Abstract
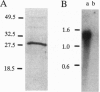
Expansins are unusual proteins discovered by virtue of their ability to mediate cell wall extension in plants. We identified cDNA clones for two cucumber expansins on the basis of peptide sequences of proteins purified from cucumber hypocotyls. The expansin cDNAs encode related proteins with signal peptides predicted to direct protein secretion to the cell wall. Northern blot analysis showed moderate transcript abundance in the growing region of the hypocotyl and no detectable transcripts in the nongrowing region. Rice and Arabidopsis expansin cDNAs were identified from collections of anonymous cDNAs (expressed sequence tags). Sequence comparisons indicate at least four distinct expansin cDNAs in rice and at least six in Arabidopsis. Expansins are highly conserved in size and sequence (60-87% amino acid sequence identity and 75-95% similarity between any pairwise comparison), and phylogenetic trees indicate that this multigene family formed before the evolutionary divergence of monocotyledons and dicotyledons. Sequence and motif analyses show no similarities to known functional domains that might account for expansin action on wall extension. A series of highly conserved tryptophans may function in expansin binding to cellulose or other glycans. The high conservation of this multigene family indicates that the mechanism by which expansins promote wall extensin tolerates little variation in protein structure.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1992 May 11;20 (Suppl):2013–2018. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N. C., Gibeaut D. M. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993 Jan;3(1):1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cosgrove D. J. Characterization of long-term extension of isolated cell walls from growing cucumber hypocotyls. Planta. 1989;177:121–130. [PubMed] [Google Scholar]
- Cosgrove D. J. How do plant cell walls extend? Plant Physiol. 1993 May;102(1):1–6. doi: 10.1104/pp.102.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. J. Wall extensibility: its nature, measurement and relationship to plant cell growth. New Phytol. 1993 May;124(1):1–23. doi: 10.1111/j.1469-8137.1993.tb03795.x. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto T., Foong F., Shoseyov O., Doi R. H. Analysis of functional domains of endoglucanases from Clostridium cellulovorans by gene cloning, nucleotide sequencing and chimeric protein construction. Mol Gen Genet. 1992 Feb;231(3):472–479. doi: 10.1007/BF00292718. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991 Dec 11;19(23):6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y., Rasheed S. A simple procedure for maximum yield of high-quality plasmid DNA. Biotechniques. 1990 Dec;9(6):676–679. [PubMed] [Google Scholar]
- McCaldon P., Argos P. Oligopeptide biases in protein sequences and their use in predicting protein coding regions in nucleotide sequences. Proteins. 1988;4(2):99–122. doi: 10.1002/prot.340040204. [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S. J., Cosgrove D. J. Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 1995 Jan;107(1):87–100. doi: 10.1104/pp.107.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S., Cosgrove D. J. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci U S A. 1994 Jul 5;91(14):6574–6578. doi: 10.1073/pnas.91.14.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S., Durachko D. M., Cosgrove D. J. Two endogenous proteins that induce cell wall extension in plants. Plant Cell. 1992 Nov;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- Poole D. M., Hazlewood G. P., Huskisson N. S., Virden R., Gilbert H. J. The role of conserved tryptophan residues in the interaction of a bacterial cellulose binding domain with its ligand. FEMS Microbiol Lett. 1993 Jan 1;106(1):77–83. doi: 10.1111/j.1574-6968.1993.tb05938.x. [DOI] [PubMed] [Google Scholar]
- Rayle D. L., Cleland R. E. The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992 Aug;99(4):1271–1274. doi: 10.1104/pp.99.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim D. F., Speicher D. W. Microsequence analysis of electroblotted proteins. II. Comparison of sequence performance on different types of PVDF membranes. Anal Biochem. 1992 Nov 15;207(1):19–23. doi: 10.1016/0003-2697(92)90493-q. [DOI] [PubMed] [Google Scholar]
- Wright H. T., Sandrasegaram G., Wright C. S. Evolution of a family of N-acetylglucosamine binding proteins containing the disulfide-rich domain of wheat germ agglutinin. J Mol Evol. 1991 Sep;33(3):283–294. doi: 10.1007/BF02100680. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]