wnt5a mAb | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 8B | ||||||
| 1 | 2 | − 71.83333* | 3.48250 | .000 | − 82.4257 | − 61.2410 |
| 3 | 1.00000 | 3.48250 | 1.000 | − 9.5923 | 11.5923 | |
| 4 | − 65.33333* | 3.48250 | .000 | − 75.9257 | − 54.7410 | |
| 5 | − 1.00000 | 3.48250 | 1.000 | − 11.5923 | 9.5923 | |
| 6 | − 9.33333 | 3.48250 | .109 | − 19.9257 | 1.2590 | |
| 2 | 1 | 71.83333* | 3.48250 | .000 | 61.2410 | 82.4257 |
| 3 | 72.83333* | 3.48250 | .000 | 62.2410 | 83.4257 | |
| 4 | 6.50000 | 3.48250 | .441 | − 4.0923 | 17.0923 | |
| 5 | 70.83333* | 3.48250 | .000 | 60.2410 | 81.4257 | |
| 6 | 62.50000* | 3.48250 | .000 | 51.9077 | 73.0923 | |
| 3 | 1 | − 1.00000 | 3.48250 | 1.000 | − 11.5923 | 9.5923 |
| 2 | − 72.83333* | 3.48250 | .000 | − 83.4257 | − 62.2410 | |
| 4 | − 66.33333* | 3.48250 | .000 | − 76.9257 | − 55.7410 | |
| 5 | − 2.00000 | 3.48250 | .992 | − 12.5923 | 8.5923 | |
| 6 | − 10.33333 | 3.48250 | .059 | − 20.9257 | .2590 | |
| 4 | 1 | 65.33333* | 3.48250 | .000 | 54.7410 | 75.9257 |
| 2 | − 6.50000 | 3.48250 | .441 | − 17.0923 | 4.0923 | |
| 3 | 66.33333* | 3.48250 | .000 | 55.7410 | 76.9257 | |
| 5 | 64.33333* | 3.48250 | .000 | 53.7410 | 74.9257 | |
| 6 | 56.00000* | 3.48250 | .000 | 45.4077 | 66.5923 | |
| 5 | 1 | 1.00000 | 3.48250 | 1.000 | − 9.5923 | 11.5923 |
| 2 | − 70.83333* | 3.48250 | .000 | − 81.4257 | − 60.2410 | |
| 3 | 2.00000 | 3.48250 | .992 | − 8.5923 | 12.5923 | |
| 4 | − 64.33333* | 3.48250 | .000 | − 74.9257 | − 53.7410 | |
| 6 | − 8.33333 | 3.48250 | .191 | − 18.9257 | 2.2590 | |
| 6 | 1 | 9.33333 | 3.48250 | .109 | − 1.2590 | 19.9257 |
| 2 | − 62.50000* | 3.48250 | .000 | − 73.0923 | − 51.9077 | |
| 3 | 10.33333 | 3.48250 | .059 | -.2590 | 20.9257 | |
| 4 | − 56.00000* | 3.48250 | .000 | − 66.5923 | − 45.4077 | |
| 5 | 8.33333 | 3.48250 | .191 | − 2.2590 | 18.9257 | |
|
VAR00001 Tukey HSD | ||||||
|
Multiple Comparisons | ||||||
|---|---|---|---|---|---|---|
| (I) 1 = 0nm,2 = 20nm,3 = 0nm+NSC23766,4 = 20nm+NSC23766,5 = 0nm+Y-27632,6 = 20nm+Y-27632 | (J) 1 = 0nm,2 = 20nm,3 = 0nm+NSC23766,4 = 20nm+NSC23766,5 = 0nm+Y-27632,6 = 20nm+Y-27632 | Mean Difference (I-J) | Std. Error | Sig. | 95% Confidence Interval |
|
| Lower Bound | Upper Bound | |||||
| Figure 8D | ||||||
| 1 | 2 | − 78.00000* | 3.68053 | .000 | − 89.1947 | − 66.8053 |
| 3 | − 4.16667 | 3.68053 | .864 | − 15.3613 | 7.0280 | |
| 4 | − 5.50000 | 3.68053 | .670 | − 16.6947 | 5.6947 | |
| 5 | 3.16667 | 3.68053 | .953 | − 8.0280 | 14.3613 | |
| 6 | − 11.00000 | 3.68053 | .056 | − 22.1947 | .1947 | |
| 2 | 1 | 78.00000* | 3.68053 | .000 | 66.8053 | 89.1947 |
| 3 | 73.83333* | 3.68053 | .000 | 62.6387 | 85.0280 | |
| 4 | 72.50000* | 3.68053 | .000 | 61.3053 | 83.6947 | |
| 5 | 81.16667* | 3.68053 | .000 | 69.9720 | 92.3613 | |
| 6 | 67.00000* | 3.68053 | .000 | 55.8053 | 78.1947 | |
| 3 | 1 | 4.16667 | 3.68053 | .864 | − 7.0280 | 15.3613 |
| 2 | − 73.83333* | 3.68053 | .000 | − 85.0280 | − 62.6387 | |
| 4 | − 1.33333 | 3.68053 | .999 | − 12.5280 | 9.8613 | |
| 5 | 7.33333 | 3.68053 | .370 | − 3.8613 | 18.5280 | |
| 6 | − 6.83333 | 3.68053 | .447 | − 18.0280 | 4.3613 | |
| 4 | 1 | 5.50000 | 3.68053 | .670 | − 5.6947 | 16.6947 |
| 2 | − 72.50000* | 3.68053 | .000 | − 83.6947 | − 61.3053 | |
| 3 | 1.33333 | 3.68053 | .999 | − 9.8613 | 12.5280 | |
| 5 | 8.66667 | 3.68053 | .205 | − 2.5280 | 19.8613 | |
| 6 | − 5.50000 | 3.68053 | .670 | − 16.6947 | 5.6947 | |
| 5 | 1 | − 3.16667 | 3.68053 | .953 | − 14.3613 | 8.0280 |
| 2 | − 81.16667* | 3.68053 | .000 | − 92.3613 | − 69.9720 | |
| 3 | − 7.33333 | 3.68053 | .370 | − 18.5280 | 3.8613 | |
| 4 | − 8.66667 | 3.68053 | .205 | − 19.8613 | 2.5280 | |
| 6 | − 14.16667* | 3.68053 | .007 | − 25.3613 | − 2.9720 | |
| 6 | 1 | 11.00000 | 3.68053 | .056 | -.1947 | 22.1947 |
| 2 | − 67.00000* | 3.68053 | .000 | − 78.1947 | − 55.8053 | |
| 3 | 6.83333 | 3.68053 | .447 | − 4.3613 | 18.0280 | |
| 4 | 5.50000 | 3.68053 | .670 | − 5.6947 | 16.6947 | |
| 5 | 14.16667* | 3.68053 | .007 | 2.9720 | 25.3613 | |
|
VAR00001 Tukey HSD | ||||||
The mean difference is significant at the 0.05 level.
References
- 1.Joensuu H., Hohenberger P., Corless C.L. Gastrointestinal stromal tumour. Lancet. 2013;382:973–983. doi: 10.1016/S0140-6736(13)60106-3. [DOI] [PubMed] [Google Scholar]
- 2.Yang H., Shen C., Zhang B., Chen H., Chen Z., Chen J. Expression and clinicopathological significance of CD9 in gastrointestinal stromal tumor. J Korean Med Sci. 2013;28:1443–1448. doi: 10.3346/jkms.2013.28.10.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terada T. Smooth muscles and stem cells of embryonic guts express KIT, PDGFRRA, CD34 and many other stem cell antigens: suggestion that GIST arise from smooth muscles and gut stem cells. Int J Clin Exp Pathol. 2013;6:1038–1045. [PMC free article] [PubMed] [Google Scholar]
- 4.Simon S., Grabellus F., Ferrera L., Galietta L., Schwindenhammer B., Muhlenberg T., Taeger G., Eilers G., Treckmann J., Breitenbuecher F. DOG1 regulates growth and IGFBP5 in gastrointestinal stromal tumors. Cancer Res. 2013;73:3661–3670. doi: 10.1158/0008-5472.CAN-12-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirano K., Shishido-Hara Y., Kitazawa A., Kojima K., Sumiishi A., Umino M., Kikuchi F., Sakamoto A., Fujioka Y., Kamma H. Expression of stem cell factor (SCF), a KIT ligand, in gastrointestinal stromal tumors (GISTs): a potential marker for tumor proliferation. Pathol Res Pract. 2008;204:799–807. doi: 10.1016/j.prp.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Lv A., Li Z., Tian X., Guan X., Zhao M., Dong B., Hao C. SKP2 high expression, KIT exon 11 deletions, and gastrointestinal bleeding as predictors of poor prognosis in primary gastrointestinal stromal tumors. PLoS One. 2013;8:e62951. doi: 10.1371/journal.pone.0062951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubota D., Yoshida A., Tsuda H., Suehara Y., Okubo T., Saito T., Orita H., Sato K., Taguchi T., Yao T. Gene expression network analysis of ETV1 reveals KCTD10 as a novel prognostic biomarker in gastrointestinal stromal tumor. PLoS One. 2013;8:e73896. doi: 10.1371/journal.pone.0073896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinomura Y., Kinoshita K., Tsutsui S., Hirota S. Pathophysiology, diagnosis, and treatment of gastrointestinal stromal tumors. J Gastroenterol. 2005;40:775–780. doi: 10.1007/s00535-005-1674-0. [DOI] [PubMed] [Google Scholar]
- 9.Joensuu H., Eriksson M., Sundby Hall K., Hartmann J.T., Pink D., Schutte J., Ramadori G., Hohenberger P., Duyster J., Al-Batran S.E. One vs three years of adjuvant imatinib for operable gastrointestinal stromal tumor: a randomized trial. JAMA. 2012;307:1265–1272. doi: 10.1001/jama.2012.347. [DOI] [PubMed] [Google Scholar]
- 10.Demetri G.D., Reichardt P., Kang Y.K., Blay J.Y., Rutkowski P., Gelderblom H., Hohenberger P., Leahy M., von Mehren M., Joensuu H. Efficacy and safety of regorafenib for advanced gastrointestinal stromal tumours after failure of imatinib and sunitinib (GRID): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:295–302. doi: 10.1016/S0140-6736(12)61857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miettinen M., Lasota J. Gastrointestinal stromal tumors–definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Arch. 2001;438:1–12. doi: 10.1007/s004280000338. [DOI] [PubMed] [Google Scholar]
- 12.Mucciarini C., Rossi G., Bertolini F., Valli R., Cirilli C., Rashid I., Marcheselli L., Luppi G., Federico M. Incidence and clinicopathologic features of gastrointestinal stromal tumors A population-based study. BMC Cancer. 2007;7:230. doi: 10.1186/1471-2407-7-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y.L., Wang T.H., Hsu H.C., Yuan R.H., Jeng Y.M. Overexpression of CTHRC1 in Hepatocellular Carcinoma Promotes Tumor Invasion and Predicts Poor Prognosis. PloS One. 2013;8:e70324. doi: 10.1371/journal.pone.0070324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu P., Weaver V.M., Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen-Ngoc K.V., Cheung K.J., Brenot A., Shamir E.R., Gray R.S., Hines W.C., Yaswen P., Werb Z., Ewald A.J. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc Natl Acad Sci U S A. 2012;109:E2595–2604. doi: 10.1073/pnas.1212834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pyagay P., Heroult M., Wang Q., Lehnert W., Belden J., Liaw L., Friesel R.E., Lindner V. Collagen triple helix repeat containing 1, a novel secreted protein in injured and diseased arteries, inhibits collagen expression and promotes cell migration. Circ Res. 2005;96:261–268. doi: 10.1161/01.RES.0000154262.07264.12. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto S., Nishimura O., Misaki K., Nishita M., Minami Y., Yonemura S., Tarui H., Sasaki H. Cthrc1 selectively activates the planar cell polarity pathway of Wnt signaling by stabilizing the Wnt-receptor complex. Dev Cell. 2008;15:23–36. doi: 10.1016/j.devcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Park E.H., Kim S., Jo J.Y., Kim S.J., Hwang Y., Kim J.M., Song S.Y., Lee D.K., Koh S.S. Collagen triple helix repeat containing-1 promotes pancreatic cancer progression by regulating migration and adhesion of tumor cells. Carcinogenesis. 2013;34:694–702. doi: 10.1093/carcin/bgs378. [DOI] [PubMed] [Google Scholar]
- 19.Wang P., Wang Y.C., Chen X.Y., Shen Z.Y., Cao H., Zhang Y.J., Yu J., Zhu J.D., Lu Y.Y., Fang J.Y. CTHRC1 is upregulated by promoter demethylation and transforming growth factor-beta1 and may be associated with metastasis in human gastric cancer. Cancer Sci. 2012;103:1327–1333. doi: 10.1111/j.1349-7006.2012.02292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan F., Liu F., Liu H., Hu Y., Liu D., Li G. CTHRC1 is associated with peritoneal carcinomatosis in colorectal cancer: a new predictor for prognosis. Med Oncol. 2013;30:473. doi: 10.1007/s12032-013-0473-3. [DOI] [PubMed] [Google Scholar]
- 21.Joensuu H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum Pathol. 2008;39:1411–1419. doi: 10.1016/j.humpath.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Rutkowski P., Bylina E., Wozniak A., Nowecki Z.I., Osuch C., Matlok M., Switaj T., Michej W., Wronski M., Gluszek S. Validation of the Joensuu risk criteria for primary resectable gastrointestinal stromal tumour - the impact of tumour rupture on patient outcomes. Eur J Surg Oncol. 2011;37:890–896. doi: 10.1016/j.ejso.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z.G., Lambert C., Servotte S., Chometon G., Eckes B., Krieg T., Lapiere C., Nusgens B., Aumailley M. Effects of constitutively active GTPases on fibroblast behavior. Cell Mol Life Sci. 2006;63:82–91. doi: 10.1007/s00018-005-5416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steigen S.E., Eide T.J. Gastrointestinal stromal tumors (GISTs): a review. APMIS. 2009;117:73–86. doi: 10.1111/j.1600-0463.2008.00020.x. [DOI] [PubMed] [Google Scholar]
- 25.Pidhorecky I., Cheney R.T., Kraybill W.G., Gibbs J.F. Gastrointestinal stromal tumors: current diagnosis, biologic behavior, and management. Ann Surg Oncol. 2000;7:705–712. doi: 10.1007/s10434-000-0705-6. [DOI] [PubMed] [Google Scholar]
- 26.Lee E.J., Kang G., Kang S.W., Jang K.T., Lee J., Park J.O., Park C.K., Sohn T.S., Kim S., Kim K.M. GSTT1 copy number gain and ZNF overexpression are predictors of poor response to imatinib in gastrointestinal stromal tumors. PLoS One. 2013;8:e77219. doi: 10.1371/journal.pone.0077219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fletcher C.D., Berman J.J., Corless C., Gorstein F., Lasota J., Longley B.J., Miettinen M., O'Leary T.J., Remotti H., Rubin B.P. Diagnosis of gastrointestinal stromal tumors: A consensus approach. Hum Pathol. 2002;33:459–465. doi: 10.1053/hupa.2002.123545. [DOI] [PubMed] [Google Scholar]
- 28.Fletcher C.D., Berman J.J., Corless C., Gorstein F., Lasota J., Longley B.J., Miettinen M., O'Leary T.J., Remotti H., Rubin B.P. Diagnosis of gastrointestinal stromal tumors: a consensus approach. Int J Surg Pathol. 2002;10:81–89. doi: 10.1177/106689690201000201. [DOI] [PubMed] [Google Scholar]
- 29.Miao C.G., Yang Y.Y., He X., Li X.F., Huang C., Huang Y., Zhang L., Lv X.W., Jin Y., Li J. Wnt signaling pathway in rheumatoid arthritis, with special emphasis on the different roles in synovial inflammation and bone remodeling. Cell Signal. 2013;25:2069–2078. doi: 10.1016/j.cellsig.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Rabelo Fde S., da Mota L.M., Lima R.A., Lima F.A., Barra G.B., de Carvalho J.F., Amato A.A. The Wnt signaling pathway and rheumatoid arthritis. Autoimmun Rev. 2010;9:207–210. doi: 10.1016/j.autrev.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Spano D., Zollo M. Tumor microenvironment: a main actor in the metastasis process. Clin Exp Metastasis. 2012;29:381–395. doi: 10.1007/s10585-012-9457-5. [DOI] [PubMed] [Google Scholar]
- 32.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 33.Zeller J., Turbiak A.J., Powelson I.A., Lee S., Sun D., Showalter H.D., Fearon E.R. Investigation of 3-aryl-pyrimido[5,4-e][1,2,4]triazine-5,7-diones as small molecule antagonists of beta-catenin/TCF transcription. Bioorg Med Chem Lett. 2013;23:5814–5820. doi: 10.1016/j.bmcl.2013.08.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi S.S., Witek R.P., Yang L., Omenetti A., Syn W.K., Moylan C.A., Jung Y., Karaca G.F., Teaberry V.S., Pereira T.A. Activation of Rac1 promotes hedgehog-mediated acquisition of the myofibroblastic phenotype in rat and human hepatic stellate cells. Hepatology. 2010;52:278–290. doi: 10.1002/hep.23649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X., McGann J.C., Liu B.Y., Hannoush R.N., Lill J.R., Pham V., Newton K., Kakunda M., Liu J., Yu C. Phosphorylation of Dishevelled by protein kinase RIPK4 regulates Wnt signaling. Science. 2013;339:1441–1445. doi: 10.1126/science.1232253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Y., Niu C.C., Deng G., Li Z.Q., Pan J., Zhao C., Yang Z.L., Si W.K. The Wnt5a/Ror2 noncanonical signaling pathway inhibits canonical Wnt signaling in K562 cells. Int J Mol Med. 2011;27:63–69. doi: 10.3892/ijmm.2010.560. [DOI] [PubMed] [Google Scholar]
- 37.Wada H., Okamoto H. Roles of noncanonical Wnt/PCP pathway genes in neuronal migration and neurulation in zebrafish. Zebrafish. 2009;6:3–8. doi: 10.1089/zeb.2008.0557. [DOI] [PubMed] [Google Scholar]
- 38.Gao B. Wnt regulation of planar cell polarity (PCP) Curr Top Dev Biol. 2012;101:263–295. doi: 10.1016/B978-0-12-394592-1.00008-9. [DOI] [PubMed] [Google Scholar]
- 39.Miyoshi H., Ajima R., Luo C.T., Yamaguchi T.P., Stappenbeck T.S. Wnt5a potentiates TGF-beta signaling to promote colonic crypt regeneration after tissue injury. Science. 2012;338:108–113. doi: 10.1126/science.1223821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh M. WNT/PCP signaling pathway and human cancer (review) Oncol Rep. 2005;14:1583–1588. [PubMed] [Google Scholar]
- 41.Otey C., Goicoechea S., Garcia-Mata R. Roles of the small GTPases RhoA and Rac1 in cell behavior. F1000 Biol Rep. 2009;1:4. doi: 10.3410/B1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeda K., Kobayashi Y., Udagawa N., Uehara S., Ishihara A., Mizoguchi T., Kikuchi Y., Takada I., Kato S., Kani S. Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat Med. 2012;18:405–412. doi: 10.1038/nm.2653. [DOI] [PubMed] [Google Scholar]
- 43.Kamai T., Yamanishi T., Shirataki H., Takagi K., Asami H., Ito Y., Yoshida K. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin Cancer Res. 2004;10:4799–4805. doi: 10.1158/1078-0432.CCR-0436-03. [DOI] [PubMed] [Google Scholar]
- 44.Deakin N.O., Ballestrem C., Turner C.E. Paxillin and Hic-5 interaction with vinculin is differentially regulated by Rac1 and RhoA. PLoS One. 2012;7:e37990. doi: 10.1371/journal.pone.0037990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chauhan B.K., Lou M., Zheng Y., Lang R.A. Balanced Rac1 and RhoA activities regulate cell shape and drive invagination morphogenesis in epithelia. Proc Natl Acad Sci U S A. 2011;108:18289–18294. doi: 10.1073/pnas.1108993108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill C.S., Wynne J., Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 47.El-Sibai M., Pertz O., Pang H., Yip S.C., Lorenz M., Symons M., Condeelis J.S., Hahn K.M., Backer J.M. RhoA/ROCK-mediated switching between Cdc42- and Rac1-dependent protrusion in MTLn3 carcinoma cells. Exp Cell Res. 2008;314:1540–1552. doi: 10.1016/j.yexcr.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nethe M., Hordijk P.L. The role of ubiquitylation and degradation in RhoGTPase signalling. J Cell Sci. 2010;123:4011–4018. doi: 10.1242/jcs.078360. [DOI] [PubMed] [Google Scholar]
- 49.Patel S., Takagi K.I., Suzuki J., Imaizumi A., Kimura T., Mason R.M., Kamimura T., Zhang Z. RhoGTPase activation is a key step in renal epithelial mesenchymal transdifferentiation. J Am Soc Nephrol. 2005;16:1977–1984. doi: 10.1681/ASN.2004110943. [DOI] [PubMed] [Google Scholar]
- 50.Bravo-Cordero J.J., Moshfegh Y., Condeelis J., Hodgson L. Live cell imaging of RhoGTPase biosensors in tumor cells. Methods Mol Biol. 2013;1046:359–370. doi: 10.1007/978-1-62703-538-5_22. [DOI] [PMC free article] [PubMed] [Google Scholar]