Abstract
Recent progress in molecular engineering of genetically-encoded probes whose spectral properties are controlled with light, such as photoactivatable, photoswitchable and reversibly switchable fluorescent proteins, have brought the new possibilities to bioimaging and super-resolution microscopy. The development of modern photoconvertible proteins is linked to the studies of light-induced chromophore transformations. Here, we summarize the current view on the chromophore chemistry in the photocontrollable fluorescent proteins. We describe both the fundamental principles and specific molecular mechanisms underlying the irreversible and reversible chromophore photoconversions. We discuss advancements in super-resolution microscopy that became possible due to the new protein phenotypes and understanding of their chromophore transformations.
Introduction
The progress in biological imaging is tightly linked to the development of molecular probes. Among different probes, genetically-encoded fluorescent proteins (FPs) of Aequoria victoria green fluorescent protein (GFP) family enable specific labeling of cells and molecules. Of particular interest are FPs whose fluorescence is regulated by light irradiation of specific wavelengths. These photocontrollable FPs are indispensable tools for monochrome and multicolor super-resolution imaging [1]. Specifically, new probes have advanced different types of super-resolution techniques, including single molecule based photoactivated localization microscopy (PALM) [2], and ensemble imaging based reversible saturable optical fluorescence transition (RESOLFT) [3]. In addition, modern light controllable FPs are used in photochomic Fluoresence resonance energy transfer (FRET) [4], photolabelling of tissues in live animals [5] and optical manipulation of processes in a cell [6].
The light-controllable FPs can be classified in three groups: photoactivatable FPs (PAFPs), photoswitchable FPs (PSFPs) and reversibly photoswitchable FPs (rsFPs). PAFPs undergo activation from a dark to a fluorescent state. PSFPs can be photoconverted from one fluorescent state (color) to another. In contrast to PAFPs and PSFPs, which could be photoactivated only once, fluorescence of rsFPs can be photoswitched on and off repeatedly.
Engineering of FPs controllable by light was accompanied by studies of their chromophore chemistry and rearrangements in the protein structure. These studies laid the basis for development of FPs with improved characterstics and FPs with new photochemical phenotypes.
In this review we focus on the chromophore chemistry of light-controllable FPs. We first summarize the basic principles of chromophore formation and spectral properties of FPs. Then we describe various irreversible and reversible chromophore phototransformations in FPs. Finally, we discuss how new photocontrollable FPs and understanding of mechanistic basis of their photoconversion provide the new imaging applications.
Principles of chromophore chemistry
FPs form a chromophore without enzymes or cofactors except for molecular oxygen. In most FPs a chromophore-forming tripeptide consists of invariant Tyr66 and Gly67 and a variable residue at position 65 [7]. The chromophore is positioned inside a β-barrel protein fold. In a variety of chromophores several core structures can be defined (Figure 1a). Transformations of these structures, such as oxidation, cyclization, protonation-deprotonation, formation of hydrogen bonds and stacking with surrounding residues, determine the spectral properties of specific FPs [1, 8]. Red shift of FP spectra correlates with the increased number of conjugated double bonds in a chromophore and its planarity [8]. Protonation of the chromophores results in a blue shift of their absorbance. In most cases, protonated forms do not emit light, and their fluorescence can be observed only at low temperatures [9, 10]. Upon light absoption they either quickly undergo excited state proton transfer (ESPT) [9, 11], dissipate the excited-state energy [12] or exhibit cis-trans isomerization [13, 14].
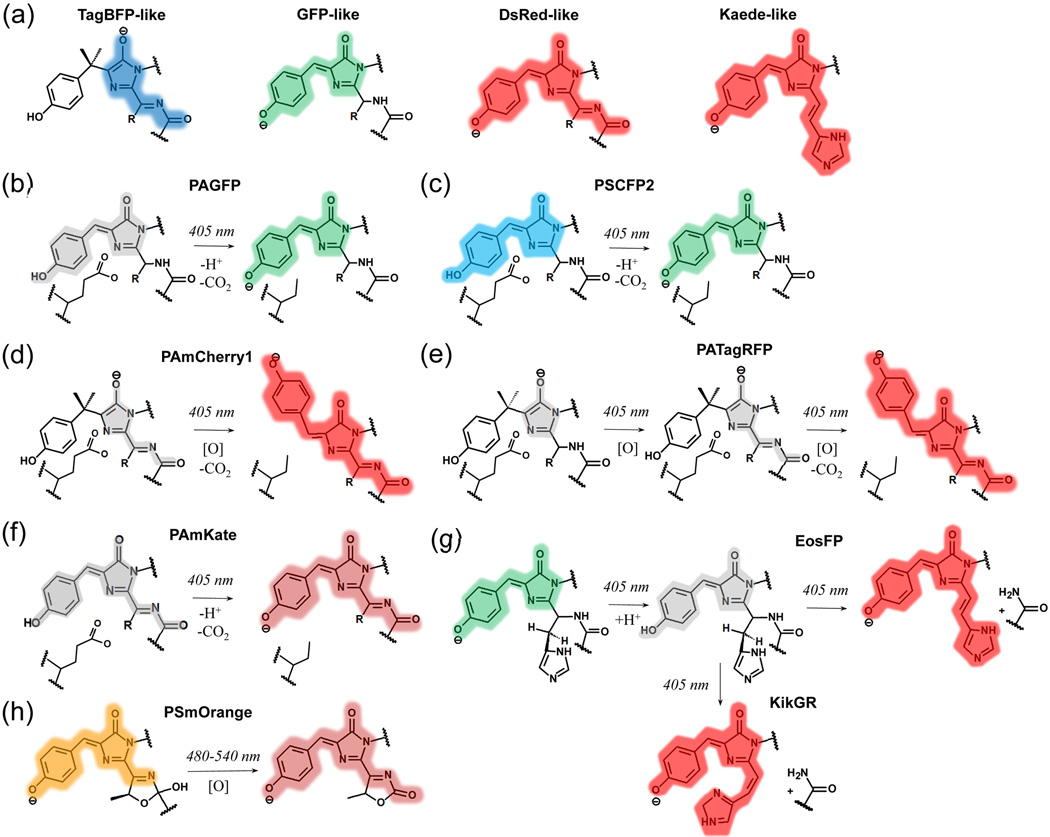
Figure 1. Light induced chromophore transformations in irreversibly photoswitchable FPs.
Colors highlighting chromophores correspond to the spectral range of observed fluorescence. Non-fluorescent chromophores are shown in gray. [O] denotes an oxidation reaction. (a) Core chromophores of GFP-like proteins, formed by invariant Tyr66, Gly67, and variable residue 65 (the numbering follows that for GFP). (b-h) Chromophore transformations are shown for PAGFP. (b), PAmCherry1 (d), EosFP (g), KikGR (g), and PSmOrange (h) were confirmed by structural data and/or mass-spectrometry. Chromophore transformations shown for PSCFP2 (c), PATagRFP (e), and PAmKate (f) are proposed based on spectral studies. (b-f) The conservative Glu222 residue, which is decarboxylated upon photoactivation, is shown.
Green and some cyan FPs share a core green GFP-like chromophore, which is 4-(p-hydroxybenzylidene)-5-imidazolinone (Figure 1a). Its anionic form absorbs at 470–510 nm and emits at 500–530 nm, while in the protonated form the spectra shift to 390–400 nm and 460–470 nm, respectively.
There are two types of core red chromophores: a DsRed-like and a Kaede-like (Figure 1a) [15, 16]. The DsRed-like chromophore can be formed either autocatalytically or by a photochemical transformation, while the Kaede-like chromophore appears only photochemically from the His65-Tyr66-Gly67 tripeptide. Anionic forms of both chromophores absorb at 540–570 nm and emit at 570–630 nm. Protonated DsRed-like chromophore absorbs at 440–460 nm. Emission of the protonated DsRed-like chromophore observed at low temperatures peaked at 530 nm [9].
The DsRed-like chromophore forms via a core TagBFP-like chromophore (Figure 1a) through oxidation of the Cα-Cβ bond in the Tyr66 side chain [17, 18]. The TagBFP-like chromophore absorbs at 390–410 nm and emits at 450–470 nm [18].
Light can cause reversible and irreversible chromophore transformations. The reversible photoconversions include cis-trans isomerization, protonation-deprotonation, and hydration-dehydration [19, 20]. The irreversible transformations include extension of conjugated π-system in a chromophore [16, 21], decarboxilation of neighboring Glu222 [21, 22], and polypeptide backbone break [5, 16]. The residues at positions 69 and 203 are minimally required for irreversibly photoactivatable phenotype, as well as conservative Glu222 in some FPs [7]. Reversibly switching phenotype depends on residues at positions 148, 165, 167, and 203 [7]. The light-induced chromophore transformations in photocontrollable FPs are discussed below.
Irreversible chromophore phototransformations
Dark-to-green PAFP PAGFP and cyan-to-green PSFPs PSCFP and PSCFP2 (Table 1) share a common mechanism of photoactivation, which was first studied in wild-type GFP (Figure 1b). Initially the protonated chromophore is stabilized by the Glu222 [23, 24]. Violet light absorption induces ESPT, which transforms the chromophore into intermediate anionic excited states, and leads to subsequent Glu222 decarboxylation. The decarboxylation causes rearrangement of hydrogen bonding network and stabilization of the anionic GFP-like chromophore. PAGFP differs from wild-type GFP by a single Thr203His mutation [25], which stabilizes protonated form in its initial state and lowers the energy of the excited state via stacking with the chromophore [22]. Similar mechanism was suggested for PSCFP and PSCFP2 with the difference in cyan emission of the protonated GFP-like chromophore before photoactivation (Figure 1c).
Table 1.
Spectral and biochemical properties of monomeric photocontrollable fluorescent proteins
| Fluorescent Protein |
Excitation (Absorbance) maximum, nm |
Emission maximum, nm |
Photoconversion light, nm |
Extinction coefficient, M−1 cm−1 |
Quantum yield |
pKa | Reference | |
|---|---|---|---|---|---|---|---|---|
| Irreversibly photoswitchable FPs | ||||||||
| PAGFP | 400 | 515 | 405 | 20,700 | 0.13 | ND | [25] | |
| 504 | 517 | 17,400 | 0.79 | 6.3 | ||||
| PAmCherry1 | (404) | ND | 405 | 6,500 | ND | ND | [45] | |
| 564 | 595 | 18,000 | 0.46 | 6.3 | ||||
| PATagRFP | (278) | ND | 405 | ND | ND | ND | [26] | |
| 562 | 595 | 66,000 | 0.38 | 5.3 | ||||
| PAmKate | (442) | ND | 405 | ND | ND | ND | [27] | |
| 586 | 628 | 25,000 | 0.18 | 5.6 | ||||
| PSCFP2 | 400 | 470 | 405 | 43,000 | 0.20 | 4.3 | [46] | |
| 490 | 511 | 47,000 | 0.23 | 6.1 | ||||
| Dendra2 | 490 | 507 | 405/480 | 45,000 | 0.50 | 6.6 | [47] | |
| 553 | 573 | 35,000 | 0.55 | 6.9 | ||||
| tdEosFP a | 506 | 516 | 405 | 84,000 | 0.66 | 5.7 | [48] | |
| 569 | 581 | 33,000 | 0.60 | ND | ||||
| mEos2 | 506 | 519 | 405 | 56,000 | 0.84 | 5.6 | [49] | |
| 573 | 584 | 46,000 | 0.66 | 6.4 | ||||
| mEos3.2 | 507 | 516 | 405 | 63,400 | 0.84 | 5.4 | [50] | |
| 572 | 580 | 32,200 | 0.55 | 5.8 | ||||
| mKikGR | 505 | 515 | 405 | 49,000 | 0.69 | 6.6 | [51] | |
| 580 | 591 | 28,000 | 0.63 | 5.2 | ||||
| mClavGR2 | 488 | 504 | 405 | 19,000 | 0.77 | 8.0 | [52] | |
| 566 | 583 | 32,000 | 0.53 | 7.3 | ||||
| mMaple | 489 | 505 | 405 | 15,000 | 0.74 | 8.2 | [53] | |
| 566 | 583 | 30,000 | 0.56 | 7.3 | ||||
| PSmOrange | 548 | 565 | 480/540 | 113,300 | 0.51 | 6.2 | [5] | |
| 636 | 662 | 32,700 | 0.28 | 5.6 | ||||
| PSmOrange2 | 546 | 561 | 480/540 | 51,000 | 0.61 | 6.6 | [44] | |
| 619 | 651 | 18,900 | 0.38 | 5.4 | ||||
| FPs with complex photoconversion phenotype | ||||||||
| mIrisFP | (386) | ND | 405 | 405 | 12,000 | ND | ND | [43] |
| 486 | 516 | 473 | 47,000 | 0.54 | 5.4 | |||
| (446) | ND | 473 | 21,000 | ND | ND | |||
| 546 | 578 | 561 | 33,000 | 0.59 | 7.6 | |||
| NijiFP | (~375) | ND | 405 | 405 | ND | ND | ND | [32] |
| 469 | 507 | 488 | 41,100 | 0.64 | 7.0 | |||
| (~440) | ND | 440 | ND | ND | ND | |||
| 526 | 569 | 561 | 42,000 | 0.65 | 7.3 | |||
| Reversibly switchable FPs | ||||||||
| Dronpa | (392) | ND | 405 | ND | ND | ND | [54] | |
| 503 | 518 | 488 | 95,000 | 0.85 | 5.0 | |||
| Dronpa3 | (ND) | ND | 405 | ND | ND | ND | [55] | |
| 487 | 514 | 488 | 58,000 | 0.33 | ND | |||
| rsFastLime | (384, 496) | ND | 405 | ND | ND | ND | [34] | |
| 496 | 518 | 488 | 39,100 | 0.77 | ND | |||
| Padron b | (505) | ND | 488 | ND | ND | ND | [35] | |
| (396), 503 | 522 | 405 | 43,000 | 0.64 | ND | |||
| bsDronpa | (385) | ND | 405 | ND | ND | ND | [35] | |
| 460 | 504 | 488 | 45,000 | 0.50 | ND | |||
| mGeosM | (390) | ND | 405 | ND | ND | ND | [56] | |
| 503 | 514 | 488 | 51,600 | 0.85 | 5.0 | |||
| rsEGFP | (396) | ND | 405 | ND | ND | ND | [3] | |
| 493 | 519 | 488 | 47,000 | 0.36 | 6.5 | |||
| rsEGFP2 | (408) | ND | 405 | ND | ND | ND | [57] | |
| 478 | 503 | 488 | 61,300 | 0.30 | 5.8 | |||
| rsCherry | 572 | 610 | 550 | 81,000 | 0.009 | ND | [37] | |
| 572 | 610 | 450 | 80,000 | 0.02 | 6.0 | |||
| rsCherryRev | 572 | 608 | 450 | 41,800 | 0.0017 | ND | [37] | |
| 572 | 608 | 550 | 42,300 | 0.0051 | 5.5 | |||
| rsTagRFP | 440 | 585 | 440 | 15,300 | 0.0013 | ND | [13] | |
| 567 | 585 | 570 | 36,800 | 0.11 | 6.6 | |||
| Dreiklang | (340, 511) | ND | 365 | ND | ND | ND | [20] | |
| (412), 511 | 529 | 405 | 83,000 | 0.41 | 7.2 | |||
For each FP, the upper row describes its parameters before photoconversion and the lower row summarizes the parameters after photoconversion. mIrisFP and NijiFP exhibit complex photoswitching behavior, which involves two states for both green and red fluorescent colors. Maturation half-times are indicated for purified FPs.
tdEosFP is a tandem dimeric fusion of the doubled size.
Padron was shown to form dimers at 4°C while being monomeric at 37°C [35]. ND, not determined.
Photoactivation of all dark-to-red PAFPs (Table 1), such as PAmCherry1, PATagRFP, and PAmKate, is based on photoinduced formation of the anionic DsRed-like chromophore from its different precursors. The dark form of PAmCherry1 corresponds to non-fluorescent TagBFP-like chromophore [21]. In the photoactivated form PAmCherry1 adopts the anionic DsRed-like chromophore in the trans conformation (Figure 1d). The photoactivation causes decarboxylation of Glu222, which leads to the oxidation of Tyr67 Cα-Cβ bond and formation of the DsRed-like chromophore [21]. In contrast to PAmCherry1, spectroscopic data suggests that the dark state of PATagRFP has a structure of cyclized dehydrated α-enolate, the mTagBFP-like chromophore precursor (Figure 1e) absorbing at about 350 nm [26]. Spectral data for PAmKate suggest that the dark state of PAmKate likely corresponds to the protonated DsRed-like chromophore. A conjugation of the anionic chromophore with a side chain of the Arg203 residue possibly causes its far-red shift (Figure 1f) [27].
Kaede-like green-to-red PSFPs include a large number of FPs (Table 1), all of which share common His65-Tyr66-Gly67 anionic GFP-like chromophore in their initial state (Figure 1g). Irradiation with violet light initiates a β-elimination reaction, which results in the Kaede-like red chromophore. In this process His65 Nα-Cα bond is cleaved and a Cα-Cβ double bond in the His65 side chain is subsequently formed, thus, extending the π-conjugated chromophore system [16]. While red chromophores of EosFP and Dendra2 have the His65 Cα-Cβ double bond in the trans conformation [28, 29], KikGR adopts the cis chromphore (Figure 1g). To account for this difference, a two-step elimination E1 mechanism, involving a loss of the leaving group followed by a deprotonation of a hydrogen nearby the carbocation, has been proposed for KikGR [30].
IrisFP and NijiFP possess unique ability to undergo both irreversible and reversible chromophore photoconversions. The irreversible chromophore photoconversion is analogous to those observed in their parental EosFP and Dendra2 [31, 32]. Both green and red forms of these proteins can be reversibly switched on and off similar to negative switching green and red rsFPs, see Table 1 and below. In both FPs this phenotype is induced by F181S mutation, which leads to rearrangements of the residues surrounding the chromophore and enables reversible photoswitching via cis-trans chromophore isomerization.
Phototransformation of the most red shifted PSFPs orange-to-farred PSmOrange and PSmOrange2 (Table 1) results in the unique far-red chromophore. Blue-green light irradiation of the mOrange-like chromophore causes its oxidation and cleavage of the polypeptide backbone. The formed far-red PSmOrange-like chromophore contains N-acylimine with the C=O group in the third five-member dihydrooxazole ring, which is co-planar with the chromophore core (Figure 1h) [5]. The greater co-planarity of the C=O group in PSmOrange-like chromophore than in the DsRed-like chromophore results in the most far-red shifted absobance of this chromophore among currently available red FPs.
Reversible chromophore photoconversions
All rsFPs can be classified in three groups on the basis on their photoswitching behavior (Table 1). Negative rsFPs are switched off by the light of the same wavelength that excites fluorescence in their on state. In contrast, positive switching rsFPs are activated by the same wavelength that excites fluorescence. A third group is represented by a single rsFP Dreiklang, whose on-off photoswitching is controlled by light at wavelengths that do not correspond to its excitation maximum.
Structural studies uncovered a similar mechanism of reversible photoconversion in all rsFPs, except Dreiklang [33, 34]. The light induces cis-trans isomerization of the p-hydroxybenzylidene chromophore moiety between two states. In negative switching green rsFPs, including Dronpa, rsFastlime, Dronpa3, and possibly mGeosM and rsEGFP, the anionic fluorescent cis GFP-like chromophore undergoes a cis-trans isomerization to the dark protonated trans GFP-like chromophore (Figure 2a). The protonation, loss of planarity, and flexibility in the rearranged protein cavity are responsible for non-radiative dissipation of excitation energy by the chromophore in the trans conformation [14, 33]. The chromophore pocket preference for the coplanar cis or the noncoplanar trans configuration determines the rsFP steady state [33]. The similar chromophore transformation also occurs during a reversible photoswitching of green and red forms of IrisFP [31]. In contrast, in positive switching rsFPs, such as Padron, the anionic cis chromophore in the on state does not undergo photoisomerization. This fluorescent anionic chromophore is in a thermal equilibrium with the protonated non-fluorescent chromophore. The latter protonated cis chromophore is the one, which undergoes photoisomerization into the anionic non-fluorescent trans chromophore under violet light irradiation (Figure 2b) [35, 36]. Interestingly, the difference in photobehavior for negative and positive switching rsFPs results from amino-acid changes at key positions 148, 165, and 167 only [35, 37].
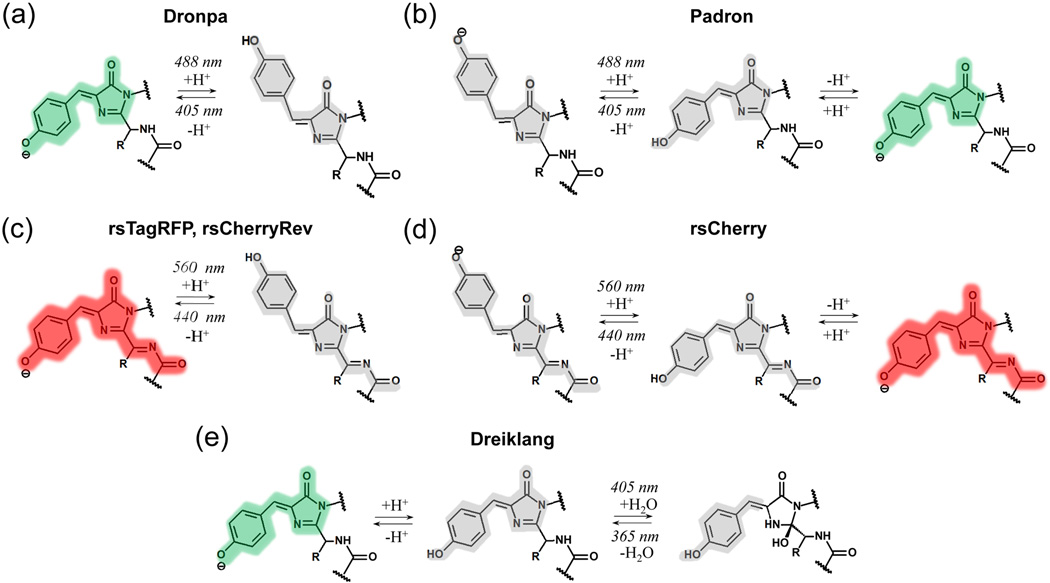
Figure 2. Light induced chromophore transformations in reversibly photoswitchable FPs.
Colors highlighting chromophores correspond to the spectral range of observed fluorescence. Non-fluorescent chromophores are shown in gray. (a-e) Chromophore transformations shown for Dronpa (a), Padron (b), rsTagRFP (c), and Dreiklang (e) were confirmed by both structural and spectral data. Chromophore transformations for rsCherryRev (c) and rsCherry (d) are proposed based on their spectral properties.
The NMR analysis of Dronpa photoswitching proposes that the on-to-off transition starts with a protonation of the chromophore and a loss of its hydrogen bond with the adjacent Ser148 residue. These events cause increased chromophore flexibility, conformational changes of the surrounding amino acids, and chromophore isomerization [14]. Overall, the flexibility of the β-barrel during photoswitching of Dronpa was revealed. The processes of chromophore cis-trans isomerization and protonation might be also concerted during the on-to-off photoconversion [36]. The opposite off-to-on switching of Dronpa was found to include the ESPT from the protonated dark chromophore [11]. In contrast to negative switching rsFPs, structural studies of positive switching rsFPs revealed rather limited rearrangement of the chromophore pockets during photoswitching [38, 39].
In red rsFPs, rsCherryRev and rsCherry the similar cis-trans isomerization might also occur (Figure 2c,d). However, a lack of an absorbance peak corresponding to the neutral DsRed-like chromophore suggests that the non-fluorescent anionic DsRed-like chromophores (trans in rsCherryRev and cis in rsCherry) may be in the equilibrium with a small fraction of the protonated chromophore, which undergoes photoconversion under blue light illumination.
Spectral and structural studies of rsTagRFP support a switching mechanism similar to that in Dronpa [40]. In contrast to rsCherryRev, the presense of 440 nm absorbance peak in the dark state of rsTagRFP indicates that in the off state the majority of rsTagRFP molecules contain the protonated trans DsRed-like chromophore capable to photoisomerization (Figure 2c) [13].
Dreiklang has a unique photoswitching mechanism [20]. In the fluorescent state its GFP-like chromophore is in the anionic cis form. Spectral data suggest that the anionic chromophore is in equilibrium with the protonated chromophore, which is capable to undergo phototransformation. Violet light causes a hydration reaction, converting the imidazolinone ring of the chromophore into a 2-hydroxyimidazolidinone (Figure 2e). UV light reverses the hydration reaction. Thus, decoupling of the wavelengths for fluorescence excitation and photoswitching occurs because none of the chromophore states, which undergo photoconversion, absorb at the same wavelengths as the fluorescent cis anionic GFP-like chromophore.
Conclusions
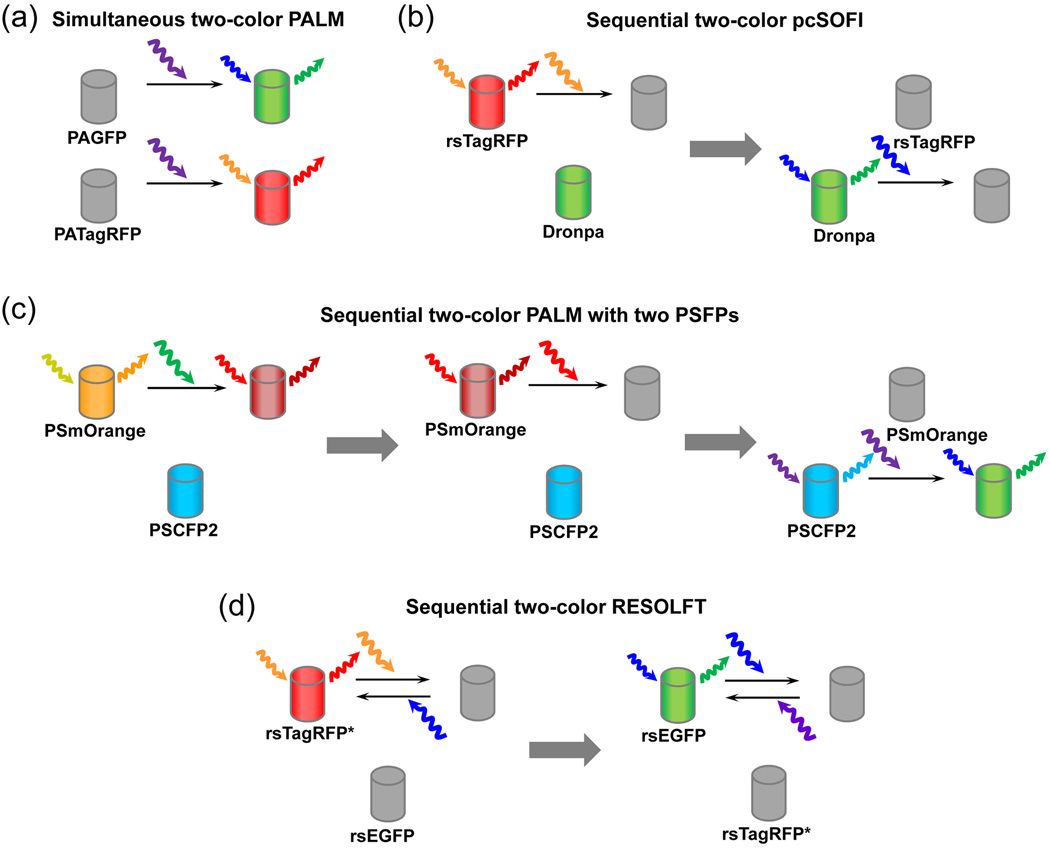
Recent studies refined our understanding and uncovered new mechanisms of the light-induced chromophore transformations. This knowledge led to engineering of advanced photocontrollable FPs, which enabled multicolor super-resolution imaging. Possible combinations of FPs that have been applied or potentially can be applied in two-color super-resolution microscopy are shown in Figure 3.
Figure 3. Proposed combinations of photocontrollable FPs for super-resolution microscopy.
(a) Green PAGFP and red PATagRFP or PAmCherry1 are suitable for simultaneous two-color PALM. Both FPs are activated by violet light and fluorescence signals of activated FPs are detected in separate channels. Simultaneous imaging is possible because the activated states of two FPs can be dicriminated from the initial states and between each other. (b) Green rsFP Dronpa and red rsTagRFP can be imaged together in a sequential pcSOFI. First, fluctuations of fluorescence from individual rsTagRFP molecules in the off state are detected and analyzed. Then rsTagRFP is bleached to prevent its activation with blue light used for switching off Dronpa. Next, fluctuations of fluorescence from individual Dronpa molecules in the off state are detected and analyzed. (c) Two PSFPs, PSmOrange or PSmOrange2 and PSCFP2, can potentially be imaged sequentially in two-color PALM with no interference of non-activated signal of one FP into the channel of another FP. First, PSmOrange molecules are activated, imaged, localized, and bleached. Orange and far-red fluorescence signals of PSmOrange are distinguishable from PSCFP2 signal. Then PSCFP2 are stochastically activated, imaged and localized. (d) Combinations of currently available green rsFP, such as rsEGFP, with future red rsFP, like rsTagRFP with improved fatigue resistance (denoted as rsTagRFP*), for use insequential two-color RESOLFT. rsTagRFP* is imaged first with rsEGFP in the dark state. Then rsTagRFP* may be bleached to avoid activation of rsTagRFP by blue light. Next, rsEGFP is imaged.
Some questions about the mechanistic basis of photoconversion are yet to be answered. For example, chemical basis for a fatigue resistance (number of switching cycles required to decrease fluorescence of rsFP in half) is unclear. This property is crucial to use rsFPs in live cell RESOLFT microscopy [3]. Until the engineering of a fatigue resistant rsEGFP variant [3], RESOLFT with rsFPs was suboptimal, and implementation of the related technique, termed stimulated emission depletion (STED), was performed with permanently fluorescent far-red FP, TagRFP657, in fixed cells [41]. Understanding of fatigue resistance will aid in development of an improved version of rsTagRFP for two-color RESOLFT (denoted as rsTagRFP* in Figure 3d). Interestingly, rsTagRFP have already shown its usefulness to complement Dronpa in a single-molecule based super-resolution technique, called photochromic stochastic optical fluctuation imaging (pcSOFI) [42] (Figure 3). These rsFPs delivered robust single-molecule fluorescence intensity fluctuations observable over extended periods, which is required for pcSOFI,
Photocontrollable FPs with unique photochemistry have found their specific applications. For example, mIrisFP with complex photobehavior was used in super-resolution imaging with selective pulse-chase photolabeling [43]. rsTagRFP has been applied to photochromic FRET in which reversible switching on and off allows the accurate FRET quantification in a live cell. [4]. Another FRET phenomenon, termed FRET-facilitated photoswitching, was demonstrated using PSmOrange2 [44]. The red-shifted spectra of PSmOrange, particularly a possibility to use green light for its photoswitching, enabled its use for labeling and tracking the specific cell population in a living animal [5]. Flexibility of the β-barrel of Dronpa mutants led to their light-inducible oligomerization and allowed to regulate intracellular processes [6].
Future efforts in understanding of the chromophore chemistry in light-controllable FPs should eventually result in new probes and advanced imaging technologies. Novel FP phenotypes can be obtained by combining different independent pathways of chromophore transformations in a single protein. These future photocontrollable FPs should advance multicolor photolabelling in super-resolution imaging of live cells.
Highlights.
Basic principles and specific photochemical reactions in fluorescent proteins are described
Irreversible and reversible light-induced chromophore transformations are discussed
Advanced bioimaging and super-resolution microscopy applications are outlined
Acknowledgments
This work was supported by grants from the Academy of Finland (263371 and 266992) and the US National Institutes of Health (GM073913, CA164468 and EB013571).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
* of special interest
** of outstanding interest
- 1.Shcherbakova DM, Subach OM, Verkhusha VV. Red fluorescent proteins: advanced imaging applications and future design. Angew Chem Int Ed Engl. 2012;51:10724–10738. doi: 10.1002/anie.201200408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betzig E, Patterson GH, Sougrat R, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 3. Grotjohann T, Testa I, Leutenegger M, et al. Diffraction-unlimited all-optical imaging and writing with a photochromic GFP. Nature. 2011;478:204–208. doi: 10.1038/nature10497. The paper reports the reversibly switchable variant of EGFP with a high fatigue resistance. rsEGFP was used in super-resolution RESOLFT microscopy of live cells.
- 4. Subach FV, Zhang L, Gadella TW, et al. Red fluorescent protein with reversibly photoswitchable absorbance for photochromic FRET. Chem Biol. 2010;17:745–755. doi: 10.1016/j.chembiol.2010.05.022. Engineered in this paper rsTagRFP is the only red reversily switchable fluorescent protein, which changes its absoption upon switching. This property enabled its use in photochromic fluorescence resonance energy transfer (pcFRET).
- 5. Subach OM, Patterson GH, Ting LM, et al. A photoswitchable orange-to-far-red fluorescent protein, PSmOrange. Nat Methods. 2011;8:771–777. doi: 10.1038/nmeth.1664. This paper reports the first orange-to-far-red PSFP and uncoveres the nature of its unique far-red chromophore. The applications of PSmOrange include photolabeling in vivo, multicolor imaging and super-resolution microscopy.
- 6. Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. The phenomenon of chromophore pocket flexibility of Dronpa and the ability to induce light dependent oligomerization of its mutants were employed in the design of the new approach to opticallly control cellular proteins.
- 7.Subach FV, Piatkevich KD, Verkhusha VV. Directed molecular evolution to design advanced red fluorescent proteins. Nat Methods. 2011;8:1019–1026. doi: 10.1038/nmeth.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Subach FV, Verkhusha VV. Chromophore transformations in red fluorescent proteins. Chem Rev. 2012;112:4308–4327. doi: 10.1021/cr2001965. This review provides the detailed description of both autocatalytic and light-induced chemical transformations of chromophores in FPs of various phenotypes.
- 9.Henderson JN, Osborn MF, Koon N, et al. Excited state proton transfer in the red fluorescent protein mKeima. J Am Chem Soc. 2009;131:13212–13213. doi: 10.1021/ja904665x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu X, Kallio K, Shi X, et al. Ultrafast excited-state dynamics in the green fluorescent protein variant S65T/H148D. 1. Mutagenesis and structural studies. Biochemistry. 2007;46:12005–12013. doi: 10.1021/bi7009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fron E, Flors C, Schweitzer G, et al. Ultrafast excited-state dynamics of the photoswitchable protein Dronpa. J Am Chem Soc. 2007;129:4870–4871. doi: 10.1021/ja069365v. [DOI] [PubMed] [Google Scholar]
- 12.Elsliger MA, Wachter RM, Hanson GT, et al. Structural and spectral response of green fluorescent protein variants to changes in pH. Biochemistry. 1999;38:5296–5301. doi: 10.1021/bi9902182. [DOI] [PubMed] [Google Scholar]
- 13.Subach FV, Zhang L, Gadella TW, et al. Red fluorescent protein with reversibly photoswitchable absorbance for photochromic FRET. Chem. Biol. 2010;17:745–755. doi: 10.1016/j.chembiol.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizuno H, Mal TK, Walchli M, et al. Light-dependent regulation of structural flexibility in a photochromic fluorescent protein. Proc Natl Acad Sci U S A. 2008;105:9227–9232. doi: 10.1073/pnas.0709599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross LA, Baird GS, Hoffman RC, et al. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizuno H, Mal TK, Tong KI, et al. Photo-induced peptide cleavage in the green-to-red conversion of a fluorescent protein. Mol Cell. 2003;12:1051–1058. doi: 10.1016/s1097-2765(03)00393-9. [DOI] [PubMed] [Google Scholar]
- 17.Subach OM, Malashkevich VN, Zencheck WD, et al. Structural characterization of acylimine-containing blue and red chromophores in mTagBFP and TagRFP fluorescent proteins. Chem. Biol. 2010;17:333–341. doi: 10.1016/j.chembiol.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subach OM, Gundorov IS, Yoshimura M, et al. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Innovation: Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005;6:885–891. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- 20. Brakemann T, Stiel AC, Weber G, et al. A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching. Nat Biotechnol. 2011;29:942–947. doi: 10.1038/nbt.1952. Dreiklang is the only rsFP, which is not turned on or off during excitation of fluorescence. The paper explains the chemical nature of chromophore transformations, and demonstrates applicability of Dreiklang in both single-molecule and ensemble based super-resolution microscopy.
- 21.Subach FV, Malashkevich VN, Zencheck WD, et al. Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states. Proc Natl Acad Sci U S A. 2009;106:21097–21102. doi: 10.1073/pnas.0909204106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson JN, Gepshtein R, Heenan JR, et al. Structure and mechanism of the photoactivatable green fluorescent protein. J Am Chem Soc. 2009;131:4176–4177. doi: 10.1021/ja808851n. [DOI] [PubMed] [Google Scholar]
- 23.van Thor JJ, Gensch T, Hellingwerf KJ, Johnson LN. Phototransformation of green fluorescent protein with UV and visible light leads to decarboxylation of glutamate 222. Nat Struct Biol. 2002;9:37–41. doi: 10.1038/nsb739. [DOI] [PubMed] [Google Scholar]
- 24.Bell AF, Stoner-Ma D, Wachter RM, Tonge PJ. Light-driven decarboxylation of wild-type green fluorescent protein. J Am Chem Soc. 2003;125:6919–6926. doi: 10.1021/ja034588w. [DOI] [PubMed] [Google Scholar]
- 25.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 26.Subach FV, Patterson GH, Renz M, et al. Bright monomeric photoactivatable red fluorescent protein for two-color super-resolution sptPALM of live cells. J. Am. Chem. Soc. 2010;132:6481–6491. doi: 10.1021/ja100906g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunewardene MS, Subach FV, Gould TJ, et al. Superresolution imaging of multiple fluorescent proteins with highly overlapping emission spectra in living cells. Biophys J. 2011;101:1522–1528. doi: 10.1016/j.bpj.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nienhaus K, Nienhaus GU, Wiedenmann J, Nar H. Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP. Proc Natl Acad Sci U S A. 2005;102:9156–9159. doi: 10.1073/pnas.0501874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adam V, Nienhaus K, Bourgeois D, Nienhaus GU. Structural basis of enhanced photoconversion yield in green fluorescent protein-like protein Dendra2. Biochemistry. 2009;48:4905–4915. doi: 10.1021/bi900383a. [DOI] [PubMed] [Google Scholar]
- 30.Tsutsui H, Shimizu H, Mizuno H, et al. The E1 mechanism in photo-induced beta-elimination reactions for green-to-red conversion of fluorescent proteins. Chem. Biol. 2009;16:1140–1147. doi: 10.1016/j.chembiol.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Adam V, Lelimousin M, Boehme S, et al. Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc. Natl. Acad. Sci. USA. 2008;105:18343–18348. doi: 10.1073/pnas.0805949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adam V, Moeyaert B, David CC, et al. Rational design of photoconvertible and biphotochromic fluorescent proteins for advanced microscopy applications. Chem Biol. 2011;18:1241–1251. doi: 10.1016/j.chembiol.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Henderson JN, Ai HW, Campbell RE, Remington SJ. Structural basis for reversible photobleaching of a green fluorescent protein homologue. Proc Natl Acad Sci U S A. 2007;104:6672–6677. doi: 10.1073/pnas.0700059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stiel AC, Trowitzsch S, Weber G, et al. 1.8 A bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants. Biochem J. 2007;402:35–42. doi: 10.1042/BJ20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andresen M, Stiel AC, Folling J, et al. Photoswitchable fluorescent proteins enable monochromatic multilabel imaging and dual color fluorescence nanoscopy. Nat. Biotechnol. 2008;26:1035–1040. doi: 10.1038/nbt.1493. [DOI] [PubMed] [Google Scholar]
- 36.Bourgeois D, Adam V. Reversible photoswitching in fluorescent proteins: a mechanistic view. IUBMB life. 2012;64:482–491. doi: 10.1002/iub.1023. [DOI] [PubMed] [Google Scholar]
- 37.Stiel AC, Andresen M, Bock H, et al. Generation of monomeric reversibly switchable red fluorescent proteins for far-field fluorescence nanoscopy. Biophys. J. 2008;95:2989–2997. doi: 10.1529/biophysj.108.130146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faro AR, Carpentier P, Jonasson G, et al. Low-temperature chromophore isomerization reveals the photoswitching mechanism of the fluorescent protein Padron. J Am Chem Soc. 2011;133:16362–16365. doi: 10.1021/ja207001y. [DOI] [PubMed] [Google Scholar]
- 39.Brakemann T, Weber G, Andresen M, et al. Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron. J Biol Chem. 2010;285:14603–14609. doi: 10.1074/jbc.M109.086314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pletnev S, Subach FV, Dauter Z, et al. A structural basis for reversible photoswitching of absorbance spectra in red fluorescent protein rsTagRFP. J Mol Biol. 2012;417:144–151. doi: 10.1016/j.jmb.2012.01.044. The paper demonstrates that reversible switching of rsTagRFP is accompanied by cis–trans isomerization and protonation/deprotonation of the chromophore, with the deprotonated cis- and protonated trans-isomers corresponding to its on and off states, respectively.
- 41.Morozova KS, Piatkevich KD, Gould TJ, et al. Far-red fluorescent protein excitable with red lasers for flow cytometry and superresolution STED nanoscopy. Biophys J. 2010;99:L13–L15. doi: 10.1016/j.bpj.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dedecker P, Mo GC, Dertinger T, Zhang J. Widely accessible method for superresolution fluorescence imaging of living systems. Proc Natl Acad Sci U S A. 2012;109:10909–10914. doi: 10.1073/pnas.1204917109. The paper describes the pcSOFI superresolution technique. In this technique, irradiating rsFP at an appropriate wavelength produces single-molecule intensity fluctuations, from which a superresolution picture can be extracted by a statistical analysis of the fluctuations in each pixel as a function of time. The two-color pcSOFI was demonstrated using Dronpa and rsTagRFP.
- 43.Fuchs J, Bohme S, Oswald F, et al. A photoactivatable marker protein for pulse-chase imaging with superresolution. Nat Methods. 2010;7:627–630. doi: 10.1038/nmeth.1477. [DOI] [PubMed] [Google Scholar]
- 44. Subach OM, Entenberg D, Condeelis JS, Verkhusha VV. A FRET-facilitated photoswitching using an orange fluorescent protein with the fast photoconversion kinetics. J Am Chem Soc. 2012;134:14789–14799. doi: 10.1021/ja3034137. Compared to PSmOrange, the orange-to-far-red PSFP, called PSmOrange2, reported in this paper has blue-shifted photoswitching action spectrum, 9-fold higher photoconversion contrast and 10-fold faster photoswitching kinetics. This results in 4-fold more PSmOrange2 molecules being photoconverted in mammalian cells.
- 45.Subach FV, Patterson GH, Manley S, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chudakov DM, Verkhusha VV, Staroverov DB, et al. Photoswitchable cyan fluorescent protein for protein tracking. Nat Biotechnol. 2004;22:1435–1439. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- 47.Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 48.Wiedenmann J, Ivanchenko S, Oswald F, et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci U S A. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKinney SA, Murphy CS, Hazelwood KL, et al. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang M, Chang H, Zhang Y, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods. 2012;9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 51.Habuchi S, Tsutsui H, Kochaniak AB, et al. mKikGR, a monomeric photoswitchable fluorescent protein. PLoS One. 2008;3:e3944. doi: 10.1371/journal.pone.0003944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoi H, Shaner NC, Davidson MW, et al. A monomeric photoconvertible fluorescent protein for imaging of dynamic protein localization. J Mol Biol. 2010;401:776–791. doi: 10.1016/j.jmb.2010.06.056. [DOI] [PubMed] [Google Scholar]
- 53.McEvoy AL, Hoi H, Bates M, et al. mMaple: a photoconvertible fluorescent protein for use in multiple imaging modalities. PLoS One. 2012;7:e51314. doi: 10.1371/journal.pone.0051314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 55.Flors C, Hotta J, Uji-i H, et al. A stroboscopic approach for fast photoactivation-localization microscopy with Dronpa mutants. J Am Chem Soc. 2007;129:13970–13977. doi: 10.1021/ja074704l. [DOI] [PubMed] [Google Scholar]
- 56.Chang H, Zhang M, Ji W, et al. A unique series of reversibly switchable fluorescent proteins with beneficial properties for various applications. Proc Natl Acad Sci U S A. 2012;109:4455–4460. doi: 10.1073/pnas.1113770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grotjohann T, Testa I, Reuss M, et al. rsEGFP2 enables fast RESOLFT nanoscopy of living cells. eLife. 2012;1:e00248. doi: 10.7554/eLife.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]