Abstract
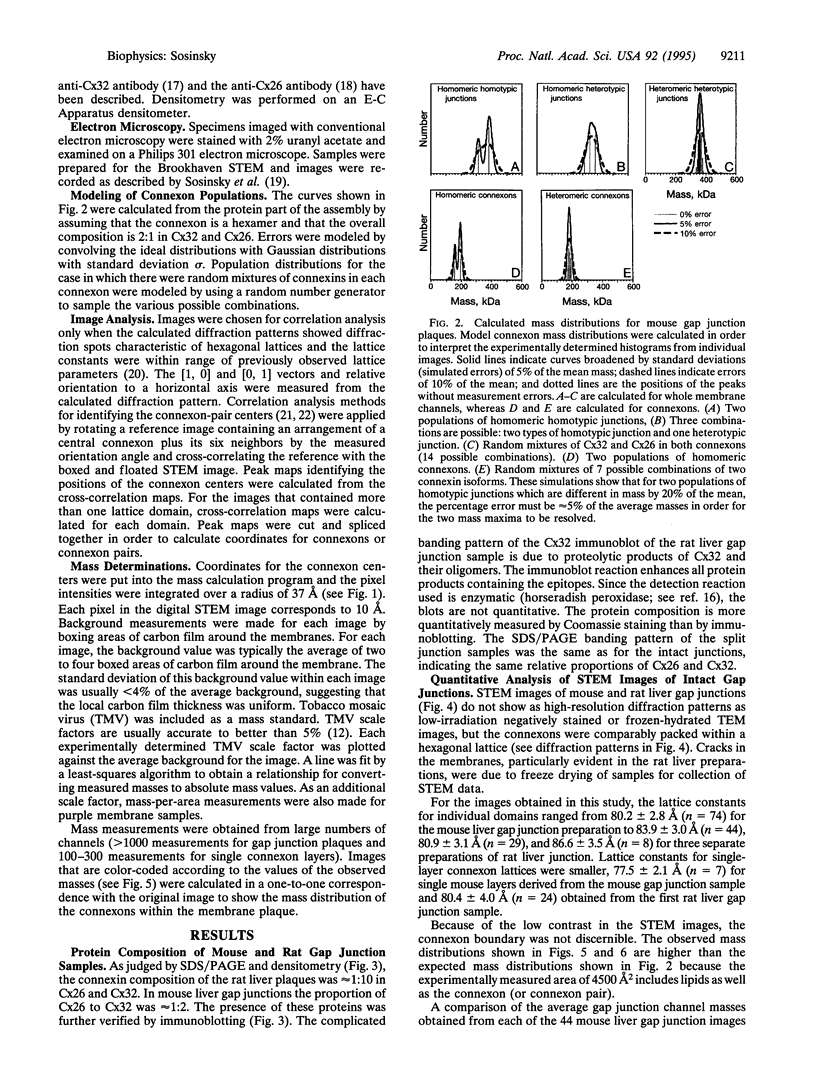
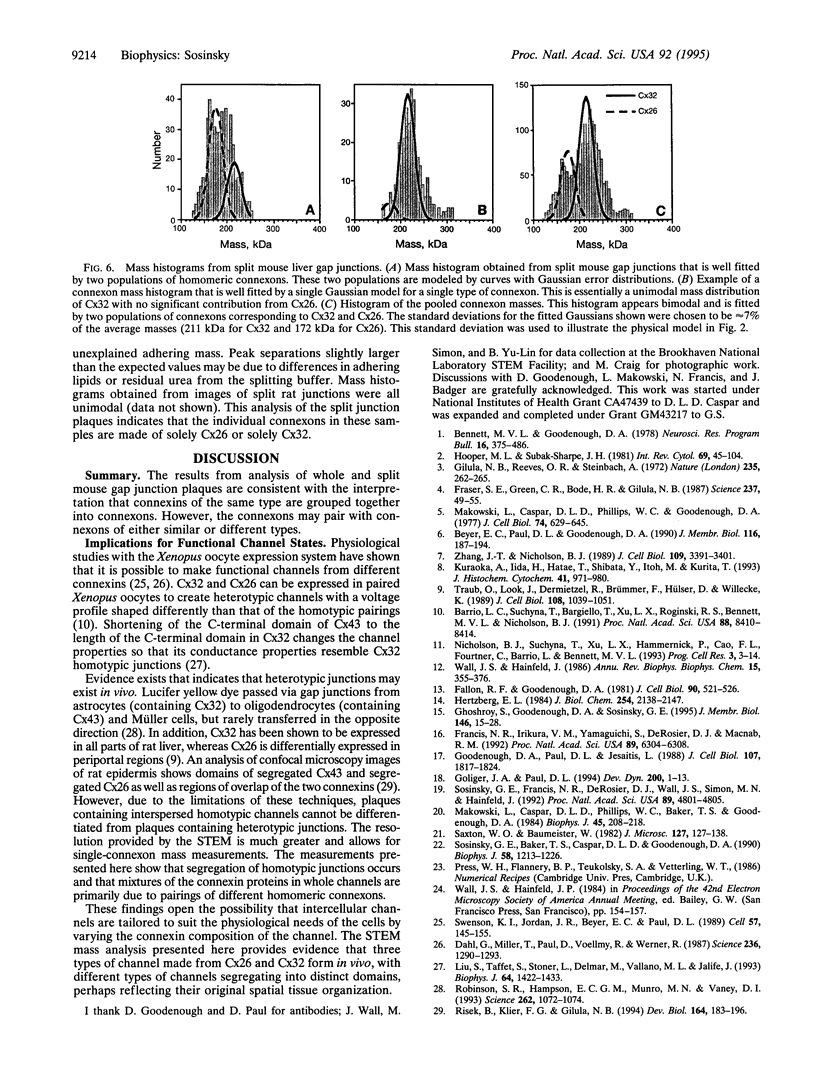
Gap junctions are plaque-like clusters of intercellular channels that mediate intercellular communication. Each of two adjoining cells contains a connexon unit which makes up half of the whole channel. Gap junction channels are formed from a multigene family of proteins called connexins, and different connexins may be coexpressed by a single cell type and found within the same plaque. Rodent gap junctions contain two proteins, connexins 32 and 26. Use of a scanning transmission electron microscope for mass analysis of rodent gap junction plaques and split gap junctions prvided evidence consistent with a model in which the channels may be made from (i) solely connexin 26, (ii) solely connexin 32, or (iii) mixtures of connexin 26 and connexin 32 in which the two connexons are made entirely of connexin 26 and connexin 32. The different types of channels segregate into distinct domains, implying tha connexon channels self-associate to give a non-random distribution within tissues. Since each connexin confers distinct physiological properties on its membrane channels, these results imply that the physiological properties of channels can be tailored by mixing the constituent proteins within these macromolecular structures.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrio L. C., Suchyna T., Bargiello T., Xu L. X., Roginski R. S., Bennett M. V., Nicholson B. J. Gap junctions formed by connexins 26 and 32 alone and in combination are differently affected by applied voltage. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8410–8414. doi: 10.1073/pnas.88.19.8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. V., Goodenough D. A. Gap junctions, electrotonic coupling, and intercellular communication. Neurosci Res Program Bull. 1978 Sep;16(3):1–486. [PubMed] [Google Scholar]
- Beyer E. C., Paul D. L., Goodenough D. A. Connexin family of gap junction proteins. J Membr Biol. 1990 Jul;116(3):187–194. doi: 10.1007/BF01868459. [DOI] [PubMed] [Google Scholar]
- Dahl G., Miller T., Paul D., Voellmy R., Werner R. Expression of functional cell-cell channels from cloned rat liver gap junction complementary DNA. Science. 1987 Jun 5;236(4806):1290–1293. doi: 10.1126/science.3035715. [DOI] [PubMed] [Google Scholar]
- Fallon R. F., Goodenough D. A. Five-hour half-life of mouse liver gap-junction protein. J Cell Biol. 1981 Aug;90(2):521–526. doi: 10.1083/jcb.90.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis N. R., Irikura V. M., Yamaguchi S., DeRosier D. J., Macnab R. M. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser S. E., Green C. R., Bode H. R., Gilula N. B. Selective disruption of gap junctional communication interferes with a patterning process in hydra. Science. 1987 Jul 3;237(4810):49–55. doi: 10.1126/science.3037697. [DOI] [PubMed] [Google Scholar]
- Ghoshroy S., Goodenough D. A., Sosinsky G. E. Preparation, characterization, and structure of half gap junctional layers split with urea and EGTA. J Membr Biol. 1995 Jul;146(1):15–28. doi: 10.1007/BF00232677. [DOI] [PubMed] [Google Scholar]
- Gilula N. B., Reeves O. R., Steinbach A. Metabolic coupling, ionic coupling and cell contacts. Nature. 1972 Feb 4;235(5336):262–265. doi: 10.1038/235262a0. [DOI] [PubMed] [Google Scholar]
- Goliger J. A., Paul D. L. Expression of gap junction proteins Cx26, Cx31.1, Cx37, and Cx43 in developing and mature rat epidermis. Dev Dyn. 1994 May;200(1):1–13. doi: 10.1002/aja.1002000102. [DOI] [PubMed] [Google Scholar]
- Goodenough D. A., Paul D. L., Jesaitis L. Topological distribution of two connexin32 antigenic sites in intact and split rodent hepatocyte gap junctions. J Cell Biol. 1988 Nov;107(5):1817–1824. doi: 10.1083/jcb.107.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg E. L., Gilula N. B. Isolation and characterization of gap junctions from rat liver. J Biol Chem. 1979 Mar 25;254(6):2138–2147. [PubMed] [Google Scholar]
- Hooper M. L., Subak-Sharpe J. H. Metabolic cooperation between cells. Int Rev Cytol. 1981;69:45–104. doi: 10.1016/s0074-7696(08)62320-7. [DOI] [PubMed] [Google Scholar]
- Kuraoka A., Iida H., Hatae T., Shibata Y., Itoh M., Kurita T. Localization of gap junction proteins, connexins 32 and 26, in rat and guinea pig liver as revealed by quick-freeze, deep-etch immunoelectron microscopy. J Histochem Cytochem. 1993 Jul;41(7):971–980. doi: 10.1177/41.7.8390496. [DOI] [PubMed] [Google Scholar]
- Liu S., Taffet S., Stoner L., Delmar M., Vallano M. L., Jalife J. A structural basis for the unequal sensitivity of the major cardiac and liver gap junctions to intracellular acidification: the carboxyl tail length. Biophys J. 1993 May;64(5):1422–1433. doi: 10.1016/S0006-3495(93)81508-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L., Caspar D. L., Phillips W. C., Baker T. S., Goodenough D. A. Gap junction structures. VI. Variation and conservation in connexon conformation and packing. Biophys J. 1984 Jan;45(1):208–218. doi: 10.1016/S0006-3495(84)84149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L., Caspar D. L., Phillips W. C., Goodenough D. A. Gap junction structures. II. Analysis of the x-ray diffraction data. J Cell Biol. 1977 Aug;74(2):629–645. doi: 10.1083/jcb.74.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risek B., Klier F. G., Gilula N. B. Developmental regulation and structural organization of connexins in epidermal gap junctions. Dev Biol. 1994 Jul;164(1):183–196. doi: 10.1006/dbio.1994.1190. [DOI] [PubMed] [Google Scholar]
- Robinson S. R., Hampson E. C., Munro M. N., Vaney D. I. Unidirectional coupling of gap junctions between neuroglia. Science. 1993 Nov 12;262(5136):1072–1074. doi: 10.1126/science.8093125. [DOI] [PubMed] [Google Scholar]
- Saxton W. O., Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J Microsc. 1982 Aug;127(Pt 2):127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Sosinsky G. E., Baker T. S., Caspar D. L., Goodenough D. A. Correlation analysis of gap junction lattice images. Biophys J. 1990 Nov;58(5):1213–1226. doi: 10.1016/S0006-3495(90)82462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky G. E., Francis N. R., DeRosier D. J., Wall J. S., Simon M. N., Hainfeld J. Mass determination and estimation of subunit stoichiometry of the bacterial hook-basal body flagellar complex of Salmonella typhimurium by scanning transmission electron microscopy. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):4801–4805. doi: 10.1073/pnas.89.11.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson K. I., Jordan J. R., Beyer E. C., Paul D. L. Formation of gap junctions by expression of connexins in Xenopus oocyte pairs. Cell. 1989 Apr 7;57(1):145–155. doi: 10.1016/0092-8674(89)90180-3. [DOI] [PubMed] [Google Scholar]
- Traub O., Look J., Dermietzel R., Brümmer F., Hülser D., Willecke K. Comparative characterization of the 21-kD and 26-kD gap junction proteins in murine liver and cultured hepatocytes. J Cell Biol. 1989 Mar;108(3):1039–1051. doi: 10.1083/jcb.108.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J. S., Hainfeld J. F. Mass mapping with the scanning transmission electron microscope. Annu Rev Biophys Biophys Chem. 1986;15:355–376. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- Zhang J. T., Nicholson B. J. Sequence and tissue distribution of a second protein of hepatic gap junctions, Cx26, as deduced from its cDNA. J Cell Biol. 1989 Dec;109(6 Pt 2):3391–3401. doi: 10.1083/jcb.109.6.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]