Abstract
Aims
To determine whether testing for isolated 1p or 19q losses, or as a codeletion, has any significance in the workup of glioblastomas (GBMs).
Methods
Upfront 1p/19q testing by fluorescence in situ hybridization (FISH) and/or polymerase chain reaction (PCR)-based loss of heterozygosity (LOH) was done in 491 gliomas that were histologically diagnosed as GBMs. Outcomes were determined and measured against 1p/19q results.
Results
Twenty-eight showed apparent 1p/19q codeletion by either FISH and/or PCR-based LOH, but only 1/26 showed codeletion by both tests. Over 90% of tumours with apparent codeletion by either FISH or LOH also had 10q LOH and/or EGFR amplification, features inversely related to true whole-arm 1p/19q codeletion. Furthermore, only 1/28 tumours demonstrated an R132H IDH1 mutation. Neither 1p/19q codeletion by FISH nor LOH had an impact on GBM survival. Isolated losses of 1p or 19q also had no impact on survival.
Conclusions
These data suggest that (i) 1p/19q testing is not useful on gliomas that are histologically GBMs; (ii) codeletion testing should be reserved only for cases with compatible morphology; and (iii) EGFR, 10q, and IDH1 testing can help act as safeguards against a false-positive 1p/19q result.
Keywords: 1p/19q, FISH, glioblastoma, LOH
Introduction
In diffusely infiltrative glial neoplasms, codeletion of the short arm of chromosome 1 and the long arm of chromosome 19 has been known to be a marker of oligodendroglial lineage for nearly 20 years [1,2]. This codeletion is the result of an unbalanced translocation between the two chromosomes with subsequent loss of der(1;19)(p10;q10) [3,4]. An oligodendroglioma that shows classic morphology such as small round nuclei with perinuclear cytoplasmic clearing, delicate branching vasculature, and microcalcifications has a greater than 80% likelihood of carrying the codeletion, while gliomas demonstrating mixed morphology (oligoastrocytomas) harbour the deletion less often [5–8]. Because of the strong association between 1p/19q and oligodendroglial lineage, testing for codeletion can also be very useful in differentiating oligodendroglioma from histological mimics that lack codeletion, including dysembryoplastic neuroepithelial tumour, clear cell ependymoma, and central neurocytoma [9,10]. Furthermore, mixed oligoastrocytomas with codeletion seem to behave more like oligodendrogliomas than comparable tumours lacking the codeletion [11–13].
1p/19q codeletion has more than just diagnostic relevance. Oligodendrogliomas with codeletion also demonstrate improved response to adjuvant radiochemotherapy and longer survival, especially at the grade III level [14–20]. Recently, both EORTC and RTOG clinical trials showed that codeletion in grade III oligodendroglial tumours is not just prognostic, but specifically predictive of better response to procarbazine, lomustine and vincristine (PCV) chemotherapy [21,22]. Interestingly, though, 1p/19q codeletion may not be a useful prognostic marker in the absence of adjuvant therapy [23].
While testing for 1p/19q codeletion is understandably an integral component of molecular neuro-oncology, there is still some heterogeneity in (i) selection of cases for testing, (ii) the variety of assays in use, and (iii) interpretation criteria. Two of the more common methods are fluorescence in situ hybridization (FISH) and polymerase chain reaction (PCR)-based microsatellite loss of heterozygosity (LOH). The former targets 1p36/1p25 and 19q13/19p13 via fluorophore-labelled DNA probes in tissue sections. These loci were initially selected because, as minimally deleted regions in gliomas, they are very sensitive [19,24,25]. However, they are not quite as specific for true whole-arm codeletion, especially if cut-off criteria are too inclusive [26]. Another widely used tool is PCR-based analysis of LOH in microsatellites scattered throughout chromosomes 1p and 19q, thereby providing a more accurate measure of copy number status along the entire lengths of both chromosome arms. Not surprisingly, microsatellite LOH analysis is slightly better than FISH at prognostic stratification [26].
Because of the aforementioned diagnostic and prognostic value of 1p/19q, it is widely used in the workup of gliomas. Such routine testing inevitably identifies cases that have either partial or total isolated losses at either 1p or 19q, yet cannot be histologically classified as oligodendroglial tumours. This raises the question as to whether such isolated partial or total deletions or codeletions have any prognostic significance. Some have suggested 19q copy number status is a significant prognostic stratifier for glioblastomas (GBMs) [27,28], though not all have found such an association [29–34]. Herein we studied 1p/19q testing in a large cohort of GBMs to prove whether 1p/19q testing has any value at all in GBMs.
Materials and methods
Cohort
Detailed treatment and survival data from 532 GBMs from 2002 to 2010 were retrieved from the Hillman Cancer Registry at the University of Pittsburgh (Table 1). Tumour location was available in 484 cases, surgical procedure for 493, and adjuvant therapy for 465. Regarding outcome, 470 (88.3%) of the patients were deceased at the time of analysis, with an overall median survival of 8.7 months. Cases of recurrent and/or treated gliomas were excluded from analysis. Diagnoses were made according to standard WHO criteria at the time of initial biopsy (i.e. astrocytic nuclear morphology, pleomorphism, endothelial proliferation, mitoses, palisading necrosis) [35]. All molecular data (see below) were generated at the time of initial diagnosis in each case. All data collection and analyses were done in accordance with the University of Pittsburgh and the University of Kentucky committees on human subjects.
Table 1.
Cohort characteristics
| Age | Median | 63 years |
| Range | 18–89 years | |
| Gender | Male (%) | 311 (58.5) |
| Female (%) | 221 (41.5) | |
| Surgery | Biopsy only (%) | 168 (34.1) |
| STR (%) | 187 (37.9) | |
| GTR (%) | 138 (28.0) | |
| Adjuvant therapy | None (%) | 82 (17.6) |
| Adjuvant with TMZ (%) | 312 (67.1) | |
| Adjuvant without TMZ (%) | 71 (15.3) | |
| Location | Frontal (%) | 143 (29.5) |
| Temporal (%) | 133 (27.5) | |
| Parietal (%) | 85 (17.6) | |
| Occipital (%) | 17 (3.5) | |
| Multilobe (%) | 59 (12.2) | |
| Other (%) | 47 (9.7) |
A total of 532 cases comprised the complete GBM cohort, 491 of which had upfront 1p/19q testing, 484 had known tumour location, 493 had known surgery type, and 465 had known information regarding adjuvant therapy. A total of 470 patients (88.3%) were deceased at the time of analysis. Mean survival was 8.7 months. TMZ, temozolomide; STR, subtotal resection; GTR, gross total resection.
Fluorescence in situ hybridization
Formalin-fixed paraffin-embedded blocks were analysed via FISH techniques using probes for 1p36, 19q13 and EGFR (7p12) (Abbott Molecular, Des Plaines, IL, USA), as previously described [36]. A minimum of 60 neoplastic nuclei were analysed in each tumour. For ploidy control, locus-specific probes were used for chromosomes 1 (1q25), 19 (19p13) and centromeric probe for chromosome 7 (CEP7). Codeletion was counted if the 1p36/1q25 and 19q13/19p13 ratios were both below 0.87 and at least 20% of tumour nuclei showed relative deletion. For EGFR, amplification was defined as an EGFR/CEP7 signal ratio >2.0. These cut-off points exceeded 3 SDs from the mean target: ploidy control ratio of 20 non-neoplastic autopsy brain tissue specimens.
PCR-based microsatellite LOH analysis
DNA from formalin-fixed paraffin-embedded tissue was probed as described previously [36]. Chromosome 1p was interrogated with seven microsatellite markers (D1S1172, D1S226, D1S162, D1S1161, D1S199, D1S407, D1S171). Prior to 2007, two markers were used on 19q (D19S112 and D19S206) and two were used on 10q (D10S1173 and D10S520); from 2007 onward (comprising 75% of the total cohort), additional microsatellites on 19q (D19S559) and 10q (D10S1171) were targeted, bringing the total microsatellites on those arms to three. PCR was performed, and the products were analysed using capillary gel electrophoresis on GeneMapper ABI 3730 (Applied Biosystems, Foster City, CA, USA). In most cases, patient-matched germline DNA from a peripheral blood sample was used as a control. When normal tissue was not available, peak height ratios falling outside 2 SDs beyond the mean of previously validated normal values for each polymorphic allele paring were assessed as showing LOH. Loss of heterozygosity regions were defined by bracketing only with informative markers. Noninformative loci were mapped and included within preserved or lost regions. To be considered code-leted the majority of informative microsatellite loci on both 1p and 19q had to show LOH. Any LOH on 10q was counted as 10q LOH.
IDH1 immunohistochemistry
Analysis of the R132H mutant isocitrate dehydrogenase 1 (mtIDH1) was performed using a mutation-specific monoclonal antibody by standard immunohistochemical methods [37,38].
Statistics
Univariate analyses were performed using log-rank tests on Kaplan–Meier curves; multivariate analysis was performed via Cox proportional hazards survival regression. Statistical analyses were performed using GraphPad software (La Jolla, CA, USA) and http://statpages.org/prophaz.html. Differences were considered significant if P < 0.05.
Results
Cohort characteristics
The male : female ratio in the GBM cohort was 1.4:1 and a median age of 63 years (range 18–89 years) (Table 1). Nearly 60% of all GBMs arose in the frontal or temporal lobes, 17.6% localized to a parietal lobe, only 3.5% were in an occipital lobe, 12.2% involved at least two lobes, and 9.7% were in other sites (e.g. basal ganglial). Two-thirds of patients received either gross total or subtotal surgical resection, with the other third having only computerized tomography (CT)-guided stereotactic biopsies. About 67% of patients received temozolomide (TMZ) and radio-therapy, with the remainder split between adjuvant therapies lacking TMZ or no therapy at all – reasons for these include pre-2005 cases where TMZ was not yet part of routine care, adverse side effects of TMZ, electing to pursue alternative treatment, too ill to pursue any additional therapy, or refusing treatment altogether.
1p/19q status and FISH-LOH concordance in GBMs
Four hundred and ninety-one GBMs were tested for 1p/19q codeletion by either FISH alone (44), LOH alone (1), or both (446); only 28 (5.7%) showed apparent 1p/19q codeletion by either FISH and/or PCR-based LOH (Table 2). In this subset of 28 GBMs there was a strong inverse correlation between the two methods (Spearman r = −0.91, P < 0.0001), meaning an extremely poor concordance between FISH and PCR-based LOH when either test suggested codeletion. For example, 18/28 had codeletions detected only by FISH, with negative results by matched LOH testing. Seven had 1p/19q codeletions detected only by LOH, with concomitant negative FISH results. Two GBMs showed codeletions by FISH but did not have LOH data. Only 1 of the 28 GBMs showed 1p/19q codeletion by both methods.
Table 2.
GBMs with 1p/19q codeletion by FISH or LOH analysis
| Case | Age (years) |
Gender | Primary site |
1p/19q codeletion by FISH |
1p36/ 1q25 ratio |
% 1p36 deleted |
19q13 /19p13 ratio |
% 19q deleted |
1p/19q codeletion by LOH |
10q LOH |
EGFR amp |
MGMT status |
R132H IDH1 status |
Therapy | Survival (months) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 72 | Male | Frontal | Yes | 0.48 | 90 | 0.45 | 95 | No | Yes | Yes | N/A | neg | Resection, no adjuvant | 0.76 |
| 2 | 72 | Female | Parietal | Yes | 0.5 | 95 | 0.63 | 75 | No | N/A | Yes | N/A | neg | Biopsy, no adjuvant | 0.23 |
| 3 | 57 | Male | Temporal | Yes | 0.58 | 80 | 0.84 | 40 | No | Yes | Yes | pos | neg | Resection, TMZ/XRT, NovoTTF-100A, bevacizumab/irinotecan | 8.0 |
| 4 | 80 | Male | Parietal | Yes | 0.59 | 82 | 0.64 | 72 | No | Yes | No | N/A | neg | Resection, TMZ/XRT | 9.4 |
| 5 | 51 | Male | Temporal | Yes | 0.59 | 87 | 0.82 | 45 | No | Yes | No | neg | neg | Resection, TMZ/XRT, bevacizumab/irinotecan | 63.5 |
| 6 | 49 | Female | frontal | Yes | 0.6 | 80 | 0.55 | 90 | No | Yes | Yes | N/A | neg | Resection, TMZ/XRT | 10.6 |
| 7 | 62 | Female | Parietal | Yes | 0.64 | 71 | 0.75 | 55 | No | Yes | No | pos | neg | Resection, TMZ/XRT, bevacizumab/irinotecan | 26.1 |
| 8 | 58 | Male | Temporal | Yes | 0.65 | 74 | 0.73 | 65 | No | No | No | Low-level | neg | Resection, TMZ/XRT, carboplatin/etoposide | 27.3 |
| 9 | 69 | Male | Parietal | Yes | 0.73 | 58 | 0.70 | 67 | No | Yes | No | pos | neg | Biopsy, no adjuvant | 8.8 |
| 10 | 65 | Female | Temporal | Yes | 0.75 | 52 | 0.68 | 64 | No | Yes | No | pos | neg | Resection, TMZ/XRT | 6.7 |
| 11 | 77 | Male | N/A | Yes | 0.77 | 52 | 0.60 | 79 | No | Yes | Yes | pos | neg | Resection, adjuvant unknown | 4.1 |
| 12 | 33 | Male | Brainstem | Yes | 0.77 | 47 | 0.82 | 37 | No | Yes | No | N/A | neg | Biopsy, XRT only | 11.9 |
| 13 | 63 | Male | Temporal | Yes | 0.78 | 44 | 0.85 | 35 | No | Yes | Yes | neg | neg | Biopsy, TMZ and irinotecan with XRT | 11.7 |
| 14 | 52 | Male | Occipital | Yes | 0.78 | 59 | 0.89 | 25 | No | No | No | Low-level | neg | Resection, TMZ/XRT | 12.3 |
| 15 | 52 | Female | Temporal | Yes | 0.79 | 41 | 0.74 | 49 | N/A | Yes | No | N/A | pos | Biopsy, XRT only | 6.0 |
| 16 | 61 | Male | Frontal | Yes | 0.8 | 46 | 0.75 | 48 | No | Yes | Yes | N/A | neg | Resection, TMZ/XRT | 22.7 |
| 17 | 48 | Male | Frontal | Yes | 0.81 | 38 | 0.67 | 65 | No | No | Yes | neg | neg | Resection, adjuvant unknown | 24.7 |
| 18 | 60 | Male | Frontal | Yes | 0.82 | 35 | 0.55 | 82 | Yes | Yes | No | pos | neg | Biopsy, TMZ/XRT | 9.9 |
| 19 | 52 | Male | Occipital | Yes | 0.85 | 29 | 0.69 | 68 | No | Yes | Yes | Low-level | neg | Resection, TMZ/XRT | 12.3 |
| 20 | 57 | Female | Frontal | Yes | 0.85 | 28 | 0.74 | 63 | No | Yes | Yes | Low-level | neg | Resection, TMZ/XRT | 8.7 |
| 21 | 61 | Female | N/A | Yes | N/A | N/A | N/A | N/A | N/A | N/A | No | N/A | neg | Unknown | 21.0 |
| 22 | 75 | Male | Parietal | No | 0.58 | 76 | 1.1 | 19 | Yes | Yes | Yes | N/A | neg | Resection, TMZ/XRT, bevacizumab/irinotecan | 14.7 |
| 23 | 59 | Male | N/A | No | 0.92 | 18 | 0.75 | 59 | Yes | Yes | Yes | N/A | neg | Unknown | 42.0 |
| 24 | 47 | Male | Parietal | No | 0.96 | 7 | 1.0 | 2 | Yes | Yes | No | N/A | neg | Resection, TMZ/XRT, bevacizumab/irinotecan | 76.2 |
| 25 | 85 | Male | Parietal | No | 0.97 | 7 | 0.95 | 11 | Yes | Yes | No | N/A | neg | Resection, no adjuvant | 3.4 |
| 26 | 76 | Female | Frontal | No | 0.99 | 16 | 0.99 | 12 | Yes | Yes | No | pos | neg | Biopsy, XRT only | 2.8 |
| 27 | 57 | Male | N/A | No | 0.89 | 22 | 0.98 | 5 | Yes | Yes | No | neg | neg | Unknown | 22.1 |
| 28 | 59 | Male | N/A | No | 0.94 | 18 | 0.97 | 7 | Yes | Yes | No | Low-level | neg | Unknown | 5.6 |
Of the entire cohort, 21 tumours showed apparent 1p/19q codeletion by FISH. Only one of those tumours was also codeleted by PCR-based LOH analysis. In the majority of cases, patient survival was more consistent with a GBM than a 1p/19q-codeleted anaplastic oligodendroglioma. Survival was also generally poor in the 8 cases called codeleted by LOH analysis. In all but 3 cases either 10q LOH and/or EGFR amplification was detected. Representative virtual whole-slide images are available online for Cases 2 (http://image.upmc.edu:8080/NeuroPathology/GlialTumors/GlialTumor4/GT.114.svs/view.apml?), 5 (http://image.upmc.edu:8080/NeuroPathology/GlialTumors/GlialTumor4/GT.116.svs/view.apml?), 8 (http://image.upmc.edu:8080/NeuroPathology/GlialTumors/GlialTumor4/GT.117.svs/view.apml?) and 18 (http://image.upmc.edu:8080/NeuroPathology/GlialTumors/GlialTumor4/GT.115.svs/view.apml?).
Amp, amplification; neg, negative; pos, positive; TMZ, temozolomide; XRT, radiotherapy; N/A, not available; FISH, fluorescence in situ hybridization; LOH, loss of heterozygosity; GBM, glioblastoma; PCR, polymerase chain reaction.
Tumour location in this subset of 28 GBMs was similar to the cohort as a whole, with about 57% being located in either the frontal or temporal lobes. This subset was also similar to the whole cohort regarding age (median 59.5 years) and male : female ratio (1.4:1).
Histological characteristics of GBMs with apparent 1p/19q codeletion
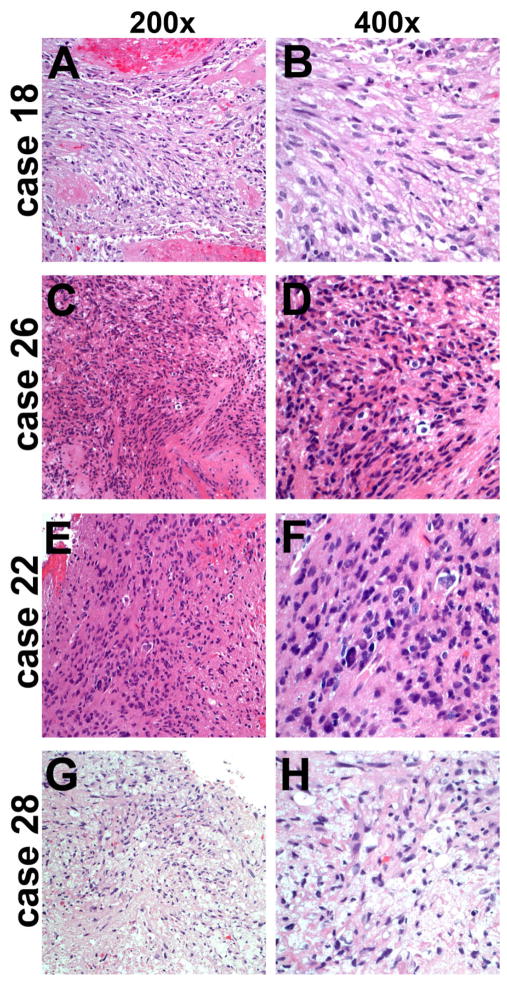
The morphologic characteristics of GBMs with 1p/19q codeletion by FISH were studied in detail. Four representative cases are shown in Figure 1, and web links to virtual slides of four other representative cases are embedded in Table 2. All cases had strong astrocytic nuclear morphology – i.e. irregular angulated nuclei – as well as microvascular proliferation and/or necrosis. No significant oligodendroglial components were present in any of the cases. Likewise, GBMs showing codeletion by LOH analysis were strongly astrocytic. Even the single case showing codeletion by both assays was more astrocytic than oligodendroglial and had 10q LOH (Figure 2A,B, Case 18 in Table 2); that patient died within 10 months of initial diagnosis.
Figure 1.
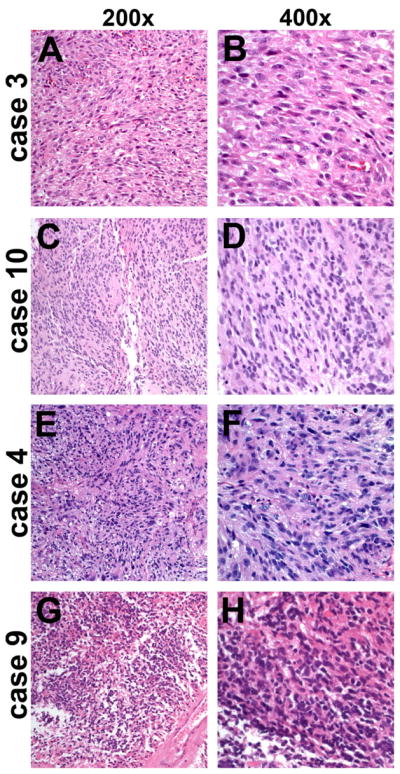
GBMs with false 1p/19q codeletion by FISH. Despite strong astrocytic morphology, all four cases of GBM showed 1p/19q codeletion by FISH (case numbers correspond to Table 2). However, none were codeleted by PCR-based LOH analysis and all had 10q LOH. Case 3 (A, B) also had EGFR amplification. None of the four patients survived beyond 10 months after initial surgery. FISH, fluorescence in situ hybridization; LOH, loss of heterozygosity; GBM, glioblastoma; PCR, polymerase chain reaction.
Figure 2.
Glioblastomas with false 1p/19q codeletion by LOH. Analogous to Figure 1, these four histologically unequivocal GBMs showed 1p/19q codeletion by LOH (case numbers correspond to Table 2). Only case 18 appeared codeleted by FISH and all had 10q LOH. Case 22 (E, F) also had EGFR amplification. None of the four patients survived beyond 15 months after initial surgery. FISH, fluorescence in situ hybridization; LOH, loss of heterozygosity; GBM, glioblastoma.
Genetic characteristics of GBMs with apparent 1p/19q codeletion
In the entire GBM cohort, 43.6% had EGFR amplification and 82.2% had 10q LOH. Both genetic lesions are known to be strongly associated with astrocytic rather than oligodendroglial tumours, and inversely correlate with true whole-arm 1p/19q codeletion. Interestingly, 25 of 27 GBMs (92.6%) with apparent 1p/19q codeletion also had either EGFR amplification and/or 10q LOH (Table 2). (One tumour did not have EGFR amplification and did not have available 10q data.)
Several groups have shown a strong correlation between whole-arm 1p/19q codeletion and a concomitant IDH1/2 mutation, the latter occurring in the form of R132H IDH1 about 90% of the time [39–41]. In other words, whole-arm codeletion only occurs in conjunction with an IDH1/2 mutation (although most IDH1/2-mutant gliomas do not have codeletions). In this subset of 28 GBMs, only 1 showed R132H IDH1 immunoreactivity, a temporal lobe tumour with 10q LOH in a 52 year-old woman who survived 6 months after diagnosis (Table 2, Case 15).
Outcomes of GBMs with apparent 1p/19q codeletion
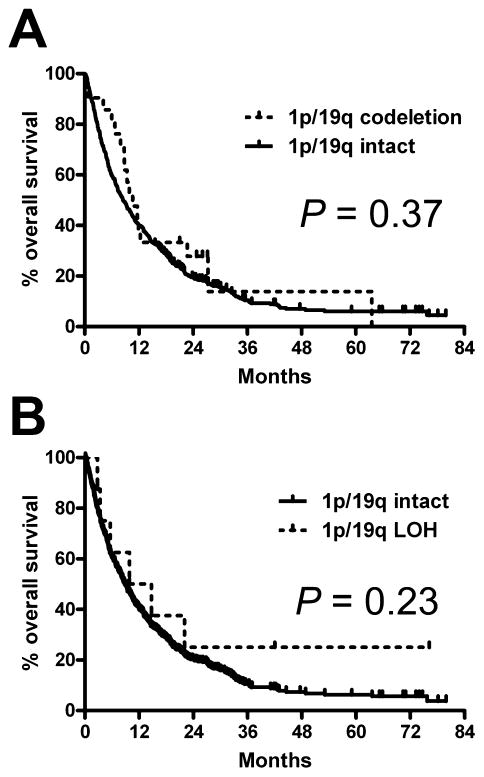
The main objective in 1p/19q testing is to identify gliomas that will respond better to adjuvant therapy. In the current subset of gliomas that were morphologically GBMs (usually having other genetic lesions typically discordant with codeletion), cases identified as 1p/19q-codeleted showed no difference in survival, either by FISH (Figure 3A) or LOH (Figure 3B). Via multivariate analysis, neither test showed even a weak trend towards independent prognostic value (Table 3). On the other hand, well-known variables such as patient age, bulk resection and use of TMZ all showed significance as expected.
Figure 3.
1p/19q data lacks prognostic significance in histological GBMs. On univariate analysis, neither 1p/19q FISH (A) nor PCR-based LOH data (B) had any relevance concerning overall survival. FISH, fluorescence in situ hybridization; LOH, loss of heterozygosity; GBM, glioblastoma; PCR, polymerase chain reaction.
Table 3.
Multivariate analysis of clinical and molecular factors in GBM
| Variable | HR | 95% CI | P |
|---|---|---|---|
| Age 45+ vs. <45 years | 2.7 | 1.8–4.1 | <0.0001 |
| Resection vs. biopsy alone | 0.53 | 0.41–0.68 | <0.0001 |
| TMZ vs. no TMZ | 0.35 | 0.27–0.46 | <0.0001 |
| 1p/19q codeletion by FISH | 0.87 | 0.50–1.5 | 0.61 |
| 1p/19q codeletion by LOH | 0.73 | 0.27–2.0 | 0.54 |
| 10q LOH | 0.97 | 0.70–1.3 | 0.85 |
| EGFR amplification | 1.0 | 0.79–1.3 | 0.91 |
Patient age, surgery type and adjuvant therapy with TMZ were all independent prognostic variables in tumours that were histological GBMs. In contrast, neither 1p/19q codeletion by FISH or LOH analysis had any impact on survival; neither did 10q LOH or EGFR amplification.
HR, hazard ratio; CI, confidence interval; FISH, fluorescence in situ hybridization; LOH, loss of heterozygosity; GBM, glioblastoma.
Often on 1p/19q testing, isolated losses of 1p or 19q are seen in GBMs. In this cohort, besides the cases with apparent codeletion, 58/456 GBMs (12.7%) showed isolated 1p36 loss via FISH and 41/453 (9.1%) had isolated 19q13 loss. Via PCR, 82/423 (19.4%) had subtotal 1p LOH, while 5 (1.2%) had total 1p LOH. Likewise, 76/424 (17.9%) showed subtotal 19q LOH and 28 (6.6%) had complete isolated 19q LOH. Neither total nor subtotal isolated 1p or 19q deletions had any correlation with survival on univariate (Figure S1) or multivariate analyses (not shown).
Discussion
Inherent in any test is knowing when to use that test and how to interpret results that conflict with other pieces of data. While not common, a 1p/19q codeletion result can be seen in the occasional GBM [42,43] and begs the question as to whether the molecular test or the histological appearance of the tumour should take precedence. Based on a large cohort of GBMs with 1p/19q data, our conclusions are that diagnostic and prognostic weight should be given to tumour morphology over an apparent codeletion, and that additional molecular testing for 10q LOH, EGFR amplification and/or IDH1 mutations could help triage high-grade gliomas for additional 1p/19q analysis.
A key issue in 1p/19q testing is what cut-off criteria to adopt. At the moment there is a great deal of cut-off heterogeneity for the most widely used 1p36/1q25 and 19q13/19p13 FISH probes. Previously, we determined that each probe pair ratio should be <0.75 or have relative loss in greater than 40% tumour nuclei to produce maximal prognostic stratification, although the longstanding institutional cut-offs, established using non-neoplastic control tissue, were <0.87 and 20% [26]. But in this dataset, even adopting those more stringent cut-offs still yielded false-positive codeletions, with 8 tumours showing ratios below 0.75 on both 1p and 19q and 15 tumours showing codeletion in greater than 40% of cells (Table 2). Thus, although rare, such cases will occur if upfront reflex 1p/19q testing is done on all diffusely infiltrative gliomas.
In the 8 cases in this subset that met our initial LOH codeletion criteria, usually only a single 1p or 19q micro-satellite remained intact. For there to be true whole-arm codeletion, all microsatellites should be lost on 1p and 19q. Because none of the tumours in this cohort showed complete loss of microsatellites on both arms, and none had other molecular signatures compatible with a codeleted tumour (see below), it is highly probable that none truly had oligodendroglioma-like whole-arm codeletion. Thus, anything less than complete loss of all microsatellite loci on both arms should be viewed with scepticism.
Going forward, more advanced assays like array comparative genomic hybridization or multiplex ligation assays are likely to be better at prognostic stratification, specifically avoiding false-positive cases. For now such platforms generally remain the purview of larger centres, and FISH and PCR-based LOH will likely remain popular in many laboratories for some time. Nevertheless, their technical limitations must be recognized; specifically, that neither assay can always reliably distinguish partial from whole-arm losses.
Other molecular tests can identify gliomas that would not benefit from focused 1p/19q analysis. For example, LOH on 10q is associated with high-grade astrocytic tumours [12,44]. We recently showed that, even in oligodendroglial-appearing tumours with 1p/19q codeletion by FISH, the presence of 10q LOH predicted a discrepant negative 1p/19q LOH result and survival more like GBM than anaplastic oligodendroglioma [26]. Similarly, EGFR amplification is a hallmark of GBMs and is inversely correlated with true whole-arm 1p/19q codeletion [44,45]. This is consistent with prior work showing that tumours resembling anaplastic oligodendroglioma, but with EGFR amplification or 10q LOH, are better classified as small cell GBMs [46]. In our cohort, none of the tumours with amplification and 1p/19q codeletion by FISH also had codeletion via LOH, and their median survival was typical of GBMs (Table 2). And two recent large studies have shown that true whole-arm 1p/19q codeletion is virtually never seen in the absence of an IDH1/2 mutation [39,40]. Our data are consistent with those observations, and suggests upfront IDH1/2 screening is useful before testing for 1p/19q codeletion analysis.
Regarding GBMs with an oligodendroglial component (GBM-O), some have shown that such tumours have a slightly better prognosis, though the presence of necrosis negates that effect [47]. One study showed no difference in survival [48], although many of the ‘GBM-O’ cases in that cohort had EGFR amplification and may have been small cell GBMs [46]. Nevertheless, none of the codeleted GBMs in this cohort showed appreciable oligodendroglial morphology (see Table 2 hyperlinks for representative virtual slides), or if even some of the tumour was equivocal for oligodendroglial morphology (e.g. case 18 virtual slide in Table 2), the tumour was negative for R132H IDH1 and had 10q LOH, both of which are more consistent with GBM than an oligodendroglioma [26,46,49].
Taken together, our data suggest that upfront reflex 1p/19q testing of all gliomas is unhelpful, insofar as even the rare ‘positive’ result in a GBM is unlikely to have clinically relevant whole-arm codeletion, and a change in diagnosis or prognosis is not warranted. In questionable cases where codeletion testing needs to be done, assessment of 10q and EGFR copy number and/or IDH1/2 mutations can act as a check on 1p/19q results.
EORTC and RTOG studies showed 1p/19q codeletion to be predictive of PCV response in anaplastic oligodendrogliomas [21,22]. However, only one case in this entire current cohort involved adjuvant PCV, as second-line therapy in a GBM with no sign of 1p/19q codeletion (not shown), and the patient survived 10 months after surgery.
From a broader perspective, molecular testing in gliomas is still the way of the future, but results cannot be interpreted in isolation. Clinical trial assignment often depends on specific molecular results, and the field as a whole seems to be heading towards reclassification of gliomas based on upfront genetic testing. However, this study demonstrates that, because 1p/19q testing has no utility in a histologically unequivocal GBM, light microscopic examination will likely remain a primary component of glioma diagnostics for quite some time.
Supplementary Material
Isolated 1p or 19q have no significant effect on GBM response according to adjuvant therapy. Neither 1p deletion by FISH (A–C) nor LOH (G–I), nor 19q3 deletion by FISH (D–F) nor LOH (J–L), correlated with any difference in GBM survival, regardless of whether there was no adjuvant therapy (A, D, G, J), adjuvant therapy without TMZ (B, E, H, K), or adjuvant therapy with TMZ (C, F, I, L). FISH, fluorescence in situ hybridization; LOH, loss of heterozygosity; TMZ, temozolomide.
Acknowledgments
The authors thank the In Situ Hybridization Laboratory at the University of Pittsburgh for their assistance with the FISH data and the Molecular Anatomic Pathology Laboratory at the University of Pittsburgh for their assistance with the LOH data. The authors also acknowledge the assistance of Sharon Winters of the Hillman Cancer Registry in providing clinical follow-up data.
C.H. was supported by K08 CA155764-01A1, 2P20 RR020171, and the University of Kentucky College of Medicine Physician Scientist Program.
Footnotes
Author contributions
Kenneth H. Clark – histological imaging and manuscript preparation.
John L. Villano – manuscript preparation.
Marina N. Nikiforova – molecular analyses.
Ronald L. Hamilton – collection of cases and clinical data.
Craig Horbinski – project conception, data analysis, and manuscript preparation.
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
References
- 1.Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W, Collins VP. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol. 1994;145:1175–90. [PMC free article] [PubMed] [Google Scholar]
- 2.Kraus JA, Koopmann J, Kaskel P, Maintz D, Brandner S, Schramm J, Louis DN, Wiestler OD, von Deimling A. Shared allelic losses on chromosomes 1p and 19q suggest a common origin of oligodendroglioma and oligoastrocytoma. J Neuropathol Exp Neurol. 1995;54:91–5. doi: 10.1097/00005072-199501000-00011. [DOI] [PubMed] [Google Scholar]
- 3.Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, Flynn H, Passe S, Felten S, Brown PD, Shaw EG, Buckner JC. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–61. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 4.Griffin CA, Burger P, Morsberger L, Yonescu R, Swierczynski S, Weingart JD, Murphy KM. Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol. 2006;65:988–94. doi: 10.1097/01.jnen.0000235122.98052.8f. [DOI] [PubMed] [Google Scholar]
- 5.Jeuken JW, von Deimling A, Wesseling P. Molecular pathogenesis of oligodendroglial tumors. J Neurooncol. 2004;70:161–81. doi: 10.1007/s11060-004-2748-1. [DOI] [PubMed] [Google Scholar]
- 6.Reifenberger G, Louis DN. Oligodendroglioma: toward molecular definitions in diagnostic neuro-oncology. J Neuropathol Exp Neurol. 2003;62:111–26. doi: 10.1093/jnen/62.2.111. [DOI] [PubMed] [Google Scholar]
- 7.Giannini C, Burger PC, Berkey BA, Cairncross JG, Jenkins RB, Mehta M, Curran WJ, Aldape K. Anaplastic oligodendroglial tumors: refining the correlation among histopathology, 1p 19q deletion and clinical outcome in Intergroup Radiation Therapy Oncology Group Trial 9402. Brain Pathol. 2008;18:360–9. doi: 10.1111/j.1750-3639.2008.00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald JM, See SJ, Tremont IW, Colman H, Gilbert MR, Groves M, Burger PC, Louis DN, Giannini C, Fuller G, Passe S, Blair H, Jenkins RB, Yang H, Ledoux A, Aaron J, Tipnis U, Zhang W, Hess K, Aldape K. The prognostic impact of histology and 1p/19q status in anaplastic oligodendroglial tumors. Cancer. 2005;104:1468–77. doi: 10.1002/cncr.21338. [DOI] [PubMed] [Google Scholar]
- 9.Fouladi M, Helton K, Dalton J, Gilger E, Gajjar A, Merchant T, Kun L, Newsham I, Burger P, Fuller C. Clear cell ependymoma: a clinicopathologic and radiographic analysis of 10 patients. Cancer. 2003;98:2232–44. doi: 10.1002/cncr.11783. [DOI] [PubMed] [Google Scholar]
- 10.Tong CY, Ng HK, Pang JC, Hu J, Hui AB, Poon WS. Central neurocytomas are genetically distinct from oligodendrogliomas and neuroblastomas. Histopathology. 2000;37:160–5. doi: 10.1046/j.1365-2559.2000.00977.x. [DOI] [PubMed] [Google Scholar]
- 11.Eoli M, Bissola L, Bruzzone MG, Pollo B, Maccagnano C, De Simone T, Valletta L, Silvani A, Bianchessi D, Broggi G, Boiardi A, Finocchiaro G. Reclassification of oligoastrocytomas by loss of heterozygosity studies. Int J Cancer. 2006;119:84–90. doi: 10.1002/ijc.21759. [DOI] [PubMed] [Google Scholar]
- 12.Fuller CE, Schmidt RE, Roth KA, Burger PC, Scheithauer BW, Banerjee R, Trinkaus K, Lytle R, Perry A. Clinical utility of fluorescence in situ hybridization (FISH) in morphologically ambiguous gliomas with hybrid oligodendroglial/astrocytic features. J Neuropathol Exp Neurol. 2003;62:1118–28. doi: 10.1093/jnen/62.11.1118. [DOI] [PubMed] [Google Scholar]
- 13.Wick W, Weller M. Classification and management of anaplastic gliomas. Curr Opin Neurol. 2009;22:650–6. doi: 10.1097/WCO.0b013e3283329ce7. [DOI] [PubMed] [Google Scholar]
- 14.Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, Ramsay DA, Louis DN. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90 :1473–9. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 15.Smith JS, Perry A, Borell TJ, Lee HK, O’Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, Jenkins RB. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–45. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 16.Perry A, Fuller CE, Banerjee R, Brat DJ, Scheithauer BW. Ancillary FISH analysis for 1p and 19q status: preliminary observations in 287 gliomas and oligodendroglioma mimics. Front Biosci. 2003;8:a1–9. doi: 10.2741/896. [DOI] [PubMed] [Google Scholar]
- 17.van den Bent MJ, Looijenga LH, Langenberg K, Dinjens W, Graveland W, Uytdewilligen L, Sillevis Smitt PA, Jenkins RB, Kros JM. Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer. 2003;97:1276–84. doi: 10.1002/cncr.11187. [DOI] [PubMed] [Google Scholar]
- 18.Fallon KB, Palmer CA, Roth KA, Nabors LB, Wang W, Carpenter M, Banerjee R, Forsyth P, Rich K, Perry A. Prognostic value of 1p, 19q, 9p, 10q, and EGFR-FISH analyses in recurrent oligodendrogliomas. J Neuropathol Exp Neurol. 2004;63:314–22. doi: 10.1093/jnen/63.4.314. [DOI] [PubMed] [Google Scholar]
- 19.Felsberg J, Erkwoh A, Sabel MC, Kirsch L, Fimmers R, Blaschke B, Schlegel U, Schramm J, Wiestler OD, Reifenberger G. Oligodendroglial tumors: refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol. 2004;14:121–30. doi: 10.1111/j.1750-3639.2004.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouwenhoven MC, Gorlia T, Kros JM, Ibdaih A, Brandes AA, Bromberg JE, Mokhtari K, van Duinen SG, Teepen JL, Wesseling P, Vandenbos F, Grisold W, Sipos L, Mirimanoff R, Vecht CJ, Allgeier A, Lacombe D, van den Bent MJ. Molecular analysis of anaplastic oligodendroglial tumors in a prospective randomized study: a report from EORTC study 26951. Neuro Oncol. 2009;11:737–46. doi: 10.1215/15228517-2009-011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J, Fink K, Souhami L, Laperriere N, Curran W, Mehta M. Phase III trial of chemoradiotherapy for ana-plastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31:337–43. doi: 10.1200/JCO.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kouwenhoven MC, Delattre JY, Bernsen HJ, Frenay M, Tijssen CC, Grisold W, Sipos L, Enting RH, French PJ, Dinjens WN, Vecht CJ, Allgeier A, Lacombe D, Gorlia T, Hoang-Xuan K. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC Brain Tumor Group Study 26951. J Clin Oncol. 2013;31:344–50. doi: 10.1200/JCO.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- 23.Weller M, Berger H, Hartmann C, Schramm J, Westphal M, Simon M, Goldbrunner R, Krex D, Steinbach JP, Ostertag CB, Loeffler M, Pietsch T, von Deimling A. Combined 1p/19q loss in oligodendroglial tumors: predictive or prognostic biomarker? Clin Cancer Res. 2007;13:6933–7. doi: 10.1158/1078-0432.CCR-07-0573. [DOI] [PubMed] [Google Scholar]
- 24.Kitange G, Misra A, Law M, Passe S, Kollmeyer TM, Maurer M, Ballman K, Feuerstein BG, Jenkins RB. Chromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas. Genes Chromosomes Cancer. 2005;42:68–77. doi: 10.1002/gcc.20108. [DOI] [PubMed] [Google Scholar]
- 25.Smith JS, Tachibana I, Lee HK, Qian J, Pohl U, Mohrenweiser HW, Borell TJ, Hosek SM, Soderberg CL, von Deimling A, Perry A, Scheithauer BW, Louis DN, Jenkins RB. Mapping of the chromosome 19 q-arm glioma tumor suppressor gene using fluorescence in situ hybridization and novel microsatellite markers. Genes Chromosomes Cancer. 2000;29:16–25. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1007>3.3.co;2-9. [DOI] [PubMed] [Google Scholar]
- 26.Horbinski C, Nikiforova MN, Hobbs J, Bortoluzzi S, Cieply K, Dacic S, Hamilton RL. The importance of 10q status in an outcomes-based comparison between 1p/19q fluorescence in situ hybridization and polymerase chain reaction-based microsatellite loss of heterozygosity analysis of oligodendrogliomas. J Neuropathol Exp Neurol. 2012;71:73–82. doi: 10.1097/NEN.0b013e318240fa65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneshiro D, Kobayashi T, Chao ST, Suh J, Prayson RA. Chromosome 1p and 19q deletions in glioblastoma multiforme. Appl Immunohistochem Mol Morphol. 2009;17:512–6. doi: 10.1097/PAI.0b013e3181a2c6a4. [DOI] [PubMed] [Google Scholar]
- 28.Korshunov A, Sycheva R, Golanov A. The prognostic relevance of molecular alterations in glioblastomas for patients age < 50 years. Cancer. 2005;104:825–32. doi: 10.1002/cncr.21221. [DOI] [PubMed] [Google Scholar]
- 29.Batchelor TT, Betensky RA, Esposito JM, Pham LD, Dorfman MV, Piscatelli N, Jhung S, Rhee D, Louis DN. Age-dependent prognostic effects of genetic alterations in glioblastoma. Clin Cancer Res. 2004;10:228–33. doi: 10.1158/1078-0432.ccr-0841-3. [DOI] [PubMed] [Google Scholar]
- 30.Felsberg J, Rapp M, Loeser S, Fimmers R, Stummer W, Goeppert M, Steiger HJ, Friedensdorf B, Reifenberger G, Sabel MC. Prognostic significance of molecular markers and extent of resection in primary glioblastoma patients. Clin Cancer Res. 2009;15:6683–93. doi: 10.1158/1078-0432.CCR-08-2801. [DOI] [PubMed] [Google Scholar]
- 31.Houillier C, Lejeune J, Benouaich-Amiel A, Laigle-Donadey F, Criniere E, Mokhtari K, Thillet J, Delattre JY, Hoang-Xuan K, Sanson M. Prognostic impact of molecular markers in a series of 220 primary glioblastomas. Cancer. 2006;106:2218–23. doi: 10.1002/cncr.21819. [DOI] [PubMed] [Google Scholar]
- 32.Pinto LW, Araujo MB, Vettore AL, Wernersbach L, Leite AC, Chimelli LM, Soares FA. Glioblastomas: correlation between oligodendroglial components, genetic abnormalities, and prognosis. Virchows Arch. 2008;452:481–90. doi: 10.1007/s00428-007-0562-9. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt MC, Antweiler S, Urban N, Mueller W, Kuklik A, Meyer-Puttlitz B, Wiestler OD, Louis DN, Fimmers R, von Deimling A. Impact of genotype and morphology on the prognosis of glioblastoma. J Neuropathol Exp Neurol. 2002;61:321–8. doi: 10.1093/jnen/61.4.321. [DOI] [PubMed] [Google Scholar]
- 34.Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, Westphal M, Schackert G, Simon M, Tonn JC, Heese O, Krex D, Nikkhah G, Pietsch T, Wiestler O, Reifenberger G, von Deimling A, Loeffler M. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27:5743–50. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 35.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO Classification of Tumors of the Central Nervous System. 4. Lyon: IARC; 2007. [Google Scholar]
- 36.Horbinski C, Hamilton RL, Nikiforov Y, Pollack IF. Association of molecular alterations, including BRAF, with biology and outcome in pilocytic astrocytomas. Acta Neuropathol. 2010;119:641–9. doi: 10.1007/s00401-009-0634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capper D, Weissert S, Balss J, Habel A, Meyer J, Jager D, Ackermann U, Tessmer C, Korshunov A, Zentgraf H, Hartmann C, von Deimling A. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2009;20:245–54. doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009;118:599–601. doi: 10.1007/s00401-009-0595-z. [DOI] [PubMed] [Google Scholar]
- 39.Labussiere M, Idbaih A, Wang XW, Marie Y, Boisselier B, Falet C, Paris S, Laffaire J, Carpentier C, Criniere E, Ducray F, El Hallani S, Mokhtari K, Hoang-Xuan K, Delattre JY, Sanson M. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74:1886–90. doi: 10.1212/WNL.0b013e3181e1cf3a. [DOI] [PubMed] [Google Scholar]
- 40.Yip S, Butterfield YS, Morozova O, Chittaranjan S, Blough MD, An J, Birol I, Chesnelong C, Chiu R, Chuah E, Corbett R, Docking R, Firme M, Hirst M, Jackman S, Karsan A, Li H, Louis DN, Maslova A, Moore R, Moradian A, Mungall KL, Perizzolo M, Qian J, Roldan G, Smith EE, Tamura-Wells J, Thiessen N, Varhol R, Weiss S, Wu W, Young S, Zhao Y, Mungall AJ, Jones SJ, Morin GB, Chan JA, Cairn-cross JG, Marra MA. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol. 2012;226:7–16. doi: 10.1002/path.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 42.von Deimling A, Fimmers R, Schmidt MC, Bender B, Fass-bender F, Nagel J, Jahnke R, Kaskel P, Duerr EM, Koopmann J, Maintz D, Steinbeck S, Wick W, Platten M, Muller DJ, Przkora R, Waha A, Blumcke B, Wellenreuther R, Meyer-Puttlitz B, Schmidt O, Mollenhauer J, Poustka A, Stangl AP, Lenartz D, von Ammon K. Comprehensive allelotype and genetic anaysis of 466 human nervous system tumors. J Neuropathol Exp Neurol. 2000;59:544–58. doi: 10.1093/jnen/59.6.544. [DOI] [PubMed] [Google Scholar]
- 43.Brat DJ, Seiferheld WF, Perry A, Hammond EH, Murray KJ, Schulsinger AR, Mehta MP, Curran WJ. Analysis of 1p, 19q, 9p, and 10q as prognostic markers for high-grade astrocytomas using fluorescence in situ hybridization on tissue microarrays from Radiation Therapy Oncology Group trials. Neuro Oncol. 2004;6:96–103. doi: 10.1215/S1152851703000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Idbaih A, Marie Y, Lucchesi C, Pierron G, Manie E, Raynal V, Mosseri V, Hoang-Xuan K, Kujas M, Brito I, Mokhtari K, Sanson M, Barillot E, Aurias A, Delattre JY, Delattre O. BAC array CGH distinguishes mutually exclusive alterations that define clinicogenetic subtypes of gliomas. Int J Cancer. 2008;122:1778–86. doi: 10.1002/ijc.23270. [DOI] [PubMed] [Google Scholar]
- 45.Horbinski C. Practical molecular diagnostics in neuropathology: making a tough job a little easier. Semin Diagn Pathol. 2010;27:105–13. doi: 10.1053/j.semdp.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 46.Perry A, Aldape KD, George DH, Burger PC. Small cell astrocytoma: an aggressive variant that is clinicopathologically and genetically distinct from anaplastic oligodendroglioma. Cancer. 2004;101:2318–26. doi: 10.1002/cncr.20625. [DOI] [PubMed] [Google Scholar]
- 47.Miller CR, Dunham CP, Scheithauer BW, Perry A. Significance of necrosis in grading of oligodendroglial neoplasms: a clinicopathologic and genetic study of newly diagnosed high-grade gliomas. J Clin Oncol. 2006;24:5419–26. doi: 10.1200/JCO.2006.08.1497. [DOI] [PubMed] [Google Scholar]
- 48.Hegi ME, Janzer RC, Lambiv WL, Gorlia T, Kouwenhoven MC, Hartmann C, von Deimling A, Martinet D, Besuchet Schmutz N, Diserens AC, Hamou MF, Bady P, Weller M, van den Bent MJ, Mason WP, Mirimanoff RO, Stupp R, Mokhtari K, Wesseling P. Presence of an oligodendroglioma-like component in newly diagnosed glioblastoma identifies a pathogenetically heterogeneous subgroup and lacks prognostic value: central pathology review of the EORTC_26981/NCIC_CE. 3 trial. Acta Neuropathol. 2012;123:841–52. doi: 10.1007/s00401-011-0938-4. [DOI] [PubMed] [Google Scholar]
- 49.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Isolated 1p or 19q have no significant effect on GBM response according to adjuvant therapy. Neither 1p deletion by FISH (A–C) nor LOH (G–I), nor 19q3 deletion by FISH (D–F) nor LOH (J–L), correlated with any difference in GBM survival, regardless of whether there was no adjuvant therapy (A, D, G, J), adjuvant therapy without TMZ (B, E, H, K), or adjuvant therapy with TMZ (C, F, I, L). FISH, fluorescence in situ hybridization; LOH, loss of heterozygosity; TMZ, temozolomide.