Abstract
Alzheimer’s disease (AD) is characterized by multiple, intertwined pathological features, including amyloid-β (Aβ) aggregation, metal ion dyshomeostasis, and oxidative stress. We report a novel compound (ML) prototype of a rationally designed molecule obtained by integrating structural elements for Aβ aggregation control, metal chelation, reactive oxygen species (ROS) regulation, and antioxidant activity within a single molecule. Chemical, biochemical, ion mobility mass spectrometric, and NMR studies indicate that the compound ML targets metal-free and metal-bound Aβ (metal–Aβ) species, suppresses Aβ aggregation in vitro, and diminishes toxicity induced by Aβ and metal-treated Aβ in living cells. Comparison of ML to its structural moieties (i.e., 4-(dimethylamino)phenol (DAP) and (8-aminoquinolin-2-yl)methanol (1)) for reactivity with Aβ and metal–Aβ suggests the synergy of incorporating structural components for both metal chelation and Aβ interaction. Moreover, ML is water-soluble and potentially brain permeable, as well as regulates the formation and presence of free radicals. Overall, we demonstrate that a rational structure-based design strategy can generate a small molecule that can target and modulate multiple factors, providing a new tool to uncover and address AD complexity.
Alzheimer’s disease (AD) is characterized by a loss of brain function which affects memory, cognition, and behavior.1 Development of a cure for AD has been hindered by a lack of understanding of both the causes and mechanisms of disease onset and progression.2–6 The AD brain exhibits several characteristic pathological features, such as accumulation of misfolded amyloid-β (Aβ), metal ion dyshomeostasis, and elevated oxidative stress.3–12 Two amyloidogenic peptides, Aβ40 and Aβ42, present in the brain at ca. 90% and 9%, respectively, are primarily produced upon cleavage of amyloid precursor protein (APP) by β- and γ-secretases.3–6 Both peptides tend to aggregate, generating oligomers and fibrils.3–6,8,12,13 Although Aβ is proposed to be a causative agent in AD, a relationship between specific peptide oligomers and toxicity remains unclear despite recent findings indicating soluble Aβ oligomers as possible neurotoxic species.3–6,8,12–15 In addition to Aβ imbalance, high levels of metal ions (Cu, ca. 0.4 mM; Zn, ca. 1 mM; Fe, ca. 0.9 mM) have been found in Aβ plaques of AD brains.3,5–12 These metals, particularly Cu and Zn, bind to Aβ peptides facilitating their aggregation. Moreover, dysregulated redox active metal ions, Cu(i/ii) and Fe(ii/iii), both unbound and bound to Aβ peptides, are observed to promote overproduction of reactive oxygen species (ROS) that damage biological molecules, such as proteins, DNA, and lipids.3,5–12,16–18 Overall, because of the involvement of numerous factors (e.g., metal-free/-associated Aβ species, metals, free radicals) and their potential interconnection in AD pathogenesis, the causative agents in this multifaceted disease remain to be unambiguously identified.
Chemical reagents to target and regulate these multiple factors in AD are desirable to advance our understanding of AD complexity and offer possible answers for remediation. Toward this effort, small molecules have been developed via a rational structure-based incorporation approach by integrating an Aβ interacting framework with a metal chelation moiety into a single molecule designed to target and modulate metal–associated Aβ (metal–Aβ) species.8,9,12,18–26 These compounds were observed to control metal-induced Aβ aggregation, attenuate ROS formation by metal–Aβ, or regulate metal–Aβ toxicity in vitro and in living cells.21–26 In addition, interaction and reactivity of natural products, such as the green tea extract, (−)-epigallocatechin-3-gallate, and myricetin, with metal–Aβ species have also been investigated showing distinct reactivity with metal–Aβ over metal-free Aβ.27,28 To the best of our knowledge, however, a single designed compound, targeting all these factors (i.e., Aβ, metal–Aβ, metal ions, free radicals, Figure 1) and regulating their reactivities, has not been reported to date.
Figure 1.
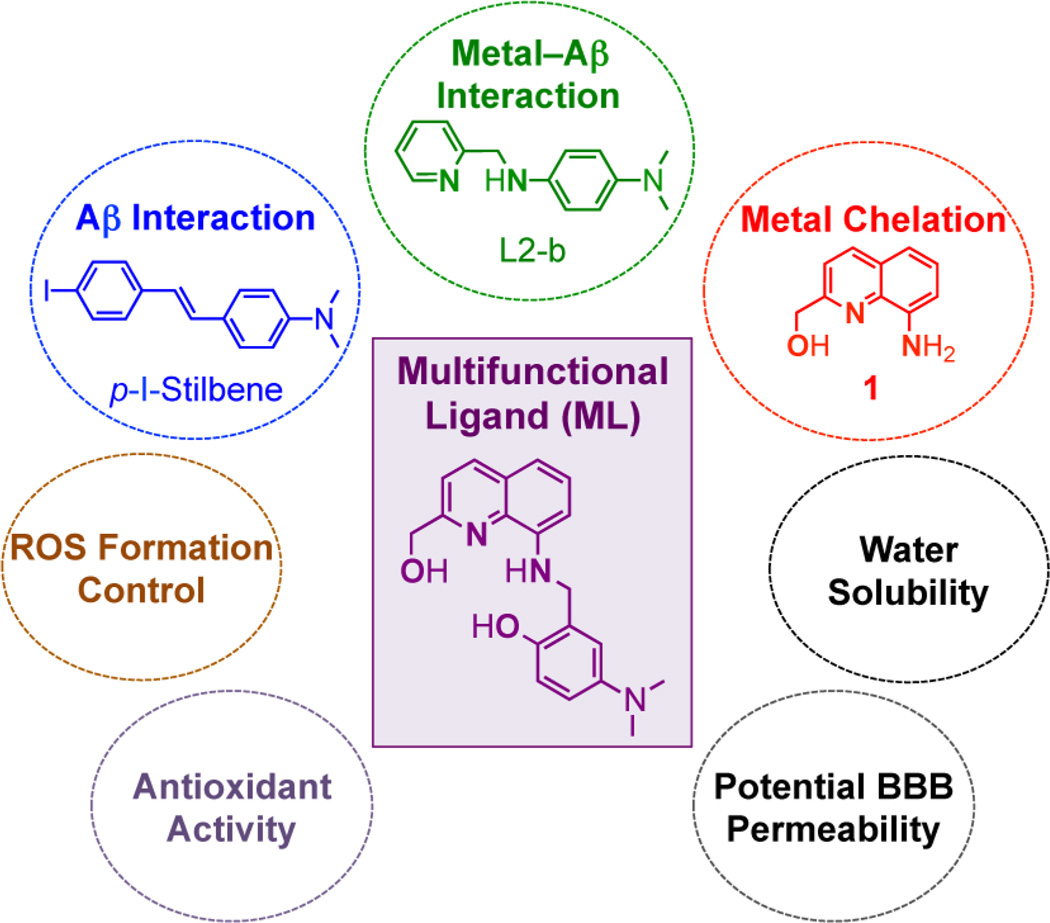
Rational structure-based design principle (incorporation approach) of a multifuncitonal ligand (ML). Atoms responsible for metal binding are in bold. Chemical structures: ML = 4-(dimethylamino)-2-(((2-(hydroxymethyl)quinolin-8-yl)-amino)-methyl)phenol; p-I-stilbene = (E)-4-(4-iodostyryl)-N,N-dimethylaniline); L2-b = N1,N1-dimethyl-N4-(pyridin-2-ylmeth-yl)benzene-1,4-diamine; 1 = (8-aminoquinolin-2-yl)methanol.
Herein, we present a novel ligand (ML) as the first example of a rationally designed molecule to afford multiple properties within a single entity (Figure 1). Our investigations of ML’s activity toward Aβ, metal–Aβ, metal ions, and free radicals, as well as its potential blood-brain barrier (BBB) permeability confirm that careful selection and consideration of molecular properties can result in the design of a molecule to target and modulate multiple pathological features of AD. The compound 1 (Figure 1 for structure), without an Aβ interacting moiety, was also studied in parallel to demonstrate that ML’s reactivity toward Aβ and metal–Aβ could arise from the synergy of its metal chelation and Aβ interaction properties.
RESULTS AND DISCUSSION
Design Consideration for a Multifunctional Ligand (ML)
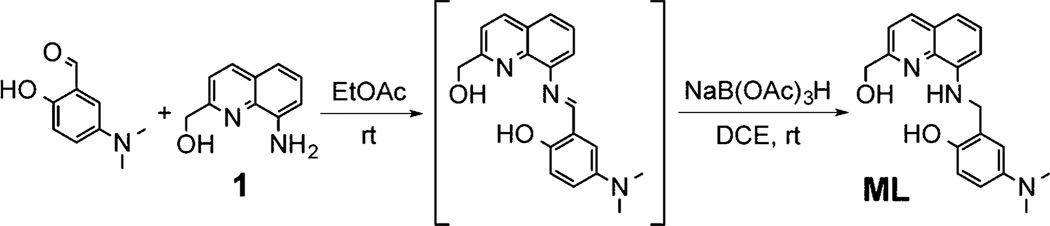
To develop a chemical tool capable of both targeting and modulating the reactivity of multiple AD pathological factors in biological systems, we designed a novel molecule (ML) with the potential for interaction with Aβ and metal–Aβ, metal chelation, control of ROS generation, antioxidant activity, water solubility, and BBB permeability (Figure 1). For Aβ/metal–Aβ interactions and metal chelation, ML was constructed by combining p-I-stilbene, a known Aβ imaging agent,29 with L2-b, a molecule previously reported to target and regulate metal–Aβ22 and 1, a metal chelator30 (Figure 1). For enhanced metal binding properties, an additional hydroxyl group, along with nitrogen and oxygen donor atoms from 1, was incorporated into ML affording a tetradentate ligand for Cu(ii) with 1:1 metal-to-ligand stoichiometry.30 ML was constructed to accommodate a slightly distorted square planar geometry for Cu(ii) similar to 2-[(8-quinolinylamino)methyl]phenol.30 In this conformation, the ligand cannot easily accommodate the preferred tetrahedral geometry of Cu(i) for redox cycles of Cu(i/ii) and, thus, is able to inhibit ROS generation. For antioxidant activity, substituents (i.e., both quinoline and phenolic groups, Figure 1)31–33 known to have antioxidant capability were integrated into ML. Lastly, polar functionalities (e.g., hydroxyl and amino groups) were introduced into the backbone for water solubility. All structural elements were selected to adhere to values of Lipinski’s rules and logBB for possible drug-likeness and BBB penetration (Table 1).22,25,34,35 ML was synthesized by modifications to previously reported procedures (Schiff base condensation followed by reduction of imine to amine, ca. 50% yield) as shown in Scheme 1.30
Table 1.
Values (MW, clogP, HBA, HBD, PSA, logBB, and −logPe)a for ML and 1
| Compound | MW | clogP | HBA | HBD | PSA (Å) | logBB | −logPe | CNS± predictionb |
|---|---|---|---|---|---|---|---|---|
| ML | 323 | 2.57 | 5 | 3 | 68.1 | −0.478 | 4.49 (±0.01) | CNS+ |
| 1 | 174 | 0.889 | 3 | 3 | 58.6 | −0.593 | 4.70 (±0.01) | CNS+ |
| Lipinski’s rules and others | ≤ 450 | ≤ 5.0 | ≤ 10 | ≤ 5 | ≤ 90 | > 3.0 (readily); < −1.0 (poorly) | < 5.4; > 5.7 | CNS+; CNS− |
MW, molecular weight; clogP, calculated logarithm of the octanol–water partition coefficient; HBA, hydrogen bond acceptor atoms; HBD, hydrogen bond donor atoms; PSA, polar surface area; logBB = −0.0148 × PSA + 0.152 × clogP + 0.139 (logBB > 3.0, readily crosses BBB; logBB < −1.0, poorly distributed to the brain); −logPe values were determined using the Parallel Artificial Membrane Permeability Assay (PAMPA), and average −logPe values were then calculated by the PAMPA 9 Explorer software v. 3.5.
Prediction of a compound’s ability to penetrate the central nervous system (CNS) on the basis of literature values. Compounds categorized as CNS+ possess the ability to penetrate the BBB and are available in the CNS. Compounds assigned as CNS− have poor permeability through the BBB; therefore, their bioavailability into the CNS is considered to be minimal.
Scheme 1.
Synthetic Route to ML
Direct Interactions of ML with Soluble Forms of Aβ and Metal–Aβ
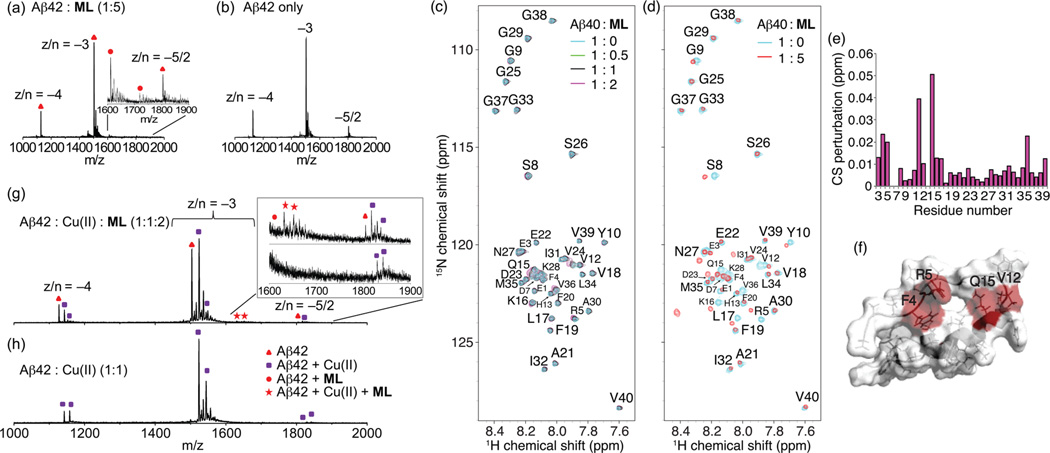
To determine whether ML binds to Aβ40 and Aβ42, mixtures of the compound with either peptide were first monitored by mass spectrometry (MS). In the mass spectrum of a 1:5 mixture of Aβ42 and ML (Figure 2a), there were three peaks representing Aβ42 with charge (z) to oligomer number (n) ratio z/n = −4, −3, and −5/2, similar to the mass spectrum of pure Aβ42 without ML (Figure 2b). Moreover, there were two tailing peaks (m/z = 1611.3 and 1718.7, respectively) corresponding to z/n = −3 complexes of Aβ42 with one and two ML molecules bound, respectively. In the mass spectrum of the mixture of Aβ40 and ML (Supporting Information Figure S1), a tailing peak indicating to the z/n = −3 complex of Aβ40 and ML was also observed. These results suggest that ML can directly bind to both Aβ40 and Aβ42 with either a 1:1 or 1:2 Aβ:ML stoichiometry.
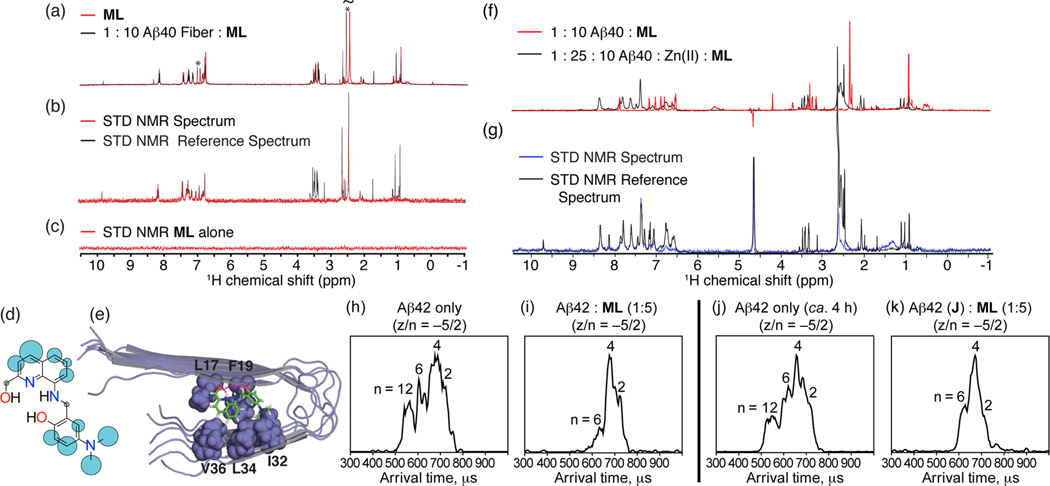
Figure 2.
Interactions of ML with soluble metal-free or Cu(ii)-treated Aβ species monitored by mass spectrometry or SOFAST-HMQC NMR. Mass spectra of (a) 1:5 mixture of Aβ42 and ML and (b) pure Aβ42 (z/n = charge/oligomer number). (c and d) 2D SOFAST-HMQC NMR spectra of ML-titrated metal-free monomeric Aβ40. Freshly dissolved Aβ40 (80 µM) in 50 mM Tris-DCl (pD 7.3) was titrated to (c) 40–160 µM or (d) 400 µM ML at 4 °C. The contour level of (d) has been adjusted to clearly show ligand-bound resonances. (e) Chemical shift perturbations within Aβ40 following addition of ML (Aβ:ML = 1:2). (f) Residues with largest changes in chemical shift at 1:1 Aβ40:ML mapped onto the NMR structure of the helical conformer of the Aβ40 structural ensemble (PDB 2LFM). Mass spectra of (g) Aβ42, Cu(ii), and ML (1:1:2) and of (h) Aβ42 and Cu(ii) (1:1). Peaks for pure Aβ42, Cu(ii)-bound Aβ42, ML-bound Aβ42, and Aβ42–Cu(ii)–ML complexes are noted with triangles, rectangles, circles, and stars, respectively.
Direct binding of ML to monomeric Aβ40 was supported by NMR experiments, which showed either a small but detectable broadening or a chemical shift change for freshly prepared Aβ40 with a stoichiometric amount of ML, particularly for Aβ residues F4, R5, V12, and Q15 (Figure 2c–f). These residues form an apparent binding pocket in the aqueous NMR structure of Aβ40 (Figure 2f).36 Larger changes in chemical shift were observed with an excess of ML possibly due to a change in the conformation or oligomerization state of Aβ40 at this concentration of ML (Figure 2d). The chemical shift of methionine 35 (M35) did not change appreciably, indicating that ML does not cause oxidation of Aβ40.37
To investigate the interaction of ML with metal-bound Aβ, samples of Cu(ii)–Aβ40/42 or Zn(ii)–Aβ40/42 with and without ML were analyzed by MS (Figure 2g and h; Supporting Information Figures S2 and S3). The mass spectrum of a 1:1 mixture of Aβ42 and Cu(ii) without ML displayed two sets of peaks for each charge state z/n = −4, −3, and −5/2, corresponding to Aβ42 with one and two Cu(ii) binding, respectively (Figure 2h). When ML was added to preincubated Cu(ii)–Aβ42, a mixture of species was observed (Figure 2g). In addition to the peaks observed in ML-free samples corresponding to Aβ42 with one or two Cu(ii), three additional peaks representing metal-free Aβ42 with z/n = −4, −3, and −5/2 were also detected, indicating that ML can competitively chelate Cu(ii) from Aβ42. Note that peak intensities of Aβ42 with two Cu(ii) bound decreased dramatically upon ML addition. A tailing peak corresponding to a complex of Aβ42 with one ML bound was observed. Moreover, peaks indicating complexes of Aβ42, ML, and one or two Cu(ii) (m/z = 1633 and 1653, respectively) were present, indicating ML bound to Aβ42 and metal–Aβ42.
Similar to Aβ42, the mass spectrum of preincubated Cu(ii)–Aβ40 treated with ML displayed peaks indicative of both Aβ40–Cu(ii)–ML ternary complexes and copper-liberated Aβ40 (Supporting Information Figure S2). Zn(ii)-associated Aβ40/42 were also investigated with ML. The mass spectrum of a 1:1:2 mixture of Aβ40/42:Zn(ii):ML showed peaks corresponding to Aβ40/42–Zn(ii)–ML complexes (z/n = −3 and −5/2) as well as slight increased intensity of peaks corresponding to metal-free Aβ compared to those without ML (Supporting Information Figure S3). Overall, these results indicate that ML not only forms complexes with both Cu(ii)–Aβ and Zn(ii)–Aβ but also competitively chelates metal ions from metal–Aβ generating metal-free Aβ species.
Interactions of ML with Aβ Fibers
To probe the interaction of ML with larger aggregates, we measured the interaction of ML with metal-free Aβ40 fibers by 1H NMR (Figure 3a). Even at low fiber concentrations (Aβ40:ML, 1:10), chemical shift perturbations were detected within the quinoline and aniline rings and for the dimethylamino group of ML. The monomeric Aβ signal was not observed, indicating that ML does not completely convert metal-free Aβ fibers into the monomeric form, in agreement with results from aggregation experiments (vide infra). The interaction of ML with Aβ40 fibers was probed more directly by saturation transfer difference (STD) NMR experiments (Figure 3b and c). Signals in STD NMR are proportional to the proximity of each ligand atom to its macromolecular binding partner, allowing atomic-level mapping of ligand binding interactions to be made.38 Relatively strong saturation effects can be seen throughout ML (Figure 3d), suggesting that it can pack tightly against fibers. This binding mode is supported by docking simulations (Figure 3e and Supporting Information Figures S4 and S5) showing intimate interaction of ML with the side-chains and backbone of the unpaired β sheet at the end of the Aβ fiber. The lowest energy conformations were stabilized by hydrogen bonding of the hydroxyl and amino moieties of ML to the peptide backbone, π–π stacking of ML’s quinoline ring with the phenyl ring of F19, and Van der Waals interactions of ML’s dimethylamino group with the side chains of I32 and L34 on the opposing β sheet (Figure 3e and Supporting Information Figures S4 and S5). Preservation of Van der Waals interactions between the dimethylamino moiety and Aβ fibers suggests that this structural group in a meta position to the bridgehead could also be effective for Aβ interaction compared to that in a para position, as for p-I-stilbene (Figure 1).29 In this conformation, most of ML was in contact with the fiber with the exception of the bridge-head CH2, in agreement with the STD results. Some features of the Aβ fiber–ML interaction were dependent on the specific Aβ fiber model used in the simulations and also varied among the low energy conformations obtained for each model (Supporting Information Figures S4 and S5). In docking investigations, ML would consistently intercalate between the “steric zipper” of the exposed β strands at the end of the Aβ fiber, most likely disrupting the potential hydrogen bonding network for incoming Aβ40 monomers and preventing fiber extension. Strong chemical shift perturbations in ML upon treatment with Zn(ii) in the presence of Aβ40 fibers demonstrate that ML interacts with Zn(ii) (Figure 3f), as confirmed by metal binding studies (vide infra). STD NMR of Zn(ii)–Aβ40 fibers showed a similar saturation pattern (Figure 3g) as the metal-free samples (Figure 3b) but with reduced intensity, indicating that ML could bind to Zn(ii)–Aβ fibers in a similar manner, although somewhat less effectively.
Figure 3.
(a–g) Interaction of ML with fibrillar Aβ40 species by saturation transfer difference (STD) NMR and (h–k) influence of ML on early Aβ42 oligomerization monitored by mass spectrometry and ion mobility studies. (a) Chemical shift changes in the 1H spectra of ML upon the addition of 10 mol % metal-free Aβ40 fibers in 100% D2O (20 mM deuterated Tris–DCl, pD 7.4). Large chemical shift changes can be seen in the aniline ring and dimethylamino groups (marked with an asterisk). (b) 1H STD NMR spectra of ML with Aβ40 fibers (Aβ:ML = 1:10). Comparison of STD signal intensity (red) to the STD reference (black) reflects the relative proximity of the corresponding proton to the Aβ40 fiber. (c) 1H STD NMR spectra of ML alone showing the absence of an STD signal in the absence of Aβ40 fibers. (d) Normalized STD intensities mapped to ML’s structure. Larger blue circles indicate a more intense STD effect; gray circles indicate the absence of an STD signal. (e) Lowest energy docked conformation of ML to Aβ40 fibers (PDB 2LMO). Other docked conformations and a cluster analysis can be found in Supporting Information Figures S4 and S5. (f) Comparison of the 1H spectra of ML (200 µM) with Aβ40 fibers (20 µM) in 100% D2O (20 mM deuterated Tris–DCl, pD 7.4) with (black) and without (red) 500 µM ZnCl2. The large chemical shift changes are evidence of binding of Zn(ii) to ML. (g) 1H STD NMR spectra of ML with Aβ40 fibers in a ratio of 10:1 in the presence of ZnCl2 (500 µM). Arrival time distriubtions (ATDs) for the z/n = −5/2 peak of (h) pure Aβ42 and (i) 1:5 mixture of Aβ42 and ML sample, respectively. (j) ATD for the −5/2 peak of the Aβ42 sample prepared and placed on ice for ca. 4 h. (k) ATD for the −5/2 peak of the preincubated Aβ42 sample immediately following the addition of ML (ca. 5 min).
Metal Binding Properties of ML
Cu(ii) and Zn(ii) binding to ML was measured by UV–visible (UV–vis) spectroscopy (Supporting Information Figure S6). Upon addition of one equivalent of CuCl2 or ZnCl2 to a solution of ML at pH 7.4, a new absorption band at ca. 457 nm (for CuCl2) or ca. 450 nm (for ZnCl2) was observed, indicative of metal binding to the ligand. Moreover, ML’s selectivity toward Cu(ii) or Zn(ii) over other biologically relevant divalent metal ions [Mg(ii), Ca(ii), Mn(ii), Fe(ii), Co(ii), and Ni(ii)] was investigated by competition experiments. ML was bound to Cu(ii) selectively, even at 20-fold higher concentrations (1 mM) of other divalent ions (Supporting Information Figure S7). ML was shown to be relatively selective for Zn(ii) over Mg(ii), Ca(ii), or Mn(ii) at stoichiometric ratios, whereas it could not bind Zn(ii) over Fe(ii), Co(ii), Ni(ii), or Cu(ii). ML could interact with Zn(ii) over excess Mg(ii) and Ca(ii) (Supporting Information Figure S7). Metal binding and Cu(ii) selectivity of 1 (see Figure 1 for structure) were also confirmed (Supporting Information Figure S7).
Solution speciation and metal binding affinities of ML were determined by UV–vis variable-pH (2–8) titration experiments. Three acidity constants (pKa) were obtained (2.628, 3.971, and 6.230), suggesting the presence of mono-, di-, and triprotonated ligand forms in the pH range of 2–8 (Supporting Information Figure S8). The solution speciation diagram based on the acidity constants indicates that ML exists mainly in neutral form (ca. 94%) at physiological pH (i.e., 7.4). Stability constants for ML with Cu(ii) and Zn(ii) were identified by solution speciation studies of Cu(ii)–ML and Zn(ii)–ML complexes. Dissociation constants (Kd) for ML with Cu(ii) (picomolar range) and Zn(ii) (nanomolar range) based on concentrations of unchelated Cu(ii) or Zn(ii) at pH 7.4 (Supporting Information Figure S8) demonstrate that ML coordinates Cu(ii) more strongly than Zn(ii). ML chelates Cu(ii) and Zn(ii) more effectively than 1 (micromolar range for both metal ions, pH 7.4; Supporting Information Figure S9) and preferentially forms 1:1 metal:ML complexes, whereas 1 generates a mixture of 1:1 and 1:2 complexes of metal:1. Moreover, Kd of ML values for Cu(ii) and Zn(ii) are comparable to those of Cu(ii)–Aβ (picomolar to nanomolar) and Zn(ii)–Aβ (nanomolar to micromolar),5–9,12,16–18 implying that ML could interact with metal ions surrounded by soluble Aβ species, which supports the outcomes from MS (vide supra) and reactivity studies with metal–Aβ species (vide infra).
Modulation of the Early Oligomerization of Aβ by ML
Ion mobility–mass spectrometry (IM–MS)39 was employed to investigate the influence of ML on early Aβ aggregation. IM–MS is capable of separating species with the same mass-to-charge ratio, but different oligomer conformations and sizes, and has been successfully applied to studying Aβ structure and screening for small molecule inhibitors of Aβ aggregation.40–43 The arrival time distributions (ATDs) for Aβ42 peaks at z/n = −5/2 with and without ML are shown in Figures 3h–k. The ATD of the −5/2 peak of Aβ42 without ML (Figure 3h) exhibited four features with arrival times at ca. 720, 680, 620, and 540 µs, previously assigned41 as the −5 dimer, − 10 tetramer, −15 hexamer, and −30 dodecamer, respectively. Dodecamers, potentially associated with memory impairment in mice,41,44 may be of particular interest to AD pathology. The ATD of −5/2 peak for ML-treated Aβ42 displayed three features that were assigned to the −5 dimer, −10 tetramer, and −15 hexamer based on their cross section measurements (Figure 3i). Notably, the feature representing Aβ42 dodecamer was not observed in the presence of ML. The intensity of the −15 hexamer feature is lower compared to that of the Aβ42 sample without ML (Figure 3h and i). These results indicate that the formation of hexamer and dodecamer is partially and completely inhibited, respectively, by ML in solution. In the presence of ML, the largest Aβ species detected are hexamers because no peaks arrive at shorter arrival times (Aβ40 system was also studied; results given in Supporting Information Figure S1).
The ability of ML to disaggregate preformed Aβ42 aggregates was also explored. The ATD for pure Aβ42 incubated on ice for ca. 4 h showed four features corresponding to Aβ42 dimers, tetramers, hexamers, and dodecamers (Figure 3j). Concentrated ML was added to the Aβ42 sample to prepare a 1:5 mixture of Aβ42 and ML. The −5/2 ATD of the mixture (Figure 3k) exhibited three features corresponding to Aβ42 dimers, tetramers, and hexamers. The dodecamers disappeared after the addition of ML, which presents that the compound disaggregates preformed dodecamers in solution. Taken together, these IM–MS studies indicate that ML not only inhibits dodecamer formation but also disaggregates preformed dodecamers in the early oligomerization of Aβ in solution under the MS conditions. ML likely continues to interact with a fraction of the smaller oligomers, possibly redirecting them into conformations of lower toxicity (vide infra).
Control of Metal-Free and Metal-Induced Aβ Aggregation by ML
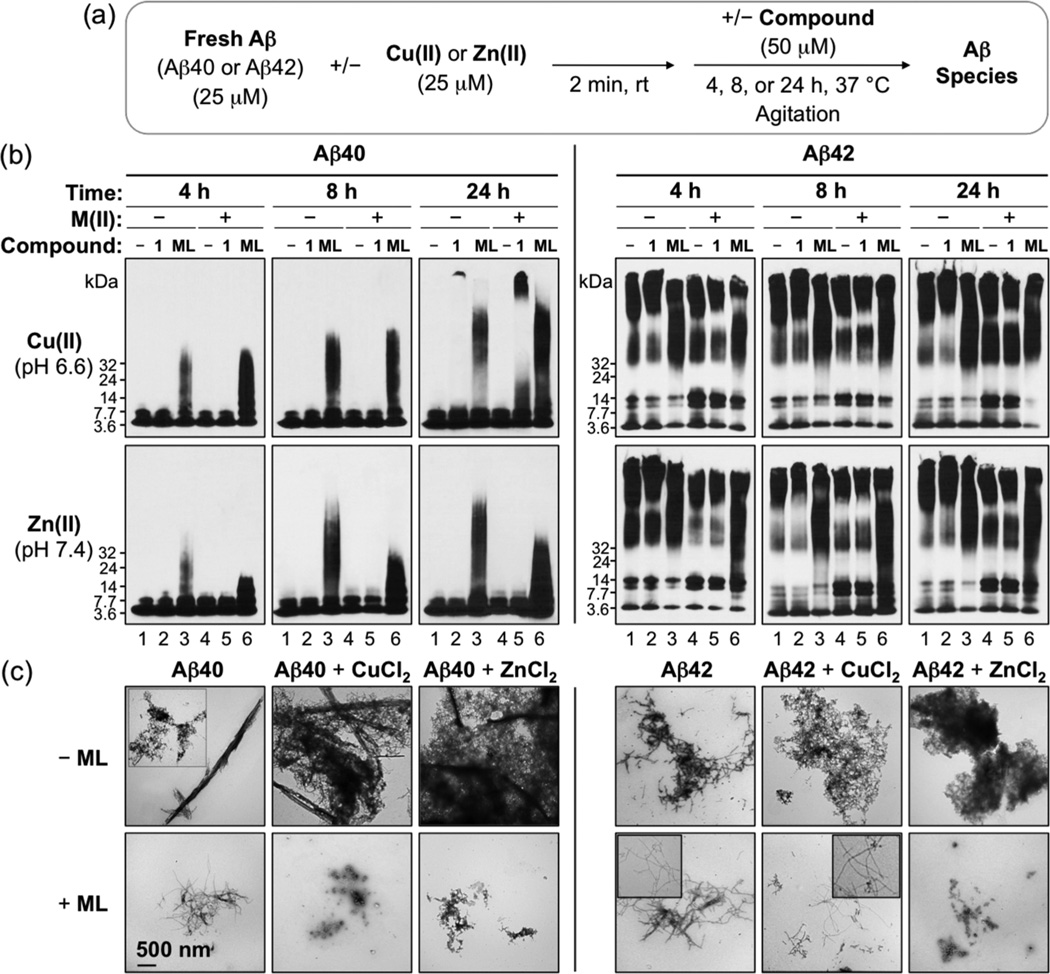
In addition to IM–MS, ML’s ability to inhibit the formation of Aβ aggregates was evaluated in the absence and presence of metal ions by gel electrophoresis/Western blot (to visualize the size distribution of Aβ species) and transmission electron microscopy (TEM) (to examine the morphological change of Aβ species) (Figure 4).21–28 As shown in Figure 4b, the smeared band from ML-treated metal-free Aβ40/Aβ42 samples (lane 3 in all gels) was indicated and compared to lanes from samples of Aβ only or 1-added Aβ (lanes 1 and 2 in all gels). This smeared band is composed of a distribution of Aβ species having various molecular weight (MW) and demonstrates that ML could direct the production of Aβ species with a wide range of MW. ML also exhibited noticeable effects on the modulation of metal-induced Aβ aggregation. Aβ species with a diverse MW distribution (smearing) were presented for samples of ML-incubated Cu(ii)–Aβ or Zn(ii)–Aβ species (lane 6 in all gels) over compound-free or 1-treated analogues (lanes 4 and 5 in all gels).45 Aβ aggregates were shorter in ML-treated metal-free Aβ samples than in nontreated samples as observed by TEM; upon incubation of ML with metal–Aβ species, unstructured Aβ aggregates were mainly visible (Figure 4c).
Figure 4.
Influence of ML or 1 on the formation of metal-free and metal-induced Aβ40/42 aggregates. (a) Scheme of the inhibition experiment. (b) Aβ species were visualized by gel electrophoresis using immunoblotting with an anti-Aβ antibody (6E10). Experimental conditions: Aβ (25 µM); CuCl2 or ZnCl2 (25 µM); ML or 1 (50 µM); 4, 8, or 24 h; pH 6.6 (for metal-free and Cu(ii) experiments) or 7.4 (for metal-free and Zn(ii) experiments); 37 °C; constant agitation. Lanes: (1) Aβ; (2) Aβ + 1; (3) Aβ + ML; (4) Aβ + [CuCl2 or ZnCl2]; (5) Aβ + [CuCl2 or ZnCl2] + 1; (6) Aβ + [CuCl2 or ZnCl2] + ML. (c) TEM images of the 24 h incubated samples from (b).
Furthermore, to understand the structural aspect of ML’s control on Aβ aggregation (Figure 4), the inhibition experiment was performed employing individual structural components of ML, 4-(dimethylamino)phenol (DAP), and 1. As depicted in Supporting Information Figure S10, the effect of ML on metal-free and Cu(ii)-induced Aβ aggregation was more noticeable overall than that of individual structural moieties, DAP and 1 (which were relatively less reactive than ML with a different MW distribution of resulting Aβ species or showed no significant reactivity). In addition, reactivity of a mixture of DAP and 1 (DAP + 1) with metal-free Aβ and Cu(ii)–Aβ was indicated to be similar to DAP only and relatively less reactive than ML showing distinct-sized Aβ species. With Zn(ii) present, DAP and the mixture (DAP + 1) influenced Zn(ii)-triggered Aβ aggregation, similar to ML (Supporting Information Figure S10). Thus, the DAP moiety, the structural portion of ML that contains the dimethylamino functionality believed to be important for Aβ interaction,21–26,29 could interact and react with metal-free and metal-associated Aβ species either less than ML (presenting different-sized Aβ) or similar to ML. Therefore, the reactivity of ML with Aβ species is proposed to derive mainly from its overall framework rather than from individual structural moieties.
We further investigated ML’s impact on the disaggregation of preformed aggregates of Aβ and metal–Aβ (Supporting Information Figure S11). The MW distributions for both Aβ40 and Aβ42 aggregates, with and without metal ions upon incubation with ML (Supporting Information Figure S11b, lanes 3 and 6) were broadly similar to those in the inhibition experiments (Figure 4b). Metal-free and metal-treated Aβ aggregates with and without 1 presented no discernible difference. TEM revealed that ML could transform preformed aggregates into relatively smaller, amorphous conformations that were more apparent for the Zn(ii)–Aβ aggregates relative to the Cu(ii)–Aβ aggregates (Supporting Information Figure S11c). Taken together, our inhibition and disaggregation results demonstrate that ML modulates metal-free and metal-induced Aβ aggregation to different extents. This modulatory activity of ML on Aβ aggregation may occur via direct interaction with metal-free or metal-bound Aβ species to form complexes, chelation of metal ions from metal-bound Aβ species, or both, as supported by MS/IM–MS and NMR results (vide supra).
Regulation of Metal-Free and Metal-Associated Aβ-Induced Toxicity by ML in Living Cells
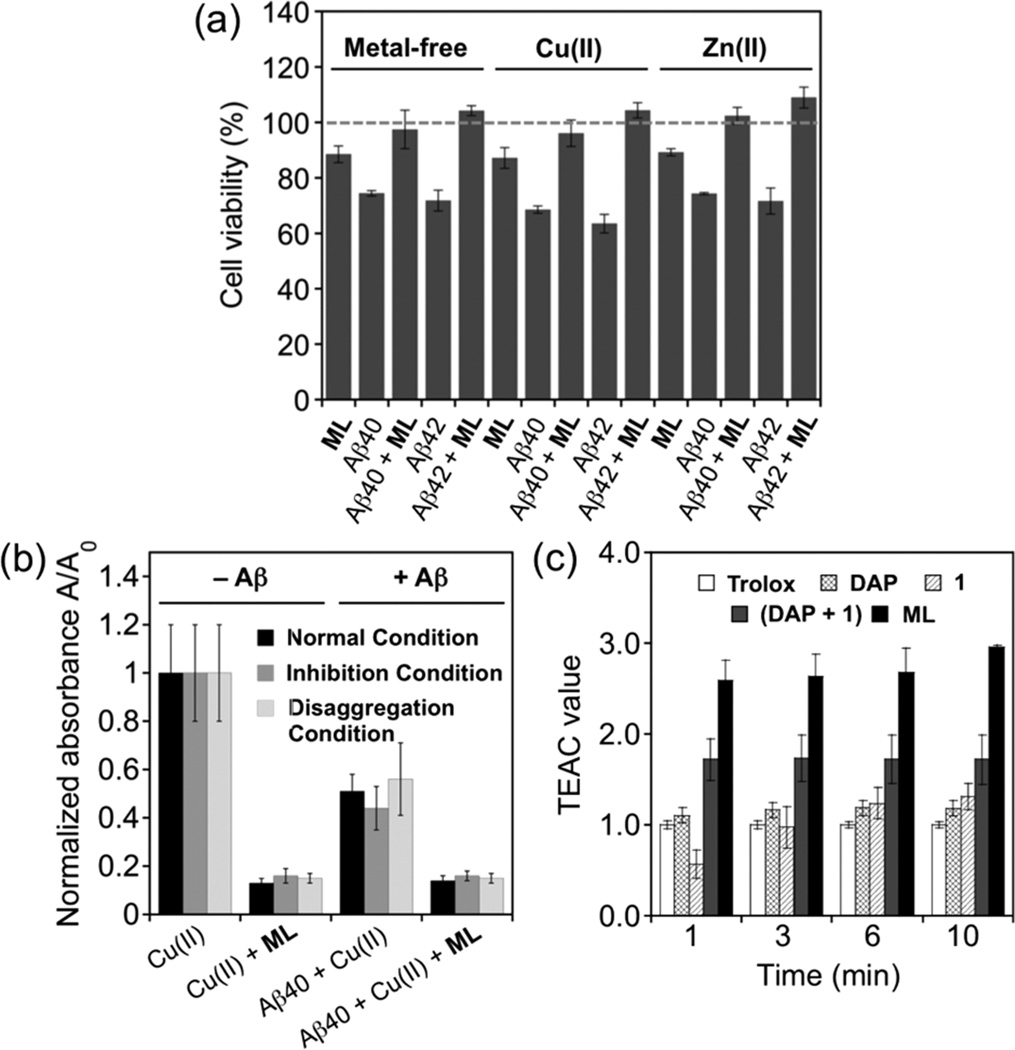
We examined the neuroprotective properties of ML toward Aβ- or metal–Aβ-induced toxicity in murine Neuro-2a neuroblastoma cells with and without overexpression of the Swedish mutant human APP (N2aAPPswe AD cell line;46 both Aβ40 and Aβ42 were employed). Cells incubated with Aβ (10 µM) for 24 h in the absence and presence of metal ions (Cu(ii) or Zn(ii), 10 µM) showed viability of ca. 70% (Aβ), ca. 60–70% (Aβ with Cu(ii)), or ca. 70% (Aβ with Zn(ii)) (Figure 5a and Supporting Information Figure S12). Upon addition of ML (10 µM) to Aβ-treated N2aAPPswe or N2a cells, ca. 90–100% cell survival was observed with and without metal ions (Figure 5a and Supporting Information Figure S12).47 Compound 1, compared to ML, was not shown to improve cell viability significantly in N2a cells incubated by both metal-free Aβ and metal–Aβ (Supporting Information Figure S12). Overall, our cell studies suggest that ML may regulate Aβ/metal–Aβ-induced toxicity in living cells.
Figure 5.
Biological activities of ML. (a) Effect of ML on toxicity triggered by metal-free Aβ and metal–Aβ species in N2aAPPswe cells. Cells treated with Aβ40/42 (10 µM), a metal chloride salt (CuCl2 or ZnCl2; 10 µM), or ML (10 µM) were incubated for 24 h at 37 °C. Cell viability (%) was determined by the MTT assay compared to cells treated with DMSO only (0–1%, v/v) (MTT = 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide). Data are mean ± SEM, P < 0.05, n = 3. (b) Inhibitory activity toward ROS formation in the absence and presence of freshly prepared Aβ40 (normal condition) and Aβ40 aggregates (inhibition and disaggregation conditions), determined by the 2-deoxyribose assay. The absorbance values are normalized compared to ligand-free condition (Aβ/CuCl2/ML = 25/10/125 µM). (c) Antioxidant activity of ML, DAP, 1, and a mixture of DAP and 1 (DAP + 1) identified by the TEAC assay. The TEAC values are relative to a vitamin E analogue, Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid).
ROS Formation Control, Antioxidant Capacity, and BBB Permeability of ML
From a biological perspective, inhibiting ROS formation by binding and constraining Cu(ii) from redox cycling is an attractive feature. Accordingly, we explored the inhibitory ability of ML toward ROS production by the 2-deoxyribose assay.48 As shown in Figure 5b, copper-mediated generation of hydroxyl radicals was significantly reduced upon treatment with ML, in the absence and presence of Aβ (both freshly prepared and aggregated Aβ species). In addition, ML, compared to its individual structural components, DAP and 1, and the mixture (DAP + 1), inhibited the production of hydroxyl radicals by ca. 2- to 4-fold (Supporting Information Figure S13). This result suggests that the overall structure of ML, particularly its metal binding site that is not preferable for redox cycling of Cu(i/ii) (Figure 1),30 guides its capability of controlling Cu-triggered formation of hydroxyl radicals.
The antioxidant activity of ML was also evaluated by the Trolox equivalent antioxidant capacity (TEAC) assay which measures a compound’s ability to quench ABTS cation radicals (ABTS•+; ABTS = 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)) in solution49,50 and in cell lysates. As depicted in Figure 5c, ML scavenged free radicals more effectively in solution than DAP, 1, and Trolox (vitamin E analogue) by a factor of ca. 2.6. Additionally, the mixture (DAP + 1) was observed to scavenge free radicals better than the individual structural component, DAP or 1, but less than ML. The antioxidant capacity of ML was relatively greater than 1 and Trolox within M17 human neuroblastoma cell lysates (1.41 ± 0.15 for ML; 0.86 ± 0.10 for 1; 1.00 ± 0.08 for Trolox). Overall, the studies of ML’s antioxidant activity demonstrate that the presence of both phenolic and quinoline groups within one framework could enhance antioxidant capability.
Lastly, for potential brain applications, the BBB permeability of ML, predicted by Lipinski’s rules and logBB (Table 1), was first examined by the Parallel Artificial Membrane Permeability Assay adapted for BBB (PAMPA-BBB).22,25,26,34,35,51 These values (Table 1), when compared to previously reported BBB permeable molecules,22,25,26,34,35,51 indicate that ML may cross the BBB. The brain uptake of ML was further investigated using male CD1 mice. The brain and plasma concentrations of ML at 5 min52 after its administration to mice (10 mg/kg, n = 3) by oral gavage were 14.3 ± 4.0 ng/g and 5.91 ± 1.24 ng/mL, respectively. The brain-to-plasma ratio of ML was approximately 2.4. These overall in vivo results in conjunction with the in vitro data (Table 1) suggest that ML is able to cross the BBB and to become available within the CNS. Taken together, other biological properties of ML (e.g., ROS clearance, ROS formation control, BBB permeability) demonstrate this structural framework could be valuable for potential biological applications (particularly, in the brain).
CONCLUSIONS
The complexity of AD is suggested to arise from multiple pathological factors, such as Aβ, metal–Aβ, metal ions, and free radicals; however, the roles of individual elements and, more importantly, their interconnection in disease development remain unclear. To advance our understanding of this aspect and target and control all these features, we have rationally designed a novel molecule, ML, by incorporating structural moieties for Aβ/metal–Aβ interactions, metal chelation, ROS generation control, and antioxidant activity into a single framework. Water solubility and BBB permeability were considered for potential biological applications, particularly in the brain, as part of our design approach. To the best of our knowledge, ML is the first example of a single designed molecule that can control multiple reactivities, including metal-free and metal-induced Aβ aggregation, toxicity induced by Aβ and metal–Aβ, ROS generation, and free radical reactions. ML’s properties validates our rational structure-based design strategy and supports the idea that a molecule can be tailored to a specific purpose despite the challenges and complexity of the pathological features of the disease it is intended to examine. There is great heterogeneity in the toxicity landscape of AD.13 We demonstrated ML’s ability to interact with a broad spectrum of Aβ species (i.e., soluble monomers and oligomers, insoluble fibers). Derivatization of ML to a more lipophilic form may further extend its use to interact and react with membrane-bound Aβ or Aβ oligomer channels within membranes. In our future efforts, we intend to extend this work and build a foundation toward the development of chemical tools for uncovering complex AD pathogenesis that will form the basis for the discovery of effective therapeutics for this disease.
EXPERIMENTAL SECTION
Materials and Methods
All reagents were purchased from commercial suppliers and used as received unless otherwise noted. Aβ40 and Aβ42 were purchased from Anaspec (Aβ42 = DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA; Fremont, CA, U. S. A.). The compound DAP was obtained from Ark Pharm., Inc. (Libertyville, IL, U. S. A.). The compounds 130,53 and 5-(dimethylamino)-2-hydroxybenzaldehyde54 were prepared following previously reported procedures. NMR and mass spectrometric analyses of small molecules were conducted on a 400 MHz Varian NMR spectrometer and a Micromass LCT Electrospray Time-of-Flight (TOF) mass spectrometer, respectively. Trace metal contamination was removed from buffers and solutions used for metal binding and Aβ experiments (vide infra) by treating with Chelex overnight (Sigma-Aldrich, St. Louis, MO, U. S. A.). Optical spectra were recorded on an Agilent 8453 UV–visible (UV–vis) spectrophotometer. Transmission electron microscopic (TEM) images were taken using a Philips CM-100 transmission electron microscope (Microscopy and Image Analysis Laboratory, University of Michigan, Ann Arbor, MI, U. S. A.). Absorbance values for biological assays, including cell viability assay, PAMPA-BBB, 2-deoxyribose assay, and TEAC assay, were measured on a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA, U. S. A.). Mass spectra for investigating the interaction of Aβ with ML in the absence and presence of Cu(ii) and Zn(ii) were acquired on a traveling-wave Quadrupole TOF (Q-TOF) mass spectrometer (Waters Synapt Prototype, Milford, MA, U. S. A.)55 and a home-built electrospray ionization (ESI) ion mobility–mass spectrometer.56 NMR studies of Aβ with ML or Zn(ii) were carried out on a 900 MHz Bruker spectrometer equipped with a cryogenic probe at Michigan State University in Lansing, MI, U. S. A.
Synthesis of 4-(Dimethylamino)-2-(((2-(hydroxymethyl)-quinolin-8-yl)amino)methyl)phenol (ML)
A solution (dry ethyl acetate (EtOAc), 8.0 mL) of 130,53 (174 mg, 0.99 mmol) and 5-(dimethylamino)-2-hydroxybenzaldehyde54 (164 mg, 0.99 mmol) was stirred overnight at room temperature. After removing the solvent, the resulting solid material was dissolved in dichloroethane (DCE, 8.0 mL) followed by addition of sodium triacetoxyborohydride (NaB-(OAc)3H, 420 mg, 2.0 mmol). After stirring for 24 h at room temperature, the crude product was purified by column chromatography (SiO2, 1:5 hexanes/EtOAc, Rf = 0.47). The final product (orange powder, HCl salt form) was obtained by recrystallization (upon addition of 1:1 HCl/H2O to a MeOH solution of crude products) (198 mg, 0.50 mmol, 51%). 1H NMR (400 MHz, CD3OD, δ (ppm)): 8.73 (1H, d, J = 8.4 Hz), 7.92 (1H, d, J = 8.8 Hz), 7.80 (1H, d, J = 8.0 Hz), 7.67 (1H, t, J = 8.0 Hz), 7.61 (1H, d, J = 2.8 Hz), 7.55 (2H, m), 7.03 (1H, d, J = 8.8 Hz), 5.12 (2H, s), 4.74 (2H, s), 3.16 (6H, s). 13C NMR (100 MHz, DMSO-d6, δ (ppm)): 159.2, 156.0, 141.8, 139.8, 134.5, 133.7, 127.9, 127.8, 126.2, 121.4, 120.7, 119.6, 115.8, 115.1, 106.2, 63.2, 46.0, 42.2. HRMS: [M + H]+ calcd, 324.1707; found, 324.1697. Anal. Calcd for C19H23Cl2N3O2 (ML· 2HCl·H2O): C, 55.08; H, 6.08; N, 10.14. Found: C, 54.68; H, 5.96; N, 9.80.
Ion Mobility–Mass Spectrometry (IM–MS)
Lyophilized Aβ40 and Aβ42 were dissolved in 10 mM ammonium acetate buffer (pH 7.4) to generate a final peptide concentration of 10 µM for all mass spectrometry experiments. Mass spectra were recorded on a prototype of the commercial Waters Synapt instrument (Milford, MA, U. S. A.)55 and a home-built ESI ion mobility–mass spectrometer.56 Briefly, for ion mobility measurements, ions were generated continuously by a nano-ESI source, focused, and stored in the ion funnel. The ions were then pulsed into a temperature-controlled drift cell filled with 3–5 torr helium gas, where they gently pass through under the influence of a weak electric field. The ions exiting the drift cell were mass analyzed with a quadrupole mass filter, detected by a conversion dynode and channel electron multiplier, and recorded as a function of time to obtain the arrival time distributions (ATDs).
The velocity of the ions in the drift cell vd is proportional to the electric field E
| (1) |
Here, the proportionality constant K is termed ion mobility. The absolute ion mobility is dependent on the temperature (T) and the pressure (P) of the buffer gas (He), so it is typically converted to the reduced mobility K0
| (2) |
The ions exiting the drift cell are mass analyzed and detected as a function of the arrival time, tA. The reduced mobility K0 can be determined from the instrument parameters by using eq 3 and plotting tA versus P/V57
| (3) |
In eq 3, l is the length of the drift cell (4.503 cm), V is the voltage across the drift cell, and t0 is the time the ions spend outside the drift cell before hitting the detector. All of these quantities are either known constants or are measured for each experiment.
The reduced ion mobility K0 can be related to the collision cross section Ω using kinetic theory58
| (4) |
Here, q is the ion charge, N is the buffer gas number density at STP, μ is the reduced mass of the ion–He collision, and kB is the Boltzmann constant. The measured reduced mobility (K0) and the collision cross section (Ω) provide information about the three-dimensional configurations of the ions. For peptide and protein ions, the secondary/tertiary structural information and the oligomerization states can be identified by comparison with modeling.41
2D NMR Spectroscopy
The interaction of prefibrillar Aβ40 with ML was determined by a series of 2D band-Selective Optimized Flip-Angle Short Transient Heteronuclear Multiple Quantum Correlation (SOFAST-HMQC) experiments by titrating a 80 µM solution of Aβ40 with a 40 mM stock solution of ML in DMSO.59 The influence of DMSO on the spectrum of Aβ40 is minor at the maximum concentration of DMSO used (1% v/v final concentration).23 NMR samples were prepared from 15N-labeled Aβ40 (rPeptide, Bogart, GA, U. S. A.) by first dissolving the peptide in 1% NH4OH, lyophilizing, and then resuspending in 150 µL of 1 mM NaOD (pH 10). The peptide was then diluted 1:1 with deuterated Tris–DCl for a final buffer concentration of 50 mM Tris–DCl, verified to be pD 7.3 before the start of each titration using the relation pD = pH meter reading +0.4.60 Trace metals were removed by treating all buffers and solutions with Chelex (Sigma-Aldrich, St. Louis, MO, U. S. A.) prior to the experiment. Each spectrum was obtained from 256 t1 experiments, 16 transients, and a 100 ms recycle delay on a Bruker Avance 900 MHz spectrometer at 4 °C. 2D data were processed using TOPSPIN 2.1 (from Bruker). Resonance assignment and volume fit calculations were performed with SPARKY 3.113 using published assignments for Aβ40 as a guide.36,61,62
Saturation Transfer Difference (STD) NMR Spectroscopy
For the STD NMR experiments, an 80 µM solution of fibrillar Aβ40 was prepared by incubating for 24 h at 37 °C with constant agitation in 50 mM deuterated Tris–DCl, 95% D2O with or without 80 µM ZnCl2 at pD 7.4 (corrected for the isotope effect). To minimize the aggregation of ML that occurs at high concentration at neutral pH, ML was added to fibrillar Aβ40 by dilution from a 3.1 mM stock solution in acidic 1 mM DCl (pD 4) for a final ML:Aβ40 molar ratio of 10:1 (200 µM ML:20 µM Aβ40) in 50 mM deuterated Tris–DCl, 95% D2O with or without 80 µM ZnCl2 at pD 7.4. STD experiments were acquired with a train of 60 dB Gaussian-shaped pulses of 50 ms duration at centered at either −2.0 ppm (on resonance63–65) or 100 ppm (off resonance) with a total saturation time of 3 s on a Bruker 500 MHz (metal-free samples) or 900 MHz spectrometer (Zn(ii) samples).66 A total of 2048 transients with a 2 s recycle delay were collected for all spectra. Chemical shifts are referenced to water due to the potential interaction of Aβ40 with the chemical shift standard sodium 4,4-dimethyl-4-silapentane-1-sulfonate (DSS).67
Docking of ML with Fibrillar Aβ40
Flexible ligand docking studies of ML against the Aβ40 fiber were conducted using AutoDock4.2.68 The starting conformations of the fiber were obtained from previous solid-state NMR models of Aβ40 fibers (PDB LMO and LMN); current structural constraints are consistent with both models for Aβ40 fibers formed under agitation.69 A model for ML was first constructed and energy minimized using the PRODRG server.70 Prior to docking, hydrogen atoms were added to ML and the Aβ40 fiber using AutoDock Tools.68 Kollman charges71 were used for the Aβ40 fiber and Gasteiger charges72,73 were introduced to ML. An electrostatics grid map of the system and atomic affinity grid maps for each atom type were then set up using AutoGrid 4.2, using 126 points in each dimension centered on the Aβ40 fiber with a grid spacing of 0.636 Å.74 Docking was performed using a Lamarckian Genetic Algorithm to search the conformation space of ML for low energy binding orientations.74 An initial random population of 200 individuals was used to start the docking run, which was set to have a maximum of 25 × 106 energy evaluations and a maximum of 270 000 generations. The resulting structures from docking were clustered using an RMSD cutoff of 2.0 Å.
Metal Binding Experiments
Unless otherwise stated, metal binding properties of ML (50 µM, 1% v/v DMSO) and 1 (50 or 500 µM, 1% DMSO) were investigated in a Chelex-treated buffered solution containing 20 mM 2-[4-(2-hydroxyethyl)piperazin-1-yl]-ethanesulfonic acid (HEPES), pH 7.4, and 150 mM NaCl. To a solution of ML or 1, 1 equiv of CuCl2 or ZnCl2 was treated and incubated for 30 min (for ML with CuCl2), 1 h (for ML with ZnCl2), or 10 min (for 1 with CuCl2 or ZnCl2) at room temperature. To examine the metal selectivity of ML or 1, 1 or 20 equiv of MgCl2, CaCl2, MnCl2, FeCl2, CoCl2, NiCl2, and ZnCl2 (for Cu(ii) selectivity) or CuCl2 (for Zn(ii) selectivity) was first treated to a solution containing 50 µM of ligand (ML or 1). The spectra were recorded after an additional 5 min incubation at room temperature. The Fe(ii) samples were maintained anaerobically by purging the solutions with N2. CuCl2 (for Cu(ii) selectivity experiments) or ZnCl2 (for Zn(ii) selectivity experiments) (50 µM) was then added to a solution of compound (ML or 1) and a divalent metal chloride salt. The spectra were taken after an additional 5 min incubation at room temperature. Quantification of metal selectivity was calculated by comparing and normalizing the absorption values of metal–ligand complexes at 485 nm (Cu(ii) selectivity of ML), 291 nm (Cu(ii) selectivity of 1), and 425 nm (Zn(ii) selectivity of ML) to the absorption at this wavelength before and after the addition of CuCl2 (AM/ACu) or ZnCl2 (AM/AZn).
Solution Speciation Studies for ML, 1, Cu(ii)–ML/1, and Zn(ii)–ML/1 Complexes
The pKa values for ML and 1 were determined by UV–vis variable-pH titrations based on a previously reported procedure.22–26 To obtain pKa values for the ligands, a solution (100 mM NaCl, 10 mM NaOH, pH 12) of ML or 1 (50 µM) was titrated with small aliquots of HCl to obtain at least 30 spectra in the range of pH 2–8 (for ML) or pH 2–9 (for 1). In addition, to investigate Cu(ii) or Zn(ii) binding to ligand at various pHs, solutions containing a ligand (ML or 1) and a metal chloride salt ([M(ii)]:[L] = 1:2; [CuCl2] = 25 µM; [ZnCl2] = 50 µM (for ML) or 250 µM (for 1)) were prepared. The solution was titrated with small aliquots of HCl. At least 30 spectra were measured over the range of pH 2–9 for both Cu(ii) and Zn(ii) systems. The acidity and stability constants were calculated by using the HypSpec program (Protonic Software, Leeds, UK).75 Speciation diagrams of ligands and their corresponding metal complexes were modeled using the HySS2009 program (Protonic Software, Leeds, UK).76
Aβ Aggregation Experiments
Aβ experiments were performed according to previously published methods.21–28,77 Prior to experiments, Aβ40 or Aβ42 was dissolved in ammonium hydroxide (NH4OH, 1% v/v, aq), aliquoted, lyophilized overnight, and stored at −80 °C. For experiments described herein, a stock solution of Aβ was prepared by dissolving lyophilized peptide in 1% NH4OH (10 µL) and diluting with ddH2O. The concentration of the solution was determined by measuring the absorbance of the solution at 280 nm (ε = 1450 M−1cm−1 for Aβ40; ε = 1490 M−1cm−1for Aβ42). The peptide stock solution was diluted to a final concentration of 25 µM in Chelex-treated buffered solution containing HEPES (20 µM, pH 6.6 for metal-free and Cu(ii) samples; pH 7.4 for metal-free and Zn(ii) samples) and NaCl (150 µM). For the inhibition studies,21–28,77 a compound (final concentration 50 µM, 1% v/v DMSO) was added to the sample of Aβ (25 µM) in the absence and presence of a metal chloride salt (CuCl2 or ZnCl2, 25 µM) followed by incubation at 37 °C with constant agitation for 4, 8, and 24 h. For the disaggregation studies,21–28,77 Aβ with and without metal ions was incubated for 24 h at 37 °C with constant agitation prior to treatment with a compound (50 µM). The resulting samples containing Aβ, a metal chloride salt, and a compound were incubated at 37 °C with constant agitation for 4, 8, and 24 h.
Gel Electrophoresis
Samples from the inhibition and disaggregation experiments were analyzed by gel electrophoresis with Western blot using anti-Aβ antibody (6E10).21–28,77 Each sample (10 µL) was separated on a 10–20% Tris–tricine gel (Invitrogen, Grand Island, NY, U. S. A.). Following separation, the proteins were transferred onto nitrocellulose which was blocked with bovine serum albumin (BSA, 3% w/v, Sigma-Aldrich, St. Louis, MO, U. S. A.) in Tris-buffered saline (TBS) containing 0.1% Tween-20 (TBS-T) for 2 or 3 h at room temperature. The membranes were incubated with antibody (6E10, 1:2000, Covance, Princeton, NJ, U. S. A.) in a solution of 2% BSA (w/v in TBS-T) overnight at 4 °C. After washing, the horseradish peroxidase-conjugated goat antimouse secondary antibody (1:5000) in 2% BSA was added for 1 h at room temperature. The ThermoScientific SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL, U. S. A.) was used to visualize protein bands.
Transmission Electron Microscopy (TEM)
Samples for TEM were prepared according to previously reported methods.21–28,77 Glow-discharged grids (Formar/Carbon 300-mesh, Electron Microscopy Sciences, Hatfield, PA, U. S. A.) were treated with Aβ samples from the inhibition and disaggregation experiments (5 µL) for 2 min at room temperature. Excess sample was removed using filter paper followed by washing twice with ddH2O. Each grid was incubated with uranyl acetate (1% w/v ddH2O, 5 µL, 1 min). Upon removal of excess uranyl acetate, the grids were dried for 15 min at room temperature. Images from each sample were taken on a Philips CM-100 transmission electron microscope (80 kV, 25 000× magnification).
Cell Viability Measurements
The N2a cell line was purchased from the American Type Cell Collection (ATCC, Manassas, VA, U. S. A.). N2a cells stably overexpressing the Swedish mutant (K670N and M671L) human APP (N2aAPPswe)46 were the generous gift of Professor Gopal Thinakaran (University of Chicago). Both cell lines ware maintained in media containing 45% DMEM, 50% OPTI-MEM, 5% fetal bovine serum (FBS, Atlanta Biologicals, Lawrenceville, GA, U. S. A.), 100 U/mL penicillin (GIBCO, Grand Island, NY, U. S. A.), and 100 mg/mL streptomycin (GIBCO). For the N2aAPPswe cell line, 0.2 mg/mL G418 (Geneticin, GIBCO) was added to the culture medium. The cells were grown in a humidified atmosphere with 5% CO2 at 37 °C. For the MTT assay (MTT = 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazoliu bromide, Sigma-Aldrich), cells were seeded in a 96 well plate (15 000 cells/100 µL). The cells were then treated with Aβ (10 µM) with or without CuCl2 or ZnCl2 (10 µM) followed by the addition of ML or 1 (final concentration 10 µM, 1% v/v final DMSO concentration). After 24 h incubation, 25 µL MTT (5 mg/mL in phosphate buffered saline (PBS, pH 7.4, GIBCO) was added to each well and the plate was incubated for 4 h at 37 °C. Formazan produced by the cells was solubilized by addition of an acidic solution of N,N-dimethylformamide (50%, v/v, aq) and sodium dodecyl sulfate (SDS, 20%, w/v) overnight at room temperature in the dark. The absorbance was measured at 600 nm using a microplate reader. The concentrations (0 to 10 µM) of ML or 1, which do not interfere with the analysis of MTT assay, were selected for the cell studies.
2-Deoxyribose Assay
The ability of ML, DAP, 1, and a mixture of DAP and 1 (DAP + 1) to suppress free radical Fenton chemistry was determined by the 2-deoxyribose assay. The assay was performed based on previously established methods with some modifications.48 Chelexed solutions were used, and reactions (total volume, 200 µL) were setup by mixing, in the following order, buffer (50 mM NaH2PO4, pH 7.2), ligand (125 µM), CuCl2 (10 µM), 2-deoxy-d-ribose (15 mM), H2O2 (200 µM), and sodium ascorbate (2 mM) and allowed to react for 1 h at 37 °C with constant agitation. The reactions were quenched upon addition of trichloroacetic acid (200 µL of 2.8% w/v) and 2-thiobarbituric acid (200 µL of 1% w/v) and heated at 100 °C for 20 min, cooled for 5 min, and their absorbance values at 532 nm measured immediately afterward. In addition, samples without ligand were prepared as a control. Experiments were performed in triplicates. Normalized absorbance values (A/A0) were calculated by taking the absorbance (A) and dividing by the absorbance of the control (A0).
Experiments in the presence of Aβ (25 µM) were carried out in three formats. (a) Experiments with freshly prepared Aβ40 were conducted in the “normal condition” as for Aβ-free samples in which all reagents were added simultaneously, shaken for 2 h at 37 °C, and heated to develop the pink chromogen at 532 nm. (b) Experiments were also performed in the “inhibition condition” in which Aβ/CuCl2/ML (25/10/125 µM) samples were mixed and incubated for 24 h, then assay reagents were added (2-deoxyribose, H2O2, and ascorbate), and the assay was carried out. (c) Experiments were also completed in the “disaggregation condition” in which ML (125 µM) was added to Aβ aggregates generated by treating Aβ (25 µM) with CuCl2 (10 µM) for 24 h and the resulting solution was incubated for 24 h, at which point the assay was carried out. Copper-only and copper-ML-only controls were prepared for comparison. Data was analyzed as for Aβ-free experiments, by normalizing the absorbance at 532 nm against copper-only controls.
Trolox Equivalent Antioxidant Capacity (TEAC) Assay
The antioxidant activity of ML and 1 was determined by the TEAC assay in EtOH and with M17 human neuroblastoma cell lysates. (a) The assay in solution was performed according to a previously reported method with slight modifications.49,50 To generate blue ABTS cation radicals (ABTS•+; ABTS = 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt; Sigma-Aldrich), a solution of ABTS (7.0 mM) and potassium persulfate (2.5 mM; Sigma-Aldrich) was prepared in 5 mL of water and incubated for 16 h at room temperature in the dark. The resulting solution was diluted with EtOH to an absorbance of ca. 0.7 at 734 nm. ABTS•+ solution (200 µL) was added to the wells of a clear 96 well plate and incubated for 5 min at 30 °C. Various final concentrations (0, 1, 2.5, 5, 7.5, 10, and 15 µM) of ML, DAP, 1, (DAP + 1) or Trolox (Trolox = 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid; Sigma-Aldrich; dissolved in EtOH) were added and incubated with the ABTS•+ solution at 30 °C for different time periods (1, 3, 6, and 10 min). The percent inhibition was calculated according to the measured absorbance at 734 nm (% inhibition =100 × (A0 − A)/A0) and was plotted as a function of ligand concentration. The TEAC value of compounds for each time point was calculated as a ratio of the slope of the compound to that of Trolox. The measurements were carried out in triplicate. (b) The assay employing cell lysates was conducted following the protocol of the antioxidant assay kit purchased from Cayman Chemical Company (Ann Arbor, MI, U. S. A.), with minor modifications. The human neuroblastoma SK-N-BE(2)-M17 (M17) cells were used for this assay. This cell line purchased from ATCC (Manassas, VA, U. S. A.) was maintained in media containing 1:1 minimum Essential Media (MEM, GIBCO) and Ham’s F12K Kaighn’s Modification Media (F12K, GIBCO), 10% (v/v) fetal bovine serum (FBS, GIBCO), 100 U/mL penicillin (GIBCO), and 100 mg/mL streptomycin (GIBCO). The cells were grown and maintained at 37 °C in a humidified atmosphere with 5% CO2. For the antioxidant assay using cell lysates, cells were seeded in a 6 well plate and grown to approximately 80–90% confluence. Cell lysates were prepared following a previously reported method with modifications.78 M17 cells were washed once with cold PBS (pH 7.4, GIBCO) and harvested by gently pipetting off adherent cells with cold PBS. The cell pellet was generated by centrifugation (2000g for 10 min at 4 °C). This cell pellet was sonicated on ice (5 s pulses five times with 20 s intervals between each pulse) in 2 mL of cold Assay Buffer (5 mM potassium phosphate, pH 7.4, containing 0.9% NaCl and 0.1% glucose). The cell lysates were centrifuged at 10 000g for 10 min at 4 °C. The supernatant was removed and stored on ice until use. To standard and sample 96 wells, 10 µL of the supernatant of cell lysates was delivered followed by addition of compound, metmyoglobin, ABTS, and H2O2 in order. After 5 min incubation at room temperature on a shaker, absorbance values at 750 nm were recorded. The final concentrations (0.045, 0.090, 0.135, 0.180, 0.225, and 0.330 mM) of ML, 1, and Trolox were used. The percent inhibition was calculated according to the measured absorbance (% inhibition = (A0 − A)/A0, where A0 is absorbance of the supernatant of cell lysates) and was plotted as a function of compound concentration. The TEAC value of ligands was calculated as a ratio of the slope of the standard curve of the compound to that of Trolox. The measurements were conducted in triplicate.
Parallel Artificial Membrane Permeability Adapted for the Blood-Brain Barrier (PAMPA-BBB) Assay
PAMPA-BBB experiments were carried out using the PAMPA Explorer kit (pION Inc., Billerica, MA, U. S. A.) with modification to previously reported protocols.22,25,26,34,35,51 Each stock solution was diluted with Prisma HT buffer (pH 7.4, pION) to a final concentration of 25 µM (1% v/v final DMSO concentration). The resulting solution was added to wells of the donor plate (200 µL, number of replicates = 12 (for ML) and 11 (for 1)). BBB-1 lipid formulation (5 µL, pION) was used to coat the polyvinylidene fluoride (PVDF, 0.45 µM) filter membrane on the acceptor plate. This acceptor plate was placed on top of the donor plate forming a sandwich. Brain sink buffer (BSB, 200 µL, pION) was added to each well of the acceptor plate. The sandwich was incubated for 4 h at ambient temperature without stirring. UV–vis spectra of the solutions in the reference, acceptor, and donor plates were measured using a microplate reader. The PAMPA Explorer software v. 3.5 (pION) was used to calculate −log Pe for each compound. CNS+/− designations were assigned by comparison to compounds that were identified in previous reports.34,35,51
Brain Uptake Studies
The experiments of ML’s brain uptake were carried out using male CD1 mice (purchased from Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China)) by Contract Research Organization, Shanghai ChemPartner Co., Ltd. (Shaghai, China). The studies reported here adhere to the principles of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International. ML (10 mg/kg; single dose; sterile water) was administrated to mice by oral gavage. At 5 min postdose52 (n = 3 at each time point), 150 µL of blood was withdrawn via retro orbital puncture or cardiac puncture and transferred into tubes with spray-coated K2EDTA as anticoagulant. Blood samples were put on ice and centrifuged to obtain plasma samples (2000g, 5 min, 4 °C). Immediately following blood collection, mice were euthanized by pure CO2 inhalation and the whole brain was collected, rinsed with cold saline, dried on filter paper, and weighed. The brain samples were immediately homogenized with three volumes (v/w) of homogenizing solution (PBS). Both plasma and brain samples were added with an internal standard (propranolol) in acetonitrile (CH3CN; protein precipitation). The mixture was vortexed for 2 min and centrifuged at 14 000 rpm for 5 min and an aliquot of the supernatant was analyzed for concentration of ML by LC-MS/MS (UPLC/MS-MS API-5500, Framingham, MA, U. S. A.), with analytical lower limit of quantitation (LLOQ) values for ML at 1 ng/mL (plasma) and 4 ng/mL (brain). The supernatant was stored at −80 °C prior to analysis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the National Institutes of Health to A.R. (GM084018, GM095640, and RR023597) and to M.T.B. (AG047116), the American Heart Association, Ruth K. Broad Biomedical Foundation, Alfred P. Sloan Fellowship, Rush Alzheimer’s Disease Core Center (P30AG010161), and the National Science Foundation (CHE-1253155) (to M.H.L.). This work was also supported by the 2013 Research Fund (Project Number 1.130068.01) of UNIST (Ulsan National Institute of Science and Technology) (to M.H.L.). C.K. thanks the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2012001725) for support. We are grateful to Professor Gopal Thinakaran (University of Chicago) for the generous supply of the N2aAPPswe cell line. We also thank Alaina DeToma for experimental assistance on the PAMPA assay.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Additional mass spectra, UV–vis spectra, docking diagrams, cluster energy analyses, electrophoresis visualizations, TEM images, and selectivity, speciation, and cytotoxicity, and inhibitory activity studies. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Thies W, Bleiler L. Alzheimer’s Association. Alzheimer’s Dementia. 2013;9:208–245. doi: 10.1016/j.jalz.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Corbett A, Pickett J, Burns A, Corcoran J, Dunnett SB, Edison P, Hagan JJ, Holmes C, Jones E, Katona C, Kearns I, Kehoe P, Mudher A, Passmore A, Shepherd N, Walsh F, Ballard C. Nat. Rev. Drug Discov. 2012;11:833–846. doi: 10.1038/nrd3869. [DOI] [PubMed] [Google Scholar]
- 3.Jakob-Roetne R, Jacobsen H. Angew. Chem. Int. Ed. 2009;48:3030–3059. doi: 10.1002/anie.200802808. [DOI] [PubMed] [Google Scholar]
- 4.Hamley IW. Chem. Rev. 2012;112:5147–5192. doi: 10.1021/cr3000994. [DOI] [PubMed] [Google Scholar]
- 5.Kepp KP. Chem. Rev. 2012;112:5193–5239. doi: 10.1021/cr300009x. [DOI] [PubMed] [Google Scholar]
- 6.Savelieff MG, Lee S, Liu Y, Lim MH. ACS Chem. Biol. 2013;8:856–865. doi: 10.1021/cb400080f. [DOI] [PubMed] [Google Scholar]
- 7.Duce JA, Bush AI. Prog. Neurobiol. 2010;92:1–18. doi: 10.1016/j.pneurobio.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 8.DeToma AS, Salamekh S, Ramamoorthy A, Lim MH. Chem. Soc. Rev. 2012;41:608–621. doi: 10.1039/c1cs15112f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott LE, Orvig C. Chem. Rev. 2009;109:4885–4910. doi: 10.1021/cr9000176. [DOI] [PubMed] [Google Scholar]
- 10.Que EL, Domaille DW, Chang CJ. Chem. Rev. 2008;108:1517–1549. doi: 10.1021/cr078203u. [DOI] [PubMed] [Google Scholar]
- 11.Zatta P, Drago D, Bolognin S, Sensi SL. Trends Pharmacol. Sci. 2009;30:346–355. doi: 10.1016/j.tips.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Pithadia AS, Lim MH. Curr. Opin. Chem. Biol. 2012;16:67–73. doi: 10.1016/j.cbpa.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Miller Y, Ma B, Nussinov R. Chem. Rev. 2010;110:4820–4838. doi: 10.1021/cr900377t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haass C, Selkoe DJ. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 15.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farreel MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Nat. Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faller P, Hureau C. Dalton Trans. 2009:1080–1094. doi: 10.1039/b813398k. [DOI] [PubMed] [Google Scholar]
- 17.Faller P. Chem Bio Chem. 2009;10:2837–2845. doi: 10.1002/cbic.200900321. [DOI] [PubMed] [Google Scholar]
- 18.Telpoukhovskaia M, Orvig C. Chem. Soc. Rev. 2013;42:1836–1846. doi: 10.1039/c2cs35236b. [DOI] [PubMed] [Google Scholar]
- 19.Rodríguez-Rodríguez C, Telpoukhovskaia M, Orvig C. Coord. Chem. Rev. 2012;256:2308–2332. [Google Scholar]
- 20.Braymer JJ, DeToma AS, Choi J-S, Ko KS, Lim MH. Int. J. Alzheimer’s Dis. 2011;2011:623051. doi: 10.4061/2011/623051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hindo SS, Mancino AM, Braymer JJ, Liu Y, Vivekanandan S, Ramamoorthy A, Lim MH. J. Am. Chem. Soc. 2009;131:16663–16665. doi: 10.1021/ja907045h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi J-S, Braymer JJ, Nanga RPR, Ramamoorthy A, Lim MH. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21990–21995. doi: 10.1073/pnas.1006091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braymer JJ, Choi J-S, DeToma AS, Wang C, Nam K, Kampf JW, Ramamoorthy A, Lim MH. Inorg. Chem. 2011;50:10724–10734. doi: 10.1021/ic2012205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi J-S, Braymer JJ, Park SK, Mustafa S, Chae J, Lim MH. Metallomics. 2011;3:284–291. doi: 10.1039/c0mt00077a. [DOI] [PubMed] [Google Scholar]
- 25.Pithadia AS, Kochi A, Soper MT, Beck MW, Liu Y, Lee S, DeToma AS, Ruotolo BT, Lim MH. Inorg. Chem. 2012;51:12959–12967. doi: 10.1021/ic302084g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Kochi A, Pithadia AS, Lee S, Nam Y, Beck MW, He X, Lee D, Lim MH. Inorg. Chem. 2013;52:8121–8130. doi: 10.1021/ic400851w. [DOI] [PubMed] [Google Scholar]
- 27.Hyung SJ, DeToma AS, Brender JR, Lee S, Vivekanandan S, Kochi A, Choi JS, Ramamoorthy A, Ruotolo BT, Lim MH. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3743–3748. doi: 10.1073/pnas.1220326110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeToma AS, Choi J-S, Braymer JJ, Lim MH. Chem Bio Chem. 2011;12:1198–1201. doi: 10.1002/cbic.201000790. [DOI] [PubMed] [Google Scholar]
- 29.Kung HF, Lee C-W, Zhuang Z-P, Kung M-P, Hou C, Plössl KJ. Am. Chem. Soc. 2001;123:12740–12741. doi: 10.1021/ja0167147. [DOI] [PubMed] [Google Scholar]
- 30.Lim MH, Wong BA, Pitcock WH, Mokshagundam D, Baik M-H, Lippard SJ. J. Am. Chem. Soc. 2006;128:14364–14373. doi: 10.1021/ja064955e. [DOI] [PubMed] [Google Scholar]
- 31.Orhan Puskullu M, Tekiner B, Suzen S. Mini Rev. Med. Chem. 2013;13:365–372. doi: 10.2174/138955713804999793. [DOI] [PubMed] [Google Scholar]
- 32.Havsteen BH. Pharmacol. Ther. 2002;96:67–202. doi: 10.1016/s0163-7258(02)00298-x. [DOI] [PubMed] [Google Scholar]
- 33.Rice-Evans CA, Miller NJ, Paganga G. Free Radical Biol. Med. 1996;20:933–956. doi: 10.1016/0891-5849(95)02227-9. [DOI] [PubMed] [Google Scholar]
- 34.Di L, Kerns EH, Fan K, McConnell OJ, Carter GT. Eur. J. Med. Chem. 2003;38:223–232. doi: 10.1016/s0223-5234(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 35.Avdeef A, Bendels S, Di L, Faller B, Kansy M, Sugano K, Yamauchi YJ. Pharm. Sci. 2007;96:2893–2909. doi: 10.1002/jps.21068. [DOI] [PubMed] [Google Scholar]
- 36.Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A. Biochem. Biophys. Res. Commun. 2011;411:312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou L, Shao H, Zhang Y, Li H, Menon NK, Neuhaus EB, Brewer JM, Byeon I-JL, Ray DG, Vitek MP, Iwashita T, Makula RA, Przybyla AB, Zagorski MG. J. Am. Chem. Soc. 2004;126:1992–2005. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- 38.Mayer M, Meyer B. J. Am. Chem. Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 39.Wyttenbach T, Bowers MT. In: Modern Mass Spectrometry, Topics in Current Chemistry. Christoph A, Schalley, editors. Vol. 225. Berlin: Springer; 2003. pp. 207–232. [Google Scholar]
- 40.Bernstein SL, Wyttenbach T, Baumketner A, Shea J-E, Bitan G, Teplow DB, Bowers MT. J. Am. Chem. Soc. 2005;127:2075–2084. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- 41.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea J-E, Ruotolo BT, Robinson CV, Bowers MT. Nat. Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gessel MM, Wu C, Li H, Bitan G, Shea J-E, Bowers MT. Biochemistry. 2012;51:108–117. doi: 10.1021/bi201520b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng X, Gessel MM, Wisniewski ML, Viswanathan K, Wright DL, Bahr BA, Bowers MT. J. Biol. Chem. 2012;287:6084–6088. doi: 10.1074/jbc.C111.328575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ash KH. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 45.Upon 24 h incubation of Cu(ii)-treated Aβ42 with ML, small sized Aβ species disappeared.
- 46.Thinakaran G, Teplow DB, Siman R, Greenberg B, Sisodia SS. J. Biol. Chem. 1996;271:9390–9397. doi: 10.1074/jbc.271.16.9390. [DOI] [PubMed] [Google Scholar]
- 47.The concentration of ML with and without metal ions used for Figure 5a is relatively nontoxic in both N2a and N2aAPPswe cells (ca. 85% cell survival for 24 h; Supporting Information Figure S12). The concentrations (0 to 10 µM) of ML, which do not interfere with the analysis of MTT assay, were selected for cell studies.
- 48.Charkoudian LK, Pham DM, Franz KJ. J. Am. Chem. Soc. 2006;128:12424–12425. doi: 10.1021/ja064806w. [DOI] [PubMed] [Google Scholar]
- 49.Re R, Pellegrini N, Proteggente A, Pannala A, Yang M, Rice-Evans C. Free Radical Biol. Med. 1999;26:1231–1237. doi: 10.1016/s0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- 50.Schugar H, Green DE, Bowen ML, Scott LE, Storr T, Böhmerle K, Thomas F, Allen DD, Lockman PR, Merkel M, Thompson KH, Orvig C. Angew. Chem., Int. Ed. 2007;46:1716–1718. doi: 10.1002/anie.200603866. [DOI] [PubMed] [Google Scholar]
- 51.BBB Protocol and Test Compounds. Woburn, MA: pION Inc.; 2009. [Google Scholar]
- 52.Due to limited stability of ML, confirmed employing human, rat, and mouse liver microsomes (half time (t1/2) < 30 min, susceptible to metabolism in liver microsomes; [ML] = 1 µM, 37 °C, ketanserin as a reference compound; this in vitro metabolic stability study was performed by a Contract Research Organization, Shaghai Chem-Partner Co., Ltd.), the brain and plasma concentrations were measured at a short incubation time point (i.e., 5 min).
- 53.Roth R, Erlenmeyer H. Helv. Chim. Acta. 1954;37:1064–1068. [Google Scholar]
- 54.Waibel M, Hasserodt J. Tetrahedron Lett. 2009;50:2767–2769. [Google Scholar]
- 55.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. Int. J. Mass Spectrom. 2007;261:1–12. [Google Scholar]
- 56.Wyttenbach T, Kemper PR, Bowers MT. Int. J. Mass Spectrom. 2001;212:13–23. [Google Scholar]
- 57.Gidden J, Baker ES, Ferzoco A, Bowers MT. Int. J. Mass Spectrom. 2005;240:183–193. [Google Scholar]
- 58.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. New York: Wiley; 1988. [Google Scholar]
- 59.Schanda P, Brutscher BJ. Am. Chem. Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 60.Glasoe PK, Long FA. J. Phys. Chem. 1960;64:188–190. [Google Scholar]
- 61.Yoo SI, Yang M, Brender JR, Vivekanandan S, Sun K, Joo NE, Jeong S-H, Ramamoorthy A, Kotov NA. Angew. Chem., Int. Ed. 2011;50:5110–5115. doi: 10.1002/anie.201007824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fawzi NL, Ying J, Torchia DA, Clore GM. J. Am. Chem. Soc. 2010;132:9948–9951. doi: 10.1021/ja1048253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soong R, Brender JR, Macdonald PM, Ramamoorthy AJ. Am. Chem. Soc. 2009;131:7079–7085. doi: 10.1021/ja900285z. [DOI] [PubMed] [Google Scholar]
- 64.Huang R, Vivekanandan S, Brender JR, Abe Y, Naito A, Ramamoorthy AJ. Mol. Biol. 2012;416:108–120. doi: 10.1016/j.jmb.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narayanan S, Reif B. Biochemistry. 2005;44:1444–1452. doi: 10.1021/bi048264b. [DOI] [PubMed] [Google Scholar]
- 66.Airoldi C, Sironi E, Dias C, Marcelo F, Martins A, Rauter AP, Nicotra F, Jimenez-Barbero J. Chem. Asian J. 2013;8:596–602. doi: 10.1002/asia.201201063. [DOI] [PubMed] [Google Scholar]
- 67.Laurents DV, Gorman PM, Guo M, Rico M, Chakrabartty A, Bruix MJ. Biol. Chem. 2005;280:3675–3685. doi: 10.1074/jbc.M409507200. [DOI] [PubMed] [Google Scholar]
- 68.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petkova AT, Yau W-M, Tycko R. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schüttelkopf AW, van Aalten DMF. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 71.Singh UC, Kollman PA. J. Comput. Chem. 1984;5:129–145. [Google Scholar]
- 72.Gasteiger J, Marsili M. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 73.Gasteiger J, Marsili M. Tetrahedron Lett. 1978;19:3181–3184. [Google Scholar]
- 74.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 75.Gans P, Sabatini A, Vacca A. Ann. Chim. 1999;89:45–49. [Google Scholar]
- 76.Alderighi L, Gans P, Ienco A, Peters D, Sabatini A, Vacca A. Coord. Chem. Rev. 1999;184:311–318. [Google Scholar]
- 77.Mancino AM, Hindo SS, Kochi A, Lim MH. Inorg. Chem. 2009;48:9596–9598. doi: 10.1021/ic9014256. [DOI] [PubMed] [Google Scholar]
- 78.Spencer VA, Sun J-M, Li L, Davie JR. Methods. 2003;31:67–75. doi: 10.1016/s1046-2023(03)00089-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.