Abstract
Background
Pharmacogenomics (PGx) is positioned to have a widespread impact on the practice of medicine, yet physician acceptance is low. The presentation of context-specific PGx information, in the form of clinical decision support (CDS) alerts embedded in a computerized provider order entry (CPOE) system, can aid uptake. Usability evaluations can inform optimal design, which, in turn, can spur adoption.
Objectives
The study objectives were to: 1) evaluate an early prototype, commercial CPOE system with PGx-CDS alerts in a simulated environment, 2) identify potential improvements to the system user interface, and 3) understand the contexts under which PGx knowledge embedded in an electronic health record is useful to prescribers.
Methods
Using a mixed methods approach, we presented seven cardiologists and three oncologists with five hypothetical clinical case scenarios. Each scenario featured a drug for which a gene encoding drug metabolizing enzyme required consideration of dosage adjustment. We used Morae® to capture comments and on-screen movements as participants prescribed each drug. In addition to PGx-CDS alerts, ‘Infobutton®’ and ‘Evidence’ icons provided participants with clinical knowledge resources to aid decision-making.
Results
Nine themes emerged. Five suggested minor improvements to the CPOE user interface; two suggested presenting PGx information through PGx-CDS alerts using an ‘Infobutton’ or ‘Evidence’ icon. The remaining themes were strong recommendations to provide succinct, relevant guidelines and dosing recommendations of phenotypic information from credible and trustworthy sources; any more information was overwhelming. Participants’ median rating of PGx-CDS system usability was 2 on a Likert scale ranging from 1 (strongly agree) to 7 (strongly disagree).
Conclusions
Usability evaluation results suggest that participants considered PGx information important for improving prescribing decisions; and that they would incorporate PGx-CDS when information is presented in relevant and useful ways.
Keywords: Clinical decision support systems, Clinical knowledge resources (not a MeSH term), Medical order entry systems, Pharmacogenetics, User-Computer Interface
1. Introduction
Mapping of the human genome in 2003 opened new avenues for research [1]. Pharmacogenomics (PGx), the study of how genetic variations affect differences in drug response, is positioned to be the first genomic advance to have a widespread impact on the practice of medicine [2]. Already, the FDA includes information about 43 PGx biomarkers in the labels of 155 drugs [3].
Although surveys of US physicians indicate wide acceptance of the concept of PGx, these surveys also suggest the adoption rate remains low [4–6]. Given the growing number of biomarkers and the complexity of combinatory relationships of disease, gene, PGx test, drug, and dose, clinical decision support (CDS) will be needed to facilitate diffusion of PGx information to inform clinical practice [7]. Presentation of PGx context-specific information, such as alerts embedded in computerized provider order entry (CPOE) systems, can aid uptake of PGx knowledge.
CPOE with PGx-CDS alerts will be useful to classify drug responders, tailor drug therapy, optimize drug response, and reduce adverse drug events [8]. Yet, poorly designed alerts could introduce errors, cause alert fatigue, or hinder adoption of CPOE systems [9]. Usability evaluations inform proper design of clinical information systems, and can facilitate adoption [10,11]. To date, usability studies focus on CDS alerts in the context of CPOE systems [10], however the usability of PGx-CDS alerts has not yet been studied. The objectives of this study were to: 1) Evaluate the usability of an early prototype, commercial CPOE system with PGx-CDS alerts in a simulated work environment (from the prescriber perspective), 2) Identify improvements to the CPOE user interface, and 3) Understand the contexts under which PGx knowledge embedded in an EHR could be useful to clinicians.
2. Methods
2.1 Study Design
Ours was a usability study of a CPOE system with PGx-CDS alerts that was comprised of a heuristic evaluation, supplemented by a participant satisfaction survey. We employed two sets of heuristics. The first set used fourteen principles customized by Zhang for the health domain [11], and is based on usability heuristics and rules from Neilsen [12] and Shneiderman [13]. The second set used seven clinical knowledge heuristics designed to measure knowledge utilization in healthcare [14–16] (Appendix A). Our data collection method centered on a concurrent ‘think-aloud’ process, by which our aim was to identify possible improvements that could be made to the prototype CPOE system, and understand under what circumstances prescribers accessed knowledge to aid prescribing decisions.
2.2 Study Set-Up
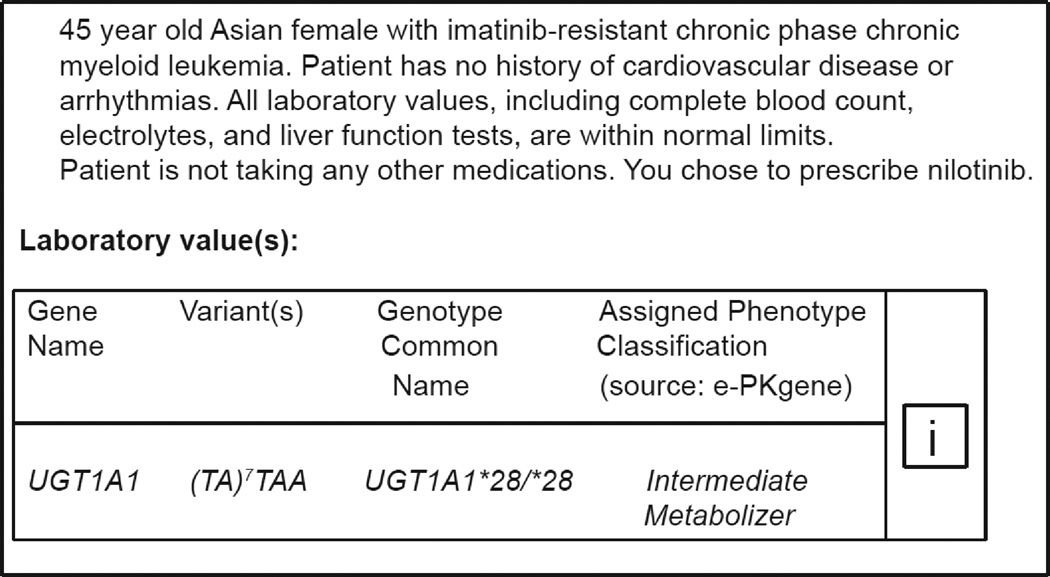
The study was conducted using a prototype version of PowerChart® (Cerner Millennium®), the University of Washington’s (UW) inpatient EHR application. The CPOE system provides auto-fill selection of generic drugs, and standard doses, routes and frequencies. We employed Cerner’s Discern Expert® rules engine to provide real-time CDS alerts. The CPOE and Discern Expert modules of PowerChart had not been implemented at UW when our study began. From within PowerChart, participants could access both the UW Health Sciences Library (‘Links and Reports’) and the internet, and therefore were accustomed to certain available clinical knowledge resources. However, none provided focused genomic or PGx recommendations. To address this, each scenario was comprised of two parts. First, each participant read a short clinical case including the indicated drug and patient-specific PGx allelic variants with corresponding phenotype. Participants could then click an ‘InfoButton®’ linking to curated genomic knowledge resources that further described the relevant gene and laboratory value (Figure 1a; Appendix B).
Figure 1.
a: Case Scenario, PGx Allelic Variants, Phenotype, and Infobutton®
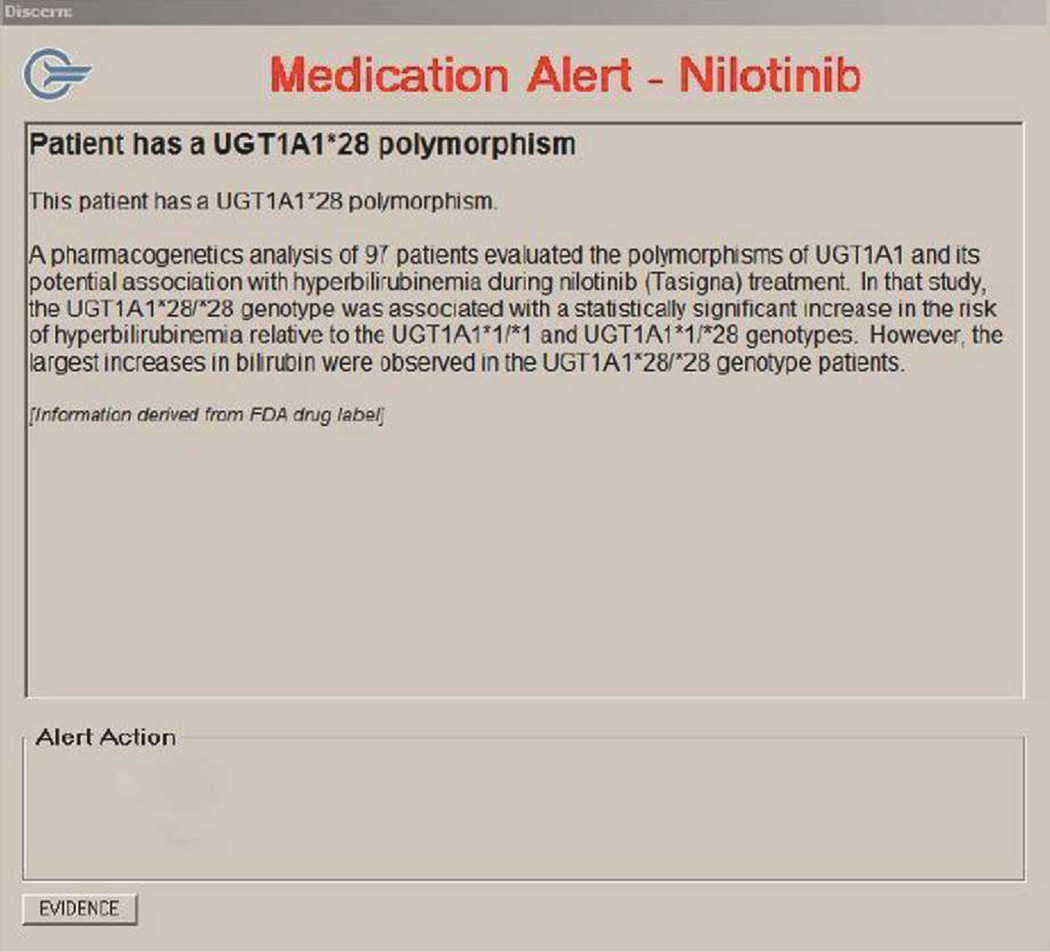
b: Clinical Decision Support Alert in Discern® System
Once satisfied, participants were directed to the second part of the exercise, using the CPOE system to prescribe the drug for the hypothetical patient. In this context, when the PGx-CDS alert popped up, the participant was provided an opportunity to view additional evidence -curated PGx knowledge resources specific to the drug-gene pair. Participants accessed these resources by clicking on an ‘Evidence’ button in the alert (Figure 1b; Appendix B).
2.3 Drugs Selected for Study
In previous work Overby designed 565 decision rules for CDS, specific to the 71 drugs for which biomarkers were codified in the FDA label as of May 2011 [17]. Fifty-five percent of these drugs involved genes encoding cytochrome P-450 (CYP-450) drug metabolism; 42% of these defined cardiology or oncology drugs. Thus, for the usability study, we created CDS alerts around the five cardiology (carvedilol, clopidogrel, metoprolol, propafenone, warfarin) and six oncology (capecitabine, irinotecan, nilotinib, tamoxifen, 6-mercaptopurine (6-MP), 6-thioguanine (6-TG)) drugs for which a gene encoding a drug-metabolizing enzyme was listed in the label. (As 6-MP and 6-TG are identically metabolized, oncology participants were randomized to complete a scenario using one or the other.) Our previous work categorized rules as either low or highly-actionable and, accordingly, created two levels of CDS alerts per drug.
2.4 Participants
Participants were cardiology or oncology fellows, recruited through email by program coordinators not involved in the study. Prospective participants were directed to a website describing the project. Participants signed the online consent form and scheduled a time to complete simulations in a computer laboratory on campus. Each participant who completed the study received a $100 gift card. The UW Human Subjects Committee approved all study procedures.
2.5 Scenarios and Tasks
We developed hypothetical patients and scenarios, crafted such that participants were asked to prescribe drugs, and then to consider modifying the appropriate dose, based on alert messages triggered by the medication ordered and patient-specific laboratory data [18]. Each participant completed the tasks associated with five clinical case scenarios featuring five unique drugs in their clinical discipline. We created discrete prescribing tasks (e.g. ‘find the drug you wish to order in the CPOE system’) for which we provided instructions (Appendices C and D). The first scenario and drug served as the ‘orientation’ scenario and was the same for all participants within each discipline (metoprolol/ tamoxifen). The facilitator guided each participant through each task as a mock-up of a PGx-CDS alert was presented. The second scenario was also the same within each discipline; only the drug with a low-actionable alert was offered (clopidogrel/ nilotinib). We used a pseudo-randomization process to randomize participants to the order of the remaining scenarios and actionability level of alerts.
2.6 Conducting Simulations
Two investigators were present at each simulation, all conducted between April and August 2011. (Appendix C) The facilitator (CLO) read instructions to participants and facilitated completion of all tasks. We used Morae®, a commercially available software package (TechSmith®) [19] to capture audio and on-screen participant activity. The software captured the start and stop time for each task. Each participant was asked to ‘think-aloud’ as they completed the tasks. The observer (EBD) recorded observations and statements made by each participant throughout. We assessed post-simulation satisfaction through online administration of the Post-Study System Usability Questionnaire (PSSUQ; Task 6) [20–23]. This 19-item instrument is comprised of three domains: system usefulness, information quality and interface quality. Finally, we asked participants to record their year in clinical training and their previous experience and comfort using computers, EHRs, and PowerChart.
2.7 Mixed Methods Analyses
We employed the convergent, parallel, mixed methods design, integrating both quantitative and qualitative analyses [24]. For the qualitative analysis, each audio-video recorded simulation provided us with images of all tasks completed, time to completion, types of clinical knowledge resources accessed, participants’ statements and the tasks associated therewith. Two coders (CLO, CJL) mapped statements made by participants and observations recorded by the investigator-observer to evaluation heuristics. Unlike traditional usability studies, we identified ‘positive’ as well as ‘negative’ heuristics; the former demonstrated the features that satisfied the needs of clinicians, while the latter informed possible enhancements to the CPOE system. When concordance between the two coders was not achieved, a third investigator served as arbiter (EBD). All data were downloaded from Morae® and analyzed in Microsoft Excel® (Redmond, WA). The quantitative analysis employed descriptive statistics (medians and interquartile ranges for continuous, and proportions for categorical data). As the PSSUQ is scored on a 7-point Likert scale (1=strongly agree; 7=strongly disagree), we considered it a continuous outcome. Analyses were performed in Excel or Stata 11® (Stata Corp., College Station, TX).
3. Results
3.1 Participants
Seven cardiologists and three oncologists completed the study. All felt comfortable using EHRs and PowerChart and had experience using other EHRs. One-third had previously worked or had a serious hobby in a technology-related field (Table).
Table.
Results - Participant Characteristics, Heuristic Evaluation and Satisfaction
| Cardiology (n=7) |
Oncology (n=3) |
||
|---|---|---|---|
| Participant Characteristics | |||
| Year in fellowship, n (%) | |||
| First | 1/7 (14%) | 0 | |
| Second | 1/7 (14%) | 2/3 (67%) | |
| Third | 3/7 (43%) | 1/3 (33%) | |
| Fourth | 2/7 (29%) | 0 | |
| How comfortable are you using computers?*, median (IQR) | 1.0 (1.0 to 2.0) | 1.0 (1.0 to 1.0) | |
| How comfortable are you using EHRs?*, median (IQR) | 1.0 (1.0 to 1.0) | 1.0 (1.0 to 1.0) | |
| How comfortable are you using PowerChart?*,median (IQR) | 1.0 (1.0 to 2.0) | 1.0 (1.0 to 1.0) | |
| Have used EHRs other than PowerChart, n (%) | 7/7 (100%) | 3/3 (100%) | |
| EHRs previously used** | EpicCare®, CPRS®, Gopher® | EpicCare®, CASMED®, CPRS®, Massachusetts LMR®, Ameritech® | |
| Ever worked or had serious hobby in a technology-related field, n (%) | 2/7 (29%) | 1/3 (33%) | |
| Heuristic Evaluation | |||
| Total (positive, neutral, negative) | Total (positive, neutral, negative) | ||
| Usability heuristics, counts | 117 (38, 42, 37) | 80 (21, 34, 25) | |
| Clinical Knowledge Characteristic heuristics, counts | 132 (33, 63, 36) | 56 (9, 34, 13) | |
| Satisfaction - Post-Study System Usability Questionnaire(PSSUQ) | |||
| Median (IQR) | Median (IQR) | ||
| Total Score (Q1-Q19)± | 1.9 (1.3 to 2.2) | 2.1 (1.3 to 2.4) | |
| System Usefulness (Q1-Q8) | 1.8 (1.1 to 2.0) | 2.0 (1.3 to 2.1) | |
| Information Quality (Q9-Q15) | 2.3 (1.3 to 2.7) | 2.4 (1.3 to 2.7) | |
| Interface Quality (Q16-Q18) | 2.0 (1.0 to 2.7) | 1.7 (1.3 to 2.7) | |
1=very comfortable, 5=not at all comfortable;
Types of EHRs Previously Used: CASMED®: http://www.casmed.com/about-us/partners.cfm; EpicCare®: http://www.epic.com/software-ambulatory.php; Gopher, Regenstrief Medical Record System®: http://clinfowiki.org/wiki/index.php/Regenstrief_Medical_Record_System_(RMRS); Massachusetts General Longitudinal Medical Record (LMR)®: http://www.massgeneral.org/stoecklecenter/programs/patient_exper/lmr.aspx; Meditech®: http://home.meditech.com/en/d/home/; Veterans Affairs Computerized Patient Record System (CPRS)®: http://www.ehealth.va.gov/EHEALTH/CPRS_Demo.asp;
Task 1=Orientation Task; Task 2=Low-actionable alert only; Tasks 3–5=Randomization to low- or high-actionable alert;
Likert Scale for PSSUQ: 1–7, where 1=Strongly agree; 7=Strongly disagree
EHR = electronic health record; IQR = interquartile range; MP = mercaptopurine; Q = question; SD = standard Deviation; TG = thioguanine
3.3 Tasks
The study was comprised of six tasks, each consisting of several steps. As the orientation task, Task 1 consisted of 14 steps. Tasks 2 through 5 consisted of the same five tasks each. Task 6 required completion of the PSSUQ. (Appendix D) Each physician spent an average of between 3.6 to 4.9 minutes per prescribing task exploring the user interface and PGx resources and providing concurrent think-aloud feedback.
3.4 Heuristic evaluation
3.4.1 Quantitative heuristic evaluation
Of the 197 usability heuristics identified, approximately equal numbers were positive, neutral and negative for both specialties, with a few more categorized as neutral (Table). As expected, the greatest numbers of heuristics were attributed to ordering tasks (Appendix D). Eighty-seven percent (171/197) of the usability heuristics were attributed to Visibility of System State (39%), Flexibility and Efficiency (27%), and Match between System and Real World (20%). Example quotes are presented in Appendix E. Of the 188 clinical knowledge characteristics heuristics, approximately equal numbers were positive and negative, with a much greater number being categorized as neutral. Sixty-nine percent (130/188) of these heuristics were attributed to Availability (27%), Trialability (25%), and Relevance to Practice (18%). Example quotes are presented in Appendix F. Based on participant feedback, we were able to identify useful clinical knowledge resources that were not provided in the current study but could possibly be added to the Infobutton or EVIDENCE icon in the future. These were UpToDate®, [25] Micromedex®, [26] Chemoregimen.com®, [27] and ePocrates® [28].
3.4.2 Qualitative heuristic evaluation
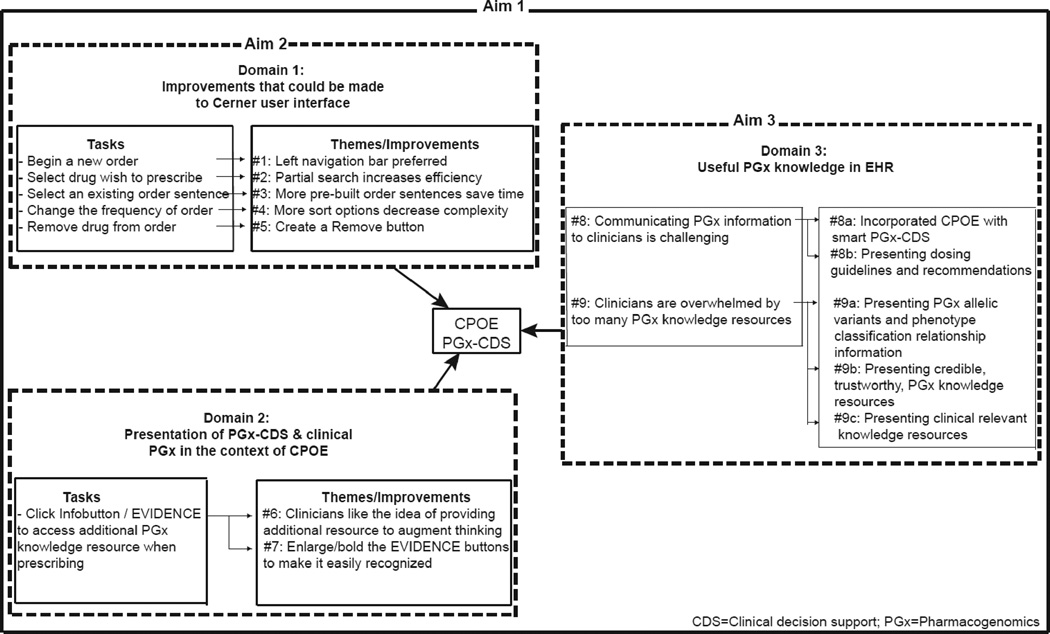
Nine themes and corresponding improvements emerged from the heuristic evaluation. Each theme describes participants’ perceptions, and is grouped into one of three domains described below: 1) Potential improvements to the Cerner CPOE user interface; 2) Presentation of PGx-CDS and clinical PGx in the context of a CPOE system; and 3) Useful PGx knowledge embedded in an EHR (Figure 3).
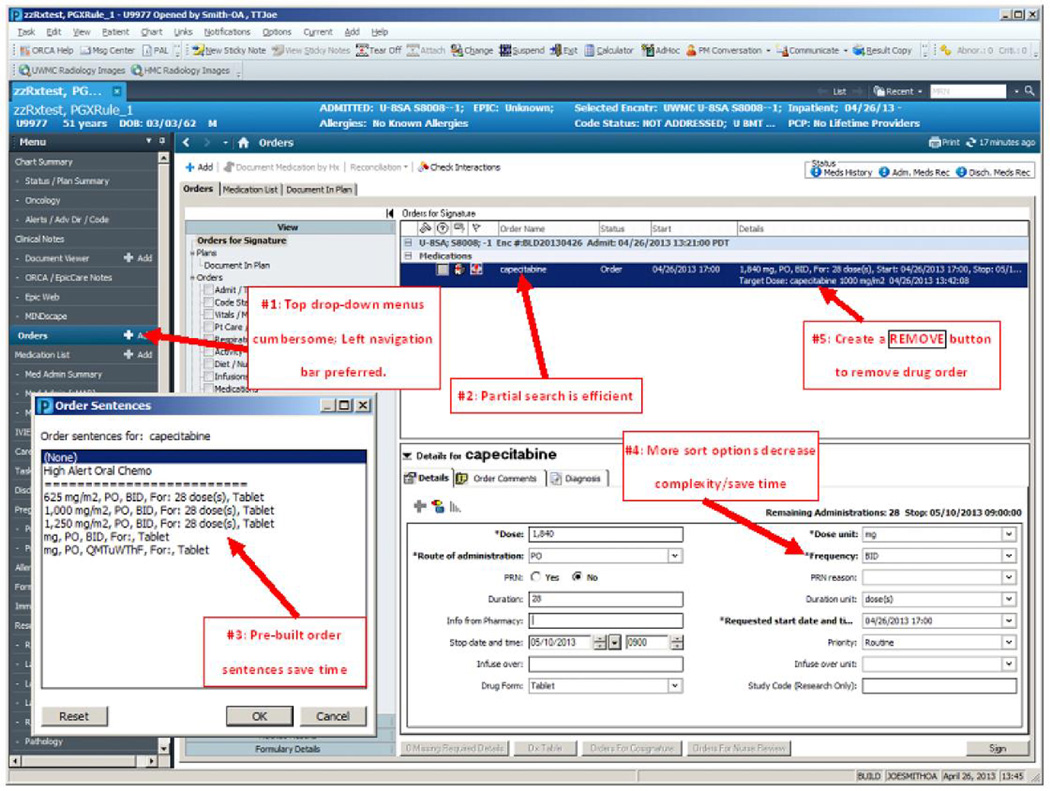
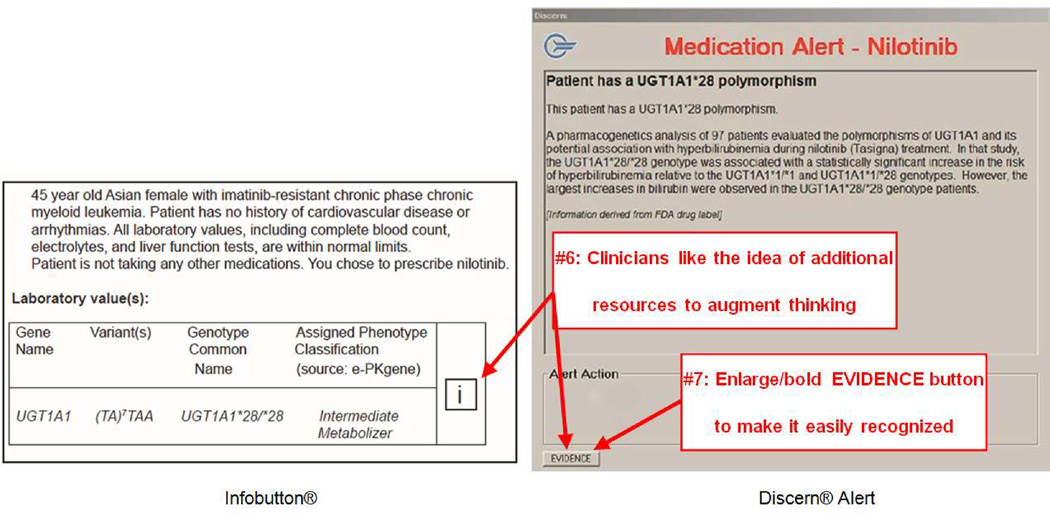
Figure 3.
a: Results – Domain 1: Improvements that could be made to the CPOE user interface PowerChart® – Orders Tab and Orders Details
b: Results – Domain 2: Improvements that could be made to the PGx-CDS presentation
Domain 1:Potential improvements to Cerner® CPOE system user interface (Figure 4a)
Theme #1: Drop-down menus are cumbersome to navigate; left navigation menu bar preferred
All participants were able to quickly select ‘Orders’ and add an order in the CPOE test environment. Some participants browsed and selected functions from the top drop-down navigation menu bar; others from the left menu bar. Prescribing could be more easily and quickly accomplished if the commonly used features/functions were displayed on the left menu bar.
“I usually don’t use these pull-down menus as much, for things that I do rapidly. If every time I had to write an order I had to pull down, it might get cumbersome.” [Cardiologist #5; Metoprolol]
“Order on the left side is pretty intuitive.” [Cardiologist #6; Metoprolol]
Theme #2: Partial drug name search function in ordering system increases efficiency and reduces errors
Participants frequently did not key-in the complete drug name during a search. The drug search function in the CPOE test environment was able to display matches, which alleviated some effort required of participants. However, the system only matched generic names, while participants preferred to type brand names.
“Our order is Plavix®, hopefully they have trade names of the medication… and I don’t think they do. Clopidogrel, I found that. They don’t have the trade name, which stinks because I don’t like typing in clopidogrel.”[Cardiologist #6; Clopidogrel]
Theme #3: More pre-built order sentences save time; avoid free text
Participants were positive about the function that presented standard doses within the “Order Sentences” pop-up box after they selected the drug they wished to prescribe. They stated they would like to have even more pre-built order sentence options pop-up, as long as they fit on one screen. Simply clicking on the dose, rather than typing the dose, or using the “Dose Change” box saved time.
“I like that the standard doses come up [looking at the Order Sentences pop-up box].” [Oncologist #3; Tamoxifen]
Theme #4: Too many dosage change options created complexity when changing dose frequency. More sort options would be helpful to speed up the search process
Almost all participants were unable to change the dose frequency quickly, and were frustrated that too many options were listed. Some participants simply scrolled through all the options and went directly to “Special Instructions”, which allowed them to type in frequencies using free text. This was especially problematic for oncologists. Sorting on specialty or a list of ‘favorites’ would be useful.
“For chemotherapy drugs, it would be nice for a totally different box to come up for the frequencies. Obviously oral chemo wouldn’t utilize that but IV, I think, would. I would worry about this being missed” [Oncologist #3; Irinotecan]
Theme #5: Removing a drug from an order was not always intuitive. Clicking a Remove button to remove the order would be more straightforward
Many participants felt that removing the drug from the system was not straightforward. [It was removed by right clicking on it.]
“I’m not exactly sure how to do that [remove the order]; There might need to be a button explaining that, but a good default is a right click” [Oncologist #3; Tamoxifen]
Domain 2: Presentation of PGx-CDS and clinical PGx in the context of a CPOE system (Figure 4b)
Theme #6: Clinicians liked the knowledge resources
Participants spoke favorably about the value these tools provided in offering a rich and comprehensive set of PGx knowledge resources to augment their current thinking.
“How the drug is metabolized…this is good. I don’t think enough about this in clinical medicine”[Cardiologist #1; Metoprolol; ePKGene]
Theme #7: Icons that identify knowledge resources are not always easily recognized and intuitively presented; icons should be made bold or bigger
The Infobutton and EVIDENCE icons were designed to facilitate access to curated, genomic knowledge resources providing additional information about the PGx information described in the scenario. Several participants did not see the EVIDENCE icon right away.
“Oh, EVIDENCE. I didn’t see the EVIDENCE button. Maybe make it bold or something.” [Cardiologist #5; Metoprolol]
But all participants understood that the Infobutton and EVIDENCE icons were tools by which they could access additional knowledge when needed.
“Honestly, good to know evidence screen is here, but I already have enough information. I don't think I will use this button every time I have a clinical alert [Evidence button]. I would use it in clinical scenarios when I was more unsure.” [Cardiologist #1; Warfarin]
Domain 3: Useful PGx knowledge embedded in an EHR
Theme #8: Communicating genomics and PGx information to clinicians is challenging because clinicians are not trained to interpret genomics and PGx information
Participants were sometimes confused and didn’t know how to interpret genomics and PGx data, which could have impacted patient safety.
“I don’t know how to read this table. I’m confused. It’s confusing because it’s almost indicating that there are 3 alleles. I have no idea how to read this table. So it’s not very helpful to me [referring to the Warfarin dosing table]” [Cardiologist #6; Warfarin; PGx-CDS alert]
To address this challenge, participants identified two potential improvements to the presentation of information: smart PGx-CDS alerts, those that use dosing guidelines and recommendations.
Theme #8a: Smart PGx-CDS alerts facilitate clinical PGx adoption and improve care
Participants felt confident and commonly overrode the CDS alert when prescribing a familiar drug or regimen. But all agreed that PGx-CDS alerts can be very helpful and can positively affect patient care in many ways, especially when clinicians incorrectly interpret genomics and PGx information.
“The alert is good. It raises something I didn’t know about the association with hyperblirubinemia” [Oncologist #3; Irinotecan; PGx-CDS alert]
Many participants talked about alert fatigue when PGx-CDS alerts popped out during the simulations. They expressed that it was overwhelming and annoying when alerts appear for so many medications. They stated that ‘smart’ PGx-CDS alerts should contain not only genetically but also clinically relevant information. Moreover, they felt it would be more convincing if alerts summarized the medical literature and indicated which changes will be important to improve patient outcomes, increase safety, or reduce costs.
“I work with systems where there were a number of pop out text boxes with different medication in order entry. The number of them that pop out will impact how seriously I take them.” [Oncologist #3; Tamoxifen; PGx-CDS alert]
Theme #8b: Busy clinicians want information primarily about dosing guidelines and recommendations
Participants commonly sought additional information when prescribing a medication or regimen with which they were not familiar. Some stated they would talk with more experienced colleagues or pharmacists before prescribing. Others said they would browse resources such as UpToDate®, PubMed®, Micromedex®, and Chemoregimen.com®.
“How often should I dose him if I want to give him Plavix®? That information would be helpful” [Cardiologist #3; Clopidogrel; PGx-CDS alert]
Theme #9: Participants were overwhelmed by too many PGx knowledge resources
After completing the scenarios, participants seemed overwhelmed when they were asked to browse additional information about genomics and PGx knowledge resources.
“This is one big long abstract. Way too much information for a busy day. And so, the summary should be, like, one line” [Cardiologist #4; Warfarin; PLOS]
To address this concern, participants identified three potential improvements in the presentation of information: presentation of phenotype information, credible/trustworthy information, and only clinically relevant information.
Theme #9a: Participants were most interested in the relationship between PGx allelic variants and phenotype classification, as the latter was more helpful to them as they prescribed
Participants were most interested in the phenotype classification for genetic variants and easily interpreted these, even though they were not trained to interpret genomics and PGx data. The following quotes refer to phenotype classifications from a knowledge resource and the phenotype classification listed along with patient laboratory values, respectively.
“Poor metabolizer [reading ‘Assigned Phenotype Classification’ on lab values] Gene name means nothing to me, variants mean nothing to me, genome means nothing to me, poor metabolizer suggests that less of the drug is going to have more of an effect” [Cardiologist #2; Carvedilol, lab value]
Theme #9b: Clinicians want to be able to assess the credibility of knowledge resources
Clinicians were not familiar with all of the knowledge resources that were presented, and indicated they needed to know the source of each type of information in order to assess its credibility and trustworthiness.
“I’m not going to look up all of this information. Usually, maybe in the future, but at this point, I don’t have much of an interest. As long as the information is trustworthy, that’s the biggest thing for making clinical decisions. There’s so much information, it has to be accurate or it’s gonna cause a lot of problems” [Cardiologist #6; Carvedilol; ePKGene]
Theme#9c: Clinicians want solely information that is clinically relevant
Busy clinicians at the point of care indicated they wished to view solely information that is clinically relevant to prescribing the drug at hand.
“The knowledge resources were bad, interesting scientifically but not clinically.” [Cardiologist #4; Warfarin; Survey]
3.6 Satisfaction
For both cardiologists and oncologists, the median total PSSUQ score was approximately 2.0, corresponding with strong agreement that the participants were satisfied with the CPOE system with PGx-CDS alerts. There was less variability in domain responses for cardiologists (1.8–2.3) than oncologists (1.7–2.4). Both cardiologists and oncologists rated system usefulness and interface quality higher than information quality. (Table)
4. Discussion
With the rapid evolution of personalized medicine and increasing reliance on EHRs to reduce medical errors, incorporation of PGx knowledge into decision support systems is a timely challenge. However, the complexity of these systems, the clinical decisions they influence, and the varying skill sets of CPOE users will complicate efforts to properly evaluate the features that lead to effective decision support. Here, we have presented a mixed methods approach to provide a heuristic evaluation of such a system. In a simulated environment, using a commercially available CPOE system with CDS capabilities, we designed and tested an early prototype of patient-specific PGx-CDS alerts in which we embedded a rich collection of PGx clinical knowledge resources. We evaluated the usability of the system, identifying potential improvements to the CPOE system user interface, and understanding contexts in which PGx knowledge could be found useful to clinicians.
Participants were seasoned EHR users and prescribers, although unfamiliar with the Cerner system. The amount of time they spent per task suggests they were genuinely interested in exploring the user interface and took seriously their charge of evaluating the available PGx resources. Spending between 3–5 minutes per prescription order on average is not realistic in clinical practice, but did enable us to gain valuable feedback about both the user interface and the PGx resources. When considering both categories of heuristics (usability and clinical knowledge resources), participants found both positive and negative aspects to each.
Participants identified five potential improvements to the ordering process in the CPOE user interface (domain 1), despite the fact that the system is a commercially available product that is widely used. These improvements would aid in prescribing the correct drug and dose; provide greater flexibility when searching and navigating medications by brand/generic names; and provide a more standardized selection of dosing sentences (drug + dosage form + route), thereby reducing the number of mouse clicks and free text typing. We noted that oncologists had a more difficult time prescribing within the constraints of the existing system than did cardiologists. This makes intuitive sense, as the prescribing of chemotherapy regimens is frequently more complex, and is best facilitated by an oncology-specific CPOE module. (Ours has since been implemented.)
Participants agreed with the idea of providing additional resources to guide drug dosing, and made use of the clinical knowledge resources behind the Infobutton and EVIDENCE icons to achieve this end (domain 2) [29]. Dissatisfaction with the size or placement of icons shows the importance of user-centered design to minimize errors and slips in utilization of the interface, particularly outside of a controlled environment.
The vision of PGx is to prescribe the right drug in the right dose to the right patient at the right time. To do so, PGx information must be presented in a useful fashion (domain 3) and on demand [29,30]. Although our results suggest that incorporating PGx resources into smart PGx-CDS alerts is key to increasing PGx adoption into daily practice and improving patient safety [31], we learned that presenting PGx information to clinicians is challenging, and that presenting too much information overwhelms participants. Clinically relevant PGx knowledge must be represented in language the physicians find intuitive and easily interpretable [32,33]. In order to avoid alert fatigue, rules for alerts must also be carefully developed and triggered such that recommended actions are clear and facilitate a rapid response [32,34]. PGx data are complex; creating the appropriate infrastructure to communicate this information is crucial. [35] Our results suggest that the use of simple dosing guidelines and recommendations would be well received.
Participants indicated they had not been trained to interpret genomic data and infrequently experienced PGx-based drug prescribing; all indicated they were most interested in the relationship between genotype and phenotype, as the phenotype was more helpful to them as they prescribed. Indeed, others have shown that the overall knowledge level of genetics of many health care providers is low. [36] A survey conducted by the American Medical Informatics Association (AMIA), in collaboration with Medco Health Solutions, Inc.® (now Express Scripts®), found that although 98% of provider participants agreed about the utility of genetic testing in drug therapy, only 10% actually felt that they had been adequately informed about the process. [34] Results suggest that prescribers would benefit from education focused on presentation, interpretation, and clinical applicability of PGx information. Further, from the physician’s point of view, it is important that the selected PGx knowledge is from credible, trustworthy and familiar sources. Separately, in taking an important step to optimize patient safety and quality of care, an AMIA task force has recently published ten recommendations, in four domains, for improving the usability of EHRs. The first domain is the usability and human factors research agenda. [37] Our usability study contributes to this research agenda.
A unified framework to assess EHR usability exists, and consists of four components: Task, User, Representation, and Function – TURF. [38] However, this framework is intended for use in a more general way, and at a more conceptual level, rather than in an individual study. For this reason, we used the TURF framework as background information but focused on our evaluation of heuristics and assessment of participant satisfaction with the existing CPOE system.
Ours was a pilot study, utilizing one EHR in the inpatient setting to study participant responses, in depth. The literature suggests that ten is a sufficient number of participants to adequately identify up to 80% of usability issues. [39,40] Our participants did identify the same issues repeatedly, although we cannot be totally confident that we reached saturation. A greater number and richer mix of participants might have revealed additional insights. Despite the rigorous, repeated recruitment and motivational ($100) efforts we made over a number of months, no additional participants volunteered. As the CPOE system is already in use, we were unable to influence its design, but we are able to forward recommendations to medical center information technology professionals who are equipped to modify features and functionality of the system.
To our knowledge, this is the first usability study to evaluate PGx-CDS alerts in a commercial CPOE system. In evaluating five clinical case scenarios in each of two medical disciplines, we had an opportunity to explore and understand physicians’ expectations of the provision of PGx-CDS alerts presented in a CPOE system, and their perspective on PGx-based drug prescribing. In time, using PGx data at the point of prescribing will improve medication safety and overall quality of care. Correctly configuring functionality and selectively presenting the most important information in a readily interpretable format will be required. Our work has helped to inform this nascent field.
Supplementary Material
Figure 2.
Heuristic Evaluation Results
Highlights.
CPOE with PGx-CDS can improve medication safety.
Usability can drive adoption; few evaluations have been completed.
We conducted a usability study with cardiologists and oncologists.
Results suggest design improvements could be made to the user interface.
Provision of PGx information is important; must be presented in intuitive ways.
Summary Points.
What was already known on the topic
The mapping of the human genome in 2003 opened new avenues of research and launched the era of personalized medicine, including pharmacogenomics (PGx).
Health information technology is an indispensable support to facilitate diffusion of information to inform clinical practice. Specifically, computerized prescriber order entry (CPOE) with clinical decision support (CDS) is a sophisticated technology that can improve practitioner performance and medication safety.
Although increasing evidence has demonstrated that PGx-based drug dosing is useful in improving safety, the rate of adoption has been low.
Usability is one of the key factors driving adoption and appropriate utilization of electronic health records, and can be applied to development of CPOE/CDS systems.
What this study adds to our knowledge
In this study we evaluate the usability of PGx-CDS alerts in the context of a commercially available CPOE system. [11] We create clinical scenarios and tasks featuring the prescribing of cardiology drugs and oncology drugs by physician participants. Results suggest:
Design improvements could be made to the user interface of the existing CPOE system, in order to facilitate the incorporation of PGx dosing into clinical care.
Both cardiologists and oncologists agree that it is important to provide information about the association between genetic variants and potential drug dose modifications.
Communicating genomics and PGx information to clinicians is challenging and can be overwhelming. Clinically relevant PGx information in CDS alerts should be presented as simple dosing guidelines and recommendations from credible sources, and communicated by phenotype (e.g. poor metabolizer) rather than genotype.
Acknowledgements
This work was sponsored by AHRQ 5K08 HSO14739 (PI: Devine);
NIH/NLM T15 LM07442; NIH/NHGRI T15 Hg000035; NIH NCCR UL1RR025014 (Overby). The funders played no role in the study design, data collection, analysis and interpretation of data, writing the manuscript, or decision to submit the manuscript for publication.
The authors wish to thank Mervin Johnsingh for his aid in setting up the simulations; Bernardo Goulart and Veena Shankaran for pilot testing the oncology simulations; Lingtak-Neandar Chan and Daniel Capurro for pilot testing the cardiology simulations; Isabelle Ragueneau-Majlessi, Cathy Yeung, and Sophie Argon for providing pilot feedback on genetic laboratory values used in the simulations; Isabell Ragueneau-Majlessi (UW Pharmaceutics, PI: e-PKgene Project) for allowing us to use e-PKgene in our research; Cathy Yeung (UW Pharmaceutics e-PKGene) for assisting with developing ePKgene resources specific to this project; Pamela Wareham (UW Medicine Pharmacy Informatics, Associate Director, Pharmacy) for allowing members of her group to contribute time to this project; and Guilherme Del Fiol (University of Utah and Duke) for his assistance with OpenInfobutton® configuration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors Contributions
Emily Beth Devine: Conceived and designed the study, collected, analyzed and interpreted the data, wrote the manuscript, and jointly with Dr. Overby decided on journal for submission.
Casey L. Overby: Conceived and designed the study, collected, analyzed and interpreted the data, edited the manuscript for critically important content, and jointly with Dr. Devine decided on journal for submission.
Chia-Ju Lee: Analyzed and interpreted the data, drafted the manuscript and revised it for critically important content, and approved the final version.
Neil Abernethy: Assisted in study design, interpreted the data, critically revised the manuscript for critically important content, and approved the final version.
Jeannine McCune: Designed the study, interpreted the data, revised the manuscript for critically important content, and approved the final version.
Joe W. Smith: Designed the study, revised the manuscript for critically important content, and approved the final version.
Peter Tarczy-Hornoch: Contributed to conception and design of the study, revised the manuscript for critically important content, and approved the final version.
Conflict of Interest Statement
Emily Beth Devine: has no conflicts to declare
Casey L. Overby: has no conflicts to declare
Chia-Ju Lee: has no conflicts to declare
Neil Abernethy: has no conflicts to declare
Jeannine McCune: has no conflicts to declare
Joe W. Smith: has no conflicts to declare Peter
Tarczy-Hornoch: has no conflicts to declare
References
- 1.Green ED, Guyer MS. National Human Genome Research Institute. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470(7333):204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 2.Realizing the Potential of Pharmacogenomics. [Accessed: March 14, 2014];Opportunities and Challenges. Report of the Secretary’s Advisory Committee on Genetics, Health, and Society. 2008 May; Available from: http://osp.od.nih.gov/sites/default/files/SACGHS_PGx_report.pdf.
- 3.United States Food and Drug Administration. [Accessed: March 14, 2014];Table of Pharmacogenomic Biomarkers in Drug Labels. Available from: http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm.
- 4.Stanek EJ, Sanders CL, Teagarden JR, Johansen KA, Aubert RE, Agatep BC, Khalid M, Patel A, Frueh FW, Epstein RS. Presented at the 59th Annual Meeting of the American Society of Human Genetics. Honolulu, Hawaii: 2009. Oct, Who are adopters of pharmacogenomics among U.S. physicians? abstract. [Google Scholar]
- 5.Sanders CL, Stanek EJ, Teagarden JR, Agatep BC, Johansen KA, Aubert RE, Khalid M, Patel A, Frueh FW, Epstein RS. Honolulu, Hawaii: Presented at the 59th Annual Meeting of the American Society of Human Genetics; Oct, 2009. Putting knowledge into practice: well informed physicians are twice as likely to order or recommend pharmacogenetic testing. abstract. [Google Scholar]
- 6.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawamoto K, Lobach DF, Willard HF, Ginsburg GS. A national clinical decision support infrastructure to enable the widespread and consistent practice of genomic and personalized medicine. BMC Med Inform Decis Mak. 2009 Mar 23;9:17. doi: 10.1186/1472-6947-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott GP, Shah P, Wyatt JC, Makubate B, Cross FW. Making electronic prescribing alerts more effective: scenario-based experimental study in junior doctors. J Am Med Inform Assoc. 2011;18:789–798. doi: 10.1136/amiajnl-2011-000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horsky J, Phansalkar S, Desai A, Bell D, Middleton B. Design of decision support interventions for medication prescribing. Int J Med Inform. 2013;82:492–503. doi: 10.1016/j.ijmedinf.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Chan J, Shojania KG, Easty AC, Etchells EE. Does user-centered design affect the efficiency, usability and safety of CPOE order sets? J Am Med Inform Assoc. 2011;18:276–281. doi: 10.1136/amiajnl-2010-000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Johnson TR, Patel VL, Paige DL, Kubose T. Using usability heuristics to evaluate patient safety of medical devices. J Biomed Inform. 2003;36:23–30. doi: 10.1016/s1532-0464(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen J. Usability engineering. Boston: AP Professional; 1994. [Google Scholar]
- 13.Shneiderman B. Designing the user interface. 3rd ed. Reading, Massachusetts: Addison-Wesley; 1998. [Google Scholar]
- 14.Curran JA. Development of a knowledge exchange and utilization model for emergency practice [dissertation] Halifax, NS, Canada: Dalhousie University; 2009. [Google Scholar]
- 15.Rogers EM. Diffusion of Innovations. 5th ed. New York: Free Press; 2003. Note: describes 3 of the 7 characteristics – complexity, trialability and compatibility. [Google Scholar]
- 16.Estabrooks CA, Wallin L, Milner M. Measuring knowledge utilization in health care. Int J Policy Eval Manage. 2003;1:3–36. [Google Scholar]
- 17.Overby CL, Devine EB, Tarczy-Hornoch P, Kalet Deriving rules and assertions from pharmacogenomic knowledge resources in support of patient drug exposure predictions. J Am Med Inform Assoc. 2012;19:840–850. doi: 10.1136/amiajnl-2011-000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overby CL, Devine EB, Abernethy N, McCune JS, Tarczy-Hornoch P. Making pharmacogenomic-based prescribing alerts more effective: a scenario-based experimental study with physicians. Int J Med Inform. doi: 10.1016/j.jbi.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morae®, by TechSmith. [Accessed: March 14, 2014]; Available from URL: http://techsmith.com/morae.html.
- 20.Lewis JR. Psychometric evaluation of the Post-Study System Usability Questionnaire: The PSSUQ; Proceedings of the Human factors Society 36th Annual Meeting; 1992. [Google Scholar]
- 21.Lewis JR. IBM computer usability satisfaction questionnaires: Psychometric evaluation and instructions for use. [Accessed: March 14, 2014];Technical report 54.786. IBM Corporation. 1993 Available from URL: http://drjim.0catch.com/usabqtr.pdf.
- 22.Lewis JR. Psychometric evaluation of the PSSUQ using data from five years of usability studies. [Accessed: March 14, 2014];International Journal of Human-Computer Interaction. 2002 14:463–488. Available from URL: http://drjim.0catch.com/PsychometricEvaluationOfThePSSUQ.pdf. [Google Scholar]
- 23.Fruhling A, Lee S. Assessing the reliability, validity and adaptability of the PSSUQ; AMCIS 2005 Proceedings; [Accessed: March 14, 2014]. Available from URL: http://aisel.aisnet.org/amcis2005/378/. [Google Scholar]
- 24.Creswell JW, Klassen AC, Clark VL, Smith KC. Office of Behavioral and Social Sciences Research. National Institutes of Health; [Accessed: March 14, 2014]. Best practices for mixed methods research in the health sciences. Available from URL: http://obssr.od.nih.gov/mixed_methods_research/pdf/Best_Practices_for_Mixed_Methods_Research.pdf. [Google Scholar]
- 25.UpToDate®. [Accessed: March 14, 2014];Walters Kluwer Health. Available from URL: http://www.uptodate.com/home/about-us.
- 26.Micromedex 2.0®. [Accessed: March 14, 2014];Truven Health Analytics.©2013. Available from URL: http://www.micromedex.com/
- 27.Chemoregimen.com®. [Accessed: March 14, 2014];PO Box 3852. South Pasadena, CA, USA 91031. Available from URL: http://chemoregimen.com/Home-28.html.
- 28.ePocrates® Inc. [Accessed: March 14, 2014];Copyright © 2013. Available from URL: http://www.epocrates.com/mobile?CID=PPC-Brand-IMEpocratesPPC-Brand-Content12&gclid=CNvinOy787UCFag7MgodpAoAPA.
- 29.Chaffee BW, Zimmerman CR. Developing and implementing clinical decision support for use in a computerized prescriber-order-entry system. Am J Health-Syst Pharm. 2010;67(5):391–400. doi: 10.2146/ajhp090153. [DOI] [PubMed] [Google Scholar]
- 30.Crockett DK, Ridge PG, Wilson AR, Lyon E, Williams MS, Narus SP, Facelli JC, Mitchell JA. Consensus: a framework for evaluation of uncertain gene variants in laboratory test reporting. Genome Med. 2012 May 28;4(5):48. doi: 10.1186/gm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheuner MT, de Vries H, Kim B, Meili RC, Olmstead SH, Teleki S. Are electronic health records ready for genomic medicine? Genet Med. 2009;11:510–517. doi: 10.1097/GIM.0b013e3181a53331. [DOI] [PubMed] [Google Scholar]
- 32.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, Delaney JT, Bowton E, Brothers K, Johnson K, Crawford DC, Schildcrout J, Masys DR, Dilks HH, Wilke RA, Clayton EW, Shultz E, Laposata M, McPherson J, Jirjis JN, Roden DM. Operational Implementation of Prospective Genotyping for Personalized Medicine: The design of the Vanderbilt PREDICT Project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li AC, Kannry JL, Kushniruk A, Chrimes D, McGinn TG, Edonyabo D, Mann DM. Integrating usability testing and think-aloud protocol analysis with “near-live” clinical simulations in evaluating clinical decision support. Int J Med Inform. 2012;81:761–772. doi: 10.1016/j.ijmedinf.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Wilke RA, Xu H, Denny JC, Roden DM, Krauss RM, McCarty CA, Davis RL, Skaar T, Lamba J, Savova G. The emerging role of electronic medical records in pharmacogenomics. Clin Pharmacol Ther. 2011;89:379–386. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neri PM, Pollard SE, Volk LA, Newmark LP, Varugheese M, Baxter S, Aronson SJ, Rehm HL, Bates DW. Usability of a novel clinician interface for genetic results. J Biomed Inform. 2012;45:950–957. doi: 10.1016/j.jbi.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baars MJ, Henneman L, Ten Kate LP. Deficiency of knowledge of genetics and genetic tests among general practitioners, gynecologists, and pediatricians: a global problem. Genet Med. 2005;7:605–610. doi: 10.1097/01.gim.0000182895.28432.c7. [DOI] [PubMed] [Google Scholar]
- 37.Middleton B, Bloomrosen M, Dente MA, Hasmat B, Koppel R, Overhage JM, Payne TH, Rosenbloom ST, Weaver C, Zhang J. Enhancing patient safety and quality by improving the usability of electronic health record systems: recommendations from AMIA. J Am Med Inform Assoc. 2013;0:1–7. doi: 10.1136/amiajnl-2012-001458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Walji MJ. TURF: Toward a unified framework of EHR usability. J Biomed Inform. 2011;44:1056–1067. doi: 10.1016/j.jbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Virzi RA. Refining the test phase of usability evaluation: How many subjects is enough? Human Factors: The Journal of the Human Factors and Ergonomics Society. 1992;34:457–468. [Google Scholar]
- 40.Johnson CD, Zeiger RF, Das AK, Goldstein MK. Task analysis of writing hospital admission orders: evidence of a problem-based approach. AMIA Symposium Proceedings. 2006:389–393. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.