Abstract
Ovarian cancer is an extremely aggressive disease associated with a high percentage of tumor recurrence and chemotherapy resistance. Understanding the underlying mechanism of tumor relapse is crucial for effective therapy of ovarian cancer. DNA damage-binding protein 2 (DDB2) is a DNA repair factor mainly involved in nucleotide excision repair. Here, a novel role was identified for DDB2 in the tumorigenesis of ovarian cancer cells and the prognosis of patients with ovarian cancer. Overexpressing DDB2 in human ovarian cancer cells suppressed its capability to recapitulate tumors in athymic nude mice. Mechanistic investigation demonstrated that DDB2 is able to reduce the cancer stem cell (CSC) population characterized with high aldehyde dehydrogenase activity in ovarian cancer cells, probably through disrupting the self-renewal capacity of CSCs. Low DDB2 expression correlates with poor outcomes among patients with ovarian cancer, as revealed from the analysis of publicly available gene expression array datasets. Given the finding that DDB2 protein expression is low in ovarian tumor cells, enhancement of DDB2 expression is a promising strategy to eradicate CSCs and would help to halt ovarian cancer relapse.
Introduction
Epithelial ovarian cancer is the fifth leading cause of cancer-related deaths in women in the United States and the leading cause of gynecologic cancer deaths. Most of the tumors are initially responsive to platinum-based chemotherapy and the patients enter into clinical remission after initial treatment. However, recurrence occurs in more than 70% of patients despite treatment (1). The high relapse rate in ovarian cancer results in greater mortality and is estimated to account for 5% of all deaths by cancer in women for 2013 (2). Therefore, reducing ovarian cancer relapse is especially important to prolonging progression-free survival and decreasing the mortality in patients with ovarian cancer.
Over the past several years, it has been increasingly evident that a small population of cancer cells, referred to as “cancer stem cells (CSC),” is the most important trigger of tumor progression (3, 4). The CSC theory suggests that tumor cells are organized hierarchically with a small self-renewing population of stem cells generating a large population of proliferative cells to maintain the tumors. These CSCs have been identified in a variety of solid tumors including ovarian cancers (5–8). Each type of CSC has a distinctive pattern of surface markers (i.e., CD44, CD133, and CD117) and nonsurface markers [i.e., aldehyde dehydrogenase (ALDH) activity] that can be targeted for CSC isolation (9). In addition, CSCs can also be isolated by detection of side-population (SP) phenotypes with Hoechst 33342 dye efflux technique (10) and their ability to grow as floating spheres in serum-free medium (11). Ovarian CSCs have been successfully isolated based on the expression of distinctive cell surface markers CD44, CD117, MyD88, and CD133 (5, 12, 13), as well as the activity of ALDH (13). All isolated ovarian CSCs fulfill all currently accepted criteria of the existence of a subpopulation of tumor-initiating cells.
CSCs possess several key properties, including (i) self-renewal, (ii) multipotent differentiation into nontumorigenic cells, (iii) resistance to toxic xenobiotics, and (iv) the ability to induce tumors when transplanted into immunodeficient mice (14). A number of reports support the presence of rare CSCs that are resistant to chemotherapy and radiotherapy. These resistant CSCs are believed to be the main source of tumor relapse (15). Thus, there is an urgent need for detailed characterization of these CSCs to device new treatment modalities.
DDB2 is a 48-kDa protein originally identified as a component of the damage-specific DNA-binding heterodimeric complex DDB (16). DNA damage-binding protein 2 (DDB2) is able to bind UV-damaged DNA and serves as the initial damage recognition factor during nucleotide excision repair (NER; ref. 17). The low expression of DDB2 in cisplatin-resistant ovarian cancer cell lines (18) and high-grade colon cancer (19) and skin cancer (20) indicates a link between DDB2 expression and tumor progression. Recently, new functions of DDB2 beyond its role in DNA repair have been identified, e.g., inhibiting cellular apoptosis through downregulation of Bcl-2 (18, 21) and p21 (22), suppressing colon tumor metastasis through blocking epithelial–mesenchymal transition (EMT; ref. 19), and limiting the motility and invasiveness of invasive human breast tumor cells by regulating NF-κB activity (23), as well as mediating premature senescence (24). In this study, we reveal a novel role of DDB2 in the inhibition of tumorigenesis. DDB2 overexpression resulted in a reduction of the CSC population associated with repression of the tumorigenicity of ovarian cancer cells, whereas DDB2 knockdown resulted in an expansion of the CSC population.
Material and Methods
Cell culture
Human ovarian cancer cell line A2780 and its derived cisplatin-resistant cell line CP70 (25) were kindly provided by Dr. Paul Modrich (Duke University, Durham, NC). Ovarian cancer cell line 2008 and its resistant cell line 2008C13 (26) were kindly provided by Dr. Francois X. Claret (University of Texas M.D. Anderson Cancer Center, Houston, TX). The A2780 derivative and 2008 derivative cisplatin-resistant cell lines were produced by intermittent, incremental exposure of the sensitive parental cell line to various concentrations of cisplatin. SKOV3 ovarian cancer cell line was kindly provided by Dr. Thomas C. Hamilton (Fox Chase Cancer Center, Philadelphia, PA). CP70 cells with overexpression of DDB2 (CP70-DDB2) were established in our laboratory (18). All cell lines were authenticated by DNA (STR) profiling, and maintained in RPMI 1640 supplemented with 10% FBS, 100 μg/mL streptomycin, and 100 units/mL penicillin. Cells were grown at 37°C in humidified atmosphere of 5% CO2 in air.
Tissue microarray and immunohistochemistry
Confirmed, formalin-fixed, paraffin-embedded human ovarian normal tissue and epithelial tumors were obtained from the Department of Pathology, The Ohio State University (Columbus, OH). Cores were obtained from the most viable/nonnecrosis areas of the tissue. Each sample had 2 independent cores. A tissue microarray (TMA) was constructed from 16 normal tissues and 43 patients with ovarian cancer (grade 1, 4; grade 2, 14; grade 3, 25). TMA sections were subjected to deparaffinization and rehydration. The endogenous peroxidase was quenched by 3% (v/v) hydrogen peroxide. The epitope retrieval was performed using Dako TRS solution (Dako) for 25 minutes at 96°C in a vegetable steamer. Primary mouse antibody against DDB2 (ab51017; 1:10; Abcam) was incubated for 1 hour at room temperature, and detected using a Mach 3 Mouse HRP-Polymer kit (Biocare Medical) and diaminobenzidine teterhydroxychloride (Dako). Tissues were counterstained with Richard Allen hematoxylin. Intensity of staining was blind scored from 1 (no staining) to 4 (highest intensity of staining).
Plasmids and gene transfection
pReceiver-Lv105-DDB2 (GeneCopoeia) and pcDNA3.1-His-DDB2 plasmids were introduced into cells by using either lentivirus infection or electroporation with NEPA-21 Electroporator (Nepa Gene Co., Ltd). DDB2 expression lentiviruses were generated as described before (21). To establish shDDB2 stably transfected cell lines, MISSION shDDB2 (TRCN0000083993) plasmids (Sigma) were transfected into 2008 cells using eletroporation. The transfected cells were selected in the medium containing 2 μg/mL Puromycin, and the transfectant lines with stable DDB2 downregulation were confirmed by Western blotting.
Immunoblotting
Whole cell lysates were prepared by boiling cell pellets for 10 minutes in SDS lysis buffer [2% SDS, 10% Glycerol, 62 mmol/L Tris-HCl, pH 6.8 and a complete mini-protease inhibitor cocktail (Roche Applied Science)]. After protein quantification with Bio-Rad Dc Protein Assay (Bio-Rad Laboratories), equal amounts of proteins were loaded, separated on a polyacrylamide gel, and transferred to a nitro-cellulose membrane. Protein bands were immunodetected with appropriate antibodies, e.g., goat anti-DDB2 (R&D Systems), rabbit anti-Nanog (Cell Signaling Technology), mouse anti-Tubulin (Millipore), and mouse anti-IκBα (Cell Signaling Technology).
Semisolid colony-forming assay
Cells were trypsinized and counted. A total of 1,000 cells were mixed with semisolid media (MethoCult H4100; STEMCELL Technologies Inc.) containing serum-free DMEM/F12 (Life Technologies) supplemented with 20% KnockOut Serum Replacement (Life Technologies), 20 ng/mL EGF (Life Technologies), 10 ng/mL basic fibro-blast growth factor (bFGF; Life Technologies), 100 μg/mL streptomycin, and 100 units/mL penicillin (Life Technologies), and seeded in 6-well Ultra-Low Attachment plates (Corning). The number of larger (more than 50 cells), symmetric, and prototypical colonies was counted after 6 days.
Flow cytometry analysis and cell sorting
Anti–CD117-PE and anti–CD44-FITC (BD Pharmingen) were used for flow cytometric analysis and cell sorting. Detection of ALDH activity was conducted using the ALDEFLUOR assay (STEMCELL Technologies) according to the manufacturer’s instruction. For each sample, one half of cells was treated with 50 mmol/L diethylaminobenzaldehyde to define negative gates. Flow cytometric analysis and sorting were performed on a BD FACS Aria III at The Ohio State University Analytical Cytometry Shared Resource. CP70-ALDH+, 2008-CD44+CD117+, and 2008C13-CD44+CD117+ cells were maintained in Ultra-Low Attachment plates in KnockOut DMEM/F12 medium supplemented with 20% KnockOut Serum Replacement, 20 ng/mL EGF, 10 ng/mL bFGF, 100 μg/mL streptomycin, and 100 units/mL penicillin. Cells were grown at 37°C in humidified atmosphere of 5% CO2 in air.
Isolation of putative CSCs based on sphere formation
CSCs are able to form colonies from a single cell more efficiently than their progeny (27) and to grow as spheres in nonadherent culture conditions (28). We seeded SKOV3 cells in Ultra-Low Attachment 96-well plate by serial dilution, and cultured them in 100 μL KnockOut DMEM/F12 medium supplemented with 20% KnockOut Serum Replacement, 20 ng/mL EGF, and 10 ng/mL bFGF. A total of 10 μL of fresh medium was added every 2 days. After 1 month, wells that contain just a single colony were marked. These SKOV3 spheroids were then subcultured from the wells into larger vessels for further growing and defined as putative CSCs.
Sphere-forming assay
A total of 1,000 of CSCs were plated in triplicate in Ultra-Low Attachment plates in serum-free DMEM/F12 medium supplemented with serum replacement, EGF, and bFGF. Sphere formation was assessed 2 weeks after cell seeding. Spheres are defined as floating nonadherent multicellular cell aggregates.
Quantitative real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen), and the first strand cDNA was generated by the Reverse Transcription System (Promega) in a 20-μL reaction containing 1 μg of total RNA. A 0.5 μL aliquot of cDNA was amplified by Fast SYBR Green PCR Master Mix (Life Technologies) in each 20 μL reaction with the following primers: DDB2, forward, 5′-CTCCTCAATGGAGGGAA-CAA-3′, reverse, 5′-GTGACCACCATTCGGCTACT-3′; Nanog, forward, 5′-GTCCCAAAGGCAAACAACCC-3′, reverse, 5′-TTGACCGGGACCTTGTCTTC-3′; GAPDH, forward, 5′-GAAGGTGAAGGTCGGAGT-3′, reverse, 5′-GAAGATGGTGATGGGATTTC-3′. PCR reactions were run on the ABI 7900 Fast Real-Time PCR system in the OSUCCC Nucleic Acid Core Facility.
Xenograft tumor growth
Athymic NCr-nu/nu mice and nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (6–8 weeks, female, 20 to 25 g body weight) were obtained from National Cancer Institute. Animals were maintained in accordance with institutional policies, and all studies were performed with approval of the Institutional Animal Care and Use Committee of The Ohio State University. To assess the tumorigenicity of DDB2-overexpressing CP70 cells, 5 × 106 cells were resuspended (1:1) in PBS:Matrigel (BD Biosciences) and injected subcutaneously into the flank of nude mice. To evaluate the effect of DDB2 on the tumorigenicity of SKOV3 spheroids, pcDNA3.1-His-DDB2 plasmids or empty vectors were cotransfected with pcDNA3.1-GFP plasmids into SKOV3 spheroids by electroporation. Cells were cultured for 2 days, and viable cells with GFP expression were sorted by fluorescence-activated cell sorting (FACS). A total of 1 × 104 of these cells were mixed (1:1) with Matrigel and injected subcutaneously into the axillae of NOD/SCID mice. Tumor growth was measured using calipers, and volumes were calculated based on the formula V = (a × b2)/2, in which a is the longest and b is the shortest diameter of the tumor. Tumor growth curves were compared using a Student t test.
Statistical analysis
Student t test and nonparametrics Mann–Whitney test were performed for data analysis by using the Minitab software. Log-rank analysis was used to determine statistical significance of the Kaplan–Meier survival curve. For all statistical methods, P < 0.05 was considered statistically significant.
Results
DDB2 is downregulated in ovarian cancers
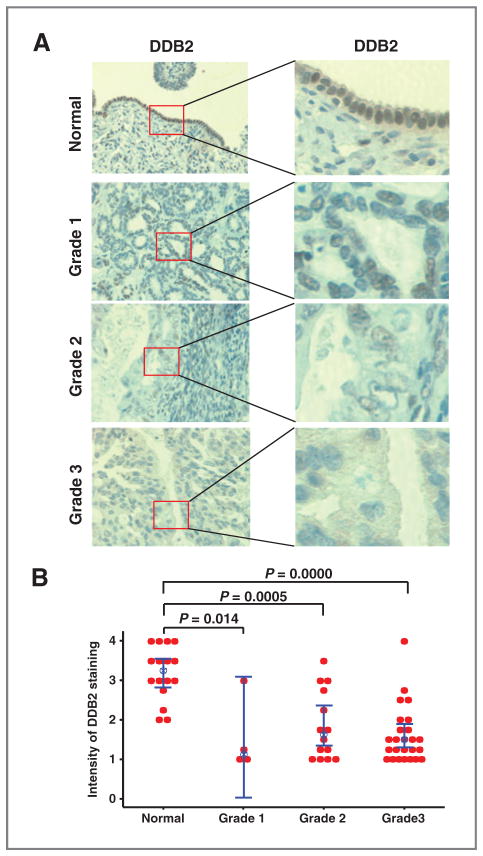
It has been reported that DDB2 protein expression is downregulated in human colon cancer and skin cancer demonstrated by immunohistochemical (IHC) staining (19, 20). To determine the DDB2 protein expression level in primary ovarian carcinoma, we constructed TMA and analyzed DDB2 expression using IHC with an antibody specific for DDB2. IHC staining of the samples showed that normal ovary epithelial cells exhibit strong DDB2 staining, whereas ovarian carcinoma cells in most of tumor tissue samples display low DDB2 expression (Fig. 1A). Statistical analysis revealed a significant decrease of DDB2 expression in ovarian carcinomas regardless the grade (Fig. 1B), indicating a strong correlation between the loss of DDB2 expression and the ovarian carcinoma.
Figure 1.
Loss of DDB2 expression in ovarian cancer. DDB2 IHC of human TMA of normal ovary (n = 16), ovarian serous adenocarcinoma grade 1 (n = 4), grade 2 (n = 14), and grade 3 (n = 25). A, representative images of sections of each group are shown. B, intensity of DDB2 staining was scored from 1 to 4, and an individual value plot was generated to display the distribution of the intensity of DDB2 staining for normal ovary and ovarian cancer grade 1, 2, and 3. Bar, 95% confidence interval for the mean. Blue dot, median value. P value is calculated by the nonparametrics Mann–Whitney test.
DDB2 represses tumorigenicity of ovarian cancer cells in vivo and in vitro
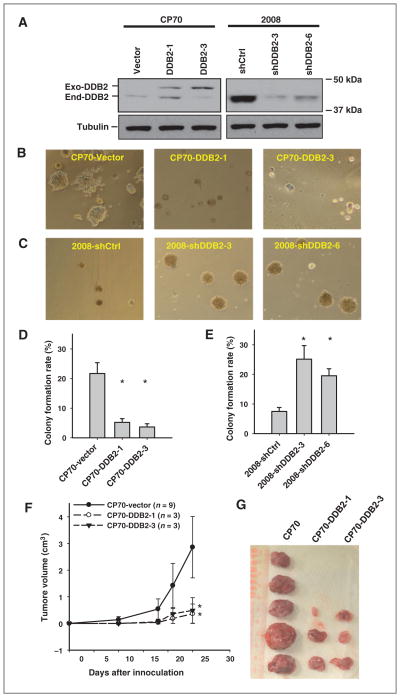
To further investigate the role of DDB2 in the tumorigenicity of ovarian cancer cells, we overexpressed DDB2 in cisplatin-resistant ovarian cancer cell line CP70, which expresses low level of endogenous DDB2 (18), and established two stable cell lines with DDB2 overexpression (Fig. 2A, left). We also transfected DDB2 short hairpin RNA (shRNA) into another ovarian cancer cell line 2008 and selected two stable DDB2-knockdown cell lines (Fig. 2A, right). We then assessed the anchorage-independent growth of these cells in vitro by culturing them in semisolid serum-free medium. As shown in Fig. 2B–E, DDB2 over-expression significantly inhibited anchorage-independent growth of CP70 cells, whereas DDB2 knockdown promoted anchorage-independent growth of 2008 cells.
Figure 2.
DDB2 represses tumorigenicity of ovarian cancer cells in vitro and in vivo. A, two clones of DDB2 stably overexpressing CP70 cells and two clones of DDB2 stably knockdown 2008 cells were selected, and DDB2 expression was determined. Exo-DDB2, Exogenous His-Xpress-tagged DDB2; End-DDB2, Endogenous DDB2. B to E, the DDB2-manipulated CP70 cells (B and D) and 2008 cells (C and E) were seeded in semisolid media containing serum-free DMEM/F12 supplemented with EGF and bFGF in Ultra-Low Attachment plate and allowed to grow for 6 days. Representative images of colonies were shown (B and C). The number of colonies were counted and plotted (D and E). Bar, SD, n = 6; *, P < 0.01 compared with control cells. F and G, CP70-vector and two DDB2-overexpressing CP70 cell lines were injected subcutaneously into nude mice and the tumors formed were counted after 4 weeks. Tumor sizes were measured (F), and tumors were removed from mice after 4 weeks (G). Bar, SD; *, P < 0.01 compared with CP70-vector.
We next sought to determine the effects of DDB2 over-expression on the tumorigenicity of CP70 cells in vivo in immunocompromised mice. CP70-vector cells and two clones of CP70-DDB2 cells were injected separately into nude mice subcutaneously. Tumors were formed in all mice injected with CP70-vector cells within 1 month, whereas tumors could only be found in 3 of 9 mice injected with two different CP70 cell lines stably overexpressing DDB2 up to 3 months. The tumor growth curves and the final tumor sizes also indicate that DDB2 overexpression suppresses the growth of implanted human ovarian cancer cells (Fig. 2F and G). Taken together, these in vitro and in vivo data indicate that DDB2 is able to suppress tumorigenicity of ovarian cancer cells.
DDB2 overexpression reduces the CSC subpopulation in ovarian cancer cells
It has recently been reported that DDB2 suppresses the tumorigenicity and invasiveness of colon cancer cells through suppressing EMT (19). We then analyzed the EMT markers in CP70 cells with or without overexpression of DDB2. We were unable to find any effect of DDB2 overexpression on the cellular levels of epithelial marker E-cadherin and mesenchymal marker vimentin (Supplementary Fig. S1A). In addition, we failed to see any change in morphology of the cells with DDB2 overexpression (Supplementary Fig. S1B). Similarly, we did not find any change of EMT in various clones of ovarian cancer cell line 2008 with stable DDB2 knockdown (Supplementary Fig. S1C). These results indicate that DDB2 may not suppress EMT of ovarian cancer cells.
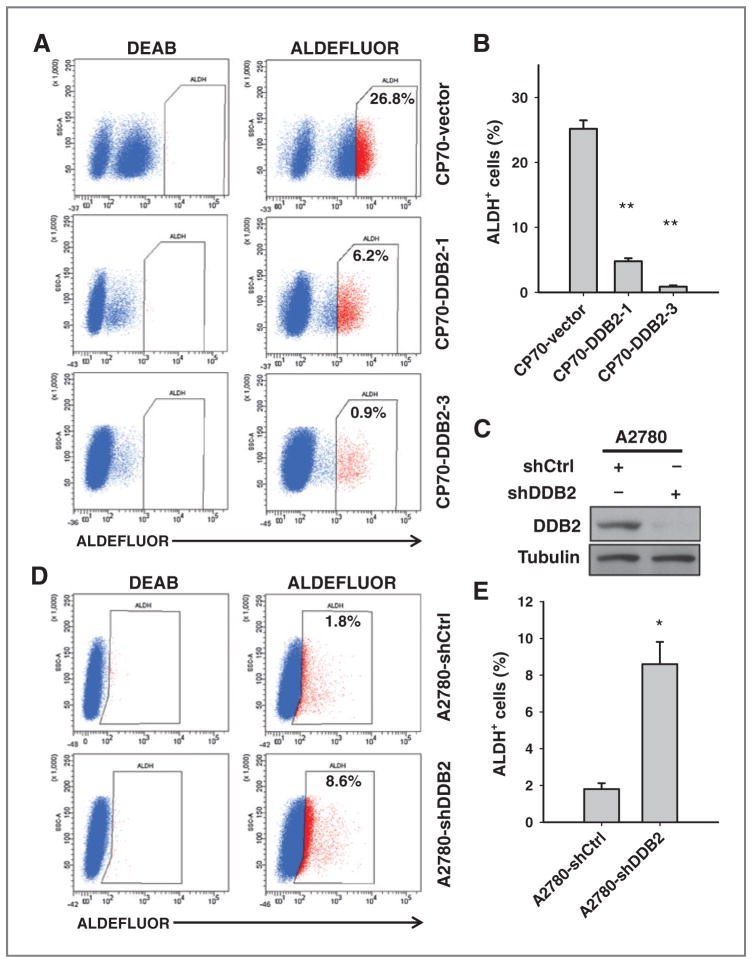
CSCs are thought to contribute to the initiation of the tumor (15). We thus hypothesized that DDB2 may regulate the amount of CSC subpopulation in cancer cells. ALDH has been proposed to be a marker of both normal and cancer stem cells (29) and has been used to identify CSCs from ovarian cancers (13). We thus analyzed the abundance of putative CSC subpopulations characterized with ALDH+ in CP70 and DDB2-overexpressing CP70 cells using FACS. It is clear that two DDB2-overexpressing CP70 cell lines exhibit reduced ALDH+ subpopulation, in comparison with CP70-vector cells (Fig. 3A and B, Supplementary Fig. S2A and S2B). Our immunoblotting analysis also revealed diminished ALDH1A1 expression in both DDB2-overexpressing CP70 cell lines (Supplementary Fig. S3). In contrast, downregulation of DDB2 expression in A2780 cells increased the percentage of ALDH+ cells (Fig. 3C–E), further supporting that DDB2 is able to decrease the abundance of ALDH+ cells in ovarian cancer cells. In addition, we also analyzed ALDH+ cells in DDB2-knockdown 2008 cells and demonstrated the similar result (Supplementary Fig. S4A and S4B). Besides ALDH activity, CD44+CD117+ was also used as a phenotypic marker of ovarian CSCs (5). We then sought to determine whether DDB2 expression also affects the percentage of CD44+ CD117+ fraction. Indeed, we found a decrease in CD44+ CD117+ population in CP70 cells with DDB2 overexpression, and an increase in CD44+CD117+ population in 2008 cells with DDB2 knockdown (Supplementary Fig. S5A–S5D). SP cells, identified based on their ability to efflux the Hoechst 33342 fluorscent dye, have been shown to possess CSC properties (10). We then attempted to determine the effect of DDB2 on the percentage of SP cells. Different from above-detected ALDH+ and CD44+ CD117+ populations, SP cells only account for a very small fraction of the total cancer cell population (less than 0.1%). Nevertheless, the overexpression of DDB2 reduced the SP fraction in one cell line (Supplementary Fig. S6A and S6B). Collectively, these results indicate that DDB2 is able to reduce the CSCs pool existing in ovarian cancer cells.
Figure 3.
DDB2 reduces the abundance of putative CSCs in ovarian cancer cell lines. A, ALDEFLUOR staining of CP70-vector and two DDB2-overexpressing CP70 cell lines were conducted and analyzed by FACS. Number, percentage of ALDH+ cells. B, average percentage of ALDH+ cells in indicated cells. C, A2780 cells were transfected with DDB2 shRNA for 2 days. D, ALDEFLUOR staining of A2780 cells transiently transfected with either shControl (shCtrl) or shDDB2 analyzed by FACS. E, average percentage of ALDH+ cells in indicated cells. Bar, SD, n = 3; *, P < 0.05; **, P < 0.01 compared with A2780-shControl.
Ovarian CSCs exhibit low DDB2 expression level
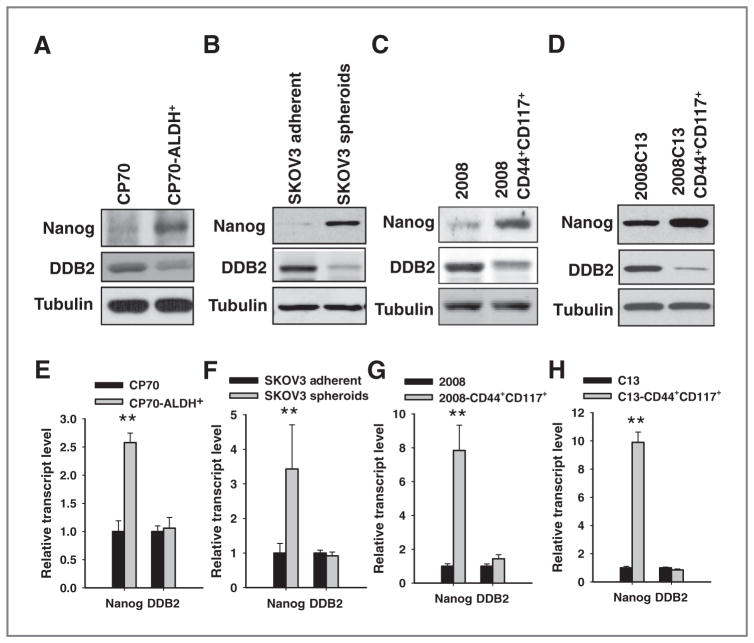
To elucidate the mechanism of the DDB2-mediated regulation of the CSC population, we isolated the putative CSCs from various ovarian cancer cell lines, e.g., CP70, SKOV3, 2008, and 2008C13 based on the activity of ALDH, expression of specific surface markers CD44 and CD117, as well as the ability to form spheres and grow in nonadherent serum-free culture condition, respectively (9). The characteristics of isolated CSCs, including the ability to form tumor spheres in ultra-low attachment plates and the ability to form xenografts in immunodeficient mice at lower cell numbers, have been authenticated (Supplementary Figs. S7A–S7C; S8A–S8D; S9A–S9G). Both immunoblotting (Fig. 4A–D) and real-time (RT) PCR (Fig. 4E–F) analyses showed enhanced expression of Nanog, one of the self-renewal markers of stem cells and upregulated in various ovarian CSCs (5, 30), in these CSC populations in comparison with their parent bulk cancer cells, further supporting the stemness of these CSC populations. Interestingly, DDB2 protein level was found to decrease, while its mRNA level did not change in all CSC populations compared with their parent bulk cells (Fig. 4A–H). These data indicate that low DDB2 protein expression might be required for the maintenance of CSCs, whereas cellular DDB2 expression in CSCs is most likely regulated at posttranscription level.
Figure 4.
The expression of DDB2 protein, but not DDB2 mRNA, is reduced in ovarian CSCs. A to D, the protein level of DDB2 in various CSCs and their parental bulk cancer cells was determined using immunoblotting with anti-DDB2 antibody. The stem cell marker Nanog was also detected to show the CSC property. E to H, the mRNA level of DDB2 in various CSCs and their parental bulk cancer cells was determined using quantitative RT-PCR. The stem cell marker Nanog was also detected to show the CSC property. Bar, SD; n = 3; **, P < 0.01 compared with bulk cancer cells.
DDB2 inhibits the self-renewal property and tumorigenicity of ovarian CSCs through suppressing the NF-κB pathway
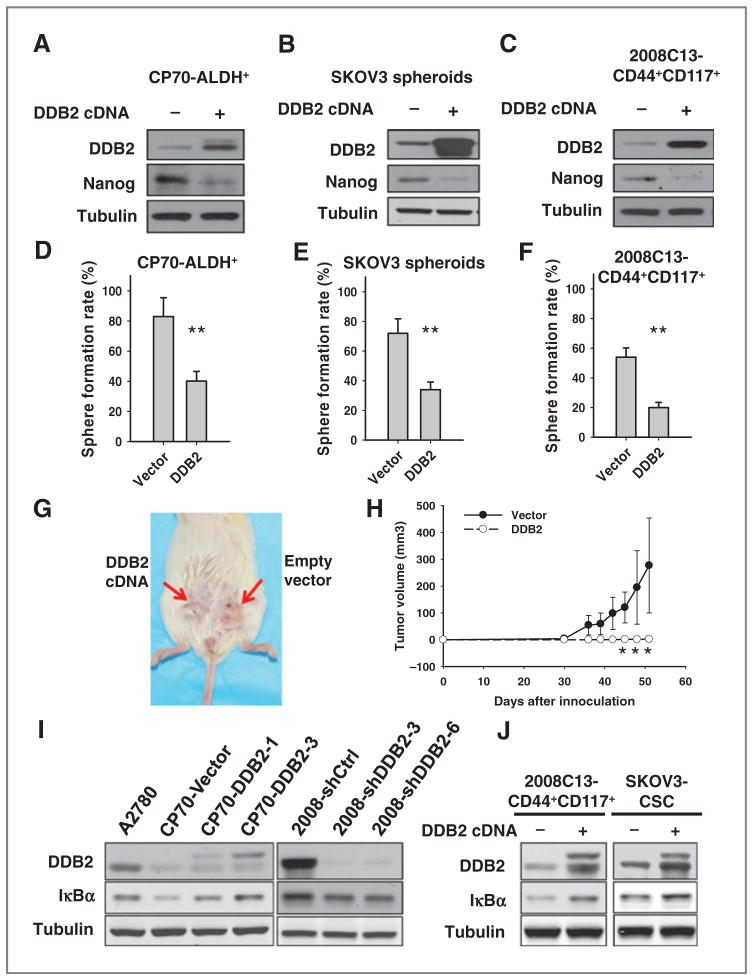
To further investigate whether low DDB2 expression is required to maintain the stem cell properties of ovarian CSCs, we transfected DDB2-expressing plasmids into three ovarian CSC populations and analyzed the expression of stem cell marker Nanog and their capability to form spheres in nonadherent culture condition. As shown in Fig. 5A–C, overexpression of DDB2 was accompanied by a reduced Nanog expression in all three CSCs. In addition, DDB2 overexpression significantly reduced the capability of CSCs to form floating spheres in serum-free medium (Fig. 5D–F).
Figure 5.
Overexpression of DDB2 in ovarian CSCs alters the properties of CSCs. CP70-ALDH+ (A and D), SKOV3 spheroids (B and E), and 2008C13-CD44+CD117+ (C and F) cells were transfected with DDB2-expressing vector for 2 days. The expression of DDB2 and CSC marker Nanog were determined using immunoblotting (A to C). The sphere formation was assessed in nonadherent conditions (D to F). Bar, SD; n = 3; **, P < 0.01. G and H, NOD/SCID mice were subcutaneously injected with 1 × 104 SKOV3 spheroids transiently transfected with either empty vectors or DDB2-expressing vectors. Representative image for tumor growth is shown (G). Tumor sizes were measured and the tumor growth was plotted (H); n = 5; bar, SD; *, P < 0.05 compared with the vector group at the same time point. I, the expression of IκBα was determined in various ovarian cancer cell lines with either stable overexpression or knockdown of DDB2. J, 2008C13-CD44+CD117+ cells and SKOV3 spheroids were transiently transfected with DDB2-expressing plasmids for 2 days and the expression of IκBα was detected using immunoblotting.
These data indicate that DDB2 is able to disrupt the self-renewal property of ovarian CSCs. Furthermore, we assessed the effect of DDB2 overexpression on the capability of CSCs to form xenografts in vivo. SKOV3 spheroids were transfected with either empty vectors or DDB2 expression plasmids, and 1 × 104 of these cells were injected into NOD/SCID mice subcutaneously. As shown in Fig. 5G and H, at 50 days after injection, tumors can be found in five of five sites injected with vector-transfected SKOV3 spheroids, whereas only two of five sites injected with DDB2-over-expressing SKOV3 spheroids display visible tumor. In addition, the mean volumes of xenograft tumors generated from DDB2-overexpressing SKOV3 spheroids were significantly smaller than those arising from empty vector–transfected SKOV3 spheroids. Thus, DDB2 overexpression significantly inhibited the tumorigenicity of ovarian CSCs.
It has been recently reported that DDB2 is able to attenuate the activity of NF-κB through upregulating expression of IκBα in invasive breast cancer cells (23). Through the analysis of publicly available datasets (www.cbioportal.org; Supplementary Materials and Methods), we found that DDB2 mRNA expression level positively correlates with IκBα mRNA expression in patients with ovarian cancer (Supplementary Fig. S10). Given NF-κB activation is required for the maintenance of CSCs in breast tumor (31–34), this prompted us to test whether DDB2 inhibits ovarian CSC population through enhancing IκBα expression. We thus determined the IκBα expression in DDB2-overexpressing and DDB2-downregulating ovarian cancer cell lines. As shown in Fig. 5I, DDB2 overexpression enhanced the protein level of IκBα, whereas DDB2 downregulation reduced the protein level of IκBα in the ovarian cancer cell lines. We then transiently transfected DDB2 into 2008C13-CD44+CD117+ cells and SKOV3 spheroids, and found that overexpression of DDB2 also increased the expression of IκBα in both ovarian CSC populations (Fig. 5J). These data indicate that DDB2-mediated reduction of ovarian CSC population could be attributed, at least in part, to the DDB2-induced upregulation of IκBα, which results in the inactivation of the NF-κB pathway.
Low DDB2 mRNA expression level correlates with poor prognosis of patients with ovarian cancer
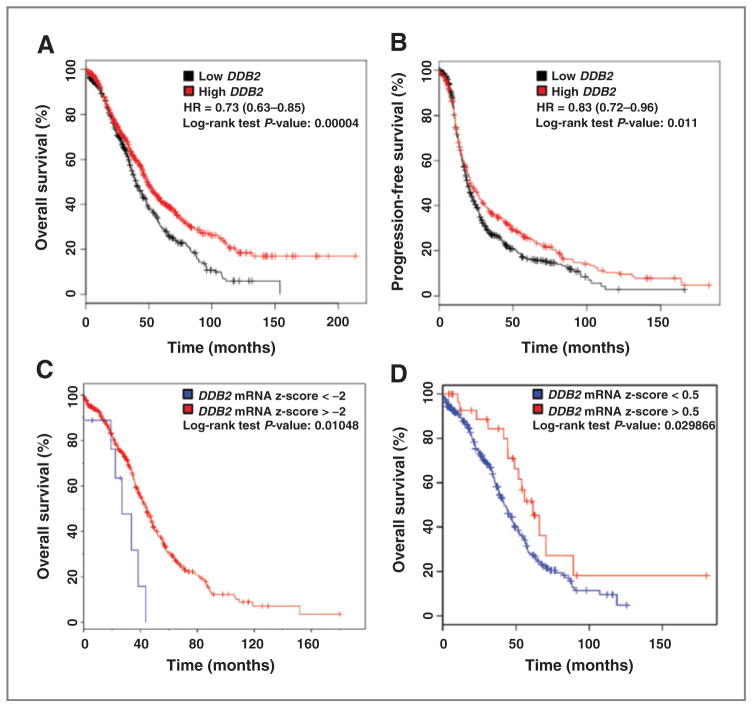
Our data from the studies in vitro and in vivo indicate that loss of DDB2 is associated with highly tumorigenic potential in ovarian cancer cells. To understand whether there is any relationship between the level of DDB2 expression and the prognosis in human patients with ovarian cancer, we evaluated publicly available datasets for DDB2 mRNA expression (Supplementary Materials and Methods). First, we used an online tool (http://kmplot.com) to assess the prognostic value of the microarray-quantified DDB2 expression level in patients with ovarian cancer included in 10 datasets (35). Kaplan–Meier survival curves showed that low DDB2 mRNA expression correlates with poor overall and progression-free survival among patients with ovarian cancer (Fig. 6A and B). We then used another online tool cBioPortal (http://cbioportal.org) to analyze the effect of extremely low DDB2 expression or high DDB2 expression on the overall survival of patients with ovarian cancer from The Cancer Genome Atlas (TCGA) data portal (36–38). Similarly, the Kaplan–Meier survival analyses demonstrated that patients with ovarian cancer with DDB2 mRNA expression Z-score less than −2 exhibited poor prognosis compared with those patients with DDB2 expression Z-score greater than −2 (median survival months, 26.94 vs. 44.29), while the patients with DDB2 expression Z-score higher than 0.5 exhibited better prognosis than those with DDB2 expression Z-score less than 0.5 (median survival months, 55.88 vs. 41.53; Fig. 6C and D). However, although the TCGA dataset demonstrates that overall survival is different, there is no difference in progression-free survival. These analyses suggest that DDB2 low expression is associated with poor outcome in patients with ovarian cancer.
Figure 6.
Prognostic significance of DDB2 in ovarian cancer. A and B, the effect of DDB2 mRNA expression level on the overall survival (A) and progression-free survival (B) in 1,464 patients with ovarian cancer was analyzed and the Kaplan–Meier plots were generated by the Kaplan–Meier Plotter (http://www.kmplot.com). C and D, TCGA data were also analyzed to reveal the effect of extremely low DDB2 mRNA level (C) and high DDB2 mRNA level (D) on the overall survival of patients with ovarian serous cystadenocarcinoma by using cBioPortal (http://cbioportal.org).
Discussion
Despite significant advances in diagnosing and treating ovarian cancer, one of the major clinical and scientific problems that remain unresolved is the prediction and inhibition of ovarian tumor recurrence after clinical remission. In the present study, we provide evidence showing that low DDB2 expression is correlated with poor outcome of patients with ovarian cancer, overexpression of DDB2 in ovarian cancer cells reduces their tumorigenicity through limiting the CSC population, thus defining a novel function for DDB2 in the control of tumor relapse. DDB2 has been considered a tumor suppressor based on the findings that DDB2−/− mice were not only susceptible to UV-induced carcinogenesis, but also developed spontaneous malignant tumors at a high rate (39, 40). The analysis of publicly available datasets in this study indicates that low DDB2 mRNA expression correlates with poor outcome of patients with ovarian cancer. Indeed, this kind of correlation can also be found in patients with breast (23) and lung cancer (http://www.kmplot.com). Therefore, in combination with our findings in this study and others (19), we believe that DDB2 plays an important role in impeding tumor progression and tumor relapse.
DDB2 has been reported to inhibit metastasis of colon cancer (19) and limit the invasiveness of breast cancer (23). Mechanistically, DDB2 constitutively represses genes that are the key activators of EMT through its transcriptional regulation function in various colon cancer cell lines (19). In addition, DDB2 attenuates the activity of NF-κB by upregulating expression of IκB in breast cancer cells. EMT is believed to be a crucial mechanism for tumor metastatic progression (41), whereas NF-κB plays a causal role in migration and invasion of tumor cells and is required for maintenance of the malignant phenotype (42). Thus, the DDB2-dependent decrease of EMT and NF-κB activity could explain in part the correlation between high DDB2 expression and an improvement of the outcome in patients with cancer. Besides these known mechanisms, we demonstrated for the first time in this study that DDB2 is able to reduce the abundance of CSCs in the bulk ovarian cancer cells, which provide a novel mechanism to explain the DDB2-mediated suppression of tumorigenicity. Our data demonstrated that the low expression of DDB2 is required for the maintenance of CSCs. Given that CSCs are believed to be the source of tumor recurrence and metastasis, enhancement of DDB2 expression in ovarian tumors bears great potential in the improvement of the prognosis of patients with tumor.
Mechanistic investigations demonstrated that overexpression of DDB2 reduces the expression of several CSCs self-renewal markers and their capability to form spheres in nonadherent condition. The maintenance and survival of CSCs are controlled by many pathways, such as Wnt, Notch, and Hedgehog (43). It has been reported that the Notch pathway in glioblastoma CSCs involves the constitutive activation of NF-κB signaling, which upregulates Notch pathway genes, and promotes the survival of CSCs (44). In addition, mammary epithelial NF-κB is able to regulate the self-renewal of breast CSCs in Her2-dependent tumorigenesis (45). Given that DDB2 is able to upregulate the expression of IκBα, the inhibitor of NF-κB, in breast tumor cells (23) and ovarian cancer cells, as well as ovarian CSCs, as demonstrated in the present study, we reason that DDB2 could suppress the survival of CSCs through downregulating the NF-κB signaling. Moreover, NF-κB is required for the induction and maintenance of the EMT (42), a process that can be used by cancer cells to reacquire “stemness.” Thus, it seems possible that DDB2 reduces the abundance of CSC through inhibiting NF-κB–mediated EMT. Indeed, it has been reported that DDB2 is able to suppress EMT by constitutively repressing genes that are the key activators of EMT in colon cancer (24). However, we failed to find the alterations of epithelial marker E-cadherin and mesenchymal marker vimentin in ovarian cancer cells after DDB2 over-expression or knockdown. Therefore, we incline to the mechanism that DDB2 reduces the CSC subpopulation through shutting down the pathways required for the maintenance of CSCs.
The cellular level of DDB2 could be regulated at both transcript and protein levels. According to the publicly available TCGA datasets, 16.5% (52/316) patients with ovarian cancer display downregulated DDB2 mRNA (Z-score < −1), whereas 8.5% (27/316) display DDB2 mRNA upregulation (Z-score > 1; www.cbiopor-tal.org). Similarly, downregulation of the DDB2 mRNA can also be found in majority of the colon carcinoma datasets (24). In ovarian cancer datasets, heterozygous deletion contributes partially to the downregulation of DDB2 mRNA, while promoter CpG methylation does not seem to be a determinant of DDB2 mRNA expression (Supplementary Fig. S11; www.cbioportal.org). At the protein level, DDB2 can be ubiquitylated by DDB-Cul4A ubiquitin ligase, and its ubiquitylation and subsequent degradation are essential for its functions in NER (46, 47). DDB2 is also ubiquitylated in human cells in the absence of exogenous DNA damage (48), indicating that the steady-state level of DDB2 can be regulated by posttranslational modification. Furthermore, DDB2 protein stability is reported to be regulated by USP24, a deubiquitylating enzyme, by removing the ubiquitin moiety from modified DDB2, thereby preventing DDB2 degradation (49). We found in this study that CSCs display reduced DDB2 protein level but not mRNA level in comparison with their parental bulk cancer cells. Thus, it is more likely that ovarian CSCs regulate the cellular DDB2 expression by decreasing its protein stability, and the underlying mechanisms warrant a further investigation in the future.
In conclusion, we report here a novel function of DDB2 in limiting tumorigenicity of ovarian cancer cells. Overexpression of DDB2 is able to inhibit the self-renewal ability of ovarian CSCs, probably through inhibiting the NF-κB pathway, leading to a reduction of CSC subpopulation, and finally results in an impediment of tumor progression.
Supplementary Material
Acknowledgments
The authors thank Dr. Paul Modrich (Duke University), Dr. Francois X. Claret (University of Texas M.D. Anderson Cancer Center), and Dr. Thomas C. Hamilton (Fox Chase Cancer Center) for kindly providing cell lines; Jason Bacher and Kristin Kovach in the Department of Pathology, the Ohio State University, for their invaluable helps with TMA construction and IHC staining; and the Cytometry Core Facility at the Ohio State University for FACS analysis. They also thank Dr. Roberts Snapka (the Ohio State University) for critical reading of the article, and Dr. Altaf Wani (OSU) for the selfless supports of this project.
Grant Support
This work was supported by the NIH (grant CA151248, to Q.-E. Wang).
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
ImplicationsDDB2 status has prognostic potential, and elevating its expression eradicates CSCs and could reduce ovarian cancer relapse
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Authors’ Contributions
Conception and design: C. Han, Q.-E. Wang
Development of methodology: L. Gong, W. Zhao, Q.-E. Wang
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): R. Zhao, A. Srivastava, M. Qu, W. Zhao, Q.-E. Wang
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): H. Mao, W. Zhao, Q.-E. Wang
Writing, review, and/or revision of the manuscript: R. Zhao, X. Liu, W. Zhao, J. Yu, Q.-E. Wang
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): H. Mao
Study supervision: Q.-E. Wang
References
- 1.Hennessy BT, Coleman RL, Markman M. Ovarian cancer. Lancet. 2009;374:1371–82. doi: 10.1016/S0140-6736(09)61338-6. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Cancer Facts & Figures 2013. Atlanta, GA: American Cancer Society; 2013. [Google Scholar]
- 3.Hermann PC, Huber SL, Heeschen C. Metastatic cancer stem cells: a new target for anti-cancer therapy? Cell Cycle. 2008;7:188–93. doi: 10.4161/cc.7.2.5326. [DOI] [PubMed] [Google Scholar]
- 4.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–20. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baba T, Convery PA, Matsumura N, Whitaker RS, Kondoh E, Perry T, et al. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene. 2009;28:209–18. doi: 10.1038/onc.2008.374. [DOI] [PubMed] [Google Scholar]
- 7.Curley MD, Therrien VA, Cummings CL, Sergent PA, Koulouris CR, Friel AM, et al. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem Cells. 2009;27:2875–83. doi: 10.1002/stem.236. [DOI] [PubMed] [Google Scholar]
- 8.Bapat SA, Mali AM, Koppikar CB, Kurrey NK. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65:3025–9. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- 9.Tirino V, Desiderio V, Paino F, De RA, Papaccio F, La NM, et al. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB. 2013;27:13–24. doi: 10.1096/fj.12-218222. [DOI] [PubMed] [Google Scholar]
- 10.Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, Dinulescu DM, Connolly D, Foster R, et al. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian inhibiting substance responsiveness. Proc Natl Acad Sci U S A. 2006;103:11154–9. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rybak AP, He L, Kapoor A, Cutz JC, Tang D. Characterization of sphere-propagating cells with stem-like properties from DU145 prostate cancer cells. Biochim Biophys Acta. 2011;1813:683–94. doi: 10.1016/j.bbamcr.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Alvero AB, Chen R, Fu HH, Montagna M, Schwartz PE, Rutherford T, et al. Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle. 2009;8:158–66. doi: 10.4161/cc.8.1.7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71:3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea–a paradigm shift. Cancer Res. 2006;66:1883–90. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–43. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 16.Dualan R, Brody T, Keeney S, Nichols AF, Admon A, Linn S. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics. 1995;29:62–9. doi: 10.1006/geno.1995.1215. [DOI] [PubMed] [Google Scholar]
- 17.Tang J, Chu G. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair (Amst) 2002;1:601–16. doi: 10.1016/s1568-7864(02)00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barakat BM, Wang QE, Han C, Milum K, Yin DT, Zhao Q, et al. Over-expression of DDB2 enhances the sensitivity of human ovarian cancer cells to cisplatin by augmenting cellular apoptosis. Int J Cancer. 2009;127:977–88. doi: 10.1002/ijc.25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy N, Bommi PV, Bhat UG, Bhattacharjee S, Elangovan I, Li J, et al. DDB2 suppresses epithelial-to-mesenchymal transition in colon cancer. Cancer Res. 2013;73:3771–82. doi: 10.1158/0008-5472.CAN-12-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoyanova T, Roy N, Bhattacharjee S, Kopanja D, Valli T, Bagchi S, et al. p21 cooperates with DDB2 protein in suppression of ultraviolet ray-induced skin malignancies. J Biol Chem. 2012;287:3019–28. doi: 10.1074/jbc.M111.295816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao R, Han C, Eisenhauer E, Kroger J, Zhao W, Yu J, et al. DNA damage-binding complex recruits HDAC1 to repress Bcl-2 transcription in human ovarian cancer cells. Mol Cancer Res. 2013 doi: 10.1158/1541-7786.MCR-13-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoyanova T, Roy N, Kopanja D, Bagchi S, Raychaudhuri P. DDB2 decides cell fate following DNA damage. Proc Natl Acad Sci U S A. 2009;106:10690–5. doi: 10.1073/pnas.0812254106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ennen M, Klotz R, Touche N, Pinel S, Barbieux C, Besancenot V, et al. DDB2: a novel regulator of NF-kappaB and breast tumor invasion. Cancer Res. 2013;73:5040–52. doi: 10.1158/0008-5472.CAN-12-3655. [DOI] [PubMed] [Google Scholar]
- 24.Roy N, Stoyanova T, Dominguez-Brauer C, Park HJ, Bagchi S, Raychaudhuri P. DDB2, an essential mediator of premature senescence. Mol Cell Biol. 2010;30:2681–92. doi: 10.1128/MCB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Behrens BC, Hamilton TC, Masuda H, Grotzinger KR, Whang-Peng J, Louie KG, et al. Characterization of a cis-diamminedichloroplatinum (II)-resistant human ovarian cancer cell line and its use in evaluation of platinum analogues. Cancer Res. 1987;47:414–8. [PubMed] [Google Scholar]
- 26.Andrews PA, Velury S, Mann SC, Howell SB. cis-Diamminedichloroplatinum(II) accumulation in sensitive and resistant human ovarian carcinoma cells. Cancer Res. 1988;48:68–73. [PubMed] [Google Scholar]
- 27.Franken NA, Rodermond HM, Stap J, Haveman J, van BC. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–9. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 28.Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–98. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McAuliffe SM, Morgan SL, Wyant GA, Tran LT, Muto KW, Chen YS, et al. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc Natl Acad Sci U S A. 2012;109:E2939–48. doi: 10.1073/pnas.1206400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Y, Luo JL, Karin M. IkappaB kinase alpha kinase activity is required for self-renewal of ErbB2/Her2-transformed mammary tumor-initiating cells. Proc Natl Acad Sci U S A. 2007;104:15852–7. doi: 10.1073/pnas.0706728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu M, Sakamaki T, Casimiro MC, Willmarth NE, Quong AA, Ju X, et al. The canonical NF-kappaB pathway governs mammary tumorigenesis in transgenic mice and tumor stem cell expansion. Cancer Res. 2010;70:10464–73. doi: 10.1158/0008-5472.CAN-10-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Zhang H, Gu P, Bai J, Margolick JB, Zhang Y. NF-kappaB pathway inhibitors preferentially inhibit breast cancer stem-like cells. Breast Cancer Res Treat. 2008;111:419–27. doi: 10.1007/s10549-007-9798-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto M, Taguchi Y, Ito-Kureha T, Semba K, Yamaguchi N, Inoue J. NF-kappaB non-cell-autonomously regulates cancer stem cell populations in the basal-like breast cancer subtype. Nat Commun. 2013;4:2299. doi: 10.1038/ncomms3299. [DOI] [PubMed] [Google Scholar]
- 35.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 36.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Consortium TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon T, Chakrabortty A, Franks R, Valli T, Kiyokawa H, Raychaudhuri P. Tumor-prone phenotype of the DDB2-deficient mice. Oncogene. 2005;24:469–78. doi: 10.1038/sj.onc.1208211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itoh T, Iwashita S, Cohen MB, Meyerholz DK, Linn S. Ddb2 is a haploinsufficient tumor suppressor and controls spontaneous germ cell apoptosis. Hum Mol Genet. 2007;16:1578–86. doi: 10.1093/hmg/ddm107. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Huber MA, Azoitei N, Baumann B, Grunert S, Sommer A, Pehamberger H, et al. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–81. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res. 2010;16:3113–20. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 44.Garner JM, Fan M, Yang CH, Du Z, Sims M, Davidoff AM, et al. Constitutive activation of signal transducer and activator of transcription 3 (STAT3) and nuclear factor kappaB signaling in glioblastoma cancer stem cells regulates the Notch pathway. J Biol Chem. 2013;288:26167–76. doi: 10.1074/jbc.M113.477950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu M, Sakamaki T, Casimiro MC, Willmarth NE, Quong AA, Ju X, et al. The canonical NF-kappaB pathway governs mammary tumorigenesis in transgenic mice and tumor stem cell expansion. Cancer Res. 2010;70:10464–73. doi: 10.1158/0008-5472.CAN-10-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, Zhang Y, Douglas L, Zhou P. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J Biol Chem. 2001;276:48175–82. doi: 10.1074/jbc.M106808200. [DOI] [PubMed] [Google Scholar]
- 47.El-Mahdy MA, Zhu Q, Wang QE, Wani G, Praetorius-Ibba M, Wani AA. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem. 2006;281:13404–11. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- 48.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L, Lubin A, Chen H, Sun Z, Gong F. The deubiquitinating protein USP24 interacts with DDB2 and regulates DDB2 stability. Cell Cycle. 2012;11:4378–84. doi: 10.4161/cc.22688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.