Abstract
Broadly neutralizing monoclonal antibodies (bnMAbs) that target the high-mannose patch centered around the glycan at position 332 on HIV Env are promising vaccine leads and therapeutic candidates as they effectively protect against mucosal SHIV challenge and strongly suppress SHIV viraemia in established infection in macaque models. However, these antibodies demonstrate varying degrees of dependency on the N332 glycan site and the origins of their neutralization breadth are not always obvious. By measuring neutralization on an extended range of glycan site viral variants, we found that some bnMAbs can utilize alternate N-linked glycans in the absence of the N332 glycan site and therefore neutralize a substantial number of viruses lacking the site. Furthermore, many of the antibodies can neutralize viruses in which the N332 glycan site is shifted to the 334 position. Finally, we found that a combination of three antibody families that target the high-mannose patch can lead to 99% neutralization coverage of a large panel of viruses containing the N332/334 glycan site and up to 66% coverage for viruses that lack the N332/334 glycan site. The results indicate that a diverse response against the high-mannose patch may provide near equivalent coverage as a combination of bnMAbs targeting multiple epitopes. Additionally, the ability of some bnMAbs to utilize other N-linked glycan sites can help counter neutralization escape mediated by shifting of glycosylation sites. Overall, this work highlights the importance of promiscuous glycan binding properties in bnMAbs to the high-mannose patch for optimal anti-viral activity either in protective or therapeutic modalities.
Introduction
Broadly neutralizing HIV antibodies provide important leads for vaccine design and may be valuable in therapy (1–6). They define sites that are both conserved and accessible to antibodies and their characteristics, such as germline gene usage and degree of somatic hypermutation, can help inform the choice of immunogens and immunization strategies most likely to induce broadly neutralizing antibodies though vaccination. The value of such antibodies to the HIV vaccine design and therapy efforts has greatly increased with the isolation, in the last few years, of many potent, broadly neutralizing human monoclonal antibodies (bnMAbs) from infected individuals (7–14). The most potent neutralizers of this new generation of bnMAbs appear to be a group that targets and penetrates the glycan shield of HIV Env to recognize both glycans and protein surface of the V3 and V4 regions underneath (8, 15–17). A number of the glycans involved are of the high-mannose variety and form a patch (“the high-mannose patch”), (18–21) centered on a glycan at Asn 332 (N332), although some complex glycans appear to be interspersed at the edges of this patch. The bnMAbs are often described as “N332-dependent”. The prototype antibodies in this class, each isolated from individual donors, are PGT121, PGT128 and PGT135, but many somatic variants have also been generated (8, 22). An earlier isolated bnMAb, 2G12, is often included with this set of antibodies since its binding is N332-dependent but it is less potent and broad in neutralization, recognizes glycans solely, and has a unique domain-exchanged structure (23–26).
The outstanding in vitro neutralization potency of at least one of the bnMAbs targeting the high-mannose patch, PGT121, has been shown to translate into in vivo efficacy (27). Passively administered PGT121 protects against high-dose vaginal SHIV challenge in macaques at relatively low serum antibody titers (6, 27). In addition, the antibody given alone strongly suppresses SHIV replication in chronically infected macaques (5). Similarly, a close variant of PGT121, 10-1074, has also been shown to suppress viral load in combination with other bnMAbs in humanized mouse models (4) and macaques (6).
The bnMAbs targeting the high-mannose patch have other features that make them particularly interesting from a vaccine design standpoint. Of note, these antibodies appear to arise with a relatively high frequency in comparison to other broadly neutralizing specificities in infected individuals and at a relatively early stage (28, 29). The latter point is emphasized by the development of a broadly neutralizing N332-dependent serum antibody response in a SHIV-infected macaque in only about 9 months (30). The early development of Abs targeting the high-mannose patch is also consistent with the somewhat lower level of somatic hypermutation typically found among bnMAbs of this specificity as compared to, for example, VRC01-like antibodies that target the CD4 binding site (14). One factor favoring the relatively high frequency of broadly neutralizing responses against the high-mannose patch may be the relatively unrestricted antibody gene requirements for recognition of this region. Thus, whereas the CD4bs-directed antibodies of the VRC01 family all use a single VH germline gene, VH1-2, the antibodies that target the high-mannose patch derive from several VHDHJH and VLJL recombinations and pairings (8, 15–17, 24, 31, 32). The usage of different antibody germline genes by the N332-dependent MAbs likely reflects the different modes of binding of these antibodies as determined structurally and hence different points of contact with gp120 (15–17).
Interestingly, a recent longitudinal study in HIV-infected individuals found that the N-glycan site at position 332 is somewhat under-represented in founder subtype C viruses (45/68, 66%) relative to unmatched chronic viruses (52/62, 84%) (33). The study also monitored two HIV-infected individuals for whom a N-glycan site was identified at position 334 early in infection that shifted to 332 immediately before the emergence of neutralization breadth, and then shifted back to the position at 334 to permit neutralization escape. The study has vaccine implications in that the proportion of isolates whose transmission could be blocked by antibodies targeting the high-mannose patch may be lower than previously thought. Another recent study showed that a bnMAb recognizing the N332 glycan region (PGT121) gains some of its breadth by a promiscuous ability to recognize both high-mannose and complex-type carbohydrates (17, 22). Here we extend the concept of promiscuity and show that, depending on isolate context, a single MAb to the high-mannose patch can neutralize viruses using differing proximal glycan sites to differing degrees. Our results demonstrate a notable complexity in how HIV glycan-dependent antibodies recognize their epitopes and provide important considerations for their use in therapy and in the design of immunogens to elicit this class of antibodies that are distinct from approaches for the elicitation of bnMAbs targeting other sites like the CD4bs.
Results
bnMAbs to the high-mannose patch can neutralize many viruses in which the N332 glycan site is shifted to the 334 position
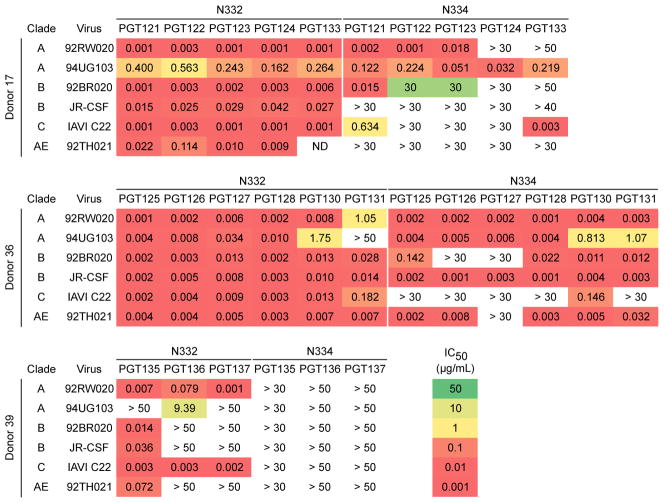
Moore et al. recently identified two HIV-infected donors in which a shift of the N332 glycan site to the 334 position resulted in escape from serum neutralizing antibodies with N332-dependent specificity (33). We sought to determine the effects of such a shift on a panel of bnMAbs similarly targeting the high-mannose patch by using a modified 6-virus indicator panel (34, 35). Notably, one of the viruses in the panel, 92TH021, naturally harbors a N-glycan site at position N334. We tested neutralization of three families of N332-dependent bnMAbs isolated from three elite neutralizers in the IAVI Protocol G cohort; PGT121 and variants PGT122, PGT123, PGT124 and PGT133 (from donor 17); PGT128 and variants PGT125, PGT126, PGT127, PGT130 and PGT131 (from donor 36); PGT135 and variants PGT136 and PGT137 (from donor 39) (8, 34–36). A summary of the donors and a list of the somatic variants isolated from each donor are tabulated in table S1.
As shown in Fig. 1, a sizable fraction of the bnMAbs were capable of neutralizing this indicator panel of viruses when the N332 glycan site was shifted to the 334 position (34, 35). These results suggest that for many antibodies targeting the high-mannose patch, escape does not necessarily follow a shift in the glycan site from N332 to N334. There were, however, notable differences in the ability of bnMAbs to recognize the N334 glycan site. First, there were overall differences between antibody families. The PGT128 family of antibodies was the most effective at neutralizing viruses with the N334 glycan site, while the PGT135 family was completely unable to neutralize these variants. The PGT121 family of antibodies was able to neutralize the N334 viruses to an intermediate degree. Second, there were more subtle differences among the somatic variants of an antibody family. For the PGT121 family of antibodies, for example, PGT121 was able to neutralize all the N334 glycan site variants in the panel except for the isolates JR-CSF and 92TH021. Conversely, the PGT124 somatic variant of PGT121 was unable to neutralize any of the viruses in the panel except for the isolate 94UG103. Differences between somatic variants were also found among the PGT128 family of antibodies. Within this family, PGT130 was capable of neutralizing all of the isolates in the panel, while PGT127 was only capable of neutralizing half of the isolates. Given the notable differences between bnMAbs in recognizing N334 glycan site variants, we were next interested in evaluating the significance of these differences on a larger virus panel.
Figure 1. Neutralization of viruses by bnMAbs to the high-mannose patch with the N332 glycan site shifted to the 334 position.
Neutralization potency was measured on a 6-virus indicator panel for which the glycan site at N332 was shifted to the 334 position. Presented values are neutralization IC50 in μg/ml and colored according to the listed scale. Values listed as “ND” were not determined.
The bnMAbs display differences in the ability to neutralize viruses naturally expressing the N332 or N334 glycan site
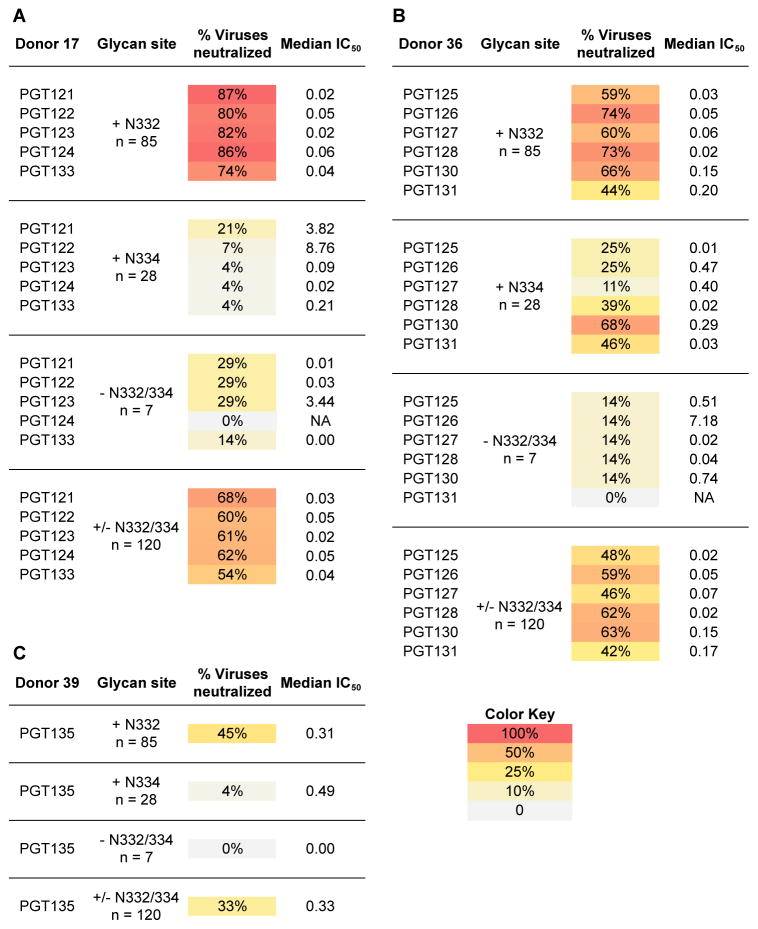
We wanted to compare differences in neutralization breadth and potency of bnMAb somatic variants on viruses naturally expressing the N332 glycan site, the N334 site or neither. To this end, we tested neutralization activity against a cross-clade 120-virus panel, which contained 85 viruses with a N332 glycan site, 28 viruses with a N334 glycan site, and 7 viruses without either N332 or N334 glycan sites (Fig. 2A–C and table S2). To evaluate neutralization profiles, we measured neutralization breadth and potency (IC50 cutoffs of 50 μg/mL) for each antibody (Fig. 2A–C). Given the poor activity of the somatic variants from the PGT135 family, we only tested bnMAb PGT135 and not PGT136 or PGT137 on this large panel.
Figure 2. BnMAbs to the high-mannose patch neutralize many isolates with the glycan site at the 332 position but also some bnMAbs can neutralize isolates with the site at the 334 position and some with no glycan site at either position.
Percent neutralization breadth and median IC50 were determined at an IC50 cutoff of 50 μg/mL for the (A) PGT121 antibody family (B) PGT128 antibody family and (C) PGT135. Viruses were separated into those naturally containing a glycan site at N332 or N334 or without a glycan site at either position.
These results revealed differences between antibody families in the neutralization of viruses with a glycan site naturally present at the N332 position (Fig. 2A–C). The PGT121 family of antibodies neutralized N332 glycan site viruses more effectively overall than the PGT128 family or PGT135 (Fig. 2A–C). Nevertheless, no somatic variants from either of these antibody families were able to neutralize all 85 viruses, indicating that the glycan at the N332 site is often necessary, but not sufficient for neutralization by this class of antibodies.
Interestingly, there were marked differences in breadth and potency among somatic variants within a clonal lineage in the ability to neutralize viruses with a glycan at the N334 site (fig. S1). For the PGT121 antibody family, for example, mostly PGT121 showed (although limited) activity against the N334 glycan site virus panel (Fig. 2A and fig. S1). On the other hand, PGT124, which is homologous to the recently described variant 10-1074, containing only one residue difference in CDRH3, appeared to be especially ineffective against viruses naturally containing a N334 glycan site, as similarly noted for 10–1074 (22). Similarly, for the PGT128 antibody family, PGT130 was able to neutralize 68% of the N334 glycan site viruses compared to 11% for PGT127 (Fig. 2B). Indeed, of all the bnMAbs tested, PGT130 was the most effective at neutralizing viruses with a glycan site at N334, suggesting that immunogens designed to accommodate a PGT130 mode of Env recognition could provide additional neutralization coverage against N334 glycan site viruses (Fig. 2B). PGT135, meanwhile, was only able to neutralize a single virus in this N334 panel (Fig. 2C), which is consistent with the results from the indicator panel.
Having demonstrated some degree of flexibility in recognizing isolates with a glycan at either N332 or N334 sites, we surprisingly found somatic variants from both PGT121 and PGT128 antibody families that were able to neutralize viruses naturally lacking a glycan at either the 332 or 334 positions. To explore the degrees of dependency of bnMAbs on the N332/334 glycan sites, we created a panel of virus mutants entirely lacking these sites and tested neutralization by the PGT121 and PGT128 family of antibodies.
Some of the bnMAbs to the high-mannose patch neutralize viruses despite removal of N332 and N334 glycan sites by alanine mutagenesis
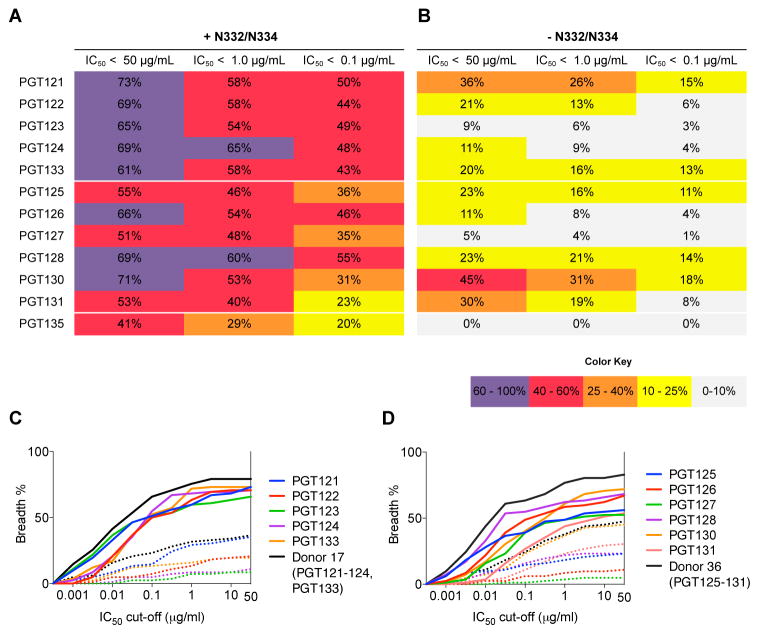
We took a panel of 80 viruses consisting of 62 N332 and 18 N334 glycan site viruses, eliminated the glycan sites in each case by alanine substitution (table S3 and S4), measured neutralization breadth and potency of the PGT121 and PGT128 antibody families and then tabulated percent viruses neutralized at different IC50 cutoffs (Fig. 3 and fig. S2).
Figure 3. A substantial fraction of isolates in a large panel are neutralized by high-mannose patch bnMAbs when the N332 or N334 glycan sites are eliminated by alanine mutagenesis.
Antibody families were tested on an 80-virus panel, whose members naturally include a glycan site at N332/334 (A) and on the same panel with the glycan site at N332/334 removed by alanine mutagenesis (B). Listed are percent viruses neutralized at the indicated neutralization IC50 cutoffs. Cumulative distribution frequency (CFD) of percent viruses neutralized in the 80-virus panel with and without the N332/334 glycan site present at different IC50 cutoffs for the (C) PGT121 antibody family and the (D) PGT128 antibody family.
In contrast to previous interpretations (8, 17, 22), somatic variants from both PGT121 and PGT128 antibody families were capable of neutralizing a number of viruses lacking both N332 and N334 glycan sites. PGT121 and PGT130 were the most effective antibodies against those viruses and maintained a remarkable neutralization breadth of 36% and 45%, respectively (Fig. 3 and fig. S2 and table S5–6). The potency of PGT130 against the N332A and N334A variants was almost identical to the wild-type viruses while the potency of PGT121 was about 10 fold lower (fig. S2). The ability to recognize Env despite lacking a key portion of the epitope as suggested from structural studies, (contact to the N332 glycan comprises 449 Å2 out of 1081 Å2 of the epitope in the case of PGT128 bound to an outer domain construct (15)), is remarkable and may be due to the ability to utilize other glycan sites in place of the N332 site.
Alternate glycan sites can be utilized by some bnMAbs targeting the high-mannose patch
Given that antibodies targeting the high-mannose patch can neutralize some virus isolates in the absence of N-glycan sites at either position 332 or 334, we investigated whether alternate glycan sites may be used in place of N332/334 for neutralization by the antibodies. We thus chose isolates for which loss of the N332 or N334 glycan site did not have a demonstrable impact on neutralization and then removed putative alternate glycan sites, either singly or in conjunction with N332 or N334 glycan sites, and subsequently tested neutralization. Figure S3 summarizes the sensitivities of various mAbs targeting the high-mannose patch to the presence of a number of key glycan sites identified previously (8, 15–17, 24) and below.
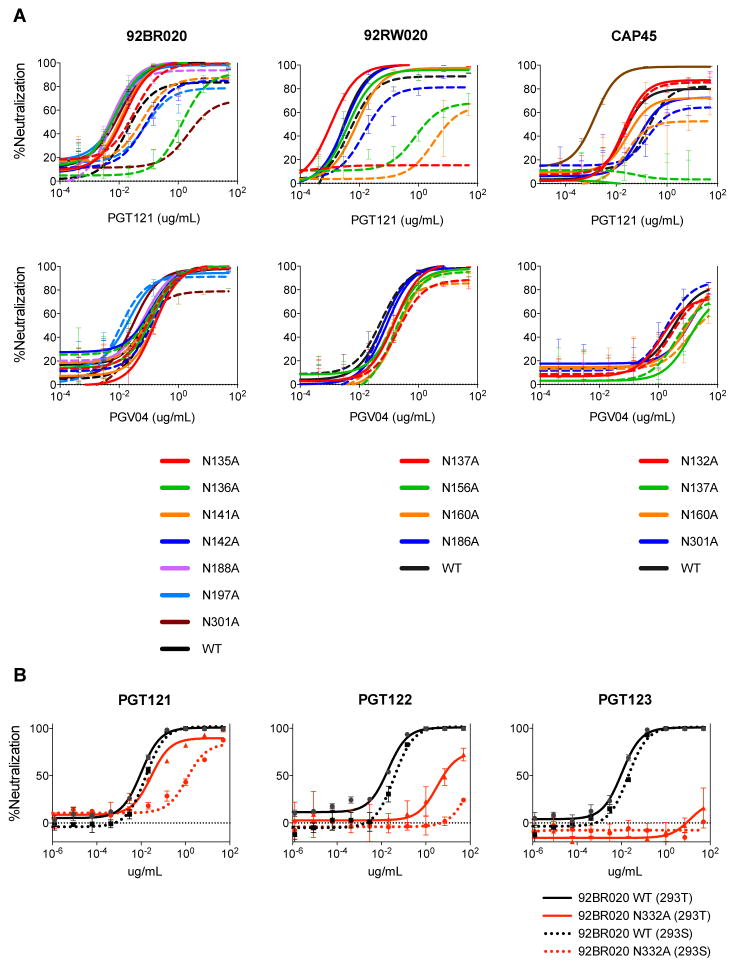
We first studied the PGT121 family of antibodies and focused on isolates 92BR020 and 92RW020, which naturally have a glycan site at N332, isolate CAP45, which naturally has a glycan site at N334, and isolate ZM53, which does not have a glycan at either N332 or N334 sites (Figure 4A). Based on recent crystal and EM structures of recombinant gp140 trimer suggesting PGT121/PGT122-glycan contacts with V1/V2 (17) and on reduced PGT121 binding to JR-CSF ΔV1/V2 Env by flow cytometry (fig. S4A), we introduced alanine substitutions at glycan sequons in the V1/V2 loops in combination with the N332A substitution. PGT121 neutralization was greatly reduced for isolate 92BR020 by removal of the N136 and N332 glycan site combination as well as by the N301 and N332 combination, whereas no large effects were observed for single substitution variants. To demonstrate that these substitutions do not result in global neutralization sensitivity, we also tested the CD4bs antibody PGV04 and demonstrated that loss of these glycan sites did not have an effect on overall antigenicity (Fig. 4A and fig. S5). Similarly, for isolate 92RW020, removal of glycan sites at both N137 and N332 resulted in complete loss of PGT121 neutralization, with large reductions in neutralization titers following removal of the N156 and N332 as well as the N160 and N332 glycan site combinations. For isolate CAP45, for which we eliminated the N334 site, we also noted a co-dependency on the N137 glycan site for neutralization by PGT121. The results show that, for the isolates described, PGT121 can tolerate the loss of the N332 glycan site provided the glycan sites in the V1/V2 loops are present. For isolate ZM53, we found that removal of the glycan site at position N136 reduces PGT121-123 neutralization but interestingly has the opposite effect for PGT124 and PGT133 (Fig. 6A–C and fig. S6). These results indicate a different mode of binding within the somatic variants of this donor and underline the divergent evolution between antibodies of the PGT121 family (35).
Figure 4. PGT121 can utilize glycans in the V1/V2 loops to neutralize isolates when the glycan site at N332 or N334 is removed by alanine mutagenesis.
(A) Neutralization potency of PGT121 was measured on 92BR020, 92RW020 and CAP45 glycan mutant viruses for which N-linked glycosylation sites have been removed singly (solid line) or in combination with N332 (dashed line) by alanine mutagenesis. PGV04 was included as a control. (B) Neutralization potency of PGT121-123 was measured on virus isolate 92BR020 N332A mutant virus (red) made in HEK 293T cells (solid line) and a GnT1−/− deficient HEK 293S cell line (dotted line) and the potency compared to WT virus (black).
Figure 6. bnMAbs to the high-mannose patch can utilize alternate glycan sites to neutralize viruses that naturally do not have a glycan site present at the N332/334 position.
(A) PGT121, (B) PGT123, and (C) PGT124 were tested for neutralization of ZM53 glycan site mutants. (D) PGT125, (E) PGT127, and (F) PGT128 were tested for neutralization of BG505 glycan site mutants. Mutants were generated by site-directed mutagenesis.
Considering that glycans in the V1/V2 loops are often complex-type and that PGT121-123 are capable of interacting with complex glycans (17, 22), we wanted to determine the glycoform of the glycan sites that PGT121 utilizes in V1/V2. To this effect, we produced viruses in the GnT1−/− deficient 293S cell line (37, 38), which results in Env bearing only Man5–9GlcNAc2 glycoforms. In the presence of a glycan at the N332 site, the 293S-grown viruses were sensitive to PGT121 neutralization indicating no or little dependence on complex-type glycans for neutralization and indicating that the critical glycan site(s) in the V1/V2 loops are likely complex-type (Fig. 4B). Further, the results on PGT121 neutralization taken as a whole imply that the antibody makes critical use of the N332 glycan site when it is present, but if it is absent, then greater use can be made of the V1/V2 complex glycan sites to enhance the neutralization breadth of the antibody. This idea is further corroborated by removal of the glycan site combinations N136 + N156 and N136 + N301 (fig. S7), which show little effect on neutralization potency, suggesting that while the antibody may be making contacts to these alternate glycans, reliance on these glycans only becomes important in the absence of the N332 glycan.
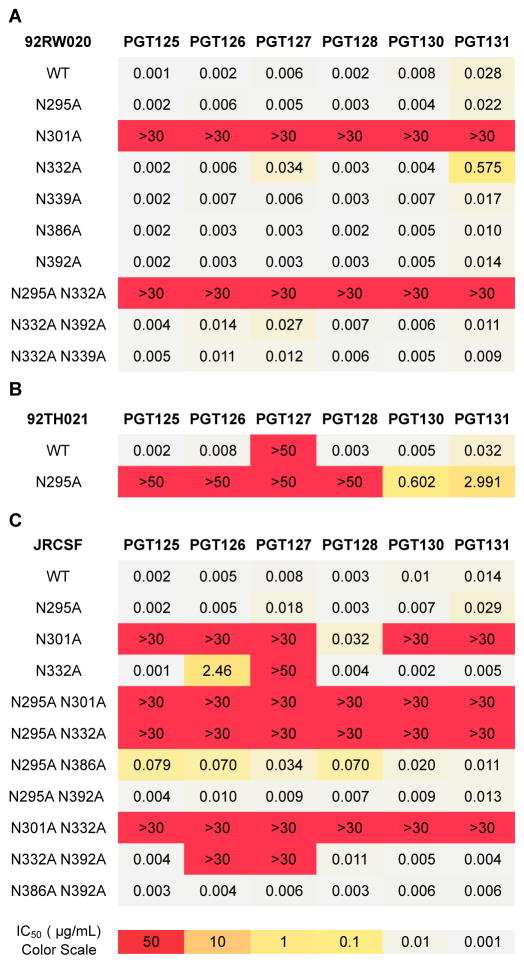
We next performed a similar analysis for the PGT128 family of antibodies and focused on isolates 92RW020 and JR-CSF, which naturally have a glycan site at N332, on 92TH021, which has a glycan site at N334, and on BG505, which does not have a glycan at either N332 or N334 sites. To better determine which alternate glycan sites may be involved in the PGT128 family epitope, we first tested binding to cell surface JR-CSF ΔV1/V2 and ΔV3 Env constructs and saw enhanced binding of ΔV1/V2 relative to wild-type and a large decrease in binding to the ΔV3 Env construct (fig. S4B). Accordingly, we decided to focus our mutagenesis on the glycan sites that comprise the canonical high-mannose glycan patch: N295, N332, N339, N386, and N392. We also included the N301 glycan, located adjacent to the high-mannose patch. Looking first at the isolate 92RW020, we only observed loss of neutralization with the N301A substitution alone as well as for the N295A + N332A combination (Fig. 5A). These results suggest that there is little or no flexibility in dependency on the N301 glycan site, but that the glycans at the N295 and N332 sites are close enough in proximity to substitute for each other when one of the two sites is missing (15). For 92TH021, which naturally contains a glycan at the N334 site, loss of the N295 glycan site alone was able to abrogate neutralization or reduce potency for the PGT128 antibody family. This result suggests that, in some cases, the PGT128 family antibodies depend more on the N295 glycan site for neutralization (Fig. 5B). For the isolate JR-CSF, we see a similar result as for 92RW020. However, we do see some effects following removal of the N295 and N386 glycan sites in combination as well as for the N332 and N392 glycan site combination (Fig. 5C) in neutralization of JR-CSF by some PGT128 family antibodies. This result suggests that the N295/N386 glycan sites may also be utilized by PGT128 family antibodies to some degree and further highlights the complexity of glycan site promiscuity of some bnMAbs. For BG505, removal of N295A and N301A glycan sites generally abrogated neutralization for the PGT128 family, but interestingly, there was variability in the sensitivity of different antibody variants to the removal of other glycan sites. Neutralization by PGT125 (Fig. 6D–F), for example, was not affected by any glycan site mutations other than N295A and N301A, while the remaining antibodies within the clonal lineage demonstrated lower neutralization plateaus for other glycan mutants within the high-mannose patch (fig. S8). Further, similar to observations for PGT124 and PGT133 on isolate ZM53, removal of the V1 glycan site at position N137 resulted in enhanced neutralization IC50 and/or neutralization plateaus for all somatic variants except PGT125, perhaps by increasing accessibility of the N295 and N301 glycans or protein contacts (fig. S8).
Figure 5. PGT128 family antibodies can utilize alternate glycan sites in the high-mannose patch to neutralize isolates when the N332 or N334 glycan sites are removed by alanine mutagenesis.
Neutralization potency of the PGT128 antibody family was measured on a panel of (A) 92RW020, (B) 92TH021 and (C) JR-CSF glycan mutant viruses for which N-linked glycan sites in the V3 region were removed alone or in combination with other N-linked glycan sites.
Glycan modeling demonstrates close proximity of terminal mannose residues of glycans at N295, N332 and N334
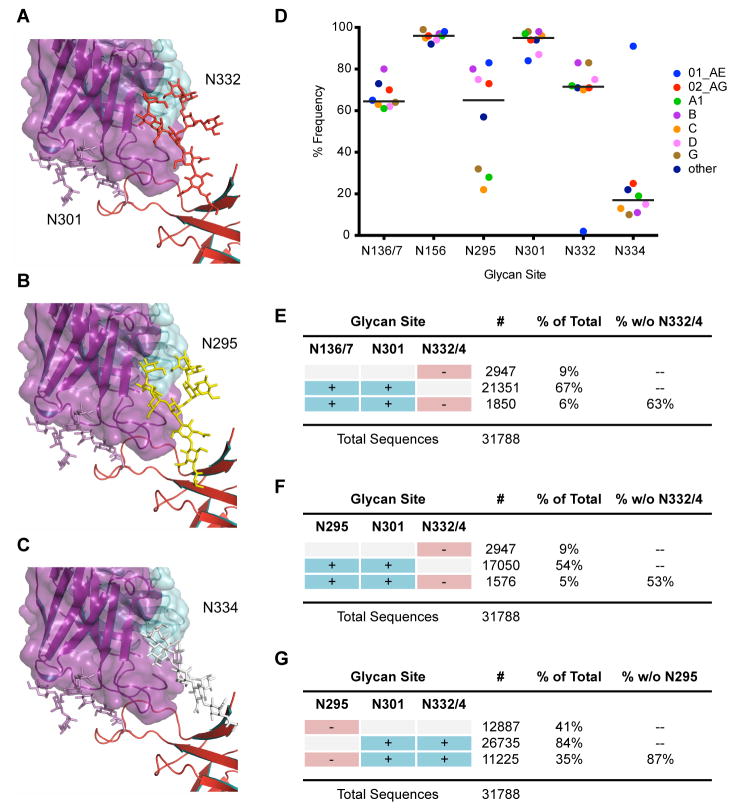
To further understand how the N-glycan sites at positions 332, 334, and 295 appear to have some degree of interchangeability for binding by PGT128-like antibodies, we performed ensemble modeling of glycan conformations at the N295 and N334 sites (Fig. 7A–C). We used two computational methods to sample glycan conformations at N295 and N334 glycan sites and evaluated root mean square deviation (RMSD) between the crystallized N332 glycan and the computationally generated N295 and N334 glycans.
Figure 7. Structural modeling of high-mannose patch glycan sites and calculations of glycan site frequencies.
Glycan models to rationalize PGT128 promiscuity. (A) Close-up view of PGT128 binding to eODmV3 including the glycans at position N301 and N332, as defined by the crystal structure of eODmV3+PGT128. (B) Model for an N-linked Man8GlcNAc2 glycan at position N295 on eODmV3. (C) Model for an N-linked Man8GlcNAc2 glycan at position N334 on eODmV3. Coloring is as follows: PGT128, magenta; eODmV3, red; N301 glycan, magenta; N332 glycan, red; N295 glycan, yellow; N334 glycan, white. Cross-clade frequency of glycans at different positions on Env. (D) Frequency of critical N-linked glycan sites in 31,788 viruses in the Los Alamos database. A glycan site was determined as being present accordingly to the Nx(T or S) motif, where x is not a proline. Only clades for which > 100 sequences were available are included in the graph. The line represents the mean of the frequency for each glycan site. (E–F) Frequency of combinations of the N-linked glycan sites shown to be important for bnMAb neutralization in the same set of 31,788 virus sequences.
From the GlycanRelax ensembles, the lowest RMSDs were 4.9 Å at N295 and 4.2 Å at N334, while from the GlycanTransfer simulations, the lowest RMSDs were 1.8 Å at 295 and 3.1 Å at 334. The lowest RMSD structures from GlycanTransfer are shown in Figure 7B and 7C. These results support the structural plausibility of glycans at position N295 or N334 making productive contacts to PGT128, and suggest that, compared to the glycan at position N334, the glycan at position N295 might more readily mimic the N332 glycan. Further, the results also suggest that in order for glycans at N295 or N334 sites to fully recapitulate the interactions made by the N332 glycan, small protein backbone conformational changes may be required in gp120, possibly at the base of the V3 loop.
These analyses offer a potential explanation for the reduced potency of bnMAbs against viruses with a N334 glycan site compared to viruses naturally harboring the N332 glycan site (Fig. 2A–C). Overall, these analyses support the notion that while moving the glycan sites from N332 to N334 negatively impacts the binding of antibodies that form critical contacts with the base of the glycan, antibodies that form contacts to the terminal carbohydrates of the glycan remain capable of neutralization due to the inherent flexibility of the glycan arms.
Glycan sites that are important for neutralization are highly conserved among isolates
We were interested in the frequency of the critical glycan sites in global isolates. Accordingly, we analyzed over 30,000 sequences in the Los Alamos database for the frequency of not only single glycan sites, but also the frequency of glycan site pairs among cross-clade Env isolates.
First, given that previous work demonstrated a higher frequency of the N334 glycan site among clade C founder viruses, which might confer resistance against PGT128-like antibodies, (33), we wanted to determine the frequency of the N334 glycan site among cross-clade isolates. The results revealed that a glycan site is present at the N332 position far more often than at the N334 position, with 73% and 17% frequency among Env isolates, respectively (Fig. 7D and table S7). Strikingly, only approximately 10% of Env sequences contain no glycan site at either position N332 or N334, highlighting the high conservation of the N332/334 glycan site and strongly suggesting a role for the presence of a glycan at this site for replication fitness.
We were next interested in determining the frequency of glycan sites that are important in bnMAb neutralization targeting the high-mannose patch. Strikingly, the N301 glycan site is conserved in 96% of isolates, while the N136/7 glycan site is as highly conserved as the N332 glycan site, with 72% frequency among isolates (Fig. 7D and table S7). The N295 glycan site is also highly represented, although less than the other glycan sites, with 58% frequency among global isolates.
Considering that two or more glycan sites are important for forming most bnMAb epitopes (15, 16), we next analyzed the frequency of combinations of glycan sites (Fig. 7E–7G). The combination of N332/334 and N301 sites was present in 84% of isolates in the 31,788-virus dataset (Fig. 7E). Of note, for isolates lacking N332/334, the N136/137 and N301 glycan site pair is present at a frequency of 63% and the N295 and N301 glycan site pair at 53% (Fig. 7F and 7G). Therefore, for over half of those sequences that do not have N332 or N334 glycan sites, bnMAbs that also recognize the N136/137 or N295 glycans may remain capable of recognizing Env. The high frequency of these alternative glycan sites, individually and in pairs, suggests that the elicitation or delivery of antibodies with promiscuous glycan site recognition will greatly improve protection coverage and may limit virus escape during antibody therapy.
Discussion
The study of a subset of broadly neutralizing HIV antibodies that recognize the high-mannose patch of gp120 has provided new insights into antibody recognition of a glycoprotein. These insights include: 1) the ability of antibodies to penetrate between relatively tightly packed glycans and access the protein surface underneath as described only recently (15, 16, 39), 2) the potential of antibodies to form extended contacts over many glycan residues (15, 16, 39), and 3) the very high affinities of the antibodies involved despite predominant interaction with glycans (24, 40). These antibodies also have very high potencies of neutralization in vitro and an antiviral activity in vivo that is exceptional (5, 6, 8, 27). Initially, a set of these antibodies were grouped together and were assumed to recognize a very similar epitope, as they often lost neutralization activity against isolates that do not contain a glycan at the N332 site (8, 28, 36). Here, we describe a new level of complexity in antibody recognition of the mixed glycan-protein epitopes of the N332 region of HIV gp120. We show that the same antibody can target differently positioned clusters of neighboring glycan sites on different HIV isolates and that closely related antibody variants, originating from the same HIV-infected individual, can bind to differing glycan arrangements. It is instructive to consider the quantitative conclusions that can be drawn from the results presented here that may influence vaccine design and antibody therapy strategies.
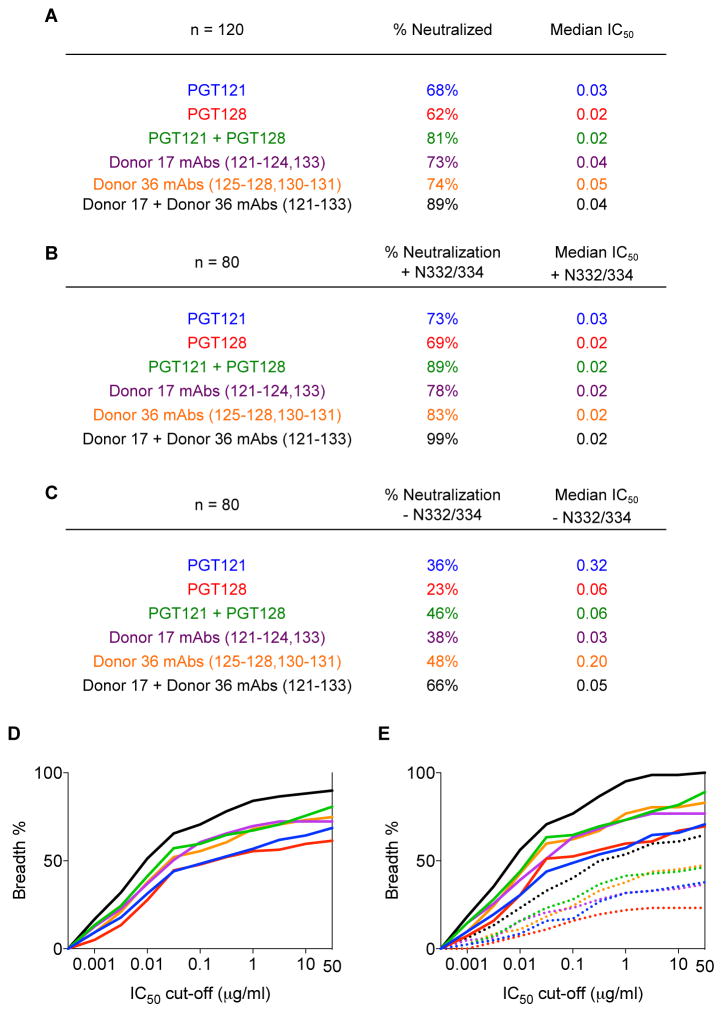
We consider first the implications for vaccine efficacy. The data presented highlights differing modes of N332-epitope recognition by bnAbs from both within and between donors. One effect of this complexity and diversity is that polyclonal antibody responses with promiscuous glycan binding behavior will be notably more effective at neutralizing a diversity of HIV isolates than single bnMAbs lacking promiscuous behavior. To evaluate quantitatively, we plotted a cumulative frequency distribution of percent viruses neutralized at different IC50 cutoffs for individual bnMAbs of the PGT121 and PGT128 antibody families on the cross-clade 120-virus panel (Fig. 8A–E). The elicitation of single PGT121 or PGT128 specificities with no promiscuous capacity (i.e. N332 recognition only) would lead to up to 62% and 52% neutralization coverage, respectively (table S8). A theoretical combination of PGT121 and PGT128, however, would enhance overall neutralization coverage to 81% (Fig. 8A and table S8). Strikingly, the elicitation or delivery of diverse variants from both antibody families with promiscuous capacity would lead to neutralization coverage reaching close to 90% for wild-type viruses (Fig. 8A and 8D). Although we have noted previously the highly broad neutralization coverage of antibody combinations targeting non-overlapping epitopes (8), these results indicate that the elicitation or delivery of antibodies with overlapping or partially overlapping epitopes on just the high-mannose patch could potentially provide up to 90% cross-clade coverage.
Figure 8. Combinations of antibodies targeting the high-mannose patch increase neutralization coverage compared to single antibodies.
(A) Summary of neutralization breadth and potency on the 120-virus panel for individual antibodies and theoretical antibody combinations. (B) Summary of neutralization breadth and potency on an 80 wild-type virus subset of the 120-virus panel and (C) on the same 80-virus panel with the glycan site at N332/334 removed by alanine mutagenesis. The analysis includes individual antibodies and theoretical antibody combinations. (D) Cumulative frequency distribution at different IC50 cutoffs for individual antibodies and theoretical antibody combinations on the 120-virus panel. (E) Cumulative frequency distribution at different IC50 cutoffs for individual antibodies and theoretical antibody combinations on the 80-virus subset of the 120-virus panel with (solid) and without (dotted) the glycan site at N332/334.
In terms of implications of this study to potential antibody therapy, though also applicable to vaccine efficacy, it is of interest to consider the breadth and potency of neutralization against the panel of 80 viruses with and without the N332/334 glycan site. Against isolates with the N332/334 glycan site present, PGT121 and PGT128 were able to neutralize 73% and 69% of the panel, respectively, but this was reduced to 36% and 23% against the same viruses lacking the N332/334 glycan site (Fig. 3 and Fig. 8B–C). However, a theoretical combination of PGT121 and PGT128 antibodies would increase neutralization coverage against those viruses to 46% (Fig. 8B–C). Further, a theoretical combination of all somatic variants from all donors would extend neutralization coverage even further to 66%, which presents an astounding recovery in neutralization breadth (Fig. 8B–C, Fig. 8E, and table S5–6). Thus, antibody therapy with diverse antibodies targeting the high-mannose patch would be more effective than single monoclonal therapy in limiting virus escape.
Finally, immunogen design strategies targeting the high-mannose patch will undoubtedly begin with single well-explored specificities such as PGT128, but this design should probably be extended rapidly to cover other specificities as molecular details emerge. In particular, the observation that PGT121 and PGT128 are able to utilize alternate glycan sites to the N332 site, namely N136/137 and N295 respectively, suggests that immunogens incorporating an extended region of N-glycans beyond the N332 and N301 sites might be necessary to elicit antibodies with this promiscuity of binding. Additionally, given the enhanced neutralization potency or binding upon removal of certain glycan sites for PGT128 and PGT124, the need to accommodate potentially obstructive glycans may also be a key part of the antibody maturation process and should be considered during immunogen design. Further, immunogens that reflect the natural glycan heterogeneity at individual sites may also be required to generate bnMAbs that are promiscuous in their recognition of the core-component of both high-mannose-type and complex-type N-linked glycans (Fig. 4B) (17, 22). The extent of promiscuity that emerges in antibodies induced through immunization will depend upon the immunogens and immunization protocols used and should be carefully monitored.
The elicitation of antibodies to the high-mannose patch presents many challenges. A substantial obstacle is the general lack of immunogenicity of glycans. The frequency of chronically infected donors who develop antibody responses targeting the high-mannose patch, however, suggests that this region can be immunogenic. Another obstacle is to avoid eliciting anti-glycan antibodies that are self-reactive. We note that a number of isolates that contain a glycan site at the 334 position are in fact highly resistant to all known high-mannose patch targeting antibodies, but become sensitive upon shifting the glycan site to the 332 position (fig. S9). Thus, while our results suggest that not all viruses with a N334 glycan site are resistant, there are additional nuances in the recognition of this site that would be greatly informed by additional structural studies as well as by the isolation of new bnMAbs that specifically target the glycan at the N334 position. Finally, the data presented here uses a readily available large cross-clade panel of isolates. Similar studies could be fruitfully carried out on recently isolated founder viruses for comparative studies.
In summary, broadly neutralizing antibodies targeting the high-mannose patch of HIV gp120 such as PGT121 and PGT128, exhibit outstanding in vitro and in vivo anti-viral activity. Although generally considered as N332 glycan site dependent, these antibodies and their somatic variants show a marked ability to retain neutralization potency despite the loss of this glycan site through amino acid substitution. An important factor here appears to be the propensity of the antibodies to make use of neighboring glycans. The antibodies also show the ability to neutralize many viruses isolated from natural infection in which the glycan site is located at the N334 rather than the N332 position. In terms of antibody therapy, the promiscuous behavior described may impact upon and limit neutralization escape. In terms of vaccine design, immunogens and immunization strategies will need to be carefully chosen to maximize the potential of induced antibodies targeting the high-mannose patch.
Materials and Methods
Study Design
The objective of the study was to better define the high-mannose patch epitope cluster on HIV Env and to identify key glycan components that would inform immunogen design for the elicitation of broadly neutralizing antibodies by vaccination. To this end, known broadly neutralizing antibodies that target the epitope cluster were extensively tested in neutralization assays to identify the glycan sites that are important for neutralization. Removal of glycan sites by alanine substitution was performed by site-directed mutagenesis and variants evaluated in neutralization assays. HIV Env sequences from the Los Alamos database were analyzed computationally to determine the frequency of single and pairwise glycan sites that were identified to be important for neutralization. Finally, glycans at different glycan sites on gp120 were modeled by the programs GlycanRelax and GlycanTransfer to determine the spatial dynamics between glycan sites. The viruses tested in the study were selected because they are publically available and are routinely used by other groups to evaluate neutralization breadth and potency. Antibodies were chosen because they are also publically available and demonstrate the greatest neutralization breadth and potency among this class of antibodies. All experiments were performed in duplicate unless otherwise noted.
Antibodies and antigens
The following antibodies and reagents were procured by the IAVI Neutralizing Antibody Consortium: antibody 2G12 (Polymun Scientific), PGT121-128, PGT130-131, PGT133, PGT135-137, and PGV04.
Pseudovirus production and neutralization assays
To produce pseudoviruses, plasmids encoding Env were co-transfected with an Env-deficient genomic backbone plasmid (pSG3ΔEnv) in a 1:2 ratio with the transfection reagent Fugene 6 (Promega). Pseudoviruses were harvested 72 hours post transfection for use in neutralization assays. Neutralizing activity was assessed using a single round of replication pseudovirus assay and TZM-bl target cells, as described previously (8). Briefly, TZM-bl cells were seeded in a 96-well flat bottom plate at a concentration of 20,000 cells/well. The serially diluted virus/antibody mixture, which was pre-incubated for 1 hr, was then added to the cells and luminescence was quantified 48 hrs following infection via lysis and addition of Bright-Glo™ Luciferase substrate (Promega). To determine IC50 values, serial dilutions of mAbs were incubated with virus and the dose-response curves were fitted using nonlinear regression.
Antibody expression and purification
Antibody plasmids were co-transfected at a 1:1 ratio in 293 FreeStyle cells using 293fectin (Invitrogen). Transfections were performed according to the manufacturer’s protocol and antibody supernatants were harvested four days following transfection. Antibody supernatants were purified over a protein A column, eluted with 0.1M citric acid (pH 3.0), and dialyzed against phosphate-buffered saline.
Envelope mutations
Mutations were introduced by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene) and mutants were verified by Sanger DNA sequencing.
ELISA assays
Ninety-six-well ELISA plates were coated with 50 uL PBS containing 100 ng of goat anti-human IgG Fc (Pierce) or 100 ng of gp120 per well. The wells were washed four times with PBS containing 0.05% Tween 20 and blocked with 3% BSA at room temperature for 1 h. Serial dilutions or mAb were then added to the wells, and the plates were incubated at room temperature for 1 hour. After washing four times, goat anti-human IgG Fab′2 conjugated to alkaline phosphatase (Pierce) was diluted 1:1000 in PBS containing 1% BSA and 0.025% Tween 20 and added to the wells. The plate was incubated at room temperature for 1 h, washed four times, and the plate was developed by adding 50 uL of alkaline phosphatase substrate (Sigma) to 5 mL alkaline phosphatase staining buffer (pH 9.8), according to the manufacturer’s instructions. The optical density at 405 nm was read on a microplate reader (Molecular Devices).
Cell surface binding assays
To produce cell surface Env trimer, plasmids encoding Env were co-transfected with an Env-deficient genomic backbone plasmid (pSG3ΔEnv) in a 1:2 ratio with the transfection reagent Fugene 6 (Promega). The cells were then harvested 48 hrs following transfection and the supernatant discarded. Titrating amounts of mAbs were added to the transfected cells and incubated for 1h at 4°C in 1x PBS. The cells were washed three times with 1x PBS and fixed with 2% PFA (PolySciences) for 20 min at RT. Following three washes with 1x PBS, the cells were then stained with a 1:200 dilution of goat anti-human IgG F(ab′)2 conjugated to phycoerythrin (Jackson) for 1h at RT. Binding was analyzed using flow cytometry, and binding curves were generated by plotting the mean fluorescence intensity of antigen binding as a function of antibody concentration. FlowJo software was used for data interpretation.
Statistics
Statistical analyses were done with Prism 6.0 for Mac (GraphPad). Viruses that are not neutralized at an IC50 or IC90 were given a value of 50 μg/ml for median calculations. For combinations of antibodies, a virus was counted as covered if at least one of the monoclonal antibodies was neutralized depending on individual concentrations (IC50). This approach does not take additivity into account and therefore underestimates the neutralization potency of antibody combinations.
Glycan Modeling
GlycanRelax (41), was employed to generate 1000 different conformations of Man8 at position 295 and 334 of the eODmV3 in PDB:3TYG but in the absence of PGT128. In addition, a new biased glycan sampling program, GlycanTransfer, was written in MSL (42) to explore alternative glycan conformations capable of mimicking the interaction of the N332 glycan with PGT128. Brief details of the GlycanTransfer program are as follows. The N-linked Man8 from position 332 in PDB:3TYG was transferred to position 295 and 334 by aligning the backbone atoms of the asparagine to the backbone atoms of N295/S334. Next, 22 dihedral angles were allowed to rotate within confined angular ranges above and below the values in PDB:3TYG or the mean values defined by Petrescu et al (43) and Wormald et al (44). The allowed range was +/− 1.5 standard deviations according to Petrescu et al and Wormald et al (43, 44). To effectively explore the conformational space, systematic sampling (angles closest to protein first) and random sampling protocols were utilized independently and in combination. Throughout the trajectories, models were selected when a set of dihedral angles produced glycan conformations with a low root-mean-square-deviation (RMSD) to a specified sugar ring, either the beta-D-mannose (BMA) group or one of the terminal mannose (MAN) groups of Man8. RMSDs reported in the main text are for BMA.
Supplementary Material
Fig. S1. Neutralization of viruses with a natural N334 glycan site by bnMAbs targeting the high-mannose patch.
Fig. S2. Neutralization breadth and potency on an 80-virus panel with and without the glycan site at N332/334 present.
Fig. S3. Summary of dependency on different glycan sites for bnMAbs targeting the high-mannose patch.
Fig. S4. Binding to cell surface Env by bnMAbs to the high-mannose patch shows differing sensitivity to the presence of V1/V2 and V3 loops.
Fig. S5. Loss of the N332 glycan site does not affect overall neutralization sensitivity of pseudoviruses.
Fig. S6. PGT121-123 can utilize the N136 glycan site for neutralization when no glycan site at position N332 or N334 is naturally present.
Fig S7. Loss of glycan site combinations N136 + N301 and N136 + N156 does not greatly affect neutralization for PGT121
Fig. S8. PGT125-131 can utilize the N295 and N301 glycan sites when no glycan site at position N332 or N334 is naturally present.
Fig. S9. Some resistant viruses naturally containing a glycan at the N334 become sensitive to neutralization upon shifting the glycan to the N332 position.
Table S1. Summary of key features of bnMAbs targeting the high-mannose patch.
Table S2. Neutralization breadth and potency of bnMAbs to the high-mannose patch on a cross-clade 120-virus panel.
Table S3. Neutralization breadth and potency of bnMAbs to the high-mannose patch on an 80-virus subset of the 120-virus panel.
Table S4. Neutralization breadth and potency of bnMAbs to the high-mannose patch on the 80-virus panel with the glycan site at N332/334 removed by alanine mutagenesis.
Table S5. Summary of neutralization breadth and potency on the 80-virus wild-type panel.
Table S6. Summary of neutralization breadth and potency on the 80-virus panel with the glycan site at N332/334 removed by alanine mutagenesis.
Table S7. Frequency of critical N-linked glycan sites in 31,788 viruses in the Los Alamos database.
Table S8. Neutralization breadth and potency of bnMAbs to the high-mannose patch on a cross-clade 120-virus panel.
Acknowledgments
We would like to thank all the study participants and research staff at each of the Protocol G clinical centers and all of the Protocol G team members. In addition, we would like to thank all of the IAVI Protocol G project, clinical and site team members, the IAVI Human Immunology Laboratory and all of the Protocol G clinical investigators, specifically, Melissa Simek, George Miiro, Anton Pozniak, Dale McPhee, Olivier Manigart, Etienne Karita, Andre Inwoley, Walter Jaoko, Jack DeHovitz, Linda-Gail Bekker, Punnee Pitisuttithum, Robert Paris, Jennifer Serwanga, and Susan Allen.
Funding: This work was supported by the International AIDS Vaccine Initiative through the Neutralizing Antibody Consortium SFP1849 (D.R.B., P.P., I.A.W.), NIH R01 AI033292 (D.R.B.), AI84817 (I.A.W.) and 1U19AI090970 (P.P.), Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery Grant UM1AI100663 (D.R.B., I.A.W.); NIH Interdisciplinary Training Program in Immunology 5T32AI007606-10 (D.S.), Canadian Institutes of Health Research fellowship (J.-P.J.). A portion of the neutralization experiments were done by Michael Seaman’s group (Beth Israel Deaconess Medical Center, Harvard Medical School) and this work was funded through the Bill and Melinda Gates foundation (grant no. 38619).
Footnotes
Author contributions: DS, KJD, PP, and DRB planned and designed the experiments. DS, KJD, KML, KFS-F, AR, LW, and MSS performed the experiments. BB, DWK, SM, and WRS designed and performed computational analyses. DS, KJD, and DRB composed the first draft. J-PJ, BB, DWK, WRS, IAW, and PP contributed manuscript revisions. WRS, IAW, PP, and DRB supervised research.
Competing interests: The IAVI holds U.S. patent 61/515,528 on the PGT antibodies. All other authors declare no competing interests.
Data and materials availability: Gene sequences of the reported antibodies and the primers used for antibody isolation have been deposited under GenBank accession numbers JN201894–JN201927.
References and Notes
- 1.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. A Blueprint for HIV Vaccine Discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 3.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 4.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu RB, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang HW, Shekhar K, Gupta S, Nkolola JP, Seaman MS, Smith KM, Borducchi EN, Cabral C, Smith JY, Blackmore S, Sanisetty S, Perry JR, Beck M, Lewis MG, Rinaldi W, Chakraborty AK, Poignard P, Nussenzweig MC, Burton DR. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M, Lifson JD, Dimitrov DS, Nussenzweig MC, Martin MA. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Investigators PGP, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Miiro G, Serwanga J, Pozniak A, McPhee D, Manigart O, Mwananyanda L, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Allen S, Kaminsky S, Zamb T, Moyle M, CKoff W, Poignard P, Burton DR Protocol G Principal Investigators. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao H-X, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Becker J, Benjamin B, Blakesley R, Bouffard G, Brooks S, Coleman H, Dekhtyar M, Gregory M, Guan X, Gupta J, Han J, Hargrove A, Ho S-L, Johnson T, Legaspi R, Lovett S, Maduro Q, Masiello C, Maskeri B, McDowell J, Montemayor C, Mullikin J, Park M, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Thomas J, Thomas P, Vemulapalli M, Young A, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia S-M, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BTM, Kwong PD, Mascola JR, Haynes BF NISC Comparative Sequencing Program. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013 doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 15.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong L, Lee JH, Doores KJ, Murin CD, Julien J-P, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS ONE. 2011;6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci USA. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Go EP, Irungu J, Zhang Y, Dalpathado DS, Liao HX, Sutherland LL, Alam SM, Haynes BF, Desaire H. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes’ accessibility. J Proteome Res. 2008;7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantophlet R, Burton DR. GP120: Target for Neutralizing HIV-1 Antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 22.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PNP, Spencer DIR, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci USA. 2012;109:E3268–77. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 25.Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moldt B, Rakasz EG, Schultz N, Chan-Hui PY, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, Burton DR. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci USA. 2012;109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L. CAPRISA002 Study Team, The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch RM, Tran L, Louder MK, Schmidt SD, Cohen M, Dersimonian R, Euler Z, Gray ES, Abdool Karim S, Kirchherr J, Montefiori DC, Sibeko S, Soderberg K, Tomaras G, Yang Z-Y, Nabel GJ, Schuitemaker H, Morris L, Haynes BF, Mascola JR CHAVI 001 Clinical Team Members. The development of CD4 binding site antibodies during HIV-1 infection. J Virol. 2012;86:7588–7595. doi: 10.1128/JVI.00734-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker LM, Sok D, Nishimura Y, Donau O, Sadjadpour R, Gautam R, Shingai M, Pejchal R, Ramos A, Simek MD, Geng Y, Wilson IA, Poignard P, Martin MA, Burton DR. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc Natl Acad Sci USA. 2011;108:20125–20129. doi: 10.1073/pnas.1117531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pancera M, Yang Y, Louder MK, Gorman J, Lu G, McLellan JS, Stuckey J, Zhu J, Burton DR, Koff WC, Mascola JR, Kwong PD. N332-Directed broadly neutralizing antibodies use diverse modes of HIV-1 recognition: inferences from heavy-light chain complementation of function. PLoS ONE. 2013;8:e55701. doi: 10.1371/journal.pone.0055701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber M, Le KM, Doores KJ, Fulton Z, Stanfield RL, Wilson IA, Burton DR. Very few substitutions in a germ line antibody are required to initiate significant domain exchange. J Virol. 2010;84:10700–10707. doi: 10.1128/JVI.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, Lambson BE, Ranchobe N, Ping L, Ngandu N, Karim QA, Karim SSA, Swanstrom RI, Seaman MS, Williamson C, Morris L. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, Julien JP, Briney B, Ramos A, Saye KF, Le K, Mahan A, Wang S, Kardar M, Yaari G, Walker LM, Simen BB, St John EP, Chan-Hui PY, Swiderek K, Kleinstein SH, Alter G, Seaman MS, Chakraborty AK, Koller D, Wilson IA, Church GM, Burton DR, Poignard P. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog. 2013;9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi BK, Bobrowicz P, Davidson RC, Hamilton SR, Kung DH, Li H, Miele RG, Nett JH, Wildt S, Gerngross TU. Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. Proc Natl Acad Sci USA. 2003;100:5022–5027. doi: 10.1073/pnas.0931263100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang VT, Crispin M, Aricescu AR, Harvey DJ, Nettleship JE, Fennelly JA, Yu C, Boles KS, Evans EJ, Stuart DI, Dwek RA, Jones EY, Owens RJ, Davis SJ. Glycoprotein structural genomics: solving the glycosylation problem. Structure/Folding and Design. 2007;15:267–273. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O’Dell S, Patel N, Shahzad-ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang G-Y, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang Z-Y, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang L-X, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scanlan CN, Offer J, Zitzmann N, Dwek RA. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- 41.Pancera M, Majeed S, Ban YEA, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kulp DW, Subramaniam S, Donald JE, Hannigan BT, Mueller BK, Grigoryan G, Senes A. Structural informatics, modeling, and design with an open-source Molecular Software Library (MSL) J Comput Chem. 2012;33:1645–1661. doi: 10.1002/jcc.22968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrescu AJ, Milac AL, Petrescu SM, Dwek RA, Wormald MR. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 44.Wormald MR, Petrescu AJ, Pao YL, Glithero A, Elliott T, Dwek RA. Conformational studies of oligosaccharides and glycopeptides: complementarity of NMR, X-ray crystallography, and molecular modelling. Chem Rev. 2002;102:371–386. doi: 10.1021/cr990368i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Neutralization of viruses with a natural N334 glycan site by bnMAbs targeting the high-mannose patch.
Fig. S2. Neutralization breadth and potency on an 80-virus panel with and without the glycan site at N332/334 present.
Fig. S3. Summary of dependency on different glycan sites for bnMAbs targeting the high-mannose patch.
Fig. S4. Binding to cell surface Env by bnMAbs to the high-mannose patch shows differing sensitivity to the presence of V1/V2 and V3 loops.
Fig. S5. Loss of the N332 glycan site does not affect overall neutralization sensitivity of pseudoviruses.
Fig. S6. PGT121-123 can utilize the N136 glycan site for neutralization when no glycan site at position N332 or N334 is naturally present.
Fig S7. Loss of glycan site combinations N136 + N301 and N136 + N156 does not greatly affect neutralization for PGT121
Fig. S8. PGT125-131 can utilize the N295 and N301 glycan sites when no glycan site at position N332 or N334 is naturally present.
Fig. S9. Some resistant viruses naturally containing a glycan at the N334 become sensitive to neutralization upon shifting the glycan to the N332 position.
Table S1. Summary of key features of bnMAbs targeting the high-mannose patch.
Table S2. Neutralization breadth and potency of bnMAbs to the high-mannose patch on a cross-clade 120-virus panel.
Table S3. Neutralization breadth and potency of bnMAbs to the high-mannose patch on an 80-virus subset of the 120-virus panel.
Table S4. Neutralization breadth and potency of bnMAbs to the high-mannose patch on the 80-virus panel with the glycan site at N332/334 removed by alanine mutagenesis.
Table S5. Summary of neutralization breadth and potency on the 80-virus wild-type panel.
Table S6. Summary of neutralization breadth and potency on the 80-virus panel with the glycan site at N332/334 removed by alanine mutagenesis.
Table S7. Frequency of critical N-linked glycan sites in 31,788 viruses in the Los Alamos database.
Table S8. Neutralization breadth and potency of bnMAbs to the high-mannose patch on a cross-clade 120-virus panel.