Abstract
The high-mobility group–box transcription factor sex-determining region Y–box 2 (Sox2) is essential for the maintenance of stem cells from early development to adult tissues. Sox2 can reprogram differentiated cells into pluripotent cells in concert with other factors and is overexpressed in various cancers. In glioblastoma (GBM), Sox2 is a marker of cancer stemlike cells (CSCs) in neurosphere cultures and is associated with the proneural molecular subtype. Here, we report that Sox2 expression pattern in GBM tumors and patient-derived mouse xenografts is not restricted to a small percentage of cells and is coexpressed with various lineage markers, suggesting that its expression extends beyond CSCs to encompass more differentiated neoplastic cells across molecular subtypes. Employing a CSC derived from a patient with GBM and isogenic differentiated cell model, we show that Sox2 knockdown in the differentiated state abolished dedifferentiation and acquisition of CSC phenotype. Furthermore, Sox2 deficiency specifically impaired the astrocytic component of a biphasic gliosarcoma xenograft model while allowing the formation of tumors with sarcomatous phenotype. The expression of genes associated with stem cells and malignancy were commonly downregulated in both CSCs and serum-differentiated cells on Sox2 knockdown. Genes previously shown to be associated with pluripontency and CSCs were only affected in the CSC state, whereas embryonic stem cell self-renewal genes and cytokine signaling were downregulated, and the Wnt pathway activated in differentiated Sox2-deficient cells. Our results indicate that Sox2 regulates the expression of key genes and pathways involved in GBM malignancy, in both cancer stemlike and differentiated cells, and maintains plasticity for bidirectional conversion between the two states, with significant clinical implications.
Abbreviations: α-SMA, actin alpha 2 smooth muscle (ACTA2); CNS, central nervous system; CSC, cancer stem-like cell; DEG, differentially expressed gene; ESC, embryonic stem cell; GFAP, glial fibrillary acidic protein; GBM, glioblastoma; GS, gliosarcoma; IF, intermediate filament protein; IHC, immunohistochemistry; NM, neurosphere media; NSC, neural stem cell; Sox2, SRY (sex determining region Y)-box 2; SVZ, subventricular zone; SDC, serum differentiated cell; TCGA, the Cancer Genome Atlas; TMA, tissue microarray; WHO, World Health Organization
Introduction
In a variety of tumor types, a worse prognosis is associated with increased plasticity, loss of differentiation makers, and induction of stem cell phenotypes [1]. One key regulator of stemness is the high-mobility group (HMG)–box transcription factor sex-determining region Y (SRY)–box 2 (Sox2), essential in early embryonic development [2]. Sox2 activity is critical for the maintenance of mouse embryonic stem cells (ESCs) [3] and is associated with the promoter region of 7% of protein-coding genes in human ESCs [4]. Sox2 activates and represses the expression of different gene sets in various tissues during development through HMG-box domain-mediated DNA binding. Cooperative binding of other transcription factors contributes to target specificity [5]. Sox2 is highly expressed in neural stem cells (NSC), neural progenitors, and immature astrocytes in the subventricular zone (SVZ) of developing rodent brains and is required for maintenance and identity of NSC in the adult mouse brain [6]. Sox2 is re-expressed in astrocytes undergoing cell division in the mouse neocortex in response to brain injury [7]. In the adult human brain, Sox2 expression is restricted to regions with neurogenic potential, such as the SVZ [8]. Outside of normal development, Sox2 is an essential component of the transcription factor cocktail for reprogramming differentiated cells into iPS cells [9]. SOX2 gene is amplified or otherwise overexpressed in several tumor types [5]. Transcriptional regulation by Sox2 is context dependent, and its impact on malignancy is cancer-type specific. For example, Sox2 functions as an oncogene in lung and esophagus squamous cell carcinomas [10], whereas in ovarian cancer, its expression is restricted to cancer stemlike cells (CSCs) [11].
Glioblastoma (GBM), a WHO grade IV astrocytoma, is the most aggressive primary central nervous system tumor. Sox2 transcript and protein are upregulated in GBM tumors in relation to nontumor brain tissues [12,13], gene amplification is observed in 4% of GBM tumors profiled by The Cancer Genome Atlas (TCGA) [14], and promoter hypomethylation occurs with high frequency [12]. Sox2 is a component of the GBM proneural molecular subclass signature, along with other regulators of neural stem/progenitor cell fate [15]. Notable phenotypic plasticity of GBMs contributes to intratumor heterogeneity, encompassing cells in a range of developmental states, from CSCs to more differentiated neoplastic cells. These various developmental states can be partially captured ex vivo. Culturing GBM cells in serum-free medium supplemented with epidermal growth factor (EGF) and fibroblast growth factor (FGF) developed for NSCs allows the selection and propagation of neurospheres, a population enriched in CSC, defined by the ability to self-renew and differentiate into cells forming the bulk of the tumor, phenocopying the parental tumor in mouse orthotopic xenografts [16,17]. Sox2, ubiquitously expressed in GBM neurosphere cell cultures [16–21], has been shown to function in the proliferation of GBM CSC population and tumorigenicity [20]. Neurosphere cultures derived from adult mouse SVZ also express Sox2, and differentiation induced by FBS is accompanied by down-regulation of Sox2 mRNA and protein [22]. Similarly, culturing dissociated GBM tumors in FBS-containing media typically results in loss of the CSC phenotype and impaired tumorigenicity associated with down-regulation of NSC markers, including Sox2, and gain of more mature astrocytic markers [16,17,19].
Here, we verify that the endogenous expression of Sox2 in GBM biopsies and xenograft tumors is more widespread than expected for a marker of CSCs, raising the question as to whether in GBM tumors Sox2 performs distinct functions in differentiated and CSCs. Although few long-term serum-cultured GBM cells can regain expression of Sox2 [23], these cells accumulate genomic abnormalities leading to considerably divergence from the original tumor [16]. We have previously described a patient-derived low -passage serum-differentiated cell (SDC) line, which, contrary to typical GBM samples, retains Sox2 expression, along with the ability to dedifferentiate into neurospheres in vitro and to phenocopy the parental tumor in orthotopic xenografts [19]. These SDCs and CSCs originating from the same tumor specimen [19] constitute an adequate model to study Sox2 function in the two developmental states under an isogenic background. Gliosarcoma (GS), a WHO histologic variant [24] comprising approximately 1.8% to 2.8% of all GBMs, is characterized by biphasic malignant glial and sarcomatous components, resulting from mesenchymal metaplasia [25]. GSs are not distinguished from GBMs regarding clinical management and large-scale molecular profiling efforts, such as the TCGA, and are assigned to multiple molecular subclasses [26]. The availability of a GS model [19] presented an opportunity to contrast Sox2 functions in the glial and mesenchymal compartments, further highlighting its role in astrocytic tumorigenicity.
Materials and Methods
Tumor Samples and Cell Culture
Resected brain tumors were collected at Henry Ford Hospital (Detroit, MI) with written consent from patients in accordance with institutional guidelines and graded pathologically according to the WHO criteria. GBM tumors were dissociated, as previously described [27]. Dissociated cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies) supplemented with 10% FBS (HyClone) for traditional monolayer cultures or in neurosphere medium (NM), composed of DMEM/F-12 supplemented with N2 (Gibco), 0.5 mg/ml BSA (Sigma), 25 μg/ml gentamicin (Gibco), 0.5% antibiotic/antimycotic (Invitrogen), 20 ng/ml basic fibroblast growth factor, and 20 ng/ml EGF (Peprotech). Cells were maintained in culture for up to passage 10 (low passage).
Immunohistochemistry
Immunohistochemistry was performed as described previously [19]. The following primary antibodies were used: anti-Sox2 (Cell Signaling Technology), anti–glial fibrillary acidic protein (anti-GFAP) (Abcam), anti-nestin (Millipore), anti-vimentin (Santa Cruz Biotechnology), anti–α-smooth muscle actin (anti–α-SMA) (Abcam), and anti–major histocompatibility complex I (anti-MHC I) (Abcam). Anti-Sox2 antibody specificity was verified by the absence of signal on Sox2 knockdown, as shown in Figure 4. Reticulin staining was performed with a kit (DakoCytomation). Images were captured using Nikon E800M microscope and DXM1200C digital camera. For immunofluorescence, secondary antibodies conjugated to DyLight 488 and DyLight 649 (Jackson ImmunoResearch Laboratories) were employed, nuclei were stained with DAPI (Invitrogen), and images captured using Nikon Eclipse 80i microscope equipped with epifluorescence.
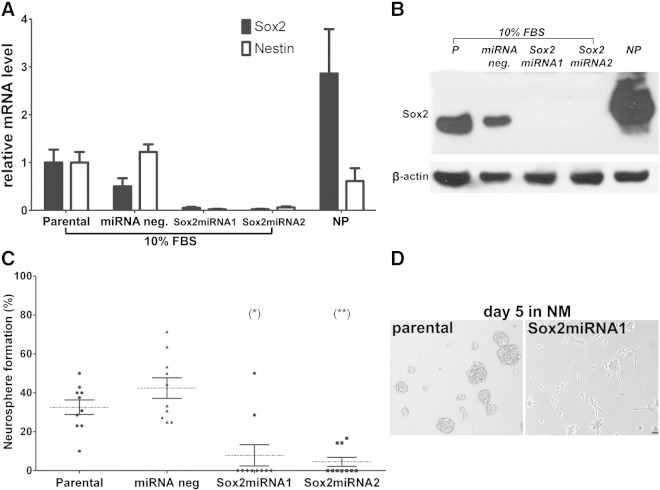
Figure 4.
Sox2 knockdown prevents dedifferentiation of HF2303 GBM cells. (A) Sox2 and nestin mRNA levels in HF2303 cells (parental) and cells transfected with miRNA constructs targeting Sox2 (Sox2miRNA1 and Sox2miRNA2) or nonsilencing miRNA (miRNAneg) was evaluated by quantitative RT-PCR. HF2303 neurosphere cells (NP) were included for comparison. Results represent means ± SEM; n = 6. (B) Sox2 protein expression in whole-cell lysates determined by Western blot analysis confirms Sox2 knockdown in Sox2miRNA1 and Sox2miRNA2 lines. β-Actin was used as loading control. (C) The effect of Sox2 knockdown on dedifferentiation of HF2303 monolayers into neurospheres. Cells were seeded at low density in 96-well plates and cultured in NM. Neurosphere formation was monitored and quantified at day 12. Results represent means ± SEM, for n = 10 (*P < .05 and **P < .01), in relation to parental cells after Dunn multiple comparison test. (D) Representative images contrast Sox2-expressing and Sox2-deficient cells in NM culture. Bar, 20 μm.
Sox2 Knockdown and Ectopic Expression
Sox2 knockdown in low-passage 10% FBS cells was achieved by oligonucleotides targeting human SOX2-coding or nonsilencing control sequences cloned into BLOCK-iT Pol II miR RNAi expression vectors (Life Technologies) before cell transfection with Lipofectamine 2000 (Life Technologies). Sox2 knockdown in neurospheres was achieved using GIPZ Lentiviral shRNAmir (clones V3LHS_404430 and V3LHS_404432) and nonsilencing control (RHS4346) (Thermo Scientific Open Biosystems). Sox2 ectopic expression was achieved by subcloning Sox2 cDNA from pCMV6-XL5-NM_002106.2 (OriGene) into pcDNA 3.1 mammalian expression vector (Invitrogen), under control of constitutive CMV promoter; the empty vector was used as control. Plasmid DNA constructs were verified by DNA sequencing and stably transfected into GBM monolayer cells using Lipofectamine 2000 (Invitrogen).
Neurosphere Formation Assay
Monolayer cells were harvested by trypsin treatment, washed three times in serum-free DMEM/F-12 medium, and plated in NM in regular tissue culture–treated flasks at a density of 10 cells per well. Neurosphere formation was monitored for 2 weeks, and neurospheres containing more than 32 cells were counted.
Cell Proliferation Assay
Cells were harvested and plated onto 96-well plates (500 cells per well for 10% FBS cultures and 1000 cells per well for neurosphere cultures). Cell viability was quantified for 7 days by intracellular ATP level measurements using CellTiter-Glo Luminescent Cell Viability Assay (Promega).
Xenograft Tumors
Following IACUC guidelines in an institutionally approved animal use protocol, dissociated GBM cells were implanted into 8-week-old female nude mice (NCRNU; Taconic Farms). Animals were anesthetized with a mixture of ketamine and xylazine. The injection site was manually drilled 2.5 mm to the right of the bregma and 1 mm posterior to the coronal suture. Dissociated cells (3 × 105 per mouse) were injected at a depth of 3 mm using a Hamilton syringe. Animals were monitored daily and killed on the first signs of neurologic deficit or weight loss greater than 20%. Xenograft tissue microarray (TMA) slides were obtained through participation in the Ivy GBM consortium.
Proteasome Inhibition
Exponentially growing cells were plated at a density of 1 × 105 cells/ml in growth medium. After 24 hours, medium was replaced with growth medium containing 10 μM MG132 (Sigma). Cells were incubated for 4 to 24 hours at 37°C (5% CO2) before whole-cell lysates were obtained.
Western Blot Analysis
Fifteen micrograms of total protein from whole-cell lysates were denatured in sodium dodecyl sulfate–gel loading buffer and separated by NuPAGE SDS-PAGE gel system (Life Technologies). Proteins were electrophoretically transferred to PVDF membranes (Life Technologies). Primary antibodies used were Sox2 (Chemicon) and β-actin (Sigma-Aldrich). Immobilon Western Chemiluminescent HRP substrate (Millipore) was used for detection.
Quantitative Real-Time Polymerase Chain Reaction Analysis
Total RNA was extracted with RNeasy Mini Kit (Qiagen), eluted in RNase-free water, and stored at − 80°C. cDNA was prepared from 1 μg of DNAse I–treated RNA using SuperScript III and oligo(dT) (Invitrogen). Relative quantification of gene expression was performed by real-time polymerase chain reaction (PCR) with SYBR Green and ViiA 7 detection system (Applied Biosystems). β-Actin was used as internal reference, and relative mRNA levels were quantified by the 2(− ΔΔCt) method. DNA oligo sequences are available on request.
Global Gene Expression Analysis
RNA samples were analyzed by Illumina Human HT12v4.0 Expression BeadChip, at the Wayne State University Applied Genomics Technology Center. The gene expression image files obtained from the Illumina iScan were uploaded to GenomeStudio (version 2010.3) using the Gene Expression module. Illumina array data were normalized using the rank invariant method, and the background was subtracted in BeadStudio. Normalization artifacts of negative expression values were set to a minimum null value. A single probe with the largest expression range across the samples was selected for genes with multiple probes. Fold change was calculated on the basis of the mean values for Sox2-expressing cells (HF2303 parental and miRNAneg) and Sox2-deficient cells (Sox2miR1 and Sox2miR2). Genes with mean Illumina intensity levels lower than 20 for both groups were excluded. A stringent fold-change cutoff of 3.5 was used to generate the differentially expressed gene (DEG) lists. Global gene expression data files are available at GEO. MetaCore (Thomson Reuters) and IPA (Ingenuity Systems) software applications were used to further analyze the data sets. Microarray data are available through GEO (GSE51441).
TCGA Data Analysis
Preprocessed level 2 Agilent Human Gene Expression Microarray data were obtained from the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/, July 2, 2012) for 517 GBM cases with no prior cancer diagnoses. Analysis was conducted at probe level. Pearson correlation coefficient between each of two Sox2 open reading frame probes (A_23_P159606, A_24_0379969, r = 0.96) and all other probes was conducted. A gene was considered to have consistent correlation if it showed correlation of ρ > 0.4 (positive) or ρ < − 0.4 (negative) for at least 50% of its probes with either of the Sox2 probes. For visualization, consistently correlated probes were z score transformed and averaged to give a single value per gene. Hierarchical clustering of the genes was performed using the Pearson correlation and complete linkage. Molecular class was predicted from the metagene signatures for the four classes. Analysis was performed using R Software.
Statistical Analysis
Results are represented as means ± SEM. Differences were analyzed by unpaired two-tailed Student's t test or one-way analysis of variance using Prism 5 (GraphPad Software). For Figures 4C and W1C, Dunn multiple-comparison test was used. ANCOVA was used for age-adjusted tests of means, and Cox regression was used for survival analysis (R Software). P < .05 was considered significant.
Results
Sox2 is Widely Expressed in GBM Tumors
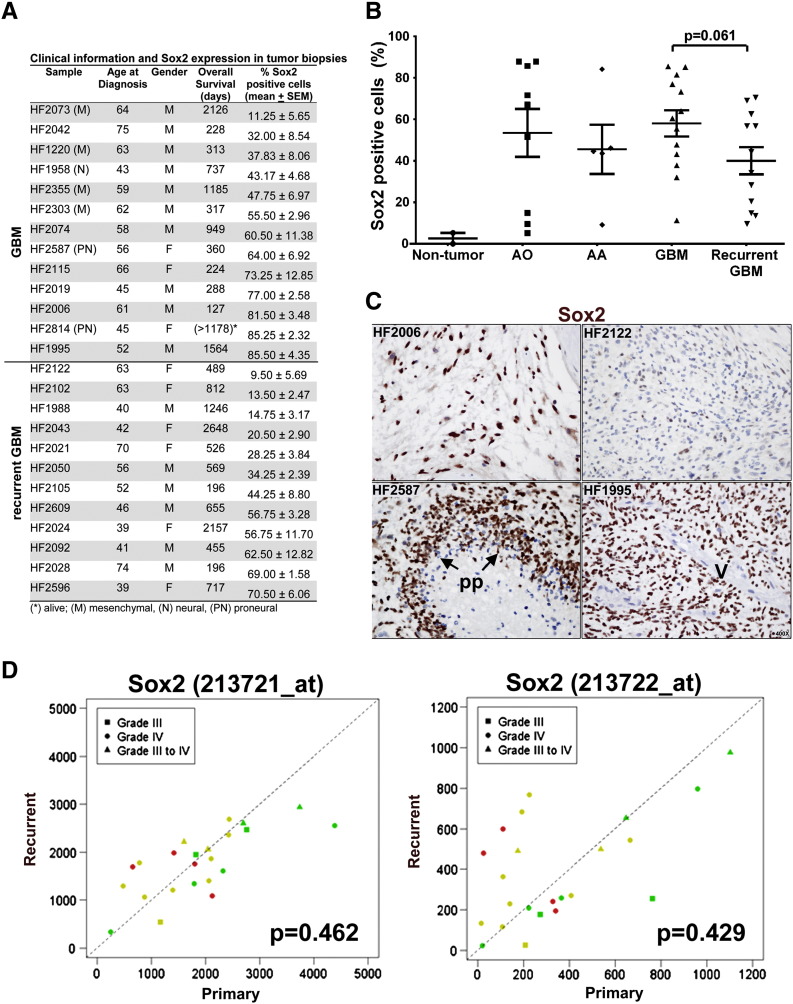
Sox2 protein expression was assessed in newly diagnosed (n = 13) and recurrent (n = 12) GBMs and in grade III anaplastic oligodendroglioma (n = 9) and anaplastic astrocytomas (n = 5). The percentage of Sox2-positive cells in GBM tumors ranged from 9% to 85% (Figure 1, A and B). Sox2 was similarly highly expressed in the grade III gliomas (Figure 1B), in agreement with previous reports [28,29]. Representative images are shown in Figure 1C. The subclass assignment [15] was available for seven GBM samples (Figure 1A). In the proneural samples, 64% and 85% of cells were positive for Sox2 (Figure 1A), consistent with Sox2 being a component of the proneural subclass signature [15]. Sox2-positive cells ranged from 11.2% to 55.5% in the mesenchymal tumors. An 18% decrease in the mean percentage of Sox2-positive cells in the recurrent GBM group relative to newly diagnosed was observed (P = .0609; 95% confidence interval = − 0.9%- 36.9%) (Figure 1B). When we adjusted for patient age at diagnosis (Figure 1A), a significant decrease in Sox2 expression was observed in the recurrent tumor group (21.7%, ANCOVA P = .0266; 95% confidence interval = 2.8%- 40.6%). To further investigate how Sox2 expression changes on recurrence, transcriptome data for 23 paired primary and recurrent grade III and grade IV gliomas [30] were analyzed. Sox2 expression obtained from two Affymetrix U133 probes were not significantly different between primary and recurrent GBMs (213721_at, P = .4618; 213722_at, P = .4293) (Figure 1D). Sox2 expression tended to decrease on recurrence for the younger patients and for the grade III tumors (Figure 1D).
Figure 1.
Sox2 expression in GBM tumors. Sox2 expression in 25 GBM biopsy specimens was assessed by immunohistochemistry. Sox2-positive nuclei were quantified in four randomly selected × 400 optical fields per specimen. (A) Clinical information. (B) Scatterplot represents the percentage of Sox2-positive cells for each specimen (mean ± SEM). Anaplastic oligodendroglioma (AO; n = 9), anaplastic astrocytoma (AA; n = 5), and nontumor brain (n = 2) samples are included for reference. (C) Representative images show nuclear Sox2 expression in GBM biopsies; pseudopalisating necrosis (pp) and vessels (V) are indicated. (D) Graphical representation of Sox2 mRNA expression in 23 paired primary (horizontal axis) and recurrent (vertical axis) gliomas. Square, WHO grade III primary and recurrent gliomas (n = 3); circle, primary and recurrent (grade IV) GBMs (n = 16); triangle, primary grade III and recurrent GBM (n = 4). Patients grouped by age (years): Green, < 40; yellow, 40 to 60; red, > 60.
Together, these observations indicate that Sox2 expression is not restricted to a small subpopulation of undifferentiated neoplastic cells, suggesting that Sox2 could regulate different programs in CSCs and more differentiated GBM cells.
Sox2 Expression is Conserved in Patient-Derived GBM Orthotopic Xenografts
To enrich for cells with CSC phenotype, neurosphere cultures from two newly diagnosed GBMs (HF2303 and HF2587) and two recurrent GBMs (HF2609 and HF2596) were implanted intracranially in immunocompromised mice. Sox2 was expressed in 55% to 70% of the nuclei for these biopsies (Figures 1 A and 2A). As expected, matched neurosphere cells were highly enriched in Sox2-positive cells, and biopsy expression pattern was replicated in the xenograft tumors (Figure 2A).
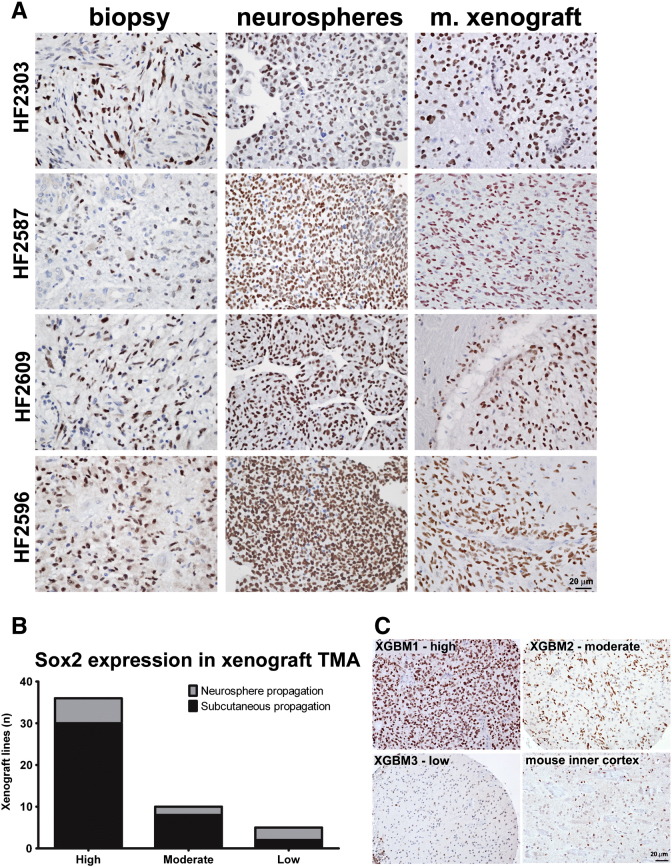
Figure 2.
Sox2 expression in GBM neurospheres and orthotopic xenografts. (A) Sox2 expression in four sets of matched GBM biopsy, cultured neurosphere cells, mouse xenograft, two newly diagnosed (HF2303 and HF2587), and two recurrent tumors (HF2609 and HF2596). (B) Sox2-positive cells were quantified in TMA containing samples of xenograft tumors obtained from 51 GBM specimens propagated in vitro as neurosphere cultures or subcutaneously in nude mouse flanks, before intracranial implant in nude mice. Xenograft lines from subcutaneous and neurosphere propagation were ranked for Sox2 expression as high (> 65% positive cells), moderate (between 20% and 65% positive cells), and low (< 20% cells). (C) Representative images for each xenograft category in B and normal mouse brain (inner cortex). Scale, 20 μm.
Sox2 expression was then evaluated in a TMA composed of samples from 51 mouse orthotopic GBM xenografts, including 40 originating from patient tumors propagated subcutaneously and 11 from neurosphere cultures. As shown in Figure 2B, 36 (71%) of the xenograft lines presented high percentage (> 65%) of Sox2-positive nuclei, whereas only 5 (9.8%) of the xenografts displayed low (< 20%) Sox2-positive nuclei. Representative images are shown (Figure 2C). These results indicate that robust Sox2 protein expression observed for the majority of the patient-derived orthotopic xenograft tumors is independent of the use of neurosphere culture or subcutaneous tumors as the preimplantation propagation method.
The intermediate filament (IF) protein nestin is a direct target of Sox2 regulation in neural stem and progenitor cells [31] and also commonly expressed in a large percentage of cells in GBM tumors [19]. As expected, in neurosphere-derived GBM xenografts, Sox2 coexpressed with nestin (Figure 3A). Vimentin, the main IF protein of mesenchymal cells, also expressed in neural stem and progenitor cells, was coexpressed with Sox2 in the GBM xenografts (Figure 3A). GFAP, a major IF of mature astrocytes, also expressed in astrocytic progenitors, coexpressed with Sox2 (Figure 3A), as previously reported [32]. Sox2 coexpression with IF proteins markers of diverse lineages is in agreement with Sox2 not being restricted to undifferentiated tumor cells, underlying the plasticity of high-grade astrocytomas.
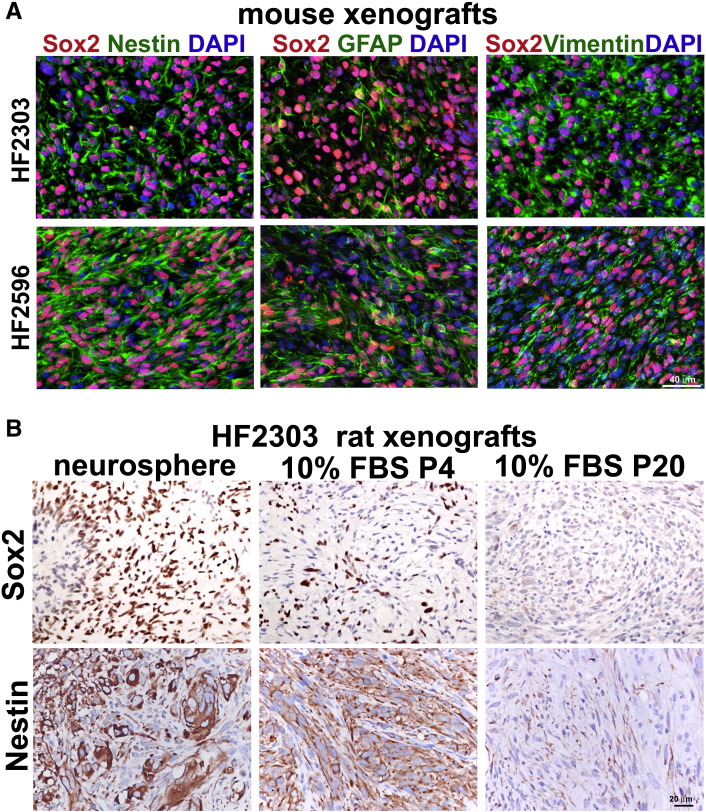
Figure 3.
Sox2 is coexpressed with GFAP, nestin, and vimentin in neurosphere-derived GBM xenografts. (A) Coexpression of Sox2 (red) with the IF proteins (green) nestin, GFAP, and vimentin in HF2303 and HF2596 GBM mouse xenografts. Nuclei counterstained with DAPI. Scale, 40 μm. (B) Sox2 and nestin are expressed in xenografts formed from HF2303 neurospheres or low-passage 10% FBS cultures (P4) and downregulated in higher passage (P20) 10% FBS serum cultures. Scale, 20 μm.
Whereas Sox2 is downregulated in the majority of GBM cells exposed to 10% FBS, we have previously characterized a GBM specimen, HF2303 (GS1), which retains Sox2 expression in low-passage SDCs [19]. Here, we show that Sox2 expression in xenograft tumors are consistent with the in vitro expression: Sox2-positive cells are observed in xenografts derived from HF2303 neurosphere/CSCs and low-passage SDCs (10% FBS P4) and not in higher passage SDCs (10% FBS P20, Figure 3B). Nestin expression in the tumors follows the Sox2 pattern (Figure 3B).
Sox2 Function in Plasticity of Differentiated GBM Cells
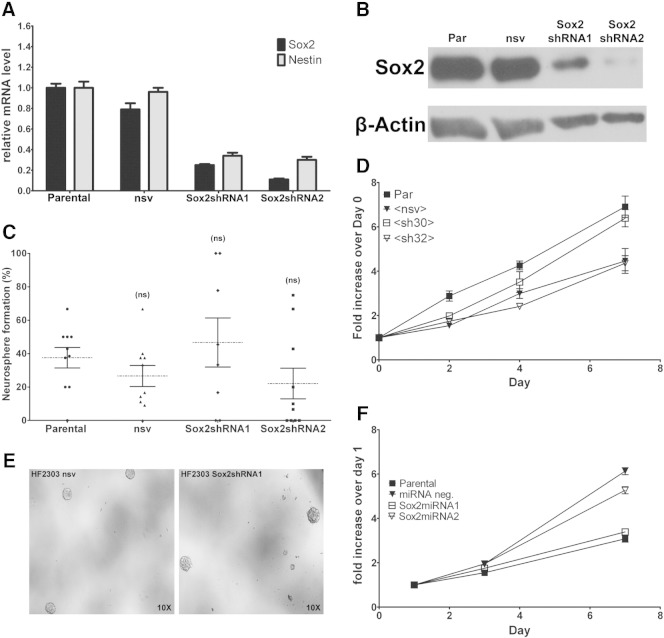
To study Sox2 function in differentiated patient-derived GBM cells, low-passage HF2303 SDCs were transfected with miRNA constructs targeting the Sox2-coding region (Sox2miRNA1 and Sox2miRNA2) or with a nonsilencing control (miRNAneg). Sox2 knockdown in HF2303_Sox2miRNA1 and HF2303_Sox2miRNA2 was verified by quantitative PCR (Figure 4A) and Western blot analysis (Figure 4B). Knocking down Sox2 expression in HF2303 cells significantly decreased the level of nestin transcript, a direct target of Sox2 (Figure 4A). Expression of Sox2 in isogenic HF2303 neurospheres (NP) are shown for comparison (Figure 4, A and B). We have previously demonstrated that HF2303 SDCs not only retain Sox2 expression but also the ability to dedifferentiate, forming neurospheres in a clonogenic assay [19]. Sox2 knockdown significantly impaired neurosphere formation in NM (Figure 4, C and D), whereas no effect in proliferation was observed (Figure W1).
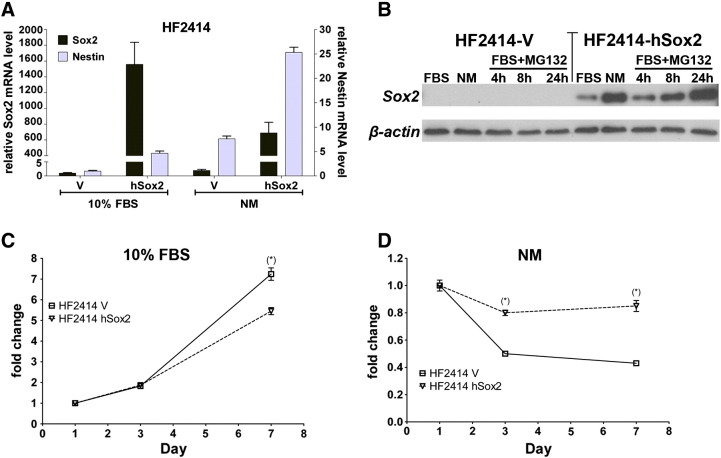
Sox2 Expression is not Sufficient to Restore Plasticity and Tumorigenicity to Serum-Cultured Primary GBM Cells
We have previously described neurosphere/CSCs and SDCs from another GBM specimen (HF2414/GBM2) [19], demonstrating that the CSCs express Sox2 and are tumorigenic, whereas the low-passage SDCs lose Sox2 expression, along with the ability to dedifferentiate in vitro, and to form tumors [19], typical observations for GBM SDCs [16]. To determine whether Sox2 ectopic expression was sufficient to restore plasticity and tumorigenicity to HF2414 SDCs, cells were stably transfected with Sox2 cDNA construct. Detectable levels of Sox2 protein expression were only attained in clones expressing very high levels of Sox2 transcript (HF2414-hSox2) in relation to clones expressing the empty vector (HF2414 V) (Figure 5, A and B). Endogenous Sox2 expression was negligible in HF2414 V cells and was not upregulated when cells were cultured in serum-free NM for 48 hours (Figure 5, A and B). Nestin mRNA was upregulated on ectopic Sox2 expression and further upregulated when the cells were transferred to NM (Figure 5A). The discrepancy between mRNA and protein levels (Figure 5, A and B) suggests that the posttranslational regulation of Sox2 expression is affected by culture conditions, because increased protein levels were observed when HF2414-hSox2 was transferred from 10% FBS to NM for 48 hours (Figure 5B), which was not mediated by an increase in mRNA expression (Figure 5A). Treatment with proteasome inhibitor MG132 (10 μM) increased Sox2 protein levels in HF2414-hSox2 cells (Figure 5B). Although predicted ubiquitin-binding sites exist in Sox2 (http://ubpred.org), we found no evidence of direct ubiquitination of Sox2 (not shown). Sox2 protein was not detected in HF2414-V cells, cultured under the same conditions (Figure 5B), in agreement with irreversible loss of endogenous Sox2 commonly observed for GBM SDCs. Ectopic expression of Sox2 had a negative effect in HF2414 cell proliferation in 10% FBS (Figure 5C) and was not sufficient to induce CSC phenotype or restore tumorigenicity to HF2414 SDCs (not shown) but led to significant increase in cell viability in NM (Figure 5D).
Figure 5.
Sox2 ectopic expression in low-passage primary GBM monolayer line HF2414. HF2414 cells constitutively expressing Sox2 (HF2414-hSox2) or the empty vector (HF2414-V) were maintained in 10% FBS culture or transferred to NM for 48 hours. (A) Sox2 and nestin mRNA levels were determined by quantitative RT-PCR. Results represent means ± SEM, for n = 6. (B) Sox2 protein expression in whole-cell lysates was determined by Western blot analysis. Sox2 protein was detected in the HF2414-hSox2 grown in serum, and protein expression was increased when cells were transferred to NM or when proteasome inhibitor MG132 was added to the 10% FBS serum containing medium in a time-dependent manner. No Sox2 protein was observed in the HF2414-V cell line under the same culture conditions. (C) Cell proliferation in 10% FBS medium measured by CellTiterGlo was decreased for HF2414-hSox2 cells relative to HF2414-V cells. Mean ± SE (n = 10) is shown in the graph (*P < .05). (D) When the cells were plated in low density in NM, no neurospheres formed, and there was a reduction in cell viability for 7 days, significantly more accentuated for HF2414-V. Mean ± SE (n = 10) is shown in the graph (*P < .05).
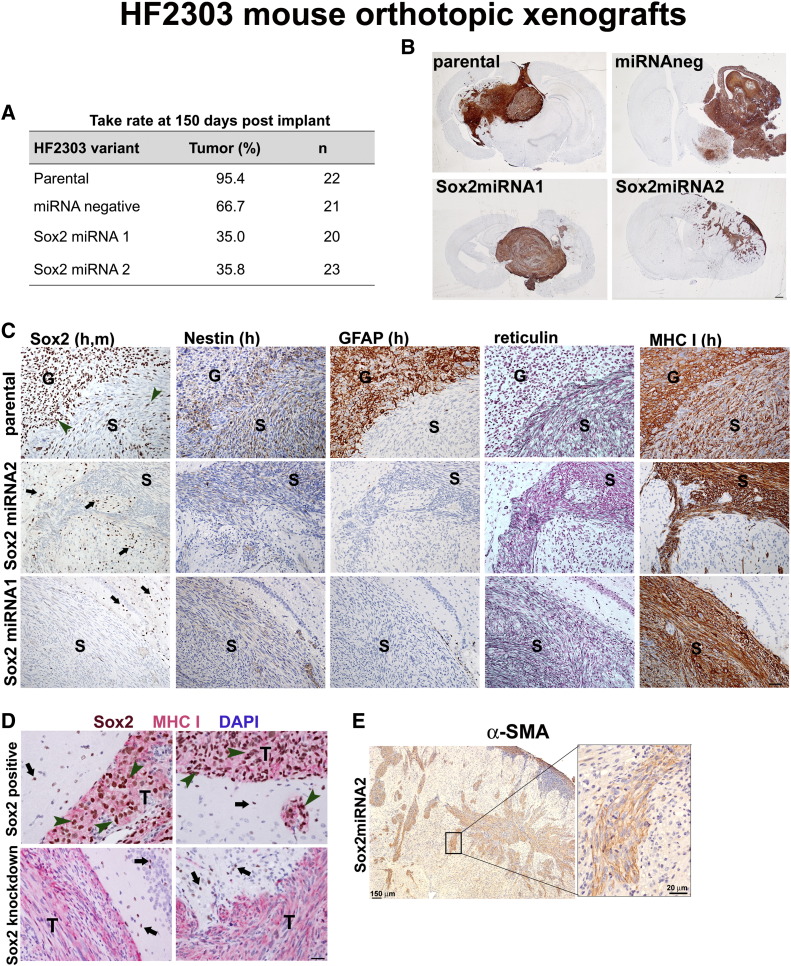
Sox2 Knockdown Alters Tumor Formation and Abolishes the Astrocytic Phenotype in Biphasic GS Tumor
We have previously demonstrated that CSCs and low-passage SDCs from HF2303 GS tumor retain the intrinsic ability to generate the biphasic glial and sarcomatous tumor in orthotopic xenografts [19]. Sox2 deficiency in HF2303 SDCs reduced tumorigenicity, as observed by a 50% decrease intake rate relative to the control group (miRNAneg) (Figure 6A). GBM xenograft tumors were labeled with a human-specific marker (MHC I, Figure 6B). Distinct GFAP and reticulin-positive regions were observed in the Sox2-positive tumors (Figure 6C), as previously demonstrated [19]. Sox2 was expressed in both the glial and sarcomatous compartments of the xenograft tumors, although fewer positive cells were observed in the sarcomatous compartment (Figure 6C). No Sox2-positive tumor cells or GFAP expression was observed in the Sox2miRNA xenografts, and the entire tumor consisted of the sarcomatous component, as evidenced by the reticulin staining (Figure 6C). Murine and human Sox2 proteins share 99.4% amino acid identity, leading to cross-species antibody labeling. Because Sox2-positive cells exist in the adult mouse brain (e.g., Figure 2C), colabeling of the human marker and Sox2 was used to distinguish between Sox2 expression in the human tumor (Figure 6, C and D, green arrowheads) and in mouse host brain tissue (Figure 6, C and D, black arrows), demonstrating that Sox2 protein was not detected in neoplastic cells of xenograft tumors from Sox2-deficient HF2303 cells. GS sarcomatous compartment can display diverse mesenchymal phenotypes. HF2303 mesenchymal component in the biopsy and xenografts is positive for α-SMA, a marker of myogenic lineage [19]. Sox2-deficient xenografts expressed α-SMA throughout the tumor (Figure 6E), further verifying the sarcomatous nature of the tumors.
Figure 6.
Sox2 loss affects the astrocytic compartment of GS. HF2303-parental, HF2303-mirRNAneg, HF2303-Sox2miRNA1, and HF2303-Sox2miRNA2 cells were implanted intracranially in immunocompromised mice (n = 20-23).( A) Percentage of animals in each group that succumbed to tumor burden within 150 days post-implant. (B) Representative images of the xenograft tumors labeled with the human cell marker MHC I. Scale, 300 μm. (C) Sequential coronal sections of FFPE xenograft brains were labeled with anti-Sox2 antibody recognizing both human and mouse (h,m) proteins, with human-specific antibodies against nestin, GFAP, and MHC I, and stained with reticulin. GFAP and reticulin are markers of the glial (G) and sarcomatous (S) phases of the biphasic GS tumors. Representative images for Sox2-positive and Sox2-negative tumors are shown. Scale, 40 μm. (D) Colabeling of the human marker MHC I (red) and Sox2 (brown). Human cells expressing Sox2 in the tumor (T) (green arrowhead) and mouse brain cells expressing Sox2 (black arrow) are shown for representative images of Sox2-expressing and Sox2-deficient xenograft tumors. Scale, 20 μm. (E) α-SMA is expressed throughout the tumor from Sox2-deficient HF2303 cells.
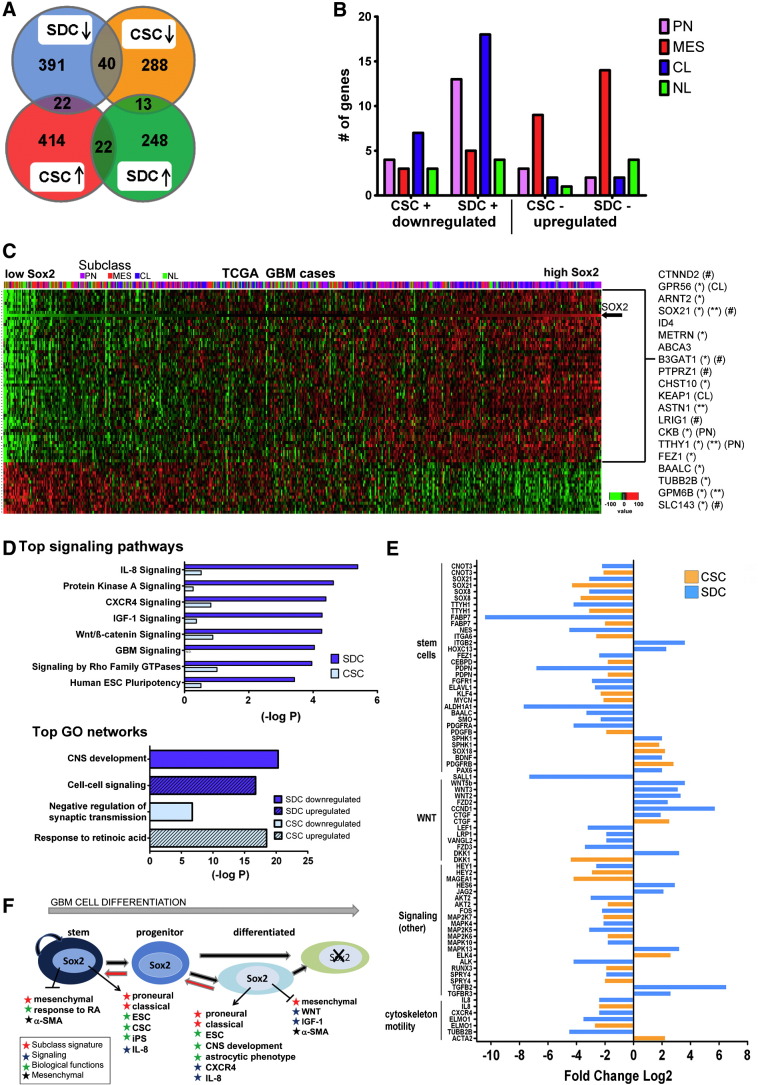
Sox2 Regulates the Expression of Distinct Sets of Genes in Differentiated and CSC States
To better understand transcriptional regulation by Sox2 in differentiated GBM cells, genes differentially expressed between Sox2-positive HF2303 low-passage SDCs (parental and HF2303miRneg) and cells where Sox2 has been knocked down (HF2303_Sox2miRNA1 and HF2303_Sox2miRNA2) were derived from genome-wide transcriptome data (Illumina HT12v4.0). To compare these results with Sox2 activity in isogenic GBM CSCs, lentivirus-shRNA–mediated Sox2 knockdown in HF2303 neurospheres was attained (Figure W1), and differential gene expression between Sox2-positive (parental and NSV-shRNA) and Sox2-deficient (Sox2shRNA1 and Sox2shRNA2) was similarly analyzed. Applying a 3.5-fold change cutoff (1.8 log2), Sox2 knockdown affected the expression of 736 genes in SDCs (453 downregulated and 283 upregulated) and 799 genes in CSCs (341 downregulated and 458 upregulated) (Figure 7A). Table W1 contains the complete DEG lists. Only 62 transcripts were commonly regulated in SDCs and CSCs, whereas 35 presented opposite change in expression between the two groups (Figure 7A and Table W1). Overlap with Sox2 binding to cis-regulatory sequences was observed for 25.1% and 15.1% of DEGs in SDC and CSC, respectively (Table W1), suggesting a direct regulation by Sox2 for this subset. A total of 94 DEGs overlapped with GBM molecular subclass signature genes [15]. In Sox2-deficient cells, proneural and classic signature genes were notably downregulated in SDCs and to a lesser extent in CSCs, whereas mesenchymal signature genes were upregulated in both groups (Figure 7B and Table W1).
Figure 7.
Sox2 regulates distinct gene sets in GBM stem and differentiated cells. (A) Venn diagram shows the overlap of genes differentially expressed on Sox2 knockdown in HF2303 SDC and CSC. Down arrow, downregulated, positive correlation with Sox2 expression; up arrow, upregulated, negative correlation with Sox2 expression. (B) Genes differentially regulated in HF2303 CSC and SDC that overlap with the subclass signature. On Sox2 knockdown, proneural (PN) and classic (CL) signature genes are downregulated in SDC and modestly in CSC [positive (+) correlation], whereas mesenchymal (MES) signature genes are upregulated [negative (−) correlation] in both groups. The pattern of expression of neural (NL) signature genes did not correlate with Sox2 expression. (C) Sox2 expression in the TCGA GBM data set was significantly correlated with 73 genes. The 517 GBM cases are ordered according to the level of Sox2 expression and colored by their molecular class (red, mesenchymal; blue, classic; green, neural; purple, proneural). The mRNA expression data were converted to z scores, averaged across probes for each gene, and then ordered by hierarchical clustering. The heat map displays the standardized intensity with red high and green low. Select genes positively correlated with Sox2 expression are noted. Overlap with genes downregulated on Sox2 knockdown in HF2303 SDC and CSC are indicated by (*) and (**), respectively. Genes that are direct targets of Sox2 (#) or part of subclass signatures are indicated. (D) Signaling pathway analysis of the DEGs in CSC and SDC shows the top signaling pathways (IPA) and top GO networks (GeneGo/MetaCore). (E) Graph with selected DEGs between Sox2-expressing and Sox2-deficient HF2303 cells, represented as log2 fold change. (F) Schematic representation of Sox2 role in GBM cell plasticity and transcriptional regulation in stemlike and differentiated GBM cells.
To compare the above results with genes correlated with Sox2 expression in tumor tissues, gene expression data for 517 GBMs in TCGA were analyzed to identify 56 and 17 genes presenting positive and negative correlation with Sox2 expression, respectively (Table W2 and Figure 7C). Significantly, 28.6% of the positively correlated genes overlapped with the DEGs in HF2303: 3 genes in both CSC and SDC states, 1 in CSCs, and the remaining 12 in the SDC state (Table W2 and Figure 7C), suggesting that genes correlated with Sox2 expression in the tumors are better represented in SDCs than in CSCs.
Top canonical signaling pathways and top GO network pathways representing the DEGs on Sox2 knockdown in CSCs and SDCs were identified using IPA and GeneGO/MetaCore, respectively (Figure 7D). Genes altered in SDCs were more connected in the pathway analyses than those in CSCs. Chemokine signaling was significantly affected by Sox2 knockdown in HF2303 cells. IL-8 expression was downregulated in both CSC and SDCs (Figure 7E), and DEG genes were enriched in IL-8 signaling components, especially in SDCs (Figure 7D). Chemokine (C-X-C motif) receptor 4 (CXCR4), a chemokine receptor involved in glioma cell invasion, was downregulated in SDC (Figure 7E), affecting CXCR4 signaling (Figure 7D).
Wnt receptors Frizzled homolog 2, as well as ligands Wnt2, Wnt3, and Wnt5B, were upregulated, whereas low-density lipoprotein–related protein 1, a negative regulator, and lymphoid enhancer-binding factor 1, a transcription mediator of Wnt signaling, were downregulated (Figure 7E). Up-regulation of Wnt target genes connective tissue growth factor, cyclin D1, and Dickkopf homolog 1 (Figure 7E) provide additional evidence that Wnt signaling is activated on Sox2 knockdown in SDCs (Figure 7D). Genes involved in GBM, insulin-like growth factor 1 (IGF-1), protein kinase A, and ρ GTPase signaling were also significantly enriched in the SDC data set (Figure 7D).The top GO networks indicate that Sox2 loss in SDCs results in down-regulation of genes involved in central nervous system development and up-regulation of genes involved in cell-cell signaling (Figure 7D). Sox2 knockdown in CSCs led to up-regulation of genes associated with response to retinoic acid (Figure7D). Conversely, retinoic acid decreased Sox2 expression in GBM neurosphere cells [33].
On Sox2 knockdown, α-SMA (actin, α 2) was upregulated in CSCs (Figure 7E) and in SDCs (by 3.2-fold, not shown), in agreement with the widespread expression of α-SMA observed for the Sox2-deficient GS xenografts (Figure 6E) and with the lower expression of Sox2 in the sarcomatous phase observed for Sox2-positive tumors (Figure 6C). Genes commonly altered on Sox2 knockdown in both SDC and CSC states are enriched in ESC genes, including Sox21 and Tweety 1, both positively correlated with Sox2 expression in the TCGA data set (Figure 7C), CCR4-NOT transcription complex subunit 3 (CNOT3), fatty acid binding protein 7 (FABP7), and podoplanin (PDPN) (Figure 7E). Several genes associated with stem/progenitor cells were exclusively downregulated in SDCs: Brain and acute leukemia, cytoplasmic (BAALC), smoothened homolog (SMO), Sal-like 1 (SALL1), nestin (NES), and platelet-derived growth factor receptor α (PDGFRα) (Figure 7E), in agreement with enrichment of ESC self-renewal signaling genes in SDCs (Figure 7D). Genes downregulated in Sox2-deficient CSCs include n-Myc (MYCN), Krüppel-like factor 4 (KLF4), integrin-α6 (ITGA6), and aldehyde dehydrogenase 1 family, member A1 (ALDH1A1) (Figure 7E). A schematic model summarizing the main findings reported here is shown in Figure 7F.
Discussion
Sox2 is considered a CSC marker for GBMs due in part to its ubiquitous expression in tumorigenic neurosphere cultures [16,18–21] and analogous role in the determination of NSC identity [6]. However, Sox2 expression in 9% to 85% of GBM tumor cells reported here, and comparable to previous reports [13,28,29], indicates that Sox2 expression encompasses more differentiated neoplastic cells as well. To investigate Sox2 function in GBM differentiated cells, we used a rare low-passage SDC line derived from a patient with GBM that retains Sox2 expression, along with dedifferentitation and tumorigenic potential [19]. Tissue culture manipulation, such as the use of FBS to induce astrocytic differentiation, has provided insights into the differentiation potential of both NSCs [34] and GBM CSCs [35]. High-passage 10% FBS cell cultures incur accumulation of genomic abnormalities leading to a deviation of the GBM phenotype, even when these cells are tumorigenic [16,36]. Low-passage 10% FBS cultures commonly present down-regulation of NSC markers, including Sox2, and loss of tumorigenic potential [16,19], thus only partially representing the molecular profile of differentiated cells in the tumor.
Despite the caveats for the use of FBS as differentiation stimulus for GBM cells, we report a significant overlap of genes affected by Sox2 knockdown in SDCs and genes highly correlated with Sox2 expression in GBM tumor tissue profiled by TCGA. Several genes associated with ESCs and cancer were commonly downregulated in both CSCs and SDCs on Sox2 knockdown: Sox21, a known Sox2 target in GBMs [32] and mediator of Sox2 activity in several cell types, including colon cancer [37]; Tweety 1, a proneural subclass signature member and ER protein also required in embryonic and brain development [38]; CNOT3, involved in ESC self-renewal [39]; FABP7, expressed in radial glia cells and associated with GBM malignancy and cell migration [40]; and PDPN, involved in tumor invasion [41]. Several other genes associated with stem and progenitor cells were exclusively downregulated in Sox2-deficient SDCs: BAALC, restricted to neuroectoderm tissues and overexpressed in acute leukemia and GBM [42]; Shh receptor SMO; SALL1, a zinc finger transcription factor activator of NANOG in ESCs [43]; and the known Sox2 targets nestin and PDGFRα [32]. Wnt pathway activation on Sox2 knockdown, reported here for GBM SDCs, has been observed in osteosarcomas [44]. Interestingly, Wnt activation has been shown to induce differentiation of GBM CSCs [45]. Activation of CXCR4 expression by Sox2 is in agreement with previous reports [21]. Paracrine IL-8 in perivascular niches has been recently shown to regulate GBM CSCs [46], and here, we report for the first time a strong correlation of Sox2 expression in GBM SDCs and CSCs with IL-8 expression and signaling. Activation of IGF-1 signaling on Sox2 knockdown in SDCs may explain the reported sensitivity of Sox2-deficient GBM cells to the IGF-1R inhibition [21]. Loss of Sox2 led to down-regulation of several genes exclusively in CSCs, in particular transcription factors that act in concert with Sox2 to reprogram somatic cells into iPS cells (MYCN and KLF4) [47] and other genes previously associated with CSCs (ALDH1A1 and ITGA6) [48].
Sox2 is part of the gene signature for the GBM proneural molecular subclass [15]. Here, we show that Sox2 was correlated with the expression of both proneural and classic signature genes in the TCGA data set and in vitro functional studies. Furthermore, loss of Sox2 resulted in up-regulation of mesenchymal signature genes in vitro. No significant difference in Sox2 expression between recurrent and newly diagnosed GBMs was observed at protein level in this study or at transcript level in paired samples [30]. Mechanisms leading to the up-regulation of Sox2 in GBM tumors include gene amplification [12,14,18], promoter hypomethylation [12], translational regulation [49], and activated signaling pathways [50]. Posttranslational regulation of Sox2 level by proteolysis has been previously observed in committed neuronal precursor cells [7]. Sox2 protein stabilization by MG132 treatment reported here has also been observed in mouse ESCs [51], and although the mechanism remains to be determined, it has possible implications for the clinical use of proteasome inhibitors.
Sox2 expression is retained in patient-derived orthotopic xenografts generated from neurosphere cells, in agreement with previous studies [52], and also from subcutaneously propagated tumors. It has been previously reported that Sox2 loss in GBM neurospheres impairs tumorigenicity [20,50]. Here, we show that Sox2 knockdown in GBM SDCs abolished dedifferentiation and acquisition of CSC phenotype in vitro and decreased tumorigenicity. Ectopic Sox2 expression was not sufficient to reprogram typical Sox2-deficient low-passage GBM SDCs to regain plasticity and tumorigenicity, possibly due to insufficient levels of required cofactors, similar to what has been observed in a high-passage GBM cell line [12]. Furthermore, we show that in the context of biphasic GSs, Sox2 specifically regulates the glial component, which displays histologic features typical of GBMs, whereas the loss of Sox2 did not affect the sarcomatous compartment, identified by reticulin staining of the dense extracellular matrix and composed of GFAP-negative spindle cells [24]. These findings have potential clinical implications in that any intervention aimed at decreasing Sox2 activity or downstream pathways in GS could favor metaplastic sarcomatous tumor growth.
We propose that Sox2 function in GBMs resemble not only its role in ESC and NSC identity, maintenance, and fate but also its function in committed progenitors and proliferating astrocytes [7]. As evidences continue to point to a “plastic CSC model” [53] for GBMs, our results indicate that Sox2 may be central to the maintenance of developmental plasticity during glial tumor progression, regulating dedifferentiation and acquisition of CSC properties. Furthermore, Sox2 regulates gene expression and signaling critical to malignancy in more differentiated cells.
Acknowledgments
The authors thank Enoch Carlton and Kevin Nelson for immunohistology and management of tumor samples, respectively. The GBM xenograft TMA slides were obtained through the Ivy GBM consortium, supported by the Ben and Catherine Ivy Foundation.
Footnotes
This work was supported by the LIGHT Research Program at the Hermelin Brain Tumor Center.
This article refers to supplementary materials, which are designated by Figure W1 and Tables W1 and W2 and are available online at www.neoplasia.com.
Appendix A. Supplementary materials
Figure W1.
Sox2 knockdown in HF2303 neurospheres cells. (A) Sox2 and nestin mRNA levels in HF2303 (parental) cells and cells transfected with shRNA constructs targeting Sox2 (Sox2shRNA1 and Sox2shRNA2) or nonsilencing miRNA (NSV) were evaluated by quantitative reverse transcription (RT)–PCR. Results represent means ± SEM, n = 3. (B) Sox2 protein expression in whole-cell lysates determined by Western blot analysis confirms Sox2 knockdown in Sox2shRNA1 and Sox2shRNA2 lines; α-actin was used as loading control. (C) The effect of Sox2 knockdown on clonogenicity. Cells were seeded at low density in 96-well plates and cultured in NM. Neurosphere formation was monitored and quantified at day 11 (mean ± SEM). ns, nonsignificant; P > .05 in relation to parental cells. (D) The effect of Sox2 knockdown on proliferation of HF2303 neurospheres. Cells were seeded at low density in 96-well players and cultured in NM. Cell viability was quantified by intracellular ATP level measured using CellTiter-Glo Luminescent Cell Viability Assay (Promega). (E) Representative images contrast Sox2-expressing and Sox2-deficient cells in NM. (F) The effect of Sox2 knockdown on proliferation of HF2303 monolayers. Cells were seeded at low density in 96-well players and cultured in FBS. Viability was measured on days 1, 3, and 7.
Table W1.
DEGs in HF2303 GBM Cells on Sox2 Knockdown.
| SDC | ||||
|---|---|---|---|---|
| Description | Symbol | Fold Change (Log2) | Sox2 Target (Ref) | Subclass Signature |
| ATP-binding cassette, subfamily A (ABC1), member 8 | ABCA8 | − 4.3 | ||
| ATP-binding cassette, subfamily A (ABC1), member 9 | ABCA9 | − 1.9 | ||
| ATP-binding cassette, subfamily B (MDR/TAP), member 9 | ABCB9 | − 1.8 | ||
| ATP-binding cassette, subfamily D (ALD), member 1 | ABCD1 | − 1.9 | ||
| Abhydrolase domain containing 11 | ABHD11 | − 2.9 | ||
| Abhydrolase domain containing 3 | ABHD3 | − 2.0 | ||
| ACN9 homolog (Saccharomyces cerevisiae) | ACN9 | − 2.3 | ||
| Acyl-CoA synthetase bubblegum family member 1 | ACSBG1 | − 5.8 | CL | |
| Acyl-CoA synthetase family member 2 | ACSF2 | − 1.8 | ||
| ADAM metallopeptidase with thrombospondin type 1 motif, 1 | ADAMTS1 | − 2.2 | ||
| Adenosine A1 receptor | ADORA1 | − 2.1 | ||
| AE binding protein 1 | AEBP1 | − 2.3 | ||
| Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | AGT | − 5.6 | ||
| Adenosylhomocysteinase-like 2 | AHCYL2 | − 3.0 | ||
| Allograft inflammatory factor 1-like | AIF1L | − 3.9 | ||
| AKNA domain containing 1 | AKNAD1 | − 2.2 | ||
| Aldo-keto reductase family 1 member 15 (ALD2) | AKR1B15 | − 3.1 | ||
| V-akt murine thymoma viral oncogene homolog 2 | AKT2 | − 3.0 | CL | |
| Aldehyde dehydrogenase 1 family, member A1 | ALDH1A1 | − 7.7 | (1) | |
| Anaplastic lymphoma kinase (Ki-1) | ALK | − 4.2 | (1) | |
| Ankyrin 2, neuronal | ANK2 | − 4.1 | ||
| Ankyrin repeat domain 20 family, member A4 | ANKRD20A4 | − 3.8 | ||
| Ankyrin repeat domain 9 | ANKRD9 | − 1.9 | ||
| Ankyrin repeat and sterile α motif domain containing 1B | ANKS1B | − 2.3 | (1) | NL |
| Adaptor-related protein complex 2, α 1 subunit | AP2A1 | − 2.1 | (1) | |
| Adenomatosis polyposis coli down-regulated 1 | APCDD1 | − 2.6 | ||
| Amyloid β (A4) precursor-like protein 1 | APLP1 | − 2.2 | ||
| Arf-GAP with ρ-GAP domain, ANK repeat and PH domain-containing protein 3 | ARAP3 | − 1.9 | ||
| ADP-ribosylation factor-like 4A | ARL4A | − 2.3 | (1) | |
| Aryl-hydrocarbon receptor nuclear translocator 2 | ARNT2 | − 3.2 | ||
| Acetylserotonin O-methyltransferase-like | ASMTL | − 1.8 | ||
| ATPase, class V, type 10B | ATP10B | − 2.5 | ||
| Β-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) | B3GAT1 | − 2.8 | (1) | |
| Brain and acute leukemia, cytoplasmic | BAALC | − 3.3 | ||
| Brain-specific angiogenesis inhibitor 3 | BAI3 | − 3.3 | (1) | PN |
| Bardet-Biedl syndrome 2 | BBS2 | − 1.9 | ||
| Butyrylcholinesterase | BCHE | − 4.0 | (1) | |
| Brain expressed, X-linked 1 | BEX1 | − 3.1 | PN | |
| Bloom syndrome | BLM | − 2.2 | (1) | CL |
| Bone marrow stromal cell antigen 2 | BST2 | − 2.5 | ||
| BTB (POZ) domain containing 11 | BTBD11 | − 3.7 | (1) | |
| BTG family, member 2 | BTG2 | − 3.0 | ||
| Chromosome 15 open reading frame 59 | C15orf59 | − 2.6 | ||
| Chromosome 18 open reading frame 56 | C18orf56 | − 3.8 | ||
| Chromosome 1 open reading frame 54 | C1orf54 | − 3.2 | MES | |
| Chromosome 1 open reading frame 61 | C1orf61 | − 3.5 | (1) | |
| Chromosome 2 open reading frame 40 | C2orf40 | − 2.1 | ||
| Chromosome 2 open reading frame 76 | C2orf76 | − 1.9 | ||
| Chromosome 3 open reading frame 70 | C3orf70 | − 4.9 | ||
| Chromosome 7 open reading frame 43 | C7orf43 | − 2.1 | ||
| Chromosome 8 open reading frame 4 | C8orf4 | − 3.4 | (1) | |
| Carbonic anhydrase II | CA2 | − 5.3 | ||
| Calcium channel, voltage-dependent, P/Q type, α 1A subunit | CACNA1A | − 2.1 | (1) | |
| Calcium channel, voltage-dependent, β 2 subunit | CACNB2 | − 2.1 | (1) | |
| Cell adhesion molecule 4 | CADM4 | − 3.7 | ||
| Ca2+−dependent activator protein for secretion 2 | CADPS2 | − 2.5 | (1) | |
| Calcitonin receptor-like | CALCRL | − 4.9 | (1) | |
| Calcium/calmodulin-dependent protein kinase (CaM kinase) II β | CAMK2B | − 2.0 | (1) | CL |
| Calcium/calmodulin-dependent protein kinase II inhibitor 1 | CAMK2N1 | − 2.7 | (1) | |
| Caspase recruitment domain family, member 8 | CARD8 | − 2.7 | ||
| Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, β, convertase) | CASP1 | − 2.1 | MES | |
| Coiled-coil domain containing 74B | CCDC74B | − 2.0 | ||
| CD276 molecule | CD276 | − 1.8 | ||
| CD83 molecule | CD83 | − 2.4 | (1) | |
| Cat eye syndrome chromosome region, candidate 1 | CECR1 | − 4.2 | ||
| Cell cycle exit and neuronal differentiation 1 | CEND1 | − 3.4 | ||
| Centrosomal protein 112kDa (CCDC46) | CEP112 | − 2.0 | ||
| Complement factor I | CFI | − 2.9 | ||
| Coiled-coil-helix-coiled-coil-helix domain containing protein 10, mitochondrial | CHCHD10 | − 2.2 | ||
| Cell adhesion molecule with homology to L1CAM (close homolog of L1) | CHL1 | − 7.9 | (1) | |
| Choline phosphotransferase 1 | CHPT1 | − 1.9 | ||
| Cholinergic receptor, nicotinic, α 9 | CHRNA9 | − 2.8 | ||
| Carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | CHST1 | − 3.8 | ||
| Carbohydrate sulfotransferase 10 | CHST10 | − 2.5 | ||
| Carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 | CHST2 | − 2.8 | ||
| Creatine kinase, brain | CKB | − 1.9 | ||
| Chloride channel 2 | CLCN2 | − 2.7 | ||
| Claudin 23 | CLDN23 | − 2.6 | ||
| CAP-GLY domain containing linker protein 2 | CLIP2 | − 2.8 | CL | |
| Calsyntenin 2 | CLSTN2 | − 2.5 | (1) | |
| CCR4-NOT transcription complex, subunit 3 | CNOT3 | − 2.2 | ||
| Canopy 4 homolog (zebrafish) | CNPY4 | − 2.5 | ||
| Coagulation factor C homolog, cochlin (Limulus polyphemus) | COCH | − 2.1 | ||
| Collagen, type XXII, α 1 | COL22A1 | − 2.2 | (1) | |
| Collagen, type IV, α 5 (Alport syndrome) | COL4A5 | − 2.3 | (1) | |
| Collagen, type IV, α 6 | COL4A6 | − 4.8 | (1) | |
| Collagen, type IX, α 2 | COL9A2 | − 2.2 | ||
| Coronin, actin binding protein, 2B | CORO2B | − 2.0 | (1) | |
| DNA-directed RNA polymerase III subunint RPC9 | CRCP | − 1.9 | ||
| Cysteine-rich protein 1 (intestinal) | CRIP1 | − 3.0 | ||
| Crystallin, γ S | CRYGS | − 4.5 | ||
| Cysteine and glycine-rich protein 2 | CSRP2 | − 2.9 | ||
| Chemokine (C-X3-C motif) ligand 1 | CX3CL1 | − 2.4 | (1) | |
| Coxsackie virus and adenovirus receptor | CXADR | − 2.3 | (1) | |
| Chemokine (C-X-C motif) ligand 14 | CXCL14 | − 3.3 | ||
| Chemokine (C-X-C motif) receptor 4 | CXCR4 | − 2.4 | (1) (3) | |
| Cytochrome b5 reductase 2 | CYB5R2 | − 3.5 | ||
| Cytochrome c, somatic | CYCS | − 1.9 | (1) | |
| Cytochrome P450, family 51, subfamily A, polypeptide 1 | CYP51A1 | − 2.2 | ||
| Dysbindin (dystrobrevin binding protein 1) domain containing 1 | DBNDD1 | − 2.2 | ||
| Dehydrogenase/reductase (SDR family) member 3 | DHRS3 | − 2.2 | ||
| DIRAS family, GTP-binding RAS-like 3 | DIRAS3 | − 2.1 | ||
| Discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | DLG3 | − 2.6 | ||
| DLGAP1 antisense RNA 2 | DLGAP1-AS2 | − 2.3 | ||
| Distal-less homeobox 1 | DLX1 | − 1.8 | ||
| DNA-directed RNA polymerase III subunit RPC9 | DNAJC30 | − 1.8 | ||
| Dynamin 3 | DNM3 | − 1.8 | (1) | PN |
| Dipeptidyl-peptidase 4 (CD26, adenosine deaminase complexing protein 2) | DPP4 | − 2.1 | ||
| Dorsal inhibitory axon guidance protein | DRAXIN | − 4.0 | ||
| Dystrobrevin, α | DTNA | − 2.8 | (1) | |
| Dual specificity phosphatase 4 | DUSP4 | − 2.8 | ||
| Dual specificity phosphatase 6 | DUSP6 | − 2.9 | ||
| Endothelin receptor type B | EDNRB | − 5.0 | ||
| Endonuclease/exonuclease/phosphatase family domain containing 1 | EEPD1 | − 1.8 | ||
| Eukaryotic translation initiation factor 1A, Y-linked | EIF1AY | − 4.2 | ||
| ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R) | ELAVL1 | − 2.7 | ||
| Engulfment and cell motility 1 | ELMO1 | − 3.5 | (1) | |
| Elastin (supravalvular aortic stenosis, Williams-Beuren syndrome) | ELN | − 2.8 | ||
| Elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 | ELOVL2 | − 8.3 | (1) | CL |
| ER membrane protein complex subunit 10 | EMC10 | − 1.9 | ||
| Energy homeostasis associated | ENHO | − 2.1 | ||
| Enolase superfamily member 1 | ENOSF1 | − 1.8 | ||
| Epoxide hydrolase 4 | EPHX4 | − 3.1 | ||
| Estrogen-related receptor γ | ESRRG | − 2.3 | (1) | |
| Fatty acid binding protein 5 (psoriasis-associated) | FABP5 | − 1.9 | (1) | |
| Fatty acid binding protein 7, brain | FABP7 | − 10.4 | ||
| Family with sequence similarity 196, member A | FAM196A | − 3.7 | ||
| Family with sequence similarity 212, member B | FAM212B | − 2.4 | ||
| Family with sequence similarity 43, member A | FAM43A | − 4.7 | (1) | |
| Family with sequence similarity 69, member C | FAM69C | − 5.8 | ||
| Family with sequence similarity 84, member B | FAM84B | − 2.6 | (1) | |
| Fasciculation and elongation protein ζ 1 (zygin I) | FEZ1 | − 2.4 | ||
| Fin bud initiation factor | FIBIN | − 4.9 | ||
| Hypothetical gene supported by AK094963 | FLJ37644 | − 2.5 | ||
| Fibronectin leucine rich transmembrane protein 3 | FLRT3 | − 3.3 | ||
| V-fos FBJ murine osteosarcoma viral oncogene homolog | FOS | − 2.2 | ||
| Forkhead box S1 | FOXS1 | − 2.1 | ||
| FERM and PDZ domain containing 3 | FRMPD3 | − 3.2 | ||
| Frizzled homolog 3 (Drosophila) | FZD3 | − 3.4 | CL | |
| Γ-aminobutyric acid (GABA) A receptor, β 1 | GABRB1 | − 2.9 | ||
| Galactose-3-O-sulfotransferase 4 | GAL3ST4 | − 2.0 | ||
| UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) | GALNT1 | − 1.8 | (1) | |
| UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) | GALNT4 | − 1.8 | CL | |
| UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1 | GALNTL1 | − 4.5 | ||
| Growth associated protein 43 | GAP43 | − 5.2 | (1) | |
| Growth arrest-specific 8 | GAS8 | − 3.0 | ||
| Gap junction protein, β 2, 26kDa | GJB2 | − 2.5 | ||
| Glycosyltransferase 25 domain containing 2 | GLT25D2 | − 2.0 | ||
| Guanine nucleotide binding protein (G protein), α inhibiting activity polypeptide 1 | GNAI1 | − 2.1 | (1) | NL |
| Guanine nucleotide binding protein (G protein), γ 11 | GNG11 | − 3.2 | (1) | |
| Guanine nucleotide binding protein (G protein), γ 2 | GNG2 | − 5.2 | (1) | |
| Guanine nucleotide binding protein (G protein), γ 7 | GNG7 | − 2.1 | CL | |
| Glycoprotein M6A | GPM6A | − 4.8 | PN | |
| Glycoprotein M6B | GPM6B | − 3.7 | ||
| G protein–coupled receptor 162 | GPR162 | − 2.0 | ||
| G protein–coupled receptor 56 | GPR56 | − 3.2 | ||
| Growth factor receptor-bound protein 10 | GRB10 | − 1.9 | ||
| Hyaluronan synthase 3 | HAS3 | − 4.9 | ||
| Histone deacetylase 4 | HDAC4 | − 3.0 | (1) | |
| Hepatocyte cell adhesion molecule | HEPACAM | − 3.9 | ||
| Hairy/enhancer-of-split related with YRPW motif 1 | HEY1 | − 2.6 | (1) | |
| HHIP antisense RNA 1 | HHIP-AS1 | − 4.8 | ||
| Huntingtin interacting protein 1 | HIP1 | − 3.0 | ||
| High-mobility group AT-hook 1 | HMGA1 | − 1.8 | ||
| Homer homolog 1 (Drosophila) | HOMER1 | − 6.5 | ||
| Hippocalcin-like 1 | HPCAL1 | − 2.2 | (1) | |
| Heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | HS3ST1 | − 2.7 | (1) | |
| Heat shock 70kDa protein 6 (HSP70B′) | HSPA6 | − 2.1 | ||
| Heat shock protein, α-crystallin-related, B6 | HSPB6 | − 4.1 | ||
| Indoleamine 2,3-dioxygenase 1 | IDO1 | − 2.8 | ||
| Iduronate 2-sulfatase (Hunter syndrome) | IDS | − 1.9 | ||
| Intraflagellar transport 57 homolog (Chlamydomonas) | IFT57 | − 2.4 | ||
| Intraflagellar transport 81 homolog (Chlamydomonas) | IFT81 | − 1.8 | ||
| Insulin-like growth factor 2 mRNA binding protein 3 | IGF2BP3 | − 5.7 | ||
| Insulin-like growth factor binding protein 2, 36kDa | IGFBP2 | − 6.0 | ||
| Insulin-like growth factor binding protein 3 | IGFBP3 | − 1.9 | (1) | |
| Insulin-like growth factor binding protein 5 | IGFBP5 | − 2.0 | ||
| Immunoglobulin superfamily, member 1 | IGSF1 | − 2.0 | ||
| Immunoglobulin superfamily, member 5 | IGSF5 | − 2.1 | ||
| Interleukin 17 receptor B | IL17RB | − 2.0 | ||
| Interleukin-1 receptor accessory protein-like 1 | IL1RAPL1 | − 8.5 | PN | |
| Interleukin-27 receptor, α | IL27RA | − 2.2 | (1) | |
| Interleukin-33 | IL33 | − 5.0 | ||
| Interleukin-8 | IL8 | − 2.4 | (1) | |
| Inositol 1,4,5-triphosphate receptor, type 2 | ITPR2 | − 2.4 | (1) | |
| Potassium voltage-gated channel, subfamily F, member 1 | KCNF1 | − 2.0 | (1) | CL |
| Kv channel interacting protein 4 | KCNIP4 | − 2.5 | (1) | |
| Potassium inwardly-rectifying channel, subfamily J, member 2 | KCNJ2 | − 2.0 | (1) | |
| Potassium channel, subfamily K, member 12 | KCNK12 | − 2.8 | ||
| Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | KCNN4 | − 2.5 | ||
| Potassium channel tetramerisation domain containing 12 | KCTD12 | − 2.4 | ||
| Potassium channel tetramerisation domain containing 13 | KCTD13 | − 2.6 | ||
| Potassium channel tetramerisation domain containing 17 | KCTD17 | − 2.2 | ||
| Lysine (K)-specific demethylase 5D | KDM5D | − 2.8 | ||
| KIAA0408 | KIAA0408 | − 2.3 | ||
| KIAA1147 | KIAA1147 | − 1.8 | ||
| KIAA1211 | KIAA1211 | − 2.6 | ||
| Kinesin family member 13B | KIF13B | − 2.0 | (1) | |
| Kinesin family member 26B | KIF26B | − 2.5 | ||
| Kinesin family member 5C | KIF5C | − 5.0 | ||
| Kelch domain containing 9 | KLHDC9 | − 2.6 | ||
| Kelch-like 29 (Drosophila) | KLHL29 | − 2.5 | ||
| Kynureninase (l-kynurenine hydrolase) | KYNU | − 2.6 | (1) | MES |
| Laminin, α 1 | LAMA1 | − 3.1 | ||
| Lymphoid enhancer-binding factor 1 | LEF1 | − 3.2 | ||
| LEM domain containing 1 | LEMD1 | − 5.2 | ||
| Lectin, galactoside-binding, soluble, 9B | LGALS9B | − 1.9 | ||
| Leucine-rich repeat LGI family, member 2 | LGI2 | − 3.9 | ||
| Leukemia inhibitory factor receptor α | LIFR | − 2.3 | ||
| LIM domain kinase 1 | LIMK1 | − 2.9 | ||
| Long intergenic nonprotein coding RNA 341 | LINC00341 | − 2.1 | ||
| Long intergenic nonprotein coding RNA 623 | LINC00623 | − 3.2 | ||
| Lipase, endothelial | LIPG | − 3.1 | ||
| LOC100128288 | − 1.8 | |||
| LON peptidase N-terminal domain and ring finger 1 | LONRF1 | − 2.7 | ||
| LON peptidase N-terminal domain and ring finger 2 | LONRF2 | − 2.9 | (1) | |
| Lysophosphatidic acid receptor 1 | LPAR1 | − 1.9 | ||
| Latrophilin 3 | LPHN3 | − 2.0 | (1) | PN |
| Lipoprotein lipase | LPL | − 4.2 | ||
| Lipid phosphate phosphatase-related protein type 5 | LPPR5 | − 4.4 | ||
| Low-density lipoprotein–related protein 1 (α-2-macroglobulin receptor) | LRP1 | − 1.9 | ||
| Low-density lipoprotein–related protein 1B (deleted in tumors) | LRP1B | − 2.5 | (1) | |
| Low-density lipoprotein receptor–related protein 4 | LRP4 | − 3.0 | ||
| Leucine rich repeat containing 4C | LRRC4C | − 3.4 | (1) | |
| Leucine rich repeat neuronal 2 | LRRN2 | − 4.5 | ||
| Leucine rich repeat neuronal 3 | LRRN3 | − 2.5 | ||
| LY6/PLAUR domain containing 1 | LYPD1 | − 2.7 | ||
| Leucine zipper, putative tumor suppressor 1 | LZTS1 | − 4.8 | ||
| Mal, T cell differentiation protein | MAL | − 4.8 | ||
| MAM domain containing 2 | MAMDC2 | − 3.2 | (1) | |
| Mannosidase, α, class 1C, member 1 | MAN1C1 | − 3.9 | ||
| Mitogen-activated protein kinase kinase 5 | MAP2K5 | − 3.1 | ||
| Mitogen-activated protein kinase 10 | MAPK10 | − 1.8 | (1) | |
| Mitogen-activated protein kinase 4 | MAPK4 | − 2.1 | ||
| Mitogen-activated protein kinase 8 interacting protein 1 | MAPK8IP1 | − 2.8 | ||
| MARCKS-like 1 | MARCKSL1 | − 2.1 | PN | |
| Midkine (neurite growth-promoting factor 2) | MDK | − 2.4 | ||
| Mesenchyme homeobox 2 | MEOX2 | − 2.0 | (1) | CL |
| Meteorin, glial cell differentiation regulator | METRN | − 3.1 | ||
| Major facilitator superfamily domain containing 6 | MFSD6 | − 1.9 | ||
| Matrix Gla protein | MGP | − 1.9 | ||
| MKL/myocardin-like 2 | MKL2 | − 2.9 | ||
| Megalencephalic leukoencephalopathy with subcortical cysts 1 | MLC1 | − 2.4 | ||
| Musashi homolog 1 (Drosophila) | MSI1 | − 2.1 | ||
| Microseminoprotein, prostate associated | MSMP | − 4.6 | ||
| Metallothionein 1X | MT1X | − 2.2 | ||
| Myosin binding protein C, slow type | MYBPC1 | − 5.0 | NL | |
| Myosin VB | MYO5B | − 1.9 | ||
| Neurocan | NCAN | − 2.9 | ||
| Noncompact myelin associated protein | NCMAP | − 2.5 | ||
| NDRG family member 4 | NDRG4 | − 3.2 | ||
| NEL-like 2 (chicken) | NELL2 | − 2.5 | ||
| Nestin | NES | − 4.5 | (5) | CL |
| Nuclear factor I/B | NFIB | − 3.0 | (1) | |
| Nuclear factor of κ light polypeptide gene enhancer in B cells inhibitor, β | NFKBIB | − 2.7 | ||
| Neuroligin 4, X-linked | NLGN4X | − 1.8 | (1) | |
| Neuroligin 4, Y-linked | NLGN4Y | − 7.3 | ||
| Nicotinamide nucleotide adenylyltransferase 2 | NMNAT2 | − 3.6 | ||
| Nicotinamide nucleotide adenylyltransferase 3 | NMNAT3 | − 2.1 | (1) | |
| Neuro-oncological ventral antigen 1 | NOVA1 | − 2.7 | (1) | |
| Neuronal PAS domain protein 2 | NPAS2 | − 3.9 | ||
| Neuronal PAS domain protein 3 | NPAS3 | − 3.3 | (1) | CL |
| Neuropeptide B | NPB | − 2.6 | ||
| Neuropeptide Y receptor Y5 | NPY5R | − 6.6 | ||
| Notch-regulated ankyrin repeat protein | NRARP | − 3.2 | ||
| Neuronal cell adhesion molecule | NRCAM | − 4.3 | ||
| Neurogranin (protein kinase C substrate, RC3) | NRGN | − 2.8 | (1) | |
| Neuropilin 2 | NRP2 | − 1.8 | (1) | |
| Netrin 1 | NTN1 | − 2.2 | (1) | |
| OAF homolog (Drosophila) | OAF | − 2.4 | ||
| Olfactomedin-like 3 | OLFML3 | − 2.6 | ||
| Oxysterol binding protein-like 6 | OSBPL6 | − 2.4 | (1) | |
| Orthopedia homeobox | OTP | − 2.2 | ||
| Prostate androgen-regulated mucin-like protein 1 | PARM1 | − 4.2 | ||
| Protocadherin 20 | PCDH20 | − 3.4 | ||
| Protocadherin 9 | PCDH9 | − 6.4 | (1) | |
| Protocadherin γ subfamily A, 5 | PCDHGA5 | − 2.5 | ||
| Protocadherin γ subfamily C, 3 | PCDHGC3 | − 2.0 | ||
| Phosphodiesterase 1B, calmodulin-dependent | PDE1B | − 5.2 | ||
| Phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4 dunce homolog, Drosophila) | PDE4B | − 2.1 | (1) | |
| Phosphodiesterase 8B | PDE8B | − 3.3 | (1) | |
| Platelet-derived growth factor receptor, α polypeptide | PDGFRA | − 4.2 | (2) | |
| PDZ and LIM domain 4 | PDLIM4 | − 2.4 | ||
| Podoplanin | PDPN | − 6.8 | MES | |
| Paternally expressed 10 | PEG10 | − 2.6 | ||
| Pellino homolog 1 (Drosophila) | PELI1 | − 2.9 | PN | |
| GPI deacylase | PGAP1 | − 2.8 | (1) | |
| PiggyBac transposable element derived 5 | PGBD5 | − 2.9 | NL | |
| Pleckstrin homology-like domain, family A, member 1 | PHLDA1 | − 2.0 | ||
| Phosphatidylinositol glycan anchor biosynthesis, class F | PIGF | − 3.8 | ||
| Protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) | PIN4 | − 2.1 | ||
| Pigeon homolog (Drosophila) | PION | − 2.2 | ||
| Phosphatidylinositol transfer protein, cytoplasmic 1 | PITPNC1 | − 3.2 | ||
| Protein kinase (cAMP-dependent, catalytic) inhibitor α | PKIA | − 2.6 | (1) | |
| Protein kinase (cAMP-dependent, catalytic) inhibitor β | PKIB | − 3.3 | ||
| Phospholipase C, γ 1 | PLCG1 | − 3.1 | ||
| Phospholipase D family, member 3 | PLD3 | − 3.0 | ||
| Pleckstrin homology domain containing, family B (evectins) member 1 | PLEKHB1 | − 3.1 | (1) | |
| Pleckstrin homology domain containing, family G (with ρGef domain) member 1 | PLEKHG1 | − 2.4 | ||
| Prostate transmembrane protein, androgen induced 1 | PMEPA1 | − 4.5 | ||
| Paraneoplastic antigen MA2 | PNMA2 | − 3.4 | (1) | |
| Podocalyxin-like 2 | PODXL2 | − 1.9 | PN | |
| POM (POM121 homolog, rat) and ZP3 fusion | POMZP3 | − 4.4 | ||
| Periostin, osteoblast specific factor | POSTN | − 5.7 | (1) | |
| Phosphatidic acid phosphatase type 2C | PPAP2C | − 2.5 | ||
| Protein phosphatase 1, catalytic subunit, β isoform | PPP1CB | − 1.8 | ||
| Protein kinase, cAMP-dependent, catalytic, β | PRKACB | − 1.8 | ||
| Protein kinase C, θ | PRKCQ | − 2.2 | ||
| Proline rich 7 (synaptic) | PRR7 | − 2.2 | ||
| Prostaglandin F2 receptor negative regulator | PTGFRN | − 2.3 | (1) | |
| Prothymosin, α (gene sequence 28) | PTMA | − 1.9 | ||
| Pleiotrophin (heparin binding growth factor 8, neurite growth-promoting factor 1) | PTN | − 2.4 | ||
| Protein tyrosine phosphatase, nonreceptor type 12 | PTPN12 | − 1.9 | ||
| Protein tyrosine phosphatase, nonreceptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) | PTPN13 | − 2.4 | (1) | |
| Protein tyrosine phosphatase, receptor type, D | PTPRD | − 6.4 | (1) | |
| RAB36, member RAS oncogene family | RAB36 | − 3.5 | ||
| RAN binding protein 2 | RANBP2 | − 2.3 | ||
| Rap guanine nucleotide exchange factor (GEF) 5 | RAPGEF5 | − 5.4 | ||
| RAS, dexamethasone-induced 1 | RASD1 | − 1.9 | ||
| Ras interacting protein 1 | RASIP1 | − 4.1 | ||
| Ras association (RalGDS/AF-6) domain family 2 | RASSF2 | − 3.0 | ||
| Retinol binding protein 7, cellular | RBP7 | − 2.5 | ||
| Regulator of calcineurin 2 | RCAN2 | − 2.2 | ||
| Receptor accessory protein 1 | REEP1 | − 2.1 | (1) | PN |
| Replication factor C (activator 1) 2, 40kDa | RFC2 | − 1.9 | ||
| Raftlin family member 2 | RFTN2 | − 4.9 | ||
| Regulator of G-protein signaling 2, 24kDa | RGS2 | − 3.2 | (1) | |
| Rhomboid domain containing 3 | RHBDD3 | − 2.2 | ||
| Ras homolog gene family, member U | RHOU | − 3.0 | (1) | |
| Regulating synaptic membrane exocytosis 4 | RIMS4 | − 2.6 | ||
| Ρ family GTPase 2 | RND2 | − 1.9 | ||
| Ring finger protein 112 | RNF112 | − 2.0 | ||
| Reticulon 1 | RTN1 | − 4.2 | ||
| Sal-like 1 (Drosophila) | SALL1 | − 7.3 | (1) | |
| Sterile α motif domain containing 13 | SAMD13 | − 2.7 | ||
| Sterile α motif domain containing 5 | SAMD5 | − 3.4 | ||
| Secretogranin II (chromogranin C) | SCG2 | − 3.0 | ||
| Sodium channel, voltage-gated, type IV, β | SCN4B | − 2.4 | ||
| Sodium channel, voltage-gated, type IX, α subunit | SCN9A | − 1.9 | (1) | |
| Scrapie responsive protein 1 | SCRG1 | − 5.0 | (1) | |
| Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | SEMA3A | − 2.5 | (1) | |
| Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E | SEMA3E | − 5.1 | ||
| Sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A | SEMA6A | − 2.5 | (1) | CL |
| Selenoprotein N, 1 | SEPN1 | − 2.2 | ||
| Septin 3 | SEPT3 | − 2.2 | ||
| Septin 5 | SEPT5 | − 2.0 | ||
| SET binding protein 1 | SETBP1 | − 2.0 | (1) | |
| Sideroflexin 5 | SFXN5 | − 3.7 | (1) | |
| Sarcoglycan, ε | SGCE | − 6.4 | ||
| Homolog of rat pragma of Rnd2 | SGK223 | − 2.4 | ||
| SH3 and PX domains 2A | SH3PXD2A | − 2.1 | (1) | |
| SHC (Src homology 2 domain containing) family, member 4 | SHC4 | − 6.8 | (1) | |
| SLAIN motif family, member 1 | SLAIN1 | − 3.9 | ||
| Solute carrier family 10 (sodium/bile acid cotransporter family), member 4 | SLC10A4 | − 3.3 | ||
| Solute carrier family 1 (glial high affinity glutamate transporter), member 3 | SLC1A3 | − 1.9 | (1) | |
| Solute carrier family 25, member 13 (citrin) | SLC25A13 | − 2.0 | (1) | |
| Solute carrier family 25 (mitochondrial carrier), member 18 | SLC25A18 | − 3.6 | ||
| Solute carrier family 26, member 7 | SLC26A7 | − 2.4 | ||
| Solute carrier family 27 (fatty acid transporter), member 1 | SLC27A1 | − 2.0 | ||
| Solute carrier family 35, member B4 | SLC35B4 | − 2.1 | ||
| Solute carrier family 35, member F1 | SLC35F1 | − 3.1 | ||
| Solute carrier family 39 (zinc transporter), member 12 | SLC39A12 | − 3.0 | ||
| Solute carrier family 45, member 3 | SLC45A3 | − 5.6 | ||
| Solute carrier family 4, anion exchanger, member 3 | SLC4A3 | − 3.8 | (1) | |
| Solute carrier organic anion transporter family, member 2A1 | SLCO2A1 | − 3.7 | ||
| Solute carrier organic anion transporter family, member 5A1 | SLCO5A1 | − 2.5 | PN | |
| SLIT and NTRK-like family, member 3 | SLITRK3 | − 4.1 | ||
| Sarcolipin | SLN | − 6.4 | ||
| SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily d, member 3 | SMARCD3 | − 1.9 | (1) | |
| Structural maintenance of chromosomes 6 | SMC6 | − 2.6 | ||
| Smoothened homolog (Drosophila) | SMO | − 2.3 | (1) | CL |
| Sine oculis binding protein homolog (Drosophila) | SOBP | − 1.9 | ||
| Suppressor of cytokine signaling 2 | SOCS2 | − 2.5 | (1) | CL |
| Sortilin-related receptor, L(DLR class) A repeats-containing | SORL1 | − 3.1 | ||
| SRY (sex-determining region Y)-box 15 | SOX15 | − 3.5 | ||
| SRY (sex-determining region Y)-box 2 | SOX2 | − 4.0 | PN | |
| SRY (sex-determining region Y)-box 21 | SOX21 | − 3.1 | (1) (2) | |
| SRY (sex-determining region Y)-box 3 | SOX3 | − 4.4 | ||
| SRY (sex-determining region Y)-box 8 | SOX8 | − 3.1 | ||
| SPARC-like 1 (mast9, hevin) | SPARCL1 | − 2.4 | ||
| Sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 | SPOCK2 | − 4.0 | ||
| Spondin 1, extracellular matrix protein | SPON1 | − 4.9 | ||
| Secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | SPP1 | − 5.2 | ||
| Sprouty homolog 1, antagonist of FGF signaling (Drosophila) | SPRY1 | − 3.1 | (1) | |
| Sprouty homolog 2 (Drosophila) | SPRY2 | − 2.2 | (1) | CL |
| Sprouty homolog 4 (Drosophila) | SPRY4 | − 1.9 | (1) | |
| SLIT-ROBO Ρ GTPase activating protein 3 | SRGAP3 | − 2.0 | (1) | PN |
| Sorcin | SRI | − 2.8 | (1) | |
| SFRS protein kinase 2 | SRPK2 | − 4.0 | (1) | |
| Sushi-repeat-containing protein, X-linked | SRPX | − 2.6 | ||
| Single stranded DNA binding protein 4 | SSBP4 | − 2.5 | (1) | |
| ST6 β-galactosamide α-2,6-sialyltranferase 1 | ST6GAL1 | − 3.9 | ||
| Stanniocalcin 1 | STC1 | − 2.3 | ||
| Serine/threonine kinase 32A | STK32A | − 2.2 | ||
| Storkhead box 2 | STOX2 | − 3.2 | ||
| Syntaxin 1A (brain) | STX1A | − 2.1 | ||
| Syntabulin (syntaxin-interacting) | SYBU | − 2.1 | ||
| Synemin, intermediate filament protein | SYNM | − 2.2 | ||
| TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa | TAF6 | − 3.4 | ||
| Transgelin 3 | TAGLN3 | − 3.4 | ||
| Transcobalamin II; macrocytic anemia | TCN2 | − 2.8 | ||
| Tyrosyl-DNA phosphodiesterase 1 | TDP1 | − 2.4 | ||
| Transforming growth factor, α | TGFA | − 2.5 | (1) | |
| Thrombospondin 2 | THBS2 | − 2.6 | ||
| TIMP metallopeptidase inhibitor 4 | TIMP4 | − 4.5 | ||
| Tousled-like kinase 1 | TLK1 | − 2.0 | (1) | |
| Transmembrane 7 superfamily member 2 | TM7SF2 | − 2.1 | ||
| Transmembrane protein 108 | TMEM108 | − 2.3 | (1) | |
| Transmembrane protein 132A | TMEM132A | − 2.2 | ||
| Transmembrane protein 163 | TMEM163 | − 3.2 | (1) | |
| Transmembrane protein 255A | TMEM255A | − 2.6 | ||
| Transmembrane protein 26 | TMEM26 | − 2.3 | ||
| Transmembrane protein 55A | TMEM55A | − 1.9 | ||
| Transmembrane protein 86A | TMEM86A | − 4.0 | ||
| Transmembrane and tetratricopeptide repeat containing 2 | TMTC2 | − 5.2 | (1) | |
| Tumor necrosis factor, α-induced protein 6 | TNFAIP6 | − 4.6 | ||
| Tumor necrosis factor (ligand) superfamily, member 15 | TNFSF15 | − 2.6 | ||
| Tumor necrosis factor (ligand) superfamily, member 4 (tax-transcriptionally activated glycoprotein 1, 34kDa) | TNFSF4 | − 6.9 | ||
| TP53 target 1 (nonprotein coding) | TP53TG1 | − 1.8 | ||
| Tumor protein D52 | TPD52 | − 1.8 | (1) | |
| Tubulin polyglutamylase complex subunit 2 | TPGS2 | − 2.7 | ||
| Thiamin pyrophosphokinase 1 | TPK1 | − 6.3 | (1) | |
| Tribbles homolog 2 (Drosophila) | TRIB2 | − 2.0 | (1) | CL |
| TLR4 interactor with leucine-rich repeats | TRIL | − 2.9 | ||
| Tripartite motif-containing 36 | TRIM36 | − 1.9 | (1) | |
| Tripartite motif-containing 47 | TRIM47 | − 3.1 | ||
| Tripartite motif-containing 9 | TRIM9 | − 3.3 | ||
| Trichorhinophalangeal syndrome I | TRPS1 | − 2.2 | (1) | |
| Thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 | TSTD1 | − 4.6 | ||
| Tubulin tyrosine ligase-like family, member 4 | TTLL4 | − 2.8 | ||
| Tweety homolog 1 (Drosophila) | TTYH1 | − 4.2 | ||
| Tweety homolog 3 (Drosophila) | TTYH3 | − 2.4 | (1) | |
| Tubulin, β 2B | TUBB2B | − 4.5 | ||
| Ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) | USP14 | − 2.1 | ||
| Vang-like 2 (van gogh, Drosophila) | VANGL2 | − 1.9 | ||
| VGF nerve growth factor inducible | VGF | − 2.0 | ||
| Visinin-like 1 | VSNL1 | − 1.9 | (1) | |
| V-set and transmembrane domain containing 2 like | VSTM2L | − 1.9 | ||
| Von Willebrand factor A domain containing 1 | VWA1 | − 3.0 | ||
| Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | WAS | − 2.5 | ||
| WAS/WASL interacting protein family, member 1 | WIPF1 | − 4.0 | MES | |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, γ polypeptide | YWHAG | − 1.9 | ||
| Zinc finger and BTB domain containing 7C | ZBTB7C | − 6.5 | (1) | |
| Zic family member 2 (odd-paired homolog, Drosophila) | ZIC2 | − 3.3 | ||
| Zinc finger, matrin type 4 | ZMAT4 | − 4.2 | (1) | |
| Zinc finger protein 226 | ZNF226 | − 1.9 | ||
| Zinc finger protein 3 | ZNF3 | − 2.1 | ||
| Zinc finger protein 331 | ZNF331 | − 2.0 | ||
| Zinc finger protein 573 | ZNF573 | − 1.8 | ||
| Zinc finger protein 615 | ZNF615 | − 2.0 | ||
| Zinc finger protein 679 | ZNF679 | − 4.4 | ||
| Zinc finger protein 680 | ZNF680 | − 2.4 | (1) | |
| Zinc finger protein 681 | ZNF681 | − 2.0 | ||
| ABI gene family, member 3 (NESH) binding protein | ABI3BP | 2.1 | (1) | |
| Ankyrin repeat and BTB (POZ) domain containing 1 | ABTB1 | 2.5 | ||
| Adenosine A2b receptor | ADORA2B | 2.0 | ||
| Adrenergic, α-1B-, receptor | ADRA1B | 2.0 | (1) | |
| Adrenergic, β-2-, receptor, surface | ADRB2 | 4.0 | (1) | |
| AGSK1 | 1.8 | |||
| Ajuba LIM protein | AJUBA | 2.4 | ||
| Adenylate kinase 4 | AK4 | 1.9 | ||
| Aldo-keto reductase family 1, member C1 | AKR1C1 | 6.3 | ||
| Aldo-keto reductase family 1, member C3 (3-α hydroxysteroid dehydrogenase, type II) | AKR1C3 | 2.2 | ||
| Aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-α hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4) | AKR1C4 | 3.7 | ||
| Activated leukocyte cell adhesion molecule | ALCAM | 2.2 | (1) | |
| Angiopoietin-like 2 | ANGPTL2 | 2.0 | ||
| Ankyrin repeat domain 1 (cardiac muscle) | ANKRD1 | 4.0 | ||
| Alanyl (membrane) aminopeptidase (aminopeptidase N, aminopeptidase M, microsomal aminopeptidase, CD13, p150) | ANPEP | 2.3 | ||
| Annexin A11 | ANXA11 | 2.7 | ||
| Annexin A3 | ANXA3 | 4.9 | NL | |
| Annexin A6 | ANXA6 | 2.0 | ||
| Adenomatosis polyposis coli down-regulated 1-like | APCDD1L | 2.3 | (1) | |
| Ρ GTPase activating protein 30 | ARHGAP30 | 1.8 | ||
| Ρ GDP dissociation inhibitor (GDI) β | ARHGDIB | 2.8 | ||
| Ρ guanine nucleotide exchange factor (GEF) 10 | ARHGEF10 | 2.5 | ||
| Aspartate β-hydroxylase | ASPH | 2.2 | (1) | |
| Argininosuccinate synthetase 1 | ASS1 | 2.1 | ||
| Arginine vasopressin-induced 1 | AVPI1 | 2.1 | ||
| AXL receptor tyrosine kinase | AXL | 2.3 | (1) | |
| Brain abundant, membrane attached signal protein 1 | BASP1 | 2.6 | (1) | NL |
| Breast cancer anti-estrogen resistance 3 | BCAR3 | 2.9 | ||
| Brain-derived neurotrophic factor | BDNF | 2.0 | ||
| BEN domain containing 7 | BEND7 | 2.1 | ||
| Chromosome 11 open reading frame 75 | C11orf75 | 3.2 | ||
| Chromosome 11 open reading frame 87 | C11orf87 | 1.9 | ||
| Calmodulin-like 4 | CALML4 | 3.0 | ||
| Caspase 4, apoptosis-related cysteine peptidase | CASP4 | 2.3 | MES | |
| Castor zinc finger 1 | CASZ1 | 2.9 | ||
| Caveolin 1, caveolae protein, 22kDa | CAV1 | 1.9 | ||
| Coiled-coil domain containing 147 | CCDC147 | 2.3 | ||
| Coiled-coil domain containing 80 | CCDC80 | 2.8 | ||
| Cyclin D1 | CCND1 | 5.7 | (1) | |
| CD163 molecule-like 1 | CD163L1 | 3.0 | ||
| CD24 molecule | CD24 | 4.9 | ||
| CDC14 cell division cycle 14 homolog B (Saccharomyces cerevisiae) | CDC14B | 2.3 | ||
| Cadherin 11, type 2, OB-cadherin (osteoblast) | CDH11 | 2.0 | (1) | |
| Cadherin 13, H-cadherin (heart) | CDH13 | 4.4 | (1) | |
| Complement factor B | CFB | 2.9 | ||
| Chorionic gonadotropin, β polypeptide | CGB | 2.2 | ||
| Chorionic gonadotropin, β polypeptide 1 | CGB1 | 4.3 | ||
| Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | CITED2 | 2.1 | (1) | |
| Cytidine monophospho-N-acetylneuraminic acid hydroxylase, pseudogene | CMAHP | 2.2 | ||
| CKLF-like MARVEL transmembrane domain containing 7 | CMTM7 | 2.5 | ||
| Calponin 1, basic, smooth muscle | CNN1 | 3.8 | ||
| Calponin 2 | CNN2 | 2.0 | MES | |
| Collagen, type XI, α 1 | COL11A1 | 1.8 | (1) | |
| Collagen, type XII, α 1 | COL12A1 | 2.9 | (1) | |
| Collagen, type V, α 1 | COL5A1 | 2.9 | (1) | MES |
| Collagen, type VI, α 2 | COL6A2 | 1.8 | ||
| Collagen, type VIII, α 1 | COL8A1 | 1.9 | ||
| Coronin, actin binding protein, 2A | CORO2A | 2.0 | ||
| Carboxypeptidase A4 | CPA4 | 3.9 | (1) | |
| Cadherin-like and PC-esterase domain containing 1 | CPED1 | 2.2 | ||
| Colony stimulating factor 1 receptor, formerly McDonough feline sarcoma viral (v-fms) oncogene homolog | CSF1R | 2.9 | ||
| Chondroitin sulfate proteoglycan 4 | CSPG4 | 3.7 | ||
| C-terminal binding protein 2 | CTBP2 | 2.6 | (1) | |
| Connective tissue growth factor | CTGF | 1.9 | ||
| Cathepsin H | CTSH | 2.2 | ||
| Chemokine (C-X-C motif) receptor 7 | CXCR7 | 2.8 | (1) | |
| Cytochrome P450, family 26, subfamily B, polypeptide 1 | CYP26B1 | 2.4 | (1) | |
| Cysteine-rich, angiogenic inducer, 61 | CYR61 | 2.7 | ||
| Cytohesin 3 | CYTH3 | 1.8 | ||
| Disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | DAB2 | 3.4 | MES | |
| DEP domain containing 7 | DEPDC7 | 2.7 | ||
| Deiodinase, iodothyronine, type II | DIO2 | 2.0 | ||
| DIX domain containing 1 | DIXDC1 | 1.9 | ||
| Dickkopf homolog 1 (Xenopus laevis) | DKK1 | 3.2 | (4) | |
| DnaJ (Hsp40) homolog, subfamily B, member 12 | DNAJB12 | 2.1 | ||
| Dedicator of cytokinesis 2 | DOCK2 | 1.9 | (1) | |
| Dermatan sulfate epimerase | DSE | 2.2 | MES | |
| Deltex 3-like (Drosophila) | DTX3L | 1.9 | ||
| Dual specificity phosphatase 1 | DUSP1 | 2.0 | (1) | |
| Endothelin converting enzyme 2 | ECE2 | 3.7 | (1) | |
| EH-domain containing 4 | EHD4 | 2.2 | (1) | |
| Elongation factor, RNA polymerase II, 2 | ELL2 | 1.8 | (1) | |
| ELM2 and Myb/SANT-like domain containing 1 | ELMSAN1 | 1.9 | ||
| Ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | ENPP2 | 2.8 | NL | |
| V-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | ETS2 | 4.2 | (1) | |
| Eyes absent homolog 1 (Drosophila) | EYA1 | 4.0 | ||
| Coagulation factor III (thromboplastin, tissue factor) | F3 | 2.6 | ||
| Family with sequence similarity 133, member A | FAM133A | 2.3 | ||
| Family with sequence similarity 24, member B | FAM24B | 3.3 | ||
| Fibrillin 1 | FBN1 | 1.9 | ||
| Fibroblast growth factor 13 | FGF13 | 2.4 | (1) | |
| Filamin A interacting protein 1-like | FILIP1L | 1.9 | ||
| Hypothetical LOC642477 | FLJ39632 | 3.4 | ||
| Fibronectin leucine rich transmembrane protein 2 | FLRT2 | 3.0 | (1) | |
| Folate receptor 1 (adult) | FOLR1 | 3.5 | ||
| Forkhead box C1 | FOXC1 | 2.9 | ||
| Fraser syndrome 1 | FRAS1 | 3.4 | ||
| Follistatin | FST | 3.5 | ||
| FXYD domain containing ion transport regulator 5 | FXYD5 | 3.3 | MES | |
| Frizzled homolog 2 (Drosophila) | FZD2 | 2.4 | (1) | |
| Growth arrest-specific 6 | GAS6 | 4.1 | ||
| Guanylate binding protein 2, interferon-inducible | GBP2 | 2.7 | ||
| Growth differentiation factor 15 | GDF15 | 1.8 | ||
| Glucose-fructose oxidoreductase domain containing 1 | GFOD1 | 2.8 | ||
| GDNF family receptor α 1 | GFRA1 | 2.6 | ||
| Gap junction protein, γ 12, 47kDa | GJC2 | 2.3 | ||
| G protein-coupled receptor 1 | GPR1 | 2.4 | (1) | |
| G protein-coupled receptor, family C, group 5, member C | GPRC5C | 2.3 | (1) | |
| Glutathione peroxidase 3 (plasma) | GPX3 | 2.3 | ||
| GRAM domain containing 3 | GRAMD3 | 2.7 | ||
| Gasdermin A | GSDMA | 2.1 | ||
| H2A histone family, member Y2 | H2AFY2 | 2.2 | ||
| HLA complex P5 | HCP5 | 2.3 | ||
| Hairy and enhancer of split 6 (Drosophila) | HES6 | 2.9 | ||
| Homeobox C13 | HOXC13 | 2.3 | ||
| Homeobox C6 | HOXC6 | 2.1 | ||
| Heat shock 27kDa protein 3 | HSPB3 | 2.1 | ||
| Heat shock 27kDa protein family, member 7 (cardiovascular) | HSPB7 | 6.0 | ||
| Immediate early response 3 | IER3 | 2.2 | ||
| Interferon induced transmembrane protein 1 | IFITM1 | 2.5 | ||
| Insulin-like growth factor binding protein 4 | IGFBP4 | 3.5 | ||
| Interleukin 13 receptor, α 2 | IL13RA2 | 3.4 | ||
| Interleukin 15 | IL15 | 2.1 | (1) | |
| Interleukin 7 receptor | IL7R | 2.4 | ||
| Integrin, α 11 | ITGA11 | 2.2 | ||
| Integrin, β 2 (complement component 3 receptor 3 and 4 subunit) | ITGB2 | 3.6 | MES | |
| Jagged 2 | JAG2 | 2.1 | ||
| Junctophilin 2 | JPH2 | 2.0 | ||
| Kazrin, periplakin interacting protein | KAZN | 3.0 | ||
| Potassium voltage-gated channel, Shab-related subfamily, member 1 | KCNB1 | 3.4 | ||
| Potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 | KCNS1 | 1.8 | ||
| KIAA1522 | KIAA1522 | 2.0 | (1) | |
| Kruppel-like factor 11 | KLF11 | 1.9 | ||
| Killin, p53-regulated DNA replication inhibitor | KLLN | 1.9 | ||
| Keratin 18 pseudogene 55 | KRT18P55 | 3.0 | ||
| Keratin 80 | KRT80 | 3.8 | ||
| Keratin 81 | KRT81 | 6.8 | ||
| Lactamase, β | LACTB | 2.0 | ||
| Layilin | LAYN | 1.9 | ||
| Leprecan-like 1 | LEPREL1 | 3.2 | ||
| LIM and senescent cell antigen-like domains 2 | LIMS2 | 4.6 | ||
| Long intergenic nonprotein coding RNA 263 | LINC00263 | 2.8 | ||
| LOC400464 | 2.0 | |||
| LOC400879 | 2.0 | |||
| LOC645638 | 3.6 | |||
| Lysyl oxidase | LOX | 3.0 | MES | |
| Leucine rich repeat neuronal 1 | LRRN1 | 1.9 | (1) | |
| Melanoma antigen family C, 2 | MAGEC2 | 2.4 | (1) | |
| Mitogen-activated protein kinase 13 | MAPK13 | 3.2 | MES | |
| Membrane-associated ring finger (C3HC4) 3 | MARCH3 | 1.9 | ||
| Mab-21 domain containing 2 | MB21D2 | 3.4 | ||
| Meis homeobox 3 pseudogene 1 | MEIS3P1 | 1.8 | ||
| Microfibrillar-associated protein 4 | MFAP4 | 1.8 | ||
| Microtubule associated monoxygenase, calponin and LIM domain containing 2 | MICAL2 | 3.0 | (1) | |
| MHC class I polypeptide-related sequence B | MICB | 2.1 | ||
| Mohawk homeobox | MKX | 3.3 | ||
| Melanophilin | MLPH | 2.9 | ||
| Matrix metallopeptidase 3 (stromelysin 1, progelatinase) | MMP3 | 3.2 | ||
| Mesothelin | MSLN | 3.8 | ||
| Methionine sulfoxide reductase B3 | MSRB3 | 3.7 | ||
| Mucin 1, cell surface associated | MUC1 | 1.9 | ||
| Matrix-remodelling associated 8 | MXRA8 | 2.2 | ||
| Myosin, light chain 1, alkali; skeletal, fast | MYL1 | 3.7 | (1) | |
| Myosin, light chain 9, regulatory | MYL9 | 3.4 | ||
| Myosin IC | MYO1C | 2.9 | ||
| Nucleosome assembly protein 1-like 3 | NAP1L3 | 1.8 | ||
| Neuroblastoma, suppression of tumorigenicity 1 | NBL1 | 3.4 | ||
| Neutral cholesterol ester hydrolase 1 | NCEH1 | 2.0 | ||
| Neutrophil cytosolic factor 2 (65kDa, chronic granulomatous disease, autosomal 2) | NCF2 | 3.6 | MES | |
| NIMA (never in mitosis gene a)-related kinase 7 | NEK7 | 2.8 | ||
| Nexilin (F actin binding protein) | NEXN | 2.5 | (1) | |
| NK2 homeobox 2 | NKX2-2 | 1.9 | PN | |
| Neuromedin U | NMU | 2.1 | ||
| N-terminal Xaa-Pro-Lys N-methyltransferase 1 | NTMT1 | 1.9 | ||
| Netrin G1 | NTNG1 | 3.8 | (1) | |
| Nuclear casein kinase and cyclin-dependent kinase substrate 1 | NUCKS1 | 4.7 | ||
| Opioid growth factor receptor-like 1 | OGFRL1 | 2.4 | ||
| Oxytocin receptor | OXTR | 1.9 | ||
| Procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), α polypeptide II | P4HA2 | 2.0 | MES | |
| P antigen family, member 5 (prostate associated) | PAGE5 | 4.2 | ||
| Palmdelphin | PALMD | 3.1 | ||
| 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | PAPSS2 | 3.5 | ||
| Paired box 6 | PAX6 | 2.0 | ||
| Platelet-derived growth factor receptor-like | PDGFRL | 2.3 | ||
| PDZ and LIM domain 1 (elfin) | PDLIM1 | 2.9 | ||
| Phosphoglucomutase 3 | PGM3 | 1.8 | ||
| Pleckstrin homology-like domain, family B, member 2 | PHLDB2 | 2.3 | ||
| Paired-like homeodomain 2 | PITX2 | 4.7 | (1) | |
| Placenta-specific 8 | PLAC8 | 2.9 | ||
| Pleiomorphic adenoma gene-like 1 | PLAGL1 | 2.0 | ||
| Pleckstrin homology domain containing, family F (with FYVE domain) member 1 | PLEKHF1 | 2.7 | ||
| Phospholipid scramblase 4 | PLSCR4 | 2.6 | ||
| Plexin A2 | PLXNA2 | 1.9 | (1) | |
| Phosphatidic acid phosphatase type 2 domain containing 1A | PPAPDC1A | 2.4 | ||
| Prickle homolog 1 (Drosophila) | PRICKLE1 | 2.9 | ||
| Proline rich 16 | PRR16 | 2.6 | (1) | |
| Protease, serine, 23 | PRSS23 | 2.6 | ||
| Pregnancy specific β-1-glycoprotein 4 | PSG4 | 5.0 | ||
| Protein tyrosine phosphatase, receptor type, F | PTPRF | 2.4 | ||
| Protein tyrosine phosphatase, receptor type, M | PTPRM | 1.9 | (1) | |
| RAB32, member RAS oncogene family | RAB32 | 2.1 | MES | |
| RAB3B, member RAS oncogene family | RAB3B | 5.1 | ||
| Retinoic acid early transcript 1G | RAET1G | 2.2 | ||
| Retinoic acid receptor, β | RARB | 3.9 | ||
| Retinoic acid receptor responder (tazarotene induced) 2 | RARRES2 | 2.6 | ||
| Ras association (RalGDS/AF-6) domain family 7 | RASSF7 | 1.8 | ||
| RNA binding motif, single stranded interacting protein | RBMS3 | 1.9 | (1) | |
| RNA binding protein with multiple splicing | RBPMS | 2.5 | (1) | |
| Reticulocalbin 3, EF-hand calcium binding domain | RCN3 | 1.9 | ||
| Reversion-inducing-cysteine-rich protein with kazal motifs | RECK | 1.9 | ||
| RAS-like, estrogen-regulated, growth inhibitor | RERG | 4.1 | ||
| Raftlin, lipid raft linker 1 | RFTN1 | 3.7 | ||
| RGM domain family, member B | RGMB | 1.9 | ||
| Regulator of G-protein signaling 10 | RGS10 | 1.8 | ||
| Rhomboid 5 homolog 2 (Drosophila) | RHBDF2 | 2.4 | ||
| Ribonuclease, RNase A family, 4 | RNASE4 | 2.4 | ||
| Ring finger protein 150 | RNF150 | 2.7 | ||
| Arginyl aminopeptidase (aminopeptidase B) | RNPEP | 1.8 | ||
| Ribosomal protein S6 kinase, 90kDa, polypeptide 2 | RPS6KA2 | 2.2 | ||
| Related RAS viral (r-ras) oncogene homolog 2 | RRAS2 | 2.1 | (1) | |
| Sodium channel, nonvoltage-gated 1 α | SCNN1A | 3.5 | ||
| Serologically defined colon cancer antigen 3 | SDCCAG3 | 2.3 | ||
| Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B | SEMA3B | 3.1 | ||
| Serpin peptidase inhibitor, clade B (ovalbumin), member 7 | SERPINB7 | 3.1 | (1) | |
| SERTA domain containing 4 | SERTAD4 | 3.2 | ||
| SERTAD4 antisense RNA 1 | SERTAD4-AS1 | 3.3 | ||
| Surfactant associated 1, pseudogene | SFTA1P | 2.3 | ||
| SH2 domain containing 4A | SH2D4A | 2.4 | ||
| SH3-domain GRB2-like 3 | SH3GL3 | 7.2 | (1) | NL |
| SH3 domain containing ring finger 2 | SH3RF2 | 2.8 | ||
| Solute carrier family 1 (neutral amino acid transporter), member 5 | SLC1A5 | 2.0 | ||
| Solute carrier family 22 (organic cation transporter), member 4 | SLC22A4 | 1.8 | ||
| Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 | SLC25A24 | 1.9 | ||
| Solute carrier family 25, member 43 | SLC25A43 | 2.0 | ||
| Solute carrier family 38, member 1 | SLC38A1 | 3.2 | (1) | |
| Solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | SLC6A9 | 2.0 | CL | |
| Solute carrier family 9 (sodium/hydrogen exchanger), member 7 | SLC9A7 | 2.3 | ||
| SMAD family member 6 | SMAD6 | 3.1 | (1) | |
| Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) | SMG6 | 2.1 | ||
| Sosondowah ankyrin repeat domain family member A | SOWAHA | 2.5 | ||
| SPANX family, member C | SPANXC | 2.9 | (1) | |
| SPANX family, member E | SPANXE | 4.9 | ||
| SPATA31 subfamily C, member 2 | SPATA31C2 | 2.2 | ||
| Sphingosine kinase 1 | SPHK1 | 2.0 | ||
| Serine palmitoyltransferase, long chain base subunit 3 | SPTLC3 | 3.0 | ||
| Serglycin | SRGN | 3.1 | ||
| Slingshot homolog 3 (Drosophila) | SSH3 | 2.0 | CL | |
| Sjogren syndrome/scleroderma autoantigen 1 | SSSCA1 | 2.2 | (1) | |
| ST6 (α-N-acetyl-neuraminyl-2,3-β-galactosyl-1,3)-N-acetylgalactosaminide α-2,6-sialyltransferase 3 | ST6GALNAC3 | 2.0 | (1) | |
| StAR-related lipid transfer (START) domain containing 5 | STARD5 | 1.9 | (1) | |
| StAR-related lipid transfer (START) domain containing 8 | STARD8 | 2.2 | (1) | |
| Stanniocalcin 2 | STC2 | 3.5 | ||
| Syntaxin binding protein 5 (tomosyn) | STXBP5 | 2.7 | (1) | |
| Syncoilin, intermediate filament protein | SYNC | 2.3 | ||
| Transgelin | TAGLN | 3.3 | ||
| TEA domain family member 4 | TEAD4 | 3.1 | ||
| Teneurin transmembrane protein 4 | TENM4 | 2.2 | ||
| Transforming growth factor, β 2 | TGFB2 | 6.5 | ||
| Transforming growth factor, β receptor III | TGFBR3 | 2.6 | ||
| Thrombospondin 1 | THBS1 | 2.7 | (1) | MES |
| TIMP metallopeptidase inhibitor 3 (Sorsby fundus dystrophy, pseudoinflammatory) | TIMP3 | 4.6 | ||
| Transmembrane protein 255B | TMEM255B | 2.0 | ||
| Transmembrane protein 44 | TMEM44 | 1.9 | ||
| Transmembrane protein 45A | TMEM45A | 2.3 | (1) | |
| Tumor protein p53 inducible protein 11 | TP53I11 | 2.8 | (1) | |
| Two pore segment channel 2 | TPCN2 | 2.0 | ||
| Tropomyosin 1 (α) | TPM1 | 2.0 | ||
| Tropomyosin 2 (β) | TPM2 | 1.8 | ||
| Tetraspanin 10 | TSPAN10 | 2.9 | ||
| Tetraspanin 4 | TSPAN4 | 2.1 | ||
| Tuftelin 1 | TUFT1 | 2.5 | ||
| Thioredoxin interacting protein | TXNIP | 1.8 | ||
| Urothelial cancer associated 1 | UCA1 | 2.4 | ||
| Vascular endothelial growth factor C | VEGFC | 1.8 | ||
| WAS protein family, member 3 | WASF3 | 1.9 | ||
| Wingless-type MMTV integration site family member 2 | WNT2 | 3.3 | ||
| Wingless-type MMTV integration site family, member 3 | WNT3 | 3.1 | ||
| Wingless-type MMTV integration site family, member 5B | WNT5B | 3.6 | ||
| WW and C2 domain containing 1 | WWC1 | 2.1 | ||
| Yip1 interacting factor homolog A (Saccharomyces cerevisiae) | YIF1A | 1.9 | ||
| Zinc finger, CCHC domain containing 5 | ZCCHC5 | 3.1 | (1) | |
| Zinc finger protein 618 | ZNF618 | 2.2 | ||
| Zinc finger protein 730 | ZNF730 | 1.8 | ||
| CSC | ||||
|---|---|---|---|---|
| Description | Symbol | Fold Change (Log2) | Sox2 Target (Ref) | Subclass Signature |
| AP2 associated kinase 1 | AAK1 | − 2.5 | (1) | |
| ATP-binding cassette, subfamily A (ABC1), member 2 | ABCA2 | − 2.5 | ||
| Acetylcholinesterase (Yt blood group) | ACHE | − 2.2 | ||
| Acid phosphatase 6, lysophosphatidic | ACP6 | − 2.7 | ||
| Acyl-CoA synthetase short-chain family member 1 | ACSS1 | − 2.0 | ||
| Activin A receptor, type IIA | ACVR2A | − 3.5 | (1) | |
| Adenosine deaminase | ADA | − 2.1 | ||
| ADAM metallopeptidase domain 15 | ADAM15 | − 2.1 | ||
| ADAM metallopeptidase with thrombospondin type 1 motif, 6 | ADAMTS6 | − 2.2 | (1) | |
| ADAM metallopeptidase with thrombospondin type 1 motif, 8 | ADAMTS8 | − 3.9 | (1) | |
| Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | AGT | − 2.1 | ||
| Allograft inflammatory factor 1-like | AIF1L | − 2.3 | ||
| Adenylate kinase 4 | AK4 | − 2.5 | ||
| A kinase (PRKA) anchor protein 10 | AKAP10 | − 1.8 | ||
| V-akt murine thymoma viral oncogene homolog 2 | AKT2 | − 1.8 | CL | |
| Aldehyde dehydrogenase 4 family, member A1 | ALDH4A1 | − 2.1 | ||
| Aldehyde dehydrogenase 6 family, member A1 | ALDH6A1 | − 2.7 | ||
| Antagonist of mitotic exit network 1 homolog (Saccharomyces cerevisiae) | AMN1 | − 2.1 | ||
| Angiopoietin 2 | ANGPT2 | − 4.3 | ||
| Ρ guanine nucleotide exchange factor (GEF) 39 | ARHGEF39 | − 1.8 | ||
| ADP-ribosylation factor-like 4A | ARL4A | − 2.9 | (1) | |
| Arylsulfatase G | ARSG | − 1.8 | ||
| Ankyrin repeat and SOCS box-containing 9 | ASB9 | − 3.6 | (1) | |
| Asparaginase like 1 | ASRGL1 | − 2.1 | ||
| Astrotactin 1 | ASTN1 | − 2.0 | ||
| Autophagy related 13 | ATG13 | − 3.6 | ||
| ATPase type 13A4 | ATP13A4 | − 3.6 | ||
| ATPase, Na +/K + transporting, β 2 polypeptide | ATP1B2 | − 2.3 | ||
| ATPase, class II, type 9B | ATP9B | − 2.1 | (1) | |
| ATP binding domain 4 | ATPBD4 | − 1.9 | ||
| UDP-GlcNAc:βGal β-1,3-N-acetylglucosaminyltransferase 2 | B3GNT2 | − 2.0 | (1) | |
| Bardet-Biedl syndrome 9 | BBS9 | − 3.1 | ||
| B cell receptor-associated protein 29 | BCAP29 | − 6.0 | ||
| B-box and SPRY domain containing | BSPRY | − 2.1 | ||
| Βcellulin | BTC | − 2.2 | (1) | |
| Chromosome 10 open reading frame 12 | C10orf12 | − 2.4 | ||
| Chromosome 11 open reading frame 63 | C11orf63 | − 2.4 | ||
| Chromosome 19 open reading frame 6 | C19orf6 | − 1.8 | ||
| Chromosome 2 open reading frame 80 | C2orf80 | − 5.8 | ||
| Chromosome 6 open reading frame 15 | C6orf15 | − 2.0 | ||
| Chromosome 6 open reading frame 170 | C6orf170 | − 1.9 | ||
| Carbonic anhydrase II | CA2 | − 4.3 | ||
| Carbonic anhydrase IV | CA4 | − 2.4 | (1) | NL |
| Calcium channel, voltage-dependent, P/Q type, α 1A subunit | CACNA1A | − 2.8 | (1) | |
| Calcium channel, voltage-dependent, β 1 subunit | CACNB1 | − 2.6 | ||
| Cell adhesion molecule 1 | CADM1 | − 4.5 | ||
| Ca2+−dependent activator protein for secretion 2 | CADPS2 | − 2.0 | (1) | |
| Calpain 9 | CAPN9 | − 2.8 | (1) | |
| Calcyphosine | CAPS | − 2.3 | ||
| Coiled-coil domain containing 103 | CCDC103 | − 4.3 | ||
| Coiled-coil domain containing 94 | CCDC94 | − 2.2 | ||
| CD14 molecule | CD14 | − 2.2 | MES | |
| CD276 molecule | CD276 | − 2.4 | ||
| CD79b molecule, immunoglobulin-associated β | CD79B | − 2.1 | ||
| Cadherin 6, type 2, K-cadherin (fetal kidney) | CDH6 | − 3.1 | (1) | CL |
| Carcinoembryonic antigen-related cell adhesion molecule 21 | CEACAM21 | − 2.2 | ||
| CCAAT/enhancer binding protein (C/EBP), δ | CEBPD | − 1.8 | (1) | |
| CUGBP, Elav-like family member 1 | CELF1 | − 2.4 | ||
| Centrosomal protein 152kDa | CEP152 | − 1.8 | ||
| Centrosomal protein 170kDa | CEP170 | − 2.7 | (1) | |
| Centrosomal protein 70kDa | CEP70 | − 2.1 | ||
| Complement factor I | CFI | − 2.2 | ||
| Chorionic gonadotropin, β polypeptide | CGB | − 2.1 | ||
| Chromogranin B (secretogranin 1) | CHGB | − 2.3 | (1) | |
| C-type lectin domain family 4, member A | CLEC4A | − 1.9 | ||
| C-type lectin domain family 9, member A | CLEC9A | − 2.0 | ||
| CCR4-NOT transcription complex, subunit 3 | CNOT3 | − 2.1 | ||
| Cannabinoid receptor 1 (brain) | CNR1 | − 2.1 | ||
| Cordon-bleu WH2 repeat protein | COBL | − 3.5 | (1) | |
| Component of oligomeric golgi complex 5 | COG5 | − 2.3 | ||
| Collagen, type XI, α 2 | COL11A2 | − 2.0 | (1) | |
| Collagen, type V, α 1 | COL5A1 | − 1.8 | (1) | MES |
| Carnitine palmitoyltransferase 1A (liver) | CPT1A | − 1.9 | ||
| Crystallin, μ | CRYM | − 4.5 | NL | |
| Cold shock domain containing C2, RNA binding | CSDC2 | − 3.0 | (1) | |
| CTAGE family, member 11, pseudogene | CTAGE11P | − 2.2 | ||
| CTS telomere maintenance complex component 1 | CTC1 | − 2.4 | ||
| Chemokine (C-X-C motif) ligand 3 | CXCL3 | − 1.8 | ||
| Dachsous 1 (Drosophila) | DCHS1 | − 2.1 | ||
| Doublecortin-like kinase 2 | DCLK2 | − 2.6 | ||
| Decorin | DCN | − 3.0 | (1) | |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 | DDX51 | − 1.9 | ||
| DENN/MADD domain containing 1A | DENND1A | − 2.5 | (1) | |
| Der1-like domain family, member 3 | DERL3 | − 8.2 | ||
| Dehydrogenase/reductase (SDR family) member 12 | DHRS12 | − 3.7 | ||
| DEXH (Asp-Glu-X-His) box polypeptide 58 | DHX58 | − 2.3 | ||
| Diaphanous homolog 3 (Drosophila) | DIAPH3 | − 2.1 | (1) | |
| DIS3 mitotic control homolog (Saccharomyces cerevisiae) | DIS3 | − 1.8 | ||
| Dickkopf homolog 1 (Xenopus laevis) | DKK1 | − 4.4 | (4) | |
| Dynein, axonemal, assembly factor 3 | DNAAF3 | − 5.2 | ||
| DnaJ (Hsp40) homolog, subfamily C, member 12 | DNAJC12 | − 2.5 | ||
| Estrogen receptor binding site associated, antigen, 9 | EBAG9 | − 2.0 | ||
| Extracellular matrix protein 2, female organ and adipocyte specific | ECM2 | − 3.0 | ||
| EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | − 1.9 | (1) | |
| Engulfment and cell motility 1 | ELMO1 | − 2.7 | (1) | |
| Exoribonuclease 1 | ERI1 | − 2.1 | ||
| ERI1 exoribonuclease family member 2 | ERI2 | − 2.1 | ||
| Ets variant gene 3 | ETV3 | − 2.9 | (1) | |
| Fatty acid binding protein 7, brain | FABP7 | − 2.0 | ||
| Family with sequence similarity 153, member B | FAM153B | − 3.7 | ||
| Family with sequence similarity 5, member B | FAM5B | − 2.6 | (1) | |
| Fanconi anemia, complementation group A | FANCA | − 2.7 | ||
| Fatty acyl CoA reductase 2 | FAR2 | − 2.0 | ||
| Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | FGFR1 | − 2.9 | ||
| Filamin A interacting protein 1-like | FILIP1L | − 2.1 | ||
| Hypothetical protein FLJ22184 | FLJ22184 | − 2.8 | ||
| Fibronectin leucine rich transmembrane protein 3 | FLRT3 | − 1.9 | ||
| Fibromodulin | FMOD | − 2.0 | ||
| Forkhead box A2 | FOXA2 | − 2.0 | (1) | |
| Forkhead box R1 | FOXR1 | − 8.3 | ||
| FRAS1 related extracellular matrix protein 2 | FREM2 | − 3.8 | ||
| FRMD6 antisense RNA 1 | FRMD6-AS1 | − 2.1 | ||
| GRB2-associated binding protein 1 | GAB1 | − 4.6 | (1) | |
| GRB2-associated binding protein 2 | GAB2 | − 1.9 | (1) | |
| UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) | GALNT13 | − 2.9 | (1) | |
| UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) | GALNT3 | − 2.8 | ||
| Growth associated protein 43 | GAP43 | − 4.2 | (1) | |
| Gap junction protein, β 6, 30kDa | GJB6 | − 3.0 | ||
| Guanine nucleotide binding protein (G protein), γ 7 | GNG7 | − 2.5 | CL | |
| G patch domain containing 2 | GPATCH2 | − 2.2 | ||
| Glycoprotein M6B | GPM6B | − 3.2 | ||
| Glutathione peroxidase 3 (plasma) | GPX3 | − 2.0 | ||
| Glutamate receptor, ionotropic, kainate 1 | GRIK1 | − 1.8 | CL | |
| G protein-coupled receptor kinase 5 | GRK5 | − 1.8 | (1) | |
| Germ cell associated 1 | GSG1 | − 2.0 | ||
| H1 histone family, member N, testis-specific | H1FNT | − 2.8 | ||
| HemK methyltransferase family member 1 | HEMK1 | − 2.6 | ||
| Hairy/enhancer-of-split related with YRPW motif 2 | HEY2 | − 2.9 | (1) | |
| Hypoxia inducible factor 3, α subunit | HIF3A | − 2.2 | ||
| Major histocompatibility complex, class II, DM β | HLA-DMB | − 5.2 | ||
| Heat shock 27kDa protein 3 | HSPB3 | − 2.1 | ||
| HUS1 checkpoint homolog b (S. pombe) | HUS1B | − 3.4 | ||
| Huntingtin interacting protein K | HYPK | − 1.9 | ||
| Islet cell autoantigen 1, 69kDa | ICA1 | − 3.0 | ||
| Interferon, α-inducible protein 27 | IFI27 | − 2.3 | ||
| Insulin-like growth factor binding protein 7 | IGFBP7 | − 3.1 | (1) | |
| IGF-like family member 3 | IGFL3 | − 4.8 | ||
| Immunoglobulin superfamily, member 11 | IGSF11 | − 3.2 | (1) | |
| Interleukin 8 | IL8 | − 2.4 | (1) | |
| InaD-like (Drosophila) | INADL | − 2.9 | ||
| Insulinoma-associated 1 | INSM1 | − 2.2 | ||
| Inositol hexakisphosphate kinase 3 | IP6K3 | − 1.8 | ||
| Interferon regulatory factor 4 | IRF4 | − 1.9 | ||
| Integrin, α 6 | ITGA6 | − 2.6 | ||
| Kelch repeat and BTB (POZ) domain containing 3 | KBTBD3 | − 2.0 | ||
| Potassium channel tetramerisation domain containing 21 | KCTD21 | − 5.2 | ||
| KIAA0319-like | KIAA0319L | − 3.4 | (1) | |
| Kruppel-like factor 4 (gut) | KLF4 | − 2.3 | ||
| Kruppel-like factor 7 (ubiquitous) | KLF7 | − 1.8 | ||
| Kelch domain containing 8A | KLHDC8A | − 2.1 | CL | |
| Kelch-like 23 (Drosophila) | KLHL23 | − 2.0 | ||
| Kelch-like 5 (Drosophila) | KLHL5 | − 3.8 | ||
| Killer cell lectin-like receptor subfamily F, member 1 | KLRF1 | − 4.5 | ||
| Like-glycosyltransferase | LARGE | − 5.1 | (1) | |
| Lysocardiolipin acyltransferase 1 | LCLAT1 | − 1.8 | ||
| Leptin (obesity homolog, mouse) | LEP | − 1.9 | (1) | |
| Leprecan-like 1 | LEPREL1 | − 3.0 | ||
| Leucine-rich repeat LGI family, member 2 | LGI2 | − 3.5 | ||
| Long intergenic nonprotein coding RNA 261 | LINC00261 | − 3.1 | ||
| Long intergenic nonprotein coding RNA 282 | LINC00282 | − 2.1 | ||
| Long intergenic nonprotein coding RNA 324 | LINC00324 | − 1.9 | ||
| Long intergenic nonprotein coding RNA 478 | LINC00478 | − 2.2 | ||
| Long intergenic nonprotein coding RNA 612 | LINC00612 | − 1.8 | ||
| Long intergenic nonprotein coding RNA 623 | LINC00623 | − 2.9 | ||
| LOC100129502 | − 1.9 | |||
| LOC100130276 | − 2.8 | |||
| LOC100131096 | − 2.3 | |||
| LOC100132354 | − 2.2 | |||
| LOC255167 | − 2.1 | |||
| LOC283710 | − 2.9 | |||
| LOC285696 | − 3.7 | |||
| LOC389765 | − 2.5 | |||
| LOC441155 | − 2.9 | |||
| LOC645355 | − 2.1 | |||
| LOC653562 | − 7.3 | |||
| LOC728755 | − 2.2 | |||
| LOC730441 | − 3.4 | |||
| Lysyl oxidase-like 3 | LOXL3 | − 2.3 | ||
| Plasticity related gene 1 | LPPR4 | − 2.7 | ||
| Leucine rich repeat containing 37, member A3 | LRRC37A3 | − 3.1 | ||
| Leucine rich repeat neuronal 1 | LRRN1 | − 3.9 | (1) | |
| Leucine rich repeat neuronal 3 | LRRN3 | − 3.0 | ||
| Lumican | LUM | − 2.2 | ||
| Lymphocyte antigen 6 complex, locus G5C | LY6G5C | − 2.2 | ||
| Lymphocyte antigen 6 complex, locus H | LY6H | − 3.8 | ||
| Melanoma antigen family A, 11 | MAGEA11 | − 4.2 | ||
| Mal, T cell differentiation protein | MAL | − 2.1 | ||
| MANSC domain containing 1 | MANSC1 | − 2.0 | ||
| Microtubule-associated protein 2 | MAP2 | − 2.9 | (1) | PN |
| Mitogen-activated protein kinase kinase 6 | MAP2K6 | − 1.8 | ||
| Mitogen-activated protein kinase kinase 7 | MAP2K7 | − 2.1 | (1) | |
| Microtubule-associated protein 6 | MAP6 | − 2.6 | ||
| Microtubule-associated protein τ | MAPT | − 2.7 | (1) | PN |
| Membrane-associated ring finger (C3HC4) 2 | MARCH2 | − 1.8 | ||
| Microtubule associated serine/threonine kinase family member 4 | MAST4 | − 3.4 | ||
| MEF2B neighbor | MEF2BNB | − 2.7 | ||
| Malignant fibrous histiocytoma amplified sequence 1 | MFHAS1 | − 2.8 | (1) | |
| Myocardial infarction associated transcript (nonprotein coding) | MIAT | − 3.1 | ||
| MLLT4 antisense RNA 1 (head to head) | MLLT4-AS1 | − 1.9 | ||
| Metallophosphoesterase 1 | MPPE1 | − 2.0 | ||
| Mucin 12, cell surface associated | MUC12 | − 1.9 | ||
| Myeloid-associated differentiation marker | MYADM | − 2.1 | ||
| Myosin binding protein C, slow type | MYBPC1 | − 2.3 | NL | |
| V-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) | MYCN | − 2.1 | (1) | |
| Myosin IE | MYO1E | − 1.8 | (1) | |
| Myosin VC | MYO5C | − 2.2 | (1) | CL |
| N-myc downstream regulated gene 1 | NDRG1 | − 1.8 | ||
| Nicotinamide nucleotide transhydrogenase | NNT | − 3.3 | (1) | |
| NADPH oxidase activator 1 | NOXA1 | − 2.3 | (1) | |
| Nuclear pore complex interacting protein family, member B15 | NPIPL2 | − 2.6 | ||
| Neuropeptide Y receptor Y5 | NPY5R | − 2.5 | ||
| Neuronal cell adhesion molecule | NRCAM | − 2.2 | ||
| Nuclear receptor binding SET domain protein 1 | NSD1 | − 2.2 | ||
| Neurotensin | NTS | − 7.2 | ||
| 2′-5′-oligoadenylate synthetase 2, 69/71kDa | OAS2 | − 1.8 | ||
| Protein kinase C and casein kinase substrate in neurons 2 | PACSIN2 | − 2.0 | ||
| Peptidyl arginine deiminase, type II | PADI2 | − 5.2 | ||
| Protocadherin γ subfamily A, 5 | PCDHGA5 | − 2.2 | ||
| Protocadherin γ subfamily B, 6 | PCDHGB6 | − 2.4 | ||
| Pecanex-like 4 (Drosophila) | PCNXL4 | − 5.6 | ||
| Phosphodiesterase 8B | PDE8B | − 6.0 | (1) | |
| Platelet-derived growth factor β polypeptide (simian sarcoma viral (v-sis) oncogene homolog) | PDGFB | − 1.9 | (1) | |
| PDZ and LIM domain 5 | PDLIM5 | − 2.3 | (1) | |
| Podoplanin | PDPN | − 1.8 | MES | |
| PDZ domain containing 2 | PDZD2 | − 1.9 | ||
| PDZ domain containing RING finger 3 | PDZRN3 | − 3.0 | ||
| PHD finger protein 8 | PHF8 | − 3.4 | ||
| Phosphatidylinositol-4-phosphate 5-kinase-like 1 | PIP5KL1 | − 2.1 | ||
| Phospholipase A2, group IID | PLA2G2D | − 2.3 | ||
| Polo-like kinase 2 (Drosophila) | PLK2 | − 1.8 | ||
| Phospholipid transfer protein | PLTP | − 3.2 | ||
| Postmeiotic segregation increased 2 pseudogene 1 | PMS2P1 | − 2.0 | ||
| POM121 transmembrane nucleoporin-like 8 pseudogene | POM121L8P | − 4.3 | ||
| Protein phosphatase 2 (formerly 2A), regulatory subunit B″, α | PPP2R3A | − 2.2 | ||
| Proline rich 13 | PRR13 | − 2.1 | ||
| Protein tyrosine phosphatase type IVA, member 2 | PTP4A2 | − 2.5 | ||
| Poliovirus receptor-related 2 (herpesvirus entry mediator B) | PVRL2 | − 4.4 | ||
| Pyrroline-5-carboxylate reductase 1 | PYCR1 | − 4.2 | ||
| Pyrin and HIN domain family, member 1 | PYHIN1 | − 3.8 | ||
| RAB24, member RAS oncogene family | RAB24 | − 1.9 | ||
| RAB3A, member RAS oncogene family | RAB3A | − 1.9 | ||
| RAB3A interacting protein (rabin3) | RAB3IP | − 3.5 | (1) | |
| Retinoic acid receptor responder (tazarotene induced) 1 | RARRES1 | − 2.6 | ||
| RNA binding motif (RNP1, RRM) protein 3 | RBM3 | − 3.2 | ||
| Retinol binding protein 7, cellular | RBP7 | − 2.6 | ||
| Regulator of calcineurin 2 | RCAN2 | − 3.0 | ||
| Replication factor C (activator 1) 5, 36.5kDa | RFC5 | − 2.8 | ||
| Raftlin family member 2 | RFTN2 | − 2.4 | ||
| Regulator of cell cycle | RGCC | − 2.3 | ||
| Rhomboid, veinlet-like 2 (Drosophila) | RHBDL2 | − 2.3 | ||
| Ρ family GTPase 2 | RND2 | − 2.3 | ||
| Ring finger protein 14 | RNF14 | − 2.0 | ||
| Ring finger protein 148 | RNF148 | − 1.8 | ||
| RNA, Ro-associated Y4 | RNY4 | − 2.3 | ||
| Ribosomal protein L37a | RPL37A | − 2.8 | ||
| Ribosomal protein S6 kinase, 90kDa, polypeptide 2 | RPS6KA2 | − 2.4 | ||
| Radical S-adenosyl methionine domain containing 2 | RSAD2 | − 3.4 | ||
| Runt-related transcription factor 3 | RUNX3 | − 1.9 | ||
| Scavenger receptor class A, member 3 | SCARA3 | − 1.8 | ||
| Secretogranin III | SCG3 | − 7.1 | PN | |
| Secretagogin, EF-hand calcium binding protein | SCGN | − 2.1 | ||
| Sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F | SEMA4F | − 2.6 | ||
| Serine incorporator 2 | SERINC2 | − 3.4 | ||
| Surfactant associated 1, pseudogene | SFTA1P | − 2.8 | ||
| SH2B adaptor protein 2 | SH2B2 | − 2.7 | (1) | |
| SH3-domain kinase binding protein 1 | SH3KBP1 | − 2.2 | ||
| SH3 domain containing ring finger 2 | SH3RF2 | − 3.1 | ||
| Signal-induced proliferation-associated gene 1 | SIPA1 | − 2.8 | (1) | |
| Signal-regulatory protein α | SIRPA | − 4.8 | (1) | |
| Solute carrier family 16, member 4 (monocarboxylic acid transporter 5) | SLC16A4 | − 1.8 | ||
| Solute carrier family 2 (facilitated glucose transporter), member 12 | SLC2A12 | − 2.6 | ||
| Solute carrier family 30 (zinc transporter), member 1 | SLC30A1 | − 3.6 | (1) | |
| Solute carrier family 35, member F1 | SLC35F1 | − 4.9 | ||
| Solute carrier family 38, member 8 | SLC38A8 | − 3.0 | ||
| Solute carrier family 39 (zinc transporter), member 4 | SLC39A4 | − 2.6 | ||
| Solute carrier family 5 (iodide transporter), member 8 | SLC5A8 | − 4.4 | ||
| Solute carrier family 6 (proline IMINO transporter), member 20 | SLC6A20 | − 2.2 | ||
| Solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | SLC6A9 | − 5.3 | CL | |
| Solute carrier organic anion transporter family, member 1C1 | SLCO1C1 | − 2.4 | ||
| Secretory leukocyte peptidase inhibitor | SLPI | − 3.7 | ||
| Small integral membrane protein 1 | SMIM1 | − 2.2 | ||
| Small nucleolar RNA, H/ACA box 28 | SNORA28 | − 1.8 | ||
| Small nucleolar RNA, C/D box 114-3 | SNORD114-3 | − 1.8 | ||
| Small nucleolar RNA, C/D box 14C | SNORD14C | − 2.1 | ||
| Small nucleolar RNA, C/D box 6 | SNORD6 | − 5.9 | ||
| Small nucleolar RNA, C/D box 95 | SNORD95 | − 2.0 | ||
| Sorbin and SH3 domain containing 1 | SORBS1 | − 1.8 | (1) | |
| SRY (sex-determining region Y)-box 2 | SOX2 | − 1.6 | PN | |
| SRY (sex-determining region Y)-box 21 | SOX21 | − 4.3 | (1)(2) | |
| SOX2 overlapping transcript (nonprotein coding) | SOX2-OT | − 2.0 | ||
| SRY (sex-determining region Y)-box 8 | SOX8 | − 3.7 | ||
| Sperm associated antigen 1 | SPAG1 | − 2.0 | ||
| SPARC-like 1 (mast9, hevin) | SPARCL1 | − 2.2 | ||
| Spermatogenesis and centriole associated 1 | SPATC1 | − 2.2 | ||
| Speedy/RINGO cell cycle regulator family member E3 | SPDYE3 | − 2.7 | ||
| Sprouty homolog 4 (Drosophila) | SPRY4 | − 2.0 | (1) | |
| SLIT-ROBO Ρ GTPase activating protein 1 | SRGAP1 | − 2.1 | (1) | |
| ST8SIA6 antisense RNA 1 | ST8SIA6-AS1 | − 1.9 | ||
| STAU2 antisense RNA 1 | STAU2-AS1 | − 3.2 | ||
| Synapse differentiation inducing 1 | SYNDIG1 | − 2.2 | ||
| TBC1 domain family, member 17 | TBC1D17 | − 2.5 | ||
| Transducin (β)-like 3 | TBL3 | − 1.8 | ||
| Tyrosyl-DNA phosphodiesterase 1 | TDP1 | − 1.9 | ||
| Trefoil factor 3 (intestinal) | TFF3 | − 3.0 | ||
| Transmembrane protein 100 | TMEM100 | − 1.9 | ||
| Transmembrane protein 106A | TMEM106A | − 1.9 | (1) | |
| Transmembrane protein 156 | TMEM156 | − 3.1 | ||
| Transmembrane protein 161B | TMEM161B | − 2.2 | ||
| Transmembrane protein 164 | TMEM164 | − 3.0 | ||
| Transmembrane protein 178A | TMEM178A | − 1.9 | ||
| Tumor protein p63 | TP63 | − 5.4 | ||
| Tumor protein D52 | TPD52 | − 3.8 | (1) | |
| Tripartite motif-containing 24 | TRIM24 | − 1.8 | (1) | |
| tRNA methyltransferase 44 homolog (Saccharomyces cerevisiae) | TRMT44 | − 2.8 | ||
| Transient receptor potential cation channel, subfamily M, member 3 | TRPM3 | − 2.8 | (1) | |
| Tetraspanin 14 | TSPAN14 | − 2.1 | ||
| Testis specific protein, Y-linked 1 | TSPY1 | − 2.4 | ||
| Tweety homolog 1 (Drosophila) | TTYH1 | − 3.1 | ||
| Taxilin γ 2, pseudogene | TXLNG2P | − 2.0 | ||
| UDP glucuronosyltransferase 2 family, polypeptide B11 | UGT2B11 | − 4.6 | ||
| Urotensin 2 | UTS2 | − 2.3 | ||
| Vaccinia related kinase 3 | VRK3 | − 4.9 | ||
| WD repeat domain 49 | WDR49 | − 2.0 | (1) | |
| WD repeat domain 74 | WDR74 | − 3.0 | ||
| WD repeat domain 86 | WDR86 | − 2.4 | ||
| WD repeat domain 89 | WDR89 | − 3.3 | ||
| X antigen family, member 1D | XAGE1D | − 6.3 | ||
| Zinc finger and BTB domain containing 40 | ZBTB40 | − 7.8 | ||
| Zinc finger with KRAB and SCAN domains 5 | ZKSCAN5 | − 3.2 | ||
| Zinc finger protein 182 | ZNF182 | − 5.5 | ||
| Zinc finger protein 519 | ZNF519 | − 2.3 | ||
| Zinc finger protein 555 | ZNF555 | − 1.9 | ||
| Zinc finger protein 608 | ZNF608 | − 2.0 | (1) | |
| Zinc finger protein 678 | ZNF678 | − 2.3 | ||
| Zinc finger protein 717 | ZNF717 | − 2.3 | ||
| Zinc finger protein 761 | ZNF761 | − 1.8 | ||
| Zinc finger protein 880 | ZNF880 | − 2.4 | ||
| Zinc finger protein 890, pseudogene | ZNF890P | − 2.0 | ||
| Zinc finger and SCAN domain containing 12 | ZSCAN12 | − 1.8 | ||
| Zinc finger and SCAN domain containing 16 | ZSCAN16 | − 2.4 | ||
| Zinc finger, X-linked, duplicated B | ZXDB | − 1.8 | ||
| Aminoadipate aminotransferase | AADAT | 5.0 | (1) | |
| Acyl-Coenzyme A dehydrogenase family, member 8 | ACAD8 | 1.8 | ||
| Actin, α 2, smooth muscle, aorta | ACTA2 | 2.2 | ||
| ADAM metallopeptidase domain 28 | ADAM28 | 2.3 | (1) | |
| ADAM metallopeptidase with thrombospondin type 1 motif, 18 | ADAMTS18 | 1.8 | ||
| Adenylate cyclase 9 | ADCY9 | 3.3 | ||
| Alcohol dehydrogenase 6 (class V) | ADH6 | 2.3 | ||
| AF4/FMR2 family, member 3 | AFF3 | 2.7 | (1) | |
| 1-acylglycerol-3-phosphate O-acyltransferase 3 | AGPAT3 | 2.5 | ||
| Aldo-keto reductase family 1 member 15 (ALD2) | AKR1B15 | 1.9 | ||
| Aminolevulinate, δ-, synthase 2 (sideroblastic/hypochromic anemia) | ALAS2 | 2.9 | ||
| Angiopoietin-like 3 | ANGPTL3 | 2.3 | ||
| Angiopoietin-like 4 | ANGPTL4 | 1.9 | ||
| Ankyrin 1, erythrocytic | ANK1 | 2.4 | ||
| Anoctamin 2 | ANO2 | 2.1 | ||
| Annexin A8-like 2 | ANXA8L2 | 2.5 | ||
| Adaptor-related protein complex 1 associated regulatory protein | AP1AR | 3.3 | ||
| Amyloid β (A4) precursor protein-binding, family B, member 2 (Fe65-like) | APBB2 | 8.0 | ||
| Adenomatosis polyposis coli down-regulated 1-like | APCDD1L | 1.9 | (1) | |
| Apolipoprotein E | APOE | 1.9 | ||
| Ρ GDP dissociation inhibitor (GDI) β | ARHGDIB | 2.2 | ||
| ADP-ribosylation factor-like 9 | ARL9 | 1.8 | ||
| Armadillo repeat containing 8 | ARMC8 | 1.9 | ||
| Achaete-scute complex homolog 2 (Drosophila) | ASCL2 | 2.8 | ||
| Aspartic peptidase, retroviral-like 1 | ASPRV1 | 2.1 | ||
| Argininosuccinate synthetase 1 | ASS1 | 2.7 | ||
| ATPase, class V, type 10B | ATP10B | 2.4 | ||
| Attractin | ATRN | 2.1 | ||
| BAI1-associated protein 2-like 1 | BAIAP2L1 | 2.1 | (1) | |
| Biglycan | BGN | 2.5 | ||
| Βine-homocysteine methyltransferase 2 | BHMT2 | 1.9 | ||
| BR serine/threonine kinase 2 | BRSK2 | 2.1 | ||
| Bone marrow stromal cell antigen 1 | BST1 | 2.0 | ||
| Biotinidase | BTD | 2.4 | ||
| Blood vessel epicardial substance | BVES | 2.1 | ||
| Chromosome 11 open reading frame 31 | C11orf31 | 4.5 | ||
| Chromosome 17 open reading frame 99 | C17orf99 | 3.2 | ||
| Chromosome 1 open reading frame 115 | C1orf115 | 2.1 | ||
| Chromosome 1 open reading frame 141 | C1orf141 | 3.4 | ||
| Chromosome 1 open reading frame 170 | C1orf170 | 1.9 | ||
| Chromosome 20 open reading frame 201 | C20orf201 | 2.3 | ||
| Chromosome 21 open reading frame 49 | C21orf49 | 2.0 | ||
| Chromosome 21 open reading frame 67 | C21orf67 | 2.2 | ||
| Chromosome 2 open reading frame 40 | C2orf40 | 5.2 | ||
| Chromosome 2 open reading frame 81 | C2orf81 | 2.7 | ||
| Chromosome 3 open reading frame 52 | C3orf52 | 2.2 | ||
| Chromosome 8 open reading frame 46 | C8orf46 | 4.7 | ||
| Chromosome 9 open reading frame 106 | C9orf106 | 2.0 | ||
| Calcium binding protein 7 | CABP7 | 3.9 | ||
| Calcium channel, voltage-dependent, L type, α 1C subunit | CACNA1C | 2.3 | (1) | |
| Cancer antigen 1 | CAGE1 | 3.5 | ||
| Caspase 6, apoptosis-related cysteine peptidase | CASP6 | 2.8 | ||
| Coiled-coil domain containing 163, pseudogene | CCDC163P | 2.3 | ||
| Coiled-coil domain containing 42B | CCDC42B | 1.8 | ||
| Coiled-coil domain containing 57 | CCDC57 | 3.2 | (1) | |
| Chemokine (C-C motif) receptor 2 | CCR2 | 2.1 | ||
| CD163 molecule-like 1 | CD163L1 | 2.2 | ||
| CD302 molecule | CD302 | 2.4 | ||
| CDC14 cell division cycle 14 homolog A (Saccharomyces cerevisiae) | CDC14A | 5.0 | ||
| CDC42 effector protein (Ρ GTPase binding) 5 | CDC42EP5 | 2.1 | ||
| Cadherin 15, M-cadherin (myotubule) | CDH15 | 6.0 | (1) | |
| CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) | CDIPT | 1.9 | ||
| Cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | CDKN2D | 2.2 | ||
| Cdon homolog (mouse) | CDON | 1.9 | ||
| Chromodomain protein, Y-linked, 1 | CDY1 | 2.0 | ||
| Carcinoembryonic antigen-related cell adhesion molecule 2, pseudogene | CEACAM22P | 3.5 | ||
| Centrosomal protein 112kDa (CCDC46) | CEP112 | 2.2 | ||
| Centrosomal protein 85kDa-like | CEP85L | 6.7 | ||
| Cripto, FRL-1, cryptic family 1 | CFC1 | 3.6 | ||
| Chorionic gonadotropin, β polypeptide 1 | CGB1 | 1.9 | ||
| ChaC, cation transport regulator homolog 2 (E. coli) | CHAC2 | 2.7 | ||
| Carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 | CHST15 | 2.9 | ||
| C-type lectin domain family 4, member C | CLEC4C | 4.7 | ||
| Clavesin 2 | CLVS2 | 4.8 | ||
| CKLF-like MARVEL transmembrane domain containing 4 | CMTM4 | 2.7 | ||
| Cannabinoid receptor interacting protein 1 | CNRIP1 | 3.0 | ||
| Collagen, type I, α 1 | COL1A1 | 2.6 | MES | |
| Collagen, type I, α 2 | COL1A2 | 2.9 | (1) | MES |
| Collagen, type IV, α 6 | COL4A6 | 2.5 | (1) | |
| Collagen, type V, α 3 | COL5A3 | 3.9 | ||
| Collagen, type VI, α 2 | COL6A2 | 2.8 | ||
| Collagen, type IX, α 2 | COL9A2 | 1.8 | ||
| COMM domain containing 8 | COMMD8 | 2.1 | ||
| Coronin, actin binding protein, 1A | CORO1A | 2.0 | ||
| Carboxypeptidase B1 (tissue) | CPB1 | 3.3 | ||
| Cytokine receptor-like factor 2 | CRLF2 | 2.8 | ||
| Crystallin, γ S | CRYGS | 2.9 | ||
| Chondrosarcoma associated gene 1 | CSAG1 | 2.9 | ||
| Connective tissue growth factor | CTGF | 2.5 | ||
| Cathepsin H | CTSH | 5.2 | ||
| Cytokine-like 1 | CYTL1 | 2.5 | ||
| d-2-hydroxyglutarate dehydrogenase | D2HGDH | 2.6 | ||
| DAB2 interacting protein | DAB2IP | 3.0 | ||
| Death associated protein-like 1 | DAPL1 | 2.2 | ||
| Deleted in azoospermia 4 | DAZ4 | 3.1 | ||
| DCN1, defective in cullin neddylation 1, domain containing 2 (Saccharomyces cerevisiae) | DCUN1D2 | 2.5 | ||
| Defensin, β 106A | DEFB106A | 5.9 | ||
| Defensin, β 128 | DEFB128 | 2.7 | ||
| DEP domain containing 7 | DEPDC7 | 2.1 | ||
| DiGeorge syndrome critical region gene 5 (noncoding) | DGCR5 | 4.0 | ||
| Disrupted in renal carcinoma 3 | DIRC3 | 2.0 | ||
| DKFZP434L187 protein | DKFZP434L187 | 1.9 | ||
| Δ-like 2 homolog (Drosophila) | DLK2 | 2.9 | ||
| Dmx-like 1 | DMXL1 | 2.0 | ||
| DNA (cytosine-5-)-methyltransferase 3 β | DNMT3B | 2.5 | ||
| Deoxynucleotidyltransferase, terminal, interacting protein 2 | DNTTIP2 | 3.2 | ||
| Dipeptidyl-peptidase 8 | DPP8 | 5.4 | ||
| Dermatopontin | DPT | 2.0 | ||
| Dpy-19-like 2 pseudogene 2 (C. elegans) | DPY19L2P2 | 3.1 | ||
| Dorsal inhibitory axon guidance protein | DRAXIN | 2.1 | ||
| Dopamine receptor D2 | DRD2 | 3.5 | ||
| Dystonin | DST | 1.9 | ||
| E2F transcription factor 8 | E2F8 | 1.9 | ||
| Epstein-Barr virus induced gene 3 | EBI3 | 2.1 | ||
| Ephrin-B1 | EFNB1 | 2.4 | ||
| ELK4, ETS-domain protein (SRF accessory protein 1) | ELK4 | 2.6 | ||
| EP400 N-terminal like | EP400NL | 2.2 | ||
| Envoplakin | EVPL | 2.3 | ||
| Coagulation factor VIII, procoagulant component (hemophilia A) | F8 | 6.1 | ||
| Fatty acid binding protein 4, adipocyte | FABP4 | 2.0 | ||
| Family with sequence similarity 110, member D | FAM110D | 2.5 | ||
| Family with sequence similarity 159, member B | FAM159B | 2.7 | ||
| Family with sequence similarity 174, member A | FAM174A | 2.1 | ||
| Family with sequence similarity 179, member A | FAM179A | 2.5 | ||
| Family with sequence similarity 196, member A | FAM196A | 1.8 | ||
| Family with sequence similarity 27-like | FAM27L | 2.1 | ||
| Family with sequence similarity 65, member C | FAM65C | 4.1 | ||
| Family with sequence similarity 71, member C | FAM71C | 5.8 | ||
| Family with sequence similarity 72, member A | FAM72A | 1.8 | ||
| Fibulin 1 | FBLN1 | 2.6 | (1) | |
| FBXO22 antisense RNA 1 | FBXO22-AS1 | 3.2 | ||
| Fc receptor-like 3 | FCRL3 | 2.0 | ||
| FLJ31945 | 6.2 | |||
| FLJ37786 | 2.6 | |||
| FLJ45248 | 1.8 | |||
| Filamin C, γ (actin binding protein 280) | FLNC | 2.4 | ||
| Fibronectin type III domain containing 5 | FNDC5 | 2.6 | ||
| Folate receptor 1 (adult) | FOLR1 | 2.0 | ||
| Forkhead box K2 | FOXK2 | 5.2 | (1) | |
| Forkhead box protein O6 | FOXO6 | 4.8 | ||
| Forkhead box S1 | FOXS1 | 3.2 | ||
| Fraser syndrome 1 | FRAS1 | 2.6 | ||
| FSHD region gene 2 family, member C | FRG2C | 2.6 | ||
| Frizzled-related protein | FRZB | 2.4 | ||
| FtsJ methyltransferase domain containing 2 | FTSJD2 | 2.5 | ||
| FXYD domain containing ion transport regulator 5 | FXYD5 | 1.9 | ||
| Glucose-6-phosphatase, catalytic subunit | G6PC | 2.7 | ||
| Γ-aminobutyric acid (GABA) receptor, ρ 1 | GABRR1 | 5.2 | ||
| G antigen 1 | GAGE1 | 6.9 | ||
| GRINL1A combined protein | GCOM1 | 3.2 | ||
| Glutamine-fructose-6-phosphate transaminase 2 | GFPT2 | 2.6 | ||
| Glycoprotein, α-galactosyltransferase 1 (GGTA1), noncoding RNA. | GGTA1P | 2.6 | ||
| GTPase, IMAP family member 4 | GIMAP4 | 2.1 | ||
| Gap junction protein, β 2, 26kDa | GJB2 | 2.0 | ||
| GLI pathogenesis-related 1 (glioma) | GLIPR1 | 3.0 | ||
| Guanine nucleotide binding protein (G protein), α 14 | GNA14 | 1.8 | ||
| Golgin A6 family, member C | GOLGA6C | 3.5 | ||
| Golgin A6 family-like 9 | GOLGA6L9 | 2.1 | ||
| Golgin A8 family, member I | GOLGA8I | 3.5 | ||
| Glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 | GPIHBP1 | 2.0 | ||
| Glycoprotein M6A | GPM6A | 2.2 | PN | |
| G protein-coupled receptor 128 | GPR128 | 2.3 | ||
| G protein-coupled receptor 18 | GPR18 | 1.9 | ||
| Growth factor receptor-bound protein 14 | GRB14 | 1.9 | ||
| GREB1 protein | GREB1 | 2.2 | ||
| Glutamate receptor, ionotropic, N-methyl d-aspartate 2B | GRIN2B | 3.0 | ||
| General transcription factor IIH, polypeptide 2, 44kDa | GTF2H2 | 3.8 | ||
| Glycogenin 2 | GYG2 | 2.0 | ||
| Hyaluronan synthase 3 | HAS3 | 3.4 | ||
| HAUS augmin-like complex, subunit 5 | HAUS5 | 2.2 | ||
| Hemoglobin, α 1 | HBA1 | 1.9 | ||
| Hyperpolarization activated cyclic nucleotide-gated potassium channel 1 | HCN1 | 2.3 | ||
| Histone deacetylase 7 | HDAC7 | 1.9 | ||
| High density lipoprotein binding protein (vigilin) | HDLBP | 6.8 | (1) | |
| Hairy and enhancer of split 2 (Drosophila) | HES2 | 2.7 | ||
| Histone cluster 1, H2ag | HIST1H2AG | 3.0 | ||
| Major histocompatibility complex, class I, A | HLA-A | 3.4 | ||
| Major histocompatibility complex, class II, DR α | HLA-DRA | 2.2 | ||
| H2.0-like homeobox | HLX | 3.6 | ||
| Histamine N-methyltransferase | HNMT | 2.0 | (1) | |
| Homeobox B4 | HOXB4 | 2.2 | ||
| Harakiri, BCL2 interacting protein (contains only BH3 domain) | HRK | 4.2 | ||
| Heat shock transcription factor, Y-linked 1 | HSFY1 | 5.1 | ||
| Heat shock protein 90kDa α (cytosolic), class B member 4 (pseudogene) | HSP90AB4P | 1.8 | ||
| Histatin 3 | HTN3 | 2.4 | ||
| Hydrocephalus inducing homolog (mouse) | HYDIN | 3.5 | ||
| Insulin-like growth factor binding protein 3 | IGFBP3 | 2.3 | (1) | |
| Interleukin 15 receptor, α | IL15RA | 2.1 | MES | |
| Interleukin 17 receptor B | IL17RB | 4.4 | ||
| Interleukin 1 receptor, type II | IL1R2 | 2.1 | (1) | |
| Interleukin 1 receptor accessory protein | IL1RAP | 1.9 | (1) | |
| Interleukin 2 receptor, β | IL2RB | 2.4 | ||
| Interleukin 5 (colony-stimulating factor, eosinophil) | IL5 | 2.9 | ||
| Inhibitor of growth family, member 1 | ING1 | 1.9 | ||
| Inversin | INVS | 2.4 | ||
| Interferon regulatory factor 5 | IRF5 | 2.3 | ||
| Interferon regulatory factor 6 | IRF6 | 1.8 | ||
| KAT8 regulatory NSL complex subunit 1-like | KANSL1L | 2.2 | ||
| Potassium inwardly-rectifying channel, subfamily J, member 4 | KCNJ4 | 2.6 | ||
| Potassium large conductance calcium-activated channel, subfamily M β member 3 | KCNMB3 | 4.6 | ||
| Potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 | KCNS3 | 1.9 | (1) | |
| Potassium channel, subfamily V, member 2 | KCNV2 | 2.5 | ||
| KIAA1033 | KIAA1033 | 1.8 | ||
| KIAA1211-like | KIAA1211L | 2.2 | ||
| Killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 | KIR3DL1 | 2.5 | ||
| Klotho | KL | 2.6 | ||
| Kallikrein-related peptidase 14 | KLK14 | 4.6 | ||
| Laminin, β 3 | LAMB3 | 2.3 | (1) | |
| Leukocyte cell derived chemotaxin 1 | LECT1 | 1.8 | ||
| LIM homeobox 1 | LHX1 | 8.0 | (1) | |
| LIM homeobox 6 | LHX6 | 2.2 | ||
| Long intergenic nonprotein coding RNA 265 | LINC00265 | 2.2 | ||
| Long intergenic nonprotein coding RNA 426 | LINC00426 | 1.9 | ||
| Long intergenic nonprotein coding RNA 427 | LINC00427 | 2.4 | ||
| Long intergenic nonprotein coding RNA 545 | LINC00545 | 1.8 | ||
| Lines homolog (Drosophila) | LINS | 2.0 | ||
| LIM homeobox transcription factor 1, α | LMX1A | 3.2 | (1) | |
| LOC100128239 | 3.2 | |||
| LOC100128292 | 2.1 | |||
| LOC100131551 | 1.9 | |||
| LOC100132418 | 1.8 | |||
| LOC100506060 | 1.8 | |||
| LOC148696 | 1.9 | |||
| LOC286083 | 2.0 | |||
| LOC402160 | 3.6 | |||
| LOC440895 | 2.3 | |||
| LOC644248 | 2.4 | |||
| LOC645177 | 3.1 | |||
| LOC728724 | 2.6 | |||
| LOC93432 | 2.2 | |||
| Leucine-rich repeats and guanylate kinase domain containing | LRGUK | 2.7 | ||
| Low density lipoprotein-related protein 12 | LRP12 | 3.3 | (1) | |
| Leucine rich repeat containing 32 | LRRC32 | 4.5 | (1) | |
| LY6/PLAUR domain containing 6B | LYPD6B | 8.0 | ||
| Mab-21-like 1 (C. elegans) | MAB21L1 | 3.1 | CL | |
| MAFF interacting protein (pseudogene) | MAFIP | 2.8 | ||
| Melanoma antigen family A, 1 (directs expression of antigen MZ2-E) | MAGEA1 | 2.7 | (1) | |
| Melanoma antigen family A, 3 | MAGEA3 | 2.4 | (1) | |
| Melanoma antigen family B, 1 | MAGEB1 | 2.1 | ||
| Melanoma antigen family C, 2 | MAGEC2 | 1.9 | (1) | |
| Mago-nashi homolog B (Drosophila) | MAGOHB | 4.3 | ||
| Mannosidase, β A, lysosomal-like | MANBAL | 2.2 | (1) | |
| Microtubule-associated protein 1 light chain 3 γ | MAP1LC3C | 3.1 | ||
| Membrane-associated ring finger (C3HC4) 1 | MARCH1 | 3.3 | ||
| MARVEL domain containing 1 | MARVELD1 | 2.4 | ||
| Myoglobin | MB | 3.7 | ||
| MCF.2 cell line derived transforming sequence | MCF2 | 7.3 | ||
| Minichromosome maintenance domain containing 2 | MCMDC2 | 2.0 | ||
| Mucolipin 3 | MCOLN3 | 2.2 | ||
| Methyltransferase like 2B | METTL2B | 2.3 | ||
| Microfibrillar-associated protein 4 | MFAP4 | 7.1 | ||
| MGC72080 | 2.5 | CL | ||
| Matrix Gla protein | MGP | 2.0 | ||
| MER1 repeat containing imprinted transcript 1 | MIMT1 | 2.6 | ||
| MKL/myocardin-like 2 | MKL2 | 2.0 | ||
| Myeloid/lymphoid or mixed-lineage leukemia 3 | MLL3 | 2.1 | (1) | |
| Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 | MLLT10 | 3.3 | (1) | |
| Motilin | MLN | 3.0 | ||
| Membrane metallo-endopeptidase | MME | 3.8 | ||
| Matrix metallopeptidase 13 (collagenase 3) | MMP13 | 3.1 | (1) | |
| Matrix metallopeptidase 23A (pseudogene) | MMP23A | 2.2 | ||
| Membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) | MPP4 | 2.8 | ||
| Mitochondrial ribosomal protein L3 | MRPL3 | 1.9 | ||
| Mitochondrial ribosomal protein S12 | MRPS12 | 3.0 | ||
| Membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; β polypeptide) | MS4A2 | 2.2 | ||
| Mucin 19, oligomeric | MUC19 | 5.0 | ||
| MAX interactor 1 | MXI1 | 5.0 | ||
| V-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian) | MYCL1 | 2.0 | (1) | |
| Myosin, light chain 10, regulatory | MYL10 | 1.8 | ||
| Myosin IB | MYO1B | 4.6 | ||
| Myosin IC | MYO1C | 2.3 | ||
| NLR family, apoptosis inhibitory protein | NAIP | 2.4 | ||
| Neuroblastoma breakpoint family, member 15 | NBPF15 | 2.8 | ||
| Neurocalcin δ | NCALD | 1.8 | PN | |
| Neutrophil cytosolic factor 1, (chronic granulomatous disease, autosomal 1) | NCF1 | 2.6 | ||
| Nonprotein coding RNA 185 | NCRNA00185 | 1.8 | ||
| NEBL antisense RNA 1 | NEBL-AS1 | 1.8 | ||
| NEL-like 1 (chicken) | NELL1 | 2.1 | (1) | |
| Nucleolar complex associated 4 homolog (Saccharomyces cerevisiae) | NOC4L | 2.1 | ||
| NADPH oxidase organizer 1 | NOXO1 | 2.2 | (1) | |
| Neuropeptide S | NPS | 2.2 | (1) | |
| Neuronal pentraxin II | NPTX2 | 3.1 | (1) | |
| Neurexin 2 | NRXN2 | 2.1 | PN | |
| Neurotensin receptor 1 (high affinity) | NTSR1 | 2.8 | ||
| Optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) | OPA3 | 2.7 | ||
| Olfactory receptor, family 11, subfamily A, member 1 | OR11A1 | 2.1 | ||
| Olfactory receptor, family 2, subfamily A, member 9 pseudogene | OR2A9P | 4.6 | ||
| Olfactory receptor, family 51, subfamily E, member 2 | OR51E2 | 3.6 | ||
| OTU domain containing 1 | OTUD1 | 2.1 | ||
| Purinergic receptor P2X, ligand-gated ion channel, 3 | P2RX3 | 3.5 | ||
| P antigen family, member 5 (prostate associated) | PAGE5 | 1.9 | ||
| Paralemmin 3 | PALM3 | 4.7 | ||
| Par-3 partitioning defective 3 homolog (C. elegans) | PARD3 | 3.0 | ||
| Protocadherin 11 X-linked | PCDH11X | 3.5 | ||
| Protocadherin α 12 | PCDHA12 | 5.5 | ||
| Protocadherin γ subfamily B, 7 | PCDHGB7 | 2.9 | ||
| Phosducin-like 3 pseudogene 4 | PDCL3P4 | 2.2 | ||
| Platelet-derived growth factor receptor, β polypeptide | PDGFRB | 2.8 | ||
| Pyridoxal-dependent decarboxylase domain containing 2, pseudogene | PDXDC2P | 1.9 | ||
| PDZ domain containing 1 | PDZK1 | 3.1 | ||
| Platelet/endothelial cell adhesion molecule (CD31 antigen) | PECAM1 | 1.9 | ||
| Pepsinogen 3, group I (pepsinogen A) | PGA3 | 3.5 | ||
| Placental growth factor, vascular endothelial growth factor-related protein | PGF | 2.7 | ||
| Pleckstrin homology-like domain, family A, member 3 | PHLDA3 | 2.0 | ||
| Phosphatidylinositol 4-kinase type 2 β | PI4K2B | 2.3 | ||
| Phospholipase A2, group IIC | PLA2G2C | 2.5 | ||
| Plasminogen activator, urokinase | PLAU | 1.8 | MES | |
| Plasminogen activator, urokinase receptor | PLAUR | 2.5 | MES | |
| Phospholipase C-like 2 | PLCL2 | 2.4 | ||
| Pleckstrin homology domain containing, family G (with ΡGef domain) member 3 | PLEKHG3 | 2.3 | ||
| Premelanosome protein | PMEL | 2.6 | ||
| POM121 transmembrane nucleoporin-like 1, pseudogene | POM121L1P | 2.6 | ||
| Proopiomelanocortin (adrenocorticotropin/ β-lipotropin/ α-melanocyte stimulating hormone/ β-melanocyte stimulating hormone/ β-endorphin) | POMC | 2.0 | ||
| Peptidylprolyl isomerase E (cyclophilin E) | PPIE | 1.9 | ||
| Protein phosphatase 1, regulatory (inhibitor) subunit 9B | PPP1R9B | 2.0 | ||
| PR domain containing 1, with ZNF domain | PRDM1 | 4.4 | ||
| Protein kinase C, ε | PRKCE | 2.2 | (1) | |
| Proline rich 5 (renal) | PRR5 | 4.2 | (1) | |
| Prostaglandin D2 synthase 21kDa (brain) | PTGDS | 5.5 | ||
| Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | PTGS1 | 5.2 | ||
| Protein tyrosine phosphatase, receptor type, E | PTPRE | 1.8 | ||
| Protein tyrosine phosphatase, receptor type, O | PTPRO | 1.9 | (1) | |
| Pyridine nucleotide-disulphide oxidoreductase domain 2 | PYROXD2 | 1.8 | ||
| RAB20, member RAS oncogene family | RAB20 | 2.2 | (1) | |
| RAB30, member RAS oncogene family | RAB30 | 1.8 | ||
| Retinoic acid early transcript 1G | RAET1G | 6.2 | ||
| RAN guanine nucleotide release factor | RANGRF | 1.9 | ||
| Rap guanine nucleotide exchange factor (GEF) 5 | RAPGEF5 | 2.2 | ||
| Retinoic acid receptor, α | RARA | 2.2 | ||
| RAS protein activator like 2 | RASAL2 | 2.7 | ||
| RASD family, member 2 | RASD2 | 2.6 | (1) | |
| Ras association (RalGDS/AF-6) domain family 6 | RASSF6 | 4.1 | ||
| Retinoblastoma binding protein 6 | RBBP6 | 2.4 | ||
| RCC1 domain containing 1 | RCCD1 | 2.0 | ||
| RELT tumor necrosis factor receptor | RELT | 3.0 | ||
| Regulatory factor X, 7 | RFX7 | 1.8 | ||
| Rhomboid 5 homolog 2 (Drosophila) | RHBDF2 | 2.1 | ||
| Ripply1 homolog (zebrafish) | RIPPLY1 | 2.2 | ||
| Ring finger protein 186 | RNF186 | 2.1 | ||
| Retinitis pigmentosa 1 (autosomal dominant) | RP1 | 2.4 | (1) | |
| Reprimo-like | RPRML | 2.4 | ||
| Ras-related associated with diabetes | RRAD | 2.2 | ||
| Runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | RUNX1T1 | 2.1 | (1) | |
| Sterile α motif domain containing 14 | SAMD14 | 2.3 | ||
| SET binding factor 1 pseudogene 1 | SBF1P1 | 2.9 | ||
| Small Cajal body-specific RNA 13 | SCARNA13 | 2.3 | ||
| Small Cajal body-specific RNA 9-like | SCARNA9L | 2.6 | ||
| Secretogranin V (7B2 protein) | SCG5 | 2.6 | ||
| Sodium channel, voltage-gated, type I, β | SCN1B | 2.9 | ||
| Sidekick homolog 2 (chicken) | SDK2 | 2.2 | (1) | |
| Selectin E (endothelial adhesion molecule 1) | SELE | 1.9 | ||
| Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D | SEMA3D | 3.2 | (1) | |
| Serpin peptidase inhibitor, clade A (α-1 antiproteinase, antitrypsin), member 3 | SERPINA3 | 4.9 | ||
| Serpin peptidase inhibitor, clade I (neuroserpin), member 1 | SERPINI1 | 4.0 | NL | |
| SH3 and PX domains 2A | SH3PXD2A | 6.2 | (1) | |
| Src homology 2 domain containing E | SHE | 3.2 | ||
| Src homology 2 domain containing F | SHF | 2.1 | ||
| SIX homeobox 4 | SIX4 | 1.8 | ||
| Solute carrier family 16, member 10 (aromatic amino acid transporter) | SLC16A10 | 2.5 | (1) | |
| Solute carrier family 16, member 14 (monocarboxylic acid transporter 14) | SLC16A14 | 1.9 | ||
| Solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 | SLC17A7 | 2.5 | ||
| Solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | SLC25A15 | 2.3 | ||
| Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 | SLC25A24 | 7.4 | ||
| Solute carrier family 25, member 52 | SLC25A52 | 2.5 | ||
| Solute carrier family 26, member 4 | SLC26A4 | 2.5 | ||
| Solute carrier family 27 (fatty acid transporter), member 4 | SLC27A4 | 3.5 | ||
| Solute carrier family 39 (zinc transporter), member 12 | SLC39A12 | 3.0 | ||
| Solute carrier family 39 (zinc transporter), member 3 | SLC39A3 | 1.8 | ||
| Solute carrier family 39 (zinc transporter), member 8 | SLC39A8 | 2.0 | ||
| Solute carrier family 45, member 3 | SLC45A3 | 2.2 | ||
| Solute carrier family 46, member 2 | SLC46A2 | 2.3 | ||
| Solute carrier family 5 (sodium/glucose cotransporter), member 2 | SLC5A2 | 5.7 | ||
| SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | SMARCC2 | 2.5 | ||
| Small integral membrane protein 8 | SMIM8 | 2.6 | ||
| Snail homolog 2 (Drosophila) | SNAI2 | 2.0 | ||
| Small nuclear RNA activating complex, polypeptide 3, 50kDa | SNAPC3 | 3.9 | ||
| Stannin | SNN | 2.0 | ||
| Small nucleolar RNA, H/ACA box 72 | SNORA72 | 2.6 | ||
| Small nucleolar RNA, C/D box 101 | SNORD101 | 1.9 | ||
| Small nucleolar RNA, C/D box 113-6 | SNORD113-6 | 3.4 | ||
| Sorting nexin family member 21 | SNX21 | 2.2 | ||
| Sosondowah ankyrin repeat domain family member A | SOWAHA | 3.2 | ||
| SRY (sex-determining region Y)-box 18 | SOX18 | 2.2 | ||
| SP100 nuclear antigen | SP100 | 2.2 | MES | |
| Sperm associated antigen 16 | SPAG16 | 2.0 | ||
| SPATA31 subfamily D, member 5, pseudogene | SPATA31D5P | 2.3 | ||
| Speedy homolog A (Drosophila) | SPDYA | 2.5 | ||
| Speedy/RINGO cell cycle regulator family member E6 | SPDYE6 | 3.3 | ||
| Sphingosine kinase 1 | SPHK1 | 1.8 | ||
| Spondin 1, extracellular matrix protein | SPON1 | 2.0 | ||
| Steroidogenic acute regulatory protein | STAR | 3.5 | ||
| Serine/threonine kinase 16 | STK16 | 4.3 | ||
| Serine/threonine kinase 17b | STK17B | 2.1 | (1) | |
| Suppressor of G2 allele of SKP1 (Saccharomyces cerevisiae) pseudogene 3 | SUGT1P3 | 2.4 | ||
| Sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 | SULT1A3 | 2.1 | ||
| Sad1 and UNC84 domain containing 3 | SUN3 | 5.2 | ||
| Sushi domain containing 2 | SUSD2 | 2.9 | ||
| Supervillin | SVIL | 6.8 | (1) | |
| Synaptonemal complex central element protein 3 | SYCE3 | 2.0 | ||
| Synaptotagmin III | SYT3 | 2.0 | ||
| Transgelin | TAGLN | 2.6 | ||
| T cell acute lymphocytic leukemia 2 | TAL2 | 4.3 | ||
| TBC1 domain family, member 26 | TBC1D26 | 2.7 | ||
| Transducin (β)-like 1Y-linked | TBL1Y | 3.9 | ||
| T cell, immune regulator 1, ATPase, H + transporting, lysosomal V0 subunit A3 | TCIRG1 | 1.9 | MES | |
| Testis development related 1 (nonprotein coding) | TDRG1 | 3.9 | ||
| TEK tyrosine kinase, endothelial (venous malformations, multiple cutaneous and mucosal) | TEK | 2.0 | (1) | |
| Testis-specific kinase 2 | TESK2 | 7.3 | ||
| Tet methylcytosine dioxygenase 3 | TET3 | 5.4 | ||
| THAP domain containing 4 | THAP4 | 2.5 | ||
| Tight junction protein 3 | TJP3 | 3.2 | ||
| Transmembrane 4 L six family member 19 | TM4SF19 | 2.0 | ||
| Transmembrane channel-like 4 | TMC4 | 4.9 | ||
| Transmembrane protein with EGF-like and two follistatin-like domains 2 | TMEFF2 | 2.3 | (1) | |
| Transmembrane protein 107 | TMEM107 | 1.8 | ||
| Transmembrane protein 110 | TMEM110 | 2.2 | ||
| Transmembrane protein 120B | TMEM120B | 2.0 | ||
| Transmembrane protein 191B | TMEM191B | 2.7 | ||
| Transmembrane protein 65 | TMEM65 | 1.8 | ||
| Thioredoxin-related transmembrane protein 1 | TMX1 | 5.5 | ||
| Tumor necrosis factor, α-induced protein 2 | TNFAIP2 | 1.9 | ||
| Tumor necrosis factor, α-induced protein 8 | TNFAIP8 | 1.9 | MES | |
| Tumor necrosis factor receptor superfamily, member 18 | TNFRSF18 | 2.5 | ||
| Tumor necrosis factor (ligand) superfamily, member 12 | TNFSF12 | 2.8 | ||
| TNNI3 interacting kinase | TNNI3K | 2.7 | ||
| Topoisomerase (DNA) I pseudogene 1 | TOP1P1 | 2.9 | ||
| TOX high-mobility group box family member 2 | TOX2 | 2.6 | ||
| Tumor protein p53 regulated apoptosis inducing protein 1 | TP53AIP1 | 2.1 | ||
| Trafficking protein particle complex 6B | TRAPPC6B | 1.9 | ||
| TRNA aspartic acid methyltransferase 1 | TRDMT1 | 2.4 | ||
| Tripartite motif-containing 51E, pseudogene | TRIM51EP | 2.7 | ||
| Transient receptor potential cation channel, subfamily M, member 1 | TRPM1 | 3.5 | ||
| Testis-specific transcript, Y-linked 5 | TTTY5 | 2.4 | ||
| Tubulin, α 8 | TUBA8 | 2.3 | ||
| Tubulin, β pseudogene 5 | TUBBP5 | 4.4 | ||
| Urocortin 2 | UCN2 | 3.1 | ||
| Vestigial like 2 (Drosophila) | VGLL2 | 1.9 | ||
| Vitelline membrane outer layer 1 homolog (chicken) | VMO1 | 2.0 | (1) | |
| Vanin 2 | VNN2 | 4.8 | ||
| WASH and IL9R antisense RNA 1 | WASIR1 | 4.0 | ||
| WD repeat domain 63 | WDR63 | 4.1 | ||
| WD repeat domain 87 | WDR87 | 1.8 | ||
| WAP four-disulfide core domain 2 | WFDC2 | 2.9 | ||
| YME1-like 1 (Saccharomyces cerevisiae) | YME1L1 | 2.2 | ||
| YPLR6490 | 2.4 | |||
| Zinc finger, CCHC domain containing 12 | ZCCHC12 | 2.1 | ||
| Zinc finger, FYVE domain containing 27 | ZFYVE27 | 2.4 | ||
| ZMIZ1 antisense RNA 1 | ZMIZ1-AS1 | 2.0 | ||
| Zinc finger, MYND-type containing 15 | ZMYND15 | 2.7 | ||
| Zinc finger protein 138 | ZNF138 | 2.9 | ||
| Zinc finger protein 48 | ZNF48 | 2.2 | ||
| Zinc finger protein 493 | ZNF493 | 2.3 | ||
| Zinc finger protein 570 | ZNF570 | 3.8 | ||
| Zinc finger protein 607 | ZNF607 | 2.0 | ||
| Zinc finger protein 793 | ZNF793 | 3.6 | ||
| Zinc finger protein 815, pseudogene | ZNF815P | 2.9 | ||
| Zinc finger protein 847, pseudogene | ZNF847P | 2.7 | ||
| Zona pellucida-like domain containing 1 | ZPLD1 | 2.1 | ||
Bold, genes commonly regulated in SCS and CSC; italic, genes regulated in oposite directions in SDC and CSC.
References
1. Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, Zheng S, Hood L, Goodlett DR, Foltz G,et al. (2011). The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics12,11.
2. Ferletta M, Caglayan D, Mokvist L, Jiang Y, Kastemar M, Uhrbom L, Westermark B (2011). Forced expression of Sox21 inhibits Sox2 and induces apoptosis in human glioma cells. Int J Cancer129,45–60.
3. Hägerstrand D, He X, Bradic Lindh M, Hoefs S, Hesselager G, Ostman A, Nistér M (2011). Identification of a SOX2-dependent subset of tumor- and sphere-forming glioblastoma cells with a distinct tyrosine kinase inhibitor sensitivity profile. Neuro Oncol13,1178–1191.
4. Park SB, Seo KW, So AY, Seo MS, Yu KR, Kang SK, Kang KS (2012). SOX2 has a crucial role in the lineage determination and proliferation of mesenchymal stem cells through Dickkopf-1 and c-MYC. Cell Death Differ 19, 534–545.
5. Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H (2004) Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol24, 8834–8846.
Table W2.
Genes Highly Correlated with Sox2 mRNA Expression in the TCGA Data.
| Gene ID | Correlation | In Vitro Sox2 Knockdown DEG | Sox2 Target (Ref) | Verhaak Subclass |
|---|---|---|---|---|
| ABCA3 | Positive | |||
| ARNT2 | Positive | SDC | ||
| ASTN1 | Positive | CSC | ||
| B3GAT1 | Positive | SDC | (1) | |
| BAALC | Positive | SDC | ||
| BAI2 | Positive | |||
| BCAN | Positive | PN | ||
| C1orf61 | Positive | SDC | (1) | PN |
| C3orf39 | Positive | |||
| CHST10 | Positive | SDC | ||
| CKB | Positive | SDC | PN | |
| CLIP3 | Positive | |||
| CTNND2 | Positive | |||
| DBI | Positive | |||
| DCLK2 | Positive | |||
| DDR1 | Positive | |||
| DENND5A (RAB61P1) | Positive | (1) | ||
| DNAJB5 | Positive | |||
| DTX1 | Positive | |||
| EBF4 | Positive | |||
| FAM181B (MGC33846) | Positive | (1) | ||
| FEZ1 | Positive | SDC | (1) | |
| FXYD6 | Positive | (1) | PN | |
| GDF1 | Positive | |||
| GNA11 | Positive | |||
| GPM6B | Positive | SDC, CSC | ||
| GPR56 | Positive | SDC | CL | |
| HEPN1 | Positive | |||
| ID4 | Positive | |||
| JAKMIP2 | Positive | |||
| KEAP1 | Positive | CL | ||
| LRIG1 | Positive | (1) | ||
| METRN | Positive | SDC | ||
| MLC1 | Positive | CL | ||
| MT3 | Positive | |||
| NFIX | Positive | |||
| NLRP1 | Positive | |||
| NR2F1 | Positive | (1) | ||
| OLIG1 | Positive | |||
| OSBPL6 | Positive | SDC | (1) | |
| PCDHGA1 | Positive | |||
| POU3F2 | Positive | |||
| PTPRZ1 | Positive | (1) | ||
| RGMA | Positive | |||
| SHC3 | Positive | |||
| SLC1A3 | Positive | SDC | (1) | |
| SOX2 | Positive | SDC,CSC | (1) | PN |
| SOX21 | Positive | SDC,CSC | (1) (2) | |
| TBCB | Positive | |||
| TCEA2 | Positive | |||
| TTYH1 | Positive | SDC, CSC | PN | |
| TUBA4A | Positive | |||
| TUBB2A | Positive | |||
| TUBB2B | Positive | SDC | ||
| TUBB6 | Positive | |||
| TUBB8 | Positive | |||
| ZBTB47 | Positive | |||
| ANXA3 | Negative | SDC | NL | |
| ATF1 | Negative | |||
| B3GNT2 | Negative | (1) | ||
| C4orf32 | Negative | (1) | ||
| C6orf211 | Negative | |||
| FEM1B | Negative | |||
| ITGA4 | Negative | MES | ||
| MOB1A | Negative | |||
| MST4 | Negative | |||
| NUDT15 | Negative | |||
| PCYOX1 | Negative | |||
| PTPN2 | Negative | |||
| RALB | Negative | (1) | ||
| STT3B | Negative | |||
| TBC1D8B | Negative | |||
| THOC7 | Negative | |||
| TMF1 | Negative |
References
1. Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, Zheng S, Hood L, Goodlett DR, Foltz G,et al. (2011). The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics12,11.
2. Ferletta M, Caglayan D, Mokvist L, Jiang Y, Kastemar M, Uhrbom L, Westermark B (2011). Forced expression of Sox21 inhibits Sox2 and induces apoptosis in human glioma cells. Int J Cancer129,45–60.
References
- 1.Ben-Porath I., Thomson M.W., Carey V.J., Ge R., Bell G.W., Regev A., Weinberg R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avilion A.A., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A.A. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 4.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarkar A., Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferri A.L., Cavallaro M., Braida D., Di Cristofano A., Canta A., Vezzani A., Ottolenghi S., Pandolfi P.P., Sala M., DeBiasi S. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 7.Bani-Yaghoub M., Tremblay R.G., Lei J.X., Zhang D., Zurakowski B., Sandhu J.K., Smith B., Ribecco-Lutkiewicz M., Kennedy J., Walker P.R. Role of Sox2 in the development of the mouse neocortex. Dev Biol. 2006;295:52–66. doi: 10.1016/j.ydbio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Baer K., Eriksson P.S., Faull R.L.M., Rees M.I., Curtis M.A. Sox-2 is expressed by glial and progenitor cells and Pax-6 is expressed by neuroblasts in the human subventricular zone. Exp Neurol. 2007;204:828–831. doi: 10.1016/j.expneurol.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 10.Bass A.J., Watanabe H., Mermel C.H., Yu S., Perner S., Verhaak R.G., Kim S.Y., Wardwell L., Tamayo P., Gat-Viks I. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bareiss P.M., Paczulla A., Wang H., Schairer R., Wiehr S., Kohlhofer U., Rothfuss O.C., Fischer A., Perner S., Staebler A. SOX2 expression associates with stem cell state in human ovarian carcinoma. Cancer Res. 2013;73:5544–5555. doi: 10.1158/0008-5472.CAN-12-4177. [DOI] [PubMed] [Google Scholar]
- 12.Alonso M.M., Diez-Valle R., Manterola L., Rubio A., Liu D., Cortes-Santiago N., Urquiza L., Jauregi P., Lopez de Munain A., Sampron N. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLoS One. 2011;6:e26740. doi: 10.1371/journal.pone.0026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz M., Temme A., Senner V., Ebner R., Schwind S., Stevanovic S., Wehner R., Schackert G., Schackert H.K., Fussel M. Identification of SOX2 as a novel glioma-associated antigen and potential target for T cell-based immunotherapy. Br J Cancer. 2007;96:1293–1301. doi: 10.1038/sj.bjc.6603696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 17.Galli R., Binda E., Orfanelli U., Cipelletti B., Gritti A., De Vitis S., Fiocco R., Foroni C., Dimeco F., Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 18.Annovazzi L., Mellai M., Caldera V., Valente G., Schiffer D. SOX2 expression and amplification in gliomas and glioma cell lines. Cancer Genomics Proteomics. 2011;8:139–147. [PubMed] [Google Scholar]
- 19.deCarvalho A.C., Nelson K., Lemke N., Lehman N.L., Arbab A.S., Kalkanis S., Mikkelsen T. Gliosarcoma stem cells undergo glial and mesenchymal differentiation in vivo. Stem Cells. 2010;28:181–190. doi: 10.1002/stem.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gangemi R.M., Griffero F., Marubbi D., Perera M., Capra M.C., Malatesta P., Ravetti G.L., Zona G.L., Daga A., Corte G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 21.Hagerstrand D., He X., Bradic Lindh M., Hoefs S., Hesselager G., Ostman A., Nister M. Identification of a SOX2-dependent subset of tumor- and sphere-forming glioblastoma cells with a distinct tyrosine kinase inhibitor sensitivity profile. Neuro Oncol. 2011;13:1178–1191. doi: 10.1093/neuonc/nor113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavallaro M., Mariani J., Lancini C., Latorre E., Caccia R., Gullo F., Valotta M., DeBiasi S., Spinardi L., Ronchi A. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development. 2008;135:541–557. doi: 10.1242/dev.010801. [DOI] [PubMed] [Google Scholar]
- 23.Fang X., Yoon J.G., Li L., Yu W., Shao J., Hua D., Zheng S., Hood L., Goodlett D.R., Foltz G. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics. 2011;12:11. doi: 10.1186/1471-2164-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louis D.N., Ohgaki H., Wiestler O.D., Cavenee W.K. IARC; Lyon, France: 2007. WHO Classification of Tumours of the Central Nervous System. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meis J.M., Ho K.L., Nelson J.S. Gliosarcoma: a histologic and immunohistochemical reaffirmation. Mod Pathol. 1990;3:19–24. [PubMed] [Google Scholar]
- 26.deCarvalho A.C., Mikkelsen T. Gliosarcoma stem cells: glial and mesenchymal differentiation. In: Hayat M.A., editor. Stem cells and Cancer Stem Cells: Therapeutic Applications in Disease and Injury. Springer; Dordrecht: 2012. pp. 75–81. [Google Scholar]
- 27.Hasselbach L.A., Irtenkauf S.M., Lemke N.W., Nelson K.K., Berezovsky A.D., Carlton E.T., Transou A.D., Mikkelsen T., Decarvalho A.C. Optimization of high grade glioma cell culture from surgical specimens for use in clinically relevant animal models and 3D immunochemistry. J Vis Exp. 2014;83:e51088. doi: 10.3791/51088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmberg J., He X., Peredo I., Orrego A., Hesselager G., Ericsson C., Hovatta O., Oba-Shinjo S.M., Marie S.K., Nister M. Activation of neural and pluripotent stem cell signatures correlates with increased malignancy in human glioma. PLoS One. 2011;6:e18454. doi: 10.1371/journal.pone.0018454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phi J.H., Park S.H., Kim S.K., Paek S.H., Kim J.H., Lee Y.J., Cho B.K., Park C.K., Lee D.H., Wang K.C. Sox2 expression in brain tumors: a reflection of the neuroglial differentiation pathway. Am J Surg Pathol. 2008;32:103–112. doi: 10.1097/PAS.0b013e31812f6ba6. [DOI] [PubMed] [Google Scholar]
- 30.Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka S., Kamachi Y., Tanouchi A., Hamada H., Jing N., Kondoh H. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol. 2004;24:8834–8846. doi: 10.1128/MCB.24.20.8834-8846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferletta M., Caglayan D., Mokvist L., Jiang Y., Kastemar M., Uhrbom L., Westermark B. Forced expression of Sox21 inhibits Sox2 and induces apoptosis in human glioma cells. Int J Cancer. 2011;129:45–60. doi: 10.1002/ijc.25647. [DOI] [PubMed] [Google Scholar]
- 33.Ying M., Wang S., Sang Y., Sun P., Lal B., Goodwin C.R., Guerrero-Cazares H., Quinones-Hinojosa A., Laterra J., Xia S. Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition. Oncogene. 2011;30:3454–3467. doi: 10.1038/onc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gage F.H., Ray J., Fisher L.J. Isolation, characterization, and use of stem cells from the CNS. Annu Rev Neurosci. 1995;18:159–192. doi: 10.1146/annurev.ne.18.030195.001111. [DOI] [PubMed] [Google Scholar]
- 35.Singh S.K., Clarke I.D., Terasaki M., Bonn V.E., Hawkins C., Squire J., Dirks P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 36.Li A., Walling J., Kotliarov Y., Center A., Steed M.E., Ahn S.J., Rosenblum M., Mikkelsen T., Zenklusen J.C., Fine H.A. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res. 2008;6:21–30. doi: 10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- 37.Kuzmichev A.N., Kim S.K., D'Alessio A.C., Chenoweth J.G., Wittko I.M., Campanati L., McKay R.D. Sox2 acts through Sox21 to regulate transcription in pluripotent and differentiated cells. Curr Biol. 2012;22:1705–1710. doi: 10.1016/j.cub.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Kumada T., Yamanaka Y., Kitano A., Shibata M., Awaya T., Kato T., Okawa K., Abe T., Oshima N., Nakahata T. Ttyh1, a Ca2 +-binding protein localized to the endoplasmic reticulum, is required for early embryonic development. Dev Dyn. 2010;239:2233–2245. doi: 10.1002/dvdy.22348. [DOI] [PubMed] [Google Scholar]
- 39.Hu G., Kim J., Xu Q., Leng Y., Orkin S.H., Elledge S.J. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Rosa A., Pellegatta S., Rossi M., Tunici P., Magnoni L., Speranza M.C., Malusa F., Miragliotta V., Mori E., Finocchiaro G. A radial glia gene marker, fatty acid binding protein 7 (FABP7), is involved in proliferation and invasion of glioblastoma cells. PLoS One. 2012;7:e52113. doi: 10.1371/journal.pone.0052113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wicki A., Christofori G. The potential role of podoplanin in tumour invasion. Br J Cancer. 2007;96:1–5. doi: 10.1038/sj.bjc.6603518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanner S.M., Austin J.L., Leone G., Rush L.J., Plass C., Heinonen K., Mrózek K., Sill H., Knuutila S., Kolitz J.E. BAALC, the human member of a novel mammalian neuroectoderm gene lineage, is implicated in hematopoiesis and acute leukemia. Proc Natl Acad Sci U S A. 2001;98:13901–13906. doi: 10.1073/pnas.241525498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karantzali E., Lekakis V., Ioannou M., Hadjimichael C., Papamatheakis J., Kretsovali A. Sall1 regulates embryonic stem cell differentiation in association with nanog. J Biol Chem. 2011;286:1037–1045. doi: 10.1074/jbc.M110.170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basu-Roy U., Seo E., Ramanathapuram L., Rapp T.B., Perry J.A., Orkin S.H., Mansukhani A., Basilico C. Sox2 maintains self renewal of tumor-initiating cells in osteosarcomas. Oncogene. 2012;31:2270–2282. doi: 10.1038/onc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rampazzo E., Persano L., Pistollato F., Moro E., Frasson C., Porazzi P., Della Puppa A., Bresolin S., Battilana G., Indraccolo S. Wnt activation promotes neuronal differentiation of glioblastoma. Cell Death Dis. 2013;4:e500. doi: 10.1038/cddis.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Infanger D.W., Cho Y., Lopez B.S., Mohanan S., Liu S.C., Gursel D., Boockvar J.A., Fischbach C. Glioblastoma stem cells are regulated by interleukin-8 signaling in a tumoral perivascular niche. Cancer Res. 2013;73:7079–7089. doi: 10.1158/0008-5472.CAN-13-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abad M., Mosteiro L., Pantoja C., Canamero M., Rayon T., Ors I., Grana O., Megias D., Dominguez O., Martinez D. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- 48.Lathia J.D., Gallagher J., Heddleston J.M., Wang J., Eyler C.E., Macswords J., Wu Q., Vasanji A., McLendon R.E., Hjelmeland A.B. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge Y., Zhou F., Chen H., Cui C., Liu D., Li Q., Yang Z., Wu G., Sun S., Gu J. Sox2 is translationally activated by eukaryotic initiation factor 4E in human glioma-initiating cells. Biochem Biophys Res Commun. 2010;397:711–717. doi: 10.1016/j.bbrc.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 50.Lee C., Fotovati A., Triscott J., Chen J., Venugopal C., Singhal A., Dunham C., Kerr J.M., Verreault M., Yip S. Polo-like kinase 1 inhibition kills glioblastoma multiforme brain tumor cells in part through loss of SOX2 and delays tumor progression in mice. Stem Cells. 2012;30:1064–1075. doi: 10.1002/stem.1081. [DOI] [PubMed] [Google Scholar]
- 51.Jeong C.H., Cho Y.Y., Kim M.O., Kim S.H., Cho E.J., Lee S.Y., Jeon Y.J., Lee K.Y., Yao K., Keum Y.S. Phosphorylation of Sox2 cooperates in reprogramming to pluripotent stem cells. Stem Cells. 2010;28:2141–2150. doi: 10.1002/stem.540. [DOI] [PubMed] [Google Scholar]
- 52.Chen R., Nishimura M.C., Bumbaca S.M., Kharbanda S., Forrest W.F., Kasman I.M., Greve J.M., Soriano R.H., Gilmour L.L., Rivers C.S. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 53.Marjanovic N.D., Weinberg R.A., Chaffer C.L. Cell plasticity and heterogeneity in cancer. Clin Chem. 2013;59:168–179. doi: 10.1373/clinchem.2012.184655. [DOI] [PMC free article] [PubMed] [Google Scholar]