Abstract
X-linked spinal & bulbar muscular atrophy (SBMA) is characterized by adult-onset muscle weakness and lower motor neuron degeneration. SBMA is caused by CAG-polyglutamine (polyQ) repeat expansions in the androgen receptor (AR) gene. Pathological findings include motor neuron loss, with polyQ-AR accumulation in intranuclear inclusions. SBMA patients exhibit myopathic features, suggesting a role for muscle in disease pathogenesis. To determine the contribution of muscle, we developed a BAC mouse model featuring a floxed first exon to permit cell-type-specific excision of human AR121Q. BAC fxAR121 mice develop systemic and neuromuscular phenotypes, including shortened survival. After validating termination of AR121 expression and full rescue with ubiquitous Cre, we crossed BAC fxAR121 mice with Human Skeletal Actin-Cre mice. Muscle-specific excision prevented weight loss, motor phenotypes, muscle pathology, and motor neuronopathy, and dramatically extended survival. Our results reveal a crucial role for muscle expression of polyQ-AR in SBMA, and suggest muscle-directed therapies as effective treatments.
INTRODUCTION
X-linked spinal and bulbar muscular atrophy (SBMA, Kennedy’s disease) is an inherited neuromuscular disorder characterized by adult onset proximal muscle weakness due to lower motor neuron degeneration. SBMA patients also display signs of androgen insensitivity, including gynecomastia, reduced fertility, and testicular atrophy (Katsuno et al., 2012b). This finding, together with the X-linked inheritance, led to analysis of the androgen receptor (AR) gene as the potential cause of SBMA. While a CAG repeat in the first exon of the AR gene varies in length from 5 – 34 triplets in normals, SBMA patients were found to harbor repeats ranging from 37 – 66 CAG repeats (La Spada et al., 1991). As CAG encodes the amino acid glutamine (Q), SBMA was the first disorder identified to result from expansion of a CAG – polyQ repeat tract. Eight other inherited neurodegenerative disorders were subsequently found to be caused by expanded CAG repeats; hence, in addition to SBMA, the CAG – polyQ repeat disease category includes Huntington’s disease (HD), dentatorubral pallidoluysian atrophy (DRPLA), and six forms of spinocerebellar ataxia: SCA1, 2, 3, 6, 7 and 17 (La Spada and Taylor, 2010).
For decades, research into the basis of neurological disease focused upon the contribution of neuronal dysfunction to disease pathogenesis. However, over the last 15 years, there has been a growing appreciation of the importance of non-neuronal cells in maintaining neuron function and contributing to neurological disease pathogenesis (Garden and La Spada, 2012). In amyotrophic lateral sclerosis (ALS), the inability to recapitulate SOD1 neurotoxicity in transgenic mice upon expression of mutant SOD1 in motor neurons argued against cell autonomous degeneration (Lino et al., 2002; Pramatarova et al., 2001). Chimeric expression of mutant and normal SOD1 in the spinal cords of mice demonstrated a role for non-neuronal cells in ALS motor neuron degeneration, and revealed that astrocytes and microglia are key determinants of disease onset and disease progression (Clement et al., 2003). Subsequently, conditional gene silencing of the ubiquitously expressed SOD1 mutant within astrocytes and microglia indicated that mutant SOD1 within either glial cell type is a key determinant of disease progression (Boillee et al., 2006; Yamanaka et al., 2008), while similar mutant gene silencing in NG2+ precursor cells of oligodendrocytes is a key contributor to disease onset (Kang et al., 2013). In the CAG – polyQ repeat disease field, careful study of a line of SCA7 transgenic mice revealed that Purkinje cell neurodegeneration occurred even when the polyQ-ataxin-7 transgene was not expressed in Purkinje cells, leading to a hypothesis of non-cell autonomous SCA7 neurodegeneration (Garden et al., 2002). As Purkinje cell neurons are intimately associated with a specialized astroglial cell type known as the Bergmann glia, SCA7 transgenic mice were engineered to express mutant ataxin-7 in only Bergmann glial cells in the cerebellum. These animals developed cerebellar ataxia and Purkinje cell degeneration, demonstrating the non-cell autonomous nature of polyQ neurodegeneration (Custer et al., 2006). Studies in HD and in Parkinson’s disease mouse models have similarly shown that expression of mutant disease protein in one cell type is capable of producing dysfunction and demise of a different neuronal cell type, especially for cells in direct communication with one another (reviewed in (Ilieva et al., 2009)). All of this preceding work suggests an overarching theme in neurodegenerative disease pathogenesis: preferential degeneration of select neuron populations does not necessarily stem from intrinsic molecular pathology restricted to the neuron subtype of interest (i.e. cell autonomous toxicity), but rather often results from pathological processes occurring in one or more neighboring cell types that perform crucial functions upon which the exquisitely vulnerable neuron subtype stalwartly depends (i.e. non-cell autonomous toxicity).
Skeletal muscle is a major source of trophic support for innervating motor neurons, and has been shown to contribute not only to neuron survival during development, but also to synaptic activity and axonal function (Funakoshi et al., 1995). SBMA patients often exhibit features of myopathy, as progressive muscle weakness occurs in the context of elevated serum creatine kinase levels (Katsuno et al., 2012b). Muscle biopsies of SBMA patients reveal mixed pathological findings, with both myopathy and neurogenic atrophy features (Soraru et al., 2008). Knock-in mice expressing AR with 113 glutamines (AR113Q) develop early myopathy findings with little or no significant motor neuron loss until late in their disease course (Yu et al., 2006), consistent with muscle as a key site for SBMA disease pathogenesis. Moreover, while widespread transgenic expression of human AR20Q at levels comparable to endogenous AR does not produce a neuromuscular phenotype (Sopher et al., 2004), skeletal muscle-specific over-expression of wild-type AR (AR22Q) in mice is sufficient to produce SBMA-like neuromuscular disease, accompanied by denervation of target muscle and motor neuron axon degeneration, complete with androgen dependence, gender bias, axonopathy and muscle wasting (Monks et al., 2007). Additionally, testosterone treatment of asymptomatic female transgenic mice over-expressing the AR22Q transgene in muscle yielded pronounced neuromuscular deficits, but without detectable motor neuron pathology (Johansen et al., 2009). Although development of SBMA-like disease phenotypes upon skeletal-muscle specific expression of AR22Q may simply stem from the very high level of AR transgene over-expression in this model (Monks et al., 2007), the SBMA-like phenotype in males can be reversed upon cessation of testosterone treatment. These findings indicate that muscle-restricted AR toxicity may underlie SBMA disease pathogenesis, and therapies targeting skeletal muscle may prove beneficial for patients. In support of this thesis, transgenic expression of anabolic insulin growth factor-1 (IGF-1) directed to muscle can rescue nerve pathology in SBMA transgenic mice, producing a significant extension in lifespan (Palazzolo et al., 2009).
Although the preceding studies suggest a role for muscle dysfunction as a component of SBMA motor neuronopathy, the necessity of polyQ-AR expression in muscle for SBMA disease pathogenesis is yet to be investigated. To directly examine the role of muscle expression of AR in SBMA pathogenesis, we developed a BAC transgenic mouse model featuring a floxed first exon to permit cell-type specific excision of the human AR gene. We engineered the human AR transgene to carry 121 CAG repeats (BAC fxAR121), and found that BAC fxAR121 mice develop a gender-restricted, progressive neuromuscular phenotype, characterized by weight loss, motor deficits, muscle atrophy, myopathy, and shortened lifespan. By conditionally terminating expression of mutant polyQ-AR in the skeletal muscles of BAC fxAR121 male mice, we document a crucial role for muscle expression of mutant polyQ-AR in SBMA disease pathogenesis and predict that muscle-directed therapies hold great promise as definitive treatments for SBMA motor neuron degeneration.
RESULTS
Generation and expression analysis of BAC fxAR121 transgenic mice
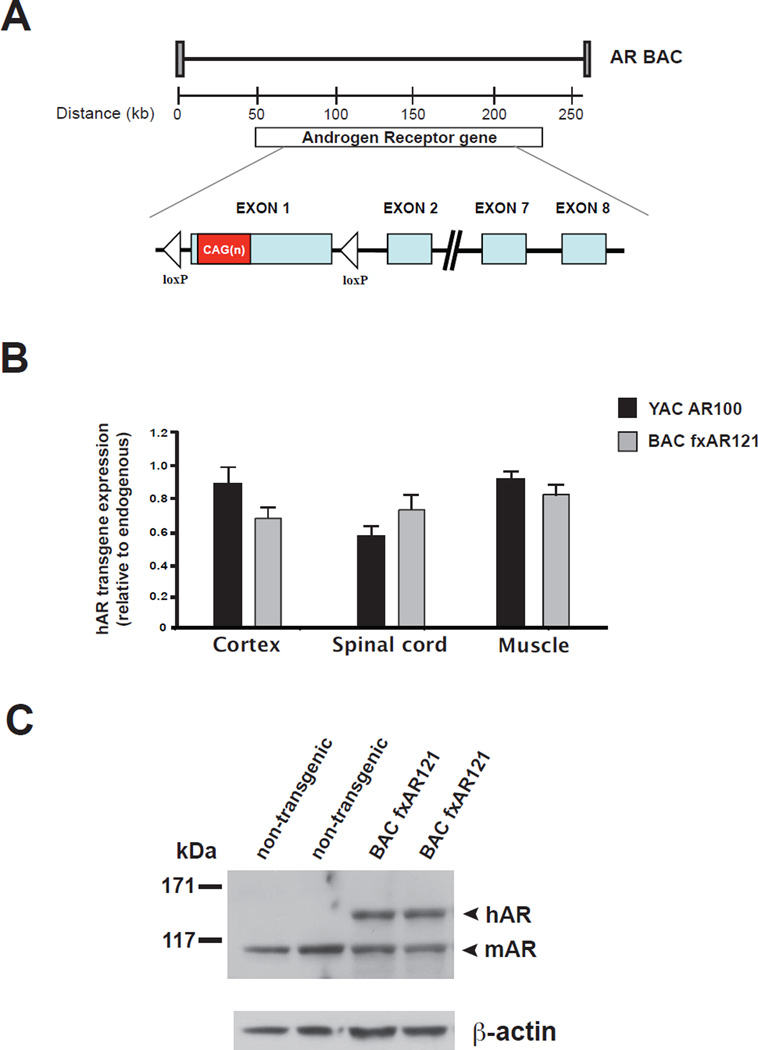
The human AR gene is composed of eight exons that span ~180 kb of DNA. Using the Human Genome Browser Gateway (http://genome.ucsc.edu/cgi-bin/hgGateway), we identified two overlapping BACs that span the entire length of the AR gene. Through a novel recombineering strategy, we fused these two BACs to create an AR BAC construct that, in addition to all eight AR exons, includes ~50 kb of DNA 5’ to the first AR exon and ~30 kb of DNA 3’ to the last AR exon (Sopher and La Spada, 2006). With this recombineering approach, we also introduced a 121 CAG repeat tract and engineered two loxP sites flanking AR exon 1, to create a floxed AR CAG121 BAC (BAC fxAR121) transgenic construct (Figure 1A). We then derived BAC fxAR121 transgenic mice, and when we performed RT-PCR analysis of human AR (hAR) transgene expression, we determined that hAR RNA expression levels in the cortex, spinal cord, and muscle of BAC fxAR121 mice are comparable to endogenous mouse AR (mAR) RNA expression levels and to hAR RNA expression levels in AR YAC CAG100 (YAC AR100) mice (Figure 1B). We measured protein expression in brain and spinal cord, and similarly found that hAR protein levels in the CNS of the BAC fxAR121 mice are comparable to mAR endogenous protein levels (Figure 1C) and to YAC AR100 transgene expression levels (Figure S1). These are encouraging results, as YAC AR100 mice develop a highly representative SBMA motor neuronopathy (Sopher et al., 2004).
Figure 1. Generation and expression analysis of BAC fxAR121 transgenic mice.
(A) Diagram of BAC fxAR121 transgenic construct. After we generated a BAC containing all eight exons of the human Androgen Receptor (hAR) gene, we introduced a CAG121 repeat and two loxP sites flanking exon 1.
(B) RT-PCR analysis of cortex, spinal cord, and quadriceps muscle hAR transgene RNA revealed expression levels that are slightly less than endogenous mouse AR RNA expression levels for BAC fxAR121 mice, and comparable to previously published YAC AR100 SBMA mice (Sopher et al., 2004).
(C) Western blot analysis of whole brain protein lysates confirmed expression of full-length human AR protein (arrowhead, hAR) in the CNS of BAC fxAR121 mice. In this experiment, we used an anti-AR antibody that cross-reacts with mouse AR protein, which is detected at a lower molecular mass (arrowhead, mAR). β-actin immunoblotting serves as a loading control.
Male BAC fxAR121 mice display progressive neuromuscular disease and premature death
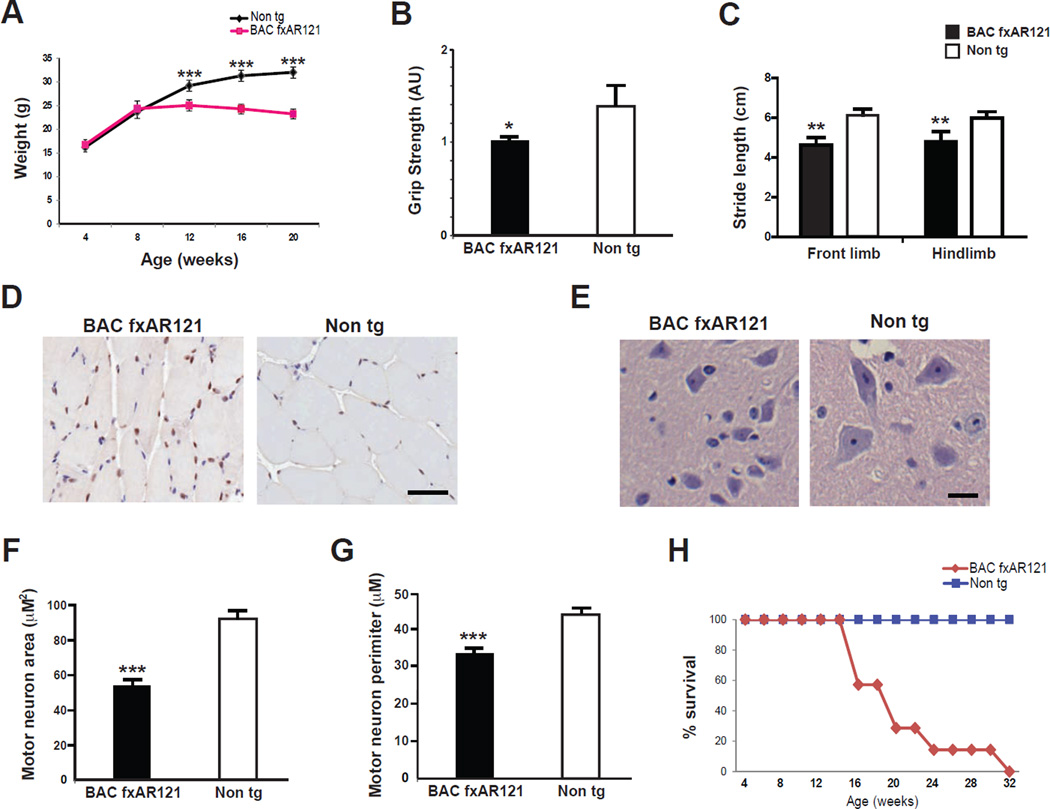
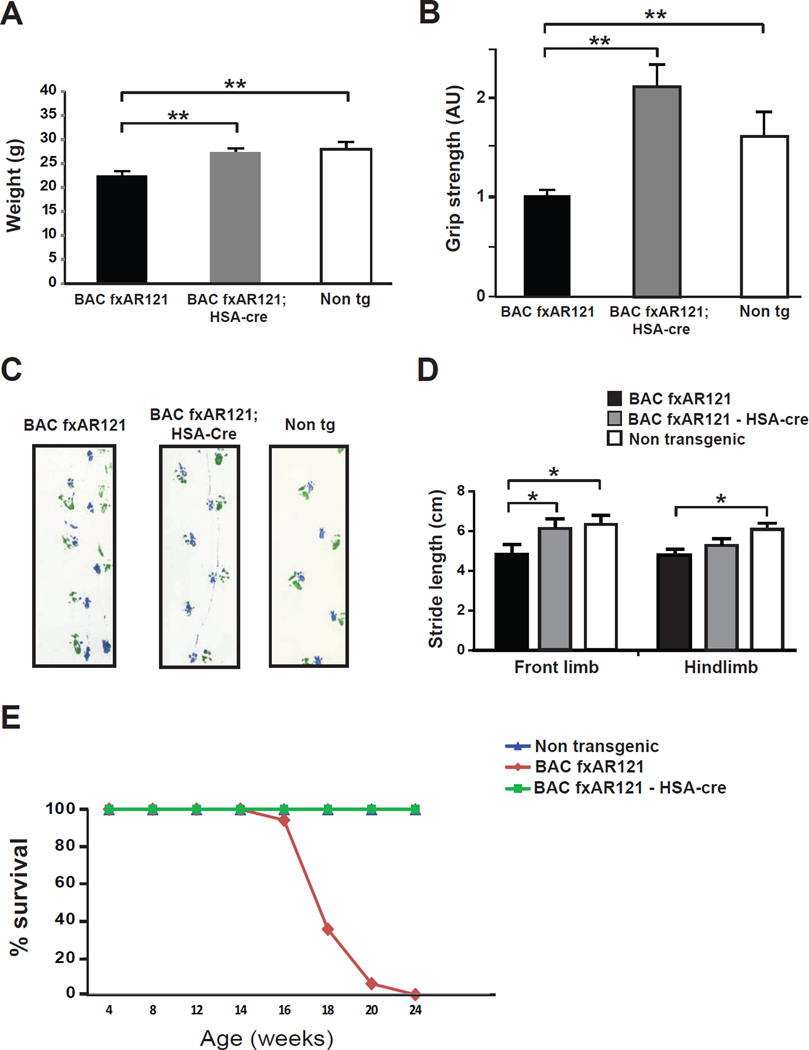
We then inspected the BAC fxAR121 transgenic mice for visible phenotypes and behavioral impairments. Beginning at 12 weeks of age, we observed a significant decrease in the weight of male BAC fxAR121 mice, and this weight loss phenotype steadily progressed as the mice aged (Figure 2A). By the time of significant weight loss, BAC fxAR121 mice exhibited onset of a motor phenotype characterized by weakness, as 13 week-old male BAC fxAR121 mice displayed significantly reduced grip strength (Figure 2B). With disease progression, male BAC fxAR121 mice also exhibited gait abnormalities, based upon impaired performance on stride length testing (Figure 2C). Muscle sections from male BAC fxAR121 mice contained AR protein aggregates (Figure 2D), and NADH staining of such muscle sections revealed muscle fiber atrophy (Figure S2A), consistent with an ongoing process of muscle atrophy due to AR polyQ neurotoxicity, as previously reported in the YAC AR100 mice (Sopher et al., 2004). When we examined ventral horn neurons from lumbar spinal cord sections, we did not observe any changes in motor neuron numbers between male BAC fxAR121 mice and non-transgenic littermate controls (Figure S2B), but noted that ventral horn neurons from BAC fxAR121 mice appear smaller than ventral horn neurons in non-transgenic littermate controls (Figure 2E). We therefore measured ventral horn neuron soma size and perimeter in lumbar spinal cord sections, and documented marked reductions in neuron soma size and perimeter in BAC fxAR121 mice (Figure 2F, 2G). Although BAC fxAR121 mice develop neuromuscular disease phenotypes reminiscent of YAC AR100 mice (Sopher et al., 2004), the disease phenotype in BAC fxAR121 is more progressive than in the YAC AR100 model, as lifespan was dramatically shortened in male BAC fxAR121 mice (Figure 2H). While male BAC fxAR121 mice display prominent neuromuscular disease and premature death, female BAC fxAR121 mice appear phenotypically normal on behavioral testing, and do not suffer weight loss or reduced survival (Figure S2C and S2D).
Figure 2. BAC fxAR121 transgenic mice display systemic and neuromuscular phenotypes.
(A) By 12 weeks of age, male BAC fxAR121 mice no longer continue to gain weight, unlike their non-transgenic littermates (n = 6 – 14 / group). ***P < .001, t-test.
(B) Combined grip strength analysis at 13 weeks of age reveals significant weakness in male BAC fxAR121 mice in comparison to non-transgenic littermate controls (n = 3 / group). Grip strength is given in arbitrary units, with BAC fxAR121 performance set to 1. *P < .05, t-test.
(C) Measurement of front limb and hindlimb stride length was performed for 20 week-old male BAC fxAR121 mice and non-transgenic littermate controls (n = 5 – 6 / group). Significant decreases in mean stride length were observed for BAC fxAR121 mice in comparison to controls. **P < .01, t-test.
(D) AR immunostaining of quadriceps muscle sections from 20 week-old male BAC fxAR121 mice and non-transgenic littermate controls reveals focal AR protein inclusions at the muscle cell periphery in BAC fxAR121 mice. These inclusions appear only occasionally in muscle sections from non-transgenic mice. Scale bar = 50 µm.
(E) H&E-stained spinal cord sections from 28 week-old male BAC fxAR121 mice and non-transgenic littermate controls demonstrate that ventral horn neurons in the lumbar spinal cord are reduced in size in BAC fxAR121 males. Scale bar = 20 µm.
(F) Quantification of ventral horn neuron soma size shown in (E), (n = 3 / group). ***P < .001, t-test.
(G) Quantification of ventral horn neuron perimeter shown in (E), (n = 3 / group). ***P < .001, t-test.
(H) Kaplan-Meier plot of male BAC fxAR121 mice and non-transgenic littermate controls (n = 9 / group) reveals a dramatic reduction in lifespan for BAC fxAR121 mice. P < .0001, Log-rank test. Error bars = s.e.m.
Ubiquitous excision of floxed hAR transgene prevents SBMA disease phenotypes in BAC fxAR121 – CMV-Cre bigenic mice
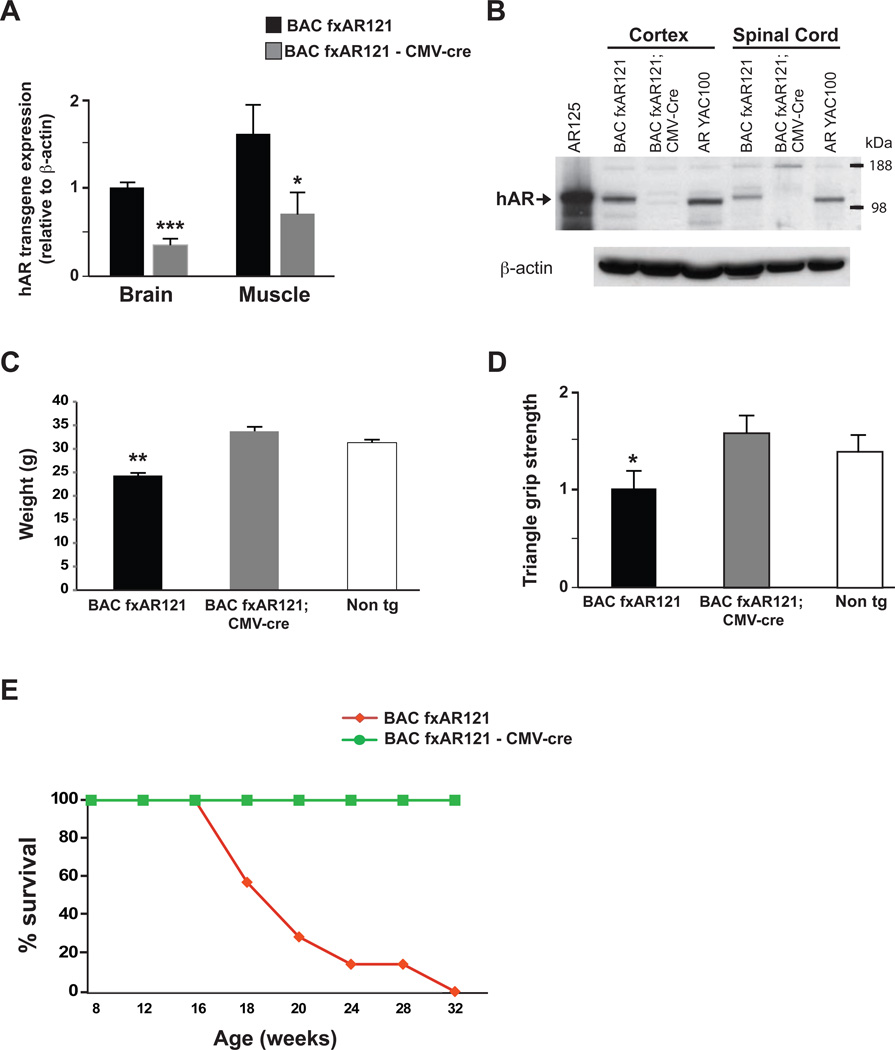
To confirm that efficient in vivo excision of the first exon of the hAR gene and corresponding inactivation of hAR gene expression in BAC fxAR121 mice were feasible, we crossed BAC fxAR121 mice with CMV-Cre transgenic mice. In the resulting BAC fxAR121 – CMV-Cre bigenic progeny, excision of transgenic hAR exon 1, which includes the translational start site, should occur in all tissues, yielding inactivation of mutant hAR expression and prevention of neuromuscular disease. RT-PCR analysis confirmed that BAC fxAR121 – CMV-Cre bigenic mice display markedly reduced hAR RNA expression levels in brain and muscle (Figure 3A). Furthermore, immunoblot analysis yielded no detectable hAR protein in brain or spinal cord protein lysates isolated from BAC fxAR121 – CMV-Cre bigenic mice (Figure 3B), confirming high-level excision of hAR exon 1 and corresponding inactivation of hAR transgene expression. Phenotype analysis of BAC fxAR121 – CMV-Cre bigenic mice indicated normal weight gain, grip strength, and lifespan (Figure 3C, 3D and 3E), thereby validating the utility of the floxed first hAR exon as a target for Cre-mediated excision and hAR transgene inactivation.
Figure 3. Ubiquitous Cre-mediated excision of the human AR121 transgene rescues systemic and neuromuscular phenotypes in BAC fxAR121 mice.
(A) We measured hAR transgene expression in 6 week-old male BAC fxAR121 – CMV-Cre bigenic mice in comparison to singly transgenic male BAC fxAR121 mice (n = 3 / group) by qRT-PCR analysis. We observed marked reductions in hAR transgene expression in the brain and quadriceps muscle of BAC fxAR121 – CMV-Cre bigenic mice. Results are normalized to hAR transgene expression in BAC fxAR121 brain, which was set to 1. ***P < .001, *P < .05, t-test.
(B) Western blot analysis of cortex and spinal cord for 6 week-old male BAC fxAR121 – CMV-Cre bigenic mice and littermate male BAC fxAR121 mice indicates that hAR protein expression is no longer detectable in BAC fxAR121 – CMV-Cre bigenic mice, when immunoblotted with human-specific anti-AR antibody. The first lane contains protein isolated from HEK293 cells transfected with a hAR125Q expression construct, and YAC AR100 protein lysates are included as an additional positive control. β-actin immunoblotting serves as a loading control.
(C) Ubiquitous Cre expression in BAC fxAR121 mice rescues weight loss in 16 week-old male BAC fxAR121 – CMV-Cre bigenic mice (n = 4 – 10 / group). **P < .01, ANOVA with post-hoc Tukey test.
(D) Ubiquitous Cre expression in BAC fxAR121 mice rescues grip strength in 13 week-old BAC fxAR121 – CMV-Cre bigenic mice (n = 4 / group). Grip strength is given in arbitrary units, with BAC fxAR121 performance set to 1. *P < .05, ANOVA with post-hoc Tukey test.
(E) Kaplan-Meier plot of male BAC fxAR121 mice and male BAC fxAR121 – CMV-Cre bigenic mice (n = 9 / group) reveals significantly extended lifespan in BAC fxAR121 – CMV-Cre mice. P < .0001, Log-rank test. BAC fxAR121 – CMV-Cre bigenic mice are still alive and well beyond 24 months of age. Error bars = s.e.m.
BAC fxAR121 – HSA-Cre bigenic mice exhibit muscle-restricted inactivation of hAR transgene expression
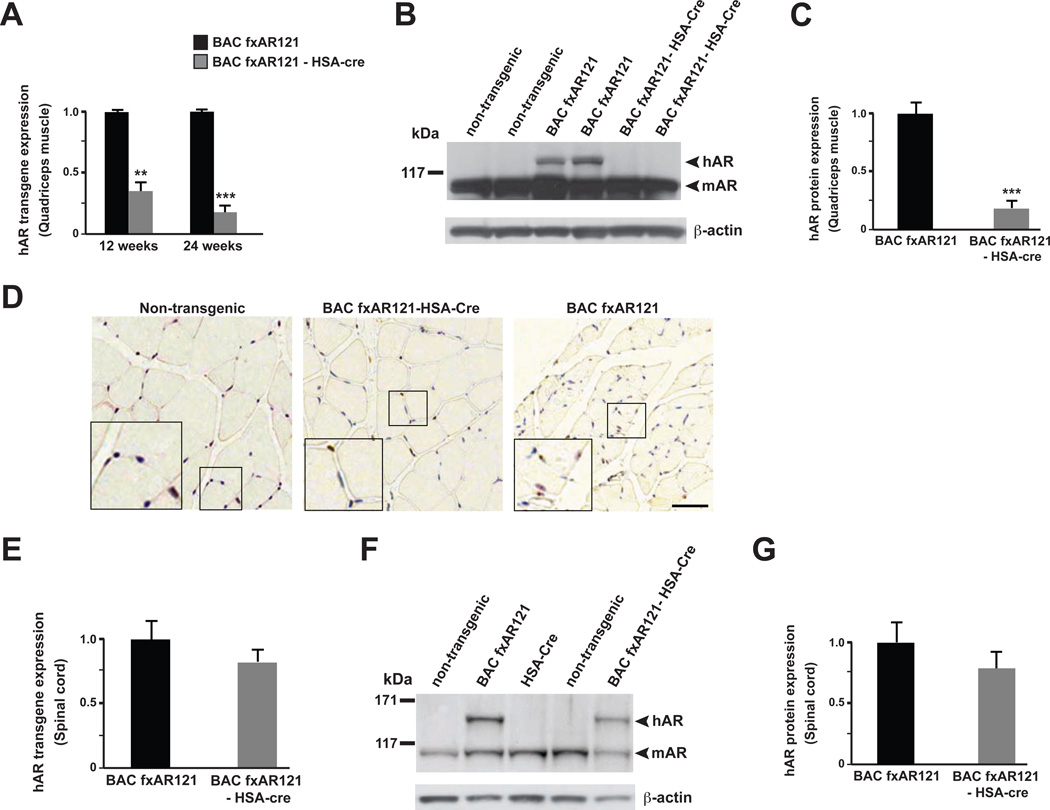
To evaluate the contribution of polyQ-AR muscle expression to SBMA disease phenotypes, we obtained mice carrying the Cre-recombinase gene placed under the control of the Human Skeletal Actin (HSA) promoter (Brennan and Hardeman, 1993). The HSA-Cre mice are known to direct efficient Cre-mediated excision of floxed transgenes in the skeletal muscle lineage only, with no ectopic Cre-excision in the spinal cord or in other CNS regions (Miniou et al., 1999). After crossing BAC fxAR121 mice with HSA-Cre mice, we obtained cohorts of mice with the following genotypes: BAC fxAR121 – HSA-Cre bigenic mice; BAC fxAR121 singly transgenic mice; HSA-Cre singly transgenic mice; and non-transgenic mice. RT-PCR analysis of RNAs isolated from quadriceps muscle obtained from the various progeny of this cross revealed marked reductions in the level of hAR transgene expression in BAC fxAR121 – HSA-Cre bigenic mice in comparison to BAC fxAR121 singly transgenic mice (Figure 4A). Immunoblot analysis corroborated the RNA expression data, as we observed minimal amounts of hAR121Q protein in muscle samples obtained from BAC fxAR121 – HSA-Cre bigenic mice (Figure 4B, 4C). AR immunostaining of quadriceps muscle sections independently confirmed a dramatic reduction in AR protein aggregates in BAC fxAR121 – HSA-Cre mice (Figure 4D). To assure the specificity of muscle excision by the HSA-Cre driver, we obtained RNA and protein lysates from spinal cord, and noted that hAR transgene RNA expression levels and hAR121Q protein levels are similar in the CNS of BAC fxAR121 – HSA-Cre mice and BAC fxAR121 singly transgenic mice (Figure 4E, 4F and 4G). These results confirmed muscle-restricted excision of the hAR transgene in BAC fxAR121 – HSA-Cre bigenic mice, indicating that BAC fxAR121 – HSA-Cre mice can be used to assess the role of muscle expression of polyQ-AR in SBMA neuromuscular disease and cellular pathology. Furthermore, Cre recombinase expression in skeletal muscle and the accompanying marked reduction of hAR transgene expression in BAC fxAR121 – HSA-Cre mice did not alter expression of endogenous mouse AR in the muscle, brain, or spinal cord of BAC fxAR121 – HSA-Cre bigenic mice (Figure S3).
Figure 4. Muscle-restricted inactivation of human AR transgene expression in BAC fxAR121 – HSA-Cre mice.
(A) We measured hAR transgene expression in quadriceps muscle in male BAC fxAR121 – HSA-Cre bigenic mice in comparison to singly transgenic male BAC fxAR121 mice (n = 3 / group) by qRT-PCR analysis. We documented a marked reduction in hAR transgene expression in BAC fxAR121 – HSA-Cre mice at 12 weeks and 24 weeks of age. Transgene expression is given in arbitrary units, with BAC fxAR121 set to 1. **P < .01, ***P < .001, t-test.
(B) Western blot analysis of quadriceps muscle for 12 week-old male BAC fxAR121 – HSA-Cre bigenic mice and littermate male BAC fxAR121 mice indicates that hAR protein expression (arrowhead) is extremely reduced in muscle from BAC fxAR121 – HSA-Cre mice, when immunoblotted with anti-AR antibody, which cross-reacts with mAR protein (arrowhead). β-actin immunoblotting serves as a loading control.
(C) Densitometry quantification of muscle protein immunoblot data shown in (B), n = 4 separate experiments. ***P < .001, t-test.
(D) AR immunostaining of quadriceps muscle sections from 28 week-old male BAC fxAR121 mice, BAC fxAR121 – HSA-Cre mice, and non-transgenic littermate controls reveals a marked reduction in AR protein inclusions at the muscle cell periphery in BAC fxAR121 – HSA-Cre mice. Scale bar = 50 µm.
(E) We measured hAR transgene expression in spinal cord from 12 week-old male BAC fxAR121 – HSA-Cre bigenic mice and BAC fxAR121 mice (n = 3 / group) by qRT-PCR analysis, and observed similar hAR expression levels. P = n.s., t-test. Transgene expression is given in arbitrary units, with BAC fxAR121 set to 1.
(F) Western blot analysis of spinal cord for 12 week-old male BAC fxAR121 – HSA-Cre bigenic mice and littermate male BAC fxAR121 mice indicates that hAR protein expression (arrowhead) is not reduced, relative to mouse AR protein expression (arrowhead) in BAC fxAR121 – HSA-Cre mice, when immunoblotted with anti-AR antibody, which cross-reacts with mouse AR protein. β-actin immunoblotting serves as a loading control.
(G) Densitometry quantification of spinal cord protein immunoblot data shown in (F), n = 4 separate experiments. P = n.s., t-test. Error bars = s.e.m.
Inactivation of hAR transgene expression in muscle prevents SBMA disease phenotypes and neuromuscular pathology without reducing AR protein aggregation in the CNS
To determine the role of polyQ-AR muscle expression in SBMA disease pathogenesis, we performed a thorough phenotyping analysis on BAC fxAR121 – HSA-Cre bigenic mice, BAC fxAR121 singly transgenic mice, and non-transgenic controls. Elimination of AR expression in muscle prevented weight loss in BAC fxAR121 – HSA-Cre bigenic mice (Figure 5A). We also documented normal motor phenotypes in BAC fxAR121 – HSA-Cre mice, as BAC fxAR121 – HSA-Cre mice displayed significantly improved grip strength, gait performance, and front limb stride length, in comparison to singly transgenic BAC fxAR121 mice (Figure 5B, 5C, and 5D). Indeed, for all tested measures, BAC fxAR121 – HSA-Cre bigenic mice performed comparably to non-transgenic controls. Lastly, although BAC fxAR121 mice had reduced survival, there was not a single instance of premature death in the BAC fxAR121 – HSA-Cre cohort (Figure 5E). Indeed, all BAC fxAR121 – HSA-Cre mice were still alive beyond one year of age, without visible phenotypes or evidence of muscle weakness.
Figure 5. Cre-mediated excision of the human AR121 transgene from skeletal muscle rescues systemic and neuromuscular phenotypes in BAC fxAR121 mice.
(A) Cre expression in skeletal muscle rescues weight loss in 20 week-old male BAC fxAR121 mice (n = 5 – 10 / group). **P < .01, ANOVA with post-hoc Tukey test.
(B) Cre expression in skeletal muscle rescues grip strength in 20 week-old male BAC fxAR121 mice (n = 5 – 10 / group). Grip strength is given in arbitrary units, with BAC fxAR121 performance set to 1. **P < .01, ANOVA with post-hoc Tukey test.
(C) Footprint analysis of 20 week-old male BAC fxAR121 mice, BAC fxAR121 – HSA-Cre mice, and littermate non-transgenic controls reveal normal stride in non-transgenic individuals (right), but obviously shortened strides in BAC fxAR121 mice (left). The stride of BAC fxAR121 – HSA-Cre mice is visibly improved (middle), more closely resembling the performance of non-transgenic mice. Fore paw prints are green, and hind paw prints are blue.
(D) Quantification of stride length analysis shown in (C), (n = 6 – 8 / group). *P < .05, ANOVA with post-hoc Tukey test, front limb. *P < .05, ANOVA with post-hoc Tukey test, hindlimb: Non tg vs. BAC fxAR121, P < .05. BAC fxAR121-HSA-Cre vs. BAC fxAR121, P = n.s.
(E) Kaplan-Meier plot of male BAC fxAR121 mice, male BAC fxAR121 – HSA-Cre bigenic mice, and male non-transgenic littermates (n = 9 – 13 / group) reveals significantly extended lifespan in BAC fxAR121 – HSA-Cre mice. P < .0001, Log-rank test. BAC fxAR121 – HSA-Cre bigenic mice are still alive and well beyond 16 months of age. Error bars = s.e.m.
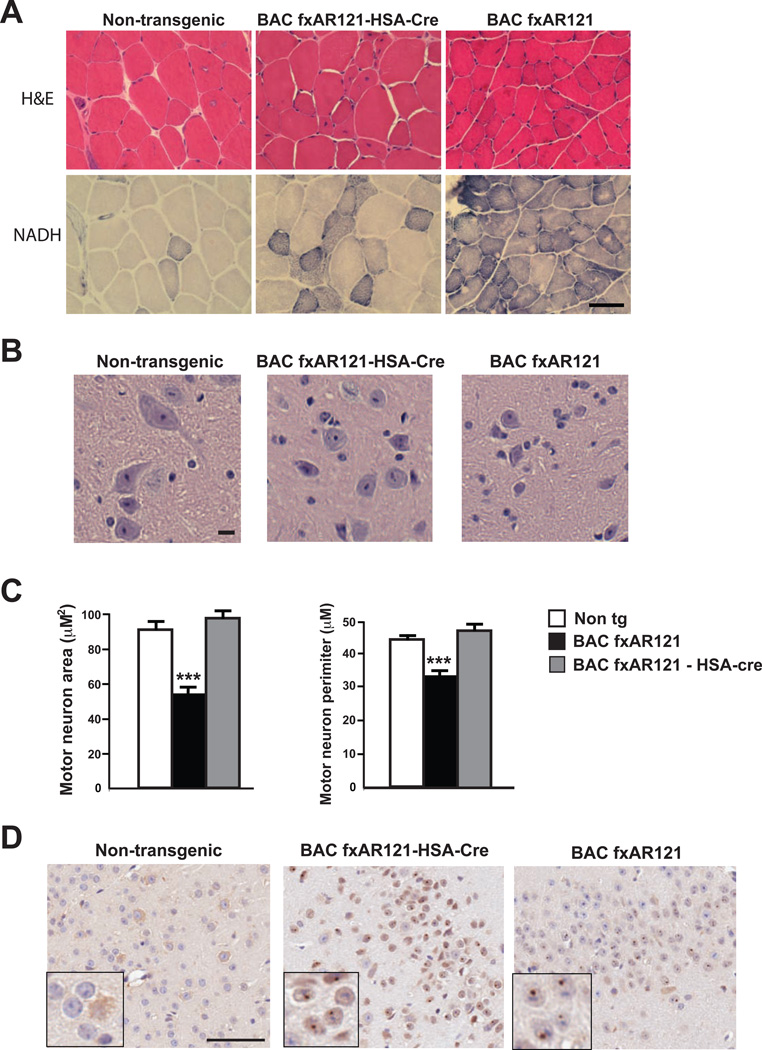
After completing behavioral paradigms for testing motor function, we considered the role of muscle expression of polyQ-AR protein in SBMA-associated neuromuscular disease pathology. Staining of muscle sections with hematoxylin and eosin revealed marked muscle fiber atrophy and increased connective tissue in BAC fxAR121 mice compared to age-matched controls, and NADH staining demonstrated considerable areas of muscle fiber atrophy in BAC fxAR121 mice (Figure 6A). Evidence of neuromuscular pathology in BAC fxAR121 – HSA-Cre bigenic mice, however, was minimal and limited (Figure 6A), consistent with a predominant role for polyQ-AR muscle expression in SBMA neuromuscular disease. To further characterize the extent of motor neuronopathy in the BAC fxAR121 mice and the role of polyQ-AR muscle expression in the production of this phenotype, we compared ventral horn regions of lumbar spinal cord sections obtained from BAC fxAR121 mice, BAC fxAR121 – HSA-Cre mice, and non-transgenic littermate controls. We noted that ventral horn neurons from BAC fxAR121 mice are reduced in size, but that ventral horn neurons in BAC fxAR121 – HSA-Cre mice appear comparable to controls (Figure 6B). Quantification of ventral horn neuron soma size and perimeter indicated that excision of mutant AR transgene expression in skeletal muscle was sufficient to completely rescue this motor neuron degeneration phenotype in BAC fxAR121 – HSA-Cre mice (Figure 6C). RT-PCR analysis and immunoblot analysis of BAC fxAR121 – HSA-Cre brain RNA and protein samples confirmed that skeletal muscle excision of hAR transgene expression in BAC fxAR121 mice does not reduce the level of hAR transgene expression in the brain (Figure S4A and S4B), as expected. Moreover, despite rescuing spinal cord neurodegeneration, excision of hAR transgene expression in muscle did not result in an appreciable reduction in AR nuclear inclusion formation in frontal cortex neurons of BAC fxAR121 – HSA-Cre mice (Figure 6D), and immunoblot analysis of brain protein lysates from BAC fxAR121 – HSA-Cre mice corroborated this finding, as insoluble, aggregated AR protein persists in brain samples from BAC fxAR121 – HSA-Cre mice (Figure S4C).
Figure 6. Excision of human AR transgene from skeletal muscle rescues neuromuscular pathology without reducing AR protein aggregate burden in CNS of BAC fxAR121 mice.
(A) Quadriceps muscle sections from 18 week-old male non-transgenic littermate controls, BAC fxAR121 – HSA-Cre mice, and BAC fxAR121 mice were stained with hematoxylin and eosin (H&E) and NADH. H&E staining of non-transgenic muscle (left) shows normal fibers of similar shape and size, but BAC fxAR121 muscle (right) reveals significant atrophy, with most fibers of markedly reduced caliber. Sections of BAC fxAR121 – HSA-Cre muscle (middle) appear relatively normal, with most fibers of large caliber, though one area of smaller fibers is present. NADH staining of non-transgenic muscle (left) yields occasional dark-staining fibers, while NADH staining of BAC fxAR121 sections (right) uncovers collections of very dark-staining fibers, undergoing atrophy. Evidence for muscle fiber atrophy is only occasionally present in BAC fxAR121 – HSA-Cre mice (middle). Scale bar = 50 µm.
(B) H&E-stained spinal cord sections from 28 week-old male BAC fxAR121 mice, BAC fxAR121 – HSA-Cre mice, and non-transgenic littermate controls reveals that ventral horn neurons in the lumbar spinal cord are reduced in size in BAC fxAR121 males, and that this size reduction phenotype is rescued in BAC fxAR121 – HSA-Cre mice. Scale bar = 50 µm.
(C) Quantification of ventral horn neuron soma size and perimeter shown in (B), n = 3 / group. ***P < .001, ANOVA with post-hoc Tukey test. Error bars = s.e.m.
(D) AR immunostaining of frontal cortex from 28 week-old male BAC fxAR121 mice, BAC fxAR121 – HSA-Cre mice, and non-transgenic littermates reveals prominent nuclear inclusions in the cortex of both BAC fxAR121 and BAC fxAR121 – HSA-Cre mice (see inset for AR nuclear aggregates, which are visualized as dark brown puncta). Scale bar = 50 µm.
Excision of hAR transgene expression in skeletal muscle prevents motor neuron axon pathology in SBMA transgenic mice
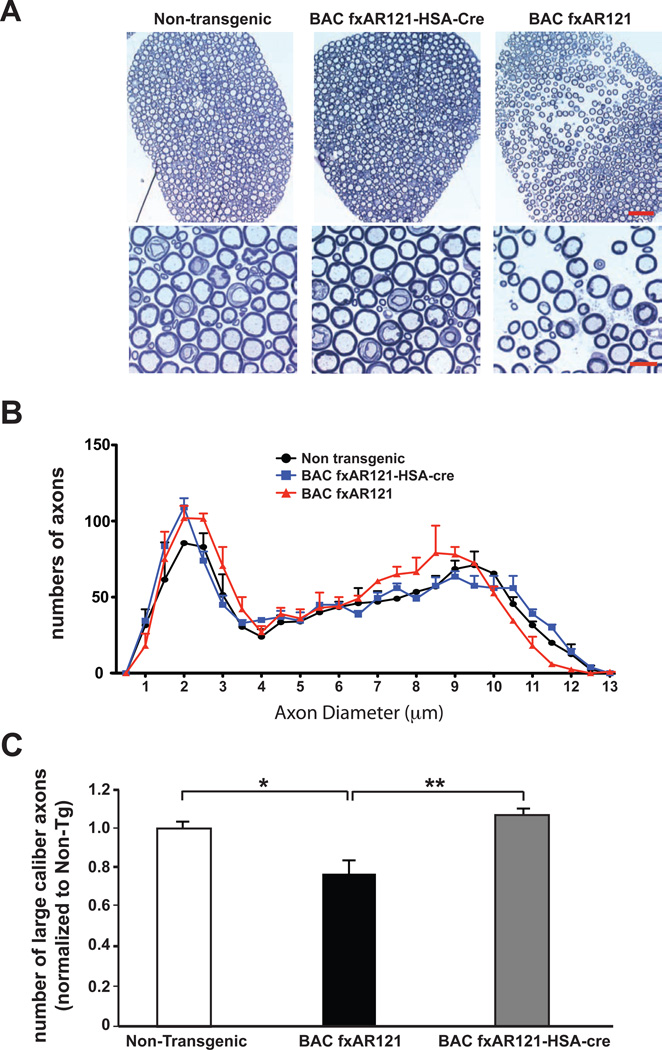
To further evaluate the effect of skeletal muscle expression of mutant AR protein on SBMA motor neuron disease, we generated sections of spinal cord motor axons from the L5 root, and observed excessive spacing of axons in spinal cord sections from end-stage BAC fxAR121 mice (Figure 7A). This aberrant spacing phenotype was absent from L5 root spinal cord motor axons in littermate BAC fxAR121 – HSA-Cre bigenic mice (Figure 7A). When we measured axon diameters in L5 motor roots and counted axon numbers based upon diameter, we noted that BAC fxAR121 mice exhibit a shift towards smaller-sized axons, but that BAC fxAR121 – HSA-Cre mice retain a distribution of motor axons that is not shifted towards smaller-sized axons and thus resemble non-transgenic controls (Figure 7B). Indeed, we documented a significant decrease in the number of large caliber axons (i.e. > 9 µm in diameter) in the BAC fxAR121 mice, and found that such large caliber axons were not reduced in L5 roots from BAC fxAR121 – HSA-Cre mice (Figure 7C), confirming that skeletal muscle excision of hAR transgene expression can prevent this SBMA motor neuronopathy phenotype in AR BAC transgenic mice. Importantly, evaluation of L5 motor roots from younger BAC fxAR121 mice, BAC fxAR121 – HSA-Cre mice, and non-transgenic controls revealed comparable numbers of motor axons and roughly equivalent numbers of large caliber axons prior to the onset of motor neuronopathy (Figure S5A and S5B), ruling out developmental variation in motor axon size selection as the basis for differences in large caliber axon numbers in older mice.
Figure 7. Excision of human AR transgene from skeletal muscle rescues motor neuron axon degeneration in BAC fxAR121 mice.
(A) We sectioned L5 spinal cord root motor axons, and observed increased spacing in 28 week-old male BAC fxAR121 mice (right). No evidence of axon spacing was present in micrographs from non-transgenic mice (left) or BAC fxAR121 – HSA-Cre littermates (middle). Scale bar, top panel = 50 µm. Scale bar, bottom = 10 µm.
(B) Mean diameters of L5 spinal cord root motor axons in 28 week-old male non-transgenic mice, BAC fxAR121 – HSA-Cre mice, and BAC fxAR121 mice.
(C) Counting of motor axons revealed a significant reduction in large caliber (> 9 µm diameter) axons in BAC fxAR121 mice. This phenotype is rescued in BAC fxAR121 – HSA-Cre bigenic mice. **P < .01, ANOVA with post-hoc Tukey test: P < .01 for BAC fxAR121-HSA-Cre vs. BAC fxAR121, P < .05 for Non-transgenic vs. BAC fxAR121. Error bars = s.e.m.
DISCUSSION
Motor neuron diseases comprise a group of neurodegenerative disorders characterized by progressive degeneration of motor neurons, accompanied by muscle weakness and wasting. Despite decades of research, the mechanistic basis of the observed motor neuron loss remains enigmatic. Although motor neuron demise is the cardinal feature of motor neuron disorders, recent investigations into the cellular basis of disease pathogenesis in ALS and autosomal recessive spinal muscular atrophy (SMA) indicate that the pathogenic cascade is not restricted to an intrinsic pathological process playing out in the motor neuron (Bricceno et al., 2012; Ilieva et al., 2009). Rather, considerable evidence indicates that many motor neuron diseases display non-cell autonomous degeneration, and for SMA, muscle cell pathology appears to significantly contribute to disease pathogenesis (Bricceno et al., 2012).
In the case of SBMA, certain features of the disease, together with previous studies in mouse models, suggested that muscle might play a key role in this motor neuronopathy (Katsuno et al., 2012b; Palazzolo et al., 2009; Yu et al., 2006); however, none of these studies definitively addressed whether muscle expression of the disease-causing polyQ-expanded AR is required for SBMA neuromuscular phenotypes. Here we provide evidence for the necessity of polyQ-AR muscle expression for the full spectrum of SBMA motor neuron disease. We find that excision of a human AR121Q transgene from muscle in BAC fxAR121 mice prevents the development of both systemic and neuromuscular SBMA disease phenotypes. Importantly, we document that BAC fxAR121 – HSA-Cre mice retain strong expression of mutant AR121Q protein in the spinal cord and brain with continued production of AR protein aggregates, indicating that CNS expression is not sufficient to yield motor neuronopathy in SBMA transgenic mice. As BAC fxAR121 – HSA-Cre mice are still healthy without signs of neuromuscular disease beyond 12 months of age, our findings indicate that expression of polyQ-AR protein in muscle is a primary driver of disease pathogenesis and progression in SBMA. Although the BAC fxAR121 mouse model recapitulates key features of SBMA disease in human patients, BAC fxAR121 mice do not exhibit motor neuron loss prior to their demise. Hence, we cannot completely exclude a role for motor neuron expressed AR in the motor neuron death occurring in SBMA patients. Nonetheless, in a concurrent study, peripheral delivery of an antisense oligonucleotide (ASO) directed against AR prevented systemic and neuromuscular phenotypes in two different SBMA mouse models – BAC fxAR121 and AR113 knock-in mice (Lieberman et al., 2014). As peripheral delivery of anti-AR ASO yielded dramatic reductions in polyQ-AR expression in skeletal muscle but did not produce knock-down of mutant AR gene expression in the CNS (Lieberman et al., 2014), this work corroborates a principal role for muscle in SBMA disease pathogenesis.
If muscle expression of polyQ-AR protein is a key site for motor neuron disease pathogenesis in SBMA, as both this genetic rescue study and the pharmacological rescue study demonstrate, could muscle also be a primary site for disease pathogenesis in related motor neuronopathies? SMA is an autosomal recessive lower motor neuron disease caused by loss-of-function mutations in the survival motor neuron (SMN) gene (Lefebvre et al., 1995), and while SMA shares certain features with SBMA, SMA patients are much more severely affected than SBMA patients. Numerous lines of investigation indicate that there are muscle abnormalities in SMA, including deficits in myoblast fusion, delayed muscle maturation, and onset of weakness before frank motor neuron loss (Murray et al., 2008; Mutsaers et al., 2011). Indeed, early studies of SMA in mouse models confirmed that loss-of-function of SMN protein restricted to muscle resulted in a severe neuromuscular phenotype, most reminiscent of muscular dystrophy (Cifuentes-Diaz et al., 2001). Furthermore, muscle-specific conditional rescue in SMA mice yields significant improvement in weight, survival, and motor behavior, but without any appreciable effect on synaptic function (Martinez et al., 2012). As gene therapy approaches relying upon transduction of muscle as a means to achieve motor neuron delivery simultaneously target muscle, the beneficial effects of SMN restoration in muscle and in motor neuron cells in mice have been difficult to separate (Bosch-Marce et al., 2011; Martinez et al., 2012). However, peripheral delivery of an ASO capable of restoring normal levels of SMN expression, by correcting the SMN2 gene splicing defect, yielded dramatic rescue of disease phenotypes in SMA mice, and far exceeded the efficacy of CNS-only delivery (Hua et al., 2011). Although enhanced liver production of IGF-1, which targets muscle and thereby affects motor neuron health, was implicated in this peripheral treatment intervention, this result nonetheless underscores the importance of non-CNS cell types in SMA disease pathogenesis and highlights the potential importance of muscle as a disease-relevant cell type in SMA (Duan et al., 2010).
The other major motor neuron disease is ALS, a combined upper motor neuron and lower motor neuron disorder. Many investigations into the cellular basis of familial ALS1 caused by SOD1 mutations have been performed in SOD1 mouse models, and these studies have firmly established that SOD1-mediated disease in ALS1 mice is non-cell autonomous, with predominant roles delineated for astrocytes, microglia, and oligodendrocytes (Boillee et al., 2006; Kang et al., 2013; Yamanaka et al., 2008). However, while overexpression of mutant SOD1 within skeletal muscle can damage it (Dobrowolny et al., 2008), excision of mutant SOD1 transgene expression in muscle in SOD1 conditional mouse models does not produce any appreciable effect on disease course (Miller et al., 2006; Towne et al., 2008), arguing against a primary role for muscle in ALS1. With the discovery of TDP-43 aggregate pathology in ALS, and subsequent realization that familial ALS can be caused by mutations in TDP-43, FUS/TLS, and most frequently GGGGCC repeat expansions in the C9orf72 gene (DeJesus-Hernandez et al., 2011; Renton et al., 2011), the cellular basis of ALS disease pathogenesis deserves further study. Indeed, recent work on multi-system proteinopathy pedigrees indicates that accumulation of misfolded proteins in muscle cells falls within the phenotype spectrum of observed histopathology in such patients (Benatar et al., 2013), suggesting that muscle pathology could be contributing to motor neuronopathy findings in certain cases.
Skeletal muscle provides trophic factor support for innervating motor neurons, and functionally interfaces with motor neurons at the neuromuscular junction, where muscle cells receive electrical impulses and modulate electrical activity through post-synaptic regulation (Tabebordbar et al., 2013). If expression of polyQ-AR protein in skeletal muscle is contributing to SBMA motor neuron degeneration, then what aspects of muscle dysfunction promote the disease process? A variety of studies, in both SBMA patients and mouse models, strongly suggest that loss of trophic factor support from muscle accounts for SBMA motor neuron degeneration. Indeed, motor neurons rely upon a number of growth factors, including vascular endothelial growth factor (VEGF) and IGF-1, which both can function as neurotrophic factors. Analysis of YAC AR100 transgenic mice revealed a marked reduction in the expression of VEGF (Sopher et al., 2004), a neurotrophic factor which, when over-expressed in muscle, can delay motor neuron disease in SOD1-expressing ALS1 transgenic mice (Azzouz et al., 2004). IGF-1, another neurotrophic growth factor shown to promote motor neuron survival in both ALS and SMA (Bosch-Marce et al., 2011; Dobrowolny et al., 2005), appears to be particularly important in SBMA muscle, as muscle-directed expression of IGF-1 can rescue systemic and neuromuscular phenotypes in SBMA mice (Palazzolo et al., 2009). Other growth factors implicated in SBMA disease pathogenesis include type II transforming growth factor-β (TGF-β), glial-derived neurotrophic factor (GDNF), and neurotrophin-4 (Katsuno et al., 2010; Yu et al., 2006). Hence, elimination of polyQ-AR expression from muscle in BAC fxAR121 – HSA-Cre mice may promote motor neuron survival by restoring the ability of skeletal muscle cells to produce normal levels of neurotrophic growth factors and then deliver them to innervating motor neurons.
Although our discovery of muscle as a crucial site for initiation of SBMA motor neuron disease illustrates that skeletal muscle cells and innervating motor neurons are inextricably linked, the most compelling aspect of this work is its immediate potential for therapeutic impact. Current therapies for SBMA are aimed at targeting the mutant AR for degradation or preventing its full activation (Katsuno et al., 2012a), and these approaches are fraught with potential for side effects. Of the most attractive therapeutic options available for patients with SBMA or patients suffering from related neurodegenerative proteinopathies, dosage reduction shows the greatest promise and is typically achieved via targeting the disease-causing RNA transcript for destruction or translation repression (La Spada, 2009). For SBMA, the prospect of targeting the AR gene in the CNS, though feasible, could be problematic, as reduced AR action in the CNS is associated with malaise, lack of focus, listlessness, and loss of libido upon pharmacological castration (Tammela, 2012). If peripheral ASO delivery, targeting muscle, offers the potential for stemming SBMA motor neuron degeneration, muscle weakness, and muscle atrophy, this would represent a dramatic paradigm shift for SBMA therapeutics. While the utility of peripheral therapy directed to muscle for related motor neuron diseases such as SMA and ALS remains controversial, further development of peripheral delivery strategies for these disorders may deserve consideration. Indeed, even neurodegenerative proteinopathies such as HD might benefit from peripheral dosage reduction therapies, as such disorders involve non-CNS metabolic pathology (Mochel et al., 2007), and in the case of PD, may stem from the production of misfolded protein species that form in the enteric nervous system and then propagate to the CNS (Wakabayashi et al., 1989). As therapies directed at knocking down disease-associated genes in the periphery are less invasive and thus potentially much safer than treatments requiring CNS delivery, our findings suggest that peripheral dosage reduction should be explored as a potential treatment option for neurodegenerative disorders.
EXPERIMENTAL PROCEDURES
Mouse studies
All animal experiments adhered to NIH guidelines and were approved by the University of Washington IACUC and UCSD IACUC. Construction of the BAC fxAR121 transgene, and derivation of BAC fxAR121 mice have been described (Sopher and La Spada, 2006). Blinded observers visually inspected all mice for general and neurological phenotypes starting at 8 weeks of age and weekly thereafter. Behavioral testing to evaluate grip length and stride length was performed as described previously (Sopher et al., 2004). Stride length measurements were obtained for each animal by a blinded investigator.
RT-PCR analysis
Total muscle, brain, or spinal cord RNA samples were isolated using the Trizol method (Life Technologies). Quantification of mRNA was performed using an Applied Biosystems 7500 Real Time Sequence Detection System with ABI Assays-on-Demand primers and TaqMan® based probes. ABI TaqMan primer and probe set designations are available upon request. PGK1 was used as the internal control to normalize results. Relative expression levels were calculated via the Standard Curve method (Livak et al., 1995).
Western Blot analysis
Protein lysates from whole brain, spinal cord, or muscle tissue were prepared as previously described (Sopher et al., 2004). We loaded 50 µg of homogenized proteins per lane, and after running 3–8% Tris-Acetate gels (Invitrogen), samples were transferred to PVDF membranes (Millipore), which were blocked in 5% milk in PBS at RT for 1 hr. Membranes were incubated with an anti-AR antibody (H280, sc-13062; AR441, Abcam) or beta-actin (ab8226, Abcam) in PBS-T with 3% BSA at 4°C overnight. The primary antibody was visualized with horseradish-peroxidase conjugated anti-rabbit or anti-mouse IgG (Santa Cruz) at 1:5,000 dilution and Enhanced Chemiluminescence (Amersham). Densitometry analysis for human and mouse AR protein quantification was performed using the NIH ImageJ software application, and normalized to β-actin signal intensity.
Histopathology studies
Quadriceps, gastrocnemius, and triceps muscles were collected immediately after euthanasia and then flash-frozen in isopentane pre-cooled in liquid nitrogen and stored at −80°C until further processing. Cryostat sections (8 µm in thickness) were stained or reacted with a standard panel of histochemical stains and reactions as previously described (Dubowitz and Shewry, 2007). AR immunostaining was performed, as previously described (Sopher et al., 2004).
For motor neuron counts, serial 20 µm transverse sections of the lumbar cord (L3–L5) were cut and Nissl stained. A blinded investigator counted the number of positively stained motor neurons in the ventral horn region on each section (n ≥ 4 sections/spinal cord). For motor neuron soma size and perimeter quantification, serial 20 µm transverse sections of the lumbar cord (L3–L5) were cut and stained with H&E. A blinded investigator traced motor neuron soma on lumbar ventral horns on each section (n ≥ 4 sections/spinal cord). Only polygonal neurons with a clearly identifiable nucleus were counted (n ≥ 100 motor neurons/genotype). Motor neuron soma area and perimeter were determined using ImageJ measurement tools, and pixel number was converted to µm or µm2, based upon the magnification scale.
Our technique for preparation of semi-thin sections of lumbar level 5 (L5) roots has been described (Arnold et al., 2013). Briefly, anesthetized mice were transcardially perfused with PBS, followed by 4% paraformaldehyde in phosphate buffer for fixation. L5 roots were collected and incubated in 2% osmium tetroxide in 0.05 M cacodylate buffer. The roots were then washed, dehydrated, and embedded for sectioning in Epon-Araldite resin (Electron Microscopy Sciences). After generating 1 µm-thick sections, L5 root sections were stained with 1% toluidine blue for 30 sec. Entire roots were imaged and cross-sectional diameter of each axon was measured with Bioquant software. Axonal diameters of motor axons from L5 roots were recorded and then subdivided into 0.5 µm bins.
Statistical analysis
All data were prepared for analysis with standard spread sheet software (Microsoft Excel). Statistical analysis was done using Microsoft Excel, Prism 4.0 (Graph Pad), or the VassarStats website < http://faculty.vassar.edu/lowry/VassarStats.html>. For ANOVA, if statistical significance (P < 0.05) was achieved, we performed post-hoc analysis to account for multiple comparisons. The level of significance (alpha) was always set at 0.05.
Supplementary Material
Acknowledgments
This work was supported by funding from the N.I.H. (R01 NS041648 to A.R.L. and R01 NS027036 to D.W.C.) and from the Muscular Dystrophy Association (Basic Research Grant to A.R.L. and Development Award to C.J.C.). D.W.C. receives salary support from the Ludwig Institute. S.-C. L. was a recipient of a National Institute of Aging training grant (T32 AG000216).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arnold ES, Ling SC, Huelga SC, Lagier-Tourenne C, Polymenidou M, Ditsworth D, Kordasiewicz HB, McAlonis-Downes M, Platoshyn O, Parone PA, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci U S A. 2013;110:E736–E745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouz M, Ralph GS, Storkebaum E, Walmsley LE, Mitrophanous KA, Kingsman SM, Carmeliet P, Mazarakis ND. VEGF delivery with retrogradely transported lentivector prolongs survival in a mouse ALS model. Nature. 2004;429:413–417. doi: 10.1038/nature02544. [DOI] [PubMed] [Google Scholar]
- Benatar M, Wuu J, Fernandez C, Weihl CC, Katzen H, Steele J, Oskarsson B, Taylor JP. Motor neuron involvement in multisystem proteinopathy: Implications for ALS. Neurology. 2013;80:1874–1880. doi: 10.1212/WNL.0b013e3182929fc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillee S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, Kassiotis G, Kollias G, Cleveland DW. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- Bosch-Marce M, Wee CD, Martinez TL, Lipkes CE, Choe DW, Kong L, Van Meerbeke JP, Musaro A, Sumner CJ. Increased IGF-1 in muscle modulates the phenotype of severe SMA mice. Hum Mol Genet. 2011;20:1844–1853. doi: 10.1093/hmg/ddr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan KJ, Hardeman EC. Quantitative analysis of the human alpha-skeletal actin gene in transgenic mice. J Biol Chem. 1993;268:719–725. [PubMed] [Google Scholar]
- Bricceno KV, Fischbeck KH, Burnett BG. Neurogenic and myogenic contributions to hereditary motor neuron disease. Neurodegener Dis. 2012;9:199–209. doi: 10.1159/000335311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Diaz C, Frugier T, Tiziano FD, Lacene E, Roblot N, Joshi V, Moreau MH, Melki J. Deletion of murine SMN exon 7 directed to skeletal muscle leads to severe muscular dystrophy. J Cell Biol. 2001;152:1107–1114. doi: 10.1083/jcb.152.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement AM, Nguyen MD, Roberts EA, Garcia ML, Boillee S, Rule M, McMahon AP, Doucette W, Siwek D, Ferrante RJ, et al. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- Custer SK, Garden GA, Gill N, Rueb U, Libby RT, Schultz C, Guyenet SJ, Deller T, Westrum LE, Sopher BL, et al. Bergmann glia expression of polyglutamine-expanded ataxin-7 produces neurodegeneration by impairing glutamate transport. Nat Neurosci. 2006;9:1302–1311. doi: 10.1038/nn1750. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolny G, Aucello M, Rizzuto E, Beccafico S, Mammucari C, Boncompagni S, Belia S, Wannenes F, Nicoletti C, Del Prete Z, et al. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Dobrowolny G, Giacinti C, Pelosi L, Nicoletti C, Winn N, Barberi L, Molinaro M, Rosenthal N, Musaro A. Muscle expression of a local Igf-1 isoform protects motor neurons in an ALS mouse model. J Cell Biol. 2005;168:193–199. doi: 10.1083/jcb.200407021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C, Ren H, Gao S. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins: roles in skeletal muscle growth and differentiation. Gen Comp Endocrinol. 2010;167:344–351. doi: 10.1016/j.ygcen.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Dubowitz V, Shewry CA. Histochemical stains and reactions. Muscle Biopsy: A Practical Approach. (3rd ed.) 2007:21–37. [Google Scholar]
- Funakoshi H, Belluardo N, Arenas E, Yamamoto Y, Casabona A, Persson H, Ibanez CF. Muscle-derived neurotrophin-4 as an activity-dependent trophic signal for adult motor neurons. Science. 1995;268:1495–1499. doi: 10.1126/science.7770776. [DOI] [PubMed] [Google Scholar]
- Garden GA, La Spada AR. Intercellular (mis)communication in neurodegenerative disease. Neuron. 2012;73:886–901. doi: 10.1016/j.neuron.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden GA, Libby RT, Fu YH, Kinoshita Y, Huang J, Possin DE, Smith AC, Martinez RA, Fine GC, Grote SK, et al. Polyglutamine-expanded ataxin-7 promotes non-cell-autonomous purkinje cell degeneration and displays proteolytic cleavage in ataxic transgenic mice. J Neurosci. 2002;22:4897–4905. doi: 10.1523/JNEUROSCI.22-12-04897.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen JA, Yu Z, Mo K, Monks DA, Lieberman AP, Breedlove SM, Jordan CL. Recovery of function in a myogenic mouse model of spinal bulbar muscular atrophy. Neurobiol Dis. 2009;34:113–120. doi: 10.1016/j.nbd.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Li Y, Fukaya M, Lorenzini I, Cleveland DW, Ostrow LW, Rothstein JD, Bergles DE. Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis. Nat Neurosci. 2013;16:571–579. doi: 10.1038/nn.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Minamiyama M, Waza M, Doi H, Kondo N, Mizoguchi H, Nitta A, Yamada K, Banno H, et al. Disrupted transforming growth factor-beta signaling in spinal and bulbar muscular atrophy. J Neurosci. 2010;30:5702–5712. doi: 10.1523/JNEUROSCI.0388-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno M, Tanaka F, Adachi H, Banno H, Suzuki K, Watanabe H, Sobue G. Pathogenesis and therapy of spinal and bulbar muscular atrophy (SBMA) Prog Neurobiol. 2012a doi: 10.1016/j.pneurobio.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Katsuno M, Tanaka F, Adachi H, Banno H, Suzuki K, Watanabe H, Sobue G. Pathogenesis and therapy of spinal and bulbar muscular atrophy (SBMA) Prog Neurobiol. 2012b;99:246–256. doi: 10.1016/j.pneurobio.2012.05.007. [DOI] [PubMed] [Google Scholar]
- La Spada AR. Getting a handle on Huntington's disease: silencing neurodegeneration. Nat Med. 2009;15:252–253. doi: 10.1038/nm0309-252. [DOI] [PubMed] [Google Scholar]
- La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010 doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lieberman A, Yu Z, Murray S, Peralta R, Low A, Shuling G, Yu XX, Cortes CJ, Bennett CF, Monia BP, et al. Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell Reports. 2014 doi: 10.1016/j.celrep.2014.02.008. [submitted]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino MM, Schneider C, Caroni P. Accumulation of SOD1 mutants in postnatal motoneurons does not cause motoneuron pathology or motoneuron disease. J Neurosci. 2002;22:4825–4832. doi: 10.1523/JNEUROSCI.22-12-04825.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- Martinez TL, Kong L, Wang X, Osborne MA, Crowder ME, Van Meerbeke JP, Xu X, Davis C, Wooley J, Goldhamer DJ, et al. Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy. J Neurosci. 2012;32:8703–8715. doi: 10.1523/JNEUROSCI.0204-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TM, Kim SH, Yamanaka K, Hester M, Umapathi P, Arnson H, Rizo L, Mendell JR, Gage FH, Cleveland DW, et al. Gene transfer demonstrates that muscle is not a primary target for non-cell-autonomous toxicity in familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2006;103:19546–19551. doi: 10.1073/pnas.0609411103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miniou P, Tiziano D, Frugier T, Roblot N, Le Meur M, Melki J. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res. 1999;27:e27. doi: 10.1093/nar/27.19.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochel F, Charles P, Seguin F, Barritault J, Coussieu C, Perin L, Le Bouc Y, Gervais C, Carcelain G, Vassault A, et al. Early energy deficit in Huntington disease: identification of a plasma biomarker traceable during disease progression. PLoS One. 2007;2:e647. doi: 10.1371/journal.pone.0000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks DA, Johansen JA, Mo K, Rao P, Eagleson B, Yu Z, Lieberman AP, Breedlove SM, Jordan CL. Overexpression of wild-type androgen receptor in muscle recapitulates polyglutamine disease. Proc Natl Acad Sci U S A. 2007;104:18259–18264. doi: 10.1073/pnas.0705501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray LM, Comley LH, Thomson D, Parkinson N, Talbot K, Gillingwater TH. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17:949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- Mutsaers CA, Wishart TM, Lamont DJ, Riessland M, Schreml J, Comley LH, Murray LM, Parson SH, Lochmuller H, Wirth B, et al. Reversible molecular pathology of skeletal muscle in spinal muscular atrophy. Hum Mol Genet. 2011;20:4334–4344. doi: 10.1093/hmg/ddr360. [DOI] [PubMed] [Google Scholar]
- Palazzolo I, Stack C, Kong L, Musaro A, Adachi H, Katsuno M, Sobue G, Taylor JP, Sumner CJ, Fischbeck KH, et al. Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron. 2009;63:316–328. doi: 10.1016/j.neuron.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramatarova A, Laganiere J, Roussel J, Brisebois K, Rouleau GA. Neuron-specific expression of mutant superoxide dismutase 1 in transgenic mice does not lead to motor impairment. J Neurosci. 2001;21:3369–3374. doi: 10.1523/JNEUROSCI.21-10-03369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopher BL, La Spada AR. Efficient recombination-based methods for bacterial artificial chromosome fusion and mutagenesis. Gene. 2006;371:136–143. doi: 10.1016/j.gene.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Sopher BL, Thomas PS, Jr, LaFevre-Bernt MA, Holm IE, Wilke SA, Ware CB, Jin LW, Libby RT, Ellerby LM, La Spada AR. Androgen receptor YAC transgenic mice recapitulate SBMA motor neuronopathy and implicate VEGF164 in the motor neuron degeneration. Neuron. 2004;41:687–699. doi: 10.1016/s0896-6273(04)00082-0. [DOI] [PubMed] [Google Scholar]
- Soraru G, D'Ascenzo C, Polo A, Palmieri A, Baggio L, Vergani L, Gellera C, Moretto G, Pegoraro E, Angelini C. Spinal and bulbar muscular atrophy: skeletal muscle pathology in male patients and heterozygous females. J Neurol Sci. 2008;264:100–105. doi: 10.1016/j.jns.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Tabebordbar M, Wang ET, Wagers AJ. Skeletal muscle degenerative diseases and strategies for therapeutic muscle repair. Annu Rev Pathol. 2013;8:441–475. doi: 10.1146/annurev-pathol-011811-132450. [DOI] [PubMed] [Google Scholar]
- Tammela TL. Endocrine prevention and treatment of prostate cancer. Mol Cell Endocrinol. 2012;360:59–67. doi: 10.1016/j.mce.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Towne C, Raoul C, Schneider BL, Aebischer P. Systemic AAV6 delivery mediating RNA interference against SOD1: neuromuscular transduction does not alter disease progression in fALS mice. Mol Ther. 2008;16:1018–1025. doi: 10.1038/mt.2008.73. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K, Takahashi H, Takeda S, Ohama E, Ikuta F. Lewy bodies in the enteric nervous system in Parkinson's disease. Arch Histol Cytol. 1989;52(Suppl):191–194. doi: 10.1679/aohc.52.suppl_191. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Chun SJ, Boillee S, Fujimori-Tonou N, Yamashita H, Gutmann DH, Takahashi R, Misawa H, Cleveland DW. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Dadgar N, Albertelli M, Gruis K, Jordan C, Robins DM, Lieberman AP. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J Clin Invest. 2006;116:2663–2672. doi: 10.1172/JCI28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.