Summary
The emergent discipline of metabolomics has attracted considerable research effort in hepatology. Here we review the metabolomic data for nonalcoholic fatty liver disease (NAFLD), nonalcoholic steatohepatitis (NASH), cirrhosis, hepatocellular carcinoma (HCC), cholangiocarcinoma (CCA), alcoholic liver disease (ALD), hepatitis B and C, cholecystitis, cholestasis, liver transplantation and acute hepatotoxicity in animal models. A metabolomic window has permitted a view into the changing biochemistry occurring in the transitional phases between a healthy liver and hepatocellular carcinoma or cholangiocarcinoma. Whether provoked by obesity and diabetes, alcohol use or oncogenic viruses, the liver develops a core metabolomic phenotype (CMP) that involves dysregulation of bile acid and phospholipid homeostasis. The CMP commences at the transition between the healthy liver (Phase 0) and NAFLD/NASH, ALD or viral hepatitis (Phase 1). This CMP is maintained in the presence or absence of cirrhosis (Phase 2) and whether or not either HCC or CCA (Phase 3) develop. Inflammatory signalling in the liver triggers the appearance of the CMP. Many other metabolomic markers distinguish between Phases 0, 1, 2 and 3. A metabolic remodelling in HCC has been described but metabolomic data from all four Phases demonstrate that the Warburg shift from mitochondrial respiration to cytosolic glycolysis foreshadows HCC and may occur as early as Phase 1. The metabolic remodelling also involves an upregulation of fatty acid β-oxidation, also beginning in Phase 1. The storage of triglycerides in fatty liver provides high energy-yielding substrates for Phases 2 and 3 of liver pathology. The metabolomic window into hepatobiliary disease sheds new light on the systems pathology of the liver.
Metabolomics and the liver in brief
Over the past decade or more, many authors have defined the terms metabolomics and metabonomics. It is unproductive and unnecessary to add further to these definitions here. All the reader needs to know from the point of view of hepatobiliary disease, is that metabolomics is a window that offers a view distinct from the lenses of genomics, transcriptomics and proteomics. There can be no other organ where such a plethora of both lipids and water-soluble metabolites are metabolically interchanged. No other organ exceeds the rates of metabolism and energy production and consumption as found in the liver. Not only is the liver the source of myriad endogenous metabolites and precursors used by other organs, but also houses a vast array of detoxication enzymes that are crucial for rendering less toxic, more water-soluble and readily excretable the 1–3 million xenobiotics that we are exposed to in our lifetimes [1]. The hepatic metabolome is therefore a highly complex and dynamic flux of small metabolites (say, <1.5 kDa, to include the larger phospholipid species, such as cardiolipins). Metabolomics in its practice combines high-throughput analytical chemistry, typically, methodologies based upon mass spectrometry or nuclear magnetic resonance spectroscopy, with multivariate data analysis. These technologies permit comparison of “global” metabolite profiles in an “unbiased” fashion for two or more groups of samples. Of course, no metabolomic investigation has ever delivered a global metabolite profile for a sample set, as this would require employment of multiple analytical platforms and several sample preparation protocols that performed from millimolar down to sub-picomolar concentrations. Moreover, different analytical platforms combined with specific sample preparation procedures each provide a different metabolomic window in the metabolic life of the liver. Accordingly, metabolomic findings reported are always biased by the laboratory analytical procedures employed, often highly so.
This notwithstanding, many metabolomic investigators in recent years have entered the field of hepatobiliary disease and a considerable volume of publications has appeared. This review is therefore timely and we will attempt to make sense of a large and heterogeneous set of published studies concerning the varied hepatobiliary elements of pathophysiology where metabolomics has had something to say. This metabolomic window on hepatobiliary disease has furnished an overabundance of potential disease biomarkers. More importantly, in our view, the metabolomic lens has begun to provide new insights into liver disease mechanisms, new understandings that may unmask potential therapeutic targets and, one day, new treatment modalities.
The metabolomic window into nonalcoholic diseases of the liver
Overview
In this review and as depicted in Fig. 1, we will describe the extent to which metabolomics has informed on the progression from the healthy liver to hepatocellular carcinoma (HCC) through the various phases of nonalcoholic fatty liver disease (NAFLD), nonalcoholic steatohepatitis (NASH) and liver cirrhosis. We will also examine what metabolomics has taught about the various influencing factors and putative risk factors for these diseases, such as obesity, diabetes, alcohol, hepatitis B and C virus (HBV, HCV) infection. In addition, we will also review what metabolomics has contributed to the understanding of the change in hepatic function after liver transplantation.
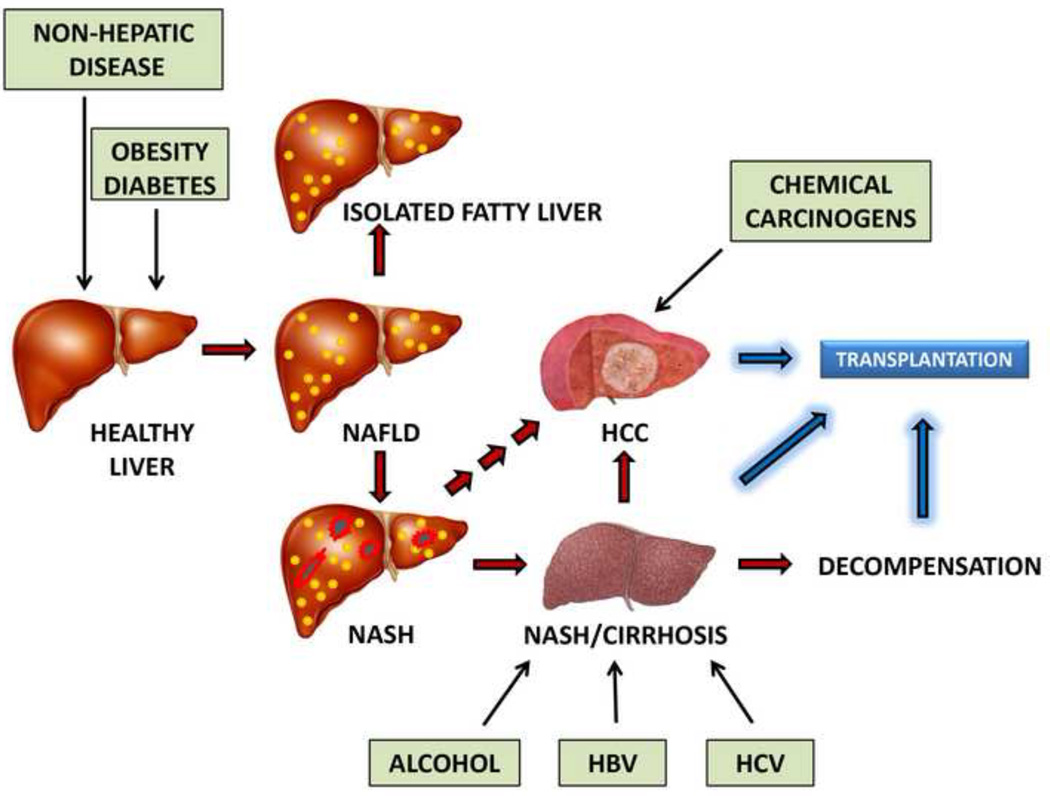
Fig. 1. Major liver diseases and potential influencing factors.
This schematic shows the development of NAFLD from a healthy liver various influencing factors (green boxes). Steatosis is shown in yellow. NAFLD mostly becomes isolated fatty liver, but some cases progress to NASH, showing both steatosis and inflammatory necrosis (shown in red and black). NASH may progress to cirrhosis and then to HCC or to HCC directly. HCC, cirrhosis and decompensated cirrhosis may all be treated by liver transplantation. Chemical carcinogens, such as aflatoxin B1, together with alcohol and HBV and HCV infection, are all potential influencing factors (green boxes).
Abbreviations used: NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; HCC, hepatocellular carcinoma; HBV, hepatitis B virus; HCV, hepatitis C virus.
Nonalcoholic fatty liver disease (NAFLD)
Nonalcoholic fatty liver disease (NAFLD) is a highly prevalent condition that affects 15% to 45% persons in developed nations [2] and in both children and adults from all ethnic groups [3]. A diagnosis of NAFLD implies an increased risk of such diseases as cardiovascular disease, diabetes, colonic adenomas, hypothyroidism and polycystic ovary syndrome [3]. NAFLD is generally considered to be the hepatic manifestation of metabolic syndrome [4]. The reference standard for diagnosing hepatic steatosis remains liver biopsy [3]. Investigators have employed metabolomic protocols in an attempt to define biomarkers that might replace this invasive procedure for a disease of such high prevalence. Table 1 shows a summary of 11 studies with metabolomic components that inform regarding the formation of hepatic steatosis. Animal models and studies in living human subjects and human tissues have been employed. One common finding is that of increased lipid species in the liver and serum/plasma, including cholesterol esters [5, 6], triacylglycerols [4–7], diacylglycerols [4], sphingomyelins [4], various bile salts [8–10], together with lactate [9, 11, 12] and glutamate [11, 13]. In addition, cysteine-glutathione disulfide and both oxidized and reduced glutathione were all reported to be depressed in the liver and serum/plasma [8, 9]. Finally, where diets that instigate fatty liver had been used, depressed concentrations of glucose were reported both in rat liver [14] and mouse serum [11], but in one study, elevated plasma glucose was reported [12]. Taken together with elevated mouse serum/plasma lactate [11, 12], pyruvate and alanine [12], and human plasma lactate [9], these results would suggest that NAFLD engages in cytosolic glycolysis. NAFLD is frequently associated with insulin resistance and insulin has been reported in mice to activate pyruvate kinase M2 [15], the enzyme switch to glycolysis involved in the Warburg effect and thus the production of lactate and alanine from glucose via pyruvate. Furthermore, the reduction in glutathione derivatives in human liver [8] and plasma [9] in NAFLD are clear signs of active oxidative stress in the liver.
Table 1.
Summary of metabolomic studies examining the development of NAFLD
| species [ref] | tissue | platform | up-regulated | down-regulated | conclusions |
|---|---|---|---|---|---|
|
Human [9 obese with normal liver; 24 NAFLD] GNMT−/− mice [10] |
serum | UPLC- ESI- QTOFMS |
Common to NAFLD & GNMT−/− mice: DCA | Common to NAFLD & GNMT−/− mice: FAs 20:2n6, 18:3n3/18:3n6, LPC(20:2), LPC(20:1), SM(36:3), SM(d18:2/16:0), SM(18:2/14:0), SM(d18:1/12:0), LPC(20:2), LPC(20:1), TDCA, TCDCA, DG(18:0/22:6) | |
|
Human [50 HV; 25 NAFLD] [7] |
plasma | HPTLC GCFID LCMS |
Triacylglycerols, FAs: 16:0, 14:1n5, 16:1n7, 18:1n9, 18:1n7, 18:3n6, 18:4n3, 20:3n6, 22:5n3, 15-HETE | - | lipogenesis↑ |
|
Human [25 HV; 11 NAFLD] [9] |
plasma | UPLC- ESI- TQMS GCMS |
GCA, TCA, GCDCA, 4:0-carnitine, glutamate, tyrosine, lactate | cysteine-glutathione disulfide, LPC(18:1), cortisone, uridine | oxidative stress↑ Bile salts↑ |
|
Human [23 non-steatotic; 23 steatotic] [8] |
liver | UPLC- ESI- QTOFMS |
Dextrin, GCDCA-3-sulfate, TCDCA, glycerophosphocholine, LPC(16:0), LPC(18:3), LPE(16:0), LPE(18:0), LPE(18:3), PC(36:5), PC(36:2), PC(36:4) | GSH, GSSG, L-glutamyl-L-lysine, L-leucyl-L-proline, glutamate | Oxidative stress↑ Bile salts↑ Phospholipid synthesis↑ Hepatic glucose catabolism↑ |
|
Mouse fed methionine and choline deficient diet vs. control mice Human [28 HV; 15 NAFLD; 11 NAFLD with necro-inflammation [12] |
serum | NMR | Mice: lactate NAFLD: lactate, glutamate |
Mice: glucose, choline, TMAO, betaine, VLDL | Glycolysis↑ |
| Mouse [fed high-fat diet] [12] | plasma | NMR | lactate, pyruvate, glucose, fucose, phosphatidylcholine, TMAO, alanine | albumin | glucose uptake/mobilization↑ glycolysis↑ phospholipid synthesis↑ |
| Mouse [fed high-fat diet] [12] | plasma | NMR | lactate, pyruvate, glucose, fucose, phosphatidylcholine, TMAO, alanine | albumin | glucose uptake/mobilization↑ glycolysis↑ phospholipid synthesis↑ |
| Mouse [24h starvation] [5] | liver | FPLC HPTLC LCMS |
Cholesterol esters, triacylglycerols TG(49:4), TG(44:2), TG(48:3) | phosphatidylcholine | mobilization of TGs from adipose to liver↑ |
| Mouse (LDLr−/−) fed high-fat diet ± cholesterol [4] | liver | NMR | Triacylglycerols, diacylglycerols, sphingomyelins, G-6-P, G-1-P, glycerol | PUFA/MUFA, fumaric acid | cholesterol is influencing factor SCD1↓ |
| Rat H4IIEC3 hepatoma cells treated with PA (apoptosis) ± OA(steatosis) [14] | cells | GCMS | Associated with PA + OA (steatosis): fructose, gluconate, glutamate, desmosterol | Associated with PA alone (apoptosis): adenosine, malate, serine, citrate, aspartate, C16 ceramide, diacylglycerol | None for NAFLD, but several for NASH (lipoapoptosis phenotype) |
| Piglet [Caesarean section] [6] | liver | NMR | Total lipid (>5 mg/g liver), cholesterol esters, triacylglycerols, glycerol | glycerol phosphate | gluconeogenesis from glycerol↓ |
Abbreviations used: HV, healthy volunteers; PA, palmitic acid; OA, oleic acid; NMR, nuclear magnetic resonance spectroscopy; FPLC, fast performance liquid chromatography; HPTLC, high performance thin-layer chromatography; LCMS, liquid chromatography-mass spectrometry; GCFID, gas chromatography with flame ionization detection; UPLC, ultraperformance liquid chromatography; ESI, electrospray ionization; TQMS, triple quadrupole mass spectrometry; QTOFMS quadrupole time-of-flight mass spectrometry; TMAO, trimethylamine N-oxide; TG, triacylglycerol (triglyceride); FA, fatty acid; 15-HETE, (±)-15-hydroxy-5Z,8Z,11Z,13E-eicosatetraenoic acid (non-enzymic oxidation product of arachidonic acid); DCA, deoxycholic acid; GCA, glycocholic acid; TCA, taurocholic acid; GCDCA, glycochenodeoxycholic acid; TCDCA, taurochenodeoxycholic acid; G-6-P, glucose-6-phosphate; G-1-P, glucose-1-phosphate; GNMT, glycine N-methyltransferase; PUFA, polyunsaturated fatty acids; MUFA, monounsaturated fatty acids; LPC, lysophosphocholine; SCD1, stearoyl-CoA desaturase-1
The lipidomic component of the observations summarized in Table 1 is of interest. Firstly, it has been reported that phosphocholine, choline, betaine and trimethylamine N-oxide (TMAO) were up-regulated metabolites in both liver and plasma of rodents fed diets that provoked fatty liver [11, 12]. This is a clear indication of an increased turnover of phosphatidylcholine and phosphatidylethanolamine species in the liver, thus releasing free fatty acids through the action of phospholipases A1 and A2. These fatty acids, if not catabolized by β-oxidation, will be stored in the liver as triacylglycerols. This is what was observed in the metabolomic studies of animals with fatty liver [4–6]. Therefore, fatty liver is not just a deposition of fat in the liver but rather a rearrangement and repartitioning of lipid stores as has been proposed [5]. Using a mouse 24h starvation protocol, it was observed that the triacylglycerols TG(44:2) and TG(48:3) massively increased in the liver by 2427% and 1198%, respectively. These are the most abundant triacylglycerols in adipose tissue and these findings suggest that adipose may be a source of triacylglycerols deposited in the liver in NAFLD [5]. Secondly, elevated hepatic concentrations of various lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE) and phosphatidylcholine (PC) species have been reported for human steatotic vs. non-steatotic livers [8]. These molecules are obvious candidates for the elevated choline and choline metabolites discussed above. Finally, three studies in humans reported elevated bile salts in the liver [8] that spilled over to elevated bile acids in serum/plasma [9, 10]. Bile acids act as signaling molecules in the liver that regulate lipid and glucose homeostasis [3, 16]. Certain bile acids, in particular, chenodeoxycholic acid (CDCA) and deoxycholic acid (DCA) are endogenous ligands that activate the farnesoid X receptor (FXR) [17]. The nuclear receptor FXR modulates conversion of cholesterol to bile acids by the regulation of the expression of CYP7A1 [3]. Moreover, FXR reduces lipogenesis by downregulating expression of SREBP-1, activates the nuclear receptor PPARα causing an increase in β-oxidation of free fatty acids (FFA), both of which processes reduce hepatic FFA levels [3, 16]. There is a single report of elevated hepatic levels of the bile salts glycochenodeoxycholate 3-sulfate (GCDCA-3S) and taurochenodeoxycholate (TCDCA) in human fatty liver [8]. TCDCA is a relatively weak activator of FXR [17] and GCDCA-3S appears not to have been studied in this regard. It is curious that NAFLD existed in the presence of increased serum/plasma concentrations of glycocholate, taurocholate, glycochenodeoxycholate [9] and deoxycholate [10], which may not reflect hepatic concentrations of the FXR activators CDCA and DCA. This theme will be returned to in the next section.
Nonalcoholic steatohepatitis (NASH)
NASH is a more advanced stage of NAFLD with a major inflammatory component [2]. NAFLD may progress to NASH, but >80% of cases remain as isolated fatty liver (IFL) with no or minimal progression to cirrhosis and no increased risk of death relative to the general population [3]. It has been estimated that ~11% NASH develop cirrhosis over 15 years and ~7% progress to hepatocellular carcinoma (HCC) over 6.5 years, either via cirrhosis or sometimes directly [3] (see Fig. 1). The origins of the hepatic inflammation in NASH continues to involve a major research effort and one theory posits that the hepatitis originates in visceral adipose, which is intrinsically pro-inflammatory [2]. A study in mice fed a high-fat diet supports this theory [18].
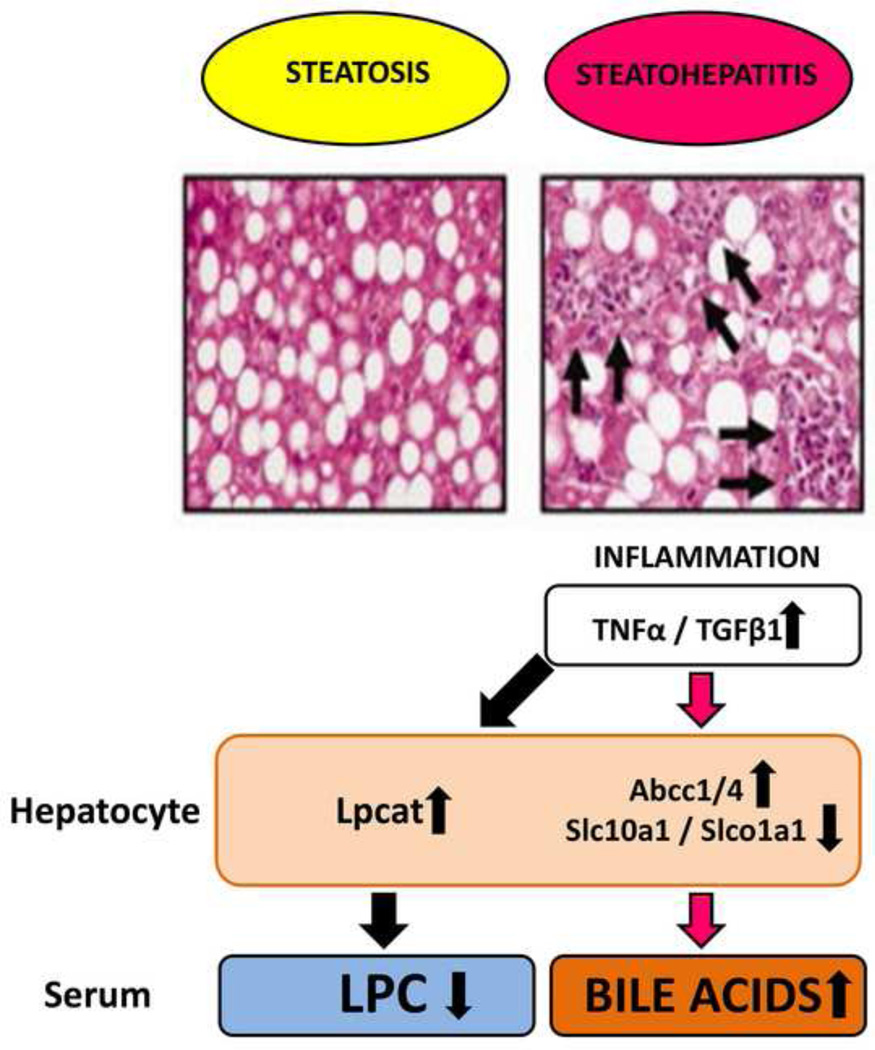
There have been relatively few metabolomic studies addressing the pathobiology of NASH and its progression from simple NAFLD and all these have examined serum/plasma only. Five studies are summarized in Table 2. As with NAFLD, triacylglycerols and several fatty acids were elevated in plasma [7] and like NAFLD, several other fatty acids and LPCs were attenuated in plasma [9]. When a small series of NASH was compared with NAFLD, significant changes in serum concentrations of only three phospholipids were reported [10]. A study using NMR, which, unlike mass-spectrometry-based platforms, does not have the power of detecting a large range of molecules [19], contributed raised serum concentrations of glucose, glutamate and taurine [11]. The greatest metabolomic insights into NASH pathogenesis come from a recent study that combined high-end analytics and targeted gene expression by qPCR [20]. These workers generated NASH in mice fed a methionine- and choline-deficient (MCD) diet. UPLC-ESI-TOFMS metabolomics revealed a statistically significant depression of LPC(16:0), LPC(18:0) and LPC(18:1) in serum with a significant rise in tauro-β-muricholate, taurocholate and 12-HETE for MCD fed mice compared with mice on a normal diet. As a positive control, genetically obese ob/ob mice with severe steatosis were administered galactosamine (GalN), which provoked severe inflammation and hepatocyte injury with marked upregulation of hepatic mRNAs coding for TNFα and TGFβ1. Serum of GalN-injected ob/ob steatotic mice compared with saline-injected ob/ob steatotic mice displayed the same changes in LPCs and bile acids as the MCD fed mice. Thus, the decline in serum LPC and the rise in serum bile acids is a signature of the inflammatory component of NASH, rather than the steatotic component. To investigate further the mechanisms involved in these perturbations of LPC and bile acid homeostasis in the NASH model, hepatic mRNA levels were determined by qPCR for genes involved in the metabolism and transport of LPC, bile acids and 12-HETE. Lysophosphatidylcholine acyltransferases (LPCAT) that convert LPC to PC [21] were all upregulated with two- to four-fold elevations in hepatic Lpcat1, Lpcat2 and Lpcat3 mRNAs in the NASH model. Additionally, the transporters SLC10A1 and SLCO1A1 that uptake bile salts into hepatocytes and the transporters ABCC1 and ABCC4 that export bile acids from the liver were highly downregulated and upregulated, respectively [20]. Taken together, these observations explain how the inflammatory phenotype of NASH in a mouse model results in the changes in serum metabolites described in Table 2 and this is shown in Fig. 2. Importantly, similar perturbations have been observed in NASH patients [9], suggesting that similar mechanisms may operate in humans. Finally, it should be stated that biomarkers for NASH are limited and therapeutic options are poorly developed, which serves to emphasize the need for further metabolomic research in this area.
Table 2.
Summary of metabolomic studies examining the development of NASH
| species [ref] | tissue | platform | up-regulated | down-regulated | conclusions |
|---|---|---|---|---|---|
| Human [9 obese with normal liver; 24 NAFLD; 9 NASH] [10] |
serum | UPLC- ESI- QTOFMS |
NASH vs. NAFLD: PC(14:0/20:4), LPC(18:1) | NASH vs. NAFLD: LPC(24:0) | A few lipid changes between NAFLD and NASH of uncertain origin. |
| Human 28 HV; 6 NASH |
serum | NMR | Glucose, glutamate, taurine | - | Increased glucose mobilization. |
| Humans [HV 50; NASH 50] [7] |
plasma | HPTLC GCFID LCMS |
Triacylglycerols, FAs: 14:1n5, 16:1n7, 18:1n9, 18:1n7, 18:3n6, 20:3n6, 22:6n3, 5-HETE, 8-HETE, 15-HETE, 11-HETE | lipogenesis↑ | |
| Human [25 HV; 24 NASH] [9] |
plasma | UPLC- ESI- TQMS GCMS |
N-Acetylthreonine, aspartate, glutamate, phenylalanine, tyrosine, 3-(4-hydroxyphenyl)-lactate, kynurenine, isoleucine, leucine, valine, ornithine, glutamylvaline, γ-glutamyl-leucine, γ-glutamyl-phenylalanine, γ-glutamyl-tyrosine, erythronate, mannose, glucose, pyruvate, lactate,2-oxoglutarate, carnitine, propionylcarnitine, butyrylcarnitine, 2-methybutyroylcarnitine, GCA, TCA, GCDCA, xanthine, urate, pseudouridine, erithritol | N-Acetylglycine, betaine, histidine, phenylacetate, indolepropionate, 2-aminobutyrate, cysteine-glutathione disulfide, glycerate, 20:5n3, 22:6n3, 11:1n1, 20:4n6, 2-hydroxypalmitate, 3-carboxy-4-methyl-5-propyl-2-furanpropanoate, glycerophosphocholine, LPC(18:1), LPC(18:2), LPC(20:4), cortisone, threonate, hippurate, catechol sulfate, indoleacrylate, 3-phenylpropionate | Elevated bile acids GCA, TCA and GCDCA a sign of liver injury or insulin resistance. High rate of GSH turnover reflective of oxidative stress. Origin of certain depressed PUFAs and LPCs uncertain. Increased glucose mobilization and formation of pyruvate and lactate suggests cytosolic glycolysis. Elevation of essential amino acids suggests increased protein turnover. |
| Mouse fed methionine- and choline-deficient diet [11] | serum | UPLC- ESI- QTOFMS |
Tauro-β-muricholate, TCA, 12-HETE | LPC(16:0), LPC(18:0), LPC(18:1) | Disruption of bile acid and phospholipid homeostasis due to hepatic inflammatory signaling. |
For abbreviations, see Table 1 footnotes.
Fig. 2. Mechanisms leading to lowered LPCs and elevated bile acids in serum in NASH.
Reproduced with permission from Tanaka et al. Disruption of phospholipid and bile acid homeostasis in mice with nonalcoholic steatohepatitis. Hepatology 2012;56:118–129.
Abbreviations used: LPC, lysophosphatidylcholine.
Fibrosis and cirrhosis
Liver fibrosis is a scarring process involving the deposition of excess connective tissue in response to injury. Cirrhosis may be considered as the end-stage of this reaction, comprising formation of fibrous septa and hepatocyte nodules. Oxidative stress provokes the inflammatory reactions and apoptosis involved in the generation of cirrhosis [22]. It is now clear that NAFLD/NASH may develop into cirrhosis, although the histological features of precursor NASH in the cirrhotic liver may be challenging to diagnose [23]. Cirrhosis may arise due to a large number of causes, principal among which are not only NAFLD/NASH but also alcoholic fatty liver disease and viral hepatitis B or C (see Fig. 1). There are no cut-off values for laboratory analyses that give a diagnosis of cirrhosis [24] and so the generation of novel metabolomic biomarkers to detect early cirrhosis has become a justifiable aim. Table 3 summarizes such studies.
Table 3.
Summary of metabolomic studies examining the development of hepatic fibrosis and cirrhosis
| species [ref] | tissue | platform | up-regulated | down-regulated | conclusions |
|---|---|---|---|---|---|
| Human [63 HV; 36 LC] [27] |
serum | NMR | Acetate, pyruvate, glutamine, "N-acetylglycoproteins", 2-oxoglutarate, taurine, glycerol, tyrosine, 1-methylhistidine, phenylalanine | LDL, VLDL, leucine, isoleucine, valine, acetoacetate, choline, unsaturated lipid | Downregulation of essential amino acids suggests depressed protein turnover. |
| Human [16 HV; 25 LC] [28] |
serum | GCxGC- TOFMS |
D-Alanine, D-proline | L-Alanine, L-valine, L-isoleucine, L-leucine, L-serine, L-asparagine | Targeted amino acid analysis reveals loss of ability by the cirrhotic liver to metabolize D-amino acids |
| Human [22 HV; 18 LC (alcohol); 19 LC (HBV) [29] |
serum | UPLC-ESI- QTOFMS |
GCDCA, GCA, L-acetylcarnitine, myristamide, oleamide (only in alcohol cirrhosis) | LPC(16:0), LPC(18:2), LPC(18:0), LPC(20:3), LPC(20:5) myristamide, oleamide (only in HBV cirrhosis) | Pattern very similar to NASH and so NASH signature dominates serum picture in cirrhosis, irrespective of origin as alcohol or HBV. |
| Human [HV and LC (HBV)] [30] |
serum | LCMS | GCDCA | LPCs | Confirmatory of bile acid and phospholipid perturbations. |
| Human [30 HV; 30 LC (compensated); 30 LC (decompensated) [31] |
serum | NMR | - | In both compensated and decompensated LC: isoleucine, valine | Many other changes recorded, but with OPLS-DA correlations <0.85 |
| Human [30 HV; 30 CHB; 30 LC] [32] |
serum | LCMS | Relative to HV: 16:1-carnitine, 18:1-carnitine | Relative to HV: carnitine, pimeloylcarnitine Also relative to CHB: PE(22:6/16:0), PE(20:4/18:0) | Elevation of two MUFA-carnitines suggests reduced β-oxidation of these two fatty acids. |
| Human [24 HV; 17 LC] [34] |
faeces | UPLC-ESI- QTOFMS |
LPC(16:0), LPC(18:0), LPC(18:1), LPC(18:2) | CDCA, 7-ketolithocholic acid, urobilin, urobilinogen | Increased LPCs in faeces consistent with lower serum LPCs in LC. Biliary excretion of bile acids known to be reduced in LC |
| Human [57 non-LC; 11 LC] [33] | liver | NMR | UFA, phosphocholine, glutamate, phosphoethanolamine | Choline, TMAO, α-glucose, glutamine, aspartate, β-glucose | Shift from glutamine and glucose to glutamate suggests a net release of ammonium and impaired ammonium detoxication. |
| Rat treated with CCl4, then with Xia Yu Xue Decoction [25] | urine | GCMS | Apart from propionate and leucine, all changes due to CCl4 were reversed by Xia Yu Xue Decoction. | Effect of CCl4: Propionate, benzoate, leucine, octanoate, phenol, glycine, indole, oleic acid, lysine | Some metabolic changes of uncertain origin that may be associated with fibrosis |
| Rat treated with CCl4 [26] | urine | UPLC-ESI- QTOFMS |
Glycocholate | Hard to evaluate in this model if GCA is really a marker for fibrosis | |
| Rat treated with thioacetamide [22] | liver | NMR | Lactate, choline, proline, "glutamine/glutamate" | TMA, glycogen, inosine, fumarate | Raw data very poor and these NMR findings have uncertain validity |
Abbreviations used: LC, liver cirrhosis; CHB, chronic hepatitis B; TMA, trimethylamine; CCl4, carbon tetrachloride; UFA, unsaturated fatty acid units (-CH=CH-CH2-). GCxGC-TOFMS, Two-dimensional gas chromatography time-of-flight mass spectrometry. OPLS-DA, orthogonal partial least squares projection to latent structures-discriminant analysis, PE, phosphatidylethanolamine. For other abbreviations, see footnotes to Table 1
Three studies have been conducted in rats administered hepatotoxins to provoke fibrosis and cirrhosis. Histopathology confirmed that rats exposed to thioacetamide in their drinking water developed hepatic fibrosis after one month and cirrhosis after three months. Liver extracts examined by NMR had higher levels of lactate [22], suggesting a degree of anaerobic metabolism within the fibrotic liver. Two studies treated rats with carbon tetrachloride (CCl4), which induced fibrosis [25, 26] and the authors evaluated treatment with the Chinese medicine Xia Yu Xue Decoction [25] or scoparone, a drug isolated from a medicinal plant [26]. Many metabolomic signals were reported after CCl4 administration, including decreases in the urinary excretion of certain amino acids and gut flora metabolites (which were mostly reversed by Xia Yu Xue Decoction) [25] and an increased urinary excretion of glycocholate [26]. Neither serum nor liver tissue was examined in these studies. Thus, hepatic fibrosis provoked in a normal, rather than fatty, rat liver, is associated with somewhat minor changes in the urinary metabolome.
Eight metabolomic investigations of hepatic cirrhosis have all been performed on human materials, six on serum [27–32], one on liver biopsies [33], and one on faeces [34]. No clear picture emerges from these studies. An increased serum concentration of non-essential amino acids [27] and certain D-amino acids [28], a decreased serum concentration of essential amino acids [27, 28, 31], suggests that the cirrhotic liver has an impaired ability to metabolize both protein and D-amino acids. Other notable observations include the decrease in several LPCs in serum of cirrhotics versus healthy volunteers, whether the cirrhosis was due to alcohol or hepatitis B [29]. This pattern is similar to that observed for NASH (Table 2), although the cirrhotic patients studied had a background of alcohol abuse or hepatitis B. Moreover, glycochenodeoxycholic acid and glycocholic acid concentrations were also elevated in serum [29]. Clearly, the mechanism proposed by Gonzalez and colleagues [20] shown in Fig. 2 may apply not only to NASH but other inflammatory liver diseases.
Selective impairment of hepatic β-oxidation was apparent from a reduced serum carnitine and increased serum palmitoleoylcarnitine (16:1) and oleoylcarnitine (18:1) concentrations [32]. Impaired ammonium detoxication in cirrhosis is implied from a reported shift from hepatic levels of glutamine and glucose to glutamate [33]. Finally, a very interesting report catalogued changes in the faecal metabolome between 24 healthy volunteers and 17 cirrhotics [34]. In faeces from cirrhotic patients, there was an increased concentration of the major LPCs (16:0, 18:0, 18:1, 18:2) and a decreased faecal excretion of chenodeoxycholic acid and 7-ketolithocholic acid, the latter reported as a gut flora metabolite of the former by Bacteroides intestinalis [35]. The data on the faecal excretion of LPCs and bile acids further supports and enhances the mechanism outlined in Fig. 2.
Hepatocellular carcinoma (HCC)
More than half a million people are diagnosed each year with hepatocellular carcinoma (HCC). The disease has a poor prognosis, generally because of its late presentation and its incidence is growing in developed countries. There has been considerable research effort to try to define biomarkers that would aid earlier detection and thus improve patient outcomes. Many researchers, particularly in China, have employed metabolomic protocols towards this end. Table 4 contains details of 24 metabolomic investigations of human HCC [27, 32, 36–57], three of chemically-induced rat HCC [42, 46, 58] and two of hepatocellular adenomas in the flatfish Limanda limanda [59, 60]. Many investigators of human HCC employed healthy volunteers as a control group, especially for the collection of serum/plasma or urine [27, 32, 36, 37, 39–41, 43–45, 47–50, 53, 57], others used cirrhotics as a comparator group [36, 37, 39, 45–48, 50–52, 54], while others included acute hepatitis [36, 37], chronic hepatitis [36, 37, 46, 48, 50], benign liver tumours [43] and acute myeloid leukemia [45] as comparator groups. These metabolomic comparisons have permitted insights into the biochemical transitions to HCC from various precursor states, at least as viewed through serum/plasma or urine. A relatively few studies have addressed the hepatic metabolome directly by interrogating tumour tissue and paired uninvolved liver, for human HCC [38, 55, 56], chemically-induced rat HCC [42] and fish hepatocellular adenoma [59, 60]. Two recent reports also combined transcriptomic and metabolomic analyses of human HCC [55, 56]. As will be demonstrated below, comparison of the outputs of metabolomic investigations of NAFLD/NASH, cirrhosis and HCC will permit a new understanding of the chain of biochemical events that lead from a healthy liver to HCC.
Table 4.
Summary of metabolomic studies examining the development of hepatocellular carcinoma
| species [ref] | tissue | platform | up-regulated | down-regulated | conclusions |
|---|---|---|---|---|---|
| Human [63 HV; 39 HCC] [27] |
serum | NMR | Acetate, "N-acetylglyco-proteins", pyruvate, glutamine, 2-oxoglutarate, glycerol, tyrosine, 1-methylhistidine, phenylalanine | LDL, VLDL, valine, acetoacetate, choline, taurine, "unsaturated lipid" | Increased lipid catabolism because of LDL/VLDL↓ and acetate↑. Many signs that TCA cycle is impaired – pyruvate↑, acetoacetate↓, glutamine/2-oxoglutarate↑ Essential amino acids and metabolites elevated due to increased protein turnover. |
| Human [25 HV; 25 LC(HBV); 24 HCC] [39] |
serum | HPLC- ESI- TOFMS |
TCA, GCA, bilirubin, TCDCA, GCDCA, FA(18:1), carnitine, acetylcarnitine | Hypoxanthine, phytosphingosine, dihydrosphingosine, LPC(18:2), LPC(18:3), LPC(16:1), LPC(18:0), taurine, 6-methyl-nicotinic acid | HCC/HV ratios much smaller than LC/HC ratios, suggesting that HCC diminishes the LC metabolomic phenotype. Bile acid and LPC findings consistent with other data. Data suggest reduced β-oxidation of FAs. Reduced sphingosines suggest increased ceramide synthesis and thus, increased death signalling. |
| Human [38 HV; 41 HCC] [44] |
serum | UPLC- ESI- TQMS |
1-Methyladenosine | - | A biomarker study that compared 1-methyladenosine ± AFP |
| Human [90 HV; 48 LC; 82 HCC] [47] |
serum | UPLC- ESI- QTOF- MS |
Canavaninosuccinate, phenylalanine, GCDCA, oleamide | LPC(16:0), LPC(18:0), PC(16:0/22:6), PC(16:0)/20:4), PC(18:0/18:2) | Canavaninosuccinate synthesized by argininosuccinate synthase, presumably induced in HCC. Decreased PCs as well as LPCs suggests increased biliary excretion of phospholipids, rather than increased synthesis of PCs from LPCs by Lpcat (Fig. 2). |
| Human [30 HV; 30 CHB; 30 LC; 30 HCC] [48] |
serum | UPLC- ESI- QTOF- MS |
GCA, GCDCA, 16:1-carnitine, | Tryptophan, LPC(14:0), 10:0-carnitine, 10:1-carnitine, 8:0-carnitine, 6:0-carnitine | Increased β-oxidation of short- to medium-chain FAs. |
| Human [30 HV; 30 CHB; 30 LC; 30 HCC] [50] |
serum | UPLC- ESI- QTOF- MS |
16:1-carnitine, 18:1-carnitine, PE(22:6/16:0) | carnitine, pimeloylcarnitine | Reduced β-oxidation of MUFAs. |
| Human [184 LC; 78 HCC] [51] |
serum | UPLC- ESI- QTOF- MS |
- | Relative to LC: GCA, GDCA, TCA | Bile acid export to blood mostly a feature of LC not HCC. |
| Human [49 LC; 40 HCC] [52] |
serum | UPLC- ESI- QTOF- MS |
Relative to LC: PhePhe | Relative to LC: TCDCA, GDCA, 3β,6β-dihydroxy-5β-cholan-24-oic acid, 18:1-carnitine, 18:2-carnitine | Bile acid export to blood mostly a feature of LC not HCC. Increased β-oxidation of FA(18:1) AND FA(18:2) in HCC. |
| Human [93 LC; 28 small HCC; 33 large HCC] [54] |
serum | NMR | Glutamate, acetate | Glutamine | Shift from glutamine in LC to glutamate in HCC suggests defect in ammonium detoxication in HCC. Acetate↑ suggests increased β-oxidation of FAs. |
| Human [6 HV; 22 AML; 7 LC; 20 HCC] [45] |
plasma | UPLC- ESI- QTOF- MS |
Bilirubin, biliverdin, GDCA, DCA 3-sulfate, 7α-hydroxy-3-oxochol-4-en-24-oic acid, 3-oxachol-4,6-dien-24-oic acid, LPC(24:0) | LPC(14:0), LPC(16:0), LPC(18:0), LPC(18:1), LPC(18:2), LPC(18:3), LPC(20:2), LPC(20:3), LPC(20:4), LPC(20:5), LPC(22:6), FA(24:0), FA(24:1) | Increased bile acid transport into blood, including fetal bile acids. Increased metabolism of LPCs and/or biliary excretion. |
| Human [30 HV; 28 HCC] [32] |
plasma | GCTOF- MS |
No significantly elevated molecules in HCC vs. HV | No significantly depressed molecules in HCC vs. HV | Multiple comparisons not allowed for. |
| Human [71 HV; 24 BLT; 82 HCC] [43] |
serum urine | UPLC- ESI- QTOF- MS GCTOF- MS |
Serum: GCDCA, GCA, cysteine, fumarate, 2-oxoglutarate, lactate, pyruvate, inosine, erythronate, carnitine Urine: GCA, dopamine, adenosine, urate, xanthine, phenylalanine, dihydrouracil, hypotaurine, threonine, N-acetylneuraminic acid |
Serum: AA, EPA, DHA, glycerol, FA(14:0), FA(24:1), glycine, serine, aspartate, citrulline, ornithine, kynurenine, tryptophan, lysine, glucosamine, 5-oxo-proline, phenylalanine, β-alanine, α-tocopherol, glycerate, 3-amino-2-piperidone, D-arabino-hexos-2-ulose, arabinose, creatinine, oleamide, phosphate Urine: Cysteine, TMAO, homovanillate, normetanephrine, adenine, cysteic acid, 6-aminohexanoate, creatine |
Reduced ammonium detoxication through the urea cycle. Metabolic reprogramming to glycolysis. Increased export of bile acids into blood and then to urine. Reduced serum free carnitine consistent with increased β-oxidation of FAs. |
| Human [50 HV; 27 LC; 30 acute hepatitis; 20 chronic hepatitis; 48 HCC] [36] |
urine | HPLC | Assay specific for cis-diols and so nucleosides detected: pseudouridine, 1-methyladenosine, xanthosine, 1-methylinosine, 1- and 2-methylguanosine, N4-acetylcytidine, adenosine | - | Probably a sign of increased tRNA turnover and of inflammation rather than HCC. |
| Human [50 HV; 27 LC; 30 acute hepatitis; 20 chronic hepatitis; 48 HCC] [37] |
urine | LCMS | - | - | Reanalysis by LCMS of same samples as [232.] with no further information. |
| Human [17 HV; 16 HCC (11 with HCV] [57] |
urine | NMR | No elevated molecules. | Glycine, TMAO, hippurate, citrate | Decreased glycine may drive decreased urinary hippurate. |
| Human [20 HV; 19 paired pre- and post-operative HCC patients; patients divided into 7 recurrent; 11 nonrecurrent; one unknown] [49] |
urine | GC- TOFMS |
Adenine (RPost/NRPost), threonine (RPost/RPre) | - | Not corrected for multiple comparisons. Most findings not statistically significant. |
| Human [12 HV; 25 HCC] [53] |
urine | UPLC- ESI- QTOF- MS |
GCA | - | Increased bile acid export into blood in HCC vs. HV. |
| Human [20 HV; 20 HCC] [40] |
urine | GCMS | A total of 18 metabolites listed as different between HV and HCC, but no correction for multiple comparisons was made. Therefore, only xylitol and urea elevated. | - | Xylitol a sign of a switch from the TCA cycle to the pentose phosphate pathway and thus from catabolism to anabolism, e.g. synthesis of nucleic acids |
| Human [24 HV; 21 HCC] [41] |
urine | UPLC- ESI- QTOF- MS |
A total of 15 metabolites listed as different between HV and HCC, but no correction for multiple comparisons was made. Therefore, no metabolites elevated. | Three metabolites significantly reduced: carnitines 4:0, 8:1 and 9:0 | Authors claim the decline of acylcarnitines in urine is a sign of reduced β-oxidation in HCC. It is surely a sign of increased β-oxidation on HCC. |
| Human [31 HCC from 17 patients; 14 adjacent nontumour livers] [38] |
liver | NMR | Glutamine, glutamate | α-glucose, β-glucose | Increased glycolysis. |
| Human [30 pairs HCC and nontumour tissue; 356 HCC] [55] |
liver | LCMS GCMS |
55 Annotated metabolites upregulated in HCC, of which 5-methylthioadenosine, 4:0-carnitine, 6:0-carnitine, 16:0-carnitine, 18:0-carnitine and ophthalmate had greatest fold change | 103 Annotated metabolites downregulated in HCC, of which NAD+, glycerol 3-phosphate, LPC(18:2), GCA and xanthosine had greatest fold change | Consistent with shift to glycolysis. Decreased β-oxidation of short- and long-chain FAs. |
| Human [31 pairs HCC and nontumour tissue; 59 HCC typed by transcriptomics as G1 to G6] [56] |
liver | GCMS | - | Downregulated in HCC: glucose, glycerol 2- and 3-phosphate, malate, alanine, myo-inositol, FA(18:2) Downregulated in transcriptomic groups G1 and G3: palmitate, 1-palmitoylglycerol, 1-stearoylglycerol |
Consistent with shift to glycolysis in HCC. Consistent with increased β-oxidation of long-chain FAs in groups G1 and G3. |
| Rat [20 DEN-treated HCC; 28 control rats] Human 262 HCC sera; 76 LC sera; 74 hepatitis sera] [46] |
serum | UPLC- ESI- QTOF- MS |
Three molecules elevated in HCC rat serum – LPC(22:5), LPE(16:0), TCA Same three molecules used as biomarkers to distinguish HCC, LC and hepatitis |
- | Only elevated bile acid consistent with other data. |
| Rat [5 control; 5 DEN-treated HCC; 5 DEN-treated HCC with lung metastases (HLM)] [58] |
serum urine | GCTOF- MS |
Serum: A total of 47 metabolites listed, of which 18 had P<0.05 by ANOVA for control, HCC and HLM. No correction for multiple comparisons was made. Nine remained significant after Bonferroni correction. Elevated: lactate, tyrosine Urine: 13 metabolites had P<0.05 by ANOVA for control, HCC and HLM. Only one remained significant after Bonferroni correction. No metabolites elevated. |
Serum: Oxalate, glutamate, arabinose, glucose, stearate, adipate, phosphoinositide Urine: hippurate |
Decreased hippurate may be a sign of decreased glycine or CoA availability. Glycine appeared to rise >4-fold in rat HCC. Therefore, most likely explanation, other than an effect on the gut flora, is decreased CoA availability due to enhanced β-oxidation of FAs. |
| Rat [5 control; 5 DEN-treated HCC; 5 DEN-treated HCC with lung metastases (HLM)] [42] |
liver | NMR | A total of 15 metabolites listed as different between control and HCC rats, but no correction for multiple comparisons was made. Six remained significant after Bonferroni correction. Elevated: Leucine, acetate, glutamine | Three metabolites significantly reduced: TMAO, glucose, glycogen | Increased glycolysis and β-oxidation of FAs. |
| Dab (Limanda limanda) [9 hepatocellular adenomas; 9 paired nontumour samples] [60] |
liver | FTICR- MS |
MR = 521.3500 (probably, LPC(18:1) [3 ppm error]) | MR = 495.3343 (probably, LPC(16:0) [13 ppm error]) MR = 533.2902 (?) MR = 591.2066 (?) |
Possible changes in LPCs in this liver tumour. |
| Dab (Limanda limanda) [10 hepatocellular adenomas; 10 paired nontumour samples] [59] |
liver | NMR | Propionate, succinate, lactate | Lysine, phosphocholine | Switch to glycolysis in the tumour. |
Abbreviations used: FTICR, Fourier transform ion cyclotron resonance mass spectrometry; MR, relative molecular mass (molecular weight); GCTOFMS, gas chromatography time-of-flight mass spectrometry; BLT, benign liver tumour; AA, arachidonic acid; EPA, 5Z,8Z,11Z,14Z,17Z-eicosapentaenoic acid; DHA, 4Z,7Z,10Z,13Z,16Z,19Z-docosahexaenoic acid; TQMS, triple quadrupole mass spectrometry; AFP, α-fetoprotein; LPE, lysophosphoethanolamine; TCA, taurocholic acid; CHB, chronic hepatitis B; RPost, recurrent HCC post-surgery; NRPost, nonrecurrent HCC post-surgery; RPre, recurrent HCC pre-surgery; For other abbreviations, see footnotes to Table 1
Disease progression from fatty liver to hepatocellular carcinoma
The metabolomic observations encompassed in Tables 1–4 have been combined into a visual format (Fig. 3) which permits a biochemical view of the changes occurring from fatty liver through cirrhosis to HCC. Only observations reported in at least two independent human studies have been entered into this Figure. The paramount conclusion is that elevated bile acids and lowered LPCs are common across all three groups of pathology. The bile acids affected include GCA, TCA, GDCA and GCDCA. A whole range of LPCs, comprising saturated, monounsaturated and polyunsaturated long-chain and very long-chain fatty acids are affected. The probable mechanisms by which these metabolic perturbations have occurred were discussed above and are shown, in part, in Fig. 2. Increased biliary excretion of phospholipids is an additional factor already discussed above. Of importance is that these alterations in hepatic metabolism would appear to occur very early in the chain of events leading from the normal liver to HCC (Fig. 1) and therefore must be maintained throughout the progression to HCC.
Fig. 3. Venn diagram showing the up- and down-regulated metabolites in NAFLD/NASH, cirrhosis and HCC.
Elevated bile acids and lowered lysophosphatidylcholines are common across the pathological evolution in humans and comprise a core metabolomic phenotype.
For abbreviations, see Fig. 1.
As shown in Fig. 3, NAFLD/NASH (see Tables 1, 2) is characterized by upregulation of lactate, glucose, glutamate and tyrosine, together with the downregulation of cortisone. This would suggest that, in the fatty liver states, hepatic glucose is mobilized from glycogen almost certainly due to insulin resistance [61]. The rise in lactate may be a sign of a degree of metabolic remodelling to aerobic glycolysis in response to elevated glucose, although there was little evidence of the other glycolytic metabolites, pyruvate and alanine [56], being elevated in NAFLD/NASH. The rise in glutamate is a sign of reduced cytosolic glutamine synthesis and thus an impairment of ammonium detoxication [62]. This is usually associated with cirrhosis and liver failure, but may also be manifest in NASH [63]. The upregulation of tyrosine in NAFLD/NASH is at odds with a single report in which plasma tyrosine was lower in NAFLD than controls [64]. Elevated tyrosine is more likely to be correct as the metabolomic report [9] employed a more specific analysis. Finally, the metabolomic investigations of NAFLD and NASH [9] have reported that plasma cortisone is downregulated. This is consistent with the upregulation of 11β-hydroxysteroid dehydrogenase type 1 (HSD11B1) in visceral obesity, metabolic syndrome and type 2 diabetes [65, 66] and its role in NAFLD [67]. HSD11B1 activates cortisone to cortisol in liver and adipose tissue. Thus, the aforementioned metabolomic signals are consistent with known biochemical characteristics of fatty liver. In particular, elevated serum bile acids and reduced LPCs are in accord with known changes in gene expression in NASH (Fig. 2).
As shown in Table 3, a relatively small number of metabolomic studies have addressed the conversion of either normal or fatty human liver states to cirrhosis. Only two metabolomic markers specific to cirrhosis could therefore be defined, downregulation of the branched-chain amino acids (BCAAs) valine and isoleucine. Lowered plasma BCAAs in cirrhosis was first observed almost six decades ago [68] and is due to hepatic metabolism of BCAAs to provide carbon skeletons for the TCA cycle [69]. Noteworthy is the carry forward from NAFLD/NASH into cirrhosis of elevated bile acids and reduced LPCs (Fig. 3).
The greatest number of human metabolomic studies was conducted in HCC and, not surprisingly, there occur a large number of metabolomic changes in HCC relative to cirrhosis or to control subjects (Table 4). As shown in Fig. 3, there are many signs of a metabolic reprogramming in the livers of HCC patients, detected by metabolomics. For example, the decrease in glucose, citrate and glycerol 3-phosphate coupled with an increase in pyruvate are all signs of the Warburg effect [70], a switch from mitochondrial respiration to cytosolic aerobic glycolysis [71, 72]. By a metabolomic comparison of paired HCC biopsies and uninvolved liver tissues, we have calculated that the switch to the aerobic glycolysis in HCC is no more than four-fold [56]. Although tumours are generally considered to synthesize fatty acids de novo from citrate via acetyl-CoA [72], the accumulated metabolomic data in HCC (Fig.3) tend to point to increased fatty acid β-oxidation, with elevated acetate and 2-oxoglutarate (immediate precursor of carnitine) and reduced free fatty acids, carnitine and carnitine esters. Furthermore, some transcriptomic types of HCC, in particular G1 and G3, displayed markedly reduced 1-palmitoylglycerol, 1-stearoylglycerol and palmitate compared with surrounding uninvolved liver tissue [56]. Thus, metabolic reprogramming in HCC appears to comprise a modest Warburg shift to glycolysis and a major upregulation of fatty acid catabolism in some tumour types.
The metabolomic window into other hepatobiliary diseases
Alcoholic liver disease
The consumption of alcoholic beverages leads to exposure of the liver to ethanol. While many consider the pharmacological effects of ethanol consumption enjoyable, ethanol is nevertheless a solvent that can exhibit potent toxicological effects, in particular, on the liver. Alcohol exposure to laboratory animals can provoke a range of pathologies that parallels nonalcoholic liver disease. For example, 20 to 40 kg micropigs voluntarily consume an ethanol-supplemented diet (40% daily energy needs), developing peak blood ethanol levels >200 mg/dl and, within 6 months, hepatic steatosis, inflammation and fibrosis. Alcoholic micropigs displayed increased hepatic TG levels relative to controls with elevated fatty acid ratios of 16:1n7/16:0 and 18:1n9/18:0, due to increased stearoyl-CoA desaturase activity. The authors concluded that increased de novo lipogenesis and reduced LPC synthesis and export were responsible for the accumulation of TG during alcoholic steatohepatitis (ASH) [73]. Athymic nude mice gavaged with ethanol solutions from 5% gradually to 40%, developed mild hepatic hemorrhage, with elevated serum PC, decreased saturated and monounsaturated LPC, and elevated polyunsaturated LPC levels [74]. Similarly, rats fed 5% ethanol developed fatty infiltration after 2 months with mild inflammation and oxidative stress after 3 months. NMR metabolomics suggested that hepatic fatty acids and TG increased and plasma fatty acids and PC decreased [75]. These contradictions may reflect a species difference but more likely underscore the relative weakness of NMR as a lipidomic tool.
Another approach to study alcohol-induced liver disease (ALD) has been to employ the Ppara-null mouse, since the nuclear receptor PPARα is a master regulator of hepatic lipid metabolism whose biochemical effects can be detected through metabolomics, both in humans [76] and in the mouse [77]. Ppara-null and control mice were fed a 4% ethanol-containing liquid diet and an isocaloric control diet, respectively. After one month, steatosis with elevated hepatic TGs was observed for the Ppara-null mice only. Metabolomic analysis revealed elevated indole-3-lactic acid associated with the development of ALD in ethanol-treated Ppara-null mice [78]. In an enlarged study, these authors reported that indole-3-lactic acid and phenyllactic acid were potential biomarkers for early ALD [79]. CYP2E1 is the principal ethanol-inducible hepatic enzyme responsible for ethanol metabolism and hepatotoxicity [80]. A metabolomic study in Cyp2e1-null and control mice reported that the ethanol metabolite acetate can acetylate taurine in the liver, leading to ethanol-dose-dependent production of N-acetyltaurine [81], a potential biomarker of ethanol hepatotoxicity. This reaction was found only in wild-type animals with hepatic CYP2E1.
Viral hepatitis B and C
Evaluation of liver disease in patients with hepatitis B or C is essential to identify patients who require antiviral therapy and to determine prognosis. Staging of liver fibrosis and the occurrence of cirrhosis associated with HBV or HCV infection is traditionally done by biopsy, but now there has been a move towards the use of noninvasive biomarkers [82]. None of the serum biomarkers that were originally developed for hepatitis C involves small molecules. Metabolomic studies in hepatitis B and C patients are very timely. The first study of its kind to evaluate deteriorating liver function in chronic hepatitis B using metabolomics was conducted in China, where HBV infection occurs in 80–90% of HCC cases [39]. Using LCMS, they established a decline in serum LPC(16:0), LPC(18:0), LPC(18:1) and LPC(18:2), together with an elevation of GCDCA (or its isomer GDCA) [83]. Another Chinese study reported similar results when examining the progression of chronic hepatitis B to cirrhosis [84]. This, of course, is the same fingerprint as seen in NALFD/NASH, cirrhosis and HCC (Figs. 2, 3). It was also reported that serum GCA, GCDCA and TCA were elevated in hepatitis B-induced cirrhosis [39]. There do not appear to be metabolomic studies comparing HBV-positive and HBV-negative subjects. It should also be pointed out that HBV may cause HCC in the absence of cirrhosis. Currently, there are no biomarkers for predicting HCC development in HBV-positive patients without cirrhosis and this should be a priority for metabolomic research.
HCV infection accounts for 70% of chronic hepatitis and 30% of liver transplants in developed countries [85–87]. Regarding HCV, atomic emission spectroscopy on scalp hair has been performed in 73 HCV-positive and 82 HCV-negative subjects, the hair concentrations of Ca, Cu, Fe, Mg, Mn and Zn determined and the data analyzed by multivariate data analysis [88]. This metallomics [89] study showed that Mg, Ca and Zn were most closely associated with HCV infection. No biological discussion of the findings was made. There has been a claim that NMR metabolomics on urine can distinguish HCV-infected from uninfected persons [90], although little data were provided. A metabolomic comparison of HCV-infected and mock-infected hepatocytes revealed small but significant increases in alanine, tyrosine and adenosine with HCV infection [91]. Interestingly, similar elevations have been recorded for NAFLD/NASH (tyrosine) and HCC (adenosine) (see Fig. 3). Preliminary findings in HCV-infected tree shrews (Tupaia belangeri chinensis) suggested that HCV affects many pathways in the liver, with alterations in LPCs and bile acids (as for other liver diseases, Fig.3), carnitine esters, fatty acids and LPEs [92]. It is clear, therefore, that both HBV and HCV infection, together with NASH, trigger similar molecular events represented by the mechanisms shown in Fig. 2. Moreover, both alcohol- and HBV-induced cirrhosis displayed higher bile acids and lower LPCs than healthy controls in an almost identical manner [29]. It would appear that the depressed LPCs and elevated bile acids in serum represent a phenotype of hepatitis and cirrhosis independent of etiological origin, and that this phenotype is carried forward into any resultant HCC.
Cholangiocarcinoma
Cholangiocarcinoma (CCA) is an aggressive cancer originating from the biliary tract. It would appear that obesity, diabetes, hepatitis B and C, alcohol use and cirrhosis are all major risk factors for CCA, suggesting a common pathogenesis with HCC [93]. It has also been proposed that genetically impaired biliary excretion of phospholipids underlies CCA [94, 95]. Metabolomic investigations support this view, with lower phosphatidylcholine and elevated glycine- and taurine-conjugated bile acids reported in the bile of CCA patients [96, 97].
Cholestasis and cholecystitis
Interruption of bile flow may have an extrahepatic and obstructive or an intrahepatic and biochemical basis. An NMR metabolomic study has been performed in rats in an attempt to use urinary biomarkers to distinguish the two mechanisms [98]. Metabolomics revealed that cholestasis induced in Fxr-null mice by a cholic acid diet resulted in increased urinary excretion of bile salt tetrols, predominantly 3α,6,7α,12α-tetrahydroxy-5β-cholestan-26-oyltaurine, due to an adaptive upregulation of the steroid-hydroxylating cytochrome P450 CYP3A11 in these mice [99]. An adaptive response was also characterized in a rat cholestasis model, with a shift from cytotoxic to cytoprotective bile acids in plasma and urine [100].
Injection of Escherichia coli into the rabbit gallbladder produces a model for acalculous cholecystitis (AAC). Compared to saline-injected controls, AAC animals displayed increased serum LDL and VLDL, with decreased serum phospholipids, lactate, 3-hydroxybutyrate, citrate, lysine, asparagine, histidine and glucose as demonstrated by NMR metabolomics [101]. These observations need to be refined with the use of LCMS-based metabolomics.
Liver transplantation
As shown in Fig. 1, several end-stage liver diseases require transplantation. A metabolomic study of single patient with hepatitis B and HCC who underwent two consecutive liver transplants showed that the first failed graft was associated with elevated blood lactate, uric acid, citrate, glutamine and methionine, diagnostic of dysfunctional hepatic metabolic fluxes [102]. A series of 15 HCC patients displayed increased valine, alanine, acetone, succinate, glutamine, choline, lactate and glucose one day after transplantation. After 7 days, lipids and choline increased while glucose and amino acids decreased [31]. The metabolomic window appears to offer new insights into specific hepatic metabolic changes in the transplantation perioperative period.
Miscellaneous other hepatobiliary diseases
Metabolomic studies have been reported that are of relevance to Wilson's disease [103, 104], primary biliary cirrhosis [105], primary sclerosing cholangitis [105], the hepatic stage of malaria [106–108], as well as various aspects of hepatic encephalopathy [109–112].
The metabolomic window into acute liver toxicity in animal models
High-throughput metabolomic screening of hepatotoxins in laboratory animals first used NMR and pattern recognition algorithms [113–115] but, in early studies, also employed Fourier-transform infrared spectroscopy [116]. Metabolomic profiles of numerous hepatotoxins in laboratory animals have been described, and include hydrazine [117], bromobenzene [118, 119], methapyrilene [120], methylenedianiline [121], D-galactosamine [121–123], clofibrate [121], allyl formate [124], the anti-HBV compound Bay41-4109 [125], paracetamol [126–133], isoniazid [134, 135], carbon tetrachloride [131, 136–138], α-naphthylisothiocyanate [137], perfluorododecanoic acid [139], valproate [140], Huang-yao-zi [141], dimethylnitrosamine [142], polychlorinated biphenyls [143, 144], 2,3,7,8-tetrachlorodibenzo-p-dioxin [143], methamphetamine [145], (+)-usnic acid [146], pentamethychromanol [147] and methotrexate [131]. Detailed analysis of these drug-induced liver injury (DILI) studies falls beyond the scope of this review. However, the reader is directed to The Liver Toxicity Biomarker Study on DILI and closely related topics that have been reviewed [148–153].
A proposed metabolomics-based model for major liver disease
Based upon a review of the available literature, we propose a three-stage progression from hepatic insult of the healthy liver to carcinoma (Fig. 4). A core metabolomic phenotype (CMP) arises early in this progression and comprises readily discernible changes in bile acids and phospholipids (Tables 1–4, Fig. 3). The CMP is maintained whether or not cirrhosis arises and/or HCC or CCA develop (Stages 2 and 3, respectively). This CMP is common to all etiologies in Stage 1, including NAFLD/NASH, ALD and viral hepatitis. Other metabolomic perturbations distinguish the different stages (Fig. 3). We also propose that the metabolic remodelling described for HCC [56] begins at the transition from Phase 0 to Phase 1 as a consequence of the presence of inflammatory signalling in the liver, as outlined in Fig. 2. Thus, this body of accumulated metabolomic data may begin to cast further light on hepatobiliary diseases.
Fig. 4. Diverse hepatic insults leading to a core metabolomic phenotype en route from the healthy liver to HCC and CCA.
Elevated serum bile acids and urinary bile salts together with decreased serum lysophosphatidylcholines represent the core metabolomic phenotype (CMP). A metabolic remodelling begins in the transition from the healthy liver (Phase 0) to NAFLD/NASH, ALD or viral hepatitis (Phase 1). During Stage 1 there occurs a Warburg shift from mitochondrial respiration to cytosolic glycolysis, together with an increase in fatty acid β-oxidation. This metabolic remodelling persists through cirrhosis (Phase 2) and into carcinoma (Phase 3). Note that the sum of the carcinoma energy production D+E+F is greater than the summed energy production A+B+C in the healthy liver.
Abbreviations used: CCA, cholangiocarcinoma; CMP, core metabolomic phenotype; ALD, alcoholic liver disease. For other abbreviations, see Fig. 1.
Acknowledgements
We would like to thank our colleague Professor Jean-François Dufour for encouraging us to write this review. The authors wish to acknowledge the financial support of the National Institutes of Health/National Institute of Allergy and Infectious Diseases (grant U19 AI067773-07/08), the Hassan Badawi Foundation Against Liver Cancer and Imperial Tobacco Limited, UK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Idle JR, Gonzalez FJ. Metabolomics. Cell Metab. 2007;6:348–351. doi: 10.1016/j.cmet.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farrell GC, van Rooyen D, Gan L, Chitturi S. NASH is an Inflammatory Disorder: Pathogenic, Prognostic and Therapeutic Implications. Gut Liver. 2012;6:149–171. doi: 10.5009/gnl.2012.6.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres DM, Williams CD, Harrison SA. Features, diagnosis, and treatment of nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2012;10:837–858. doi: 10.1016/j.cgh.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Vinaixa M, Rodriguez MA, Rull A, Beltran R, Blade C, Brezmes J, et al. Metabolomic assessment of the effect of dietary cholesterol in the progressive development of fatty liver disease. J Proteome Res. 2010;9:2527–2538. doi: 10.1021/pr901203w. [DOI] [PubMed] [Google Scholar]
- 5.van Ginneken V, Verhey E, Poelmann R, Ramakers R, van Dijk KW, Ham L, et al. Metabolomics (liver and blood profiling) in a mouse model in response to fasting: a study of hepatic steatosis. Biochim Biophys Acta. 2007;1771:1263–1270. doi: 10.1016/j.bbalip.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Hyde MJ, Griffin JL, Herrera E, Byrne CD, Clarke L, Kemp PR. Delivery by Caesarean section, rather than vaginal delivery, promotes hepatic steatosis in piglets. Clin Sci (Lond) 2010;118:47–59. doi: 10.1042/CS20090169. [DOI] [PubMed] [Google Scholar]
- 7.Puri P, Wiest MM, Cheung O, Mirshahi F, Sargeant C, Min HK, et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 2009;50:1827–1838. doi: 10.1002/hep.23229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Canaveras JC, Donato MT, Castell JV, Lahoz A. A comprehensive untargeted metabonomic analysis of human steatotic liver tissue by RP and HILIC chromatography coupled to mass spectrometry reveals important metabolic alterations. J Proteome Res. 2011;10:4825–4834. doi: 10.1021/pr200629p. [DOI] [PubMed] [Google Scholar]
- 9.Kalhan SC, Guo L, Edmison J, Dasarathy S, McCullough AJ, Hanson RW, et al. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism. 2011;60:404–413. doi: 10.1016/j.metabol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barr J, Vazquez-Chantada M, Alonso C, Perez-Cormenzana M, Mayo R, Galan A, et al. Liquid chromatography-mass spectrometry-based parallel metabolic profiling of human and mouse model serum reveals putative biomarkers associated with the progression of nonalcoholic fatty liver disease. J Proteome Res. 2010;9:4501–4512. doi: 10.1021/pr1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Wang L, Yan X, Liu Q, Yu C, Wei H, et al. A proton nuclear magnetic resonance metabonomics approach for biomarker discovery in nonalcoholic fatty liver disease. J Proteome Res. 2011;10:2797–2806. doi: 10.1021/pr200047c. [DOI] [PubMed] [Google Scholar]
- 12.Toye AA, Dumas ME, Blancher C, Rothwell AR, Fearnside JF, Wilder SP, et al. Subtle metabolic and liver gene transcriptional changes underlie diet-induced fatty liver susceptibility in insulin-resistant mice. Diabetologia. 2007;50:1867–1879. doi: 10.1007/s00125-007-0738-5. [DOI] [PubMed] [Google Scholar]
- 13.Noguchi Y, Young JD, Aleman JO, Hansen ME, Kelleher JK, Stephanopoulos G. Tracking cellular metabolomics in lipoapoptosis- and steatosis-developing liver cells. Mol Biosyst. 2011;7:1409–1419. doi: 10.1039/c0mb00309c. [DOI] [PubMed] [Google Scholar]
- 14.Griffin JL, Scott J, Nicholson JK. The influence of pharmacogenetics on fatty liver disease in the wistar and kyoto rats: a combined transcriptomic and metabonomic study. J Proteome Res. 2007;6:54–61. doi: 10.1021/pr0601640. [DOI] [PubMed] [Google Scholar]
- 15.Hines IN, Hartwell HJ, Feng Y, Theve EJ, Hall GA, Hashway S, et al. Insulin resistance and metabolic hepatocarcinogenesis with parent-of-origin effects in AxB mice. Am J Pathol. 2011;179:2855–2865. doi: 10.1016/j.ajpath.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei J, Qiu de K, Ma X. Bile acids and insulin resistance: implications for treating nonalcoholic fatty liver disease. J Dig Dis. 2009;10:85–90. doi: 10.1111/j.1751-2980.2009.00369.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 18.Stanton MC, Chen SC, Jackson JV, Rojas-Triana A, Kinsley D, Cui L, et al. Inflammatory Signals shift from adipose to liver during high fat feeding and influence the development of steatohepatitis in mice. J Inflamm (Lond) 2011;8:8. doi: 10.1186/1476-9255-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beyoglu D, Idle JR. Metabolomics and its potential in drug development. Biochem Pharmacol. 2013;85:12–20. doi: 10.1016/j.bcp.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka N, Matsubara T, Krausz KW, Patterson AD, Gonzalez FJ. Disruption of phospholipid and bile acid homeostasis in mice with nonalcoholic steatohepatitis. Hepatology. 2012;56:118–129. doi: 10.1002/hep.25630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, Chen YQ, Bonacci TM, Bredt DS, Li S, Bensch WR, et al. Identification and characterization of a major liver lysophosphatidylcholine acyltransferase. J Biol Chem. 2008;283:8258–8265. doi: 10.1074/jbc.M710422200. [DOI] [PubMed] [Google Scholar]
- 22.Constantinou MA, Theocharis SE, Mikros E. Application of metabonomics on an experimental model of fibrosis and cirrhosis induced by thioacetamide in rats. Toxicol Appl Pharmacol. 2007;218:11–19. doi: 10.1016/j.taap.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Yeh MM, Brunt EM. Pathology of nonalcoholic fatty liver disease. Am J Clin Pathol. 2007;128:837–847. doi: 10.1309/RTPM1PY6YGBL2G2R. [DOI] [PubMed] [Google Scholar]
- 24.Wiegand J, Berg T. The etiology, diagnosis and prevention of liver cirrhosis: part 1 of a series on liver cirrhosis. Dtsch Arztebl Int. 2013;110:85–91. doi: 10.3238/arztebl.2013.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gou X, Tao Q, Feng Q, Peng J, Sun S, Cao H, et al. Urinary metabonomics characterization of liver fibrosis induced by CCl(4) in rats and intervention effects of Xia Yu Xue Decoction. J Pharm Biomed Anal. 2013;74:62–65. doi: 10.1016/j.jpba.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 26.Zhang A, Sun H, Dou S, Sun W, Wu X, Wang P, et al. Metabolomics study on the hepatoprotective effect of scoparone using ultra-performance liquid chromatography/electrospray ionization quadruple time-of-flight mass spectrometry. Analyst. 2013;138:353–361. doi: 10.1039/c2an36382h. [DOI] [PubMed] [Google Scholar]
- 27.Gao H, Lu Q, Liu X, Cong H, Zhao L, Wang H, et al. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009;100:782–785. doi: 10.1111/j.1349-7006.2009.01086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldhier MC, Almstetter MF, Nurnberger N, Gruber MA, Dettmer K, Oefner PJ. Improved enantiomer resolution and quantification of free D-amino acids in serum and urine by comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry. J Chromatogr A. 2011;1218:4537–4544. doi: 10.1016/j.chroma.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 29.Lian JS, Liu W, Hao SR, Guo YZ, Huang HJ, Chen DY, et al. A serum metabonomic study on the difference between alcohol- and HBV-induced liver cirrhosis by ultraperformance liquid chromatography coupled to mass spectrometry plus quadrupole time-of-flight mass spectrometry. Chin Med J (Engl) 2011;124:1367–1373. [PubMed] [Google Scholar]
- 30.Du Z, Zhang L, Liu S. Application of liquid chromatography-mass spectrometry in the study of metabolic profiling of cirrhosis in different grades. Se Pu. 2011;29:314–319. doi: 10.3724/sp.j.1123.2011.00314. [DOI] [PubMed] [Google Scholar]
- 31.Qi SW, Tu ZG, Peng WJ, Wang LX, Ou-Yang X, Cai AJ, et al. (1)H NMR-based serum metabolic profiling in compensated and decompensated cirrhosis. World J Gastroenterol. 2012;18:285–290. doi: 10.3748/wjg.v18.i3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin X, Zhang Y, Ye G, Li X, Yin P, Ruan Q, et al. Classification and differential metabolite discovery of liver diseases based on plasma metabolic profiling and support vector machines. J Sep Sci. 2011;34:3029–3036. doi: 10.1002/jssc.201100408. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Granados B, Morales JM, Rodrigo JM, Del Olmo J, Serra MA, Ferrandez A, et al. Metabolic profile of chronic liver disease by NMR spectroscopy of human biopsies. Int J Mol Med. 2011;27:111–117. doi: 10.3892/ijmm.2010.563. [DOI] [PubMed] [Google Scholar]
- 34.Huang HJ, Zhang AY, Cao HC, Lu HF, Wang BH, Xie Q, et al. Metabolomic analyses of faeces reveals malabsorption in cirrhotic patients. Dig Liver Dis. 2013 doi: 10.1016/j.dld.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Fukiya S, Arata M, Kawashima H, Yoshida D, Kaneko M, Minamida K, et al. Conversion of cholic acid and chenodeoxycholic acid into their 7-oxo derivatives by Bacteroides intestinalis AM-1 isolated from human feces. FEMS Microbiol Lett. 2009;293:263–270. doi: 10.1111/j.1574-6968.2009.01531.x. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Xu G, Zheng Y, Kong H, Pang T, Lv S, et al. Diagnosis of liver cancer using HPLC-based metabonomics avoiding false-positive result from hepatitis and hepatocirrhosis diseases. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;813:59–65. doi: 10.1016/j.jchromb.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Xu G, Zheng Y, Kong H, Wang C, Zhao X, et al. Strategy for metabonomics research based on high-performance liquid chromatography and liquid chromatography coupled with tandem mass spectrometry. J Chromatogr A. 2005;1084:214–221. doi: 10.1016/j.chroma.2004.10.100. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Li C, Nie X, Feng X, Chen W, Yue Y, et al. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J Proteome Res. 2007;6:2605–2614. doi: 10.1021/pr070063h. [DOI] [PubMed] [Google Scholar]
- 39.Yin P, Wan D, Zhao C, Chen J, Zhao X, Wang W, et al. A metabonomic study of hepatitis B-induced liver cirrhosis and hepatocellular carcinoma by using RP-LC and HILIC coupled with mass spectrometry. Mol Biosyst. 2009;5:868–876. doi: 10.1039/b820224a. [DOI] [PubMed] [Google Scholar]
- 40.Wu H, Xue R, Dong L, Liu T, Deng C, Zeng H, et al. Metabolomic profiling of human urine in hepatocellular carcinoma patients using gas chromatography/mass spectrometry. Anal Chim Acta. 2009;648:98–104. doi: 10.1016/j.aca.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Wang W, Lv S, Yin P, Zhao X, Lu X, et al. Metabonomics study of liver cancer based on ultra performance liquid chromatography coupled to mass spectrometry with HILIC and RPLC separations. Anal Chim Acta. 2009;650:3–9. doi: 10.1016/j.aca.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Zhang S, Li Z, Yang J, Huang C, Liang R, et al. (1)H-NMR-based metabolomics of tumor tissue for the metabolic characterization of rat hepatocellular carcinoma formation and metastasis. Tumour Biol. 2011;32:223–231. doi: 10.1007/s13277-010-0116-7. [DOI] [PubMed] [Google Scholar]
- 43.Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004945. M110 004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen F, Xue J, Zhou L, Wu S, Chen Z. Identification of serum biomarkers of hepatocarcinoma through liquid chromatography/mass spectrometry-based metabonomic method. Anal Bioanal Chem. 2011;401:1899–1904. doi: 10.1007/s00216-011-5245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patterson AD, Maurhofer O, Beyoglu D, Lanz C, Krausz KW, Pabst T, et al. Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer Res. 2011;71:6590–6600. doi: 10.1158/0008-5472.CAN-11-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan Y, Yin P, Tang L, Xing W, Huang Q, Cao D, et al. Metabolomics study of stepwise hepatocarcinogenesis from the model rats to patients: potential biomarkers effective for small hepatocellular carcinoma diagnosis. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.010694. M111 010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang B, Chen D, Chen Y, Hu Z, Cao M, Xie Q, et al. Metabonomic profiles discriminate hepatocellular carcinoma from liver cirrhosis by ultraperformance liquid chromatography-mass spectrometry. J Proteome Res. 2012;11:1217–1227. doi: 10.1021/pr2009252. [DOI] [PubMed] [Google Scholar]
- 48.Zhou L, Wang Q, Yin P, Xing W, Wu Z, Chen S, et al. Serum metabolomics reveals the deregulation of fatty acids metabolism in hepatocellular carcinoma and chronic liver diseases. Anal Bioanal Chem. 2012;403:203–213. doi: 10.1007/s00216-012-5782-4. [DOI] [PubMed] [Google Scholar]
- 49.Ye G, Zhu B, Yao Z, Yin P, Lu X, Kong H, et al. Analysis of urinary metabolic signatures of early hepatocellular carcinoma recurrence after surgical removal using gas chromatography-mass spectrometry. J Proteome Res. 2012;11:4361–4372. doi: 10.1021/pr300502v. [DOI] [PubMed] [Google Scholar]
- 50.Lin X, Yang F, Zhou L, Yin P, Kong H, Xing W, et al. A support vector machine-recursive feature elimination feature selection method based on artificial contrast variables and mutual information. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;910:149–155. doi: 10.1016/j.jchromb.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 51.Ressom HW, Xiao JF, Tuli L, Varghese RS, Zhou B, Tsai TH, et al. Utilization of metabolomics to identify serum biomarkers for hepatocellular carcinoma in patients with liver cirrhosis. Anal Chim Acta. 2012;743:90–100. doi: 10.1016/j.aca.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao JF, Varghese RS, Zhou B, Nezami Ranjbar MR, Zhao Y, Tsai TH, et al. LC-MS based serum metabolomics for identification of hepatocellular carcinoma biomarkers in Egyptian cohort. J Proteome Res. 2012;11:5914–5923. doi: 10.1021/pr300673x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang A, Sun H, Yan G, Han Y, Ye Y, Wang X. Urinary metabolic profiling identifies a key role for glycocholic acid in human liver cancer by ultra-performance liquid-chromatography coupled with high-definition mass spectrometry. Clin Chim Acta. 2013;418:86–90. doi: 10.1016/j.cca.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 54.Nahon P, Amathieu R, Triba MN, Bouchemal N, Nault JC, Ziol M, et al. Identification of serum proton NMR metabolomic fingerprints associated with hepatocellular carcinoma in patients with alcoholic cirrhosis. Clin Cancer Res. 2012;18:6714–6722. doi: 10.1158/1078-0432.CCR-12-1099. [DOI] [PubMed] [Google Scholar]
- 55.Budhu A, Roessler S, Zhao X, Yu Z, Forgues M, Ji J, et al. Integrated Metabolite and Gene Expression Profiles Identify Lipid Biomarkers Associated With Progression of Hepatocellular Carcinoma and Patient Outcomes. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beyoglu D, Imbeaud S, Maurhofer O, Bioulac-Sage P, Zucman-Rossi J, Dufour JF, et al. Tissue metabolomics of hepatocellular carcinoma: Tumor energy metabolism and the role of transcriptomic classification. Hepatology. 2013 doi: 10.1002/hep.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shariff MI, Gomaa AI, Cox IJ, Patel M, Williams HR, Crossey MM, et al. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: a validation study. J Proteome Res. 2011;10:1828–1836. doi: 10.1021/pr101096f. [DOI] [PubMed] [Google Scholar]
- 58.Li ZF, Wang J, Huang C, Zhang S, Yang J, Jiang A, et al. Gas chromatography/time-of-flight mass spectrometry-based metabonomics of hepatocarcinoma in rats with lung metastasis: elucidation of the metabolic characteristics of hepatocarcinoma at formation and metastasis. Rapid Commun Mass Spectrom. 2010;24:2765–2775. doi: 10.1002/rcm.4703. [DOI] [PubMed] [Google Scholar]
- 59.Southam AD, Easton JM, Stentiford GD, Ludwig C, Arvanitis TN, Viant MR. Metabolic changes in flatfish hepatic tumours revealed by NMR-based metabolomics and metabolic correlation networks. J Proteome Res. 2008;7:5277–5285. doi: 10.1021/pr800353t. [DOI] [PubMed] [Google Scholar]
- 60.Stentiford GD, Viant MR, Ward DG, Johnson PJ, Martin A, Wenbin W, et al. Liver tumors in wild flatfish: a histopathological, proteomic, and metabolomic study. OMICS. 2005;9:281–299. doi: 10.1089/omi.2005.9.281. [DOI] [PubMed] [Google Scholar]
- 61.Pessayre D. Role of mitochondria in non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2007;22(Suppl 1):S20–S27. doi: 10.1111/j.1440-1746.2006.04640.x. [DOI] [PubMed] [Google Scholar]
- 62.Adeva MM, Souto G, Blanco N, Donapetry C. Ammonium metabolism in humans. Metabolism. 2012;61:1495–1511. doi: 10.1016/j.metabol.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 63.Felipo V, Urios A, Montesinos E, Molina I, Garcia-Torres ML, Civera M, et al. Contribution of hyperammonemia and inflammatory factors to cognitive impairment in minimal hepatic encephalopathy. Metab Brain Dis. 2012;27:51–58. doi: 10.1007/s11011-011-9269-3. [DOI] [PubMed] [Google Scholar]
- 64.Mukherjee S, Vaidyanathan K, Vasudevan DM, Das SK. Role of plasma amino acids and gaba in alcoholic and non-alcoholic fatty liver disease-a pilot study. Indian J Clin Biochem. 2010;25:37–42. doi: 10.1007/s12291-010-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seckl JR, Walker BR. Minireview: 11beta-hydroxysteroid dehydrogenase type 1- a tissue-specific amplifier of glucocorticoid action. Endocrinology. 2001;142:1371–1376. doi: 10.1210/endo.142.4.8114. [DOI] [PubMed] [Google Scholar]
- 66.Cooper MS, Stewart PM. 11Beta-hydroxysteroid dehydrogenase type 1 and its role in the hypothalamus-pituitary-adrenal axis, metabolic syndrome, and inflammation. J Clin Endocrinol Metab. 2009;94:4645–4654. doi: 10.1210/jc.2009-1412. [DOI] [PubMed] [Google Scholar]
- 67.Moon SS, Lee YS, Kim JG, Lee IK. Association of 11beta-hydroxysteroid dehydrogenase type 1 gene polymorphisms with serum alanine aminotransferase activity. Diabetes Res Clin Pract. 2013 doi: 10.1016/j.diabres.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 68.Muting D, Wortmann V. Amino acid metabolism in liver diseases. Dtsch Med Wochenschr. 1956;81:1853–1856. doi: 10.1055/s-0028-1115247. [DOI] [PubMed] [Google Scholar]
- 69.Dam G, Ott P, Aagaard NK, Vilstrup H. Branched-chain amino acids and muscle ammonia detoxification in cirrhosis. Metab Brain Dis. 2013 doi: 10.1007/s11011-013-9377-3. [DOI] [PubMed] [Google Scholar]
- 70.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 71.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 72.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zivkovic AM, Bruce German J, Esfandiari F, Halsted CH. Quantitative lipid metabolomic changes in alcoholic micropigs with fatty liver disease. Alcohol Clin Exp Res. 2009;33:751–758. doi: 10.1111/j.1530-0277.2008.00892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li S, Liu H, Jin Y, Lin S, Cai Z, Jiang Y. Metabolomics study of alcohol-induced liver injury and hepatocellular carcinoma xenografts in mice. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:2369–2375. doi: 10.1016/j.jchromb.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 75.Fernando H, Bhopale KK, Kondraganti S, Kaphalia BS, Shakeel Ansari GA. Lipidomic changes in rat liver after long-term exposure to ethanol. Toxicol Appl Pharmacol. 2011;255:127–137. doi: 10.1016/j.taap.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patterson AD, Slanar O, Krausz KW, Li F, Hofer CC, Perlik F, et al. Human urinary metabolomic profile of PPARalpha induced fatty acid beta-oxidation. J Proteome Res. 2009;8:4293–4300. doi: 10.1021/pr9004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhen Y, Krausz KW, Chen C, Idle JR, Gonzalez FJ. Metabolomic and genetic analysis of biomarkers for peroxisome proliferator-activated receptor alpha expression and activation. Mol Endocrinol. 2007;21:2136–2151. doi: 10.1210/me.2007-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manna SK, Patterson AD, Yang Q, Krausz KW, Li H, Idle JR, et al. Identification of noninvasive biomarkers for alcohol-induced liver disease using urinary metabolomics and the Ppara-null mouse. J Proteome Res. 2010;9:4176–4188. doi: 10.1021/pr100452b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manna SK, Patterson AD, Yang Q, Krausz KW, Idle JR, Fornace AJ, et al. UPLC-MS-based urine metabolomics reveals indole-3-lactic acid and phenyllactic acid as conserved biomarkers for alcohol-induced liver disease in the Ppara-null mouse model. J Proteome Res. 2011;10:4120–4133. doi: 10.1021/pr200310s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lieber CS. Metabolism of alcohol. Clin Liver Dis. 2005;9:1–35. doi: 10.1016/j.cld.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 81.Shi X, Yao D, Chen C. Identification of N-acetyltaurine as a novel metabolite of ethanol through metabolomics-guided biochemical analysis. J Biol Chem. 2012;287:6336–6349. doi: 10.1074/jbc.M111.312199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Castera L. Noninvasive methods to assess liver disease in patients with hepatitis B or C. Gastroenterology. 2012;142:1293 e1294–1302 e1294. doi: 10.1053/j.gastro.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 83.Yang J, Zhao X, Liu X, Wang C, Gao P, Wang J, et al. High performance liquid chromatography-mass spectrometry for metabonomics: potential biomarkers for acute deterioration of liver function in chronic hepatitis B. J Proteome Res. 2006;5:554–561. doi: 10.1021/pr050364w. [DOI] [PubMed] [Google Scholar]
- 84.Zhang L, Jia X, Peng X, Ou Q, Zhang Z, Qiu C, et al. Development and validation of a liquid chromatography-mass spectrometry metabonomic platform in human plasma of liver failure caused by hepatitis B virus. Acta Biochim Biophys Sin (Shanghai) 2010;42:688–698. doi: 10.1093/abbs/gmq078. [DOI] [PubMed] [Google Scholar]
- 85.Sherman M, Shafran S, Burak K, Doucette K, Wong W, Girgrah N, et al. Management of chronic hepatitis C: consensus guidelines. Can J Gastroenterol. 2007;21(Suppl C):25C–34C. [PMC free article] [PubMed] [Google Scholar]
- 86.Sherman M, Shafran S, Burak K, Doucette K, Wong W, Girgrah N, et al. Management of chronic hepatitis B: consensus guidelines. Can J Gastroenterol. 2007;21(Suppl C):5C–24C. [PMC free article] [PubMed] [Google Scholar]
- 87.Davis GL, Albright JE, Cook SF, Rosenberg DM. Projecting future complications of chronic hepatitis C in the United States. Liver Transpl. 2003;9:331–338. doi: 10.1053/jlts.2003.50073. [DOI] [PubMed] [Google Scholar]
- 88.Lloyd GR, Ahmad S, Wasim M, Brereton RG. Pattern recognition of inductively coupled plasma atomic emission spectroscopy of human scalp hair for discriminating between healthy and hepatitis C patients. Anal Chim Acta. 2009;649:33–42. doi: 10.1016/j.aca.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 89.Sperling M, Karst U. Metallomics: an emerging interdisciplinary science. Anal Bioanal Chem. 2013;405:1789–1790. doi: 10.1007/s00216-012-6619-x. [DOI] [PubMed] [Google Scholar]
- 90.Godoy MM, Lopes EP, Silva RO, Hallwass F, Koury LC, Moura IM, et al. Hepatitis C virus infection diagnosis using metabonomics. J Viral Hepat. 2010;17:854–858. doi: 10.1111/j.1365-2893.2009.01252.x. [DOI] [PubMed] [Google Scholar]
- 91.Roe B, Kensicki E, Mohney R, Hall WW. Metabolomic profile of hepatitis C virus-infected hepatocytes. PLoS One. 2011;6:e23641. doi: 10.1371/journal.pone.0023641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun H, Zhang A, Yan G, Piao C, Li W, Sun C, et al. Metabolomic analysis of key regulatory metabolites in HCV-infected tree shrews. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M112.019141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Palmer WC, Patel T. Are common factors involved in the pathogenesis of primary liver cancers? A meta-analysis of risk factors for intrahepatic cholangiocarcinoma. J Hepatol. 2012;57:69–76. doi: 10.1016/j.jhep.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet. 2005;366:1303–1314. doi: 10.1016/S0140-6736(05)67530-7. [DOI] [PubMed] [Google Scholar]
- 95.Komichi D, Tazuma S, Nishioka T, Hyogo H, Chayama K. Glycochenodeoxycholate plays a carcinogenic role in immortalized mouse cholangiocytes via oxidative DNA damage. Free Radic Biol Med. 2005;39:1418–1427. doi: 10.1016/j.freeradbiomed.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 96.Sharif AW, Williams HR, Lampejo T, Khan SA, Bansi DS, Westaby D, et al. Metabolic profiling of bile in cholangiocarcinoma using in vitro magnetic resonance spectroscopy. HPB (Oxford) 2010;12:396–402. doi: 10.1111/j.1477-2574.2010.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hashim Abdalla MS, Taylor-Robinson SD, Sharif AW, Williams HR, Crossey MM, Badra GA, et al. Differences in phosphatidylcholine and bile acids in bile from Egyptian and UK patients with and without cholangiocarcinoma. HPB (Oxford) 2011;13:385–390. doi: 10.1111/j.1477-2574.2011.00296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ishihara K, Katsutani N, Asai N, Inomata A, Uemura Y, Suganuma A, et al. Identification of urinary biomarkers useful for distinguishing a difference in mechanism of toxicity in rat model of cholestasis. Basic Clin Pharmacol Toxicol. 2009;105:156–166. doi: 10.1111/j.1742-7843.2009.00410.x. [DOI] [PubMed] [Google Scholar]
- 99.Cho JY, Matsubara T, Kang DW, Ahn SH, Krausz KW, Idle JR, et al. Urinary metabolomics in Fxr-null mice reveals activated adaptive metabolic pathways upon bile acid challenge. J Lipid Res. 2010;51:1063–1074. doi: 10.1194/jlr.M002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aoki M, Konya Y, Takagaki T, Umemura K, Sogame Y, Katsumata T, et al. Metabolomic investigation of cholestasis in a rat model using ultra-performance liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2011;25:1847–1852. doi: 10.1002/rcm.5072. [DOI] [PubMed] [Google Scholar]
- 101.Li Z, Shen H, Zhang Y, Lu M, Qiao X, Meng X, et al. Metabolomic study of serum from rabbits with acute acalculous cholecystitis. Inflamm Res. 2012;61:987–995. doi: 10.1007/s00011-012-0491-1. [DOI] [PubMed] [Google Scholar]
- 102.Serkova NJ, Zhang Y, Coatney JL, Hunter L, Wachs ME, Niemann CU, et al. Early detection of graft failure using the blood metabolic profile of a liver recipient. Transplantation. 2007;83:517–521. doi: 10.1097/01.tp.0000251649.01148.f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wilmarth PA, Short KK, Fiehn O, Lutsenko S, David LL, Burkhead JL. A systems approach implicates nuclear receptor targeting in the Atp7b(−/−) mouse model of Wilson's disease. Metallomics. 2012;4:660–668. doi: 10.1039/c2mt20017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Santos EM, Ball JS, Williams TD, Wu H, Ortega F, van Aerle R, et al. Identifying health impacts of exposure to copper using transcriptomics and metabolomics in a fish model. Environ Sci Technol. 2010;44:820–826. doi: 10.1021/es902558k. [DOI] [PubMed] [Google Scholar]
- 105.Trottier J, Bialek A, Caron P, Straka RJ, Heathcote J, Milkiewicz P, et al. Metabolomic profiling of 17 bile acids in serum from patients with primary biliary cirrhosis and primary sclerosing cholangitis: a pilot study. Dig Liver Dis. 2012;44:303–310. doi: 10.1016/j.dld.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 106.Sengupta A, Basant A, Ghosh S, Sharma S, Sonawat HM. Liver Metabolic Alterations and Changes in Host Intercompartmental Metabolic Correlation during Progression of Malaria. J Parasitol Res. 2011;2011:901854. doi: 10.1155/2011/901854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sengupta A, Ghosh S, Basant A, Malusare S, Johri P, Pathak S, et al. Global host metabolic response to Plasmodium vivax infection: a 1H NMR based urinary metabonomic study. Malar J. 2011;10:384. doi: 10.1186/1475-2875-10-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ghosh S, Sengupta A, Sharma S, Sonawat HM. Metabolic fingerprints of serum, brain, and liver are distinct for mice with cerebral and noncerebral malaria: a (1)H NMR spectroscopy-based metabonomic study. J Proteome Res. 2012;11:4992–5004. doi: 10.1021/pr300562m. [DOI] [PubMed] [Google Scholar]
- 109.Barba I, Chatauret N, Garcia-Dorado D, Cordoba J. A 1H nuclear magnetic resonance-based metabonomic approach for grading hepatic encephalopathy and monitoring the effects of therapeutic hypothermia in rats. Liver Int. 2008;28:1141–1148. doi: 10.1111/j.1478-3231.2008.01801.x. [DOI] [PubMed] [Google Scholar]
- 110.Williams HR, Cox IJ, Taylor-Robinson SD. Metabonomics in hepatic encephalopathy: lucidity emerging from confusion. Liver Int. 2008;28:1050–1051. doi: 10.1111/j.1478-3231.2008.01817.x. [DOI] [PubMed] [Google Scholar]
- 111.Jimenez B, Montoliu C, MacIntyre DA, Serra MA, Wassel A, Jover M, et al. Serum metabolic signature of minimal hepatic encephalopathy by (1)H-nuclear magnetic resonance. J Proteome Res. 2010;9:5180–5187. doi: 10.1021/pr100486e. [DOI] [PubMed] [Google Scholar]
- 112.Bajaj JS, Gillevet PM, Patel NR, Ahluwalia V, Ridlon JM, Kettenmann B, et al. A longitudinal systems biology analysis of lactulose withdrawal in hepatic encephalopathy. Metab Brain Dis. 2012;27:205–215. doi: 10.1007/s11011-012-9303-0. [DOI] [PubMed] [Google Scholar]
- 113.Robertson DG, Reily MD, Sigler RE, Wells DF, Paterson DA, Braden TK. Metabonomics: evaluation of nuclear magnetic resonance (NMR) and pattern recognition technology for rapid in vivo screening of liver and kidney toxicants. Toxicol Sci. 2000;57:326–337. doi: 10.1093/toxsci/57.2.326. [DOI] [PubMed] [Google Scholar]
- 114.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 115.Wang Y, Bollard ME, Keun H, Antti H, Beckonert O, Ebbels TM, et al. Spectral editing and pattern recognition methods applied to high-resolution magic-angle spinning 1H nuclear magnetic resonance spectroscopy of liver tissues. Anal Biochem. 2003;323:26–32. doi: 10.1016/j.ab.2003.07.026. [DOI] [PubMed] [Google Scholar]
- 116.Harrigan GG, LaPlante RH, Cosma GN, Cockerell G, Goodacre R, Maddox JF, et al. Application of high-throughput Fourier-transform infrared spectroscopy in toxicology studies: contribution to a study on the development of an animal model for idiosyncratic toxicity. Toxicol Lett. 2004;146:197–205. doi: 10.1016/j.toxlet.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 117.Bollard ME, Keun HC, Beckonert O, Ebbels TM, Antti H, Nicholls AW, et al. Comparative metabonomics of differential hydrazine toxicity in the rat and mouse. Toxicol Appl Pharmacol. 2005;204:135–151. doi: 10.1016/j.taap.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 118.Heijne WH, Lamers RJ, van Bladeren PJ, Groten JP, van Nesselrooij JH, van Ommen B. Profiles of metabolites and gene expression in rats with chemically induced hepatic necrosis. Toxicol Pathol. 2005;33:425–433. doi: 10.1080/01926230590958146. [DOI] [PubMed] [Google Scholar]
- 119.Waters NJ, Waterfield CJ, Farrant RD, Holmes E, Nicholson JK. Integrated metabonomic analysis of bromobenzene-induced hepatotoxicity: novel induction of 5-oxoprolinosis. J Proteome Res. 2006;5:1448–1459. doi: 10.1021/pr060024q. [DOI] [PubMed] [Google Scholar]
- 120.Craig A, Sidaway J, Holmes E, Orton T, Jackson D, Rowlinson R, et al. Systems toxicology: integrated genomic, proteomic and metabonomic analysis of methapyrilene induced hepatotoxicity in the rat. J Proteome Res. 2006;5:1586–1601. doi: 10.1021/pr0503376. [DOI] [PubMed] [Google Scholar]
- 121.Ishihara K, Katsutani N, Aoki T. A metabonomics study of the hepatotoxicants galactosamine, methylene dianiline and clofibrate in rats. Basic Clin Pharmacol Toxicol. 2006;99:251–260. doi: 10.1111/j.1742-7843.2006.pto_455.x. [DOI] [PubMed] [Google Scholar]
- 122.Feng B, Wu SM, Lv S, Liu F, Chen HS, Gao Y, et al. A novel scoring system for prognostic prediction in d-galactosamine/lipopolysaccharide-induced fulminant hepatic failure BALB/c mice. BMC Gastroenterol. 2009;9:99. doi: 10.1186/1471-230X-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Coen M, Goldfain-Blanc F, Rolland-Valognes G, Walther B, Robertson DG, Holmes E, et al. Pharmacometabonomic investigation of dynamic metabolic phenotypes associated with variability in response to galactosamine hepatotoxicity. J Proteome Res. 2012;11:2427–2440. doi: 10.1021/pr201161f. [DOI] [PubMed] [Google Scholar]
- 124.Yap IK, Clayton TA, Tang H, Everett JR, Hanton G, Provost JP, et al. An integrated metabonomic approach to describe temporal metabolic disregulation induced in the rat by the model hepatotoxin allyl formate. J Proteome Res. 2006;5:2675–2684. doi: 10.1021/pr0601584. [DOI] [PubMed] [Google Scholar]
- 125.Shi C, Wu CQ, Cao AM, Sheng HZ, Yan XZ, Liao MY. NMR-spectroscopy-based metabonomic approach to the analysis of Bay41-4109, a novel anti-HBV compound, induced hepatotoxicity in rats. Toxicol Lett. 2007;173:161–167. doi: 10.1016/j.toxlet.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 126.Sun J, Schnackenberg LK, Holland RD, Schmitt TC, Cantor GH, Dragan YP, et al. Metabonomics evaluation of urine from rats given acute and chronic doses of acetaminophen using NMR and UPLC/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:328–340. doi: 10.1016/j.jchromb.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 127.Chen C, Krausz KW, Shah YM, Idle JR, Gonzalez FJ. Serum metabolomics reveals irreversible inhibition of fatty acid beta-oxidation through the suppression of PPARalpha activation as a contributing mechanism of acetaminophen-induced hepatotoxicity. Chem Res Toxicol. 2009;22:699–707. doi: 10.1021/tx800464q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cheng J, Ma X, Krausz KW, Idle JR, Gonzalez FJ. Rifampicin-activated human pregnane X receptor and CYP3A4 induction enhance acetaminophen-induced toxicity. Drug Metab Dispos. 2009;37:1611–1621. doi: 10.1124/dmd.109.027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Winnike JH, Li Z, Wright FA, Macdonald JM, O'Connell TM, Watkins PB. Use of pharmaco-metabonomics for early prediction of acetaminophen-induced hepatotoxicity in humans. Clin Pharmacol Ther. 2010;88:45–51. doi: 10.1038/clpt.2009.240. [DOI] [PubMed] [Google Scholar]
- 130.Fukuhara K, Ohno A, Ando Y, Yamoto T, Okuda H. A 1H NMR-based metabolomics approach for mechanistic insight into acetaminophen-induced hepatotoxicity. Drug Metab Pharmacokinet. 2011;26:399–406. doi: 10.2133/dmpk.dmpk-11-rg-005. [DOI] [PubMed] [Google Scholar]
- 131.Kumar BS, Chung BC, Kwon OS, Jung BH. Discovery of common urinary biomarkers for hepatotoxicity induced by carbon tetrachloride, acetaminophen and methotrexate by mass spectrometry-based metabolomics. J Appl Toxicol. 2012;32:505–520. doi: 10.1002/jat.1746. [DOI] [PubMed] [Google Scholar]
- 132.Prot JM, Bunescu A, Elena-Herrmann B, Aninat C, Snouber LC, Griscom L, et al. Predictive toxicology using systemic biology and liver microfluidic "on chip" approaches: application to acetaminophen injury. Toxicol Appl Pharmacol. 2012;259:270–280. doi: 10.1016/j.taap.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 133.Shintu L, Baudoin R, Navratil V, Prot JM, Pontoizeau C, Defernez M, et al. Metabolomics-on-a-chip and predictive systems toxicology in microfluidic bioartificial organs. Anal Chem. 2012;84:1840–1848. doi: 10.1021/ac2011075. [DOI] [PubMed] [Google Scholar]
- 134.Liao Y, Peng SQ, Yan XZ, Zhang LS. Metabonomics profile of urine from rats administrated with different treatment period of isoniazid. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2007;29:730–737. [PubMed] [Google Scholar]
- 135.Li F, Lu J, Cheng J, Wang L, Matsubara T, Csanaky IL, et al. Human PXR modulates hepatotoxicity associated with rifampicin and isoniazid co-therapy. Nat Med. 2013 doi: 10.1038/nm.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang X, Shao L, Gong Y, Mao Y, Liu C, Qu H, et al. A metabonomic characterization of CCl4-induced acute liver failure using partial least square regression based on the GC/MS metabolic profiles of plasma in mice. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;870:178–185. doi: 10.1016/j.jchromb.2008.05.049. [DOI] [PubMed] [Google Scholar]
- 137.Yang L, Xiong A, He Y, Wang Z, Wang C, Li W, et al. Bile acids metabonomic study on the CCl4- and alpha-naphthylisothiocyanate-induced animal models: quantitative analysis of 22 bile acids by ultraperformance liquid chromatography-mass spectrometry. Chem Res Toxicol. 2008;21:2280–2288. doi: 10.1021/tx800225q. [DOI] [PubMed] [Google Scholar]
- 138.Lin Y, Si D, Zhang Z, Liu C. An integrated metabonomic method for profiling of metabolic changes in carbon tetrachloride induced rat urine. Toxicology. 2009;256:191–200. doi: 10.1016/j.tox.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 139.Ding L, Hao F, Shi Z, Wang Y, Zhang H, Tang H, et al. Systems biological responses to chronic perfluorododecanoic acid exposure by integrated metabonomic and transcriptomic studies. J Proteome Res. 2009;8:2882–2891. doi: 10.1021/pr9000256. [DOI] [PubMed] [Google Scholar]
- 140.Lee MS, Jung BH, Chung BC, Cho SH, Kim KY, Kwon OS, et al. Metabolomics study with gas chromatography-mass spectrometry for predicting valproic acid-induced hepatotoxicity and discovery of novel biomarkers in rat urine. Int J Toxicol. 2009;28:392–404. doi: 10.1177/1091581809340329. [DOI] [PubMed] [Google Scholar]
- 141.Liu Y, Huang R, Liu L, Peng J, Xiao B, Yang J, et al. Metabonomics study of urine from Sprague-Dawley rats exposed to Huang-yao-zi using (1)H NMR spectroscopy. J Pharm Biomed Anal. 2010;52:136–141. doi: 10.1016/j.jpba.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 142.Sun C, Teng Y, Li G, Yoshioka S, Yokota J, Miyamura M, et al. Metabonomics study of the protective effects of Lonicera japonica extract on acute liver injury in dimethylnitrosamine treated rats. J Pharm Biomed Anal. 2010;53:98–102. doi: 10.1016/j.jpba.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 143.Lu C, Wang Y, Sheng Z, Liu G, Fu Z, Zhao J, et al. NMR-based metabonomic analysis of the hepatotoxicity induced by combined exposure to PCBs and TCDD in rats. Toxicol Appl Pharmacol. 2010;248:178–184. doi: 10.1016/j.taap.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 144.Shi X, Wahlang B, Wei X, Yin X, Falkner KC, Prough RA, et al. Metabolomic analysis of the effects of polychlorinated biphenyls in nonalcoholic fatty liver disease. J Proteome Res. 2012;11:3805–3815. doi: 10.1021/pr300297z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shima N, Miyawaki I, Bando K, Horie H, Zaitsu K, Katagi M, et al. Influences of methamphetamine-induced acute intoxication on urinary and plasma metabolic profiles in the rat. Toxicology. 2011;287:29–37. doi: 10.1016/j.tox.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 146.Lu X, Zhao Q, Tian Y, Xiao S, Jin T, Fan X. A metabonomic characterization of (+)- usnic acid-induced liver injury by gas chromatography-mass spectrometry-based metabolic profiling of the plasma and liver in rat. Int J Toxicol. 2011;30:478–491. doi: 10.1177/1091581811414436. [DOI] [PubMed] [Google Scholar]
- 147.Parman T, Bunin DI, Ng HH, McDunn JE, Wulff JE, Wang A, et al. Toxicogenomics and metabolomics of pentamethylchromanol (PMCol)-induced hepatotoxicity. Toxicol Sci. 2011;124:487–501. doi: 10.1093/toxsci/kfr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McBurney RN, Hines WM, Von Tungeln LS, Schnackenberg LK, Beger RD, Moland CL, et al. The liver toxicity biomarker study: phase I design and preliminary results. Toxicol Pathol. 2009;37:52–64. doi: 10.1177/0192623308329287. [DOI] [PubMed] [Google Scholar]
- 149.Watkins PB. Biomarkers for the diagnosis and management of drug-induced liver injury. Semin Liver Dis. 2009;29:393–399. doi: 10.1055/s-0029-1240008. [DOI] [PubMed] [Google Scholar]
- 150.Beger RD, Sun J, Schnackenberg LK. Metabolomics approaches for discovering biomarkers of drug-induced hepatotoxicity and nephrotoxicity. Toxicol Appl Pharmacol. 2010;243:154–166. doi: 10.1016/j.taap.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 151.O'Connell TM, Watkins PB. The application of metabonomics to predict drug-induced liver injury. Clin Pharmacol Ther. 2010;88:394–399. doi: 10.1038/clpt.2010.151. [DOI] [PubMed] [Google Scholar]
- 152.Ellinger-Ziegelbauer H, Adler M, Amberg A, Brandenburg A, Callanan JJ, Connor S, et al. The enhanced value of combining conventional and "omics" analyses in early assessment of drug-induced hepatobiliary injury. Toxicol Appl Pharmacol. 2011;252:97–111. doi: 10.1016/j.taap.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 153.Gonzalez E, van Liempd S, Conde-Vancells J, Gutierrez-de Juan V, Perez-Cormenzana M, Mayo R, et al. Serum UPLC-MS/MS metabolic profiling in an experimental model for acute-liver injury reveals potential biomarkers for hepatotoxicity. Metabolomics. 2012;8:997–1011. doi: 10.1007/s11306-011-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]