Abstract
The past decade has seen explosive growth in new high speed imaging methods. These can broadly be classified as either point-scanning (which offer better depth penetration) or parallelized systems (which offer higher speed). We discuss each class generally, and cover specific advances in diffraction-limited microscopes (laser-scanning confocal, spinning-disk, and light-sheet) and super-resolution microscopes (single-molecule imaging, stimulated emission-depletion, and structured illumination). A theme of our review is that there is no free lunch: each technique has strengths and weaknesses, and an advance in speed usually comes at the expense of either spatial resolution or depth penetration.
Introduction
Fluorescence microscopy provides a unique combination of high contrast and molecular specificity that is well suited to a wide array of research areas, from cell biology [1] to neuroscience [2]. The last decade has seen explosive growth in new imaging techniques, tremendously improving the performance of fluorescence microscopes. These new methods make it challenging to determine which technique is appropriate for a given experiment, as many factors--including spatial resolution, imaging speed and the desired sample penetration--must be considered.
Although developments in hardware and brighter, more photostable fluorophores continue to result in faster and more sensitive imaging, there are still inherent speed limitations in fluorescence microscopy (Fig. 1). Existing fluorescence microscopes can be broadly divided into two classes – point-scanning and parallelized systems. Point-scanning microscopes (such as laser-scanning confocal microscopy, LSCM) scan a single excitation focus through the sample, mapping the resulting fluorescence from each scan position to a unique pixel in the image. It is often assumed that the speed of point-scanning systems can be improved by simply increasing the scan speed, yet the resulting decrease in per-pixel dwell time lowers the total signal and degrades the image’s signal-to-noise ratio (SNR). Increasing the illumination intensity compensates for this effect, but can also result in higher levels of photodamage and photobleaching (and at high intensities these processes can scale nonlinearly with intensity). Also, given the finite pool of fluorophores in the sample, above a certain illumination intensity effectively all fluorophores are excited and further increases in intensity are of no benefit. Higher speed, or higher SNR at the same speed, can be achieved by parallelizing excitation (i.e. using multiple simultaneous excitation foci to illuminate the sample). Widefield microscopy (illuminating the entire sample volume at once) exemplifies the highest degree of parallelization, thus offering the fastest image acquisition rates. However, this increased acquisition speed comes at a price, as any degree of parallelization results in ‘crosstalk’ between spatially distinct points in the sample, degrading optical sectioning and contaminating the in-focus signal with scattered light.
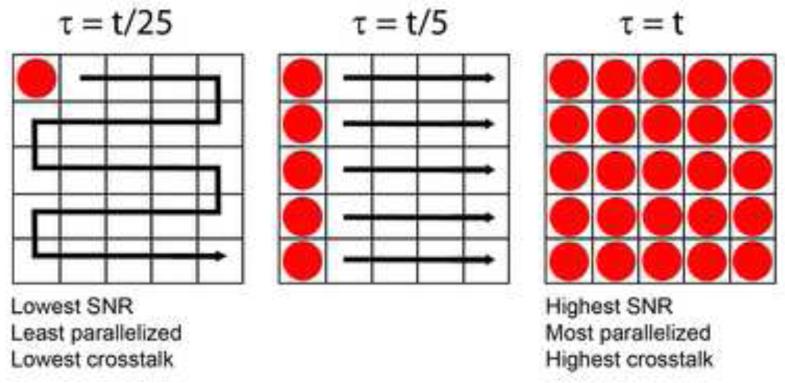
Fig. 1. Effects of parallelizing excitation.
Left: a single excitation spot (red) is scanned through a 5 × 5 = 25 pixel grid, illuminating one pixel at a time. This serial illumination is reminiscent of point-scanning confocal microscopy, and implies that for total frame exposure time t (the time required to illuminate all pixels), the per-pixel dwell time is only τ = t/25. Note that in this case, there is no crosstalk as pixels are illuminated sequentially. Middle: excitation has been partially parallelized, so that an entire column of pixels is excited at once (similar to line-scanning confocal microscopy). The per-pixel dwell time is now τ = t/5 for total exposure time t, increasing the signal-to-noise ratio (SNR) relative to the point-scanning case at left. Since multiple pixels are illuminated simultaneously, emission originating from spatially distinct regions in the sample (especially from outside the focal plane) may leak over to neighboring pixels, resulting in crosstalk and degrading optical sectioning relative to the point-scanning case. Another example of partially parallelized excitation is the spinning disk confocal microscope. Right: all pixels are simultaneously illuminated (similar to widefield microscopy), so for frame exposure time t, each pixel is also exposed for time τ = t. SNR is maximized, but so is pixel crosstalk, and optical sectioning is worse than either the point- or line-scanning case.
High speed imaging at the diffraction-limit
Point-scanners image large volumes much more slowly than parallelized systems, but in certain applications they are preferred. For example, when imaging deep into samples (especially when coupled with multiphoton excitation), robust performance in the presence of scattering is often as desirable as imaging fast. Additionally, when recording from multiple sites in live samples (as in functional imaging), scanning the entire volume is unnecessary and point-scanners can be advantageously used to sample arbitrary regions of interest (‘random access scanning’). A major limitation of these systems has been slow scan speed in the axial direction, resulting from the need to move a relatively massive objective or sample chamber during refocusing.
One solution is to use a customized, light-weight mirror to rapidly translate the excitation at a location upstream of the sample, and then refocus this excitation at the sample plane [3]. Such ‘remote refocusing’ permits kHz scan rates over hundreds of microns in all three dimensions, enabling, for example, the study of neuronal activity in populations of neurons (Fig. 2a-c). Other routes to high speed 3D scanning are to use acousto-optic scanning technology to rapidly move the excitation focus [4] or to increase the number of excitation foci (i.e. by parallelization). For example, multiplexing 4 pulsed two-photon (2P) beams that are offset spatially and temporally yields a 4× increase in speed, and was used to image neural activity in intact mouse brains [5].
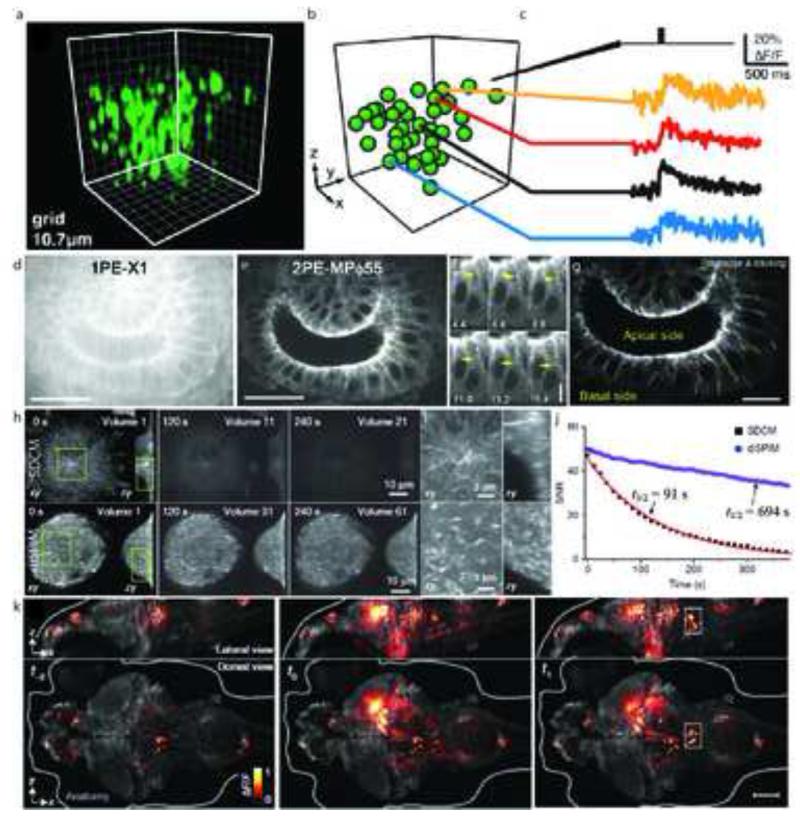
Fig. 2. High speed imaging at the diffraction-limit.
(a) Functional imaging of cortical neurons was performed in tissue slabs with a temporal resolution of 1kHz, along a cyclic trajectory. The image shows a 3D rendering of neurons of interest in the scan volume (100×100×100μm) loaded with OGB1-AM. (b) Segmentation of (a) representing neurons as spheres. (c) Fluorescence measurements from selected neurons in response to stimulation with an extracellular electrical burst. (d) 1PE and (e) 2PE excitation images of a live drosophila embryo expressing EB1-GFP. Images were collected approximately 25 μm into the sample and consisted of horizontal sections of the hindgut as observed from the dorsal side. EB1 comets, indicated by yellow arrows (f) are observed in 2PE, but not 1PE. (g) Tracked EB1 comet paths are shown with colored lines. (h) Bleaching comparison between SDCM and diSPIM, on cells expressing EB3-GFP. At similar initial SNR, cells imaged with SDCM were sampled 3× less frequently than diSPIM and with 3.2× fewer planes per volume, but exhibited greatly increased bleaching rates. (i) Higher magnification views of the rectangular subregions indicated in (h), emphasizing the superior axial resolution obtained in diSPIM compared to SDCM. (j) Bleaching rates were calculated from a 20×20 μm area inside each cell; SNR was calculated as the ratio of the measured area inside the cell to an area outside the cell. Data were fitted to single exponentials; bleaching half times are indicated on the graph. (k) Whole brain imaging of neuronal activity in live Zebrafish embryos reported by GCaMP5G, a genetically encoded calcium indicator, in elavl3:GCaMP5G fish. Changes in GCaMP5G fluorescence are superimposed over the reference anatomy. Panels represent whole-brain volumes recorded at time intervals of 1.39 s. Nearly simultaneous activation of numerous neurons occurs at t0 spanning both the midbrain and hindbrain. Scalebars: (d,e): 20 μm; (f): 5 μm; (g): 10 μm; (k): 100 μm. Panels (a-c) are adapted from Botcherby et al. [3] with permission from PNAS. Panels (d-g) are adapted from Shimozawa et al. [6] with permission from PNAS. Panels (h-j) are adapted from Wu et al. [9] with permission from Nature. Panel (k) is adapted from Ahrens et al. [13] with permission from Nature.
Improvements have also been made to more highly paralellized systems, such as spinning disk confocal microscopy (SDCM). By increasing the interpinhole distance and utilizing two photon excitation, pinhole crosstalk can be reduced, permitting imaging at depths greater than 100 μm and enabling the tracking of microtubule plus-end binding protein EB1 25 μm into live drosophila embryos (Fig. 2d-g) [6]. Another parallelized technique that can achieve high imaging rates is temporal focusing, in which a laser pulse is scanned across the sample on a picosecond timescale. In conjunction with an image-intensifier and a fast, scientific complementary metal-oxide-semiconductor (sCMOS) camera, this technique has enabled measurement of calcium activity at 80 Hz throughout 99 neurons in the brain of an adult nematode [7].
Most fluorescence microscopes, even those using multiphoton excitation, dose the imaging volume outside the focal plane causing unnecessary photodamage. Light sheet fluorescence microscopy (LSFM) solves this problem by illuminating only the sample plane currently in focus. Since an entire line- or plane within the sample is illuminated simultaneously, excitaiton is parallelized and imaging rates and SNR are much improved relative to point-scanning systems.
LSFM has proved especially useful in developmental- and neurobiology. For example, LSFM on an inverted microscope base was used to follow the development of individual neurons at volumetric imaging rates of 0.5 Hz for periods exceeding 8 hrs in C. elegans embryos [8]. The spatial and temporal resolution of this inverted design has been recently improved by rapidly capturing and registering two perpendicular specimen views and performing joint-deconvolution on the result (Figure 2h-j) [9]. LSFM has also been applied to embryogenesis studies in zebrafish and drosophila embryos [10-12] at speeds sufficient for following cellular development and neural activity, including a recent implementation that used multiple illumination objectives to obtain whole brain functional imaging of zebrafish larvae with cellular resolution at 0.8 s/volume (Figure 2k) [13].
Faster super-resolution imaging
Super-resolution microscopes extract higher resolution spatial information than would be possible in standard ‘diffraction-limited’ systems (where resolution is typically limited to ~250 nm laterally, and ~500 nm axially). Regardless of their operating principle, all super-resolution microscopes come with a hefty price. Since the pixel size in an image must be at least 2× as fine as the desired resolution (the Nyquist criterion), an N-fold increase in image resolution in D-dimensions requires an ND-fold decrease in pixel size. In order to achieve this pixel size without comprising SNR or imaging rate, the signal collection rate must increase by at least ND. A microscope which doubles resolution in 3D must therefore collect at least ~8× as much signal in the same time as a conventional microscope for the same SNR. For point-scanning systems, the signal collection rate must be increased by N2D to maintain the same imaging speed, due to the required reduction in pixel dwell time. These rather stringent requirements can be met by increasing the density of available fluorophores or by increasing the excitation intensity. In practice, finite label density, photodamage and sample perturbation make it difficult to maintain either the SNR or imaging speed available in conventional microscopy. For these reasons, images acquired at high speed by super-resolution microscopy are often quite noisy.
One super-resolution method, single-molecule imaging (SMI), sequentially excites random subsets of photoswitchable fluorophores, localizes the centers of these molecules (typically to a precision ~10× better than the diffraction limit), and constructs a single super-resolved image from the aggregate localizations. Extracting the best possible resolution in SMI relies critically on the choice of the dye [14, 15], and to a lesser extent on the algorithms used in constructing the super-resolved images. Even though brighter and faster switching fluorophores continue to improve the imaging rate [16], and these techniques are inherently parallelized due to widefield illumination and detection, imaging speed is still inherently limited by the number of molecules that may be switched on and localized at once. A partial remedy is using multi-emitter fitting algorithms that permit greater numbers of molecules to be localized in any given frame [17, 18]. When used with sCMOS detectors, these algorithms allow super-resolved images to be obtained at rates as high as ~30 frames per second [19]. Other strategies do not attempt to localize individual emitters; instead they rely on iterative deconvolution [20] or compressed sensing [21] to increase the limits on active emitter density.
A particularly fast SMI technique is Bayesian analysis of blinking and bleaching (3B). In 3B, an analysis of the fluctuations of all fluorophores in the dataset is used to model the most likely distribution of fluorophore locations. The practical benefit is that many highly overlapping fluorophores can be turned on in an individual frame, greatly reducing the number of raw images required to construct a super-resolved image (Figure 3a-e) [22, 23]. The greatest bottleneck to 3B microscopy is the excessive time required to process the raw data (a 15 × 15 pixel region comprising 200 raw frames requires 6 hours on a 3.3 GHz single core machine). However, this limitation can be ameliorated by paralellization on cluster or cloud computing systems [24].
Fig. 3. Fast super-resolution imaging.
Comparative 3B (a) and widefield (b) images of immunolabeled, fixed podosomes. Images were acquired in 6 s. (c-e) Higher magnification views of white, blue, and green rectangles in (a) to emphasize resolution. (f) Widefield and (g) parallelized RESOLFT images of rsEGFP-labeled keratin in a live cell, acquired in 0.4 s. Insets represent higher magnification views of rectangle in (g). The marked fiber has an apparent diameter of 95 nm in the RESOLFT image. (h-j) Instant SIM images of GFP-labeled endoplasmic reticulum in a live cell. (h): overview; (i, j): higher magnification views of white rectangle and square in (h), emphasizing fast temporal dynamics. Arrows in (j) highlight growth and remodeling of individual ER tubules. Images were acquired at 100 Hz. Scalebars: (a,b): 2 μm; (c-e): 500 nm; (f): 10 μm; (g): 5 μm; (h): 200 nm; (i, j): 10 μm (insets: 1 μm). Panels (a-e) are adapted from Cox et al. [22] with permission from Nature. Panels (f-g) are adapted from Chmyrov et al. [34] with permission from Nature. Panels (h-j) are adapted from York et al. [37] with permission from Nature.
As noted above, increasing imaging speed, especially for super-resolution imaging, requires an accompanying increase in light dose. In SMI, where hundreds or thousands of raw frames are usually required for a single super-resolution image, photobleaching and photodamage are serious concerns (especially when imaging in 3D or over extended durations). Limiting the excitation to the vicinity of the focal plane with multiphoton illumination [25] or LSFM [26, 27] can help in these applications.
Another widely utilized super-resolution technique is stimulated emission depletion (STED) microscopy. STED forces fluorophores at the periphery of a point of illumination to undergo stimulated emission instead of fluorescence, creating a subdiffractive excitation focus that can be scanned throughout the sample. This scanning makes STED inherently slower than widefield techniques, especially over large areas. As such, STED benefits from parallelization, and a STED with 4 beams has been developed that improves speed 4-fold [28]. Although the tremendous peak power intensities required for STED (MW/cm2) limit its effectiveness in some live applications, it has been successfully used to image neuronal morphology in living mice [29, 30].
Instead of stimulated emission, the contrast between bright and dark states in reversibly-switchable fluorescent proteins can be used to generate a subdiffractive focus in the sample (reversible saturable optical fluorescence transitions, RESOLFT). RESOLFT can be performed at much lower intensities (1-10 kW/cm2) than in STED [31], and has been used to study morphological changes in neurons with a time resolution of several seconds, over many hours, 10-50 μm into living hippocampal brain slices [32]. The development of faster switching fluorescent proteins improves image acquisition rates [33], as does parallelization (Fig. 3f-g) [34].
The fastest method of super-resolution microscopy is currently linear structured illumination microscopy (SIM). This technique doubles the resolution of a widefield microscope without increasing excitation intensities significantly and can be used with conventional (non-switchable) fluorophores. SIM uses contributions from both excitation and emission point spread functions to improve spatial resolution, employing sharply patterned light for illumination while collecting fluorescence on a camera and using mathematical processing and deconvolution to enhance resolution. Recent implementations have achieved resolutions of 120 nm laterally and 360 nm axially with volumetric temporal resolution approaching 1 Hz for >100 cellular volumes [35, 36]. Other SIM designs use point-based illumination patterns to confer better sectioning and to improve depth penetration, and have been implemented in point-scanning [37], and parallelized modes [38, 39].
Very recently, the mathematical post-processing operations required in most SIM have been eliminated entirely, enabling a 10-100 fold improvement in imaging rates (Figure 3 h-j) [40]. As with other super-resolution microscopy, the duration of SIM is ultimately limited by photobleaching and photodamage. Implementing SIM in an LSFM geometry can mitigate this issue, albeit with diminished resolution [41]. Finally, use of switchable fluorescent proteins can increase the resolution of SIM (‘nonlinear SIM’) in a manner similar to RESOLFT, by creating effectively finer regions of excitation in the sample. Like RESOLFT, the speed of these techniques is primarily limited more by available switchable fluorophores than by hardware [42].
Alternative high speed imaging strategies
A few more ‘exotic’ imaging methods improve speed through creative use of hardware. An alternative parallelization scheme uses acousto-optical devices to simultaneously excite multiple points in the sample at distinct radiofrequencies (Fig. 4a), capturing the fluorescence on a point detector and using software post-processing to enable an effective imaging rate of up to 4.4 kHz [43]. While, another route to high speed imaging trades spatial resolution and field of view for temporal resolution by using a fast digital micromirror device (DMD) to rapidly and sequentially shutter subsets of pixels during a single camera exposure (Fig. 4b). Such ‘temporal pixel multiplexing’ has been used to record calcium transients in cardiac myocytes [44]. However, the current implementation of this technology is inefficient, as the DMD deflects the majority of emission light away from the camera. Nevertheless, for sufficiently bright samples where spatial resolution is not paramount, temporal pixel multiplexing provides a straightforward means of converting excess detector pixels into increased temporal resolution.
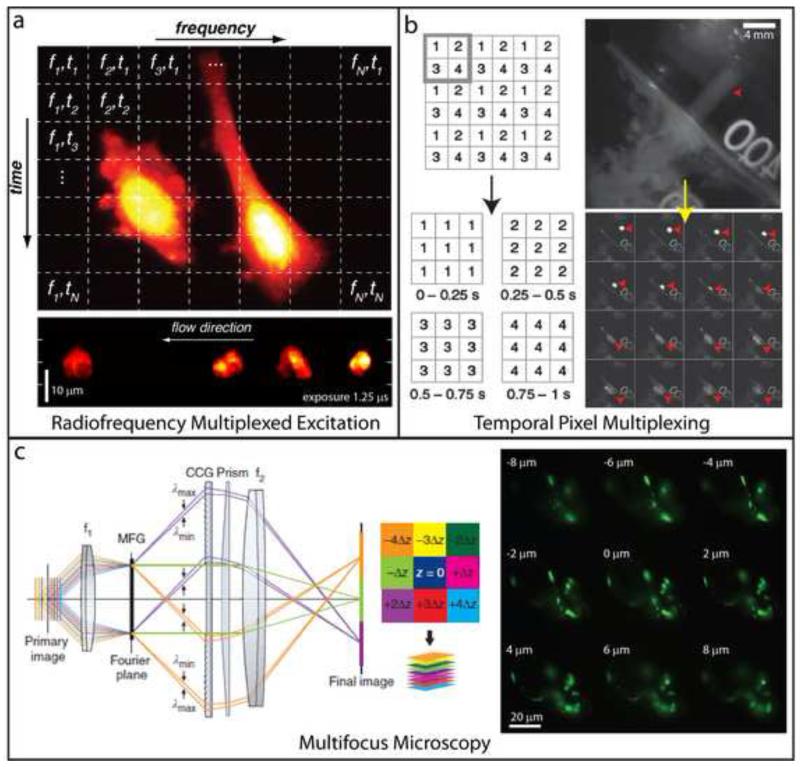
Fig. 4. Alternative strategies for high-speed fluorescence imaging.
(a) Top: Radiofrequency multiplexed excitation concept. Each pixel in the horizontal direction is illuminated at a different radiofrequency. Scanning the excitation in the vertical direction ensures that successive rows of pixels are illuminated sequentially, thus resulting in a unique ‘tag’ for each pixel in the image. Emission is collected on a fast point-detector and post-processing algorithms are applied to reconstruct an image. The method attains kHz or faster imaging rates, sufficient to ‘freeze’ flowing cells moving at 1 m/s (bottom). (b) Left: Temporal pixel multiplexing (TPM) concept. An image is divided into subregions (e.g. gray square), and neighboring pixels in each subregion are illuminated sequentially during image acquisition. Combining all pixels from a given illumination period (blocks of 1s, 2s, 3s, and 4s in bottom graphic) results in an image sequence with reduced spatial resolution, but increased temporal resolution (here a high resolution image acquired in one second is converted into a sequence of 4 lower resolution images, each acquired in 0.25 s). Right: An example highlighting a high spatial resolution scene of a milk droplet falling into water, acquired in 40 ms. Motion blur obscures the droplet (red arrowhead). Applying the TPM concept subdivides the image into a series of 16 lower resolution images, each spanning a 2.5 ms interval, which clearly resolve the milk droplet. (c) Left: Placing a suitably engineered diffraction grating (MFG) one Fourier plane away from the primary image plane spatially segregates light corresponding to different focal planes in the sample. In combination with color-correction optics, an instant focal stack is obtained on a widefield detector. Right: A multifocus stack of neurons in a developing C. elegans embryo. Nine planes were simultaneously acquired with an exposure time of 0.11 s. Panel (a) is adapted from Diebold et al. [42] with permission from Nature. Panel (b) is adapted from Bub et al. [43] with permission from Nature. Panel (c) is adapted from Abrahamsson et al. [44] with permission from Nature.
All of the methods described above require some form of axial refocusing in order to acquire volumes, and are thus limited by the inertia of the refocusing element. Multifocus microscopy, eliminates axial refocusing by exciting the sample with widefield illumination and using a diffraction grating to record multiple, focus-shifted images simultaneously onto a single camera (Fig. 4c) [45]. In its current implementation, 9 images are corrected for spherical aberration and simultaneously recorded onto a sensitive camera; redesigning the grating element would allow more (~25) images to be recorded onto a proportionately large detector (such as an sCMOS).
Outlook
We have emphasized advances in the speed of fluorescence microscopy, but other areas of research will prove equally important. The speed and imaging duration of all fluorescence techniques are ultimately limited by the fluorescent probe; to this end, brighter and more photostable fluorophores [46] and improved buffer conditions [47] (especially for super-resolution imaging) are essential. We also see promise in recent denoising [48] and deconvolution [49] methods that help in extracting more information from limited fluorescence; freely available, open-source image processing packages would help these tools reach their full potential.
Highlights.
We describe recent advances in high speed fluorescence microscopy.
Modern microscopes are dividable into two classes – point-scanning and parallelized.
Advances in microscope technology are enabling improvements in imaging rate, spatial resolution and sample penetration.
Improvements in imaging rate often come at the expense of spatial resolution or sample penetration.
Acknowledgements
We thank H. Eden, Y. Wu, R. Christensen and P. Chandris for critical feedback on this manuscript. This work was supported by the Intramural Research Program of the NIH - National Institute of Biomedical Imaging and Bioengineering.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particlular interest, published within the period of review, have been highlighted as:
*of special interest
**of outstanding interest
- [1].Fischer RS, Wu Y, Kanchanawong P, et al. Microscopy in 3D: a biologist’s toolbox. Trends Cell Biol. 2011;21:682–691. doi: 10.1016/j.tcb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wu Y, Christensen R, Colon-Ramos D, Shroff H. Advanced optical imaging techniques for neurodevelopment. Current Opinion in Neurobiology. 2013;23:1090–1097. doi: 10.1016/j.conb.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Botcherby EJ, Smith CW, Kohl MM, et al. Aberration-free three-dimensional multiphoton imaging of neuronal activity at kHz rates. PNAS. 2012;109:2919–2924. doi: 10.1073/pnas.1111662109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4*].Katona G, Szalay G, Maak P, et al. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nature Methods. 2012;9:201–208. doi: 10.1038/nmeth.1851. Acousto-optic scanning technology permits high speed 3D imaging hundreds of times faster than galvanometric scanners, enabling 50 distinct points to be interrogated across nearly cm sized volumes every ms.
- [5].Cheng A, Goncalves JT, Golshani P, et al. Simultaneous two-photon calcium imaging at different depths with spatiotemporal multiplexing. Nature Methods. 2011;8:139–142. doi: 10.1038/nmeth.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shimozawa T, Yamagata K, Kondo T, et al. Improving spinning disk confocal microscopy by preventing pinhole cross-talk for intravital imaging. PNAS. 2013;110:3399–3404. doi: 10.1073/pnas.1216696110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schrodel T, Prevedal R, Aumayr K, et al. Brain-wide 3D imaging of neuronal activity in Caenorhabditis elegans with sculpted light. Nature Methods. 2013;10:1013–1020. doi: 10.1038/nmeth.2637. [DOI] [PubMed] [Google Scholar]
- [8].Wu Y, Ghitani A, Christensen R, et al. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2011;108:17708–17713. doi: 10.1073/pnas.1108494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9**].Wu Y, Wawrzusin P, Senseney J, et al. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat Biotechnol. 2013;31:1032–1038. doi: 10.1038/nbt.2713. Dual-view LSFM in conjunction with joint deconvolution provides ~350 nm isotropic spatial resolution at volumetric imaging rates up to 2 volumes/second.
- [10].Krzic U, Gunther S, Saunders TE, et al. Multiview light-sheet microscope for rapid in toto imaging. Nat Methods. 2012;9:730–733. doi: 10.1038/nmeth.2064. [DOI] [PubMed] [Google Scholar]
- [11].Tomer R, Khairy K, Amat F, K. PJ. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods. 2012;9:755–763. doi: 10.1038/nmeth.2062. [DOI] [PubMed] [Google Scholar]
- [12].Truong TV, Supatto W, Koos DS, et al. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat. Methods. 2011;8:757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- [13].Ahrens MB, Orger MB, Robson DN, et al. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- [14].Dempsey GT, Vaughan JC, Chen KH, et al. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Durisic N, Laparra-Cuervo L, Sandoval-Alvarez A, et al. Single-molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate. Nature Methods. 2014;11:156–162. doi: 10.1038/nmeth.2784. [DOI] [PubMed] [Google Scholar]
- [16].Jones SA, Shim S-H, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods. 2011;8:499–505. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huang F, Schwartz SL, Byars JM, Lidke KA. Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomed. Opt Express. 2011;2:1377–1393. doi: 10.1364/BOE.2.001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Holden S, Uphoff S, Kapanidis AN. DAOSTORM: an algorithm for high-density super-resolution microscopy. Nat Methods. 2011;8:279–280. doi: 10.1038/nmeth0411-279. [DOI] [PubMed] [Google Scholar]
- [19].Huang F, Hartwich TMP, Rivera-Molina F, et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nature Methods. 2013;10:653–658. doi: 10.1038/nmeth.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mukamel EA, Babcock H, zhuang X. Statistical deconvolution for superresolution fluorescence microscopy. Biophys. J. 2012;102:2391–2400. doi: 10.1016/j.bpj.2012.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhu L, Zhang W, Elnatan D, Huang B. Faster STORM using compressed sensing. Nat. Methods. 2012;9:721–723. doi: 10.1038/nmeth.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22**].Cox S, Rosten E, Monypenny J, et al. Bayesian localization microscopy reveals nanoscale podosome dynamics. Nat Methods. 2011;9:195–200. doi: 10.1038/nmeth.1812. Analysis of the fluctuations of all fluorophores in the dataset is used to model the most likely distribution of fluorophore locations. This reduces the number of raw images required to construct a super-resolved image by an order of magnitude (~200 instead of ~2000), allowing imaging of cellular dynamics with ~50 nm spatial resolution at 4 second temporal resolution.
- [23].Rosten E, Jones GE, Cox S. ImageJ plug-in for Bayesian analysis of blinking and bleaching. Nature Methods. 2013;10:97–98. doi: 10.1038/nmeth.2342. [DOI] [PubMed] [Google Scholar]
- [24].Hu YS, Nan X, Sengupta P, et al. Accelerating 3B single-molecule super-resolution microscopy with cloud computing. Nature Methods. 2013;10:96–97. doi: 10.1038/nmeth.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].York AG, Ghitani A, Vaziri A, et al. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nature Methods. 2011;8:327–333. doi: 10.1038/nmeth.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zanacchi FC, Lavagnino Z, Donnorso MP, et al. Live-cell 3D super-resolution imaging in thick biological samples. Nat. Methods. 2011;8:1047–1049. doi: 10.1038/nmeth.1744. [DOI] [PubMed] [Google Scholar]
- [27].Hu YS, Zhu Q, Elkins K, et al. Light-sheet Bayesian microscopy enables deep-cell super-resolution imaging of heterochromatin in live human embryonic stem cells. Optical Nanoscopy. 2013:2. doi: 10.1186/2192-2853-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bingen P, Reuss M, Engelhardt J, Hell SW. Parallelized STED fluorescence nanoscopy. Optics Express. 2011;19:23716–23726. doi: 10.1364/OE.19.023716. [DOI] [PubMed] [Google Scholar]
- [29*].Berning S, Willig KI, Steffens H, et al. Nanoscopy in a living mouse brain. Science. 2012;335:551. doi: 10.1126/science.1215369. STED was used to visualize changes in neuronal morphology at sub-100 nm resolution on the minute time-scale in living mice.
- [30].Willig KI, Steffens H, Gregor C, et al. Nanoscopy of filamentous actin in cortical dendrites of a living mouse. Biophys. J. 2014;106:L01–03. doi: 10.1016/j.bpj.2013.11.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Grotjohann T, Testa I, Leutenegger M, et al. Diffraction-unlimited all-optical imaging and writing with a photochromic GFP. Nature. 2011;478:204–208. doi: 10.1038/nature10497. [DOI] [PubMed] [Google Scholar]
- [32].Testa I, Urban NT, Jakobs S, et al. Nanoscopy of living brain slices with low light levels. Neuron. 2012;75:992–1000. doi: 10.1016/j.neuron.2012.07.028. [DOI] [PubMed] [Google Scholar]
- [33].Grotjohann T, Testa I, Reuss M, et al. rsEGFP2 enables fast RESOLFT nanoscopy of living cells. eLIFE. 2012;1:e00248. doi: 10.7554/eLife.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34*].Chmyrov A, Keller J, Grotjohann T, et al. Nanoscopy with more than 100,000 ‘doughnuts’. Nat Methods. 2013;10:737–740. doi: 10.1038/nmeth.2556. Incoherently superimposing two standing light waves enables massive parallelization of RESOLFT allowing super-resolution imaging of 100×100 μm fields of view in living cells at <1 s temporal resolution.
- [35].Shao L, Kner P, Rego EH, Gustafsson MGL. Super-resolution 3D microscopy of live whole cells using structured illumination. Nat. Methods. 2011;8:1044–1046. doi: 10.1038/nmeth.1734. [DOI] [PubMed] [Google Scholar]
- [36].Lesterlin C, Ball G, Schermelleh L, Sherratt D. RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature. 2014;506:249–253. doi: 10.1038/nature12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].De Luca GM, Breedijk RM, Brandt RA, et al. Re-scan confocal microscopy: scanning twice for better resolution. Biomed Opt Express. 2013;4:2644–2656. doi: 10.1364/BOE.4.002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].York AG, Parekh SH, Dalle Nogare D, et al. Resolution Doubling in Live, Multicellular Organisms via Multifocal Structured Illumination Microscopy. Nature Methods. 2012;9:749–754. doi: 10.1038/nmeth.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Schulz O, Pieper C, Clever M, et al. Resolution doubling in fluorescence microscopy with confocal spinning-disk image scanning microscopy. PNAS. 2013;110:21000–21005. doi: 10.1073/pnas.1315858110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40**].York AG, Chandris P, Nogare DD, et al. Instant super-resolution imaging in live cells and embryos via analog image processing. Nat Methods. 2013;10:1122–1126. doi: 10.1038/nmeth.2687. Elimination of mathematical post-processing operations using optics allows SIM imaging at frame rates of up to 100 Hz over 100 × 100 μm2 fields of view.
- [41].Gao L, Shao L, Higgins CD, et al. Noninvasive Imaging beyond the Diffraction Limit of 3D Dynamics in Thickly Fluorescent Specimens. Cell. 2012;151:1370–1385. doi: 10.1016/j.cell.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rego EH, Shao L, Macklin JJ, et al. Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proc. Natl. Acad. Sci. USA. 2011;109:E135–E143. doi: 10.1073/pnas.1107547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43*].Diebold ED, Buckley BW, Gossett DR, Jalali B. Digitally synthesized beat frequency multiplexing for sub-millisecond fluorescence microscopy. Nature Photonics. 2013;7:806–810. Parallelization with acousto-optical devices simultaneously excites multiple points in the sample at distinct radiofrequencies. Imaging rates of 4.4 kHz enable blur-free observation of rapidly moving (1 m/s or ~50,000 cells/s) cells in flow cytometry.
- [44*].Bub G, Tecza M, Helmes M, et al. Temporal pixel multiplexing for simultaneous high-speed, high-resolution imaging. Nature Methods. 2010;7:209–211. doi: 10.1038/nmeth.1429. Temporal pixel multiplexing was used to convert a 10 Hz CCD into a 250 Hz detector, at the expense of spatial resolution and light efficiency.
- [45*].Abrahamsson S, Chen J, Hajj B, et al. Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nature Methods. 2012;10:60–63. doi: 10.1038/nmeth.2277. A custom diffraction grating is incorporated into the imaging path of an epifluorescence microscope, recording 9 simultaneous images onto a high speed camera.
- [46].Vaughan JC, Jia S, Zhuang X. Ultrabright photoactivatable fluorophores created by reductive caging. Nature Methods. 2012;9:1181–1184. doi: 10.1038/nmeth.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Olivier N, Keller D, Gonczy P, Manley S. Resolution doubling in 3D-STORM imaging through improved buffers. PLoS One. 2013;8:e69004. doi: 10.1371/journal.pone.0069004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Carlton PM, Boulanger J, Kervrann C, et al. Fast live simultaneous multiwavelength four-dimensional optical microscopy. PNAS. 2010;107:16016–16022. doi: 10.1073/pnas.1004037107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Arigovindan M, Fung JC, Elnatan D, et al. High-resolution restoration of 3D structures from widefield images with extreme low signal-to-noise-ratio. PNAS. 2013;110:17344–17349. doi: 10.1073/pnas.1315675110. [DOI] [PMC free article] [PubMed] [Google Scholar]