Abstract
Huntington’s disease (HD) is caused by CAG / polyglutamine repeat expansions in the huntingtin (htt) gene, yielding proteins that misfold and resist degradation. HD belongs to a large class of neurodegenerative proteinopathies including Alzheimer’s disease, Parkinson’s disease, and tauopathies. Previous studies demonstrated that mutant htt interferes with transcriptional programs coordinated by PPARγ co-activator 1α (PGC-1α), a regulator of mitochondrial biogenesis and oxidative stress. To test if restoration of PGC-1α could treat HD, we attempted an in vivo genetic rescue in mice. We found that PGC-1α induction ameliorates HD neurodegeneration and virtually eliminates htt protein aggregation, in part by attenuating oxidative stress. Further studies revealed that PGC-1α promotes htt turnover and aggregate elimination by transactivation of TFEB, a master regulator of the autophagy-lysosome pathway, and that TFEB alone is capable of reducing htt aggregation and neurotoxicity, placing PGC-1α upstream of TFEB. PGC-1α and TFEB thus hold great promise as therapies for HD and other neurodegenerative proteinopathies.
Introduction
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder in which patients develop involuntary movements (“chorea”), suffer cognitive decline, and experience psychiatric illness (1). The disorder is relentlessly progressive, and patients succumb to their disease usually 10 to 30 years after onset. Neuropathology studies established that a region of the midbrain, known as the striatum, is principally involved in HD (2). In classic HD, significant cerebral cortex degeneration and atrophy also occur, while cerebellar, thalamic and spinal cord neuron populations are spared. HD displays anticipation, which is defined as an earlier age of onset and more rapid disease progression in successive generations within an affected pedigree. The cause of the disease is expansion of a CAG trinucleotide repeat within the first exon of the huntingtin (htt) gene (3). The CAG repeat is translated into an expanded polyglutamine (polyQ) tract in the amino-terminal region of the htt protein, and once the polyQ tract is expanded, it misfolds to adopt a pathogenic conformation. HD is one of nine inherited neurodegenerative disorders that are all caused by CAG repeats located within the coding regions of their genes (4). A considerable body of work has shown that polyQ disease proteins undergo a conformational change when the glutamine tract exceeds a certain length threshold, typically in the mid-30’s range (5). Misfolding of the polyQ disease protein is the crux of its molecular pathology, as polyQ expansion tracts from the different disease proteins can all be detected by specific antibodies, such as 1C2 (6). Although polyQ disease proteins undergo structural transformations driven by a common mutational motif, each disorder is characterized by a distinct pattern of neuropathology. As the different polyQ disease proteins exhibit widespread and overlapping patterns of expression, the mechanistic basis of this selective vulnerability remains unclear (4).
Prior to the discovery of the HD gene, several lines of evidence implicated mitochondrial dysfunction in this disorder (7), including especially studies of the mitochondrial toxin 3- nitropropionic acid (3-NP) in rodents (8). Since the characterization of the htt gene, numerous studies have extended these findings. Weight loss, despite increased caloric intake, has been documented in HD patients and mouse models (9, 10), suggestive of negative energy balance. Bioenergetics studies of striatal neurons from late-stage HD patients revealed reduced activities for key components of the oxidative phosphorylation pathway, including complexes II, III, and IV of the electron transport chain (11). Analysis of adenine nucleotide ratios strongly supports these findings, as ATP production is decreased as a function of CAG repeat length in human HD lymphoblastoid cell lines (12).
As data for mitochondrial dysfunction in HD accumulated, investigators sought a mechanistic basis for these findings. Studies of mitochondria isolated from HD patients and mice indicated that HD mitochondria depolarize at decreased calcium levels, and mutant htt protein may directly interact with mitochondria to yield this effect (13–15). However, after the discovery of the HD gene, investigators soon realized that entry of mutant htt protein to the nucleus is a crucial step in disease pathogenesis, and assembled considerable evidence for transcription dysregulation (16). While evaluating the HD N171-82Q mouse model (17) for metabolic abnormalities, we uncovered a phenotype of profound hypothermia and deranged body temperature regulation (18). This finding led us to consider a role for the transcription factor PPARγ co-activator 1α (PGC-1α) in HD, as PGC-1α is principally responsible for coordinating the adaptive thermogenesis response in rodents (19). Furthermore, PGC-1α stimulates the expression of genes required for mitochondrial energy production, while concomitantly inducing genes dedicated to countering reactive oxygen species (ROS) generated as by-products of oxidative metabolism (20, 21). PGC-1α is thus the key regulatory node in a complex network of transcription programs that culminate in mitochondrial biogenesis and enhanced mitochondrial function, making it a strong candidate for involvement in HD. Indeed, earlier work had shown that PGC-1α knock-out mice develop neurological abnormalities and show prominent neurodegeneration (22, 23). Based upon in vitro and in vivo studies of PGC-1α function in HD mice, and upon striatal RNA expression array data from patients, we and others demonstrated a central role for PGC-1α transcription interference in HD (18, 24). Taken together, these studies, and subsequent work done by another group (25), support a model for HD pathogenesis in which htt interference with PGC-1α transcription co-activation is a major contributor to the mitochondrial dysfunction (26).
To test the hypothesis that PGC-1α is a major factor in HD neurological dysfunction and neurodegeneration, we set out to determine if genetic over-expression of PGC-1α could compensate for the documented interference with PGC-1α function. We established an inducible transgenic system for PGC-1α, and used this approach to create HD transgenic mice that express increased levels of PGC-1α. We report here that not only does PGC-1α ameliorate HD neurological phenotypes, but PGC-1α also virtually eradicates htt protein aggregates in the brains of HD mice. Our studies indicate that PGC-1α’s ability to co-activate the expression of reactive oxygen species (ROS) defense genes, diminishing levels of oxidative stress, promotes ubiquitin-proteasome system function, and this contributes to htt aggregate reduction. However, further investigation of PGC-1α’s ability to enhance proteostasis led us to identify TFEB, a master regulator of the autophagy-lysosomal pathway (27), as a key target of PGC-1α. When we examined the mechanistic basis of htt protein aggregate reduction, we determined that TFEB alone is capable of preventing htt aggregation, even without PGC-1α induction, placing TFEB downstream to PGC-1α in the prevention of htt aggregation and neurotoxicity. Our findings indicate that PGC-1α and TFEB are potent therapeutic targets for HD, and likely for other neurodegenerative proteinopathies.
Results
Induction of PGC-1α expression rescues neurological phenotypes in HD transgenic mice
To determine if increased expression of PGC-1α can ameliorate HD, we developed a system to induce PGC-1α expression in transgenic mice. As over-expression of PGC-1α during development can produce disease phenotypes and result in lethality, we obtained a tet-responsive element (TRE)-PGC-1α transgenic line (28). We also obtained a line of mice genetically engineered to contain the reverse-tet transactivator (rtTA) cDNA downstream of the Rosa26 gene promoter, with a floxed STOP cassette placed between the promoter and rtTA coding sequence (29). After crossing the Rosa26-floxed STOP-rtTA line with mice carrying male germline-specific protamine 1-Cre recombinase (30), we derived F1 males with the STOP cassette excised in haploid germ cells (i.e. Rosa26-rtTA). These males were crossed to TRE-PGC-1α females to generate Rosa26-rtTA – TRE-PGC-1α bigenic mice. When Rosa26-rtTA – TRE-PGC-1α bigenic mice receive doxycycline (Dox), the rtTA becomes activated and should promote the expression of PGC-1α (fig. S1, A). To validate our induction system, we derived Rosa26-rtTA – TRE-PGC-1α bigenic mice. These mice were fed Dox for six weeks beginning at weaning, and we then measured the expression levels of PGC-1α by RT-PCR. We observed marked induction of PGC-1α in cortex and striatum, but only a moderate increase of PGC-1α in muscle and brown adipose tissue (BAT) (Fig. 1,A).
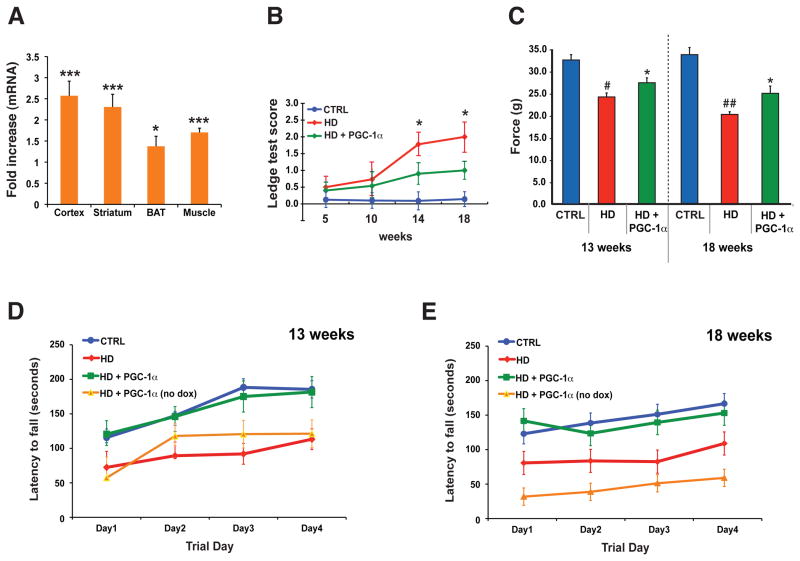
Figure 1. PGC-1α expression rescues HD neurological phenotypes.
A) PGC-1α RNA expression upon doxycycline induction in 10 week-old bigenic Rosa26-rtTA – TRE- PGC-1α mice (n = 6 / group). Fold increase is normalized to endogenous PGC-1α in controls. (* P < .05; *** P < .001). Error bars = s.e.m.
B) The ledge test, a direct measure of coordination, was performed on cohorts (n = 10 – 15 / group) of littermate controls (CTRL), HD N171-82Q mice (HD), and HD N171-82Q mice induced to express PGC-1α (HD + PGC-1α) at 14 and 18 weeks of age (* P < .05). Errors bar = s.d.
C) Grip strength analysis in HD transgenic mice (n = 13 – 15 / group). HD mice display reduced forepaw grip strength at 13 weeks of age (# P < .05), worsening further at 18 weeks of age (## P < .01). Induction of PGC-1α yields a significant improvement at 13 and 18 weeks of age (* P < .05). Each score is the mean of three tests / time point per mouse, and errors bar = s.e.m.
D) Rotarod analysis on 13 week-old cohorts (n = 10 – 12 / group), including a cohort (n = 3) of HD + PGC-1α mice that did not receive doxycycline (no dox). The HD + PGC-1α group performed comparably to controls, but significantly better than the HD and HD + PGC-1α (no dox) groups (P < .01). Error bars = s.d.
E) Rotarod analysis on 18 week-old cohorts (n = 10 – 12 / group). The HD + PGC-1α group performed comparably to controls, but significantly better than the HD and HD + PGC-1α (no dox) groups (P < .01). Error bars = s.d.
The N171-82Q mouse model recapitulates phenotype abnormalities representative of human HD (17). To test if restoration of PGC-1α function can ameliorate neurological disease in HD, we crossed N171-82Q mice with inducible PGC-1α bigenic mice, utilizing a breeding scheme that yielded three different cohorts: triple transgenic mice, HD mice (no rtTA and PGC-1α transgenes), and non-HD controls (fig. S1, B). We subjected the cohorts to behavioral testing, and noted that expression of PGC-1α, at levels consistent with prior induction, significantly improved performance on the ledge test, once HD mice become symptomatic (Fig. 1, B). PGC-1α expression improved forepaw grip strength in the HD mice (Fig. 1, C), and rectified foreleg stride length distances on gait analysis. PGC-1α expression also enabled HD triple transgenic mice to perform comparably to control mice on the rotarod, and HD triple transgenic mice, not treated with Dox, did as poorly as the HD cohort (Fig. 1, D–E).
PGC-1α prevents huntingtin protein aggregation and rescues HD neurodegeneration
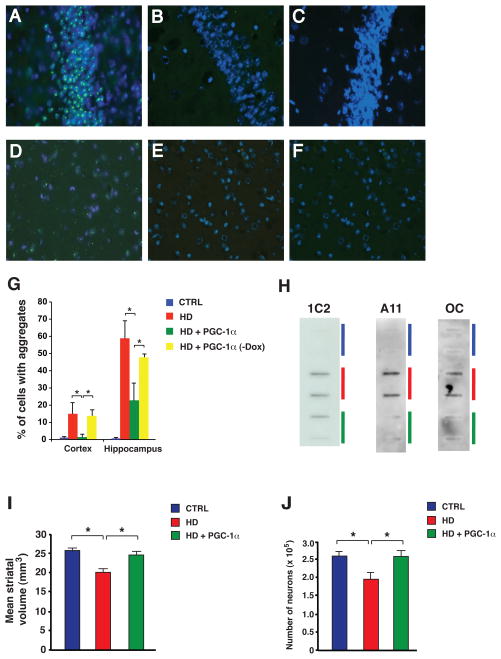
The formation of protein aggregates, visible at the light microscope level, is an established pathological hallmark in HD (62, 63). Although initial studies suggested that htt aggregates are toxic, it is now generally accepted that misfolded htt protein conformers, either in monomeric form or as soluble oligomers or fibrils, are the toxic species (31, 32). Nonetheless, while aggregates are not the toxic species, their production requires high concentrations of misfolded htt; hence, their elimination correlates with marked reductions in pathogenic htt protein (33). When we examined the brains of 18 week-old HD mice induced to express PGC-1α, we observed a dramatic reduction in htt protein aggregation in the hippocampus and cortex (Fig. 2, A–F). Quantification of neurons containing htt protein aggregates in HD, triple transgenic, and control mice confirmed this observation, and demonstrated that induction of PGC-1α in triple transgenic mice is required to achieve this outcome (Fig. 2, G). We also noted a significant reduction in htt protein aggregation in the striatum, though there were fewer cells with aggregates there. We then evaluated the brains of younger mice, and determined that prominent htt protein aggregation is apparent at 10 weeks of age, but already dramatically reduced in HD mice induced to express PGC-1α. Filter trap assay is a widely used method to measure the level of SDS-insoluble misfolded proteins (34), and a variety of antibodies are available to detect different species of amyloidogenic proteins, including 1C2 (for expanded polyQ tracts), A11 (for prefibrillar oligomers), and OC (for fibrils) (35). Using each of these antibodies, we performed filter trap assays on protein lysates isolated from the striatum of HD, triple transgenic, and control mice, and noted an obvious reduction in the level of SDS-insoluble polyQ-expanded htt protein, oligomeric htt protein, and fibrillar htt protein in HD mice induced to express PGC-1α (Fig. 2, H). We measured the levels of SDS-insoluble polyQ-expanded htt protein, oligomeric htt protein, and fibrillar htt protein, and also quantified the levels of htt transgene mRNA and soluble htt protein (fig. S2). Observed reductions in the levels of different misfolded, insoluble htt amyloidogenic species could not be attributed to an effect of PGC-1α upon HD transgene expression, as real-time RT-PCR analysis revealed similar levels of htt transgene mRNA in HD transgenic mice expressing PGC-1α and in HD transgenic mice lacking both transgenes (fig. S2). As expected, reductions in soluble htt protein levels corresponded to reductions in insoluble htt protein conformers in HD transgenic mice expressing PGC-1α (fig. S2). To determine if improved behavior and reduced htt aggregation in HD triple transgenic mice were accompanied by an amelioration of neurodegeneration, we completed a stereological assessment of the striatum, and found that induction of PGC-1α significantly increases both striatal volume and neuron number (Fig. 2, I-J; fig. S3). We also charted survival, comparing HD mice induced to express PGC-1α with their HD littermates, and did not detect any extension in overall lifespan (fig. S4, A). Closer examination of the Kaplan-Meier plot, however, revealed that there was a significant extension in lifespan for Rosa26-rtTA – HD – TRE-PGC-1α transgenic mice, when the first 50% of deaths are considered (fig. S4, A). Analysis of 20 week-old mice induced to express PGC-1α indicated reduced RNA expression, such that the level of PGC-1α in BAT was roughly equivalent to the PGC-1α level in non-induced littermates by this age. As HD N171- 82Q mice may die because of impaired thermoregulation (18), we measured the body temperature of HD mice induced to express PGC-1α and noted that body temperature did not improve in these mice (fig. S4, B). However, as CNS-restricted rescue of HD neurodegeneration can extend lifespan (36), we considered other possible explanations and found that after 20 weeks of age, HD triple transgenic mice also exhibit a modest reduction (~ 25%) in PGC-1α induction in the brain. We thus attribute the lack of lifespan extension in Rosa26-rtTA – HD – TRE-PGC-1α transgenic mice to inadequate induction of PGC-1α in BAT, combined with diminished induction of PGC-1α in the CNS.
Figure 2. PGC-1α expression prevents htt aggregate formation and rescues HD neurodegeneration.
A–F) Sections from the frontal cortex (A–C) and hippocampus CA3 region (D–F) of 18 week-old HD mice (A,D), non-HD littermate controls (B,E), and HD mice induced to express PGC-1α (C,F). Anti-htt antibody EM48 (green); DAPI (blue).
G) Quantification of htt aggregate formation in 18 week-old HD mice (* P < 0.05). Error bars = s.d.
H) Filter trap assays were performed using different antibodies that detect alternative misfolded species of htt protein. 1C2 reveals a reduction in SDS-insoluble htt protein for 18 week-old HD transgenic mice expressing PGC-1α (green), compared to HD mice lacking the PGC-1α transgene (red). A11 reveals a reduction in oligomeric htt protein for 18 week-old HD transgenic mice expressing PGC-1α (green), compared to HD mice (red). OC reveals a reduction in fibrillar htt protein for 18 week-old HD transgenic mice expressing PGC-1α (green), compared to HD mice (red). Non-HD littermate controls do not exhibit appreciable levels of SDS-insoluble, oligomeric, or fibrillar htt protein (blue).
I) Mean striatum volume in 18 week-old non-HD (CTRL), HD transgenic (HD) and HD mice induced to express PGC-1α (HD + PGC-1α). (* P < .05). Error bars = s.e.m.
J) Number of neurons in 18 week-old non-HD (CTRL), HD transgenic (HD) and HD mice induced to express PGC-1α (HD + PGC-1α). (* P < .05). Error bars = s.e.m.
PGC-1α boosts mitochondrial function and inhibits oxidative damage in HD mice
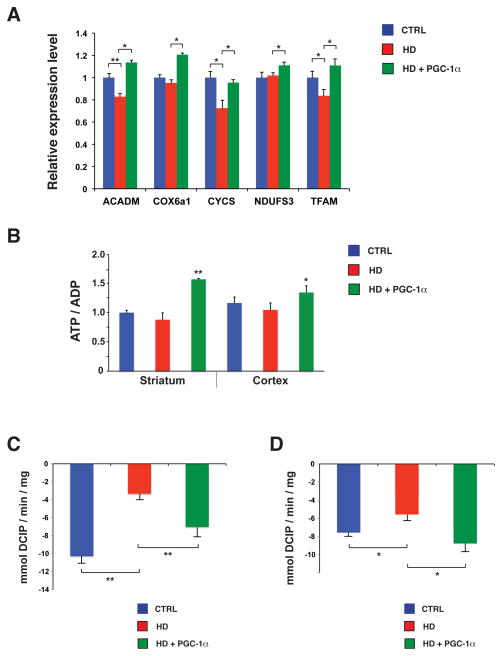
PGC-1α is a master regulator of mitochondrial biogenesis and respiration, and acts as a transcriptional co-activator that transduces physiological stimuli into specific metabolic programs (21). We, and others, have documented marked reductions in mitochondrial function in HD (18, 24, 25). To determine if PGC-1α over-expression can rescue this transcription interference, we assayed the expression levels of PGC-1α target genes in the striatum of 13 week-old HD mice, non-HD littermates, and HD mice induced to express PGC-1α. RT-PCR analysis indicated that PGC-1α induction significantly increases the expression of mitochondrial genes required for increased biogenesis and enhanced energy production (Fig. 3, A). To measure ATP content, we performed HPLC analysis of adenine nucleotides from extracts of striatum and cortex dissected from 13 week-old mice. Although the ATP/ADP ratio was only slightly reduced in HD mice, the ATP/ADP ratio was markedly increased in triple transgenic mice in striatum and cortex (Fig. 3, B). We then measured complex I activity and complex II activity, which were both decreased in HD mice, and found that PGC-1α induction significantly rescued these enzymatic activities (Fig. 3, C-D), confirming that impaired mitochondrial function can be rescued by increased PGC-1α activation. We also quantified mitochondrial DNA copy number, and found that PGC-1α significantly increased mitochondrial genome number in HD mice induced to express PGC-1α (fig. S5, A). Mitochondrial DNA copy number was reduced in ST-Hdh striatal-like cells heterozygous or homozygous for the Q111 htt allele (fig. S5, B), but transfection of a PGC-1α expression construct into ST-Hdh Q111/Q111 striatal-like cells yielded a significant increase in mitochondrial DNA copy number in this system (fig. S5, C).
Figure 3. PGC-1α expression restores mitochondrial function in HD transgenic mice.
A) Real-time RT-PCR analysis of striatal RNAs, obtained from sets (n = 4 – 6 / group) of 13 week-old HD mice, are significantly decreased in HD mice, but all five mitochondrial PGC-1α target genes are markedly increased in HD mice expressing PGC-1α (** P < .01; * P < .05). Error bars = s.e.m.
B) ATP/ADP ratios are significantly elevated upon PGC-1α induction in the striatum (** P < .01) and cortex (* P < .05). Error bars = s.e.m.
C) We measured mitochondrial complex I activity in the striatum of 13 week-old HD mice, and and observed a significant rescue of complex I activity in HD mice expressing PGC-1α (** P < .01). Error bars = s.e.m.
D) Mitochondrial complex II activity is markedly improved in the striatum of 13 week-old HD transgenic mice expressing PGC-1α (* P < .05). Error bars = s.e.m.
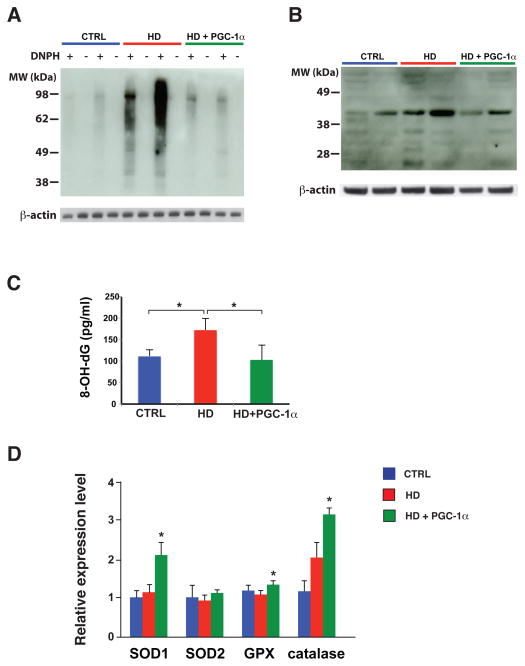
In parallel with its induction of genes that promote mitochondrial biogenesis and respiration, PGC-1α concomitantly drives the expression of genes whose products counter oxidative stress (20). Thus, PGC-1α-regulated induction of ROS defense genes prevents oxidative stress in the face of increased mitochondrial respiration. An important set of questions is whether oxidative stress contributes to HD neuronal demise, and whether PGC-1α transcription interference promotes oxidative stress in HD. To address these questions, we pursued a series of assays designed to measure oxidative protein damage, lipid peroxidation, and oxidative DNA damage. We began by performing immunoblot analysis to measure carbonyl adduct formation in striatum, and noted high levels of protein carbonyls in HD mice (Fig. 4, A). Protein carbonyl content was dramatically reduced in striatal samples from HD mice induced to express PGC-1α, however (Fig. 4, A). We then assessed lipid peroxidation by immunoblot analysis of 4-hydroxynonenal (4-HNE) adduct formation on proteins isolated from the striatum of HD, triple transgenic, and control mice. Western blot analysis revealed strong immunoreactivity for 4-HNE-containing proteins in HD striatal samples, but yielded weaker signals for HD mice induced to express PGC-1α and for non-HD controls (Fig. 4, B). Quantification of Thio-Barbituric Acid Reactive Species (TBARS) in striatal samples from HD, triple transgenic, and control mice corroborated the lipid peroxidation data obtained in the 4-HNE assay. We examined DNA oxidative damage in HD mice by measuring the levels of 8-hydroxy-2-deoxyguanosine (8-OH-dG), and noted much higher levels of 8-OH-dG in the striatum of HD mice, which were reduced to normal in HD mice over-expressing PGC-1α (Fig. 4, C). To determine if the amelioration of oxidative stress observed in Dox-treated Rosa26-rtTA – HD – TRE-PGC-1α transgenic mice stems from an increase in the expression of PGC-1α-regulated ROS defense genes, we performed RT-PCR analysis, and noted significantly increased levels of expression for three of four targets (Fig. 4, D). These findings suggest that the brains of HD mice exhibit markedly increased levels of oxidative stress, whereas induction of PGC-1α in triple transgenic mice yields levels of oxidative stress akin to normal controls, and this protection against oxidative stress correlates with recovery of PGC-1α-regulated ROS defense genes.
Figure 4. PGC-1α expression protects against HD oxidative damage by inducing reactive oxygen species defense genes.
A) Immunoblot analysis for protein carbonyl content in the striatum of 13 week-old HD transgenic mice was performed by preparing DNPH-derivatized protein lysates, and comparing with non-derivatized protein lysates in alternate lanes, as shown. We reprobed for β-actin to confirm equivalent protein loading.
B) Immunoblot analysis for lipid peroxidation via 4-hydroxynonenal (4-HNE) adduct formation in the striatum of 13 week-old HD transgenic mice. We reprobed for β-actin to confirm equivalent protein loading.
C) We measured the level of 8-OH-deoxyguanosine (8-OH-dG) in the striatum of 13 week-old HD mice, and observed a significant reduction in 8-OH-dG levels in HD mice induced to express PGC-1α (* P < .05). Error bars = s.e.m.
D) Real-time RT-PCR analysis of striatal RNAs, obtained from sets (n = 4 – 6 / group) of 13 week-old HD mice. The expression levels of most key reactive oxygen species defense genes subject to PGC-1α regulation are significantly increased in HD mice induced to express PGC-1α (* P < .05). Error bars = s.e.m.
PGC-1α amelioration of htt protein aggregation correlates with reduced oxidative stress
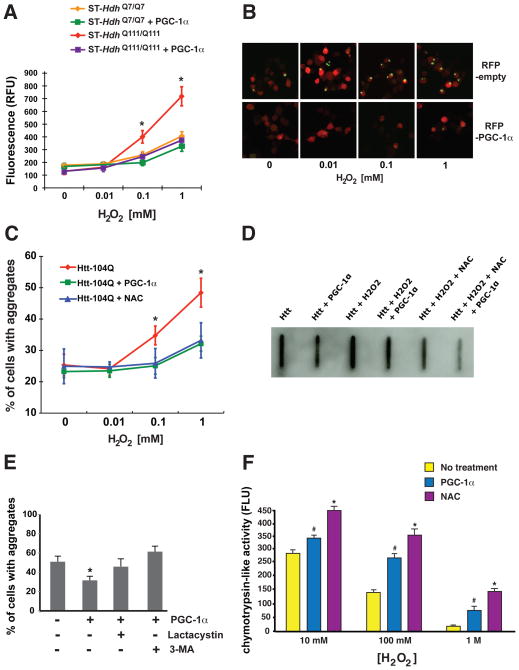
As our studies of HD mice revealed a role for PGC-1α in reducing htt aggregate formation, we investigated whether PGC-1α mediates this effect by countering oxidative stress. To test this hypothesis, we began by examining the levels of oxidative stress in ST-Hdh striatal-like cells derived from a knock-in HD mouse model that features expression of full-length htt protein (37). Quantification of oxidative stress levels indicated that ST-HdhQ111/Q111 striatal-like cells have significantly increased ROS levels compared to ST-HdhQ7/Q7 striatal-like cells (Fig. 5, A). Co-expression of PGC-1α completely normalized ROS levels in ST-HdhQ111/Q111 striatal-like cells (Fig. 5, A). To gauge the ability of oxidative stress to promote htt aggregate formation, we transfected Neuro2a cells with a GFP-tagged exon 1-htt-104Q expression construct, along with RFP-PGC-1α or RFP empty vector. After differentiating Neuro2a cells into neuron-like cells, we supplemented the media with increasing concentrations of hydrogen peroxide, and noted concentration-dependent production of protein aggregates in htt-104Q expressing cells (Fig. 5, B). Co-expression of PGC-1α, however, blocked htt aggregate formation (Fig. 5, B–C). To determine if inhibition of aggregate formation reflected reduced oxidative stress, we established a third set of Neuro2a cells in which we added the ROS scavenger N-acetyl-cysteine (NAC), and observed a significant reduction in htt aggregates, akin to the reduction achieved with PGC-1α (Fig. 5, C). We examined the effect of various conditions and treatments upon htt aggregation in transfected Neuro2a cells by filter trap assay, and found that oxidative stress promoted production of SDS-insoluble htt, and that PGC-1α and NAC reduced the level of SDS-insoluble protein (Fig. 5, D). Indeed, when expression of PGC-1α was combined with NAC treatment, we observed a marked reduction in insoluble htt protein (Fig. 5, D). To rule out an effect of oxidative stress upon CMV-driven expression of htt-104Q, we measured htt RNA levels by realtime RT-PCR analysis under different conditions, and noted levels of expression consistent with transfection efficiency.
Figure 5. PGC-1α expression counters htt protein aggregate formation induced by oxidative stress.
A) At increasing hydrogen peroxide concentrations, ST-Hdh Q111/Q111 cells exhibited significantly greater levels of ROS formation compared to ST-Hdh Q7/Q7 cells; however, over-expression of PGC-1α in ST-Hdh Q111/Q111 cells prevented ROS formation (* P < .05).
B) Neuro2a cells were transfected with an htt-104Q-eGFP expression construct, co-transfected with either an empty vector (RFP-empty) or a PGC-1α expression construct (RFP-PGC-1α). As the hydrogen peroxide concentration increased, we noted greater numbers of cells containing punctate htt-104Q staining. Co-expression of PGC-1α markedly diminished the frequency of cells containing aggregated htt.
C) Quantification of the effect of PGC-1α upon htt protein aggregate formation in Neuro2a cells exposed to oxidative stress. There is a significant reduction in the % of Neuro2a cells with htt aggregates upon PGC-1α over-expression, or when cultured in the presence of the ROS scavenger, N-acetylcysteine (NAC) (* P < .05). Error bars = s.d.
D) Filter trap assay on Neuro2a cells expressing polyQ-expanded htt protein under different. Oxidative stress promoted the formation of insoluble htt protein, while PGC-1α expression or NAC supplementation reduced insoluble htt protein. Combining PGC-1α and NAC together yielded the greatest reduction in insoluble htt protein.
E) We measured htt protein aggregate formation in Neuro2a cells expressing htt-Q82 in 1 mM hydrogen peroxide in the absence or presence of PGC-1α, lactacystin, and 3-methyladenine (3- MA). Proteasome inhibition or macroautophagy inhibition prevented PGC-1α from reducing htt protein aggregation (* P < .05). Error bars = s.e.m.
F) In the absence of oxidative stress, chymotrypsin-like activity levels are identical for untreated Neuro2a cells and PGC-1α-expressing or NAC-treated Neuro2a cells. Significant oxidative stress yields marked reductions in chymotrypsin-like activity for untreated cells (# P < .0001), but under such oxidative stress conditions, Neuro2a cells that express PGC-1α, or are exposed to NAC, retain much higher levels of chymotrypsin-like activity (** P < .01). Error bars = s.e.m.
PGC-1α amelioration of huntingtin neurotoxicity is protein-turnover pathway dependent
Although PGC-1α-mediated reduction of htt protein aggregation in Neuro2a cells is dramatic, an important question is whether decreased htt aggregate formation has any effect upon polyQ-htt neurotoxicity. This issue is crucial, as aggregate formation can be protective – if associated with a reduction in the concentration of toxic htt monomers and oligomers (38). To address this, we measured the level of apoptotic cell death in Neuro2a cells by immunostaining for activated caspase-3, as apoptotic cell death is an accepted measure of polyQ neurotoxicity (39). When we compared Neuro2a cells expressing GFP-htt-25Q or GFP-htt-104Q, we observed numerous htt- 104Q expressing cells that exhibited caspase-3 activation, while almost all htt-25Q expressing cells were negative for activated caspase-3 (fig. S6, A). We then counted htt-104Q expressing cells that were positive for activated caspase-3 upon increasing levels of oxidative stress, when treated with NAC or co-expressing PGC-1α. We found that PGC-1α over-expression or NAC treatment prevented htt-104Q dependent cell death, most significantly under maximal oxidative stress conditions (fig. S6, B). As previous work has drawn a distinction between the toxicity of diffuse htt protein vs. aggregated htt protein (31), we quantified the extent of caspase-3 activation in htt-104Q expressing Neuro2a cells that lacked visible aggregates, and in htt-104Q expressing Neuro2a cells that contained visible aggregates. When we considered htt-104Q cells lacking visible aggregates, we noted significant protection against cell death by both PGC-1α and NAC (fig. S6, C). Interestingly, we also observed significant protection against cell death by PGC-1α and NAC in htt-104Q expressing cells that contained visible aggregates (fig. S6, D), suggesting that reduced oxidative stress has neuroprotective effects, regardless of whether diffuse or aggregated forms of htt predominate.
The two major pathways for protein turnover in the CNS are the ubiquitin-proteasome system (UPS) and the macroautophagy pathway (hereafter referred to as simply autophagy). To delineate the pathway by which PGC-1α promotes htt protein turnover, we examined the role of the UPS and autophagy in htt protein aggregation. To do this, we quantified htt protein aggregation in Neuro2a cells exposed to oxidative stress, and treated PGC-1α-transfected cells with lactacystin to inhibit the UPS, or with 3-methyladenine (3-MA) to inhibit autophagosome formation. Htt-104Q expressing Neuro2a cells treated with lactacystin or 3-MA no longer displayed a reduction in htt aggregate formation when co-transfected with PGC-1α, indicating that PGC-1α-mediated htt aggregate elimination requires the UPS and autophagy pathway to be functional (Fig. 5, E). Numerous studies of polyQ neurotoxicity have shown that modulating the UPS can reduce polyQ aggregate formation and toxicity (40, 41). To test if PGC-1α-mediated aggregate reduction might involve modulation of UPS degradation function, we assayed the chymotrypsin-like activity of the proteasome in Neuro2a cells under baseline and oxidative stress conditions. Upon exposure to high concentrations of hydrogen peroxide, chymotrypsin-like activity markedly decreased; however, when Neuro2a cells expressed PGC-1α or were exposed to NAC, significantly higher levels of chymotrypsin-like activity were retained (Fig. 5, F). These findings suggest that PGC-1α-mediated mitigation of oxidative stress may promote htt protein turnover and aggregate reduction by supporting enhanced UPS activity.
PGC-1α induction of TFEB prevents htt protein aggregation
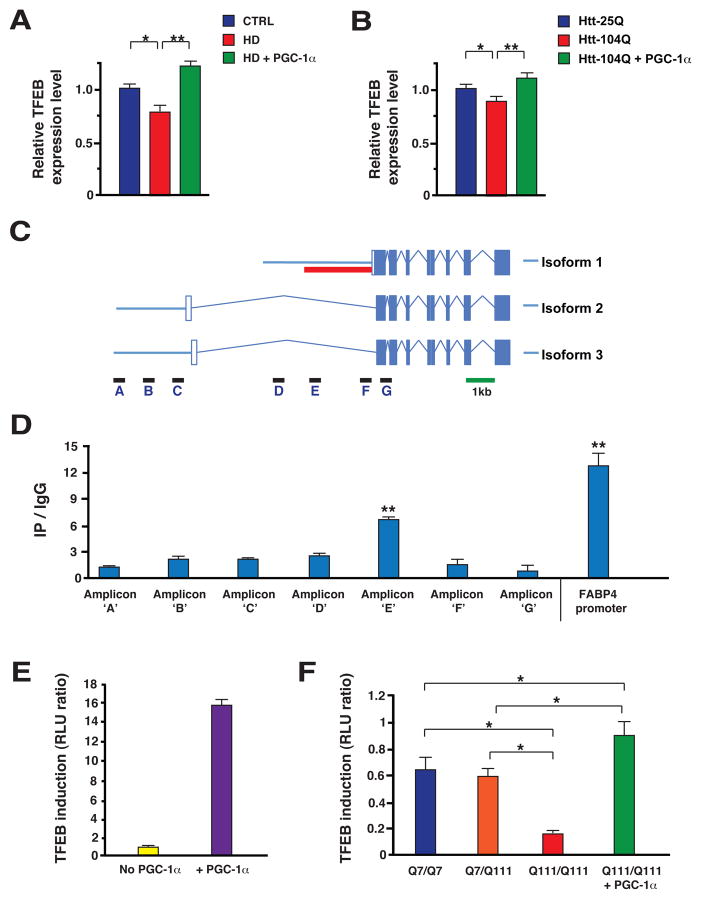
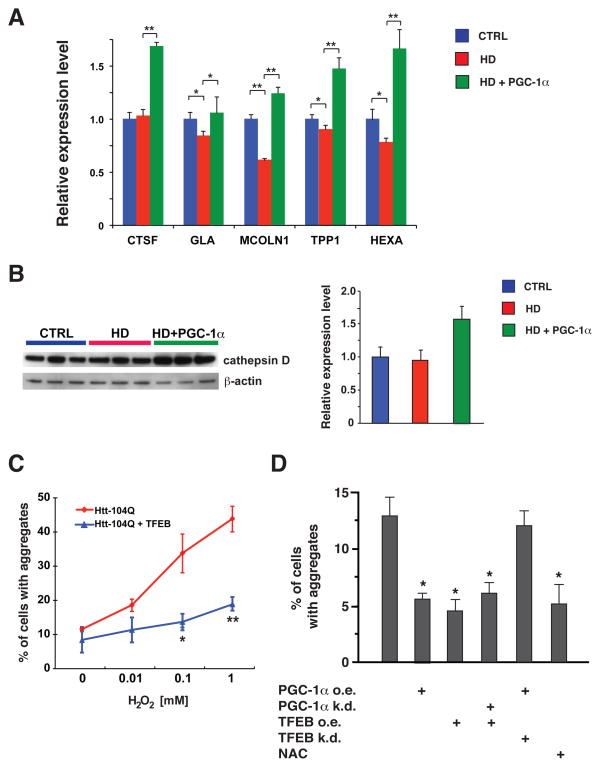
In light of the importance of autophagy for PGC-1α-mediated turnover of htt protein aggregates, we considered possible mechanisms by which PGC-1α might be promoting autophagy pathway function. Based upon very recent work, transcription factor EB (TFEB) has emerged as a master regulator of both the autophagy pathway and lysosomal biogenesis (27), leading us to test if TFEB gene expression is regulated by PGC-1α. We began our studies by measuring TFEB RNA levels, and noted a significant reduction in TFEB expression in HD mice (Fig. 6, A). This reduction in TFEB gene expression was strongly rescued in HD transgenic mice expressing PGC-1α (Fig. 6, A). We obtained corroborating results when we transfected Neuro2a cells with htt exon 1 expression constructs of different glutamine lengths, noting again that PGC-1α cotransfection strongly rescued endogenous TFEB expression in the presence of htt-104Q (Fig. 6, B). To delineate the basis of PGC-1α regulation of TFEB, we analyzed the TFEB promoter region, and found that the TFEB gene possesses at least three different transcription start sites, giving rise to three different isoforms, all with same coding exons but different 5’ UTRs (Fig. 6, C). Chromatin immunoprecipitation (ChIP) analysis revealed that PGC-1α primarily occupies the proximal promoter region for the TFEB isoform with the most 3’ transcription start site (Fig. 6, D). Based upon this finding, we generated a TFEB promoter-reporter construct linked to luciferase and then performed transactivation assays in Neuro2a cells. We found that transfection of PGC-1α into Neuro2a cells expressing this reporter construct yielded a robust induction of luciferase activity (Fig. 6, E), confirming the utility of this TFEB promoter-reporter construct for measuring PGC-1α-regulated transcription. We then performed TFEB promoter-reporter assays in ST-Hdh striatal-like cells, and observed significant reductions in TFEB transactivation in Q111/Q111 cells, compared to Q7/Q7 cells and Q7/Q111 cells (Fig. 6, F). Q111/Q111 cells transfected with PGC-1α, however, displayed a pronounced rescue of luciferase activity (Fig. 6, F), confirming a key transcription regulatory role for PGC-1α. Further studies in Neuro2a cells reinforced the importance of PGC-1α for transactivation of TFEB expression, as PGC-1α knock-down under baseline conditions, or upon sucrose treatment to induce lysosomal stress, yielded significant reductions in TFEB expression levels (fig. S7). As PGC-1α can positively regulate the expression of TFEB, we wondered if TFEB target gene expression would be altered in HD mice subject to PGC-1α rescue. To evaluate this hypothesis, we selected a subset of TFEB target genes, and we measured their RNA expression levels in the striatum of HD mice. We found that four of these five TFEB target genes displayed significantly reduced expression in HD mice, and all five TFEB target genes were induced, most markedly, in HD mice expressing PGC-1α (Fig. 7, A). Western blot analysis of cathepsin D, another TFEB target, indicated that HD mice expressing PGC-1α show increased protein levels of this TFEB target (Fig. 7, B). As TFEB has been proposed to be a master regulator of the autophagy-lysosomal pathway, we next tested if TFEB over-expression could reduce htt protein aggregate formation in Neuro2a cells subjected to increasing levels of oxidative stress. These studies revealed that TFEB over-expression is capable of dramatically reducing htt protein aggregation (Fig. 7, C), akin to what we had observed with PGC-1α over-expression (Fig. 5, C). This finding raised an important question: Does PGC-1α depend upon TFEB induction to prevent htt protein aggregation? To address this, we again measured htt aggregation in Neuro2a cells exposed to oxidative stress, but transfected the htt-104Q-expressing Neuro2a cells with a PGC-1α expression construct, alone or in combination with a TFEB shRNA knock-down construct (Fig. 7, D). In a separate set of experiments, htt-104Q-expressing Neuro2a cells were transfected with a TFEB expression construct, alone or in combination with a PGC-1α shRNA construct (Fig. 7, D). As expected, PGC-1α over-expression or TFEB over-expression each reduced htt protein aggregation; however, while TFEB over-expression reduced htt aggregates despite co-expression of a PGC-1α shRNA, PGC-1α over-expression could not significantly reduce htt protein aggregation when combined with TFEB knock-down (Fig. 7, D). These findings place PGC-1α upstream of TFEB and autophagy-lysosome pathway activation.
Figure 6. PolyQ-expanded htt interferes with PGC-1α transactivation of TFEB expression.
A) Real-time RT-PCR analysis of striatal RNAs, obtained from sets (n = 4 – 6 / group) of 13 week-old HD mice, reveals that TFEB expression is significantly decreased in HD mice (* P < .05), but markedly increased in HD mice expressing PGC-1α (** P < .01). Error bars = s.e.m.
B) TFEB expression is significantly decreased in htt-104Q-expressing Neuro2a cells (* P < .05); however, co-transfection with a PGC-1α expression construct strongly rescues polyQ-htt repression of TFEB expression (** P < .01). Error bars = s.e.m.
C) Diagram of the TFEB transcription start sites and promoter region. Boxes represent exons (with coding exons filled), solid lines correspond to 5’ and 3’ UTRs, and tented lines indicate introns. Positions of amplicons employed in the chromatin immunoprecipitation (ChIP) analysis in panel D are shown, as is the ~2 kb fragment (red solid line) that was cloned into a luciferase reporter vector to yield the TFEB promoter-reporter construct used in panels E and F.
D) Results of ChIP analysis for PGC-1α occupancy of the TFEB promoter. Isolated DNAs were subjected to qPCR analysis for a series of amplicons in the TFEB promoter to exon 1 region (panel C). PGC-1α occupancy was greatest for amplicon ‘E’ (** P < .01). Error bars = s.e.m.
E) PGC-1α transactivation of TFEB gene expression using a TFEB luciferase promoter-reporter construct containing ~2 kb of the isoform 1 proximal promoter (panel C).
F) Htt polyQ length-dependent repression of TFEB transactivation in ST-Hdh striatal-like cells. We performed transactivation assays with the TFEB promoter-reporter construct, and noted a significant repression of TFEB transactivation in Q111/Q111 cells (* P < .05). However, when we co-transfected Q111/Q111 cells with the PGC-1α expression construct, we observed a marked rescue (* P < .05). Error bars = s.e.m.
Figure 7. PGC-1α promotes TFEB target gene induction and TFEB-mediated htt aggregate reduction.
A) Real-time RT-PCR analysis of striatal RNAs, obtained from sets (n = 4 – 6 / group) of 13 week-old HD mice, reveals that the expression levels of four out of five TFEB target genes are significantly decreased in HD mice, while all five TFEB target genes are markedly increased in HD mice expressing PGC-1α (* P < .05; ** P < .01). Error bars = s.e.m.
B) Western blot analysis of cathepsin D, a TFEB target gene, reveals an increased expression of the fully processed ~32 kDa mature form in the striatum of HD mice expressing PGC-1α. Blots were reprobed for β-actin to permit normalization upon Image J analysis of band intensities, which is shown to the right.
C) Neuro2a cells were transfected with an htt-104Q-eGFP expression construct, and co-transfected with either an empty vector or a TFEB expression construct. As the hydrogen peroxide concentration increased, we detected greater numbers of cells containing punctate htt- 104Q staining. Co-expression of TFEB markedly diminished the % of cells containing aggregated htt (* P < .05; ** P < .01). Error bars = s.e.m.
D) Neuro2a cells were cultured at 0.1 mM hydrogen peroxide, transfected with a htt-104Q-eGFP expression construct, and co-transfected with a PGC-1α expression construct (PGC-1α o.e.), PGC-1α shRNA knock-down construct (PGC-1α k.d), TFEB expression construct (TFEB o.e.), or TFEB shRNA knock-down construct (TFEB k.d.), as indicated. We noted a significant reduction in htt aggregates upon PGC-1α over-expression, TFEB over-expression, and TFEB over-expression despite simultaneous PGC-1α knock-down (* P < .05). However, PGC-1α over-expression in the presence of TFEB knock-down did not reduce htt aggregate formation. N-acetylcysteine (NAC) is a positive control for aggregate reduction (* P < .05). Error bars = s.e.m.
Discussion
The recognition of protein misfolding as a shared feature of all neurodegenerative diseases, inherited or sporadic, represented a fundamental advance in our understanding of these disorders. HD is but one member of a large class of neurodegenerative proteinopathies that includes Alzheimer’s disease (AD), Parkinson’s disease (PD), prion diseases, amyotrophic lateral sclerosis, and tauopathies (42). These disorders are all characterized by an age-dependent process of neuronal dysfunction and demise, presumably initiated by the cell’s inability to deal with a specific misfolded protein stress. Why are neurons, and other highly specialized CNS cell types, exquisitely susceptible to degeneration in these different diseases? An answer to this question may lie in the fact that neurons, and other specialized CNS cells, are unique because such cells: 1) are post-mitotic; 2) constantly demand high levels of energy; and 3) need to maintain protein quality control throughout a bipolar, elongated cell body. For these reasons, any process that disrupts mitochondrial function, either at the level of bioenergetics capacity, or organelle / protein quality control, tends to preferentially compromise neurological function, resulting in neurodegeneration (43, 44).
HD is a neurodegenerative disorder characterized by selective vulnerability of the striatum and cortical projection neurons. An extensive literature has established that impaired energy metabolism and mitochondrial dysfunction are prominent features of HD pathogenesis (18, 24, 25, 45). Here we evaluated the contribution of impaired PGC-1α function to HD pathogenesis by attempting a genetic rescue using an inducible, bigenic PGC-1α expression system. After validating the system, we performed a behavioral and neuropathological analysis of HD mice induced to express PGC-1α. Our results indicate that PGC-1α up-regulation can restore normal motor and coordination function, and prevent neuron loss in the striatum. Analysis of PGC-1α-regulated genes and energy production revealed a recovery of target gene expression, complex I and II activities, and ATP generation. These compelling findings strongly support a role for PGC-1α transcription interference in HD, and confirm that PGC-1α deserves to be considered as a therapeutic target in HD. As PGC-1α boosted mitochondrial function and reduced oxidative stress, our results suggest that PGC-1α may also have therapeutic application in related neurodegenerative proteinopathies, such as PD. This view is supported by a study in which a meta-GSEA analysis of 17 microarray data sets from PD brains identified coordinate down-regulation of 425 PGC-1α target genes as a significantly shared feature in PD, and showed that PGC-1α over-expression could mitigate α-synuclein and rotenone toxicity in neurons (46). Moreover, another group has documented diminished PGC-1α function in PD, and linked the PGC-1α dysfunction to altered degradation of a novel parkin substrate, known as PARIS (47).
While we anticipated that PGC-1α expression might reverse HD neurological phenotypes and neurodegeneration, our analysis of HD neuropathology yielded an unexpected result – PGC-1α virtually eliminated htt protein aggregates. We hypothesized that one possible explanation for this effect is PGC-1α’s ability to reduce oxidative stress. Human HD brains exhibit signs of oxidative damage, consistent with high levels of oxidative stress (48). To determine if such oxidative damage is recapitulated in the striatum of HD N171-82Q mice, we measured protein carbonyl adduct formation, lipid peroxidation, and oxidative DNA damage, and observed strong evidence of oxidative damage. In the striatum of HD mice induced to express PGC-1α, markers of oxidative damage were not increased, but instead were similar to levels detected in non-HD controls. Real-time RT-PCR analysis indicated that the expression of ROS defense genes, subject to PGC-1α induction, was significantly increased. We also measured ROS levels in ST-Hdh striatal-like cells from HD knock-in mice, and observed a roughly three-fold increase in ROS levels in ST-HdhQ111/Q111 cells. Transfection of a PGC-1α expression construct into ST-HdhQ111/Q111 cells normalized ROS levels, confirming that PGC-1α is capable of countering oxidative stress associated with htt neurotoxicity. While reduced oxidative stress is likely neuroprotective, we tested if a connection exists between oxidative stress and htt protein aggregation. We observed a marked increase in htt-104Q aggregate formation in differentiated Neuro2a cells under conditions of increasing oxidative stress, and noted that co-expression of PGC-1α prevented htt protein aggregation. When we tested if the ROS scavenger NAC could block oxidative stress-dependent htt-104Q aggregate formation, we observed strong inhibition of htt aggregation. Filter trap analysis corroborated these results, and support the conclusion that htt protein aggregation is favored by oxidative stress conditions. As a pre-clinical trial of NAC in α-synuclein mice markedly reduced α-synuclein accumulation and prevented loss of dopaminergic nerve terminals and striatal neurodegeneration (49), attenuation of oxidative stress may have the potential to ameliorate neurodegenerative proteinopathies.
One of the major challenges for post-mitotic neurons is to maintain protein quality control. To eliminate misfolded proteins, neurons rely upon a properly functioning UPS and autophagy-lysosomal pathway. If either of these protein turnover pathways is incapacitated, then neuronal dysfunction and neurodegeneration will result (50). When we evaluated the role of the UPS and the autophagy pathway in PGC-1α-dependent amelioration of htt protein aggregation, we found that a fully functional UPS and autophagy pathway are necessary. As oxidative stress can inhibit UPS function by inactivation of proteasome subunits through direct oxidative modification (51–53), or impair the UPS by presenting it with an excessive load of oxidatively damaged substrates (54), we hypothesized that PGC-1α-mediated mitigation of oxidative stress might enhance UPS function, thereby contributing to htt protein turnover and aggregate reduction. When we directly studied the effect of oxidative stress upon one key measure of UPS enzymatic activity, we noted that oxidative stress dramatically inhibited this proteasomal activity, but expression of PGC-1α or treatment with NAC provided a significant rescue of the enzymatic activity in Neuro2a cells.
Although our findings implicate reduced oxidative stress as a factor in the elimination of htt protein aggregates by PGC-1α, it seemed unlikely that this alone could account for PGC-1α’s profound anti-aggregation effect. As a normally functioning autophagy pathway is critical for maintaining protein quality control in the CNS (50), we hypothesized that a potential link might exist between PGC-1α transactivation and enhanced autophagy-lysosome pathway activity. In 2009, a transcription factor known as TFEB was identified as a key regulator of lysosome biogenesis (55, 56). Further studies then demonstrated that TFEB promotes the expression of genes in the autophagy pathway in addition to genes encoding components of the lysosome (27), indicating that TFEB is a major node in the regulation of the entire autophagy-lysosome pathway. To evaluate the role of TFEB in PGC-1α-mediated htt protein turnover, we measured TFEB expression levels in HD in vitro and in vivo models, and found that polyQ-expanded htt repressed the levels of TFEB gene expression – an effect that was rescued in both cases by PGC-1α. ChIP analysis of TFEB promoter regions associated with different transcription start sites localized PGC-1α occupancy to a proximal region of one of these promoters, enabling us to derive a TFEB promoter-reporter construct. When we studied TFEB transactivation in ST-Hdh striatal-like cells with this promoter-reporter construct, we observed significant repression of TFEB promoter activity in Q111/Q111 homozygous cells, and again documented that PGC-1α expression could dramatically rescue this repression. We then tested if TFEB expression could prevent htt protein aggregation in Neuro2a cells expressing htt-104Q protein under conditions of oxidative stress, and observed marked TFEB-dependent reductions in htt aggregate formation. To clarify the pathway relationship between PGC-1α and TFEB in the suppression of htt protein aggregation, we tested if PGC-1α required TFEB to limit htt aggregate formation or if TFEB required PGC-1α to limit htt aggregate formation. Importantly, we found that TFEB is capable of reducing htt aggregation when PGC-1α is knocked down, but that PGC-1α in the presence of TFEB knock-down no longer reduced htt aggregation. Our findings indicate that PGC-1α promotes htt protein turnover and aggregate suppression by co-activating the expression of TFEB, and places PGC-1α upstream of TFEB in the transcriptional regulation of the autophagy-lysosome pathway.
Why should PGC-1α co-activation be linked to enhanced autophagy pathway function? As PGC-1α promotes mitochondrial biogenesis and increased mitochondrial metabolic activity, the up-regulation of mitochondrial number and mass required to achieve higher energy production likely results in a proportionately greater accumulation of damaged mitochondria that need to be turned over. Consequently, enhanced autophagy-lysosome pathway function would be required to accommodate this increased need for mitochondrial turnover via autophagy – a process known as mitophagy (57). Although the transcription factor(s) that PGC-1α co-activates to promote autophagy-lysosome pathway function remain unknown, the PPARs are likely candidates, since they are potent inducers of mitochondrial biogenesis and mitochondrial activity in a wide range of cell types (58). As neurons are continually on the edge for energy production and protein quality control, our findings highlight the importance of PGC-1α function and TFEB action in neurodegenerative disease, and establish PGC-1α and TFEB as attractive therapeutic targets with the potential for treating a broad range of neurodegenerative disorders characterized by protein misfolding, including HD.
Materials and Methods
Mouse breeding and behavioral studies
All animal experiments adhered to NIH guidelines and were approved by the University of Washington IACUC and UCSD IACUC. Rosa26-floxed STOP-rtTA mice and protamine-1 Cre transgenic mice were obtained from the Jackson laboratory. The TRE-PGC-1α mice were obtained from the Kelly lab (28). All transgenic and gene-targeted lines were crossed onto the C57BL/6J strain background for more than 10 generations before directed breeding experiments. Behavioral studies and survival analysis were performed as previously described (18, 59, 60).
Neuropathology analysis
Mice were euthanized and perfused as previously described (59, 60), and frozen parasagittal or coronal sections were cut at 10 μm thickness on a sliding microtome. After permeabilization with 0.05% TritonX/PBS for 10 min, slides were blocked with 5% normal goat serum (Vector lab, Burlingame, CA) and 1% fetal bovine serum (Sigma) for 1 hr and then incubated with primary antibody EM-48 (1:50, Millipore) for one hour, washed with PBS 3x and incubated with secondary antibody for 1 hr. Nuclear staining was achieved with Hoechst 33342 (Molecular Probes, Eugene, OR). Confocal imaging analysis was performed on a Zeiss Axiovert 200M inverted microscope (Carl Zeiss, Inc., Thornwood, NY). Quantification of aggregation was performed by counting the number of cells in the field with punctate EM-48 staining, and dividing by the total number of cells in the field. For each brain region, we analyzed five sections / individual, and studied at least six mice / cohort. For stereology analysis, we used the Cavalieri method (61). Briefly, the region of interest in each section was selected with a 4x objective on an Olympus BX55 microscope (Olympus, Denmark). Each region was then divided into randomly selected squares by Stereo-Investigator software (MBF bioscience). An average of 25 squares / area in each section was used to count the total number of nucleoli-containing neurons using a 100x oil objective. Total number of neurons was calculated according to the optical fractionator (62). Volume of the striatum was determined from serial section analysis using point counting and Cavalieri’s rule. Images from sections were captured at 12.5x and projected onto a video monitor. Point counting was performed as above. Volume was computed separately for the right and left sides and corrected for shrinkage (56). For striatal volume, sections were stained with cresyl violet as previously described, and neurons were differentiated from glial cells by size (63). In all cases, the scorer was blinded to the cohort status of the mice.
Vector constructs
The htt expression constructs have been previously described (18). For expression of PGC-1α, we cloned a mouse PGC-1α cDNA (kind gift of D.E. Kelly) into the multiple cloning site of the pTAG-RFP vector (Evrogen; Moscow, Russia). We then inserted the CMV promoter with a Kozak sequence between the stop codon of RFP and the PGC-1α cDNA. We confirmed RFP and PGC-1α expression by Western blot analysis. The TFEB expression construct was obtained from Origene (Rockville, MD). To derive the TFEB promoter-report construct, we PCR-amplified a 2 kb proximal promoter fragment from mouse BAC RP23-205M10 DNA (BACPAC Resources Center; Oakland, CA), and inserted this fragment into the Nhe I and Hind III restriction sites in the pGL3-Basic vector (Promega). For PGC-1α and TFEB knock-down, we screened a series of PGC-1α and TFEB shRNA’s (MISSION® shRNA, Sigma), and noted superior knock-down with PGC-1α shRNA TRCN0000095313 and TFEB shRNA TRCN0000085548, which were used for all subsequent experimentation.
Statistical analysis
All data were prepared for analysis with standard spread sheet software (Microsoft Excel). Statistical analysis was done using Microsoft Excel, Prism 4.0 (Graph Pad), or the VassarStats website <http://faculty.vassar.edu/lowry/VassarStats.html>. For ANOVA, if statistical significance (p < 0.05) was achieved, we performed post-hoc analysis to account for multiple comparisons. The level of significance (alpha) was always set at 0.05.
Supplementary Material
Acknowledgments
We thank G. MacDonald, A.C. Smith, K. Saijo, C.K. Glass, and B.L. Sopher for technical assistance, D.E. Kelly for the PGC-1α expression construct and the TRE-PGC-1α transgenic mice, and C.G. Glabe for the A11 and OC antibodies. This work was supported by funding from the Hereditary Disease Foundation, the CHDI, and grants from the N.I.H. (R01 AG033082 and R01 NS065874 to A.R.L., P01 HL034322 to E.R.L., and R01 AG018440, R01 NS057096 and R01 AG022074 to E.M.)
Footnotes
The authors declare no competing interests.
A.R.L. designed experiments, provided funding, and wrote the paper. T.T. designed and performed all experiments, and wrote the paper. T.D.A., B.E.M., K.R.S., J.A., and R.A.V.R. assisted with the performance of the experiments. V.A.D. assisted with mouse experiment performance and design. E.R.L. and E.M. performed experiments, analyzed results, and contributed data.
References
- 1.Nance MA. Clinical aspects of CAG repeat diseases. Brain Pathol. 1997;7:881–900. doi: 10.1111/j.1750-3639.1997.tb00892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Huntington’s, Disease, Collaborative, Research, Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 4.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paulson HL. Protein fate in neurodegenerative proteinopathies: polyglutamine diseases join the (mis)fold. Am J Hum Genet. 1999;64:339–345. doi: 10.1086/302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, Saudou F, Weber C, David G, Tora L, et al. Polyglutamine expansion as a pathological epitope in Huntington’s disease and four dominant cerebellar ataxias. Nature. 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- 7.Beal MF, Hyman BT, Koroshetz W. Do defects in mitochondrial energy metabolism underlie the pathology of neurodegenerative diseases? Trends Neurosci. 1993;16:125–131. doi: 10.1016/0166-2236(93)90117-5. [DOI] [PubMed] [Google Scholar]
- 8.Beal MF, Brouillet E, Jenkins BG, Ferrante RJ, Kowall NW, Miller JM, Storey E, Srivastava R, Rosen BR, Hyman BT. Neurochemical and histologic characterization of striatal excitotoxic lesions produced by the mitochondrial toxin 3-nitropropionic acid. J Neurosci. 1993;13:4181–4192. doi: 10.1523/JNEUROSCI.13-10-04181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aziz NA, van der Burg JM, Landwehrmeyer GB, Brundin P, Stijnen T, Roos RA. Weight loss in Huntington disease increases with higher CAG repeat number. Neurology. 2008;71:1506–1513. doi: 10.1212/01.wnl.0000334276.09729.0e. [DOI] [PubMed] [Google Scholar]
- 10.Menalled LB, Chesselet MF. Mouse models of Huntington’s disease. Trends Pharmacol Sci. 2002;23:32–39. doi: 10.1016/s0165-6147(00)01884-8. [DOI] [PubMed] [Google Scholar]
- 11.Browne SE, Beal MF. The energetics of Huntington’s disease. Neurochem Res. 2004;29:531–546. doi: 10.1023/b:nere.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- 12.Seong IS, Ivanova E, Lee JM, Choo YS, Fossale E, Anderson M, Gusella JF, Laramie JM, Myers RH, Lesort M, MacDonald ME. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 13.Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 14.Orr AL, Li S, Wang CE, Li H, Wang J, Rong J, Xu X, Mastroberardino PG, Greenamyre JT, Li XJ. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 16.Riley BE, Orr HT. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 2006;20:2183–2192. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- 17.Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, Slunt HH, Ratovitski T, Cooper JK, Jenkins NA, Copeland NG, Price DL, Ross CA, Borchelt DR. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 18.Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, Gilbert ML, Morton GJ, Bammler TK, Strand AD, Cui L, Beyer RP, Easley CN, Smith AC, Krainc D, Luquet S, Sweet IR, Schwartz MW, La Spada AR. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A coldinducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 20.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 21.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 22.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Chaturvedi RK, Adhihetty P, Shukla S, Hennessy T, Calingasan N, Yang L, Starkov A, Kiaei M, Cannella M, Sassone J, Ciammola A, Squitieri F, Beal MF. Impaired PGC-1alpha function in muscle in Huntington’s disease. Hum Mol Genet. 2009;18:3048–3065. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenamyre JT. Huntington’s disease--making connections. N Engl J Med. 2007;356:518–520. doi: 10.1056/NEJMcibr067022. [DOI] [PubMed] [Google Scholar]
- 27.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, Medeiros DM, Valencik ML, McDonald JA, Kelly DP. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- 29.Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci U S A. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 32.Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol. 2004;11:1215–1222. doi: 10.1038/nsmb860. [DOI] [PubMed] [Google Scholar]
- 33.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 34.Muchowski PJ, Ning K, D’Souza-Schorey C, Fields S. Requirement of an intact microtubule cytoskeleton for aggregation and inclusion body formation by a mutant huntingtin fragment. Proc Natl Acad Sci U S A. 2002;99:727–732. doi: 10.1073/pnas.022628699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayed R, Head E, Sarsoza F, Saing T, Cotman CW, Necula M, Margol L, Wu J, Breydo L, Thompson JL, Rasool S, Gurlo T, Butler P, Glabe CG. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beal MF, Ferrante RJ. Experimental therapeutics in transgenic mouse models of Huntington’s disease. Nat Rev Neurosci. 2004;5:373–384. doi: 10.1038/nrn1386. [DOI] [PubMed] [Google Scholar]
- 37.Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto S, Pouladi MA, Talantova M, Yao D, Xia P, Ehrnhoefer DE, Zaidi R, Clemente A, Kaul M, Graham RK, Zhang D, Vincent Chen HS, Tong G, Hayden MR, Lipton SA. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat Med. 2009;15:1407–1413. doi: 10.1038/nm.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young JE, Garden GA, Martinez RA, Tanaka F, Sandoval CM, Smith AC, Sopher BL, Lin A, Fischbeck KH, Ellerby LM, Morrison RS, Taylor JP, La Spada AR. Polyglutamine-expanded androgen receptor truncation fragments activate a Bax-dependent apoptotic cascade mediated by DP5/Hrk. J Neurosci. 2009;29:1987–1997. doi: 10.1523/JNEUROSCI.4072-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cummings CJ, Reinstein E, Sun Y, Antalffy B, Jiang Y, Ciechanover A, Orr HT, Beaudet AL, Zoghbi HY. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron. 1999;24:879–892. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 41.Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 42.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 43.Kaplan J. Spinocerebellar ataxias due to mitochondrial defects. Neurochem Int. 2002;40:553–557. doi: 10.1016/s0197-0186(01)00127-9. [DOI] [PubMed] [Google Scholar]
- 44.Schon EA, Santra S, Pallotti F, Girvin ME. Pathogenesis of primary defects in mitochondrial ATP synthesis. Semin Cell Dev Biol. 2001;12:441–448. doi: 10.1006/scdb.2001.0281. [DOI] [PubMed] [Google Scholar]
- 45.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 46.Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA, Grunblatt E, Moran LB, Mandel SA, Riederer P, Miller RM, Federoff HJ, Wullner U, Papapetropoulos S, Youdim MB, Cantuti-Castelvetri I, Young AB, Vance JM, Davis RL, Hedreen JC, Adler CH, Beach TG, Graeber MB, Middleton FA, Rochet JC, Scherzer CR. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) Repression of PGC-1alpha Contributes to Neurodegeneration in Parkinson’s Disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Browne SE, Ferrante RJ, Beal MF. Oxidative stress in Huntington’s disease. Brain Pathol. 1999;9:147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clark J, Clore EL, Zheng K, Adame A, Masliah E, Simon DK. Oral N-acetyl-cysteine attenuates loss of dopaminergic terminals in alpha-synuclein overexpressing mice. PLoS One. 2010;5:e12333. doi: 10.1371/journal.pone.0012333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friguet B, Stadtman ER, Szweda LI. Modification of glucose-6-phosphate dehydrogenase by 4-hydroxy-2-nonenal. Formation of cross-linked protein that inhibits the multicatalytic protease. J Biol Chem. 1994;269:21639–21643. [PubMed] [Google Scholar]
- 52.Okada K, Wangpoengtrakul C, Osawa T, Toyokuni S, Tanaka K, Uchida K. 4- Hydroxy-2-nonenal-mediated impairment of intracellular proteolysis during oxidative stress. Identification of proteasomes as target molecules. J Biol Chem. 1999;274:23787–23793. doi: 10.1074/jbc.274.34.23787. [DOI] [PubMed] [Google Scholar]
- 53.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276:30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 54.Malkus KA, Tsika E, Ischiropoulos H. Oxidative modifications, mitochondrial dysfunction, and impaired protein degradation in Parkinson’s disease: how neurons are lost in the Bermuda triangle. Mol Neurodegener. 2009;4:24. doi: 10.1186/1750-1326-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 56.Rosen GD, Williams RW. Complex trait analysis of the mouse striatum: independent QTLs modulate volume and neuron number. BMC Neurosci. 2001;2:5. doi: 10.1186/1471-2202-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Batlevi Y, La Spada AR. Mitochondrial autophagy in neural function, neurodegenerative disease, neuron cell death, and aging. Neurobiol Dis. 2011;43:46–51. doi: 10.1016/j.nbd.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 59.Garden GA, Libby RT, Fu YH, Kinoshita Y, Huang J, Possin DE, Smith AC, Martinez RA, Fine GC, Grote SK, Ware CB, Einum DD, Morrison RS, Ptacek LJ, Sopher BL, La Spada AR. Polyglutamine-expanded ataxin-7 promotes non-cell-autonomous purkinje cell degeneration and displays proteolytic cleavage in ataxic transgenic mice. J Neurosci. 2002;22:4897–4905. doi: 10.1523/JNEUROSCI.22-12-04897.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.La Spada AR, Fu Y, Sopher BL, Libby RT, Wang X, Li LY, Einum DD, Huang J, Possin DE, Smith AC, Martinez RA, Koszdin KL, Treuting PM, Ware CB, Hurley JB, Ptacek LJ, Chen S. Polyglutamine-expanded ataxin-7 antagonizes CRX function and induces cone-rod dystrophy in a mouse model of SCA7. Neuron. 2001;31:913–927. doi: 10.1016/s0896-6273(01)00422-6. [DOI] [PubMed] [Google Scholar]
- 61.Sonmez OF, Odaci E, Bas O, Colakoglu S, Sahin B, Bilgic S, Kaplan S. A stereological study of MRI and the Cavalieri principle combined for diagnosis and monitoring of brain tumor volume. J Clin Neurosci. 2010;17:1499–1502. doi: 10.1016/j.jocn.2010.03.044. [DOI] [PubMed] [Google Scholar]
- 62.Ubhi K, Rockenstein E, Mante M, Inglis C, Adame A, Patrick C, Whitney K, Masliah E. Neurodegeneration in a transgenic mouse model of multiple system atrophy is associated with altered expression of oligodendroglial-derived neurotrophic factors. J Neurosci. 2010;30:6236–6246. doi: 10.1523/JNEUROSCI.0567-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Everall IP, DeTeresa R, Terry R, Masliah E. Comparison of two quantitative methods for the evaluation of neuronal number in the frontal cortex in Alzheimer disease. J Neuropathol Exp Neurol. 1997;56:1202–1206. doi: 10.1097/00005072-199711000-00004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.