Abstract
Neuroblastoma is a common childhood tumor and accounts for 15% of pediatric cancer deaths. To investigate the microRNA (miRNA) profile and role of Dicer and Drosha in neuroblastoma, we assessed the expression of 162 human miRNAs, Dicer and Drosha in 66 neuroblastoma tumors by using real-time PCR methods. We found global downregulation of miRNA expression in advanced neuroblastoma and identified 27 miRNAs that can clearly distinguish low- from high-risk patients. Furthermore, expression levels of Dicer or Drosha were low in high-risk neuroblastoma tumors, which accounted for global downregulation of miRNAs in advanced disease and correlated with poor outcome. Notably, for patients with non–MYCN-amplified tumors, low expression of Dicer can serve as a significant and independent predictor of poor outcome (hazard ratio, 9.6; P = 0.045; n = 52). Using plausible neural networks to select a combination of 15 biomarkers that consist of 12 miRNAs' signature, expression levels of Dicer and Drosha, and age at diagnosis, we were able to segregate all patients into four distinct patterns that were highly predictive of clinical outcome. In vitro studies also showed that knockdown of either Dicer or Drosha promoted the growth of neuroblastoma cell lines. Our results reveal that a combination of 15 biomarkers can delineate risk groups of neuroblastoma and serve as a powerful predictor of clinical outcome. Moreover, our findings of growth promotion by silencing Dicer/Drosha implied their potential use as therapeutic targets for neuroblastoma.
Introduction
microRNAs (miRNAs) are a class of small, evolutionarily conserved, endogenous noncoding RNAs that act as negative regulators of gene expression by inhibiting translation or promoting RNA degradation. miRNAs are transcribed by RNA polymerase II to generate the primary miRNA transcripts, which are processed into the ~70-nt hairpin structured pre-miRNA by Drosha, a RNase III endonuclease in the nucleus. After being transported to cytoplasm by exportin 5, pre-miRNA is further processed by another RNase III endonuclease, Dicer, to generate the ~22-nt mature miRNA (1). Recent studies have shown that miRNAs are involved in the regulation of multiple physiologic processes, including apoptosis, proliferation, and differentiation, and are implicated in the pathogenesis of various diseases. In cancer, miRNA have been shown to function as both tumor suppressors and oncogenes by targeting genes that are critical regulators in cancer development, and are also shown to be useful for cancer classification and prognostication (2). Furthermore, the components of miRNA biogenesis machinery, including Drosha and Dicer, have been implicated in tumorigenesis (3–10). These findings suggest that miRNAs may serve as both prognostic markers and therapeutic targets.
Neuroblastoma is a common childhood tumor derived from primitive sympathetic neuroblasts and accounts for 15% of pediatric cancer deaths. The hallmark of this tumor is its plethoric clinical behavior, ranging from malignant tumor progression to spontaneous regression or differentiation to benign ganglioneuroma (11). The biological variables, such as genomic amplification of the MYCN oncogene, allelic loss of chromosome 1p, 3p, 11q, and 14q, gain of 17q, and differential expression of neurotrophin receptors TrkA/TrkB have been associated with tumor behavior (12). However, the key prognostic indicator in neuroblastoma is MYCN amplification, which occurs in about 20% of cases and is associated with high tumor stage and poor outcome (13, 14). Currently, treatment of neuroblastoma patients is tailored to risk group assignment according to well-known prognostic factors in addition to MYCN amplification, such as patient age at diagnosis, International Neuroblastoma Staging System (INSS) stage, tumor histopathology, and DNA index (15, 16). Although risk group assignment is useful for therapeutic stratification, there is still a lack of reliable markers for predicting treatment failure in neuroblastoma patients. Thus, it will be important to search for neuroblastoma-specific prognostic markers that can provide better prediction of tumor behavior and refine risk assessment.
Recent microarray studies of neuroblastomahave shown that gene expression profile may be useful for molecular classification and clinical prognostication (17–19). Emerging evidence suggests that miRNA expression profiling may add another dimension to sort out subtypes of various cancers (20). In this study, we delineated miRNA expression profiling of neuroblastoma and assessed its usefulness in clinical prognostication, as well as its role in tumorigenesis of neuroblastoma.
Materials and Methods
Neuroblastoma samples and cell lines
A total of 66 neuroblastoma primary tumors with a tumor cell content of ≥80%were obtained from three sources: the Children's Oncology Group, the Pediatric Oncology Group, and the Cooperative Human Tissue Network collected from 1986 to 1995. The samples were fully encoded and examined under a protocol approved by the Institutional Review Board of Human Subjects Research Ethics Committee (University of California San Diego, La Jolla, CA). The clinicopathologic information is listed in Table 1 and Supplementary Table S1. The cohort was representative of the population with neuroblastoma in general (21). The 5-year event-free survival was 50.8% with a median follow-up time of 5.33 years, and the 5-year overall survival rate was 56.9% with a median follow-up time of 6.08 years. The neuroblastoma cell lines Be2C, NMB7, and NB5 were maintained as described previously (22, 23). Total RNA was isolated using TRIZOL reagent (Invitrogen) according to the manufacturer's instructions.
Table 1.
Relationships between expression levels of Dicer and Drosha and various clinicopathologic characteristics
| Characteristics | Cases | Dicer | Drosha | ||||
|---|---|---|---|---|---|---|---|
| High | Low | P* | High | Low | P* | ||
| Sex | |||||||
| Male | 35 | 23 | 12 | 1.0 | 17 | 18 | 1.0 |
| Female | 23 | 15 | 8 | 12 | 11 | ||
| Age at diagnosis | |||||||
| <1.5 years | 43 | 36 | 7 | <0.001 | 22 | 21 | 0.301 |
| ≧1.5 years | 22 | 8 | 14 | 8 | 14 | ||
| INSS stage | |||||||
| 1 | 7 | 4 | 3 | 0.038† | 5 | 2 | 0.006† |
| 2 | 17 | 14 | 3 | 10 | 7 | ||
| 3 | 12 | 9 | 3 | 6 | 6 | ||
| 4 | 22 | 10 | 12 | 4 | 18 | ||
| 4S | 7 | 7 | 0 | 5 | 2 | ||
| Risk | |||||||
| Low and intermediate | 40 | 32 | 8 | 0.013 | 26 | 14 | <0.001 |
| High | 25 | 12 | 13 | 4 | 21 | ||
| Histology | |||||||
| Favorable | 29 | 24 | 5 | 0.004 | 16 | 13 | 0.079 |
| Unfavorable | 18 | 7 | 11 | 5 | 13 | ||
| MYCN | |||||||
| Nonamplified | 52 | 37 | 15 | 0.321 | 28 | 24 | 0.015 |
| Amplified | 13 | 7 | 6 | 2 | 11 | ||
Two-sided Fisher's exact test.
Comparing stage 1, 2, and 4S with stage 3 and 4.
Real-time PCR quantification of miRNAs and computational analysis
Expressions of 162 mature miRNAs in 66 human neuroblastoma patients were analyzed by TaqMan miRNA Assays Human Panel-Early Access Kit (Applied Biosystems) according to the manufacturer's protocol. Briefly, gene-specific reverse transcription was performed for each miRNA from 2.5 ng of total RNA in 15 µL reaction volume by using Taq-Man miRNA Reverse Transcription Kit, followed by q-PCR amplification using sequence-specific primers from TaqMan miRNA assays Human Panel on the 7300 Sequence Detection System (Applied Biosystems). Total RNA input was normalized based on threshold cycle (Ct) values of common internal control for miRNA quantification assays, U6 rRNA (24, 25), and all Ct values ≥36, which were considered as not expressed, were adjusted to 36.
Data analysis was performed by using GENESPRING software (version 7.2, Silicon Genetics). First, data transformation was set to real-time PCR and then normalization was performed by using U6, allowing comparison among samples. To highlight miRNAs that characterize each clinicopathologic factors, a per-gene on median normalization was first performed with subsequent statistical comparisons performed by ANOVA with the Benjamin and Hochberg correction for false positive reduction. Hierarchical clustering for both miRNAs and conditions was generated by using standard correlation as a measure of similarity.
Further analyses to identify differentially expressed miRNAs among risk and classified samples were performed by using the prediction analysis of microarray (PAM) algorithm (26). We identified the miRNAs that result in best-risk tumor classification across all samples by using the method of the nearest shrunken centroids, as implemented in PAM. The prediction error was calculated by means of 10-fold cross-validation and then the miRNAs were selected yielding the minimum misclassification error.
Real-time reverse transcriptase-PCR of Dicer and Drosha
Total RNA was reverse transcribed to cDNA using Super-Script First-Strand Synthesis System with random hexamer primers (Invitrogen). Real-time quantitative PCR was performed using cDNA transcribed from 10 ng total RNA, 1× SYBR Green Master Mix (Applied Biosystems), and either Dicer, Drosha, or GAPDH primers as previously described (4) on an Applied Biosystems PRISM 7300-HT. All reactions were run in triplicate with expression levels normalized against GAPDH.
Plausible neural network analysis
Plausible neural networks (PNN; refs. 27, 28) is an intelligent self-organizing neural networks system. PNN performs unsupervised learning, associative memory, clustering, classification, function approximation, and belief judgment in single-network architecture. One specific advantage of PNN is the capability to deal with all kinds of measurement simultaneously. PNN was trained using all of the neuroblastoma patient information and biomarkers including Dicer, Drosha, and miRNAs to form multivariate patterns. PNN performs feature selection by calculating the joint mutual information relationship between the biomarkers and the patterns. With these features, PNN was used to perform cluster analysis and predict the patients' clinical outcome.
Short hairpin RNA design and transfection
All the short hairpin RNA (shRNA) clones were obtained from National RNAi Core Facility (Genomics Research Center, Academia Sinica). shRNA against target sequence was 5 ′-GCTGGCTGTAAAGTACGACTA-3′ (clone ID TRCN0000051262) for human Dicer (shDicer), 5′-GCCAGATGAGACTGAAGACAT-3′ (TRCN0000022253) for Drosha (shDrosha), and 5′- TCCTAAGGTTAAGTCGCCCTCG-3′ (TRCN0000072243) for firefly luciferase as the negative control (shLuc). The shRNAs plasmids were transfected into neuroblastoma cell lines with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Knockdown efficiency of Dicer and Drosha was confirmed by quantitative reverse transcriptase-PCR and Western blot analysis at 48 to 72 hours posttransfection.
Cell proliferation and soft-agarose colony forming assay
Cells were seeded at a density of 5 × 103 in a 96-well plate 24 hours before transfection with different shRNAs or negative control shLuc plasmids using Lipofectamine 2000 (Invitrogen). Forty-eight hours posttransfection, proliferation rates were determined daily by incubation in 10% alamarBlue for 4 hours and followed by measurement of fluorescence with excitation at 544 nm and emission at 590 nm by spectrophotometer (Spectramax 190, Molecular Devices).
For soft-agarose colony-forming assay, 60-mm dishes were coated with 0.6% agarose in RPMI-1640 supplemented with 10% fetal bovine serum (FBS). This layer was then overlaid with a suspension of 1,000 cells, which were transfected with shRNA for 48 hours, in 0.36% agarose/RPMI/FBS and allowed to grow for two weeks at 37°C. The cell colonies >1 mm in diameter were counted after staining with 5% MTT for 15 minutes. Experiments were performed in triplicate using three neuroblastoma cell lines (Be2C, NMB7, and NB5).
Statistical analysis
The best cutoff value for separating two groups in terms of gene expression levels (Dicer, Drosha) were determined by Student's t-test; these values were found to be −4.5 and −5.13 for Dicer and Drosha, respectively, which are close to their mean values (−4.2 and −5.13, respectively). The association between various clinical characteristics and expression levels of Dicer and Drosha were examined by Fisher's exact test. Event-free survival (EFS) and overall survival (OS) were estimated with the Kaplan-Meier method and were compared by log-rank test using GraphPad Prism software (version 4, GraphPad Software). Cox regression model was used for analysis of factors potentially related to EFS or OS. All statistical analyses were performed with SAS software (version9.0, SAS Institute Inc). A P value <0.05 was considered as significant.
Results
Global downregulation of miRNA expression profile in advanced neuroblastoma tumors
The expression levels of 162 miRNAs in 66 primary neuroblastoma samples were quantified by real-time PCR method using TaqMan MiRNA Assay kit. Global miRNA expression profiles were generated using unsupervised agglomerative hierarchical clustering and are shown in Supplementary Fig. S1. Notably, 23 of the 162 miRNAs were not detectable in any neuroblastoma sample. In stage 4 tumors, most of the remaining miRNAs were also downregulated, especially in those with MYCN amplification. Overall 33 miRNAs were differentially expressed among different stages (P < 0.01, Supplementary Table S2), and miRNA profiles could discriminate tumors with low- or intermediate-risk features from most of stage 4 MYCN-amplified tumors, which were clustered together (Supplementary Fig. S1).
miRNA signature can discriminate high-risk neuroblastoma patients from low-risk group
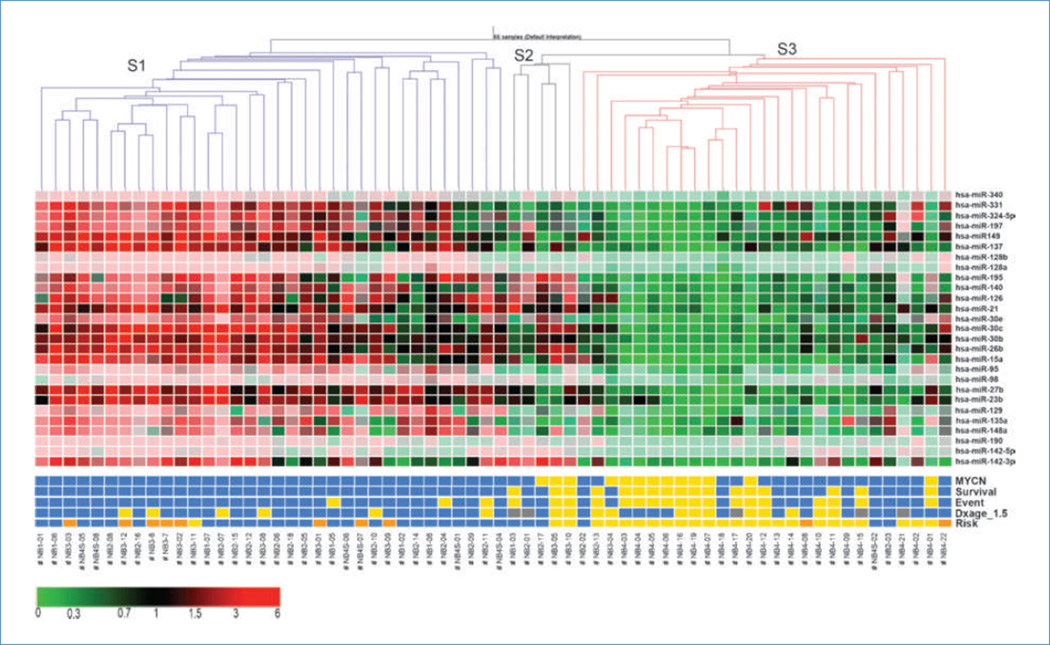
To identify the smallest set of miRNAs predictive of neuroblastoma risk group, we used the PAM algorithms to find 27 miRNAs that could discriminate between high- and low-risk groups. These miRNAs also have ANOVA P value of <0.001 in risk examination (Supplementary Table S3). Based on these 27 selected miRNAs, PAM correctly classified 23 of the 25 (92%) high-risk samples and 26 of the 31 (84%) low-risk samples. Using this signature, 9 of 10 intermediate-risk samples were classified to low risk and one stage 4 sample was grouped to high risk, and all 10 patients with intermediate risk had good outcome. In addition, cluster analysis based on these 27 selected miRNAs generated a tree that could discriminate risk groups and predict clinical outcome (Fig. 1). The tree consisted of a left branch (S1) and a right branch which could be subdivided into S2 and S3. The S1 branch was composed almost exclusively of cases without features of aggressive neuroblastoma and all survived (n = 34). None of these patients harbored MYCN-amplified tumors and 28 of the 34 cases were diagnosed at <1.5 years. The patients in this cluster belonged to stage 1, 2, 3, and 4S; all of them except for those stage 3 were classified as low risk by the current Children's Oncology Group system. Among 12 stage 3 patients, 7 were classified as intermediate risk and 1 patient with high risk fell into this cluster, whereas the other 4 cases classified as high risk did not. Furthermore, six of the seven stage 4S fell into S1 and one into S3 group, but all survived.
Figure 1.
Hierarchical clustering of 66 primary neuroblastoma samples for 27 miRNAs selected, showing three main samples groups, S1 (blue), S2 (black), and S3 (red), using standard correlation as measure of similarity. Columns, samples; rows, miRNAs. Color scale, expression level relative to mean expression of a miRNA across all samples (red, high expression; green, low expression). Clinical characteristics: yellow, MYCN amplification, dead, event, age at diagnosis (Dxage) = 1.5–5 years, and high-risk; blue, MYCN not amplified, alive, no event, age at diagnosis <1.5 years, and low risk; gray, age at diagnosis >5 years; orange, intermediate risk. Neuroblastoma samples were coded as #NBx-01, NBx-02, etc., where × refers to INSS stage.
On the other hand, the right branch (S2 + S3) consisted of cases that were nearly all associated with more advanced disease and poor outcome. The smaller subbranch S2 (n = 5) included three cases with MYCN amplification at stage 2 or 3 and one case at stage 1 who died. The other subbranch S3 (n = 27) consisted of all stage 4 patients (n = 22). Remarkably, seven of the nine samples with stage 4 and MYCN amplification status were exclusively clustered together under this subbranch.
Our findings suggest that miRNA expression profile is significantly correlated with clinical and biological features of neuroblastoma.
Downregulation of Dicer and Drosha in advanced neuroblastoma
From the above cluster tree view and statistical analysis, the most prominent feature of miRNA expression profile was a widespread downregulation in high-risk samples, especially stage 4 tumors. Because Dicer and Drosha are essential for miRNA biogenesis and alterations of their expression levels have been reported in some human tumors (4, 6, 7, 29), we explored the possibility that global miRNA downregulation in stage 4 neuroblastoma could be due to reduced expression of Dicer and/or Drosha. With the use of real-time reverse transcriptase-PCR, Dicer and Drosha expression were determined in 65 of 66 neuroblastoma samples (one sample had insufficient RNA). As expected, expression levels of Dicer and Drosha were significantly lower in stage 4 (P < 0.05) than in other stages (except Dicer expression in stage 1), with the most striking difference in Dicer expression between stage 4 and 4S (P < 0.001), whereas no statistically significant difference among neuroblastoma tumors of other stages was noted (Supplementary Fig. S2). By probing the GSE13136 microarray dataset, we also observed lower expression of Drosha and Dicer in stage 4 neuroblastoma, although significant difference was only noted for Drosha comparing stage 4 versus non–stage 4 samples (Supplementary Fig. S3).
For the purpose of clinical correlative analyses, we used Student's t-test to select a cutoff value that could best discriminate high- and low-expression groups for Dicer and Drosha. As shown in Table 1, Dicer and Drosha expression were in general well correlated with various characteristics except for gender. Low Drosha expression was frequently seen in tumors of stage 4 (18 of 22, 82%), tumors of high risk (21 of 25, 84%), and MYCN-amplified tumors (11 of 13, 85%). Low Dicer expression was also significantly correlated with unfavorable age at diagnosis, stage, risk, and Shimada histology (P < 0.001, 0.038, 0.013, and 0.004, respectively; Table 1). Thus, low expression of Dicer and/or Drosha was significantly associated with high-risk phenotype.
Lower expression of Dicer or Drosha is associated with shorter survival
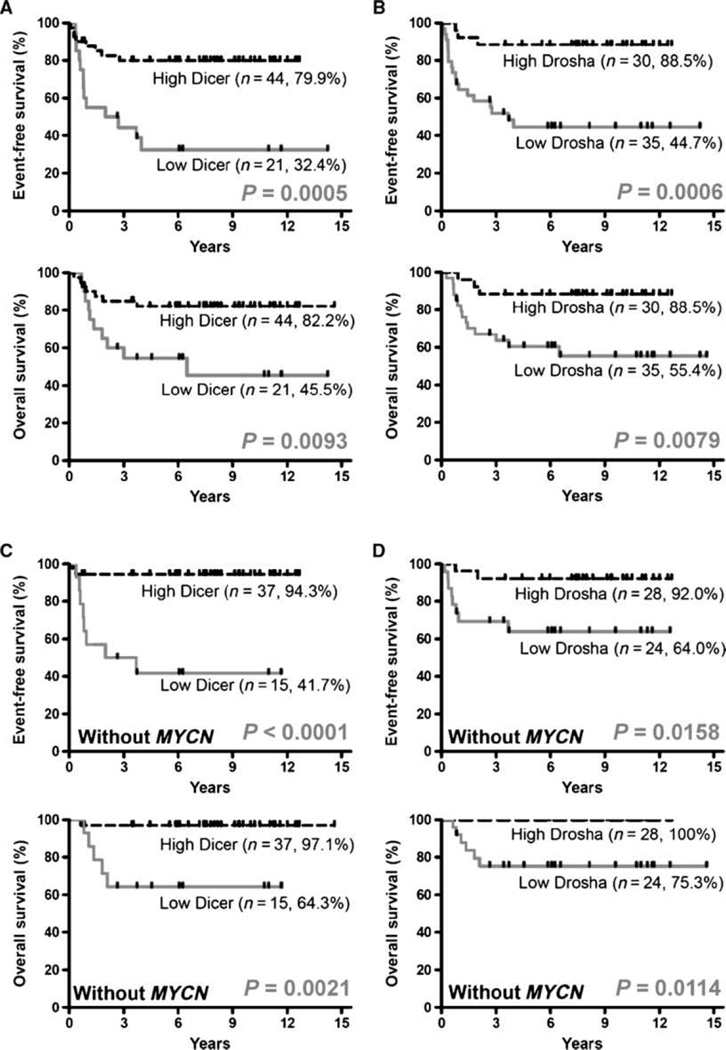
Next, we examined the prognostic value of Dicer and Drosha expression. The Kaplan-Meier survival analyses showed that neuroblastoma patients with low expression levels of Dicer had a significantly shorter EFS (32.4% versus 79.9%; P = 0.0005) and OS (45.5 versus 82.2%; P = 0.0093) than those with high levels of Dicer, respectively (Fig. 2A). Similarly, low Drosha expression was significantly associated with shorter EFS (44.7% versus 88.5%, P = 0.0006) and OS (55.4% versus 88.5%; P = 0.0079) than high Drosha expression (Fig. 2B). Furthermore, univariate Cox regression analysis of various parameters with OS showed that in addition to the well-known prognostic factors for neuroblastoma such as clinical stage, risk, and MYCN status, low expression of Dicer (P = 0.0142) or Drosha (P = 0.0158) was another significant predictive factor for poor outcome (Supplementary Table S4). However, when further analyzed by multivariate Cox regression, only MYCN amplification (P = 0.0077) and stage (P = 0.035) remained independent predictors. This is also true for the correlation between EFS and expression of Dicer and Drosha (Supplementary Table S4). Further supporting evidence was obtained by analysis of two large and independent cohorts of microarray database. A similar trend of association was noted between lower Dicer and decreased EFS, as determined by Kaplan-Meier survival estimates (Supplementary Fig. S7) and Cox regression analysis (Supplementary Table S6), with hazard ratios of 1.24 and 1.29 for E-TABM-38 (n = 251) and E-MTAB-16 (n = 262), respectively.
Figure 2.
Expression levels of Dicer and Drosha were significantly correlated with survival of neuroblastoma patients as a whole and those without MYCN amplification. Kaplan-Meier estimates of EFS and OS for neuroblastoma patients are shown according to the expression levels of Dicer (A and C) and Drosha (B and D) for 65 neuroblastoma patients (A and B) and for 52 neuroblastoma patients without MYCN amplification (C and D). P values were obtained using the log-rank test.
Expression of Dicer is an independent predictor for survival in neuroblastoma patients without MYCN amplification
In the above analyses of all 66 patients, MYCN is such a strong prognostic factor that it may have masked the effect of other factors in subgroup of patients. Analysis of neuroblastoma patients without MYCN amplification revealed that the low expression levels of Dicer or Drosha were significantly correlated with shorter EFS and OS (Fig. 2C and D). Notably, further analysis by univariate and multivariate Cox regression model identified Dicer expression as a significant and independent predictor for overall survival (hazard ratio, 9.6; P = 0.045; Table 2) in this subgroup.
Table 2.
Cox regression analyses of the various factors associated with event-free survival and overall survival in neuroblastoma patients without MYCN (n = 52)
| Event-free survival | |||
|---|---|---|---|
| Variables | HR (95% CI) | Favorable/Unfavorable | P |
| Univariate analysis | |||
| Stage | 2.66 (0.77–9.17) | 1, 2, 3, 4S/4 | 0.123 |
| Gender | 3.66 (0.78–7.26) | Female/Male | 0.102 |
| Risk | 4.45 (1.25–15.77) | Low, Middle/High | 0.021 |
| Age at diagnosis_1.5 | 14.03 (2.97–66.36) | <1.5 years/≧1.5 years | 0.001 |
| Dicer | 12.20 (2.57–57.82) | High/Low | 0.002 |
| Drosha | 5.48 (1.16–25.84) | High/Low | 0.032 |
| Multivariate analysis | |||
| Age at diagnosis_1.5 | 9.72 (1.37–69.10) | <1.5 years/≧1.5 years | 0.023 |
| Dicer | 1.88 (0.22–16.23) | High/Low | 0.568 |
| Drosha | 4.12 (0.69–24.68) | High/Low | 0.121 |
| Overall survival | |||
| Variables | HR (95% CI) | Favorable/Unfavorable | P |
| Univariate analysis | |||
| Stage | 5.86 (1.07–32.06) | 1, 2, 3, 4S/4 | 0.041 |
| Gender | 4.06 (0.47–34.81) | Female/Male | 0.648 |
| Risk | 16.32 (1.90–139.92) | Low, Middle/High | 0.011 |
| Age at diagnosis* | 594.6 (0.01–∞) | <1.5 years/≧1.5 years | 0.242 |
| Dicer | 13.39 (1.56–114.77) | High/Low | 0.018 |
| Drosha | 2.51 (0.46–13.74) | High/Low | 0.288 |
| Multivariate analysis | |||
| Stage | 3.22 (0.56–18.55) | 1, 2, 3, 4S/4 | 0.190 |
| Dicer | 9.60 (1.05–87.44) | High/Low | 0.045 |
Abbreviations: HR, hazard ratio; 95% CI, 95% confidence interval.
Because there was no death among the patients with diagnosis less than 1.5 years or 1 year.
These findings suggest that low expression of Dicer and Drosha may account for global reduction of miRNAs in advanced neuroblastoma and may significantly impact on patient outcome. Notably, expression levels of Dicer can serve as an important and independent predictor in patients without MYCN amplification.
Identification of a unique signature of 12 miRNAs, Dicer, Drosha, and age at diagnosis that can delineate clinical risk group by PNN analysis
Next, we investigated whether a combination of all biomarkers including Dicer, Drosha, and miRNAs can provide a better prediction for clinical outcome. To facilitate multivariate biomarker analysis, we utilize the theory of PNN (27), which is a self-organizing neural network model with plausible reasoning capability.
Using PNN Solution software, we trained a PNN on all the biomarkers and clinical information of the neuroblastoma samples except outcome information. During training, PNN uses an unsupervised learning algorithm that makes use of each biomarker to provide partial-weighted evidence to form multivariate patterns. The network then uses these patterns to assign the patient samples into different natural clusters.
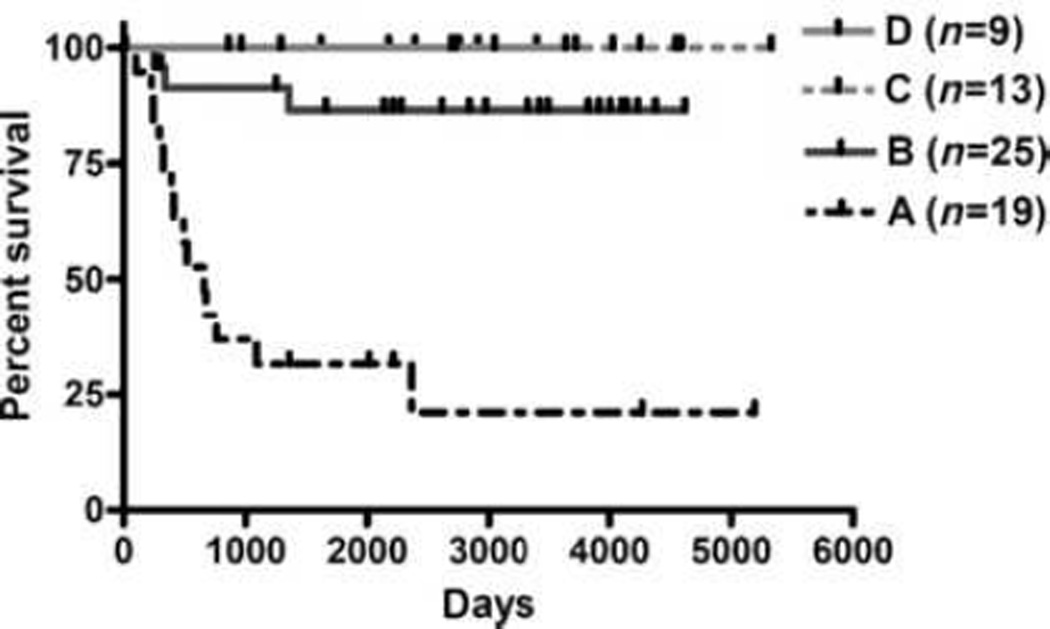
Then we used PNN Solution to calculate mutual information content to measure the joint relationship between the biomarkers and the patterns. Because of their strong mutual information relationship, the top-ranked biomarkers provide the strongest information to distinguish the patterns. Thus, we kept only the top 15 biomarkers for further PNN analysis; these consisted of 12 miRs' signature, expression of Dicer and Drosha, and age at diagnosis. We then used the 15 biomarkers to train the network again to form new clearer patterns, and let PNN assign the neuroblastoma patients into four different natural clusters (Supplementary Table S5). Figure 3 shows the Kaplan-Meier survival curve of OS for neuroblastoma patients according to their cluster (group) assignment. It reveals that group A contains mostly high-risk patients (18 of 19) with a fatal outcome (74%); in group B, 76% (19 of 25) of the patients are of low/intermediate risk, and 12% are high-risk patients that have fatality. Groups C and D consist of low/intermediate-risk patients that are alive, except for one patient. It shows that multivariate patterns found by PNN analysis can distinguish between short-term and long-term neuroblastoma patient survival, better than using Dicer and Drosha expression levels alone to determine neuroblastoma patient survival. These results were further confirmed by assigning neuroblastoma patients into training and test set to correlate with patient survival (see details in Supplementary Results, and Supplementary Fig. S4).
These findings imply that a combination of these 15 biomarkers may serve as a powerful predictor of neuroblastoma clinical outcome.
Knockdown of Dicer or Drosha can promote neuroblastoma cell proliferation and transformation
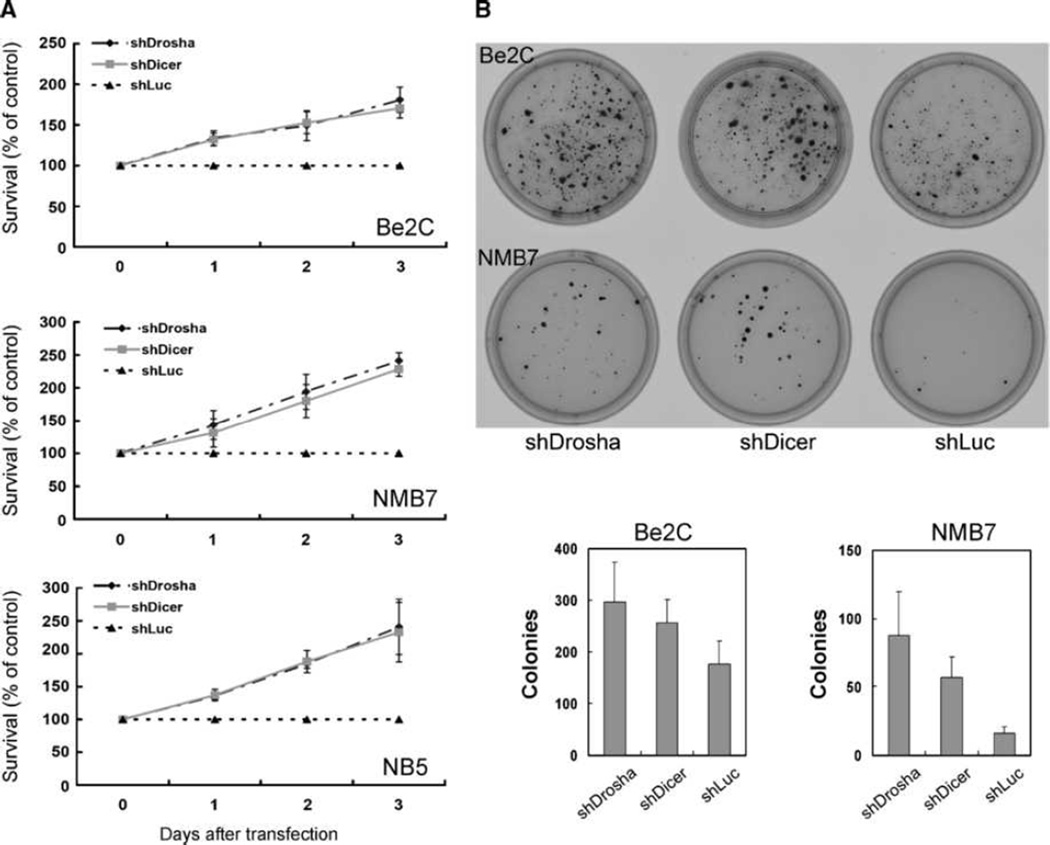
To elucidate the role of Dicer and Drosha on neuroblastoma tumorigenesis, we repressed the expression of Dicer and Drosha in three neuroblastoma cell lines (Be2C, NMB7, and NB5) with shRNA. The extent of knockdown at mRNA and protein levels varied from 33% to 81% inhibition (Supplementary Fig. S5). As expected, knockdown of either Dicer or Drosha led to a reduction in expression of miRNAs as reflected by qPCR detection of miR-17-5p and let-7a, and miRNA array (Supplementary Figs. S5 and S6). As compared with the negative control (shLuc), Drosha knockdown not only enhanced cell proliferation (Fig. 4A), but also induced greater number and size of colonies when cultured in soft agarose (Fig. 4B). Similar results were obtained with knockdown of Dicer (Fig. 4). These results were also confirmed by other independent shRNA clones against each gene (data not shown). These in vitro findings coupled with the above-mentioned clinical correlative studies suggest that Dicer and Drosha deregulate expression of miRNAs and promote tumor progression in neuroblastoma.
Figure 4.
Knockdown of Dicer or Drosha enhances cell proliferation of neuroblastoma cell lines. Neuroblastoma cell lines Be2C, NMB7, and NB5 were transfected with shRNA against Drosha (shDrosha), Dicer (shDicer), or luciferase (shLuc). Cell proliferation was determined at indicated day after transfection using alamarBlue assay (A) or colony formation assay (B) in soft agar and expressed as percentage of growth as compared with negative control, shLuc. Values are mean ± SE from at least three independent experiments.
Discussion
In this study, we determined the expression profile of miRNAs, Dicer, and Drosha in primary neuroblastoma samples and identified 27 miRNAs that can clearly distinguish low- from high-risk patients, using the PAM algorithm. Furthermore, using the PNN system to select a combination of 15 biomarkers that consist of 12 miRNAs, expression levels of Dicer and Drosha, and age at diagnosis, we were able to separate all patients into four distinct patterns that correlated well with clinical-pathologic characteristics of patients, and more importantly, with patient survival. If proven in future prospective study, this means that a combination of 12 miRNAs with Dicer and Drosha and age at diagnosis may serve as neuroblastoma-specific biomarkers for risk-group assignment and selection of optimal therapy, as well as a powerful predictor of clinical outcome.
Our finding of downregulation of miRNAs in neuroblastoma is consistent with several recent reports showing that the expression of most miRNAs is markedly reduced in a variety of human tumors, including breast cancer (30), thyroid anaplastic carcinoma(31), and prostate cancer (32). Notably, we found striking global downregulation of miRNAs in high-risk tumors, especially those with stage 4 and MYNC amplification. Similar findings were reported in a study of 157 miRNAs in 35 primary neuroblastoma tumors by Chen and Stalling (24). Ten of the 20 miRNAs associated with MYCN in their study are among the 27 miRNAs we identified to correlate with tumor risk (Supplementary Table S3). Thus, downregulation of miRNAs was associated with more advanced neuroblastoma, suggesting that overall repression of miRNAs may play a role in tumor progression.
Recent evidence suggested that expression of certain miRNAs was regulated by MYCN (33, 34). Furthermore, a widespread dysregulation of miRNA expression in neuroblastoma caused by overexpression of MYCN and large-scale chromosomal imbalances was noted, and a 15-miRNA signature was found to be predicative of clinical outcome (33). Although there are some overlapping in the selected miRNAs with those noted in our study, the contribution of Dicer and Drosha was not addressed in these studies. We showed that lower expression of Dicer and Drosha was responsible for global downregulation of miRNAs, and expression levels of Dicer and Drosha were significantly lower in stage 4 than in other stages, with a striking dichotomy in Dicer expression between stage 4 and 4S. Importantly, lowexpression of Dicer and Drosha significantly correlated with shorter EFS and OS of neuroblastoma patients. Thus, expression of Dicer and Drosha seemed to be another significant prognostic factor for neuroblastoma. Moreover, in cases without MYCN amplification, low expression levels of Dicer and Drosha correlated with worse patient survival, and multivariate analysis showed that low expression of Dicer could serve as a significant and independent prognostic factor for non– MYCN-amplified patients. Until now, no single marker or gene-expression profile can reliably predict outcome in tumor without MYCN amplification, which represents 80% of neuroblastoma(35, 36). If further validated in a larger and prospective cohort study, lower expression of Dicer may aid the prognostication of this large group of neuroblastoma patients.
Emerging evidence has linked the miRNA processing machinery to cancer. It has been reported that reduced expression of Dicer is associated with poor prognosis in non–small cell lung carcinomas (4) and ovarian cancer (10). This is consistent with our finding in neuroblastoma, but our data showed further that low expression of either Dicer or Drosha seemed to have prognostic impact. On the other hand, upregulation of Dicer has also been reported in prostate adenocarcinoma with correlation to more advanced clinical stage and lymph node status (7). Similarly, upregulation of Drosha was reported to be associated with increased cell proliferation and poor prognosis in esophageal cancer (6). In cervical cancer, copy number–driven overexpression of Drosha and changes in miRNA profile seem to be important for cervical cancer progression (9). In addition, Dicer1 mutation (missense mutation, L1583R) with truncation of a functional domain of RNase III has recently been identified in familial pleuropulmonary blastoma (37). These studies revealed that dysregulation of Dicer and Drosha exists in many types of human cancer, which may affect tumor progression and prognosis by altering miRNA expression profile.
Until now, the mechanism that regulates expression of Dicer and Drosha remains unclear. Zhang et al. have reported that copy number of Dicer and Ago2 at genomic locus are abnormal in breast cancer, ovarian cancer, and melanoma (38). In addition, deletion of Dicer locus at chr14q32.13 in lung cancer (29) and genomic amplification of Drosha locus at chr5p13.3 leading to Drosha overexpression in cervical squamous cell carcinoma (9) have been reported. As to neuroblastoma, loss of heterogeneity for 14q 23–32 (which may encompass Dicer locus) occurs in 22% of primary neuroblastoma tumors and is more frequently associated with presence of 11q loss of heterogeneity and inversely correlated with MYCN amplification (39). However, a conflicting report showed that deletions of 14q32 were only seen in the low-and intermediate-risk group (40). It is not clear whether the deletion encompassed the Dicer gene in the abovementioned reports and whether the locus of Dicer at 14q32.13 is aberrant in our samples set. The possibility of genetic abnormalities at Dicer and Drosha loci and the existence of other mechanisms such as epigenetic regulation of Dicer and Drosha in neuroblastoma await further investigation.
Supplementary Material
Figure 3.
The 15 biomarkers can delineate clinical risk groups and refine risk assessment in neuroblastoma. Based on PNN analysis, 15 biomarkers, including 12 miRNAs, Dicer, Drosha, and age at diagnosis (Dxage), were self-organized into four cluster patterns. Kaplan-Meier estimates of OS for neuroblastoma patients according to their pattern assignment by PNN analysis.
Acknowledgments
We thank the Children's Oncology Group, the Pediatric Oncology Group, and the Cooperative Human Tissue Network for providing neuroblastoma samples, and the National RNAi Core of Academia Sinica for providing RNAi for Dicer and Drosha.
Grant Support
Academia Sinica and grant FD-R-002319 from the Orphan Products Development Office of the U. S. Food and Drug Administration.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 2.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Gregory RI, Shiekhattar R. MicroRNA Biogenesis and Cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 4.Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of micro-RNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugito N, Ishiguro H, Kuwabara Y, et al. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006;12:7322–7328. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- 7.Chiosea S, Jelezcova E, Chandran U, et al. Up-regulation of dicer, a component of the microRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 9.Muralidhar B, Goldstein L, Ng G, et al. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212:368–377. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- 10.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 12.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 13.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 14.Tang XX, Zhao H, Kung B, et al. The MYCN enigma: significance of MYCN expression in neuroblastoma. Cancer Res. 2006;66:2826–2833. doi: 10.1158/0008-5472.CAN-05-0854. [DOI] [PubMed] [Google Scholar]
- 15.Castleberry RP, Pritchard J, Ambros P, et al. The International Neuroblastoma Risk Groups (INRG): a preliminary report. Eur J Cancer. 1997;33:2113–2116. doi: 10.1016/s0959-8049(97)00202-5. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka T, Iehara T, Sugimoto T, et al. Diversity in neuroblastomas and discrimination of the risk to progress. Cancer Lett. 2005;228:267–270. doi: 10.1016/j.canlet.2004.12.051. [DOI] [PubMed] [Google Scholar]
- 17.Schramm A, Schulte JH, Klein-Hitpass L, et al. Prediction of clinical outcome and biological characterization of neuroblastoma by expression profiling. Oncogene. 2005;24:7902–7912. doi: 10.1038/sj.onc.1208936. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Diskin S, Rappaport E, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 19.Oberthuer A, Berthold F, Warnat P, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- 20.Tricoli JV, Jacobson JW. MicroRNA: potential for cancer detection, diagnosis, and prognosis. Cancer Res. 2007;67:4553–4555. doi: 10.1158/0008-5472.CAN-07-0563. [DOI] [PubMed] [Google Scholar]
- 21.Attiyeh EF, London WB, Mosse YP, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 22.Diccianni MB, Omura-Minamisawa M, Batova A, Le T, Bridgeman L, Yu AL. Frequent deregulation of p16 and the p16/G1 cell cycle-regulatory pathway in neuroblastoma. Int J Cancer. 1999;80:145–154. doi: 10.1002/(sici)1097-0215(19990105)80:1<145::aid-ijc26>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 23.Lin YC, Diccianni MB, Kim Y, et al. Human p16γ, a novel transcriptional variant of p16(INK4A), coexpresses with p16(INK4A) in cancer cells and inhibits cell-cycle progression. Oncogene. 2007;26:7017–7027. doi: 10.1038/sj.onc.1210507. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67:976–983. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- 25.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen YY, Chen JJ. Neural networks and belief logic. Proceedings of the Fourth International Conference on Hybrid Intelligent Systems (HIS04); IEEE Computer Society; Washington, DC. 2004. pp. 460–461. [Google Scholar]
- 28.Wei-Chao C, Ping IH, Yuan-Yan C, Michael H, Pei-Jung L, Chung-Hsuan C. Observation of peptide differences between cancer and control in gastric juice. Proteomics Clin Appl. 2008;2:55–62. doi: 10.1002/prca.200780066. [DOI] [PubMed] [Google Scholar]
- 29.Chiosea S, Jelezcova E, Chandran U, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 30.Iorio MV, Ferracin M, Liu C-G, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 31.Visone R, Pallante P, Vecchione A, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 32.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 33.Bray I, Bryan K, Prenter S, et al. Widespread dysregulation of MiRNAs by MYCN amplification and chromosomal imbalances in neuroblastoma: association of miRNA expression with survival. PLoS ONE. 2009;4:e7850. doi: 10.1371/journal.pone.0007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fontana L, Fiori ME, Albini S, et al. Antagomir-17–5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS ONE. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schleiermacher G, Michon J, Huon I, et al. Chromosomal CGH identifies patients with a higher risk of relapse in neuroblastoma without MYCN amplification. Br J Cancer. 2007;97:238–246. doi: 10.1038/sj.bjc.6603820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oberthuer A, Warnat P, Kahlert Y, et al. Classification of neuroblastoma patients by published gene-expression markers reveals a low sensitivity for unfavorable courses of MYCN non-amplified disease. Cancer Lett. 2007;250:250–267. doi: 10.1016/j.canlet.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Hill DA, Ivanovich J, Priest JR, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Huang J, Yang N, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson PM, Seifried BA, Kyemba SK, et al. Loss of heterozygosity for chromosome 14q in neuroblastoma. Med Pediatr Oncol. 2001;36:28–31. doi: 10.1002/1096-911X(20010101)36:1<28::AID-MPO1008>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 40.Mosse YP, Diskin SJ, Wasserman N, et al. Neuroblastomas have distinct genomic DNA profiles that predict clinical phenotype and regional gene expression. Genes Chromosomes Cancer. 2007;46:936–949. doi: 10.1002/gcc.20477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.