Abstract
Despite recent evidences suggesting that agents inducing endoplasmic reticulum (ER) stress could be exploited as potential antitumor drugs in combination with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), the mechanisms of this anticancer action are not fully understood. Moreover, the effects of ER stress and TRAIL in nontransformed cells remain to be investigated. In this study we report that ER stress-inducing agents sensitizes both transformed and nontransformed cells to TRAIL-induced apoptosis. In addition, glucose-regulated protein of 78 kDa (GRP78) knockdown by RNA interference induces ER stress and facilitates apoptosis by TRAIL. We demonstrate that TRAIL death-inducing signaling complex (DISC) formation and early signaling are enhanced in ER stressed cells. ER stress alters the cellular levels of different apoptosis-related proteins including a decline in the levels of FLIP and Mcl-1 and the up-regulation of TRAIL-R2. Up-regulation of TRAIL-R2 following ER stress is dependent on the expression of PKR-like ER kinase (PERK) and independent of CAAT/enhancer binding protein homologous protein (CHOP) and Ire1α. Silencing of TRAIL-R2 expression by siRNA blocks the ER stress-mediated sensitization to TRAIL-induced apoptosis. Furthermore, simultaneous silencing of cFLIP and Mcl-1 expression by RNA interference results in a marked sensitization to TRAIL-induced apoptosis. Finally, in FLIP-overexpressing cells ER stress-induced sensitization to TRAIL-activated apoptosis is markedly reduced. In summary, our data reveal a pleiotropic mechanism involving both apoptotic and anti-apoptotic proteins for the sensitizing effect of ER stress on the regulation of TRAIL receptor-mediated apoptosis in both transformed and non-transformed cells.
Keywords: ER stress, TRAIL, Death-inducing signaling complex, TRAIL-R2, FLIP, Mcl-1
Introduction
TRAIL/Apo2L is a cytokine of the tumor necrosis factor (TNF) gene superfamily that selectively induces apoptosis in many tumor cells while leaving normal cells intact and thus is an attractive candidate for antitumor therapies [1]. TRAIL induces apoptosis through its binding to membrane bound death domain (DD)-containing receptors TRAIL-R1/DR4 and TRAIL-R2/DR5. This interaction induces the recruitment of the intracellular adaptor molecule FADD (FAS-associated death domain protein), that concurrently engages procaspase-8 at the death-inducing signaling protein complex (DISC) [2]. Within the DISC, caspase-8 is activated by transcatalytic and autocatalytic cleavage and released into the cytoplasm, initiating the protease cascade that leads to the effector caspases activation, thereby triggering the execution of steps that induce apoptosis (extrinsic apoptotic pathway). In addition, activated cas-pase-8 is able to cleave Bid, a BH3-only proapoptotic member of the Bcl-2 family protein, releasing a truncated protein (tBid) that translocates to the mitochondrial external membrane and, in concert with other proapoptotic Bcl-2 family proteins, as Bax and Bak, induces the release of apoptogenic factors, thereby amplifying caspase activation [3]. The apoptotic signal from the DISC may be inhibited by the cellular FLICE-inhibitory protein (FLIP) [4]. In most cells, two alternatively spliced isoforms of cFLIP exist: a caspase-8 homologue cFLIPL that lacks the amino acids critical for proteolytic caspase activity; and cFLIPS, which is comprised of the two death effector domains alone [4]. Although the role of cFLIP in apoptotic signaling remains controversial, there is strong evidence that it displays antiapoptotic activity [5–7]. Despite the ubiquitous expression of TRAIL receptors, some tumor cells, including most breast cancer cells, show either partial or complete resistance to the apoptotic effects of TRAIL [8, 9]. To overcome this resistance, TRAIL-induced signaling is being studied alone or in combination with different sensitizing agents, such as chemotherapeutic drugs, cytokines and inhibitors of survival pathways [10–12].
The ER is a central cellular organelle that is responsible for the folding and maturation of most secreted and transmembrane proteins. In response to different environmental and physiological stress conditions that increase the load of unfolded proteins in the ER, protein sensors located in the luminal face of the ER membrane activate an intracellular signaling mechanism called the unfolded protein response (UPR) to enhance the expression of ER-resident chaperones [13]. Moreover, insufficient vascularization of solid tumors and the increased production of immunoglobulins in multiple myeloma cells are conditions that activate the UPR, to promote tumor survival and progression [14]. Furthermore, UPR activation in tumor cells has been shown to induce apoptosis, at least in part, through enhancing TRAIL-R2 expression [15–17]. Interestingly, ER stress and UPR activation sensitize tumor cells to exogenous TRAIL-induced apoptosis, although the molecular mechanism of this synergistic action remains unclear [18–20]. In addition, a cross-talk between the UPR and the TRAIL system has been reported. Thus, activation of TRAIL receptors induces the translocation of pro-apoptotic Par-4/GRP78 complex to the cell surface of tumor cells [17]. However, it is unknown whether ER stress has any impact on the sensitivity of nontransformed cells to TRAIL. We hereby report that UPR activation sensitizes both nontransformed and transformed cells to TRAIL-induced apoptosis. We also demonstrate that GRP78 plays a survival role in TRAIL-induced apoptosis because GRP78 knockdown by siRNA induces ER stress and markedly sensitizes cells to TRAIL. Furthermore, we show that ER stress up-regulates TRAIL-R2 expression through a PERK-dependent, CHOP and Ire1α-independent mechanism. Finally, we demonstrate that anti-apoptotic FLIP and Mcl-1 proteins are down-regulated in cells undergoing ER-stress and this contribute to the sensitization observed to TRAIL.
Materials and methods
Cell culture and reagents
Soluble human His-tagged recombinant TRAIL (residues 95–281) was produced as described [21]. Tunicamycin and thapsigargin were purchased from Sigma Chemical Corp (St. Louis, MO). zVAD-fmk was from Bachem AG (Bachem, Bubendorf, Switzerland). The human breast epithelial cell line MCF-10A was maintained in DMEM/F12 supplemented with 5% donor horse serum, 2 mM L-glutamine, 20 ng/ml epidermal growth factor (EGF), 10 μg/ml insulin, 100 ng/ml cholera toxin/ml, 0.5 μg/ml hydrocortisone, 50 U/ml penicillin, and 50 μg/ml streptomycin. The human hTERT-immortalized retinal pigment epithelial cell line hTERT RPE-1 was grown in DMEM/F12 medium with 10% foetal bovine serum, 2 mM glutamine and antibiotics. The human tumor cell lines MDA-MB231 (breast), MDA-MB231FLIPL [22], HeLa (cervix) and MM1s (multiple myeloma) were cultured in RPMI medium containing 10% foetal bovine serum, 2 mM glutamine and antibiotics. All cell lines were maintained at 37°C in a humidified 5% CO2, 95% air incubator.
Analysis of cell surface proteins by flow cytometry
Cells were detached from the culture dish with RPMI 1640 medium containing 3 mM EDTA and cytofluorimetric analysis of proteins was performed as described [22] using the CellQuest software (Becton–Dickinson, Mountain View, CA). Anti-TRAIL-R2 antibody was from Abcam (Cambridge, UK). FITC-conjugated affinity-purified rabbit anti-mouse immunoglobulins was purchased from DAKO (Glostrup, Denmark).
Isolation of the TRAIL DISC
DISC precipitation was performed using biotin-tagged recombinant TRAIL (bio-TRAIL) [21]. Cells were incubated for 15 h in the presence or absence of tunicamycin as indicated in the figure legend. After this incubation, cells were treated with bio-TRAIL for 30 min. DISC formation was determined as reported [12].
RNA interference
siRNAs against Bid (5′-GAAGACAUCAUCCGGAAUAdT dT-3′), TRAIL-R1 (5′-CACCAAUGCUUCCAACAAUdTd T-3′), TRAIL-R2 (5′-GACCCUUGUGCUCGUUGUCdTd T-3′), GRP78 (5′-GGAGCGCAUUGAUUGAUACUAGA dTdT-3′), CHOP (5′AAGAACCAGCAGAGGUCACAAd TdT-3′), Ire1alpha (5′GCGUCUUUUACUACGUAAUdTd T-3′), PERK (5′ CAAACUGUAUAACGGUUUAdTdT-3′) and a scrambled control (5′-GAGCGCUAGACAAUGAAGd TdT-3′) were from SIGMA-Proligo (Boulder, CO). Cells were transfected with siRNAs using DharmaFECT 1 (Dharmacon, Lafayette, CO) as described by the manufacturer.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was isolated from cells with Trizol reagent (Life Technologies, Inc.) as recommended by the supplier. cDNAs were synthesized from 2 μg of total RNA using a RT-PCR kit (PerkinElmer Life Sciences) with the supplied oligo(dT) primer under conditions described by the manufacturer. PCR reactions were performed using the following primers: XBP1s (forward, 5′CCTTGTAGTTGAGA CCCAGG-3′; reverse, 5′-GGGCTTGGTATATATGTG G-3′), CHOP (forward, 5′-GGTGGCAGCGCGACA GACCAAAAAT-3′; reverse 5′-TCCAAGCCTTCCCCCT GCGTA-3′), TRAIL-R2 (forward, 5′-GCCTCATGGACA ATGAGATAAAGGTGGCT-3′; reverse, 5′-CCAAACT CAAAGTACGCACAAACGG-3′). PCR cycle conditions were 95°C for 5 min followed either by 30 cycles of 95°C for 45 s, 55°C for 30 s, and 72°C for 30 s (CHOP) or 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for 30 cycles (TRAIL-R2). XBP1s mRNA was measured as described [23].
Analysis of apoptosis
Cells (5 × 105/well) were treated in 6-well plates as indicated in the figure legends. After treatment, hypodiploid apoptotic cells were detected by flow cytometry according to published procedures [24]. Briefly, cells were washed with phosphate buffered saline (PBS), fixed in cold 70% ethanol and then stained with propidium iodide while treating with RNAse. Quantitative analysis of cell cycle and hypodiploid cells was carried out in a FACSCalibur cytometer using the Cell Quest software (Becton–Dickinson, Mountain View, CA).
Chromatin condensation and fragmentation in apoptotic cells was assessed after staining of cellular DNA with DAPI by viewing the cell preparations under a Leica fluorescent microscope.
Immunoblot analysis of proteins
Proteins were resolved on SDS–polyacrylamide minigels and detected as described previously [12]. The following antibodies were used in our studies: anti-FLIP monoclonal antibody NF6 from Alexis Corp. (San Diego, CA), anti-caspase-8 generously provided by Dr. Gerald Cohen (Leicester University, UK), anti-FADD obtained from BD Bioscience (Erembodegem, Belgium), anti-GAPDH, myeloid cell leukemia-1 (Mcl-1), CHOP and GRP78 were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), anti-Bid, a kind donation of Dr. X Wang (Howard Hughes Institute, Dallas, Texas), antibodies against TRAIL-R1/DR4, and TRAIL-R2/DR5 from R&D Systems (Minneapolis, USA), Ire1α, p-eIF2α and anti-cleaved caspase-3 were from Cell Signaling (MA, USA), anti-caspase-9 monoclonal antibody was purchased from MBL International, Woburn, MA) and poly(ADP-ribose) polymerase (PARP) monoclonal antibody was from Roche Molecular Biochemicals (Monza, Italy).
Statistical analysis
All data are presented as the mean ± SD of at least three independent experiments. The differences among different groups were determined by the Student’s t test. P value <0.05 was considered significant.
Results
ER stress sensitizes both transformed and nontransformed cells to TRAIL-induced apoptosis
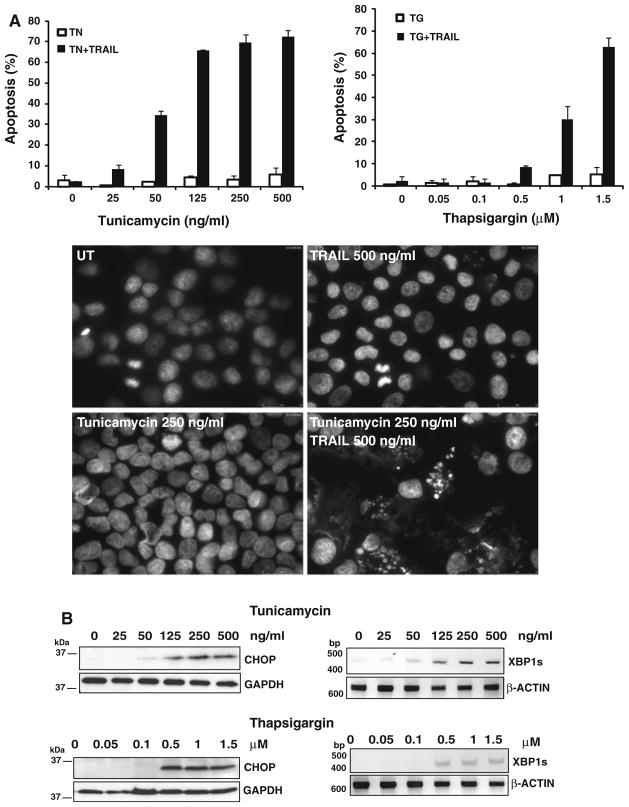
It has been suggested that agents that induce ER stress may increase the therapeutic response to TRAIL in tumor cells [18–20]. However, little is known about the impact that ER stress may have on the sensitivity to TRAIL in nontransformed cells. Hereby, we have addressed this issue by studying the effect of ER stress in the apoptotic response to TRAIL of different transformed and nontransformed cell lines. To examine the effect of ER stress on the apoptotic response to TRAIL, the breast cancer cell line MDA-MB231 was treated with different concentrations of tunicamycin to determine the sub-toxic dose capable of sensitizing these cells to TRAIL-induced apoptosis (Fig. 1a). Tunicamycin at doses between 25 and 500 ng/ml markedly sensitized MDA-MB231 cells to TRAIL. Lower panels show chromatin condensation and fragmentation only in cells treated with tunicamycin (15 h) followed by incubation in the presence of TRAIL (6 h). To further substantiate these observations we also determined the effect of the ER calcium pump inhibitor thapsigargin, a well known ER stress inducer, in the sensitivity of cancer cells to TRAIL. Treatment of MDA-MB231 cells with the ER calcium pump inhibitor thapsigargin clearly sensitized MDA-MB231 cells to the apoptotic action of TRAIL (Fig. 1a). Strikingly, the dose–response of the various ER stress inducers in the sensitization of cells to TRAIL-induced apoptosis closely correlated with the activation of the UPR as determined by the induction of the CCAAT/enhancer binding protein homologous protein (CHOP) as well as formation of the spliced mRNA form of the X-box binding protein-1 (XBP-1) XBP-1s (Fig. 1b).
Fig. 1.
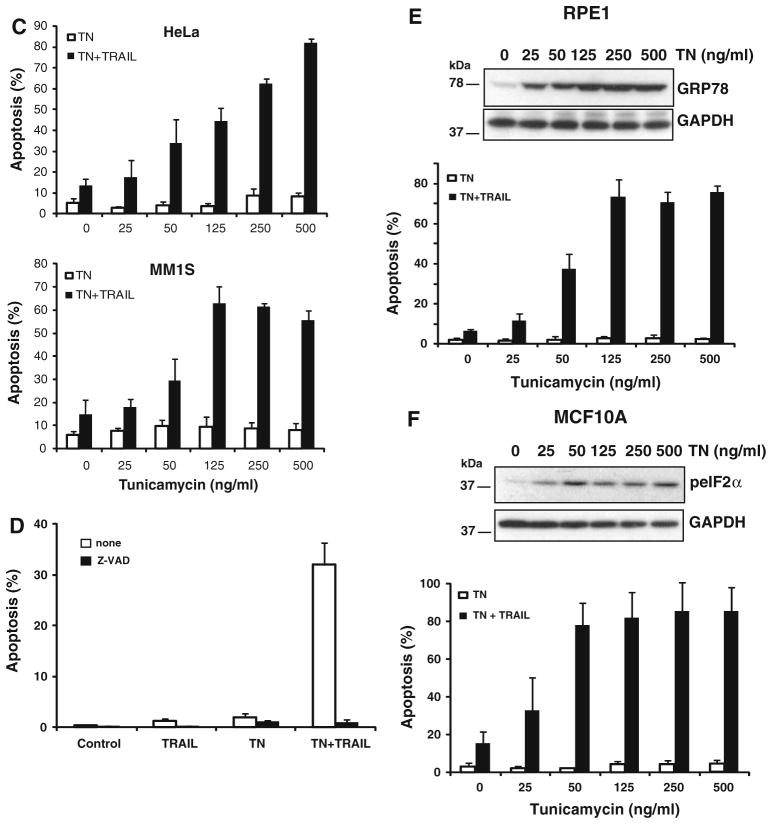
ER stress sensitizes both transformed and nontransformed cells to TRAIL-induced apoptosis. a MDA-MB231 cells (upper panels) were treated for 15 h with a range of concentrations of tunicamycin or thapsigargin prior to incubation in the presence or absence TRAIL (500 ng/ml) for 6 h. Apoptosis was assessed as described under “Materials and methods” section. Error bars represent S.D. from three different experiments. Lower panels show a picture of apoptotic and non-apoptotic cells in untreated cultures (UT) or cultures of MDA-MB231 cells treated as indicated. b Activation of the UPR was determined by immunoblotting (CHOP) or RT-PCR (XBP1s) after 15 h-treatment with tunicamycin or thapsigargin. GAPDH was used as a protein loading control. RT-PCR product of β-actin was used as a control for mRNA input. Results are representative of at least two independent experiments. c Hypodiploid cells were determined in HeLa (upper panel) or MM.1S (lower panel) cell cultures treated as described in a. Error bars represent S.D. from three different experiments. d MDA-MB231 cells were treated for 15 h with tunicamycin (250 ng/ml) prior to incubation with or without Z-VAD-fmk (50 μM) in the presence or absence TRAIL (500 ng/ml) for 6 h. Apoptosis was assessed as described under “Materials and methods” section. Error bars represent S.D. from three different experiments. Induction of apoptosis in RPE1 (e) and MCF-10A (f) cells treated as described in a. Error bars represent S.D. from three different experiments. Activation of the UPR was determined by western blot analysis of GRP78 expression (RPE1) and eIF2α phosphorylation (MCF-10A)
Subsequently, other cancer cell lines of different origin were tested to determine whether similar doses of tunicamycin also induced sensitization to TRAIL-induced apoptosis (Fig. 1c). In cervical carcinoma HeLa and multiple myeloma MM.1S cells, tunicamycin alone induced little cell death at the concentration indicated. However, if the cells were exposed to tunicamycin for 15 h before adding TRAIL, apoptotic cell death was markedly increased in both cancer cell lines. Caspase activation was required for the sensitization observed as the pan-caspase inhibitor Z-VAD-fmk completely abrogated the cell death induced by the combination of tunicamycin and TRAIL (Fig. 1d). We also investigated the effect of ER stress in the sensitivity of nontransformed cells to TRAIL. Breast (MCF10A) and retinal pigment (hTERT-RPE1) epithelial cells lines were treated with tunicamycin prior to the addition of TRAIL and apoptosis was determined. Results shown in Fig. 1e and f clearly indicate that tunicamycin activated the UPR and markedly promoted TRAIL-induced apoptosis in these cells. Collectively, these results demonstrated that the ER is a key regulator of the sensitivity of both transformed and nontransformed cells to TRAIL.
ER stress promotes TRAIL-induced activation of the mitochondrial apoptotic pathway
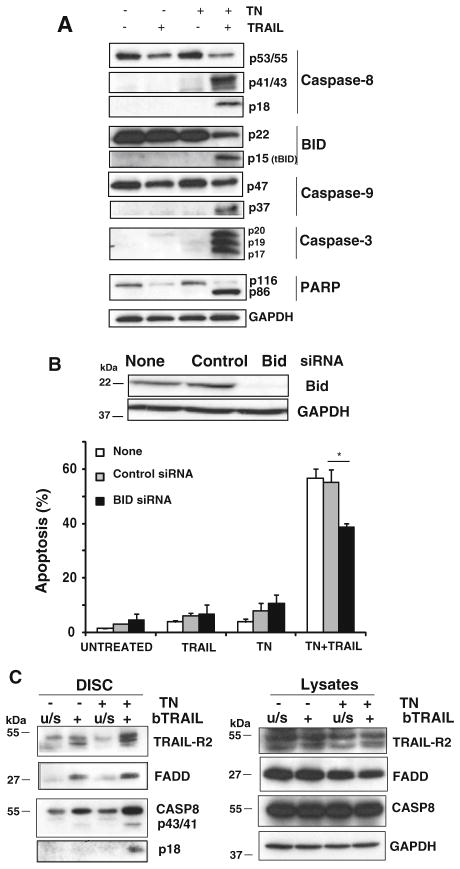
To further establish the mechanism of ER stress-promoted, TRAIL-induced apoptosis, we examined caspases activation during this cell death process. In TRAIL-induced apoptosis, procaspase-8 is recruited and processed at the DISC in a FADD- and TRAIL receptor-dependent manner. Procaspase-8 is first cleaved to the p43/41 intermediate fragments releasing the small subunit, p12, and then subsequently processed to generate the large catalytically active p18 subunit. To test if caspase-8 activation by TRAIL was blocked in the TRAIL resistant cell line MDA-MB231, processing of procaspase-8 was determined by western blotting in cells treated with TRAIL following ER stress. As shown in Fig. 2a, TRAIL-induced processing of procaspase-8 to its 43–41 kDa intermediate fragments was observed only in the cells previously treated with tunicamycin. Furthermore, full processing of caspase-8 to generate the p18 mature subunit only took place in ER stressed cells treated with TRAIL.
Fig. 2.
DISC formation and activation of a mitochondria-operated pathway of apoptosis by TRAIL in ER stressed cells. a MDA-MB231 cells were incubated with tunicamycin (250 ng/ml) for 15 h prior to treatment with TRAIL (500 ng/ml) for 4 h. Caspase-8 activation, Bid cleavage, caspase-9 processing, caspase-3 activation and PARP-1 cleavage were assessed by immunoblotting. GAPDH was used as a protein loading control. Results are representative of at least two independent experiments. b MDA-MB231 were transfected with 50 nM of either Bid or scrambled (control) siRNA for 48 h as described in “Materials and methods” section. After this incubation cells were treated with or without tunicamycin for 15 h prior to incubation with or without TRAIL (500 ng/ml) for 6 h and apoptosis was determined as percentage of subG1 cells. Error bars represent S.D. from three independent experiments. *P <0.05 as compared with scrambled siRNA. Immunoblot analysis was also performed in siRNA transfected cells to verify protein knockdown. c MDA-MB231 cells were either treated or not with tunicamycin (250 ng/ml) for 15 h before the incubation with biotin-labelled TRAIL (bTRAIL, 1 μg/ml) for 30 min. Unstimulated receptor controls (u/s) represent the addition of bTRAIL to an equivalent volume of lysate isolated from TRAIL-unstimulated cells. DISC was isolated as described in “Materials and methods” section, and its components TRAIL-R2, caspase-8 and FADD, analyzed by western blotting. Lysates are included as a positive control for the expression of these proteins in MDA-MB231 cells. Data shown are representative of three independent experiments
Activation of caspase-8 leads to the processing of its substrate Bid generating a 15 kDa fragment which translocates to mitochondria. Thus, we examined the activation of the mitochondria-controlled apoptotic pathway by TRAIL in tunicamycin-treated cells by determining the loss of intact Bid and the generation of truncated Bid (tBid). Results shown in Fig. 2a indicate that the amount of intact Bid was clearly diminished in the cells treated with tunicamycin and TRAIL in comparison with the cultures treated only with TRAIL or tunicamycin. Accordingly, formation of tBid was only observed in cells treated with tunicamycin and TRAIL. Similarly, caspase-9 and caspase-3 processing were also stimulated by TRAIL in ER stressed cells (Fig. 2a). In contrast, in the absence of tunicamycin, processing of caspases was barely detected. To further confirm that the apoptosis cascade was fully active in MDA-MB231 cells treated with both tunicamycin and TRAIL, we analyzed the processing of the nuclear protein PARP, a substrate of executioner caspases. As shown in Fig. 2a, PARP cleavage was only induced in cells pre-treated with tunicamycin and subsequently treated with TRAIL.
Ligation of pro-apoptotic TRAIL receptors by their ligand activates both mitochondria-dependent and independent mechanisms of apoptosis in various cell types [25, 26]. We then asked whether apoptosis induced by TRAIL in ER stressed cells required the activation of a mitochondria-operated pathway. For this purpose, we silenced the expression of the BH3-only protein Bid, a protein substrate of caspase-8 involved in the mitochondrial pathway activated by death receptors and determined apoptosis in MDA-MB231 cells treated with tunicamycin and TRAIL. Results shown in Fig. 2b demonstrate that Bid is at least partially involved in the apoptosis induced by ER stress and TRAIL which suggested a certain role of the mitochondria in this apoptotic pathway.
ER stress facilitates TRAIL DISC formation
In MDA-MB231 cells resistance to TRAIL occurred at a step prior to caspase-8 activation (Fig. 2a). In non-stressed cells, TRAIL induced the formation of a death-inducing signaling complex (DISC) with recruitment of procaspase-8 and FADD to the TRAIL-R2 (Fig. 2c). Caspase-8 processing to its p43/41 kDa forms was also observed at 30 min after TRAIL addition. Interestingly, ER stress clearly enhanced TRAIL DISC formation, as indicated by the higher amounts of TRAIL-R2, procaspase-8 and FADD in the DISC. Most importantly, in ER stressed cells but not in control cells, activation of procaspase-8 processing at the DISC following TRAIL addition proceeded to completion at 30 min upon TRAIL stimulation (Fig. 2c).
CHOP and Ire1α-independent, PERK-dependent up-regulation of TRAIL-R2 is involved in sensitization to TRAIL-induced apoptosis by ER stress
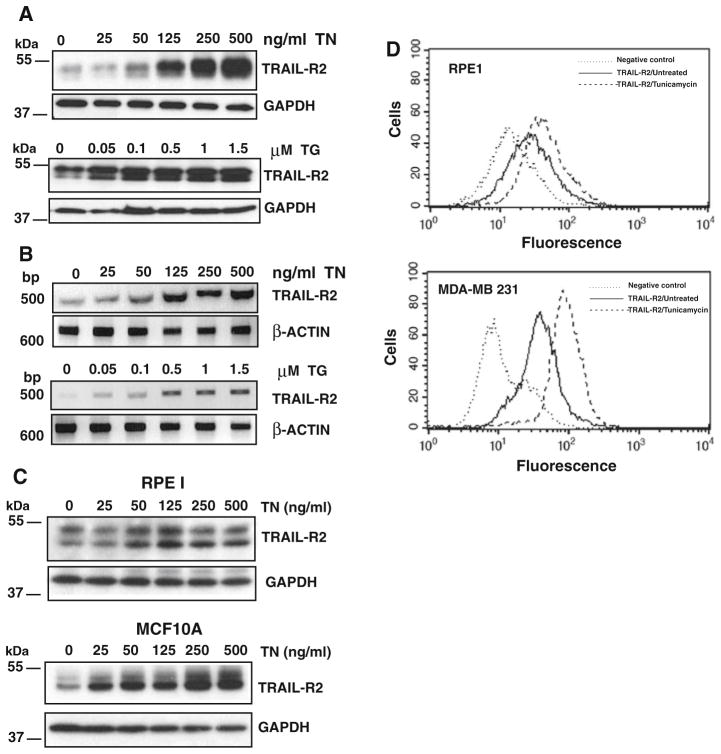
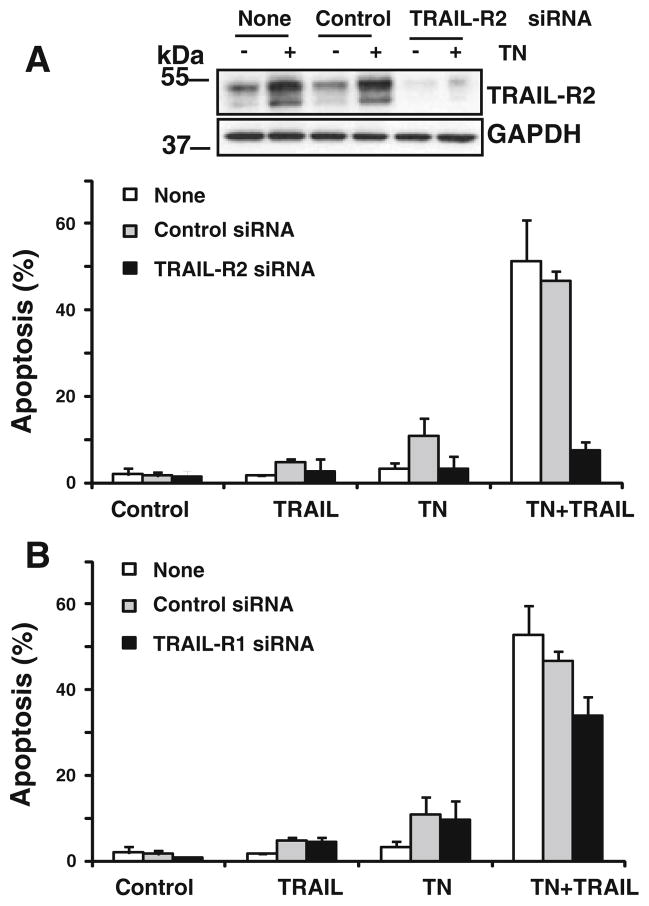
It has been reported that ER stress induces the expression of TRAIL-R2/DR5 [15, 16]. To elucidate the mechanism of the sensitization to TRAIL observed in our studies following ER stress, we first determined the expression of proapoptotic TRAIL-R2 in MDA-MB231 cells subjected to ER stress inducers. In these experiments, tunicamycin and thapsigargin induced a dose-dependent up-regulation of TRAIL-R2 protein and mRNA expression as measured by western blot and RT-PCR analysis, respectively (Fig. 3a, b). Strikingly, the dose–response relationship of TRAIL-R2 increase in ER stressed cells closely correlated with the sensitization data (Fig. 1a). Importantly, ER stress also induced TRAIL-R2 up-regulation in nontransformed cells (Fig. 3c). Moreover, ER stress-mediated increase in TRAIL-R2 expression was also observed at the cell surface, in both transformed and nontransformed cells (Fig. 3d). Next, we examined the importance of TRAIL-R2 up-regulation in the sensitization to TRAIL by silencing TRAIL-R2 expression with a siRNA duplex specific for TRAIL-R2 mRNA prior to ER stress. Results of Fig. 4a demonstrate that TRAIL-R2 siRNA efficiently down-regulated TRAIL-R2 protein expression and prevented TRAIL-R2 increase by tunicamycin. Interestingly, apoptosis induced by the combination of tunicamycin and TRAIL was completely prevented in cells transfected with siRNA to TRAIL-R2 (Fig. 4a).
Fig. 3.
ER stress induces TRAIL-R2 up-regulation in both transformed and nontransformed cells. Breast tumor cells MDA-MB231 were treated with the different ER stress inducers at the indicated concentrations for 15 h. TRAIL-R2 protein (a) and mRNA (b) levels were determined by immunoblotting and RT-PCR, respectively. c Nontransformed cells were incubated for 15 h in the presence or absence of the indicated concentrations of tunicamycin and TRAIL-R2 protein expression was assessed by western blot. d Nontransformed (RPE1) and transformed (MDA-MB231) cells were incubated for 15 h in the absence or presence of tunicamycin (250 ng/ml). Cell surface expression of TRAIL-R2 was determined by flow cytometry as described in “Materials and methods” section. Data shown are representative of three independent experiments
Fig. 4.
TRAIL-R2 plays a major role in apoptosis induced by TRAIL in ER stressed cells. MDA-MB231 cells were transfected with 50 nM TRAIL-R2 (a), TRAIL-R1 (b) or scrambled (control) siRNA (a, b) for 48 h prior to incubation in the absence or presence of tunicamycin (250 ng/ml) for 15 h. After this incubation cells were treated with or without TRAIL (500 ng/ml) for 6 h and apoptosis was determined as described in “Materials and methods” section. Error bars represent S.D. from three independent experiments. Protein knockdown was assessed by western blotting and GAPDH was used as a control for protein loading. Results are representative of at least three independent experiments
It has been reported that inhibition of glycosylation by tunicamycin facilitates TRAIL-R1 transport to the cell surface and overcome resistance to TRAIL [27]. We have examined the role of TRAIL-R1 in the sensitization of MDA-MB231 cells to TRAIL by tunicamycin. As shown in Fig. 4b silencing the expression of TRAIL-R1 partially inhibited apoptosis induced by the combination of tunicamycin and TRAIL. Together, all these results suggest that a certain amount of heteromeric complexes TRAIL-R1/R2 [28] may be involved in the induction of apoptosis by TRAIL in ER stressed cells.
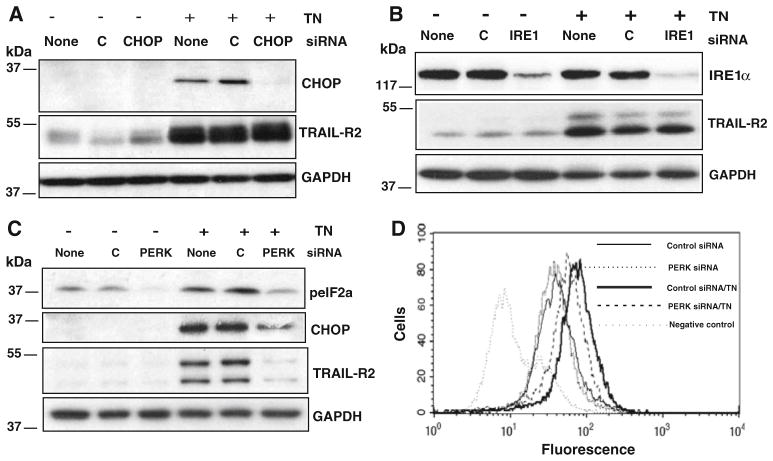
The C/EBP homologous protein (CHOP) has a dual role both as an inhibitor of C/EBPs function and as a transcriptional activator of other genes [29]. CHOP-dependent transcriptional up-regulation of TRAIL-R2 expression following ER stress has been reported in different cell types [16, 18]. We explored this issue in MDA-MB231 cells by silencing CHOP expression with siRNA prior to the activation of ER stress with tunicamycin. As can be observed in Fig. 5a RNA interference efficiently reduced cellular levels of CHOP protein. Rather unexpectedly, CHOP silencing did not prevent ER stress-mediated TRAIL-R2 up-regulation (Fig. 5a), suggesting that the role of this transcription factor in the regulation of TRAIL-R2 expression upon ER stress could be cell type-dependent.
Fig. 5.
ER stress up-regulates TRAIL-R2 through a PERK-dependent, CHOP and Ire1-independent mechanism. MDA-MB231 cells were transfected with scrambled (c) or CHOP (a), IRE1α (b) or PERK (c, d) siRNAs for 48 h to silence protein expression. Transfected cells were then treated with tunicamycin for 15 h prior to the analysis of protein knockdown and TRAIL-R2 expression by western blotting (a, b, c) or cell surface levels of TRAIL-R2 by flow cytometry (d). Results are representative of at least three independent experiments
It has been reported that the Ire1α/XBP-1 pathway of the UPR plays a certain role in TRAIL-R2 up-regulation following ER stress by 2-deoxy-D-glucose [20]. To further elucidate the role of the Ire1α arm of the UPR in the regulation of TRAIL-R2 expression by ER stress in MDA-MB231 cells Ire1α expression was silenced by RNA interference prior to ER stress induction with tunicamycin. In Ire1α-depleted cells we assessed the impact of ER stress on TRAIL-R2 expression. Knockdown of IRE1α did not prevent ER stress-induced TRAIL-R2 up-regulation as measured either in whole cell lysates (Fig. 5b) or cell surface expression (not shown).
An important ER stress sensor and UPR mediator is the PKR-like ER kinase (PERK), a transmembrane protein that is kept in an inactive state through its interaction with the ER chaperone GRP78/BIP [30]. Upon ER stress, GRP78/BIP dissociates from PERK leading to activation of this kinase. We examined the role of PERK in the regulation of TRAIL-R2 expression in MDA-MB231 cells treated with tunicamycin. Results shown in Fig. 5c indicate that silencing PERK expression clearly inhibited PERK signaling as demonstrated by the reduced phosphorylation of the PERK substrate eIF2α and the inhibition of ER stress-mediated up-regulation of the transcriptional regulator CHOP. Interestingly, knockdown of PERK markedly inhibited TRAIL-R2 up-regulation in cells treated with tunicamycin (Fig. 5c) and significantly reduced cell surface expression (Fig. 5d). The different effects of PERK silencing on total and cell surface expression of TRAIL-R2 in tunicamycin-treated cells can be explained by the UPR-independent modulation of TRAIL-R2 transport to the cell surface following inhibition of protein glycosylation by tunicamycin, as previously reported [27].
Survival role of GRP78 in TRAIL-induced apoptosis
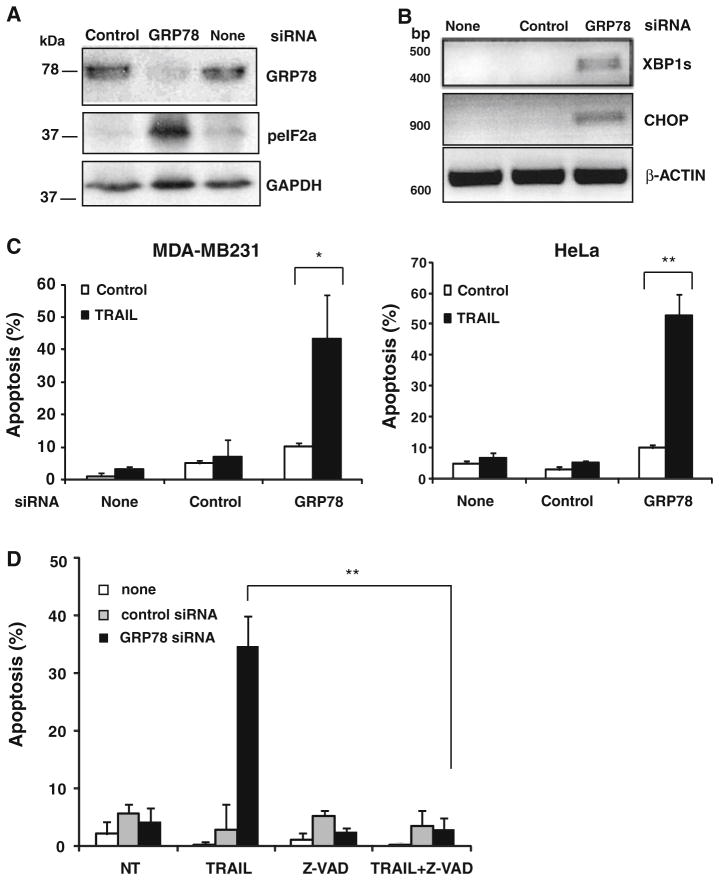
Under ER homeostasis, the ER chaperone GRP78/BIP directly interacts with all three ER stress sensors, PERK, ATF6 and IRE1, and keeps them in inactive forms in non-stressed cells [13, 31]. Upon ER stress, misfolded proteins accumulate in the lumen of the ER and GRP78 is released from the ER stress sensors. Release from GRP78 allows the activation and transduction of the unfolded protein signals across the ER membrane to the cytosol and the nucleus. We reasoned that reducing GRP78 levels by RNA interference should lead to ER stress and UPR activation. As shown in Fig. 6a, silencing GRP78 expression in MDA-MB231 cells induced the phosphorylation of the α subunit of eukaryotic translation initiation factor 2 (eIF2α), a substrate of ER resident PKR-like kinase. Furthermore, GRP78 knockdown also induced CHOP and XBP1s expression, two markers of activated UPR (Fig. 6b). We then determined whether sensitivity to TRAIL was affected in cells in which ER stress had been induced by GRP78 knockdown. Strikingly, following GRP78 silencing there was a clear sensitization to TRAIL-induced apoptosis in both MDA-MB231 and HeLa cells (Fig. 6c). Furthermore, cell death induced by TRAIL in GRP78-depleted cells was dependent on caspase activation as it was completely inhibited in the presence of the pancaspase inhibitor Z-VAD-fmk (Fig. 6d).
Fig. 6.
GRP78 knockdown activates the UPR and sensitizes cells to TRAIL-induced apoptosis. a Cell lysates from MDA-MB231 cells transfected with siRNAs (50 nM) for 48 h were analyzed for protein expression as described in “Materials and methods” section. b MDA-MB231 cells were transfected with 50 nM GRP78 siRNA or a scrambled (control) siRNA for 48 h as described in “Materials and methods” section. XBP1s and CHOP mRNA levels were determined by RT-PCR and β-actin was used as a control for mRNA input. Results are representative of at least two independent experiments. c MDA-MB231 or HeLa cells were transfected with siRNAs for 48 h and treated with TRAIL (500 ng/ml) for 24 h. Apoptosis was assessed as the percentage of hypodiploid cells. Error bars represent S.D. from three independent experiments. *P <0.05, **P <0.01. d MDA-MB231 cells transfected with siRNAs (50 nM) for 48 h were incubated with or without TRAIL for 24 h in the presence or absence of Z-VAD-fmk. Hypodiploid cells were determined as described under “Materials and methods” section. Error bars represent S.D. from three independent experiments. **P <0.01
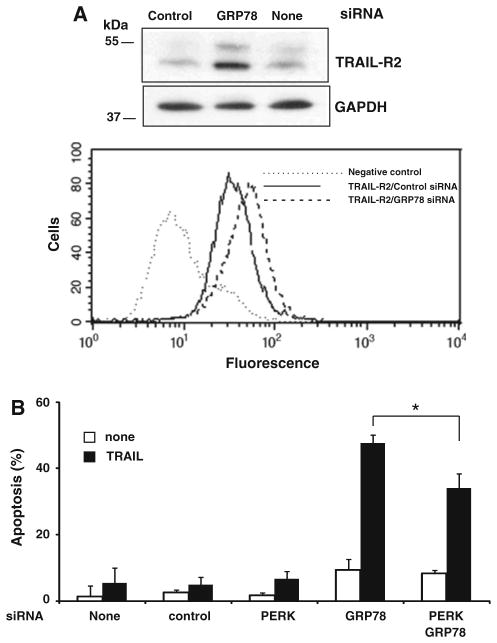
We next examined whether ER stress induced following GRP78 knockdown affected TRAIL-R2 levels and TRAIL-induced apoptosis in MDA-MB231 cells. As shown in Fig. 7a, GRP78 silencing also caused an increase in both total and cell surface levels of TRAIL-R2. Furthermore, silencing PERK expression partially reduced sensitization to TRAIL in GRP78-depleted cells (Fig. 7b), suggesting a certain role of TRAIL-R2 up-regulation in the sensitization to TRAIL in these cells. These results also suggested that other regulators of apoptosis signaling by TRAIL were probably involved in the sensitization of MDA-MB231 cells to TRAIL by ER stress.
Fig. 7.
GRP78 knockdown up-regulates TRAIL-R2 expression and sensitizes cells to TRAIL-induced apoptosis through a PERK-dependent mechanism. a Cell lysates from MDA-MB231 cells transfected with siRNAs (50 nM) for 48 h were analyzed for protein expression by western blotting (upper panel) or cell surface expression by flow cytometry (lower panel) as described in “Materials and methods” section. b MDA-MB231 were transfected with siRNAs for 48 h and treated with TRAIL (500 ng/ml) for 24 h. Apoptosis was assessed as the percentage of cells with sub-G1 DNA content. Error bars represent S.D. from three independent experiments. * Indicate a statistically significant difference in sensitivity to TRAIL when comparing GRP78 siRNA-transfected cells with cells transfected with GRP78/PERK siRNAs. (P <0.05)
Role of FLIP and Mcl-1 in the sensitization of cells to TRAIL upon ER stress
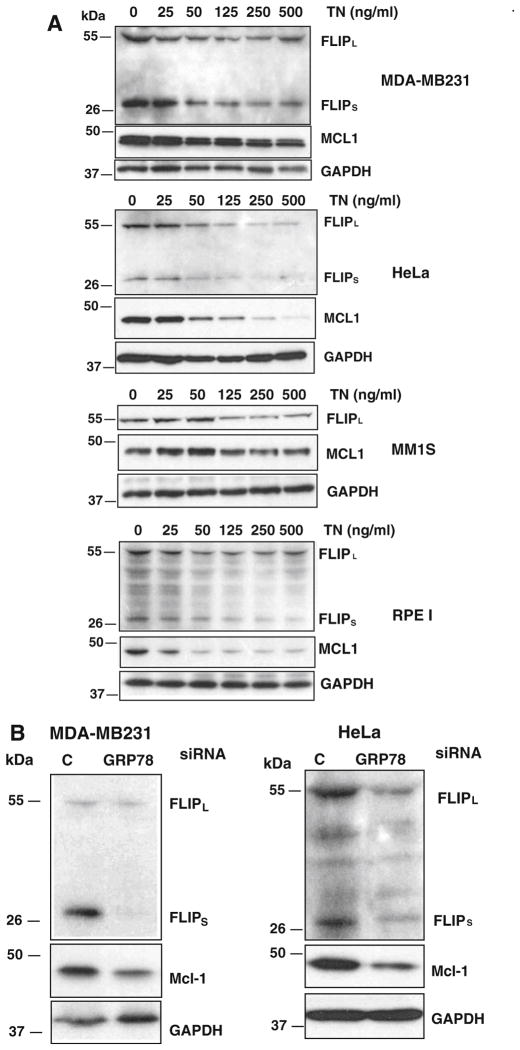
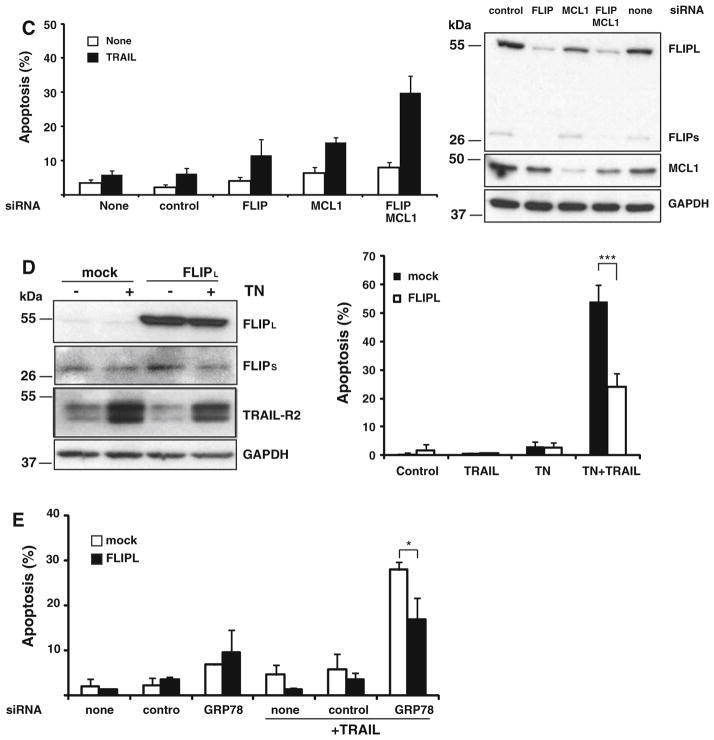
We have recently reported that simultaneous down-regulation of anti-apoptotic Mcl-1 and FLIP proteins following treatment of breast tumor cells with microtubules interfering agents promotes sensitization to TRAIL [22]. To further investigate the mechanisms underlying the sensitization by ER stress to TRAIL we examined the expression of these anti-apoptotic proteins in both transformed and nontransformed cells treated with tunicamycin. As can be observed in Fig. 8a, 15 h-treatment with tunicamycin caused a dose-dependent down-regulation of FLIP and Mcl-1 proteins in all cell lines. Remarkably, the dose–response of tunicamycin in the down-regulation of FLIP and Mcl-1 (Fig. 8a) strongly correlated with the sensitization to TRAIL (Fig. 1) in the various cell lines tested. Likewise, GRP78 knockdown also induced the down-regulation of Mcl-1 and cFLIP proteins (Fig. 8b), further suggesting that ER stress is an important regulator of the expression of both anti-apoptotic proteins. To determine whether down-regulation of these anti-apoptotic proteins in ER stressed cells was a crucial event in the sensitization to TRAIL, Mcl-1 and FLIP expression were silenced prior to TRAIL treatment. As shown in Fig. 8c simultaneous knockdown of both cFLIP and Mcl-1 significantly augmented the sensitization caused by either siRNA alone. These data further support our hypothesis that down-regulation of cFLIP and Mcl-1 expression by ER stress is critical for the sensitization of cells to TRAIL-induced apoptosis.
Fig. 8.
Role of FLIP and Mcl-1 in ER-induced sensitization to TRAIL. a Cells were treated for 15 h with the indicated concentrations of tunicamycin and protein expression was determined in cell lysates by western blotting with specific antibodies. b MDA-MB231 or HeLa cells were transfected with 50 nM GRP78 siRNA or a control siRNA for 48 h as described in “Materials and methods” section. Cell lysates from transfected cells were analyzed for protein expression. Results are representative of at least three independent experiments. c MDA-MB231 cells were transfected with siRNAs for 48 h and treated with TRAIL (500 ng/ml) for 6 h. Apoptosis was assessed as the percentage of hypodiploid cells. Error bars represent S.D. from three independent experiments. Cell lysates from MDA-MB231 cells transfected with siRNAs (50 nM) for 48 h were analyzed for protein expression as described in “Materials and methods” section. d Mock-transfected and MDA-MB231 cells transfected with the pCR3.V64-Met-Flag-FLIPL vector were treated with or without tunicamycin (250 ng/ml) for 15 h prior to the incubation with or without TRAIL (500 ng/ml) for 6 h. Protein expression was determined by western blotting (left panel). Apoptosis was measured as the percentage of cells with sub-G1 DNA content (right panel). Error bars represent S.D. from three independent experiments. ***P <0.001. e Mock or FLIPL-overexpressing MDA-MB231 cells were transfected with siRNAs (50 nM) for 48 h and then treated with TRAIL for 24 h. Apoptosis was assessed as the percentage of cells with sub-G1 DNA content. Error bars represent S.D. from three independent experiments. *P <0.05
To further assessed the role of cFLIP in the sensitization observed we determined the sensitizing effect of tunicamycin in TRAIL-induced apoptosis in MDA-MB231 cells over-expressing cFLIPL. Results shown in Fig. 8d demonstrate that cells over-expressing cFLIPL were clearly more resistant to ER stress-induced sensitization to TRAIL than cells expressing normal cFLIP levels. Similarly, sensitization to TRAIL-induced apoptosis following GRP78 knockdown was significantly reduced in cells over-expressing cFLIPL (Fig. 8e). In summary, our data indicate that down-regulation of anti-apoptotic proteins cFLIP and Mcl-1 together with the up-regulation of TRAIL-R2 expression are key events in the sensitization of ER stressed cells to TRAIL-induced apoptosis.
Discussion
In response to a protein overload in the ER, cells activate the complex signaling of the UPR to increase the folding capacity of the ER and restore homeostasis. These adaptive mechanisms are sometimes insufficient to maintain homeostasis in the ER and an apoptotic cell death process is then induced [32, 33]. Although the activation of the intrinsic apoptotic pathway has been reported in ER stressed cells [34], a number of evidences support the involvement of death receptors and in particular TRAIL-R2/DR5 in the death of cells undergoing ER stress [15–17]. In addition, TRAIL-R2 up-regulation by ER stress treatments has been suggested to play a prominent role in the sensitization of different tumor cell lines to exogenous TRAIL by ER stress treatments [18–20]. However, the molecular mechanisms of the sensitization to TRAIL have not been fully elucidated. Regarding the mechanism underlying the up-regulation of TRAIL-R2 expression following ER stress, the involvement of the transcription factor CHOP has been previously reported [16, 18, 19, 35]. In contrast, studies in melanoma cells have indicated that stable knockdown of x-box-binding protein-1 (XBP-1) with shRNA inhibited ER stress-induced up-regulation of TRAIL-R2, by a CHOP-independent mechanism [20]. Ire1-dependent splicing of the XBP-1 mRNA generate the XBP-1s transcription factor, a potent activator of UPR target genes [36]. Our data indicate that in MDA-MB231 cells up-regulation of TRAIL-R2 expression and sensitization to TRAIL upon ER stress occurs through a CHOP and Ire1-independent process. Our results also demonstrate that ER stress-mediated up-regulation of TRAIL-R2 was inhibited by blocking the ER stress transducer PERK by RNA interference. Although the axis ATF4-CHOP is an important signaling pathway in the PERK arm of the UPR, it is known that approximately half of the PERK-dependent UPR target genes are ATF4-independent [37], suggesting that other PERK effectors should be involved in the regulation of TRAIL-R2 expression in MDA-MB231 cells. In mammalian cells, the three ER transmembrane components, IRE1, PERK, and ATF6, initiate distinct UPR signaling branches and display distinct sensitivities toward different forms of ER stress [38]. Moreover, cross-talking and functional redundancy are important characteristic of the three arms of the UPR [13]. Furthermore, there are marked differences in terms of tissue-specific regulation of the UPR [39]. Taken together, these features of the UPR may explain the differences between our results and others in term of the regulation by ER stress of TRAIL-R2 expression.
Despite a reduction in TRAIL-R2 expression in PERK-silenced cells following ER stress, sensitization to TRAIL-induced apoptosis was only partially reduced. These results suggested that other apoptosis regulators of TRAIL signaling should be also modulated by ER stress. In this respect, we observed a decreased expression of FLIP and Mcl-1 in different cell lines undergoing ER stress. There are conflicting results on the role of Mcl-1 in the resistance of cells to TRAIL. Down-regulating Mcl-1 expression has been reported to sensitize tumor cells to TRAIL-induced apoptosis [40, 41]. However, our previous results indicated that Mcl-1 knockdown by RNA interference only weakly facilitated activation of apoptosis by TRAIL in MDA-MB231 cells [22]. Taken together, these data suggest that involvement of Mcl-1 in the resistance of cells to TRAIL might be cell type-dependent. Interestingly, simultaneous down-regulation of Mcl-1 and cFLIP significantly enhanced sensitization to apoptosis, suggesting the cooperation between Mcl-1 and cFLIP in the mechanism of cell resistance to TRAIL [22]. On the other hand, down-regulation of FLIP is sufficient to sensitize tumor cells to TRAIL [12, 42, 43]. Mcl-1 and cFLIP are short half-life proteins which are subject to constitutive polyubiquitination and subsequent degradation by the proteasome [44, 45]. It has been demonstrated that c-Jun N-terminal kinase (JNK) activation by TNF-α reduces cFLIPL stability by a mechanism involving the JNK-mediated phosphorylation and activation of the E3 ubiquitin ligase Itch, which ubiquitinates cFLIPL and induces its proteasomal degradation [46]. Although in ER-stressed cells JNK could be activated by the Ire1/TRAF2/Ask1 pathway [47], our unpublished results indicate that silencing Ire1 expression by RNA interference did not prevent ER stress-induced FLIP down-regulation. However, we can not exclude a role of the JNK/Itch pathway in the down-regulation of FLIP observed in our studies as it has been recently reported that ER stress induced by a vitamin E derivative down-regulates FLIPL expression through a TRAIL-R2/JNK-dependent Itch E3 ligase-mediated ubiquitination and degradation by the proteasome [48]. Proteasome-mediated degradation of Mcl-1 is regulated by phosphorylation [49]. It has been proposed that JNK could be responsible for the priming phosphorylation of Mcl-1 necessary for the docking of GSK-3 to Mcl-1 [50]. On the other hand, Mcl-1 is a target for the BH3-containing E3 ubiquitin ligase Mule [51]. It remains to be determined whether the proteasome-mediated degradation of Mcl-1 through the JNK/GSK-3/Mule pathway is also involved in the down-regulation of Mcl-1 in ER stressed cells.
GRP78 is a major ER chaperone involved in protein folding and assembly and a key regulator of the UPR. Our data show for the first time that silencing GRP78 expression by RNA interference activates the UPR and markedly sensitizes cells to TRAIL further supporting the anti-apoptotic function of GRP78. We also demonstrate that TRAIL-R2 up-regulation and down-regulation of FLIP and Mcl-1 induced by GRP78 knockdown are important events in the sensitization to TRAIL. Several evidences have shown that GRP78 can also be present outside the ER [52]. In tumor cells, GRP78 is frequently overexpressed at the cell surface where it could bind to various extracellular ligands to result either in cell death or cell survival [17, 53]. In this respect, it has been recently reported that prostate apoptosis response-4 (PAR-4) activates an extrinsic pathways for apoptosis upon binding to GRP78 at the cell surface [17]. In contrast to these results, our data support a survival role of GRP78 preventing the activation of apoptosis by TRAIL. Differences in the cell surface levels of GRP78 may explain these apparently discrepant results, with cells expressing low levels of GRP78 being more resistant to TRAIL. In these cells, ER stress induced by GRP78 down-regulation would activate the pro-apoptotic UPR and sensitize cells to TRAIL.
The findings that different tumor cells can be sensitized to TRAIL by agents that induce ER stress have lead other authors to suggest that these treatments could be potential antitumor therapies in combination with TRAIL [18–20]. However, our data also demonstrate that not only tumor cell lines but also nontransformed cells are markedly sensitized to TRAIL-induced apoptosis by ER stress. In the nontransformed cell lines, TRAIL-R2 up-regulation and down-regulation of FLIP and Mcl-1 are also observed, suggesting that these changes in apoptosis-related proteins belong to a preemptive response of mammalian cells to ER stress. These results imply that targeting ER stress could also affect the sensitivity of normal cells to TRAIL and hence this therapeutic strategy may not be acceptable. On the other hand, tumor cells are often subject to ER stress in vivo. Tumor microenvironment is characterized by glucose deprivation, acidosis, and severe hypoxia. These combined factors leads to the accumulation of misfolded proteins in the ER, triggering the UPR to facilitate tumor survival and growth [14]. Besides the survival role of the UPR, our in vitro results suggest that activating the UPR in tumors by microenvironmental cues may modulate the expression of proteins of the extrinsic pathway of apoptosis, like TRAIL-R2, FLIP and Mcl-1, sensitizing these cells to TRAIL. This may contribute to explain the differential sensitivity of normal and cancer cells to intraperitoneally administered TRAIL observed in in vivo studies [54, 55].
Collectively, our data reveal a pleiotropic mechanism involving both apoptotic and anti-apoptotic proteins for the sensitizing effect of ER stress on the regulation of TRAIL receptor-mediated apoptosis in both transformed and non-transformed cells. These data further highlight the central role of the UPR to maintain protein folding homeostasis and to facilitate the activation of the extrinsic apoptotic pathway when homeostasis can not be restored following sustained ER stress. As we show that ER stress also sensitizes nontransformed cells to TRAIL, a more complete understanding of how UPR does this could provide insights into the role of UPR signaling in normal physiology and regulation of TRAIL action.
Acknowledgments
We thank C. Palacios, R. Yerbes, T. Sánchez-Pérez and A. Cano for stimulating scientific discussions and feedback. This work was supported by grants from Ministerio de Educación y Ciencia (SAF2006-00633 and SAF2009-07163), Red Temática de Investigación Cooperativa en Cáncer (RTICC: RD06/0020/0068) and Junta de Andalucía (CTS-211 and CVI-4497) (to AL-R) and NIH (RO1GM087415) and American Cancer Society (RSG-10-027-01-CSM) (to M.N.). RMP was supported by a contract from Instituto de Salud Carlos III.
Contributor Information
Rosa Martín-Pérez, Centro Andaluz de Biología Molecular y Medicina Regenerativa (CABIMER), Consejo Superior de Investigaciones Científicas, Avenida Americo Vespucio s/n, 41092 Sevilla, Spain.
Maho Niwa, Division of Biological Sciences, Section of Molecular Biology, University of California, San Diego, La Jolla, CA 92093, USA.
Abelardo López-Rivas, Email: abelardo.lopez@cabimer.es, Centro Andaluz de Biología Molecular y Medicina Regenerativa (CABIMER), Consejo Superior de Investigaciones Científicas, Avenida Americo Vespucio s/n, 41092 Sevilla, Spain.
References
- 1.Ashkenazi A. Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat Rev Drug Discov. 2008;7:1001–1012. doi: 10.1038/nrd2637. [DOI] [PubMed] [Google Scholar]
- 2.Sprick MR, Weigand MA, Rieser E, et al. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity. 2000;12:599–609. doi: 10.1016/s1074-7613(00)80211-3. [DOI] [PubMed] [Google Scholar]
- 3.Deng Y, Lin Y, Wu X. TRAIL-induced apoptosis requires Bax-dependent mitochondrial release of Smac/DIABLO. Genes Dev. 2002;16:33–45. doi: 10.1101/gad.949602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irmler M, Thome M, Hahne M, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 5.Krueger A, Schmitz I, Baumann S, Krammer PH, Kirchhoff S. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J Biol Chem. 2001;276:20633–20640. doi: 10.1074/jbc.M101780200. [DOI] [PubMed] [Google Scholar]
- 6.Sharp DA, Lawrence DA, Ashkenazi A. Selective knockdown of the long variant of cellular FLICE inhibitory protein augments death receptor-mediated caspase-8 activation and apoptosis. J Biol Chem. 2005;280:19401–19409. doi: 10.1074/jbc.M413962200. [DOI] [PubMed] [Google Scholar]
- 7.Yeh WC, Itie A, Elia AJ, et al. Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity. 2000;12:633–642. doi: 10.1016/s1074-7613(00)80214-9. [DOI] [PubMed] [Google Scholar]
- 8.Keane MM, Ettenberg SA, Nau MM, Russell EK, Lipkowitz S. Chemotherapy augments TRAIL-induced apoptosis in breast cell lines. Cancer Res. 1999;59:734–741. [PubMed] [Google Scholar]
- 9.Wang S, El-Deiry WS. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene. 2003;22:8628–8633. doi: 10.1038/sj.onc.1207232. [DOI] [PubMed] [Google Scholar]
- 10.Chinnaiyan AM, Prasad U, Shankar S, et al. Combined effect of tumor necrosis factor-related apoptosis-inducing ligand and ionizing radiation in breast cancer therapy. Proc Natl Acad Sci USA. 2000;97:1754–1759. doi: 10.1073/pnas.030545097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frew AJ, Lindemann RK, Martin BP, et al. Combination therapy of established cancer using a histone deacetylase inhibitor and a TRAIL receptor agonist. Proc Natl Acad Sci USA. 2008;105:11317–11322. doi: 10.1073/pnas.0801868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palacios C, Yerbes R, Lopez-Rivas A. Flavopiridol induces cellular FLICE-inhibitory protein degradation by the proteasome and promotes TRAIL-induced early signaling and apoptosis in breast tumor cells. Cancer Res. 2006;66:8858–8869. doi: 10.1158/0008-5472.CAN-06-0808. [DOI] [PubMed] [Google Scholar]
- 13.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 14.Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–10634. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- 15.He Q, Lee DI, Rong R, et al. Endoplasmic reticulum calcium pool depletion-induced apoptosis is coupled with activation of the death receptor 5 pathway. Oncogene. 2002;21:2623–2633. doi: 10.1038/sj.onc.1205345. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi H, Wang HG. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J Biol Chem. 2004;279:45495–45502. doi: 10.1074/jbc.M406933200. [DOI] [PubMed] [Google Scholar]
- 17.Burikhanov R, Zhao Y, Goswami A, Qiu S, Schwarze SR, Rangnekar VM. The tumor suppressor Par-4 activates an extrinsic pathway for apoptosis. Cell. 2009;138:377–388. doi: 10.1016/j.cell.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiraishi T, Yoshida T, Nakata S, et al. Tunicamycin enhances tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in human prostate cancer cells. Cancer Res. 2005;65:6364–6370. doi: 10.1158/0008-5472.CAN-05-0312. [DOI] [PubMed] [Google Scholar]
- 19.Jiang CC, Chen LH, Gillespie S, et al. Tunicamycin sensitizes human melanoma cells to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by up-regulation of TRAIL-R2 via the unfolded protein response. Cancer Res. 2007;67:5880–5888. doi: 10.1158/0008-5472.CAN-07-0213. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Jiang CC, Lavis CJ, et al. 2-Deoxy-D-glucose enhances TRAIL-induced apoptosis in human melanoma cells through XBP-1-mediated up-regulation of TRAIL-R2. Mol Cancer. 2009;8:122. doi: 10.1186/1476-4598-8-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harper N, Hughes MA, Farrow SN, Cohen GM, MacFarlane M. Protein kinase C modulates tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by targeting the apical events of death receptor signaling. J Biol Chem. 2003;278:44338–44347. doi: 10.1074/jbc.M307376200. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Perez T, Ortiz-Ferron G, Lopez-Rivas A. Mitotic arrest and JNK-induced proteasomal degradation of FLIP and Mcl-1 are key events in the sensitization of breast tumor cells to TRAIL by antimicrotubule agents. Cell Death Differ. 2010;17:883–894. doi: 10.1038/cdd.2009.176. [DOI] [PubMed] [Google Scholar]
- 23.Hirota M, Kitagaki M, Itagaki H, Aiba S. Quantitative measurement of spliced XBP1 mRNA as an indicator of endoplasmic reticulum stress. J Toxicol Sci. 2006;31:149–156. doi: 10.2131/jts.31.149. [DOI] [PubMed] [Google Scholar]
- 24.Gong J, Traganos F, Darzynkiewicz Z. A selective procedure for DNA extraction from apoptotic cells applicable for gel electrophoresis and flow cytometry. Anal Biochem. 1994;218:314–319. doi: 10.1006/abio.1994.1184. [DOI] [PubMed] [Google Scholar]
- 25.Walczak H, Bouchon A, Stahl H, Krammer PH. Tumor necrosis factor-related apoptosis-inducing ligand retains its apoptosis-inducing capacity on Bcl-2- or Bcl-xL-overexpressing chemotherapy-resistant tumor cells. Cancer Res. 2000;60:3051–3057. [PubMed] [Google Scholar]
- 26.Ruiz de Almodovar C, Ruiz-Ruiz C, Munoz-Pinedo C, Robledo G, Lopez-Rivas A. The differential sensitivity of Bc1-2-overexpressing human breast tumor cells to TRAIL or doxorubicin-induced apoptosis is dependent on Bc1-2 protein levels. Oncogene. 2001;20:7128–7133. doi: 10.1038/sj.onc.1204887. [DOI] [PubMed] [Google Scholar]
- 27.Jin Z, McDonald ER, 3rd, Dicker DT, El-Deiry WS. Deficient tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor transport to the cell surface in human colon cancer cells selected for resistance to TRAIL-induced apoptosis. J Biol Chem. 2004;279:35829–35839. doi: 10.1074/jbc.M405538200. [DOI] [PubMed] [Google Scholar]
- 28.Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 2000;12:611–620. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 29.Ubeda M, Wang XZ, Zinszner H, Wu I, Habener JF, Ron D. Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol Cell Biol. 1996;16:1479–1489. doi: 10.1128/mcb.16.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 31.Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 32.Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004;11:372–380. doi: 10.1038/sj.cdd.4401378. [DOI] [PubMed] [Google Scholar]
- 33.Zinszner H, Kuroda M, Wang X, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masud A, Mohapatra A, Lakhani SA, Ferrandino A, Hakem R, Flavell RA. Endoplasmic reticulum stress-induced death of mouse embryonic fibroblasts requires the intrinsic pathway of apoptosis. J Biol Chem. 2007;282:14132–14139. doi: 10.1074/jbc.M700077200. [DOI] [PubMed] [Google Scholar]
- 35.Zou W, Yue P, Khuri FR, Sun SY. Coupling of endoplasmic reticulum stress to CDDO-Me-induced up-regulation of death receptor 5 via a CHOP-dependent mechanism involving JNK activation. Cancer Res. 2008;68:7484–7492. doi: 10.1158/0008-5472.CAN-08-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calfon M, Zeng H, Urano F, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 37.Harding HP, Zhang Y, Zeng H, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 38.DuRose JB, Tam AB, Niwa M. Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress. Mol Biol Cell. 2006;17:3095–3107. doi: 10.1091/mbc.E06-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meng XW, Lee SH, Dai H, et al. Mcl-1 as a buffer for proapoptotic Bcl-2 family members during TRAIL-induced apoptosis: a mechanistic basis for sorafenib (Bay 43–9006)-induced TRAIL sensitization. J Biol Chem. 2007;282:29831–29846. doi: 10.1074/jbc.M706110200. [DOI] [PubMed] [Google Scholar]
- 41.Ricci MS, Kim SH, Ogi K, et al. Reduction of TRAIL-induced Mcl-1 and cIAP2 by c-Myc or sorafenib sensitizes resistant human cancer cells to TRAIL-induced death. Cancer Cell. 2007;12:66–80. doi: 10.1016/j.ccr.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Hietakangas V, Poukkula M, Heiskanen KM, Karvinen JT, Sistonen L, Eriksson JE. Erythroid differentiation sensitizes K562 leukemia cells to TRAIL-induced apoptosis by downregulation of c-FLIP. Mol Cell Biol. 2003;23:1278–1291. doi: 10.1128/MCB.23.4.1278-1291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y, Suh N, Sporn M, Reed JC. An inducible pathway for degradation of FLIP protein sensitizes tumor cells to TRAIL-induced apoptosis. J Biol Chem. 2002;277:22320–22329. doi: 10.1074/jbc.M202458200. [DOI] [PubMed] [Google Scholar]
- 44.Nijhawan D, Fang M, Traer E, et al. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poukkula M, Kaunisto A, Hietakangas V, et al. Rapid turnover of c-FLIPshort is determined by its unique C-terminal tail. J Biol Chem. 2005;280:27345–27355. doi: 10.1074/jbc.M504019200. [DOI] [PubMed] [Google Scholar]
- 46.Chang L, Kamata H, Solinas G, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 47.Urano F, Wang X, Bertolotti A, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 48.Tiwary R, Yu W, Li J, Park SK, Sanders BG, Kline K. Role of endoplasmic reticulum stress in alpha-TEA mediated TRAIL/DR5 death receptor dependent apoptosis. PLoS One. 2010;5:e11865. doi: 10.1371/journal.pone.0011865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Opferman JT. Unraveling MCL-1 degradation. Cell Death Differ. 2006;13:1260–1262. doi: 10.1038/sj.cdd.4401978. [DOI] [PubMed] [Google Scholar]
- 50.Inoshita S, Takeda K, Hatai T, et al. Phosphorylation and inactivation of myeloid cell leukemia 1 by JNK in response to oxidative stress. J Biol Chem. 2002;277:43730–43734. doi: 10.1074/jbc.M207951200. [DOI] [PubMed] [Google Scholar]
- 51.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Papandreou I, Denko NC, Olson M, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Misra UK, Deedwania R, Pizzo SV. Activation and crosstalk between Akt, NF-kappaB, and unfolded protein response signaling in 1-LN prostate cancer cells consequent to ligation of cell surface-associated GRP78. J Biol Chem. 2006;281:13694–13707. doi: 10.1074/jbc.M511694200. [DOI] [PubMed] [Google Scholar]
- 54.Walczak H, Miller RE, Ariail K, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 55.Ashkenazi A, Pai RC, Fong S, et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]