Abstract
Charcot-Marie-Tooth disease (CMT) is genetically heterogeneous and classification based on motor nerve conduction velocity and inheritance is used to direct genetic testing. With the less common genetic forms of CMT, identifying the causative genetic mutation by Sanger sequencing of individual genes can be time-consuming and costly. Next-generation sequencing technologies show promise for clinical testing in diseases where a similar phenotype is caused by different genes. We report the unusual occurrence of CMT4J, caused by mutations in FIG4, in a apparently dominant pedigree. The affected proband and her mother exhibit different disease severities associated with different combinations of compound heterozygous FIG4 mutations, identified by whole exome sequencing. The proband was also shown to carry a de novo nonsense mutation in the dystrophin gene, which may contribute to her more severe phenotype. This study is a cautionary reminder that in families with two generations affected, explanations other than dominant inheritance are possible, such as recessive inheritance due to three mutations segregating in the family. It also emphasizes the advantages of next-generation sequencing approaches that screen multiple CMT genes at once for patients in whom the common genes have been excluded.
Keywords: Charcot-Marie-Tooth, FIG4, CMT4J, autosomal recessive CMT, whole exome sequencing
Introduction
Charcot-Marie-Tooth disease (CMT) is a genetically heterogeneous group of peripheral nerve disorders characterised by progressive distal weakness, deformity and sensory loss. Dominant, X-linked and recessive forms occur and CMT can be further classified into CMT1 (upper limb motor conduction velocity [MCV] <38m/s) and CMT2 (MCV > 38m/s) [1]. There are currently at least 36 genes described to cause CMT, in addition to those that cause motor or sensory predominant forms of inherited neuropathy, and this number continues to grow rapidly [2]. Classification based on MCV and inheritance is typically used to direct genetic testing [3]. With the less common genetic forms of CMT, attempting to identify the causative genetic mutation by Sanger sequencing of individual genes can be time-consuming and costly. Approaches in which multiple genes are sequenced at once using next-generation sequencing (NGS) techniques like whole exome sequencing, whole genome sequencing and sequencing of disease gene panels have proven effective research tools and show promise as alternatives to Sanger sequencing for clinical testing, particularly in diseases where a similar phenotype is caused by many different genes [4].
CMT4J is a rare cause of recessive CMT caused by mutations in the FIG4 gene, which codes for a phosphatase of particular phosphatidylinositol species involved in the cycling of intracellular organelles [5]. Though classified as a demyelinating CMT, mouse models of the disease show evidence of a neuronopathy affecting the anterior horn cell and sensory ganglia, and an axonal neuropathy, all of which contribute to ongoing neurodegeneration and progression of disease [5–7]. Complete inactivation of FIG4 results in Yunis-Varon Syndrome, an autosomal-recessive disorder with structural brain abnormalities and developmental delay [8]. We report the unusual occurrence of CMT4J in a pedigree that appeared to exhibit dominant inheritance. The proband was found to have a de novo nonsense mutation in the DMD gene, indicating she is also a carrier of Duchenne muscular dystrophy. The proband and her affected mother exhibit different disease severities associated with different combinations of compound heterozygous FIG4 mutations that were identified by exome sequencing. The DMD mutation in the proband may also contribute to her more severe phenotype.
Case Report
Patient II.1
The proband, a 13 year old girl, born to unrelated Caucasian parents of English and Irish heritage, presented with frequent falls and ankle contractures. Concerns were first raised at 2 years of age due to frequent falls and she had difficulties with her gait, climbing stairs and handwriting during childhood. Examination (Table 1) revealed mild weakness of hip flexion and ankle dorsiflexion. She was areflexic in the upper and lower limbs. Nerve conduction studies at 14 years of age (Table 2) showed a length-dependent sensorimotor neuropathy with motor conduction velocities between 18–20m/s. Previous creatine kinase levels had ranged between 500–1660 U/L (normal < 200U/L) and a previous muscle biopsy at 4 years of age had shown minor type 2 fibre atrophy and mild variation in fibre diameter. Testing for the CMT1A duplication was negative.
Table 1.
Phenotype of proband (II.1) and her mother (I.1)
| I.1 | II.1 | |
|---|---|---|
|
| ||
| Genotype | l41T/F98fsX38 | l41T/exon 2 deletion |
|
| ||
| Age at diagnosis (years) | 14 years | 41 years |
|
| ||
| Age at symptom onset | 2 years | Teens |
|
| ||
| Strength | ||
|
| ||
| Small muscles of the hand (MRC) | 5/5 (14 years) 4/5 (15 years) |
5/5 |
|
| ||
| Hip flexion (MRC) | 4+/5 | 5/5 |
|
| ||
| Ankle dorsiflexion (MRC) | 4/5 | 5/5 |
| Sensation | ||
| Vibration | reduced to left ankle and right knee | normal |
|
| ||
| Proprioception | reduced to both ankles | normal |
|
| ||
| Pinprick | reduced to base of toes | normal |
|
| ||
| Reflexes | generalised areflexia | present in UL, absent in LL |
Table 2.
Nerve conduction studies in the proband (II.1) and her mother (I.1)
| II.1 | I.1 | |||
|---|---|---|---|---|
|
| ||||
| Right | Left | Right | Left | |
|
| ||||
| Sensory conduction | ||||
| Median | absent | absent | ||
| Ulnar | absent | |||
| Sural | absent | absent | absent | |
| Motor conduction | ||||
| Median - APB | ||||
| DML (≤4.4) | 7.4 ms | 6.5 ms | 3.3 ms | |
| CMAP - wrist (≥4) | 4.9 mV | 4.6 mV | 4.5 mV | |
| CMAP - elbow | 1.8 mV | 2.1 mV (dispersed) | 3.7 mV | |
| CV (≥49) | 18 ms | 20m/s | 34 m/s | |
| Ulnar - ADM | ||||
| DML (≤3.3) | 5.4 ms | 3.2 ms | ||
| CMAP - wrist (≥6) | 2.5 mV | 4.6 mV | ||
| CMAP - elbow | 2.0 mV | 3.1 mV | ||
| CV (≥49) | 20m/s | 31 m/s | ||
| Posterior Tibial - AH | ||||
| DML (≤5.8) | 5.0 ms | |||
| CMAP - ankle (≥4.0) | absent | 2.2 mV | ||
| CMAP - knee | 0.7 mV (dispersed) | |||
| CV (≥41) | 28m/s | |||
| Common Peroneal - EDB | ||||
| CMAP (ankle) | absent | absent | absent | |
Patient I.1
The proband's 41 year old mother reported frequent ankle sprains and pain and intermittent paresthesiae in her hands since her twenties. She had bilateral surgery for carpal tunnel syndrome. On examination, she was able to walk on her heels and had no proximal or distal weakness though reflexes were absent at the knees and ankles (Table 1). Nerve conduction tests (Table 2) showed upper limb motor conduction velocities between 31–34 m/s. Both individuals met clinical criteria for diagnosis of CMT.
Whole exome sequencing methods
This study was approved by the Sydney Children's Hospitals Network Human Research Ethics Committee (10/CHW/45). Written informed consent was obtained from all patients. Exome capture was performed on genomic DNA with Agilent Whole Exome SureSelect v2 kit according to manufacturer's instructions. Captured exome DNA was subjected to Illumina sequencing; on average across samples, 91% of exome target bases were covered to a total depth of >20X with high quality (Q20) reads. Reads were processed by Picard and aligned to the human reference genome hg19 [9] with Burrows-Wheeler Aligner [10], and single nucleotide variant (SNV) and small insertion/deletion (indel) calling on the exomes was performed by using the Genome Analysis Toolkit (GATK) [11]. Variants were annotated using a modified version of the Ensembl Variant Effect Predictor [12] and filtered for inheritance patterns and predicted functional severity using the xBrowse web server (http://atgu.mgh.harvard.edu/xbrowse). The population frequency of candidate variants was determined by comparison with an in-house database of 25,991 reference exomes at the Broad Institute of Harvard and MIT, as well as the 1000 Genomes phase 2 and Exome Sequencing Project data-sets. To identify genes satisfying possible recessive inheritance, we searched for homozygous or compound heterozygous variants in Patient II.1 where one allele had been transmitted from each parent; variants were further filtered based on potential functional impact (including predicted missense, nonsense, and essential splice site SNPs, and all coding region indels), and variants with a frequency >1% in any of our reference populations were excluded.
Sanger sequencing confirmation
Exons 2 and 4 of FIG4 and exon 39 of the DMD gene were sequenced in all family members by Sanger sequencing using standard methods (see Supplementary Information for details). FIG4 exon 2 copy number was measured in genomic DNA with a quantitative Taqman polymerase chain reaction assay using using primers flanking exon 2 (Hs02702611_cn) with internal reference RNase P [13].
Results
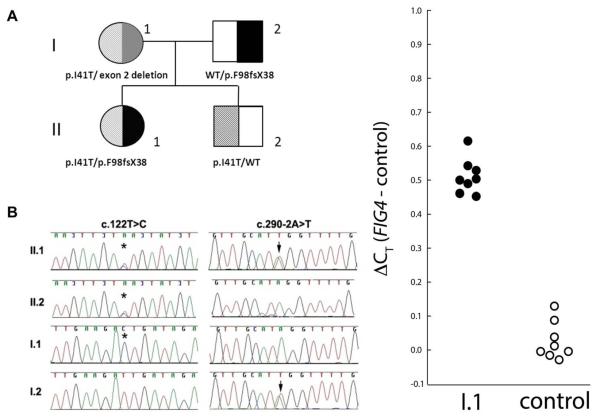
Whole exome sequencing for the proband II.1 detected two mutations in the FIG4 gene. She was heterozygous for a previously described missense mutation c.122T>C in exon 2 which results in the amino acid substitution p.Ile41Thr. She was also heterozygous for a novel mutation (c.290-2A>T), which alters the invariant splice site consensus sequence and is predicted to prevent normal splicing of exon 4, likely resulting in an out-of-frame deletion that markedly truncates the protein (p.F98fsX38) (Fig.1A, B). The combination of a heterozygous missense p.Ile41Thr mutation with a protein truncation mutation is characteristic of patients with recessive CMT4J [13]. The proband's asymptomatic father (I.2) carried the exon 4 splice site mutation (c.290-2A>T) in a heterozygous state. The proband's mother (I.1) appeared homozygous for the c.122T>C missense variant in exon 2 on Sanger sequencing. To determine whether she was truly homozygous for the variant or carried a deletion of exon 2 on her other FIG4 allele, we performed quantitative PCR (qPCR) from genomic DNA. The results demonstrate the presence of a single copy of exon 2. Therefore individual I.1 is compound heterozygous for the c.122T>C missense variant and an exon 2 deletion (Fig.1C). A comparative genomic hybdridization (CGH) array showed no evidence of a larger deletion involving FIG4 or surrounding DNA. The proband's asymptomatic brother was heterozygous for only the c.122T>C mutation. In addition, WES identified a heterozygous nonsense mutation in exon 39 of the DMD gene in Patient II.1 (c.5530C>T, p.R1844X – reference sequence NM_004006.2). Sanger sequencing confirmed the mutation in Patient II.1 but in neither parent, indicating it had arisen de novo.
Figure 1.
Pedigree, genotypes and results of quantitative Taqman polymerase chain reaction
(A) Pedigrematograms of family members showing those with the c.122T>C (★) and c.290-2A>T (⬇) mutations. I.1 appears homozygous for the c.122T>C mutation. (C) qRTPCR with intronic primers flanking exon 2, as per Figure 4 in Nicholson et al [9]. An additional cycle is required for detection of FIG4 in patient I-1 compared with the control, indicative of reduced abundance of the exon 2 sequence in the patient DNA. The difference is a little less than one cycle, as also seen in Nicholson et al, Figure 4.
Discussion
Historically, providing all families with a genetic diagnosis for CMT has been a considerable challenge. Currently, the causative gene mutation can be identified in only 60% or so of individuals with CMT, with dominantly inherited rearrangements or point mutations in PMP22, GJB1, MPZ and MFN2 accounting for 92% of the genetic diagnoses [14, 15]. The clinical challenge is to predict the causative gene if these first four genes are negative for mutations, since each of the other known genetic causes accounts for <1% of CMT. While phenotypic clues to specific forms of CMT have been reported, they are often common to multiple forms of CMT and are not universally present, making them of limited value for directing genetic testing [16]. In addition, as our family shows, clues from phenotype or family history may be misleading. With evidence of CMT in both mother and daughter, the dominant forms of CMT would have been prioritised for traditional Sanger sequencing. One of the benefits of multi-gene NGS is that erroneous assumptions about inheritance pattern or phenotype are less likely to derail the diagnostic process, compared to traditional approaches. NGS may also identify mutations in other disease causing genes that may modify the phenotype and, as in the case of our family, have implications for future pregnancies. Even with NGS approaches, filtering of data based on the likely inheritance patterns is commonly performed and it is important to remember that assumptions about the inheritance pattern are sometimes incorrect. With a rapid decline in the laboratory and interpretation costs, NGS looks highly promising as a cheaper and more effective alternative to traditional Sanger sequencing of individual genes, which can cost hundreds to thousands of dollars per gene.
There is unexplained variability in the clinical phenotypes associated with FIG4 mutations. A comprehensive description of reported patients with FIG4 mutations has previously been published [13]. Patients with genotype I41T/null may have onset in early childhood or as late as the 6th decade, and may show gradual deterioration or rapid progression. Unlike other forms of CMT, proximal weakness is seen early and may result in wheelchair dependence as early as 3rd decade of life. Asymmetric involvement of the extremities and rapidly progressive involvement of a single limb resembling chronic inflammatory demyelinating neuropathy have also been described [17, 18]. Nerve conduction studies show combined axonal and demyleinating features, often with MCV less than 20 m/s, though a single patient with MCV in the axonal range has previously been reported [13]. Electromyography (EMG) shows signs of active and chronic denervation in proximal and distal muscles [13].
The proband's mother shows features that are much milder than previously described with CMT4J. A deletion of exon 2 alone would keep the FIG4 protein in-frame but missing 33 amino acids from the internal N-terminal region. The effect of this is not known, but if the mutant protein retains some function, this could explain why the mother's phenotype is milder than her daughter's. One other patient with the I41T/ exon deletion FIG4 genotype has been previously reported but their phenotype and nerve conduction velocity were not described [13]. The de novo null mutation in the DMD gene carried by the daughter may also contribute to her muscle weakness, as is seen in a small number of female DMD mutation carriers [19].
No patients homozygous for the I41T FIG4 mutation have been reported even though the expected frequency for this genotype is 1/1,000,000 in Northern Europeans [13]. A possible reason is that this genotype causes subclinical disease. We report the second patient heterozygous for an exon deletion involving FIG4. It is important for clinicians to consider this class of mutations when an individual with CMT has only a single heterozygous change in FIG4, such as the I41T substitution, since deletions are routinely missed by Sanger sequencing and other tests are required.
Currently, the conventional approach of classifying CMT based on phenotypic markers, nerve conduction findings and pedigree is useful for the common genetic causes of CMT and leads to a diagnosis in more than half of patients. For those without an early diagnosis, this study emphasises the advantages of NGS since the number of possible genetic causes is large and digenic inheritance is possible. It is also a cautionary reminder that while families in which two generations are affected are usually affected due to dominantly inherited forms of CMT, other explanations are possible, such as recessive inheritance due to three mutations segregating within the family. The clinician is wise to remain alert to this possibility.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NHMRC grants 1022707 and 1031893 (NFC), NIH grant R01 GM24872 (MHM), and a grant from the Thyne Reid Foundation (MPM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Harding AE, Thomas PK. The clinical features of hereditary motor and sensory neuropathy types I and II. Brain. 1980;103:259–80. doi: 10.1093/brain/103.2.259. [DOI] [PubMed] [Google Scholar]
- [2].Kaplan J-C, Hamroun D. The 2013 version of the gene table of neuromuscular disorders (nuclear genome) Neuromuscul Disord. 2012;22:1108–1135. doi: 10.1016/j.nmd.2012.10.021. [DOI] [PubMed] [Google Scholar]
- [3].Reilly MM, Murphy SM, Laura M. Charcot-Marie-Tooth disease. J Peripher Nerv Syst. 2011;16:1–14. doi: 10.1111/j.1529-8027.2011.00324.x. [DOI] [PubMed] [Google Scholar]
- [4].Rossor AM, Polke JM, Houlden H, Reilly MM. Clinical implications of genetic advances in Charcot-Marie-Tooth disease. Nat Rev Neurol. 2013;9:562–571. doi: 10.1038/nrneurol.2013.179. [DOI] [PubMed] [Google Scholar]
- [5].Chow CY, Zhang Y, Dowling JJ, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Katona I, Zhang X, Bai Y, et al. Distinct pathogenic processes between Fig4-deficient motor and sensory neurons. Eur J Neurosci. 2011;33:1401–10. doi: 10.1111/j.1460-9568.2011.07651.x. [DOI] [PubMed] [Google Scholar]
- [7].Ferguson CJ, Lenk GM, Jones JM, et al. Neuronal expression of Fig4 is both necessary and sufficient to prevent spongiform neurodegeneration. Hum Mol Genet. 2012;21:3525–34. doi: 10.1093/hmg/dds179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Campeau PM, Lenk GM, Lu JT, et al. Yunis-Varon syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am J Hum Genet. 2013;92:781–91. doi: 10.1016/j.ajhg.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- [10].Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nicholson G, Lenk GM, Reddel SW, et al. Distinctive genetic and clinical features of CMT4J: a severe neuropathy caused by mutations in the PI(3,5)P phosphatase FIG4. Brain. 2011;134:1959–71. doi: 10.1093/brain/awr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Murphy SM, Laura M, Fawcett K, et al. Charcot-Marie-Tooth disease: frequency of genetic subtypes and guidelines for genetic testing. J Neurol Neurosurg Psychiatry. 2012;83:706–10. doi: 10.1136/jnnp-2012-302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Saporta AS, Sottile SL, Miller LJ, Feely SM, Siskind CE, Shy ME. Charcot-Marie-Tooth disease subtypes and genetic testing strategies. Ann Neurol. 2011;69:22–33. doi: 10.1002/ana.22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wilmshurst JM, Ouvrier R. Hereditary peripheral neuropathies of childhood: an overview for clinicians. Neuromuscul Disord. 2011;21:763–75. doi: 10.1016/j.nmd.2011.05.013. [DOI] [PubMed] [Google Scholar]
- [17].Zhang X, Chow CY, Sahenk Z, Shy ME, Meisler MH, Li J. Mutation of FIG4 causes a rapidly progressive, asymmetric neuronal degeneration. Brain. 2008;131:1990–2001. doi: 10.1093/brain/awn114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cottenie E, Menezes MP, Rossor AM, et al. Rapidly progressive asymmetrical weakness in Charcot-Marie-Tooth disease type 4J resembles chronic inflammatory demyelinating polyneuropathy. Neuromuscul Disord. 2013;23:399–403. doi: 10.1016/j.nmd.2013.01.010. [DOI] [PubMed] [Google Scholar]
- [19].Soltanzadeh P, Friez MJ, Dunn D, et al. Clinical and genetic characterization of manifesting carriers of DMD mutations. Neuromuscul Disord. 2010;20:499–504. doi: 10.1016/j.nmd.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.