Abstract
CXCL12 and its unique receptor CXCR4, is critical for the homing of a variety of cell lineages during both development and tissue repair. CXCL12 is particularly important for the recruitment of hemato/lymphopoietic cells to their target organs. In conjunction with the damage-associated alarmin molecule HMGB1, CXCL12 mediates immune effector and stem/progenitor cell migration towards damaged tissues for subsequent repair. Previously, we showed that cell migration to HMGB1 simultaneously requires both IKKβ and IKKα-dependent NF-κB activation. IKKβ-mediated activation maintains sufficient expression of HMGB1's receptor RAGE, while IKKα-dependent NF-κB activation ensures continuous production of CXCL12, which complexes with HMGB1 to engage CXCR4. Here using fibroblasts and primary mature macrophages, we show that IKKβ and IKKα are simultaneously essential for cell migration in response to CXCL12 alone. Non-canonical NF-κB pathway subunits RelB and p52 are also both essential for cell migration towards CXCL12, suggesting that IKKα is required to drive non-canonical NF-κB signaling. Flow cytometric analyses of CXCR4 expression show that IKKβ, but not IKKα, is required maintain a critical threshold level of this CXCL12 receptor. Time-lapse video microscopy experiments in primary MEFs reveal that IKKα is required both for polarization of cells towards a CXCL12 gradient and to establish a basal level of velocity towards CXCL12. In addition, CXCL12 modestly up-regulates IKKα-dependent p52 nuclear translocation and IKKα-dependent expression of the CXCL12 gene. On the basis of our collective results we posit that IKKα is needed to maintain the basal expression of a critical protein co-factor required for cell migration to CXCL12.
Keywords: CXCL12, CXCR4, cell migration, NF-κB, IKKα, IKKβ
INTRODUCTION
The chemokine CXCL12, also known as stromal cell derived factor 1 (SDF-1), regulates the migration and homing of stem/progenitor cells. Along with its unique receptor, CXCR4, CXCL12 exerts a pivotal role during embryogenesis for lymphopoiesis, myelopoiesis, cardiogenesis, angiogenesis and neurogenesis (1). In postnatal life, CXCL12/CXCR4 signaling plays an important role in the homing of hematopoietic and lymphopoietic cells, and in the targeted trafficking of stem/progenitor cells during tissue repair and/or regeneration. Moreover, since CXCR4 is expressed by several types of cancer cells, CXCL12 exerts pleiotropic effects regulating processes essential to tumor cell metastasis {reviewed in (2)}. Binding of CXCL12 to CXCR4 activates multiple signaling pathways including MAPK p42/44, Jak/STAT and the PI-3K-AKT-NF-κB axes (3-7). CXCR4 expression is regulated by NF-κB and the CXCR4 promoter contains p50/p65 binding sites (6, 8), Moreover, CXCL12 stimulation of mesoangioblasts (blood vessel stem cells) triggers NF-κB p65 nuclear translocation, which is required for mesoangioblast migration in response to CXCL12 (9).
NF-κB transcription factors mediate stress-like inflammatory responses, participate in developmental programming and regulate normal and malignant cell growth and survival (10-12). In the canonical NF-κB signaling pathway, specific NF-κB subunits are released from their inhibitory IκBs by virtue of the serine-threonine kinase activities of the IKK signalosome. In response to extracellular stress signals, IκBα is phosphorylated by the IKK complex thereby targeting it for ubiquitination and subsequent proteasomal destruction. The removal of IκB results in exposure of the nuclear localization sequence of NF-κB, leading to nuclear translocation of NF-κB hetero-dimers (including p65/RelA:p50 and c-Rel:p50) and the subsequent activation of their target genes (13). The IKK complex consists of two serine-threonine kinases, IKKα and IKKβ, and NEMO/IKKγ, a regulatory or docking protein that facilitates IKK complex assembly and the regulated transmission of upstream activating signals to IKKα and IKKβ (13-15). Most extracellular stimuli cause only transient IKK activation and mechanisms to limit IKK activity are physiologically important because persistent NF-κB activity is associated with numerous pathological conditions.
IKKβ is almost always the IκBα kinase in vivo, although IKKα can infrequently assume this role (16, 17). In contrast to IKKβ, IKKα functions to attenuate or resolve acute inflammatory responses by more than one mechanism (18-20) and is the essential kinase activator of the non-canonical NF-κB pathway (21). Upon activation by the NF-κB inducing kinase (NIK), IKKα phosphorylates specific serine residues in the carboxy-terminal domain of NFκB2/p100, leading to its proteasomal processing into the mature NF-κB p52 subunit and subsequent nuclear translocation of p52-RelB hetero-dimers (22). Non-canonical NF-κB activation also requires new protein synthesis and thus is not activated as rapidly as the classical IKKβ/NEMO dependent pathway. Moreover, only a few extracellular signals have been identified that can activate the NIK-IKKα non-canonical NF-κB pathway (including CD40L, LTβ and BAFF) (11). Interestingly, extracellular stimuli resulting in cellular responses that require sustained NF-κB induction appear to activate both the IKKβ/NEMO and NIK/IKKα signaling pathways (23-26). In addition to driving nuclear translocation of RelB/p52 heterodimers, the IKKα dependent non-canonical pathway has also been reported to activate p65/p52 (25) and a subset of p50/p65 heterodimers (sequestered in the cytoplasm in p100 complexes) (26), suggesting that IKKα can also contribute to certain pro-inflammatory responses.
Although a host of cytokines and chemokines are targets of NF-κB transcription factors, only a paucity of data (and only in either immortalized or engineered cell lines in vitro) has suggested that canonical NF-κB activation in migrating cells may contribute to their chemotactic responses (27-29). We have previously shown that both the IKKβ-driven canonical and the IKKα-dependent p52/RelB non-canonical NF-κB pathways are simultaneously critical for cell migration to HMGB1 (30, 31). Even though it is well established that HMGB1 (32-34) and CXCL12 (6, 8, 35-38) both activate the canonical NF-κB pathway, until our recent published work, it was not known if their unique chemotactic properties require cells to express specific NF-κB target genes needed for cells to migrate towards these two chemoattractants. Here we show that IKKβ and IKKα mediated canonical and non-canonical NF-κB signaling pathways are essential for the migration of fibroblasts and macrophages in response to CXCL12. IKKβ, but not IKKα, is required to maintain a threshold level of cell surface CXCR4, which is needed to maintain CXCL12-elicited chemotaxis. In conjunction with the latter functional role of IKKβ, IKKα, (via its unique function to activate the RelB/p52 non-canonical NF-κB pathway), is critically important for the initial polarization and velocity of cell movement towards a CXCL12 gradient.
MATERIALS AND METHODS
1.1 Ethics Statement
All animal work was approved by the IACUC committee of Stony Brook University in accordance with USA NIH guidelines for the use of animals in biomedical research. These studies utilized only in vitro experiments with primary embryonic fibroblasts (MEFs) or bone marrow progenitors (BMPs) isolated from the femurs of adult mice and subsequently differentiated to mature macrophages in vitro. Mouse pups or adult mice were euthanized by an IACUC approved protocol prior to the isolation of MEFS or BMPs.
1.2 Conditional and inducible IKKα KO mice
Mice with IKKα alleles flanked by LoxP recombination sites (IKKαf/f mice) that express Cre recombinase under the control of the macrophage lysozyme (MLys) promoter only in mature macrophages (MΦ) and neutrophils (IKKαf/f:MLysCre mice) (30). Alternatively, other mice express a tamoxifen (4-OHT) inducible Cre gene (IKKαf/f:CreERT2 mice) that have been previously described (30). All animal work was approved by Stony Brook University's IACUC committee in accordance with NIH guidelines.
1.3 Reagents
Recombinant murine CXCL12/SDF-1 was obtained from PeproTech (Rocky Hill, NJ). Human recombinant PDGF and human recombinant complement C5a were purchased from R&D Systems (Minneapolis, MN); purified fibronectin was obtained from Roche (Indianapolis, IN). Tamoxifen (4-hydroxytamoxifen, 4-OHT) was obtained from Sigma-Aldrich (St. Louis, MO); Alexafluor 647-conjugated anti-mouse CXCR4 antibody was purchased from Biolegend (San Diego, CA). All materials for the in vitro cell migration assays were obtained from Neuroprobe (Cabin John, MD) and included 48 well microchemotaxis chamber and 8 μm pore size cellulose nitrate filters (for macrophages) and 8 μm pore size PVP-free polycarbonate filters (for fibroblasts).
1.4 Cells and tissue culture
Immortalized WT, IKKα KO, p52 KO and RelB KO MEFs were maintained as previously described in Dulbecco's Modified Eagle's Medium (DMEM) with 10% Fetal Bovine Serum (FBS), 100 units/ml penicillin and 100 μg/ml streptomycin. Bone marrow progenitors from the femurs of IKKα WT (IKKαf/f), IKKαf/f:MLysCre and IKKβf/f:MLysCre adult mice were differentiated to MΦ in M-CSF conditioned DMEM/10%FBS for 7 days as previously described (30); and the loss of IKKα or IKKβ in myeloid cell progenitors does not affect the efficiency of their differentiation to mature macrophages or neutrophils (30). Primary MEFs were isolated from 5-6 day old IKKαf/f:CreERT2 mouse embryos also as described in prior reports (30, 31).
1.5 Retroviral transduction
IKKβ and IKKα KO MEFs were stably transduced with a Moloney murine retroviral vector containing a murine CXCR4 cDNA expressed as part of a bi-cistronic IRES-Puromycin expression cassette (39). Murine CXCR4 cDNA was subcloned upstream of an IRES-puromycin cassette in the BIP murine Moloney retroviral vector (40, 41). The generation of amphotyped viruses, infection of cells and selection of stable puromycin resistant cell populations have been previously described (30, 40, 41).
1.6 In vitro chemotaxis assays
Chemotaxis assays with MEFs and MΦ were performed as previously described (30, 31). MEF and MΦ migration assays were performed with 5 × 104 and 1 × 105 cells respectively per well of a 48 well microchemotaxis (Boyden-type) chamber. Chemotaxis of MEFs utilized 8 μm pore size PVP-free polycarbonate filters pre-coated with fibronectin (50 μg/ml) and chambers were incubated for 3 hours at 37°C. MEFs were quantified as the number of migrated cells per high power field (400×), and the background control (serum-free media) was subtracted so data are expressed as net migrated cells. Macrophage chemotaxis employed cellulose nitrate filters (8 μm pore size) in chambers incubated for 3 hours at 37°C. Distances of macrophages migrated into the nitrocellular filters were measured by the leading front method as previously described (42). Macrophage migration data is presented as net cell movement per 3 hours. In our experience and prior work (30, 31), nitrocellulose filters, which measure the leading front distance of migrating cells, is a superior and more sensitive technique for myeloid cells (including macrophages, monocytes and neutrophils) than counting cells per high power field with polycarbonate filters. Mesenchymal cells such as fibroblasts do not migrate into the nitrocellulose filters, even when employing very large pore size (12 μm) filters. For fibroblast migration assays we used thin (10 μm depth) polycarbonate filters with straight 8 μm holes to count cells adhering to the underside of the filter.
For IBIDI time-lapse chemotaxis experiments, cells of different genotypes were distinguished from each other by staining with 500 nM Green CMFDA or 500 nM Orange CMRA CellTracker™(Invitrogen) for 15 min in a humidified tissue culture incubator. Labeled cells were washed, resuspended at 4 × 105 cells/ml in DMEM (phenol red free) +0.1% BSA, mixed and placed (300 μl) in a μ-Slide (IBIDI/Integrated BioDiagnostics Inc.) pre-coated with 50 μg/ml fibronectin. After cells had firmly attached to the substratum, slides were laid on a 37°C humidified stage of an UltraVIEW ERS Spinning Disk confocal microscope (Perkin Elmer). A 0-30 μg/ml CXCL12 gradient, mixed with fluorescent beads (Molecular Probes) for microscope viewing, was allowed to form in the chamber's channel as described (43). Pictures of cells at the edge of the gradient were captured every 2 minutes for up to 3 h (microscope objective Zeiss, 5X magnification, numerical aperture 0.15; EM-CCD Hamamatsu C9100 Camera; UltraVIEW ERS acquisition software). Directional tracks are defined as those with ending points closer to the CXCL12 source compared to their starting points, whereas non-directional tracks are the opposite. Indeterminate tracks starting and ending at the same distance from the CXCL12 gradient (moving laterally) are also considered non-directional tracks. Cell tracks were analyzed with ImageJ software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, MD, USA, http://rsb.info.nih.gov/ij/, 1997-2008).
1.7 Flow cytometry
Primary mature macrophages were detached from cell culture dishes by incubation in DPBS containing 10% EDTA for 10 minutes at 4°C. Cells were resuspended at 5 × 106 cells per ml in flow cytometry staining buffer (DPBS + 1% BSA + 0.02% NaN3); and 5 × 105 cells were blocked with 2 μg of mouse IgG for 15 minutes on ice and subsequently stained with 1 μg of Alexafluor 647-conjugated anti-mouse CXCR4 antibody (Biolegend) for an additional 15 minutes on ice. Cells were washed twice with flow cytometry staining buffer and then fixed in 2 % paraformaldehyde prior to analysis. For intracellular staining, macrophages were permeabilized by washing twice with 1× BD perm/wash buffer (BD Biosciences, San Jose, CA). Next, cells were resuspended in 100 μl of perm/wash buffer and blocked with 2 μg of mouse IgG for 30 minutes on ice. Cells were stained with 1 μg of fluorophore conjugated anti-CXCR4 antibody for 30 minutes on ice and then washed twice with perm/wash buffer and fixed with 2% paraformaldehyde. Flow cytometric data were collected using a BD FACS Calibur II flow cytometer and the data were analyzed using FlowJo (Tree Star Inc., Ashland, OR). As a negative control for the specificity of the anti-CXCR4 antibody, cells were stained with an isotype control antibody conjugated with the same fluorophore.
1.8 Immunoblotting and cell fractionation
Immunoblotting and the preparation of nuclear and cytoplasmic fractions were performed as previously described (30, 41). HRP conjugated secondary antibody signals were detected with an enhanced chemiluminescent detection kit (GE Healthcare, Piscataway, NJ). Nuclear p52 levels were normalized with respect to nuclear lamin B1. Film images were acquired with a Fluor-S Multimager (BioRad). Scanned protein bands were quantified with Quantity One 4.5.0 software (BioRad) and processed with Adobe Creative Suite.
1.9 Sybr green real time RT-PCR assays
Total cell RNAs were prepared and Sybr green real time RT-PCR was performed and quantified as previously described (30) Forward (F) and reverse (R) PCR primer sequences were as follows:
Gapdh F (GCTCACTGGCATGGCCTTC),
Gapdh R (CCTTCTTGATGTCATCATACTTGGC);
Cxcl12 F (GCACGGCTGAAGAACAACAAC),
Cxcl12 R (TTCCTCGGGCGTCTGACTC).
1.10 Statistical analysis
P values were determined with either Prism V4.0 Software or InStat (both obtained from GraphPad Inc., San Diego, CA) to 4 significant figures by two tailed Student's t test, one way or two way ANOVA with Tukey's multiple comparisons post test as indicated in the figure legends of specific experiments.
RESULTS
2.1 The IKKα and IKKβ driven NF-κB pathways are both required for cell migration in response to CXCL12
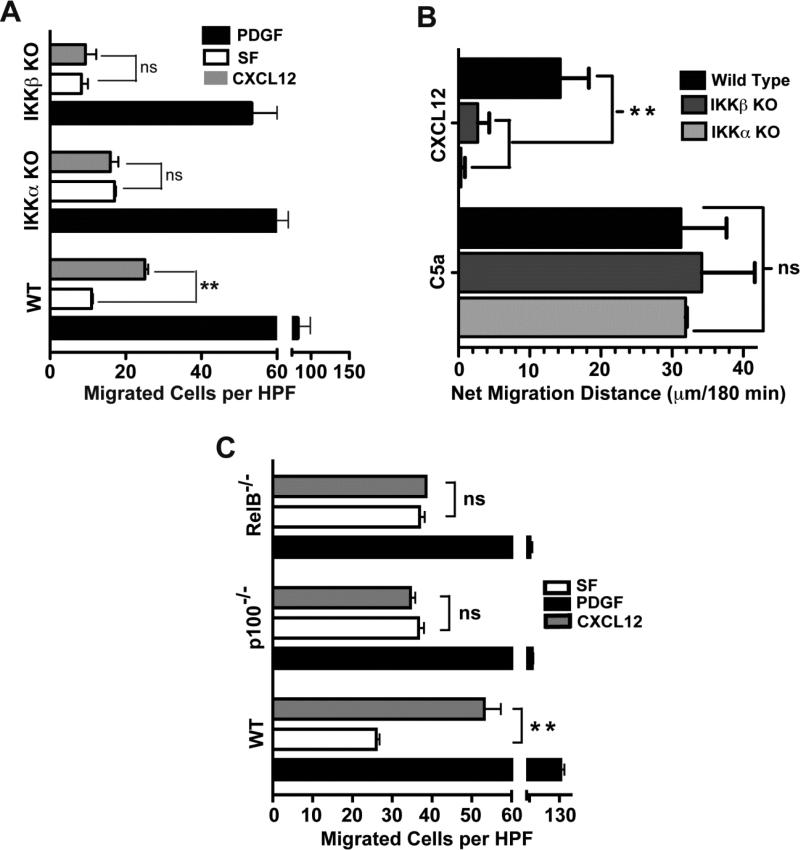
We have previously shown that the NF-κB canonical pathway is required for cell migration to HMGB1 and CXCL12 but not to other cytokines (9), and that the IKKβ and IKKα-driven canonical and non-canonical NF-κB pathways are both essential for cell migration to HMGB1 (30, 31). Akin to our findings with HMGB1-mediated cell migration, we find that IKKβ and IKKα are essential for the migration of immortalized MEFs (Figure 1A) but not to a PDGF positive control. To determine if this was also a property of other cell types lacking IKKα or IKKβ, we performed analogous experiments with mature primary macrophages differentiated from bone marrow progenitors of IKKβf/f;MLysCre and IKKαf/f;MLysCre mice, in which IKKβ or IKKα respectively are absent in mature myeloid cells. Immunoblots demonstrating that IKKβ and IKKα proteins are not expressed by mature myeloid cells derived from these mice were reported previously (30) and other representative examples are shown in Suppl. Figure 1A. Indeed, akin to IKKβ and IKKα KO MEFs, mature macrophages derived from these mice also are unable to migrate to CXCL12 as compared to mature macrophages from WT mice. In contrast, macrophages from each of these 3 mouse strains migrate equally well to the positive control, complement activation fragment C5a (Figure 1B). The essential IKKα requirement for chemotaxis in response to CXCL12 is functionally linked to the non-canonical NF-κB pathway, because MEFs lacking either the non-canonical NF-κB subunits p52 or RelB also failed to migrate towards CXCL12, while their ability to respond to PDGF was similar to that of WT MEFs (Figure 1C).
Figure 1. IKKα, IKKβ and NF-κB p52 and RelB are each required for cell migration in response to CXCL12.
(A) Chemotaxis of immortalized WT, IKKβ KO and IKKα KO MEFs (5 × 104 cells) was evaluated using a 48 well microchemotaxis chambers as previously described (30). Cells were allowed to respond to CXCL12 (50 ng/ml), serum-free media (SF) as the negative control and PDGF (10 ng/ml) as the positive control for robust cell migration. The number of cells that traversed an 8 μm pore-size polycarbonate filter after a 3 hour incubation at 37°C were counted using a microscope at 400× magnification. Numbers represent mean ± SEM of cells per high power field (HPF), n = 4, ** indicates p<0.01, ns = not significant. PDGF served as a positive migration control for all 3 cell backgrounds but only to illustrate that each cell type migrate towards PDGF but in contrast to WT MEFs, IKKα KO and IKKβ KO cells do not migrate towards CXCL12. The absolute degrees of migration of WT, IKKα KO and IKKβ KO MEFs towards PDF are somewhat differ from each other, which likely reflects intrinsic properties of these different cell lines. Thus, statistical analyses of the results in Figure 1A were done for WT MEFs exposed to media vs. CXCL12: IKKα KO MEFs exposed to media vs. CXCL12; and IKKβ KO MEFs exposed to media vs. CXCL12. (B) Primary mature WT and conditional IKKα and IKKβ KO macrophages (105 cells) were exposed to CXCL12 (50 ng/ml) or C5a (2 nM) as a positive control in 48 well microchemotaxis chambers for 3 hrs as previously described (30). Data is presented as net migration distance per 400× field after subtracting basal migration in serum free media (n = 3-4). Mean value of WT macrophage basal migration towards serum free media control was 35 ± 2.6 μm. Statistical significance is indicated, ns = not significant.
(C) Chemotaxis of WT, NF-κB p100/p52−/−and NF-κB RelB−/− MEFs (5 × 104 cells) was evaluated using a 48 well microchemotaxis chambers as previously described (30). Cells were allowed to respond to CXCL12 (50 ng/ml), serum-free media (SF) as the negative control and PDGF (10 ng/ml) as the positive control for robust cell migration. The number of cells that traversed an 8 μm pore-size polycarbonate filter after a 3 hour incubation at 37°C were counted using a microscope at 400× magnification. Numbers represent mean ± SEM of cells per high power field (HPF), n = 4, ** indicates p<0.01, ns = not significant.
2.2 IKKβ is required to maintain sufficient levels of CXCR4
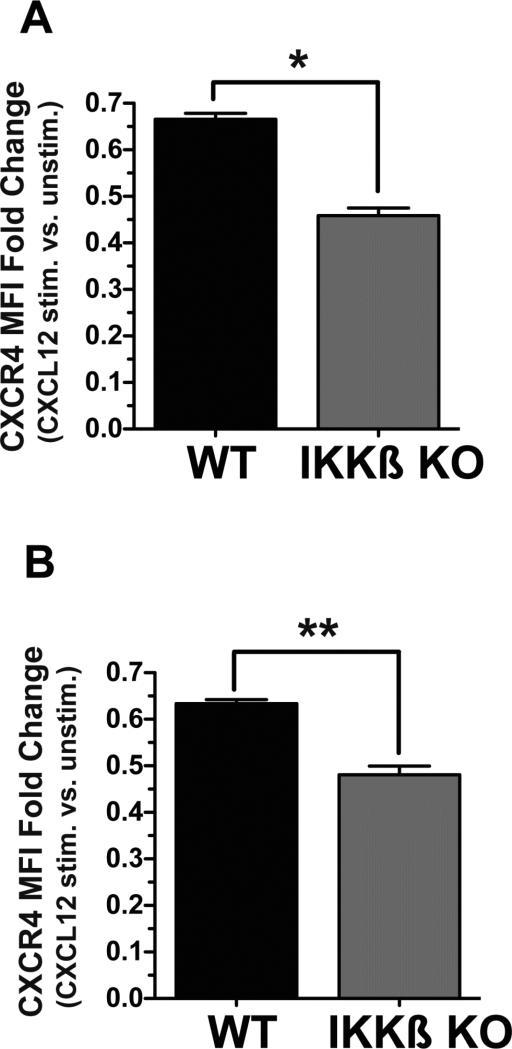
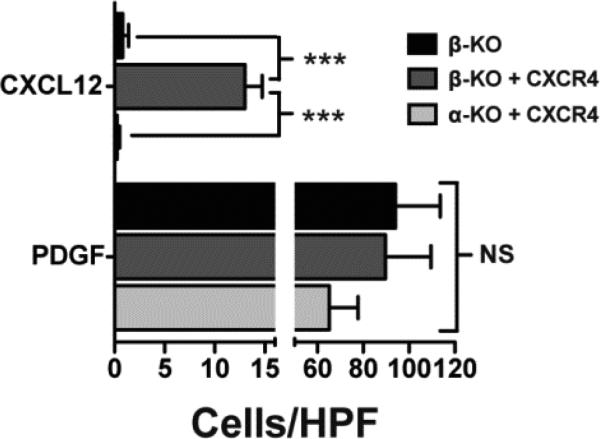
Since the CXCL12 receptor CXCR4 is a direct target of the canonical NF-κB pathway (6, 8), we performed experiments to investigate if the essential requirement for IKKβ was linked to significant differences of CXCR4 expression levels in WT and IKKβ KO cells. To this end, we analyzed the relative expression levels of total cellular and cell surface CXCR4 protein in WT and conditional IKKβ KO IKKβf/f;MLysCre mature macrophages by quantitative flow cytometry. We performed this analysis by flow cytometry, and not by immunobloting nor by immunofluoresence staining of cells, because only flow cytometry provides a quantitative assessment of CXCR4 expression on a single cell basis. This analysis of total cellular and cell surface levels of CXCR4 in multiple batches of WT and IKKβ KO macrophages revealed that the expression levels of CXCR4 were significantly reduced in IKKβ KO cells relative to their WT counterparts (Figure 2A). Since cell surface CXCR4 is internalized upon binding to its ligand CXCL12 (44, 45), we observed a significantly greater reduction of cell surface CXCR4 with IKKβ KO Macs compared to WT Macs after 2 hr of exposure to CXCL12 (see results shown as fold change in MFI with an without CXCL12 treatment for total and cell surface levels of CXCR4 in Figure 2A&B respectively). Similar to our observations with primary mature macrophages, we also noted reduced cell surface CXCR4 levels in IKKβ KO MEFs compared to WT or NF-κB p52 KO MEFs when cells were treated with CXCL12 (Supplementary Figure 2). Representative quantitative flow cytometry experiments comparing cell surface and total CXCR4 expression in un-stimulated versus CXCL12-treated cells are shown in Supplementary Figure 3. In addition, these quantitative experiments demonstrate that total levels of CXCR4 in unstimulated IKKβ KO primary mature macrophages are significantly reduced compared to their WT counterparts (Supplementary Figure 3C). To investigate if the IKKβ-dependent maintenance of a critical, threshold level of cell surface CXCR4 is functionally linked to IKKβ's essential requirement for cell migration to CXCL12, we enforced the expression of murine CXCR4 in both IKKβ and IKKα KO MEFs by stable retroviral transduction and evaluated their migration in response to CXCL12 by quantitative flow cytometry analysis. The enhanced cell surface expression of CXCR4 in these cells, as well as the specificity of the anti-CXCR4 antibody (31), was verified as shown in Supplementary Figure 4. Indeed, the CXCL12-induced migration of IKKβ KO MEFs was rescued by the enforced expression of exogenous murine CXCR4; in contrast, IKKα KO MEFs expressing similar levels of exogenously introduced CXCR4 still failed to migrate towards CXCL12 (Figure 3).
Figure 2. CXCR4 protein expression levels in Wt versus IKKβ conditional KO macrophages.
Mature macrophages derived from WT and myeloid cell conditional IKKβf/f:MLysCre mice were stained with anti-CXCR4 antibody after cell permeabilization to reveal total cellular CXCR4 protein expression (A), or without permeabilization to quantity cell surface CXCR4 levels (B) and then analyzed by flow cytometry. Data are shown as mean fluorescence intensities (MFI) from 4 independent experiments and are expressed as fold change of cells stimulated with CXLC12 (50 ng/ml) for 2 hr vs. unstimulated cells. *p<0.05 and **p<0.01 by one way ANOVA with Tukey's multiple comparisons post test.
Figure 3. Enforcing CXCR4 expression rescues IKKβ but not IKKα cell migration towards CXCL12.
IKKβ and IKKα KO MEFs were stably transduced with a Moloney murine retroviral vector harboring a murine CXCR4 cDNA in a bi-cistronic IRES-Puromycin expression cassette (MuCXCR4-BIP). Cell surface CXCR4 expression was verified by flow cytometry. IKKα and IKKβ KO MEFs stably transduced with MuCXCR4-BIP and IKKβ KO MEFs as a negative control were exposed for 3 hrs to CXCL12 (50 ng/ml) in 48 well microchemotaxis chamber. PDGF (10 ng/ml) served as the positive control for cell migration. Numbers represent mean ± SEM of cells per high power field (n = 5), *** = p<0.001, NS = not significant.
2.3 IKKα is required at the earliest phase of CXCL12-mediated cell migration
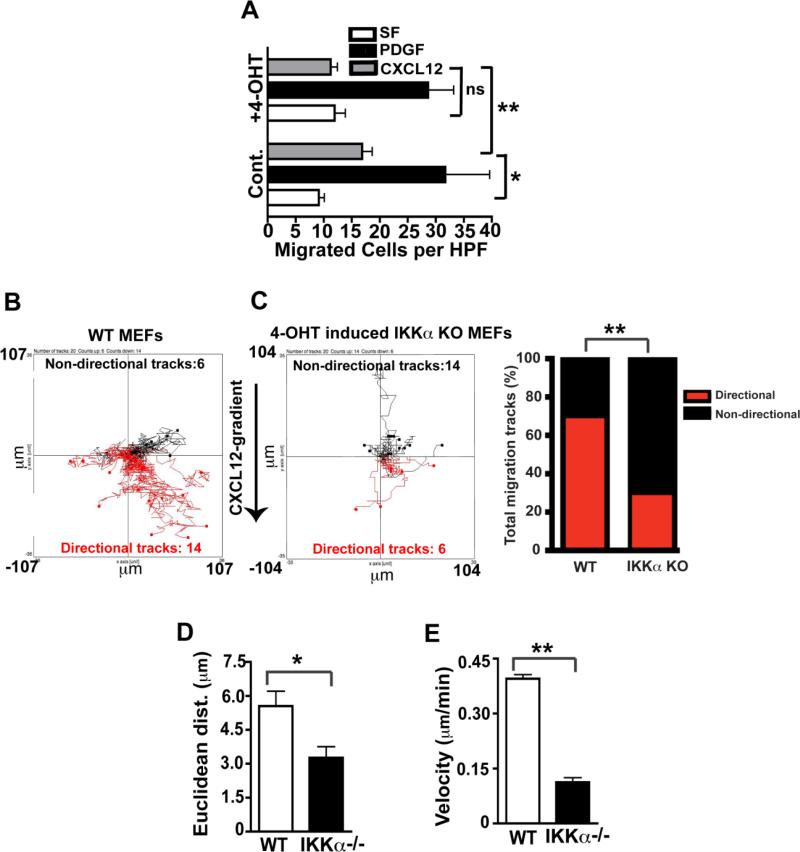
To begin to explore the mechanism of action of IKKα in CXCL12 induced cell migration, we dissected the migration parameters of WT vs. induced IKKα KO primary MEFs in time-lapse microscopy experiments employing IBIDI cell migration slides (30, 43). In vitro migration assays were performed with primary IKKαf/f;CreERT2 MEFs sham-treated (WT control) or treated with 4-OHT (IKKα induced deletion) in IBIDI μ slides as previously described (30). Immunoblotting confirmed the ablation of IKKα protein expression in IKKαf/f;CreERT2 MEFs exposed to 4-OHT (30) (Supplementary Figure 1B). As shown in Figure 4A, IKKα is required in primary MEFs for migration towards CXCL12, as it is in immortalized MEFs (shown above in Figure 1). The functional impact of IKKα for directional chemotaxis is best determined at the earliest phase of the migration response, which defines cellular polarization/orientation towards the CXCL12 gradient. In this analysis, we defined directional tracks as those with ending points closer to the higher CXCL12 concentration in the gradient, compared to their starting points, whilst non-directional tracks are the opposite; and indeterminate tracks starting and ending at the same distance from the CXCL12 gradient (moving laterally) are also considered non-directional tracks {see detailed description of how the IBIDI experiments were performed in Materials and Methods} (30, 43). These time lapse time video microscopy experiments revealed that IKKα is critical for determining initial cellular orientation/polarization towards a CXCL12 gradient (compare directions of migration tracks in WT vs, induced IKKα KO cells in Figure 4B and 4C respectively and see statistically analyzed bar graph results summary in Figure 4C). The Euclidean distance (e.g. the straight line distance between starting and arriving points) and velocity of all tracks were compared to each other to ensure that all cells in the population were analyzed equally for these two migration parameters independent of their directionality to avoid biasing the results solely on the cells migrating directly towards the CXCL12 gradient. Importantly, Figures 4D and 4E reveal that IKKα is also required for optimal Euclidean migration distance and for the degree of cellular velocity within the initial 60 minutes of the chemotactic response to CXCL12.
Figure 4. Functional consequences of IKKα ablation on specific CXCL12 migration parameters.
(A): IKKαf/f:CreERT2 MEFs were incubated with or without 4-OHT (100 nM) for 36 hours, and migration assays were performed using serum free medium (SF), or in response to the same media containing CXCL12 (30 ng/ml) or the positive control PDGF (10 ng/ml). Numbers represent mean ± SEM of cells per high power field (n = 3), ** = p<0.01, * = p <0.05, NS = not significant. (B and C): Cells were exposed CXCL12 gradients (0-30 ng/ml) in μ slides for 1 hour, and their movement was recorded by time-lapse microscopy. Red lines are tracks of cells moving in the direction of the CXCL12 gradient, and black lines are cell tracks moving in other directions. The difference in the number of directional and non-directional tracks between WT and the IKKα ablated MEFs is statistically significant (χ2 test **p<0.01); and bar graphs summarizing these results are shown to the right of panel C. (D and E): WT and IKKα KO (4-OHT treated) cell tracks in Panels B & C were analyzed for their relative Euclidean distances and velocities, *p<0.05 and **p<0.01 by two-tailed Student's t tests.
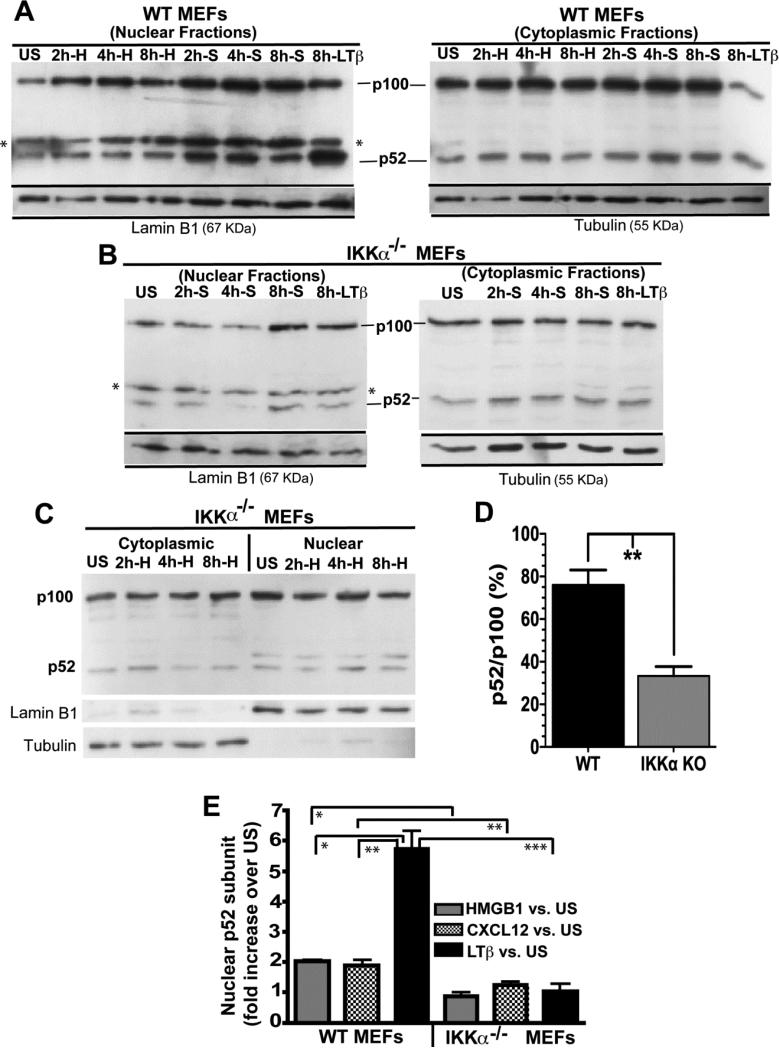
2.4 CXCL12 up-regulates the NF-κB non-canonical pathway
Because the IKKα dependent NF-κB non-canonical pathway is required for the earliest phase of migration in response to CXCL12 (polarization and initial velocity), these results suggest that the IKKα /p52/RelB axis could be necessary to maintain a sufficient basal level of a critical protein co-factor. In conjunction with such a mechanism, CXCL12 may also stimulate IKKα dependent p52 nuclear translocation to up-regulate the expression of a subset of IKKα/RelB/p52-dependent genes. To explore the latter possibility, MEFs were exposed to CXCL12 for up to 8 hours and cytoplasmic and nuclear cell fractions were scored for their levels of p100 (p52's precursor) and the processed p52 NF-κB subunit. Representative immunoblots are shown in Figure 5A-C with lamin-B1 and β-tubulin serving as normalization/reference controls for nuclear and cytoplasmic proteins respectively. Cells treated for a similar duration with anti-murine lymphotoxin β receptor (mLTβR) antibody, an agonist which is known to robustly activate IKKα-dependent non-canonical NF-κB signaling (46), served as a positive control. Quantitative densitometric analysis of multiple immunoblots of nuclear and cytoplasmic extracts of WT vs. IKKα−/− cells indicate that CXCL12, as we previously had shown for HMGB1 (30), modestly induces p52 nuclear translocation in WT but not in IKKα−/− MEFs (Figure 5E). In contrast, and as expected, LTβR stimulation induced robust p100>p52 processing in WT MEFs, while no effect was seen in IKKα null cells (Figure 5E). The reduced levels of cytoplasmic and nuclear p52 (in comparison to its p100 precursor) observed in IKKα null MEFs in Figure 5 are in agreement with prior reports showing that IKKα is required to maintain a constitutive, basal level of cytoplasmic and nuclear p52 (46, 47). Indeed, quantitative densitometric analysis of immunoblots from multiple independent experiments show that unstimulated IKKα KO MEFs present a reduced degree of p100 > p52 processing (revealed by their relative p52/p100 ratios) in comparison to WT MEFs (Figure 5D). In contrast to IKKα KO MEFs, similar independent experiments show that unstimulated IKKβ KO MEFs present p52/p100 ratios of 85±12.5%, which are comparable to WT MEFs
Figure 5. CXCL12 is a modest, IKKα-dependent inducer of NF-kB p52 nuclear translocation.
(A) Immortalized WT or (B and C) IKKα−/− MEFs were stimulated with either 50 ng/ml CXCL12, 100 ng/ml HMGB1 or 10 μg/ml of an antagonistic anti-LTβR antibody for the indicated times. S refers to CXCL12/SDF-1 and H denotes HMGB1. Nuclear and cytoplasmic protein fractions were analyzed by western blotting as shown with p100 and p52 identified by an anti-p100/p52 antibody. The asterisk in the immunoblot denotes a cross-reactive artifact band in the anti-p52 nuclear extracts. Immunoblots were stripped and re-probed with antibodies against Lamin B1 or β-Tubulin as protein loading controls for nuclear and cytoplasmic cell fractions, respectively. (D) Comparative analysis of p52:p100 levels in un-stimulated WT vs. unstimulated IKKα KO MEFs from 3 independent experiments (one of which is shown in panels A-C). E) Densitometric quantification of p52 nuclear import in WT and IKKα−/− cells in response to HMGB1, CXCL12/SDF-1 or LTβR stimulations at the 8 hr time point. Numbers represent 3 independent experiments. p52 signals were all normalized to lamin B1 as a nuclear protein reference control. Error bars are standard error of the mean. *P<0.05; **P<0.01; ***P<0.001 by two-tailed Student's t tests.
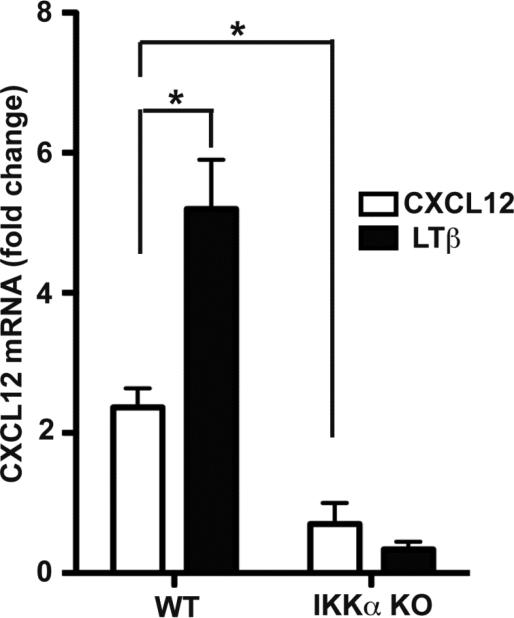
In light of the above biochemical observations, we next investigated if IKKα was also required for the induction of RelB/p52 target gene expression in response to CXCL12. Because the CXCL12 gene is well known direct target of RelB/p52 NF-κB heterodimers, we analyzed CXCL12 mRNA expression levels by qRT-PCR in WT and IKKα KO MEFs in response to CXCL12 with LTβ receptor activation serving as a positive control. Indeed as shown in Figure 6, CXCL12 is a modest IKKα-dependent inducer of its own expression.
Figure 6. CXCL12 up-regulated the level of its mRNA with a dependency on IKKα.
Real time RT-PCR analysis of CXCL12 mRNA expression in WT and IKKα−/− MEFs in response to CXCL12 or LTβR stimulation. Data were normalized to GAPDH mRNA qRT-PCRs as a reference control. +P<0.05.
DISCUSSION
Cell migration is a sophisticated, multi-step process critical for animal development, innate and adaptive immunity and the recognition and repair of damaged tissue. Inappropriate and/or dysregulated cell migration contributes to the progression of cancers, poor wound healing, inflammatory-induced tissue injury and other maladies {reviewed in (48, 49)}.
3.1 Cell migration responses towards CXCL12 are mechanistically related to chemotactic responses to HMGB1
CXCL12 (SDF-1), along with HMGB1, belong to a small group of chemoattractants that are essential for stem cell trafficking (50-54). Consequently, these molecules are critically important for the recognition and repair of damaged tissue in vivo (50, 53, 54). Extracellular HMGB1 engages the cell surface Receptor for Advanced Glycation End-products (RAGE) and thereby elicits a range of inflammatory reactions by acting as a cytokine and chemoattractant (33, 34, 55, 56). The pertussis toxin sensitivity of HMGB1 chemotactic responses suggested early on that at least one G-protein coupled receptor is also required for HMGB1-mediated cell migration responses (57); and recent work by our lab and others has shown that CXCR4 functions as this essential receptor for HMGB1-mediated cell migration responses (31, 58). HMGB1 forms functional complexes with a variety of endogenous and exogenous effectors (including CpG-ODNS, LPS, IL-1β and nucleosomes) to dramatically enhance inflammatory responses (59-62); and HMGB1-partner complexes signal through the specific partner molecule's receptor, independent of HMGB1 receptors, to dramatically enhance the production of cytokines associated with inflammatory responses (63). Recently, we reported that the HMGB1-mediated cell migration responses require extracellular CXCL12, which is secreted by cells migrating towards HMGB1 (31, 64). HMGB1 forms functional complexes with CXCL12 (64); and the mechanism of cell migration towards HMGB1 involves HMGB1-CXCL12 complexes signaling via CXCR4 (31, 58), which is believed to be sufficient to drive cell migration towards HMGB1 in the absence of other HMGB1 receptors (58). Interestingly, CXCL12 ligation of CXCR4 activates some of the same pathways as HMGB1 including canonical NF-κB signaling, moreover, CXCR4 (6, 8) and its ligand CXCL12 (46, 65), are direct targets of the IKKβ-dependent canonical and IKKα-dependent non-canonical NF-κB pathways, respectively. Importantly and unlike other cell migratory responses, we have previously shown that chemotaxis to HMGB1 or CXCL12 in vitro and in vivo uniquely requires activating of the canonical NF-κB pathway (9, 57). Furthermore, we have also previously shown that the IKKβ/canonical and IKKα/non-canonical NF-κB signaling pathways are simultaneously required for HMGB1 chemotactic responses in vitro and in vivo (30, 31), with IKKβ signaling necessary to maintain sufficient levels of RAGE (30); and IKKα signaling essential for maintaining a sufficient expression of its direct target CXCL12 for cells to migrate towards HMGB1/CXCL12 complexes (31, 58). In this new study we present evidence that the IKKβ and IKKα-driven NF-κB signaling pathways are also both essential for cell migration to CXCL12 alone, independent of HMGB1, thereby linking the regulation of cell migration to the two unique chemoattractants mediating cell recruitment to damaged tissues.
3.2 IKKβ and IKKα-dependent NF-κB signaling are simultaneously essential for directed cell movement towards CXCL12/SDF-1
Our data collectively points to constitutive IKKβ-dependent canonical NF-κB signaling as essential to maintain a specific threshold level of cell surface CXCR4, the sole receptor that positively mediates directed cell migration to CXCL12. Thus, on the basis of the new data presented here and our prior work (30), IKKβ-dependent canonical NF-κB signaling works by a common mechanism of action to regulate chemotactic responses to HMGB1 and CXCL12 by maintaining sufficient expression levels of the receptors for these two cytokines.
In contrast, the critical need for IKKα-dependent non-canonical NF-κB p52/RelB activation for cell migration towards CXCL12 is not linked to CXCR4 expression levels. Although the precise downstream target of the non-canonical NF-κB IKKα/p52/RelB signaling axis needed for cell migration to CXCL2 remains to be identified, our time lapse video microscopy experiments reveal that IKKα is required for the initial cell orientation/polarization and velocity towards a CXCL12 gradient. Taken collectively, along with our cell fractionation experiments showing that IKKα up-regulates the basal level of p100>p52 processing, our results herein provide evidence that cells migrating towards CXCL12 require the IKKα/p52/RelB pathway to maintain a sufficient level of a critical effector/adaptor of CXCR4-mediated CXCL12 signaling that is specifically needed for cell migration to CXCL12 alone. In contrast, cell migration towards HMGB1/CXCL12 complexes are believed to signal via an alternate conformation of the CXCR4 receptor (58).
In summary, we posit that the IKKα/p52/RelB pathway functions in a constitutive fashion to maintain basal levels of this putative CXCR4 signaling effector/co-factor; and our future work will in part be directed to identify this potentially novel regulatory protein factor.
CONCLUSIONS
Our experiments show that IKKβ and IKKα are each essential for cell migration towards CXCL12 for different reasons. The results demonstrate that IKKβ is needed to maintain a threshold level of cell surface CXCR4, the CXCL12 receptor, while the dual requirement for IKKα is not linked to CXCR4 expression. Time lapse video microscopy analysis performed with inducible IKKα KO primary fibroblasts reveal that the essential need for IKKα becomes apparent within the initial 30-60 minutes of a CXCL12 chemotactic response, as IKKα appears to be critical for both initial cell polarization and velocity towards CXCL12. We also present evidence that CXCL12 is a modest iIKKα-dependent inducer of NF-κB p52 nuclear translocation and also up-regulates its own IKKα/RelB/p52-dependent expression. Because IKKα up-regulates the basal level of p100>p52 processing and is essential at the earliest phase of CXCL12-mediated cell migration, taken together our data suggests that IKKα is required to maintain the basal expression of a critical NF-κB p52/RelB target gene, which is required for CXCL12-mediated cell migration.
Supplementary Material
Cell migration to CXCL12 requires IKKβ- and IKKα-dependent NF-κB signaling
IKKβ is required to maintain a sufficient threshold level of CXCR4
IKKα is required for cell polarization and velocity towards CXCL12
IKKα is needed to maintain a sufficient level of active NF-κB p52/RelB heterodimers
CXCL12 modestly induces IKKα-dependent p52 nuclear translocation.
ACKNOWLEDGEMENTS
MP gratefully acknowledges the generous hospitality of Prof. Ruggero Pardi and the expert assistance of Dr. Raffaella Molteni (S. Raffaele Institute, Milan, Italy) for the IBDI chamber/time-lapse migration experiments. The authors declare no direct financial interests.
Grant support: The initial phase of this work was supported by the MAIN FP6 European Union Network of Excellence (MP and KBM) and subsequently by USA NIH grants GM066882 awarded to KBM and GM063769 to RRK and 5T32 GM008468 (for support of DMH) awarded to Stony Brook University's Molecular and Cellular Biology graduate training program.
Non-standard abbreviations
- C5a
complement component 5a
- HMGB1
High Mobility Group Box 1
- HPF
high power field
- IKK
Inhibitor of NF-κB kinase
- MEFs
Mouse Embryo Fibroblasts
- MΦ
mature macrophages
- PDGF
platelet derived growth factor
- RAGE
receptor for advanced glycation end products
- SDF-1/CXCL12
Stromal Cell Derived Factor 1/C-X-C motif ligand 12
- SF
serum free media
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
We the authors declare no conflicts of interest.
REFERENCES
- 1.Miller RJ, Banisadr G, Bhattacharyya BJ. CXCR4 signaling in the regulation of stem cell migration and development. Journal of Neuroimmunology. 2008;198:31–38. doi: 10.1016/j.jneuroim.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kucia M, Jankowski K, Reca R, Wysoczynski M, Bandura L, Allendorf DJ, Zhang J, Ratajczak J, Ratajczak MZ. CXCR4-SDF-1 signaling, locomotion, chemotaxis and adhesion. J. Mol. Histol. 2004;35:233–245. doi: 10.1023/b:hijo.0000032355.66152.b8. [DOI] [PubMed] [Google Scholar]
- 3.Ganju RK, Brubaker SA, Meyer J, Dutt P, Yang Y, Qin S, Newman W, Groopman JE. The α-Chemokine, Stromal Cell-derived Factor-1α, Binds to the Transmembrane G-protein-coupled CXCR-4 Receptor and Activates Multiple Signal Transduction Pathways. Journal of Biological Chemistry. 1998;273:23169–23175. doi: 10.1074/jbc.273.36.23169. [DOI] [PubMed] [Google Scholar]
- 4.Tilton B, Ho L, Oberlin E, Loetscher P, Baleux F, Clark-Lewis I, Thelen M. Signal Transduction by Cxc Chemokine Receptor 4: Stromal Cell–Derived Factor 1 Stimulates Prolonged Protein Kinase B and Extracellular Signal–Regulated Kinase 2 Activation in T Lymphocytes. The Journal of Experimental Medicine. 2000;192:313–324. doi: 10.1084/jem.192.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chernock RD, Cherla RP, Ganju RK. SHP2 and cbl participate in α-chemokine receptor CXCR4–mediated signaling pathways. Blood. 2001;97:608–615. doi: 10.1182/blood.v97.3.608. [DOI] [PubMed] [Google Scholar]
- 6.Helbig G, Christopherson KW, Bhat-Nakshatri P, Kumar S, Kishimoto H, Miller KD, Broxmeyer HE, Nakshatri H. NF-{kappa} B Promotes Breast Cancer Cell Migration and Metastasis by Inducing the Expression of the Chemokine Receptor CXCR4. J. Biol. Chem. 2003;278:21631–21638. doi: 10.1074/jbc.M300609200. [DOI] [PubMed] [Google Scholar]
- 7.Neuhaus T, Stier S, Totzke G, Gruenewald E, Fronhoffs S, Sachinidis A, Vetter H, Ko YD. Stromal cell-derived factor 1α (SDF-1α) induces gene-expression of early growth response-1 (Egr-1) and VEGF in human arterial endothelial cells and enhances VEGF induced cell proliferation. Cell Proliferation. 2003;36:75–86. doi: 10.1046/j.1365-2184.2003.00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kukreja P, Abdel-Mageed AB, Mondal D, Liu K, Agrawal KC. Up-regulation of CXCR4 expression in PC-3 cells by stromal-derived factor-1alpha (CXCL12) increases endothelial adhesion and transendothelial migration: role of MEK/ERK signaling pathway-dependent NF-kappaB activation. Cancer Res. 2005;65:9891–9898. doi: 10.1158/0008-5472.CAN-05-1293. [DOI] [PubMed] [Google Scholar]
- 9.Palumbo R, Galvez BG, Pusterla T, De Marchis F, Cossu G, Marcu KB, Bianchi ME. Cells migrating to sites of tissue damage in response to the danger signal HMGB1 require NF-{kappa}B activation. J. Cell Biol. 2007;179:33–40. doi: 10.1083/jcb.200704015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldwin A., Jr. The NF-kappaB and IkappaB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 11.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 13.May MJ, Ghosh S. IkappaB kinases: kinsmen with different crafts. Science. 1999;284:271–273. doi: 10.1126/science.284.5412.271. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto Y, Gaynor RB. IkappaB kinases: key regulators of the NF-kappaB pathway. Trends Biochem Sci. 2004;29:72–79. doi: 10.1016/j.tibs.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 16.Cao Y, Bonizzi G, Seagroves TN, Greten FR, Johnson R, Schmidt EV, Karin M. IKKalpha Provides an Essential Link between RANK Signaling and Cyclin D1 Expression during Mammary Gland Development. Cell. 2001;107:763–775. doi: 10.1016/s0092-8674(01)00599-2. [DOI] [PubMed] [Google Scholar]
- 17.Hansberger MW, Campbell JA, Danthi P, Arrate P, Pennington KN, Marcu KB, Ballard DW, Dermody TS. IkappaB kinase subunits alpha and gamma are required for activation of NF-kappaB and induction of apoptosis by mammalian reovirus. J Virol. 2007;81:1360–1371. doi: 10.1128/JVI.01860-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 19.Li Q, Lu Q, Bottero V, Estepa G, Morrison L, Mercurio F, Verma IM. Enhanced NF-{kappa}B activation and cellular function in macrophages lacking I{kappa}B kinase 1 (IKK1). PNAS. 2005;102:12425–12430. doi: 10.1073/pnas.0505997102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Yang Y, Chernishof V, Loo RRO, Jang H, Tahk S, Yang R, Mink S, Shultz D, Bellone CJ, Loo JA, Shuai K. Proinflammatory Stimuli Induce IKK[alpha]-Mediated Phosphorylation of PIAS1 to Restrict Inflammation and Immunity. Cell. 2007;129:903–914. doi: 10.1016/j.cell.2007.03.056. [DOI] [PubMed] [Google Scholar]
- 21.Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, Karin M. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 22.Xiao G, Harhaj EW, Sun S-C. NF-[kappa]B-Inducing Kinase Regulates the Processing of NF-[kappa]B2 p100. Molecular Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 23.Yang CH, Murti A, Pfeffer SR, Kim JG, Donner DB, Pfeffer LM. Interferon alpha /beta promotes cell survival by activating nuclear factor kappa B through phosphatidylinositol 3-kinase and Akt. J Biol Chem. 2001;276:13756–13761. doi: 10.1074/jbc.M011006200. [DOI] [PubMed] [Google Scholar]
- 24.Saitoh T, Nakayama M, Nakano H, Yagita H, Yamamoto N, Yamaoka S. TWEAK Induces NF-{kappa}B2 p100 Processing and Long Lasting NF-{kappa}B Activation. J. Biol. Chem. 2003;278:36005–36012. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- 25.Yang CH, Murti A, Pfeffer LM. Interferon Induces NF-{kappa}B-inducing Kinase/Tumor Necrosis Factor Receptor-associated Factor-dependent NF-{kappa}B Activation to Promote Cell Survival. J. Biol. Chem. 2005;280:31530–31536. doi: 10.1074/jbc.M503120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basak S, Kim H, Kearns JD, Tergaonkar V, O'Dea B, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, Hoffmann A. A Fourth IkappaB protein within the NF-kappaB Signaling Module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko J, Kim IS, Jang SW, Lee YH, Shin SY, Min DS, Na DS. Leukotactin-1/CCL15-induced chemotaxis signaling through CCR1 in HOS cells. FEBS Lett. 2002;515:159–164. doi: 10.1016/s0014-5793(02)02465-1. [DOI] [PubMed] [Google Scholar]
- 28.Ko J, Jang SW, Kim YS, Kim IS, Sung HJ, Kim HH, Park JY, Lee YH, Kim J, Na DS. Human LZIP binds to CCR1 and differentially affects the chemotactic activities of CCR1-dependent chemokines. Faseb J. 2004;18:890–892. doi: 10.1096/fj.03-0867fje. [DOI] [PubMed] [Google Scholar]
- 29.Jang S-W, Kim YS, Kim YR, Sung HJ, Ko J. Regulation of Human LZIP Expression by NF-{kappa}B and Its Involvement in Monocyte Cell Migration Induced by Lkn-1. J. Biol. Chem. 2007;282:11092–11100. doi: 10.1074/jbc.M607962200. [DOI] [PubMed] [Google Scholar]
- 30.Penzo M, Molteni R, Suda T, Samaniego S, Raucci A, Habiel DM, Miller F, Jiang H.-p., Li J, Pardi R, Palumbo R, Olivotto E, Kew RR, Bianchi ME, Marcu KB. Inhibitor of NF-{kappa}B Kinases {alpha} and {beta} Are Both Essential for High Mobility Group Box 1-Mediated Chemotaxis. J Immunol. 2010;184:4497–4509. doi: 10.4049/jimmunol.0903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kew RR, Penzo M, Habiel DM, Marcu KB. The IKKα-Dependent NF-κB p52/RelB Noncanonical Pathway Is Essential To Sustain a CXCL12 Autocrine Loop in Cells Migrating in Response to HMGB1. J Immunology. 2012;188:2380–2386. doi: 10.4049/jimmunol.1102454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huttunen HJ, Fages C, Rauvala H. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-kappaB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J Biol Chem. 1999;274:19919–19924. doi: 10.1074/jbc.274.28.19919. [DOI] [PubMed] [Google Scholar]
- 33.Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B, Stern DM, Nawroth PP. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med. 2005;83:876–886. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 34.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 35.Tang C-H, Chuang J-Y, Fong Y-C, Maa M-C, Way T-D, Hung C-H. Bone-derived SDF-1 stimulates IL-6 release via CXCR4, ERK and NF-{kappa}B pathways and promotes osteoclastogenesis in human oral cancer cells. Carcinogenesis. 2008;29:1483–1492. doi: 10.1093/carcin/bgn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu D-Y, Tang C-H, Yeh W-L, Wong K-L, Lin C-P, Chen Y-H, Lai C-H, Chen Y-F, Leung Y-M, Fu W-M. SDF-1alpha up-regulates interleukin-6 through CXCR4, PI3K/Akt, ERK, and NF-kappaB-dependent pathway in microglia. European Journal of Pharmacology. 2009;613:146–154. doi: 10.1016/j.ejphar.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Chu C-Y, Cha S-T, Lin W-C, Lu P-H, Tan C-T, Chang C-C, Lin B-R, Jee S-H, Kuo M-L. Stromal cell-derived factor-1{alpha} (SDF-1{alpha}/CXCL12)-enhanced angiogenesis of human basal cell carcinoma cells involves ERK1/2-NF-{kappa}B/interleukin-6 pathway. Carcinogenesis. 2009;30:205–213. doi: 10.1093/carcin/bgn228. [DOI] [PubMed] [Google Scholar]
- 38.Huang CY LC, Chen MY, Yang WH, Chen YH, Chang CH, Hsu HC, Fong YC, Tang CH. Stromal cell-derived factor-1/CXCR4 enhanced motility of human osteosarcoma cells involves MEK1/2, ERK and NF-kappaB-dependent pathways. J Cell Physiol. 2009;221:204–212. doi: 10.1002/jcp.21846. [DOI] [PubMed] [Google Scholar]
- 39.Diamond P, Labrinidis A, Martin SK, Farrugia AN, Gronthos S, To LB, Fujii N, O'Loughlin PD, Evdokiou A, Zannettino ACW. Targeted Disruption of the CXCL12/CXCR4 Axis Inhibits Osteolysis in a Murine Model of Myeloma-Associated Bone Loss. Journal of Bone and Mineral Research. 2009;24:1150–1161. doi: 10.1359/jbmr.090210. [DOI] [PubMed] [Google Scholar]
- 40.Massa PE, Li X, Hanidu A, Siamas J, Pariali M, Pareja J, Savitt AG, Catron KM, Li J, Marcu KB. Gene expression profiling in conjunction with physiological rescues of IKKalpha null cells with wild type or mutant IKKalpha reveals distinct classes of IKKalpha/NF-kappaB-dependent genes. J. Biol. Chem. 2005;280:14057–14069. doi: 10.1074/jbc.M414401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penzo M, Massa PE, Olivotto E, Bianchi F, Borzi RM, Hanidu A, Li X, Li J, Marcu KB. Sustained NF-kappaB activation produces a short-term cell proliferation block in conjunction with repressing effectors of cell cycle progression controlled by E2F or FoxM1. Journal of Cellular Physiology. 2009;218:215–227. doi: 10.1002/jcp.21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zigmond SH, Hirsch JG. Leukocyte locomotion and chemotaxis. New methods for evaluation, and demonstration of a cell-derived chemotactic factor. J Exp Med. 1973;137:387–410. doi: 10.1084/jem.137.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zantl R, Rädler U, Horn E. Chemotaxis in mu channels. Imaging & Microscopy. 2006;8:30–32. [Google Scholar]
- 44.Amara A, Gall SL, Schwartz O, Salamero J, Montes M, Loetscher P, Baggiolini M, Virelizier J-L, Arenzana-Seisdedos F. HIV Coreceptor Downregulation as Antiviral Principle: SDF-1alpha -dependent Internalization of the Chemokine Receptor CXCR4 Contributes to Inhibition of HIV Replication. J. Exp. Med. 1997;186:139–146. doi: 10.1084/jem.186.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orsini MJ, Parent JL, Mundell SJ, Benovic JL, Marchese A. Trafficking of the HIV coreceptor CXCR4. Role of arrestins and identification of residues in the c-terminal tail that mediate receptor internalization. J Biol Chem. 1999;274:31076–31086. doi: 10.1074/jbc.274.43.31076. [DOI] [PubMed] [Google Scholar]
- 46.Dejardin E, Droin NM, Delhase M, Haas E, Ca Y, Makris C, Karin M, Ware CF, Green DR. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 47.Qing G, Xiao G. Essential Role of I{kappa}B Kinase {alpha} in the Constitutive Processing of NF-{kappa}B2 p100. J. Biol. Chem. 2005;280:9765–9768. doi: 10.1074/jbc.C400502200. [DOI] [PubMed] [Google Scholar]
- 48.Horwitz R, Webb D. Cell migration. Curr Biol. 2003;13:R756–759. doi: 10.1016/j.cub.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 49.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 50.Imitola J, Raddassi K, Park KI, Mueller F-J, Nieto M, Teng YD, Frenkel D, Li J, Sidman RL, Walsh CA, Snyder EY, Khoury SJ. Directed migration of neural stem cells to sites of CNS injury by the stromal cell-derived factor 1alpha/CXC chemokine receptor 4 pathway. Proceedings of the National Academy of Sciences. 2004;101:18117–18122. doi: 10.1073/pnas.0408258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palumbo R, Bianchi ME. High mobility group box 1 protein, a cue for stem cell recruitment. Biochemical Pharmacology. 2004;68:1165–1170. doi: 10.1016/j.bcp.2004.03.048. [DOI] [PubMed] [Google Scholar]
- 52.Guo Y, Hangoc G, Bian H, Pelus LM, Broxmeyer HE. SDF-1/CXCL12 enhances survival and chemotaxis of murine embronic stem cells and production of primitive and definitive hematopoietic progenitor cells. Stem Cells. 2005;23:1324–1332. doi: 10.1634/stemcells.2005-0085. [DOI] [PubMed] [Google Scholar]
- 53.Galvez BG, Sampaolesi M, Brunelli S, Covarello D, Gavina M, Rossi B, Costantin G, Torrente Y, Cossu G. Complete repair of dystrophic skeletal muscle by mesoangioblasts with enhanced migration ability. J. Cell Biol. 2006;174:231–243. doi: 10.1083/jcb.200512085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carbajal KS, Schaumburg C, Strieter R, Kane J, Lane TE. Migration of engrafted neural stem cells is mediated by CXCL12 signaling through CXCR4 in a viral model of multiple sclerosis. Proceedings of the National Academy of Sciences. 2010;107:11068–11073. doi: 10.1073/pnas.1006375107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersson U, Erlandsson-Harris H, Yang H, Tracey KJ. HMGB1 as a DNA-binding cytokine. J Leukoc Biol. 2002;72:1084–1091. [PubMed] [Google Scholar]
- 56.Dumitriu IE, Baruah P, Manfredi AA, Bianchi ME, Rovere-Querini P. HMGB1: guiding immunity from within. Trends in Immunology. 2005;26:381–387. doi: 10.1016/j.it.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 57.Palumbo R, Sampaolesi M, De Marchis F, Tonlorenzi R, Colombetti S, Mondino A, Cossu G, Bianchi ME. Extracellular HMGB1, a signal of tissue damage, induces mesoangioblast migration and proliferation. J Cell Biol. 2004;164:441–449. doi: 10.1083/jcb.200304135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schiraldi M, Raucci A, Munoz LM, Livoti E, Celona B, Venereau E, Apuzzo T, De Marchis F, Pedotti M, Bachi A, Thelen M, Varani L, Mellado M, Proudfoot A, Bianchi ME, Uguccioni M. HMGB1 promotes recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12 and signaling via CXCR4. The Journal of Experimental Medicine. 2012;209:551–563. doi: 10.1084/jem.20111739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ivanov S, Dragoi A-M, Wang X, Dallacosta C, Louten J, Musco G, Sitia G, Yap GS, Wan Y, Biron CA, Bianchi ME, Wang H, Chu W-M. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood. 2007;110:1970–1981. doi: 10.1182/blood-2006-09-044776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sha Y, Zmijewski J, Xu Z, Abraham E. HMGB1 Develops Enhanced Proinflammatory Activity by Binding to Cytokines. J Immunol. 2008;180:2531–2537. doi: 10.4049/jimmunol.180.4.2531. [DOI] [PubMed] [Google Scholar]
- 61.Urbonaviciute V, Furnrohr BG, Meister S, Munoz L, Heyder P, De Marchis F, Bianchi ME, Kirschning C, Wagner H, Manfredi AA, Kalden JR, Schett G, Rovere-Querini P, Herrmann M, Voll RE. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: implications for the pathogenesis of SLE. J. Exp. Med. 2008;205:3007–3018. doi: 10.1084/jem.20081165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hreggvidsdottir HS, Ostberg T, Wahamaa H, Schierbeck H, Aveberger A-C, Klevenvall L, Palmblad K, Ottosson L, Andersson U, Harris HE. The alarmin HMGB1 acts in synergy with endogenous and exogenous danger signals to promote inflammation. J Leukoc Biol. 2009;86:655–662. doi: 10.1189/jlb.0908548. [DOI] [PubMed] [Google Scholar]
- 63.Hreggvidsdóttir HS, Lundberg AM, Aveberger AC, Klevenvall L, Andersson U, Harris HE. HMGB1-partner molecule complexes enhance cytokine production by signaling through the partner molecule receptor. Mol Med. 2012;18:224–230. doi: 10.2119/molmed.2011.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campana L, Bosurgi L, Bianchi ME, Manfredi AA, Rovere-Querini P. Requirement of HMGB1 for stromal cell-derived factor-1/CXCL12-dependent migration of macrophages and dendritic cells. J Leukoc Biol. 2009;86:609–615. doi: 10.1189/jlb.0908576. [DOI] [PubMed] [Google Scholar]
- 65.Bonizzi G, Bebien M, Otero DC, Johnson-Vroom KE, Cao Y, Vu D, Jegga AG, Aronow BJ, Ghosh G, Rickert RC, Karin M. Activation of IKKalpha target genes depends on recognition of specific kappaB binding sites by RelB:p52 dimers. Embo J. 2004;23:4202–4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.