Graphical abstract
Keywords: Leishmania, Quantitative proteomics, Stable isotope labeling of amino acids in cell culture (SILAC), Amphotericin B, Drug resistance
Highlights
-
•
First large-scale quantitative proteomic study of amphotericin B resistance in Leishmania.
-
•
Identification of 97 differentially expressed proteins.
-
•
Upregulation of glycolysis and TCA cycle pathways.
-
•
Upregulation in reactive oxygen species scavenging and heat-shock proteins.
Abstract
Amphotericin B (AmB) in its liposomal form is now considered as either first- or second-line treatment against Leishmania infections in different part of the world. Few cases of AmB resistance have been reported and resistance mechanisms toward AmB are still poorly understood. This paper reports a large-scale comparative proteomic study in the context of AmB resistance. Quantitative proteomics using stable isotope labeling of amino acids in cell culture (SILAC) was used to better characterize cytoplasmic and membrane-enriched (ME) proteomes of the in vitro generated Leishmania infantum AmB resistant mutant AmB1000.1. In total, 97 individual proteins were found as differentially expressed between the mutant and its parental sensitive strain (WT). More than half of these proteins were either metabolic enzymes or involved in transcription or translation processes. Key energetic pathways such as glycolysis and TCA cycle were up-regulated in the mutant. Interestingly, many proteins involved in reactive oxygen species (ROS) scavenging and heat-shock proteins were also up-regulated in the resistant mutant. This work provides a basis for further investigations to understand the roles of proteins differentially expressed in relation with AmB resistance.
1. Introduction
Protozoan parasites of the genus Leishmania cause a wide range of diseases affecting 12 million people worldwide with 1.5–2 million new cases each year (Murray et al., 2005). With no vaccine available yet, the control of these parasites relies solely on chemotherapy. The first-line of treatment relies on pentavalent antimony (SbV) compounds (Alvar et al., 2006). However, in certain regions such as Bihar state in India, resistance to SbV is now widespread and alternative drugs must be used (Lira et al., 1999; Sundar, 2001; Thakur et al., 2001). Alternative drugs such as paromomycin (Sundar et al., 2007) and the orally administered miltefosine (Sundar et al., 1998) are effective and used against Leishmania. Nowadays, liposomal AmB is considered as the best drug available against visceral leishmaniasis, the most severe form of the disease that is fatal if untreated (Bern et al., 2006; Chappuis et al., 2007). However, its high cost limits its widespread use in developing countries (Olliaro and Sundar, 2009).
Clinical resistance to AmB is rare as shown by the absence of resistance in strains isolated from HIV-1 patients treated repeatedly with AmB (Lachaud et al., 2009) although few unusual cases have been reported recently in India (Srivastava et al., 2011; Purkait et al., 2012). The molecular mechanisms of AmB resistance are not very well understood. In vitro generated AmB resistant mutants showed that ergosterol in the plasma membrane was replaced by a precursor, cholesta-5,7,24-trien-3β-ol, which prevents the binding and subsequent uptake of the drug (Mbongo et al., 1998). The same substitution was observed in clinical resistant strains along with a higher expression level of the ABC transporter MDR1 (ABCB4) and of some enzymes involved in thiol metabolism (Purkait et al., 2012).
Quantitative proteomics is now emerging as a powerful approach to study drug resistance in microorganisms. Here, we used the stable isotope labeling of amino acids in cell culture (SILAC) methodology to study AmB resistance in Leishmania. The present paper reports for the first time the use of a large-scale quantitative proteomic study to characterize the proteome of an in vitro selected AmB resistant Leishmania mutant.
2. Material and methods
2.1. Cell culture and SILAC
The Leishmania infantum (MHOM/MA/67/ITMAP-263) wild-type (WT) strain and the in vitro generated resistant mutant AmB1000.1, which is resistant to 1000 nM of AmB, were described previously (Moreira et al., 2011). The resistance phenotype to AmB was tested after 7 and 63 passages in absence of drug and has been proven to be highly stable in this mutant. For SILAC experiments, we first did an incorporation assay on WT cells to evaluate the extent of isotopic incorporation and technical noise. This control experiment allowed us to confirm that 99% of incorporation in lysine and arginine amino acids was achieved. Thus, promastigotes of WT and mutant AmB1000.1 were grown in RPMI-1640 medium for SILAC (minus l-lysine and l-arginine) (Cambridge Isotope Laboratories) supplemented with 75 μM adenosine (Sigma), 28 mM HEPES (Sigma), 40 μM biotin (Sigma), 1% penicillin–streptomycin (Wisent), 5 mg/L hemin (MP Biomedicals), 10 μM biopterin and 10% heat-inactivated dialysed foetal bovine serum (Cambridge Isotope Laboratories). L. infantum WT light medium (normal isotopic abundance) was also supplemented with 242.26 mg/L l-arginine (Sigma) and 50.03 mg/L l-lysine, while 253.68 mg/L 13C6-15N4-l-arginine and 62.21 mg/L 13C6-15N2-l-lysine were added to the resistant mutant heavy medium. Mutant cells were grown in the heavy complete medium for at least 7 passages to ensure a minimum of 99% incorporation of heavy isotopes into proteins, as previously determined in our control incorporation assay. WT and mutant cells were counted using a haemocytometer and the resistant strain was then mixed with WT cells in a 1:1 ratio.
2.2. Sample preparation
The membrane-enriched (ME) fraction was obtained by sonication, ultracentrifugation and further purification by Free flow zone electrophoresis (ZE-FFE) as described previously (Brotherton et al., 2012). The cytosolic protein extraction was performed in 2D lysis buffer as described previously (Brotherton et al., 2010). Proteins were then quantified using the 2D Quant kit (GE Healthcare).
2.3. Sodium dodecyl sulfate (SDS)-PAGE (1DE)
Protein samples (30 μg) were mixed with 4× premixed protein sample buffer (BioRad) and β-mercaptoethanol (5% final concentration, Sigma), and heated at 95 °C for 5 min. Protein mixtures were then loaded on Precast Criterion XT Bis–Tris gradient gels (4–12% polyacrylamide, BioRad) and the SDS–PAGE separation was performed on a Criterion™ gel electrophoresis cell (BioRad) using a PowerPac 200 BioRad power supply set at 200 V for 50 min. For staining, gels were fixed in a solution of 50% methanol: 7.5% acetic acid for 1 h then incubated overnight with SYPRO Ruby Protein Gel stain (BioRad). The destaining step was performed for 30 min in a solution of 15% methanol: 7.5% acetic acid. Gel images were captured on a PerkinElmer ProExpress Proteomic Imaging system. Each sample lane from the SDS–PAGE gels was cut in 40 fractions (24 fractions above 50 kDa and 16 fractions below 50 kDa) with disposable blade (MEE-1×5) mounted on a One Touch GridCutter (Gel Company Inc.).
2.4. Protein in-gel digestion
Bands of interest were extracted from SDS–PAGE gels, placed in 96-well plates and washed extensively with HPLC water. Tryptic digestion was performed on a MassPrep liquid handling robot (Waters) according to the manufacturer’s specifications and to the protocol of Shevchenko et al. (1996) with minor modifications (Havlis et al., 2003). Briefly, proteins were reduced with 10 mM DTT and alkylated with 55 mM iodoacetamide. Trypsin digestion was performed using 126 nM of modified porcine trypsin (Sequencing grade, Promega) at 58 °C for 1 h. Digestion products were then extracted using 1% formic acid and 2% acetonitrile followed by 1% formic acid and 50% acetonitrile. The recovered protein extracts were pooled, vacuum centrifuge dried and resuspended into 10 μL of 0.1% formic acid. Aliquots of 2 (for cytosolic fractions) or 5 (for ME fractions) μL were analyzed by mass spectrometry.
2.5. Mass spectrometry for the ME fraction
SILAC experiments for the ME fraction were performed on a ABI QSTAR XL QqTOF mass spectrometer equipped with a nanospray II ion source (ABSciex) coupled to an Agilent 1100 HPLC as previously described (Brotherton et al., 2013). Five microliters of each protein sample were injected by the Agilent 1100 autosampler onto a 0.075 mm (internal diameter) self-packed IntegraFrit column (New Objective) packed with an isopropanol slurry of 5 μm Jupiter C18 (Phenomenex) stationary phase using a pressure vessel set at 700 psi. The length of the column was 12 cm. Samples were run using a 75 min gradient from 10–40% solvent B (solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile) at a flow rate of 250 nL/min. An information-dependent acquisition (IDA) method was set up with the MS survey range set between 400 amu and 1600 amu (1 s) followed by dependent MS/MS scans with a mass range set between 100 and 1600 amu (3 s) of the 3 most intense ions with the enhanced all mode activated. Dynamic exclusion was set for a period of 15 s and a tolerance of 100 ppm.
2.6. Mass spectrometry for the cytosolic fraction
SILAC experiments for the cytosolic protein fraction were performed on a TripleTOF 5600 mass spectrometer equipped with a nanospray III ion source (ABSciex) coupled to an Agilent 1200 HPLC. Two microliter samples were injected by the Agilent 1200 autosampler onto a 0.075 mm (internal diameter) self-packed PicoFrit column (New Objective) packed with an isopropanol slurry of 5 μm Jupiter C18 (Phenomenex) stationary phase using a pressure vessel set at 700 psi. The length of the column was 15 cm. Samples were run using a 65 min gradient from 5–35% solvent B (solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile) at a flow rate of 300 nL/min. Data were acquired using an ion spray voltage of 2.4 kV, curtain gas of 30 psi, nebulizer gas of 8 psi and an interface heater temperature of 125 °C. An information-dependent acquisition (IDA) method was set up with the MS survey range set between 400 amu and 1250 amu (250 ms) followed by dependent MS/MS scans with a mass range set between 100 and 1800 amu (50 ms) of the 20 most intense ions in the high sensitivity mode with a 2+ to 5+ charge state. Dynamic exclusion was set for a period of 3 s and a tolerance of 100 ppm.
2.7. Interpretation of tandem mass spectra and protein identification
Raw data files (n = 40 for each sample) were submitted for simultaneous searches using the Protein Pilot version 4 software (ABSciex) utilizing the Paragon and Progroup algorithms (Shilov et al., 2007). The Protein Pilot program was set up to search the L. infantum proteins in the TriTrypDB LeishPEP database (http://tritrypdb.org/common/downloads/release-4.0/Linfantum/fasta/LinfantumAnnotatedProteins_TriTrypDB-4.0.fasta) with carbamidomethyl (C) as a fixed modification and standard SILAC (Lys +8, Arg +10) settings for QSTAR or TripleTof 5600 instruments. Proteins for which at least two fully trypsin-digested light (L) and heavy (H) peptides were detected at >99% confidence and quantitative p-value lower than 0.05 were used for subsequent comparative quantitative analysis.
3. Results and discussion
Lipid formulations of the polyene antibiotic AmB are now the mainstay for the treatment of Leishmania in the epidemic region of Bihar in India where nearly 65% of the cases are refractory to the first line antimonial drugs (Bern et al., 2006; Chappuis et al., 2007). Although some sporadic unusual cases have been reported recently (Srivastava et al., 2011; Purkait et al., 2012), clinical resistance has not yet threatened the efficacy of AmB, even when used through many courses as for treating HIV co-infected patients (Lachaud et al., 2009). Using a SILAC-based quantitative proteomic method, we probed proteome alterations induced in an in vitro selected L. infantum AmB1000.1 resistant mutant. Since membrane proteins are often underrepresented in proteomic studies, we choose to analyze SILAC-labeled membrane enriched (ME) and cytosolic proteins separately in order to increase the proteome coverage.
The L. infantum AmB1000.1 resistant mutant was grown in the presence of 13C6-15N4-l-arginine and 13C6-15N2-l-lysine while the parental L. infantum WT strain was maintained in the same medium containing normal isotopic abundance amino acids. Both cell populations were counted and mutant parasites were mixed with WT cells in a 1:1 ratio. ME and cytosolic fractions were extracted from the mixed population and both fractions were independently subjected to SDS–PAGE separation. Each sample line was further fractionated into 40 pieces of gel, proteins were gel extracted, trypsin digested and peptides were identified and quantified by mass spectrometry and isotopic quantification. Only protein identifications with differences in their expression level greater than 1.2-fold when comparing mutant and WT levels were considered as significant. This expression threshold was selected after having performed a control experiment to evaluate the extent of isotopic incorporation and technical noise. A fold change of 1.2 was considered as an expression threshold well above background. Thus, 99 protein identifications corresponding to 97 individual proteins were assigned as differentially expressed in the AmB1000.1 mutant (Table 1). Among them, 83 (84%) were up-regulated and 16 (16%) were down-regulated. Two proteins (LinJ.03.0190 and LinJ.24.1700) were identified independently in both ME and cytosolic fractions as indicated in Table 1. With only two exceptions (LinJ.23.0880 and LinJ.32.0280), all the cytosolic proteins identified in this study were predicted to be devoid of transmembrane domains (TMDs) according to the TMHMM v2.0 algorithm (Table 1). In contrast, a quarter (23%) of the proteins differentially expressed in ME fraction were predicted to have at least one TMD. The identification of proteins with no predicted TMD in ME fraction might be possibly explained by non-covalent interactions with components of membranes through hydrogen bonding, van der Waals contacts or electrostatic attractions. These types of interactions are known to play critical roles in maintaining cell membrane structure and facilitating membrane functions (reviewed in Prinz and Hinshaw, 2009).
Table 1.
MS/MS identifications of differentially expressed proteins in AmB1000.1 compared to WT in SILAC experiments.
| Systematic IDs | Putative protein name | Unique peptidesa (% sequence coverage) | Ratio AmB 1000.1/WT | p-value | Error factor | TMDsb | Fractionc |
|---|---|---|---|---|---|---|---|
| Transport | |||||||
| LinJ.18.1510 | H1A-2 P-type H+-ATPase, putative | 15 (14.48) | −2,51 | 0,0130 | 2,02 | 8 | M |
| LinJ.25.1210 | ATPase beta subunit, putative | 18 (31.62) | 1,68 | 0,0009 | 1,29 | 0 | C |
| LinJ.30.3660 | ATP synthase, epsilon chain, putative | 2 (16.57) | 2,25 | 0,0892 | 4,28 | 0 | C |
| Surface | |||||||
| LinJ.10.0500 | GP63, leishmanolysin,metallo-peptidase, Clan MA(M), Family M8 | 24 (27.38) | −4,14 | 0,0001 | 1,83 | 1 | M |
| LinJ.30.0930 | Amastin-like surface protein-like protein | 2 (4.05) | −2,61 | 0,0897 | 5,65 | 4 | M |
| Cytoskeleton | |||||||
| LinJ.19.0680 | Kinesin, putative | 9 (8.70) | 1,98 | 0,0001 | 1,25 | 0 | C |
| LinJ.23.0720 | Kinesin, putative | 5 (6.86) | 1,78 | 0,0167 | 1,50 | 0 | M |
| LinJ.30.0350 | Kinesin, putative | 4 (6.39) | 2,18 | 0,0316 | 1,84 | 0 | C |
| Metabolism | |||||||
| LinJ.03.0190 | Delta-1-pyrroline-5-carboxylate dehydrogenase, putative | 8 (13.39) | 1,26 | 0,0417 | 1,25 | 0 | M |
| LinJ.03.0190 | Delta-1-pyrroline-5-carboxylate dehydrogenase, putative | 10 (21.43) | 1,67 | 0,0045 | 1,37 | 0 | C |
| LinJ.05.0350 | Trypanothione reductase | 2 (5.50) | 2,50 | 0,0320 | 2,06 | 0 | C |
| LinJ.07.0240 | Cobalamin-dependant methionine synthase, putative | 3 (2.24) | 1,91 | 0,0334 | 1,74 | 0 | C |
| LinJ.14.0680 | Fatty acid elongase, putative | 2 (5.63) | −2,22 | 0,0936 | 4,47 | 6 | M |
| LinJ.14.1240 | Enolase | 9 (24.48) | 1,53 | 0,0706 | 1,60 | 0 | C |
| LinJ.14.1450 | Myo-inositol-1-phosphate synthase | 2 (4.56) | 1,91 | 0,0975 | 3,57 | 0 | C |
| LinJ.15.1100 | Tryparedoxin peroxidase | 10 (32.16) | 1,81 | 0,0548 | 1,86 | 0 | C |
| LinJ.18.0510 | Aconitase, putative | 26 (33.15) | 1,37 | 0,0687 | 1,41 | 0 | C |
| LinJ.21.0310 | Hexokinase, putative | 14 (31.42) | 1,85 | 0,0036 | 1,42 | 0 | C |
| LinJ.21.1490 | Adenylate kinase, putative | 2 (9.29) | −3,23 | 0,0546 | 3,55 | 0 | C |
| LinJ.22.0190 | Carnitine palmitoyltransferase-like protein | 1 (2.22) | −4,34 | 0,0560 | 5,19 | 0 | M |
| LinJ.23.0050 | Peroxidoxin, tryparedoxin peroxidase | 2 (8.41) | 2,67 | 0,0276 | 2,18 | 0 | C |
| LinJ.23.0860 | ERG10 3-ketoacyl-coa thiolase-like protein | 2 (7.26) | 2,47 | 0,0140 | 1,59 | 0 | C |
| LinJ.23.0880 | Acetyl-CoA synthetase, putative | 5 (7.63) | 2,17 | 0,0054 | 1,53 | 1 | C |
| LinJ.24.1700 | Succinate dehydrogenase flavoprotein, putative | 6 (12.03) | −1,90 | 0,0147 | 1,49 | 0 | M |
| LinJ.24.1700 | Succinate dehydrogenase flavoprotein, putative | 5 (9.56) | 1,98 | 0,0127 | 1,59 | 0 | C |
| LinJ.30.2920 | Aldehyde dehydrogenase, putative | 2 (3.59) | 1,67 | 0,0013 | 1,20 | 0 | M |
| LinJ.30.3000 | Glyceraldehyde 3-phosphate dehydrogenase, glycosomal | 18 (41.83) | 1,60 | 0,0048 | 1,35 | 0 | C |
| LinJ.30.3580 | S-adenosylmethionine synthetase | 5 (13.52) | 2,22 | 0,0780 | 2,53 | 0 | C |
| LinJ.32.1920 | SODB2 iron superoxide dismutase, putative | 4 (11.54) | 2,41 | 0,0959 | 5,46 | 0 | C |
| LinJ.32.3110 | Nucleoside diphosphate kinase b | 2 (13.91) | 1,60 | 0,0462 | 1,58 | 0 | M |
| LinJ.32.3510 | GCVL-2 dihydrolipoamide dehydrogenase, putative | 8 (17.44) | 1,62 | 0,0086 | 1,40 | 0 | C |
| LinJ.34.0150 | Malate dehydrogenase | 5 (18.30) | 1,55 | 0,0526 | 1,56 | 0 | C |
| LinJ.35.1190 | NADH-dependent fumarate reductase, putative | 25 (23.98) | 1,40 | 0,0033 | 1,23 | 0 | C |
| LinJ.36.1320 | Fructose-1,6-bisphosphate aldolase | 10 (23.99) | 1,64 | 0,0073 | 1,38 | 0 | C |
| LinJ.36.4100 | S-adenosylhomocysteine hydrolase | 12 (25.63) | 1,54 | 0,0547 | 1,55 | 0 | C |
| Protein folding | |||||||
| LinJ.21.1330 | T-complex protein 1, delta subunit, putative | 11 (15.79) | 1,86 | 0,0229 | 1,67 | 0 | C |
| LinJ.23.1460 | T-complex protein 1, gamma subunit, putative | 10 (18.51) | 1,58 | 0,0161 | 1,39 | 0 | C |
| LinJ.26.1360 | Prefoldin-like protein | 5 (27.84) | 1,62 | 0,0522 | 1,63 | 0 | C |
| LinJ.28.3060 | Heat-shock protein hsp70, putative | 45 (38.59) | 1,55 | 0,0004 | 1,24 | 0 | C |
| LinJ.30.2540 | Heat shock 70-related protein 1, mitochondrial precursor, putative | 42 (46.82) | 1,93 | 0,0097 | 1,63 | 0 | C |
| LinJ.32.1060 | Chaperonin containing t-complex protein, putative | 7 (11.71) | 1,95 | 0,0053 | 1,47 | 0 | C |
| LinJ.32.1940 | Chaperonin Hsp60, mitochondrial precursor, putative | 3 (5.05) | 2,40 | 0,0123 | 1,53 | 0 | C |
| LinJ.32.3470 | Chaperonin alpha subunit, putative | 20 (36.63) | 1,72 | 0,0004 | 1,29 | 0 | C |
| LinJ.33.0370 | Heat shock protein 83-1 | 28 (26.86) | 1,52 | 0,0042 | 1,31 | 0 | C |
| LinJ.33.2520 | Heat shock protein, putative | 16 (17.98) | 1,52 | 0,0172 | 1,39 | 0 | C |
| LinJ.36.2140 | Chaperonin Hsp60, mitochondrial precursor | 44 (54.98) | 2,14 | 4,611E-06 | 1,21 | 0 | C |
| LinJ.36.7240 | Chaperonin, putative,T-complex protein 1 (theta subunit), putative | 13 (24.77) | 1,93 | 1,289E-05 | 1,22 | 0 | C |
| Proteolysis | |||||||
| LinJ.09.0820 | Oligopeptidase b,serine peptidase, clan SC, family S9A-like protein | 4 (6.02) | 1,56 | 0,0870 | 1,72 | 0 | C |
| LinJ.11.0640 | Aminopeptidase, putative,metallo-peptidase, Clan MF, Family M17 | 7 (14.77) | 1,45 | 0,0687 | 1,50 | 0 | C |
| LinJ.20.1220 | Calpain-like cysteine peptidase, putative,cysteine peptidase, Clan CA, family C2, putative | 3 (4.38) | −1,69 | 0,0115 | 1,35 | 0 | C |
| LinJ.36.1730 | Proteasome beta 5 subunit, putative | 3 (11.92) | 1,86 | 0,0438 | 1,78 | 0 | C |
| Transcription, Translation | |||||||
| LinJ.01.0800 | Eukaryotic initiation factor 4a, putative | 10 (26.05) | 1,74 | 0,0468 | 1,72 | 0 | C |
| LinJ.06.0410 | 60S ribosomal protein L19, putative | 5 (15.92) | 1,21 | 0,0238 | 1,17 | 0 | M |
| LinJ.09.1130 | Eukaryotic translation initiation factor 2 subunit, putative | 2 (3.34) | 2,02 | 0,0191 | 1,70 | 0 | C |
| LinJ.11.0100 | Seryl-tRNA synthetase, putative | 3 (3.80) | 2,17 | 0,0064 | 1,55 | 0 | C |
| LinJ.13.0460 | 40S ribosomal protein S12, putative | 8 (29.79) | 1,93 | 0,0001 | 1,20 | 0 | C |
| LinJ.17.0110 | Elongation factor 1-alpha | 23 (38.37) | 1,65 | 0,0014 | 1,32 | 0 | C |
| LinJ.18.0740 | Elongation factor Tu, putative | 2 (5.59) | 1,82 | 0,0530 | 1,85 | 0 | C |
| LinJ.21.0600 | la RNA binding protein, putative | 5 (20.54) | 1,50 | 0,0409 | 1,46 | 0 | C |
| LinJ.21.1310 | 40S ribosomal protein S23, putative | 4 (18.88) | 1,32 | 0,0324 | 1,26 | 0 | M |
| LinJ.25.0760 | Eukaryotic initiation factor 5a, putative | 2 (8.17) | 2,27 | 0,0497 | 2,27 | 0 | C |
| LinJ.26.2360 | 60S ribosomal protein L35, putative | 4 (14.17) | 1,30 | 0,0374 | 1,26 | 0 | M |
| LinJ.27.2480 | 60S acidic ribosomal subunit protein | 3 (9.91) | 2,17 | 0,0166 | 1,55 | 0 | C |
| LinJ.29.1920 | 40S ribosomal protein S15A, putative | 3 (20.77) | 2,00 | 0,0659 | 2,18 | 0 | C |
| LinJ.30.3650 | 40S ribosomal protein S14 | 4 (20.14) | −1,43 | 0,0057 | 1,20 | 0 | C |
| LinJ.32.0410 | ATP-dependent RNA helicase, putative | 17 (27.52) | 1,56 | 0,0050 | 1,33 | 0 | C |
| LinJ.32.2850 | Ribosomal protein L27, putative | 6 (28.36) | 1,23 | 0,0630 | 1,24 | 0 | M |
| LinJ.35.0240 | 60S ribosomal protein L30 | 7 (56.73) | 1,92 | 0,0236 | 1,70 | 0 | C |
| LinJ.35.1880 | 60S ribosomal protein L5, putative | 5 (11.48) | 1,28 | 0,0194 | 1,20 | 0 | M |
| LinJ.35.2000 | 40S ribosomal protein S6, putative | 3 (4.02) | 1,22 | 0,0727 | 1,26 | 0 | M |
| LinJ.35.3840 | 60S ribosomal protein L23, putative | 3 (25.90) | 2,19 | 0,0433 | 2,07 | 0 | C |
| LinJ.35.5360 | Polyadenylate-binding protein 1, putative | 6 (9.11) | 1,77 | 0,0190 | 1,55 | 0 | C |
| LinJ.36.0020 | Histone H4 | 5 (26.00) | 1,31 | 0,0043 | 1,17 | 0 | M |
| LinJ.36.3950 | RPL10a 60S ribosomal protein L10a, putative | 6 (15.42) | −1,31 | 0,0863 | 1,39 | 0 | M |
| LinJ.36.4730 | RPL18 60S ribosomal protein L18, putative | 3 (13.13) | −1,56 | 0,0409 | 1,49 | 0 | M |
| LinJ.36.5870 | Isoleucyl-tRNA synthetase, putative | 5 (4.18) | 1,96 | 0,0566 | 2,03 | 0 | C |
| LinJ.36.7320 | Eukaryotic translation initiation factor 3 subunit 8, putative | 7 (6.16) | 1,39 | 0,0698 | 1,44 | 0 | M |
| Others | |||||||
| LinJ.14.0990 | Immunodominant antigen, putative,tc40 antigen-like | 5 (6.97) | 1,48 | 0,0707 | 1,54 | 0 | C |
| LinJ.16.1710 | Prohibitin, putative | 4 (16.42) | −1,28 | 0,0695 | 1,32 | 0 | M |
| LinJ.20.0820 | Vesicle-fusing ATPase, putative,N-ethylmaleimide-sensitive factor, putative | 5 (7.86) | 1,55 | 0,0832 | 1,68 | 0 | C |
| LinJ.23.0120 | GDP-mannose pyrophosphorylase | 3 (9.50) | 1,88 | 0,0683 | 2,06 | 0 | C |
| LinJ.28.2430 | Glycosomal membrane protein, putative | 4 (10.36) | 1,66 | 0,0016 | 1,20 | 0 | M |
| LinJ.34.0720 | Flagellar attachment zone protein | 4 (10.92) | 1,88 | 0,0676 | 2,02 | 0 | C |
| LinJ.34.2680 | Regulatory subunit of protein kinase a-like protein | 9 (12.52) | 1,83 | 0,0002 | 1,21 | 0 | C |
| LinJ.35.0070 | Prohibitin, putative | 2 (6.16) | −1,71 | 0,0182 | 1,43 | 1 | M |
| LinJ.36.3360 | 14-3-3 protein-like protein | 2 (8.53) | 1,98 | 0,0144 | 1,43 | 0 | C |
| Hypothetical | |||||||
| LinJ.08.1010 | Hypothetical protein, conserved | 3 (8.77) | 1,89 | 0,0563 | 1,94 | 0 | C |
| LinJ.09.0120 | Hypothetical protein, conserved,calmodulin-like protein containing EF hand domain | 6 (13.75) | 1,40 | 0,0382 | 1,36 | 0 | C |
| LinJ.09.1070 | Hypothetical protein, conserved | 8 (15.93) | 1,53 | 0,0699 | 1,60 | 0 | C |
| LinJ.09.1530 | Hypothetical protein, conserved | 2 (13.74) | 2,15 | 0,0395 | 1,96 | 0 | C |
| LinJ.13.0740 | Hypothetical protein, conserved | 8 (9.88) | −2,13 | 0,0222 | 1,82 | 0 | C |
| LinJ.16.1050 | Hypothetical protein, conserved | 3 (4.91) | −1,77 | 0,0802 | 1,97 | 0 | M |
| LinJ.17.0780 | Hypothetical protein, conserved | 4 (5.21) | 1,72 | 0,0840 | 2,06 | 0 | C |
| LinJ.23.0810 | Hypothetical protein, conserved | 3 (3.96) | 1,52 | 0,0010 | 1,22 | 4 | M |
| LinJ.25.2100 | Hypothetical protein, conserved | 2 (7.87) | 1,60 | 0,0235 | 1,44 | 0 | C |
| LinJ.27.1220 | Hypothetical protein, conserved | 6 (9.64) | 1,65 | 0,0431 | 1,61 | 0 | C |
| LinJ.29.1570 | Hypothetical protein, conserved | 2 (3.23) | 2,40 | 0,0717 | 3,51 | 0 | C |
| LinJ.32.0280 | Hypothetical protein, conserved | 5 (4.07) | −1,59 | 0,0650 | 1,68 | 2 | C |
| LinJ.36.5380 | Hypothetical protein, conserved; kinesin-like protein, putative | 7 (10.88) | 1,51 | 0,0108 | 1,33 | 0 | M |
Peptide identifications were accepted if they reached greater than 95% probability as specified by the Peptide Prophet algorithm.
TMDs were predicted using TMHMM v2.0.
M, membrane-enriched fraction; C, cytosolic fraction.
Differentially expressed proteins with statistical significance were sorted into functional classes according to GeneDB annotations and Gene Ontology. Metabolism (27%) and transcription/translation (26%) were the main functional classes represented in this study corresponding together to more than half of all the identified proteins (Fig. 1). Another quarter of the protein hits were represented by hypothetical proteins (13%) or involved in protein folding (12%) (Fig. 1). Other functional groups, each of which representing less than 5% of the identifications, were proteolysis, transport, cytoskeleton and surface proteins, whereas 9% of the identified proteins were not assigned to any of the aforementioned classes (Fig.1).
Fig. 1.
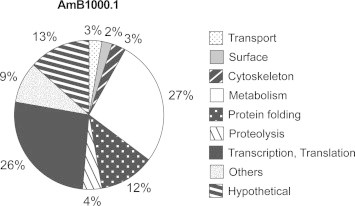
Functional assignment of proteins found differentially expressed by SILAC between L. infantum WT and AmB1000.1 mutant. Protein functional classification was based on GeneDB annotations and Gene Ontology.
Three enzymes playing a role in the conversion of S-adenosyl-l-homocysteine to S-adenosyl-l-methionine, namely the S-adenosylhomocysteine hydrolase (LinJ.36.4100), cobalamin-dependant methionine synthase (LinJ.07.0240) and S-adenosylmethionine synthetase (LinJ.30.3580), were up-regulated in the AmB resistant mutant (Table 1). Since S-adenosyl-l-methionine is involved in many cellular functions, including stress response, it may explain why this pathway was increased in the AmB mutant. Other key metabolic pathways detected in AmB1000.1 resistant mutant as being up-regulated included glycolysis with the hexokinase (LinJ.21.0310), fructose-1,6-bisphosphate aldolase (LinJ.36.1320), glyceraldehyde 3-phosphate dehydrogenase (LinJ.30.3000) and enolase (LinJ.14.1240) as well as the TCA cycle with the aconitase (LinJ.18.0510), GCVL-2 dihydrolipoamide dehydrogenase (LinJ.32.3510) and malate dehydrogenase (LinJ.34.0150) (Table 1). Curiously, the succinate dehydrogenase flavoprotein (LinJ.24.1700), another enzyme involved in the TCA cycle, was found to be up-regulated in the cytosolic protein fraction (1.98-fold) but down-regulated in the ME fraction (1.90-fold). The reason for this discrepancy is unknown. The modulation of several glycolytic enzymes in an AmB resistance context is mimicking the glycolysis state level previously reported in L. donovani antimony in vitro resistant cell lines (Biyani et al., 2011) and may thus corresponds to a general stress response more than a specific drug response. However, the TCA cycle enzymes aconitase and malate dehydrogenase as well as the inositol phosphate metabolism enzyme myo-inositol-1-phosphate synthase were also found to be up-regulated in response to AmB in a previous Candida albicans proteomic study (Hoehamer et al., 2010), similarly as we observed here and thus might be involved in the AmB resistance phenotype acquired from our in vitro Leishmania mutant.
A number of enzymes part of the reactive oxygen species (ROS) induced pathway was overproduced in our AmB mutant namely the trypanothione reductase (TR) (LinJ.05.0350) as well as two different tryparedoxin peroxidases (TPX) (LinJ.15.1100 and LinJ.23.0050) (Table 1). In addition to the tryparedoxin cascade which was previously shown to be up-regulated in AmB clinical unresponsive strain (Purkait et al., 2012), increased reduced thiols by an overproduction of TR in the AmB resistant parasites may also be involved in better antioxidant defense as it was already reported in the case of antimony resistance (Wyllie et al., 2008). Nonetheless, transfection of these individual genes (encoding TR and TXP) did not lead to a resistance phenotype to AmB (Table S1) suggesting that if they are involved in resistance it must be a more subtle role. AmB has been proposed to kill Aspergillus terreus by inducing intracellular oxidation leading to lipid peroxidation and ultimately cell death (Blum et al., 2013a). Thus, TR and TXP might possibly act by interfering somehow with this cell death pathway. It is also interesting to note that the detoxifying enzyme SODB2 iron superoxide dismutase (LinJ.32.1920) was similarly overproduced in the AmB mutant. This enzyme is known in several other organisms to be modulated in response to oxidative stress (Alscher et al., 2002). Moreover, superoxide dismutase was also shown to protect C. albicans from oxidative damage caused by AmB (Sokol-Anderson et al., 1986). However, transfection of SODB2 failed to show a direct role for resistance to AmB (Table S1) and further work would be required.
Many proteins involved in protein folding, such as heat-shock proteins and chaperonins, were also found to be up-regulated in AmB1000.1 suggesting a putative role of these proteins in AmB resistance or tolerance. In particular, the heat shock protein 83-1 (Hsp83-1, LinJ.33.0370) was found up-regulated in the AmB resistant mutant compared to WT cells (Table 1). This protein is a member of the Hsp90 family, which has been recently found to be a key player in AmB resistance in A. terreus (Blum et al., 2013b). Furthermore, Hsp60, some Hsp70 and S-adenosylmethionine synthetase were also found to be up-regulated in C. albicans in response to AmB through a proteomic screen (Hoehamer et al., 2010). The role of heat shock proteins in resistance in Leishmania has not been directly assessed by gene transfection however (Table S1).
AmB is known to bind to the membrane ergosterol of sensitive Leishmania strains, causing pore formation in the membrane and leakage of ions like K+ which results ultimately in cell death (Cohen et al., 1986; Saha et al., 1986). The H1A-2 P-type H+-ATPase (LinJ.18.1510) was down-regulated in our AmB mutant. Pma1p, the orthologous protein in yeast, is involved in the regulation of intracellular pH by hydrogen efflux (reviewed in Morsomme et al., 2000). We may therefore hypothesize that the down-regulation of this proton pump at the parasite cell surface might prevent H+ expulsion and thus might help maintaining the membrane potential gradient necessary to survive K+ leaking. Overexpression experiments did not lead to a phenotype, however (Table S1), so the gene product encoded by LinJ.18.1510 cannot be considered per se as an AmB resistance gene but still can be a contributing factor to the high level of AmB resistance since its proton activity might be a futile waste of ATP in the presence of pores in the membrane, thus unnecessarily depleting the parasite of ATP. Further experiments such as inactivation of the gene are required to assess any role of this candidate in AmB resistance in Leishmania.
Overall, the genes coding for 18 candidate proteins being detected either as up- or down-regulated from our quantitative proteomic analysis were cloned in an episomal vector and transfected in L. infantum WT and AmB resistant mutant cells, respectively (Table S1). These genes were selected because literature data were indicating a potential role in drug resistance in other organisms or because some motifs of these proteins were suggesting a possible link with chemoresistance or drug tolerance. Transfection experiments failed to show any direct link with AmB resistance, however. Since multiple mutations can co-exist and lead to drug resistance in Leishmania (Coelho et al., 2012; Ritt et al., 2013), it remains to be tested if a combination of two or more genes may lead to AmB resistance when co-transfected in the same recipient cell. A large number of protein candidates (n = 97) differentially expressed in our mutant were discovered and some might be only related to a stress response and not to the drug itself. One possible approach to decrease the false-negative discovery rate in SILAC proteomic experiments would be to study more than one mutant and concentrate on recurrent mutations in independent mutants. This strategy has indeed improved the quality of our dataset in other large genomic scale studies (Coelho et al., 2012; Ritt et al., 2013).
In conclusion, this large-scale proteomic study of an in vitro AmB Leishmania resistant mutant allowed the identification of 97 individual differentially expressed proteins when compared to the parental sensitive strain. While the candidate proteins tested by transfection were not directly involved in AmB resistance, several of the proteins identified were observed independently either in Leishmania or fungus. It is thus possible that some of these putative candidates may form protein complexes in order to sustain resistance or that their involvement in resistance is more indirect.
Acknowledgments
We would like to thank Maxim Isabelle for his help in the planning and the setting up of SILAC experiments. We also would like to thank Sandra Breuils-Bonnet and Benjamin Nehmé for sample preparation prior to MS/MS. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) and by the Ministère du Dévelopement Economique, Innovation et Exportation (MDEIE) grants to AD and by CIHR grants to GGP and MO. MO holds the Canada Research Chair in Antimicrobial Resistance.
Appendix A. Supplementary data
References
- Alscher R.G., Erturk N., Heath L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002;53:1331–1341. [PubMed] [Google Scholar]
- Alvar J., Croft S., Olliaro P. Chemotherapy in the treatment and control of leishmaniasis. Adv. Parasitol. 2006;61:223–274. doi: 10.1016/S0065-308X(05)61006-8. [DOI] [PubMed] [Google Scholar]
- Bern C., Adler-Moore J., Berenguer J., Boelaert M., den Boer M., Davidson R.N., Figueras C., Gradoni L., Kafetzis D.A., Ritmeijer K., Rosenthal E., Royce C., Russo R., Sundar S., Alvar J. Liposomal amphotericin B for the treatment of visceral leishmaniasis. Clin. Infect. Dis. 2006;43:917–924. doi: 10.1086/507530. [DOI] [PubMed] [Google Scholar]
- Biyani N., Singh A.K., Mandal S., Chawla B., Madhubala R. Differential expression of proteins in antimony-susceptible and -resistant isolates of Leishmania donovani. Mol. Biochem. Parasitol. 2011;179:91–99. doi: 10.1016/j.molbiopara.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Blum G., Hortnagl C., Jukic E., Erbeznik T., Pumpel T., Dietrich H., Nagl M., Speth C., Rambach G., Lass-Florl C. New insight into amphotericin B resistance in Aspergillus terreus. Antimicrob. Agents Chemother. 2013 doi: 10.1128/AAC.01283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G., Kainzner B., Grif K., Dietrich H., Zelger B., Sonnweber T., Lass-Florl C. In vitro and in vivo role of heat shock protein 90 in amphotericin B resistance of Aspergillus terreus. Clin. Microbiol. Infect. 2013;19:50–55. doi: 10.1111/j.1469-0691.2012.03848.x. [DOI] [PubMed] [Google Scholar]
- Brotherton M.C., Racine G., Foucher A.L., Drummelsmith J., Papadopoulou B., Ouellette M. Analysis of stage-specific expression of basic proteins in Leishmania infantum. J. Proteome Res. 2010;9:3842–3853. doi: 10.1021/pr100048m. [DOI] [PubMed] [Google Scholar]
- Brotherton M.C., Racine G., Ouameur A.A., Leprohon P., Papadopoulou B., Ouellette M. Analysis of membrane-enriched and high molecular weight proteins in Leishmania infantum promastigotes and axenic amastigotes. J. Proteome Res. 2012;11:3974–3985. doi: 10.1021/pr201248h. [DOI] [PubMed] [Google Scholar]
- Brotherton M.C., Bourassa S., Leprohon P., Legare D., Poirier G.G., Droit A., Ouellette M. Proteomic and genomic analyses of antimony resistant Leishmania infantum mutant. PLoS One. 2013;8:e81899. doi: 10.1371/journal.pone.0081899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappuis F., Sundar S., Hailu A., Ghalib H., Rijal S., Peeling R.W., Alvar J., Boelaert M. Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat. Rev. Microbiol. 2007;5:873–882. doi: 10.1038/nrmicro1748. [DOI] [PubMed] [Google Scholar]
- Coelho A.C., Boisvert S., Mukherjee A., Leprohon P., Corbeil J., Ouellette M. Multiple mutations in heterogeneous miltefosine-resistant Leishmania major population as determined by whole genome sequencing. PLoS Negl. Trop. Dis. 2012;6:e1512. doi: 10.1371/journal.pntd.0001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B.E., Ramos H., Gamargo M., Urbina J. The water and ionic permeability induced by polyene antibiotics across plasma membrane vesicles from Leishmania sp. Biochim. Biophys. Acta. 1986;860:57–65. doi: 10.1016/0005-2736(86)90498-0. [DOI] [PubMed] [Google Scholar]
- Havlis J., Thomas H., Sebela M., Shevchenko A. Fast-response proteomics by accelerated in-gel digestion of proteins. Anal. Chem. 2003;75:1300–1306. doi: 10.1021/ac026136s. [DOI] [PubMed] [Google Scholar]
- Hoehamer C.F., Cummings E.D., Hilliard G.M., Rogers P.D. Changes in the proteome of Candida albicans in response to azole, polyene, and echinocandin antifungal agents. Antimicrob. Agents Chemother. 2010;54:1655–1664. doi: 10.1128/AAC.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaud L., Bourgeois N., Plourde M., Leprohon P., Bastien P., Ouellette M. Parasite susceptibility to amphotericin B in failures of treatment for visceral leishmaniasis in patients coinfected with HIV type 1 and Leishmania infantum. Clin. Infect. Dis. 2009;48:e16–22. doi: 10.1086/595710. [DOI] [PubMed] [Google Scholar]
- Lira R., Sundar S., Makharia A., Kenney R., Gam A., Saraiva E., Sacks D. Evidence that the high incidence of treatment failures in Indian kala-azar is due to the emergence of antimony-resistant strains of Leishmania donovani. J. Infect. Dis. 1999;180:564–567. doi: 10.1086/314896. [DOI] [PubMed] [Google Scholar]
- Mbongo N., Loiseau P.M., Billion M.A., Robert-Gero M. Mechanism of amphotericin B resistance in Leishmania donovani promastigotes. Antimicrob. Agents Chemother. 1998;42:352–357. doi: 10.1128/aac.42.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira W., Leprohon P., Ouellette M. Tolerance to drug-induced cell death favours the acquisition of multidrug resistance in Leishmania. Cell Death Dis. 2011;2:e201. doi: 10.1038/cddis.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme P., Slayman C.W., Goffeau A. Mutagenic study of the structure, function and biogenesis of the yeast plasma membrane H(+)-ATPase. Biochim. Biophys. Acta. 2000;1469:133–157. doi: 10.1016/s0304-4157(00)00015-0. [DOI] [PubMed] [Google Scholar]
- Murray H.W., Berman J.D., Davies C.R., Saravia N.G. Advances in leishmaniasis. Lancet. 2005;366:1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- Olliaro P., Sundar S. Anthropometrically derived dosing and drug costing calculations for treating visceral leishmaniasis in Bihar, India. Trop. Med. Int. Health. 2009;14:88–92. doi: 10.1111/j.1365-3156.2008.02195.x. [DOI] [PubMed] [Google Scholar]
- Prinz W.A., Hinshaw J.E. Membrane-bending proteins. Crit. Rev. Biochem. Mol. Biol. 2009;44:278–291. doi: 10.1080/10409230903183472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkait B., Kumar A., Nandi N., Sardar A.H., Das S., Kumar S., Pandey K., Ravidas V., Kumar M., De T., Singh D., Das P. Mechanism of amphotericin B Resistance in Clinical Isolates of Leishmania donovani. Antimicrob. Agents Chemother. 2012;56:1031–1041. doi: 10.1128/AAC.00030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritt J.F., Raymond F., Leprohon P., Legare D., Corbeil J., Ouellette M. Gene amplification and point mutations in pyrimidine metabolic genes in 5-fluorouracil resistant Leishmania infantum. PLoS Negl. Trop. Dis. 2013;7:e2564. doi: 10.1371/journal.pntd.0002564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A.K., Mukherjee T., Bhaduri A. Mechanism of action of amphotericin B on Leishmania donovani promastigotes. Mol. Biochem. Parasitol. 1986;19:195–200. doi: 10.1016/0166-6851(86)90001-0. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Shilov I.V., Seymour S.L., Patel A.A., Loboda A., Tang W.H., Keating S.P., Hunter C.L., Nuwaysir L.M., Schaeffer D.A. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- Sokol-Anderson M.L., Brajtburg J., Medoff G. Amphotericin B-induced oxidative damage and killing of Candida albicans. J. Infect. Dis. 1986;154:76–83. doi: 10.1093/infdis/154.1.76. [DOI] [PubMed] [Google Scholar]
- Srivastava P., Prajapati V.K., Rai M., Sundar S. Unusual case of resistance to amphotericin B in visceral leishmaniasis in a region in India where leishmaniasis is not endemic. J. Clin. Microbiol. 2011;49:3088–3091. doi: 10.1128/JCM.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar S., Rosenkaimer F., Makharia M.K., Goyal A.K., Mandal A.K., Voss A., Hilgard P., Murray H.W. Trial of oral miltefosine for visceral leishmaniasis. Lancet. 1998;352:1821–1823. doi: 10.1016/S0140-6736(98)04367-0. [DOI] [PubMed] [Google Scholar]
- Sundar S. Drug resistance in Indian visceral leishmaniasis. Trop. Med. Int. Health. 2001;6:849–854. doi: 10.1046/j.1365-3156.2001.00778.x. [DOI] [PubMed] [Google Scholar]
- Sundar S., Jha T.K., Thakur C.P., Sinha P.K., Bhattacharya S.K. Injectable paromomycin for visceral leishmaniasis in India. N. Engl. J. Med. 2007;356:2571–2581. doi: 10.1056/NEJMoa066536. [DOI] [PubMed] [Google Scholar]
- Thakur C.P., Dedet J.P., Narain S., Pratlong F. Leishmania species, drug unresponsiveness and visceral leishmaniasis in Bihar, India. Trans. R. Soc. Trop. Med. Hyg. 2001;95:187–189. doi: 10.1016/s0035-9203(01)90160-9. [DOI] [PubMed] [Google Scholar]
- Wyllie S., Vickers T.J., Fairlamb A.H. Roles of trypanothione S-transferase and tryparedoxin peroxidase in resistance to antimonials. Antimicrob. Agents Chemother. 2008;52:1359–1365. doi: 10.1128/AAC.01563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


