SUMMARY
Synchrony of the mammalian circadian clock is achieved by complex transcriptional and translational feedback loops centered on the BMAL1: CLOCK heterodimer. Modulation of circadian feedback loops is essential for maintaining rhythmicity, yet the role of transcriptional coactivators in driving BMAL1:CLOCK transcriptional networks is largely unexplored. Here, we show diurnal hepatic steroid receptor coactivator 2 (SRC-2) recruitment to the genome that extensively overlaps with the BMAL1 cistrome during the light phase, targeting genes that enrich for circadian and metabolic processes. Notably, SRC-2 ablation impairs wheel-running behavior, alters circadian gene expression in several peripheral tissues, alters the rhythmicity of the hepatic metabolome, and deregulates the synchronization of cell-autonomous metabolites. We identify SRC-2 as a potent coregulator of BMAL1:CLOCK and find that SRC-2 targets itself with BMAL1:CLOCK in a feedforward loop. Collectively, our data suggest that SRC-2 is a transcriptional coactivator of the BMAL1:CLOCK oscillators and establish SRC-2 as a critical positive regulator of the mammalian circa-dian clock.
INTRODUCTION
Most physiological systems have an intrinsic rhythm centered on diurnal light and feeding cycles. The central mammalian pacemaker located within the hypothalamic suprachiasmatic nuclei (SCN) coordinates this rhythmicity with the peripheral tissues, synchronizing whole-body physiology. While the SCN can generate endogenous circadian rhythms, exogenous environmental cues, such as light and temperature, help to maintain precise synchronization of physiology with environmental changes. Disruptions in diurnal cycling are associated with increased risk for cardiovascular diseases, cancer, and several metabolic derangements (Bass and Takahashi, 2010; Takahashi et al., 2008). Molecularly, a core network of genes is responsible for circadian oscillation arising from the heterodimerization of the basic helix-loop-helix (bHLH) transcription factors Brain and Muscle ARNT-Like 1 (BMAL1, ARNTL) and Circadian Locomotor Output Cycles Kaput (CLOCK). Collectively, these transcription factors drive circadian gene expression through a common mechanism of transcriptional and translational feedback loops (Asher and Schibler, 2011). Autoregulatory feedback loops are formed by BMAL1:CLOCK binding to E box elements in promoter regions of Period (Per1/2) and Cryptochrome (Cry1/2), whose products then inhibit BMAL1:CLOCK activity. Another ancillary feedback loop involving the nuclear receptors (NRs) is formed by BMAL1:CLOCK governing the expression of Retinoic Acid Receptor-Related Orphan Receptor (ROR), Rev-erbα (Nr1d1), and Rev-erbβ (Nr1d2) expression. ROR activates Bmal1 transcription, and Rev-erbα/β proteins subsequently repress Bmal1 (Akashi and Takumi, 2005; Asher and Schibler, 2011; Bugge et al., 2012; Cho et al., 2012; Preitner et al., 2002; Sato et al., 2004; Ueda et al., 2002). Together, these feedback loops are thought to be sufficient for controlling gene expression to maintain the daily rhythmicity of most biological processes.
Oscillators in peripheral tissues (e.g., the liver) also have endogenous cycling coordinated by the central clock, but can be modulated in a tissue-specific manner by other cues, such as steroid hormones and nutrient availability. Circadian rhythm in peripheral tissues is intricately linked to metabolism, given that peripheral clock synchrony can be altered through metabolic cues and that perturbations are known to result in disruption of general energy homeostasis, glucose regulation, fatty acid metabolism, and feeding behavior (Asher and Schibler, 2011; Bass and Takahashi, 2010). Specifically, disruption of the core clock component, BMAL1, impairs glucose homeostasis, increases insulin sensitivity, and decreases whole-body adiposity after 20 weeks of age (Hatanaka et al., 2010; Kondratov et al., 2006; Rudic et al., 2004). Similarly, disruption of the heterodimeric partner of BMAL1, CLOCK, also leads to impaired glucose tolerance, increased adiposity in the CLOCK mutant, hepatic steatosis, and a broad disruption of the hepatic metabolome (Eckel-Mahan et al., 2012; Fustin et al., 2012; Marcheva et al., 2010; Rudic et al., 2004). Taken together with hepatic genome-wide localization of BMAL1 and CLOCK targets, these observations demonstrate that primary circadian transcription factors are heavily involved in the regulation of hepatic metabolism (Hatanaka et al., 2010; Kondratov et al., 2006; Rudic et al., 2004). Just as core transcriptional clock components can regulate metabolism, many hepatic metabolic transcription factors, such as the NRs ERR, PPARα, Rev-erbα/β, and ROR, have also been identified as transcriptional mediators of clock targets (Akashi and Takumi, 2005; Asher and Schibler, 2011; Bugge et al., 2012; Cho et al., 2012; Preitner et al., 2002; Sato et al., 2004; Ueda et al., 2002).
Similar crosstalk between circadian rhythm and metabolic transcriptional regulation is coordinated through the activity of transcriptional coregulators. Coregulators are utilized by NRs and other transcription factors to dynamically regulate gene transcription. In general, modulation of transcription factor activity is achieved by corepressors silencing and coactivators enhancing gene expression. As such, coregulators have broad effects on target gene expression. For example, in addition to their known role as circadian corepressors for BMAL1:CLOCK, Per2 and Cry1 have also been reported to function as corepressors for the NRs Rev-erbα and GR, respectively, to facilitate temporal cellular activity (Lamia et al., 2011; Schmutz et al., 2010). Per2 and Cry1 repression of NRs is achieved through the interaction of their LXXLL motifs with the NRs (Lamia et al., 2011; Schmutz et al., 2010). The LXXLL motif is common among NR coregulators, including the p160 family of Steroid Receptor Co-activators (SRCs). As with many other coregulators, loss of an SRC has profound and diverse physiological effects, ranging from increased tumorigenic potential through altered cell-cycle control to altered energy homeostasis through disruption of key metabolic processes. Specifically, our lab has identified Steroid Receptor Coactivator-2 (SRC-2, NCOA2, GRIP1) as an essential modulator of hepatic metabolism (Chopra et al., 2008, 2011). SRC-2 serves as a critical mediator of organism-wide energy homeostasis by regulating glucose release under fasting conditions (Chopra et al., 2008). Loss of SRC-2 is also associated with increased insulin sensitivity (Picard et al., 2002), and decreased adiposity (Hartig et al., 2011). Additionally, the activity of SRC-2 has also been shown to be directly regulated by AMP-activated protein kinase (AMPK), a key metabolic rheostat and an important sensor in circadian rhythm (Lamia et al., 2009; Um et al., 2011). AMPK activates SRC-2 as a coactivator in bile acid secretion, which is necessary for fat absorption from the gut (Chopra et al., 2011).
Given the broad metabolic impact of SRC-2, we sought to understand the previously unknown temporal metabolic effects of SRC-2 on the hepatic cistrome and metabolome. Provided with mounting evidence that metabolism and circadian rhythm are interlinked processes, and existing knowledge about the effects exerted by coactivators on transcription factors in diverse processes and tissues, we investigated the temporal role of SRC-2 through cistromic analysis of SRC-2 in the liver at two time points of the diurnal cycle. In this article, we demonstrate that SRC-2 shows preferential binding during the light phase of the circadian cycle. Additionally, we find that SRC-2 expression is rhythmic in accordance with its cistromic occupancy. Running-wheel experiments also reveal that loss of SRC-2 severely alters diurnal behavioral rhythmicity. In further exploring the temporal effects of SRC-2 on gene expression, we find that loss of SRC-2 disrupts not only a cassette of metabolic genes but also core components of the clock across several tissues, including the liver, brown adipose tissue (BAT), white adipose tissue (WAT), heart, and skeletal muscle. Likewise, loss of SRC-2 disrupts hepatic and cell-autonomous metabolite rhythmicity. Taken together, these findings suggest that SRC-2 is a transcriptional coactivator for BMAL1:CLOCK in the liver that couples metabolic gene expression with proper maintenance of the hepatic peripheral clock.
RESULTS
Differential Recruitment of SRC-2 to the Hepatic Cistrome
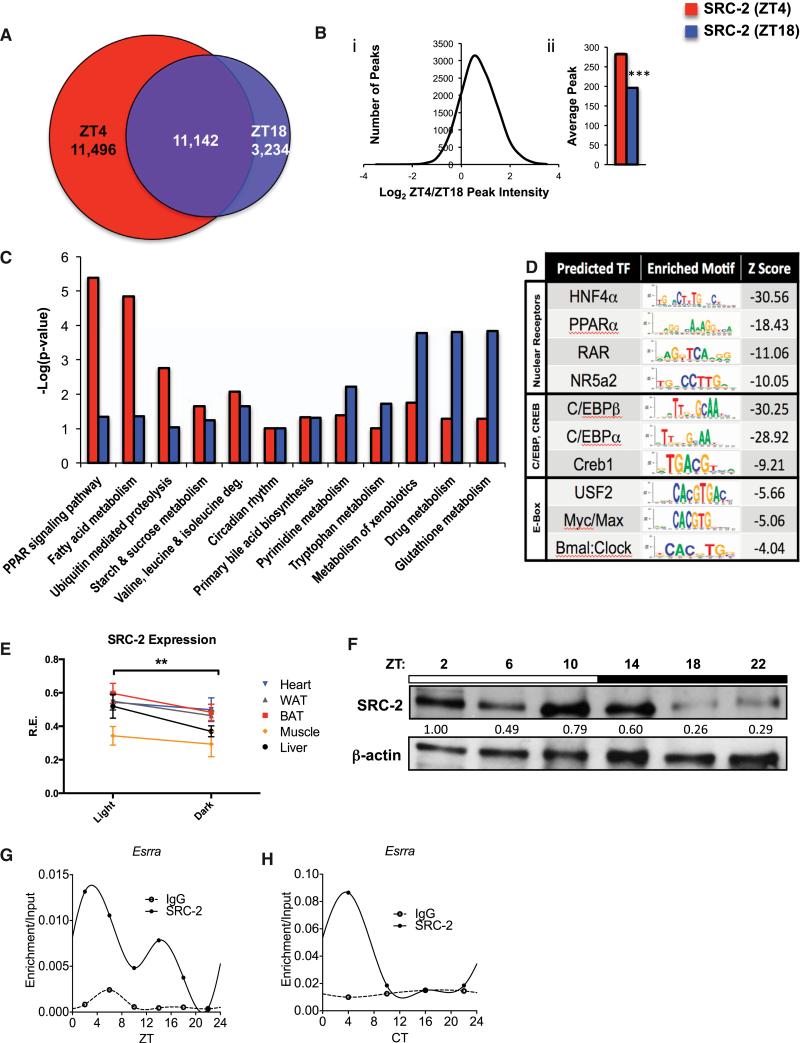
To better understand the temporal functions of SRC-2 in coordinating energy homeostasis, we analyzed its hepatic chromatin occupancy during two phases of the murine diurnal cycle, zeitgeber time 4 (ZT4) in the light phase and ZT18 in the dark phase, using chromatin immunoprecipitation followed by deep sequencing (ChIP-seq). We found an increase in total binding sites for SRC-2 at ZT4 (22,638) compared with ZT18 (14,376), but with extensive overlap (~78% of ZT18) between the two ZTs (Figure 1A). Analysis of the common binding sites revealed that SRC-2 bound more strongly at ZT4 than at ZT18, as indicated by an increase in SRC-2 peak intensity (Figure 1Bi) as well as an increase in the average number of peaks (Figure 1Bii). In accordance with a role for SRC-2 in directly controlling gene expression, SRC-2 binding sites at both ZT4 and ZT18 were strongly enriched for gene-proximal regions around the transcriptional start site (TSS) (Figures S1A–S1D). Further, in agreement with published roles for SRC-2 in regulating hepatic metabolism (Chopra et al., 2008, 2011), the functional processes associated with putative SRC-2 target genes generally clustered around major metabolic processes. In particular, ZT4 was enriched over ZT18 for PPAR signaling pathways, fatty acid metabolism, and ubiquitin-mediated proteolysis (Figure 1C, red bars). Genes called from SRC-2 binding sites at ZT18 were enriched for a different set of metabolic processes, including xeno-biotic, drug, and glutathione metabolism, suggesting distinct roles for SRC-2 in the diurnal cycle (Figure 1C, blue bars). Both the ZT4 and ZT18 genes called from SRC-2 binding sites were equally enriched for a previously reported role of SRC-2 in primary bile acid synthesis (Chopra et al., 2011) and for circadian rhythm. Analysis of select loci of metabolic targets shared by both SRC-2 binding at ZT4 and ZT18 revealed increased binding near the TSS at ZT4 of previously reported SRC-2 targets (Elovl6 and Fasn) (Jeong et al., 2006), as well as increased ZT4 peaks at new SRC-2 targets, including NRs (Nr0b2 and Shp) and metabolic targets (Ugp2 and Cyp7a1), and a circadian target (Per1) (Figure S1E). Interestingly, we also found SRC-2 targeting its own locus (Ncoa2/SRC-2) (Figure S1E). DNA sequence analyses of SRC-2 binding sites at ZT4 and ZT18 revealed the expected enrichments for NR binding motifs (HNF4α, PPARα, RAR, and NR5a2), and SRC-2 binding sites were also enriched for the E box motif, which is known to bind basic helix-loop-helix proteins such as BMAL1 and CLOCK (Figure 1D).
Figure 1. Analysis of Mouse Hepatic SRC-2 Cistromes at ZT4 and ZT18.
(A) Venn diagram comparing SRC-2 ChIP-seq intervals from murine livers at ZT4 and ZT18.
(B) Peak intensities of the overlapping binding sites plotted as log2 of SRC-2 (ZT4) over SRC-2 (ZT18) against the number of ChIP peaks (i) and in bar graph form (ii); ***p < 0.001. Data are graphed as the mean ± SEM.
(C) Comparative DAVID gene functional analyses for select enriched annotations of SRC-2 ZT4 and ZT18 targets.
(D) Top enriched SeqPos motifs common to ZT4/ZT18 SRC-2 binding regions, grouped by DNA binding domains of transcription factors.
(E) hd-qPCR expression of SRC-2 in WAT, BAT, heart, skeletal muscle, and liver in entrained WT male mice (n = 5 each) in light and dark phases. Data are graphed as the mean ± SEM. *p < 0.05.
(F) Representative immunoblot of liver SRC-2 in entrained WT mice at ZT2, ZT6, ZT10, ZT14, ZT18, and ZT22.
(G) Validation of 24 hr circadian recruitment of SRC-2 to the Esrra promoter in liver tissue assayed by ChIP-qPCR relative to input at ZT2, ZT6, ZT10, ZT14, ZT18, and ZT22.
(H) Validation of 24 hr circadian recruitment of SRC-2 to the Esrra promoter in liver tissue assayed by ChIP-qPCR relative to input at CT4, CT10, CT16, and CT22.
Given that many NRs demonstrate temporal rhythms and that SRC-2 occupancy of the cistrome at ZT4 identifies approximately twice as many peaks as at ZT18, we explored the diurnal expression of SRC-2 using high-density quantitative PCR (hdqPCR) across several key metabolic tissues (i.e., liver, BAT, WAT, heart, skeletal muscle, and SCN; Figure S1F). We identified tissue-specific differences in temporal SRC-2 expression, but generally SRC-2 had higher expression during the light phase compared with the dark phase (Figure 1E). Specifically, in liver, SRC-2 expression was found to be significantly higher at ZT2 compared with ZT18, in line with increased SRC-2 cistromic occupancy during the light phase (Figures 1A, 1E, and S1F), which coincided with cycling of hepatic SRC-2 protein. We observed the highest expression of SRC-2 at ZT2 in the light phase with an ~4-fold decrease in the dark phase at ZT18 (Figure 1F). Continuing to explore SRC-2 regulation of targets in a temporal setting, we performed ChIP-qPCR of SRC-2 over a period of 24 hr in the liver, and found that SRC-2 bound to specific promoter regions in a temporal fashion, as shown with Esrra over 24 hr in both ZT (Figure 1G) and circadian time (CT; Figure 1H) conditions (Figure S1G).
The SRC-2 and BMAL1 Cistromes Overlap Extensively
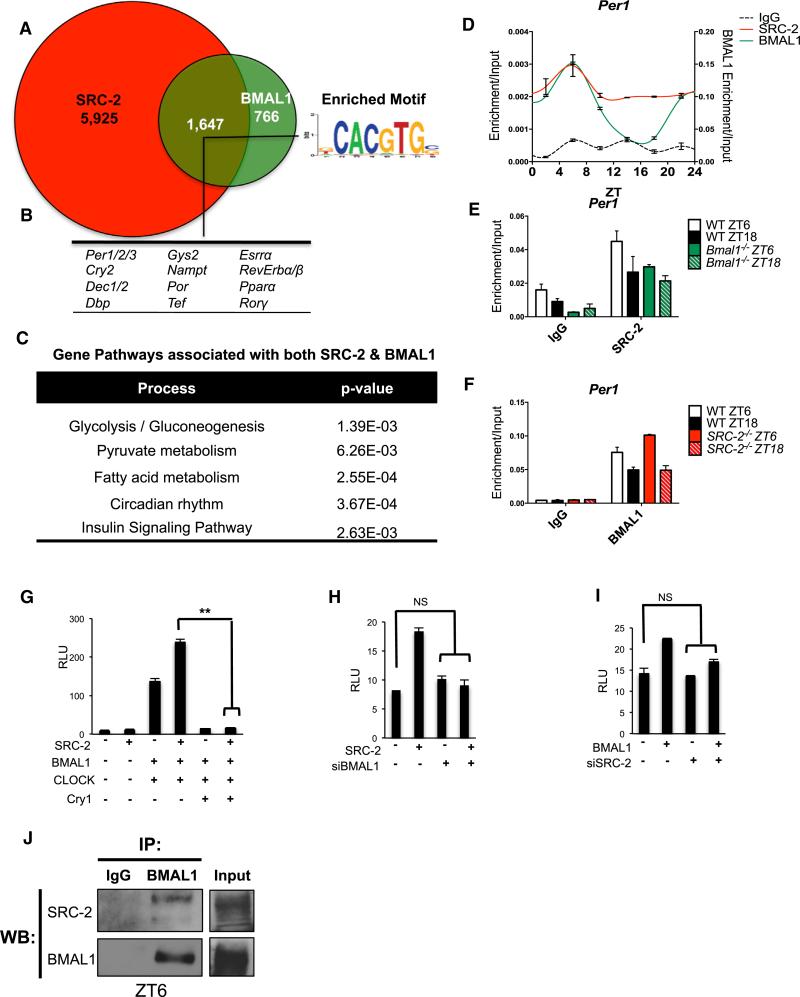
We continued to explore a role for SRC-2 in diurnal metabolic transcription, focusing on the role of SRC-2 in the light phase based on the consistency of results from the ChIP-seq data, the ChIP-qPCR of SRC-2, and light-phase expression of SRC-2. We explored a possible relationship between the SRC-2 genomic occupancy at ZT4 and transcription factors known to display rhythmic genomic recruitment with peak activity during the light phase. The results from the motif analysis of the SRC-2 cistrome (Figure 1D) led us to study the E box binding BMAL1:CLOCK (BMAL1:NPAS2) transcriptional heterodimers. Loss of any one component of the heterodimer results in metabolic derangements similar to those found upon SRC-2 ablation (Chopra et al., 2008, 2011). Because BMAL1 is the nonredundant heterodimeric partner for both CLOCK and NPAS2, we overlaid our SRC-2 ChIP-seq data with the published mouse liver cistrome for BMAL1 at CT4 (Koike et al., 2012; Rey et al., 2011), and found extensive convergence of the SRC-2 (ZT4) and BMAL1 cistromes. Analysis of these potential targets showed that ~68% of genes with BMAL1 binding sites contained at least one SRC-2 binding site within gene promoter regions (Figure 2A). Motif analysis revealed that the overlapping peaks between SRC-2 (ZT4) and BMAL1 were enriched primarily for the E box, and many SRC-2 binding sites overlapped with BMAL1 sites at known circadian genes (Per1/2/3, Cry2, Dec1/2, DBP, Dec1/2, Gys2, Nampt, Tef, Esrra, PPARα, Por, and Rev-erbα/β) (Figures 2B and S2A), suggesting that SRC-2 may serve as a transcriptional coregulator for the BMAL1:CLOCK heterodimer. Not surprisingly, the functional processes associated with the genes shared by BMAL1 and SRC-2 were highly enriched for circadian rhythm and other known BMAL1 metabolic functions, including glycolysis/gluconeogenesis, pyruvate and fatty acid metabolism, and insulin signaling (Dufour et al., 2011; Hatanaka et al., 2010; Rey et al., 2011; Figure 2C).
Figure 2. Analysis of SRC-2 Occupancy across the Hepatic Genome Shows Enrichment for Circadian Targets.
(A) Venn diagram representing the unique and overlapping genes that have SRC-2 (ZT4) and BMAL1 (CT4) (Koike et al., 2012) binding sites within the extended gene promoter region (EPR) as assayed by ChIP-seq.
(B) Circadian genes and E box sequence motif found in the SRC-2/BMAL1 overlapping portion of the data sets.
(C) Gene functional analyses of targets with shared BMAL1 and SRC-2 binding sites.
(D) Profile of 24 hr circadian recruitment of SRC-2 and BMAL1 to the Per1 promoter in liver tissue as assayed by ChIP-qPCR relative to input.
(E) Recruitment of SRC-2 to the Per1 promoter in WT and Bmal1−/− mice at ZT6 and ZT18.
(F) Recruitment of BMAL1 to the Per1 promoter in WT and SRC-2−/− mice at ZT6 and ZT18.
(G) Transactivation of cotransfected SRC-2, BMAL1, CLOCK, and Cry1 expression constructs on the Per1-promoter driving the luciferase gene in HepG2 cells. Cry1 expression was used as a control to demonstrate transcriptional repression.
(H) Transactivation assays on the Per1-Luciferase promoter with SRC-2 and siBMAL1.
(I) Transactivation assays on the Per1-Luciferase promoter with BMAL1 and siSRC-2.
(J) Coimmunoprecipitations of BMAL1 with SRC-2 at ZT6 in WT mouse liver.
To validate the temporal cistromic occupancy of SRC-2 and possible co-occupancy of SRC-2 with BMAL1 at select circadian target genes, we performed ChIP in mouse livers over a period of 24 hr. Peak SRC-2 recruitment to the Per1 promoter occurred at ZT6, when BMAL1 enrichment was also at its highest (Figures 2D and S2B). We next investigated whether SRC-2 recruitment to the Per1 promoter was BMAL1 dependent. In Bmal1−/− mice, we found that SRC-2 recruitment at ZT6 was in fact decreased compared with wild-type (WT) mice at ZT6, suggesting that BMAL1 is required for SRC-2 enrichment (Figure 2E). However, we found no significant differences in BMAL1 recruitment with the absence of SRC-2 at the Per1 promoter at ZT6 and ZT18 in SRC-2−/− mice (Figure 2F). Interestingly, hepatic BMAL1 and SRC-2 ChIP experiments showed dual recruitment on the SRC-2 promoter (Figures S2C–S2E). Again, we found that loss of Bmal1 impaired SRC-2 recruitment to its own promoter, but observed no differences in BMAL1 recruitment in SRC-2−/− livers (Figures S2C–S2E). Collectively, our ChIP-qPCR results suggest that SRC-2 and BMAL1 proteins can co-occupy the circadian target, Per1, as well as the SRC-2 promoter.
We then transiently transfected cultured cells to test SRC-2 modulation of BMAL1:CLOCK-mediated transcription on both the Per1 and SRC-2 promoters, and found that overexpression of SRC-2 enhanced BMAL1:CLOCK activity (Figures 2G and S2F). To determine whether SRC-2 transcriptional coactivation was dependent on the BMAL1:CLOCK heterodimer, we used small interfering RNA (siRNA) to deplete Bmal1. We found that SRC-2 coactivation of both the Per1 and SRC-2 promoters was impaired when compared with control levels of activity (Figures 2H, S2G, and S2H). Likewise, siRNA depletion of SRC-2 alone impaired BMAL1 activity on both the Per1 and SRC-2 promoters (Figures 2I, S2I, and S2J). Given that SRC-2 can transactivate the BMAL1:CLOCK heterodimer and the convergence of their hepatic cistromes, we investigated a possible endogenous interaction between SRC-2 and BMAL1:CLOCK in the liver. We found that endogenous hepatic SRC-2 immunoprecipitates with BMAL1 at ZT6 corresponding to peak SRC-2 protein expression (Figure 2J). We recapitulated these results in cell culture, with endogenous SRC-2 coimmunoprecipitating with both BMAL1 and CLOCK (Figure S2K). We used an in vitro pull-down to determine whether SRC-2 can directly interact with BMAL1, and found the region of SRC-2 that contains the LXXLL amino acid motifs (730–1121) to be required for BMAL1 binding (Figure S2L), suggesting that SRC-2 may interface with BMAL1 similarly to its interaction with other NRs (Li et al., 2004; Ye et al., 2011). Overall, these data suggest that SRC-2 can directly interact with BMAL1 and CLOCK, and serves as a transcriptional coactivator for the BMAL1:CLOCK heterodimer. These events may be driven by temporal interactions with the BMAL1:CLOCK heterodimer.
SRC-2 Modulates Diurnal Behavior
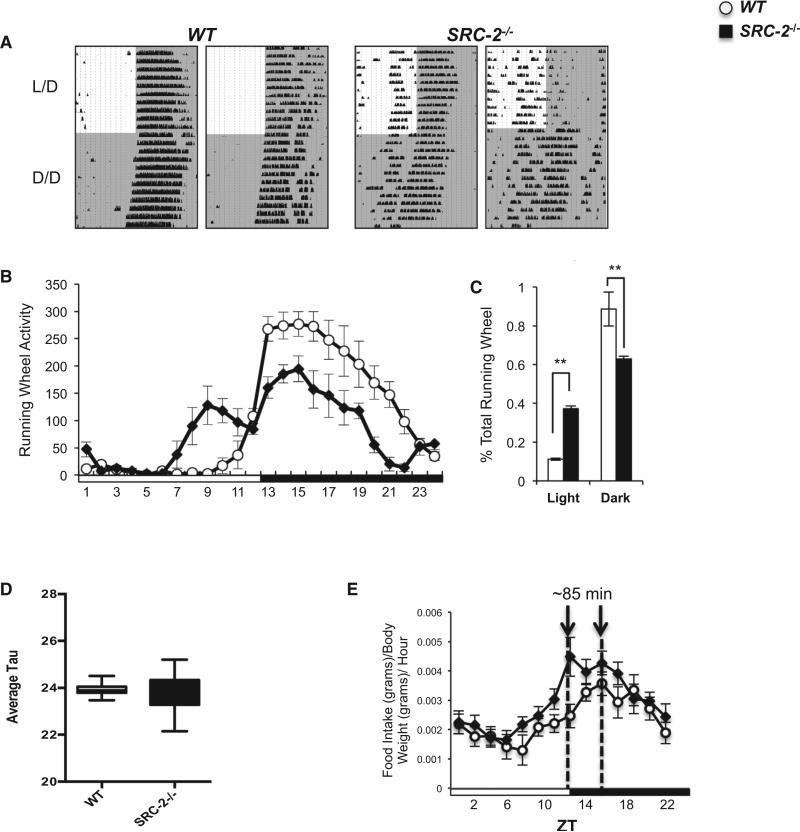
We subsequently examined the effects of genetic SRC-2 ablation on diurnal behavior by analyzing locomotor activities of WT and SRC-2−/− mice in 12 hr light/12 hr dark (24 hr L/D) cycles and in constant darkness (24 hr D/D cycles). We found that loss of SRC-2 markedly altered circadian locomotor behaviors under L/D entrained conditions (Figure 3A). The phenotypic spectrum observed in SRC-2−/− mice in L/D cycles ranged from phase-advanced behavior manifested by running activity during the light phase (SRC-2−/−, Figure 3A, left panel) to complete arrhythmic behavior (SRC-2−/−, Figure 3A, right panel). The abnormal behavior pattern for mutant mice persisted in the absence of environmental light cues (24 hr D/D cycles). Locomotor activity quantified during L/D cycles revealed that SRC-2−/− mice exhibit a bimodal running pattern, with the first minor peak of activity occurring late in the light phase and the second, major peak of activity occurring in the dark phase (Figure 3B). Interestingly, the amplitude of the major activity peak was significantly dampened in SRC-2−/− mice (Figure 3C). Therefore, unlike WT mice, in which >90% of activity occurred in the dark phase, the running activity of SRC-2−/− mice was scattered, with 40% in the light phase and only 60% in the dark phase (Figure 3C). Although SRC-2 ablation did not significantly affect the average period length for the mutant population, as shown by the near-equivalent mean tau values for WT and SRC-2−/− mice, it produced considerable daily variation in period length within individual SRC-2−/− mice (Figures 3D and S3A–S3C). Together, the above observations suggest that loss of SRC-2 disrupts the central clock, thereby altering L/D activity amplitudes.
Figure 3. Ablation of SRC-2 Disrupts Central Clock Rhythmicity.
(A) Representative actograms of male WT and SRC-2−/− (n = 6) littermate mice locomotor activity in L/D and D/D conditions.
(B) Graphical representation of locomotor activity of WT and SRC-2−/− (n = 6) mice in L/D.
(C) Percentage of time WT and SRC-2−/− (n = 6) mice spent running during a 12 hr light phase and 12 hr dark phase.
(D) Average tau for WT and SRC-2−/− mice during D/D conditions.
(E) Diurnal food consumption during L/D conditions as measured by the Columbus Instruments Comprehensive Lab Animal Monitoring System (CLAMS) in WT and SRC-2−/− (n = 9–10) mice. Data are graphed as the mean ± SEM. *p < 0.05, **p < 0.01 versus WT mice.
Circadian clock alterations are thought to promote several metabolic pathophysiologies. Using the discovered circadian defects in SRC-2−/− mice, we investigated diurnal feeding behavior. In accordance with published data (Chopra et al., 2008), we found no difference in total food consumption between WT and SRC-2−/− mice over 24 hr (data not shown). However, the peak feeding behavior of SRC-2−/− mice was shifted approximately 85 min before that of WT mice, mirroring the phase-advanced behavior seen in the actograms (Figure 3E). As in other tissues, we found cycling of SRC-2 expression in the SCN, with peak expression mirroring that in the liver (Figures S1F and S4A). To remove any effects of external light cues, we examined circadian gene expression during free-running conditions (D/D) and found significant differences in core circadian gene expression (Bmal1, Per1, and Cry1) primarily at CT22 and CT4 in the SRC-2−/− mice (Figure S3D). These results suggest that SRC-2 functions within the SCN to help maintain central clock oscillation.
Together, our data demonstrate that loss of SRC-2 results in phase-advanced phenotypes in locomotor activity and possibly in feeding behavior, and suggest a more intricate interplay of SRC-2 in coordination of circadian rhythm and metabolism.
SRC-2 Regulates Peripheral Clock Rhythmicity
Interactions between the master and peripheral clocks are necessary to maintain processes such as blood pressure, metabolism, digestion, and endocrine signaling (Mohawk et al., 2012; Ptitsyn et al., 2006). The strong behavioral defects observed with SRC-2 ablation in the running wheels led us to investigate the role of SRC-2 in several peripheral clocks.
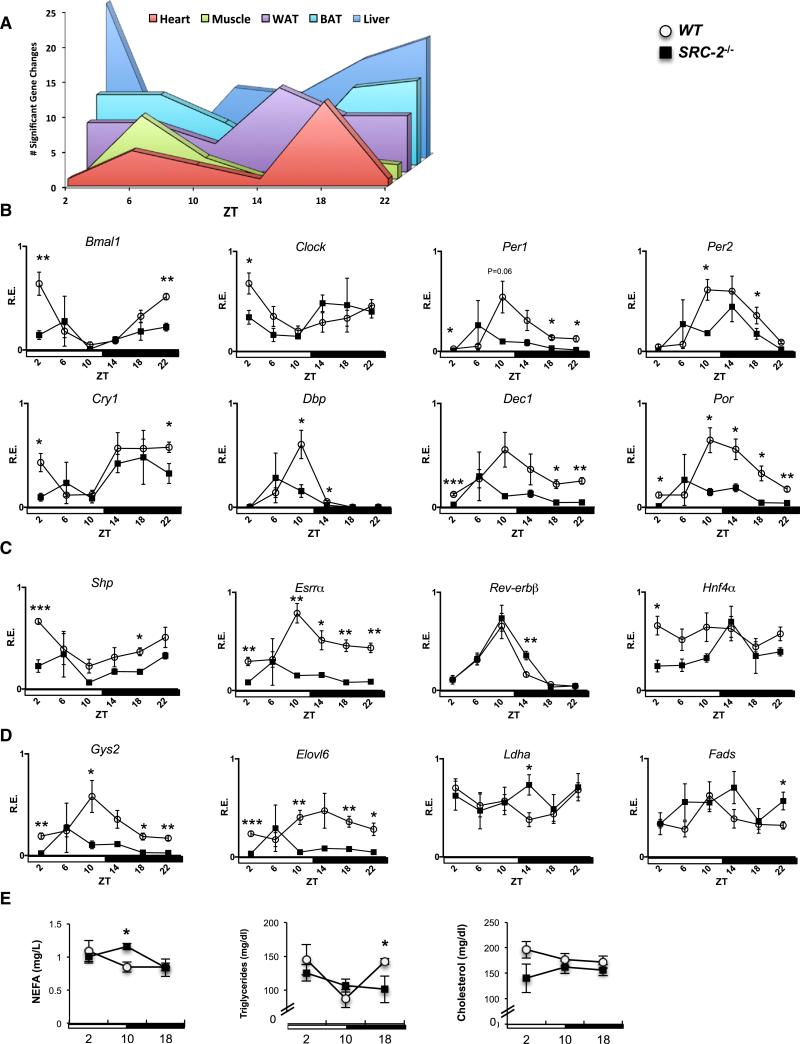
We used hd-qPCR to examine the systemic effect of SRC-2 ablation in the liver, heart, skeletal muscle, BAT, and WAT. We profiled 48 genes specifically chosen for sharing SRC-2 and BMAL1 binding sites in the ChIP-seq data sets, focusing on core circadian genes, NRs involved in coordinating metabolic cassettes, and several metabolic targets (Table S1). SRC-2 deletion significantly altered temporal expression patterns for all peripheral clocks, but in a tissue-specific manner (Figure 4A). We observed the most gene changes in the liver, with altered profiles found primarily at ZT2 and ZT22. Within the liver, altered expression between WT and SRC-2−/− mice was found in many circadian genes, including Bmal1, Clock, Per1/2, Npas2, and Cry1, as well as the clock-stabilizing genes Por, Dec1, and Dbp (Figure 4B). We also found profile changes in genes encoding the NRs (Shp [Nr0b2], Esrra, Rev-erbβ, and Hnf4a; Figure 4C) and several metabolic targets (Gys2, Elovl6, Ldha, and Fads2) (Figure 4D). We also investigated the effects of SRC-2 loss on circadian gene expression in WT and SRC-2 liver-specific knockout mice (LKO), and found that with SRC-2 absent from the liver only and an intact SCN, there was still aberrant circadian cycling (Figure S4A). Most similar to the liver was BAT, where again more gene changes occurred at ZT2-ZT6 and ZT18– ZT22, with changes in circadian genes (Por, Per1, and Cry2) and several metabolic genes (Dgat2 and Pnpla2; Figure S4B). In general, all tissues analyzed showed changes in the temporal expression of circadian oscillators. Similar to the metabolic processes revealed by hepatic ChIP-seq, we found that WAT, heart, and skeletal muscle also showed changes in genes involved in glucose metabolism (Pdk4 and Pdk1), lipid synthesis (Fads2, Elovl6, and Dgat2), and pyruvate metabolism (Pdk4 and Pdk1; Figures S4B–S4E). Overall, the fewest changes were observed in skeletal muscle and heart compared with the other tissues, and interestingly, although the heart and skeletal muscle shared similar metabolic targets, we found that the changes in these tissues occurred at different phases (ZT6 in the skeletal muscle and ZT 18 in the heart; Figure 4A).
Figure 4. Loss of SRC-2 Results in Aberrant Peripheral Circadian Clock Gene Expression in the Liver.
(A) Graphical summary representation of the gene changes observed at different ZTs in core metabolic tissues using the hd-qPCR array.
(B) hd-qPCR analysis of temporal circadian gene expression in liver from entrained WT and SRC-2−/− (n = 3–5) mice.
(C) hd-qPCR analysis of NR gene expression.
(D) hd-qPCR analysis of metabolic genes. The maximal value from each analyzed gene was normalized to one.
(E) Plasma analysis of NEFAs, triglycerides, and cholesterol at ZT2, ZT10, and ZT18 from WT and SRC-2−/− (n = 3–5) mice. Data are graphed as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 versus WT mice.
Since many of the SRC-2 targets we identified have reported functions in metabolism, we next examined whether ablation of SRC-2 could affect the oscillations of circulating plasma metabolites. Colorimetric-based assays revealed a significant increase at ZT10 in SRC-2−/− mice in plasma nonesterified fatty acids (NEFA), a significant increase in SRC-2−/− triglycerides at ZT18, and overall no change in cholesterol concentrations between SRC-2−/− and WT mice (Figure 4E). Altered diurnal cycling of plasma metabolites is likely a consequence of temporal disruption of metabolic genes targeted by SRC-2, as we have shown by hd-qPCR.
SRC-2 Ablation Alters Hepatic Metabolites
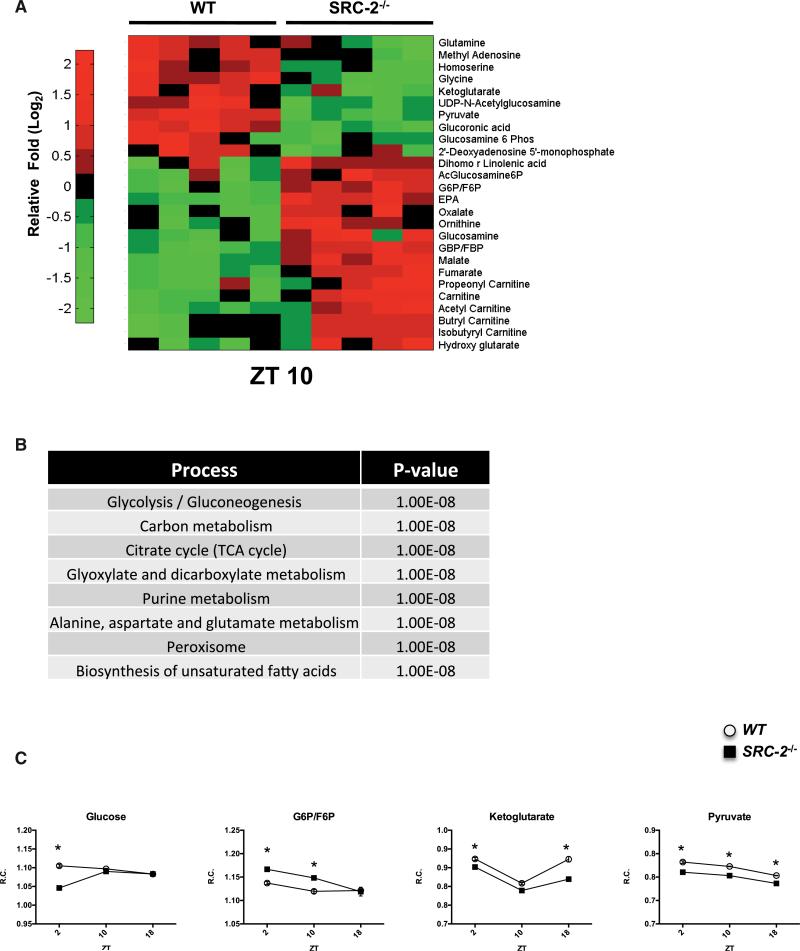
Subsequently, we went back to study the temporal regulation of metabolites in the liver of WT and SRC-2−/− mice. Our lab has previously identified that loss of SRC-2 has whole-body consequences for metabolite profiles in fed and fasted mice, but surprisingly few changes were found in the SRC-2−/− liver (York et al., 2013). Recent data suggest that in addition to the rhythmicity observed in the transcriptome, the metabolome is also subject to circadian regulation, as shown in the liver-specific Bmal1−/− mouse and Clock−/− mouse (Eckel-Mahan et al., 2012; Fustin et al., 2012). Therefore, we analyzed hepatic metabolomics over the diurnal cycle in WT and SRC-2−/− mice for metabolites in lipid/fatty acid pathways, glycolysis/gluconeogenesis, energy metabolism in the tricarboxylic acid cycle (TCA) cycle, and nucleotide synthesis. We again observed that SRC-2−/− mice showed a temporal effect with deregulation of cycling metabolites (Figures 5A, S5A, and S5B). Most of the changes observed occurred at ZT2 and ZT10, which is in agreement with our initial ChIP-seq and gene-expression studies. More specifically, the metabolite changes we observed were found to be enriched in processes associated with carbon metabolism and glycolysis/gluconeogenesis (Figure 5B). These findings support functional correlates with the processes predicted from the overlap of the SRC-2 and BMAL1 cistromes (Figure 2B). Further, analysis of selected metabolites within the glycolysis/gluconeo-genesis and the TCA cycle, including glucose, glucose-6-phosphate/fructose-6-phosphate (G6P/F6P), ketoglutarate, and pyruvate, showed that loss of SRC-2 alters the flux of hepatic metabolites in a time-dependent manner (Figure 5C).
Figure 5. Loss of SRC-2 Disrupts the Rhythmicity of the Hepatic Metabolome.
(A) Heatmap of the metabolites analyzed in WT and SRC-2−/− mice at ZT10.
(B) Functional analysis of significant metabolites altered at ZT10.
(C) Selected metabolite changes in WT and SRC-2−/− mice at ZT2, ZT10, and ZT18 (n = 3–5). Data are graphed as the mean ± SEM. *p < 0.05 versus WT mice.
SRC-2 Regulation of Metabolic Rhythmicity Is Cell Autonomous
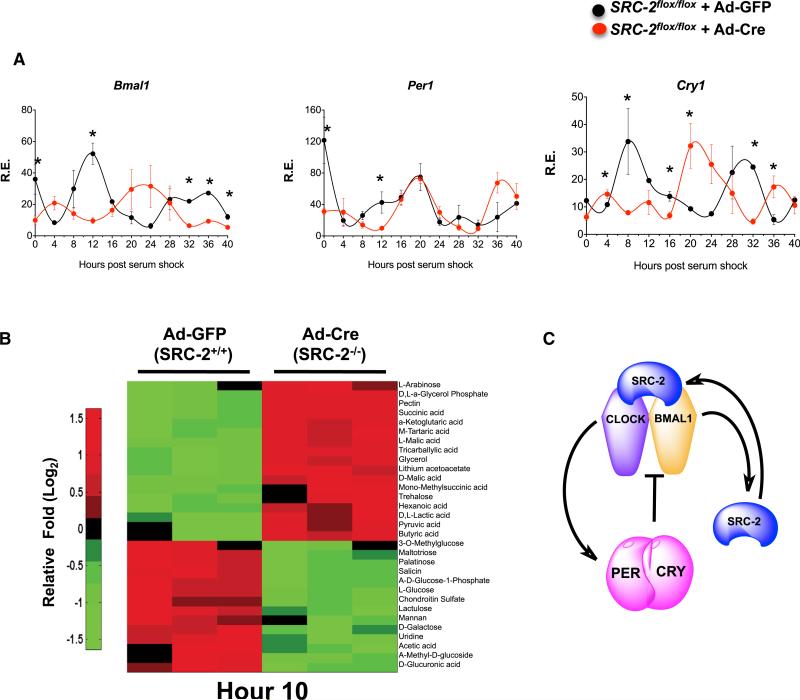
We compared systemic regulation versus cell-autonomous metabolic regulation with loss of SRC-2 by using a cell-autonomous system to remove any associated tissue-specific endogenous circadian influences. We isolated and immortalized mouse embryonic fibroblasts (MEFs) from SRC-2flox/flox animals (Figure S6A) and then infected the cells with either GFP adenovirus (Ad-GFP) or Cre-recombinase-GFP adenovirus (Ad-Cre-GFP). With SRC-2 knockout in MEFs, we still observed altered gene expression of several circadian genes, including Bmal1, Per1, and Cry1, in Ad-Cre-GFP SRC-2flox/flox MEFs (Figure 6A). Similar to the results from qPCR in cycling MEFs and liver (Figure 4B), we saw that depletion of SRC-2 in MEFs reduced the amplitude of Per2 (Figure S6B). Next, we compared primary hepatocytes from WT and SRC-2−/− mice with the circadian rhythm data obtained from whole-liver physiology. Consistent with the tissue-specific data, expression of core circadian genes was also altered in SRC-2−/− hepatocytes (Figure S6C).
Figure 6. SRC-2 Deletion Significantly Disrupts the Cell-Autonomous Clock.
(A) qPCR analysis of circadian gene expression in synchronized MEFs isolated from SRC-2flox/flox mice infected with either Ad-GFP or Ad-Cre-GFP. Data represent triplicates at each time point for maximal gene expression of Ad-GFP normalized to one, graphed as the mean ± SEM. *p < 0.05 versus WT.
(B) Heatmap of metabolic phenotype microarray of carbon metabolism in synchronized MEFs isolated from SRC-2flox/flox mice infected with either Ad-GFP or Ad-Cre-GFP at Hr10 (n = 3). Data are graphed as the mean ± SEM. *p < 0.05 versus WT mice.
(C) Schematic representation of the role of SRC-2 as a BMAL1:CLOCK transcriptional coactivator for core clock genes.
Next, we explored the cell-autonomous role of SRC-2 in the hepatic metabolic phenotypes seen in the SRC-2−/− mice. In line with the liver metabolome, quantitative analysis showed that SRC-2 deletion alters the metabolic phenotype of cells, specifically highlighting differences in carbohydrate usage. We found differences in carbon metabolism between Ad-GFP and Ad-Cre-GFP SRC-2flox/flox MEFs enriching in glycolysis and gluconeo-genesis pathways (Table S2). Kinetic analysis of metabolite uptake and utilization revealed that SRC-2 ablation imparts stronger effects at hour 10 (Hr10; Figure 6B) than at Hr2 or Hr18 (Figures S6D and S6E), suggesting that SRC-2 plays a pivotal role in cell-autonomous metabolic regulation. Collectively, these data indicate that SRC-2 is important for maintaining metabolic rhythmicity in a cell-autonomous manner.
Our model proposes that SRC-2 serves a transcriptional coactivator of the BMAL1:CLOCK heterodimer during the light phase of the circadian clock. Notably, SRC-2 also targets its own promoter in a feed-forward loop as a BMAL1:CLOCK circadian target (Figure 6C).
DISCUSSION
Our collective analysis of SRC-2 strongly indicates that it plays a systemic role in coordinating circadian and metabolic gene expression in peripheral clockworks. Circadian control depends largely on the coordinated rhythmicity of central and peripheral clocks, which at the molecular level are driven by the BMAL1: CLOCK heterodimer. We have demonstrated that SRC-2 is a transcriptional coregulator of BMAL1:CLOCK, and loss of SRC-2 disrupts diurnal rhythms stemming from the central clock, as revealed by abnormal running-wheel behaviors downstream of the transcriptome and metabolome in the liver peripheral clock. Additionally, we investigated the relevance of the overlapping SRC-2 and BMAL1 targets in human epidemiology through the HuGE Navigator, and found enrichments in human diseases such as glycogen storage disease type 1 and Von Gierke's disease (Table S3; Chopra et al., 2008), as well as several mental disorders in which we previously observed neuro-behavioral defects in the SCR-2−/− mice (Stashi et al., 2013). We also found other possible disease associations for SRC-2, including thrombosis, premature aging, and sleep disorders pertaining to circadian rhythm, all of which BMAL1 is known be associated with (Bunger et al., 2000; Kondratov et al., 2006; Somanath et al., 2011).
We have also demonstrated that SRC2 deletion disrupts metabolic rhythmicity at the cell-autonomous level. It is surprising that SRC-2 ablation has such a dramatic effect on the central circadian clock and peripheral clocks, given its previously unidentified role as a coregulator in circadian regulation.
Coactivators and corepressors are well recognized to assist in circadian transcriptional regulation by either amplifying or suppressing gene expression. Other coregulators have been described to play a role in circadian rhythm as well, including NCoR with HDAC3, PGC1-α, RIP140, JARID1a, MAP1a, TRAP150, and several others that influence the activities of both NRs and BMAL1:CLOCK (Curtis et al., 2004; DiTacchio et al., 2011; Feng et al., 2011; Lande-Diner et al., 2013; Li et al., 2013; Liu et al., 2007; Poliandri et al., 2011). We believe that the ability of SRC-2 to positively regulate circadian gene expression is largely achieved by coactivation of BMAL1:CLOCK and by SRC-2 targeting its own promoter, possibly to strengthen gene expression in a feed-forward loop. This suggests that SRC-2 serves a physiological function in modulating the dynamic multiparametric metabolic and circadian responses of living systems (Figure 6C).
The link between metabolism and circadian regulation is un-disputed. Metabolic deregulations and associated pathologies are seen in other circadian mutant mouse models. For example, analogously to SRC-2−/− mice, the Bmal1 mutant not only has gross central and peripheral clock disruptions but also shows impaired gluconeogenesis, increased insulin sensitivity, infertility, and defects in adipose accretion (Chopra et al., 2008, 2011; Hatanaka et al., 2010; Kondratov et al., 2006; Picard et al., 2002; Rudic et al., 2004). Our data reveal that SRC-2 has a surprisingly strong circadian phenotype and serves as an important coregulator for the molecular clock by positively stimulating the transcriptional activity of BMAL1:CLOCK. In addition, SRC-2 also may function with AMPK or other energy-sensing molecules to help intercalate circadian and metabolic transcriptional regulation (Lamia et al., 2009; Um et al., 2011).
Our studies highlight SRC-2 as a potent transcriptional coactivator of BMAL1:CLOCK, providing an integral link between the previously known physiological importance of SRC-2 and its newly appreciated role in circadian rhythm. Given the numerous metabolic roles of SRC-2 and its known roles in reproduction and now circadian rhythm, this work further solidifies the systemic importance of SRC-2 and aims to provide a mechanism to link these processes. In this way, SRC-2 has evolved as a master integrator of energy accretion, energy storage, reproduction, cellular proliferation, and (as shown herein) the circadian clock (O'Donnell et al., 2012; Chopra et al., 2008, 2011; Gehin et al., 2002; Hartig et al., 2011; Picard et al., 2002). The evolutionary advantages of a master regulator for these interrelated physiological processes are evident. Through loss of SRC-2 alone, we have shown a disruption of total physiology, and thus conclude that SRC-2 may play a much greater role in the regulatory axis of energy, circadian rhythm, and reproduction than previously thought.
EXPERIMENTAL PROCEDURES
Animals
The generation of SRC-2−/−, SRC-2flox/flox, SRC-2 LKO, and Bmal1−/− mice was described previously (Bunger et al., 2000; Chopra et al., 2011; Gehin et al., 2002). Congenic WT and SRC-2−/− male littermate mice (ages 10– 20 weeks) were used for all studies, except for generation of MEFs. All mice were entrained under strict temperature control and ad libitum food (normal chow, 2920X; Teklad Global) and water access under 12 hr L/D conditions for at least 2 weeks prior to tissue collection for RNA and protein. The Animal Care and Research Committee at Baylor College of Medicine approved all animal studies.
ChIP-Seq
ChIP-seq was performed by Active Motif on C57BL/6J WT liver lysates sacrificed at ZT4 and ZT18, and SRC-2−/− liver lysate at ZT4 using the mouse SRC-2 antibody 346A (Bethyl Laboratories). Approximately 24 million reads identified by Illumina's Genome Analyzer 2 were mapped to the mouse genome (Build37.1/mm9) using the ELAND software. Only the sequences that uniquely mapped with no more than two mismatches in the first 28 bp were kept and used as valid reads. Peak calling was carried out by MACS 1.3.7.1 (option: p value = 1 × 10−10 and MFOLD 30) were used.
ChIP-Seq Data Analysis
Galaxy/Cistrome's SeqPos motif tool (v1.0.0) (http://cistrome.dfci.harvard.edu/ap/root) was utilized on the entire SRC-2 ChIP-seq interval set (20,599 intervals) for de novo (MDscan algorithm) and reference-based (Transfac, JASPAR, pbm, and hPDI databases) sequence motif analyses, and Galaxy/ Cistrome's implementation of the CEAS tools was used for enrichment analyses on chromosomes and genomic annotations. DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resources v6.7 (http://david.abcc.ncifcrf.gov/) was used on protein coding genes to identify enriched biological themes. For data integration and metagenomics data analysis, the binding sites for SRC-2 and BMAL1 (Koike et al., 2012) were related to mouse reference gene annotations (ftp://ftp.ncbi.nlm.nih. gov/genomes/MapView/Mus_musculus/sequence/BUILD.37.1) and formed into a relational database system for easy querying and data sharing.
ChIP
Liver tissue was isolated and flash frozen at designated ZTs from WT or selected knockout mice as indicated. Chromatin was isolated using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling) and performed per the manufacturer's suggestion. The SRC-2 (Bethyl) and BMAL1 (Abcam) antibodies were used at 3 μg/ml. qPCR was performed with gene-specific primers and SyberGreen technology (Applied Biosystems) normalized to total input DNA. All ChIP primer sequences are available upon request.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Ljungberg and staff at the IDDRC RNA In Situ Hybridization Core for performing all in situ hybridization staining, supported by BCM IDDRC grant 5P30HD024064-23 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. We thank the MMRU staff for performing measurements of food intake in the Mouse Metabolic Research Unit at the USDA/ARS Children's Nutrition Research Center, supported by funds from the USDA ARS (https://www.bcm.edu/cnrc/mmru). We would also like to thank V. Putluri for her help with the metabolic phenotype microarray. This research was supported by grants from the NIH (NCI F31CA171350 to E.S., NIDDK PO1 P01 DK59820, and 3U19DK062434-10W1 to B.W.O'M.) and the Welch Foundation (Q1521). Additionally, funding support was provided from the Center for the Advancement of Science in Space (CASIS) Integrated OMICs Award (to B.Y., R.B.L., A.S., and B.W.O.). Partial funding support was provided by NIH grants R01 CA137019-01A (to L.F.) and P01 DK59820, CPRIT RP120092 (to A.S. and N.P.), and the Genetically Engineered Mouse Shared Resources with F. DeMayo. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
ACCESSION NUMBERS
The ChIP-seq data sets have been deposited in the NCBI Gene Expression Omnibus and are accessible under accession number GSE53039.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.01.027.
REFERENCES
- Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong L-W, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra AR, Louet JF, Saha P, An J, Demayo F, Xu J, York B, Karpen S, Finegold M, Moore D, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke's disease. Science. 2008;322:1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra AR, Kommagani R, Saha P, Louet J-F, Salazar C, Song J, Jeong J, Finegold M, Viollet B, DeMayo F, et al. Cellular energy depletion resets whole-body energy by promoting coactivator-mediated dietary fuel absorption. Cell Metab. 2011;13:35–43. doi: 10.1016/j.cmet.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis AM, Seo SB, Westgate EJ, Rudic RD, Smyth EM, Chakravarti D, FitzGerald GA, McNamara P. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J. Biol. Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J, Panda S. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour CR, Levasseur M-P, Pham NHH, Eichner LJ, Wilson BJ, Charest-Marcotte A, Duguay D, Poirier-Héon J-F, Cermakian N, Giguère V. Genomic convergence among ERRα, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS Genet. 2011;7:e1002143. doi: 10.1371/journal.pgen.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc. Natl. Acad. Sci. USA. 2012;109:5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin J-M, Doi M, Yamada H, Komatsu R, Shimba S, Okamura H. Rhythmic nucleotide synthesis in the liver: temporal segregation of metabolites. Cell Rep. 2012;1:341–349. doi: 10.1016/j.celrep.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Gehin M, Mark M, Dennefeld C, Dierich A, Gronemeyer H, Chambon P. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol. Cell. Biol. 2002;22:5923–5937. doi: 10.1128/MCB.22.16.5923-5937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig SM, He B, Long W, Buehrer BM, Mancini MA. Homeostatic levels of SRC-2 and SRC-3 promote early human adipogenesis. J. Cell Biol. 2011;192:55–67. doi: 10.1083/jcb.201004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka F, Matsubara C, Myung J, Yoritaka T, Kamimura N, Tsutsumi S, Kanai A, Suzuki Y, Sassone-Corsi P, Aburatani H, et al. Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol. Cell. Biol. 2010;30:5636–5648. doi: 10.1128/MCB.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J-W, Kwak I, Lee KY, White LD, Wang XP, Brunicardi FC, O'Malley BW, DeMayo FJ. The genomic analysis of the impact of steroid receptor coactivators ablation on hepatic metabolism. Mol. Endocrinol. 2006;20:1138–1152. doi: 10.1210/me.2005-0407. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande-Diner L, Boyault C, Kim JY, Weitz CJ. A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc. Natl. Acad. Sci. USA. 2013;110:16021–16026. doi: 10.1073/pnas.1305980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Kim JH, Koh SS, Stallcup MR. Synergistic effects of co-activators GRIP1 and beta-catenin on gene activation: cross-talk between androgen receptor and Wnt signaling pathways. J. Biol. Chem. 2004;279:4212–4220. doi: 10.1074/jbc.M311374200. [DOI] [PubMed] [Google Scholar]
- Li D-Q, Pakala SB, Reddy SDN, Peng S, Balasenthil S, Deng C-X, Lee CC, Rea MA, Kumar R. Metastasis-associated protein 1 is an integral component of the circadian molecular machinery. Nat Commun. 2013;4:2545. doi: 10.1038/ncomms3545. [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Keng VW, York B, Reineke EL, Seo D, Fan D, Silverstein KA, Schrum CT, Xie WR, Mularoni L, et al. A Sleeping Beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer. Proc. Natl. Acad. Sci. USA. 2012;109:E1377–E1386. doi: 10.1073/pnas.1115433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Géhin M, Annicotte JS, Rocchi S, Champy M-F, O'Malley BW, Chambon P, Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Poliandri AHB, Gamsby JJ, Christian M, Spinella MJ, Loros JJ, Dunlap JC, Parker MG. Modulation of clock gene expression by the transcriptional coregulator receptor interacting protein 140 (RIP140). J. Biol. Rhythms. 2011;26:187–199. doi: 10.1177/0748730411401579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Ptitsyn AA, Zvonic S, Conrad SA, Scott LK, Mynatt RL, Gimble JM. Circadian clocks are resounding in peripheral tissues. PLoS Comput. Biol. 2006;2:e16. doi: 10.1371/journal.pcbi.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis A-M, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanath PR, Podrez EA, Chen J, Ma Y, Marchant K, Antoch M, Byzova TV. Deficiency in core circadian protein Bmal1 is associated with a prothrombotic and vascular phenotype. J. Cell. Physiol. 2011;226:132–140. doi: 10.1002/jcp.22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stashi E, Wang L, Mani SK, York B, O'Malley BW. Research resource: loss of the steroid receptor coactivators confers neurobehavioral consequences. Mol. Endocrinol. 2013;27:1776–1787. doi: 10.1210/me.2013-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong H-K, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K-I, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Um JH, Pendergast JS, Springer DA, Foretz M, Viollet B, Brown A, Kim MK, Yamazaki S, Chung JH. AMPK regulates circadian rhythms in a tissue- and isoform-specific manner. PLoS ONE. 2011;6:e18450. doi: 10.1371/journal.pone.0018450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. Biochemical analysis of the canonical model for the mammalian circadian clock. J. Biol. Chem. 2011;286:25891–25902. doi: 10.1074/jbc.M111.254680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York B, Sagen JV, Tsimelzon A, Louet JF, Chopra AR, Reineke EL, Zhou S, Stevens RD, Wenner BR, Ilkayeva O, et al. Research resource: tissue- and pathway-specific metabolomic profiles of the steroid receptor coactivator (SRC) family. Mol. Endocrinol. 2013;27:366–380. doi: 10.1210/me.2012-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.