Abstract
Autophagy is the cellular process by which proteins, macromolecules, and organelles are targeted to and degraded by the lysosome. Given that neurodegenerative diseases involve the production of misfolded proteins that cannot be degraded by the protein quality-control systems of the cell, the autophagy pathway is now the focus of intense scrutiny, because autophagy is primarily responsible for maintaining normal cellular proteostasis in the central nervous system (CNS). Huntington's disease (HD) is an inherited CAG–polyglutamine repeat disorder, resulting from the production and accumulation of misfoldedhuntingtin (Htt) protein. HD shares key features with common neurodegenerative disorders, such as Alzheimer's disease (AD) and Parkinson's disease (PD) and, thus, belongs to a large class of disorders known as neurodegenerative proteinopathies. Multiple independent lines of research have documented alterations in autophagy function in HD, and numerous studies have demonstrated a potential role for autophagy modulation as a therapeutic intervention. In this review, we consider the evidence for autophagy dysfunction in HD, and delineate different targets and mechanistic pathways that might account for the autophagy abnormalities detected in HD. We assess the utility of autophagy modulation as a treatment modality in HD, and suggest guidelines and caveats for future therapy development directed at the autophagy pathway in HD and related disorders.
As postmitoticnondividing cells, neurons are susceptible to the accumulation of damaged proteins and organelles. Their complex, polarized cellular architecture and their inability to dilute insults by cell division render them particularly sensitive to the accumulation of toxic protein aggregates and defective organelles [1]. Thus, neuronal survival depends heavily on maintaining protein quality control by efficient degradation mechanisms. Autophagy, an evolutionarily conserved lysosomal degradation pathway, is active in neurons and functions to eliminate these toxic components, which are hallmarks of neurodegenerative diseases, including AD, PD and HD.
Three types of mammalian autophagy have been described: microautophagy, chaperone-mediated autophagy (CMA) and macroautophagy. The different autophagy pathways have been classified based upon the manner of cargo delivery to lysosomes, but they all culminate in cargo degradation by this organelle. Briefly, during microautophagy, the lysosomal membrane itself invaginates into the lysosomal lumen, and cargo is quickly degraded by lysosomal hydrolases. This type of autophagy is poorly understood, and its role in neurons remains unclear. During CMA, cargo is directly bound by cytosolic chaperones, and then recognized and imported into the lysosomal lumen by a receptor on the lysosomal membrane. Conversely, macroautophagy requires the formation of a double membrane-bound vesicle, the autophagosome, to isolate cargo and, after autophagosome maturation, the autophagosomes fuse with lysosomes for degradation. The importance of basal neuronal macroautophagy was demonstrated by conditional knockout of key autophagy genes autophagy protein 5 and 7 (Atg5 and Atg7), which, in the absence of any additional proteotoxic stress, resulted in neurodegeneration and the accumulation of ubiquitin-positive inclusions, which similar to what is typically observed in neurodegenerative disease [2,3].
All three types of autophagy can coexist in the same cell, and alterations in both macroautophagy and CMA have been described in many neurological disorders. Indeed, despite completely different etiologies, many neurodegenerative disorders share the common pathological finding of autophagic vacuole (AV) accumulation in degenerating neurons. Evidence now indicates that this increase in AVs is not principally the result of increased autophagy pathway activity, but rather reflects a decrease in autophagy flux; that is, the process of autophagosome maturation culminating in lysosomal fusion. Indeed, neurons are particularly sensitive to autophagy–lysosome pathway perturbations, as reflected by the high frequency of neurological disorders caused by mutations targeting the endolysosomal network [1]. Furthermore, certain disease gene mutations that cause neurodegenerative disorders [e.g. encoding presenilin-1, huntingtin, α-synuclein, parkin, Leucine-rich repeat kinase 2 (LRRK2) and dynein] directly impact proper autophagy progression at different steps [1]. This suggests that, although autophagy has powerful neuroprotective abilities, because it promotes the degradation of toxic proteins [2,3], the autophagy pathway itself might be a direct target of disease proteins in the CNS. In this review, we discuss current understanding of autophagy dysfunction in HD and the therapeutic potential for autophagy modulation in this complicated disorder and other related diseases.
Huntington's diseaseHD: a polyglutamine repeat disorder belonging to a large family of neurodegenerative proteinopathies
Huntington's disease (HD is an autosomal dominant neurodegenerative disorder characterized by involuntary motor movement, cognitive decline and psychiatric illness. The disorder is relentlessly progressive, with a median age of disease onset of approximately 40 years, and leads to death 10–30 years after the initial presentation. HD is caused by a CAG trinucleotide repeat expansion (≥36 CAG repeats) that is located in the amino-terminal region of the Htt protein and encodes an abnormally long polyglutamine (polyQ) tract. Thus, HD is one of nine inherited neurodegenerative diseases known as CAG–polyQ repeat disorders, a category that also includes six spinocerebellar ataxias (SCA1, 2, 3, 6, 7 and 17), X-linked spinobulbar muscular atrophy (SBMA) and dentatorubral-pallidoluysian atrophy (DRPLA). One of the most striking pathological hallmarks of HD is the selective degeneration of the striatum and cortical neurons that project to the striatum. Within the HD striatum, the medium spiny neurons (MSNs) turn out to be exquisitely vulnerable. Despite this pattern of selective neuronal vulnerability, Htt protein is widely expressed and readily detected in most cell types, both within and outside of the CNS. This selective neurotoxicity in the face of widespread expression is a shared feature of all polyQ disorders.
Another feature common to polyQ disorders is the production of a disease protein that misfolds, cannot be degraded and accumulates as proteinaceous aggregates. These aggregates form in the nucleus and cytoplasm as intraneuronal inclusion bodies, and are enriched in the relevant aggregation-prone polyQ-expanded disease protein. HD inclusions mostly comprise amino-terminal fragments of polyQ-expanded Htt, and are found both in neuron nuclei and dystrophic neurites throughout the cortex and striatum of patients with HD [4,5]. The extent of these inclusions correlates with the length of the polyQ expansion, suggesting that they are a feature of HD pathology [5]. Inclusions are also enriched in ubiquitin and ubiquitinated-Htt, as well as with components of the ubiquitin proteasome system and with heat-shock protein chaperones [5].
Understanding the normal function of Htt is crucial for understanding its toxicity in the context of the polyQ expansion. Complete Htt knockout in mice is embryonic lethal, suggesting that it has a nonredundant function essential for life, with a crucial role in mouse embryonic brain development, as well as being crucial for the survival of adult neurons in the forebrain [6]. The finding that normal Htt function is essential for neuronal survival in the same brain regions that are also sensitive to polyQ-Htt toxicity suggests that a loss-of-function mechanism contributes to HD pathology. In agreement with this thesis, considerable work has shown that the polyQ expansion tract alters Htt protein folding, resulting in aberrant protein conformations, which are likely to impact Htt normal function significantly [7]. However, the dominant inheritance pattern of HD supports a gain-of-function model of polyQ-Htt toxicity. In support of this model, expression of either full-length or amino-terminal truncated forms of polyQ-Htt is sufficient to induce motor abnormalities and neurodegeneration in numerous animal models, yielding phenotypes that are reminiscent of what is observed in patients with HD. The precise nature of this gain-of-function toxicity is not known; however, transcriptional dysregulation, mitochondrial dysfunction and autophagy pathology are consistent features in HD animal models [8–11]. Thus, the HD disease mechanism is likely to be complex, and might involve a combination of gain-of-function proteotoxicity and a loss-of-function of endogenous Htt protein, stemming from aberrant protein–protein interactions caused by the polyQ expansion tract [7]. The presence of misfolded, aggregate-prone proteins in the CNS of patients with HD defines HD as a member of a large family of neurodegenerative proteinopathies, which includes AD, PD, amyotrophic lateral sclerosis, prion diseases, tauopathies and synucleinopathies.
Huntington's diseaseHD: a disorder of impaired proteostasis
The finding that polyQ-Htt inclusions are positive for ubiquitin and ubiquitinated-Htt probably reflects a generalized deficiency in ubiquitin proteasome system (UPS)-mediated degradation of mutant Htt species [5]. Indeed, eukaryotic proteasomes cannot efficiently degrade long polyQ sequences, and recent work indicates that aberrant sequestration of key UPS components prevents delivery of misfolded proteins to the nuclear proteasome in HD and related disorders [12]. Failure of the UPS might lead to upregulation of autophagy via cross-talk between degradation pathways in the attempt by the cell to maintain normal proteostasis[13,14]. Whereas the proteasome has steric selectivity for its substrates and can only process those substrates that can be unfolded and passed through its core machinery, macroautophagy has no such limitation. Macroautophagy cargo can include macromolecules, entire subcellular organelles, such as mitochondria and peroxisomes, endoplasmic reticulum (ER) cisternae, or pathogenic bacteria [13]. Consequently, polyQ-expanded proteins are amenable substrates for autophagy pathway degradation [15].
The key degradation pathways of the cell (i.e. the UPS and autophagy) are significantly compromised with aging. The ‘mitochondrial–lysosomal axis’ theory of postmitotic cellular aging postulates that mitochondrial turnover, which is mediated by mitochondrial autophagy, progressively declines with age, resulting in increased oxidative cellular damage [16]. This generates a negative feedback loop, which enhances toxicity, as lysosomes (which are particularly sensitive to oxidative stress) become even more dysfunctional, and autophagy becomes less and less efficient. The gradual decline of cellular degradation pathways with age might explain why patients with HD do not present until they are middle-aged or older, owing to the ability of the UPS and autophagy to maintain neuronal homeostasis in the face of an ongoing proteotoxic insult. Once key cellular degradation pathways can no longer keep up with the burden of ongoing proteotoxic stress, polyQ-Htt aggregates accumulate in a last-ditch effort by the cell to sequester the toxic moieties away. Sometime after accumulation of protein aggregates, long-term neuronal dysfunction ensues, finally culminating in neuronal loss. Dysfunction in autophagy-mediated degradation has been described for models of HD and, over time, as quality control pathways continue to fail, accumulation of damaged organelles such as mitochondria and ER occurs, indicative of a collapse in normal proteostasis. Thus, understanding autophagy dysfunction in HD is important, because preventing or reversing this significant hit to neuronal proteostasis could represent a significant therapeutic opportunity. Interestingly, although polyQ-Htt aggregates are substrates for autophagy-mediated degradation, Htt itself might also be involved in regulating the autophagy pathway.
Macroautophagy in HD
Macroautophagy entails the packaging of cytosolic materials into a double-membrane vesicle, the autophagosome, which then fuses with components of the endolysosomal system for the ultimate degradation of the enclosed materials [1]. Macroautophagy is mediated by a series of Atg proteins, which are involved in all key steps of the pathway. In neurons of the peripheral nervous system and the CNS, autophagosomes are formed in neurites and synaptic terminal regions in the distal axon, and are then transported back to the cell soma, where active lysosomes reside. Macroautophagy is well regulated, with a key node of signal transduction being the serine/threonine kinase mammalian target of rapamycin (mTOR) in the mTORC1 complex. The mTORC1 complex is a conserved negative regulator of macroautophagy, with direct downstream effects on cellular metabolism and energetics.
Reports of biopsies of brains from patients with HD as early as the 1970s remarked on significant abnormalities in compartments of the vesicular-endocytic pathway, noting an abnormal proliferation of multivesicular bodies, endosomes and lysosomes, as well as documenting disruptions of the Golgi apparatus and disorganization of the ER [17]. Similarly, Htt staining of remaining striatal neurons in brains from patients with HD revealed dramatic increases in endosome-lysosome-like organelles and Htt-positive tubulovesicular structures, in comparison to nonHD controls [4]. Since then, increased numbers of autophagic vacuoles have been reported in many HD mouse models and in nonneuronal cells from patients with HD [18]. Furthermore, the autophagy markers p62 (a cargo-adaptor) and LC3-II (an autophagosome membrane marker) are specifically increased in the striatum of HD transgenic mice [19]. Conversely, mTOR appears to be sequestered by polyQ-Htt aggregates, inhibiting its activity and inducing autophagy [20]. Thus, various lines of investigation in the HD field implicate dysfunctional autophagy as part of HD pathology and suggest that macroautophagydysregulation contributes to neurotoxicity. However, a contradiction also quickly emerged. The observed expansion of macroautophagy compartments in HD cells was never accompanied by the expected increase in autophagy-mediated degradation [18]. Moreover, unlike other neurodegenerative disorders where autophagy flux is impaired [1], macroautophagy pathway progression appears to be comparable in HD cells and control cells [18], indicating that the defect does not lie in autophagy progression per se. Elegant work by the Cuervo group has offered one possible explanation for this conundrum, by documenting significant alterations in macroautophagy cargo recognition in HD cells, which leads to the formation of ‘empty’ autophagosomes (Figure 1). The reduced amount of cytosolic cargo inside these autophagosomes, possibly resulting from an aberrant p62–polyQ-Htt interaction, yielded a low protein degradation rate in HD samples [18]. Thus, compensatory activation of macroautophagy in response to polyQ-Htt aggregates could be detrimental to HD neuron cell survival, because resources and energy expenditure are consumed without any net removal of toxic substrates or net recycling of macromolecular building blocks for continued biosynthesis.
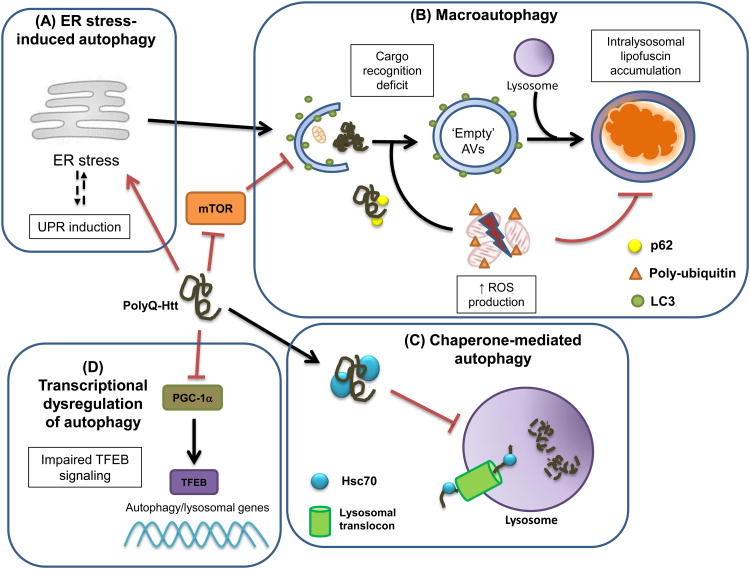
Figure 1.
Different molecular mechanisms account for autophagy pathway dysfunction in Huntington's disease (HD). In HD, a polyglutamine-expanded huntingtin (polyQ-Htt) protein is produced and accumulates in cells. The misfoldedpolyQ-htt elicits an endoplasmic reticulum (ER) stress response (A), and through direct inhibition of the ER-associated degradation pathway, impairment of ER-to-Golgi trafficking, and disruption of ER and mitochondrial calcium homeostasis, polyQ-htt ultimately yields ER stress-induced activation of the macroautophagy pathway. PolyQ-htt can sequester mammalian target of rapamycin (mTOR), resulting in disinhibition of macroautophagy; however, macroautophagy functions abnormally in HD, because the mutant polyQ-htt protein prevents recognition and loading of cargo into developing autophagosomes, resulting in an accumulation of autophagic vesicles (AVs) that are relatively empty, in terms of substrates for degradation (B). Lysosomal enzyme activity is reduced in HD, possibly because of an increased burden of reactive oxygen species (ROS), given that HD cells contain markedly increased numbers of lysosomes containing nondegradedlipofuscin (B). PolyQ-htt interacts excessively with Hsc70 and the lysosomal translocation machinery, resulting in diminished chaperone-mediated autophagy function in HD (C). As the amino-terminal fragment of polyQ-htt enters the nucleus, it has been shown to interfere with peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC-1α), a transcriptional co-regulator, recently found to promote transcription factor E-B (TFEB) expression. Thus, polyQ-htt transcriptional dysregulation leads to impaired TFEB transactivation of its target genes, which encode the proteins and enzymes required for autophagosome assembly, autophagosome-lysosome fusion and lysosomaldegradative enzyme activity (D). Hence, polyQ-htt appears capable of undermining autophagy pathway function for two branches of autophagy (macroautophagy and chaperone-mediated autophagy), and can act through multiple mechanisms for macroautophagy inhibition.
Deficient autophagic cargo recognition would also have serious deleterious effects on cell homeostasis. Cytosolic organelles are turned over by macroautophagy, a process that requires recognition of targeted membranes by the autophagy machinery. PolyQ-Htt is present at various organelle membranes [10,21] and, thus, could be directly interfering with organelle recognition by AVs. In agreement with this hypothesis, defective mitochondria accumulate in HD cells, suggesting that impaired mitochondrial recognition by autophagy contributes to the metabolic deficits and reactive oxygen species (ROS) production previously reported in HD. Protein aggregates are also targeted to macroautophagy clearance by a polyubiquitination signal. Interestingly, polyQ-Htt binds to polyubiquitinated protein aggregates [15], thereby also interfering with their recognition by the autophagy system. More work is required to understand how generalized this impaired cargo recognition is, and whether it has a substantive role in HD pathogenesis.
Experiments investigating the normal physiological function of Htt protein have highlighted a potential role for Htt in the macroautophagy process itself. Expression of either normal or mutant Htt in clonal striatal neuron-like cells can induce endolysosomal network activity, stimulating endosome tubulation and autophagy activation [10]. Endosome tubulation directly impacts important endocytic processes, such as vesicle budding, fusion, and transport. The effect of Htt on membrane dynamics is intriguing, and suggests a key role for Htt in membrane remodeling and autophagosome formation. Indeed, full-length Htt normally resides in the cytosol, and localizes to cytoplasmic vacuoles and other membranous compartments, but upon polyQ tract expansion, Htt localization is significantly altered [10,21]. Further highlighting this putative Htt function, deletion of the glutamine stretch in normal murine Htt (to create the ΔQ-Htt isoform) activated autophagy in an mTOR-independent manner in mice [22]. Heterozygous expression of the ΔQ-Htt allele intrans to the polyQ-Htt allele in Hdh140Q/ΔQ knock-in mice significantly reduced Htt aggregate load, ameliorated motor phenotypes and improved survival in this HD mouse model. Furthermore, this was accompanied by increased striatal expression of autophagy marker LC3-II. Such an autophagy phenotype was not observed when the polyQ-Htt allele was intrans to a Htt knockout allele, indicating that upregulation of macroautophagy in Hdh140Q/ΔQ knock-in mice is specifically the result of the loss of the glutamine stretch, rather than of a Htt loss-of-function effect [22].
How might Htt protein mediate macroautophagy regulation? Although this question is unresolved, it is interesting that Htt and mTOR share several structural and functional similarities. Both are protein scaffolds containing a significant number of leucine-rich HEAT repeats, which have been shown to interact specifically with membrane structures. mTOR cycles back and forth from the nucleus in response to stress, and can reversibly associate with ER and Golgi membranes. Similarly, Htt can associate with the ER and shuttle to the nucleus in response to stress [21], as well as with membranes of autophagic vesicles [10,21]. These observations suggest that Htt responds to ER stress and regulates ER-stress induced autophagy [23], as discussed in more detail below. Furthermore, several Htt domains display putative structural similarity with Atg23, vacuolar protein 8 (Vac8) and Atg11, which are all components of the yeast cytoplasm-to-vacuole-targeting pathway (reviewed in [24]), suggesting that the vertebrate Htt gene is the result of the fusion of multiple ancient yeast genes. Although this hypothesis is speculative, it deserves consideration in light of emerging evidence for the ability of Htt to perform a variety of autophagy-related functions.
Postmortem studies of HD brains have revealed a striking accumulation of lipofuscin, a nondegradable intra-lysosomal polymer, in neuronal and glial vesicular compartments [17]. Although usually considered an age-related pigment, this HD-specific lipofuscin increase exceeds that of the normal aging brain [17]. Given that intralysosomallipofuscin accumulation impairs lysosomal clearance of autophagic cargo [16], it might reflect a late-stage autophagy deficit in HD. Consistent with this notion, an increased number of lipid droplets in HD cells, in parallel with a markedly reduced association of lipid droplets with HD autophagosomes, has been reported [18]. Thus, deficient ‘lipophagy’ (i.e. macroautophagy-mediated turnover of fats and lipids) might account for this abnormal intracellular lipid storage phenotype observed in HD. In agreement with this hypothesis, Hdh140Q/ΔQ knock-in mice exhibit a complete reversal of lipofuscin accumulation in the CNS, probably because of increased autophagy in the cortex and striatum [22]. Given that Hdh140Q/ΔQ knock-in mice display a significantly improved HD disease phenotype, decreased lipofuscin accumulation might confer therapeutic benefit in HD or just correlate with phenotypic improvement, suggesting that it could serve as a valuable biomarker for HD research.
A number ofSeveral studies have shown that both UPS function and lysosome function decline with age. Macroautophagy function also diminishes with age, because of changes in both gene transcription and post-translational regulation. For example, there is a significant age-related decline in the expression of the autophagy protein Beclin-1 in human brain [25], and there is evidence for an age-related decrease in the trafficking of the lysosomal protein lysosomal associated membrane protein-2 (LAMP2) [26]. Interestingly, Beclin-1 is sequestered into polyQ-Htt inclusions in R6/2 HD transgenic mice and in the brains of patients with HD [25]. Furthermore, polyQ-Htt aggregates have also been shown to sequester mTOR[20], suggesting that an age-dependent decline of macroautophagyAtg-protein function, coupled with Htt inhibition of macroautophagy, exacerbates HD disease progression in an age-dependent fashion. Similar to other inherited polyQ disorders, polyQ expansion tract length is a major determinant of age of disease onset, but alone does not predict onset age or disease severity. Variation in HD age of onset is also influenced by environment as well as other genetic factors (and also chance), which together dictate the course of disease. Some genetic factors suggested to modify HD onset and progression include the expression levels of huntingtin-associated protein-1 (HAP-1), the adenosine A2A receptor, and peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC-1α), a master transcriptional regulator of mitochondrial biogenesis and metabolism. PGC-1α transcription interference contributes to the mitochondrial dysfunction and metabolic abnormalities in HD [27,28]. Recent work identified transcription factor E-B (TFEB), a master regulator of the autophagy–lysosome pathway, as a bona fide target of PGC-1α[11]. Importantly, impaired TFEB expression and activity was documented in the face of polyQ-expanded Htt in N171-82Q HD transgenic mice [11], implicating altered TFEB signaling in HD pathogenesis (Figure 1). Consistent with previously published work, TFEB can promote clearance of polyQ-Htt[11]. These findings highlight TFEB as a therapeutic target for HD, and for other neurodegenerative disorders characterized by accumulation of protein aggregates.
Recently, another autophagy-related genetic modifier of HD was reported. A polymorphism in Atg7, wherein a valine is substituted for an alanine at amino acid position 471 (V471A), was associated with earlier age of HD onset [29]. Although the functional significance of this polymorphism remains unknown, the valine substitution at amino acid position 471 might disrupt the hydrophobic core of Atg7. Thus, the presence of the Atg7 V471A polymorphism could further aggravate an already dysfunctional autophagy pathway in the CNS of patients with HD.
As with other protein conformation disorders, HD is characterized by the abnormal aggregation and deposition of misfolded proteins. Although polyQ-expanded Htt is a bona fide target for autophagic clearance, the wild type normal form of Htt is not [15,25], suggesting selectivity of macroautophagy towards pathogenic Htt. What makes polyQ-Htt species more susceptible to macroautophagy clearance? Htt is heavily post-translationally modified, and these biochemical changes to the Htt protein are known disease modifiers [30,31]. The IκB kinase (IKK) signaling complex has a central role in Htt post-translational modification, directly phosphorylating Htt at serine 13, which induces subsequent phosphorylation at serine 16 [31]. These phosphorylations have downstream effects leading to ubiquitination, acetylation, SUMOylation and proteolytic cleavage of Htt, all of which directly impact Htt turnover and toxicity [31]. Indeed, BAC HD mice derived with phosphomimetic (serine to aspartic acid) and phosphoresistant (serine to alanine) amino acid substitutions at Htt serine 13 and 16 confirmed the importance of these phosphorylation sites for determining polyQ-Htt turnover and neurotoxicity [32]. Another post-translational modification with relevance to HD pathogenesis is lysine 444 acetylation. HD brains are enriched in polyQ-Htt acetylated at lysine 444, and this oligomeric acetylated species was found to be preferentially targeted to autophagosomes[30], representing the first evidence for lysine acetylation as a signal for autophagosome delivery and protein clearance. Thus, acetylation might directly control pathogenicity of polyQ-Htt, because inhibitors of histone deacetylases (HDACs) reduce neurotoxicity in several models of HD [33], although this effect might involve transcription regulation in chromatin by targeting histones. However, given that increased HDAC activity is beneficial for other polyQ diseases [14], and HDACs can act as direct positive regulators of macroautophagy[34], further research into the precise regulation and role of acetylation for both autophagy pathway proteins and polyQ-Htt, and their interplay, is needed.
SUMOylation stabilizes amino-terminal Htt fragments [35], and may target proteins to PML bodies, subnuclear structures enriched for the known polyQ protein interactor CREB-binding protein (CBP). PML bodies have been proposed as sites of IKK-initiated Httcaspase cleavage, yielding toxic amino-terminal Htt fragments, underscoring the complex relation between protein trafficking, processing and degradation, and post-translational modification. Given that the sequence of events and the effects of each of these events on polyQ-Htt function and turnover are yet to be determined, understanding the (mis)regulation of post-translationally modified polyQ-Htt, and its effect on neurotoxicity, will be an important goal for future HD research.
Chaperone-mediated autophagyCMA in HD
It is well known that blockage of macroautophagy can lead to a compensatory upregulation of CMA [26], an alternative lysosome-mediated degradation pathway wherein cytosolic components are directly translocated across the lysosomal membrane [36]. CMA substrates have the pentapeptide motif KFERQ (or related degenerated sequences), and are recognized by cytosolic chaperone Hsc70. Substrate-Hsc70 complexes are then targeted to lysosomes, where they are recognized and bound by the lysosome-associated membrane protein type 2A (LAMP-2A), which mediates substrate translocation into the lysosomal lumen [36]. Thus, CMA requires soluble cargo that is amenable to unfolding and recognition by CMA machinery.
Htt has two putative KFERQ-like CMA-targeting motifs: one at amino acids 99–103 (KDRVN), and one at amino acid 248–252 (NEIKV). A third motif at the very amino-terminus (14-LKSFQ-18) is considered KFERQ-like upon phosphorylation at serine 16 [31,37]. Recent work has shown that the 99-KDRVN-103 motif modulates the interaction of Htt fragments with Hsc70, and that this interaction is probably functional. Consistent with these data, purified intact lysosomes do take up mutant Htt fragments in a LAMP2A-dependent manner [37], suggesting that polyQ-Htt is indeed a bona fide target for CMA-degradation (Figure 1). Interestingly, CMA lysosomal uptake and clearance of Htt fragments was reported to be less efficient for polyQ-expanded Htt fragments. One possible explanation for this could be that polyQ expansion in mutant Htt delays its CMA-mediated transport across the lysosomal membrane, leading to accumulation in the cytosol. In agreement with this concept, interaction with LAMP2A and Hsc70 is stronger for polyQ-Htt, resulting in a CMA pathway traffic jam [37]. Moreover, CMA appears to target selectively amino-terminal fragments of Htt, whereas full-length polyQ-Htt is primarily targeted by macroautophagy[37,38]. Indeed, Htt amino-terminal fragments co-fractionate with lysosomal fractions, a pattern distinct from full-length huntingtin[10]. Given that truncated forms of Htt are particularly cytotoxic, and are abundant in the brains of patients with HD, and other HD models, the ability of CMA to degrade them selectively offers an appealing therapeutic opportunity. Consistent with this model, one elegant study showed that specific targeting of an amino-terminal polyQ-Htt fragment to the CMA pathway greatly reduced inclusion formation and improved HD phenotypes in R6/2 mice [39]. Thus, manipulation of CMA is emerging as a strong candidate for therapy development.
Similar to macroautophagy, post-translational modification of Htt can also regulate its degradation by the CMA pathway. Clearance of phosphorylated amino-terminal fragments of polyQ-Htt depends upon LAMP2A and Hsc70 [31]. Increased CMA activity has been reported in fibroblasts from patients with HD and striatal Hdh knock-in cells [38]. Similar increases have been reported for pre-symptomatic Htt Q111 knock-in mice, but were not sustained when aged mice became symptomatic [38]. Although reductions in CMA activity have been reported in the aging brain, the observed rate of decline in HD aged mice was significantly accelerated. Although polyQ-Htt toxicity for isolated CMA-competent lysosomes has not been reported [37,38], a significant decrease in LAMP-2A staining has been recorded in older Htt Q111 knock-in mice [38]. Given that lipid composition at the lysosomal membrane regulates LAMP-2A localization and, thus, CMA activity, and altered lipid metabolism because of deficient macroautophagy occurs in HD cells [18], alterations in lipid homeostasis might cause a secondary dysfunction in CMA. This suggests a model wherein HD neurons attempt to compensate for defective macroautophagy pathway function by upregulating CMA. Over time, CMA-competent lysosomes become less and less efficient at degrading substrates, as lipids accumulate and polyQ-Htt interferes with LAMP-2a/Hsc70 cargo uptake. This proteostasis dysfunction is then compounded by a decline in CMA and macroautophagy activity in the aging brain and, ultimately, culminates in a generalized all-out collapse in autophagy pathway function, resulting in neuronal demise and death.
Endoplasmic reticulumER stress-induced autophagy in HD
In recent years, attempts at understanding the physiological function of Htt have revealed a new nexus of huntingtin-mediated neurotoxicity: the ER. The ER is a crucial site for endocytic pathway protein quality control. Normal ER function is required for folding and post-translational modification of nascent peptides, which represent fundamental steps for proper protein function and sorting. Various physiological and pathological conditions can impair the folding capacity of the ER, leading to the accumulation of misfolded proteins in the ER lumen (‘ER stress’). This triggers the unfolded protein response (UPR), an ER-to-nucleus cellular response designed to increase the capacity of the ER to fold its client proteins. UPR activation reduces global protein synthesis and upregulates chaperones and ‘foldases’. The ER compartment proliferates and ER-associated degradation (ERAD) is activated to eliminate the irreparably misfolded proteins [40]. Evidence also indicates that another consequence of ER stress and UPR activation is induction of autophagy [41], suggesting a close homeostatic balance between the autophagy and UPR pathways, with autophagy possibly operating as a survival pathway against ER stress.
ER stress has become increasingly important in the understanding of several degenerative proteinopathies, including PD and AD. Recent evidence suggests that ER stress and UPR activation are also a feature of HD neurodegeneration (Figure 1). ER stress sensors and UPR downstream proteins are upregulated and activated specifically in the striatum (but not in other brain regions) of HD transgenic mice [19,42], and in postmortem brain samples from patients with HD [43]. Furthermore, ER-stress induced autophagy has been reported in mouse models of HD [42]. However, unlike other disorders wherein the toxic protein misfolds and aggregates inside the ER lumen, directly causing ER stress, Htt has not been found inside this compartment. Despite this seeming contradiction, multiple lines of evidence suggest that polyQ-Htt could indeed be perturbing ER homeostasis. First, Htt is known to associate with various organelles of the endocytic pathway, including the ER and transport vesicles, through an amphipathic alpha-helix membrane-binding domain in its amino-terminus [10,21]. Consistent with an important role for Htt in ER function, alteration of Htt expression levels was sufficient to cause significant physical distortion of the ER network [44] and disrupted Golgi staining [10]. Furthermore, in striatal-like knock-in cells, mutant Htt perturbed ER-to-Golgi transport of signaling receptors, cytokines and enzyme delivery to lysosomes [45,46], suggesting a general dysfunction in the secretory pathway. Thus, polyQ-Htt could be disrupting proper forward vesicle transport, especially between the ER and the Golgi apparatus, which is a powerful known trigger for ER stress. Furthermore, Htt is reversibly associated with ER structures, releasing from the ER membrane and traveling to the nucleus under ER stress [23]. Given that Htt is also associated with autophagic vesicles [10,21,47], it might act as an ER sentinel, regulating autophagy in the face of ER stress [21,23]. Second, an effective UPR requires UPS degradation of irreversibly misfolded proteins via ERAD. PolyQ-Htt interferes with the function of essential ERAD factors, including Npl4, UFd1, p97 and gp78, which are all required for retrotranslocation of ERAD substrates into the cytosol and their trafficking to the proteasome [48,49]. Similarly, it has been reported that mutant Htt directly inhibits proteosomal activity [8,9], thereby directly preventing degradation of ERAD substrates.
The ER is a major calcium reservoir for the cell, and many of the UPR-responsive chaperones and foldases have calcium-dependent activities. Thus, calcium homeostasis is particularly important for efficient engagement of the UPR in the face of ER stress. Mutant Htt disrupts calcium homeostasis in many models of HD (reviewed in [50]), suggesting yet another mechanism for polyQ-Htt-mediated ER stress. Despite all of this, direct evidence of the effect of UPR and/or ER stress signaling in HD pathology remains unclear in vivo. Nonetheless, manipulation of the UPR has yielded beneficial effects in models of HD. For example, downregulation of the ER-stress sensor IRE-1α reduced mutant Htt toxicity and rescued polyQ-Htt rough-eye phenotypes in Drosophila [19]. Underscoring the importance of the IRE-1α branch of the UPR in HD, deletion of IRE-1α target X-box binding protein 1 (XBP1) in the CNS reduced polyQ-Htt levels in the brain and improved motor performance in the HD YAC128 mouse model [47]. XBP1 deficiency yielded upregulation of Forkhead box protein O1 (FOXO1), a known inducer of neuronal autophagy, which was shown previously to improve motor performance and survival in HD R6/2 mice in an autophagy-dependent manner [51]. The fact that induction of autophagy by XBP1 deficiency ameliorates HD appears contradictory, especially given previously documented abnormalities in autophagy cargo recognition in HD [18]. Hence, further research is needed to delineate the effects of XBP1 on autophagy and polyQ-Htt clearance. However, the ability of XBP1 to improve HD phenotypes suggests that combination therapies to rectify the autophagy cargo defect, reduce ER stress and enhance other protein degradation pathways could be an effective treatment strategy for HD.
Targeting autophagy for HD therapy: how and when?
The importance of neuronal autophagy and its dysregulation in neurodegenerative diseases is undeniable. Nevertheless, the precise events leading to autophagy malfunction, and the key steps affected in each disorder remain unclear. However, therapeutic manipulation of autophagy in vivo, whether through pharmacological or genetic means, has shown promise in animal models of HD, consistently resulting in an amelioration of behavioral motor abnormalities and neuropathology (Table 1). Seminal work by Ravikumar and colleagues was the first to demonstrate that macroautophagy signaling could indeed have beneficial effects in HD [15]. In this study, the rapamycin analog CCI-779, a known mTOR inhibitor, reduced polyQ-Htt aggregate load and improved motor phenotypes in HD transgenic mice by inducing autophagy activation. Trehalose, a disaccharide shown to induce macroautophagy, also alleviated toxicity, improved motor function and extended lifespan in HD R6/2 transgenic mice [52]. Similarly, rilmenidine, an mTOR-independent macroautophagy inducer, attenuated motor phenotypes and reduced levels of the toxic mutant Htt fragment in HD N171-82Q transgenic mice [53]. Recent studies also indicate that CMA might preferentially target soluble, amino-terminal fragments of polyQ-Htt[37,38]. This disease-specific species is particularly toxic, so therapies targeting upregulation of CMA might be especially beneficial in HD. Indeed, the lifespan extension obtained in R6/2 mice using a CMA-targeting adaptor molecule against polyQ-Htt has been among the most significant of all HD single-molecule preclinical trials reported so far [39].
Table 1. Preclinical trials of macroautophagy modulation in mouse models of Huntington's disease.
| Autophagy modulator | Mouse model | Outcome | Refs |
|---|---|---|---|
| CCI-779 | R6/2 | Decreased polyQ-Htt aggregate loads Improved motor performance | [20] |
| Trehalose | R6/2 | Decreased aggregates | [52] |
| Improved motor performance | |||
| Mild lifespan extension | |||
| HDAC inhibitor 4b | R6/2 | Reduced polyQ-Htt aggregates | [55] |
| Improved motor and cognitive performance | |||
| Correction of transcriptional alterations | |||
| Rilmenidine | N171-82Q | Improved HD motor phenotype (reduced tremors, increased grip strength) | [53] |
| XBP1 deficiency | YAC128 and Q111/Q7 knock-in | Decreased striatal polyQ-Htt accumulation | [47] |
| Increased striatal neuron survival | |||
| Improved motor performance | |||
| IRS2 (IGF1 signaling) | R6/2 | Decreased aggregate formation | [51] |
| Improved mitochondrial function | |||
| Slowed progression of HD motor phenotypes | |||
| QBP1 | R6/2 | Decreased aggregate formation | [39] |
| Improved motor and weight phenotypes | |||
| Significant lifespan extension |
Numerous independent lines of investigation have identified HDAC inhibitors as candidate drugs for the treatment of different neurological disorders. HDACs interact with polyQ proteins, and provide a mechanistic link between UPS impairment and macroautophagy activation [14]. HDAC inhibitor 4b ameliorated cognitive decline and motor function, and corrected transcriptional dysregulation in the striatum and cortex of R6/2 mice. Interestingly, pathways activated after treatment included post-translational modification networks (particularly ubiquitination and acetylation), processes known to affect protein turnover [54]. Inhibitor 4b also induced expression of several IKK family members, known activators of macroautophagy, and yielded increased expression of Atg4b and Map1LC3a [54]. Thus, although HDAC inhibitors exert pleiotropic effects, one possible mode of action is through macroautophagy pathway activation.
A crucial consideration for autophagy interventional therapies is the timing of treatment delivery. Early activation of autophagy should be beneficial, aiding HD neurons in the clearance of aggregate-prone polyQ-Htt. However, with more advanced disease, lysosome and proteasome abnormalities become more apparent in HD, and these defects could undermine the efficacy of autophagy induction therapies. Indeed, therapies that simultaneously activate autophagy and modulate Htt protein post-translational modifications (such as IKK signaling agonists) could yield polyQ-Htt isoforms that neurons will be incapable of turning over. Given that patients with HD can be identified at presymptomatic disease stages through genetic testing, therapeutic autophagy modulation before disease onset is theoretically possible in HD and might be necessary not only for a significant beneficial effect, but also because autophagy induction in symptomatic patients with HD could worsen disease pathology. For example, therapies that increase macroautophagy in advanced HD patients might only work if the underlying autophagic cargo recognition defects are corrected. Thus, understanding the precise molecular players underlying macroautophagy cargo recognition is an important goal of future research, with obvious implications for therapy development. Of course, it would also be ideal to advance understanding of the normal physiological function of Htt, and how the polyQ tract expansion in mutant Htt protein alters its normal function, before pushing forward with therapeutic modulation of autophagy. This is especially important for HD, because the Htt protein has been implicated in autophagy pathway regulation and membrane dynamics, and emerging data strongly point towards a possible loss of Htt normal function as a factor in the autophagy defects observed in HD. Deconstructing the mechanistic basis of Htt autophagy regulation could reveal potential therapeutic targets for modulating autophagy in HD and related neurodegenerative disorders. Ultimately, identification of the specific steps affected in the autophagy pathway in HD will be essential for the development of such rational autophagy-based therapeutic interventions.
Highlights.
The two major protein degradation pathways, the UPS and autophagy, are impaired in HD
Dysfunctional macroautophagy, including abnormal cargo recognition, is observed in HD
Huntingtin may regulate autophagy but effects of polyQ expansion on this are unclear
CMA is impaired and ER stress occurs in HD, contributing to proteostasis dysfunction
Targeting autophagy for therapy is challenging as more activation may be deleterious
Acknowledgments
Our autophagy research program and HD research are supported by grants from the National Institutes for Health (R01 AG033082 and R01 NS065874 to A.R.L.), the Muscular Dystrophy Association (Basic Research Award to A.R.L. and DG 273985 to C.J.C.), the Hereditary Disease Foundation, and the Child Health and Development Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 3.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 4.Sapp E, et al. Huntingtin localization in brains of normal and Huntington's disease patients. Ann Neurol. 1997;42:604–612. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- 5.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 6.Zeitlin S, et al. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez I, et al. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- 8.Bence NF, et al. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 9.Hipp MS, et al. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease. J Cell Biol. 2012;196:573–587. doi: 10.1083/jcb.201110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kegel KB, et al. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsunemi T, et al. PGC–1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4:142ra197. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park SH, et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154:134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 14.Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 15.Ravikumar B, et al. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 16.Terman A, et al. Mitochondrial turnover and aging of long-lived postmitotic cells: the mitochondrial-lysosomal axis theory of aging. Antioxid Redox Signal. 2010;12:503–535. doi: 10.1089/ars.2009.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tellez-Nagel I, et al. Studies on brain biopsies of patients with Huntington's chorea. J Neuropathol Exp Neurol. 1974;33:308–332. doi: 10.1097/00005072-197404000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Vicente M, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, et al. IRE1 plays an essential role in ER stress-mediated aggregation of mutant huntingtin via the inhibition of autophagy flux. Hum Mol Genet. 2012;21:101–114. doi: 10.1093/hmg/ddr445. [DOI] [PubMed] [Google Scholar]
- 20.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 21.Atwal RS, et al. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 22.Zheng S, et al. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010;6:e1000838. doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atwal RS, Truant R. A stress sensitive ER membrane-association domain in Huntingtin protein defines a potential role for Huntingtin in the regulation of autophagy. Autophagy. 2008;4:91–93. doi: 10.4161/auto.5201. [DOI] [PubMed] [Google Scholar]
- 24.Steffan JS. Does Huntingtin play a role in selective macroautophagy? CellCycle. 2010;9:3401–3413. doi: 10.4161/cc.9.17.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibata M, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 26.Kaushik S, et al. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weydt P, et al. The gene coding for PGC-1alpha modifies age at onset in Huntington's disease. Mol Neurodegener. 2009;4:3. doi: 10.1186/1750-1326-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 29.Metzger S, et al. Age at onset in Huntington's disease is modified by the autophagy pathway: implication of the V471A polymorphism in Atg7. Hum Genet. 2010;128:453–459. doi: 10.1007/s00439-010-0873-9. [DOI] [PubMed] [Google Scholar]
- 30.Jeong H, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson LM, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu X, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hockly E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci U S A. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banreti A, et al. The emerging role of acetylation in the regulation of autophagy. Autophagy. 2013;9:819–829. doi: 10.4161/auto.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steffan JS, et al. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 36.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab. 2010;21:142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi L, et al. The role of chaperone–mediated autophagy in huntingtin degradation. PLoS ONE. 2012;7:e46834. doi: 10.1371/journal.pone.0046834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koga H, et al. Constitutive upregulation of chaperone-mediated autophagy in Huntington's disease. J Neurosci. 2011;31:18492–18505. doi: 10.1523/JNEUROSCI.3219-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bauer PO, et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28:256–263. doi: 10.1038/nbt.1608. [DOI] [PubMed] [Google Scholar]
- 40.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 41.Bernales S, et al. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Martinez JM, et al. BH3-only proteins Bid and Bim(EL) are differentially involved in neuronal dysfunction in mouse models of Huntington's disease. J Neurosci Res. 2007;85:2756–2769. doi: 10.1002/jnr.21258. [DOI] [PubMed] [Google Scholar]
- 43.Carnemolla A, et al. Rrs1 is involved in endoplasmic reticulum stress response in Huntington disease. J Biol Chem. 2009;284:18167–18173. doi: 10.1074/jbc.M109.018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Omi K, et al. siRNA-mediated inhibition of endogenous Huntington disease gene expression induces an aberrant configuration of the ER network in vitro. Biochem Biophys Res Commun. 2005;338:1229–1235. doi: 10.1016/j.bbrc.2005.10.061. [DOI] [PubMed] [Google Scholar]
- 45.del Toro D, et al. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol Biol Cell. 2009;20:1478–1492. doi: 10.1091/mbc.E08-07-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeron MM, et al. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington's disease. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 47.Vidal RL, et al. Targeting the UPR transcription factor XBP1 protects against Huntington's disease through the regulation of FoxO1 and autophagy. Hum Mol Genet. 2012;21:2245–2262. doi: 10.1093/hmg/dds040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duennwald ML, Lindquist S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008;22:3308–3319. doi: 10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang H, et al. Huntingtin interacts with the cue domain of gp78 and inhibits gp78 binding to ubiquitin and p97/VCP. PLoS ONE. 2010;5:e8905. doi: 10.1371/journal.pone.0008905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Culver BP, et al. Proteomic analysis of wild-type and mutant huntingtin-associated proteins in mouse brains identifies unique interactions and involvement in protein synthesis. J Biol Chem. 2012;287:21599–21614. doi: 10.1074/jbc.M112.359307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sadagurski M, et al. IRS2 increases mitochondrial dysfunction and oxidative stress in a mouse model of Huntington disease. J Clin Invest. 2011;121:4070–4081. doi: 10.1172/JCI46305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka M, et al. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat Med. 2004;10:148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 53.Rose C, et al. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington's disease. Hum Mol Genet. 2010;19:2144–2153. doi: 10.1093/hmg/ddq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jia H, et al. Selective histone deacetylase (HDAC) inhibition imparts beneficial effects in Huntington's disease mice: implications for the ubiquitin-proteasomal and autophagy systems. Hum Mol Genet. 2012;21:5280–5293. doi: 10.1093/hmg/dds379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas EA, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington's disease transgenic mice. Proc Natl Acad Sci U S A. 2008;105:15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]