Abstract
Compounds from macro marine organisms are presumed to owe their biosynthetic origins to associated microbial symbionts, although few definitive examples exist. An upsurge in the recent literature from 2012 to 2013 has shown that four compounds previously reported from macro marine organisms are in fact biosynthesized by non-photosynthetic Gram-negative bacteria (NPGNB). Structural parallels between compounds isolated from macro marine organisms and NPGNB producers form the basis of this review. Although less attention has been given to investigating the chemistry of NPGNB sources, there exists a significant list of structural parallels between NPGNB and macro marine organism-derived compounds. Alternatively, of the thousands of compounds isolated from Gram-positive actinomycetes, few structural parallels with macro marine organisms are known. A summary of small molecules isolated from marine NPGNB sources is presented, including compounds isolated from marine myxobacteria. From this assemblage of structural parallels and diverse chemical structures, it is hypothesized that the potential for the discovery of inspirational molecules from NPGNB sources is vast and that the recent spike in the literature of macro marine compounds owing their biosynthetic origin to NPGNB producers represents a turning point in the field.
INTRODUCTION
Marine invertebrates, especially sponges and tunicates, are recognized as being prolific sources of structurally diverse biosynthetics. For decades, it has been suggested that important macro marine organism-derived natural products are biosynthesized by microorganisms. For example, in 1993, a comprehensive survey described 35 natural products in a section entitled “Marine Natural Products Assumed to Be of Microbial Origins”.1 In subsequent years, numerous remarks have been made in both research articles and reviews pointing out that the bacteria associated with sponges and tunicates seem to be the likely sources of many types of compound classes isolated from these organisms.2–10
After many decades, it appears that a new experimental-based understanding on the role of invertebrate-associated microorganisms in marine natural product biosynthesis is at hand. As an important point, it is well known that sponge-associated microorganisms may constitute over 50% of a sponge’s mass.11 Some symbionts seem to be highly sponge-specific. In fact, the novel phylum Poribacteria occurs exclusively in sponges and has the ability to produce unique polyketides.12 The potential of sponge-associated microoganisms as a source of new natural products is demonstrated by findings involving Theonella, a genus of marine sponges. An unculturable Theonella-associated microorganism, Candidatus Entotheonella palauensi, putatively produces complex cyclic polypeptides.13,14 Additionally, onnamide A, isolated from the sponge Theonella swinhoei, is considered to be produced from a sponge-associated Gram-positive bacterium.15 Similarly, a molecular genetics study of psymberin, a compound structurally related to onnamide A and isolated from the sponge Psammocinia bulbosa, led to the provisional conclusion that this compound is produced by a sponge-associated bacterium.15 Adding to this theme are reports arguing that microbes associated with sponges possess the ability to be responsible for the synthesis of structurally diverse products previously isolated from sponges.4 The most common examples include polyketides and peptides.16,17 However, even in these promising reports, long-term attempts to engage in laboratory culture to reproduce a hypothesis based on circumstantial evidence have been elusive.18–20
There are some reports involving the manzamine family of compounds that represent a possible paradigm shift. The first member of this complex group of alkaloids was isolated originally from an Okinawan Haliclona sp., and over 80 analogues have been described from more than 16 species of sponges.21–28 The ability to isolate the same biosynthetic class from numerous taxa illustrates the classic situation for presuming the biosynthetic action of a sponge-associated bacterium. Between 2004 and 2006, two reports appeared describing a Petrosiidae sp. sponge-derived actinomycete strain, Micromonospora sp. M42, which produces manzamine A.29,30 These discoveries were in a patent document and a Ph.D. dissertation, respectively, but to date there has been no follow up in the peer-reviewed literature. In 2010, however, a partial explanation was noted by a team involved with this work and consisted of a brief passage describing continuing difficulties in producing manzamines during the laboratory culture of Micromonospora sp. M42. Surprisingly, this comment was buried in the middle of a review article.7
It is well established that scanning natural products produced by the myxobacteria class of Gram-negative bacteria provides especially fertile ground for side-by-side comparison to structures reported from sponges. That there are a number of parallels in the products of sponges with those of terrestrial-derived myxobacteria is surprising, considering only 100 core structures have been isolated from this group versus the thousands of compounds isolated from Gram-positive actino-mycetes.31,32 Some additional new comparisons, based on sponge versus myxobacterial products, will also be discussed later in this report.
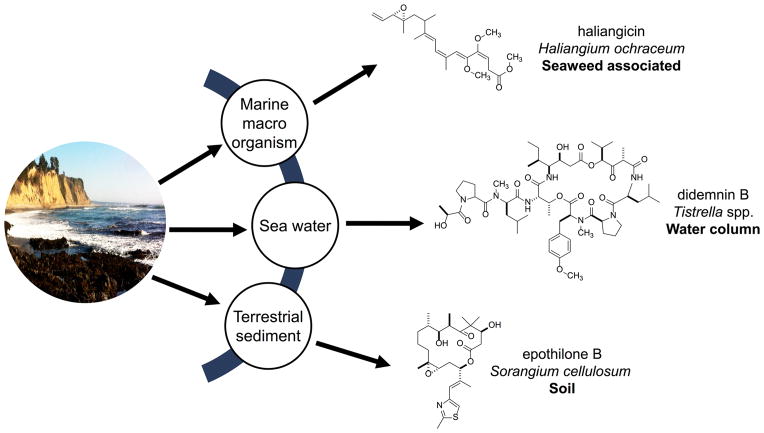
The contents of Figure 1 provide the context of both the framework and developments to be discussed in the present account. Herein, important insights will be emphasized relevant to sponge biosynthetics based on the recent literature and through a focus exclusive of the metabolites of terrestrial myxobacteria. First, an overview of compounds referred to here as non-photosynthetic Gram-negative bacteria (NPGNB) will be provided. The authors are fully aware that structural parallels between macro marine sources and Gram-negative cyanobacterial sources abound;33 however, the intention is to sharpen the scope of this review by emphasizing the emerging contribution of NPGNB-derived compounds.
Figure 1.
Prismatic view of the several dimensions of natural products derived from cultured non-photosynthetic Gram-negative bacteria as a function of their sourced environment, including terrestrial-derived sediments, oceanic water column, and macro marine organisms.
The three case examples summarized in Figure 1 provide a glimpse of the trajectory of the discussion to be provided. An important theme represented in this small collection is that significant scaffolds can be isolated through the culturing of Gram-negative microorganism strains from all possible sources: (a) the terrestrial environment, (b) the oceanic water column, and (c) in association with macro marine organisms. The key point to be made here is that each example shows a distinctive structural scaffold when the source organism was grown under laboratory conditions. The example of epothilone B is included to underscore the continued inspiration to be derived through the study of bioactive leads from terrestrial-derived myxobacteria. By contrast, the culturing of marine-derived myxobacteria has been exceedingly difficult.18,20,34,35 Nevertheless, the short list of compounds included from this source have great structural diversity,34–37 and the example of haliangicin (Figure 1) exemplifies the new outcomes now being obtained. Another ground-breaking discovery is represented by didemnin B, a clinically relevant anticancer compound first isolated from tunicates and currently obtained through culture of a marine-derived NPGNB.38–40
Finally, later in this review there will be a brief glimpse of selected, bioactive compounds from different marine-derived macroorganisms. This will be accompanied by a consideration of the emerging potential offered by NPGNB-sourced metabolites as sources of new structural complexity and clinical utility. The overall intention in mentioning the therapeutic success of compounds from marine taxa is to highlight the presently missing but promising contribution NPGNB will make.
IDENTICAL STRUCTURES FROM MACRO AND MICRO MARINE ORGANISMS
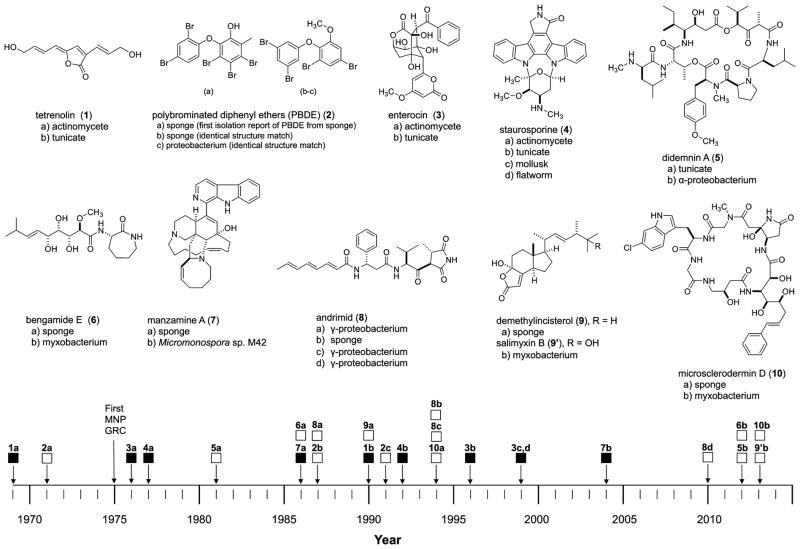
Similar natural products isolated from macro- and micro-organisms suggest a more complex origin than previously thought. Metabolites previously attributed to a macro marine organism may, in reality, be produced by microbial members of macroorganism-associated microbiomes.3–5 Increasingly, non-photosynthetic Gram-negative bacteria are being identified as the actual source of macro marine compounds.40 Figure 2 illustrates a timeline of identical carbon skeletons isolated from macro marine and microorganisms, from 1969 to 2013. There were two notable trends that emerged from the collection and organization of this information. The first is that between 2012 and 2013 there are four examples of identical carbon skeletons that have emerged from NPGNB. This upswing in discovery followed a 14-year gap (from 1999 to 2013), where to the best of the knowledge of the authors no reports, with the exception of the manzamines, of macro marine compounds having identical carbon skeletons to microorganisms occurred. The second trend of note is that while there are thousands of compounds isolated from Gram-positive microorganisms during this same time period of between 1999 and 2013, none of these compounds have been linked to a marine macroorganism. It is worth noting that, in contrast, there are approximately 100 core scaffolds isolated from Gram-negative bacteria and even less core structures from non-myxobacteria NPGNB.31 That the marine macro-microorganism connection does not involve Gram-positive bacteria has not escaped our notice and should guide further exploration in these areas.
Figure 2.
Comparative timeline, by publication date, of all structures discovered to date for which these identical carbon skeletons were derived from macro marine organisms (sponges, tunicates, mollusks, and flatworms) and also obtained through culturing of microorganisms. (Legend: Gram-positive bacteria ■; Gram-negative bacteria □; cited literature provided in the text.)
Comparative Timeline
The first Marine Natural Products Gordon Research Conference in 1975 occurred against the backdrop of pioneering work initiated in the 1950s by Bergmann, Scheuer, Pettit, Rinehart, and Moore. By the time of the conference, compounds from Gram-positive actinomycetes had already begun to be isolated in earnest. Tetrenolin (1), isolated from cultures of the actinomycete Micropolyspora venezuelensis in 1969, is the first example of a natural product with an identical structure later found in a macro marine organism.41 Compound 1 was reisolated, and its stereochemistry defined by Davidson and Ireland from a Lissoclinum sp. tunicate; it was renamed lissoclinolide.42 In 1976, enterocin (3) was isolated from two actinomycete strains, Streptomyces candidus var. enterostaticus WS-8096 and Streptomyces viridochromogenes M-127.43 Two decades later, the Fenical group also isolated enterocin from an ascidian from a Western Australian Didemnum sp.44 Adding to the list of actinomycete macro marine identical carbon skeletons is staurosporine (4), an indolo[2,3-a]carbazole first isolated from Streptomyces staurosporeus by Masuma and co-workers in Japan.45 The identical carbon skeleton was isolated subsequently from a tunicate,46 mollusk,47 and flatworm.48 Up to the turn of the 21st century, Gram-positive actinomycetes were shown to produce many macro marine natural products, in light of the identical structural matches mentioned above. Since the year 2000, there has been only one report of identical structure matches between Gram-positive actinomycetes, the manzamines, and a macro marine organism. Instead, there is accumulating evidence that macro marine compounds increasingly owe their origins to Gram-negative bacteria.
An early example of macro marine compounds found to be biosynthesized by NPGNB is the polybrominated diphenyl ethers (PBDE) (2) (Figure 2). Compounds of this type were first reported from Dysidea sp. sponges collected from the Western Caroline Islands in the early 1970s.49 Utkina and coworkers then reported an isolation of a PDBE from Dysidea fragilis and a Gram-negative Vibrio sp. associated with the sponge.50,51 However, in these reports, the authors did not specify if the PBDE was from the same sponge as the Vibrio-isolated PBDE.51 Soon afterward, the Faulkner group used flow-cytometric separation of symbionts from sponge cells and chemical analysis to demonstrate that cyanobacteria are the producers of PBDE.19,52
A flagship case of macro marine compounds having identical structures to compounds from Gram-negative organisms is the didemnins. The cyclic depsipeptide didemnin A (5) was originally isolated from a Caribbean Trididemnum sp. tunicate by Rinehart and co-workers in 1981.53 From the extract of the culture broth of Tistrella mobilis strain YIT 12409, didemnin B and nordidemnin B were isolated by Tsukimoto and coworkers.38 Following this isolation report, complete genome sequencing and analysis by Moore and co-workers determined that the Gram-negative α-proteobacteria Tistrella mobilis and Tistrella bauzanensis are the sources of the didemnins.40 The development of didemnin B, the first marine drug candidate to be clinically tested in humans, has been perturbed by supply issues. The discovery of the gene clusters responsible for the production of the didemnins in a Gram-negative α-proteobacterium provides a potential solution to the limited supply of these compounds.
As shown in Figure 2, many compounds from myxobacteria show identical carbon skeleton matches to those of macro marine organisms. The bengamides are considered to be potential antitumor agents and immunomodulators.54 A 2005 report in the patent literature indicated that the sponge-derived compound bengamide E is also produced by Gram-negative bacterium Myxococcus sp.55 Our laboratory has shown that bengamide E (6), initially isolated from a Jaspidae sp. sponge collected near the Fiji Islands, can also be isolated from Myxococcus virescens (Figure 2).54,56,57 Manzamine A (7) was discussed in the introductory paragraphs as having been isolated from both a sponge and culture broths of the actinomycete Micromonospora sp. M42.21–29
The pyrrolidinedione antibiotic andramid (8) has been isolated from a Hyatella sp. sponge and many NPGNB genera.58,59,61 The first isolation report of 8 was from an Enterobacter sp. planthopper symbiont, by Komura and coworkers.58 After the initial isolation, andrimid was isolated from a Vibrio sp. associated with a Hyatella sp. sponge.59 In this report, Oclarit and colleagues demonstrate that 8 was present in both the Vibrio sp. bacterial culture and the extract of the host sponge.59 In a separate report, the Andersen group isolated 8 from the marine bacterium Pseudomonas flurescens, adding to the indiscriminate NPGNB producers of andrimid.60 More recently, 8 was isolated from Vibrio coralliilyticus strain 2052 by Gram and co-workers, as part of a global research expedition that collected over 500 bacterial strains with antibiotic activity against pathogenic bacteria.61
The König group isolated salimyxin B (9′) from an obligate marine myxobacterium, Enhygromyxa salina, obtained from a coastal sediment sample obtained in Santa Barbara, California.34 The carbon skeleton of salimyxin B is nearly identical to demethylincisterol A3 (9), with the only difference being an additional hydroxy group. Demethylincisterol A3 had originally been isolated from the marine sponge Dictyonella incisa in 1990.62
The microsclerodermins are a family of cyclic hexapeptides originally isolated from Microscleroderma sp. sponges by Faulkner and co-workers in 1994.63 In a recent report by Müller and associates, myxobacteria in the genera Jahnella and Chondromyces were shown to produce microsclerodermin D (10).64 This compound and analogues presented are of identical structure to those isolated from Theonella sponges reported by Schmidt and Faulkner (Figure 2).65
SIMILAR STRUCTURES FROM MACRO- AND MICROORGANISMS
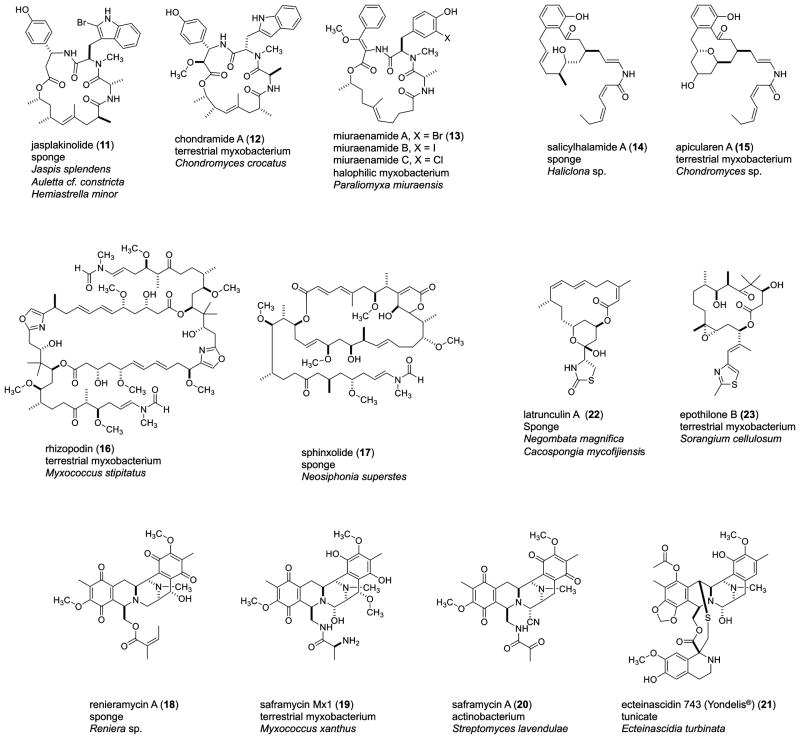
There are a number of marine-derived compounds that serve as examples of structural parallels that are not identical in carbon skeleton. Rather, these compounds have closely related chemical features. The parallelisms shown in Figure 3 imply the possibility of congruent biosynthetic machinery operating in different biota. In each of these parallelisms, an NPGNB is in some fashion implicated in the production of the compound.
Figure 3.
Molecular structures of sponge- or tunicate-derived compounds compared to those possessing similar frameworks from terrestrial- or marine-derived cultured non-photosynthetic Gram-negative bacteria.
Parallel Carbon Scaffolds
The cyclodepsipeptide jasplakinolide (11) (aka jaspamide) was isolated in 1986 from a soft-bodied Jaspis sp. sponge collected in Tonga and Fiji.66,67 Initial biological screening of 11 revealed antifungal and antihelminthic activity, with an MIC value of 25 μg/mL against Candida albicans. A separate in vivo assessment against Candida vaginal infection in mice using a 2% solution of 11 was comparable in efficacy to miconazole nitrate.62 Since its initial isolation, jasplakinolide has been the impetus for thousands of biomedicinal publications due to its F-actin effects.68,69 Structurally similar, the chondramides (12) were isolated from the myxobacterium Chondromyces crocatus by Reichenbach and colleagues and differ from jasplakinolide in the carbon count of the macrocyclic ring; the former features an 18-membered ring, while the latter possesses a 19-membered ring and different substituents around the macrocycle. Like jasplakinolide, the chondramides exert cytotoxic effects by targeting the actin cytoskeleton.70,71 In recent biological testing, members of a series of synthetic chondramide analogues were shown to block Toxoplasma gondii parasite growth on host cell monolayers with EC50 values ranging from 0.3 to 1.3 μM.72 Similar also are the miuraenamides (13), discussed under compounds from marine myxobacteria that also target the actin cytoskeleton.
Salicyclihalamide A (14) is a cytotoxic macrolide isolated from a sponge in the genus Haliclona by Boyd and co-workers in 1997.73 Structurally similar is apicularin A (15), from the myxobacterium Chondromyces robustus, isolated by the Reichenbach group in 1998.74 The principal structural difference between the salicylihalamides and apicularins is the presence of a cyclized pyran ring in the apicularins. The apicularins, like the salicylichalamides, are V-ATPase inhibitors, although they inhibit at different binding sites.75 Aplicularin has been shown to have cytotoxic effects against human cancer cell lines and causes apoptosis.76
Rhizopodin (16) is a cytostatic, antifungal macrolide, originally isolated by Reichenbach and associates from the culture broth of the myxobacterium Myxococcus stipitatus.77 The initially reported structure was corrected nearly two decades later by the Müller group through use of comprehensive NMR spectroscopy, resulting in the revision of 16 from a monomeric lactone to a symmetrical dilactone.78 The mechanism of action of 16 is through inhibition of actin polymerization.79 The structure of rhizopodin, and in particular the enamide side chain, is well adapted to the molecular surface of actin, allowing binding of the molecule to the actin and providing a significant obstacle to polymerization.79 Also sharing actin depolymerization bioactivity with rhizopodin is the macrolactone sphinoxolide (17), which was isolated shortly after rhizopodin in 1993 by D’Auria and co-workers from the New Caledonian sponge Neosiphonia superstes. Sphinoxolide shares a similar side chain to rhizopodin; however, the macrocycles are quite different.80
The tetrahydroisoquinoline compounds renieramycin A (18), saframycin Mx1 (19), saframycin A (20), and ecteinascidin (ET-743) (21) represent an interesting group of closely related structures from disparate organisms. Isolated first were the saframycins, in 1977 by Arai and co-workers, from the Gram-positive actinomycete Streptomyces lavendulae.81 The saframycins display antitumor and antimicrobial activity through DNA alkylation.82 Following the saframycins was the isolation of the dimeric quinone-bearing renieramycins in 1982, by the Faulkner group from a Mexican Reniera sp. sponge.83 In contrast to the Gram-positive origin of the saframycins, the Gram-negative myxobacterium Myxococcus xanthus is the source of saframycin Mx1 (19). Compound 19 was isolated by Reichenbach and co-workers in 1987 and differs from the saframycins in the side chain, featuring a terminal alanine instead of pyruvic acid.84 Completing this group of closely similar tetrahydroisoquinolines are the macro marine-derived ecteinascidins first isolated by Rinehart and co-workers from the Caribbean tunicate Ecteinascidia turbinata.85 Of the tetrahydroisoquinolines mentioned, the ecteinascidins have the most promising biological activity. ET-743 is by far the most prolific, with activity orders of magnitude more potent than saframycin A (20) against B16 melanoma.82 Incidentally, ET-743 is a U.S. Food and Drug Administration (FDA)-approved drug, sold under the trade name Yondelis. It has been granted orphan drug status in the U.S. for the treatment of soft tissue and ovarian cancers.86
Latrunculin A (22), a member of a group of thioimidazole bearing toxins originally isolated by Kashman in 1980 from the Red Sea sponge Negombata magnifica, and also from the Indo-Pacific sponge Cacospongia mycofijiensis, disrupts microfilament organization and has a strong anti tumor effect.87–89 Latrunculin A shares a distant structural similarity with epothilone B (23) isolated from the terrestrial myxobacterium Sorangium cellulosum by the Höfle group in 1996.90 The epothilones are microtubule stabilizers, showing nanomolar inhibition of cancer cell growth.91 The synthetic epothilone derivative Ixempra was approved by the U.S. FDA in 2007 for the treatment of taxane-resistant metastatic breast cancer and represents a “home-run” hit for terrestrial myxobacterial compounds.92
COMPOUNDS FROM NON-PHOTOSYNTHETIC, GRAM-NEGATIVE BACTERIA
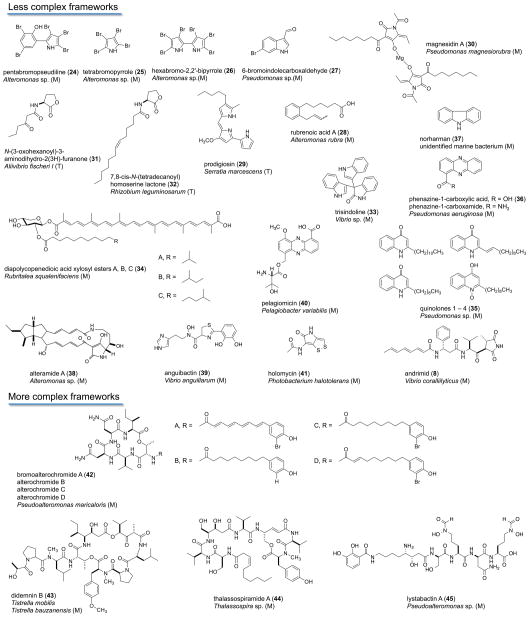
Non-photosynthetic Gram-negative bacteria are a taxonomically diverse group of organisms with the potential to contribute a broad range of molecular structures (Figure 4). However, the difficulty in culturing NPGNB under laboratory conditions, particularly marine-derived strains, has resulted in low compound titers even using relatively large-scale cultivations in the 100 L range.34,35 As a result, there are only a handful of chemically productive NPGNB genera known thus far, despite their enormous taxonomic diversity. Still, there have been focused reviews on chemically productive NPGNB genera such as Vibrio and Pseudomonas.93,94 Figure 4 groups compounds from chemically productive non-myxobacterial NPGNB genera into less complex and more complex structures from both terrestrial and marine environments.
Figure 4.
Selective view, from 1966 to 2013, of the biosynthetic products published from marine- and terrestrial-derived strains of cultured Gram-negative bacteria. Omitted from this list are marine-derived myxobacteria (shown in Figure 5) and the hundreds of metabolites (the subjects of many previous reviews) isolated from (a) terrestrial myxobacteria (gamma-proteobacteria), (b) the terrestrial genus Lysobacter (also a gamma-proteobacteria), and (c) both marine and terrestrial cyanobacteria. [Codes for the organism sources are terrestrial (T), marine (M).]
Less Complex Frameworks
Seawater-derived Gram-negative bacterial compounds were summarized by Fenical in 1993. He foreshadowed that Gram-negative bacteria are not prolific in their production of secondary metabolites and are relatively slow growing, but have the potential to produce novel chemistry.95 A brief summation of Fenical’s findings is discussed here, and many of these fall under the less complex structures category (Figure 4). Pentabromopseudiline (24), isolated from what was later classified as an Altermonas sp. by Burkholder in 1966, exhibited antibiotic activity when tested against a panel of Gram-positive bacteria and in particular Mycobacterium tuberculosis.96,97 In 1977, the Faulkner group also isolated pentabromopseudeline from a purple-pigmented bacteria (later determined to be an Altermonas sp.) purified from a seawater sample obtained in the North Pacific Gyre.98 The group also isolated the other brominated pyrroles 25 and 26.98 Compound 25 exhibited moderate antibacterial activity in a disk-diffusion assay against Staphyloccocus aureus, Escherichia coli, Pseudomonas aeruginosa, and Candida albicans. Interestingly, this compound class exhibited antibacterial activity against a panel of marine bacteria (Photobacterium fisheri, P. mandapamensis, and P. phosphoreum) as well as a degree of autotoxicity.98 In 1977, the Faulkner group continued their investigation into these compounds after obtaining the same brominated pyrroles from a yellow pseudomonad isolated from a tide pool in La Jolla, California.99 The 1977 publication describes the isolation of the then previously discussed compounds (24–26), 6-bromoindolecarboxaldehyde (27) and its debrominated analogue, the new 2-n-pentyl-4-quinolinol, and the known antibiotic 2-n-heptyl-4-quinolinol. Disk diffusion assays were used to identify particularly strong antibiotic activity from 2-n-pentyl-4-quinolinol. The publication makes a specific note of the pyrrole autotoxicity.99
Wells and co-workers at Roche Research Institute in Australia isolated three aromatic acids from the common marine bacterium Alteromonas rubra.100 Crude chloroform extracts from this bacterium exhibited in vitro bronchodilator activity as well as muscle-relaxant effects. Chromatographic purification through bioassay-guided fractionation resulted in three aromatic acids, rubrenoic acids A–C (28). Although the biological properties of the compounds described in this work did not suggest any potential clinical application, the authors concluded from their work that marine bacteria warrant further attention in biological screening programs.
An unidentified bacterial isolate (strain #157) obtained from a surface seawater sample taken at a salmon (Oncorhynus tshanytscha) net-pen farm was found to be a source of oncorhycolide. The initial publication reported no bioactiv-ities.101 However, the Andersen group, like Fenical, used this compound as a “proof of concept” structure. They concluded that by isolating this compound, it was shown that bacteria isolated from the marine environment have the potential to be the source of many novel natural products and hence are worthy of more extensive study.101
Also falling within less complex structures are derivatives of prodigiosin (29), pyrrole ring-containing pigments with many reported bioactivities including antimicrobial, cytotoxic, and antimalarial effects.102 Additionally, there is clinical promise for these compounds in the area of immunomodulation.102 Undecylprodigiosin exhibited inhibition of T-cell proliferation at nontoxic doses.103 The synthetic prodigiosin derivative PNU-15684, upon oral administration, suppressed heart allograft rejection in a rat in vivo model. Co-administration of 40 mg kg−1 with cyclosporin A caused a dramatic increase in the survival period of heart-transplanted rats.104,105 As degradation products of the prodigiosins, the magnesidins (30) are a mixture of homologous compounds bearing C5 and C7 alkyl groups present in a 1:1 ratio.106 The bacterial source of these compounds, Pseudomonas magniorubra, was obtained through washings of the marine alga Caulerpa peltata collected near Mumbai, India.107 Interestingly, prodigiosin (29), produced most notably by Serratia marcescens, has the additional distinction of being the suspected cause of a medieval-era “miracle”, since bacterial contamination and the subsequent appearance of this deep-red pigment on bread resulted in the initiation of the Feast of Corpus Christi.108
Additional less complex scaffolds from NPGNB include compounds from the marine Gram-negative bacterium Vibrio fischeri (Aliivibrio fischeri), which self-produces the chemical signal N-(3-oxohexanoyl)-3-aminodihydro-2(3H)-funanone (31), in a process known as “quorum sensing”.109 First isolated in 1981 by Eberhard and co-workers from Photobacterium fischeri, it was observed that luciferase self-expression can be turned on or off by excretion of an N-acyl homoserine lactone, 31.110 Structurally, acyl-homoserine lactones (AHL) feature a fatty acid chain and a lactone homoserine moiety, joined by an amide bond. Variations in acyl chain length are possible along with different AHLs featuring varying oxidation states at the C-3 position (i.e., carbonyl, hydroxy group).109 One example of a structural modification from 31 is 7,8-cis-N-(tetradecanoyl)-homoserine lactone (32) from Rhodobacter sphaeroides, isolated by Puskus and co-workers.111 Compound 32 features a longer acyl chain length than present in 31. Production of AHLs in Vibrio fisheri occurs through expression of lux genes that control luminescence.112,113 Euprymna scolopes and E. morsei squid species are luminescent due to colonization of the light-producing organs by V. fischeri.114,115 Vibrio fisheri from E. morsei grown in laboratory culture outside of the squid were found to produce the autoinducer 31 at levels 10-fold less than those produced by E. morsei squid isolates. The same group concluded that luminescence is a consequence of colonization of host tissues and not simply a cell-density-promoted occurrence.114,115
Kobayashi and co-workers isolated a novel indole trimer, trisindoline (33), a less complex scaffold from a Vibrio sp. that was separated from the Okinawan sponge Hyrtios altum.26 Compound 33 was shown to have antibiotic activity using disk diffusion asaays, inhibiting the growth of E. coli, B. subtilis, and S. aureus at 10 μg/disk.26 Trisindole was isolated along with known diketopiperazines. From the marine bacterium Rubritalea squalenifaciencs, Shindo and co-workers isolated the new acyl glycol-carotenoic acids diapolycopenedioic acid xylosyl esters A, B, and C.116 The source bacterium has been isolated from the sponge Halichondria okadai.116 Interestingly, the 1O2 quenching activity of diapolycopenedioic acid xylosyl esters A (34) in an 1O2 suppression model was investigated and found to have an IC50 value of 5.1 μM. The activity was more potent than those of astaxanthin or β-carotene, and the authors will further examine the role of structural features in the activity.116
A Pseudomonas sp. collected at the surface of the sponge Homophymia sp. collected in New Caledonia was found to contain a series of quinolone (35) analogues similar to the compounds reported by Faulkner and co-workers in 1977.2,99 Baker’s group, in an investigation into microorganisms associated with macro marine organisms, isolated the known phenazine alkaloid (36) from Pseudomonas aeruginosa, as well as several diketopiperazines.117 Bioactivity-guided fractionation was employed to isolate these compounds, with the phenazine alkaloids showing activity against Bacillus cereus with an MIC value of <0.5 μg/mL in disk diffusion assays.117 The sponge Isodictya setifera was also investigated for the production of phenazines; however, none were detected. The authors attributed this to the bacterium not producing the metabolite while in association with the sponge or due to insufficient amount of bacterium in the sponge for compound detection.117
Investigations of the sponge Hymeniacidon perleve collected in the East China Sea led to the isolation of 29 bacterial strains.118 Using growth inhibition against terrestrial micro-organisms, an extract from Pseudomonas piscida strain NJ6-3-1 was identified as having the broadest antimicrobial activity against a panel of microorganisms. This strain was then subjected to bioactivity-guided fractionation, using disk diffusion assays, resulting in the isolation of norharman (37). Norharman is a known antimicrobial agent first found in terrestrial plants.119
Marine Alteromonas sp. bacteria associated with the sponge Halichondria okadai are the source of alteramide A (38), isolated by Kobayashi and co-workers.120 Cytotoxic effects of 38 against murine leukemia P388, murine L1210 lymphoma, and human epidermoid carcinoma cells were determined. In this cytotoxicity screening, compound 38 exhibited IC50 values in the low micromolar range.120
A marine fish pathogen, Vibrio anguillarum strain pJM1, is the source of anguibactin (39). Structural analysis by Crosa and colleagues was supported with single-crystal X-ray diffraction of an anhydro derivative of 39.121 Anguibactin is structurally unique, having N-hydroxyhistamine, cysteine, and dihydroxybenzoic acid components. The pathogenic nature toward fish is due to the pJM1 plasmid within V. anguillarum, of which the compound is a virulence factor.121
Pelagiomicin A (40) was isolated by Imamura and coworkers from the marine halophilic Gram-negative bacterium Pelagiobacter variabilis.122 The bacterium was obtained from the macro alga Pocockiella variegata collected in Palau. Phenazine alkaloids like 40 are well known from various Streptomyces species, with the first reports dating back to the isolation of griseolutein from Streptomyces griseolutus.123,124 The cytotoxic activity of pelagiomicin was investigated against a panel of solid tumor, leukemia, and endothelial cancer cell lines, with IC50 values in the low micromolar range reported.125
Previous discussion in this review introduced the structure of andrimid (8). Of additional significance is during dereplication studies on Vibrio coralliilyticus strain 2052, it was discovered that 8 has antibiotic activity against a broad range of bacteria, due to inhibition of bacterial acetyl-CoA carboxylase.61,126 The Gram group in Denmark, along with concurrent studies by Adachi and co-workers in Japan, have spearheaded isolation work in NPGNB genera, in particular, Vibrio, Photobacterium, and Pseudoalteromonas.57,127,128 The reader is directed to a representative review entitled “Production of Bioactive Secondary Metabolites by Marine Vibrionaceae” by Mansson, which highlights the potential of Vibrio spp. as a source of bioactive small molecules.93
Interestingly, the pyrrothine antibioitic holomycin (41) was isolated from the culture of Photobacterium halotolerans strain S2753.57 Previously, compound 41 had been reported from Gram-positive Streptomyces. The authors surmise horizontal gene transfer as a reason for discovery of 41 in these disparate organisms, which represents an interesting subject for future inquiry.57,129,130
More Complex Frameworks
Under the more complex structural category, cyclic peptides are typically encountered.127 One example is the bromoalterochromides, isolated by Laatsch and co-workers from the marine bacterium Pseudoalteromonas maricaloris KMM 636T, collected in La Jolla, California.131 The antibacterial activity was also investigated and reconfirmed the results of Faulkner and colleagues with inhibition of the growth of S. aureus.99 Additionally, cytotoxicity against KB cells was observed with an IC50 value of less than 2 μg/mL.2 Bromoalterochromine A (42) is a yellow-pigmented chromo-peptide, isolated as an inseparable mixture designated A and A′.131 The amino group of the threonine residue is acylated with differing side chains, which the authors named alterochromides. The cytotoxic effects of these chromopeptides on sea urchin (Strongylocentrotus intermedius) eggs were investigated, and an MIC value of 40 μg/mL was determined; however, no antibiotic activity was observed.131
Terrestrial-derived gliding Gram-negative bacteria in the genus Lysobacter are well known and produce cyclodepsipeptides, cyclic lipodepsipeptides, cepham-type β-lactams, and polycyclic tetramate macrolactams.132,133 Most Lysobacter genera described to date occur in the terrestrial environment, but a report by Romanenko and co-workers outlined Lysobacter strain KMM 329T from a Pachastrella sp. deep-sea sponge, obtained from the Philippine Sea.134 Although no small molecules were reported by Romanenko and co-workers, this discovery of a marine-derived Lysobacter genus has the potential to provide improved knowledge of the chemistry from both terrestrial and marine-derived Lysobacter species.
As was stated earlier in this review, didemnins and thallospiramides from marine α-proteobacterial NPGNBs have been reported.39,40,135 Didemnins, a family of cyclic depsipeptides, were originally isolated from the Caribbean tunicate Trididemnum solidum.53 A dehydrodidemnin analogue, aplidine, isolated from the ascidian Aplidium albicans, is currently in phase III clinical trials initiated by PharmaMar for refractory myeloma.136 In 2003, the European Commission granted approval for use of aplidine in the treatment of acute lymphoblastic leukemia.137 Didemnin B (43) was recently isolated from the cultureable Gram-negative bacterium Tistrella mobillis, the biosynthesis of which was recently described by the Moore group.38,40
The novel cyclic peptides, thalassospiramides A (44) and B (not shown here), were isolated in 2007 by the Fenical group from the marine α-proteobacterium Thalassospira lucentensis (strain CNJ-328).135 Using an in vitro assay, the thalassospiramides showed immunosuppressive activity by inhibiting IL-5 production. Recently, the mechanisms of thallaspiramide biosynthesis have been investigated by the Moore group.138
The Butler group isolated lystabacins A–C (lystabacin A (45) shown), from the marine bacterium Pseudoalteromonas sp. strain S2B.139 As a unique observation, the strain was obtained from the Gulf of Mexico following the Deepwater Horizon oil spill that occurred in April 2010. The Pseudoalteromonas sp. strain was grown in seawater medium with crude oil added, suggesting that the bacterium could indeed degrade hydrocarbons in oil. The ability to biodegrade oil by bacteria may be facilitated by the actions of siderophores like lystabacin A (45) that sequester iron, an essential nutrient for bacterial growth.140,141
The bryostatins are well-known anticancer agents isolated initially by Pettit and co-workers from the bryozoan Bugula neritina.142,143 The cytotoxicity of the bryostatins is due to effects on protein kinase C, and numerous clinical trials have been undertaken.144–146 A Gram-negative unculturable bacterial symbiont, Candidatus Endobugula sertula, inhabiting the pallial sinus of the bryozoan B. neritina, has been implicated in the production of bryostatins.147,148 In fact, without the bacterial symbiont, the bryozoan does not produce the bryostatins.147–149 Haygood and co-workers have identified a bryostatin polyketide synthase gene cluster from E. sertula.150,151 This discovery holds enormous promise, since expression of the gene cluster could produce the bryostatins in sufficient amounts for marketing as a drug.
COMPOUNDS FROM MARINE-DERIVED MYXOBACTERIA
Myxobacteria are bacteriolytic, Gram-negative δ-proteobacteria found throughout the world. They inhabit terrestrial environments such as the soil and decaying organic matter, as well as freshwater and marine saltwater environments.152–154 The term “micropredator” has been used to describe myxobacteria, due to their physical gliding movements and lytic action on other bacteria.152–154 Two main characteristics have been ascribed to myxobacteria: formation of fruiting bodies containing propagative spores under nutrient-poor conditions, and cellular gliding over surfaces.152–154 Myxobacteria are the source of a wide array of bioactive natural products, with a relatively recent estimate in 2009 by the Müller group of 100 core structures and 500 of their derivatives having been reported in the literature.31,155–158 The majority of these compounds are from terrestrial genera such as Sorangium, Myxococcus, and Chondromyces, which is a consequence of the amenability of these species to laboratory culture conditions. For comprehensive reviews on terrestrial myxobacterial secondary metabolites, the reader is referred to excellent reviews by Reichenbach and Müller.31,155–158 Perhaps the most heralded of the terrestrial myxobacterial taxa is the genus Sorangium, the source of the epothilones. These cytotoxic macrolides were first isolated by Höfle and Reichenbach from Sorangium cellulosum strain Soce90.90 The epothilones are a distinct class of microtubule-interacting anticancer compounds from the taxanes and vinca alkaloids and often used to treat taxane-resistant tumors.92 Metabolic instability of epothilone derivatives from esterase cleavage at position 16 of the lactone was ameliorated by synthesis of the lactam-containing derivative Ixempra, approved by the U.S. FDA for treatment of refractory metastatic breast cancer in 2007.92,159
There are far fewer isolated compounds from seawater-derived myxobacterial species. The difficulty in culturing myxobacteria obtained from seawater samples has gained this taxonomically elusive group a reputation for being presently uncultureable.34,35 The number of chemical compounds from halophilic and slightly halophic, marine-derived myxobacteria can be counted on one hand. However, the compounds that have been identified illustrate the remarkable potential of marine myxobacterial taxa to produce unprecedented chemistry. In an effort to highlight this promise, the following is an update on the present literature featuring all marine-derived myxobacterial compounds shown in Figure 5.
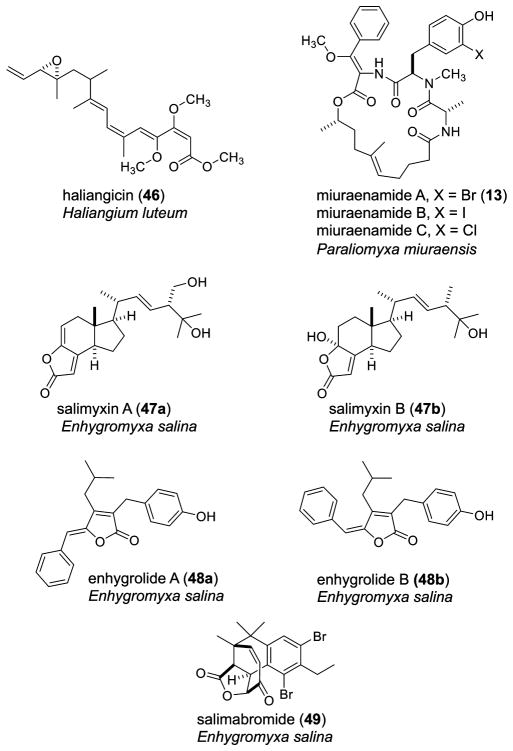
Figure 5.
Summary, from 1966 to 2013, of all biosynthetic products published from the culturing of marine-derived Gram-negative myxobacteria.
Products from Marine-Derived Myxobacteria
Starting the short list of marine-derived myxobacterial compounds is haliangicin (46), isolated by Yamanaka and co-workers in 2001 from cultures of the salt-obligate myxobacterium Haliangium luteum strain AJ-13395.36,37 This myxobacterial strain was obtained from a beach seaweed sample in Japan. Bioactivity-guided fractionation gauged by an antifungal assay was employed to isolate compound 46. The potent antifungal activity of haliangicin results from the inhibition of fungal electron transport within complex III, with an IC50 of 2.5 nM, and is comparable to the standard control amphotericin B.36,37 Structurally, haliangicin features a β-methoxyacrylate moiety with trans stereochemistry about the epoxide moiety. A cis isomer, along with geometric isomers of the polyene moiety, has also been reported, with structural disruptions in the conjugated tetraene moiety lowering antifungal activity.160
Miuraenamides A and B (13) were isolated by the Yamada group shortly following the initial report of haliangicin (46) from the slightly halophic near-shore soil myxobacterial strain SMH-27-4.161 Bioactivity-guided isolation of the miuraenamides employed a paper disk diffusion assay against phytopathogenic oomycetes, Phytophthora capsici. Given the prior knowledge of β-methoxyacrylate-containing myxobacterial compounds having activity as fungal electron transport chain inhibitors, the miuraenamides were also tested for this same activity. They were found to inhibit NADH oxidase activity at 100 μM and were compared to the positive control myxobacterial compound cystothiaxol (10 μM).161,162 Although not isolated from a truly salt-tolerant myxobacterium, the miuraenamides were obtained from a near-shore soil sample that was slightly halophilic and are therefore worthy of note. As noted earlier in Figure 3, there is a close similarity in the structure of 13 to that of jasplakinolide (11) and chondramide A (12).
Following a seven-year period with no reports of compounds from halophilic, marine-derived myxobacteria, several publications have entered the literature more recently, spearheaded by those of the König group. The compounds salimyxins A (47a) and B (47b) and enhydrolides A (48a) and B (48b) have been isolated very recently from the obligate marine myxobacterium Enhygromyxa salina. The Enhygromyxa strain was obtained from marine sediments collected from coastal Germany and California. The salimyxins and enhydrolides are tricyclic, degraded sterols and are one of the first myxobacterial compounds reported from the marine-derived taxonomic group.34 Attesting to the difficulty in culturing marine-derived myxobacteria, the isolation yield of salimyxin A (47a) was astonishing low at 0.4 mg/64 L culture.
Salimabromide (49) is another recent marine-derived compound isolated by the König group from Pleosiocystis/Enhydromyxa spp. marine myxobacteria, obtained from coastal marine sediments in Germany. Structurally, 49 is polyketide derived and represents a new carbon skeleton featuring substitution with bromine, a rare occurrence among myxobacterial secondary metabolites. Compound 49 was obtained in an equally low yield to the salimyxins and enhygrolides, with only 0.5 mg from 64 L of culture, and was the first to be isolated from the genus Enhygromyxa.35
CONCLUSIONS AND FUTURE PROSPECTS
The total number of core scaffolds discussed in this review discovered from marine-derived NPGNB is quite small in comparison with those from Gram-positive actinomycetes. Clearly, exploration of natural products from NPGNB should be intensified. As noted earlier in this review, the information base on these compounds at this point in time is sparse. In the near future, there will undoubtedly be new discoveries to add to the current knowledge of structural parallels between macro marine and Gram-negative microorganism-derived compounds.
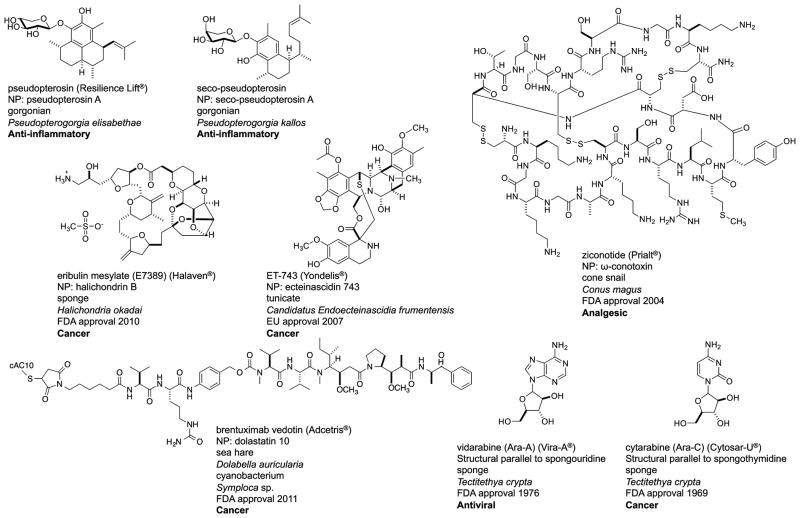
It is clear that much enthusiasm exists for the discovery and development of marine natural products as potential human therapeutic agents. Decades have been required to demonstrate the therapeutic success of compounds from marine taxa, and there are now a growing number of drugs of marine origin that have achieved U.S. FDA approval. Figure 6 shows the FDA-approved drugs that have been inspired by, or pursued in parallel with, marine invertebrate-derived natural products.163–175 Clearly missing from this collection are compounds from NPGNB. However, the potential of NPGNB compounds to gain U.S. FDA approval is great. This is highlighted by discoveries linking the didemnins and bryostatins to NPGNB producers.38,150,151 Continued attention to compound structural parallels between macro marine organisms and NPGNB organisms holds promise for the production of medically important drugs in previously unattainable yields. In this regard, NPGNB organisms have indeed entered center stage of marine natural products research.
Figure 6.
Summary of the current U.S. Federal Drug Administration (FDA)-approved drugs inspired by, or pursued in parallel with, marine invertebrate-derived natural products. The natural product (NP) compound and organism source are indicated for each. Not yet represented in this compilation are contributions from marine-derived Gram-negative bacteria.
Acknowledgments
This work was supported, in part, by grant R01-CA-047135 and a supplement to Promote Diversity in Health-Related Research. Other support was obtained from NSF CHE1214065.
Footnotes
The authors declare no competing financial interest.
DEDICATION
Dedicated to Prof. Dr. Otto Sticher, of ETH-Zurich, Zurich, Switzerland, for his pioneering work in pharmacognosy and phytochemistry.
References
- 1.Kobayashi J, Ishibashi M. Chem Rev. 1993;93:1753–1769. [Google Scholar]
- 2.Bultel-Poncé V, Berge JP, Debitus C, Nicolas JL, Guyot M. Mar Biotechnol. 1999;1:384–390. doi: 10.1007/pl00011792. [DOI] [PubMed] [Google Scholar]
- 3.Piel J. Nat Prod Rep. 2004;21:519–538. doi: 10.1039/b310175b. [DOI] [PubMed] [Google Scholar]
- 4.Piel J. Annu Rev Microbiol. 2011;65:431–453. doi: 10.1146/annurev-micro-090110-102805. [DOI] [PubMed] [Google Scholar]
- 5.Williams PG. Trends Biotechnol. 2009;27:45–52. doi: 10.1016/j.tibtech.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Gulder TAM, Moore BS. Curr Opin Microbiol. 2006;12:252–260. doi: 10.1016/j.mib.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waters AL, Hill RT, Place AR, Hamann MT. Curr Opin Biotechnol. 2010;21:780–786. doi: 10.1016/j.copbio.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radjasa OK, Vaske YM, Navarro G, Vervoort HC, Tenney K, Linington RG, Crews P. Bioorg Med Chem. 2011;19:6658–6674. doi: 10.1016/j.bmc.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerwick WH, Moore BS. Chem Biol. 2012;19:85–98. doi: 10.1016/j.chembiol.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons TL, Andrianoasolo E, McPhail K, Flatt P, Gerwick WH. Mol Cancer Ther. 2005;4:333–342. [PubMed] [Google Scholar]
- 11.Brantley SE, Molinski TF, Preston CM, DeLong EF. Tetrahedron. 1995;51:7667–7672. [Google Scholar]
- 12.Fieseler L, Horn M, Wagner M, Hentschel U. Appl Environ Microbiol. 2004;70:3724–3732. doi: 10.1128/AEM.70.6.3724-3732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt EW, Obraztsova AY, Davidson SK, Faulkner DJ, Haygood MG. Mar Biol. 2000;136:969–977. [Google Scholar]
- 14.Piel J, Butzke D, Fusetani N, Hui D, Platzer M, Wen G, Matsunaga S. J Nat Prod. 2005;68:472–479. doi: 10.1021/np049612d. [DOI] [PubMed] [Google Scholar]
- 15.Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL, Taudien S, Platzer M, Rubio BK, Robinson SJ, Crews P, Piel J. Nat Chem Biol. 2009;5:494–501. doi: 10.1038/nchembio.176. [DOI] [PubMed] [Google Scholar]
- 16.Hochmuth T, Niederkrüger H, Gernert C, Siegl A, Taudien S, Platzer M, Crews P, Hentschel U, Piel J. ChemBioChem. 2010;11:2572–2578. doi: 10.1002/cbic.201000510. [DOI] [PubMed] [Google Scholar]
- 17.Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, Sahl HG, Matsunaga S, Piel J. Science. 2012;338:387–389. doi: 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- 18.Torsvik V, Goksøyr J, Daae FL. Appl Environ Microb. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unson MD, Holland ND, Faulkner DJ. Mar Biol. 1994;119:1–11. [Google Scholar]
- 20.Amann RI, Ludwig W, Schleifer KH. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakai R, Higa T. J Am Chem Soc. 1986;108:6404–6405. [Google Scholar]
- 22.Sakai R, Kohmoto S, Higa T. Tetrahedron Lett. 1987;28:5493–5496. [Google Scholar]
- 23.Nakamura H, Deng S, Kobayashi J, Ohizumi Y. Tetrahedron Lett. 1987;28:621–624. [Google Scholar]
- 24.Kondo K, Shigemori H, Kikuchi Y, Ishibashi M, Sasaki T, Kobayashi J. J Org Chem. 1992;57:2483–2485. [Google Scholar]
- 25.Ichiba T, Corgiat JM, Scheuer PJ. J Nat Prod. 1994;57:168–170. doi: 10.1021/np50103a027. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi M, Aoki S, Gato K, Matsunami K, Kurosu M, Kitagawa I. Chem Pharm Bull. 1994;42:2449–2451. doi: 10.1248/cpb.42.2449. [DOI] [PubMed] [Google Scholar]
- 27.Peng J, Rao KV, Choo Y-M, Hamann MT. In: Modern Alkaloids: Structure, Isolation, Synthesis and Biology. Fattorusso E, Taglialatela-Scafati O, editors. Wiley-VCH Verlag GmbH & Co; Weinheim: 2008. pp. 189–232. [Google Scholar]
- 28.Kubota T, Kamijyo Y, Takahashi-Nakaguchi A, Fromont J, Gonoi T, Kobayashi J. Org Lett. 2013;15:610–612. doi: 10.1021/ol3034274. [DOI] [PubMed] [Google Scholar]
- 29.Hill RT, Peraud O, Hamann MT, Kasanah N. 10/522, 454, 2013 US Pat Appl.
- 30.Peraud O. PhD Thesis. University of Maryland; College Park, MD: 2006. Isolation and Characterization of a Sponge-Associated Actinomycete that Produces Manzamines. [Google Scholar]
- 31.Garcia RO, Krug D, Müller R. Meth Enzymol. 2009;458:59–91. doi: 10.1016/S0076-6879(09)04803-4. [DOI] [PubMed] [Google Scholar]
- 32.Fenical W, Jensen PR. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 33.Andrianasolo EH, Gross H, Goeger D, Musafija-Girt M, McPhail K, Leal RM, Mooberry SL, Gerwick WH. Org Lett. 2005;7:1375–1378. doi: 10.1021/ol050188x. [DOI] [PubMed] [Google Scholar]
- 34.Felder S, Kehraus S, Neu E, Bierbaum G, Schäberle TF, König GM. ChemBioChem. 2013;14:1363–1371. doi: 10.1002/cbic.201300268. [DOI] [PubMed] [Google Scholar]
- 35.Felder S, Dreisigacker S, Kehraus S, Neu E, Bierbaum G, Wright PR, Menche D, Schäberle TF, König GM. Chem—Eur J. 2013;19:9319–9324. doi: 10.1002/chem.201301379. [DOI] [PubMed] [Google Scholar]
- 36.Fudou R, Iizuka T, Yamanaka S. J Antibiot. 2001;54:149–152. doi: 10.7164/antibiotics.54.149. [DOI] [PubMed] [Google Scholar]
- 37.Fudou R, Iizuka T, Ando T, Shimba N, Yamanaka S. J Antibiot. 2001;54:153–156. doi: 10.7164/antibiotics.54.153. [DOI] [PubMed] [Google Scholar]
- 38.Tsukimoto M, Nagaoka M, Shishido Y, Fujimoto J, Nishisaka F, Matsumoto S, Harunari E, Imada C, Matsuzaki T. J Nat Prod. 2011;74:2329–2331. doi: 10.1021/np200543z. [DOI] [PubMed] [Google Scholar]
- 39.Lee J, Currano JN, Carroll PJ, Joullié MM. Nat Prod Rep. 2012;29:404–424. doi: 10.1039/c2np00065b. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, Kersten RD, Nam SJ, Lu L, Al-Suwailem A, Zheng H, Fenical W, Dorrestein PC, Moore BS, Qian PY. J Am Chem Soc. 2013;134:8625–8632. doi: 10.1021/ja301735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallo GG, Coronelli C, Vigevani A, Lancini GC. Tetrahedron. 1969;25:5677–5680. doi: 10.1016/s0040-4020(01)83074-x. [DOI] [PubMed] [Google Scholar]
- 42.Davidson BS, Ireland CM. J Nat Prod. 1990;53:1036–1038. doi: 10.1021/np50070a049. [DOI] [PubMed] [Google Scholar]
- 43.Miyairi N, Sakai HI, Konomi T, Imanaka H. J Antibiot. 1976;24:227–235. doi: 10.7164/antibiotics.29.227. [DOI] [PubMed] [Google Scholar]
- 44.Kang H, Jensen PR, Fenical W. J Org Chem. 1996;61:1543–1546. [Google Scholar]
- 45.Ōmura S, Iwai Y, Hirano A, Nakagawa A, Awaya J, Tsuchiya H, Takahashi Y, Masuma R. J Antibiot. 1977;30:275–282. doi: 10.7164/antibiotics.30.275. [DOI] [PubMed] [Google Scholar]
- 46.Kinnel RB, Scheuer PJ. J Org Chem. 1992;57:6327–6329. [Google Scholar]
- 47.Cantrell CL, Groweiss A, Gustafson KR, Boyd MR. Nat Prod Lett. 1999;14:39–46. [Google Scholar]
- 48.Schupp P, Eder C, Proksch P, Wray V, Schneider B, Herderich M, Paul V. J Nat Prod. 1999;62:959–962. doi: 10.1021/np980527d. [DOI] [PubMed] [Google Scholar]
- 49.Sharma GM, Vig B. Tetrahedron Lett. 1972;17:1715–1718. [Google Scholar]
- 50.Utkina NK, Kazanteseva MV, Denisenko VA. Khim Prir Soedin. 1987;4:603–605. [Google Scholar]
- 51.Elyakov GB, Kuznetsova T, Mikhailov VV, Maltsev II, Voinov VG, Fedoreyev SA. Experientia. 1991;47:632–633. [Google Scholar]
- 52.Unson MD, Faulkner DJ. Experientia. 1993;49:349–353. [Google Scholar]
- 53.Rinehart KL, Gloer JB, Hughes RG, Renis HE, McGoveren JP, Swynenberg EB, Stringfellow DA, Kuentzel SL, Li LH. Science. 1981;212:933–935. doi: 10.1126/science.7233187. [DOI] [PubMed] [Google Scholar]
- 54.Johnson TA, Sohn J, Vaske YM, White KN, Cohen TL, Vervoort HC, Tenney K, Valeriote FA, Bjeldanes LF, Crews P. Bioorg Med Chem. 2012;20:4348–4355. doi: 10.1016/j.bmc.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffman H, Haag-Richter S, Kurz M, Tiertgen H. WO2005044803. PCT Int Appl. 2005
- 56.Quiñoà E, Adamczeski M, Crews P. J Org Chem. 1986;51:4497–4498. [Google Scholar]
- 57.Adamczeski M, Quiñoà E. J Am Chem Soc. 1989;111:647–654. [Google Scholar]
- 58.Fredenhagen A, Tamura SY, Kenny PTM, Komura H, Naya Y, Nakanishi K. J Am Chem Soc. 1987;109:4409–4411. [Google Scholar]
- 59.Oclarit JM, Okada H, Ohta S, Kaminura K, Yamaoka Y, Iizuka T, Miyashiro S, Ikegami S. Microbios. 1994;314:7–16. [PubMed] [Google Scholar]
- 60.Needham J, Kelly MT, Ishige M, Andersen RJ. J Org Chem. 1994;59:2058–2063. [Google Scholar]
- 61.Wietz M, Mansson M, Gotfredsen CH, Larsen TO, Gram L. Mar Drugs. 2010;8:2946–2960. doi: 10.3390/md8122946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ciminiello E, Fattorusso E, Magno S, Mangoni A, Pansini M. J Am Chem Soc. 1990;112:3505–3509. [Google Scholar]
- 63.Bewley CA, Debitus C, Faulkner DJ. J Am Chem Soc. 1994;116:7631–7636. [Google Scholar]
- 64.Hoffmann T, Müller S, Nadmid S, Garcia R, Müller R. J Am Chem Soc. 2013;135:16904–16911. doi: 10.1021/ja4054509. [DOI] [PubMed] [Google Scholar]
- 65.Schmidt EW, Faulkner DJ. Tetrahedron. 1998;54:3043–3056. [Google Scholar]
- 66.Crews P, Manes LV, Boehler M. Tetrahedron. 1986;27:2797–2800. [Google Scholar]
- 67.Zabriskie TM, Klocke JA, Ireland CM, Marcus AH, Molinski TF, Faulkner JD, Xu C, Clardy J. J Am Chem Soc. 1986;108:3123–3124. [Google Scholar]
- 68.Bubb MR, Senderowicz AMJ, Sausville EA, Duncan KLK, Korn ED. J Biol Chem. 1994;269:14869–14871. [PubMed] [Google Scholar]
- 69.Senderowicz AMJ, Kaur G, Sainz E, Laing C, Inman WD, Rodrijuez J, Crews P, Malspeis L, Grever MR, Sausville EA, Duncan KLK. J Natl Cancer Inst. 1995;87:46–51. doi: 10.1093/jnci/87.1.46. [DOI] [PubMed] [Google Scholar]
- 70.Kunze B, Jansen R, Sasse F, Höfle G, Reichenbach H. J Antibiot. 1995;66:1262–1266. doi: 10.7164/antibiotics.48.1262. [DOI] [PubMed] [Google Scholar]
- 71.Sasse F, Kunze B, Gronewold TMA, Reichenbach H. J Natl Cancer Inst. 1998;90:1559–1563. doi: 10.1093/jnci/90.20.1559. [DOI] [PubMed] [Google Scholar]
- 72.Ma CI, Diraviyam K, Maier ME, Sept D, Sibley LD. J Nat Prod. 2013;76:1565–1572. doi: 10.1021/np400196w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Erickson KL, Beutler JA, Cardellina JH, Boyd MR. J Org Chem. 1997;62:8188–8192. doi: 10.1021/jo971556g. [DOI] [PubMed] [Google Scholar]
- 74.Kunze B, Jansen R, Sasse F, Höfle G, Reichenbach H. J Antibiot. 1998;51:1075–1080. doi: 10.7164/antibiotics.51.1075. [DOI] [PubMed] [Google Scholar]
- 75.Huss M, Sasse F, Kunze B, Jansen R, Steinmetz H, Ingenhorst G, Zeeck A, Wieczorek H. BMC Biochem. 2005;6:13–21. doi: 10.1186/1471-2091-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jansen R, Kunze B, Reichenbach H, Höfle G. Eur J Chem. 2000:913–919. [Google Scholar]
- 77.Sasse F, Steinmetz H, Höfle G, Reichenbach H. J Antibiot. 1993;46:741–748. doi: 10.7164/antibiotics.46.741. [DOI] [PubMed] [Google Scholar]
- 78.Jansen R, Steinmetz H, Sasse F, Schubert WD, Hagelüken G, Albrecht SC, Müller R. Tetrahedron Lett. 2008;49:5796–5799. [Google Scholar]
- 79.Gronewold TMA, Sasse F, Lünsdorf H, Reichenbach H. Cell Tissue Res. 1999;295:121–129. doi: 10.1007/s004410051218. [DOI] [PubMed] [Google Scholar]
- 80.D’Auria MV, Paloma LG, Mirale L, Zampella A. Tetrahedron. 1993;49:8657–8664. [Google Scholar]
- 81.Arai T, Takahashi K, Kubo A. J Antibiot. 1977;30:1015–1018. doi: 10.7164/antibiotics.30.1015. [DOI] [PubMed] [Google Scholar]
- 82.Scott JD, Williams RM. Chem Rev. 2002;102:1669–1730. doi: 10.1021/cr010212u. [DOI] [PubMed] [Google Scholar]
- 83.Frincke JM, Faulkner J. J Am Chem Soc. 1982;104:265–269. [Google Scholar]
- 84.Irschik H, Trowitzsch-Kienast W, Gerth K, Höfle G, Reichenbach H. J Antibiot. 1988;41:993–998. doi: 10.7164/antibiotics.41.993. [DOI] [PubMed] [Google Scholar]
- 85.Rinehart KL, Holt TG, Fregeau NL, Stroh JG, Keifer PA, Sun F, Li LH, Martin DG. J Org Chem. 1990;55:4512–4515. [Google Scholar]
- 86.Haefner B. Drug Discovery Today. 2003;8:536–544. doi: 10.1016/s1359-6446(03)02713-2. [DOI] [PubMed] [Google Scholar]
- 87.Kashman Y, Groweiss A, Shmueli U. Tetrahedron Lett. 1980;21:3629–3632. [Google Scholar]
- 88.Spector I, Shochet NR, Kashman Y, Groweiss A. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 89.Amagata T, Johnson TA, Cichewicz RH, Tenney K, Mooberry SL, Media J, Edelstein M, Valeriote FA, Crews PJ. Nat Prod. 2008;51:7234–7242. doi: 10.1021/jm8008585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Höfle G, Bedort N, Steinmetz H, Schomburg D, Gerth K, Reichenbach H. Angew Chem, Int Ed Engl. 1996;35:1567–1569. [Google Scholar]
- 91.Gerth K, Bedorf N, Höfle G, Irschik H, Reichenbach H. J Antibiot. 1996;49:560–563. doi: 10.7164/antibiotics.49.560. [DOI] [PubMed] [Google Scholar]
- 92.Fumoleuau P, Coudert B, Isambert N, Ferrant E. Ann Oncol. 2007;18:v9–v15. doi: 10.1093/annonc/mdm173. [DOI] [PubMed] [Google Scholar]
- 93.Mansson M, Gram L, Larsen TO. Mar Drugs. 2011;9:1440–1468. doi: 10.3390/md9091440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Isnansetyo A, Kamei Y. J Ind Microbiol Biotechnol. 2009;36:1239–1248. doi: 10.1007/s10295-009-0611-2. [DOI] [PubMed] [Google Scholar]
- 95.Fenical W. Chem Rev. 1993;93:1673–1683. [Google Scholar]
- 96.Burkholder PR, Pfister RM, Leitz FH. App Microbiol. 1966;14:649–653. doi: 10.1128/am.14.4.649-653.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lovell FM. J Am Chem Soc. 1966;88:4510. [Google Scholar]
- 98.Andersen RJ, Wolfe MS, Faulkner DJ. Mar Biol. 1974;27:281–285. [Google Scholar]
- 99.Wratten SJ, Wolfe MS, Andersen R, Faulkner DJ. Antimicrob Agents Chemother. 1977;11:411–414. doi: 10.1128/aac.11.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Holland GS, Jamieson DD, Reicheldt JL, Viset G, Wells RJ. Chem Ind. 1984 Dec 3; [Google Scholar]
- 101.Needham J, Andersen RJ, Kelly MT. Tetrahedron Lett. 1991;32:315–318. [Google Scholar]
- 102.Fürstner A. Angew Chem, Int Ed. 2003;42:3582–3603. doi: 10.1002/anie.200300582. [DOI] [PubMed] [Google Scholar]
- 103.Fürstner A, Szillat H, Gabor B, Mynott R. J Am Chem Soc. 1998;120:8305–8314. [Google Scholar]
- 104.Nakamura A, Nagai K, Ando K, Tamura G. J Antibiot. 1986;39:1155–1159. doi: 10.7164/antibiotics.39.1155. [DOI] [PubMed] [Google Scholar]
- 105.Stepkowski SM, Erwin-Cohen RA, Behbod F, Wang ME, Qu X, Tejpal N, Nagy ZS, Kahan BD, Kirken RA. Blood. 2002;15:680–689. doi: 10.1182/blood.v99.2.680. [DOI] [PubMed] [Google Scholar]
- 106.Kohl SV, Bhat JR, Patell NM, Gandhi J, Nazareth J, Divakar PV, de Sousa NJ. Tetrahedron Lett. 1974;12:983–986. [Google Scholar]
- 107.Gandhi NM, Patell JR, Gandhi J, de Souza NJ, Kohl H. Mar Biol. 1976;34:223–227. [Google Scholar]
- 108.Garlaschielli L. Chem Unserer Zeit. 1999;3:152–157. [Google Scholar]
- 109.Fuqua C, Greenberg EP. Nat Rev. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 110.Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 111.Puskas A, Greenberg EP, Kaplan S, Schaefer AL. J Bacteriol. 1997;179:7530–7537. doi: 10.1128/jb.179.23.7530-7537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nealson KH, Platt T, Hastings W. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fuqua C, Greenbery EP. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 114.McFall-Ngai MJ. Comp Biochem Phys A. 2000;126:471–480. doi: 10.1016/s1095-6433(00)00233-6. [DOI] [PubMed] [Google Scholar]
- 115.Boettcher KJ, Ruby EG. J Bacteriol. 1995;177:1053–1058. doi: 10.1128/jb.177.4.1053-1058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shindo K, Asagi E, Sano A, Hotta E, Minemura N, Mikami K, Tamesada E, Misawa N, Maoka T. J Antibiot. 2008;61:185–191. doi: 10.1038/ja.2008.28. [DOI] [PubMed] [Google Scholar]
- 117.Jayatilake GS, Thornton MP, Leonard AC, Grimwade JE, Baker B. J Nat Prod. 1996;59:292–296. doi: 10.1021/np960095b. [DOI] [PubMed] [Google Scholar]
- 118.Zheng L, Chen H, Han X, Lin W, Yan X. World J Microb Biot. 2005;21:201–206. [Google Scholar]
- 119.Agurell S, Holmstedt B, Lindgren JE, Schultes RE. Biochem Pharmacol. 1968;17:2487–2488. doi: 10.1016/0006-2952(68)90140-8. [DOI] [PubMed] [Google Scholar]
- 120.Shigemori H, Bae MA, Yazawa K, Sasaki T, Kobayashi J. J Org Chem. 1992;57:4317–4320. [Google Scholar]
- 121.Jalal MAF, Hossain MB, van der Helm D, Sanders-Loehr J, Actis LA, Crosa JH. J Am Chem Soc. 1989;111:292–296. [Google Scholar]
- 122.Imamura N, Nishijima M, Takadera T, Adachi K, Sakai M, Sano H. J Antibiot. 1997;50:8–12. doi: 10.7164/antibiotics.50.8. [DOI] [PubMed] [Google Scholar]
- 123.Umezawa H, Hayano S, Maeda K, Ogata Y, Okami Y. Jap Med J. 1950;3:111–117. doi: 10.7883/yoken1948.3.111. [DOI] [PubMed] [Google Scholar]
- 124.Fotso S, Santosa DA, Saraswati R, Yang J, Mahmud T, Zabriskie M, Proteau PJ. J Nat Prod. 2010;73:472–475. doi: 10.1021/np9005647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Singh MP, Menendez AT, Petersen PJ, Ding WD, Maiese WM, Greenstein M. J Antibiot. 1997;50:785–787. doi: 10.7164/antibiotics.50.785. [DOI] [PubMed] [Google Scholar]
- 126.Freiberg C, Pohlmann J, Nell PG, Endermann R, Schuhmacher J, Newton B, Otteneder M, Lampe T, Haebich D, Ziegelbauer K. Antimicrob Agents Chemother. 2006;50:2707–2712. doi: 10.1128/AAC.00012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Oku N, Kawabata K, Adachi K, Katsuta A, Shizuri Y. J Antibiot. 2008;61:11–17. doi: 10.1038/ja.2008.103. [DOI] [PubMed] [Google Scholar]
- 128.Mansson M, Nielsen A, Kjærulff L, Gotfredsen CH, Wietz M, Ingmer H, Gram L, Larsen TO. Mar Drugs. 2011;9:2537–2552. doi: 10.3390/md9122537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kenig M, Reading C. J Antibiot. 1979;32:549–554. doi: 10.7164/antibiotics.32.549. [DOI] [PubMed] [Google Scholar]
- 130.Hou YH, Li FC, Wang SJ, Qin S, Wang QF. Microbiol Res. 2008;163:96–104. doi: 10.1016/j.micres.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 131.Speitling M, Smetanina OF, Kuznetsova TA, Laatsch H. J Antibiot. 2007;60:36–42. doi: 10.1038/ja.2007.5. [DOI] [PubMed] [Google Scholar]
- 132.Xie Y, Wright S, Shen Y, Du L. Nat Prod Rep. 2012;29:1277–1287. doi: 10.1039/c2np20064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nett M, König GM. Nat Prod Rep. 2007;24:1245–1261. doi: 10.1039/b612668p. [DOI] [PubMed] [Google Scholar]
- 134.Romanenko LA, Uchino M, Tanaka N, Frolova GM, Mikhailov VV. Int J Syst Evol Microbiol. 2008;58:370–374. doi: 10.1099/ijs.0.65391-0. [DOI] [PubMed] [Google Scholar]
- 135.Oh DC, Strangeman WK, Kauffman CA, Jensen PR, Fenical W. Org Lett. 2007;9:1525–1528. doi: 10.1021/ol070294u. [DOI] [PubMed] [Google Scholar]
- 136.PharmaMar. Aplidin Dexamethasone in Relapsed/Refractory Myeloma (ADMYRE) In ClinicalTrials.gov [Internet] National Library of Medicine (US); Bethesda, MD: 2000. [cited Feb 8, 2014]. Available from http://clinicaltrials.gov/show/NCT01102426 NLM Identifier: NCT011102426. [Google Scholar]
- 137.Management Board Report EMEA/MB/63019/2006. European Medicines Agency; London: 2006. Annual Report of the European Medicines Agency 2005. [Google Scholar]
- 138.Ross AC, Xu Y, Lu L, Kersten RD, Shao Z, Al-Suwailem AM, Doresstein PC, Qian PY, Moore BS. J Am Chem Soc. 2013;135:1155–1162. doi: 10.1021/ja3119674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zane HK, Butler A. J Nat Prod. 2013;76:648–654. doi: 10.1021/np3008655. [DOI] [PubMed] [Google Scholar]
- 140.Bosello M, Robbel L, Linne U, Xie X, Marahiel MA. J Am Chem Soc. 2011;133:4587–4595. doi: 10.1021/ja1109453. [DOI] [PubMed] [Google Scholar]
- 141.Kreutzer MF, Kage H, Nett M. J Am Chem Soc. 2012;134:5415–5422. doi: 10.1021/ja300620z. [DOI] [PubMed] [Google Scholar]
- 142.Pettit GR, aHerald CL, Doubek DL. J Am Chem Soc. 1982;104:6846–6847. [Google Scholar]
- 143.Pettit GR. Prog Chem Org Nat Prod. 1991;57:153–195. [Google Scholar]
- 144.Mutter R, Wills M. Bioorg Med Chem. 2000;8:1841–1860. doi: 10.1016/s0968-0896(00)00150-4. [DOI] [PubMed] [Google Scholar]
- 145.Blackhall FH, Ranson M, Radford JA, Hancock BW, Soukop M, McGown AT, Robbins A, Halbert G, Jayson GC. Br J Cancer. 2001;84:465–469. doi: 10.1054/bjoc.2000.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mohammad RM, Adsay NV, Philip PA, Pettit GR, Vaitkevicius VK, Sarkar FH. Anti-Cancer Drugs. 2001;12:735–740. doi: 10.1097/00001813-200110000-00005. [DOI] [PubMed] [Google Scholar]
- 147.Davidson S, Allen SW, Lim GE, Anderson CM, Haygood MG. Appl Environ Microbiol. 2001;67:4531–4537. doi: 10.1128/AEM.67.10.4531-4537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lim GE, Haygood MG. Appl Environ Microbiol. 2004;70:4921–4929. doi: 10.1128/AEM.70.8.4921-4929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lopanik N, Lindquist N, Targett N. Oecologia. 2004;139:131–139. doi: 10.1007/s00442-004-1487-5. [DOI] [PubMed] [Google Scholar]
- 150.Haygood MG, Davidson SK. Appl Environ Microbiol. 1997;63:4612–4616. doi: 10.1128/aem.63.11.4612-4616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu H, Patel A, Sherman DH, Haygood MG. J Nat Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 152.Reichenbach H. Environ Microbiol. 1999;1:15–21. doi: 10.1046/j.1462-2920.1999.00016.x. [DOI] [PubMed] [Google Scholar]
- 153.Reichenbach H. J Ind Microbiol Biotechnol. 2001;27:149–156. doi: 10.1038/sj.jim.7000025. [DOI] [PubMed] [Google Scholar]
- 154.Singh BN. J Gen Microbiol. 1947;1:1–10. doi: 10.1099/00221287-1-1-1. [DOI] [PubMed] [Google Scholar]
- 155.Gerth K, Pradella S, Perlova O, Beyer S, Müller R. J Biotechnol. 2003;106:233–253. doi: 10.1016/j.jbiotec.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 156.Wenzel SC, Müller R. Mol Biosyst. 2009;5:567–574. doi: 10.1039/b901287g. [DOI] [PubMed] [Google Scholar]
- 157.Weissmann KJ, Müller R. Bioorg Med Chem. 2009;17:2121–2136. doi: 10.1016/j.bmc.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 158.Weissmann KJ, Müller R. Nat Prod Rep. 2010;27:1276–1295. doi: 10.1039/c001260m. [DOI] [PubMed] [Google Scholar]
- 159.Hunt JT. Mol Cancer Ther. 2009;8:275–281. doi: 10.1158/1535-7163.MCT-08-0999. [DOI] [PubMed] [Google Scholar]
- 160.Kundim BA, Itou Y, Sakagami Y, Fudou R, Iizuka T, Yamanaka S, Ojika M. J Antibiot. 2003;56:630–638. doi: 10.7164/antibiotics.56.630. [DOI] [PubMed] [Google Scholar]
- 161.Iizuka T, Fudou R, Jojima Y, Ogawa S, Yamanaka S, Inukai Y, Ojika M. J Antibiot. 2006;59:385–391. doi: 10.1038/ja.2006.55. [DOI] [PubMed] [Google Scholar]
- 162.Ojika M, Suzuki Y, Tsukamoto A, Sakagami Y, Fudou R, Yoshimura T, Yamanaka S. J Antibiot. 1998;51:275–281. doi: 10.7164/antibiotics.51.275. [DOI] [PubMed] [Google Scholar]
- 163.Wodinsky I, Kensler CJ. Cancer Chemother Rep. 1965;47:65–68. [PubMed] [Google Scholar]
- 164.Schabel FM. Chemotherapy. 1968;13:321–338. doi: 10.1159/000220567. [DOI] [PubMed] [Google Scholar]
- 165.McIntosh M, Cruz LJ, Hunkapiller MW, Gray WR, Olivera BM. Arch Biochem Biophys. 1982;218:329–334. doi: 10.1016/0003-9861(82)90351-4. [DOI] [PubMed] [Google Scholar]
- 166.Look SA, Fenical W, Jacobs RS, Clardy J. Proc Natl Acad Sci. 1986;83:6238–6240. doi: 10.1073/pnas.83.17.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Look SA, Fenical W. Tetrahedron. 1987;43:3363–3370. [Google Scholar]
- 168.Wright AE, Forleo DA, Gunawardana GP, Gunasekera SP, Koehn FE, McConnell OJ. J Org Chem. 1990;55:4508–4512. [Google Scholar]
- 169.Luesch H, Moore RE, Paul VJ, Mooberry SL, Corbett TH. J Nat Prod. 2001;64:907–810. doi: 10.1021/np010049y. [DOI] [PubMed] [Google Scholar]
- 170.Haefner B. Drug Discovery Today. 2003;8:536–544. doi: 10.1016/s1359-6446(03)02713-2. [DOI] [PubMed] [Google Scholar]
- 171.Kuznetsov G, Towle MJ, Cheng H, Kawamura T, TenDyke K, Liu D, Kishi K, Yu MJ, Littlefield BA. Cancer Res. 2004;64:5760–5766. doi: 10.1158/0008-5472.CAN-04-1169. [DOI] [PubMed] [Google Scholar]
- 172.Garber K. Nat Biotechnol. 2005;23:399–399. doi: 10.1038/nbt0405-399. [DOI] [PubMed] [Google Scholar]
- 173.Molinski TF, Dalisay DS, Lievens SL, Saludes JP. Nat Rev Drug Discovery. 2009;8:69–85. doi: 10.1038/nrd2487. [DOI] [PubMed] [Google Scholar]
- 174.Lane AL, Moore BS. Nat Prod Rep. 2011;28:411–428. doi: 10.1039/c0np90032j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Katz J, Janik JE, Younes A. Clin Cancer Res. 2011;17:6428–6436. doi: 10.1158/1078-0432.CCR-11-0488. [DOI] [PubMed] [Google Scholar]