SUMMARY
L-Asparaginase (L-ASP) is an enzyme drug that has been an asset to leukemia treatment regimens for four decades. Variability in its clinical efficacy, however, has prompted the search for biomarkers capable of distinguishing responders from non-responders. In that regard, the NCI-60 cell line panel has served as a biomarker discovery platform and has led to the identification of a correlation between L-ASP efficacy and asparagine synthetase (ASNS) expression in cultured cells. The presence of that correlation in the ovarian subpanel of the NCI-60 has made a case for repositioning L-ASP to ovarian cancer. This review presents an overview of the biomarker development process, summarizes the efforts that have been invested thus far in developing ASNS as a biomarker for ovarian cancer treatment, highlights the role of RNAi and the limitations of the NCI-60 in that process, and addresses important considerations for next steps in the development of ASNS as a predictive biomarker.
Ovarian cancer patients diagnosed at stage I or stage II have a 5-year survival rate greater than 80%. Unfortunately, however, more than 80% of patients present with stage III or IV disease, when the malignancy has spread beyond the ovaries and the 5-year survival is less than 20%. Treatment includes surgery, radiation therapy and/or chemotherapy, which is typically administered for six cycles at 3 to 4 weeks per cycle. Carboplatin is the standard first-line drug,1 and hexamethylamine and topotecan are standard second-line agents. Taxanes (e.g., paclitaxel), mustards (e.g., melphalan) and anthracyclines (e.g., doxorubicin) are also common first-line or second-line options. Despite all of those compounds, however, intrinsic and acquired drug resistance contribute to high relapse and low survival rates. Nevertheless, the postgenomic guiding principal that “all cancers are different and should be treated as such” has motivated the identification of new ovarian cancer biomarkers.
“Biomarker” generally refers to a biological measurement of one of the following five types:2–6 i) biomarkers of disease progression (referred to as “screening biomarkers” in healthy subjects and “diagnostic biomarkers” in symptomatic subjects), ii) prognostic biomarkers of outcome, iii) predictive biomarkers of therapeutic effect, iv) surrogate endpoints of therapeutic effect and v) biomarkers of toxicity or adverse events (also referred to as “safety biomarkers”). Since it is important that therapy for ovarian cancer be started in the earliest possible stage, the first biomarker category is presumably the most important. In that category, the most sensitive and specific biomarker (more properly, “biosignature”) for detecting ovarian cancer currently appears to be a panel of six serum proteins (leptin, prolactin, osteopontin, insulin-like growth factor II, macrophage inhibitory factor and CA125), which has been reported to identify ovarian cancer as early as stage I with a striking 99.3% positive predictive value and 99.2% negative predictive value.7 The second biomarker category aims to predict outcome; examples include p53, p21, Ki-67, and HOXA11 DNA methylation.8–11 The third category is similar to the second but specifically aims to predict which patients will respond to a particular treatment, as is the case for the biomarker upon which this review is focused. The fourth biomarker category provides tools for monitoring treatment response and includes the “pharmacodynamic biomarker” subcategory. Two recent examples of surrogate markers include C-reactive protein, which has demonstrated potential as a surrogate for response to platinum therapy,12 and cell-free DNA, which has demonstrated potential as a surrogate for response to antivascular therapy.13,14 In the fifth biomarker category, lipocalin-2 (LCN2) upregulation has been identified as an early marker of cisplatin-induced nephrotoxicity.15
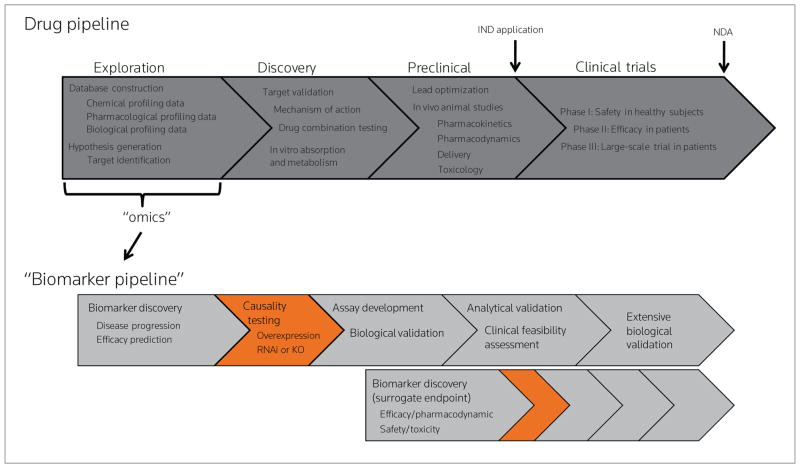
All five of the biomarker types have been invoked to curtail a substantial increase in attrition in phase II and III clinical trials—a point in the drug development process at which a significant monetary investment has been made.16 Moreover, the spike in attrition rate has prompted the call for a “biomarker pipeline” that parallels the traditional drug discovery and development pipeline (Fig. 1). In addition to curtailing the attrition rate, a biomarker pipeline would also facilitate drug repositioning (i.e., finding new uses for old drugs).17 The remainder of this article, in fact, discusses a biomarker-based rationale for repositioning the drug L-asparaginase.
Figure 1.
Drug discovery and development in the postgenomic era. Top: one rendition of the modern drug pipeline, indicating typical activities conducted during Exploration, Discovery, Preclinical Studies and Clinical Trials. Bottom: “Omic” sciences, which are typically invoked during Exploration, serve the added purpose of enabling a biomarker pipeline to parallel the drug development pipeline. The upper biomarker pipeline illustrates the typical process for biomarkers of disease progression or predictors of efficacy. The bottom biomarker pipeline illustrates that the Discovery stage for surrogate endpoint biomarkers begins much later, typically in parallel with preclinical studies.
DISCOVERY OF A PREDICTIVE L-ASPARAGINASE EFFICACY BIOMARKER
L-Asparaginase (L-ASP), the only U.S. Food and Drug Administration (FDA)-approved enzyme drug for cancer, has been used in combination with traditional chemotherapy to treat acute lymphoblastic leukemia since the early 1970s. It acts by depleting circulating asparagine and glutamine (Fig. 2),18–22 and the resulting nutrient deprivation leads to cell death via the amino acid response pathway.23–25 The endogenous enzyme asparagine synthetase (ASNS), which catalyzes asparagine synthesis from aspartate and glutamine,26,27 has long been believed to impart resistance to L-ASP. Hence, ASNS was a logical gene to investigate as an L-ASP efficacy biomarker. Toward that end, microarray-based molecular profiling of the NCI-60 cell line panel revealed a strong negative L-ASP/ASNS correlation in the leukemia subpanel of the NCI-60,28 indicating that low L-ASP efficacy (measured as reduced cellular metabolism using a tetra-zolium-formazan conversion assay) was associated with high expression of ASNS mRNA and vice-versa.
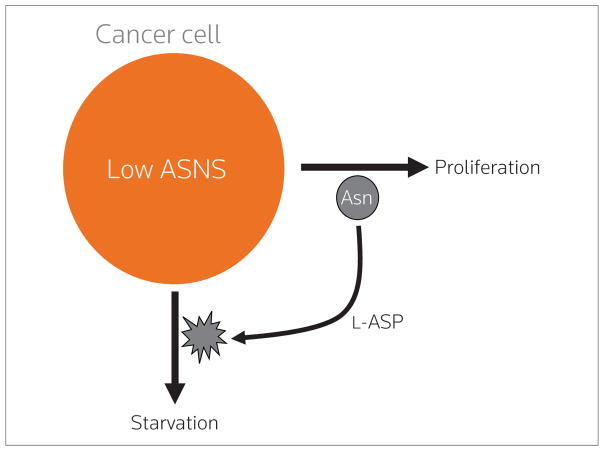
Figure 2.
Rationale for therapy with L-asparaginase (L-ASP). Cancer cells that express low asparagine synthetase (ASNS) cannot synthesize enough asparagine (Asn) to keep up with their metabolic demands. Those cells hence become dependent upon extracellular Asn, which can be depleted by the drug L-ASP to starve the cancer.
The NCI-60, originally assembled to screen for anticancer agents,29–31 has been profiled more extensively at the molecular level than any other set of cells in existence.32 The NCI-60 therefore made it possible to ask, “In addition to leukemia, do any additional cancer cell types exhibit the L-ASP/ASNS correlation?” Interestingly, the ovarian subpanel did,28 suggesting the possibility that L-ASP might be used to treat ovarian cancer and that its efficacy might be predicted by ASNS expression. The P value for the ovarian correlation, unfortunately, was not statistically significant after Bonferroni correction for multiple comparisons. Nevertheless, the correlation was later corroborated by additional mRNA expression microarray platforms33,34 and was also extended to the DNA level by comparative genomic hybridization.35 The correlation (uncorrected for multiple comparisons) was statistically significant with all of these platforms. Mechanistically, concordance between mRNA and DNA level correlations suggests that ASNS mRNA expression is strongly influenced by 7q copy number. Hence, a multi-pronged approach to biomarker discovery provided early rationale for repositioning L-ASP to possible application against a low-ASNS subset of ovarian cancer.
The observation of a statistically significant L-ASP/ASNS correlation warranted proceeding to the next phase of the biomarker development process—testing for causality (Fig. 1). Technically speaking, causality testing is not a formal requirement for the development of a biomarker, since a non-causally linked biomarker could in theory exhibit strong predictive power. Nevertheless, causality testing indicates a more direct association and, therefore, provides a stronger rationale for further development of a biomarker. Accordingly, the L-ASP/ ASNS correlation was next tested for causality using RNA interference (RNAi). Small-interfering RNA (siRNA)-mediated silencing of ASNS in three ovarian lines caused 3- to over 500-fold potentiation of L-ASP activity, indicating that the correlation was indeed causal.33 Furthermore, the L-ASP/ASNS causal link was also found to be independent of multidrug resistance as shown by experiments in a doxorubicin (Adriamycin)-selected, multidrug-resistant cell line, OVCAR-8/ADR.33 Hence, multidrug-resistant ovarian cancers with low ASNS expression might be sensitive to L-ASP. That robustness is a promising characteristic of the L-ASP/ASNS relationship considering the prominence of multidrug resistance in ovarian cancer and that clinical biomarker evaluation would probably have to be conducted, at least initially, in patients who have failed front-line therapy. These findings warranted proceeding to the next step of the biomarker development process—assay development.
ASNS ASSAY DEVELOPMENT
The assay development phase should ideally be conducted in conjunction with (continued) biological validation efforts. In that regard, since the NCI-60 panel contains only six independent ovarian cancer lines, a variety of ASNS assays were next tested in a larger set of 19 ovarian lines. In that set, the relationship between ASNS mRNA expression L-ASP efficacy did not appear as strong as it had for the NCI-60 ovarian lines. However, ASNS protein did show a persuasive association with efficacy. Using rabbit polyclonal anti-ASNS antibodies and an immunoassay (both developed for the purpose), we found that ASNS protein expression is a moderately strong predictor of L-ASP efficacy (Pearson r = −0.65, one-tailed P = 0.0014).36 In other words, the data suggest, but by no means prove, that ASNS protein is a better indicator of L-ASP efficacy in vitro than is ASNS mRNA.
PROJECTING TO THE CLINIC
Can we really expect L-ASP to benefit ovarian cancer on the basis of an in vitro correlation with ASNS expression? Single gene predictors of drug efficacy are indeed rare, and establishing the reliability of such relationships requires thorough validation.37 Several lines of evidence provide a rationale for continued development of ASNS as a predictive biomarker of L-ASP efficacy. First, the L-ASP/ASNS correlation of r = −0.65 in a diverse set of ovarian cancer lines is moderately strong.36 Second, the correlation is causal.33 Third, the causal link is unaffected by the development of multidrug resistance.33 Fourth, the L-ASP/ASNS relationship overrides numerous markers that predict poor chemotherapeutic outcome, including p53 mutation, HER2 overexpression, epidermal growth factor overexpression, CD10 negativity, platinum resistance and taxane resistance (as discussed in 33). Fifth, the correlation between L-ASP efficacy and ASNS mRNA expression in the NCI-60 is much stronger than the correlation between some FDA-approved targeted therapies and their FDA-approved clinical biomarkers (as discussed in 36). Taken together, those findings have prompted planning for a clinical trial at the National Cancer Institute to evaluate L-ASP efficacy in ovarian cancers using ASNS protein expression as a predictive biomarker.
CONCLUSIONS
Since drug resistance significantly reduces the number of available therapeutic options for ovarian cancer, adding a potential new drug to the arsenal would represent a significant advance, especially if that new drug did not exhibit cross-resistance with existing therapies. L-ASP is one such agent that could prove useful for the treatment of a low-ASNS subset of ovarian cancers.
Acknowledgments
We are grateful to our NIH collaborators Natasha Caplen, Elise Kohn, Michael Birrer, Bill Reinhold, Sudhir Varma, Jenny Llamas, Scott Martin, Paul Goldsmith, Michele Gunsior, Yves Pommier, Mark Raffeld, Laurent Ozbun, Amy Hutchinson and Stephen Chanock for their roles in making possible the work cited here and for valuable discussions. We also thank Daniel Von Hoff (TGen), Ivan Horak (Enzon Pharmaceuticals) and Aby Buchbinder (Enzon Pharmaceuticals) for valuable discussions. PLL is supported by a Pharmacology Research Associate Fellowship from NIGMS, NIH and by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The authors declare ownership interest in an NIH patent based on previously published work.
Contributor Information
Philip L. Lorenzi, Genomics & Bioinformatics Group, Laboratory of Molecular Pharmacology, Center for Cancer Research (CCR), at the National Cancer Institute (NCI) of the National Institutes of Health (NIH) in Bethesda, Maryland.
John N. Weinstein, Department of Bioinformatics and Computational Biology at M. D. Anderson Cancer Center in Houston, Texas
References
- 1.Paclitaxel plus carboplatin versus standard chemotherapy with either single-agent carbo-platin or cyclophosphamide doxorubicin and cisplatin in women with ovarian cancer: the ICON3 randomised trial. Lancet. 2002;360(9332):505–15. doi: 10.1016/S0140-6736(02)09738-6. [DOI] [PubMed] [Google Scholar]
- 2.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100(20):1432–8. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sargent DJ, Conley BA, Allegra C, Collette L. Clinical trial designs for predictive marker validation in cancer treatment trials. J Clin Oncol. 2005;23(9):2020–7. doi: 10.1200/JCO.2005.01.112. [DOI] [PubMed] [Google Scholar]
- 4.Ross JS, Symmans WF, Pusztai L, Hortobagyi GN. Pharmacogenomics and clinical biomarkers in drug discovery and development. Am J Clin Pathol. 2005;124(Suppl):S29–41. doi: 10.1309/XYQAFANAPYNC6X59. [DOI] [PubMed] [Google Scholar]
- 5.Wagner JA, Williams SA, Webster CJ. Biomarkers and surrogate end points for fit-for-purpose development and regulatory evaluation of new drugs. Clin Pharmacol Ther. 2007;81(1):104–7. doi: 10.1038/sj.clpt.6100017. [DOI] [PubMed] [Google Scholar]
- 6.Marrer E, Dieterle F. Biomarkers in oncology drug development: rescuers or troublemakers? Expert Opin Drug Metab Toxicol. 2008;4(11):1391–402. doi: 10.1517/17425255.4.11.1391. [DOI] [PubMed] [Google Scholar]
- 7.Visintin I, Feng Z, Longton G, et al. Diagnostic markers for early detection of ovarian cancer. Clin Cancer Res. 2008;14(4):1065–72. doi: 10.1158/1078-0432.CCR-07-1569. [DOI] [PubMed] [Google Scholar]
- 8.Moreno CS, Matyunina L, Dickerson EB, et al. Evidence that p53-mediated cell-cycle-arrest inhibits chemotherapeutic treatment of ovarian carcinomas. PLoS ONE. 2007;2:e441. doi: 10.1371/journal.pone.0000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singer G, Stohr R, Cope L, et al. Patterns of p53 mutations separate ovarian serous borderline tumors and low- and high-grade carcinomas and provide support for a new model of ovarian carcinogenesis: A mutational analysis with immunohistochemical correlation. Am J Surg Pathol. 2005;29(2):218–24. doi: 10.1097/01.pas.0000146025.91953.8d. [DOI] [PubMed] [Google Scholar]
- 10.Viale G, Maisonneuve P, Bonoldi E, et al. The combined evaluation of p53 accumulation and of Ki-67 (MIB1) labelling index provides independent information on overall survival of ovarian carcinoma patients. Ann Oncol. 1997;8(5):469–76. doi: 10.1023/a:1008253429700. [DOI] [PubMed] [Google Scholar]
- 11.Fiegl H, Windbichler G, Mueller-Holzner E, et al. HOXA11 DNA methylation--a novel prognostic biomarker in ovarian cancer. Int J Cancer. 2008;123(3):725–9. doi: 10.1002/ijc.23563. [DOI] [PubMed] [Google Scholar]
- 12.Hefler LA, Concin N, Hofstetter G, et al. Serum C-reactive protein as independent prognostic variable in patients with ovarian cancer. Clin Cancer Res. 2008;14(3):710–4. doi: 10.1158/1078-0432.CCR-07-1044. [DOI] [PubMed] [Google Scholar]
- 13.Kamat AA, Bischoff FZ, Dang D, et al. Circulating cell-free DNA: a novel biomarker for response to therapy in ovarian carcinoma. Cancer Biol Ther. 2006;5(10):1369–74. doi: 10.4161/cbt.5.10.3240. [DOI] [PubMed] [Google Scholar]
- 14.Kamat AA, Kim TJ, Landen CN, Jr, et al. Metronomic chemotherapy enhances the efficacy of antivascular therapy in ovarian cancer. Cancer Res. 2007;67(1):281–8. doi: 10.1158/0008-5472.CAN-06-3282. [DOI] [PubMed] [Google Scholar]
- 15.Mori K, Nakao K. Neutrophil gelatinase-associated lipocalin as the real-time indicator of active kidney damage. Kidney Int. 2007;71(10):967–70. doi: 10.1038/sj.ki.5002165. [DOI] [PubMed] [Google Scholar]
- 16.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3(8):711–5. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 17.Shah RR. Can pharmacogenetics help rescue drugs withdrawn from the market? Pharmacogenomics. 2006;7(6):889–908. doi: 10.2217/14622416.7.6.889. [DOI] [PubMed] [Google Scholar]
- 18.Fumarola C, Zerbini A, Guidotti GG. Glutamine deprivation-mediated cell shrinkage induces ligand-independent CD95 receptor signaling and apoptosis. Cell Death Differ. 2001;8(10):1004–13. doi: 10.1038/sj.cdd.4400902. [DOI] [PubMed] [Google Scholar]
- 19.Kafkewitz D, Bendich A. Enzyme-induced asparagine and glutamine depletion and immune system function. Am J Clin Nutr. 1983;37(6):1025–30. doi: 10.1093/ajcn/37.6.1025. [DOI] [PubMed] [Google Scholar]
- 20.Kitoh T, Asai S, Akiyama Y, Kubota M, Mikawa H. The inhibition of lymphocyte blastogenesis by asparaginase: critical role of glutamine in both T and B lymphocyte transformation. Acta Paediatr Jpn. 1992;34(6):579–83. doi: 10.1111/j.1442-200x.1992.tb01012.x. [DOI] [PubMed] [Google Scholar]
- 21.Reinert RB, Oberle LM, Wek SA, et al. Role of glutamine depletion in directing tissue-specific nutrient stress responses to L-asparaginase. J Biol Chem. 2006;281(42):31222–33. doi: 10.1074/jbc.M604511200. [DOI] [PubMed] [Google Scholar]
- 22.Villa P, Corada M, Bartosek I. L-asparaginase effects on inhibition of protein synthesis and lowering of the glutamine content in cultured rat hepatocytes. Toxicol Lett. 1986;32(3):235–41. doi: 10.1016/0378-4274(86)90113-x. [DOI] [PubMed] [Google Scholar]
- 23.Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2alpha. Genetics. 2000;154(2):787–801. doi: 10.1093/genetics/154.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiaville MM, Pan YX, Gjymishka A, Zhong C, Kaufman RJ, Kilberg MS. MEK signaling is required for phosphorylation of eIF2alpha following amino acid limitation of HepG2 human hepatoma cells. J Biol Chem. 2008;283(16):10848–57. doi: 10.1074/jbc.M708320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang P, McGrath BC, Reinert J, et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22(19):6681–8. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horowitz B, Meister A. Glutamine-dependent asparagine synthetase from leukemia cells. Chloride dependence, mechanism of action, and inhibition. J Biol Chem. 1972;247(20):6708–19. [PubMed] [Google Scholar]
- 27.Levintow L. Evidence that glutamine is a precursor of asparagine in a human cell in tissue culture. Science. 1957;126(3274):611–2. doi: 10.1126/science.126.3274.611. [DOI] [PubMed] [Google Scholar]
- 28.Scherf U, Ross DT, Waltham M, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24(3):236–44. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 29.Boyd MR, Paull KD. Some practical considerations and applications of the National Cancer Institute in vitro anticancer drug discovery screen. Drug Dev Res. 1995;34:91–109. [Google Scholar]
- 30.Holbeck SL. Update on NCI in vitro drug screen utilities. Eur J Cancer. 2004;40(6):785–93. doi: 10.1016/j.ejca.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 31.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6(10):813–23. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 32.Weinstein JN. Spotlight on molecular profiling: “Integromic” analysis of the NCI-60 cancer cell lines. Mol Cancer Ther. 2006;5(11):2601–5. doi: 10.1158/1535-7163.MCT-06-0640. [DOI] [PubMed] [Google Scholar]
- 33.Lorenzi PL, Reinhold WC, Rudelius M, et al. Asparagine synthetase as a causal, predictive biomarker for L-asparaginase activity in ovarian cancer cells. Mol Cancer Ther. 2006;5(11):2613–23. doi: 10.1158/1535-7163.MCT-06-0447. [DOI] [PubMed] [Google Scholar]
- 34.Shankavaram UT, Reinhold WC, Nishizuka S, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 2007;6(3):820–32. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 35.Bussey KJ, Chin K, Lababidi S, et al. Integrating data on DNA copy number with gene expression levels and drug sensitivities in the NCI-60 cell line panel. Mol Cancer Ther. 2006;5(4):853–67. doi: 10.1158/1535-7163.MCT-05-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenzi PL, Llamas J, Gunsior M, et al. Asparagine synthetase is a predictive biomarker of L-asparaginase activity in ovarian cancer cell lines. Mol Cancer Ther. 2008;7(10):3123–8. doi: 10.1158/1535-7163.MCT-08-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Covell DG. Connecting chemosensitivity, gene expression and disease. Trends Pharmacol Sci. 2008;29(1):1–5. doi: 10.1016/j.tips.2007.10.015. [DOI] [PubMed] [Google Scholar]