Graphical abstract
Keywords: Haemonchus contortus, Levamisole, Resistance, Nicotinic agonists
Highlights
-
•
A Haemonchus contortus isolate subdivided according to level of resistance to levamisole in vitro.
-
•
Increased P-glycoprotein gene expression in larvae showing low level resistance.
-
•
Decreased expression of nAChR subunit and receptor assembly genes in larvae showing higher levels of resistance.
-
•
Results suggest drug efflux mediated low level resistance, with target site changes conferring higher level resistance.
Abstract
While there is some evidence that changes in nicotinic acetylcholine receptor (nAChR) subunits confer resistance to levamisole in gastrointestinal helminth parasites, the exact nature of the resistance mechanism(s) is unclear. We utilised the presence of a resistant fraction within the Wallangra 2003 isolate of Haemonchus contortus larvae in order to subdivide the population into three subpopulations of larvae able to survive increasing concentrations of the drug. We then measured gene expression levels in the subpopulations and the larval population as a whole, focusing on genes encoding the subunit components of levamisole-sensitive receptors, genes encoding ancillary proteins involved in receptor assembly, and P-glycoprotein (P-gp) genes. The subpopulation surviving the lowest levamisole concentration showed increases of 1.5- to 3-fold in a number of P-gp genes (Hco-pgp-3, -4, -10, and -14) alongside unchanged receptor genes, compared to the whole Wallangra larval population. On the other hand, the subpopulation surviving the intermediate levamisole concentration showed an increase in only a single P-gp (Hco-pgp-14), alongside decreases in some receptor subunit (Hco-unc-63a) and ancillary protein genes (Hco-unc-50, Hco-ric-3.1 and 3.1). The subpopulation surviving the highest levamisole concentration showed further decreases in receptor subunit genes (Hco-unc-63a and Hco-unc-29 paralogs) as well as genes involved in receptor assembly (Hco-unc-74, Hco-unc-50, Hco-ric-3.1 and 3.1), alongside no increased P-gp gene levels. This suggests a biphasic pattern of drug resistance in the larvae of this worm isolate, in which a non-specific P-gp-mediated mechanism confers low levels of resistance, while higher level resistance is due to altered receptor subunit composition as a result of changes in both subunit composition and in the levels of proteins involved in receptor assembly.
1. Introduction
The use of anthelmintics for the control of intestinal parasites of livestock over the last 50 years has led to the development of resistance to each of the major chemical groups (McKellar and Jackson, 2004; Wolstenholme et al., 2004; Geary et al., 2012). The discovery of new drug compounds to facilitate the management of resistance is a slow and costly process that results in few novel compounds being released (Besier, 2007; Woods and Knauer, 2010). Hence, there is a need to manage the use of existing drugs in such a way as to minimise the impact of resistance, for example through the use of diagnostics to inform on the resistance status of worm populations present on specific properties, and hence inform on which drugs are still likely to be effective. A sensitive molecular test would be valuable for such resistance diagnostic purposes (Prichard et al., 2007). However, before such a test can be designed, an understanding of the mechanism or mechanisms of resistance to a particular anthelmintic or anthelmintic class is required, or at the very least the invariable presence of a genetic marker for that resistance. In addition, elucidating the mechanism/s of resistance may provide a better understanding of the rate at which resistance may emerge, as well as providing a tool to study parasite biology and drug targets (Sangster et al., 2005).
The cholinergenic anthelmintics, levamisole and pyrantel, act as agonists at nicotinic acetylcholine receptors (nAChRs). A subset of these receptors, known as the L-type receptors, are preferentially activated by levamisole and pyrantel (Martin et al., 2012). Activation of this L-type nAChR by the drug leads to sustained neuromuscular depolarization and spastic paralysis (Martin et al., 1997), thereby dislodging helminths from the gastrointestinal mucosa and leading to their expulsion from the host. The nAChRs are composed of five subunits forming a ring in the membrane with a central ion channel pore (Levandoski et al., 2005). The subunits are classified as subunits; those bearing two vicinal cysteines (unc-63, unc-38, acr-8) or β (non-α) subunits that lack the cysteines (unc-29, lev-1) (Martin et al., 2012). Previous reports on mechanisms of levamisole and pyrantel resistance in Haemonchus contortus, Teladorsagia circumcincta, Trichostrongylus colubriformis and Ancylostoma caninum have suggested a role for changes in the expression of a number of nAChR subunit genes: reduced transcription of unc-63a (Kopp et al., 2009; Sarai et al., 2013), reduced transcription of unc-38 (Kopp et al., 2009), reduced transcription of unc-29 (Kopp et al., 2009; Sarai et al., 2013), the presence of Hco-acr-8b (Fauvin et al., 2010; Williamson et al., 2011), and the presence of Hco-unc-63b (Neveu et al., 2010). Although not reported to date from studies of parasitic nematodes, it is possible that resistance may also be conferred by changes in ancillary proteins, RIC-3, UNC-74 and UNC-50, that are known to be essential for the proper formation, function and transport of the levamisole receptor, and for which mutations in Caenorhabditis elegans are known to confer resistance to levamisole (Gottschalk et al., 2005). Moreover, drug efflux mechanisms such as P-glycoproteins (P-gps) are involved in multidrug resistance in a variety of organisms, including helminths, and have been implicated in resistance to benzimidazoles and macrocyclic lactones (Kerboeuf et al., 2003; Blackhall et al., 2008; Lespine et al., 2012).
Sarai et al. (2013) showed that larvae of the Wallangra 2003 (WAL) isolate of H. contortus exhibited a distinctive plateau in response to levamisole in larval development assays, indicating the presence of a highly resistant fraction accounting for approximately 10–15% of the total population. While the bulk of the population behaved in a similar manner to susceptible worms in the larval development assay (equivalent IC50 values) the presence of this resistant fraction resulted in a significant shift in IC95 (13-fold higher than the susceptible isolate). The authors suggested that it might be difficult to associate gene transcription patterns with resistance in such heterogeneous isolates as the average transcription levels in a sample of worm larvae derived from the whole population might not be indicative of levels in the resistant fraction within the population. Considering that this resistant fraction, under further drug selection pressure, will most likely over time become the dominant phenotype, ascertaining the differences between the resistant fraction and the bulk of the population might give an indication of the genetic changes occurring in the early stages in the development of resistance. Such changes would therefore be the most critical to monitor in order to detect increasing levels of resistance.
A significant feature of all the parasitic nematode nAChR subunit gene expression studies described above has been the fact that they involved the comparison of genotypes or gene expression patterns between populations showing different levels of resistance, but also differing significantly in genetic background. Field-derived isolates showing resistance to the drug have most often been compared to susceptible isolates collected from different geographical regions, and/or from time periods several decades earlier than the resistant isolate. Hence, the differences seen in gene expression levels could be related to these diverse genetic backgrounds rather than being specifically associated with their level of susceptibility to the drug. The present study aimed to circumvent this issue to some degree by subdividing a single field-derived worm population solely on the basis of the ability of subpopulations to survive exposure to levamisole in vitro, and then comparing gene transcription patterns between the whole population and the various subpopulations. The heterogeneity in response to levamisole shown by WAL larvae described above provided an opportunity to subdivide the population based on in vitro levamisole sensitivity. Kotze et al. (2012) recently utilised such an experimental design in subdividing the WAL isolate on the basis of susceptibility of larvae to thiabendazole in vitro, followed by genotyping of a beta-tubulin gene in the survivors at each drug concentration. This allowed the authors to link specific SNP frequencies to relative drug resistance levels shown by the different subpopulations.
The present study therefore aimed to recover larvae from the levamisole-resistant fraction present within the WAL isolate, at three separate levels of drug exposure, and to compare these subpopulations to each other and to the WAL population as a whole. We measured gene transcription levels for putative subunit constituents of the levamisole sensitive receptor (Hco-unc-38, Hco-unc-29.1–29.4, Hco-unc-63a, Hco-unc-63b, Hco-lev-1, Hco-acr-8a and Hco-acr-8b) that have previously been implicated in resistance, as well as two further nAChR subunit genes (Hco-acr-16 and Hco-acr-26). In addition, given their potential role in resistance, we measured transcription levels of four ancillary protein genes (Hco-unc-50, Hco-unc-74, Hco-ric-3.1 and Hco-ric-3.2) (Martin et al., 2012), and nine P-gp genes.
2. Materials and methods
2.1. Collection of worms and eggs
The Kirby 1986 and Wallangra 2003 (WAL) isolates of H. contortus were maintained in sheep at the McMaster Laboratory, CSIRO Livestock Industries, Armidale, New South Wales (NSW), Australia. All animal procedures were approved by the F.D. McMaster Animal Ethics Committee, CSIRO Animal, Food and Health Sciences (Animal ethics approval number 12/18). The Kirby isolate is susceptible to all commercial anthelmintics (Albers and Burgess, 1988). The WAL isolate was derived from a worm population in the New England region of Northern NSW in 2003 (Love et al., 2003) that was resistant to levamisole (efficacy 79%), as well as benzimidazoles, closantel and short-acting macrocyclic lactones, such as ivermectin. Further selection pressure was applied to this isolate by use of a full dose of Cydectin (active ingredient moxidectin) over at least five subsequent generations. For the present study, sheep were infected with this isolate and were subsequently treated with a full dose of Cydectin 21 days after infection. After a period of at least 6 weeks had elapsed since the Cydectin dose, faeces were collected at various times for recovery of eggs.
Eggs were extracted from faeces by passage through two sieves (250 and 75 μm), followed by centrifugation within a sugar gradient (10% and 25%). Eggs were recovered from the interface between the sucrose layers and washed over a 25 μm sieve with water to remove the sucrose, before being treated with bleach briefly as described by Kotze et al. (2009). The eggs were washed thoroughly to remove the bleach and were then ready for use in larval assays or for in vitro drug selection.
2.2. Larval development assay
An in vitro larval development assay (LDA), as described by Gill et al. (1995) was used to characterize the WAL isolate with respect to levamisole response. Stock drug solution was serially diluted 2-fold in DMSO and 2 μl aliquots were added to a 96-well plate with a final drug concentration ranging from 10 to 0.0049 μg/ml. 200 μl of 2% agar (Davis Gelatine Co., powdered agar Grade J) was dispensed into each well, giving a final DMSO concentration of 1%. Plates were stored at 4 °C for up to a fortnight and re-equilibrated to room temperature before use. 30 μl of egg suspension comprising of ∼3500 eggs/mL with amphotericin B (final concentration 25 μl/mL) and tylosin tartare (final concentration 800 μl/mL) were dispensed into each well (Kotze et al., 2009). Plates were enclosed within a zip lock bag and incubated at 26 °C overnight. An aliquot of a live culture of Escherichia coli was then added to each well (as described by Kotze et al., 2009), and the plates were returned to the incubator for a further 5 days. The larvae were then killed using Lugol’s iodine solution (10 μl per well). Numbers of L3 and total larvae were counted for each well and the percentage of L3 larvae calculated for each well. These values were then expressed as a percentage of the mean of six control (no drug) wells on each assay plate (containing 1% DMSO in agar only). Three replicate assays (each with triplicate wells at each concentration) were performed for each drug with each of the H. contortus isolates. Data from the assay was analyzed using non-linear regression in GraphPad Prism 4™ (GraphPad Software Inc., La Jolla, California, USA).
2.3. Isolation of sub-populations of WAL larvae showing increasing levels of resistance
Data from larval development assays were examined in order to identify concentrations of levamisole that resulted in larval development levels of <20% compared to controls. The drug concentrations selected for use in subsequent drug exposure experiments were: 0.31, 1.25, and 2.5 μg/ml. Aliquots (29 μl) of the particular levamisole solutions (prepared earlier for LDAs) required to give these final drug concentrations were added to wells of 6-well culture plates and 2 ml of 2% agar (Davis Gelatine Co., powdered agar Grade J) was dispensed into each well (final DMSO concentration of 1.45%). All wells on each plate received the same concentration of levamisole. Control (no drug) plates were also prepared using DMSO alone (1.45%). Egg solution, 1.4 ml, containing ∼30,000 eggs with amphotericin B (final concentration 11 μl/mL) and tylosin tartare (final concentration 343 μl/mL) was then added to each well. A less concentrated egg suspension (with identical levels of amphotericin and tylosin) was added to the wells of control plates to give approximately 10,000 eggs per well. Plates were enclosed within a zip lock bag and incubated at 26 °C overnight. An aliquot of a live culture of E. coli was then added to each well (Kotze et al., 2009), and the plates were returned to the incubator for a further 5 days. The larvae from each well on a plate were then collected by flushing the contents of all 6 wells into a beaker with water. The larvae were placed onto the first of a series of sieves (40 μm followed by 25 μm followed by 20 μm) and allowed to migrate through over sequential 24 h periods in order to remove as many dead larvae as possible. After each 24 h period, the larvae that had migrated through the sieve were collected and placed onto the next sieve. Larvae were collected from below the last sieve, washed again in water, counted, and snap frozen in groups of ∼5000 in liquid nitrogen for RNA extraction.
2.4. Isolation of RNA and cDNA synthesis
RNA was extracted using Trizol LS™ (Invitrogen) as per the manufacturer’s instructions, followed by treatment with TurboDNase™ (Ambion). cDNA synthesis was performed on extracted RNA with Superscript III™ reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. cDNA was diluted to a concentration of 2.5 ng/μl for downstream applications. For each of the sub-populations, cDNA was generated from three separate samples of larvae for subsequent gene expression analysis.
2.5. Quantitative PCR
The primers used for quantitative PCR of a number of nAChR, ancillary protein and P-gp genes were as described by Sarai et al. (2013) (see Supplementary Tables 1 and 2). Three housekeeping genes (GAPDH, Actin & 18S) were employed as reference for the determination of relative expression levels. A Viia7 thermocycler (Applied Biosystems) was used, with the SYBR Green dye system (Applied Biosystems) and the following PCR cycling conditions: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles 95 °C for 15 s, 60 °C for 1 min, 95 °C for 2 min, 60 °C for 15 sec. Three separate extractions of each WAL subpopulation, as described earlier, were examined, with each PCR reaction run in quadruplicate. As reported earlier by Sarai et al. (2013), reaction efficiencies, as determined from standard curves using a series of 4, 5-fold cDNA dilutions, were in the range between 76% and 99%. Melting curve analysis of each primer pair identified the qPCR products to be homogenous and direct sequencing of these products confirmed the specific amplification of the target gene. Expression values for each gene of each sub-population were referenced to the three housekeeping genes using REST 2008 software in order to derive a value for the expression of each gene in the resistant sub-population of the WAL isolate compared to the entire WAL population. Differences between the sub-populations and the WAL isolate were then examined using a ‘ratio t-test’ on the log of the ratio as described in the GraphPad Prism handbook. Differences among the 3 subpopulations within each gene were analysed using ANOVA on the ratio data.
3. Results and discussion
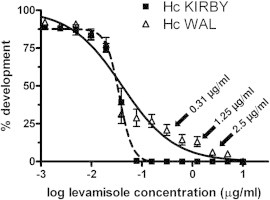
Fig. 1 shows the dose response curves describing the effects of levamisole on the development of H. contortus WAL larvae compared to the susceptible Kirby isolate. The WAL isolate showed a very low-level resistance to levamisole at the IC50 (resistance ratio 1.13). However this isolate showed the presence of a resistant fraction of approximately 10–15% as indicated by the plateau in the dose response at higher drug concentrations. This was reflected in a higher resistance ratio at the IC95 of 17-fold. These data were used to select drug concentrations for subsequent experiments to subdivide the population into groups based on their ability to survive increasing concentrations of drug. The concentrations used in these experiments are illustrated with arrows on Fig. 1.
Fig. 1.
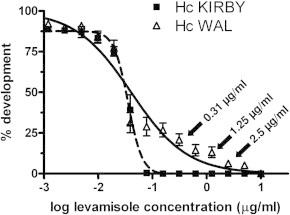
Response of Kirby and Wallangra 2003 isolates of H. contortus to levamisole in larval development assays. Each data point represents mean ± SD, n = 9 assays for levamisole (pooled data from three separate experiments, each with assays in triplicate). Arrows indicate the drug concentrations used for subsequent population subdivision experiments with the Wallangra isolate.
The isolation of resistant sub-populations of WAL larvae involved passage through three mesh sieves in order to separate live and dead larvae (based on ability to migrate through the sieves) after the 6-day larval development period in the presence of the drug. Table 1 describes the subdivision process in terms of the numbers of eggs that were incubated at the different levamisole concentrations and the numbers and percentages of larvae recovered. These numbers should be viewed as approximations only as they were based on the counting of aliquots taken from initial (pre-treatment) egg suspensions containing very large numbers of eggs (up to ∼20,000/ml). While the percentage development data from larval development assays had indicated that 6–15% of larvae should survive at the various levamisole concentrations, the process of sieving drastically reduced these numbers by nearly 15-fold to give larval recoveries of 0.7%, 0.6% and 0.2% at the three increasing levels of levamisole, respectively. A reduction in viable larvae was also observed in the control population, with losses approaching 40% (60.5% recovery of live larvae). This loss of control larvae may be expected due to failure of some eggs to hatch, some death during the development phase in the assay plates, as well as losses during the flushing of larvae from the wells and the transfer of larvae between the sequential migration steps. These factors would have been further exacerbated for drug-treated worm larvae by the presence of large numbers of dead worms during the early migration steps that may have greatly hampered migration, as well as the possibility that fully grown worms (that would have been scored as L3 stage larvae in the LDA, Fig. 1) might have been less fit after development in the presence of levamisole, and hence less able to migrate freely through the various meshes. Nevertheless, despite the lower than expected recovery of levamisole-exposed worms, it is apparent from Table 1 that a high degree of drug pressure was imposed on the population, with <1% of larvae recovered from drug assays compared to >60% for control assays. The percentage recovery of post-treatment survivors was equivalent at the concentrations of 0.31 and 1.25 μg/ml (∼0.7% and 0.6%, respectively), and then significantly lower at 2.5 μg/ml (∼0.2%, P < 0.05). Despite repeated migration steps, a significant number of dead larvae were still present in the final worm suspensions, most likely having simply ‘fallen’ through the mesh during the 24 h migration periods. It was noted that these dead larvae were mostly very small (L1 or early L2 stage). The percentages of dead larvae were approximately equal in each of the drug-pressured populations (20–29%).
Table 1.
Description of the process of recovering larvae following exposure to levamisole in larval development assays.
| Larval recovery parameter | Worm population |
||||
|---|---|---|---|---|---|
| Untreated Wallangra | Levamisole treated |
||||
| 0.31 μg/ml lev |
1.25 μg/ml lev |
2.5 μg/ml lev |
|||
| Total eggs (‘000s) | Rep1 | 12.1 | 930 | 1002 | 1682 |
| Rep2 | 10.4 | 648 | 648 | 2252 | |
| Rep3 | 15.8 | 1050 | 950 | 2722 | |
| Post-sieve survivors (‘000s) | Rep1 | 7.3 | 6.8 | 5.9 | 4.6 |
| Rep2 | 6.8 | 4.7 | 4.4 | 4.5 | |
| Rep3 | 8.4 | 7.0 | 4.8 | 5.1 | |
| Post-sieve survivors (%)⁎ | 60.43 ± 4.23a | 0.68 ± 0.02b | 0.59 ± 0.05b | 0.22 ± 0.03c | |
| Post-sieve dead larvae (%)⁎ | 8.33 ± 1.53a | 24.00 ± 3.06b | 20.00 ± 4.93b | 29.33 ± 1.20b | |
Data shown as mean ± S.E. (n = 3); values labelled with the same letter are not significantly different (P = 0.05).
It should be noted that the subdivision process resulted in three subpopulations with a degree of overlap. That is, while the survivors of 2.5 μg/ml existed as a separate subpopulation, individuals of this type also existed as a component of the population surviving 1.25 μg/ml, and, in turn, larvae that would have been able to survive of 1.25 and 2.5 μg/ml existed as a component of the subpopulation described as having survived at 0.31 μg/ml. Therefore, the transition from 0.31 to 2.5 μg/ml represented smaller and smaller subpopulations (in percentage terms) of more highly resistant worms.
The relative levels of transcription of nAChR subunits, ancillary proteins and P-gp genes in the subpopulations of worms recovered from assays at different levels of levamisole are shown in Table 2. Highlighted in colour within the Table are those genes that showed a significant change (P < 0.05) in expression in the drug-pressured populations compared to the WAL population as a whole (recovered from control assay plates). In addition, within each gene, differences in expression between worms recovered from the different drug concentrations are indicated with alphabetical notation. Hco-unc-29.3 and Hco-acr-8b were beyond the levels of detection in the quantitative PCR assays (that is, CT values >39).
Table 2.
Relative transcription of nAChR subunit, ancillary protein and P-glycoprotein genes in three resistant fractions of H. contortus Wallangra 2003 isolate compared to the whole population.
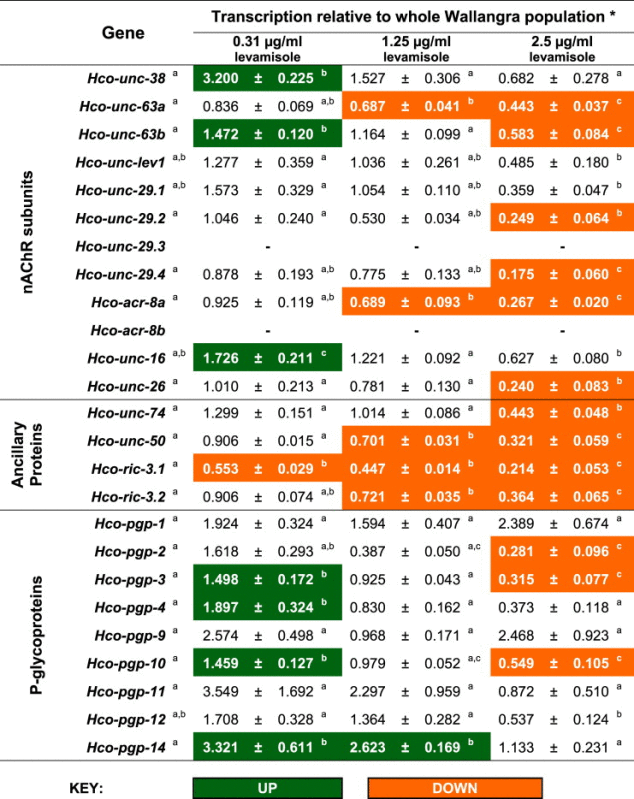 |
∗Data shown as mean ± S.E. n = 3 separate experiments; within each gene, values labelled with the same letter are not significantly different (at P = 0.05); Statistical notation for control samples is shown following the gene name; - indicates not detectable.
Among the nAChR subunit genes, there were a number of instances of reduced transcription in the drug-pressured worms compared to the controls. In each case these decreases were seen only at the highest drug concentration or in both the highest and mid concentrations. Hence, for these genes, decreasing gene expression was associated in each case with increasing drug concentration. Hco-unc-63a and Hco-acr-8a levels showed step-wise decreases at the two highest levamisole concentrations, while Hco-unc-63b, Hco-unc-29.2, Hco-unc-29.4 and Hco-unc-26 were all lower than the control population in survivors of the highest levamisole concentration. The greatest decrease was with Hco-unc-29.4 which showed an almost 6-fold reduction in the most highly drug-pressured subpopulation. Levels of Hco-unc-38 and Hco-unc-63b were elevated at the lowest drug concentration, however they were unchanged compared to controls at the other two concentrations for Hco-unc-38, and reduced at the highest concentration for Hco-unc-63b. Previous studies on mechanisms of levamisole/pyrantel resistance have focussed primarily on changes in nAChR subunits in resistant isolates: reduced transcription of unc-29 paralogs and unc-63a within H. contortus (Kopp et al., 2009; Sarai et al., 2013) as well as down-regulation of unc-38, -29 and -63 in A. caninum (Kopp et al., 2009), the presence of truncated forms of Hco-unc-63 and Hco-acr-8 (Hco-unc-63b and Hco-acr8b, respectively) within some resistant H. contortus isolates (Fauvin et al., 2010; Neveu et al., 2010; Williamson et al., 2011). The results of the present study are in agreement with some of these earlier reports in showing down-regulation of some components of the levamisole-sensitive nAChRs, namely Hco-unc-63a and the Hco-unc-29 paralogs.
The most consistent decreases in transcription in the levamisole-exposed subpopulations were observed in the ancillary protein genes (Table 2). Levels of Hco-ric-3.1 were decreased relative to the control population at all levamisole drug concentrations, while Hco-unc-50 and Hco-ric-3.2 were lower for the two highest drug concentrations, and Hco-unc-74 was reduced at the highest concentration. Studies with the free-living nematode C. elegans have shown the essential role of these ancillary proteins in the proper formation and functioning of levamisole receptors. UNC-74 encodes a thioredoxin which is closely related to the human TMX3 protein (Haugstetter et al., 2005) and is likely to be involved in the folding of AChRs. Early screens for resistance to levamisole identified unc-74 as a key component for the expression of L-type nAChRs within C. elegans (Lewis et al., 1980). UNC-50 is required for the trafficking of L-type receptors to the cell surface and the normal synaptic transmission at the neuromuscular junction and is ubiquitously expressed from early embryogenesis to adulthood, localizing mostly to the Golgi apparatus. In the absence of UNC-50, L-type nAChRs are targeted to lysozymes after they exit the endoplasmic reticulum and are degraded, thereby resulting in loss of function (Eimer et al., 2007). RIC-3 is a chaperone protein required for proper receptor assembly and functional expression in the endoplasmic reticulum. Alongside UNC-74 and -50, Boulin et al. (2011) demonstrated that co-expression with RIC-3 was required for nAChR activity in C. elegans and in Xenopus oocytes. Previous studies had identified the protein encoded by RIC-3 as essential for maturation of levamisole sensitive vulval muscle AChRs, for the maturation of the EAT-2 AChR that enables pharyngeal pumping and that RIC-3 may act at a postsynaptic site (Halevi et al., 2002). Recent immunohistochemical studies have also shown that mutations in RIC-3 caused a reduction in cell surface expression of LEV-1 (Gottschalk and Schafer, 2006) which may result in decreased sensitivity to levamisole. Further, RIC-3 has been shown to increase the functional expression of the N-type nAChR subunits such as ACR-16 within Xenopus oocytes (Bennett et al., 2012). Although the specific effects of the reduced transcript levels of these ancillary protein genes described in the present study are not known, their essential roles described above would suggest that their reduced transcription would have significant effects on the assembly and function of nAChRs in the larvae able to survive and develop in the presence of levamisole: reduced UNC-74 may interfere with the ability of nAChRs to fold correctly, reduced UNC-50 may lead to a greater targeting of receptors to the ER, followed by degradation, and finally reduced RIC-3 may lead to disruption of receptor assembly. All these effects may contribute to a reduced level of functioning levamisole-sensitive nAChRs in the drug resistant worms, hence reducing their sensitivity to the presence of levamisole during the larval development phase of the life cycle. It is not clear whether such reductions in levamisole-sensitive AChRs would have fitness costs for the resistant larvae. This was not tested in the present study as we did not assess larval fitness, except for the ability of larvae that survived the levamisole exposure to then migrate through the series of meshes.
A number of P-glycoprotein genes showed increases at the lowest drug concentration (Hco-pgp-3, -4, -10 and -14), alongside decreases or no change at the highest drug concentration (Hco-pgp-2, -3, -10) (Table 2). The increases seen at 0.31 μg/ml indicates that within the Wallangra population exists a subset of larvae able to survive exposure to this level of levamisole that also show elevated P-gp levels, suggesting that the efflux pumps may contribute to the observed drug tolerance. In addition, the lack of any increased P-gp in the worm larvae exposed to the two highest drug concentrations suggests that the larvae able to survive 1.25 and 2.5 μg/ml levamisole do not utilise P-gp efflux pumps to survive the drug exposure. Little is known about the specific interaction of levamisole with P-gps. Several imidazothiazole drugs have been shown to be reversing agents for multidrug resistances shown by mammalian cell lines suggesting that they can interact with P-gps (Naito et al., 1998). More recently, Kerboeuf and Guegnard (2011) found that levamisole stimulated P-gp activity in H. contortus eggs, suggesting that the compound may bind to ’regulatory’ sites on nematode P-gps. James and Davey (2009) reported that exposure of C. elegans to repeated selection pressure with ivermectin resulted in resistance to this drug alongside some cross-resistance to levamisole, as well as significantly increased expression of P-gp genes. This suggested that the increased transporter activity in the resistant worms had resulted in an increased ability to transport both ivermectin and levamisole, hence highlighting the latter as a likely substrate for the worm P-gps.
De Graef et al. (2013) found that in vitro exposure of L3 Cooperia oncophora to ivermectin resulted in increased expression of several P-gp genes. This raises the question as to whether the P-gp expression increases observed in the present study represent constitutive gene expression patterns in the levamisole-resistant larvae or are the result of induction following exposure to the drug. We consider that they most likely represent constitutive expression patterns as it would be likely that maintenance of the L3 worms in a drug-free environment for 72 h during the sieve migration period would have resulted in the return of any induced gene expression to baseline levels. While there is a deal of literature on the inducing effects of various agents on P-gp expression, there is very little information available on the time course of gene expression changes after removal of the inducing agents; Fardel et al (1996) found that expression of a P-gp in rat liver epithelial cells was significantly induced following exposure for 24 h to 3-methylcholanthrene, with a return to almost basal levels 72 h after removal of the inducing agent. However, the possibility remains that the increased P-gp expression levels in levamisole-exposed larvae in the present study was an induction phenomenon. This will require further experimentation to clarify.
The gene expression changes shown in Table 2 suggest the presence of a biphasic pattern of gene expression as the resistance increases in WAL larvae: increased expression of P-gps is the dominant change seen in worms able to survive the lowest drug concentration, while reduced expression of receptor subunit and assembly genes becomes significant as levamisole resistance increases. This suggests that two levels of resistance, each associated with distinct gene expression changes, are present: non-specific Pgp-mediated low-level resistance alongside target site changes conferring higher levels of resistance. This observation aligns with earlier levamisole resistance literature which reported that levamisole resistance existed in the field in Australia in two forms denoted ‘moderate level resistance’ and ‘high level resistance’ based on responses in larval development assays (Drenchrite Users Guide, 1996, Horizon Technology, Australia).
The ability of organisms to exhibit multiple resistance mechanisms which confer different levels of resistance has been documented for some time in insects (for example, Farnham, 1973). Scott and Georghiou (1986) found that three mechanism were responsible for pyrethroid resistance in a strain of housefly: enhanced metabolism via the mixed function oxidase (MFO) system, target-site insensitivity, and decreased cuticular penetration. The major mechanism of resistance was the MFO-mediated detoxification. This multi-tiered nature of resistance has also been observed with aphids (Puinean et al., 2010) and mosquitoes (Wood et al., 2010) highlighting the role of a non-specific mechanism that confers low-level resistance against a number of drug groups, alongside specific mechanisms involving target site or detoxification enzyme changes which confer higher level resistance.
Sarai et al. (2013) highlighted the difficulty of detecting gene expression changes associated with levamisole resistance when examining pooled samples of larvae from isolates displaying a degree of heterogeneity in response to the drug as shown by WAL larvae. This was because the averaging of gene expression levels across the whole population might mask changes that may confer resistance in the highly resistant subpopulation seen with this isolate. A comparison of the gene expression data from Sarai et al. (2013) with the present study is informative in this regard. The earlier study found that no nAChR subunit showed reduced transcription in pooled samples of WAL L3 stage larvae compared to the Kirby isolate. This was in contrast to larvae of the LAWES and LevR isolates in which a number of genes (most notably Hco-unc-63a in both isolates, and Hco-unc-29.2 and Hco-unc-29.4 in LAWES) were reduced compared to Kirby. Importantly, in vitro larval development assay dose responses for these two resistant isolates were well displaced from the Kirby isolate at all drug concentrations (resistance ratios at the IC50 > 650). Hence, while the earlier gene transcription measurements of WAL L3 stage worms failed to detect any changes in this isolate compared to Kirby, the present study indicates that at least some of the changes seen in LAWES and LevR do indeed occur in the most resistant individuals in the WAL isolate. This illustrates the effect of gene transcription measurements across a whole population in masking changes that may be present in a small number of resistant individuals. At the early stages of the development of resistance to an anthelmintic, the resistant genotype/phenotype is present at very low frequencies in the population, and only these few individuals are able to survive drug concentrations significantly higher than the ‘normal’ susceptible range. The present study, in examining subpopulations of worms able to survive drug concentrations significantly higher than the bulk of the population, has effectively served as a model for the detection of the early stages of resistance. It is most likely that over time and under further drug selection pressure the genotypes and phenotypes possessed by the more resistant fractions would become dominant. Hence, the present study and that of Sarai et al. (2013) illustrate a limitation in the use of gene expression-based tests to detect resistance at its early stages where only a small fraction of the population are resistant. This is in contrast to SNP-based methods which can detect changes in allele frequencies at very low levels in a population (von Samson-Himmelstjerna et al., 2009).
In conclusion, the present study suggests that reduced transcription of genes coding for structural components of levamisole-sensitive receptors (Hco-unc-63a, -63b, Hco-unc-29 paralogs, Hco-acr8a) as well as genes coding for proteins involved in the assembly and functioning of the receptors (Hco-unc-74, Hco-unc-50, Hco-ric-3.1 and 3.2) may play a significant role in levamisole resistance in larvae of a field-derived isolate of H. contortus. In addition, our data suggests a role for P-glycoproteins in worm larvae within the population that show a lower level of resistance than those characterised by the changes in receptor subunit and receptor assembly genes. Importantly, our study design has allowed us to associate gene expression patterns with levamisole sensitivity among worms with similar genetic backgrounds using subpopulations distinguishable from the whole population, and from each other only, on the basis of their ability to survive drug exposure in vitro. Though changes in expression patterns within the larvae examined here may not be indicative of changes within adult worms which will have a real impact on drug efficacy in the field, this study provides a starting point to investigate variations in gene expression patterns among adult worms differing in drug sensitivity. An investigation into gene transcription patterns and resistance status in adults cultivated from the resistant fractions examined here would provide greater insight into the relationship between resistance in this species at the larval and adult life stages.
Acknowledgements
R.S.S. is a recipient of a CSIRO Livestock Industries postgraduate scholarship. The authors have no conflicts of interest concerning the work reported in this paper.
Appendix A. Supplementary data
References
- Albers G.A.A., Burgess S.K. Serial passage of Haemonchus contortus in resistant and susceptible sheep. Vet. Parasitol. 1988;28:303–306. doi: 10.1016/0304-4017(88)90077-5. [DOI] [PubMed] [Google Scholar]
- Bennett H.M., Lees K., Harper K.M., Jones A.K., Sattelle D.B., Wonnacott S., Wolstenholme A.J. Xenopus laevis RIC-3 enhances the functional expression of the C. elegans homomeric nicotinic receptor, ACR-16, in Xenopus oocytes. J. Neurochem. 2012;123:911–918. doi: 10.1111/jnc.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besier B. New anthelmintics for livestock: the time is right. Trends Parasitol. 2007;23:21–24. doi: 10.1016/j.pt.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Blackhall W.J., Prichard R.K., Beech R.N. P-glycoprotein selection in strains of Haemonchus contortus resistant to benzimidazoles. Vet. Parasitol. 2008;152:101–107. doi: 10.1016/j.vetpar.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Boulin T., Fauvin A., Charvet C., Cortet J., Cabaret J., Bessereau J.L., Neveu C. Functional reconstitution of Haemonchus contortus acetylcholine receptors in Xenopus oocytes provides mechanistic insights into levamisole resistance. Br. J. Pharmacol. 2011;164:1421–1432. doi: 10.1111/j.1476-5381.2011.01420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Graef J., Demeler J., Skuce P., Mitreva M., von Samson-Himmelstjerna G., Vercruysse J., Claerebout E., Geldhof P. Gene expression analysis of ABC transporters in a resistant Cooperia oncophora isolate following in vivo and in vitro exposure to macrocyclic lactones. Parasitology. 2013;140:499–508. doi: 10.1017/S0031182012001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimer S., Gottschalk A., Hengartner M., Horvitz H.R., Richmond J., Schafer W.R., Bessereau J.L. Regulation of nicotinic receptor trafficking by the transmembrane Golgi protein UNC-50. Embo J. 2007;26:4313–4323. doi: 10.1038/sj.emboj.7601858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardel O., Lecureur V., Corlu A., Guillouzo A. P-glycoprotein induction in rat liver epithelial cells in response to acute 3-methylcholanthrene treatment. Biochem. Pharmacol. 1996;51:1427–1436. doi: 10.1016/0006-2952(96)00081-0. [DOI] [PubMed] [Google Scholar]
- Farnham A.W. Genetics or resistance of Pyrethroid-selected house flies, Musca domestica L. Pestic. Sci. 1973;4:513–520. [Google Scholar]
- Fauvin A., Charvet C., Issouf M., Cortet J., Cabaret J., Neveu C. CDNA-AFLP analysis in levamisole-resistant Haemonchus contortus reveals alternative splicing in a nicotinic acetylcholine receptor subunit. Mol. Biochem. Parasitol. 2010;170:105–107. doi: 10.1016/j.molbiopara.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Geary T.G., Hosking B.C., Skuce P.J., von Samson-Himmelstjerna G., Maeder S., Holdsworth P., Pomroy W.E., Vercruysse J. World Association for the Advancement of Veterinary Parasitology (W.A.A.V.P.) Guideline: Anthelmintic combination products targeting nematode infections of ruminants and horses. Vet. Parasitol. 2012;190:306–316. [Google Scholar]
- Gill J.H., Redwin J.M., van Wyk J.A., Lacey E. Avermectin inhibition of larval development in Haemonchus contortus – effects of ivermectin resistance. Int. J. Parasitol. 1995;25:463–470. doi: 10.1016/0020-7519(94)00087-5. [DOI] [PubMed] [Google Scholar]
- Gottschalk A., Schafer W.R. Visualization of integral and peripheral cell surface proteins in live Caenorhabditis elegans. J. Neurosci. Methods. 2006;154:68–79. doi: 10.1016/j.jneumeth.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Gottschalk A., Almedom R.B., Schedletzky T., Anderson S.D., Yates J.R., Schafer W.R. Identification and characterization of novel nicotinic receptor-associated proteins in Caenorhabditis elegans. Embo J. 2005;24:2566–2578. doi: 10.1038/sj.emboj.7600741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevi S., McKay J., Palfreyman M., Yassin L., Eshel M., Jorgensen E., Treinin M. The C. elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors. Embo J. 2002;21:1012–1020. doi: 10.1093/emboj/21.5.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugstetter J., Blicher T., Ellgaard L. Identification and characterization of a novel thioredoxin-related transmembrane protein of the endoplasmic reticulum. J. Biol. Chem. 2005;280:8371–8380. doi: 10.1074/jbc.M413924200. [DOI] [PubMed] [Google Scholar]
- James C.E., Davey M.W. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int. J. Parasitol. 2009;39:213–220. doi: 10.1016/j.ijpara.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Kerboeuf D., Guegnard F. Anthlemintics are substrates of nematode P glycoprotein. Antimicrob. Agents Chemother. 2011;55:2224–2232. doi: 10.1128/AAC.01477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerboeuf D., Blackhall W.J., Kaminsky R., von Samson-Himmelstjerna G. P-glycoprotein in helminths: function and perspectives for anthelmintic treatment and reversal of resistance. Int. J. Antimicrob. Agents. 2003;22:332–346. doi: 10.1016/s0924-8579(03)00221-8. [DOI] [PubMed] [Google Scholar]
- Kopp S.R., Coleman G.T., Traub R.J., McCarthy J.S., Kotze A.C. Acetylcholine receptor subunit genes from Ancylostoma caninum: altered transcription patterns associated with pyrantel resistance. Int. J. Parasitol. 2009;39:435–441. doi: 10.1016/j.ijpara.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Kotze A.C., O’Grady J., Emms J., Toovey A.F., Hughes S., Jessop P., Bennell M., Versoe P.E., Revell D.K. Exploring the anthelmintic properties of Australian native shrubs with respect to their potential role in livestock grazing systems. Parasitology. 2009;136:1065–1080. doi: 10.1017/S0031182009006386. [DOI] [PubMed] [Google Scholar]
- Kotze A.C., Cowling K., Bagnall N.H., Hines B.M., Ruffell A.P., Hunt P.W., Coleman G.T. Relative level of thiabendazole resistance associated with the E198A and F200Y SNPs in larvae of a multi-drug resistant isolate of Haemonchus contortus. Int. J. Parasitol. Drugs Drug Resist. 2012;2:92–97. doi: 10.1016/j.ijpddr.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespine A., Ménez C., Bourguinat C., Prichard R.K. P-glycoproteins and other multidrug resistance transporters in the pharmacology of anthelmintics: prospects for reversing transport-dependent anthelmintic resistance. Int. J. Parasitol. Drugs Drug Resist. 2012;2:58–75. doi: 10.1016/j.ijpddr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levandoski M.M., Robertson A.P., Kuiper S., Qian H., Martin R.J. Single-channel properties of N- and L-subtypes of acetylcholine receptor in Ascaris suum. Int. J. Parasitol. 2005;35:925–934. doi: 10.1016/j.ijpara.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Lewis J.A., Wu C.H., Berg H., Levine J.H. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics. 1980;95:905–928. doi: 10.1093/genetics/95.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love S.C.J., Neilson F.J.A., Biddle A.J., McKinnon R. Moxidectin-resistant Haemonchus contortus in sheep in northern New South Wales. Aust. Vet. J. 2003;81:359–360. doi: 10.1111/j.1751-0813.2003.tb11514.x. [DOI] [PubMed] [Google Scholar]
- Martin R.J., Robertson A.P., Bjorn H., Sangster N.C. Heterogeneous levamisole receptors: A single-channel study of nicotinic acetylcholine receptors from Oesophagostomum dentatum. Eur. J. Pharmacol. 1997;322:249–257. doi: 10.1016/s0014-2999(96)00996-x. [DOI] [PubMed] [Google Scholar]
- Martin R.J., Robertson A.P., Buxton S.K., Beech R.N., Charvet C.L., Neveu C. Levamisole receptors: a second awakening. Trends Parasitol. 2012;28:289–296. doi: 10.1016/j.pt.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKellar Q.A., Jackson F. Veterinary anthelmintics: old and new. Trends Parasitol. 2004;20:456–461. doi: 10.1016/j.pt.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Naito S., Koike K., Ono M., Machida T., Tasaka S., Kiue A., Koga H., Kumazawa J. Development of novel reversal agents, imidazothiazole derivatives, targeting MDR1- and MRP-mediated multidrug resistance. Oncol. Res. 1998;10:123–132. [PubMed] [Google Scholar]
- Neveu C., Charvet C., Fauvin A., Cortet J., Beech R.N., Cabaret J. Genetic diversity of levamisole receptor subunits in parasitic nematode species and abbreviated transcripts associated with resistance. Pharmacol. Genom. 2010;20:414–425. doi: 10.1097/FPC.0b013e328338ac8c. [DOI] [PubMed] [Google Scholar]
- Prichard R.K., von Samson-Himmelstjerna G., Blackhall W.J., Geary T.G. Foreword: towards markers for anthelmintic resistance in helminths of importance in animal and human health. Parasitology. 2007;134:1073–1076. doi: 10.1017/S0031182007000078. [DOI] [PubMed] [Google Scholar]
- Puinean A.M., Foster S.P., Oliphant L., Denholm I., Field L.M., Millar N.S., Williamson M.S., Bass C. Amplification of a Cytochrome P450 gene is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae. PLoS Genet. 2010;6:1–11. doi: 10.1371/journal.pgen.1000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster N.C., Song J., Demeler J. Resistance as a tool for discovering and understanding targets in parasite neuromusculature. Parasitology. 2005;131:S179–S190. doi: 10.1017/S0031182005008656. [DOI] [PubMed] [Google Scholar]
- Sarai R.S., Kopp S.R., Coleman G.T., Kotze A.C. Acetylcholine receptor subunit and P-glycoprotein transcription patterns in levamisole-susceptible and -resistant Haemonchus contortus. Int. J. Parasitol. Drugs Drug Resist. 2013;3:51–58. doi: 10.1016/j.ijpddr.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.G., Georghiou G.P. Mechanisms responsible for high levels of permethrin resistance in the house fly. Pestic. Sci. 1986;17:195–206. [Google Scholar]
- von Samson-Himmelstjerna G., Walsh T.K., Donnan A.A., Carriere S., Jackson F., Skuce P.J., Rohn K., Wolstenholme A.J. Molecular detection of benzimidazole resistance in Haemonchus contortus using real-time PCR and pyrosequencing. Parasitology. 2009;136:349–358. doi: 10.1017/S003118200800543X. [DOI] [PubMed] [Google Scholar]
- Williamson S.M., Storey B., Howell S., Harper K.M., Kaplan R.M., Wolstenholme A.J. Candidate anthelmintic resistance-associated gene expression and sequence polymorphisms in a triple-resistant field isolate of Haemonchus contortus. Mol. Biochem. Parasitol. 2011;180:99–105. doi: 10.1016/j.molbiopara.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Wolstenholme A.J., Fairweather I., Prichard R., von Samson-Himmelstjerna G., Sangster N.C. Drug resistance in veterinary helminths. Trends Parasitol. 2004;20:469. doi: 10.1016/j.pt.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Wood O.R., Hanrahan S., Coetzee M., Koekemoer L.L., Brooke B.D. Cuticle thickening associated with pyrethroid resistance in the major malaria vector Anopheles funestus. Parasites Vectors. 2010;3:1–7. doi: 10.1186/1756-3305-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D.J., Knauer C.S. Discovery of veterinary antiparasitic agents in the 21st Century: a view from industry. Int. J. Parasitol. 2010;40:1177–1181. doi: 10.1016/j.ijpara.2010.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


