Graphical abstract

Keywords: microRNAs, Parasite, Helminth, Caenorhabditis elegans, Gene regulation, Therapeutic
Highlights
-
•
Importance of microRNAs in helminth post-transcriptional gene regulation is reviewed.
-
•
Increasing helminth miRNA data are available from deep sequencing.
-
•
Some miRNAs are helminth-specific, many are novel to each species.
-
•
miRNAs may regulate parasite and host gene expression.
-
•
Uptake of miRNA inhibitors and mimics is feasible for functional analysis.
Abstract
microRNAs (miRNAs) are small non-coding RNAs involved in post-transcriptional gene regulation. They were first identified in the free-living nematode Caenorhabditis elegans, where the miRNAs lin-4 and let-7 were shown to be essential for regulating correct developmental progression. The sequence of let-7 was subsequently found to be conserved in higher organisms and changes in expression of let-7, as well as other miRNAs, are associated with certain cancers, indicating important regulatory roles. Some miRNAs have been shown to have essential functions, but the roles of many are currently unknown. With the increasing availability of genome sequence data, miRNAs have now been identified from a number of parasitic helminths, by deep sequencing of small RNA libraries and bioinformatic approaches. While some miRNAs are widely conserved in a range of organisms, others are helminth-specific and many are novel to each species. Here we review the potential roles of miRNAs in regulating helminth development, in interacting with the host environment and in development of drug resistance. Use of fluorescently-labeled small RNAs demonstrates uptake by parasites, at least in vitro. Therefore delivery of miRNA inhibitors or mimics has potential to alter miRNA activity, providing a useful tool for probing the roles of miRNAs and suggesting novel routes to therapeutics for parasite control.
1. Introduction
Gene expression in specific cells and tissues or at defined developmental stages can be regulated by a number of mechanisms. Transcription factor (TF) binding to specific promoter sites, usually within the 5′ upstream sequence, is currently the best understood mechanism of regulating gene expression. Presence of enhancer or suppressor DNA motifs can alter TF activity and gene expression under certain conditions. The accessibility of promoter regions to TF binding is also regulated by epigenetic modification of DNA or histones, involving methylation of DNA bases and/or methylation or acetylation of histones, resulting in changes in gene activity. Epigenetic control of gene expression is increasingly studied in cancer development and stem cell differentiation (Tarayrah and Chen, 2013), and an important role for DNA methylation in egg production and maturation of the parasitic fluke Schistosoma mansoni has also been demonstrated (Geyer et al., 2011). This review focuses on another level of gene control: post-transcriptional regulation mediated by microRNAs (miRNAs). miRNAs were first identified in Caenorhabditis elegans (Lee et al., 1993) and, with the advent of genome sequencing, have been identified in diverse organisms including humans, plants, viruses and parasitic helminths. While roles for some miRNAs have been elucidated, the functions of most are unknown. In this review we examine current data on miRNAs in helminths and their potential roles in regulating development, reproduction and survival within the host. We discuss the potential of miRNAs as therapeutic targets for parasite control and whether altered expression of specific miRNAs may play a role in anthelmintic resistance.
2. Mechanism of microRNA-mediated gene regulation
miRNAs are small non-coding RNAs of 21-25 bp in length. C. elegans genes lin-4 and let-7 were the first miRNAs to be identified and characterized (Lee et al., 1993; Reinhart et al., 2000) and both are heterochronic genes – genes which regulate the timing of developmental events (Ambros and Horvitz, 1984). Worms lacking lin-4 repeat developmental events that normally occur only in the L1 stage, in subsequent stages. Loss of another C. elegans heterochronic gene, lin-14, results in the opposite phenotype (Ambros and Horvitz, 1984). LIN-14 protein levels were shown to peak at egg hatching and decrease thereafter (Ruvkun and Giusto, 1989), while lin-4 showed the inverse expression pattern. lin-4 was cloned and found not to encode a protein, but a small RNA and the sequence of lin-4 was observed to be complementary to multiple sites in the lin-14 3′ untranslated region (UTR). From this it was concluded that lin-4 negatively regulates lin-14 post-transcriptionally, through binding sites in the 3′UTR (Lee et al., 1993; Wightman et al., 1993), thus paving the way to examine gene regulation by small RNAs. Subsequently, C. elegans let-7 was identified and shown to be a miRNA that regulated another heterochronic protein-coding gene, lin-41, through binding sites located in the 3′UTR of the lin-41 transcript (Reinhart et al., 2000). Importantly, let-7 is conserved in diverse species, establishing the importance of miRNAs as additional regulators of gene expression not only in worms, but also in higher organisms.
In animals, miRNAs bind with partial complementarity to target sites that can be present in any region of an mRNA, but are predominantly found in the 3′UTR (Bartel 2009). Each miRNA can have many target mRNAs and a single mRNA can be regulated by multiple miRNAs. miRNAs are processed from long primary transcripts (pri-miRNA) of several hundred nucleotides, by a protein complex containing the RNase Drosha to yield precursor-miRNAs (pre-miRNA) which are stem-loop structures of around 70 nucleotides (Kim et al., 2009) (Fig. 1). The enzyme Dicer then cleaves the pre-miRNA, removing the loop region to form an imperfect duplex of around 22 base pairs. One strand of the duplex (that with a weaker base paired 5′ end, referred to as the mature or guide strand) is incorporated into the RNA-induced silencing complex (RISC) and directs the complex to mRNA targets, resulting in translational repression and/or mRNA destabilization (Chekulaeva and Filipowicz, 2009). For some miRNAs, the alternative strand of the duplex (the star strand) can be functional, but is often present in lower amounts or may be degraded. The specificity of miRNA binding is determined predominantly by complementarity of nucleotides 2–7 (seed sequence) of the miRNA to the target mRNA sequence (Bartel, 2009). As discussed below, complementarity can be used to predict potential miRNA targets.
Fig. 1.
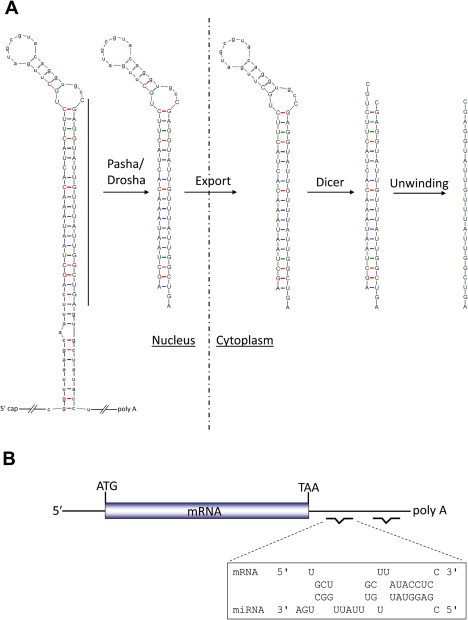
microRNA biogenesis and target binding. (A) The primary miRNA transcript (pri-miRNA) is processed in the nucleus by Drosha ribonuclease. The resulting pre-miRNA (approximately 70 nucleotides in length) is exported to the cytoplasm where it is cleaved by Dicer to form the miRNA duplex. (B) The mature strand of the miRNA duplex is incorporated into the RNA-induced silencing complex (RISC) and directs binding to mRNA target sequences, often in the 3′UTR. Binding is specified by complementarity between the target mRNA and miRNA seed sequence (nucleotides 2–7). An example of sequence complementarity is shown in (B).
3. microRNAs in helminths
3.1. Identifying microRNAs in parasitic helminths
The increasing availability of genome sequence data for parasitic helminths has paved the way to identify miRNA sequences using both computational and experimental approaches. The conservation of some miRNA sequences across species can be exploited to bioinformatically identify potential miRNAs. One approach taken previously by our lab (Winter et al., 2012) used the mature miRNA sequences present in miRBase (ftp://mirbase.org/pub/mirbase20) (Griffiths-Jones et al., 2008) to query available genome assemblies using BLASTN. The genomic regions flanking hits were excised and these candidate miRNA precursor sequences tested using RNAfold (Hofacker et al., 1994) and CIDmiRNA (Tyagi et al., 2008). A similar approach has been automated by others and is known as MapMi (Guerra-Assunção and Enright, 2010). As well as being an effective first step in miRNA identification, bioinformatic searching is particularly useful in identifying miRNAs expressed at low levels, in restricted cell types or in developmental stages not readily amenable to small RNA library preparation.
De novo identification of miRNAs is best achieved by deep sequencing of small RNA libraries. Sequences are mapped to the genome and other RNA classes including tRNAs, rRNAs and mRNAs removed from the analysis. Algorithms such as miRDeep (Friedländer et al., 2012) which incorporates RNAfold, score the likelihood of sequences representing true miRNAs based on the folding energies of genomic regions around the reads, similarity of these regions to miRNA hairpins and evidence of processing by Drosha and Dicer. In addition to miRNAs, small RNA sequencing also identifies endogenous small inhibitory (si) RNAs and PIWI-interacting RNAs (piRNAs, known as 21U RNAs in C. elegans). Each of these classes can be identified based on their different characteristic folding patterns, homology to coding sequence or for piRNAs, conserved 5′ motif sequence (Ruby et al., 2006; Friedlander et al., 2009).
Small RNA sequencing projects have been carried out for a number of helminth species (summarized in Table 1) and small RNA data are likely to increase with additional genome sequencing projects underway. In the absence of assembled genome sequence for a particular species of interest, alternative approaches have been applied to identify miRNAs. These include mapping small RNA sequence reads to genomes of closely related species, as we demonstrated for Brugia pahangi (Winter et al., 2012), where the genome data from its sister species Brugia malayi were used, and for Echinococcus granulosus (Cucher et al., 2011) where the Echinococcus multilocularis genome was utilized. Identification of miRNA/miRNA∗ duplex sequences with low RNAcofold folding energy and a 1-2nt 3′-overhang pattern, consistent with Dicer processing, has also been used to identify novel miRNAs (Huang et al., 2010). Data available so far provide a platform for detailed analysis of small RNA expression, conservation, evolution and function. For example, members of the mir-36 family have been identified in Caenorhabditis species (de Wit et al., 2009), Ascaris (Wang et al., 2011) and Brugia (Poole et al, 2010; Winter et al., 2012) as well as in platyhelminths including Schistosoma mansoni (Marco et al., 2013) and the free-living planarian Schmidtea mediterranea (Palakodeti et al., 2006; Friedlander et al., 2009). mir-36 family members appear to be helminth-specific and are expressed only in adult and embryonic stages. Deletion of the mir-36 cluster in C. elegans results in embryonic and larval lethality, indicating an essential role in early development (Alvarez-Saavedra and Horvitz, 2010). In Brugia, one highly conserved mir-36 family member was identified by deep sequencing and is enriched in adult female worms (Winter et al., 2012). Targeting this miRNA may therefore have potential in limiting microfilariae production.
Table 1.
Summary of parasitic helminth species for which microRNA sequence data is available.
| Parasite species | Developmental stage | References |
|---|---|---|
| Ascaris suum | Germline, zygote, embryo, L1, L2 | Wang et al. (2011) |
| Male and female adults | Xu et al. (2013) | |
| Brugia malayi | Mixed stage (males, females, mf) | Poole et al. (2010) |
| Brugia pahangi | Mixed sex adults, L3 | Winter et al. (2012) |
| Haemonchus contortus | Mixed sex adults, L3 | Winter et al. (2012) |
| Trichinella spiralis | Muscle stage larvae | Chen et al. (2011a) |
| Angiostrongylus cantonensis | Male and female adults | Chen et al. (2011b) |
| Young adults | Chang et al. (2013) | |
| Bursaphelenchus xylophilus | Mixed stages of different isolates | Huang et al. (2010) |
| Schistosoma japonicum | Mixed sex adults, schistosomula | Huang et al. (2009) |
| Cercariae, schistsomula, | Cai et al. (2011) | |
| Male and female adults | Cai et al. (2013) | |
| Eggs | ||
| S. mansoni | Mixed sex adults | Simoes et al. (2011) |
| Male and female adults | Marco et al. (2013) | |
| Echinococcus granulosis | Protoscolex | Cucher et al. (2011) |
| Taenia saginata | Adult worms | Ai et al. (2012) |
| Clonorchis sinensis | Adult worms | Xu et al. (2010) |
Comparative analysis has highlighted that, while some helminth miRNAs are widely conserved across diverse organisms and others are specific to helminths, many are novel. It is likely that, with additional genome data, homologues of some of these will be found in closely related helminths. C. elegans, C. remanei and C. briggsae share approximately 90% of miRNAs (de Wit et al., 2009). However, this number drops to only 27–30% when miRNAs from the necromenic nematode Pristionchus pacificus and gastrointestinal nematodes Haemonchus contortus and Ascaris suum are compared to Caenorhabditis miRNAs (de Wit et al., 2009; Wang et al., 2011; Winter et al., 2012). The diversity of miRNA sequences may indicate rapid evolution of this family of small RNAs and could reflect roles for novel miRNAs in the more complex lifecycles of parasitic species and adaptation to different environments. Comparative analysis of miRNAs identified in S. mansoni showed a similar pattern: of 112 miRNAs identified, 12% were conserved in other flatworms and 64% were unique to S. mansoni (Marco et al., 2013).
Analysis of miRNAs in P. pacificus, H. contortus, Brugia and schistosome species suggest that novel miRNAs can evolve by several mechanisms including gene duplication and arm switching. The latter results in the mature miRNA being generated from alternative arms of the precursor (5′ and 3′ mature miRNAs) and examples have been found for changes in arm usage in different lifecycle stages, suggesting functional evolution (Griffiths-Jones et al., 2011). miRNAs can be clustered in the genome and interestingly, clustering of some microRNAs is conserved across species, suggesting functional linkage. For example, clustering of mir-2 family members with mir-71 is conserved in Schistosoma species (Huang et al., 2009; de Souza Gomes et al., 2011), S. mediterranea (Palakodeti et al., 2006) and in H. contortus (Winter et al., 2012). These microRNAs have roles in suppressing apoptosis and in stress responses and their conserved linkage suggests selection for their co-ordinated expression.
Detailed analysis of small RNAs during early development of Ascaris identified some interesting features (Wang et al., 2011). De novo synthesis of miRNAs was detected in Ascaris embryos immediately following fertilization and during zygote maturation, a developmental stage considered to be transcriptionally quiescent. These miRNAs or their precursors could not be detected in oocytes or spermatids, suggesting that they are not of adult origin. This is the earliest embryonic gene expression reported in any system. Surprisingly, another class of small RNAs, piRNAs, was not found in Ascaris nor in Brugia (Wang et al., 2011; Winter et al., 2012), both clade III nematodes, but was identified in clade V nematodes for which small RNA sequence is available (Caenorhabditis, Haemonchus, Pristionchus). These RNAs are important in spermatogenesis and silencing of mobile elements in the germline and it is suggested that other small RNAs, particularly 22G- and 26G-RNAs, may carry out these key roles in Ascaris (Wang et al., 2011)
Study of miRNAs and piRNAs in the free-living planarian S. mediterranea showed that these are upregulated in neoblasts, pluripotent stem cells which proliferate and act as progenitor cells for tissue regeneration (Friedlander et al., 2009). Importantly, knockdown of the Argonaute protein Ago-2, required for miRNA and siRNA biogenesis, led to a reduction in neoblast population and defects in regeneration (Rouhana et al., 2010). Surprisingly, among the miRNAs upregulated is let-7, which in other systems promotes cell differentiation rather than proliferation. In planarians, let-7 may function differently or be involved in exiting the stem cell state. Recent studies identified neoblast-like cells in adult stage S. mansoni and it is speculated that these may contribute to the longevity of this parasite as well as to growth modulation of male-female pairs and tissue regeneration following drug treatment (Collins et al., 2013). Understanding the mechanisms regulating neoblast maintenance and differentiation and the potential involvement of miRNAs and piRNAs may identify new therapeutic targets to reduce parasite development and survival.
3.2. Roles of microRNAs in C. elegans
Currently the functions of only a few helminth miRNAs are known. C. elegans miRNAs lin-4 and let-7 were identified in genetic screens which demonstrated their essential roles in regulating hypodermal cell fates during early larval development (Lee et al., 1993) and the transition from L4 to adult stage (Reinhart et al., 2000). A previously unknown miRNA, lys-6, was identified following a genetic screen for genes regulating the asymmetrical patterning of C. elegans ASE chemoreceptors genes (Johnston and Hobert, 2003). However, even in C. elegans, determining the functions of most miRNAs has proved challenging in the absence of obvious phenotypes following gene knockout. This is thought to be due to redundancy among miRNAs; approximately 60% of miRNAs are members of a family of 2–8 genes (Ruby et al 2006). Family members share the same seed sequence, although it is not known whether they regulate the same target genes. In addition, some miRNAs may function only in certain physiological or stress conditions, or may act to maintain homeostasis. It has been shown that levels of miRNAs from multiple C. elegans families are increased on infection with pathogenic bacteria (Kudlow et al., 2012). Surprisingly, reducing miRISC activity or deleting specific miRNAs enhanced worm survival following infection. This suggested that miRNA activity, particularly in the intestine, functions to dampen pathogen responses that may be detrimental to worm physiology. This is consistent with the reduced fecundity and sterility of many C. elegans pathogen-resistant mutants (Miyata et al., 2008).
Deletion mutants of 87 C. elegans miRNAs have been generated and clear phenotypes reported for only a small group (Miska et al., 2007): deletion of the mir-35-41 gene cluster (mir-36 family) resulted in temperature sensitive embryonic and larval lethality, deletion of mir-240, mir-786 cluster increased defecation cycle lengths, while single deletion of mir-48 or mir-241 (both members of the let-7 family) affected developmental timing. A subsequent study examined the effects of deleting multiple members of miRNA families (Alvarez-Saavedra and Horvitz, 2010). This identified essential roles for mir-42-44 acting with other members of the mir-36 family, with deletion of all resulting in a penetrant embryonic and larval lethal phenotype. Deletion of the mir-51 family (mir-51-56) resulted in defects in late embryonic development, particularly a lack of pharynx attachment, while essential roles in locomotion, growth, egg-laying and dauer formation were identified for the mir-58 family (mir-58, 80,81,82). Members of the mir-58 family are orthologs of Drosophila bantam, a miRNA controlling cell proliferation and apoptosis. Re-introduction of individual C. elegans miRNAs from transgenes was sufficient to rescue the phenotypes observed following loss of all family members, supporting the idea of functional redundancy.
It was also suggested that miRNAs may function with unrelated miRNAs or with non-miRNA genes to generate robust regulatory networks. Brenner et al. (2010) examined the effect of loss of specific miRNAs in a genetic background with reduced levels of all miRNAs, using a C. elegans mutant of argonuate-like 1 (alg-1), a miRNA binding component of the silencing complex. While alg-1 mutant worms are viable, they display developmental timing defects, molting defects and early adult lethality, due mainly to reduction of let-7 family activity. In this sensitized background, enhanced or synthetic phenotypes were observed for 19 of 25 miRNA mutants tested. Brenner et al. also examined combined loss of specific miRNAs and hub genes, which encode chromatin-regulatory proteins that interact with a range of developmental signaling pathways. Combined knockdown resulted in sterile phenotypes for 4/11 miRNAs, indicating the importance of these miRNAs in regulating germline development.
Data on C. elegans miRNA expression, regulation and function are increasing and the approaches used can be applied to parasitic helminths. For some parasite miRNAs, function may be speculated based on homology to C. elegans miRNAs, from temporal or spatial expression data and from target prediction programs (see below). However, narrowing down the list of the most likely targets and testing these experimentally is a major challenge.
3.3. miRNA expression during helminth development
Small RNA library sequence read data can be used as a first indicator of miRNA abundance in particular life-cycle stages (Kato et al., 2009). Relative expression through development or in particular tissues can be confirmed by quantitative real-time PCR (qRT-PCR), miRNA microarray or Northern blot analysis. Sequence read data from the infective L3 stage and mixed adult stages of B. pahangi and H. contortus identified a number of miRNAs differentially expressed during development, while others were abundantly expressed in both stages (Winter et al., 2012). The validity of this approach was supported by expression data on homologues of C. elegans lin-4, which showed greater read numbers in the L3 stage, and let-7, which was more abundant in adult worms, consistent with expression of their C. elegans homologues.qRT-PCR confirmed the temporal expression of selected miRNAs (Winter et al., 2012) and a detailed pattern of miRNA expression through development has now been obtained for B. pahangi and H. contortus from recent microarray analyses (Winter et al., unpublished data). Arrays were designed to include all H. contortus and Brugia miRNAs identified from small RNA sequencing (mapping B. pahangi small RNA sequences to the available B. malayi genome) and computational analysis, as well as C. elegans miRNAs. Arrays were hybridised with small RNAs from a number of parasite stages including pre- and post-infective L3 larvae, L4 and adult male and female worms. One miRNA of particular interest is Brugia mir-5364, the expression of which increases significantly within 24 hours of transfer from the mosquito to the vertebrate host (Winter et al., manuscript in preparation). mir-5364 is a novel member of the let-7 family and its rapid rise suggests a possible role in adaptation to the environment of the mammalian host. A number of interesting target genes have been predicted bioinformatically and studies are underway to experimentally test these.
Temporal expression data on Ascaris miRNAs from RNA library read frequency and Northern blot analysis, showed good correlation. Of the 97 miRNAs identified, none were abundantly expressed throughout development (Wang et al., 2011). Rather, most Ascaris miRNAs seem to be expressed sequentially and could be characterized into four groups: enriched in germline; zygote, early and middle embryo stages; late embryo and larval stages; and multiple developmental stages.
Profiling of microRNAs through development of S. japonicum identified a set, including mir-1 and bantam, that are significantly down-regulated in lung-stage schistosomula compared to cercariae, although their potential targets are currently unknown (Cai et al., 2011). Analysis of small RNAs expressed in S. mansoni adult male or female worms identified 10 female enriched and 3 male enriched miRNAs (Marco et al., 2013). While the mir-2/mir-71 cluster is duplicated in schistosomes, the cluster located on chromosome 5 (autosome) shows female-biased expression while that on the sex chromosome does not. It was suggested that gene duplication into an autosome may overcome selection conflict on sex chromosomes (Gallach and Betran, 2011).
Examination of miRNA expression patterns at the developmental- and, where possible, tissue-specific levels may provide some indication as to potential biological function. As miRNAs most often negatively regulate their target genes, an inverse correlation between miRNA and mRNA expression levels may act as a filtering step in identifying likely target genes, although it should be remembered that miRNAs can have a modest effect on mRNA levels. mRNA microarray and RNA seq data are available for a number of parasitic helminths and, combined with UTR annotation and target prediction algorithms, are valuable resources in aiding target identification.
4. What are the targets of miRNA-mediated regulation?
4.1. Identification of miRNA target transcripts
Identification of biologically relevant targets is key to understanding the functions of miRNAs. A number of computational target prediction tools are available and are often used as a first step in target identification. These score the interaction of a miRNA sequence with potential binding sites, usually in the 3′UTR of mRNAs. Scoring is based on a number of features including sequence complementarity between the miRNA seed sequence (nucleotides 2–7) and the target sequence, evolutionary conservation of the predicted miRNA binding sites, free energy of the miRNA–mRNA heteroduplex, stability of the surrounding mRNA and location of binding sites, with positioning near the start or end of the 3′UTR considered favorable. TargetScan, PicTar, PITA, miRanda and mirWIP are some of the programs widely used, with each having slightly different scoring criteria (reviewed in Thomas et al. 2010). The mirWIP algorithm was developed from verified miRNA-mRNA relationships in immunoprecipitation studies in C. elegans (Hammell et al., 2008) and therefore includes experimental refinement. While these programs are available for searching mRNAs of humans and several model organisms, including C. elegans, they are not yet available for parasitic species. However, using annotated genome data, it is possible to apply these algorithms to predict parasite miRNA targets. We have used this approach to identify potential targets of Brugia miRNAs (Winter et al. in preparation), exploiting the genome data available for several filarial nematode species (Ghedin et al., 2007; Godel et al., 2012; Desjardins et al., 2013).
Experimental identification of miRNA-mRNA interactions with components of the miRISC is increasingly used to identify in vivo target genes. Proteins, such as ALG (Argonaute-Like Gene) or AIN (ALG-1 INteracting protein) tagged with GFP can be expressed in C. elegans and immunoprecipitated from miRISC using anti-GFP antibody. mRNAs and miRNAs extracted from the precipitates are then sequenced or analyzed by microarray to identify enrichment in specific developmental stages (Zhang et al., 2009). A variation of this method has been developed, referred to as high throughput sequencing by cross-linking and immunoprecipitation (HITS-CLIP), in which UV irradiation is used to cross-link miRNA-mRNAs to RISC proteins. Unbound mRNAs are trimmed with nuclease and bound complexes immunoprecipitated with antibody to RISC components. Protected fragments are PCR amplified and sequenced to identify the specific miRNA binding regions in precipitated mRNAs. This method has been used to identify Argonaute binding sites in mammalian tissues (Licatalosi et al., 2008) and has been adapted for C. elegans (Zisoulis et al., 2010). In the absence of transfection methods and use of GFP pulldowns, immunoprecipitation approaches can be adapted to identify miRNA-mRNA interactions in parasitic helminths using antibodies to specific miRISC proteins. We are currently developing this approach to identify miRNA–mRNA interactions in H. contortus (Marks et al., unpublished data).
Direct interaction between miRNA-target mRNA binding sites can be tested using reporter assays, measuring the level of luciferase or GFP expression in cells over-expressing a specific miRNA of interest. Commonly, the 3′UTR sequence is cloned downstream of the reporter gene and miRNA interaction(s) monitored by a decrease in reporter activity. Candidate binding sites can be deleted or mutated and restoration of reporter levels examined. This can be carried out in cell culture or for nematode miRNA–mRNA interactions, in transgenic C. elegans. Reporter gene analysis was used to confirm interaction of C. elegans let-7 with the 3′UTR of Ce-lin-41, resulting in a decrease in lac-Z reporter activity (Reinhart et al., 2000).
4.2. Could parasitic helminth miRNAs regulate host genes?
miRNA target gene analyses has focused predominantly on potential targets within the same organism or tissue. However miRNAs of pathogens may also have effects on host mRNA expression and be important in regulating host-parasite interactions. This has been demonstrated for viral miRNAs, particularly from human cytomegalovirus (HCMV), and may also be true of parasite miRNAs. Several HCMV miRNAs are important in the establishment and maintenance of viral infection by modulating host genes involved in cell cycle control (Grey et al., 2010) and in natural killer cell function (Stern-Ginossar et al., 2007).
The importance of miRNAs in modulating mammalian immune cell development and activity are increasingly understood (Davidson-Moncada et al., 2010). For example, the miRNA signature of T regulatory (Treg) cells has been characterized and among the miRNAs expressed are mir-21 and mir-31, which have opposing effects on the Treg TF FOXP3 (Rouas et al., 2009). In both B and T cells, mir-155 is an important regulator and loss of mir-155 can result in increased propensity for Th2 rather than Th1 responses (Rodriguez et al., 2007). In macrophages and monocytes, mir-155 expression increases on stimulation with LPS and was shown to positively regulate inflammatory responses (Tili et al., 2007). Supporting a possible role for pathogen miRNAs in modulating host immune function, recent work showed that small RNAs from a common plant fungus, Botrytis cinerea, can silence genes involved in plant immunity (Weiberg et al., 2013). The fungal small RNAs bind to plant Argonaute proteins and mediate silencing, particularly of mitogen-activated protein kinase genes, to suppress immunity.
An important finding from studies of helminth miRNAs is that many are novel and not conserved in C. elegans or other organisms. This may reflect the complex life-cycles of parasitic species, such as their ability to adapt to different temperatures and environments and longer developmental times compared to C. elegans. Interestingly, microarray analysis of H. contortus and B. pahangi miRNAs indicates enrichment of novel miRNAs in the adult stage. While these could be involved in mating and reproduction, we speculate that some may act to regulate host genes to enable parasite development and survival within the host environment. The ability of parasitic helminths to suppress host immune responses is well documented (Hewitson et al., 2009) leading to interest in their use as therapeutics for autoimmune conditions such as inflammatory bowel disease, asthma and rheumatoid arthritis (Harnett et al., 2010; Navarro et al., 2013; Weinstock and Elliott, 2013). Protein and glycan components of helminth excretory-secretory (ES) products have been studied as potential sources of immunoregulatory factors. Could miRNAs present in ES or released from dying worms also have evolved to modulate host responses to infection?
Surprisingly, miRNAs are extremely stable when released from cells and can be readily detected in serum, leading to their use as biomarkers for particular disease conditions including tissue injury and cancer (Brase et al., 2010). Recent work has also demonstrated that S. mansoni-derived miRNAs in human serum have potential as biomarkers of infection (Hoy et al., 2014). The stability of extracellular miRNAs is thought to be due to their release within microvesicles or exosomes or by complexing with proteins, including the argonaute protein Ago-2, protecting them from RNase digestion (reviewed in Hoy and Buck (2012)). We have identified several miRNAs from in vitro ES material of H. contortus adult worms (Gu et al., unpublished data). One miRNA of particular interest, Hco-mir-5352, is upregulated in adult worms compared to larval stages and, from current data, homologous sequences are present only in nematodes infecting the gastrointestinal tract (Winter et al., 2012). The T cell activation marker CD69 was identified as the highest scoring potential host target of Hco-mir-5352 and studies are currently underway to test for interaction between Hco-mir-5352 and CD69 3′UTR and any effects of this miRNA on immune cell activation and differentiation. This is potentially important for identifying novel mechanisms by which helminths may manipulate the host environment to aid survival, as well as identifying novel intervention strategies.
5. miRNAs as potential therapeutic targets
5.1. Development of miRNA inhibitors and mimics
Levels of many miRNAs, including let-7, as well as RISC activity, are in general decreased in many cancer cell lines and tumours. miRNAs are therefore receiving increasing attention as potential therapeutic targets and prognostic markers for cancer treatment. As a result, there has been significant progress made in recent years in the design and effective delivery of miRNA inhibitors and mimics. Where available, genetic knockouts, with loss of specific miRNAs or RISC components, or use of miRNA sponges (Ebert and Sharp, 2010) can also be used to determine effects of miRNA depletion. miRNA inhibitors, or anti-miRs, are modified anti-sense oligonucleotides with reverse complementary sequence to the mature endogenous miRNA to be inhibited. Anti-miRs form a duplex with the endogenous miRNA, leading to it being unable to bind to the cellular mRNA target (reviewed in van Rooij et al., 2012). miRNA inhibitors can be modified to increase resistance to nuclease digestion and enhance uptake into cells. For example, inclusion of 2′-O-methyl (OMe) group modifications, use of locked nucleic acids (LNA) with bridged 2′oxygen and 4′ carbon ribose moieties, or introduction of phosphorothioate rather than phosphodiester linkages between nucleotides, all enhance the stability of anti-miRs. Modifications are more limited with miRNA mimics, which are RNA duplex molecules, as the guide strand has to be identical to the miRNA of interest and function in the RISC. Inhibition of miRNA function was first demonstrated in C. elegans following injection of a 2′-O-methyl oligonucleotide complementary to let-7 into larval stages, resulting in the characteristic let-7 adult lethal phenotype (Hutvagner et al., 2004). Using fluorescently-labeled small RNAs, we have observed significant uptake following soaking of B. pahangi larvae and adult stages, suggesting that miRNA inhibition or mimickry is feasible in parasitic helminths (Fig. 2 and Winter et al., manuscript in preparation).
Fig. 2.
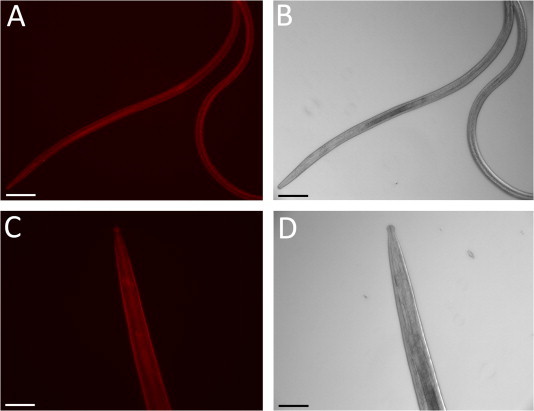
In vitro uptake of fluorescently-labeled siRNA by Brugia pahangi. 100 mosquito-derived L3 stage larvae (A and B) or 1 adult female worm (C and D) were incubated in 0.5 ml (for L3s) or 1 ml (for adults) of RPMI 1640/5% foetal calf serum/1% glucose at 37 °C/5% CO2, in the presence or absence of 0.5 μM Cy3-labeled siRNA (Ambion). After 48 h, parasites were washed three times in 1 ml PBS/0.1% Tween and pipetted (L3s) or picked (adults) onto a 2% agarose pad on microscope slides and viewed by fluorescent microscopy (A and C) or under bright field (B and D) at ×10 magnification. Images were collected using an Axioskop 2 Plus microscope (Zeiss), ORCA-ER digital camera (Hamamatsu) and Openlab (Improvision) software. No fluorescence was observed in the absence of Cy3-labeled siRNA. Scale bar = 100 μm.
Currently, one LNA-based miRNA inhibitor (miravirsen) is in human clinical trials. This is an inhibitor of mammalian mir-122, a liver-specific miRNA which, as well as being important for liver function, binds to the hepatitis C virus RNA genome and is required for HCV replication (Jopling et al., 2005). In human patients suffering from chronic HCV, miravirsen significantly reduced viral load, demonstrating the effectiveness of anti-miRs as therapeutics (Janssen et al., 2013).
5.2. Inhibition of miRNA-mediated pathway regulation
miRNAs are often considered to be fine tuners of gene expression. However as demonstrated by loss of let-7 or lin-4, specific miRNAs can clearly act as developmental switches. This is explained by the relative levels of a miRNA and target mRNA and the strength of their interaction, resulting in a molecular titration effect. For some target genes, the protein product functions only above a threshold level. If target abundance increases towards the threshold, this has the effect of titrating away the miRNAs available for binding, thus switching from gene repression to activation (Mukherji et al., 2011). miRNAs can act as master regulators of developmental transitions and responses to environmental changes/stresses, making them suitable control targets. Signaling molecules and transcription factors are particularly sensitive to changes in concentration and, as well as regulating expression of miRNAs, some TFs are also targeted by miRNAs (Leung and Sharp, 2010). Experimental and computational prediction analysis showed that TF-miRNA feedback loops occur more frequently than expected by chance (Martinez et al., 2008). Immunoprecipitation studies in C. elegans also identified enrichment for receptors, kinases and ion channels among the target mRNAs in the RISC complex (Zhang et al 2009). The important roles that these dominant miRNA targets play in helminth development suggest that changes to their regulation via miRNA inhibitors or mimics is likely to be detrimental.
Although miRNAs frequently exert only modest effects on target mRNA levels (average of 2-fold decrease), because they often regulate many genes functioning together in the same pathway or protein complex, their effect can be amplified. In addition, the effect of miRNAs on target gene level can be much greater in specific cells where the abundance of some miRNAs is high. With increasing data on temporal miRNA expression, as well as identification of many helminth-specific miRNAs, we can begin to test inhibitors and mimics as novel therapeutics. Will inhibiting parasite miRNAs upregulated on host infection, such as Bpa-mir-5364, or in adult worms limit parasite survival and reproduction? Not only are miRNAs potentially valid targets, but understanding the overall effects of miRNAs by characterizing their mRNA targets and their temporal and spatial expression, should identify co-ordinated pathways and networks required for developmental transitions or responses to stresses, which may themselves be targets for parasite control.
5.3. Involvement of host miRNAs in parasitic infections
Not only is understanding roles of parasite miRNAs important to infection control, but studying host miRNAs may also lead to novel intervention strategies. Characterizing changes in host cell miRNA profile following parasite infection may elucidate triggers and mechanisms regulating host immunity, pathology and inflammation as well as parasite-induced suppression. There is increasing evidence that miRNAs can regulate Toll-like receptor (TLR) signaling molecules, such as AP-1 and NFk-b, as well as TLR expression. It is speculated that miRNAs may provide a link between innate and adaptive immune pathways (reviewed in O′Neill et al., 2011) and are therefore relevant to understanding regulation of host immunity. A number of studies have examined changes in host miRNA levels following infection to identify parasite-induced changes and regulation of host responses. For example, infection of leukocytes with the transforming parasite Theileria results in an increase in host mir-155 which, via TFs AP-1 and c-Jun, acts to maintain the cell hyperproliferation state and parasite survival (Marsolier et al., 2013). Inhibition of mir-155 or AP-1 reverses parasite-induced transformation. Changes in miRNA expression also occur in host macrophages following implantation of B. malayi adult worms into the peritoneal cavity and this has been shown to regulate alternative macrophage proliferation (Rückerl et al., 2012). Recently it was shown that invasion and induction of innate immunity in myeloid cells by North American eastern equine encephalitis virus is blocked by a haematopoietic-cell-specific miRNA (mir-142-3p). This host miRNA binds to the 3′ end of the viral RNA genome to restrict its replication in this cell lineage, thus suppressing the immune response and aiding virus survival (Trobaugh et al., 2013).
6. microRNAs in drug resistance
miRNA expression or activity are often altered in cancer cells and some of these changes are also thought to be associated with development of drug resistance. Drug treatment of cancer cells has been shown to result in altered expression of miRNAs which target drug metabolizing enzymes and drug transporters leading to drug resistance (Yu, 2009). For example, gemcitabine treatment of xenograft tumours results in decreased expression of mir-328, a miRNA which negatively regulates the ABCG2 efflux transporter (Pan et al., 2009). Decreased mir-328 levels therefore lead to increased transporter expression and multidrug resistance.
We have speculated that the link between changes in miRNA levels and drug resistance in cancer cells may also be a feature of drug resistance in parasitic nematodes (Devaney et al., 2010). Altered levels of drug targets, including ion channels and nAChR subunits have been found in isolates of H. contortus resistant to ivermectin (Rao et al., 2009) and in A. caninum resistant to pyrantel (Kopp et al., 2009). Following drug selection, changes were also found in expression level and allele frequency of ABC transporters (Prichard and Roulet, 2007) and P-glycoproteins (Blackhall et al., 2008). Whether these changes in gene expression are mediated by miRNAs is currently unknown. In C. elegans, mutants of mir-1, which negatively regulates nAChR subunits unc-29 and unc-63, show decreased muscle sensitivity to Ach and resistance to levamisole (Simon et al., 2008). This is thought to result from changes in subunit composition affecting the biogenesis or function of heteromultimeric receptors. Recently, we have used miRNA microarray analysis to examine the expression profile of C. elegans strains and H. contortus isolates resistant to ivermectin. Our initial data have identified significant and consistent changes in expression of three miRNAs – Cel-mir-85, Cel-mir-788 and a novel H. contortus sequence Hco-mir-9551 (Gillan et al., unpublished data). Work is underway to identify alterations in mRNA levels which may be associated with these miRNA changes. Obvious targets include drug receptors, transporters and drug metabolizing enzymes, but the transcriptomic approach has the potential to identify novel mechanisms of drug resistance. Alternatively, the observed changes in miRNA expression levels may be a consequence of other changes associated with resistance. These miRNAs may also prove to be valuable and sensitive markers of anthelmintic resistance.
7. Conclusion
microRNAs of parasitic helminths are receiving increasing attention as novel regulators of development, sex differentiation, drug resistance and host-parasite interactions. The important and varied roles of miRNAs identify these as valid targets of intervention strategies. With genome sequence data available for more parasite species, miRNA identification and understanding of miRNA evolution and function will increase. Which miRNAs are conserved across species, which are specific to particular parasite clades or to modes of transmission or location within the host? Improved genome assembly/annotation together with comparative analyses will also aid target gene predictions through identification of conserved miRNA binding sites. Parasite miRNAs may also regulate host target genes and there is increasing evidence that pathogens exploit host miRNAs to aid replication and survival. This has been demonstrated for HCV, which relies on host mir-122 in liver cells, Theileria, which alters leukocyte mir-155 levels, and equine encephalitis virus where cell tropism seems to be mediated by a host miRNA. It will be important to examine effects of helminth miRNAs on both parasite and host gene expression, any parasite-induced changes in host miRNA levels and whether host miRNA expression could also influence helminth tropism. This has potential to lead to therapeutics targeting parasite pathways as well as altering host-parasite interactions that influence parasite survival.
Acknowledgements
Some of the work described here was funded by The Wellcome Trust in project grants awarded to ED and CB, by the Scottish Government’s Strategic Partnership for Animal Science Excellence and by PhD studentships funded by BBSRC/Knowledge Transfer Network (KTN)/Zoetis and by EBLEX and the University of Glasgow, UK. We acknowledge Kirsty Maitland, Neil Marks and Henry Gu for inclusion of unpublished data from our labs.
Contributor Information
Collette Britton, Email: Collette.Britton@glasgow.ac.uk.
Alan D. Winter, Email: Alan.Winter@glasgow.ac.uk.
Victoria Gillan, Email: Victoria.Gillan@glasgow.ac.uk.
Eileen Devaney, Email: Eileen.Devaney@glasgow.ac.uk.
References
- Ai L., Xu M.J., Chen M.X., Zhang Y.N., Chen S.H., Guo J., Cai Y.C., Zhou X.N., Zhu X.Q., Chen J.X. Characterization of microRNAs in Taenia saginata of zoonotic significance by Solexa deep sequencing and bioinformatic analysis. Parasitol. Res. 2012;110:2373–2378. doi: 10.1007/s00436-011-2773-x. [DOI] [PubMed] [Google Scholar]
- Alvarez-Saavedra E., Horvitz H.R. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., Horvitz H.R. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhall W.J., Prichard R.K., Beech R.N. P-glycoprotein selection in strains of Haemonchus contortus resistant to benzimidazoles. Vet. Parasitol. 2008;152:101–107. doi: 10.1016/j.vetpar.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Brase J.C., Wuttig D., Kuner R., Sultmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol. Cancer. 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner J.L., Jasiewicz K.L., Fahley A.F., Kemp B.J., Abbott A.L. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr. Biol. 2010;20:1321–1325. doi: 10.1016/j.cub.2010.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P., Hou N., Piao X., Liu S., Liu H., Yang F., Wang J., Jin Q., Wang H., Chen Q. Profiles of small non-coding RNAs in Schistosoma japonicum during development. PLoS Negl. Trop. Dis. 2011;5:e1256. doi: 10.1371/journal.pntd.0001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P., Piao X., Hao L., Liu S., Hou N., Wang H., Chen Q. A deep analysis of the small non-coding RNA population in Schistosoma japonicum eggs. PLoS One. 2013;8:e64003. doi: 10.1371/journal.pone.0064003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.H., Tang P., Lai C.H., Kuo M.L., Wang L.C. Identification and characterisation of microRNAs in young adults of Angiostrongylus cantonensis via a deep-sequencing approach. Mem. Inst. Oswaldo Cruz. 2013;108:699–706. doi: 10.1590/0074-0276108062013005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M., Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Chen M.X., Ai L., Xu M.J., Chen S.H., Zhang Y.N., Guo J., Cai Y.C., Tian L.G., Zhang L.L., Zhu X.Q., Chen J.X. Identification and characterization of microRNAs in Trichinella spiralis by comparison with Brugia malayi and Caenorhabditis elegans. Parasitol. Res. 2011;109:553–558. doi: 10.1007/s00436-011-2283-x. [DOI] [PubMed] [Google Scholar]
- Chen M.X., Ai L., Xu M.J., Zhang R.L., Chen S.H., Zhang Y.N., Guo J., Cai Y.C., Tian L.G., Zhang L.L., Zhu X.Q. Angiostrongylus cantonensis: identification and characterization of microRNAs in male and female adults. Exp. Parasitol. 2011;128:116–120. doi: 10.1016/j.exppara.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Collins J.J., 3rd, Wang B., Lambrus B.G., Tharp M.E., Iyer H., Newmark P.A. Adult somatic stem cells in the human parasite Schistosoma mansoni. Nature. 2013;494:476–479. doi: 10.1038/nature11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucher M., Prada L., Mourglia-Ettlin G., Dematteis S., Camicia F., Asurmendi S., Rosenzvit M. Identification of Echinococcus granulosus microRNAs and their expression in different life cycle stages and parasite genotypes. Int. J. Parasitol. 2011;41:439–448. doi: 10.1016/j.ijpara.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Davidson-Moncada J., Papavasiliou F.N., Tam W. MicroRNAs of the immune system. Ann. N.Y. Acad. Sci. 2010;1183:183–194. doi: 10.1111/j.1749-6632.2009.05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Gomes M., Muniyappa M.H., Carvalho S.G., Guerra-Sa R., Spillane C. Genome-wide identification of novel microRNAs and their target genes in the human parasite Schistosoma mansoni. Genomics. 2011;98:96–111. doi: 10.1016/j.ygeno.2011.05.007. [DOI] [PubMed] [Google Scholar]
- de Wit E., Linsen S.E.V., Cuppen E., Berezikov E. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res. 2009;19:2064–2074. doi: 10.1101/gr.093781.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins C.A., Cerqueira G.C., Goldberg J.M., Dunning Hotopp J.C., Haas B.J., Zucker J., Ribeiro J.M., Saif S., Levin J.Z., Fan L., Zeng Q., Russ C., Wortman J.R., Fink D.L., Birren B.W., Nutman T.B. Genomics of Loa loa, a Wolbachia-free filarial parasite of humans. Nat. Genet. 2013;45:495–500. doi: 10.1038/ng.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaney E., Winter A.D., Britton C. MicroRNAs: a role in drug resistance in parasitic nematodes. Trends Parasitol. 2010;26:428–433. doi: 10.1016/j.pt.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M.S., Sharp P.A. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander M.R., Adamidi C., Han T., Lebedeva S., Isenbarger T.A., Hirst M., Marra M., Nusbaum C., Lee W.L., Jenkin J.C., Alvarado A.S., Kim J.K., Rajewsky N. High-resolution profiling and discovery of planarian small RNAs. Proc. Natl. Acad. Sci. USA. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M., Betran E. Intralocus sexual conflict resolved through gene duplication. Trends Ecol. Evol. 2011;26:222–228. doi: 10.1016/j.tree.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer K.K., Rodriques C.M., Chalmers I.W., Munshi S.E., Truscott M., Heald J., Wilkinson M.J., Hoffmann K.F. Cytosine methylation regulates oviposition in the pathogenic blood fluke Schistosoma mansoni. Nat. Commun. 2011;2:424. doi: 10.1038/ncomms1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E., Wang S., Spiro D., Caler E., Zhao Q. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godel C., Kumar S., Koutsovoulos G., Ludin P., Nilsson D., Comandatore F., Wrobel N., Thompson M., Schmid C.D., Goto S., Bringaud F., Wolstenholme A., Bandi C., Epe C., Kaminsky R., Blaxter M., Mäser P. The genome of the heartworm, Dirofilaria immitis, reveals drug and vaccine targets. FASEB J. 2012;26:4650–4661. doi: 10.1096/fj.12-205096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F., Tirabassi R., Meyers H., Wu G., McWeeney S., Hook L., Nelson J.A. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5′UTRs. PLoS Pathog. 2010;6:e1000967. doi: 10.1371/journal.ppat.1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. MiRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Hui J.H.L., Marco A., Ronshaugen M. MicroRNA evolution by arm switching. EMBO Rep. 2011;12:172–177. doi: 10.1038/embor.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Assunção J.A., Enright A.J. MapMi: automated mapping of microRNA loci. BMC Bioinformatics. 2010;11:133. doi: 10.1186/1471-2105-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell M., Long D., Zhang L., Lee A., Carmack C.S., Han M., Ding Y., Ambros V. MirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat. Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnett M.M., Melendez A.J., Harnett W. The therapeutic potential of the filarial nematode-derived immunodulator, ES-62 in inflammatory disease. Clin. Exp. Immunol. 2010;159:256–267. doi: 10.1111/j.1365-2249.2009.04064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson J.P., Grainger J.R., Maizels R.M. Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol. Biochem. Parasitol. 2009;167:1–11. doi: 10.1016/j.molbiopara.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer L.S., Tacker M., Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- Hoy A.M., Buck A.H. Extracellular small RNAs: what, where and why? Biochem. Soc. Trans. 2012;40:886–890. doi: 10.1042/BST20120019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy A.M., Lundie R.J., Ivens A., Quintana J.F., Nausch N., Forster T., Jones F., Kabatereine N.B., Dunne D.W., Mutapi F., MacDonald A.S., Buck A.H. Parasite-derived MicroRNAs in host serum as novel biomarkers of helminth infection. PLoS Negl. Trop. Dis. 2014;8:e2701. doi: 10.1371/journal.pntd.0002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Hao P., Chen H., Hu W., Yan Q., Liu F., Han Z.-G. Genome-wide identification of Schistosoma japonicum microRNAs using a deep sequencing approach. PLoS One. 2009;4:e8206. doi: 10.1371/journal.pone.0008206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q.X., Cheng X.Y., Mao Z.C., Wang Y.S., Zhao L.L., Yan X., Ferris V.R., Xu R.M., Xie B.Y. MicroRNA discovery and analysis of pinewood nematode Bursaphelenchus xylophilus by deep sequencing. PLoS One. 2010;5:e13271. doi: 10.1371/journal.pone.0013271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., Simard M.J., Mello C.C., Zamore P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:e98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen H.L.A., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., Persson R., King B.D., Kauppinen S., Levin A.A., Hodges M.R. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kato M., de Lencastre A., Pincus Z., Slack F.J. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009;10:R54. doi: 10.1186/gb-2009-10-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kopp S.R., Coleman G.T., Traub R.J., McCarthy J.S., Kotze A.C. Acetylcholine receptor subunit genes from Ancylostoma caninum: altered transcription patterns associated with pyrantel resistance. Int. J. Parasitol. 2009;39:435–441. doi: 10.1016/j.ijpara.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Kudlow B.A., Zhang L., Han M. Systematic analysis of tissue-restricted miRISCs reveals a broad role for microRNAs in suppressing basal activity of the C. elegans pathogen response. Mol. Cell. 2012;46:530–541. doi: 10.1016/j.molcel.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Leung A.K.L., Sharp P.A. MicroRNA functions in stress responses. Mol. Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X., Darnell J.C., Darnell R.B. HITS-CLIP yields genome-wide insights into brain alternative processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A., Kozomara A., Hui J.H.L., Emery A.M., Rollinson D., Griffiths-Jones S., Ronshaugen M. Sex-biased expression of microRNAs in Schistosoma mansoni. Plos Negl. Trop. Dis. 2013;7:e2402. doi: 10.1371/journal.pntd.0002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolier J., Pineau S., Medjkane S., Perichon M., Yin Q., Flemington E., Weitzman M.D., Weitzman J.B. OncomiR addiction is generated by a miR-155 feedback loop in Theileria-transformed leukocytes. PLoS Pathog. 2013;9:e1003222. doi: 10.1371/journal.ppat.1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N.J., Ow M.C., Barrasa M.I., Hammell M., Sequerra R., Doucette-Stamm L., Roth F.P., Ambros V.R., Walhout A.J.M. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E.A., Alvarez-Saavedra E., Abbott A.L., Lau N.C., Hellman A.B., McGonagle S.M., Bartel D.P., Ambros V.R., Horvitz H.R. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:2395–2403. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S., Begun J., Troemel E.R., Ausubel F.M. DAF-16-dependent suppression of immunity during reproduction in Caenorhabditis elegans. Genetics. 2008;178:903–918. doi: 10.1534/genetics.107.083923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S., Ebert M.S., Zheng G.X.Y., Tsang J.S., Sharp P.A., van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011;43:854–860. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro S., Ferreira I., Loukas A. The hookworm pharmacopoeia for inflammatory diseases. Int. J. Parasitol. 2013;43:225–231. doi: 10.1016/j.ijpara.2012.11.005. [DOI] [PubMed] [Google Scholar]
- O’Neill L.A., Sheedy F.J., McCoy C.E. MicroRNAs: the fine tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- Palakodeti D., Smielewska M., Graveley B.R. MicroRNAs from the planarian Schmidtea mediterranea: a model system for stem cell biology. RNA. 2006;12:1640–1649. doi: 10.1261/rna.117206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.Z., Morris M.E., Yu A.M. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol. Pharmacol. 2009;75:1374–1379. doi: 10.1124/mol.108.054163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole C.B., Davis P.J., Jin J.M., McReynolds L.A. Cloning and bioinformatic identification of small RNAs in the filarial nematode, Brugia malayi. Mol. Biochem. Parasitol. 2010;169:87–94. doi: 10.1016/j.molbiopara.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Prichard R.K., Roulet A. ABC transporters and beta-tubulin in macrocyclic lactone resistance: prospects for marker development. Parasitology. 2007;134:1123–1132. doi: 10.1017/S0031182007000091. [DOI] [PubMed] [Google Scholar]
- Rao V.T., Siddiqui S.Z., Prichard R.K., Forrester S.G. A dopamine-gated ion channel (HcGGR3∗) from Haemonchus contortus is expressed in the cervical papillae and is associated with macrocyclic lactone resistance. Mol. Biochem. Parasitol. 2009;166:54–61. doi: 10.1016/j.molbiopara.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., Vetrie D., Okkenhaug K., Enright A.J., Dougan G., Turner M., Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouas R., Fayyad-Kazan H., El Zein N., Lewalle P., Rothé F., Simion A., Akl H., Mourtada M., El Rifai M., Burny A., Romero P., Martiat P., Badran B. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur. J. Immunol. 2009;39:1608–1618. doi: 10.1002/eji.200838509. [DOI] [PubMed] [Google Scholar]
- Rouhana L., Shibata N., Nishimura O., Agata K. Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev. Biol. 2010;341:429–443. doi: 10.1016/j.ydbio.2010.02.037. [DOI] [PubMed] [Google Scholar]
- Ruby J.G., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Rückerl D., Jenkins S.J., Laqtom N.N., Gallagher I.J., Sutherland T.E., Duncan S., Buck A.H., Allen J.E. Induction of IL-4Rα-dependent microRNAs identifies PI3K/Akt signaling as essential for IL-4-driven murine macrophage proliferation in vivo. Blood. 2012;120:2307–2316. doi: 10.1182/blood-2012-02-408252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G., Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989;338:313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- Simoes M.C., Lee J., Djikeng A., Cerquera G.C., Zerlotini A., da Silva-Pereira R.A., Dalby A.R., LoVerde P., El-Sayed N.M., Oliveira G. Identification of Schistosoma mansoni microRNAs. BMC Genomics. 2011;12:47. doi: 10.1186/1471-2164-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D.J., Madison J.M., Conery A.L., Thompson-Peer K.L., Soskis M., Ruvkun G.B., Kaplan J.M., Kim J.K. The MicroRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133:903–915. doi: 10.1016/j.cell.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N., Elefant N., Zimmermann A., Wolf D.G., Saleh N., Biton M., Horwitz E., Prokocimer Z., Prichard M., Hahn G., Goldman-Wohl D., Greenfield C., Yagel S., Hengel H., Altuvia Y., Margalit H., Mandelboim O. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarayrah L., Chen X. Epigenetic regulation in adult stem cells and cancer. Cell Biosci. 2013;3:41. doi: 10.1186/2045-3701-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Lieberman J., Lal A. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- Tili E., Michaille J.J., Cimino A., Costinean S., Dumitru C.D., Adair B., Fabbri M., Alder H., Liu C.G., Calin G.A., Croce C.M. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- Trobaugh D.W., Gardner C.L., Sun C., Haddow A.D., Wang E., Chapnik E., Mildner A., Weaver S.C., Ryman K.D., Klimstra W.B. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature. 2013 doi: 10.1038/nature12869. Dec 18 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S., Vaz C., Gupta V., Bhatia R., Maheshwari S., Srinivasan A., Bhattacharya A. CID-miRNA: a web server for prediction of novel miRNA precursors in human genome. Biochem. Biophys. Res. Commun. 2008;372:831–834. doi: 10.1016/j.bbrc.2008.05.134. [DOI] [PubMed] [Google Scholar]
- Van Rooij E., Purcell A.L., Levin A.A. Developing microRNA therapeutics. Circ. Res. 2012;110:496–507. doi: 10.1161/CIRCRESAHA.111.247916. [DOI] [PubMed] [Google Scholar]
- Wang J., Czech B., Crunk A., Wallace A., Mitreva M., Hannon G.J., Davis R.E. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011;21:1462–1477. doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F.-M., Zhao H., Zhang Z., Kaloshian I., Huang H.-D., Jin H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock J.V., Elliott D.E. Translatability of helminth therapy in inflammatory bowel diseases. Int. J. Parasitol. 2013;43:245–251. doi: 10.1016/j.ijpara.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Winter A.D., Weir W., Hunt M., Berriman M., Gilleard J.S., Devaney E., Britton C. Diversity in parasitic nematode genomes: the microRNAs of Brugia and Haemonchus are largely novel. BMC Genomics. 2012;13:4. doi: 10.1186/1471-2164-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M.J., Liu Q., Nisbet A.J., Cai X.Q., Yan C., Lin R.Q., Yuan Z.G., Song H.Q., He X.H., Zhu X.Q. Identification and characterization of microRNAs in Clonorchis sinensis of human health significance. BMC Genomics. 2010;2010(11):521. doi: 10.1186/1471-2164-11-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M.J., Fu J.H., Nisbet A.J., Huang S.Y., Zhou D.H., Lin R.Q., Song H.Q., Zhu X.Q. Comparative profiling of microRNAs in male and female adults of Ascaris suum. Parasitol. Res. 2013;112:I1189–I1195. doi: 10.1007/s00436-012-3250-x. [DOI] [PubMed] [Google Scholar]
- Yu A.M. Role of microRNAs in the regulation of drug metabolism and disposition. Exp. Opin. Drug Metab. Toxicol. 2009;5:1513–1528. doi: 10.1517/17425250903307448. [DOI] [PubMed] [Google Scholar]
- Zhang L., Hammell M., Kudlow B.A., Ambros V., Han M. Systematic analysis of dynamic miRNA-target interactions during C. elegans development. Development. 2009;136:3043–3055. doi: 10.1242/dev.039008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis D.G., Lovci M.T., Wilbert M.L., Hutt K.R., Liang T.Y., Pasquinelli A.E., Yeo G.W. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]


