Abstract
To determine whether initiation is rate-limiting in protein synthesis during the embryogenesis of sea urchins, polyribosome profiles of unfertilized eggs and cleavage, blastula and prism stage embryos were examined after incubation of the eggs and embryos in the presence and absence of low amounts of emetine, an inhibitor of polypeptide elongation. The ribosomes were radioactively labeled with [3H]uridine by injection of the adults during oogenesis so that we could monitor emetine-dependent shifts of monoribosomes to polyribosomes. Although initiation is not rate limiting in unfertilized eggs or 2- to 16-cell embryos of Strongylocentrotus purpuratus, it is rate limiting in blastula and prism embryos. We suggest that initiation becomes rate limiting to allow the selective translation of certain classes of mRNA during later development.
INTRODUCTION
In echinoids the rate of protein synthesis increases rapidly after fertilization (Hultin 1961a; Giudice et al., 1962; Denny and Tyler, 1964; Berg 1965; Gross et al., 1964; Epel 1967): by gastrulation it reaches a rate 115-fold greater than in the unfertilized egg (Regier and Kafatos, 1977). For polypeptide chain initiation to keep pace with this dramatic increase in the synthesis of proteins, either a very large store of preformed initiation factors, or their rapid synthesis is required. Neither the source of initiation factors during development of echinoids, nor the possible regulatory role of polypeptide chain initiation during their embryogenesis has been studied directly. It has also not been experimentally resolved whether initiation contributes to the overall activation of protein sythesis at fertilization as does polypeptide chain elongation (Hille and Albers, 1979; Brandis and Raff, 1978). Kedes et al. (1969) and Humphreys (1971) have argued that the similarity of polyribosomal profiles before and after fertilization indicates no regulation at the initiation step in unfertilized eggs and early embryos. Conversely, MacKintosh and Bell (1969) have argued that their studies with cycloheximide show that polypeptide chain initiation is limitng in the unfertilized egg.
We are investigating whether polypeptide chain initiation regulates the quantity, and hence possibly the specificity of mRNAs translated in sea urchin eggs or embryos. The studies described here resolve the qualitative question of whether polypeptide chain initiation is rate limiting in unfertilized eggs or at any stage during embryogenesis of the sea urchin Strongylocentrotus purpuratus. By polypeptide chain initiation we refer to the binding of ribosomal subunits to the AUG initiation codon on available mRNAs rather than the availability of mRNAs.
The importance of the selective translation of cytoplasmic mRNAs during development has been shown in various eukaryotes. Its mechanism can be explained by the model of Lodish (1971) which predicts that mRNAs with a high affinity for initiation factors are preferentially translated when initiation is rate limiting. Some examples of cells in which the differential translation of mRNAs have been observed are those from the early stages of the differentiating slime mold, Dictyostelium discoideum (Alton and Lodish, 1977), those from Chinese hamster cells (Walters et al., 1979), mouse sarcoma ascites cells (Geoghegan et al., 1979), and chick oviduct cells (Palmiter, 1974). The limitation in the rate of initiation at the blastula stage reported in this paper may also have significant developmental consequences: the preferential translation of poly(A)+ mRNAs observed in the blastulae of Strongylocentrotus purpuratus and Lytechinus pictus could be a result of this limitation (Nemer, 1975; Nemer et al., 1975; Rudensey and Infante, 1979).
MATERIALS AND METHODS
Eggs and embryos
Strongylocentrotus purpuratus were collected in late October from the Strait of Juan de Fuca in Northwestern Washington. Females were identified by spawning with an intracoelomic injection of 0.55 M KCl. After 4–7 days of recovering the females were injected intracoelomically with 100 µCi of [3H]uridine and fed various local kelps. To prevent the leakage of unincorporated [3H]uridine, animals were collected and handled carefully to avoid damaging the tube feet. Ninety-five percent of the label incorporated into the eggs was precipitable with TCA1 after 1 month, as observed by Piatogorsky and Tyler (1967). The animals were used for 3 to 6 months after labeling.
Adult sea urchins were spawned by the above method and eggs were fertilized in sea water which had been filtered through nitrocellulose filters with 0.45-µm pores (FSW). To prevent hardening of the fertilization membrane 0.5 mM 3-amino-1,2,4-triazole was present during fertilization (Showman and Foerder, 1979). After 30 min at 14°C with gentle stirring, embryos were washed several times in FSW containing streptomycin (100 mg/liter) by centrifugation (200gmax). Embryos were cultured at 14°C to the desired stages with paddle stirring at a concentration of 0.5% (v/v) in FSW plus streptomycin.
Inhibition of peptide synthesis with emetine
[3H]-Uridine-labeled eggs were washed with FSW containing streptomycin and were incubated at 14°C as 4.4% cultures (v/v) for 30 min. Glycylglycine was then added from a 0.5 M stock solution (pH 8.0) to a final concentration of 4.4 mM as a buffer and leucine was added to final concentrations of 8.8 µM and 9µCi/ml l-[4,5-3H]leucine. Eggs were incubated with or without 1.5 µM emetine by gentle rotation in Erlenmeyer flasks with a large surface to volume ratio for adequate aeration. At various time intervals, 1-ml samples were rapidly plunged into 30 vol of −2°C wash solution (0.21 M K+, 5 mM Mg2+, 80 mM Cl−, 130 mM acetate, 20 mM triethanolamine, 0.1 mM EDTA, 1 mM dithiothreitol). The eggs were washed free of external label by two centrifugations at 200g with 30 vol of wash solution. Washed eggs were homogenized by multiple passages through a 20-guage needle in 1.0 ml of wash solution containing 2 mg/ml yeast tRNA, 0.5% Triton X-100, 0.3% sodium deoxycholate, and 0.1 mM phenylmethylsulfonyl fluoride. Aliquots of the homogenates were precipitated with 7.5% trichloroacetic acid, heated at 90°C for 15 min to hydrolyze [3H]uridine RNA and [3H]leucyl-tRNA, cooled, collected on glass membrane filters (Gelman), and digested in 0.72 ml of 83% NCS (Amersham) before counting in 6 ml of toluene-based scintillant. Similar aliquots were precipitated with 7.5% trichloroacetic acid and analyzed for protein concentration by the modified Lowry method of Bensadoun and Weinstein (1976). Aliquots were also removed to measure total uptake of [3H]leucine. Uptake was found to be linear with time and not affected by emetine.
[3H]Uridine-labeled embryos were incubated as described for eggs except that the embryo concentration was 5% (v/v), leucine was 2.5 mM (and 9 µCi/ml [3H]leucine), and emetine, when present, was 8 µM. Processing of the samples was as for eggs except that homogenization was through a 20-guage needle fitted with 22-µm Nitex bolting cloth for embryos without spicules and 53-µm Nitex bolting cloth for prism embryos.
Polyribosome analysis
[3H]Uridine-labeled eggs and embryos were incubated in parallel with those for determining the percentage inhibition by emetine. The FSW contained unlabeled leucine, glycylglycine and, when indicated, emetine. Protein synthesis was rapidly arrested by plunging aliquots into 20 to 40 culture volumes of −2°C wash solution. The eggs and embryos were collected by centrifugation, then homogenized without Nitex in 5 vol of the same wash solution (containing no detergents). The homogenate was centrifuged twice at 9000gmax, and 0.6 ml of the postmitochondrial supernatant was layered on 12.6 ml sucrose gradients (15 to 40% (w/w) over a 1.0 ml 50% (w/w) sucrose pad). The supernatant was overlaid with 0.2 ml of wash solution. Centrifugation was for 1.25 hr at 200,000gmax in an SW 40 Beckman rotor at 2°C. The gradients were pumped from the bottom through a 0.66-cm flow cell in a Gilford spectrophotometer and the absorbance at 260 nm was continuously recorded. Fractions of 0.46 ml were collected and precipitated with 250 µg serum albumin in 7.5% TCA. Samples were collected on Gelman glass filters and counted as described in the previous section. The amount of total 260 nm absorbance in the monoribosome and polyribosomes regions were determined with a Numonics Electronic Graphics Calculator. To accurately and consistently determine the base lines in these gradients, blank gradients run with each experiment were aligned with the polyribosome gradients by using the absorption from the dense sucrose pumping solution as an internal marker.
RESULTS
Treatment with Emetine
When polypeptide chain initiation is the rate-limiting step in protein synthesis, smaller polyribosomes are formed for the same size message than when initiation is not limiting. Reducing the rate of polypeptide elongation relative to initiation with low levels of drugs such as cycloheximide or emetine allows monomeric ribosomes to accumulate on polyribosomes when polypeptide chain initiation is the rate-limiting step (Fan and Penman, 1970), without inhibiting the initiation of new polypeptide chains (Lodish, 1971). Thus, by monitoring polyribosomes in the presence and absence of these drugs, limitations at this initiation step can be identified. We used emetine to study initiation as it is more effective than cycloheximide in inhibiting protein synthesis in sea urchin embryos (Hogan and Gross, 1971) and in reticulocytes, presumably because of a difference in uptake (Lodish, 1971).
The distribution of ribosomes in polyribosomes is conveniently determined by absorption at 260 nm which measures the RNA of polyribosomes, or better, by the distribution of radio activity of labeled RNA. The values determined by measuring radioactivity are not subject to the uncertainty of base line which occurs when measuring uv absorbance Also, the monomeric ribosomes are less obscured by nonribosomal material at the top of the gradient if the fractions are filtered. Therefore, in order to measure the amounts of polyribosomes in the unfertilized eggs of S. purpuratus, we labeled the ribosomes by injecting the female adults with [3H]uridine during oogenesis.
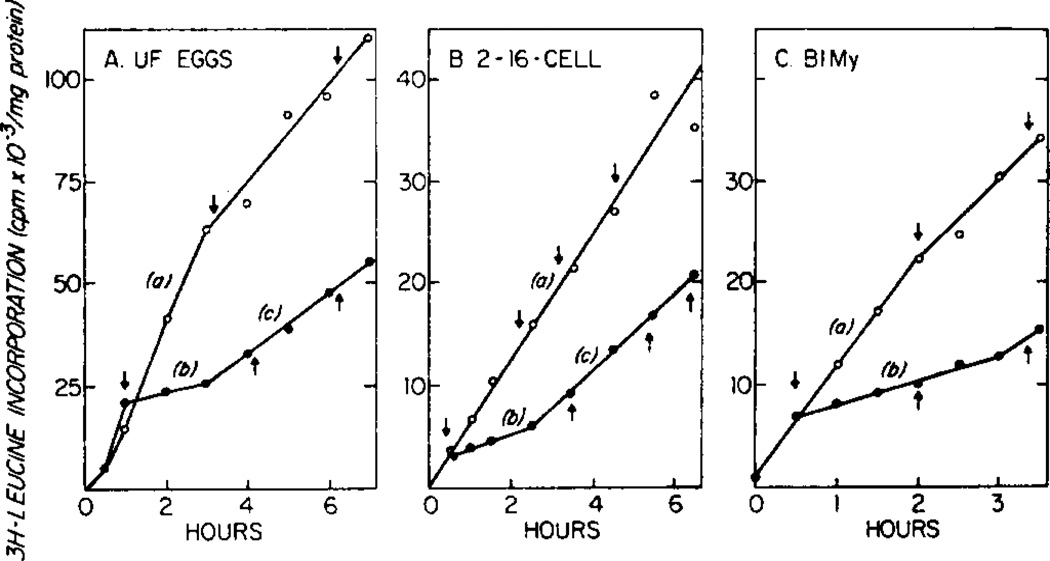
The concentrations of emetine used in these experiments inhibited protein synthesis 40 to 90%. The inhibition by emetine and the distribution of ribosomes in polyribosomes were measured with parallel cultures. In general, polyribosomes were analyzed after incubation of the eggs and embryos for longer than 1.5 complete transit times for an average sized mRNA taking into account the inhibited rate. The transit time is the length of time required, after the initiation step, for a ribosome to move along a mRNA and complete the synthesis of a peptide. Often, in our experiments inhibition by emetine decreased after 2 or 3 hr. Examples of inhibition of translation in eggs by 1.5 µM emetine and in embryos by 8µM emetine are shown in Fig. 1. In the egg culture, inhibition compared to the initial control rate, decreased from 90 to 70% after 2 hr. The inhibition in embryos, in the cases shown, was initially 75% and decreased after 2 to 2.5 hr to 40% inhibition of the initial control rate. The initial control rate was used for comparison since a decline in the rate of peptide synthesis occurred after a significant amount of leucine had been incorporated rather than after a defined period of time. The times at which control and emetine treated cultures were analyzed for polyribosome content are indicated by arrows. Polyribosome profiles of control cultures were chosen to bracket in time those with emetine to distinguish between changes in the amounts of polyribosomes due to developmental events and those due to initiation deficiencies.
FIG. 1.
Time course of inhibition of protein synthesis by emetine in unfertilized eggs (A), 2- to 16-cell embryos (B) and early mesenchyme blastula embryos (C). Unfertilized eggs and embryos were incubated and analyzed for [3H]leucine incorporation into peptides as described under Materials and Methods. At the beginning of the experiment the culture age of B was 0.7 hr and that of C 24 hr. Emetine was added at 1.0 and 0.5 hr after the addition of [3H]leucine, for eggs and embryos, respectively. Control cultures (○). Emetine-treated cultures (●). The percent inhibition was calculated by the comparison of slopes b and c with a as discussed under Results. The times at which parallel cultures were examined for polyribosome content are indicated by arrows.
The dose response of S. purpuratus embryos to emetine observed by us (data not shown) was nearly identical to that of Hogan and Gross (1971) for Arbacia punctulata embryos. However, S. purpuratus unfertilized eggs were more sensitive, requiring about four times less emetine for the same amount of inhibition as embryos.
Distribution of Polyribosomes in Unfertilized Eggs and Early Embryos
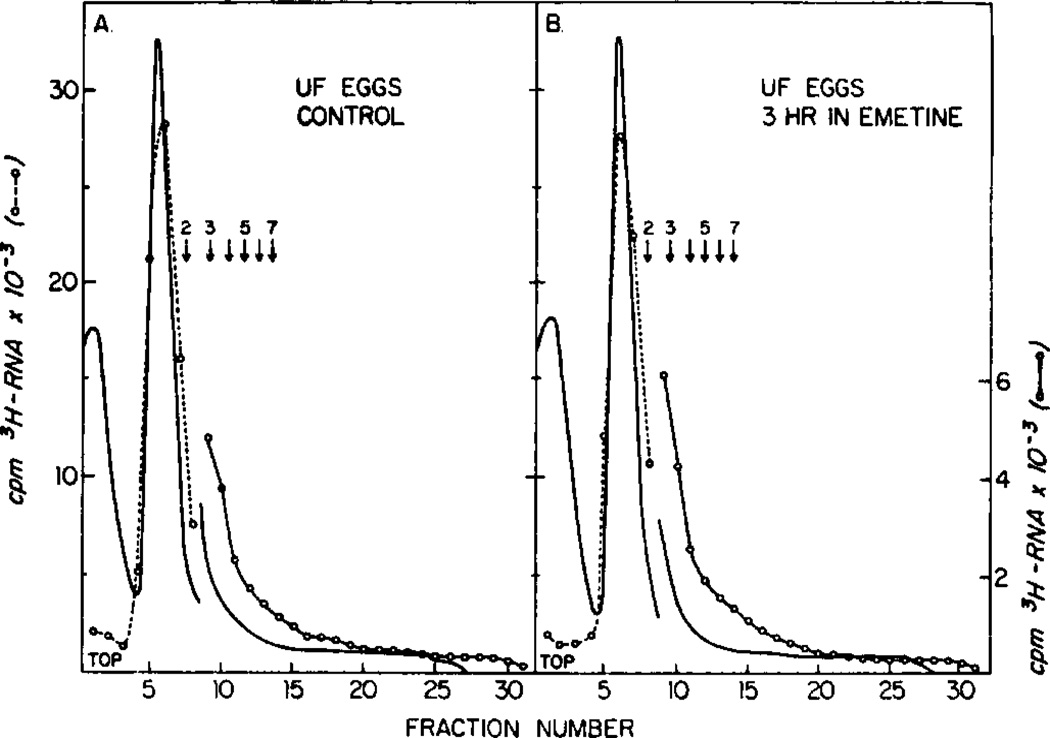
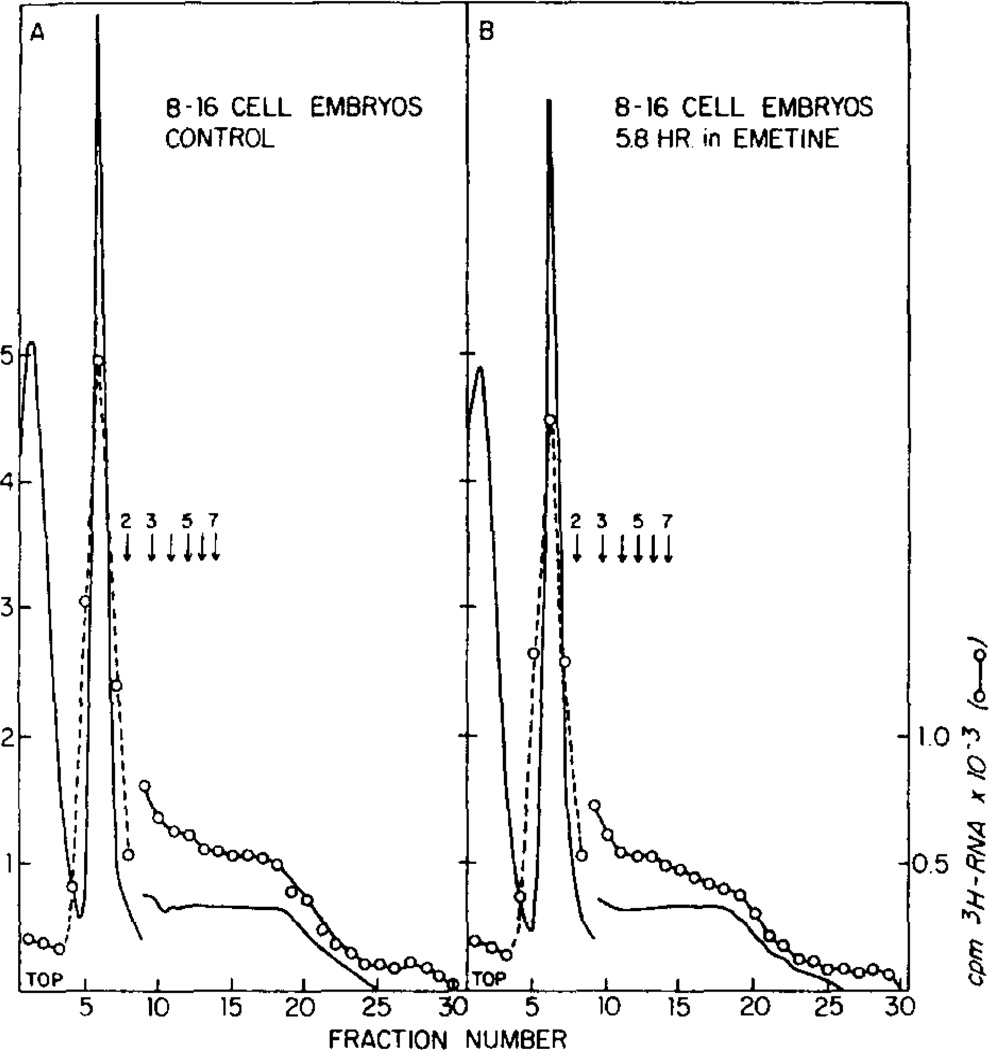
Figures 2, 3, and 4 show the distribution of polyribosomes in sucrose gradients in the cytoplasm of unfertilized eggs, 8- to 16-cell embryos and in blastula embryos in the absence and presence of emetine as described in Fig. 1. Both the amount of 3H-labeled RNA in each fraction and the absorbance at 260 nm were measured. Emetine does not appear to change the percentage of ribosomes in monoribosomes and polyribosomes, as measured by either parameter, for unfertilized eggs (Fig. 2) or 4- to 16-cell embryos (Fig. 3). However, emetine increased the amount of polyribosomes in blastula embryos (Fig. 4).
FIG. 2.
Polyribosome profiles of unfertilized egg cultures incubated in the absence (A) and presence (B) of emetine. These profiles are from eggs cultured for 3.1 hr (control) and 4.2 hr (emetine treated) in parallel with those described in Fig. 1A. No [3H]leucine was added to these cultures. Centrifugation is from left to right. The first and second peaks of absorbance at the top of the gradients are supenatant and monoribosomes peaks. The positions of polyribosomes with two to seven ribosomes are indicated by arrows. Absorbance at 260 nm (——) and [3H]RNA cpm (○ - - - ○, ○ —— ○) are indicated. There is a scale change after fraction 9. The total counts in gradients A and B are 104,970 and 102,190 cpm, respectively.
FIG. 3.
Polyribosome profiles of 8- to 16-cell embryos incubated in the absence (A) and presence (B) of emetine. Cultures were incubated in parallel with those described in Fig. 1B except that [3H]leucine was absent. The control culture was taken at the 4.5 hr point of Fig. 1B and was 32% 8-cell and 52% 16-cell. The emetine/treated culture taken at the 6.5 hr point was 83% 8-cell and 17% 16-cell. Earlier profiles were similar for both cultures. Notations are as in Fig. 2. The total counts in gradients A and B are 21,390 and 19,320 cpm, respectively.
FIG. 4.
Polyribosome profiles of late mesenchyme blastula embryos incubated in the absence (A) and presence (B) of emetine. Cultures were incubated in parallel with those described in Fig. 1C except that [3H]leucine was absent. The profile for the control culture is from the 0.5-hr point and that for the emetine-treated culture from the 3.3-hr point. The stage of development in the two cultures was similar. Total counts in the gradients are 23,830 cpm (A) and 25,391 cpm (B). Notations as in Fig. 2.
To determine if any minor shifts in polyribosome size of unfertilized eggs and cleavage embryos had occurred, the polyribosome profiles were divided into three size classes: (1) subunits, monoribosomes and polyribosomes with two ribosomes, (2) small polyribosomes containing three to seven ribosomes, and (3) large polyribosomes containing eight and more ribosomes. The dimers were included in the first size class since, under our centrifugation conditions, a large proportion of monoribosomes sedimented in the dimer region. Table 1 shows the data for the distribution of [3H]RNA in these three fractions for unfertilized eggs and early embryos (0–10 hr at 14°C). Again, no significant shifts of monoribosomes into polyribosomes or of small polyribosomes into large polyribosomes was observed for unfertilized eggs or 2- to 16-cell embryos in the presence of emetine. For the 2- to 16-cell stage the developmental changes in polyribosome distribution were small, as shown by others (Infante and Nemer, 1967; Humphreys, 1971; Goustin, 1979).
TABLE 1.
Distribution of [3H]RNA in Ribosomal Fractions of S. purpuratus eggs and early embryos
| Expt | Stage | Percentage inhibitiona with emetine |
Number of transits in emetineb |
Percentage [3H]RNAc |
||
|---|---|---|---|---|---|---|
| Polyribosomes |
||||||
| Monoribosomes | Small | Large | ||||
| 1. | UF eggs | Control (4) | 83.5 ± 0.9 | 10.1 ± 0.7 | 6.4 ± 0.7 | |
| UF eggs | 90−70 | 0.61 | 82.5 | 11.5 | 6.1 | |
| UF eggs | 90−70 | 1.63 | 82.9 | 10.8 | 6.3 | |
| 2. | Zygotesd | Control | 69.1 | 13.1 | 17.7 | |
| 2-celle | Control | 66.8 | 13.9 | 19.2 | ||
| 2-cellf | 75−40 | 2.3 | 65.8 | 17.5 | 16.6 | |
| 4-cellg | Control | 66.1 | 15.1 | 18.9 | ||
| 4-8-cellh | 75−40 | 5.0 | 66.3 | 15.1 | 18.6 | |
| 8-16-celli | Control | 65.2 | 15.1 | 19.7 | ||
| 16-cellj | 75−40 | 6.4 | 66.9 | 14.6 | 18.4 | |
| 3. | Early Blk | Control (3) | 58.9 ± 0.8 | 22.3 ± 1.0 | 18.8 ± 0.4 | |
| Early Blk | 72−49 | 1.3 | 48.0 | 26.8 | 25.2 | |
Controls had no emetine present. Emetine was added to a final concentration of 1.5 and 8.0 µM for treated cultures of unfertilized eggs (UF eggs) and embryos, respectively. The percentage inhibition was monitored during the experiments (see Figs. 1 and 2). The number of independent gradients is indicated in parentheses. Average experimental variation is given.
The number of transits was calculated from the percentage inhibition and the known transit times were adjusted for incubation at 14°C (Hille and Albers 1979; Brandis and Raff 1978).
80 S includes subunits, monoribosomes, and the dimer region. Small polyribosomes include those containing three to seven ribosomes; large polyribosomes those containing eight or more ribosomes.
The culture age at 14°C and stages at which polyribosome profiles were measured are d 1.5 hr, nuclei condensing; e 2.5 hr, 83% 2-cell, 15% 4-cell;f 4.2 hr, 65% 2-cell, 35% 4-cell; g 3.5 hr, 88% 4-cell, 12% 2-cell; h 6.1 hr, 39% 4-cell, 44% 8-cell, 17% 16-cell; i 4.7 hr, 32% 8-cell, 52% 16-cell; j 7.1 hr, 83% 16-cell, 17% 8-cell; k 10 to 12 hr, 1 hr postmorula or the earliest blastula stage detectable.
In general, when emetine inhibited protein synthesis, development was also slowed to a similar extent. This phenomenon was particularly apparent for the second division as detailed in Table 1. As the rate of cleavage is directly dependent on the rates of protein synthesis, we conclude that certain proteins must accumulate before cleavage occurs. These results support the findings of Hultin (1961b) and Black et al. (1967) that puromycin blocked cleavage in sea urchin embryos.
Table 2 shows that the data obtained by measuring absorption at 260 nm were in general less consistent than those measuring labeled RNA despite our precautions in aligning the base line. Nevertheless, the distribution of the material absorbing at 260 nm in polyribosomes always showed the same trend as that of the [3H]RNA distribution and hence confirms our conclusions.
TABLE 2.
Distribution of 260 nm absorbance in Ribosomal Fractions of S. purpuratus eggs and early embryosa
| Expt | Stage | Percentage inhibition with emetine |
Number of transits in emetine |
Percentage A260 nmb |
||
|---|---|---|---|---|---|---|
| Polyribosomes |
||||||
| Monoribosomes | Small | Large | ||||
| 1. | UF eggs | Control (2) | 86.2 ± 1.7 | 8.4 ± 0.9 | 5.4 ± 0.8 | |
| UFeggs | 90–70 (2) | 0.6/1.63 | 86.5 ± 0.8 | 7.5 ± 0.5 | 5.9 ± 0.3 | |
| 2. | Zygotes | Control | 72.0 | 12.4 | 15.6 | |
| 2-cell | Control | 68.2 | 13.6 | 18.2 | ||
| 2-cell | 75–40 | 2.3 | 69.9 | 16.8 | 13.3 | |
| 4-cell | Control | 69.5 | 13.9 | 16.6 | ||
| 4–8-cell | 75–40 | 5.0 | 70.7 | 13.5 | 15.9 | |
| 8–16-cell | Control | 69.3 | 13.9 | 16.8 | ||
| 16-cell | 75–40 | 6.4 | 69.8 | 13.5 | 16.7 | |
| 3. | Early Bl | Control (3) | 60.4 ± 3.5 | 25.5 ± 1.3 | 14.1 ± 2.3 | |
| Early Bl | 72–40 | 1.3 | 44.5 | 30.5 | 24.7 | |
Conditions and stages as in Table 1.
The distribution of material absorbing at 260 nm was determined graphically.
By early blastula stage (just 1 hr postmorula) two changes have occurred. First, about 8% more polyribosomes were present at this stage in the controls as compared to the cleavage stages (see Table 1, experiments 2 and 3, controls). This increase is presumably due to the addition of newly synthesized mRNA which accumulates after the cleavage stage (Gross and Cousineau, 1964; Wilt, 1964; Mizuno et al., 1973). Second, emetine in a concentration sufficient to inhibit protein synthesis by 72 to 49% (see Fig. 1C), shifted 10% of the monoribosomes at this stage into both small and large polyribosomes (Table 1, experiment 3). Since the number of small as well as the number of large polyribosomes increases, additional mRNA must be recruited into the polyribosomes by emetine. Thus, these results suggest that as the total protein synthesis increases in early blastulae, initiation becomes the rate-limiting step of protein synthesis.
Distribution of Polyribosomes in Late Stage Embryos
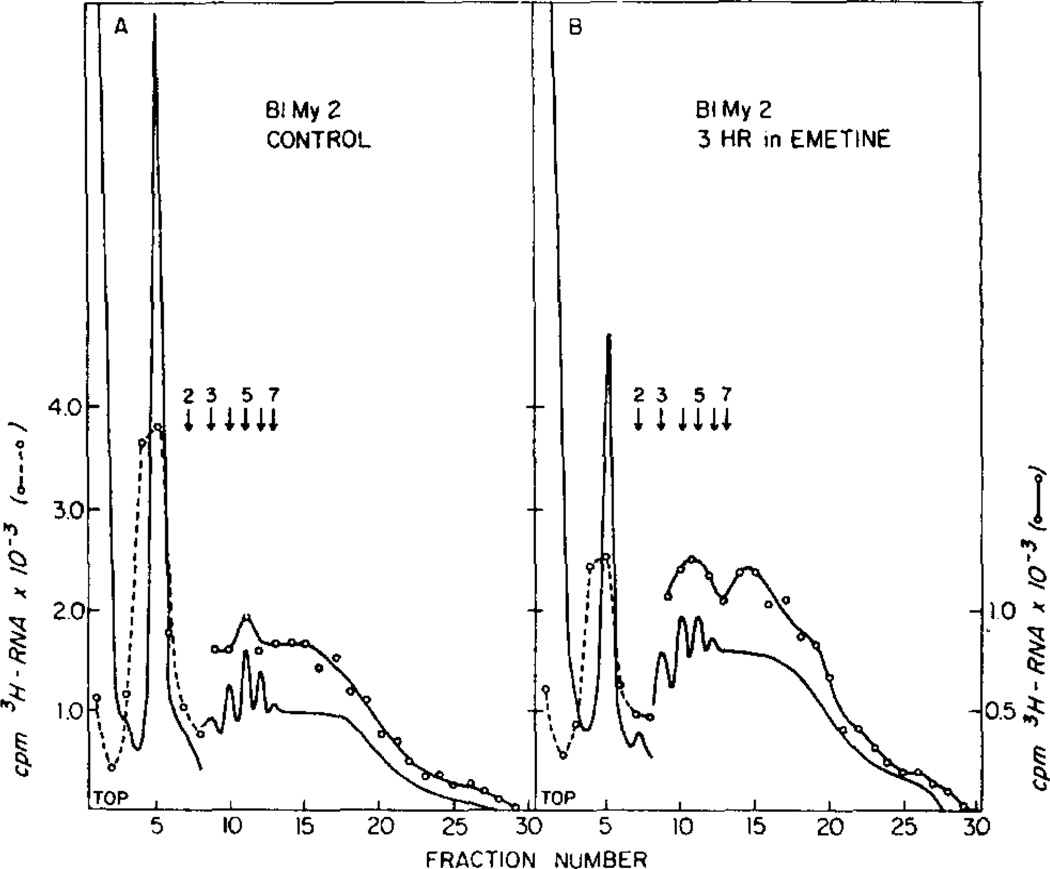
Table 3 shows data from polyribosome profiles of late stage embryos: those from the time of hatching to prism stages (16 to 52 hr at 14°C). In all of these embryos emetine increased the percentage of ribosomes in the polyribosome region. Nearly identical shifts were observed when the absorption at 260 nm was measured instead of 3H-labeled RNA (data not listed, but shown for late blastula and prism embryos in Fig. 5). The initiation step, therefore, appears to be the rate-limiting step for developmental stages from early blastula to the prism stage. In hatching blastula embryos and hatched early mesenchyme blastula embryos (BlMy1) the increase in large polyribosomes with emetine present was five times as great as that in small polyribosomes indicating that a major shift from small to large polyribosomes may have occurred as well as recruitment of mRNA into new polyribosomes.
TABLE 3.
Distribution of [3H]Rna in Ribosomal Fractions of S. purpuratus late embryosa
| Expt | Stageb | Percentage inhibition with emetine |
Number of transits in emetine |
Percentage [3H]RNA |
||
|---|---|---|---|---|---|---|
| Polyribosomes |
||||||
| Monoribosomes | Small | Large | ||||
| 1. | HatchingBlc | Control (2) | - | 42.5±0.3 | 26.4 ± 0.3 | 31.1 ± 0.6 |
| Hatching Blc | 60 | 1.5 | 34.4 | 27.9 | 37.6 | |
| 2 | BlMy-1-2d | Control | - | 49.3 | 20.7 | 30.0 |
| BlMy-1-2d | 72 | 1.05 | 31.4 | 22.8 | 45.8 | |
| 3. | BlMy-1e | Control (2) | - | 54.2 ± 0.1 | 18.9 ± 0.2 | 26.9 ± 0.2 |
| BlMy-2/Ga-1f | Control | - | 58.2 ± 0.6 | 18.5 ± 0.5 | 23.3 ± 0.1 | |
| BlMy-2f | 76 | 1.8 | 38.2 | 24.4 | 37.4 | |
| 4. | Prismg | Control (4) | - | 64.0 ± 1.7 | 20.6 ± 0.8 | 15.5 ± 1.5 |
| Prismg | 77 (2) | 0.86/1.72 | 37.3 ± 1.0 | 27.5 ± 2.2 | 35.2 ± 1.2 | |
Conditions as in Table 1.
For nomenclature of stages see Whiteley and Baltzer (1958). The culture times at 14°C at which polyribosome profiles were measured are:
16–18 hr,
23–24 hr,
24.5 hr,
27.5 hr,
48–52 hr.
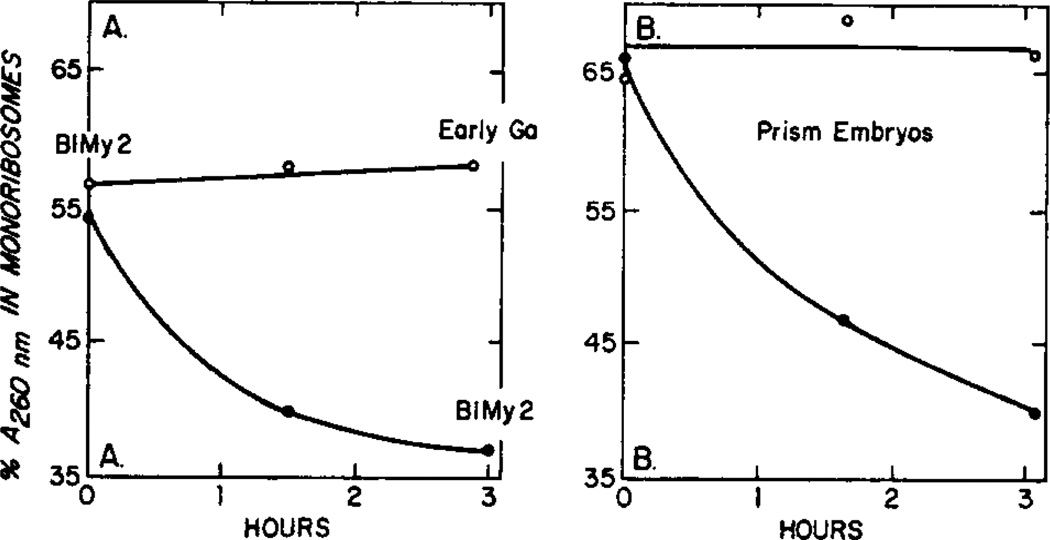
FIG. 5.
Time course of the change in monoribosome content during the incubation of late mesenchyme blastual (A) and prism (B) embryos. Control cultures (○). Emetine-treated cultures (●). Incubation conditions were as described in Table 3, experiments 3 and 4. The relative amount of material absorbing at 260 nm in the monoribosome region was graphically determined.
We attempted to determine the maximum shift of monoribosomes to polyribosomes in the presence of emetine. Figure 5 shows, however, that the shift had not reached equilibrium in 3 hr for late mesenchyme blastula or prism embryos. Two factors contribute to this difficulty. First, some mRNAs in sea urchin embryos are known to be very large and, hence, could not be loaded in 3 hr (Whiteley and Mizuno, 1981) and second, the effectiveness of emetine inhibition always decreased after 2 or 3 hr of incubation. Either emetine is inactivated or elongation factor 2 is synthesized by the embryos and competes with emetine in binding to its site on the 40 S ribosomal subunit (Vázquez, 1979; Sanchez et al., 1977; Gupta and Siminovitch, 1977).
Ribonuclease- and EDTA-Treated Polyribosomes
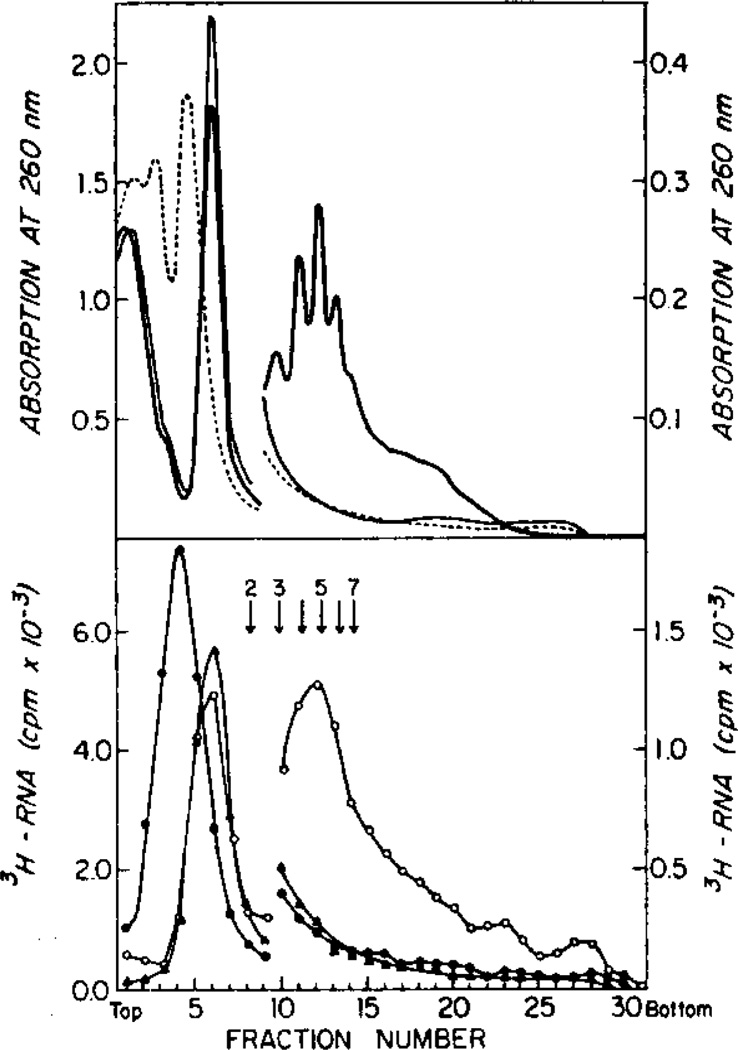
The fraction of [3H]RNA appearing in the polyribosome region of unfertilized eggs was greater than the fraction of ribosomes known to be present in polyribosomes. Infante and Nemer (1967) observed that S. purpuratus unfertilized eggs contained 5% or less of the ribosomes as polyribosomes since only that amount of the material absorbing in the region of polyribosomes shifted to the monoribosome region when digested with ribonuclease. Humphreys (1971), on the other hand, observed only 1% of the ribosomal RNA in the polyribosomes region of unfertilized L. pictus eggs. Therefore, we treated the postmitochondrial supernatant with RNase and EDTA to determine how much of the radioactivity represented polyribosomes. The results are shown in Table 4 and Fig. 6. For unfertilized eggs less than 2% of the [3H]RNA in the polyribosomes region was digested by excessive amounts of RNase A (Table 4). An additional 1% of ribosomes in the region of small polyribosomes were removed by EDTA treatment. This difference occurs because of a shift into ribosomal subunits of the monoribosomes which overlap the polyribosome region rather than an additional loss of polyribosomes. Hence, we conclude that no more than 2% of the [3H]RNA is polyribosomal in unfertilized eggs. The ribonuclease-insensitive, EDTA-insensitive material sedimenting with the polyribosomes in unfertilized eggs also occurs in extracts from embryos (Table 4, Fig. 6) and is observed by absorption at 260 nm (Fig. 6). Several other authors have described RNase-insensitive, EDTA-insensitive labeled material which cosediment with polyribosomes (Hogan and Gross, 1971; Nemer, 1975). In these cases, however, the 10% cosedimenting material was labeled with [3H]uridine during embryogenesis. Nemer showed that this material was not polyribosome like in bouyant density but heterogeneous with densities of 1.4 to 1.7 g/cm3.
TABLE 4.
Distribution of [3H]Rna in Ribosomal Fractions after ribonuclease and EDTA treatmenta
| Stage | Treatmentb | Percentage [3H]RNA |
||
|---|---|---|---|---|
| Polyribosomes |
||||
| Monoribosomes | Small | Large | ||
| UF eggs | None | 85.0 | 9.0 | 6.0 |
| +RNase | 86.9 | 7.7 | 5.4 | |
| +EDTA | 87.8 | 6.3 | 5.9 | |
| 2- to 4-cell | None | 63.3 | 15.5 | 21.2 |
| +RNase | 86.0 | 8.4 | 5.5 | |
| +EDTA | 87.2 | 7.6 | 5.2 | |
| Early Bl | None (3) | 58.9 ± 0.8 | 22.2 ± 1.0 | 18.8 ± 0.4 |
| +RNase (2) | 86.5 ± 0.1 | 8.0 ± 0.2 | 5.5 ± 0.1 | |
| +EDTA (2) | 90.7 ± 0.2 | 4.8 ± 0.1 | 4.5 ± 0.1 | |
Conditions as in Table 1.
Homogenates were treated with 29 µg/ml pancreatic ribonuclease A (Worthington) for 30–60 min at 0–4°C or made 35 mM EDTA with a 0.35 M stock just prior to loading on preformed gradients.
FIG. 6.
Polyribosome profiles of early blastula embryos and profiles of the same polyribosomes treated with 29 µg/ml pancreatic ribonuclease A (Worthington) or 35 mM EDTA. (A) Absorption at 260 nm. (B) Trichloroacetic acid precipitable [3H]RNA in the gradient fractions. Controls (——, ○ —— ○); ribonuclease-treated (——, ▲ —— ▲); EDTA treated (- - -, ● —— ●).
DISCUSSION
The experiments reported here establish that inhibition of polypeptide chain elongation by emetine in unfertilized eggs and cleavage stage embryos of sea urchins does not result in an increase in the number or size of polyribosomes. Thus, either (1) polypeptide chain initiation is not rate limiting or (2) the monoribosomes cannot be recruited onto mRNA. Bechtold and Hille have shown that in unfertilized eggs the amount of 40 S ribosomal preinitiation complex containing met-tRNAi is large compared to the rate of polypeptide synthesis that is occurring (Hille et al., 1980, and Bechtold and Hille, unpublished). Thus, the availability of active 40 S ribosomal subunits is not rate limiting and we can conclude that (1) above is correct, i.e., that the initiation step involving the binding of ribosomes to mRNA is not rate limiting in unfertilized eggs. Our conclusion is consistent with the suggesting of Kedes et al. (1969) and Humphreys (1971) that initiation is not limiting in unfertilized eggs because of a similarity in polyribosome sizes before and after fertilization. It appears, then, that the low rate of initiation in unfertilized eggs is due to a block in the availability of mRNA (Jenkins et al., 1978; Ilan and Ilan, 1978; Moon et al., 1980).
There is one observation in the literature which is contrary to our results and conclusions: MacKintosh and Bell (1969) observed an increase in the amounts of polyribosomes in unfertilized eggs and zygotes in the presence of cycloheximide. It is possible that a release of ribosomes from polyribosomes occurred in their controls during the isolation of the eggs and zygotes. When cells are slowly cooled, initiation of protein synthesis, but not elongation or termination, is impaired (Palmiter, 1973). When cycloheximide or emetine are present this release would be prevented and, hence, the cycloheximide-treated cells would have more polyribosomes than the controls. To prevent ribosomes from running off mRNA, our cultures were rapidly cooled by dilution into 20–40 culture volumes of −2°C wash Solution. Alternatively, but less likely, our disagreement could exist because Arbacia punctulata eggs are regulated differently than those of S. purpuratus.
For blastula (10 to 24 hr at 14°C) and prism embryos, we have shown that monomeric ribosomes can be mobilized into active polyribosomes by slowing the rate of mRNA translocation with low doses of emetine. Thus, we conclude that under normal conditions, the binding of ribosomes to the initiation codon of mRNA is rate limiting in blastula embryos. Hogan and Gross studied the incorporation of newly labeled RNA into polyribosomes of unhatched blastulae of the sea urchin, Arbacia punctulata, in the presence and absence of 0.02 to 1 mM emetine, concentrations which inhibit protein synthesis from 85 to nearly 100% that of normal embryos. They also observed a shift in ribosomes from monoribosomes to polyribosomes when elongation rates were inhibited less than 100%. Therefore, though Hogan and Gross interpreted their data in a different way, their data are consistent with ours and support our conclusion that polypeptide chain initiation is rate limiting in prehatched blastula embryos.
Several investigators have studied the selective translation of mRNA in sea urchin embryos. Nemer (1975) and Nemer et al. (1975) studied the size and distribution of poly(A)+ mRNA and poly(A)− nonhistone mRNA in polyribosomes from blastula and gastrula stages of the sea urchins, L. pictus and S. purpuratus. They found that these nonhistone types of mRNA had a mean S value of 19 to 22 S in 70% formamide gradients and, surprisingly, that the mean size and range of sizes of the mRNAs are essentially the same in large and small polyribosomes. They concluded that the mRNAs in the small polyribosomes are probably underloaded with ribosomes and that initiation may be the rate-limiting step of peptide synthesis at these stages. In addition they found that the poly(A)+ mRNA was on the average more fully loaded with ribosomes than poly(A)− mRNA and, therefore, reasoned that the polyadenylated segment at the 3’-terminus facilitated ribosomal loading or recycling of ribosomes. Rudensey and Infante (1979) have refined this point to suggest that some poly(A)+ mRNAs are more efficient initiators than others. Nemer and co-workers could not distinguish whether initiation was rate limiting or whether a large portion of the mRNAs were nonfunctional and not translated. Our studies now confirm that initiation is rate limiting in late stages of S. purpuratus.
What is the developmental consequence in sea urchins of the decrease in initiation rates? Nemer (1975) hypothesized that the enhancement of the poly(A)+ mRNA population between early and late blastula stages was related to preparation for the differentiation of the specialized functions of the mesenchyme cells and for gastrulation. In addition, he speculated that poly(A)+ mRNA supported specialized gene expression and that poly(A)− mRNAs were responsible for the basic cellular functions or “housekeeping.” That development arrests but general functions are maintained in the absence of the adenylation specific poly(A)+ mRNAs, has been elegantly shown by Spieth and Whiteley (1980) using cordycepin. We also suggest that the preferential translation of poly(A)+ mRNAs in late embryos when initiation is rate limiting could facilitate the differentiation of specialized structures and new functions of the developing gastrula and prism embryos. Basic cellular functions of the embryos would thus be maintained at a lower synthetic rate, without massive degradation of the mRNAs.
We conclude that the binding of 40 S ribosomal sub-units to the initiation codon is not rate limiting in unfertilized eggs or cleavage stage embryos. The availability of 40 S ribosomal preinitiation complexes, and most likely initiation factor activity, must increase during the early development of these embryos to accommodate the large increase in protein synthesis. Increase in 40 S preinitiation complexes free in the cytoplasm does occur after fertilization (Hille et al., 1980; Bechtold and Hille, unpublished). However, by the early blastula stage the amount of mRNA increases such that the components involved in peptide chain initiation are no longer sufficient. At this stage chain initiation becomes rate limiting. Regulation of translation at the level of polypeptide initiation at this and subsequent stages may be an important means of selecting the appropriate mRNAs to synthesize the proteins needed for differentiation.
Acknowledgments
We thank Dr. Charles D. Laird for the use of the Numonics Electronic Graphics Calculator and scintillation counter. This research was supported by NIH Grant H 11070, University of Washington Graduate School Research Project Fund, and Institutional Cancer Grant IN-26 from the American Cancer Society. MVD and RTM were supported by NIH National Research Service Awards GM 07270 and IT 32 HD 07183, resepctively.
Footnotes
Abbreviations used: EDTA, ethylenediaminetetraacetic acid; TCA, trichloroacetic acid.
REFERENCES
- 1.Alton TH, Lodish HF. Cell. 1977;12:301–310. doi: 10.1016/0092-8674(77)90208-2. [DOI] [PubMed] [Google Scholar]
- 2.Bensadoun A, Weinstein D. Anal. Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 3.Berg WE. Exp. Cell Res. 1965;40:469–489. doi: 10.1016/0014-4827(65)90227-2. [DOI] [PubMed] [Google Scholar]
- 4.Black RE, Babtist E, Piland J. Exp. Cell Res. 1967;48:431–439. doi: 10.1016/0014-4827(67)90366-7. [DOI] [PubMed] [Google Scholar]
- 5.Brandis JW, Raff RA. Develop. Biol. 1978;67:99–113. doi: 10.1016/0012-1606(78)90303-2. [DOI] [PubMed] [Google Scholar]
- 6.Denny PC, Tyler A. Biochem. Biophys. Res. Commun. 1964;14:245–249. doi: 10.1016/0006-291x(64)90443-7. [DOI] [PubMed] [Google Scholar]
- 7.Epel D. Proc. Nat. Acad. Sci USA. 1967;57:899–906. doi: 10.1073/pnas.57.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geoghegan T, Cereghini S, Brawerman G. Proc. Nat. Acad. Sci. USA. 1979;76:5587–5591. doi: 10.1073/pnas.76.11.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giudice G, Vitorelli ML, Monroy A. Acta Embryol. Morphol. Exp. 1962;5:113–122. [Google Scholar]
- 10.Goustin AS, Wilt FH. Develop. Biol. 1981;82:32–40. doi: 10.1016/0012-1606(81)90426-7. [DOI] [PubMed] [Google Scholar]
- 11.Gross PR, Cousineau GH. Exp. Cell Res. 1964;33:368–395. doi: 10.1016/0014-4827(64)90002-3. [DOI] [PubMed] [Google Scholar]
- 12.Gross PR, Malkin LI, Moyer WA. Proc. Nat. Acad. Sci. USA. 1964;51:407–414. doi: 10.1073/pnas.51.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta RS, Siminovitch L. Biochemistry. 1977;16:3209–3214. doi: 10.1021/bi00633a026. [DOI] [PubMed] [Google Scholar]
- 14.Hille MB, Albers AB. Nature (London) 1979;278:469–471. doi: 10.1038/278469a0. [DOI] [PubMed] [Google Scholar]
- 15.Hille MB, Bechtold MA, Hall DC, Yablonka-Reuveni Z. Amer. Zool. 1980;20:838. [Google Scholar]
- 16.Hogan B, Gross PR. J. Cell Biol. 1971;49:692–701. doi: 10.1083/jcb.49.3.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hultin T. Exp. Cell Res. 1961a;25:405–417. doi: 10.1016/0014-4827(61)90290-7. [DOI] [PubMed] [Google Scholar]
- 18.Hultin T. Experientia. 1961b;17:410–413. doi: 10.1007/BF02157974. [DOI] [PubMed] [Google Scholar]
- 19.Humphreys T. Develop. Biol. 1971;26:201–208. doi: 10.1016/0012-1606(71)90122-9. [DOI] [PubMed] [Google Scholar]
- 20.Ilan J, Ilan J. Develop. Biol. 1978;66:375–385. doi: 10.1016/0012-1606(78)90246-4. [DOI] [PubMed] [Google Scholar]
- 21.Infante AA, Nemer M. Proc. Nat. Acad. Sci. USA. 1967;58:681–688. doi: 10.1073/pnas.58.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins NA, Kaumeyer JF, Young EM, ad Raff RA. Develop. Biol. 1978;63:279–298. doi: 10.1016/0012-1606(78)90134-3. [DOI] [PubMed] [Google Scholar]
- 23.Kedes LH, Hogan B, Cognetti G, Selvig S, Yanover P, Gross PR. C.S.H.S. Quant. Biol. 1969;34:717–723. doi: 10.1101/sqb.1969.034.01.081. [DOI] [PubMed] [Google Scholar]
- 24.Lodish HF. J. Biol. Chem. 1971;246:7131–7138. [PubMed] [Google Scholar]
- 25.Lodish HF. Nature (London) 1974;251:385–388. doi: 10.1038/251385a0. [DOI] [PubMed] [Google Scholar]
- 26.MacKintosh FR, Bell E. J. Mol. Biol. 1969;41:365–380. doi: 10.1016/0022-2836(69)90282-4. [DOI] [PubMed] [Google Scholar]
- 27.Mizuno S, Whiteley HR, Whiteley AH. Differentiation. 1973;1:339–348. [Google Scholar]
- 28.Moon RT, Moe KD, Hille MB. Biochemistry. 1980;19:2723–2730. doi: 10.1021/bi00553a029. [DOI] [PubMed] [Google Scholar]
- 29.Nemer M. Cell. 1975;6:559–590. [Google Scholar]
- 30.Nemer M, Dubroff LM, Graham M. Cell. 1975;6:171–178. doi: 10.1016/0092-8674(75)90007-0. [DOI] [PubMed] [Google Scholar]
- 31.Palmiter RD. J. Biol. Chem. 1973;248:2095–2106. [PubMed] [Google Scholar]
- 32.Palmiter RD. J. Biol. Chem. 1974;249:6779–6787. [PubMed] [Google Scholar]
- 33.Piatogorsky J, Tyler A. Biol. Bull. 1967;133:229–244. doi: 10.2307/1539803. [DOI] [PubMed] [Google Scholar]
- 34.Regier JC, Kafatos FC. Develop. Biol. 1977;57:270–283. doi: 10.1016/0012-1606(77)90214-7. [DOI] [PubMed] [Google Scholar]
- 35.Rudensey LM, Infante AA. Biochemistry. 1979;18:3056–3063. doi: 10.1021/bi00581a023. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez L, Vazquez D, Jimenez A. Mol. gen. Genet. 1977;156:319–326. doi: 10.1007/BF00267188. [DOI] [PubMed] [Google Scholar]
- 37.Showman RM, Foerder CA. Exp. Cell Res. 1979;120:253–255. doi: 10.1016/0014-4827(79)90385-9. [DOI] [PubMed] [Google Scholar]
- 38.Spieth J, Whiteley AH. Develop. Biol. 1980;79:95–106. doi: 10.1016/0012-1606(80)90075-5. [DOI] [PubMed] [Google Scholar]
- 39.Vázquez D. Inhibitors of Protein Synthesis. Berlin: Springer-Verlag; 1979. [Google Scholar]
- 40.Walters RA, Yandell PM, Enger MD. Biochemistry. 1979;18:4254–4261. doi: 10.1021/bi00586a034. [DOI] [PubMed] [Google Scholar]
- 41.Whiteley AH, Baltzer F. Pubbl. Staz. Zool. Napoli. 1958;30:402–457. [Google Scholar]
- 42.Whiteley AH, Mizuno S. Roux’ Arch. 1981 doi: 10.1007/BF00848398. in press. [DOI] [PubMed] [Google Scholar]
- 43.Wilt FH. Develop. Biol. 1964;9:299–313. doi: 10.1016/0012-1606(64)90027-2. [DOI] [PubMed] [Google Scholar]