Abstract
Background
With prior studies having looked at unselected cohorts, we sought to explore the mutational landscape in a high-risk group of head and neck squamous cell carcinoma (HNSCC) tumors.
Methods
A multiplexed polymerase chain reaction (PCR) assay evaluating 68 loci in 15 genes was performed on 64 patients with high-risk HNSCC. Because of the frequent PIK3CA and AKT1 mutations in patients with oropharyngeal carcinoma, we evaluated the relationship between mutation status and both clinical/pathologic variables and tumor control in this subgroup.
Results
Seventeen of 64 patients harbored mutations in the assayed loci: 16% in PIK3CA, 9% in TP53, 2% in AKT1, and 2% in epidermal growth factor receptor (EGFR). The frequency of PIK3CA/AKT1 mutations in oropharyngeal and sinonasal primaries was increased compared to other primary sites (35% vs 6%; p = .005). There was no relationship between mutation status and overall survival (OS), disease-specific death, or progression in the oropharyngeal cohort.
Conclusion
We identified frequent PIK3CA mutations in patients with high-risk HNSCC confined predominantly to the oropharyngeal and sinonasal subsites; for the first time, mutation in AKT1 has been identified in HNSCC.
Keywords: PIK3CA, oropharyngeal, sinus, mutations, high-risk, squamous cell carcinoma
INTRODUCTION
With over 600,000 cases per year and mortality near 50%, the global burden of head and neck squamous cell carcinoma (HNSCC) is significant.1 Although the concurrent use of chemotherapy and intensity-modulated radiation therapy (IMRT) has improved survival, further advances will require the identification of novel therapeutic targets.
Toward that end, much effort has been expended in order to better characterize the molecular pathogenesis of HNSCC in the hopes of identifying potential drug targets. The discovery of frequent epidermal growth factor receptor (EGFR) overexpression in HNSCC and the development of the monoclonal antibody, cetuximab, is a notable success.2
More recently, 2 groups have looked broadly at the mutational landscape in unselected HNSCC populations using whole exome sequencing.3,4 They confirmed the presence of previously identified mutational drivers of HNSCC (TP53, cyclin-dependent kinase [CDK]N2A, phosphatase and tensin homolog [PTEN], PIK3CA, and HRAS) and, for the first time, reported inactivating NOTCH1 mutations.
Herein, we report a retrospective cohort study of 64 patients with high-risk HNSCC on whom we have genotyping data available. Although the hot-spot sequencing technique we utilized has significant limitations vis-à-vis whole exome analysis, we hoped, by using the subset of patients most likely to fail currently available treatment, to identify mutational targets of potential therapeutic value.
MATERIALS AND METHODS
Patient selection
From March 2009 to August 2011, biopsy samples from 64 patients with HNSCC treated at the Massachusetts General Hospital were selected for genotyping at the time of consultation by a multidisciplinary team of head and neck oncologists because of either (1) significant bulky nodal disease at initial presentation warranting induction chemotherapy (15 patients, 23%; 8 with N2b disease, 3 with N2c, and 4 with N3), or (2) recurrence or metastatic disease at diagnosis (49 patients; 77%). Of the 64 patients included, 23 had oropharyngeal primaries, 20 had oral cavity, 5 had hypopharyngeal, 3 had nasal cavity/sinus, 8 had laryngeal, and 5 had unknown primary. Because the number of mutations identified was highest in the oropharyngeal subset and consisted exclusively of PIK3CA/AKT1 mutations, with institutional review board approval, we then conducted a retrospective chart review to better characterize this group of 23 patients in order to examine comparative clinical outcomes. Within this subgroup, mutation analysis was conducted on 14 biopsies (61%) of primary lesions and 9 biopsies (39%) of recurrent lesions. Among the patients with oropharyngeal primaries, 7 (30%) were genotyped because of bulky nodal disease (N2b = 5; N3 = 2) that warranted induction chemotherapy and 15 (70%) were genotyped because of recurrence or metastatic disease at diagnosis.
In order to confirm the high-risk nature of the selected patients with oropharyngeal carcinoma, we compared clinical outcomes in the group of patients with oropharyngeal carcinoma selected for genotyping analysis to a representative, unselected cohort of 103 patients with locally advanced oropharyngeal carcinoma treated at the Massachusetts General Hospital during the same time period.
Genotype analysis
The assay used was a multiplexed allele-specific polymerase chain reaction (PCR) assay designed to identify somatic mutations in tumor DNA. The Diagnostic Molecular Pathology Laboratory at the Massachusetts General Hospital developed the assay using the SNaPshot method (Applied Biosystems); the assay is widely used within our hospital by clinicians working with a variety of different solid malignancies. The genotyping analysis queries 68 commonly mutated loci in 15 cancer genes (AKT1, APC, BRAF, beta-catenin, EGFR, human epidermal receptor 2 [HER2], IDH1, KIT, KRAS, MEK1, NOTCH1, NRAS, PIK3CA, phosphatase and tensin homolog [PTEN], and TP53) testing for 140 previously described somatic mutations (COSMIC database, v49 release). The PIK3CA amino acid residues evaluated include R88, E542, E545, Q546, H1047, and G1049; the AKT1 residue is E17; the TP53 residues are R175, G245, R248, R273, and R306; the PTEN residues are R130, R173, R233, and K267; and the NOTCH1 residues are L1575 and L1601.
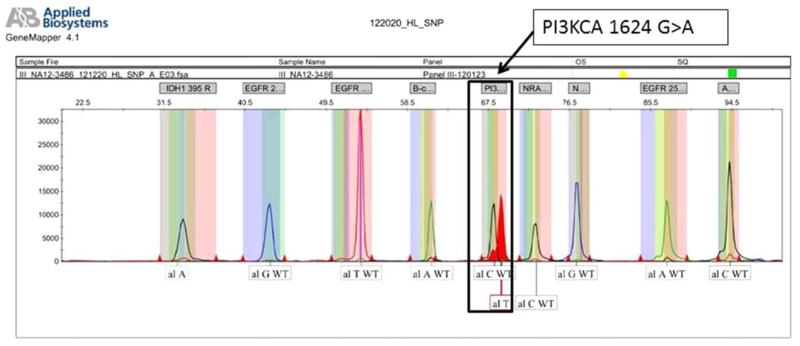
SNaPshot genotyping was performed using previously described conditions and its findings validated using direct sequencing methods in prior publications.5 Briefly, genetic regions flanking the loci of interest were co-amplified using 8 multiplexed PCR primer pools and 60 ng of total nucleic acid. PCR amplification (95 °C for 8 min followed by 45 cycles of 95 °C for 20 s, 58 °C for 30 s, and 72 °C for 1 min, and one last cylcle of 72 °C for 3 min) was followed by treatment with shrimp alkaline phosphatase and exonuclease I (USB, Cleveland, OH). Specific mutation loci were tested using 8 pools of extension primers, each annealing immediately adjacent to the nucleotide position of interest. A multiplexed single-base extension reaction that adds a fluorescently labeled ddNTP to each locus-specific probe was performed, using the ABI SNaPshot Multiplex Ready Reaction Mix (96 °C for 30 s, followed by 25 cycles of 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 30 s). Labeled extension products were resolved by capillary electrophoresis on an automatic DNA sequencer (ABI PRISM 3730 DNA Analyzer, Applied Biosystems) and data analysis was performed using GeneMapper Analysis Software version 4 (Applied Biosystems). Figure 1 shows a representative chromatogram demonstrating a PIK3CA 1624 G>A mutation leading to amino acid substitution E542K. The assay can detect mutations present at a frequency ≥5%. This technique has substantial limitations including likely underestimation of the true mutation rate within these genes (given that only certain regions were sequenced), the absence of tumor cell microdissection, and the possibility of stromal contamination because of a lack of deep sequencing.
FIGURE 1.
A representative chromatogram demonstrating a PIK3CA 1624 G>A point mutation; this leads to the substitution of glutamate for lysine at amino acid position 542. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Endpoints and statistical analyses in oropharyngeal subset
Tobacco history and alcohol use were treated as dichotomized variables; we regarded >100 cigarettes per lifetime (ever smokers) as a positive tobacco history and >14 drinks per week for men and >7 drinks per week for women (per National Institute on Alcohol Abuse and Alcoholism guidelines) as a positive alcohol history. Human papillomavirus (HPV) status was available on 19 patients (83%) and was evaluated using either p16 immunohistochemistry (n = 5) or in situ hybridization testing (n = 14) for high-risk HPV subtypes (including 16, 18, 31, and 35). Fisher’s exact test was used to evaluate associations between clinical/pathologic variables and PIK3CA/AKT mutational status. Progression was defined as the time from initial biopsy to biopsy proven recurrence or last follow-up with death from other causes as a competing risk; overall survival (OS) was defined as the time from diagnosis to death or last follow-up; time to disease-specific death was calculated from the time of initial biopsy to death because of oropharyngeal cancer or last follow-up with death from other causes as a competing risk. OS was calculated using the Kaplan-Meier product limit method; cumulative incidences of progression and disease-specific death were calculated using competing risk cumulative incidence analysis. For univariate analysis, the log-rank test was used to test for significant differences in OS between strata whereas Gray’s test was used for cumulative incidence comparisons; for multivariate analyses, we used either Cox proportional hazards regression (OS) or competing risks regression using the method of Fine and Gray (cumulative incidences of progression or disease-specific death). Mutation status was forced into all multivariate models to assess for negative confounding. In order to avoid overfitting, the size of the multivariate models were limited to 2 variables. All statistical tests were 2-sided, and a p value of < .05 was considered significant. We used R version 2.15 and STATISTICA 10.0 (Statsoft, Tulsa, OK) for all analyses.
RESULTS
Mutation frequencies in entire cohort of patients with head and neck squamous cell carcinoma
In the entire cohort, 17 of 64 patients had point mutations in the assayed loci: 16% (10) in PIK3CA, 9% (6) in TP53, 2% (1) in AKT1, and 2% (1) in EGFR. All mutations were mutually exclusive save 1 patient who had a concomitant 9 base-pair EGFR exon 20 insertion in addition to a PIK3CA H107R mutation.
The 6 mutually exclusive TP53 mutations occurred in amino acid residues R248, R306, R272, and G245 and were present in the oral cavity (3), larynx (1), and unknown primary (2) subsites. Table 1 presents relevant clinical and tumor-related data on these patients.
TABLE 1.
Clinical characteristics of patients with TP53 mutations.
| Patient no. | Mutation | Amino acid | Nucleotide | Mutation type | Biopsy@ | Subsite | T | N | Stage | HPV | ETOH | TOB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TP53 | R248L | 743G>A | Disruptive | Recurrence | Oral tongue | 3 | 1 | III | - | Neg | Neg |
| 2 | TP53 | R306* | 916C>T | Disruptive | Recurrence | Oral tongue | 4a | 0 | IVa | Neg | Neg | Neg |
| 3 | TP53 | R273H | 818G>A | Nondisruptive | Recurrence | Oral tongue | 1 | 0 | 1 | Neg | Neg | Pos |
| 4 | TP53 | R248L | 743G>T | Disruptive | Diagnosis | Larynx | 1 | 3 | IVb | Neg | Neg | Pos |
| 5 | TP53 | R248L | 743G>A | Disruptive | Diagnosis | Unknown primary | x | 3 | IVb | - | Neg | Pos |
| 6 | TP53 | G245R | 733G>C | Disruptive | Diagnosis | Unknown primary | x | 3 | IVb | - | Pos | Pos |
Abbreviations: HPV, human papillomavirus; ETOH, alcohol; TOB, tobacco; Neg, negative; Pos, positive.
Disruptive mutations were defined as those mutations resulting in a stop codon or in replacement of 1 amino acid with an amino acid of a different charge within the L2 or L3 binding domain; ETOH positivity was defined as >14 alcoholic beverages per week for men and >7 for women; TOB positivity (ever smoker) was defined as a >100 cigarettes in a lifetime.
Among the PIK3CA amino acid substitutions, 6 were E545K, 1 was H107R, 2 were E542K, and 1 was E545D; the AKT1 amino acid substitution in a patient with oropharyngeal cancer was E17K. In sum, PIK3CA/AKT1 mutations were present in 30% (7 of 23) of oropharyngeal primaries, 5% (1 of 19) of oral cavity, 66% (2 of 3) of nasal cavity/sinus, and 13% (1 of 7) of larynx; no PIK3CA/AKT1 mutations were present in hypopharyngeal (5) or unknown primaries (5). Table 2 presents tumor-related data on those with these mutations. PIK3CA/AKT1 mutations were significantly more frequent in patients with oropharyngeal and sinonasal primaries compared to other primary sites (35% vs 6%; p = .005).
TABLE 2.
Clinical characteristics of patients with PIK3CA/AKT1 mutations.
| Patient no. | Mutation | Amino acid | Nucleotide | Biopsy @ | Subsite | T | N | Stage | HPV | ETOH | TOB |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PIK3CA | E542K | 1624G>A | Diagnosis | Larynx | 4a | 0 | IVa | - | Pos | Pos |
| 2 | PIK3CA | H1047R | 3140A>G | Diagnosis | Sinonasal | 4a | 0 | IVa | - | Neg | Pos |
| 3 | PIK3CA | E545K | 1633G>A | Recurrence | Sinonasal | 2 | 0 | II | - | Neg | Pos |
| 4 | PIK3CA | E545K | 1633G>A | Diagnosis | Oral cavity | 4a | 2b | IVa | Neg | Neg | Pos |
| 5 | PIK3CA | E545K | 1633G>A | Diagnosis | Oropharynx | 4a | 2b | IVa | Pos | Neg | Pos |
| 6 | PIK3CA | E545D | 1633G>C | Diagnosis | Oropharynx | 2 | 3 | IVb | - | Neg | Pos |
| 7 | PIK3CA | E545K | 1633G>A | Recurrence | Oropharynx | 1 | 3 | IVb | Pos | Pos | Pos |
| 8 | PIK3CA | E542G | 1624G>A | Diagnosis | Oropharynx | 2 | 2b | IVa | Pos | Neg | Pos |
| 9 | AKT1 | E17K | 49G>A | Diagnosis | Oropharynx | 3 | 3 | IVb | Pos | Neg | Pos |
| 10 | PIK3CA | E545K | 1633G>A | Recurrence | Oropharynx | 3 | 1 | III | Pos | Neg | Pos |
| 11 | PIK3CA | E545K | 1633G>A | Recurrence | Oropharynx | 4a | 2b | IVa | Pos | Pos | Neg |
Abbreviations: HPV, human papillomavirus; ETOH, alcohol; TOB, tobacco; Pos, positive; Neg, negative.
HPV positive was defined as either polymerase chain reaction (PCR) amplification of a high-risk subtype or p16 positivity; ETOH positivity was defined as >14 alcoholic beverages per week for men and >7 for women; TOB positivity (ever smoker) was defined as a >100 cigarettes in a life-time.
PIK3CA-AKT1 mutations are frequent in high-risk oropharyngeal carcinoma
In order to evaluate whether the patients with oropharyngeal carcinoma selected for genotyping were truly a high-risk cohort, we compared OS and cumulative incidences of progression and disease-specific death within the genotyped cohort to an unselected group of patients with oropharyngeal squamous cell carcinoma (SCC) treated at our hospital during the same time period (2003–2010). All patients were treated with definitive, concurrent chemotherapy (either weekly cisplatinum or weekly carboplatinum and taxol) and radiation (predominantly IMRT with gross disease receiving 70 Gray). Pertinent clinicopathologic characteristics comparing the 2 groups are listed in Table 3.
TABLE 3.
Patient characteristics comparing genotyped to unselected group.
| Variable | Genotyped (n) | Unselected (n) |
|---|---|---|
| Median age | ||
| Sex | ||
| Male | 91.3% (21) | 82.5% (85) |
| Female | 8.7% (2) | 17.5% (18) |
| HPV | ||
| Positive | 79.0% (15) | 94.6% (35) |
| Negative | 21.1% (4) | 5.4% (2) |
| Tobacco status | ||
| Never | 43.5% (10) | 35.9% (37) |
| Former/active | 56.5% (13) | 64.1% (66) |
| AJCC stage | ||
| II | 4.3% (1) | 0% (0) |
| III | 47.8% (11) | 22.3% (23) |
| IVa | 26.1% (6) | 62.1% (64) |
| IVb | 17.4% (4) | 15.5% (16) |
| IVc | 4.3% (1) | 0% (0) |
| Concurrent chemotherapy | ||
| Yes | 100% (23) | 94.7% (97) |
| No | 0% (0) | 5.8% (6) |
| Induction chemotherapy | ||
| Yes | 61% (13) | 12.6% (13) |
| No | 39% (10) | 87.4% (90) |
Abbreviations: HPV, human papillomavirus; AJCC, American Joint Committee on Cancer.
Concurrent chemotherapy consisted single agent cisplatinum or carboplatinum+taxol; induction chemotherapy consisted of taxotere, cisplatin, and 5-fluorouracil.
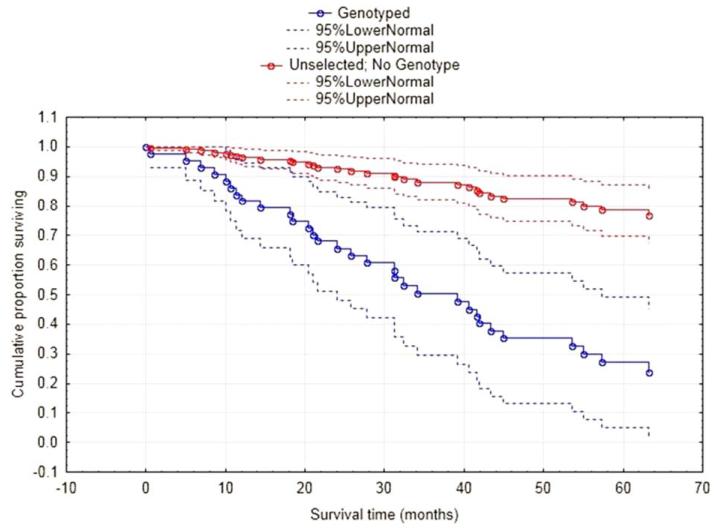
Median follow-up for all patients with oropharyngeal cancer was 58.7 months (interquartile range [IQR], 36.8–77.4); median follow-up among genotyped patients was 30.4 months (IQR, 18.9–56.8) versus 61.3 months (IQR, 43.7–61.3) for the unselected cohort. OS at 2 years in the genotyped cohort was 68.1% (95% confidence interval [CI], 51.3% to 84.8%) compared to 93.1% (95% CI, 88.7% to 97.4%) in the unselected cohort (Figure 2; p < .001). The genotyped group also had a significantly increased 2-year cumulative incidence of disease-specific death compared to the unselected cohort: 21.8% (95% CI, 6.4% to 43.1%) versus 5.0% (95% CI, 1.3% to 12.7%; p < .0001). Finally, 2-year cumulative incidence of progression was also significantly higher in the genotyped patients: 61.6% (95% CI, 36.5% to 79.1%) versus 10.0% (95% CI, 4.0% to 19.2%; p < .0001).
FIGURE 2.
Overall-survival by genotyping decision. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Among the 23 genotyped patients with oropharyngeal SCC, no mutations outside of PIK3CA or AKT1 were seen. Although 86% of genotyped patients with oropharyngeal carcinoma with PIK3CA/AKT1 mutations had a history positive for tobacco compared to 50% of wild-type, this difference was not statistically significant (p = .17). There were no statistically significant associations between either HPV status (p = .26) or alcohol history (p = 1.00) and PIK3CA/AKT1 mutational status (Table 4).
TABLE 4.
Relationship between clinical characteristics and mutation status in patients with oropharyngeal carcinoma.
| Variables | PIK3CA/AKT1 mutation (%) | Wild type (%) |
|---|---|---|
| HPV status | p = .26 | |
| Positive | 6(100) | 9 (69) |
| Negative | 0 (0) | 4(31) |
| Stage IV | p = .62 | |
| Yes | 5(71) | 13(81) |
| No | 2 (29) | 3(19) |
| Tobacco history | p = .17 | |
| Positive | 6 (86) | 8 (50) |
| Negative | 1 (14) | 8 (50) |
| Alcohol history | p = 1.0 | |
| Yes | 2 (29) | 5(31) |
| No | 5(71) | 11 (69) |
| Induction chemotherapy | p = .40 | |
| Yes | 5(71) | 8 (50) |
| No | 2 (29) | 8 (50) |
Abbreviation: HPV, human papillomavirus.
HPV positive was defined as either polymerase chain reaction (PCR) amplification of a high-risk subtype or p16 positivity; alcohol positivity was defined as >14 alcoholic beverages per week for men and >7 for women; tobacco positivity (ever smokers) was defined as >100 cigarettes in a lifetime.
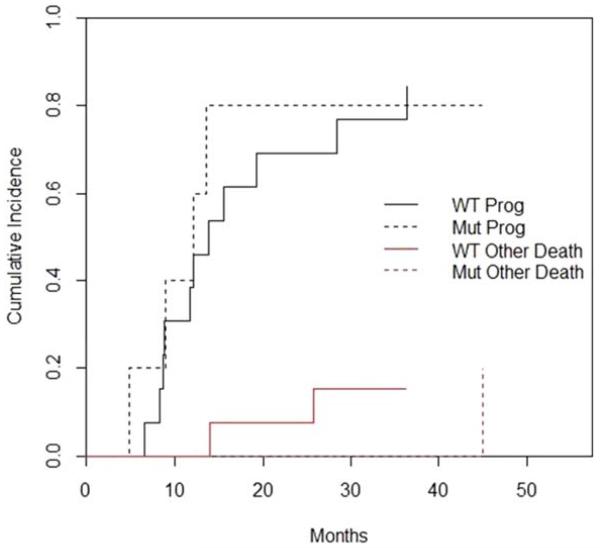
Within the genotyped cohort, 2-year cumulative incidence of progression for those with PIK3CA/AKT1 mutations was 65.7% (95% CI, 11.6% to 92.1%) versus 59.8% (95% CI, 29.7% to 80.5%) for those without (p = .96; Figure 3). Among patients with mutations, after initial treatment, 2 had locoregional failure and 1 had synchronous locoregional and distant failure; among wild-type patients, 7 had locoregional failure, 2 had distant failure, and 2 had synchronous failure. At 2 years, cumulative incidence of disease-specific death for those with mutations was 25.0% (95% CI, 0.3% to 71.3%) compared to 21.7% (95% CI, 4.8% to 46.3%) for those without (p = .89). Finally, 2-year OS for patients with mutations was 74.5% (95% CI, 46.1% to 100%) versus 78.2% (95% CI, 57.8% to 98.7%) for those without (p = .80).
FIGURE 3.
Cumulative incidence of progression and other death by mutation status. WT, wild-type; Mut, PIK3CA/AKT1 mutation; Prog, progression; other death, death from other causes without progression. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
On multivariate analysis, we controlled for tobacco history and HPV status (the 2 variables most significantly associated with mutation status on univariate testing) in order to determine whether negative confounding by these 2 variables masked any significant relationships between PIK3CA/AKT1 mutations and cumulative incidence of disease-specific death or OS. For both of these outcomes, mutation status remained nonsignificant (p = .67 and .55, respectively). For cumulative incidence of progression, because of the significant risk of overfitting, we did not conduct a multivariate analysis.
DISCUSSION
In one of the largest series to date, we found that selection for patients with high-risk HNSCC enriched for tumors with PIK3CA or AKT1 mutations (16%). To the best of our knowledge, we are the first to report both an AKT1 mutation in HNSCC and PIK3CA mutations in the sinonasal subsite.
Among patients with high-risk oropharyngeal SCC, a remarkable 30% had mutations in PIK3CA or AKT1; within this population, mutations in the PI3K-AKT pathway did not seem to impact cumulative incidence of progression, OS, or cumulative incidence of disease-specific death, even after controlling for tobacco history and HPV status in the latter 2 analyses.
We want to be up-front and clear about the potential limitations before delving into our discussion. Our study has several limitations. First, because of the sample size, more extensive multivariate modeling within the oropharyngeal subgroup risked significant overfitting. Second, there are admitted issues with selection criteria: the use of recurrence as a criterion makes prospective identification of “high risk” individuals difficult; additionally, the genotyping of those with a large disease burden for which induction chemotherapy was thought to be of potential benefit introduces significant subjectivity. Regardless, within the oropharyngeal subset, the selected patients had far worse clinical outcomes than an unselected cohort treated during the same time period at the same institution. However, the poor outcome among the genotyped oropharyngeal subgroup was largely driven by those patients with recurrent disease; those with bulky nodal disease included in the genotyped cohort had a lower absolute rate of failure. Whether this was because of the intrinsic biology of disease in these patients or to the use of induction chemotherapy in all of these patients is unclear. Regardless, if the patients with bulky nodal disease were not truly “high risk” and if PIK3CA mutations were clustered only among these patients, the hypothesized relationship between patients with high-risk oropharyngeal SCC and PIK3CA mutations may be spurious.
There is some suggestion that the PI3K pathway is upregulated in HPV-driven oropharyngeal cancer.6 The apparent association between high-risk oropharyngeal cancer and PIK3CA activating mutations could thus be completely confounded and explained by an association between PIK3CA mutations and HPV positivity, the association between HPV-driven disease and bulky lymphadenopathy, and our inclusion of patients with bulky lymph nodes in the “high risk” group. However, we found no relationship between HPV positivity and PIK3CA mutations (p = .24). Furthermore, the rate of PIK3CA mutations in only those patients with oropharyngeal carcinoma with recurrent disease (self-evidently high risk) is 33% (4 of 12). Given this, we feel confident in our conclusion that PIK3CA mutations may be more frequent in those with higher-risk oropharyngeal carcinoma. However, small numbers limit definitive conclusions regarding PIK3CA mutations and other clinical variables.
An additional limitation arises when we consider that recurrent lesions were assayed in 39% of oropharyngeal tumors sampled. In these instances, it is impossible to determine whether the mutation was present in a substantial number of clones at diagnosis or if mutation-bearing clones were selected and expanded by virtue of treatment resistance. However, if we limit our analysis only to those patients with oropharyngeal cancer who had initial biopsies genotyped, the frequency of PIK3CA or AKT1 mutations is 31% (4 of 13). The lack of normal tissue controls must also be acknowledged as a significant limitation, although in prior reports the loci assayed and the mutations identified have all been tumor-specific. We also have no downstream biological data to suggest that these mutations truly upregulate their designated pathways. Finally, the assay used examined only specific loci, did not involve the sequencing of entire genes, and did not microdissect out tumor cells; for all of these reasons, it is quite possible that we have underestimated the true rate of mutations in the genes examined.
Although the caveats above must be taken seriously, we do believe our findings help to elucidate the underlying biology of high-risk HNSCC and thus warrant further discussion.
Mutational landscape in high-risk head and neck squamous cell carcinoma
In the 17 high-risk patients with identified point mutations, 16 patients had mutually exclusive alterations in either TP53 or PIK3CA. Somatic mutations in TP53, the gene that encodes for p53 protein, a crucial cell cycle regulator, are present in approximately 40% to 80% of unselected HNSCC tumors.3,4,7 Comparatively, the frequency of TP53 missense and nonsense mutations in our own cohort was only 9%. The small number of TP53 mutations is undoubtedly because of the relatively small number of loci examined by our platform; SNaPshot analysis would have identified only 8% to 17% of the TP53 mutations picked up by the whole exome analyses. However, the absence of TP53 mutations in our oropharyngeal subset is unsurprising given the high frequency of HPV positivity; in most instances, HPV-positive oropharyngeal SCCs are TP53 wild-type.
Prior whole exome analyses of unselected HNSCC tumors also found a significant number of loss-of-function mutations in NOTCH1, a gene whose protein product plays an important role in cell fate decisions during normal development. Of interest, we did not identify any mutations in the 2 loci within the NOTCH1 gene that we examined. However, the residues we examined, L1575 and L1601, are sites of previously identified activating mutations; given the results of the whole exome analyses, we would not expect mutations at these loci.
The PI3K-AKT pathway, downstream of the ERBB tyrosine-kinase receptor family, when activated, promotes cell survival, proliferation, and radioresistance and has been previously implicated in head and neck tumorigenesis8,9; missense mutations and deletions in the primary negative regulator of this pathway, PTEN deleted on chromosome 10, have also been reported in unselected populations of patients with HNSCC. We examined several loci within AKT1, PTEN, and PIK3CA in our high-risk cohort.
Several groups have reported overexpression of phosphorylated Akt and amplification of AKT2 in HNSCC10-12; however, we are the first to identify point mutations in AKT1 in HNSCC. The specific missense mutation (E17K) identified with the plekstrin homology domain (PHD) of AKT1 results in constitutive activation of the protein.13 Previously, the mutation had been found in small numbers of breast, colorectal, prostate, ovarian, lung, and endometrial cancers.14-16 Functionally, by substituting glutamate for lysine and increasing the affinity of Akt for its phosphoinositide ligand, the mutation results in pathological localization of the protein to the plasma membrane, leading to Akt phosphorylation and thus activation.
PTEN mutations occur in HNSCC at frequencies ranging from 10% to 15%.4,17,18 We examined 4 loci in PTEN associated with inactivating missense mutations or premature stop codons and found no alterations. Although this may be because of the limited number of loci examined, there is some evidence to suggest that maintenance or overexpression of PTEN portends worse prognosis in HNSCC because of increased radioresistance.19 Although controversial,20 if true, this may mean that in our high-risk cohorts, inactivating mutations in PTEN were selected against.
Finally, copy number increases in PIK3CA (the gene that encodes the 110 kDa catalytic subunit of PI3K) are quite common in HNSCC.10,21 Several groups have investigated PIK3CA activating mutations in populations of unselected patients with HNSCC using PCR amplification of hot-spot regions within the gene; reported frequencies of these mutations range from 0% to 11%.22-25 Studies using whole exome sequencing found similarly low frequencies of PIK3CA mutations (6% to 8%).3,4 In both the targeted and whole exome analyses, the PIK3CA activating mutations were nearly all confined to residues H1047, E545, and E542. Although we evaluated additional codons, our own cohort confirms this distribution. However, we report a higher overall frequency of PIK3CA mutations (16%) in this high-risk cohort than in the unselected cohorts referenced above. More striking still is the frequency of PIK3CA mutations seen in the oropharyngeal (26%) and sinonasal (66%) subgroups.
Although our number of patients with sinonasal SCC is small (n = 3), we are the first to report the presence of PIK3CA mutations in this HNSCC subsite (both nasal cavity tumors). Because the molecular pathogenesis of sinonasal SCC is largely unexplored, with prior studies having focused largely on TP53 and EGFR mutations and amplifications,26,27 the presence of PIK3CA activating mutations in 2 of 3 patients is particularly noteworthy. A larger number of patients with sinonasal SCC must be examined before general comments on frequency can be made.
Specific investigations of PIK3CA/AKT1 mutations within oropharyngeal SCC are few. The whole exome analysis by Stransky et al4 found a frequency of PIK3CA mutations within their oropharyngeal cohort of approximately 12.5% (2 of 16). Agrawal et al3 found a similarly low number in their subpopulation of 31 patients with oropharyngeal SCC (1%; 1 of 31). Qiu et al25 identified PIK3CA activating mutations in 20.8% of pharyngeal carcinomas evaluated; however, it was impossible to determine what percentage of their subpopulation involved cancers of the oropharynx and how many of those had PIK3CA mutations.
Given the lower frequencies reported in the literature, our finding that 30% of patients with high-risk (as defined by both selection criteria and comparative clinical outcomes) oropharyngeal SCC had activating mutations in either PIK3CA or AKT1 suggested both that these mutations are enriched in this high-risk cohort and that their presence might portend a worse prognosis.
Clinical outcomes in high-risk oropharyngeal squamous cell carcinoma
Because of the high frequency of these mutations within the oropharyngeal subgroup, we then conducted further evaluation of the association between PIK3CA/AKT1 mutations and both clinicopathologic factors and outcomes. We found a trend toward a significant positive relationship between the presence of PIK3CA/AKT1 mutations and tobacco history. It is possible that, with increasing numbers of patients, this relationship would become significant. None of the other clinical variables evaluated were significantly related to PIK3CA/AKT1 mutations.
We then compared clinical outcomes in the PIK3CA-AKT1 pathway mutation-positive patients with oropharyngeal SCC to patients with high-risk oropharyngeal SCC without mutation. On univariate analyses, PIK3CA/AKT1 mutations were not significantly associated with OS, cumulative incidences of progression, or disease-specific death. We chose to conduct multivariate regression analyses controlling for the 2 clinical variables that suggested even a trend toward association with PIK3CA/AKT1 mutations: tobacco history and HPV status; we were concerned that these 2 well-established prognostic factors might mask a relationship between mutation status and clinical outcomes. However, even after controlling for HPV and tobacco status, we could discern no significant relationship between mutation status and cumulative incidence of disease-specific death or OS.
A prior report by Yu et al28 demonstrated that after multivariate adjustment, high levels of intratumoral phosphorylated Akt were associated with worse outcomes in oropharyngeal SCC. The PIK3CA/AKT1 activating mutations that we identified in our cohort should lead to a significant increase in Akt activity. Our a priori hypothesis was that such mutations might portend a worse prognosis. However, because the wild-type comparator group was also high-risk, with uniformly poor outcomes, discerning any subtle relationship between mutation status and tumor control would have been difficult. We are currently working to genotype a larger group of unselected oropharyngeal tumors in order to further evaluate the impact of PIK3CA/AKT1 mutations on clinical outcomes.
CONCLUSIONS
Ultimately, more robust, prospective data are needed to confirm our findings and determine prognostic and predictive significance of these mutations in both the patients with high-risk oropharyngeal SCC and patients with high-risk HNSCC more broadly. However, future clinical trials of PI3K-AKT pathway inhibitors should consider screening patients with HNSCC with high-risk features (eg, advanced nodal disease and large primary lesions), perhaps focusing specifically on those with sinonasal or oropharyngeal primaries, so as to select a population most likely to benefit from these targeted agents.
Acknowledgments
This work was supported by National Institute of Health/National Institute of Dental & Craniofacial Research K08DE020139 (S.M.R.).
Contract grant sponsor: National Institutes of Health; contract grant number: R01 DE015945
REFERENCES
- 1.Amonkar MM, Chastek B, Samant N, Teitelbaum A. Economic burden of resected squamous cell carcinoma of the head and neck in a US managed-care population. J Med Econ. 2011;14:421–432. doi: 10.3111/13696998.2011.584096. [DOI] [PubMed] [Google Scholar]
- 2.Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010;11:21–28. doi: 10.1016/S1470-2045(09)70311-0. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fury MG, Drobnjak M, Sima CS, et al. Tissue microarray evidence of association between p16 and phosphorylated eIF4E in tonsillar squamous cell carcinoma. Head Neck. 2011;33:1340–1345. doi: 10.1002/hed.21621. [DOI] [PubMed] [Google Scholar]
- 7.Balz V, Scheckenbach K, G€otte K, Bockm€uhl U, Petersen I, Bier H. Is the p53 inactivation frequency in squamous cell carcinomas of the head and neck underestimated? Analysis of p53 exons 2-11 and human papillomavirus 16/18 E6 transcripts in 123 unselected tumor specimens. Cancer Res. 2003;63:1188–1191. [PubMed] [Google Scholar]
- 8.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 9.Bussink J, van der Kogel AJ, Kaanders JH. Activation of the PI3-K/AKT pathway and implications for radioresistance mechanisms in head and neck cancer. Lancet Oncol. 2008;9:288–296. doi: 10.1016/S1470-2045(08)70073-1. [DOI] [PubMed] [Google Scholar]
- 10.Amornphimoltham P, Sriuranpong V, Patel V, et al. Persistent activation of the Akt pathway in head and neck squamous cell carcinoma: a potential target for UCN-01. Clin Cancer Res. 2004;10(12 Pt 1):4029–4037. doi: 10.1158/1078-0432.CCR-03-0249. [DOI] [PubMed] [Google Scholar]
- 11.Pedrero JM, Carracedo DG, Pinto CM, et al. Frequent genetic and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head and neck squamous cell carcinoma. Int J Cancer. 2005;114:242–248. doi: 10.1002/ijc.20711. [DOI] [PubMed] [Google Scholar]
- 12.Ongkeko WM, Altuna X, Weisman RA, Wang-Rodriguez J. Expression of protein tyrosine kinases in head and neck squamous cell carcinomas. Am J Clin Pathol. 2005;124:71–76. doi: 10.1309/BTLN5WTMJ3PCNRRC. [DOI] [PubMed] [Google Scholar]
- 13.Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 14.Boormans JL, Hermans KG, van Leenders GJ, Trapman J, Verhagen PC. An activating mutation in AKT1 in human prostate cancer. Int J Cancer. 2008;123:2725–2726. doi: 10.1002/ijc.23787. [DOI] [PubMed] [Google Scholar]
- 15.Malanga D, Scrima M, De Marco C, et al. Activating E17K mutation in the gene encoding the protein kinase AKT1 in a subset of squamous cell carcinoma of the lung. Cell Cycle. 2008;7:665–669. doi: 10.4161/cc.7.5.5485. [DOI] [PubMed] [Google Scholar]
- 16.Cohen Y, Shalmon B, Korach J, Barshack I, Fridman E, Rechavi G. AKT1 pleckstrin homology domain E17K activating mutation in endometrial carcinoma. Gynecol Oncol. 2010;116:88–91. doi: 10.1016/j.ygyno.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 17.Okami K, Wu L, Riggins G, et al. Analysis of PTEN/MMAC1 alterations in aerodigestive tract tumors. Cancer Res. 1998;58:509–511. [PubMed] [Google Scholar]
- 18.Shao X, Tandon R, Samara G, et al. Mutational analysis of the PTEN gene in head and neck squamous cell carcinoma. Int J Cancer. 1998;77:684–688. doi: 10.1002/(sici)1097-0215(19980831)77:5<684::aid-ijc4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 19.Pattje WJ, Schuuring E, Mastik MF, et al. The phosphatase and tensin homologue deleted on chromosome 10 mediates radiosensitivity in head and neck cancer. Br J Cancer. 2010;102:1778–1785. doi: 10.1038/sj.bjc.6605707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snietura M, Jaworska M, Mlynarczyk-Liszka J, et al. PTEN as a prognostic and predictive marker in postoperative radiotherapy for squamous cell cancer of the head and neck. PloS One. 2012;7:e33396. doi: 10.1371/journal.pone.0033396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris LG, Taylor BS, Bivona TG, et al. Genomic dissection of the epidermal growth factor receptor (EGFR)/PI3K pathway reveals frequent deletion of the EGFR phosphatase PTPRS in head and neck cancers. Proc Natl Acad Sci U S A. 2011;108:19024–19029. doi: 10.1073/pnas.1111963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenic I, Steger K, Gruber C, Arens C, Woenckhaus J. Analysis of PIK3CA and Akt/protein kinase B in head and neck squamous cell carcinoma. Oncol Rep. 2007;18:253–259. [PubMed] [Google Scholar]
- 23.Janku F, Tsimberidou AM, Garrido-Laguna I, et al. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther. 2011;10:558–565. doi: 10.1158/1535-7163.MCT-10-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murugan AK, Hong NT, Fukui Y, Munirajan AK, Tsuchida N. Oncogenic mutations of the PIK3CA gene in head and neck squamous cell carcinomas. Int J Oncol. 2008;32:101–111. [PubMed] [Google Scholar]
- 25.Qiu W, Sch€onleben F, Li X, et al. PIK3CA mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12:1441–1446. doi: 10.1158/1078-0432.CCR-05-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López F, Llorente JL, Oviedo CM, et al. Gene amplification and protein overexpression of EGFR and ERBB2 in sinonasal squamous cell carcinoma. Cancer. 2012;118:1818–1826. doi: 10.1002/cncr.26451. [DOI] [PubMed] [Google Scholar]
- 27.Holmila R, Bornholdt J, Suitiala T, et al. Profile of TP53 gene mutations in sinonasal cancer. Mutat Res. 2010;686:9–14. doi: 10.1016/j.mrfmmm.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Yu Z, Weinberger PM, Sasaki C, et al. Phosphorylation of Akt (Ser473) predicts poor clinical outcome in oropharyngeal squamous cell cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:553–558. doi: 10.1158/1055-9965.EPI-06-0121. [DOI] [PubMed] [Google Scholar]