Abstract
The metal responsive element-binding transcription factor-1 (MTF-1) responds to changes in cellular zinc levels caused by zinc exposure or disruption of endogenous zinc homeostasis by heavy metals or oxygen-related stress. Here we report the functional characterization of a complete zebrafish MTF-1 in comparison with the previously identified isoform lacking the highly conserved cysteine-rich motif (Cys-X-Cys-Cys-X-Cys) found in all other vertebrate MTF-1 orthologues. In an effort to develop novel molecular tools, a constitutively nuclear dominant-negative MTF-1 (dnMTF-1) was generated as tool for inhibiting endogenous MTF-1 signaling. The in vivo efficacy of the dnMTF-1 was determined by microinjecting in vitro transcribed dnMTF-1 mRNA into zebrafish embryos (1–2 cell stage) followed by transcriptomic profiling using an Agilent 4 × 44K array on 28- and 36-hpf embryos. A total of 594 and 560 probes were identified as differentially expressed at 28 hpf and 36 hpf, respectively, with interesting overlaps between timepoints. The main categories of genes affected by the inhibition of MTF-1 signaling were: nuclear receptors and genes involved in stress signaling, neurogenesis, muscle development and contraction, eye development, and metal homeostasis, including novel observations in iron and heme homeostasis. Finally, we investigate both the transcriptional activator and transcriptional repressor role of MTF-1 in potential novel target genes identified by transcriptomic profiling during early zebrafish development.
Keywords: MTF-1, zebrafish, zinc homeostasis, metals, embryonic development
1. Introduction
The metal response element-binding transcription factor-1 (MTF-1) is a ubiquitously expressed transcription factor most widely known for its role in the basal and metal-induced expression of metallothionein (MT) genes, and zinc (Zn) homeostasis [1]. MTF-1 is characterized by six Zn-fingers of the Cys2-His2 type which make up a DNA-binding domain that is highly conserved throughout metazoan phyla [2–5]. MTF-1 also contains three different transactivation domains, an acidic domain, a proline-rich domain and a serine/threonine-rich domain [6], all of which contribute to transcriptional activation. Another highly conserved region in vertebrate MTF-1 orthologues is a short 32-amino acid section found immediately after the serine-threonine-rich domain that contains a sumoylation motif [7] and a highly conserved cysteine-rich motif (Cys-X-Cys-Cys-X-Cys) that is required for the proper induction of MT genes [8].
MTF-1 is constitutively expressed and resides in the cytosol until activation [9]. Although multiple metals, oxidative stress and hypoxia are all capable of inducing MTF-1 target gene expression, the activation and nuclear localization of MTF-1 occurs in a Zn-dependent manner [10–12]. The six Zn-fingers are heterogeneous in function [13–15] with the first four N-terminal Zn-fingers playing a prominent role in the Zn-dependent activation of MTF-1[16]. Upon activation MTF-1 locates to the nucleus and binds to highly conserved metal-responsive promoter elements (MREs) with the core consensus sequence, TGCRCNC [14]. The two variable nucleotides at the fourth and sixth positions of the 7 base pair motif are important in determining the binding of MTF-1, and studies have indicated that MREs with an A and T at the fourth and sixth positions, respectively, have significantly greater basal activity and increased binding affinity for MTF-1 [17–19]. Basal expression of MTF-1 target genes is usually associated with minimal binding to high affinity MREs; upon exposure to various stressors, MTF-1 is recruited to the nucleus to bind additional MREs, both high and low affinity, resulting in increased transcriptional activation [20, 21]. Furthermore, Sims et al. (2012) demonstrated a role for the sixth nucleotide position in the core MRE in determining metal-specific activation of Drosophila MTF-1 [22].
Previous studies have established an essential role for the MTF-1 transcription factor as highlighted by the embryonic lethality in ‘knockout’ mice [23]. Lethality occurs by gestation day 14 and the major morphological phenotype associated with these embryos is severe liver damage characterized by enlarged, congested sinusoids, dissociation of the epithelial compartment, significantly reduced cytokeratin expression, and diffuse bleeding and edema. Although conditional ‘knockouts’ of MTF-1 in adult mice do not result in lethality, the mice are extremely susceptible to metal or oxidative stress and have significantly impaired liver regenerative capabilities [24]. An additional potential role for MTF-1 in cell differentiation has been identified by conditional ‘knockout’ in bone marrow that results in a significant reduction in leukocytes [24]. Consistent with its role in metals homeostasis, MTF-1 has been shown to regulate the expression of Zn transporter-1 (ZnT-1), ‘knockout’ of which is also embryonic lethal in mice and displays phenotypes similar to the MTF-1 ‘knockout’ [25, 26].
In recent years antisense morpholinos (MO) have become a very popular and valuable molecular tool for use in the study of gene function in zebrafish embryos. Even in the case of some essential genes, efficient use of MO ‘knockdowns’ can be used successfully to demonstrate significant changes in gene expression without producing overt abnormal phenotypes via titration of the MO used in the ‘knockdowns’. However, because of transient effects due to dilution during development they are not feasible for studies using older larvae, juveniles or adults. Previous studies have demonstrated the dominant-negative function of a C-terminal MTF-1 mutant on endogenous MTF-1 signaling in mammalian cell lines [27, 28]. Coupling this observation with the ability to create a constitutively active MTF-1 [16], use of a dominant-negative MTF-1 to inhibit endogenous in vivo signaling could be a practical approach to advancing our understanding of the multifunctional roles of MTF-1 using zebrafish as our chosen model organism. The application of a transgenic zebrafish expressing a dominant-negative bone morphogenetic protein (Bmp) under control of a heat shock-inducible promoter [29, 30] is validation of such an approach. Previous research has identified a zebrafish MTF-1 homologue that is significantly shorter than the typical vertebrate MTF-1 and missing the cysteine-rich motif [5]; although complete MTF-1 transcripts have been described other fish species [4, 31]. Therefore in our initial effort we sought to investigate the functional diversity of MTF-1 transcripts in zebrafish, followed by an investigation of the in vivo efficacy of the dominant-negative MTF-1 by microinjection of in vitro transcribed mRNA in zebrafish embryos as a precursor to the development of a transgenic zebrafish.
Here we report the functional characterization of a complete zebrafish MTF-1 in comparison with the previously identified isoform lacking the highly conserved cysteine-rich motif (Cys-X-Cys-Cys-X-Cys) found in all other vertebrate MTF-1 orthologues. In addition, we demonstrate the utility of a constitutively nuclear, dominant-negative MTF-1 construct that is capable of inhibiting both in vitro and in vivo endogenous MTF-1 signaling. Finally, we investigate the role of MTF-1 in positively and negatively regulating potential novel target genes identified by transcriptomic profiling during early zebrafish development.
2. Materials and methods
2.1 Chemicals and cell lines
Zn chloride, cadmium (Cd) chloride and copper (Cu) chloride were obtained from Sigma-Aldrich (St. Louis, MO). Cos-7 cells and the mouse cell line Hepa1c1c7 were obtained from the American Type Culture Collection (ATCC; Manassas, VA) and grown according to standard procedures. The MTF-1 null mouse embryonic fibroblast (MEF) cells were generously provided by Dr. Glen K. Andrews (University of Kansas).
2.2 Fish husbandry
The TL (Tupfel/Long fin mutations) wild-type strain of zebrafish was used for all experiments. Fertilized eggs were obtained from multiple group breedings from a Mass Embryo Production System (MEPS; Aquatic Habitats, Apopka, FL) with ~200 fish at a ratio of 2 female per 1 male fish. Procedures used in these experiments were approved by the Animal Care and Use Committee of the University of Alabama, Tuscaloosa, Alabama, USA.
2.3 Embryo development time series and cadmium treatment of adult zebrafish
A large batch (>3000) of embryos were generated by active breeding of the fish in the MEPS system for 60 minutes. The embryos were immediately rinsed and placed into large petri dishes (150 mm diameter) at a density of 100 embryos per 100 mL of 0.3× Danieau’s at 28.5°C. At ~4 hours post fertilization (hpf) the embryos were screened for normal development and 15 small petri dishes (100 mm in diameter) were setup with 20 embryos per dish in 25 mL of 0.3× Danieau’s at 28.5°C under a 14 hour light/10 hour dark cycle with water changes every 24 hours. Three replicates of 20 pooled embryos were collected at each developmental timepoint (24, 48, 72, 96 and 120 hpf), flash frozen in liquid nitrogen and stored at −80°C until RNA isolation.
Adult male and female zebrafish approximately one year in age were continuously exposed to 20 µM of Cd for a period of 96 hours. Specifically, 6 individual fish (3 male and 3 female) were place in a 4L beaker with 3L of regular fish water (360 mg Instant Ocean Sea Salt and 11 mg of NaHCO3 per liter of deionized water; 7.4 pH, 750 µ-siemens conductivity, 28.5°C) or fish water with 20 µM of Cd. The water was continuously aerated with a small airstone and the water or water with Cd was renewed (100% water change) after 48 hours. A total of 6 beakers (3 control and 3 Cd) were used in the experiment. After 96 hours, the fish were anesthetized with 150 mM of MS-222 in buffered fish water and euthanized by cervical transection. The following tissues were removed by dissection and immediately flash frozen in liquid nitrogen: brain, eye, gut, liver, heart, gill, spleen and testis. The tissues were stored at −80°C until processed for total RNA. Three replicates were collected for each sex and treatment group and consisted of equivalent amounts of total RNA pooled from three individual male or female fish.
2.4 Generation of MTF-1 constructs
All MTF-1 constructs were PCR amplified with Kapa HiFi DNA polymerase (Kapa Biosystems, Inc., Boston, MA). Primer sequences are provided in Supplementary Table 1. The full length MTF-1 was PCR amplified from embryo cDNA and cloned into the pcDNA™3.1 vector (Invitrogen, Carlsbad, CA). The MTF-1ΔCys isoform (which encodes for the first 588 amino acids of the full length MTF-1) was amplified from the full length isoform and also cloned into the pcDNA™3.1 vector. The full length MTF-1-eGFP construct and MTF-1ΔCys-eGFP construct were created by cloning the respective coding regions into the pEGFP-N1 vector (Clontech Laboratories, Inc.). To create the dnMTF-1-eGPF fusion, the MTF-1ΔTAD fragment was also amplified and cloned into the pEGFP-N1 vector. Site-directed mutagenesis was performed on the MTF-1ΔTAD/eGFP vector with the QuikChange® II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) using the following primers: MTF_mut1 5'-ACTCATCAG AAGACGCACACTGGCGAAAAGCCGTTTGTTTGCAACCAGCAA-3' and MTF_mut2 5'-TTGCTGGTTGCAAACAAACGGCTTTTCGCCAGTGTGCGTCTTCTGATGAGT-3'. The dnMTF-1-eGPF coding region and eGFP coding region were cloned into the pT7TS vector for in vitro transcription. The dnMTF-1 coding region (without the eGFP fusion) was cloned into the pT3TS vector for in vitro transcription.
2.5 Other plasmid constructs
The mouse MTF-1 expression vector was a kind gift from Dr. Glen K. Andrews [16]. The zebrafish MT luciferase construct was a kind gift from Dr. King Ming Chan [32]. The zebrafish αA crystallin and hemopexin promoters, and the human αA crystallin promoter, were amplified from genomic DNA using the Kapa HiFi DNA polymerase and cloned into the pGL4.10[luc2] vector (Promega Corporation, Madison, WI). Primer sequences are provided in Supplementary Table 1.
2.6 Transient transfections and luciferase assays
All transient transfections of the various DNA constructs were performed with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, Ca) and carried out in 48-well plates with an initial plating of 20,000 cells per well (MTF-1 null MEF cells and Hepa1c1c7 cells). 24 hours after plating 300 ng of DNA was complexed with 1 µL of Lipofectamine 2000 and then added to each well containing media with serum and incubated overnight. The total amount of DNA was kept constant by the addition of empty vector (pcDNA3.1). For luciferase assays, the Renilla luciferase (pGL4.74; Promega, Madison, WI) was used as a transfection control. The specific amounts of DNA used for each construct is listed in the figure legends. 24 hours after transfection the media was replaced with serum-free media and the cells were dose with Zn (100 µM), Cd (20 µM), or Cu (10 µM). Four hours after dosing the cells were passively lysed and used for luciferase assays. Luminescence was measured using the Dual Luciferase® Reporter Assay (Promega Corporation, Madison, WI) with a Sirius Luminometer (Titertek-Berthold, Huntsville, AL) with dual injector. The final values were expressed as a ration of firefly luciferase units to Renilla luciferase units. Each transfection experiment consisted of 6 replicates and each experiment was repeated two times.
2.7 Subcellular localization of MTF-1ΔTAD and dnMTF-1
Cos-7 cells were grown overnight on coverslips in six-well plates and the next day cells were transfected using Lipofectamine 2000 with 300 ng of either MTF-1ΔTAD or dnMTF-1 cloned into the pEGFP-N1 vector. 24 hours after transfection, the cells were dosed with Zn (100 µM) for 6 hours in serum-free media. After Zn treatment the cells were washed with 1× phosphate buffered saline and fixed in 4% formalin. The coverslips were inverted onto slides and mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Cells were visualized using a Zeiss Axio Imager Z1 fluorescence microscope with Axiovision software.
2.8 dnMTF-1 IVT mRNA microinjection experiments
2.81 Microarray experimental design
A Narishige IM-300 microinjector was used to microinject 1–2 cell embryos with ~100 pg of dnMTF-1 IVT mRNA (~2.1 nL microinjection volume with a concentration 50 ng/µL IVT mRNA), or ~100 pg of enhanced green fluorescent protein (eGFP) IVT mRNA. The dnMTF-1 is an eGFP fusion protein so proper microinjection and distribution throughout the embryo could be visualized by fluorescent microscopy. Immediately after microinjection, embryos from each treatment were divided into 4 replicates of 30 pooled embryos and held in 25 mL of 0.3× Danieau’s at 28.5°C under a 14 hour light/10 hour dark cycle. eGFP positive embryos (dnMTF-1 and eGFP) were collected at 28 and 36 hpf and flash frozen in liquid nitrogen for future RNA isolation and microarray hybridization. Total RNA was prepared using the RNA STAT60 protocol (Tel-Test, Inc., TX). RNA was quantified using a Nanodrop 2000C spectrophotometer and assessed for quality using an Agilent 2100 Bioanalyzer Lab-on-chip system. Only samples with RNA integrity number (RIN) between 9.8–10 were used for microarray analysis. Microarray hybridizations were performed by Genome Technology Core at the Whitehead Institute for Biomedical Research (Cambridge, MA) using a custom zebrafish Agilent (Agilent Technologies, Santa Clara, CA) 4×44K microarray [33]. This microarray matches Agilent’s 4×44k DNA gene expression microarray, except for addition of custom probes ensuring coverage of zebrafish cytochrome P450s and other genes involved in the chemical defensome [33]. For each RNA sample, a single microarray was hybridized with 750 ng Cy3 labeled cDNA using Agilent’s standard conditions for single-color microarrays. The Agilent Low-Input QuickAmp Labeling Kit was used for labeling, the samples were hybridized to a the custom Agilent 4×44K feature zebrafish microarray using the Agilent In situ Hybridization Kit Plus, and labeled cDNA was combined with the Agilent 10× Control Targets (to identify microarray corners). Post-hybridization, microarray slides were washed as per the Agilent In situ Hybridization Kit Plus. Arrays were scanned with an Agilent DNA Microarray Scanner.
2.82 Independent dnMTF-1 microinjection experiments for real-time RT PCR
A Narishige IM-300 microinjector was used to microinject 1–2 cell embryos with a mixture of ~40 pg of dnMTF-1 IVT mRNA and ~10 pg of eGPF IVT mRNA (~2.1 nL microinjection volume with a concentration 50 ng/µL IVT mRNA), or alone with ~50 pg of eGFP IVT mRNA. For these confirmation experiments, we chose to use dnMTF-1 without the eGFP fusion to demonstrate that the eGPF does not play an in vivo role in repression of transcription. The reduction in the microinjection concentration compared to ‘knockdowns’ used in the microarray analysis adjusts for differences in mRNA length and maintains consistency with respect to the specific number of IVT mRNA transcripts. Immediately after microinjection, embryos from each treatment were divided into 3 replicates of 20 pooled embryos and held in 25 mL of 0.3× Danieau’s at 28.5°C under a 14 hour light/10 hour dark cycle. Non-injected embryos were collected at 28 and 36 hpf, while microinjected, eGFP positive embryos were collected at 36 hpf and flash frozen in liquid nitrogen for future RNA isolation. Two independent, replicate experiments were performed to confirm the results of the gene expression profiling by microarray.
2.9 Agilent microarray data analysis
Extraction, normalization, and filtering of microarray data followed [33]. Extraction of raw microarray results was performed using Agilent’s Feature Extraction software with background detrending (spatial and multiplicative). The data were normalized using the non-linear scaling method based on rank invariant probes [34, 35] using software developed by AGM. Cy3 signal saturated probes and probes not above background in all replicates were then removed prior to statistical analysis. Statistical tests were performed using the MeV analysis suite [36]. All data were log transformed and median centered for each probe. A two-factor ANOVA was performed for time (28 vs. 36 hpf), treatment (dnMTF-1 and eGFP), and their interaction with p-value based on 1000 permutations of the data and alpha of 0.01. For post-hoc analyses to find biologically relevant results, the probes significant for treatment and time-treatment interaction were secondarily examined using Rank Product (RP) analysis [37]. Rank product analysis was performed to provide lists of probes up- and down-regulated between eGFP control and dnMTF-1 for each time point. For each test, a two-class unpaired RP analysis was performed using 1000 permutation of the data with a false discovery rate (FDR) not exceeding 5%.
Enrichment of Gene Ontology (GO; levels 3–9), KEGG Pathway, and InterPro terms for Rank Product significant probes was examined using the FatiGO+ software [38]. FatiGO+ uses GO, KEGG, and InterPro terms assigned within the Ensembl annotation of the zebrafish genome to compare lists of significant Agilent probes relative to the zebrafish genome. FatiGO+ was thus used to examine enrichment of GO, KEGG, and InterPro terms in the set of significant probes relative to the entire zebrafish gene set (less genes with probes in the significant set). FatiGO+ was performed using a two-tailed Fisher exact test and p<0.05, adjusted for multiple tests as described [38].
2.10 Gene expression assessment by real-time RT-PCR
Total RNA was prepared from whole embryos or adult tissues using the TRIzol® reagent (Life Technologies, Invitrogen) protocol. cDNA was synthesized from 1 µg total RNA using a mix of random hexamers and oligo-dT primers with the qScript™ cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD). Quantitative PCR was performed using the PerfeCTa® SYBR® Green Supermix for iQ™ (Quanta Biosciences, Gaithersburg, MD) in a MyiQ2 Two-Color Real-Time PCR Detection system (Bio-Rad, Hercules, CA). The PCR conditions used were: 95°C for 3 min, 95°C for 15 s/60°C for 30 sec (40 cycles). At the end of each PCR run, a melt curve analysis was performed to ensure that only a single product was amplified. Three technical replicates were used for each sample. For quantitation of gene expression for all zebrafish genes, a standard curve was generated by serially diluting plasmids containing a full-length copy of the target gene. No plasmid template was available for the mouse genes (18S ribosomal, ARNT, CYP1A) so relative expression was calculated as the fold change compared with the empty pcDNA vector DMSO treatment according to following equation [39]: relative mRNA expression = 2−ΔΔCt; where ΔΔCt = [Ct(Sox9b) − Ct(actin)]treatment − [Ct(Sox9b) – Ct(actin)]control. Changes in expression are reported as changes in fold induction by normalizing molecule numbers to the appropriate control. Primer sequences are provided in Supplementary Table 1.
2.11 Statistical analysis of real-time PCR and in vitro promoter assays
All statistical analyses were performed with the Prism 5 software package (GraphPad Software Inc., San Diego, CA). Data were logarithmically transformed as needed to improve equality of variances. Multifactor analyses for determining combined effects of 1) Cd-treatment in adult tissue, 2) sex of representative adult tissue samples, or 3) different developmental timepoints against multiple primer sets for the different MTF-1 isoforms were evaluated using two-way ANOVA (p-value < 0.05), with Bonferroni correction for multiple comparisons. The differences among transcript levels determined by real-time RT-PCR to confirm the microarray results were evaluated using one-way analysis of variance (ANOVA, p-value < 0.05), with Bonferroni correction for multiple comparisons. The differences among transactivation levels determined by luciferase assay were evaluated using two-way ANOVA (p-value < 0.05), with Bonferroni correction for multiple comparisons. Specific analyses are described further in the figure legends. All experiments presented in this manuscript involving real time PCR after microinjection of dnMTF-1 mRNA to confirm the microarray results or luciferase assays to characterize the different MTF-1 or promoter constructs were repeated at least twice to confirm the observations. In all cases the results were statistically consistent between replicated experiments; however, data from only one representative experiment is presented.
3. Results
3.1 Identification and cloning of a complete zebrafish MTF-1 gene
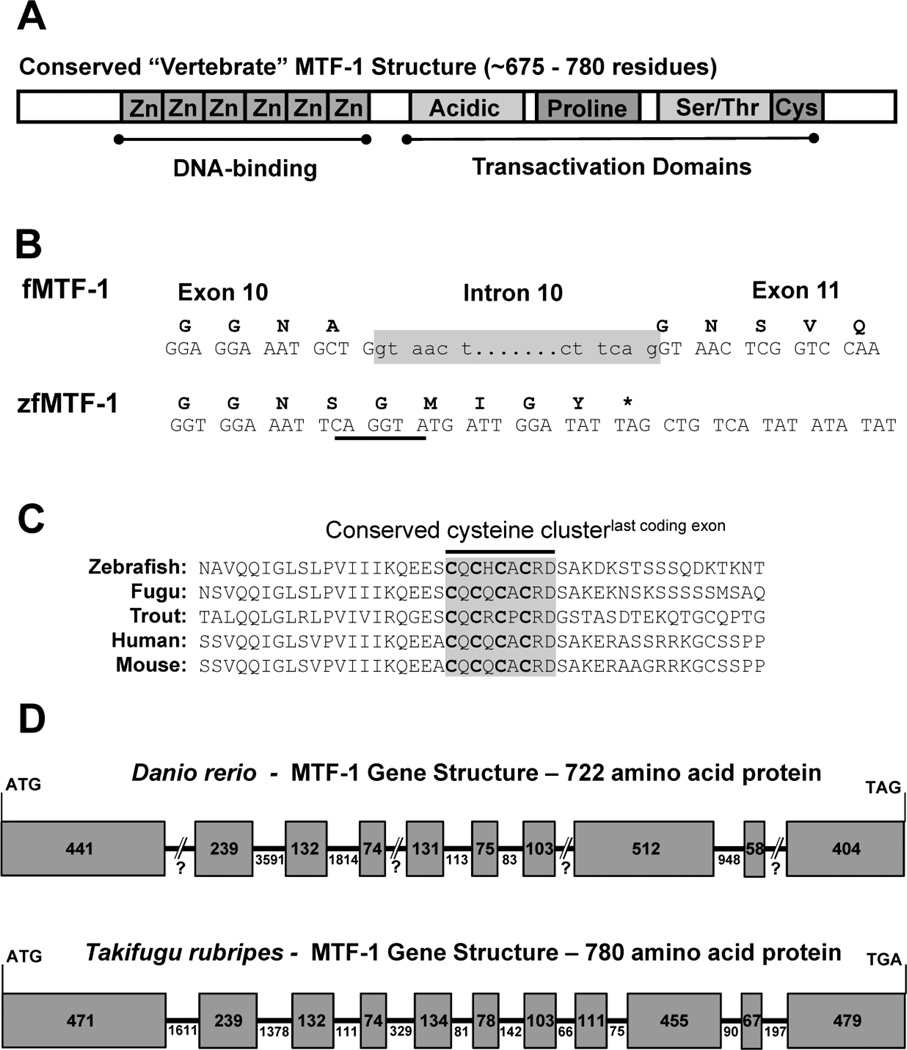
The currently published sequence for the zebrafish MTF-1 is represented by a 3,379 bp transcript, which encodes a 593 amino acid residue protein [5] that is missing the highly conserved cysteine-motif found in the C-terminal section of all other vertebrate MTF-1 proteins [8] (Figure 1A). Comparison of the truncated MTF-1 sequence with the Takifugu rubripes genomic structure for MTF-1 demonstrated that this conserved cysteine-motif occurs in the last coding exon. Close observation of the open reading frame of the truncated MTF-1 clearly shows, that prior to the last five codons of the predicted reading frame, a canonical splice site exists at the conserved intron/exon boundary of the Takifugu sequence (Figure 1B). Using the translated protein sequence from exon 11 in Takifugu, the missing exon was identified by a TBLASTN search of the zebrafish genome assembly (Zv6_scaffold 2307.4) which produced two transcripts (GENESCAN00000012450, FGENESH00000083780) that encoded the missing C-terminal end of the MTF-1 transcript (Figure 1C). Using these new sequences, primers were designed to amplify the new full length transcript of MTF-1 (herein designated as MTF-1), and additional 3' RACE PCR was used to amplify the entire 3' UTR. The new MTF-1 transcript is 2,773 nucleotides in length and encodes a 722 amino acid protein containing the Zn-finger DNA binding domain, all three transactivation domains and the highly conserved cysteine-rich motif (Supplementary Figure 1). In addition, the last exon of the zebrafish MTF-1 gene also contains the more recently characterized SUMO-interacting motif (SIM), 601VIIIKQEE608, that immediately precedes the cysteine motif and is also conserved in other vertebrate species [7].
Figure 1.
Identification of a full length zebrafish MTF-1 Transcript and Corresponding Gene Structure. A) Schematic of a typical vertebrate MTF-1 protein highlighting the various conserved domains and motifs. B) Comparison of the last Takifugu MTF-1 (fMTF-1) exon/intron boundary with zfMTF-1. fMTF-1 intron 10 is highlighted in grey. Note the underlined canonical donor splice site (CAG/GTA) five codons from the putative stop codon (*) of the zfMTF-1. C) Alignment of the primary structure of the C-terminal end of various vertebrate MTF-1’s with the putative translation of the “missing” zfMTF-1 exon. The highly conserved cysteine motif is shaded in a grey box. D) A comparison of the genetic structure of the coding exons from Takifugu and zebrafish. Exons are designated by the shaded boxes and introns are represented by the lines between boxes. The numbers in the boxes or below the lines represent length in nucleotides.
The latest assembly of the zebrafish genome (Zv9, GCA_000002035.2) contains a much more complete version of the MTF-1 gene on chromosome 16, but due to poor sequence quality or assembly challenges the current gene annotation is missing part of the 9th exon and subsequent intron. However, the overall genetic structure consisting of 10 coding exons can be ascertained by a comparison of the transcript and genomic sequences (Figure 1D). To further support the predicted genetic structure of the MTF-1 gene, we compared it to the genetic structure of the Takifugu MTF-1 gene, which contains 11 exons (Fugu 4.0, June 2005). Exons 1 thru 7, 9 and 10 of the MTF-1 gene are highly conserved with the complementary Takifugu exons (1 thru 7, 10 and 11), with Takifugu exons 8 and 9 being complementary to exon 8 of the MTF-1 gene (Figure 1D).
3.2 Expression of MTF-1 during development and in adult tissues
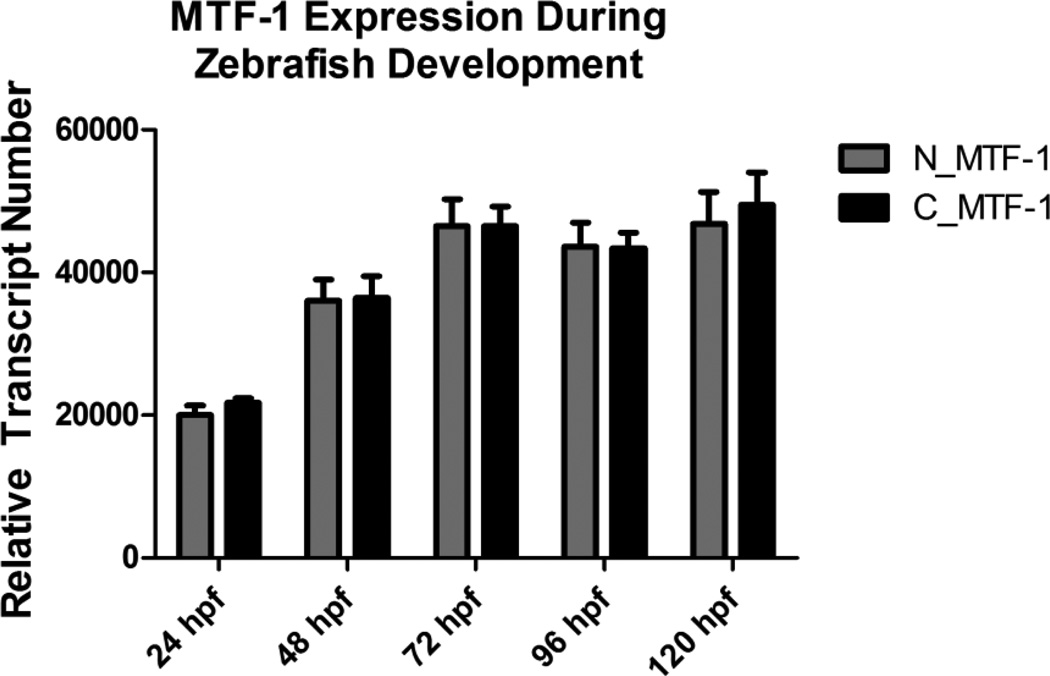
In an effort to ascertain the existence of multiple MTF-1 splice variants, we used real-time qRT-PCR to measure expression of both the full length and truncated versions of MTF-1 using two different primer sets. A primer set capable of detecting both MTF-1 isoforms was designed with a forward primer that spanned the exon/intron boundary between exons 7 and 8, and a reverse primer in exon 8 (designated as N_MTF-1). A second primer set, designed with a forward primer that spanned the exon/intron boundary between exons 9 and 10 and a reverse primer in exon 10 (designated as C_MTF-1), was capable of only detecting the full length MTF-1. Real-time qRT-PCR was performed on developing larvae starting at 24 hpf and continuing every 24 hours until 120 hpf. There was a significant increase in basal MTF-1 expression between 24 and 48 hpf, with a more modest increase and subsequent plateau in expression starting at 72 hpf. However, there was no detectable difference in expression levels between either primer set suggesting that only the full length MTF-1 is expressed during development Figure 2).
Figure 2.
Relative transcript abundance of zebrafish MTF-1 during embryonic development. Two different primer sets were used to assess relative abundance of MTF-1 transcripts in developing zebrafish embryo-larvae at five different timepoints. No statistically significant difference in transcript abundance was detected with either primer set. Error bars represent one standard deviation: n = 3 replicates of 20 pooled embryos per time point.
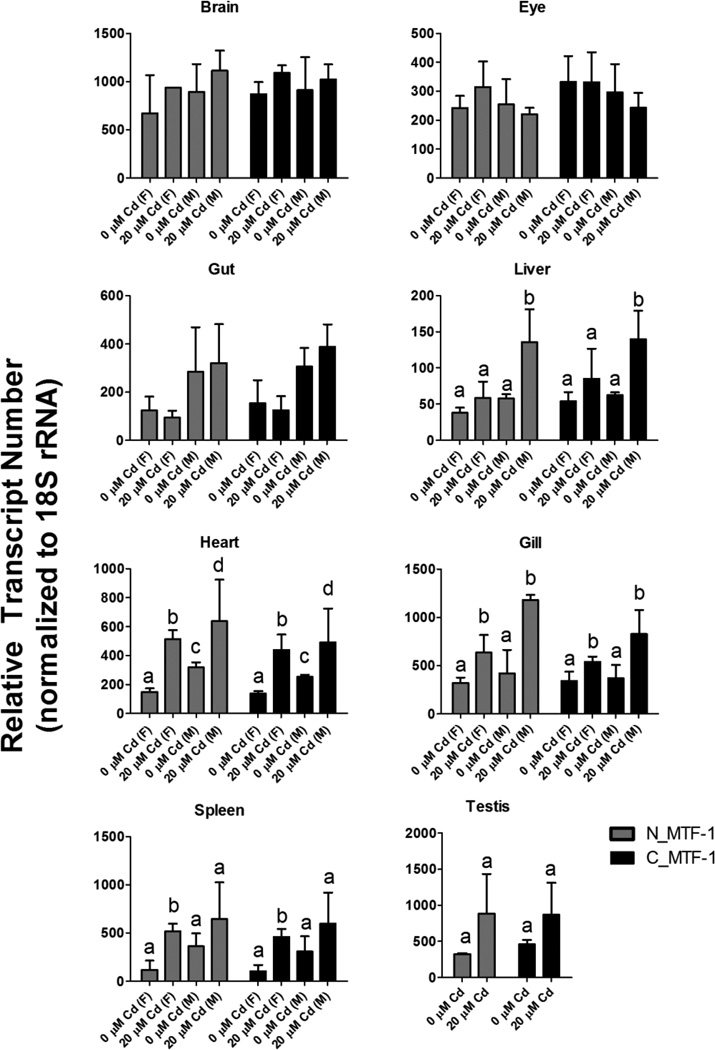
Additional expression profiling was performed on adult tissues, including brain, liver, gill, heart, eye, and spleen, from both control and Cd-exposed zebrafish. There were no statistically significant differences in MTF-1 expression between males and females, or in response to Cd treatment, in brain or eye tissues (Figure 3). Liver, gut, gill and spleen tissues did not display any significant differences in basal MTF-1 expression between males and females, whereas heart tissue did have a significant difference in basal MTF-1 expression between males and females. Both males and females had significant induction of MTF-1 expression in response to Cd treatment in both gill and heart tissues. Testis tissue had a trend toward Cd induction, and only males displayed statistically significant induction of MTF-1 in liver tissue in response to Cd In contrast, only the spleens of females had statistically significant induction of MTF-1 in response to Cd treatment. Finally, there were no statistically significant differences in expression with either primer pair (N_MTF-1 or C_MTF-1) between any of the tissues (Figure 3).
Figure 3.
Relative transcript abundance of zebrafish MTF-1 in adult tissues under basal and cadmium-induced conditions. Two different primer sets were used to assess relative abundance of MTF-1 transcripts in eight different adult tissues in control fish and fish exposed to cadmium for 96 hours. No statistically significant difference in transcript abundance was detected with either primer set. No statistically significant differences in MTF-1 expression were detected in brain, eye gut or testis. Cadmium specific effects were observed in liver, heart, gill and spleen. Sex-specific effects were observed in heart. Bars designated with the same letter represent no statistical difference. Bars designated with different letters represent statistically significant differences based on two-way ANOVA (p-value < 0.05) with Bonferroni correction for multiple comparisons. Error bars represent one standard deviation: n = 3 replicates of 3 pooled adult tissues per treatment.
3.3 Comparison of transactivation capabilities of zebrafish MTF-1 and MTF-1ΔCys
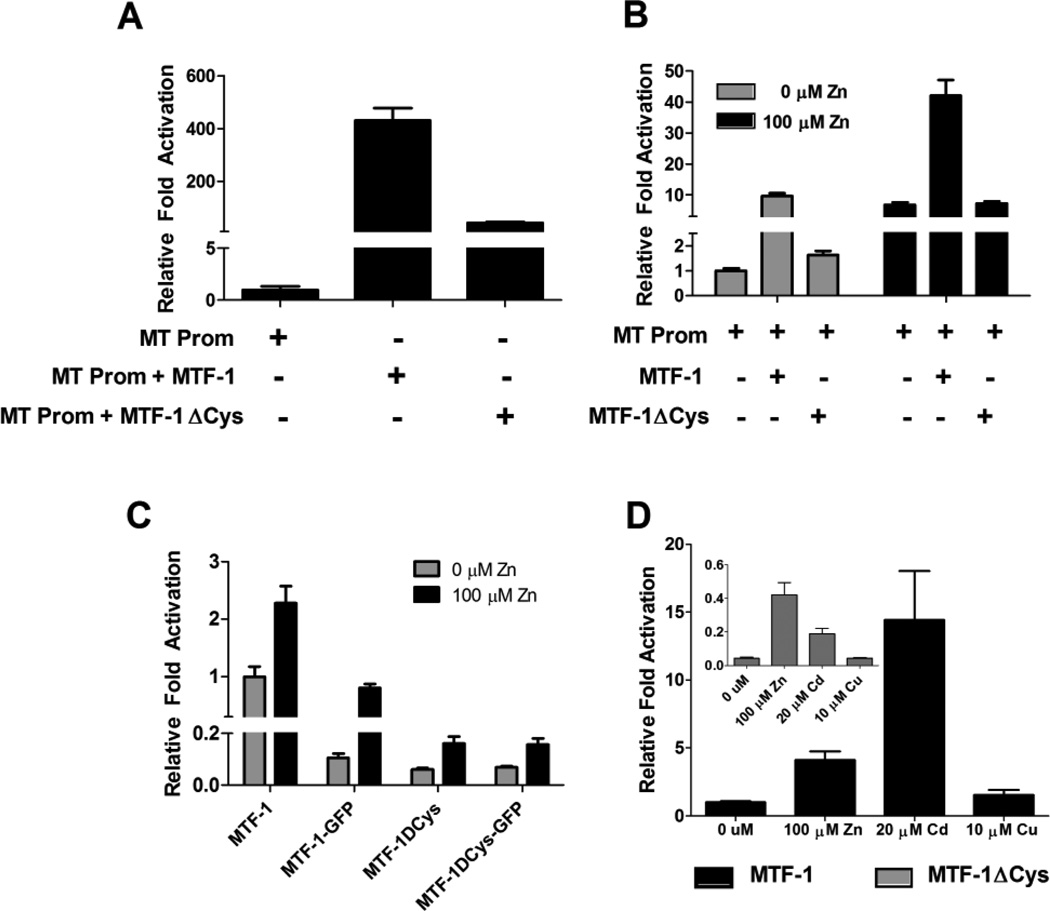
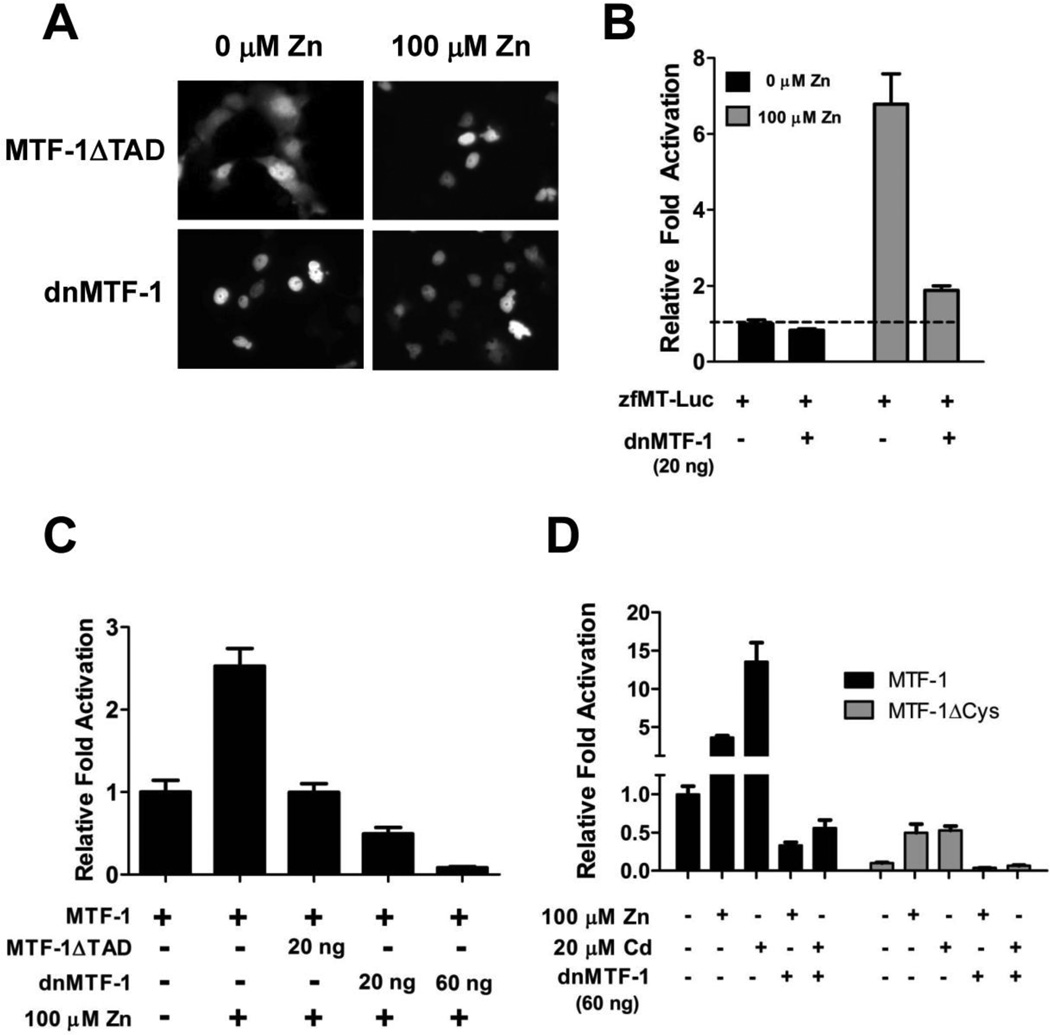
To determine any differences in transactivation potential between the two different zebrafish MTF-1 isoforms, we cloned a truncated version of the MTF-1 transcript lacking the last exon (which we designate as MTF-1ΔCys) and performed transient-transfection assays in which we measured the ability of both MTF-1 constructs to restore constitutive transcriptional activation of a zebrafish metallothionein promoter luciferase (zfMT-Luc) construct [32, 40] in the MEF MTF-1 null cell line [41, 42]. Comparison of the zfMT promoter alone to cells co-transfected with MTF-1 demonstrates an increase basal luciferase activity by 2.5 orders of magnitude (Figure 4A). In contrast, equimolar concentrations of the MTF-1ΔCys construct only increased basal luciferase activity by 1.5 orders of magnitude. To determine if the MTF-1ΔCys construct had any inhibitory effect on endogenous MTF-1 signaling, we compared overexpression of both MTF-1 and MTF-1ΔCys in Hepa1c1c7 cells using the zfMT-Luc construct. Overexpression of MTF-1 resulted in a 10-fold increase in basal luciferase activity, whereas the MTF-1ΔCys construct only produced a ~1.6 fold increase in basal luciferase activity (Figure 4B). Zn treatment resulted in ~6.8 fold increase in endogenous MTF-1 activation of the zfMT-Luc construct. Overexpression of MTF-1 resulted in a ~42-fold increase in luciferase activity compared to basal activity, whereas overexpression of MTF-1ΔCys did not appear to have any significant influence on the activation of endogenous MTF-1 by additional Zn treatment (Figure 4B).
Figure 4.
Comparison of transactivation potential of MTF-1 versus MTF-1ΔCys. A) In transient transfection assays using MEF MTF-1 null cells, MTF-1 resulted in an order of magnitude greater induction of transcriptional activation of the zfMT-Luc promoter construct compared to MTF-1ΔCys. B) In transient transfection assays using Hepa1c1c7 cells, both MTF-1 and MTF-1ΔCys significantly enhanced basal activation of the zfMT-Luc promoter construct compared to endogenous MTF-1 alone. MTF-1 significantly increased the induction of luciferase activity in response to Zn treatment. There was no difference between endogenous MTF-1 activity and transfected MTF-1ΔCys in response to additional Zn treatment. C) A comparison of the transactivation of the zfMT-Luc promoter construct by all four MTF-1 constructs in response to Zn treatment in MEF MTF-1 null cells. MTF-1 transfection resulted on an order of magnitude greater activation of the zfMT-Luc promoter construct compared to the other three constructs. D) A comparison of the transactivation of the zfMT-Luc promoter construct by either MTF-1 or MTF-1ΔCys in response different metal treatments. A-D) Amount of each construct transfected per well: 20 ng of zfMT-Luc, 3 ng of Renilla vector, 23 ng of MTF-1 (3×109 copies), 21.8 g of MTF-1ΔCys (3×109 copies), 20.9 ng of MTF-1-GFP (3×109 copies) and 19.7 ng of MTF-1ΔCys-GFP (3×109 copies). Error bars represent one standard deviation: n = 6 replicates. The results shown in each panel are representative of two independent experiments.
To further compare the MTF-1 constructs and their respective GFP-fusion constructs (MTF-1-GFP and MTF-1ΔCys-GFP), we performed additional assays with the MEF MTF-1 null cell line in which we measured the ability of each MTF-1 construct to enhance transactivation of the zfMT-Luc construct in response to metals treatment. All four MTF-1 constructs restored activation of the zfMT-Luc construct and also displayed significant induction of luciferase activity in response to 100 µM Zn treatment (Figure 4C); however, luciferase expression regulated by MTF-1 was an order of magnitude higher than the other three constructs (Figure 4C and 4D). Interestingly, the magnitude of luciferase induction by 20 µM Cd was much stronger with MTF-1 compared to MTF-1ΔCys (Figure 4D). Cu (10 µM) treatment did not have a significant effect on luciferase activity with either MTF-1 construct (Figure 4D).
3.4 Construction of a dominant-negative MTF-1
In most cases, a dominant-negative factor must be expressed at significantly higher levels than the ‘wild-type’ factor in order to competitively inhibit signaling. However, a constitutively nuclear dominant-negative MTF-1 is advantageous for creating a transgenic zebrafish strain because it should efficiently outcompete the Zn-dependent endogenous MTF-1. Using specific primers, a truncated MTF-1 isoform which encodes the first 378 amino acids of the protein and lacks all of the transactivation domains (herein designated as MTF-1ΔTAD) was amplified by PCR and cloned into the Clontech pEGFP expression vector. Using the mouse MTF-1 as a model [16], site-directed mutagenesis was performed on MTF-1ΔTAD to replace the RGEYT linker motif with TGEKP (mutant construct herein designated as dnMTF1). To compare their nuclear localization properties, both MTF-1ΔTAD and dnMTF-1 were transfected into Cos7 cells and treated with Zn. MTF-1ΔTAD is clearly localized in the cytoplasm in the absence of excess Zn and fully relocates to the nucleus a few hours after Zn treatment, whereas the dnMTF-1 is constitutively localized to the nucleus regardless of Zn treatment (Figure 5A). To test the capability of dnMTF-1 to inhibit endogenous MTF-1 signaling, mouse Hepa1c1c7 liver cells were co-transfected with the zfMT-Luc and dnMTF-1 constructs. The dnMTF-1 reduced basal and Zn-induced luciferase activity by 17% and 73%, respectively (Figure 5B).
Figure 5.
Comparison of transactivation potential of MTF-1ΔTAD versus dnMTF-1. A) Nuclear localization of MTF-1ΔTAD and dnMTF-1 in control and Zn-treated cells. dnMTF-1 is constitutively localized to the nucleus independent of metals treatment. B) In transient transfection assays in Hepa1c1c7 cells transfected with 20 ng of zfMT-Luc, the dnMTF-1 was able to significantly inhibit endogenous MTF-1 signaling in response to Zn treatment. C) In transient transfection assays in MEF MTF-1 null cells, the dnMTF-1 was much more efficient at inhibiting MTF-1 activation of the zfMT-Luc construct compared to MTF-1ΔTAD. D) In transient transfection assays in MEF MTF-1 null cells transfected with 20 ng of zfMT-Luc, the dnMTF-1 was able to significantly inhibit both Zn and Cd-induced activation of the zfMT-Luc construct by either MTF-1 and MTF-1ΔCys. Error bars represent one standard deviation: n = 6 replicates. The results shown in each panel are representative of two independent experiments.
The inhibitory capacities of MTF-1ΔTAD and dnMTF-1 were compared by assessing their ability to repress MTF-1 activation in MEF MTF-1 null cells. The cells were co-transfected with the zfMT-Luc construct, MTF-1 and either the MTF-1ΔTAD or dnMTF-1 construct. Co-transfection with MTF-1ΔTAD resulted in inhibition of Zn-dependent induction of luciferase activity (Figure 5C). In contrast, co-transfection with an equivalent amount of dnMTF-1 or a 3-fold increase in dnMTF-1 concentration resulted in 50% and 90% greater repression of luciferase activation compared to MTF-1ΔTAD, respectively (Figure 5C). To further assess the inhibitory capacity of dnMTF-1, we tested it against both the MTF-1 and MTF-1ΔCys constructs with additional Zn and Cd treatments. As seen with the previous transfection experiments, luciferase expression regulated by MTF-1 was an order of magnitude higher than MTF-1ΔCys. However, both MTF-1 and MTF-1ΔCys were equally inhibited by the dnMTF-1 construct under both control and metals treatment (Figure 5D).
3.5 Inhibition of endogenous MTF-1 signaling with the dnMTF-1 in zebrafish embryos
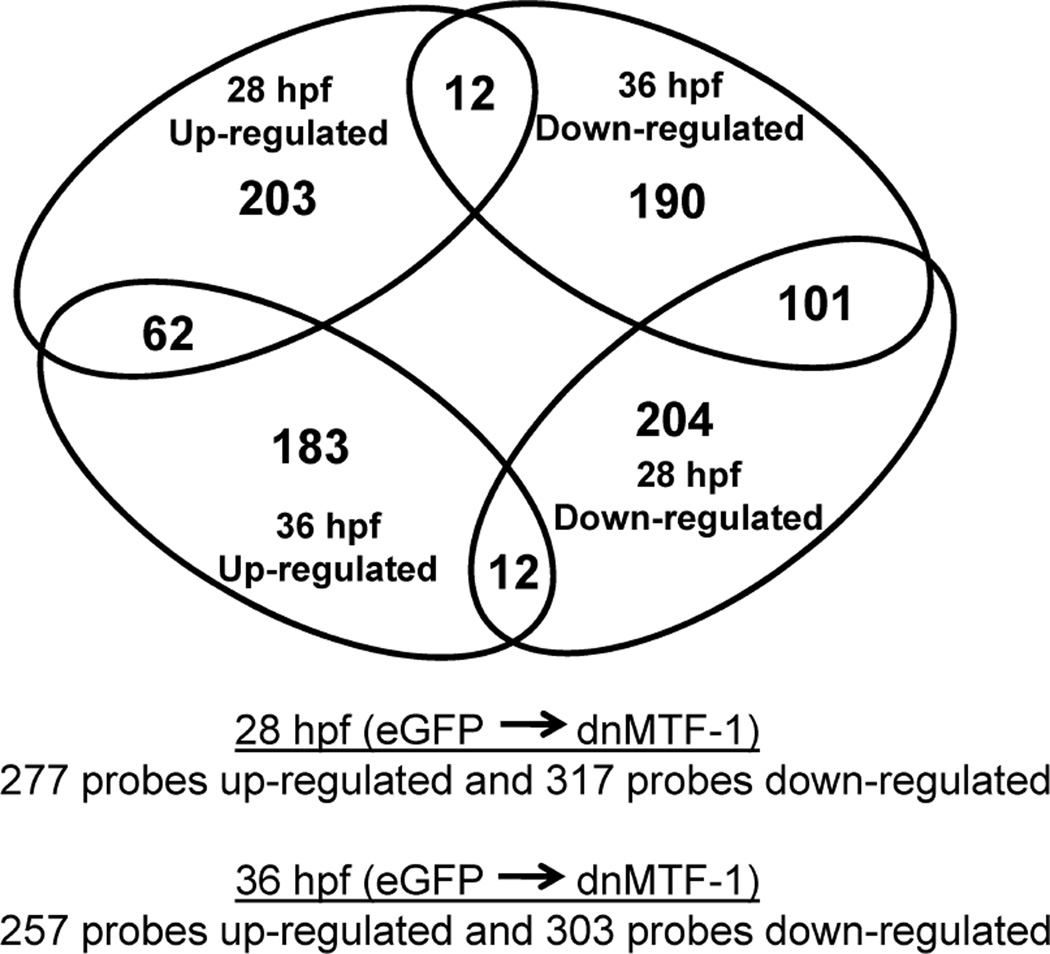
To determine the global effects of the dnMTF-1 on in vivo MTF-1 signaling, we microinjected 1–2 cell zebrafish embryos with in vitro transcribed eGFP or dnMTF-1 mRNA and assessed changes in transcript expression between dnMTF-1 and eGFP controls in embryos at 28 and 36 hpf using a custom zebrafish Agilent 4×44K microarray [33]. Successful microinjection and expression of dnMTF-1 was confirmed by fluorescence microscopy at both timepoints. After microarray normalization, probes exhibiting saturation (578 probes) or signals not above background (0 probes) were removed, leaving 21,315 probes for examination by 2-way ANOVA and Rank Product analysis. A total of 8,556 probes were found significant for treatment and/or time-treatment interaction by the two-factor ANOVA, of which a total of 594 and 560 probes were identified as differentially expressed (up or down relative to eGFP controls) by Rank Product analysis at 28 hpf and 36 hpf, respectively, with interesting overlaps between timepoints. Specifically, at 28 and 36 hpf a total of 277 and 257 probes were up-regulated, respectively; while 317 and 303 probes were down-regulated, respectively (Figure 6). In addition, there were 62 up-regulated probes overlapping between the two timepoints, while 101 down-regulated probes overlapped between the two timepoints.
Figure 6.
A representation of the differential gene expression in response to inhibition of endogenous MTF-1 signaling during early zebrafish development. Microinjection of IVT dnMTF-1 mRNA resulted in a total of 594 and 560 probes differentially expressed at 28 and 36 hpf, respectively, with interesting overlaps between timepoints.
GO terms enriched in the set of genes differentially regulated (up or down) in response to dnMTF-1 IVT mRNA (28 or 36 hpf) included four from GO Biological Processes, one from GO Molecular Function, and two from InterPro (See Table 1 for summary of enrichment). Some additional categories of genes affected by the inhibition of MTF-1 signaling included: nuclear receptors and genes involved in stress signaling, muscle development and contraction, and metal homeostasis, including novel observations in iron and heme homeostasis. Table 2 contains examples of genes differentially expressed during early zebrafish development that were identified by the gene enrichment as well as by literature review. See supplementary files for a complete list of genes differentially expressed as a result of inhibition of MTF-1 signaling as identified by microarray and gene enrichment analysis. The microarray data has additionally been deposited in the Gene Expression Omnibus (GEO) database (accession # GSE51298).
Table 1.
Gene Ontology, KEGG Pathway, and InterPro Enrichment
| Database | Term | Description | # of genes | Adj p-value |
|---|---|---|---|---|
| InterPro | IPR001064 | beta/gamma crystallin | 14 | 1.151×10−5 |
| InterPro | IPR011024 | gamma-crystallin related | 14 | 1.151×10−5 |
| GO Molecular Function | GO:0005212 | Structural constituent of eye lens | 5 | 0.0175 |
| GO Biological Process | GO:0007417 | Central nervous system development | 19 | 0.0113 |
| GO Biological Process | GO:0007420 | Brain development | 16 | 0.011 |
| GO Biological Process | GO:0001654 | Eye development | 17 | 0.011 |
| GO Biological Process | GO:0009408 | Response to heat | 6 | 0.011 |
Table 2.
Summary of Genes Differentially Expressed in Zebrafish Embyos after Inhibition of Endogenous MTF-1 Signaling
| Metal or Heme Homeostasis | |||
| Gene Symbol | Gene Name | 28 hpf | 36 hpf |
| mt2 | Metallothionein 2 | −2.2 | −6.7 |
| hamp2 | Hepcidin-2 precursor | 1.17 | −1.9 |
| hpx | Hemopexin | 2.51 | 2.04 |
| slc39a10 | Solute carrier family 39 (Zn transporter), member 10 | −1.5 | −1.05 |
| slc39a6 | Solute carrier family 39 (Zn transporter), member 6 | −1.7 | −1.3 |
| Structural Constituent of Eye Lens and Eye Development | |||
| Gene Symbol | Gene Name | 28 hpf | 36 hpf |
| aqp0a | Aquaporin 0a | −4.2 | −2.3 |
| aqp0b | Aquaporin 0b | −2.2 | −1.8 |
| atoh7 | Atonal homolog 7 | −2.7 | −2.7 |
| crx | Cone-rod homeobox | 1.0 | −7.9 |
| cryaa | Crystallin, alpha a | −2.6 | −3.3 |
| lim2.1 | Lens intrinsic membrane protein 2.1 | −1.7 | −1.7 |
| lim2.2 | Lens intrinsic membrane protein 2.2 | −2.0 | −1.9 |
| lim2.3 | Lens intrinsic membrane protein 2.3 | −1.9 | −1.6 |
| lim2.4 | Lens intrinsic membrane protein 2.4 | −2.2 | −2.1 |
| Bfsp2 | Phakinin | −2.5 | −3.7 |
| six7 | Sine oculis homeobox 7 | 1.0 | −5.2 |
| six6b | Sine oculis-related homeobox 6b | −1.6 | −1.7 |
| vsx1 | Visual system homeobox 1 protein | −1.6 | −2.3 |
| irbp | Interphotoreceptor retinoid-binding protein | −2.0 | 1.3 |
| Beta/Gamma Crystallin & Gamma Crystallin Related | |||
| crygm2c | Crystallin, gamma M2c | −3.1 | −2.8 |
| crygm2d1 | Crystallin, gamma M2d1 | −1.6 | −3.6 |
| crygm2d4 | Crystallin, gamma M2d3 | −2.1 | −2.6 |
| crygm2d6 | Crystallin, gamma M2d6 | 1.0 | −1.8 |
| crygm3 | Crystallin, gamma M3 | −1.7 | −3.5 |
| crygn2 | Crystallin, gamma N2 | −2.1 | −2.0 |
| crygc-l | Predicted: Crystallin, gamma C-like | −2.2 | −2.9 |
| cryba1b | Crystallin, beta A1b | −1.6 | −1.4 |
| cryba2a | Crystallin, beta A2a | −2.0 | −1.8 |
| cryba2b | Crystallin, beta A2b | −1.9 | −1.6 |
| cryba4 | Crystallin, beta A4 | −1.8 | −1.5 |
| crybb1 | Crystallin, beta B1 | −1.7 | −1.5 |
| Cental Nervous System and Brain Development | |||
| Gene Symbol | Gene Name | 28 hpf | 36 hpf |
| ddc | Dopa decarboxylase | −2.2 | −1.7 |
| drd3 | Dopamine receptor D3 | 1.6 | 1.33 |
| eno2 | Enolase 2 | −1.5 | −1.2 |
| hspa8l | Heat shock protein 8-like | 2.7 | 1.1 |
| neurod2 | Neurogenic differentiation 2 | 1.6 | −2.0 |
| neurog1 | Neurogenin 1 | −1.7 | −1.4 |
| olig4 | oligodendrocyte transcription factor 4 | −1.6 | −1.1 |
| oxtl | Oxytocin-like | −2.0 | −2.0 |
| syngr1a | Synaptogyrin 1 | −1.5 | −1.7 |
| synpr | Synaptoporin | −1.3 | −3.7 |
| sncb | Synuclein, beta | −1.5 | −1.8 |
| Nuclear Receptors, Transcription Factors and Signaling Proteins | |||
| Gene Symbol | Gene Name | 28 hpf | 36 hpf |
| arnt | Aryl hydrocarbon receptor nuclear translocator isoform a | 1.4 | −3.2 |
| esr1 | Estrogen receptor 1 | −1.1 | −2.4 |
| esr2a | Estrogen receptor 2a | 1.5 | 1.17 |
| esrrd | Estrogen-related receptor delta | −1.6 | −1.2 |
| esrrga | Estrogen-related receptor gamma a | 1.3 | −2.0 |
| gadd45al | Growth arrest and DNA-damage-inducible, alpha like | 1.6 | 1.4 |
| hnf4a | Hepatocyte nuclear factor 4, alpha | −1.7 | −1.4 |
| neurog3 | Neurogenin 3 | −1.7 | −1.7 |
| jak1 | Janus kinase 1 | 1.5 | 1.1 |
| junb | jun B proto-oncogene | 2.4 | 1.6 |
| nfia | Nuclear factor I/A | 1.0 | −1.9 |
| nfe2 | Nuclear factor, erythroid-derived 2 | −1.6 | 1.0 |
| rzra | Nuclear factor RZR alpha | −1.2 | −1.9 |
| nr1d1 | Nuclear receptor subfamily 1, group D, member 1 | 2.3 | 1.17 |
| nr2e3 | Nuclear receptor subfamily 2, group E, member 3 | 1.1 | −7.7 |
| socs3a | Suppressor of cytokine signaling 3a | 2.8 | 2.2 |
| socs3b | Suppressor of cytokine signaling 3b | 1.6 | 1.7 |
| fos | v-fos FBJ murine osteosarcoma viral oncogene homolog | 5.5 | 2.2 |
| p53 | p53 tumor suppressor gene | 1.8 | 1.2 |
| Muscle Development and Contraction | |||
| Gene Symbol | Gene Name | 28 hpf | 36 hpf |
| atp2a1l | ATPase, C2+ transporting, cardiac muscle, fast twitch 1 like | −1.8 | −1.5 |
| si:ch211-154h20.7 | Sarcospan | −1.5 | −1.3 |
| tagln | Transgelin | −1.5 | 1.16 |
| zgc: 162523 | Triadin | −1.5 | −1.2 |
| tnnt1 | Troponin, skeletal, slow | 3.3 | 2.8 |
| tnni2a.3 | Troponin I, skeletal, fast 2a.3 | −2.0 | −1.6 |
| tnni2b.1 | Troponin I, skeletal, fast 2b.1 | 1.1 | 1.5 |
| tnni2b.2 | Troponin I, skeletal, fast 2b.2 | −1.7 | −2.4 |
Values highlighted in grey are not statistically significant.
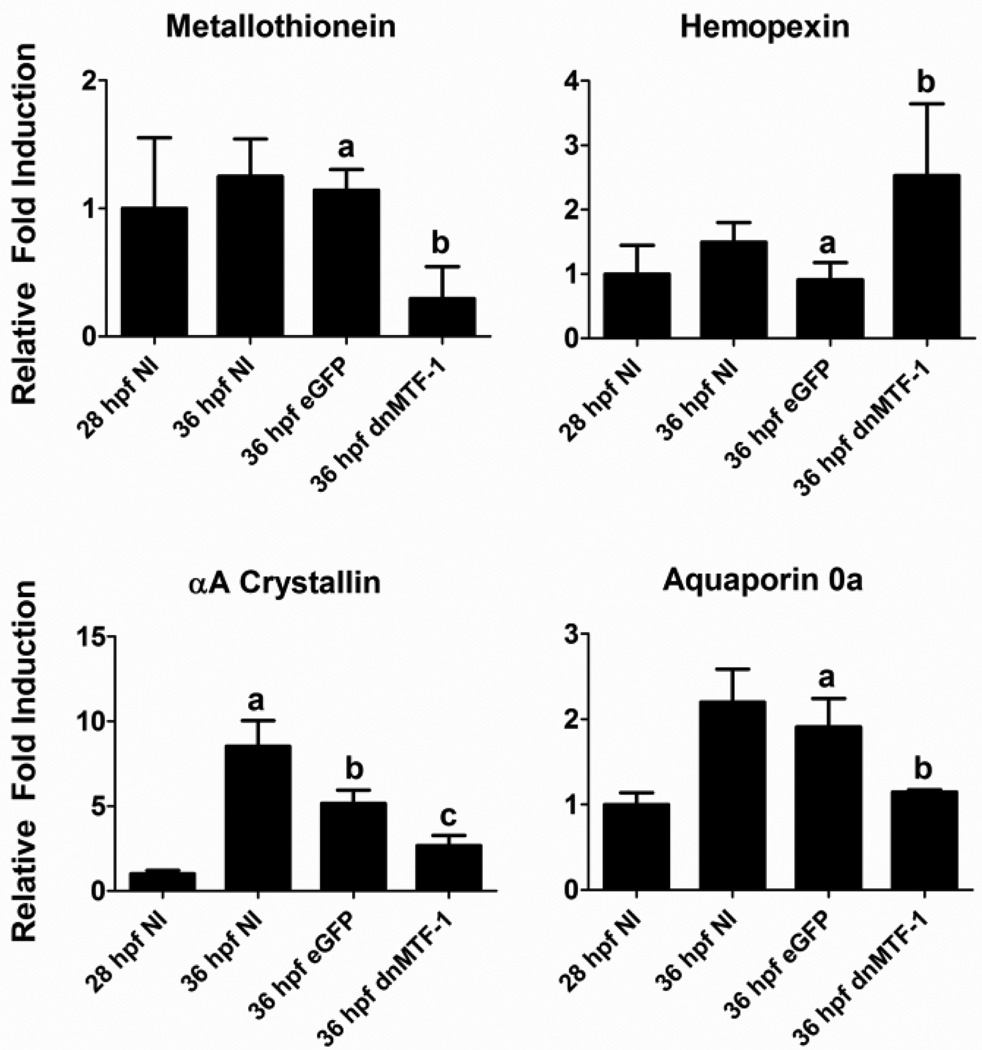
We selected three genes (metallothionein, aquaporin 0a, αA crystallin) that were down-regulated and one gene (hemopexin) that was up-regulated in the microarray experiment for confirmation by real-time RT PCR by independent dnMTF-1 mRNA microinjection experiments (Figure 7). Gene expression for eGFP and dnMTF-1 mRNA injected embryos was assessed at 36 hpf, while gene expression in non-injected embryos was assessed at both 28 and 36 hpf to provide insight into changes in basal expression at both developmental timepoints. Hemopexin was statistically up-regulated at 36 hpf in dnMTF-1 microinjected embryos, confirming the microarray results. Metallothionein, aquaporin 0a, and αA crystallin were all down-regulated in a statistically significantly manner by dnMTF-1 in comparison to the eGPF controls. While there was no significant difference in metallothionein or aquaporin 0a expression between non-injected controls and the eGPF mRNA controls, the αA crystallin expression was significantly different between the two controls, although both had significantly higher expression than the dnTMF-1 treatment. The real-time RT-PCR results were consistently observed in two independent microinjection experiments.
Figure 7.
Confirmation of microarray results with quantitative RT-PCR. Four genes that were significantly altered by inhibition of endogenous MTF-1 signaling in zebrafish embryos were selected for confirmation by qRT-PCR. Results are expressed as fold change from the 28 hpf non-injected control embryos, normalized to 18S ribosomal RNA levels. Different letters denote significant changes in response to microinjection of dnMTF-1 compared to eGPF mRNA (One-way ANOVA, p<0.05). Error bars represent one standard deviation: n = 3 replicates. The results shown in each panel are representative of two independent experiments.
3.6 Promoter analysis of putative novel MTF-1 target genes
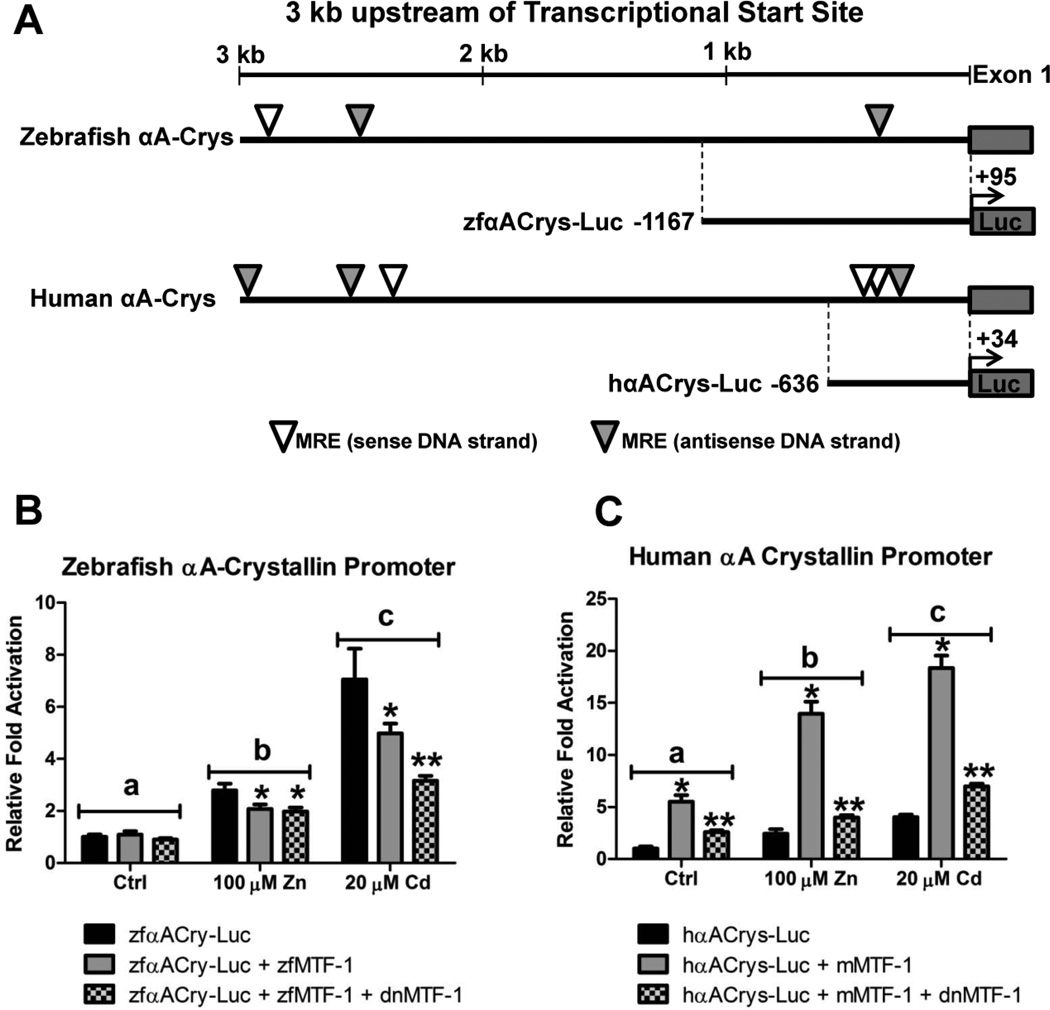
To assess the potential novel role MTF-1 in regulating some of the genes identified by microarray analysis, we sought to perform transient-transfection assays in which we measured the ability of MTF-1 to regulate the transcriptional activation or repression of two selected genes. We first assessed the ability of MTF-1 to serve as a transcriptional activator of the αA-crystallin promoter from both zebrafish and humans. A search of the first 3 kb of the upstream promoter region for zebrafish and human αA-crystallin genes using the conserved MRE motif TGCRCNC yielded three and six potential binding sites in zebrafish and human, respectively (Figure 8A). However, zebrafish only contain one potential MRE within 500 bp of the transcriptional start site, whereas the human promoter contains three potential MREs.
Figure 8.
The role of MTF-1 in regulating zebrafish and human αA crystallin gene expression. A) A schematic of the promoter region of the αA crystallin gene from both human and zebrafish displaying the location of putative MRE motifs and highlighting the regions of the promoters cloned into the pGL4.10[luc2] vector (Promega Corporation, Madison, WI). B) In transient transfection assays, MEF MTF-1 null cells were transfected with 40 ng of zfαACry-Luc, 20 ng of zebrafish MTF-1 and 60 ng of dnMTF-1. The zfαACry-Luc construct was responsive to both Zn and Cd treatment in an MTF-1 independent manner. Overexpression of MTF-1 or MTF-1 and dnMTF-1 inhibited the metal-responsiveness. C) In transient transfection assays, Hepa1c1c7 cells were transfected with 40 ng of hαACry-Luc, 20 ng of mouse MTF-1 and 60 ng of dnMTF-1. The hαACry-Luc construct was responsive to both Zn and Cd treatment in an MTF-1 dependent manner. dnMTF-1 significantly inhibited the MTF-1 mediated activation of hfαACry-Luc. Error bars represent one standard deviation: n = 6 replicates. The results shown in each panel are representative of two independent experiments.
Since an MRE motif can randomly occur about once every 1kb of DNA, transfection assays were performed to determine if MTF-1 influenced transcriptional activation of these proximal promoter regions (zfαACrys-Luc and hαA-Crys-Luc) in the MEF MTF-1 null cell line. For those assays in which we investigated the human αA-crystallin promoter, we present data using a mouse MTF-1 expression construct (generously donated by Dr. G. K. Andrews) but comparable results were obtained with the zebrafish MTF-1 (Supplementary Figure 2). The zebrafish αA-crystallin promoter was responsive to both Zn and Cd in the absence of MTF-1. Surprisingly, transfection of the zebrafish MTF-1 alone or with the dnMTF-1 actually suppressed the metals-dependent induction of luciferase (Figure 8B). Similarly, the human αA-crystallin promoter was also responsive to both Zn and Cd in the MTF-1 null cells. Furthermore, restoration of MTF-1 signaling within the cells significantly enhanced the metals-induced expression of luciferase. However, this enhanced transcriptional activation after restoration of MTF-1 was significantly inhibited by the addition of the dnMTF-1 (Figure 8C).
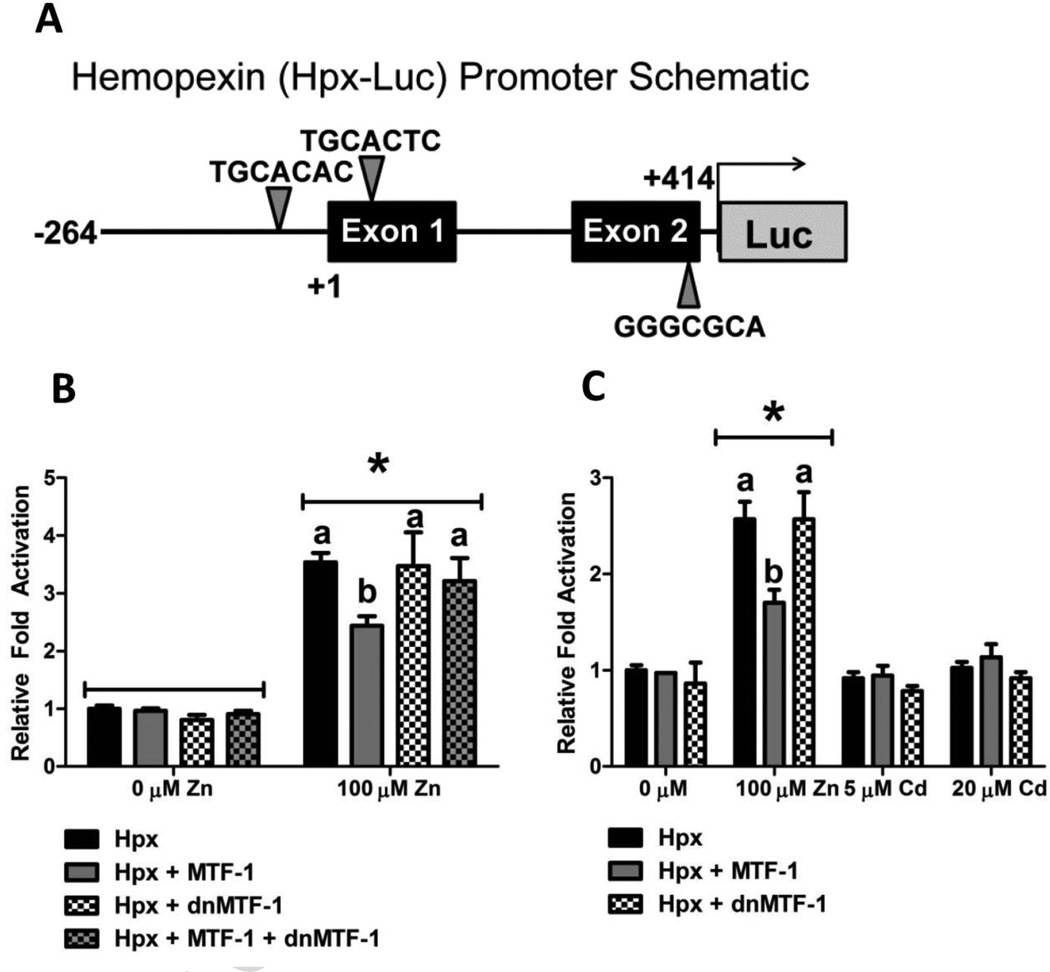
Additional transfection assays were performed to determine if MTF-1 influenced transcriptional activation of the proximal promoter region of the zebrafish hemopexin gene (zfHpx-Luc). While no conserved MRE motifs were identified in the human hemopexin gene, a search of the 3 kb upstream promoter region and first two exons of the zebrafish gene identified 3 potential MREs, one within 60 bp of the transcriptional start site, and one in each of the first two exons (Figure 9A). Initial transfection experiments in which the zfHpx-Luc construct alone or co-transfected with MTF-1 into MEF MTF-1 null cells failed to produce any significant transcriptional activation as determined by luciferase activity under basal or metals-induced conditions (data not shown). However, the zfHPX-Luc construct was able to drive basal expression of luciferase enzyme when transfected into Hepa1c1c7 cells, and displayed a significant increase in luciferase activity in response to Zn treatment (Figure 9B). Furthermore, co-transfection of MTF-1 partially suppressed the Zn-responsiveness of the hemopexin promoter, while co-transfection of dnMTF-1 had no effect on the Zn-responsiveness of the promoter (Figure 9B and 9C). Co-transfection of dnMTF-1 and MTF-1 with the zfHpx-Luc construct appears to inhibit the suppression of luciferase activity seen with co-transfection of MTF-1 alone. Finally, the zfHpx-Luc construct did not respond to Cd treatment (Figure 9C).
Figure 9.
The role of MTF-1 in regulating zebrafish hemopexin gene expression. A) Schematic of the region of the proximal zebrafish hemopexin gene promoter containing putative MRE motifs cloned into the pGL4.10[luc2] vector (Promega Corporation, Madison, WI). B and C) In transient transfection assays, Hepa1c1c7 cells were transfected with 40 ng of zfHpx-Luc, 20 ng of zebrafish MTF-1 and 60 ng of dnMTF-1. The zfHpx-Luc construct was responsive to Zn but not Cd treatment. Addition of exogenous MTF-1, but not dnMTF-1, inhibited the Zn-induced increase in luciferase activity. Co-transfection of dnMTF-1 rescued the inhibition of zfHpx-Luc activation by MTF-1. Error bars represent one standard deviation: n = 6 replicates. The results shown are representative of two independent experiments.
4. Discussion
MTF-1 is a nucleocytoplasmic shuttling protein that contains a Zn-responsive DNA-binding domain consisting of six Zn-fingers, and three distinct transcriptional activation domains that differ in their amino acid composition, the acidic domain, the proline-rich domain and the serine/threonine-rich domain, respectively. In addition, the C-terminal region of MTF-1 contains a cysteine-rich motif (C-X-C-X-C-X-C) believed to be conserved in vertebrates and has been shown to be necessary for full metal induction [8]. Recent studies have also identified a conserved SUMO-interacting motif ((V/I-X-(V/I)-(V/I)) that immediately precedes the cysteine-rich domain and appears to play important roles in regulating the subcellular localization and transcriptional activity of MTF-1 [7, 43]. A truncated MTF-1 homologue in zebrafish lacking the cysteine-rich motif typically found in vertebrates has previously been reported [5]. We report here the identification and functional characterization of the full length zebrafish MTF-1 isoform, which contains all of the domains and motifs conserved in other vertebrate species. We also provide the functional assessment of a constitutively-nuclear, dominant-negative MTF-1 isoform and demonstrate its utility as a molecular tool for inhibiting endogenous MTF-1 signaling in an in vivo model.
4.1 MTF-1 isoform diversity in fish species
Previous studies have described complete MTF-1 transcripts in two other fish species, the pufferfish (Takifugu rubripes) and tilapia (Tilapia aurea×Tilapia nilotica) [4, 31], which encode for 780 and 811 amino acid proteins, respectively. Using the Takifugu MTF-1 sequence as a guide, we were able to identify a missing exon and amplify a full zebrafish MTF-1 transcript containing an open reading frame that encodes for a 722 amino acid protein (Figure 1C). However, the identification of the complete MTF-1 transcript from zebrafish raises the question of whether the truncated MTF-1 represents an incompletely spliced mRNA sequence with a retained intron or a true alternatively spliced variant. Although there are no current publications to provide detailed descriptions, the NCBI nonredundant database does contain one full length and four putative alternatively spliced MTF-1 transcripts for the rainbow trout (Oncorhynchus mykiss; accession numbers AY586336-AY586340). In addition, earlier papers have identified potential MTF-1 splice variants in both tilapia [31] and carp [44].
The truncated zebrafish MTF-1 sequence was originally identified in a traditional phage λ cDNA library screening [5], whereas the carp and tilapia truncated MTF-1 sequences were identified by PCR amplification. Such approaches are subject to biases that could allow for the cDNA synthesis of a partially spliced transcript with a retained intron. For example, partially spliced transcripts have been identified by phage λ cDNA library screening [45] and more recent next-generation sequencing approaches regularly yield a small percentage of intron retaining transcripts [46]. Furthermore, partially spliced transcripts could easily be converted to cDNA through the use of random primers during cDNA synthesis or by the use of an oligo(dT) primer which can produce cDNAs through priming of A-rich regions of the transcript [47].
The truncated zebrafish MTF-1 transcript lacks the last coding exon (exon 10, Figure 1D) and contains an extremely long 3' UTR [5] that shares no similarity to the full length MTF-1 transcript. The 3' UTR terminates at a region within the ninth intron that contains 11 consecutive A’s, and there is no conserved polyadenylation motif (AATAAA) or any clear alternative polyadenylation signals found in other vertebrates [48] within the standard 10–30 nucleotides upstream of the cleavage/polyadenylation site [49]. The presence of a canonical 5' donor site (CAG/GT) at the end of exon 9 and preceding the predicted stop codon of the truncated zebrafish MTF-1 (Figure 1B) is inconclusive because this motif can occur at the exon/intron boundary or result from the ligation of two exons. Although the zebrafish genomic sequence assembly of exon 9 and the 5' region of the following intron is poor, the intron appears to be at least 3,600 nucleotides in length and significantly longer than the 1,520 nucleotides that make up the 3' UTR of the truncated zebrafish MTF-1 transcript. However, the last 1,440 nucleotides of the truncated zebrafish MTF-1 3' UTR sequence share ~99% similarity with the middle region of the intron that is still 1,560 nucleotides upstream of the beginning of exon 10. We cannot definitively rule out the possibility of an alternative splice variant because this discrepancy of ~1,480 nucleotides at the beginning of the intron that do not occur in the 3' UTR could result from an alternative splice site within the intron. However, there is still the strong possibility that the truncated zebrafish MTF-1 sequence results from a retained intron and the discrepancy in the intron length could just result from differences in genetic structures between individual zebrafish strains [50, 51].
A closer investigation of the tilapia and carp MTF-1 splice variants was performed to see if they provided any additional insight into the potential for alternative splicing of MTF-1 in fish species. The tilapia MTF-1 splice variant (tiMTF1-S) shows that the truncation of the transcript occurs at a point in the cDNA sequence which also contains a canonical 5' donor site (CAG/GT) upstream of the predicted stop codon [31]. Assuming the exon/intron gene structure for MTF-1 in fugu and zebrafish (Figure 1D) is conserved in tilapia, this truncation point is consistent with the exon/intron boundary at the end of the fourth coding exon. Thus, the tiMTF1-S isoform only encodes for the first five Zn-fingers in the DNA-binding domain and lacks transactivation potential. The carp MTF-1 transcripts were cloned using PCR with primers designed from an alignment that included the truncated zebrafish MTF-1; therefore, the subsequent carp MTF-1 sequences are incomplete and missing the C-terminal cysteine motif [44, 52] encoded by exon 10. However, in contrast to the tilapia MTF-1 splice variant, a comparison of the two carp MTF-1 sequences with the conserved exon/intron boundaries of the fugu and zebrafish MTF-1 gene suggests that the carp splice variant [52] is the product of a transcript missing the seventh coding exon of the MTF-1 gene but retaining exons 8 and 9. As seen with the tiMTF1-S isoform, the carp MTF-1 splice variant encodes for an MTF-1 isoform lacking any significant portion of the transactivation domain and would likely display dominant-negative activity. While an argument can be made for the identification of transcripts containing a retained intron in the case of the zebrafish and tilapia MTF-1 sequences, the carp MTF-1 splice variant is much more consistent with a properly spliced transcript missing an internal exon. Interestingly, a summary investigation of both mouse and human MTF-1 sequences within the Ensembl genome browser and AceView browser, respectively, did yield potential MTF-1 splice variant transcripts. However, despite the presence of these various truncated MTF-1 transcripts from various vertebrate species only the identification of an endogenous functional MTF-1 protein encoded from a splice variant transcript will provide definitive proof.
To fully explore the possible expression of two separate MTF-1 transcripts in zebrafish, we designed two different sets of real time PCR primers, one that would only amplify the full length MTF-1 transcript (C_MTF-1 primers) and a second set that would simultaneously amplify both MTF-1 transcripts (N_MTF-1 primers). So, if there was an abundance of both the truncated and full length MTF-1 transcripts, the N_MTF-1 primer set should show a statistically significant increase in transcript numbers compared to the C_MTF-1 primer set. We investigated the basal developmental expression at five different timepoints (24 to 120 hpf) and the sex-specific differences in basal and Cd-induced expression in eight adult tissues (Figures 2 and 3). Since the truncated MTF-1 was originally identified from a cDNA library generated from embryo RNA we took a very close look at the PCR results from the developmental time series and found on average less than a 1% difference in transcript abundance between the two primer sets. Although the sample variability was higher in the adult tissue samples, we were unable to detect any significant difference in MTF-1 transcript abundance between the two primer sets under basal conditions or after Cd treatment. Based on these observations, we concluded that the full length MTF-1 transcript is the major isoform expressed in zebrafish tissues.
4.2 Comparison of MTF-1ΔCys and MTF-1 in transactivation potential
Although our data suggests that the full length MTF-1 is the only significant transcript in zebrafish, we wanted to investigate the difference in transactivation potential between the full length MTF-1 and a truncated version lacking the last exon (designated here as MTF-1ΔCys) to determine any differences that might be relevant to biological function. This was of particular interest given that the carp and tilapia MTF-1 splice variants appear to encode for MTF-1 isoforms that would display dominant-negative function. This suggests that the expression of these splice variants could potentially serve as a feedback mechanism to regulate MTF-1 signaling. We consistently observed up to an order of magnitude greater transcriptional activation of the zfMT-Luc promoter construct with full length MTF-1 in both MTF-1 null cells and Hepa1c1c7 liver cells compared to MTF-1ΔCys (Figures 4 and 5). This is consistent with previous studies demonstrating the requirement of the cysteine-rich motif for full transcriptional activation of MTF-1 by various heavy metals [8, 53]. The inability of MTF-1ΔCys to negatively impact basal endogenous MTF-1 signaling in Hepa1c1c7 cells while still mediating an increase in fold activation in response to metal treatment (Figure 4), suggests that it is very unlikely that a MTF-1ΔCys isoform would play any significant role in altering endogenous signaling.
In addition to the conserved cysteine motif, the last exon of the zebrafish MTF-1 gene also encodes for the SUMO-interacting motif (601VIIIKQEE608) which also plays a very important role in determining the transactivation potential of MTF-1. Recent studies have demonstrated that sumoylation of MTF-1 suppresses its transcriptional activity [7] even though it does not alter the Zn-induced nuclear translocation or MRE-binding activity of MTF-1 [7, 43]. Although the exact function of sumoylated MTF-1 is not known, it is speculated that the SUMO moiety may interact with either transcriptional suppressors or activators to regulate DNA-binding activity [43]. While a mouse MTF-1 K627R mutant was no longer able to undergo sumoylation, it did not display any reduction in transcriptional activity compared to the wild-type MTF-1 [7]. Thus it is likely that the significant reduction in transcriptional activation associated with the zebrafish MTF-1ΔCys is due to the lack of the cysteine motif. However, we did observe a significant difference between our zebrafish MTF-1-eGPF fusion protein and what has been reported for the mouse MTF-1-eGPF fusion protein. Liu et al. (2011) used an MTF-1-eGFP C-terminal fusion to demonstrate that the lack of transcriptional activation of the sumoylated mouse MTF-1 was not due strictly to steric hindrance. In contrast, a C-terminal eGFP fusion to the full length zebrafish MTF-1 (MTF-1-GFP) resulted in the same significant reduction in basal transcriptional activation as seen with the MTF-1ΔCys (Figure 4C). Downstream of the cysteine-rich motif, the C-terminal region of MTF-1 is poorly conserved among vertebrate species. Thus, it is possible that structural differences between mouse and zebrafish may result in differences in steric hindrance that could interfere with zebrafish MTF-1’s potential homodimerization [54] or interaction with other co-factors [20, 55] that may be necessary for full transcriptional activation. Future studies are required for a full characterization of the role of both the sumoylation motif and the cysteine-rich motif in the transcriptional activity of the zebrafish MTF-1.
4.3 Characterization of a dominant-negative zebrafish MTF-1 construct
Although antisense morpholinos or the generation of null mutants are effective tools for investigating the function of proteins during zebrafish development, given the essential nature of MTF-1 in mouse development we wanted to determine if a dominant-negative MTF-1 construct could serve as a useful molecular tool for inhibiting endogenous MTF-1 signaling. If successful, the expression of such a repressor under the control of an inducible promoter, e.g. Tet-On system [56, 57], could be useful for studies in both embryos and adult tissue. Thus, we created a dominant-negative MTF-1 isoform by removing the entire transactivation domain (designated as MTF-1ΔTAD) from the full length MTF-1 isoform and performing site-directed mutagenesis to replace the RGEYT linker motif with TGEKP between Zn fingers 1 and 2. Using the mouse MTF-1 as a model, Li et al (2006) demonstrated that mutation of the RGEYT linker motif to TGEKP abolished the Zn-sensing capability of MTF-1 and converted MTF-1 to a constitutively nuclear transcription factor. We hypothesized that mutating this linker would produce a more efficient inhibitor of endogenous MTF-1 signaling since the cellular localization of MTF-1ΔTAD is still influenced by cellular stress. As seen in Figure 5A, mutation of this motif resulted in constitutive nuclear localization of dnMTF-1, demonstrating that the Zn-sensing capabilities of Zn fingers 1 and 2 are conserved between mouse and zebrafish. The dnMTF-1 produced similar inhibitory results as seen in a previous paper using a mouse dominant negative MTF-1 [53], and was much more successful at inhibiting MTF-1 signaling compared to MTF-1ΔTAD (Figure 5). Based on all of these observations, we can conclude that the dnMTF-1 is an effective inhibitor of MTF-1 signaling.
4.4 In vivo effects of MTF-1 inhibition in zebrafish embryos
Given the essential role for MTF-1 in mouse development, it is reasonable to assume that MTF-1 could play a similar role in zebrafish development. However, our initial goal presented here was not to perform a comprehensive investigation of the role of MTF-1 in zebrafish development. Rather, we wanted to test the utility of the dnMTF-1 as a potential alternative molecular tool to the use of antisense morpholinos or the generation of MTF-1 null mutants. Based on observations from the mouse MTF-1 “knockout” model and the significant role that liver degeneration likely played in the embryonic mortality, we chose two timepoints (28 and 36 hpf, early to mid-pharyngula stage) representing early stages of liver development [58] to test the in vivo efficacy of the dnMTF-1. In a study by Ober et al. (2006), microinjection of a wnt2bb MO significantly inhibited the development of the hepatic bud at 28 hpf and subsequent development of the liver such that normal development was delayed by at least 30 hours without any apparent negative effects on the embryo [59]. This earlier study demonstrates the resiliency of the zebrafish embryo and supports the idea that even if the role of MTF-1 in liver development is conserved among vertebrates, our early “knockdown” of MTF-1 during zebrafish development would not result in embryonic lethality due to liver degeneration. Selection of these early timepoints was also beneficial because they offered insight into novel potential roles for MTF-1 in embryonic development.
Significant changes in global transcript expression were observed at both developmental timepoints as a result of inhibition of endogenous MTF-1 signaling (Figure 6, Table 1). Significant down-regulation of endogenous MT expression and additional known MTF-1 target genes, such as hepcidin and β-synuclein [60, 61], confirm the target efficacy of the dnMTF-1. Interestingly, some known MTF-1 target genes were differentially expressed in zebrafish as a result of inhibition of endogenous MTF-1 signaling but responded in the opposite manner as expected. For example, zip10 was significantly down-regulated in zebrafish embryos at 28 hpf (Table 2) suggesting that MTF-1 is required for basal expression of Zn transporter Zip10 (solute carrier family 39, member 10; SLC39A10) during zebrafish development. Interestingly, MTF-1 has been shown to repress expression of zip10 in a Zn-specific manner in both mice and zebrafish [62, 63]. However although MTF-1 repressed zip10 expression in the presence of excess Zn, loss of the MRE in the proximal promoter of mouse zip10 also abolished transactivation of the promoter luciferase construct, suggesting that MTF-1 played a role in regulating basal expression [62]. In the case of the zebrafish zip10 gene the situation is a little more complex due to the presence of two promoter regions with differing regulatory activity. There is a proximal promoter preceding the first exon of the zebrafish gene that contains two MREs and appears to regulate zip10 induction by Zn in an MTF-1-dependent manner. In contrast, there is a second proximal promoter preceding an alternative exon 1 (exon 1’) that contains one MRE, with two additional MREs in the intron between exon 1’ and exon 2. It is this second promoter that appears to be responsible for the MTF-1-mediated repression of zip10 expression in response to high Zn levels. Interestingly, removal of the MREs in promoter 1 abolishes Zn-dependent activation of the promoter in HepG2 liver carcinoma cells, while removal of the MREs in promoter 2 abolishes basal activity of the promoter in HepG2 cells [63] again suggesting a role for MTF-1 in regulating the basal expression of the zebrafish zip10. These observations combined with the novel results represented in this paper supports a more complex mechanism of regulation in which MTF-1 might be responsible for promoting basal expression of zip10 while repressing expression in the presence of excess zinc.
A previous study demonstrated that the induction of suppressors of cytokine signaling 3 (SOCS3) by zinc occurred in an MTF-1-dependent manner in HepG2 cells [64]. In contrast, based on the microarray results both SOCS3 zebrafish genes (socs3a and socs3b) were significantly up-regulated in zebrafish embryos at both 28 and 36 hpf (Table 2). However, previous studies have demonstrated an increase in cellular oxidative stress associated with Zn deficiency which leads to DNA damage and the up-regulation of AP-1 [65, 66]. A role for AP-1 has been implicated in the regulation of SOCS3 [67], and c-Fos, a member of the heterodimeric AP-1 transcription factor, is also highly up-regulated in zebrafish embryos at both time points suggesting an alternative mechanism for induction of the SOCS genes.
Several transcription factors, including several that belong to the family of basic helix-loop-helix (bHLH) transcription factors (e.g., arnt, neurod2, neurog1, neurog3, olig4; Table 2), were differentially regulated in response to inhibition of endogenous MTF-1 signaling. Since MTF-1 is considered to be a ‘master regulator’ of Zn homeostasis, it is important to determine if the observed changes in gene expression are directly related to MTF-1 regulation or secondarily affected through disruption of normal Zn homeostasis. For example, previous studies have linked zinc homeostasis to the transcriptional regulation of cytochrome P450 1A (CYP1A), the major target gene of the aryl hydrocarbon receptor (AHR) and AHR nuclear translocator (AHR-ARNT) complex [68]. Aortic endothelial cells pretreated with N,N,N',N-tetrakis(2-pyridylmethyl) ethylenediamine (TPEN), a cell permeable, heavy metal chelator with high-affinity for Zn, displayed a significant reduction in CYP1A gene expression and EROD activity in response to PCB77 or β–naphthoflavone (β-NF) treatment [68]. We observed significant down-regulation of cyp1b1 and cyp1a1 in 36 hpf embryos microinjected with dnMTF-1 IVT mRNA (Table 2 and Supplementary Table 1), most likely due to the similar reduction of ARNT expression at 36 hpf (Table 2). To test the role of MTF-1 in direct regulation of these genes we pre-treated Hepa1c1c7 cells with Zn or TPEN and assessed the expression of arnt and cyp1a in cells further treated with either DMSO or benzo[a]pyrene (BaP). TPEN treatment resulted in significant reduction of both genes and overexpression of MTF-1 coupled with additional Zn treatment failed to rescue the TPEN-dependent inhibition (Supplementary Figure 3). Thus, at least in the case of arnt expression and its downstream targets, disruption of zinc homeostasis appears to be the causative factor leading to differential gene expression. These observations raise some interesting questions regarding the role of zinc homeostasis on bHLH transcription factor signaling pathways and highlight the importance of functional studies to confirm the direct role of MTF-1 in gene regulation.
Three additional groups of genes differentially expressed in zebrafish embryos as a result of inhibition of endogenous MTF-1 signaling were genes involved in central nervous system and brain development, eye development and muscle development. This is not surprising considering that brain and eye tissues are highly enriched in Zn. Although previous studies have demonstrated that MTF-1 is not essential for neural differentiation [23, 69], it has been acknowledged that further studies are needed to determine if MTF-1 plays a role in the structural development of the central nervous system [69]. In addition, Zn has been shown to be extremely important for both developmental and adult neurogenesis, and has been shown to play a role in synaptic signaling through glutamatergic neurons (reviewed in [70, 71]). Thus, MTF-1 could still play a significant functional role in regulating neural activity through regulation of zinc homeostasis even though it may not be essential for development or differentiation. Furthermore, Cd exposure to zebrafish during early development has been shown to disrupt neurogenesis, axon growth and muscle type development [72, 73]. Given that Cd is such a strong activator of MTF-1 signaling, these teratogenic phenotypes may be related to misregulation of several of the muscle-related genes identified in the microarray screening (Table 2).
As a first step towards confirming a novel regulatory role for MTF-1 in some of the genes identified in the microarray screening, we searched the 3kb upstream promoter region of several eye-related genes identified from our zebrafish screening results, including the humans homologues, for homology to the core consensus sequence, TGCRCNC [14]. We focused our attention on the diversity of crystallin genes, which play a very prominent role in the formation of the lens. While one or more MRE motifs were identified in crystallin genes (data not shown), we sought to further investigate the αA-crystallin gene from both zebrafish and human (Figure 8). We were particularly interested in the αA-crystallin gene because the first MRE site in the zebrafish αA-crystallin promoter occupies positions −551 to −557 within the lens-specific core promoter region as demonstrated by previous studies [74]. To determine if the MTF-1 motif played a role in determining the lens-specific expression in zebrafish, we constructed an eGFP transgenic construct containing the first 550 bp of the αA-crystallin promoter using the Tol2kit [75] and found that the motif is not required for lens-specific expression in the zebrafish eye (Supplementary Figure 4).
Although MTF-1 was not required for lens-specific expression of αA-crystallin, we still wanted to take a comparative approach to determine if MTF-1 played in role in transcriptional regulation. Additional assessments were performed using the dual luciferase assay with proximal promoter constructs from both zebrafish and humans αA-crystallin genes because typically MRE motifs within the first few hundred base pairs of the transcriptional start site play the most influential role on transcriptional activation. We successfully demonstrated that the αA-crystallin proximal promoter (Figure 8A) from both zebrafish and humans was responsive to Zn and Cd treatment in the MEF MTF-1 null cells. This observation contrasts a previous study using the human (SRA01/04) lens epithelial cell line in which the αA-crystallin gene demonstrated a Cu-responsiveness, whereas Cd or Zn treatment did not appear to have any effect on αA-crystallin expression [76]. However, it should be noted that cystallin genes are members of the heat shock protein family and may have very different functional roles depending on the cell type in which they are expressed. For example, α-crystallins (αA and αB) have been shown to play a prominent role in response to a variety of cellular stressors by inhibiting apoptosis in multiple cell types, including non-ocular tissues [77–79]. Interestingly, our data suggest that only the human αA-crystallin displayed enhanced luciferase activity in an MTF-1-dependent manner (Figure 8B and 8C). In contrast, the metal-mediated activation of the zebrafish αA-crystallin promoter was actually repressed by overexpression of MTF-1 (Figure 8). Although this was not the expected outcome, a previous study did demonstrate a reduction in expression of several crystallin genes (αB, βB1, and γM2) in response to Zn treatment that was reversed by siRNA MTF-1 knockdown in zebrafish ZF4 embryonic fibroblast cells [80], suggesting the MTF-1 could be transcriptionally repressing their expression. It is possible that the observed down-regulation of αA-crystallin in zebrafish embryos was a result of disruption of Zn homeostasis, but additional future studies are required to investigate any potential role that MTF-1 may have in regulating crystallin gene expression. However, the observations with the αA-crystallin genes suggest a similar dual role for MTF-1 in transcriptional regulation as suggested with the Zip10 expression.
Lastly, there are have been a few published papers supporting the role of MTF-1 in regulating the expression of ferroportin 1 (FPN1), an iron efflux transporter that can also transport Zn [81]. In addition, MTF-1 has been shown to regulate the Zn-dependent induction of hepcidin [61], a peptide hormone considered to be the “master regulator” of iron homeostasis by binding to FPN1 to regulate iron export. Our microarray data demonstrated a down-regulation of hepcidin 2 (hamp2) at 36 hpf in response to inhibition of endogenous MTF-1 signaling, supporting a conserved role for MTF-1 in the regulation of zebrafish hepcidin 2. Furthermore, our data suggest that MTF-1 may play a potential role as a transcriptional repressor of zebrafish hemopexin (Figure 9), a heme-binding protein that plays a major role in preserving the iron cellular content. In support of our observation, Zheng et al. (2010) identified 14 genes, differentially expressed in zebrafish gills in response to zinc-treatment, by GO enrichment that were associated with the Molecular Function term “iron ion binding” [82]. Interestingly, hemopexin was downregulated in 4 out of the 5 zinc-treatments in this study. Although we can only speculate about potential roles for MTF-1 in regulating iron and heme homeostasis, our results certainly highlight several future avenues of research.
4.5 Conclusions
The dnMTF-1 was able to successfully inhibit endogenous MTF-1 signaling during early zebrafish development, supporting the idea that a transgenic zebrafish expressing the dnMTF-1 under the control of an inducible promoter would serve as a valuable molecular tool for further investigation into the regulatory role of MTF-1. Furthermore, the research presented here highlights some important goals for future research, including the ability of MTF-1 to serve both as a transcriptional activator and a transcriptional repressor, and the distinct differences in the regulatory role of MTF-1 between zebrafish and humans that could prove useful for studies aimed at understanding the evolution of zinc homeostasis.
Supplementary Material
Highlights.
Zebrafish express a complete MTF-1 transcription factor with all conserved domains.
Dominant-negative MTF-1 suppresses in vivo signaling in zebrafish embryos.
Inhibition of MTF-1 signaling suggests novel roles in eye and brain development.
Acknowledgements
This work was supported by the National Institutes of Health Pathway to Independence grant R00ES017044 from the National Institute of Environmental Health Sciences awarded to M.J. Jenny. The authors would like to thank Drs. Jared V. Goldstone and John J. Stegeman from Woods Hole Oceanographic Institution for access to the custom zebrafish microarray design thru Agilent Technologies (Santa Clara, CA, USA). We thank Dr. Glen Andrews (University of Kansas) for providing us with the MEF MTF-1 null cell line and the mouse MTF-1 expression construct. We also thank Dr. King Ming Chan (Chinese University of Hong Kong) for the zebrafish MT promoter-luciferase construct. The U.S. Government is authorized to produce and distribute reprints for governmental purposes notwithstanding any copyright notation that may appear hereon. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interests.
References
- 1.Radtke F, Heuchel R, Georgiev O, Hergersberg M, Gariglio M, Dembic Z, Schaffner W. Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. EMBO J. 1993;12:1355–1362. doi: 10.1002/j.1460-2075.1993.tb05780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang B, Egli D, Georgiev O, Schaffner W. The Drosophila homolog of mammalian zinc finger factor MTF-1 activates transcription in response to heavy metals. Mol. Cell. Biol. 2001;21:4505–4514. doi: 10.1128/MCB.21.14.4505-4514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller HP, Brungnera E, Georgiev O, Badzong M, Muller KH, Schaffner W. Analysis of the heavy metal-responsive transcription factor MTF-1 from human and mouse. Somat. Cell. Mol. Genet. 1995;21:289–297. doi: 10.1007/BF02257464. [DOI] [PubMed] [Google Scholar]
- 4.Auf der Maur A, Belser T, Elgar G, Georgiev O, Schaffner W. Characterization of the transcription factor MTF-1 from the Japanese pufferfish (Fugu rubripes) reveals evolutionary conservation of heavy metal stress response. Biol. Chem. 1999;380:175–185. doi: 10.1515/BC.1999.026. [DOI] [PubMed] [Google Scholar]
- 5.Chen WY, John JA, Lin CH, Chang CY. Molecular cloning and developmental expression of zinc finger transcription factor MTF-1 gene in zebrafish, Danio rerio. Biochem. Biophys. Res. Commun. 2002;291:798–805. doi: 10.1006/bbrc.2002.6517. [DOI] [PubMed] [Google Scholar]
- 6.Radtke F, Georgiev O, Muller HP, Brugnera E, Schaffner W. Functional domains of the heavy metal-responsive transcription regulator MTF-1. Nucleic Acids Res. 1995;23:2277–2286. doi: 10.1093/nar/23.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu YC, Lin MC, Chen HC, Tam MF, Lin LY. The role of small ubiquitin-like modifier-interacting motif in assembly and regulation of metal-responsive transcription factor 1. J. Biol. Chem. 2011;286:42818–42829. doi: 10.1074/jbc.M111.253203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Zhang B, Harmon PM, Schaffner W, Peterson DO, Giedroc DP. A novel cysteine cluster in human metal-responsive transcription factor 1 is required for heavy metal-induced transcriptional activation in vivo. J. Biol. Chem. 2004;279:4515–4522. doi: 10.1074/jbc.M308924200. [DOI] [PubMed] [Google Scholar]
- 9.Smirnova IV, Bittel DC, Ravindra R, Jiang H, Andrews GK. Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. J. Biol. Chem. 2000;275:9377–9384. doi: 10.1074/jbc.275.13.9377. [DOI] [PubMed] [Google Scholar]
- 10.Murphy BJ, Andrews GK, Bittel D, Discher DJ, McCue J, Green CJ, Yanovsky M, Giaccia A, Sutherland RM, Laderoute KR, Webster KA. Activation of metallothionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Res. 1999;59:1315–1322. [PubMed] [Google Scholar]
- 11.Saydam N, Georgiev O, Nakano MY, Greber UF, Schaffner W. Nucleo-cytoplasmic trafficking of metal-regulatory transcription factor 1 is regulated by diverse stress signals. J. Biol. Chem. 2001;276:25487–25495. doi: 10.1074/jbc.M009154200. [DOI] [PubMed] [Google Scholar]
- 12.Stitt MS, Wasserloos KJ, Tang X, Liu X, Pitt BR, St Croix CM. Nitric oxide-induced nuclear translocation of the metal responsive transcription factor, MTF-1 is mediated by zinc release from metallothionein. Vascul. Pharmacol. 2006;44:149–155. doi: 10.1016/j.vph.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Bittel DC, Smirnova IV, Andrews GK. Functional heterogeneity in the zinc fingers of metalloregulatory protein metal response element-binding transcription factor-1. J. Biol. Chem. 2000;275:37194–37201. doi: 10.1074/jbc.M003863200. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Agarwal A, Giedroc DP. Structural and functional heterogeneity among the zinc fingers of human MRE-binding transcription factor-1. Biochemistry. 1998;37:11152–11161. doi: 10.1021/bi980843r. [DOI] [PubMed] [Google Scholar]
- 15.Koizumi S, Suzuki K, Ogra Y, Gong P, Otuska F. Roles of zinc fingers and other regions of the transcription factor human MTF-1 in zinc-regulated DNA binding. J. Cell. Physiol. 2000;185:464–472. doi: 10.1002/1097-4652(200012)185:3<464::AID-JCP18>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Li K, Kimura T, Laity JH, Andrews GK. The zinc-sensing mechanism of mouse MTF-1 invovles linker peptides between the zinc fingers. Mol. Cell. Biol. 2006;26:5580–5587. doi: 10.1128/MCB.00471-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Searle P, Stuart G, Palmiter RD. Metal regulatory elements of the mouse metallothionein-I gene. Experentia. 1987;52:407–414. doi: 10.1007/978-3-0348-6784-9_39. [DOI] [PubMed] [Google Scholar]
- 18.Searle PF. Zinc dependent binding of a liver nuclear factor to metal response element MRE-a of the mouse metallothionein-I gene and variant sequences. Nucleic Acids Res. 1990;19:4683–4689. doi: 10.1093/nar/18.16.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koizumi S, Suzuki K, Ogra Y, Yamada H, Otsuka F. Transcriptional activity and regulatory protein binding of metal-responsive elements of the human metallothionein-IIA gene. Eur. J. Biochem. 1999;259:635–642. doi: 10.1046/j.1432-1327.1999.00069.x. [DOI] [PubMed] [Google Scholar]
- 20.Daniels PJ, Andrews GK. Dynamics of the metal-dependent transcription factor complex in vivo at the mouse metallothionein-I promoter. Nucleic Acids Res. 2003;31:6710–6721. doi: 10.1093/nar/gkg913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samson SL, Gedamu L. Molecular analyses of metallothionein gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 1998;59:257–288. doi: 10.1016/s0079-6603(08)61034-x. [DOI] [PubMed] [Google Scholar]
- 22.Sims HI, Chirn G-W, II MTM. Single nucleotide in the MTF-1 binding site can determine metal-specific transcription activation. Proc. Nat. Acad. Sci., USA. 2012;109:16516–16521. doi: 10.1073/pnas.1207737109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunes C, Heuchel R, Georgiev O, Muller KH, Lichtlen P, Bluthmann H, Marino S, Aguzzi A, Schaffner W. Embryonic lethality and liver degeneration in mice lacking the metal-responsive transcriptional activator MTF-1. EMBO J. 1998;17:2846–2854. doi: 10.1093/emboj/17.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Wimmer U, Lichtlen P, Inderbitzin D, Stieger B, Meier PJ, Hunziker L, Stallmach T, Forrer R, Rulicke T, Georgiev O, Schaffner W. Metal-responsive transcription factor-1 (MTF-1) is essential for embryonic liver development and heavy metal detoxification in the adult liver. FASEB J. 2004;18:1071–1079. doi: 10.1096/fj.03-1282com. [DOI] [PubMed] [Google Scholar]
- 25.Langmade SJ, Ravindra R, Daniels PJ, Andrews GK. The transcription factor MTF-1 mediates metal regulation of the mouse ZnT1 gene. J. Biol. Chem. 2000;275:34803–34809. doi: 10.1074/jbc.M007339200. [DOI] [PubMed] [Google Scholar]
- 26.Andrews GK, Wang H, Dey SK, Palmiter RD. Mouse zinc transporter 1 gene provides an essential function during early embryonic development. Genesis. 2004;40:74–81. doi: 10.1002/gene.20067. [DOI] [PubMed] [Google Scholar]
- 27.Heuchel R, Radtke F, Georgiev O, Stark G, Aguet M, Schaffner W. The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J. 1994;13:2870–2875. doi: 10.1002/j.1460-2075.1994.tb06581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura T, Itoh N, Sone T, Tanaka K, Isobe M. C-terminal deletion mutant of MRE-binding transcription factor-1 inhibits MRE-driven gene expression. J. Cell. Biochem. 2004;93:609–618. doi: 10.1002/jcb.20210. [DOI] [PubMed] [Google Scholar]
- 29.Pyati UJ, Cooper MS, Davidson AJ, Nechiporuk A, Kimelman D. Sustained Bmp signaling is essential for cloaca development in zebrafish. Development. 2006;133:2275–2284. doi: 10.1242/dev.02388. [DOI] [PubMed] [Google Scholar]
- 30.Pyati UJ, Webb AE, Kimelman D. Transgenic zebrafish reveal stage-specific roles for Bmp signaling in ventral and posterior mesoderm development. Development. 2005;132:2333–2343. doi: 10.1242/dev.01806. [DOI] [PubMed] [Google Scholar]
- 31.Cheung AP-L, Au CY-M, Chan WW-L, Chan KM. Characterization and localization of metal-responsive-element-binding transcription factors from tilapia. Aquat. Toxicol. 2010;99:42–55. doi: 10.1016/j.aquatox.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 32.Yan CH, Chan KM. Characterization of zebrafish metallothionein gene promoter in a zebrafish caudal fin cell-line, SJD.1. Mar. Environ. Res. 2002;54:335–339. doi: 10.1016/s0141-1136(02)00141-1. [DOI] [PubMed] [Google Scholar]
- 33.Goldstone JV, McArthur AG, Kubota A, Zanette J, Parente T, Jonsson ME, Nelson DR, Stegeman JJ. Identification and developmental expression of the full complement of cytochrome P450 genes in zebrafish. BMC Genomics. 2010;11:643. doi: 10.1186/1471-2164-11-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schadt EE, Li C, Ellis B, Wong WH. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J. Cell. Biochem. 2001;(Supplement Suppl 37):120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- 35.Schadt EE, Li C, Su C, Wong WH. Analyzing high-density oligonucleotide gene expression array data. J. Cell. Biochem. 2000;80:192–202. [PubMed] [Google Scholar]
- 36.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltson A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 37.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerfu, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Letters. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 38.Al-Shahrour F, Minguez P, Tarraga J, Medina I, Alloza E, Montaner D, Dopazo J. FatiGO+: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007;35:W91–W96. doi: 10.1093/nar/gkm260. (Web Server Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Yan CHM, Chan KM. Cloning of zebrafish metallothionein gene and characterization of its gene promoter region in HepG2 cell line. Biochim. Biophys. Acta. 2004;1679:47–58. doi: 10.1016/j.bbaexp.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Jiang H, Daniels PJ, Andrews GK. Putative zinc-sensing zinc fingers of metal-response element-binding transcription factor-1 stabilize a metal-dependent chromatin complex on the endogenous metallothionein-I promoter. J. Biol. Chem. 2003;278:30394–30402. doi: 10.1074/jbc.M303598200. [DOI] [PubMed] [Google Scholar]
- 42.Jiang H, Fu K, Andrews G. Gene- and cell-type-specific effects of signal transduction cascades on metal-regulated gene transcription appear to be independent of changes in the phosphorylation of metal-response-element-binding transcription factor-1. Biochem J. 2004;382:33–41. doi: 10.1042/BJ20040504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin CY, Lin YC, Nguyen TT, Tam MF, Chein CY, Lin MT, Lin LY. Expression and characterization of SUMO-conjugated metal-responsive transcription factor 1: SIM-dependent cross-interaction and distinct DNA binding activity. J. Biochem. 2013;153:361–369. doi: 10.1093/jb/mvt002. [DOI] [PubMed] [Google Scholar]
- 44.Ferencz A, Hermesz E. Identification and characterization of two MTF-1 genes in common carp. Comp. Biochem. Physiol., Part C. 2008;148:238–243. doi: 10.1016/j.cbpc.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Jenny MJ, Ringwood AH, Schey K, Warr GW, Chapman RW. Diversity of metallothioneins in the American oyster, Crassostrea virginica, revealed by transcriptomic and proteomic approaches. Eur. J. Biochem. 2004;271:1702–1712. doi: 10.1111/j.1432-1033.2004.04071.x. [DOI] [PubMed] [Google Scholar]
- 46.James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, Herzyk P, Brown JWS, Nimmo HG. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. The Plant Cell. 2012;24:961–981. doi: 10.1105/tpc.111.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nam DK, Lee S, Zhou G, Cao X, Wang C, Clark T, Chen J, Rowley JD, Wang SM. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc. Nat. Acad. Sci., USA. 2002;99:6152–6156. doi: 10.1073/pnas.092140899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beaudoing E, Freier S, Wyatt JR, Claverie J-M, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes & Development. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 50.Guryev V, Koudijs MJ, Berezikov E, Johnson SL, Plasterk RHA, Eeden FJMv, Cuppen E. Genetic variation in the zebrafish. Genome Res. 2006;16:491–497. doi: 10.1101/gr.4791006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown KH, Dobrinski KP, Lee AS, Gokcumen O, Mills RE, Shi X, Chong WWS, Chen JYH, Yoo P, David S, Peterson SM, Raj T, Choy KW, Stranger BE, Williamson RE, Zon LI, Freeman JL, Lee C. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc. Nat. Acad. Sci., USA. 2012;109:529–534. doi: 10.1073/pnas.1112163109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferencz A, Hermesz E. Identification of a splice variant of the metal-responsive transcription factor MTF-1 in common carp. Comp. Biochem. Physiol., Part C. 2009;150:113–117. doi: 10.1016/j.cbpc.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 53.He X, Ma Q. Induction of metallothionein I by arsenic via metal-activated transcription factor 1: a critical role of C-terminal cysteine residues in arsenic sensing. J. Biol. Chem. 2009;284:12609–12621. doi: 10.1074/jbc.M901204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gunther V, Davis AM, Georgiev O, Schaffner W. A conserved cysteine cluster, essential for transcriptional activity, mediates homodimerization of human metal-responsive transcription factor-1 (MTF-1) Biochim. Biophys. Acta. 2012;1823:476–483. doi: 10.1016/j.bbamcr.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Kimura T, Huyck RW, Laity JH, Andrews GK. Zinc-induced formation of a coactivator complex containing the zinc-sensing transcription factor MTF-1, p300/CBP, and SP1. Mol. Cell. Biol. 2008;28:4275–4284. doi: 10.1128/MCB.00369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campbell LJ, Willoughby JJ, Jensen AM. Two types of Tet-On transgenic lines for doxycycline-inducible gene expression in zebrafish rod photoreceptors and a gateway-based Tet-On Toolkit. PLos ONE. 2012;7:e51270. doi: 10.1371/journal.pone.0051270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z, Huang X, Zhan H, Zeng Z, Li C, Spitsbergen JM, Meierjohann S, Schartl M, Gong Z. Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. J. Hepatol. 2012;56:419–425. doi: 10.1016/j.jhep.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 58.Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. liver morphogenesis. Dev. Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- 59.Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 60.McHugh PC, Wright JA, Brown DR. Transcriptional regulation of the beta-synuclein 5'-promoter metal response element by metal transcription factor-1. PLoS One. 2011;6:e17354. doi: 10.1371/journal.pone.0017354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balesaria S, Ramesh B, McArdle H, Bayele HK, Srai SK. Divalent metal-dependent regulation of hepcidin expression by MTF-1. FEBS Letters. 2010;584:719–725. doi: 10.1016/j.febslet.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 62.Lichten LA, Ryu M-S, Guo L, Embury J, Cousins RJ. MTF-1-mediated repression of the zinc transporter Zip10 is alleviated by zinc restriction. PLoS One. 2011;6:e21526. doi: 10.1371/journal.pone.0021526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng D, Feeney GP, Kille P, Hogstrand C. Regulation of ZIP and ZnT zinc transporters in zebrafish gill: zinc repression of ZIP10 transcription by an intronic MRE cluster. Physiol. Genomics. 2008;34:205–214. doi: 10.1152/physiolgenomics.90206.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liuzzi JP, Wong CP, Ho E, Tracey A. Regulation of hepatic suppressor of cytokine signaling 3 by zinc. J. Nutr. Biochem. 2013;24:1028–1033. doi: 10.1016/j.jnutbio.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 65.Oteiza PI, Clegg MS, Zago MP, Keen CL. Zinc deficiency induces oxidative stress and AP-1 activation in 3T3 cells. Free Radic. Biol. Med. 2000;28:1091–1099. doi: 10.1016/s0891-5849(00)00200-8. [DOI] [PubMed] [Google Scholar]
- 66.Ho E, Courtemanche C, Ames BN. Zinc deficiency induces oxidative DNA damage and increases p53 expression in human lung fibroblasts. J. Nutrition. 2003;133:2543–2548. doi: 10.1093/jn/133.8.2543. [DOI] [PubMed] [Google Scholar]
- 67.Barclay JL, Anderson ST, Waters MJ, Curlewis JD. Regulation of suppressor of cytokine signaling 3 (SOCS3) by growth hormone in pro-B cells. Mol. Endocrinol. 2007;21:2503–2515. doi: 10.1210/me.2006-0498. [DOI] [PubMed] [Google Scholar]
- 68.Shen H, Arzuaga X, Toborek M, Hennig B. Zinc nutritional status modulates expression of AHR-responsive P450 enzymes in vascular endothelial cells. Environ. Toxicol. Pharmacol. 2008;25:197–201. doi: 10.1016/j.etap.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lichtlen P, Georgiev O, Schaffner W, Aguzzi A, Brandner S. The heavy metalresponsive transcription factor-1 (MTF-1) is not required for neural differentiation. Biol Chem. 1999;380:711–715. doi: 10.1515/BC.1999.089. [DOI] [PubMed] [Google Scholar]
- 70.Levenson CW, Morris D. Zinc and neurogenesis: Making new neurons from development to adulthood. Adv. Nutr. 2011;2:96–100. doi: 10.3945/an.110.000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takeda A, Nakamura M, Fujii H, Tamano H. Synaptic Zn2+ homeostasis and its significance. Metallomics. 2013;5:417–423. doi: 10.1039/c3mt20269k. [DOI] [PubMed] [Google Scholar]
- 72.Chow ESH, Cheng SH. Cadmium affects muscle type development and axon growth in zebrafish embryonic somitogenesis. Toxicol. Sci. 2003;73:149–159. doi: 10.1093/toxsci/kfg046. [DOI] [PubMed] [Google Scholar]
- 73.Chow ESH, Hui MNY, Lin CC, Cheng SH. Cadmium inhibits neurogenesis in zebrafish embryonic brain development. Aquat. Toxicol. 2008;87:157–169. doi: 10.1016/j.aquatox.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 74.Kurita R, Sagara H, Aoki Y, Link BA, Arai K-i, Watanabe S. Suppression of lens growth by aA-crystallin promoter-driven expression of diphtheria toxin results in disruption of retinal cell organization in zebrafish. Dev. Biol. 2003;255:113–127. doi: 10.1016/s0012-1606(02)00079-9. [DOI] [PubMed] [Google Scholar]
- 75.Kwan KM, Fujimoto E, Grabher C, Magnum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien C-B. The Tol2Kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 76.Hawse JR, Cumming JR, Oppermann B, Sheets NL, Reddy VN, Kantorow M. Activation of metallothioneins and alpha-crystallin/sHSPs in human lens epithelial cells by specific metals and the metal content of aging clear human lenses. Invest. Ophthalmol. Vis. Sci. 2003;44:672–679. doi: 10.1167/iovs.02-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu JP, Schlosser R, Ma WY, Dong Z, Feng H, Lui L, Huang XQ, Liu Y, Li DW. Human alphaA- and alphaB-crystallins prevent UVA-induced apoptosis through regulation of PKCalpha, RAF/MEK/ERK and AKT signaling pathways. Exp. Eye Res. 2004;79:393–403. [PubMed] [Google Scholar]
- 78.Kamradt M, Chen F, Sam S, Cryns V. The small heat shock protein alphaB-cystallin negatively regulates apoptosis during myogenic differentiation by inhibiting caspase-3 activation. J. Biol. Chem. 2002;277:38731–38736. doi: 10.1074/jbc.M201770200. [DOI] [PubMed] [Google Scholar]
- 79.Pasupuleti N, Matsuyama S, Voss O, Doseff AI, Song K, Danielpour D, Nagaraj RH. The anti-apoptotic function of human alphaA-crystallin is directly related to its chaperone activity. Cell Death Dis. 2010;1:e31. doi: 10.1038/cddis.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hogstrand C, Zheng D, Feeney G, Cunningham P, Kille P. Zinc-controlled gene expression by metal-regulatory transcription factor 1 (MTF1) in a model vertebrate, the zebrafish. Biochem. Soc. Transact. 2008;36:1252–1257. doi: 10.1042/BST0361252. [DOI] [PubMed] [Google Scholar]
- 81.Troadec MB, Ward DM, Lo E, Kaplan J, Domenico ID. Induction of FPN1 transcription by MTF-1 reveals a role for ferroportin in transition metal efflux. Blood. 2010;116:4657–4664. doi: 10.1182/blood-2010-04-278614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng D, Kille P, Feeney GP, Cunningham P, Handy RD, Hogstrand C. Dynamic transcriptomic profiles of zebrafish gills in response to zinc supplementation. BMC Genomics. 2010;11:553. doi: 10.1186/1471-2164-11-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.