Abstract
Carboxylesterase-1 (CES1), the most versatile human carboxylesterase, plays critical roles in drug metabolism and lipid mobilization. This enzyme is highly induced by antioxidants and sensitizers in various cell lines. These compounds are known to activate nuclear factor-E2 related factor-2 (Nrf2) by reacting to kelch-like ECH-associated protein-1 (Keap1). The aims of this study were to determine whether antioxidant sulforaphane (SFN) and sensitizer trinitrobenzene sulfonate (TNBS) target Keap1 similarly and whether they use the same element for CES1 induction. Cells over-expressing Keap1 were treated with TNBS or SFN and the formation of disulfide bonds among Keap1 molecules were determined. SFN promoted intramolecular disulfide formation whereas TNBS promoted intermolecular disulfide formation of Keap1. Two elements, sensitizing/antioxidant response element (S/ARE) and ARE4, were identified to support Nrf2 in the regulated expression of CES1A1. Both elements were bound by Nrf2, however, the S/ARE element supported, whereas the ARE4 element repressed Nrf2 transactivation. The repression required higher amounts of Nrf2, suggesting that the transactivation through the S/ARE element dominates the trans-repression through the ARE4 element under normal antioxidative condition. These findings conclude that compounds, although triggering the Keap1-Nrf2 pathway, may differ in the mode of reacting with Keap1. These findings also conclude that both positive and negative Nrf2 elements exist even within the same gene, and such opposing mechanisms provide fine-tuning in transcriptional regulation by the Keap1-Nrf2 pathway. High levels of CES1 are linked to lipid retention. Excessive induction of CES1 by antioxidants and sensitizers likely provides a mechanism for potential detrimental effect on human health.
Keywords: Carboxylesterase, Keap1-Nrf2 pathway, negative and positive AREs, Keap1 reactivity, antioxidants, and skin sensitizers
1. Introduction
Carboxylesterases (CES, E.C.3.1.1.1) constitute a large class of enzymes that play important roles in drug metabolism and lipid mobilization [1-4]. In the human genome, seven carboxylesterase genes are identified [1]. However, only three are catalytically characterized including CES1, CES2 and CES3 [5]. CES1 is encoded by two distinct genes: CES1A1 and CES1A2 [1], but CES1A1 is normally expressed to a greater extent [6]. CES1 has a broad tissue distribution with the highest level in the liver microsomes [7]. CES1 is the most versatile human carboxylesterase and catalyzes hydrolytic, synthetic and transactivation reactions. While all carboxylesterases play roles in drug metabolism, emerging evidence links the sustained high-level expression of CES1 to the increased risk of developing cardiovascular diseases, obesity and insulin resistance [8, 9].
Antioxidants and skin sensitizers are two types of compounds recently shown to induce CES1 [10-13]. Some sensitizers induced CES1 by as much as 20-fold [11]. While the precise mechanism on the sensitizer-induction of CES1 remains to be established, the induction of this carboxylesterase by antioxidants, on the other hand, is mediated by Nrf2 (nuclear factor-E2 related factor-2) [14]. This transcription factor recognizes antioxidant response element (ARE) and confers transactivation [15]. RNA interference against Nrf2 abolished CES1 induction by antioxidants [14]. However, a native promoter reporter containing putative AREs was repressed by Nrf2 [14]. Majority of skin sensitizers, on the other hand, are sulfhydryl reactive agents and shown to react with kelch-like ECH-associated protein-1 (Keap1) [10, 13], an inhibitor of Nrf2. Interaction with Keap1 by sensitizers leads to Nrf2 activation. The magnitude of the activation was correlated with their sensitizing potency [10].
The present study was performed to test the hypothesis that both antioxidants and sensitizers react to Keap1 similarly and causes transactivation of CES1A1 via a novel Nrf2 element. To test this hypothesis, Keap1-transfected cells were treated with sensitizer trinitrobenzene sulfonate (TNBS) or antioxidant DL-sulforaphane (SFN) and various molecular species of Keap1 were determined. In contrary to the hypothesis, SFN promoted intramolecular oxidation whereas TNBS promoted intermolecular oxidation of Keap1. To locate the element supporting the transactivation, the regulatory sequence of the CES1A1 gene was dissected and tested for the responsiveness to TNBS, SFN or Nrf2. Two elements were identified and designated as S/ARE (sensitizing/antioxidant response element) and ARE4, respectively. Interestingly, the S/ARE element supported Nrf2 transactivation whereas the ARE4 repressed it although the S/ARE element was a more sensitive target of Nrf2.
2. Materials and Methods
2.1. Chemicals and supplies
Hanks balanced salt solution, TNBS and William's medium E (WME) were purchased from Sigma (St. Louis, MO). SFN was purchased from Alexis (San Diego, CA). Dulbecco's Modified Eagles Medium (DMEM), high fidelity Platinum Taq DNA polymerase, insulin-transferrin-selenium (ITS) G supplement were purchased from Invitrogen (Carlsbad, CA). Dual-Luciferase Reporter Assay System was from Promega (Madison, WI). Fetal bovine serum (FBS) was from HyClone laboratories (Logan, UT). The antibody against glyceradehyde-3-phosphate dehydrogenase (GAPDH) was from Abcam (Cambridge, UK), and the antibody against Nrf2 (C-20) was from Santa Cruz Biotechnology (Santa Cruz, CA). The goat anti-rabbit IgG conjugated with horseradish peroxidase was from Pierce (Rockford, IL). Plated human primary hepatocytes were obtained from the Liver Tissues Procurement and Distribution System (University of Minnesota) or CellzDirect (Pittsboro, NC). Human dermal fibroblasts (cryopreserved) were purchased from Cascade Biologics (Portland, OR). Unless otherwise specified, all other reagents were purchased from Fisher Scientific (Fair Lawn, NJ).
2.2. Reporter constructs and cotransfection assays
CES1A1 promoter reporters were prepared to contain various lengths of CES1A1 genomic sequence. All promoter reporters were subcloned from the CES1A1-6560-Luc reporter. This reporter was prepared by inserting the genomic fragment from -6560 to -21 (relatively to the translation initiation codon) into the pGL3 basic vector through Mlu I and Xho I sites. All cloning and subcloning were performed by PCR with high fidelity Platinum Taq DNA polymerase. To prepare the CES1A1-3582m-Luc reporter, site-directed mutagenesis was performed as described previously [16]. Complementary oligonucleotides (5′-CTCACCCATCACAATGTACTGAGGAATCATGAAGCAGAAA-3′) were synthesized to introduce substitutions (underlined). The primers were annealed to the CES1A1-3582-Luc reporter and subjected to a thermocycler for a total of 15 cycles. The resultant PCR-amplified constructs were then digested with Dpn I to remove the non-mutated parent construct. The mutated PCR-amplified constructs were used to transform XL1-Blue bacteria. To prepare element reporters, oligonucleotides (Table I) were synthesized, annealed and ligated into the pGL3 promoter vector through Nhe I and Xho I. All reporter constructs were subjected to sequence analysis. To determine the reporter activities, cotransfection in Huh7 cells (human hepatic cellular carcinoma line) was performed. Transfection mixtures contained 100 ng of a reporter plasmid and 0.2 ng of CMV-Renilla luciferase plasmid. In some cases, Nrf1 and Nrf2 constructs were included. Nrf1 construct was a gift of Dr. Jefferson Y. Chan of University of California Irvine whereas Nrf2 construct was purchased from OriGene (Rockville, MD). The corresponding vector was used to equalize the total amount of plasmid DNA. Typically, cells were transfected for 12 h and the medium was replaced with fresh medium supplemented with 1% FBS. The treatment lasted for 24 h and the cells were washed once with phosphate buffered saline and collected by scraping. The reporter enzyme activities were assayed with a Dual-Luciferase Reporter Assay System as described previously [16].
Table I. Sequences of Oligonucleotides.
| Oligonucleotide | Sequence |
|---|---|
| Native promoter reporters | |
| CES1A1-6560-MluI | 5′-CTGCAACCTTCCCATCTCAGCTGTGACCCA-3′ |
| CES1A1-6448-MluI | 5′-AGCTAATGAATAGGATCTGGGTTTATAATC-3′ |
| CES1A1-5017-MluI | 5′-CAATACCCTAATTTCGATCTCTGAATGACC-3′ |
| CES1A1-3582-MluI | 5′-TATTGCTTACAGCTGAAGTG-3′ |
| CES1A1-3432-MluI | 5′-CAGGCAAAACCTAGGAGTGG-3′ |
| CES1A1-2293-MluI | 5′-TGATTAGAATATCTTCCTGATGTATAAAAG-3′ |
| CES1A1-1426-MluI | 5′-GATGTTTTCCAGCTTCATCCACGTTGTAGC-3′ |
| CES1A1-3552-MluI | 5′-GTTATATGTATTAAGCAAAA-3′ |
| CES1A1-3532-MluI | 5′-TTAGATCTATTGTAAGGCT-3′ |
| CES1A1-3512-MluI | 5′-TGGTAGGCTCCAGCCTCACC-3′ |
| CES1A1-3492-MluI | 5′-CATCACAATCTGCAGAGTCA-3′ |
| CES1A1-3482-MluI | 5′-TGCAGAGTCATCATGAAGCA-3′ |
| CES1A1-10484XhoI | 5′-ATACACTCGAGTCGGGGCCTGCGAGGTCTCTGTGCA-GTTCA-3′ |
| Element reporters | |
| CES1A1-S/ARE | 5′-CACAATCTGCAGAGTCATCATGAAG-3′ |
| CES1A1-ARE4 | 5′-TTAAGATCGTGAGACAGCATTAATC-3′ |
| CES1A2-S/ARE | 5′-CACAATCTGCAGAATCATCATGAAC-3′ |
| AKR1C2-ARE | 5′-TTGATGCAGTCAGGGTGACTCAGCAGCT-3 |
| GCLM-ARE | 5′-GAAGACAATGACTAAGCAGAAATC-3′ |
| GCLC-ARE | 5′-CCTCCCCGTGACTCAGCGCTTTGT-3′ |
| NQO1-ARE | 5′-AGTCACAGTGACTCAGCAGAATCT-3′ |
| EMSA | |
| CES1A1-S/ARE | 5′-CACAATCTGCAGAGTCATCATGAAG-3′ |
| CES1A1-S/ARE(mutant) | 5′-CACAATGTACTGAGGAATCATGAAG-3′ |
| CES1A1-ARE4 | 5′-TTAAGATCGTGAGACAGCATTAATC-3′ |
| ChIP | |
| P1 | 5′- TATTGCTAGCCTGAAGTGTTGCAGGGGAGTT-3′ |
| P2 | 5′- ACACCTCGAGCTGGCTCTTGGCCTATGAAGA-3′ |
| P3 | 5′- ATAAGCTAGCTGAGTTGAGCCTATGTATTAG-3′ |
| P4 | 5′- CATTCTCGAGTCCTGGCTGTAATCTTGTCAG-3′ |
2.3. Electrophoretic mobility shift assay (EMSA)
The EMSA experiment was performed as described previously [17]. Nuclear extracts of Huh7 cells treated with SFN (10 μM) for 24 h were prepared with the nuclear and cytoplasmic extraction kit (Pierce, Rockford, IL). The sense and antisense oligonucleotides (Table I) were annealed by heating at 94°C for 5 min followed by gradually cooling to room temperature. The sense strand was synthesized as labeled or non-labeled form (for competition). Nuclear protein (5 μg) was incubated with a double-stranded biotinylated probe (0.1 pmol) at room temperature for 20 min. In competition assays, nuclear extracts were first incubated with an unlabeled probe at a 25× or 100× excess for 5 min before addition of the labeled probe. For antibody-disruption assay, the nuclear extracts were first incubated with an antibody against Nrf2 (C-20) on ice for 20 min and then with the labeled probe. The protein-DNA complexes were resolved by nondenaturing polyacrylamide gel electrophoresis (5%) and transferred onto a Biodyne® nylon membrane. The biotinylated probe was detected with streptavidin-conjugated horseradish peroxidase and chemiluminescent substrate (PIERCE, Rockford, IL). The chemiluminescent signal was captured by KODAK Image Station 2000, and the relative intensities were quantified by KODAK 1D Image Analysis Software (KODAK Molecular Imaging Software, Version 4.0, Rochester, NY).
2.4. Chromatin immunoprecipitation (ChIP)
ChIP experiment was performed, essentially described previously [17]. Huh7 Cells were treated with SFN (10 μM) for 24 h, washed and underwent cross-linking for 15 min by 1.0% formaldehyde at room temperature, and the cross-linking was terminated with glycine (final concentration of 125 mM). The soluble chromatins were prepared as described previously [17]. For ChIP experiment, chromatins were pre-cleared for 2 h at 4°C with protein G beads pre-treated with herring sperm DNA (0.2 mg/ml) and BSA (0.5 mg/ml). A fraction of the pre-cleared chromatins was stored at −80°C for later use as an input. An antibody against Nrf2 was added into the pre-cleared chromatins, and an overnight incubation at 4°C was performed. As a negative control, incubation was performed with pre-immune IgG. The antibody-bound chromatins and DNA input were analyzed by PCR for the presence of the genomic fragments containing the Nrf2-bound element with primers shown in Table I. The PCR was performed with Platinum Taq DNA polymerase for a total of 32 cycles at 94°C for 30 s, 58°C for 30 s and 68°C for 60 s. A 3-min initial denaturation was performed.
2.5. Non-reducing polyacrylamide gel electrophoresis coupled Western blotting
Non-reducing polyacrylamide gel electrophoresis was performed, essentially as described previously [18]. Briefly, 293T cells (human embryonic kidney expressing the simian virus 40 large T antigen) at 85% confluence in 24-well plates were transfected with Flag-tagged mouse or human keap1 (250 ng/well). The Flag tagged Keap1 constructs were kindly provided by Dr. Anil K. Jaiswail of the University of Maryland [12]. After a 24 h-incubation, the transfected cells were washed and then treated for 1 h with SFN or TNBS at various concentrations. The treated cells were extensively with PBS containing 40 mM N-ethylmaleimide (NEM) and the lysed in NEM-containing RIPA buffer. Lysates (4 μg) were mixed with an equal volume of denaturing sample buffer without reducing agent (62.5 mM Tris–HCl, pH 6.8, containing 10.8% glycerol, 0.001% bromophenol blue, and 1% SDS), and heated at 95 °C for 5 min. Samples were subjected to SDS–PAGE under non-reducing conditions [18] and subsequently transferred electrophoretically to an Immobilon-P membrane (Millipore, Bedford, MA). After non-specific binding sites were blocked with 5% non-fat milk, the blots were incubated with an anti-Flag antibody as described previously [16]. This experiment was repeated four times and consistent results were obtained.
2.6. Other analyses
Protein concentrations were determined with BCA assay (Pierce) based on albumin standard. Western analysis and the preparation of anti-CES1 antibody were described elsewhere [18-20]. RT-qPCR with Taqman probes was performed as described previously [21]. The Taqman probe identification numbers were: Hs00275607_m1 for CES1, Hs00168547_m1 for NAD(P)H:quinine oxidoreductase 1 (NQO1), 4352934E for GAPDH and Hs00172187_m1 for RNA polymerase II. Data are presented as mean ± SD of at least three separate experiments, except where results of blots are shown in which case a representative experiment is depicted in the figures. All data were analyzed for statistical significance with PASW Statistics 18. Significant differences were made according to One-way ANOVA followed by a DUNCAN's multiple comparison test (p < 0.05). Bars assigned different letters indicate statistical significance among data-points.
3. Results
3.1. CES1 induction by SFN and TNBS in primary cultures and cell lines
Humans are exposed to antioxidants and sensitizers primarily through the gastrointestinal track and skin. We first confirmed whether induction of CES1 by SFN and TNBS occurs comparably in primary cultures and cell lines (Huh7 and human fibrosarcoma cell line HT1080) from these organs. Cells were treated with SFN or TNBS and CES1 expression was determined initially by RT-qPCR with a Taqman probe. This probe recognizes both CES1A1 and CES1A2 mRNA. In addition, the mRNA level of NAD(P)H:quinone oxidoreductase (NQO1), an Nrf2 target gene [22], was determined as well. As summarized in Fig. 1, both SFN and TNBS significantly increased CES1 and NQO1 mRNA. The highest induction of CES1 mRNA was 3.9-fold and occurred in Huh7 cells treated with SFN (Fig. 1D) and the least (1.6 fold) in primary hepatocytes treated with TNBS (Fig. 1C). In contrast, the highest induction of NQO1 mRNA was 5.7-fold and occurred in primary hepatocytes (Fig. 1C), the least (1.9-fold) in primary fibroblasts treated with SFN (Fig. 1A). Except for HT1080 cells, SFN caused greater induction of CES1 than TNBS, and NQO1 mRNA was induced to a greater or comparable extent by TNBS (Fig. 1). In HT1080 cells, TNBS caused slightly higher induction of CES1 mRNA than SFN but the opposite was true on NQO1 induction (Fig. 1B). Both SFN and TNBS markedly increased CES1 protein (Fig. 1E).
Fig. 1. Induction of CES1 by SFN and TNBS.
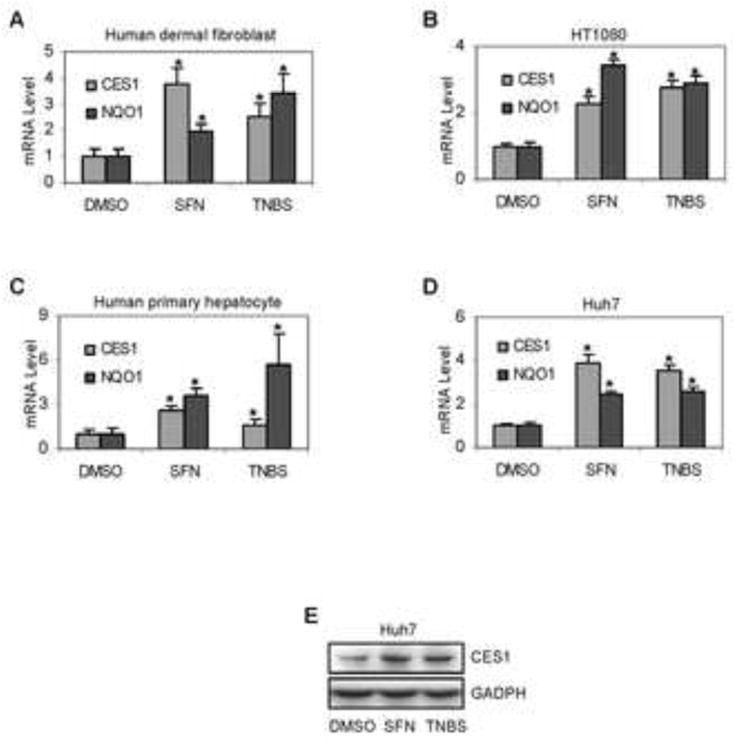
Cells were cultured in 12-well plates and were treated with SFN (10 μM), TNBS (10 μg/ml) or DMSO for 24 h. Total RNA was isolated and subjected to qRT-PCR analysis for the level of CES1 mRNA (A-D) by Taqman probes as described in the section of Materials and Methods. The signals from each target were normalized based on the signal from GAPDH and polymerase II and expressed as mean ± SD. Four individual donors were tested on human dermal fibroblasts, five on human hepatocytes and three separate experiments were performed on cell lines. The asterisk signs indicate statistical significance (P < 0.05) from DMSO-treated cells. To determine the corresponding induction at the protein level, lysates (20 μg) from control and treated Huh7 cells were subjected to SDS-PAGE and transferred electrophoretically to Trans-Blot nitrocellulose membranes. The immunoblots were blocked in 5% non-fat dry milk, incubated with the antibody against CES1 or GAPDH. The blots were detected with chemiluminescent substrate (E).
3.2. Stimulation of the CES1A1 promoter by SFN, TNBS and Nrf2
Nrf2 was implicated in the induction of CES1A1 by antioxidants including SFN [14], and many sensitizers are potent activators of Nrf2 [13]. We next tested whether TNBS and SFN induce CES1A1 via the same Nrf2 element. Reporters were prepared to contain the CES1A1 promoter and upstream sequence at varying length. As shown in Fig. 2A, reporters containing upstream sequence of 3582 bp (i.e., -3582) or further responded to both SFN and TNBS. The CES1A1-3582-Luc reporter was activated the most (Fig. 2A). In contrast, the other reporters, containing shorter sequence than the CES1A1-3582-Luc reporter, were suppressed by both chemicals, and the suppression was greater on CES1A1-3432-Luc and CES1A1-2293-Luc than CES1A1-1426-Luc (Fig. 2A).
Fig. 2. Identification of sensitizing/antioxidant response element.
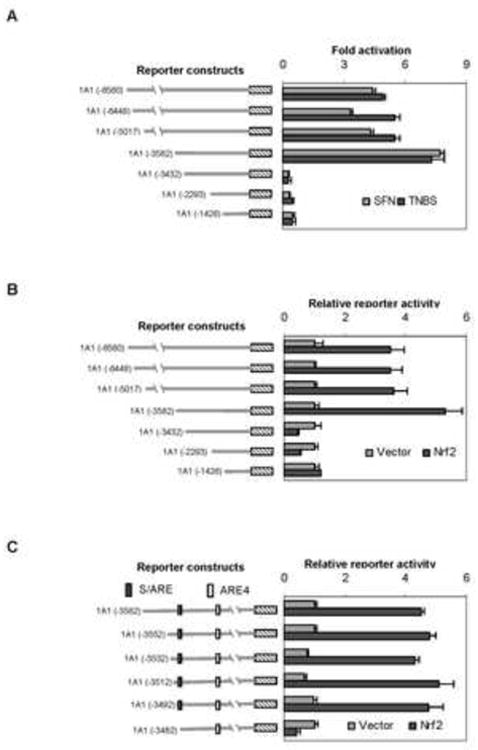
(A) Activation of CES1A1 reporters by SFN or TNBS Huh7 cells in 48 well-plates were transiently transfected by GenJet version II. For the SFN and TNBS treatment experiment, the transfection mixture contained 50 ng of a reporter and 0.2 ng of the CMV-Renilla luciferase plasmid. After incubation at 37°C for 12 h, the transfected cells were treated with SFN (10 μM), TNBS (10 μg/ml) or the same volume of DMSO for 24 h. Luciferase activities were determined with a Dual-Luciferase Reporter Assay System and the reporter activity was expressed as fold of activation. For the Nrf2 experiment, the transfection mixture contained 50 ng of a reporter and 0.2 ng of the CMV-Renilla luciferase plasmid, along with 10 ng Nrf2 expression construct or the corresponding vector. (B) Activation of CES1A1 reporters by Nrf2 Transfection was performed as described above but the the transfection mixture contained 10 ng Nrf2 expression construct or the corresponding vector. (C) Identification of CES1A1 Nrf2 response element To further narrow down the sequence that supports the responsiveness to Nrf2, the CES1A1-3582-Luc was further shortened from the 5′ end by 20 or 10 bases, and cotransfection was then performed as described above. Data were collected from three independent experiments.
Next we tested whether these reporters show a similar pattern of response to Nrf2. Huh7 cells were transfected with a reporter, along with Nrf2 or the vector. Consistent with the results on the TNBS and SFN treatment, the sequence from -3582 to -3432 was identified to support Nrf2-transactivation (Fig. 2B). Likewise, the CES1A1-3432-Luc and CES1A1-2293-Luc reporters were repressed by Nrf2 (Fig. 2B). To further narrow down the sequence supporting the action of Nrf2, deletions of the CES1A1-3582-Luc reporter were made from the 5′ end and the resultant reporters were tested for the lost responsiveness to Nrf2. As shown in Fig. 2C, the reporter 1A1-3492-Luc but not 1A1-3482-Luc was transactivated by Nrf2. Actually, the 1A1-3482-Luc reporter was repressed by Nrf2. These findings suggest that the 10-base sequence from -3492 to -3482 was critical for Nrf2-transactivation.
3.3. Characterization of the S/ARE element
The study with deletion mutants suggested that this 10-base sequence contains or is part of the Nrf2/TNBS/SFN response element: designated the S/ARE element. We next performed a set of experiments to characterize this element. We first tested whether Nrf2 binds S/ARE element and whether this binding can be competed by ARE4 (-2031), an element previously identified to support Nrf2-transactivation [14]. As shown in Fig. 3A, incubation of a biotin-labeled S/ARE probe with nuclear extracts from SFN-treated cells produced a shifted band (lane 2). This band was eliminated by an Nrf2 antibody (lane 3) and competed by non-labeled S/ARE (lanes 4 and 5), but not its mutant (lanes 6 and 7). In addition, non-labeled ARE4 element effectively competed the binding as well (lanes 8 and 9). Nevertheless, this experiment demonstrated that both the S/ARE and ARE4 were bound by Nrf2. We next tested whether the S/ARE-containing sequence is intracellularly occupied by Nrf2. ChIP analysis was performed with an Nrf2 antibody and the precipitated DNA was detected for the enriched S/ARE- or ARE4-fragment. As shown in Fig. 3B, comparable amplifications were detected with the input on both S/ARE-and ARE4-fragments, however, the ChIPed DNA produced robust amplification of the S/ARE-but not ARE4-fragment (Fig. 3B). As expected, no amplification was detected with sample precipitated with pre-immune IgG.
Fig. 3. Analyses of the S/ARE element by EMSA and ChIP.
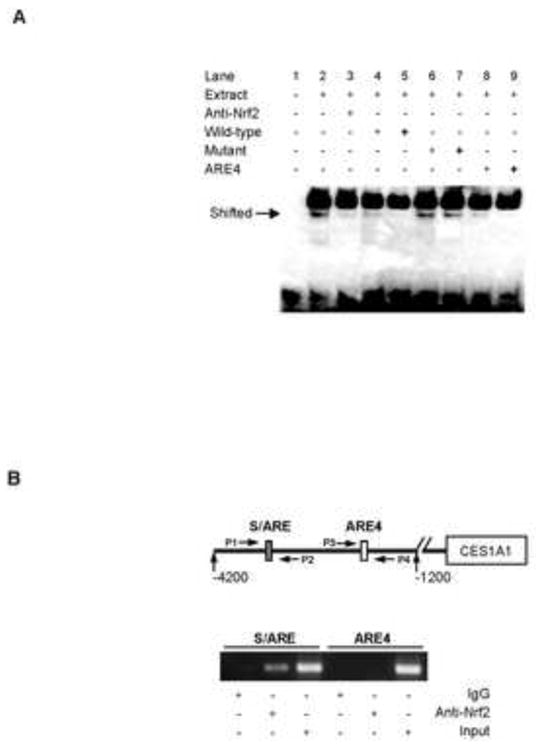
(A) EMSA analysis Nuclear extracts (5 μg) from Huh7 cells treated with SFN (10 μM) were incubated with a biotinylated probe containing the S/ARE element (0.1 pmol) for 20 min. In the competition assay, nuclear extracts were pre-incubated with the unlabeled S/ARE element, S/ARE mutant or ARE4 at 25× or 100× excess for 5 min, and then incubated with the biotinylated probe. In disruption assay, nuclear extracts were incubated first with an antibody against Nrf2 on ice for 20 min and then with the biotinylated probe. The protein-DNA complexes were electrophoretically resolved and transferred to a Biodyne® nylon membrane. The biotinylated probe was located with streptavidin-conjugated horseradish peroxidase and chemiluminescent substrate. (B) ChIP analysis Huh7 Cells were treated with SFN (10 μM) for 24 h, washed and underwent cross-linking for 15 min by 1% formaldehyde, and the cross-linking was terminated with 125 mM glycine. The soluble chromatins were prepared, pre-cleared with protein G beads and incubated with anti-Nrf2 antibody or pre-immune IgG. The antibody-bound chromatins and DNA input (1/20 of the antibody-bound chromatins) were analyzed by PCR for the presence of the genomic fragments containing the Nrf2-bound element with the primers indicated in the diagram. The primer sequences are shown in Table I.
3.4. Activation of the S/ARE element reporter by SFN, TNBS and Nrf2
The EMSA and ChIP experiments demonstrated that the S/ARE element serves as a major site for Nrf2 to interact with the CES1A1 gene. We next tested whether the interaction confers biological activities. Three reporters were tested including the S/ARE, CES1A1-3582-Luc and CES1A1-3582m-Luc reporters (Fig. 4A). The CES1A1-3582m-Luc, a mutant of the CES1A1-3582-Luc, had the S/ARE element replaced with its mutant sequence (Table I). Cotransfection was performed to test the responsiveness of these reporters to SFN, TNBS and Nrf2. As shown in Fig. 4B, SFN at 0.5 μM significantly activated the S/ARE reporter and the CES1A1-3582-Luc (bars labeled with different letters). Higher concentrations caused higher activation of both reporters except 10 μM on the S/ARE reporter (Fig. 4B). In contrast, none of the concentrations activated the CES1A1-3582m-Luc reporter. Similar responding patterns to TNBS were detected (Fig. 4C). Likewise, Nrf2 increased the activities of S/ARE and CES1A1-3582-Luc but not CES1A1-3582m-Luc reporter (Fig. 4D). Nonetheless, it was surprising that higher amounts of Nrf2 (10 and 20 ng) significantly repressed the mutant reporter CES1A1-3582m-Luc (Fig. 4D).
Fig. 4. Functional characterization of the S/ARE element.
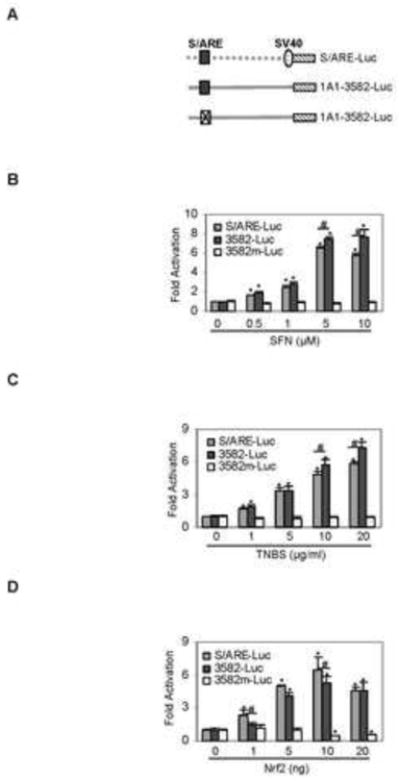
(A) Activation of S/ARE containing or disrupting reporter by SFN Huh7 cells were transiently transfected by a reporter (50 ng) of the S/ARE-Luc, CEA1A1-3582-Luc or its mutant CES1A1-3582m-Luc along with 0.2 ng of the CMV-Renilla luciferase plasmid. After incubation at 37°C for 12 h, the transfected cells were treated with SFN (0-10 μM) or the same volume of DMSO for 24 h. Luciferase activities were determined and the reporter activity was expressed as fold of activation. (B) Activation of S/ARE containing or disrupting reporter by TNBS Huh7 cells were transiently transfected as described above but treated with TNBS (0-10 μg/ml). Likewise, the reporter activities were determined. (B) Activation of S/ARE containing or disrupting reporter by Nrf2 Huh7 cells were transiently transfected with 50 ng reporter and 0.2 ng of the CMV-Renilla luciferase plasmid along with 0-20 ng Nrf2 expression construct. The corresponding vector was used to equalize the amount of total plasmid. The luciferase activities were determined 24 after the transfection. Asterisk and pound signs indicate statistical significance from the controls or between two data points linked by a line (P < 0.05).
3. 5. Activation comparison of S/ARE element with other Nrf2 elements
The EMSA, ChIP and reporter experiments established that the S/ARE element supported robust responsiveness to SFN, TNBS and Nrf2. Next, we compared the responding potential of this element with well-characterized Nrf2 elements. Reporters harboring the CES1A1-ARE4 element or the corresponding CES1A2-S/ARE were also included. In addition to Nrf2, these reporters were tested for their responsiveness to Nrf1, an Nrf2 functionally related protein [23]. For direct comparison, reporters were prepared to contain a single copy of an ARE element. As shown in Fig. 5, all reporters were transactivated by Nrf1 and Nrf2 except Nrf1 on the NQO1 reporter (Fig. 5A) and Nrf2 on the 1A1-ARE4 reporter (Fig. 5A). In both cases, their activity was actually decreased, and the decrease was statistically significant in the 1A1-ARE4 reporter (Fig. 5A). With a single exception (i.e., 1A1-ARE4), Nrf2 caused greater activation than Nrf1 on all reporters tested. The highest transactivation was detected with the CES1A1-S/ARE reporter (5.8 fold) followed by the CES1A2-S/ARE reporter (5.2 fold). The CES1A2-S/ARE reporter (Fig. 5A), compared with its CES1A2-S/ARE counterpart, was activated to a greater extent by Nrf1. In some other cases such as the reporter of GCLM (glutamate-cysteine ligase regulatory subunit), Nrf1 and Nrf2 caused comparable activation (Fig. 5A).
Fig. 5. Differential activation of various ARE reporters by Nrf1 and Nrf2.
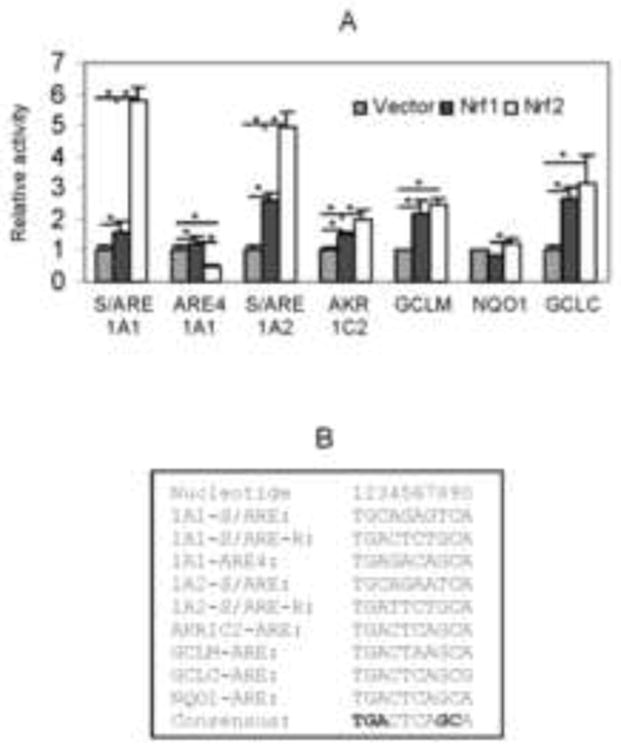
Huh7 cells were transiently transfected by a reporter (50 ng) and the CMV-Renilla luciferase plasmid (0.2 ng) along with 10 ng Nrf1, Nrf2 or the vector. The luciferase activities were determined 24 h after the transfection. The core-Nrf2 sequence from known Nrf2 target genes is shown in B. Asterisk and pound signs indicate statistical significance from the controls or between two data points linked by a line (P < 0.05).
3.6. Differential reactivity of SFN and TNBS toward Keap1
We have shown that the S/ARE element was critical for the transactivation of CES1 by Nrf2 in response to SFN and TNBS. It is well established that Nrf2 transactivation is trigged by dissociating from Keap1 [12]. In the Keap1-Nrf2 pathway, Keap1 is the initiator whereas Nrf2 is the executor [12]. Keap1 is a cysteine-rich protein and some of the cysteines serve as reactive targets for antioxidants and sensitizers. As a result, Keap1 has three forms depending on the oxidative status of cysteines: reduced Keap1, intramolecular and intermolecular Keap1. We took advantage of their differences in electrophoretic mobility and tested whether SFN and TNBS produce similar composition of these three Keap1 molecular species. Cells were transfected with Keap1 and treated with SFN or TNBS at various concentrations and the cell lysates were then separated by non-reducing electrophoresis followed by Western blotting. As shown in Fig. 6, both SFN and TNBS decreased reduced mouse Keap1. Surprisingly they differed in increasing intramolecular and intermolecular Keap1. SFN increased intramolecular Keap1 whereas TNBS increased intermolecular Keap1. Similar changes were detected with human Keap1, although to a much less extent (Fig. 6).
Fig. 6. Interaction of SFN and TNBS with kKap1.
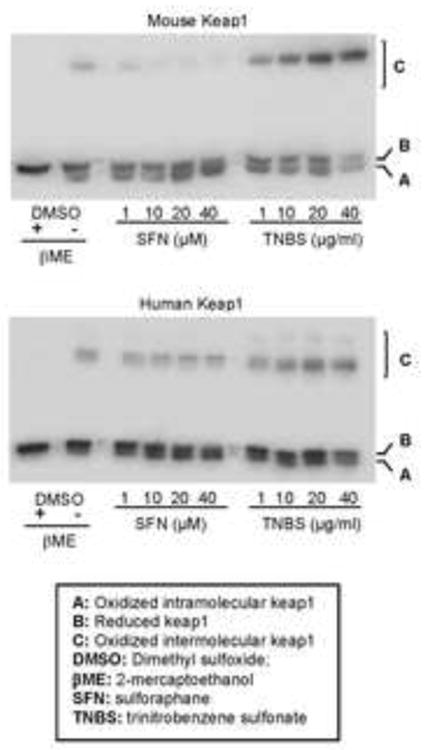
Cells (293T) at 85% confluence in 24-well plates were transfected with Flag-tagged mouse or human keap1 (250 ng/well). After a 24 h-incubation, the transfected cells were washed and then treated for 1 h with SFN or TNBS at various concentrations. Thereafter, cells were extensively with PBS containing 40 mM Nethylmaleimide (NEM) and the lysed in NEM-containing RIPA buffer. Lysates (4 μg) were analyzed by Western blotting with an anti-Flag antibody. This experiments were repeated four times and consistent results were obtained.
4. Discussion
CES1 is the most versatile human carboxylesterase and catalyzes hydrolysis, synthesis and transesterification [1, 2]. In this study, we reports that TNBS and SFN efficaciously induced CES1 through Nrf2 via two elements: S/ARE and ARE4. Nrf2 binds to both elements (Fig. 3A), although they exhibited opposite activities (Fig. 5A). S/ARE supported transactivation whereas ARE4 supported repression of Nrf2, although S/ARE was a more sensitive target (Figs. 4D and 5A). Actually, the S/ARE reporter was activated the most by Nrf2 among all ARE reporters (Fig. 5). Compared with all AREs, ARE4 differs by two nucleotides at the 4th and 5th position in the core-sequence (Fig. 5B). The ARE4 has GA whereas other AREs have CT in these positions. However, a frequency matrix analysis has predicted that 18% of ARE elements have a G and 6% an A in the 4th and 5th position, respectively [24], suggesting that this dinucleotide substitution (i.e., CT→GA) may not be entirely responsible for the observed repression by Nrf2 (Fig. 5B).
It is likely that this GA→CT substitution works with flanking nucleotides and negatively responds to Nrf2. Indeed, computer program ALGGEN-PROMO predicts that the ARE4 core-sequence overlaps with a Yin-Yang1 (YY1) element. This element, CGTGAGACA, consists of 5′ flanking dinucleotide CG and seven nucleotides of the ARE4 core sequence (in italic) including the dinucleotide GA discussed above (Fig. 6). Importantly, a recent study has shown that Nrf2 negatively regulated the transcription of the fibrosis transmembrane conductance receptor gene (CFTR) through an YY1 element overlapped with an ARE (CAAATGACA underlined) [19-25]. Although the CES1A1 YY1 element shares five nucleotides with the CFTR YY1 element, the CES1A1 YY1 element lacks the typical core nucleotides of YY1 element [19]. However, these authors detected YY1-Nrf2 complex [25], suggesting that YY1 and Nrf2 form heterodimers and bind to an YY1-Nrf2 composite site. On the other hand, Nrf2 has been established to preferably form heterodimers with small Maf proteins and confer transactivation activity [26, 27], thus dominating the repression through the YY1-Nrf2 mechanism [Fig. 6]. In support of this notion, the repression of CES1A1 reporters was evident when Nrf2 was highly expressed (Fig. 2A), or in the absence of the S/ARE element (Figs. 3D and 5B). Nevertheless, a confirmation of YY1, along with Nrf2, in the suppression of CES1 will provide an example of how a very same gene can be regulated by the same transcription factor with opposite regulatory activity.
Another interesting finding is that SFN and TNBS caused different changes in the overall conformation of Keap1. SFN promoted thiol oxidation of Keap1 within Keap1, whereas TNBS promoted the thiol oxidation between two Keap1 molecules (Fig. 5). Thus, TNBS induced the formation of large Keap1 complex. The precise mechanism on the difference remains to be determined. It is likely that TNBS activates the Keap1-Nrf2 pathway by directly reacting sulfhydryl groups of Keap1, whereas SFN activates it by altering the cellular oxidative potentials. In support of this possibility, TNBS but not SFN is potent sulfhydryl agent [28, 29]. However, it remains to be determined whether the different reactivity between SFN and TNBS represents general difference between antioxidants and skin sensitizers. Nevertheless, majority of skin sensitizers are sulfhydryl reactive agents [10, 13]. It should be emphasized that the effect on oxidative status of Keap 1 was more profound with mouse than human Keap1 (Fig. 6).
Carboxylesterases are generally considered detoxification enzymes, therefore, induction of CES1 likely represents cytoprotective response [1, 5]. On the other hand, induction of CES1 may have pharmacological significance, particularly regarding the metabolism of ester drugs. For example, the antiplatelet agent clopidogrel is activated by oxidation (5%) but inactivated by hydrolysis (95%) [2]. As a result, exposure to antioxidants or skin sensitizers may decrease the antiplatelet activity of clopidogrel. In contrast, hydrolysis of oseltamivir, a widely used anti-influenza agent, represents activation, and only the hydrolytic metabolite exerts anti-influenza activity [30]. Therefore, induction of CES1 likely leads to increased anti-viral activity of oseltamivir. We have shown that the hydrolytic metabolite, compared with the parent compound oseltamivir, is more cytotoxic [30]. However, the Nrf2-mediated induction of CES1 may cause little changes in the overall toxicity as this pathway has also been found to support the induction of multidrug resistance protein-4 [31], a transporter that effluxes the hydrolytic metabolite [32].
In addition to the pharmacological implications, induction of CES1 may have pathophysiological significance as well. CES1 hydrolyzes many endogenous compounds such as triglycerides and cholesterol esters [1, 4, 5]. Hydrolysis of cholesterol esters increases free cholesterol, leading to increased synthesis of bile acids [4]. Secretion of bile acids represents the only net elimination of excessive cholesterol [33]. On the other hand, increased free fatty acids by hydrolyzing triglycerides and cholesterol esters likely increase the synthesis and secretion of very low density lipoprotein (VLDL), a precursor that leads to the formation of low density lipoprotein (LDL) [34-36]. Indeed, high CES1 activity has been shown to facilitate VLDL maturation [35]. Transgenic expression of CES1 leads to increased secretion of apoB proteins and plasma triglycerides [34]. Elevated level of LDL increases the risk of developing atherosclerosis [37]. Therefore, excessive induction of CES1 without enhancing bile acid synthesis likely has detrimental effect.
Our study points to several important conclusions. (1) both SFN and TNBS are potent inducers of CES1 in various types of cells, establishing that induction of CES1 by antioxidants and sensitizers represents a general phenomenon; (2) the induction is achieved by transactivation through the same response element, further establishing that Nrf2 is a master sensor for functionally diverse compounds; (3) the S/ARE element, differing by a single nucleotide compared with Nrf2 consensus sequence, but is transactivated the most by Nrf2 among all element reporters tested, establishing that the consensus sequence does not represent the optimum for Nrf2 transactivation; (4) SFN and TNBS differ in inducing conformational changes of Keap1, establishing that compounds, although triggering the dissociation of Nrf2 from Keap1, use different recognition. Finally, the ARE4 element supports Nrf2 repression, establishing that flanking sequence, probably with an involvement of the native promoter, rather than the element itself specifies the nature of regulatory activities of Nrf2. Interestingly, Nrf1, commonly sharing the activity with Nrf2, surprisingly activates ARE4, providing an example of function-sharing proteins with opposite regulatory activity toward a target gene. High levels of CES1 are linked to lipid retention [7-9]. Excessive induction of CES1 by antioxidants and sensitizers may provide a mechanism for potential detrimental effect on human health [41].
Fig. 7. Diagrammatical presentation of regulated transcription of the CES1A1 through the S/ARE and ARE4 elements.
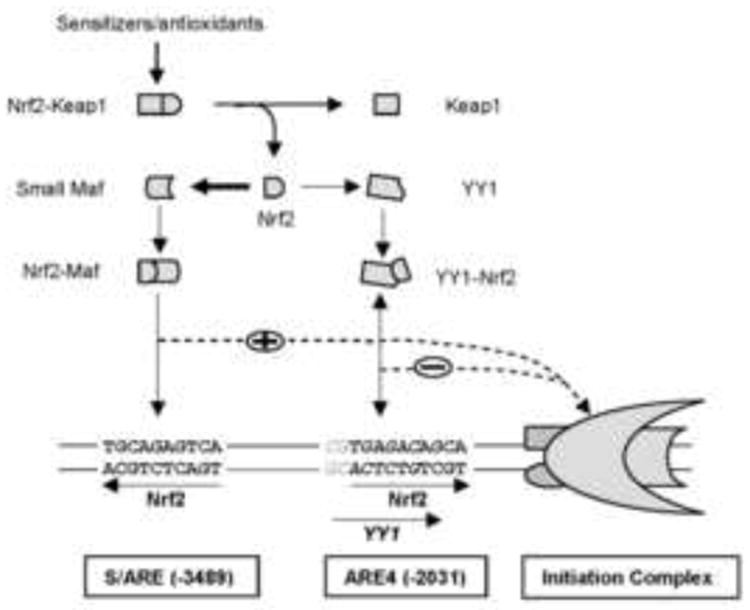
The width of the arrows between Nrf2 to small Maf and Nrf2 to YY1 suggests the dominance of the formation of Nrf2-Maf dimmers over that of Nrf2-YY1 dimers. The double ended arrow between the Nrf2-YY1 dimers and the YY1-Nrf2 composite element suggests weak interaction. The dotted lines indicate possible direct interactions.
Acknowledgments
We thank Dr. Jefferson Y. Chan of University of California Irvine the Nrf1 construct and Dr. Anil K. Jaiswail of the University of Maryland for the keap1 constructs.
This work was supported by NIH grants R01GM61988 and R01ES07965.
The authors thank Dr. Robert Rodgers of the University of Rhode Island for critical reading ot the manuscript.
Abbreviations
- AKR1C2
aldo-keto reductase-1C2
- ARE
antioxidant response element
- CES
carboxylesterase
- ChIP
chromatin-immunoprecipitation
- CYP
cytochrome P450
- DMEM
Dulbecco's Modified Eagles Medium
- EMSA
electrophoretic mobility shift assay
- FBS
fetal bovine serum
- GAPDH
glyceradehyde-3-phosphate dehydrogenase
- GCLC
glutamate-cysteine ligase catalytic subunit
- GCLM
glutamate-cysteine ligase regulatory subunit
- NQO1
NAD(P)H:quinone oxidoreductase
- Nrf2
Nuclear factor-E 2 related factor-2
- PBS
phosphate buffered saline
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- S/ARE
sensitizing/antioxidant response element
- SFN
sulforaphane
- TNBS
trinitrobenzene sulfonate
- WME
William's medium E
Footnotes
The authors indicate no potential conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Holmes R, Wright M, Laulederkind S, Cox L, Hosokawa M, Imai T, Ishibashi S, Lehner R, Miyazaki M, Potter P, Redinbo M, Robert J, Satoh T, Yamashita T, Yan B, Yokoi T, Rudolf-Zechner R, Maltais L. Recommended Nomenclature for Five Mammalian Carboxylesterase Gene Families: Human, Mouse and Rat Genes and Proteins. Mammalian Genome. 2010;21:427–441. doi: 10.1007/s00335-010-9284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang M, Mukundan M, Yang J, Charpentier N, LeCluyse EL, Black C, Yang D, Shi D, Yan B. Antiplatelet agents aspirin and clopidogrel are hydrolyzed by distinct carboxylesterases, and clopidogrel is transesterificated in the presence of ethyl alcohol. J Pharmacol Exp Ther. 2006;319:1467–1476. doi: 10.1124/jpet.106.110577. [DOI] [PubMed] [Google Scholar]
- 3.Wei E, Ben Ali Y, Lyon J, Wang H, Nelson R, Dolinsky VW, Dyck JR, Mitchell G, Korbutt GS, Lehner R. Loss of TGH/Ces3 in mice decreases blood lipids, improves glucose tolerance, and increases energy expenditure. Cell Metab. 2010;11:183–193. doi: 10.1016/j.cmet.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Zhao B, Natarajan R, Ghosh S. Human liver cholesteryl ester hydrolase: cloning, molecular characterization, and role in cellular cholesterol homeostasis. Physiol Genomics. 2005;23:304–310. doi: 10.1152/physiolgenomics.00187.2005. [DOI] [PubMed] [Google Scholar]
- 5.Sanghani SP, Sanghani PC, Schiel MA, Bosron WF. Human carboxylesterases: an update on CES1, CES2 and CES3. Protein Pept Lett. 2009;16:1207–1214. doi: 10.2174/092986609789071324. [DOI] [PubMed] [Google Scholar]
- 6.Hosokawa M, Furihata T, Yaginuma Y, Yamamoto N, Watanabe N, Tsukada E, Ohhata Y, Kobayashi K, Satoh T, Chiba K. Structural organization and characterization of the regulatory element of the human carboxylesterase (CES1A1 and CES1A2) genes. Drug Metab Pharmacokinet. 2008;23:73–84. doi: 10.2133/dmpk.23.73. [DOI] [PubMed] [Google Scholar]
- 7.Yan B. Hydrolytic Enzymes. In: Zanger, Anzenbacher, editors. Metabolism of Drugs and Other Xenobiotics. WILEY-VCH Verlag GmbH & Co. KGaA; Weinheim: 2012. pp. 165–199. [Google Scholar]
- 8.Marrades MP, González-Muniesa P, Martínez JA, Moreno-Aliaga MJ. A dysregulation in CES1, APOE and other lipid metabolism-related genes is associated to cardiovascular risk factors linked to obesity. Obes Facts. 2010;3:312–318. doi: 10.1159/000321451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagashima S, Yagyu H, Takahashi N, Kurashina T, Takahashi M, Tsuchita T, Tazoe F, Wang XL, Bayasgalan T, Sato N, Okada K, Nagasaka S, Gotoh T, Kojima M, Hyodo M, Horie H, Hosoya Y, Okada M, Yasuda Y, Fujiwara H, Ohwada M, Iwamoto S, Suzuki M, Nagai H, Ishibashi S. Depot-specific expression of lipolytic genes in human adipose tissues. J Atheroscler Thromb. 2011;18:190–9. doi: 10.5551/jat.6478. [DOI] [PubMed] [Google Scholar]
- 10.Roberts DW, Aptula AO, Patlewicz G. Electrophilic chemistry related to skin sensitization. Reaction mechanistic applicability domain classification for a published data set of 106 chemicals tested in the mouse local lymph node assay. Chem Res Toxicol. 2007;20:44–60. doi: 10.1021/tx060121y. [DOI] [PubMed] [Google Scholar]
- 11.Python F, Goebel C, Aeby P. Comparative DNA microarray analysis of human monocyte derived dendritic cells and MUTZ-3 cells exposed to the moderate skin sensitizer cinnamaldehyde. Toxicol Appl Pharmacol. 2009;239:273–283. doi: 10.1016/j.taap.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Natsch A. The Nrf2-Keap1-ARE toxicity pathway as a cellular sensor for skin sensitizers--functional relevance and a hypothesis on innate reactions to skin sensitizers. Toxicol Sci. 2010;113:284–292. doi: 10.1093/toxsci/kfp228. [DOI] [PubMed] [Google Scholar]
- 14.Maruichi T, Fukami T, Nakajima M, Yokoi T. Transcriptional regulation of human carboxylesterase 1A1 by nuclear factor-erythroid 2 related factor 2 (Nrf2) Biochem Pharmacol. 2010;79:288–295. doi: 10.1016/j.bcp.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Jung KA, Kwak MK. The Nrf2 system as a potential target for the development of indirect antioxidants. Molecules. 2010;15:7266–7291. doi: 10.3390/molecules15107266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Xie M, Song X, Gragen S, Sachdeva K, Wan Y, Yan B. DEC1 negatively regulates the expression of DEC2 through binding to the E-box in the proximal promoter. J Biol Chem. 2003;278:16899–16907. doi: 10.1074/jbc.M300596200. [DOI] [PubMed] [Google Scholar]
- 17.Liu F, Yang D, Song X, Deng R, Yan B. The far and distal enhancers in the CYP3A4 gene coordinates the proximal promoter in responding to the pregnane X receptor similarly but differentially to hepatocyte nuclear factor-4α. Biochem J. 2008;409:243–250. doi: 10.1042/BJ20070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song X, Gragen S, Li Y, Ma Y, Liu J, Yang D, Matoney L, Yan B. Intramolecular disulfide bridges are required for folding hydrolase B into a catalytically active conformation but not for maintaining it during catalysis. Biochem Biophys Res Commun. 2004;319:1072–1080. doi: 10.1016/j.bbrc.2004.05.090. [DOI] [PubMed] [Google Scholar]
- 19.Zhu W, Song L, Zhang H, Matoney L, LeCluyse E, Yan B. Dexamethasone differentially regulates the expression of carboxylesterase genes in humans and rats. Drug Metab Dispos. 2000;28:186–191. [PubMed] [Google Scholar]
- 20.Yang J, Shi D, Yang D, Song X, Yan B. Interleukin-6 suppresses the expression of carboxylesterases HCE1 and HCE2 through transcriptional repression. Mol Pharmacol. 2007;72:686–694. doi: 10.1124/mol.107.036889. [DOI] [PubMed] [Google Scholar]
- 21.Yang D, Shi D, Yang J, Deng R, Yan B. Scoparone potentially transactivates the bile salt export pump gene and the potentiation is enhanced by cytochrome P450 1A2 but inhibited by protein kinase C inhibitor. Brit J Pharmacol. 2011;164:1547–1557. doi: 10.1111/j.1476-5381.2011.01522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinkova-Kostova AT, Talalay P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch Biochem Biophys. 2010;501:116–123. doi: 10.1016/j.abb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biswas M, Chan JY. Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol Appl Pharmacol. 2010;244:16–20. doi: 10.1016/j.taap.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucl Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.René C, Lopez E, Claustres M, Taulan M, Romey-Chatelain MC. NF-E2-related factor 2, a key inducer of antioxidant defenses, negatively regulates the CFTR transcription. Cell Mol Life Sci. 2010;67:2297–309. doi: 10.1007/s00018-010-0336-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto T, Kyo M, Kamiya T, Tanaka T, Engel JD, Motohashi H, Yamamoto M. Predictive base substitution rules that determine the binding and transcriptional specificity of Maf recognition elements. Genes Cells. 2006;11:575–291. doi: 10.1111/j.1365-2443.2006.00965.x. [DOI] [PubMed] [Google Scholar]
- 27.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci U S A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maruta S, Homma K, Ohki T. Conformational changes at the highly reactive cystein and lysine regions of skeletal muscle myosin induced by formation of transition state analogues. J Biochem. 1998;124:578–584. [PubMed] [Google Scholar]
- 29.Mi L, Chung FL. Binding to protein by isothiocyanates: a potential mechanism for apoptosis induction in human non small lung cancer cells. Nutr Cancer. 2008;60(Suppl 1):12–20. doi: 10.1080/01635580802381287. [DOI] [PubMed] [Google Scholar]
- 30.Shi D, Yang J, Yang D, LeCluyse EL, Black C, You L, Akhlaghi F, Yan B. Anti-influenza prodrug oseltamivir is activated by carboxylesterase HCE1 and the activation is inhibited by anti-platelet agent clopidogrel. J Pharmacol Exp Ther. 2006;319:1477–1484. doi: 10.1124/jpet.106.111807. [DOI] [PubMed] [Google Scholar]
- 31.Xu S, Weerachayaphorn J, Cai SY, Soroka CJ, Boyer JL. Aryl hydrocarbon receptor and NF-E2-related factor 2 are key regulators of human MRP4 expression. Am J Physiol Gastrointest Liver Physiol. 2010;299:G126–35. doi: 10.1152/ajpgi.00522.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ose A, Ito M, Kusuhara H, Yamatsugu K, Kanai M, Shibasaki M, Hosokawa M, Schuetz JD, Sugiyama Y. Limited brain distribution of [3R,4R,5S]-4-acetamido-5-amino-3-(1-ethylpropoxy)-1-cyclohexene-1-carboxylate phosphate (Ro 64-0802), a pharmacologically active form of oseltamivir, by active efflux across the blood-brain barrier mediated by organic anion transporter 3 (Oat3/Slc22a8) and multidrug resistance-associated protein 4 (Mrp4/Abcc4) Drug Metab Dispos. 2009;37:315–321. doi: 10.1124/dmd.108.024018. [DOI] [PubMed] [Google Scholar]
- 33.Dawson PA, Lan T, Rao A. Bile acid transporters. J Lipid Res. 2009;50:2340–2357. doi: 10.1194/jlr.R900012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei E, Alam M, Sun F, Agellon LB, Vance DE, Lehner R. Apolipoprotein B and triacylglycerol secretion in human triacylglycerol hydrolase transgenic mice. J Lipid Res. 2007;48:2597–2606. doi: 10.1194/jlr.M700320-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Blais DR, Lyn RK, Joyce MA, Rouleau Y, Steenbergen R, Barsby N, Zhu LF, Pegoraro AF, Stolow A, Tyrrell DL, Pezacki JP. Activity-based protein profiling identifies a host enzyme, carboxylesterase 1, which is differentially active during hepatitis C virus replication. J Biol Chem. 2010;285:25602–25612. doi: 10.1074/jbc.M110.135483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Wei E, Quiroga AD, Sun X, Touret N, Lehner R. Altered lipid droplet dynamics in hepatocytes lacking triacylglycerol hydrolase expression. Mol Biol Cell. 2010;21:1991–2000. doi: 10.1091/mbc.E09-05-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siri-Tarino PW, Sun Q, Hu FB, Krauss RM. Saturated fat, carbohydrate, and cardiovascular disease. Am J Clin Nutr. 2010;91:502–509. doi: 10.3945/ajcn.2008.26285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnsen O, Murphy P, Prydz H, Kolsto AB. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res. 1998;26:512–520. doi: 10.1093/nar/26.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung KA, Kwak MK. The Nrf2 system as a potential target for the development of indirect antioxidants. Molecules. 2010;15:7266–7291. doi: 10.3390/molecules15107266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keum YS. Regulation of the Keap1/Nrf2 system by chemopreventive sulforaphane: implications of posttranslational modifications. Ann N Y Acad Sci. 2011;1229:184–189. doi: 10.1111/j.1749-6632.2011.06092.x. [DOI] [PubMed] [Google Scholar]
- 41.Yang D, Yang J, Shi D, Black C, Deng R, Yan B. Hypolipidemic agent Zguggulsterone: metabolism interplays with induction of cholesteryl ester hydrolase CES1 and bile salt export pump. J Lipid Res. 2012;53:529–539. doi: 10.1194/jlr.M014688. [DOI] [PMC free article] [PubMed] [Google Scholar]


