Abstract
The prevalence of enterococci harboring tetracycline and vancomycin-resistance genes, as well as the enterococcal surface protein (esp) has mostly been determined in clinical settings, but their prevalence in tropical recreational waters remains largely unknown. The present study determined the prevalence of tetM (tetracycline-resistance), vanA and vanB (vancomycin-resistance) in the bacterial and viral fractions, enterococci and their induced phages isolated from tropical recreational marine and fresh waters, dry and wet sands. Since lysogenic phages can act as vectors for antibiotic-resistance and virulence factors, the prevalence of the mentioned genes, as well as that of an integrase-encoding gene (int) specific for Enterococcus faecalis phages was determined. Up to 60 % and 54 % of the bacterial fractions and enterococci harbored at least one of the tested genes, respectively, suggesting that bacteria in tropical environments may be reservoirs of antibiotic-resistance and virulence genes. int was detected in the viral fractions and in one Enterococcus isolate after induction. This study opens the opportunity to determine if the presence of bacteria harboring antibiotic-resistance and virulence genes in tropical recreational waters represents a threat to public health.
Keywords: antibiotic-resistance, bacteriophages, enterococci, enterococcal surface protein
INTRODUCTION
There are increasing concerns about bacteria harboring antibiotic-resistance and virulence genes in recreational waters. One of the main reasons is that it remains unknown if microorganisms harboring these genes are a risk to bathers. Among these, enterococci have received great attention since these are ubiquitous to many aquatic ecosystems (Rivera, Hazen et al. 1988; Devriese, Laurier et al. 1992; Jett, Huycke et al. 1994; Byappanahalli, Shively et al. 2003; Roberts, Soge et al. 2009). However, certain Enterococcus spp. are also important opportunistic pathogens and infections are usually treated using tetracyclines. Because of this, some studies have focused on tetracycline-resistant enterococci and these show to harbor at least one of the tet determinants, tetM being one of the most studied in gram-positive bacteria and the most prevalent in enterococci (Murray 1998; Tannock 1999; Roberts 2005). Resistant Enterococcus infections can be treated with vancomycin, but only as a last resort due to the side effects (Rocha, Kondo et al. 2002). However, enterococci have also acquired resistance to vancomycin due to genes such as vanA and vanB, commonly described in E. faecalis and E. faecium. In addition, enterococci exhibiting resistance to antibiotics can harbor genes encoding for virulence factors, as in the case of the enterococcal surface protein (esp), described in E. faecalis and E. faecium (Shankar, Lockatell et al. 2001; Toledo-Arana, Valle et al. 2001; Hammerum and Jensen 2002; Klare, Konstabel et al. 2005). The esp variant present in E. faecium was proposed as a marker of human fecal contamination and there have been conflicting studies on the usefulness of this gene as a species-specific marker (Scott, Jenkins et al. 2005; Ahmed, Stewart et al. 2008; Byappanahalli, Przybyla-Kelly et al. 2008; Layton, Walters et al. 2009).
Another concern is the transmission of antibiotic-resistance and virulence genes by mobile elements, such as plasmids and transposons. However, such studies often ignore the potential of bacteriophages as vectors of such genes (Garcia-Vallve, Romeu et al. 2000; Oancea, Klare et al. 2004). Lysogenic phages can carry additional bacterial genes that could promote fitness and virulence in the recipient bacteria (Hendrix, Smith et al. 1999; Hendrix, Lawrence et al. 2000; Hendrix 2003). These genes are often encoded by the host and may be accidentally packaged into the phage capsid. It has been shown that the viral DNA fraction of sewage and environmental waters harbor β-lactamase-encoding genes and that lysogenic phages infecting E. faecalis may harbor genes that influence virulence in the bacterial host (Yasmin, Kenny et al. 2010; Colomer-Lluch, Jofre et al. 2011). Most lysogenic phages infecting enterococci described so far belong to the Siphoviridae family (dsDNA) and one possible approach to identify these is by amplifying integrase-encoding genes (int). Integrases are considered markers of lysogeny and are involved in the integration of the phage genome into that of the bacterial host (Groth and Calos 2004). Detection of phage integrases within an ecosystem may provide insights as to what possible risks exist within a specific ecosystem (Heinemann, Rosen et al. 2006).
The prevalence of tetM, and vanA and vanB has mostly been determined in animal husbandry facilities, and temperate and subtropical recreational waters, respectively (Agerso, Pedersen et al. 2006; Roberts, Soge et al. 2009); and to our knowledge, one study has determined the prevalence of esp in tropical regions (Betancourt and Fujioka 2009). In addition, the presence of antibiotic-resistance and virulence genes in the bacteriophage fraction of recreational waters and sand, and integrases as a way to identify these, remains largely unknown. The main reason for collecting sand samples is because it can act as a possible reservoir of antibiotic-resistance and virulence genes (Hartz, Cuvelier et al. 2008; Fernandes Cardoso de Oliveira, Ranzani de Franca et al. 2010). Therefore, the main aim of the present study was to determine the prevalence of tetM, vanA, vanB, and esp variants present in E. faecalis and E. faecium, as well as int, in the bacterial and DNA viral fractions, enterococci and their lysogenic phages in tropical recreational environments.
MATERIALS AND METHODS
Study sites
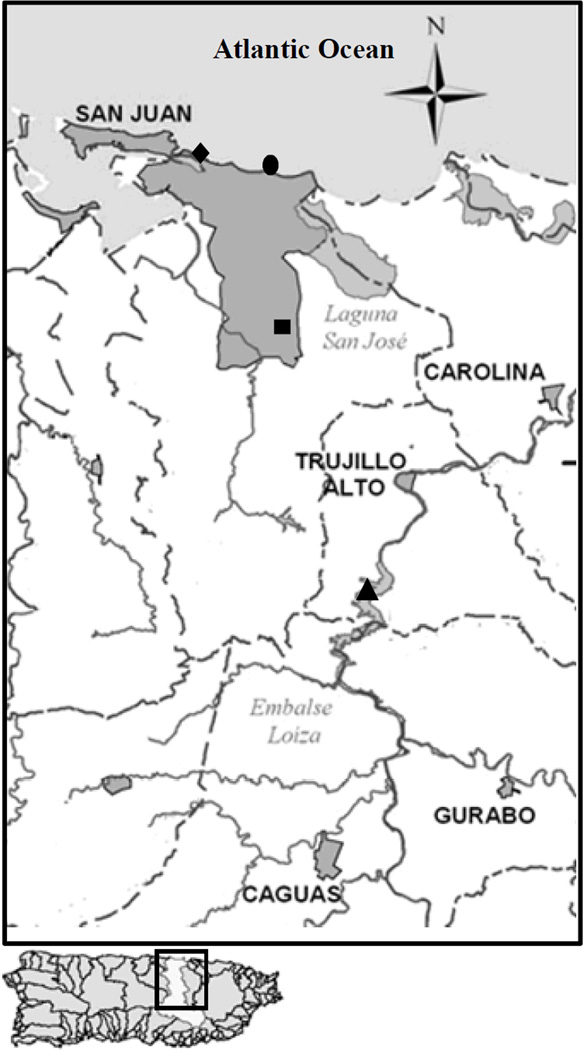
Samples were collected from several sites in Puerto Rico (Figure 1). For convenience, beaches and fresh water sources in this study will be referred as Marine water 1 (M1) and Marine water 2 (M2), and Fresh water (F1) and Fresh water (F2). M1 and M2 are beaches located in the Atlantic Ocean, alongside the northern area of the island and are heavily used by bathers year-round. Fresh water (F1) is a man-made lake whose influents are three of the island’s major rivers, serves as a major water reservoir for the San Juan metropolitan area and is also used for recreational purposes. Fresh water (F2) receives the input of minor rivers and streams used for recreational activities. One-liter grab samples of marine (M1, n=35; and M2, n=13), and fresh (F1, n=8; F2, n=10) waters were collected in sterile plastic bottles from July 2011 to May 2012. In addition, 100 g of wet and dry sands from M1 (n=16 and n=15, respectively) and M2 (n=14 and n=11, respectively). All water and sand samples were stored at 6–8 °C and processed within 6 h.
Figure 1.
Sampled sites in this study. Sites included M1(◆), M2 (●), F1 (■) F2 (▲), and in Puerto Rico. Marine water and dry and wet sands were collected from M1 and M2. Fresh water was collected from F1 and F2.
DNA extraction of the bacterial fraction and enterococci
To determine the presence of the mentioned genes in the bacterial fraction, 100 mL were processed by membrane filtration (0.4 µm pore size, 47 mm diameter) (GE Water and Process Technologies, Trevose, PA). The filter was placed in 25 mL of Azide Dextrose Broth (Difco) and incubated at 37 °C for 24 h. DNA was extracted from 1 mL by using the Fermentas GeneJet Genomic DNA Purification Kit following the manufacturer’s instructions. Alternatively, filters were placed in tubes containing 0.3 g of acid-wash glass beads (Sigma) and stored at −20 °C until processed. For this, 600 µL of sodium acetate–EDTA (AE) buffer were added, placed on a bead-beater for 60 s at maximum speed followed by centrifugation for 1 min at 1000 g. The supernatant was collected, re-centrifuged for 5 min at 10,000 g to remove any remaining debris and stored at −20 °C until processed (USEPA 2010). For the sand samples, 10 g were directly added into 20 mL of Azide Dextrose Broth, incubated at 37 °C for 24 h and 1 mL was used for DNA extraction using the Fermentas GeneJet Genomic DNA Purification Kit. Alternatively, 10 g were eluted in 120 mL of 0.1% Tween20, 100 mL of the elution were filtered and DNA from the membranes was extracted as described above.
Enumeration of enterococci was performed by using membrane filtration and m-Enterococcus agar (Difco) as previously described (USEPA 2002; Santiago-Rodriguez 2010). For the detection of enterococci in sand, 10 g were eluted in 0.1% Tween20 followed by filtration of the resulting solution and membranes were placed on m-Enterococcus agar as described. After 24–48h incubation at 37 °C, individual colonies were picked and transferred onto m-Enterococcus plates containing tetracycline (final concentration 16 µg/mL) or vancomycin (final concentration 20 µg/mL). Colonies growing in the presence of these antibiotics were picked and enriched in 1 mL of Azide Dextrose Broth for up to 48 h at 37 °C and DNA was extracted as described above. DNA quality and concentration was estimated by using a NanoDrop® (ND-1000) spectrophotometer.
Virus concentration and DNA extraction
Filtrates from above were recovered, concentrated and purified for the detection of the mentioned genes in the viral DNA fractions. as follows:, a final concentration of 0.2 % chloroform (v/v) was added to the viral suspensions and kept at room temperature for 30 min to eliminate any remaining viable bacteria. NaCl (enzyme grade >99.0 %, Fisher Scientific, NJ, USA) was added to a final concentration of 0.5 M and stored at 4 °C for 1 h. Any remaining bacterial debris was removed by centrifugation at 8000 g for 10 min at 4 °C and the supernatant was transferred to sterile Oakridge tubes. Polyethylene glycol (PEG 8000, Promega, Madison, WI, USA) was added to a final concentration of 10 % and stored at 4 °C for 24 h. Precipitates were then sedimented at 10,000 g at 4 °C for 15 min and the supernatant discarded. The phage containing pellet was resuspended in 0.5 mL of PBS (pH 7.0) overnight at 4 °C. The residual PEG and bacterial debris were removed by adding an equal volume of chloroform to a Phase Lock Gel tube (5 Prime Inc., MD, USA) and centrifuging at 4500 g for 15 min. Viral DNA was extracted by using phenol:chloroform followed by precipitation with 95 % ethanol or the Wizard DNA Clean up system following the manufacturer’s instructions (Boulanger 2009). Agarose gels (0.7% w/v) and visualized using GelStar Nucleic Acid Gel Stain (Lonza, Rockland, ME, USA) were used to confirm molecular weight and purity of the viral DNA.
Lysogen induction
One-hundred-µL of overnight culture of 50 randomly selected Enterococcus isolates harboring one or more of the genes of interest, were added to 5 mL aliquots of Azide Dextrose (Difco) containing mitomycin C (Sigma) (final concentration of 1 µg/mL). Samples were incubated in a water bath at 37 °C for 4–6 h and centrifuged at 18,000 g for 15 min at 4 °C (Raya and Hebert 2009). Supernatants were tested for the presence of lysogenic phages exhibiting the formation of viral plaques by using the double layer method and E. faecalis (ATCC 19433), E. faecium, E. gallinarum, E. hirae (ATCC 8043), E. durans (ATCC 19432), E. dispar (ATCC 51266), E. casseliflavus (ATCC 25788), E. pseudoavium (ATCC 49372) and Staphylococcus aureus (ATCC 25923) as the bacterial hosts. Briefly, 100 µL of the possible phage lysates and 1 mL of a fresh culture of each bacterial host were added to 4 mL containing Trypticase Soy Broth (TSB) (Difco) and agar (0.75 % w/v) and poured onto a Petri dish containing TSB agar containing CaCl2·2H2O (Fisher Scientific Co. NJ, USA) and NaN3 (MCB, OH, USA) (final concentrations of 2.6 mg/mL and 0.4 mg/mL, respectively). Two temperatures were tested for formation of viral plaques (22 and 37 °C) and enumerated after 24 h. Phages exhibiting the formation of viral plaques were isolated, propagated using E. faecalis (ATCC 19433) as the bacterial host and concentrated as described (Bonilla, Santiago et al. 2010). A composite of the phage lysates lacking the formation of viral plaques using the mentioned bacterial strains was concentrated as described. DNA was extracted from phages exhibiting and lacking the formation of viral plaques as described above. One of the induced phage isolates was characterized morphologically by using type-B 200 mesh copper grids placed on top of individual viral plaques and stained with uranyl acetate (UA) 2%, pH 4.5. Bacteriophages were visualized by using a Karl Zeiss Leo 922 energy filtered transmission electron microscope operated at 200 KV.
PCR amplification conditions
All primers used in the present study are described in Table 1. Primer design was performed using Primer 3 (http://frodo.wi.mit.edu/primer3/) and verified for the formation of secondary structures using NetPrimer (http://www.premierbiosoft.com/netprimer/). Detection of tetM was performed by designing primers targeting the tetM sequence found in Gene Bank (accession number AY304474.1). For the detection of the esp variant of E. faecalis, primers were designed using the gene sequence found in Gene Bank (accession number DQ845099.1). tetM and esp sequences showed a maximum identity of > 98 % (e-value 0.0) across several bacterial species and isolates. For the detection of lysogenic phages, primers were designed using the integrase gene sequence of the Enterococcus phage phiFL1B found in Gene Bank (accession number GQ478082.1).
Table 1.
Forward (F-) and reverse (R-) primers in this study. Target genes include those encoding for resistance to tetracycline (tetM) and vancomycin (vanA and vanB), the enterococcal surface protein (esp) variants found in E. faecalis and E. faecium and an integrase-encoding gene specific for E. faecalis phages (int). The 16S rRNA gene amplification was used as a control in the viral fractions and lysogenic Enterococcus phages.
| Target gene | Primer sequence | Amplicon size (bp) |
Reference |
|---|---|---|---|
| tetM | F-GGAAAATACGAAGGTGAACA | 289 | This study |
| R-GAATCCCCATTTTCCTAAGT | This study | ||
| vanA | F-GGGAAAACGACAATTGC | 732 | Roberts et. al, 2009 |
| R-GTACAATGCGGCCGTTA | Roberts et. al, 2009 | ||
| vanB | F-TTGCATGGACAAATCACTGC | 359 | Roberts et. al, 2009 |
| R-GCTCGTTTTCCTGATGGATG | Roberts et. al, 2009 | ||
| esp E. faecalis | F-CACAAATGGGTGAAGGAAGA | 490 | This study |
| R-AGACGAATTTCCCAGTTTGC | This study | ||
| esp E. faecium | F-TATGAAAGCAACAGCACAAGTT | 680 | Scott et. al, 2005 |
| R-ACGTCGAAAGTTCGATTTCC | Scott et. al, 2005 | ||
| int | F-TATTAGGAAAACCTCCGTCA | 200 | This study |
| R-ATATCTTGGGCGTAAGTGAA | This study | ||
| 16S rRNA | F-AGAGTTTGATCCTGGCTCAG | 511 | Amann et al., 1995 |
| R-ACGGGCGGTGTGTRC | Amann et al., 1995 |
Amplifications were performed in a total volume of 8µL per reaction, including 1 µL of genomic DNA at a concentration of approximately 10 ng/µL, 4 µL of Fermentas DreamTaq PCR Master Mix, 2.2 µL of milliQ water and 0.4µL of each primer at 10 µM. PCR conditions for tetM were: an initial denaturation of 94 °C for 5 min, followed by 25 cycles of 94 °C for 30 s, annealing of 55 °C for 30 s, followed by an extension of 72 °C for 30 s and a final extension of 72 °C for 7 min. Both vanA and vanB PCR conditions were: an initial denaturation of 96 °C for 3 min, followed by 35 cycles of 96 °C for 30 s, annealing of 60 °C for 1 min, extension of 72 °C for 2 min and a final extension of 72 °C for 10 min. For the detection of the esp variant found in E. faecalis the following conditions were used: an initial denaturation of 95 °C for 2 min, followed by 30 cycles of 95 °C for 45 s, annealing of 57 °C for 45 s and an extension of 72 °C for 1 min. Detection of the esp variant found in E. faecium was done by using an initial denaturation of 95 °C for 3 min, followed by 35 cycles of 94 °C for 1 min, annealing of 58 °C for 1 min and an extension of 72 °C for 1 min. int PCR parameters were: an initial denaturation of 95 °C for 2 min, followed by 35 cycles of 94 °C for 45 s, annealing of 55 °C for 45 s, followed by an extension of 72 °C for 1 min and a final extension of 72 °C for 7 min. To ensure that no bacterial DNA was present in the viral DNA, amplification of the 16S rRNA gene was performed using the following PCR conditions: an initial denaturation of 95 °C for 3 min, followed by 30 cycles of 95 °C for 30 s, annealing of 52 °C for 30 s, followed by an extension of 72 °C for 30 s and a final extension of 72 °C for 10 min. PCR products were visualized in 1.0 % agarose gels using GelStar Nucleic Acid gel stain. Positive controls were included for tetM and both esp variants and potential int products were sequenced using an ABI 3130xl Genetic Analyzer.
Statistical analyses
Fisher’s exact tests were used to determined differences in the prevalence of the mentioned genes between sample types within the sample sites. The same analysis aimed to determine significant differences in the prevalence of the esp variants between sample types. All analyses were performed with the R statistical software (v.2.11.1) (Team 2010).
RESULTS
Bacterial fraction and Enterococcus isolates
The prevalence of the tested genes in the bacterial fractions of the water and sand samples is shown in Table 2. A significant difference was noted in the prevalence of the esp variants of water samples collected from M2 (p = 0.030). The prevalence of the esp variants and int in the Enterococcus isolates is shown in Table 3 and that of tetracycline and vancomycin-resistant enterococci and tetM, vanA and vanB is shown in Table 4. Not all enterococci exhibiting resistance to either antibiotic harbored tetM or vanA and vanB. Interestingly, several of these isolates harbored two of the genes of interest. In M1 waters, 2 of the isolates were positive for both tetM and vanA, 8 harbored tetM and the esp variant present in E. faecalis and 1 harbored both vanA and the esp variant found in E. faecalis. In wet sands collected from M1 and M2, 2 and 1 of the isolates were positive for tetM and the esp variant present in E. faecium, respectively. In dry sands collected from M2, 3 of the enterococci isolated were positive for tetM and the esp variant present in E. faecium. Similarly, 1 of the isolates from F1 was positive for the latter combination of genes (data not shown). Significant differences were noted for the prevalence of tetM in M2 waters and dry sand (p = 0.0015) and M2 wet and dry sands (p = 0.0038). The prevalence of both esp variants in waters collected from M1 was significantly different as well (p = 0.033)
Table 2.
Prevalence of antibiotic-resistance and virulence-encoding genes in the bacterial fractions of tropical marine (M1, M2) and fresh waters (F1, F2) and wet and dry sands (M1, M2). Percents are included in parenthesis. Genes included those conferring resistance to tetracycline (tetM) and vancomycin (vanA and vanB), the enterococcal surface protein (esp) variants present in E. faecalis and E. faecium and an integrase-encoding gene specific for E. faecalis phages (int).
| M1 water (n=35) |
M1 wet sand (n=16) |
M1 dry sand (n=15) |
M2 water (n=13) |
M2 wet sand (n=14) |
M2 dry sand (n=11) |
F1 (n=8) |
F2 (n=10) |
|
|---|---|---|---|---|---|---|---|---|
| tetM | 6 (17) | 2 (13) | 2 (13) | 4 (31) | 1 (7) | 1 (9) | 2 (25) | 2 (20) |
| vanA | 7 (20) | 4 (25) | 2 (13) | ND* | 1 (7) | 2 (18) | 2 (25) | ND* |
| vanB | ND* | ND* | ND* | ND* | ND* | ND* | ND* | ND* |
| esp E. faecalis | 2 (5) | 2 (13) | 2 (13) | 7 (54) | 7 (50) | 4 (36) | 2 (25) | 6 (60) |
| esp E. faecium | 6 (17) | 4 (25) | 5 (33) | 1 (8) | ND* | ND* | ND* | ND* |
| int | 4 (11) | 1 (6) | 1 (7) | 2 (15) | 1 (7) | ND* | 3 (38) | 2 (20) |
ND=Not detected
Table 3.
Prevalence of the esp variants present in E. faecalis and E. faecium and int in tropical marine (M1 and M2) and fresh waters (F1 and F2) and wet and dry beach sands in the enterococci isolates. Percents are presented in parenthesis.
| M1 water (n=99) |
M1 wet sand (n=55) |
M1 dry sand (n=46) |
M2 water (n=25) |
M2 wet sand (n=21) |
M2 dry sand (n=25) |
F1 (n=19) |
F2 (n=66) |
|
|---|---|---|---|---|---|---|---|---|
| esp E. faecalis | 10 (10) | 1 (2) | 1 (2) | 2 (8) | 1 (5) | ND* | ND* | 6 (9) |
| esp E. faecium | 2 (2) | 6 (11) | 3 (7) | 2 (8) | 2 (10) | 5 (20) | ND* | 5 (8) |
| int | 3 (3) | ND* | ND* | ND* | ND* | ND* | 3 (16) | ND* |
Table 4.
Enterococcus isolates tested for tetracycline (16 µg/mL) and vancomycin (20 µg/mL) resistance isolated from tropical samples. The presence of tetracycline (tetM) and vancomycin (vanA and vanB) resistance genes was also tested in the isolates. Percents are shown in parenthesis.
| Antibiotic | Gene | ||||||
|---|---|---|---|---|---|---|---|
| Sample | Tested | Tetracycline | Vancomycin | Tested | tetM | vanA | vanB |
| M1 water | 188 | 26(14) | 18(10) | 99 | 13(13) | 2 | ND* |
| M1 wet sand | 75 | 14(19) | 10(13) | 55 | 9(16) | ND* | ND* |
| M1 dry sand | 136 | 31(23) | 31(23) | 46 | 9(20) | ND* | ND* |
| M2 water | 65 | 5(8) | 3(5) | 25 | 2(8) | ND* | ND* |
| M2 wet sand | 75 | 23(31) | 8(11) | 21 | 2(10) | ND* | ND* |
| M2 dry sand | 130 | 74(57) | 16(12) | 25 | 13(52) | ND* | ND* |
| F1 | 173 | 50(29) | 70(40) | 19 | 3(16) | ND* | ND* |
| F2 | 87 | 22(29) | 24(28) | 66 | 3(5) | ND* | ND* |
ND*= Not detected
Viral fraction and induced phages
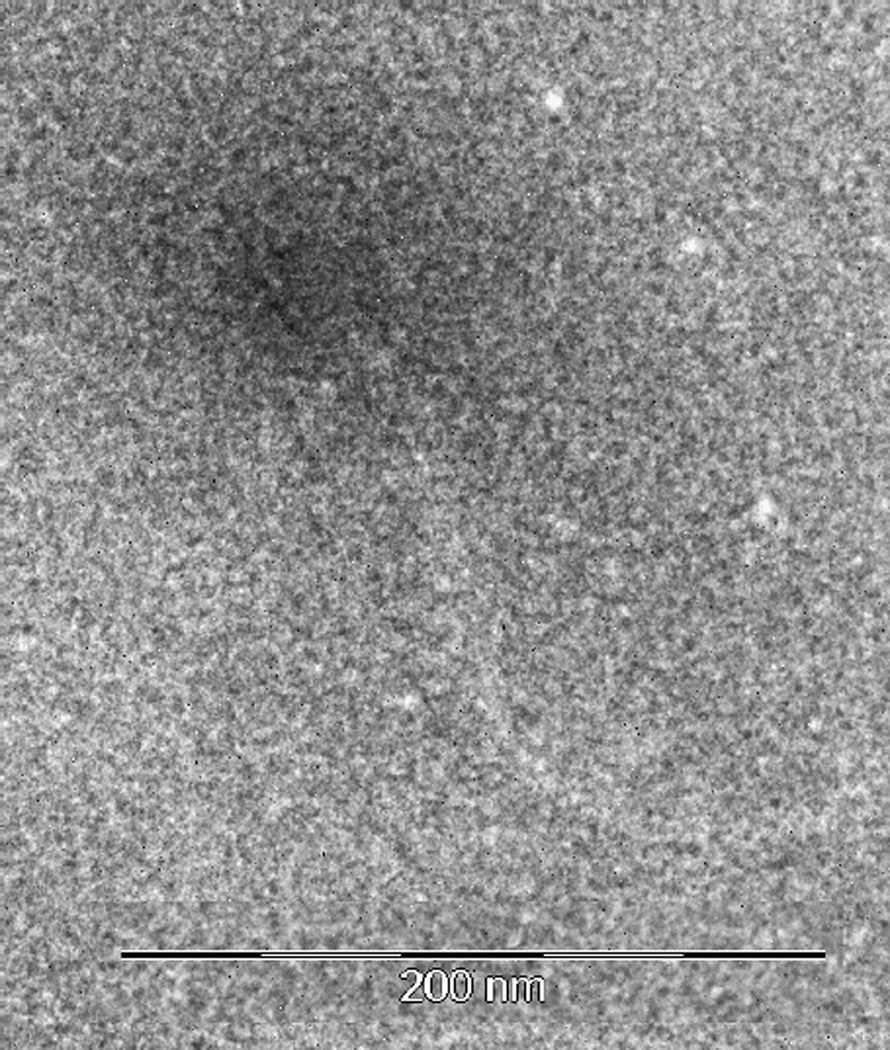
None of the antibiotic-resistance or virulence genes were detected in the viral DNA fractions. None of the samples were positive for the 16S rRNA gene and only int was detected in one of the filtrates. Similarly, the antibiotic-resistance and virulence genes tested were not detected in the induced enterococci phages. Not all induced enterococci phages were able to infect most of the Enterococcus or S. aureus type strains in this study, but those that formed viral plaques were able to replicate at 22 and 37°C in E. faecalis. In addition, enterococci phages induced from an Enterococcus isolates exhibited two different viral plaque morphologies (Data not shown). Some plaques were turbid on the edges and translucent in the center, while other plaques were completely translucent, and these phages were not positive for the integrase-encoding gene tested in this study. On the other hand, one of the induced enterococci phages that did not form viral plaques using the mentioned enterococci type strains was positive for int. Sequencing of int showed a 99 % similarity (e value=4 e-81) with int of E. faecalis phages phiFL1A, 1B, 1C, 2A, 2B, 3A and 3B. In terms of the morphology of one of the induced enterococci phage, this exhibited an icosahedral capsid and a flexible tail of approximately 80 and 180 nm, respectively (Figure 2).
Figure 2.
Characterization of an induced enterococci phage. Induced phage showed a flexible tail of approximately 180 nm and an icosahedral capsid of 80 nm (Bar=200 nm).
DISCUSSION
Many studies focusing on the prevalence of tet determinants in the environment have been done in waters associated with swine production facilities and livestock (Aminov, Garrigues-Jeanjean et al. 2001; Chee-Sanford, Aminov et al. 2001; Agerso, Pedersen et al. 2006). However, few studies that focus on the prevalence of tetracycline-resistance genes in tropical recreational waters are available. In the present study, tetM was detected in both the bacterial fractions and enterococci isolated from waters and wet and dry sands. Interestingly, significant differences were noted in the prevalence of tetM in waters and wet and dry sands collected from M2. This may suggest that the prevalence of tetM may be independent of the sample type, but this was not the case for all the sampled sites. The detection of tetracycline resistant bacteria at the sites tested may suggest that these are being introduced via human fecal material (as there are no animal husbandry facilities in proximity). Another possibility is that the constant presence of tetracycline in waters is selecting for resistant bacteria. This is reasonable since it has been shown that tetracycline can be introduced via feces and urine and detected in concentrations of approximately 0.10 µg/L (Lindsey, Meyer et al. 2001; Zhu, Snow et al. 2001). The horizontal transfer of genes conferring tetracycline-resistance via mobile elements between autochthonous and allocthonous bacteria is likely, but in tropical recreational waters it may be difficult to identify enterococci from a fecal source and those from the environment. Interestingly, some Enterococcus isolates exhibiting resistance to tetracycline were negative for tetM suggesting that these may harbor other tet determinants.
This is also one of the few reports on the detection of vanA and vanB in tropical recreational waters. Results suggest that several bacterial species in tropical environments may harbor vanA or vanA-like genes (e.g. S. aureus) (Zhu, Clark et al. 2008). The present study is comparable with previous results in which enterococci harboring vanA have been isolated from marine waters (Novais, Coque et al. 2005; Roberts, Soge et al. 2009). In contrast, vanB was not detected in any of the samples tested, but this is similar to results presented elsewhere (Schwartz, Kohnen et al. 2003). Although the reasons for this remain to be determined, it is possible that harboring vanA, instead of vanB, may be more beneficial since harboring vanA confers resistance to vancomycin and teicoplanin (Cetinkaya, Falk et al. 2000). It should be noted that most of the Enterococcus isolates that exhibited resistance to vancomycin lacked vanA or vanB. Thus, similarly to tet some of the Enterococcus isolates may harbor other genes conferring resistance to vancomycin (e.g. vanC, vanD and vanE); although it is also feasible that genes conferring resistance to other antibiotics may also be conferring resistance to vancomycin. Similar outcomes have been reported in Europe in which vancomycin-resistant enterococci have been associated with the use of avoparcin (Bager, Madsen et al. 1997). The source of vanA in tropical waters and sand remains to be determined, although the input of fecal matter remains a possibility since vancomycin-resistant bacteria have been detected in sewage effluents (Novais, Coque et al. 2005). As with tetracycline, future studies need to determine if vancomycin is being introduced into the waters tested and if it is a selective force in tropical environments.
Detection of both esp variants in the bacterial fractions and Enterococcus isolates of several of the samples may suggest an input of human and animal fecal material (Hammerum and Jensen 2002). The absence of the E. faecium variant in the bacterial fractions and in enterococci of several of the samples evaluated was not expected since these sites are visited throughout the year and may not be free from fecal contamination from non-point sources. This may suggest that the E. faecium variant may not necessarily indicate human fecal pollution in all water types, especially in the tropics, in which enterococci are naturally found in many water bodies (Muñiz, Jimenez et al. 1989). Only the bacterial fraction of waters collected from M2 and the enterococci isolated from M2 water exhibited a difference in the prevalence of both esp variants. This may suggest that, in tropical environments, similar results are obtained when detecting either esp variant.
In the present study, none of the tested genes were detected in the viral DNA fraction or the induced enterococci phages. However, the presence of other antibiotic-resistance and virulence-encoding genes in bacteriophages should not be ignored. In terms of int, although it was detected in the Enterococcus isolates and one of the induced phages, it is possible that other Enterococcus phages may harbor int sequences different from the variant tested. Phages infecting enterococci have exhibited differences in their lysogeny modules and int sequences (Yasmin, Kenny et al. 2010; Stevens, Ektefaie et al. 2011). Future studies would need to design primers targeting specific integrase genes, as integrase do not possess extensive conserved nucleotide sequences among different phage families. This study also suggests that Enterococcus isolates may harbor more than one inducible prophage, but the function of various prophages in the genomes of enterococci remains to be determined (Banks, Lei et al. 2003; Brussow, Canchaya et al. 2004).
CONCLUSIONS
This is one of the first reports which aimed to identify a battery of antibiotic-resistance and virulence genes in both the bacterial and viral fractions of tropical recreational waters and sand. The presence of antibiotic-resistance and virulence genes in the environmental microbiota of tropical samples suggests that the transmission of these genes is not restricted to strains of the clinical setting. The exact mechanisms of horizontal transfer of antibiotic-resistance and virulence genes under these environmental conditions need to be determined, as do the role of environmental microbiota as reservoirs of these genes. By detecting int in lysogenic phages, further studies could determine the presence of antibiotic-resistance and virulence factors in their genomes. Our data also point to the need to take into consideration lysogenic phages when determining the presence or absence of bacterial viruses in the environment.
Most epidemiological studies on recreational waters have focused on gastrointestinal illness; though a few recent ones have also included and eye, skin and ear infections. The present study showed a relatively high prevalence of Enterococcus spp. exhibiting resistance to antibiotics which also harbored virulence genes. It remains to be determined if these bacteria represent a risk to bathers and if these are specifically involved in skin infections. However, it is worrying that opportunistic pathogens involved in skin infections and harboring antibiotic resistance are present in recreational waters and sands, where bathers may be exposed to them.
ACKNOWLEDGEMENTS
We thank M.G. Dominguez-Bello, N. Shankar and M.N. Byappanahalli for sharing the positive controls used in this study; Miguel Urdaneta for the phage microscopy and Rita Patricio, Jean M. Carrasquillo, Roberto Lorenzini, Gabriel Vázquez and Gabriela Tirado for sample collection and processing. This research was partially supported by MBRS-RISE (NIH Grant Number 2R25GM061151-09). All sequencing was done at the Sequencing and Genotyping facility, University of Puerto Rico-Río Piedras supported in part by NCRR AABRE Grant #P20 RR16470, NIH-SCORE Grant #S06GM08102, and NSF-CREST Grant #0206200,NINDS-SNRP U54 NS39405.
REFERENCES
- Agerso Y, Pedersen AG, et al. Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J Antimicrob Chemother. 2006;57(5):832–839. doi: 10.1093/jac/dkl069. [DOI] [PubMed] [Google Scholar]
- Ahmed W, Stewart J, et al. Evaluation of the host-specificity and prevalence of enterococci surface protein (esp) marker in sewage and its application for sourcing human fecal pollution. J Environ Qual. 2008;37(4):1583–1588. doi: 10.2134/jeq2007.0474. [DOI] [PubMed] [Google Scholar]
- Aminov RI, Garrigues-Jeanjean N, et al. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol. 2001;67(1):22–32. doi: 10.1128/AEM.67.1.22-32.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bager F, Madsen M, et al. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev Vet Med. 1997;31(1–2):95–112. doi: 10.1016/s0167-5877(96)01119-1. [DOI] [PubMed] [Google Scholar]
- Banks DJ, Lei B, et al. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect Immun. 2003;71(12):7079–7086. doi: 10.1128/IAI.71.12.7079-7086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt WQ, Fujioka RS, et al. Evaluation of enterococcal surface protein genes as markers of sewage contamination in tropical recreational waters. Water Sci Technol. 2009;60(1):261–266. doi: 10.2166/wst.2009.380. [DOI] [PubMed] [Google Scholar]
- Bonilla N, Santiago T, et al. Enterophages, a group of phages infecting Enterococcus faecalis and their potential as alternate indicators of human faecal contamination. Water Sci Technol. 2010;61(2):293–300. doi: 10.2166/wst.2010.815. [DOI] [PubMed] [Google Scholar]
- Boulanger P. Purification of Bacteriophages and SDS-PAGE Analysis of Phage Structural Proteins from Ghost Particles. In: Clokie MRJ, Kropinski AM, editors. Bacteriophages: Methods and Protocols. Vol. 2. NewYork, New York: Humana Press; 2009. pp. 227–238. [DOI] [PubMed] [Google Scholar]
- Brussow H, Canchaya C, et al. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68(3):560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli MN, Przybyla-Kelly K, et al. Environmental occurrence of the enterococcal surface protein (esp) gene is an unreliable indicator of human fecal contamination. Environ Sci Technol. 2008;42(21):8014–8020. doi: 10.1021/es800481p. [DOI] [PubMed] [Google Scholar]
- Byappanahalli MN, Shively DA, et al. Growth and survival of Escherichia coli and enterococci populations in the macro-alga Cladophora (Chlorophyta) FEMS Microbiol Ecol. 2003;46(2):203–211. doi: 10.1016/S0168-6496(03)00214-9. [DOI] [PubMed] [Google Scholar]
- Cetinkaya Y, Falk P, et al. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13(4):686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee-Sanford JC, Aminov RI, et al. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl Environ Microbiol. 2001;67(4):1494–1502. doi: 10.1128/AEM.67.4.1494-1502.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer-Lluch M, Jofre J, et al. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One. 2011;6(3):e17549. doi: 10.1371/journal.pone.0017549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriese LA, Laurier L, et al. Enterococcal and streptococcal species isolated from faeces of calves, young cattle and dairy cows. J Appl Bacteriol. 1992;72(1):29–31. doi: 10.1111/j.1365-2672.1992.tb04877.x. [DOI] [PubMed] [Google Scholar]
- Fernandes Cardoso de Oliveira AJ, Ranzani de Franca PT. Antimicrobial resistance of heterotrophic marine bacteria isolated from seawater and sands of recreational beaches with different organic pollution levels in southeastern Brazil: evidences of resistance dissemination. Environ Monit Assess. 2010;169(1–4):375–384. doi: 10.1007/s10661-009-1180-6. [DOI] [PubMed] [Google Scholar]
- Garcia-Vallve S, Romeu A, et al. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 2000;10(11):1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Calos MP. Phage integrases: biology and applications. J Mol Biol. 2004;335(3):667–678. doi: 10.1016/j.jmb.2003.09.082. [DOI] [PubMed] [Google Scholar]
- Hammerum AM, Jensen LB. Prevalence of esp encoding the enterococcal surface protein, in Enterococcus faecalis and Enterococcus faecium isolates from hospital patients, poultry, and pigs in Denmark. J Clin Microbiol. 2002;40(11):4396. doi: 10.1128/JCM.40.11.4396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz A, Cuvelier M, et al. Survival potential of Escherichia coli and Enterococci in subtropical beach sand: implications for water quality managers. J Environ Qual. 2008;37(3):898–905. doi: 10.2134/jeq2007.0312. [DOI] [PubMed] [Google Scholar]
- Heinemann JA, Rosen H, et al. Environment arrays: a possible approach for predicting changes in waterborne bacterial disease potential. Environ Sci Technol. 2006;40(23):7150–7156. doi: 10.1021/es060331x. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol. 2003;6(5):506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Lawrence JG, et al. The origins and ongoing evolution of viruses. Trends Microbiol. 2000;8(11):504–508. doi: 10.1016/s0966-842x(00)01863-1. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Smith MC, et al. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci U S A. 1999;96(5):2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jett BD, Huycke MM, et al. Virulence of enterococci. Clin Microbiol Rev. 1994;7(4):462–478. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klare I, Konstabel C, et al. Spread of ampicillin/vancomycin-resistant Enterococcus faecium of the epidemic-virulent clonal complex-17 carrying the genes esp and hyl in German hospitals. Eur J Clin Microbiol Infect Dis. 2005;24(12):815–825. doi: 10.1007/s10096-005-0056-0. [DOI] [PubMed] [Google Scholar]
- Layton BA, Walters SP, et al. Distribution and diversity of the enterococcal surface protein (esp) gene in animal hosts and the Pacific coast environment. J Appl Microbiol. 2009;106(5):1521–1531. doi: 10.1111/j.1365-2672.2008.04113.x. [DOI] [PubMed] [Google Scholar]
- Lindsey ME, Meyer TM, et al. Analysis of trace levels of sulfonamide and tetracycline antimicrobials in groundwater and surface water using solid-phase extraction and liquid chromatography/mass spectrometry. Anal Chem. 2001;73(19):4640–4646. doi: 10.1021/ac010514w. [DOI] [PubMed] [Google Scholar]
- Muñiz I, Jimenez L, et al. Survival and activity of Streptococcus faecalis and Escherichia coli in tropical fresh water. Microbial Ecology. 1989;18:125–134. doi: 10.1007/BF02030121. [DOI] [PubMed] [Google Scholar]
- Murray BE. Diversity among multidrug-resistant enterococci. Emerg Infect Dis. 1998;4(1):37–47. doi: 10.3201/eid0401.980106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais C, Coque TM, et al. Environmental contamination with vancomycin-resistant enterococci from hospital sewage in Portugal. Appl Environ Microbiol. 2005;71(6):3364–3368. doi: 10.1128/AEM.71.6.3364-3368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oancea C, Klare I, et al. Conjugative transfer of the virulence gene, esp, among isolates of Enterococcus faecium and Enterococcus faecalis. J Antimicrob Chemother. 2004;54(1):232–235. doi: 10.1093/jac/dkh249. [DOI] [PubMed] [Google Scholar]
- Raya RR, Hebert EM. Isolation of Phage via Induction of Lysogens. In: Clokie MRJ, Kropinski AM, editors. Bacteriophages: Methods and Protocols. Vol. 1. New York, NY: Humana Press; 2009. pp. 23–33. [Google Scholar]
- Rivera SC, Hazen TC, et al. Isolation of fecal coliforms from pristine sites in a tropical rain forest. Appl Environ Microbiol. 1988;54(2):513–517. doi: 10.1128/aem.54.2.513-517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245(2):195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Roberts MC, Soge OO, et al. Vancomycin-resistant Enterococcus spp. in marine environments from the West Coast of the USA. J Appl Microbiol. 2009;107(1):300–307. doi: 10.1111/j.1365-2672.2009.04207.x. [DOI] [PubMed] [Google Scholar]
- Rocha JL, Kondo W, et al. Uncommon vancomycin-induced side effects. Braz J Infect Dis. 2002;6(4):196–200. doi: 10.1590/s1413-86702002000400007. [DOI] [PubMed] [Google Scholar]
- Santiago-Rodriguez TM, Davila C, Gonzalez J, Bonilla N, Marcos P, Urdaneta M, Cadete M, Monteiro S, Santos R, Santo-Domingo J, Toranzos G. Characterization of Enterococcus faecalis-infecting phages (enterophages) as markers of human fecal pollution in recreational waters. Water Research. 2010;44(16):4716–4725. doi: 10.1016/j.watres.2010.07.078. [DOI] [PubMed] [Google Scholar]
- Schwartz T, Kohnen W, et al. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol Ecol. 2003;43(3):325–335. doi: 10.1111/j.1574-6941.2003.tb01073.x. [DOI] [PubMed] [Google Scholar]
- Scott TM, Jenkins TM, et al. Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ Sci Technol. 2005;39(1):283–287. [PubMed] [Google Scholar]
- Shankar N, Lockatell CV, et al. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect Immun. 2001;69(7):4366–4372. doi: 10.1128/IAI.69.7.4366-4372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens RH, Ektefaie MR, et al. The annotated complete DNA sequence of Enterococcus faecalis bacteriophage phiEf11 and its comparison with all available phage and predicted prophage genomes. FEMS Microbiol Lett. 2011;317(1):9–26. doi: 10.1111/j.1574-6968.2010.02203.x. [DOI] [PubMed] [Google Scholar]
- Tannock GW. Medical Importance of the Normal Microflora. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. The normal microflora as a reservoir of antibiotic resistance-determinants; pp. 388–404. [Google Scholar]
- Team RDC. R: A Language and Environmeny for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- Toledo-Arana A, Valle J, et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67(10):4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA. EPA-821-R-02-022. Washington, D.C.: Office of Water; 2002. Method 1600: Membrane Filter Test Method for Enterococci in Water. [Google Scholar]
- USEPA. EPA-821-R-10-004. Washington, D.C.: 2010. Method A: Enterococci in Water by TaqMan® Quantitative Polymerase Chain Reaction (qPCR) Assay. [Google Scholar]
- Yasmin A, Kenny JG, et al. Comparative genomics and transduction potential of Enterococcus faecalis temperate bacteriophages. J Bacteriol. 2010;192(4):1122–1130. doi: 10.1128/JB.01293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Snow DD, et al. Analysis of oxytetracycline, tetracycline, and chlortetracycline in water using solid-phase extraction and liquid chromatography-tandem mass spectrometry. J Chromatogr A. 2001;928(2):177–186. doi: 10.1016/s0021-9673(01)01139-6. [DOI] [PubMed] [Google Scholar]
- Zhu W, Clark NC, et al. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob Agents Chemother. 2008;52(2):452–457. doi: 10.1128/AAC.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]