Abstract
Drug responses vary greatly among individuals due to human genetic variations, which is known as pharmacogenomics (PGx). Much of the PGx knowledge has been embedded in biomedical literature and there is a growing interest to develop text mining approaches to extract such knowledge. In this paper, we present a study to rank candidate gene-drug relations using Latent Dirichlet Allocation (LDA) model. Our approach consists of three steps: 1) recognize gene and drug entities in MEDLINE abstracts; 2) extract candidate gene-drug pairs based on different levels of co-occurrence, including abstract level, sentence level, and phrase level; and 3) rank candidate gene-drug pairs using multiple different methods including term frequency, Chi-square test, Mutual Information (MI), a reported Kullback-Leibler (KL) distance based on topics derived from LDA (LDA-KL), and a newly defined probabilistic KL distance based on LDA (LDA-PKL). We systematically evaluated these methods by using a gold standard data set of gene-drug relations derived from PharmGKB. Our results showed that the proposed LDA-PKL method achieved better Mean Average Precision (MAP) than any other methods, suggesting its promising uses for ranking and detecting PGx relations.
Keywords: Gene-drug Relation, Latent Dirichlet Allocation, Ranking, Pharmacogenomics
1. Introduction
There exists striking variability in individual responses to drug therapy as exemplified by severe adverse drug reactions which have been ranked as among the commonest causes of death in hospitalized patients.1,2 Pharmacogenomic (PGx) research is to impact this problem by linking the inherited differences to variable drug responses. However, much of our current understanding in pharmacogenomics has been dispersed across numerous journals. Hence, it is extremely important, as well as, immediately needed, to extract important facts across publications into a comprehensive knowledge base. The Pharmacogenomics Knowledge Base (PharmGKB),3 sponsored by National Institutes of Health (NIH), is such a database annotated manually by a team of curators. Compared to the vast amount of relations implicitly exist in published scientific literatures, the number of relations annotated in the PharmGKB is still limited. Thus, there is a great interest in developing automated methods to accurately detect PGx relations, such as gene-drug relations from biomedical literature.
In this paper, we present a study on detecting and ranking gene-drug relations in MED-LINE abstracts by using LDA model. Starting with MEDLINE abstracts, we first recognized gene and drug entities using existing Natural Language Processing (NLP) tools such as MetaMap. Then we extracted candidate gene-drug pairs based on different levels of co-occurrence, including abstract level (text from both titles and abstracts), sentence level, and phrase level. In order to find the most related gene-drug relations, we finally ranked candidate gene-drug pairs using different methods including three baseline methods: frequency, Chi-square test, and Mutual Information (MI), and two LDA-based methods: a previously reported Kullback-Leibler (KL) distance based on topics derived from LDA LDA-KL, and a newly defined probabilistic KL distance based on LDA LDA-PKL. The evaluation using a manually annotated data set from PharmGKB indicated that our LDA-PKL method outperformed others. To our best knowledge, this is the first attempt that applied LDA models to rank gene-drug relations for building PGx knowledge bases from biomedical literature.
2. Background
Different methods have been proposed to detect pharmacogenomic entity relations from MED-LINE abstracts, including co-occurrence based, rule based and machine learning based methods.4 The co-occurrence method is a commonly used method in literature mining, building on the assumption that two entities co-occurring in the scope of the same abstract, single sentence or single phrase,5 are likely to be related. On the other hand, the rule based methods extract relations using predefined or automatically derived patterns. Machine learning based methods train a classifier on a set of annotated relations to classify the candidate relations. Since entities co-occurring in a specific scope of text do not always define a relation, the simple co-occurrence method would generate false positives, which makes it difficult to validate the correct relations from the large numbers of candidates to integrate them into knowledge bases. Thus, other methods such as syntactic rule based methods (rule based methods using syntactic patterns) and machine leaning methods were proposed to further analyze the co-occurred relations to reduce the false positives. The syntactic rule based methods assume that the entities co-occurring in certain syntactic patterns (such as subj and obj relations) are more likely to be related.
A number of systems have been developed to extract relations among gene, drug, and diseases. Most of them, such as XplorMed, iHOP and CoPub Mapper, tried to extract relations between gene, drug and disease from abstracts of biomedical articles.6–9 Jenssen et al.9 used a co-occurrence based method to build a gene-gene relationship network by extracting all the gene-gene pairs co-occurring in the same MEDLINE abstract. The relations were then weighted by the number of times the two genes co-occurred in the same abstract. Their method achieved a precision of 60% and recall of 50%. Pharmspresso10 and Textpresso11 investigated methods to extract relationships from the full text articles, rather than abstracts or sentences. Tari et al.12 applied a rule based method using loose patterns defined by a wildcard operator (“_”) to describe syntactic relations between two entities. Recent rule-based methods, such as RelEx13 and OpenDMAP14 used dependency parse trees to detect protein-protein interactions. Coulet et al.15 presented their research on detecting PGx relations using dependency graphs generated by the Stanford Parser.16 Through manual evaluation of the randomly selected 220 raw relations, they reported precisions within the range of 70–87.7%. These automatically detected relations were normalized into a knowledge base to guide the annotation of PharmGKB.
There are also studies that have focused on detecting gene-drug relations, which is an important category of pharmacogenomic relations. For example, Chang et al.17 used a machine learning method called Maximum Entropy to classify the gene-drug relations detected by a co-occurrence based method into five categories defined in PharmGKB. Garten et al.18 created a gene-drug network from the sentence level co-occurrence over full text using Pharmspresso. Then, a logistic regression classifier was trained using these automatically extracted gene-drug relations and a group of manually curated relations. The evaluation result showed that the classifier trained from automatically detected relations was as good as, and sometimes even better than, the classifier trained from manually curated knowledge bases.
Semantic is another type of useful information to analyze co-occurred entities. The semantic meanings implicitly existed in literatures can be automatically uncovered using probabilistic models. Latent Dirichlet Allocation (LDA)19 is a widely used model to identify semantic topics from large document collections. Wang et al.20 described a method that used a variation of LDA model, named Bio-LDA model, to detect entity relationships from MEDLINE abstracts. They reported that the LDA models could detect the relationships between two bio-terms even if they did not co-occur in the same text.
In this study, we applied LDA model to rank candidate gene-drug relations. Instead of directly applying LDA to relation detection, we used it as a ranking method to prioritize candidate gene-drug relations derived from co-occurrence methods based on different levels of scope of text. Ranking is an effective way to help researchers to focus on the most related relations from large numbers of candidates mixed with false positives. We compared the LDA-based ranking methods with traditional ranking algorithms such as frequency, Chi-Square and Mutual Information, and demonstrated its usability for ranking PGx relations.
3. Methods
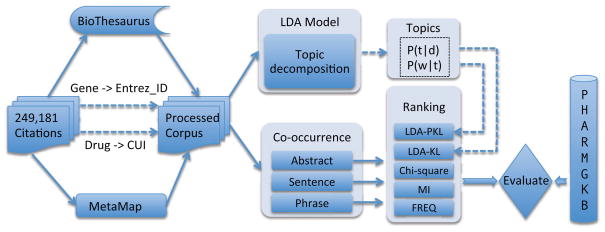
In this study, 249,181 MEDLINE abstracts listed in the gene2pubmed file from NCBI (National Center of Biotechnology Information) were used as the study corpus. Our approach consists of three steps: 1) recognize gene and drug entities in MEDLINE abstracts using natural language processing (NLP) tools; 2) extract candidate gene-drug pairs based on different levels of co-occurrence, including abstract level, sentence level, and phrase level; and 3) rank candidate gene-drug pairs using five different methods including frequency, Chi-square test, MI, LDA-KL, and LDA-PKL. These five ranking methods were then systematically evaluated using the Mean Average Precision (MAP) on a manually annotated data set derived from PharmGKB. Figure 1 shows an overview of the study design.
Fig. 1.
An overview of study design.
3.1. Data set and gene/drug recognition
In this study, we started with MEDLINE articles listed in the gene2pubmed file from NCBI, which ensures that each article in our corpus mentions at least one gene. Articles that mention many genes are often about high-throughput technologies; therefore they were removed from our study, resulting in a corpus containing 249,181 MEDLINE articles.
For each article in the corpus, its title and abstract were downloaded and processed by two NLP tools: the MetaMap program21 and a gene lookup program based on Biothesaurus.22 Instead of running MetaMap by ourselves, we downloaded the corpus of “2011 MetaMapped Medline Baseline Results”, which is a MEDLINE corpus processed by MetaMap by National Library Medicine (NLM). MetaMap outputs were used to identify drug entities - CUIs (Concept Unique Identifiers from UMLS23) from the MetaMap output with semantic types of phsu (Pharmacologic Substance) and antb (Antibiotic) were identified as drugs. One issue with this approach was that very general drug terms such as “drug” and “medicines” were also labeled as drugs. Therefore we manually reviewed top 100 frequent drugs terms from MetaMap outputs and manually removed general drug CUIs (total 15 CUIs).
To recognize genes in abstracts, we developed a gene lookup program using lexicons from Biothesaurus. To ensure the high performance of gene name recognition, we only annotated gene names that were listed in gene2pubmed and were recognized by the lookup program. If a gene name within an article was identified by the lookup program, but it was not annotated for that article according to gene2pubmed, we would ignore that gene name. An example of two processed sentences is shown in Table 1. For sentence 1, the gene entity “abcb1” as well as its alternative names: “mdr1” and “glycoprotein” were mapped to the same Entrez ID “5243”. But the ambiguous one, “impact” in sentence 2, was not considered since it could not be matched by Biothesaurus. Using sentence level co-occurrence, the candidate relation (5243, C0012265) is detected from sentence 1.
Table 1.
Example for pre-processed data.
Sentences from MEDLINE abstract 17652833:
|
|
In order to evaluate the ranking of gene-drug relations using existing annotations from PharmGKB, we further filtered extracted gene and drug entities to those that appeared in PharmGKB. For genes, PharmGKB and our gene lookup program both used Entrez Gene IDs; so it was straightforward to limit extracted genes to those in PharmGKB. However, it is more challenging to map drug entities – PharmGKB uses its own IDs for drug entities and there is no direct mapping between UMLS CUIs and PharmGKB Drug IDs. We employed KnowledgeMap,24 a general UMLS concept extraction system, to bridge the gap between UMLS drug CUIs from MetaMap and ParmGKB drug IDs. Each drug term in PharmGKB was processed by KnowledgeMap and its corresponding UMLS CUIs were identified. There were a few issues associated with the automated mapping method. First, not all drug terms in PharmGKB were mapped to UMLS CUIs by KnowledgeMap - among the 3,004 drug entities in the PharmGKB database, 2,474 of them were mapped to CUIs. Second, multiple PharmGKB drug terms might be mapped to one UMLS CUI, because of different levels of granularity between PharmGKB and UMLS drug names. For example, three PharmGKB drug terms “PA164712641 : Corticosteroids and mydriatics in combination “, “PA164712644: Corticosteroids, Combinations With Antiseptics”, and “PA164712645 : Corticosteroids, Dermatological Preparations”, were mapped to one UMLS CUI “C0001617 Corticosteroid”. As the focus of this study was the ranking methods instead of entity recognition, we removed un-mapped drug terms in PharmGKB and only kept mapped UMLS CUIs in the final list of drug entities for our evaluation. After this, a total of 9,700 distinct gene entities and 1,115 distinct drug entities were recognized from the corpus and used for our evaluation.
3.2. Detecting candidate gene-drug relation pairs
If a gene and a drug entity occur in the same scope of text, we define them as a candidate gene-drug pair. In this study, three different levels of scope were investigated: 1) abstract level, where a gene and a drug occur within the same abstract (including title); 2) sentence level, where a gene and a drug occur within the same sentence of an article (sentence boundary was determined by MetaMap program); 3) phrase level, where a gene and a drug occur within the same phrase. A phrase was defined as a fragment between any two punctuations, within one sentence.5
3.3. Ranking candidate gene-drug relations
Five different ranking methods were used to rank candidate gene-drug pairs derived from different levels of co-occurrence patterns. Three of them were baseline methods: frequency (FREQ), Chi-square test, and Mutual Information (MI). Two of them were LDA-based methods: LDA-KL (a previously reported measure) and LDA-PKL (a newly defined measure in this study). Details of each ranking algorithm were listed below.
FREQ - the frequency based method simply ranked all the relations according to the number of times that a gene and a drug entity co-occurred in the same scope of text.
-
The Chi-square test score and MI score for a gene-drug relation (wg, wd) were calculated as shown in equation 1 and equation 2, respectively.
(1) (2) Where O11 is the number of abstracts wg and wd co-occurred; O12 is the number of abstracts only wg appeared; O21 is the number of abstracts only wd appeared; O22 is the number of abstracts neither wg or wd appeared. D(wg, wd) denotes the number of abstracts containing this relation, and D(wg) denotes all the number of abstracts talking about wg.
- LDA-KL, shown in equation 4, is a score defined using Kullback-Leibler (KL) distance over all the topics derived from the LDA model, which was originally used to detect complex “bio-terms” relations from literature.20 The relations with a lower LDA-KL score are more likely to be related.
(3) (4) -
LDA-PKL - a new distance measure by combining the KL distance over different topics and the posterior probabilities of topics over MEDLINE abstracts, which is shown in equation 6. Where p(wg|tj) is the probability that gene wg appeared in topic j; T is the total number of topics automatically determined by the LDA model (T = 100 in this study); p(tj|di) is the posterior probability of topic j to MEDLINE abstract i; D is the set of MEDLINE abstracts containing relation (wg, wd).
(5) (6) In order to rank the relations using LDA latent topics, we run the LDA model with a fixed topic number of 100 on the data set containing 249,181 MEDLINE abstracts using the c-version of LDA by Blei et al.19 The data set was pre-processed before running. The gene entity and drug entity were mapped to Entrez ID and drug CUI using BioThesaurus and MetaMap, respectively. A list of 570 English stop wordsa was removed from the vocabulary. To further reduce noise, we removed 160,560 words whose frequency were less than three. The final data set contains 162,590 words.
3.4. Evaluation
To evaluate the performance of multiple ranking methods, we created a gold standard of gene-drug relations by leveraging annotations from PharmGKB. We downloaded PharmGKB files on 6/20/11 from the official website of PharmGKB. There were 11,607 gene-drug relations, associated with 3,283 MEDLINE articles. The overlap between our corpus (249,191) and PharmGKB corpus (3,283) were 898 MEDLINE articles. These 898 MEDLINE articles were associated with 1,530 gene-drug relations, according to PharmGKB.
If all the gene and drug entities were recognized correctly by our approach, candidate gene-drug relations detected at the abstract level co-occurrence should be able to cover all 1,530-candidate relations from 898 MEDLINE articles (i.e., a recall of 100%). However, there will always be errors in gene and drug entity recognition. Moreover, gene-drug relations annotated in PharmGKB were from review of full text articles; while we used only abstracts. As the purpose of this study was to evaluate relation ranking algorithms instead of entity recognition methods, we limited our evaluation data set to articles where gene/drug entities reported by PharmGKB were correctly identified by our programs. Therefore, if a PharmGKB gene-drug relation from one article contained entities (either gene or drug) that were not recognized by MetaMap and Biothesaurus from that article, we removed the relation from our evaluation. This resulted in 831 gold standard gene-drug relations from 722 MEDLINE abstracts, which served as the final evaluation data set for this study.
The candidate relations detected from the 722 MEDLINE abstracts using different co-occurrence levels were evaluated using the 831 gold standard relations. Precision and recall for detecting candidate gene-drug relations were calculated using following formulas:
| (7) |
| (8) |
Number of gold standard relations(831) To evaluate the ranking result of different algorithms, we reported the Mean Average Precision (MAP)25 as well as the precision-recall curve. The MAP score is commonly used as the standard evaluation method for ranked results from information retrieval tasks. Equation 9 shows the equation for MAP where Q is the query set, which is 1 in our case, Rjk is the set of ranked relations until you get K true relations, and {r1, …, rmj} is the set of true relation in Rjk.
| (9) |
4. Result
4.1. Detecting gene-drug relations
Using different levels of co-occurrence methods, the number of gene-drug relations detected from entire corpus and from the 722 gold standard MEDLINE abstracts were (79,885, 1,707), (43,309, 1,006), and (30,960, 692) for abstract level, sentence level and phrase level, respectively. Table 2 shows the performance of co-occurrence methods at different levels (abstract, sentence, phrase) evaluated on the 831 gold standard relations.
Table 2.
Precision and recall for detected gene-drug relations using different levels of co-occurrence.
| Method | # of extracted relations | # of extracted and correct relations | # of relations in gold standard | Precision | Recall |
|---|---|---|---|---|---|
| Abstract | 1,707 | 831 | 831 | 48.6% | 100% |
| Sentence | 1,006 | 589 | 831 | 58.5% | 70.8% |
| Clause | 692 | 447 | 831 | 64.5% | 53.7% |
Since the 831 gold standard relations only included the PharmGKB gene-drug relations between gene entity and drug entity that could be recognized using MetaMap and Biothesaurus from 722 abstracts, the abstract level co-occurrence yielded a precision of 48.6% and recall of 100%. And as expected, lower recall but higher precision was observed when we strengthen the constraint on the context level. For instance, the most strict method at the phrase level achieved a higher precision (64.5%) at the cost of lower recall (53.7%).
4.2. Ranking relations
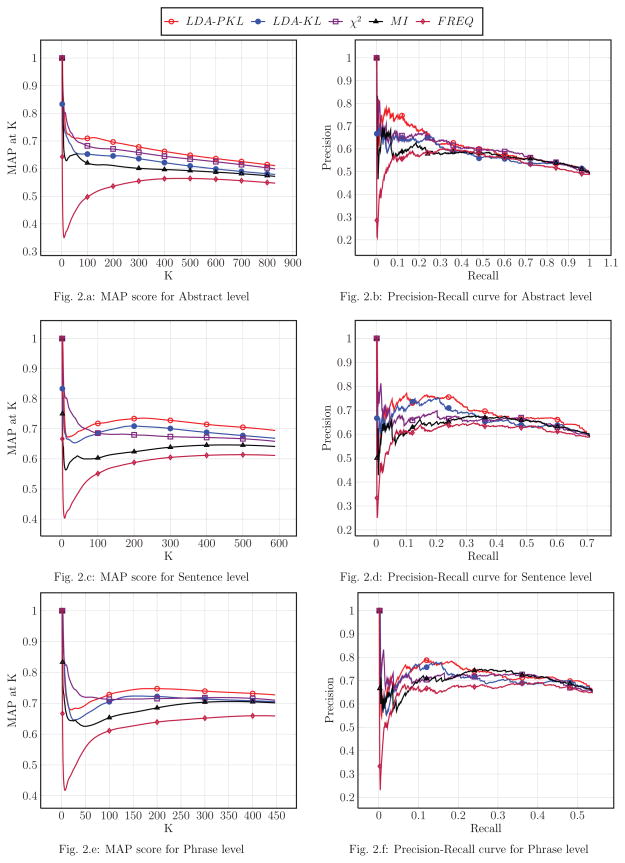
Figure 2 shows the MAP score and precision-recall curve for all the ranking algorithms over different levels of co-occurrence (Fig. 2a and Fig. 2b for abstract level, Fig. 2c and Fig. 2d for sentence level, Fig. 2e and Fig. 2f for phrase level). Among all three levels of co-occurrence based methods, our proposed LDA-PKL ranking method outperformed all others in terms of MAP score after the curves became steady. Similar results were also observed on the precision-recall curves.
Fig. 2.
MAP score and precision-recall curve for all ranking algorithms.
5. Discussion
We conducted a study on ranking candidate gene-drug relations that were extracted from MEDLINE abstracts based on simple co-occurrence methods using LDA model. Different levels of co-occurrence were used to extract candidate gene-drug relations from 249,181 MEDLINE abstracts. These candidate relations derived from literature were evaluated using a gold standard set of gene-drug relations derived from manually curated PharmGKB knowledge base. We reported the precision and recall for gene-drug relation detection at different co-occurrence levels: 48.6% and 100% for abstracts, 58.5% and 70.8% for sentences, and 64.5% and 53.7% for phrases. These results were lower than previously reported results on co-occurrence based relation detection methods for general biomedical terms by Ding et al.5 where they had 57.1% and 100% for abstracts, 63.8% and 84.9% for sentences, and 74.3% and 62.1% for phrases, respectively. This indicated gene/drug entities and their relations were more difficult to extract than general biomedical terms. We analyzed the errors and found that some gene-drug relations were missed because 1) they were not recognized by the gene and drug entity recognition programs; or 2) there were no mapping between UMLS drug CUIs and PharmGKB drug entity IDs using the KnowledgeMap.
Five different ranking methods were used to prioritize the candidate gene-drug relations detected using different levels of co-occurrence. The MAP score of proposed LDA-PKL method outperformed other methods over all the position “K” after curves became steady, which suggests that the semantic topics derived from LDA model could improve the ranking of gene-drug relations. The proposed LDA-PKL method outperformed the LDA-KL method, which only considered the KL distance over all topics, by further weighting the KL distance using the posterior probabilities of topics over MEDLINE abstracts. The Chi-square method also outperformed LDA-KL method on abstract level and part of phrase level. When considering the statistical information from the entire corpus, the Chi-square and MI methods outperformed the FREQ method that simply ranks relations by their co-occurrence frequency. The advantage of LDA-PKL on abstract level and sentence level is better than the phrase level, since the semantic topics used in ranking were derived from the abstract level using LDA model.
Although the proposed LDA-PKL algorithm outperformed other methods on MAP score after the curves became steady, in some cases from precision-recall curves, the Chi-square and MI outperformed LDA-PKL on precision. This suggested that the semantic topics derived from LDA model also introduced some noise when inferring relationships between entities (In LDA model, the PGx entities could co-occur in all topics with a probability, even if they do not co-occur in any abstract).
There are limitations in this initial study. First, better Named Entity Recognition (NER) tools are needed for identifying gene/drug entities. We analyzed the errors and found that some gene-drug relations were missed because the gene and drug entities were not recognized correctly. Although this research is not focused on NER, better gene/drug entity recognition would help detect more relations. Second, we used a fixed number of topics 100 for topic decomposition of the LDA model. It would be interesting to further analyze the performance of LDA-PKL under different numbers of topics.
The ranking algorithm proposed in this paper, LDA-PKL, could help researchers to focus on the most likely related gene-drug relations from large numbers of candidates mixed with false positives. Since LDA-PKL is not specific to gene-drug relations, our method could be extended to other types of relations between PGx entities. Our future work includes: developing more effective NER methods to accurately recognize more gene/drug entities, applying this method to mining other relations among PGx entities, and further evaluating the false positives to discover new relations that are not currently in knowledge databases.
6. Conclusion
In this paper, we presented our research on ranking gene-drug relations extracted from biomedical literature using LDA model. Our proposed method, LDA-PKL, outperformed other existing ranking methods including Chi-square, MI, and a previously reported LDA-KL method, at all co-occurrence levels, suggesting that appropriate uses of the semantic topics derived from LDA model could help analyze candidate relations that co-occur in a piece of text.
Acknowledgments
The datasets used were obtained from NLM. The evaluation data were obtained from PharmGKB website: http://www.pharmgkb.org. The authors would like to thank Dr. Josh Denny at Vanderbilt University for providing KnowledgeMap, Dr. Hongfang Liu at Mayo Clinic for providing Biothesaurus, Dr. Alan Aronson at NLM for making MetaMapped corpus available, and the PharmGKB team for the gold standard PGx relations.
This study was supported in part by grants from the NHLBI 5U19HL065962, the NLM R01LM010681, and the NCI R01CA141307.
Footnotes
The stop word list is available at ftp://ftp.cs.cornell.edu/pub/smart/english.stop
Contributor Information
YONGHUI WU, Department of Biomedical Informatics, Vanderbilt University Nashville, TN 37203, USA.
MEI LIU, Email: mei.liu@Vanderbilt.Edu, Department of Biomedical Informatics, Vanderbilt University Nashville, TN 37232, USA.
W. JIM ZHENG, Email: zhengw@musc.edu, Department of Biochemistry, Medical University of South Carolina Charleston, SC 29425, USA.
ZHONGMING ZHAO, Email: zhongming.zhao@Vanderbilt.Edu, Department of Biomedical Informatics, Vanderbilt University Nashville, TN 37232, USA.
HUA XU, Email: hua.xu@Vanderbilt.Edu, Department of Biomedical Informatics, Vanderbilt University Nashville, TN 37232, USA.
References
- 1.Roden DM, Altman RB, Benowitz NL, Flockhart DA, Giacomini KM, Johnson JA, Krauss RM, McLeod HL, Ratain MJ, Relling MV, Ring HZ, Shuldiner AR, Weinshilboum RM, Weiss ST. Pharmacogenomics: challenges and opportunities. Ann Intern Med. 2006;145:749. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giacomini KM, Krauss RM, Roden DM, Eichelbaum M, Hayden MR, Nakamura Y. When good drugs go bad. Nature. 2007;446:975. doi: 10.1038/446975a. [DOI] [PubMed] [Google Scholar]
- 3.Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, Hewett M, Lin Z, Liu Y, Liu S, Oliver DE, Rubin DL, Shafa F, Stuart JM, Altman RB. Integrating genotype and phenotype information: an overview of the pharmgkb project. pharmacogenetics research network and knowledge base. Pharmacogenomics J. 2001;1:167. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 4.Garten Y, Coulet A, Altman RB. Recent progress in automatically extracting information from the pharmacogenomic literature. Pharmacogenomics. 2010;11:1467. doi: 10.2217/pgs.10.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding J, Berleant D, Nettleton D, Wurtele E. Mining medline: abstracts, sentences, or phrases? Pac Symp Biocomput. 2002;326 doi: 10.1142/9789812799623_0031. [DOI] [PubMed] [Google Scholar]
- 6.Perez-Iratxeta C, Bork P, Andrade MA. Xplormed: a tool for exploring medline abstracts. Trends Biochem Sci. 2001;26:573. doi: 10.1016/s0968-0004(01)01926-0. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann R, Valencia A. Implementing the ihop concept for navigation of biomedical literature. Bioinformatics. 2005;21(Suppl 2):ii252. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- 8.Alako BT, Veldhoven A, van Baal S, Jelier R, Verhoeven S, Rullmann T, Polman J, Jenster G. Copub mapper: mining medline based on search term co-publication. BMC Bioinformatics. 2005;6:51. doi: 10.1186/1471-2105-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 2001;28:21. doi: 10.1038/ng0501-21. [DOI] [PubMed] [Google Scholar]
- 10.Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics. 2009;10(Suppl 2):S6. doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller HM, Rangarajan A, Teal TK, Sternberg PW. Textpresso for neuroscience: searching the full text of thousands of neuroscience research papers. Neuroinformatics. 2008;6:195. doi: 10.1007/s12021-008-9031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tari L, Hakenberg J, Gonzalez G, Baral C. Querying parse tree database of medline text to synthesize user-specific biomolecular networks. Pac Symp Biocomput. 2009:87. [PubMed] [Google Scholar]
- 13.Fundel K, Kuffner R, Zimmer R. Relex–relation extraction using dependency parse trees. Bioinformatics. 2007;23:365. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- 14.Hunter L, Lu Z, Firby J, Baumgartner J, Johnson WAHL, Ogren PV, Cohen KB. Opendmap: an open source, ontology-driven concept analysis engine, with applications to capturing knowledge regarding protein transport, protein interactions and cell-type-specific gene expression. BMC Bioinformatics. 2008;9:78. doi: 10.1186/1471-2105-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulet A, Shah NH, Garten Y, Musen M, Altman RB. Using text to build semantic networks for pharmacogenomics. J Biomed Inform. 2010;43:1009. doi: 10.1016/j.jbi.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein D, Manning CD. Accurate unlexicalized parsing. Proceedings of the 41st Annual Meeting on Association for Computational Linguistics. 2003;1:423. [Google Scholar]
- 17.Chang JT, Altman RB. Extracting and characterizing gene-drug relationships from the literature. Pharmacogenetics. 2004;14:577. doi: 10.1097/00008571-200409000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Garten Y, Tatonetti NP, Altman RB. Improving the prediction of pharmacogenes using text-derived drug-gene relationships. Pac Symp Biocomput. 2010:305. doi: 10.1142/9789814295291_0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blei DM, Ng AY, Jordan MI. Latent dirichlet allocation. J Mach Learn Res. 2003;3:993. [Google Scholar]
- 20.Wang H, Ding Y, Tang J, Dong X, He B, Qiu J, Wild DJ. Finding complex biological relationships in recent pubmed articles using bio-lda. PLoS One. 2011;6:e17243. doi: 10.1371/journal.pone.0017243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aronson AR. Effective mapping of biomedical text to the umls metathesaurus: the metamap program. Proc AMIA Symp. 2001;17 [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Torii M, Hu Z-z, Wu C. Mapping gene/protein names in free text to biomedical databases. Data Mining Workshops, International Conference on. 2007;0:101. [Google Scholar]
- 23.Bodenreider O. The unified medical language system (umls): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denny JC, Irani PR, Wehbe FH, Smithers JD, Spickard rA. The knowledgemap project: development of a concept-based medical school curriculum database. AMIA Annu Symp Proc. 2003:195. [PMC free article] [PubMed] [Google Scholar]
- 25.Sanderson M, Zobel J. Information retrieval system evaluation: effort, sensitivity, and reliability. Proceedings of the 28th annual international ACM SIGIR conference on Research and development in information retrieval; 2005. [Google Scholar]