Abstract
At macroscopic scales, the human connectome is composed of anatomically distinct brain areas, the structural pathways connecting them, and their functional interactions. Successful annotation of phenotypic associations with variation in the connectome and cataloguing of neurophenotypes promise to transform our understanding of the human brain. This review provides a survey of magnetic resonance imaging-based measurements of functional and structural connectivity. We highlight emerging areas of development and inquiry, and emphasize the importance of integrating structural and functional perspectives on brain architecture.
Introduction
First introduced in 20051, the term “connectome” embodies the advances of over a century of neuroscientific innovation and reflects an agenda for a new era. Initially defined as a complete map of neural connections within the brain, the connectome is a multiscale construct that can be examined at varying resolutions. At the extremes are the microscale, which focuses on the study of individual neurons and their synaptic connections, and the macroscale, which examines areas of cortical tissues (commonly ~1cm3or larger). Intermediate is the mesoscale, which, in humans, attends to vertical columns of 80–120 neurons (commonly referred to as micro- or mini-columns)1, 2. Although intimately associated, each resolution provides unique perspectives on the connectome. At the macroscale, and particularly in studies of the human brain, conceptualizations of the connectome have grown to also include information about function3. In this review we will use the term connectome to refer to brain areas, their anatomical connections, and their functional interactions.
Currently, macroscale methodologies are best positioned for mapping and annotating human connectomes with cognitive and behavioral associations. The higher-order, albeit lower-resolution representations, captured at the macroscale most directly relate to regulatory, cognitive, and affective processes. Interpretation of macroscale findings is most amenable to guidance from lesion and brain imaging studies. Further, comprehensive mapping and annotation of the connectome is most achievable at the lower-resolution macroscale, due to reduced computational and analytical demands. Moreover, non-invasive tools for in vivo imaging the human connectome are only available at the macroscale; in vivo microscale studies are currently limited to model organisms and neurosurgical patients.
Among the modalities used for macroconnectomics, MRI is dominant, partly because of widespread availability, safety, and spatial resolution. Diffusion-weighted MRI (dMRI) and functional MRI (fMRI) are especially popular for inferring structural and functional connectivity, respectively4. Diffusion-weighted MRI provides cubic-millimeter-resolution portrayals of white matter tracts, and insights into organizing principles guiding their orientation and trajectories; fMRI reveals a universal functional architecture, with variations among individuals meaningfully related to phenotypic variables5(e.g., behavioral, psychiatric).
The present review focuses on the mapping, characterization and analysis of macroscale connectomes. We structure our presentation in terms of a mathematical perspective that treats the connectome as a graph of interactions among brain areas. Nodes in the graph are abstract representations of brain areas and edges represent pairwise relationships between nodes. We first review approaches and challenges to parcellating the brain into discrete subunits represented by nodes, and then review the imaging and analytic methodologies used to map and quantify patterns of structural and functional connectivity that are represented by edges in the connectome.
Defining Nodes
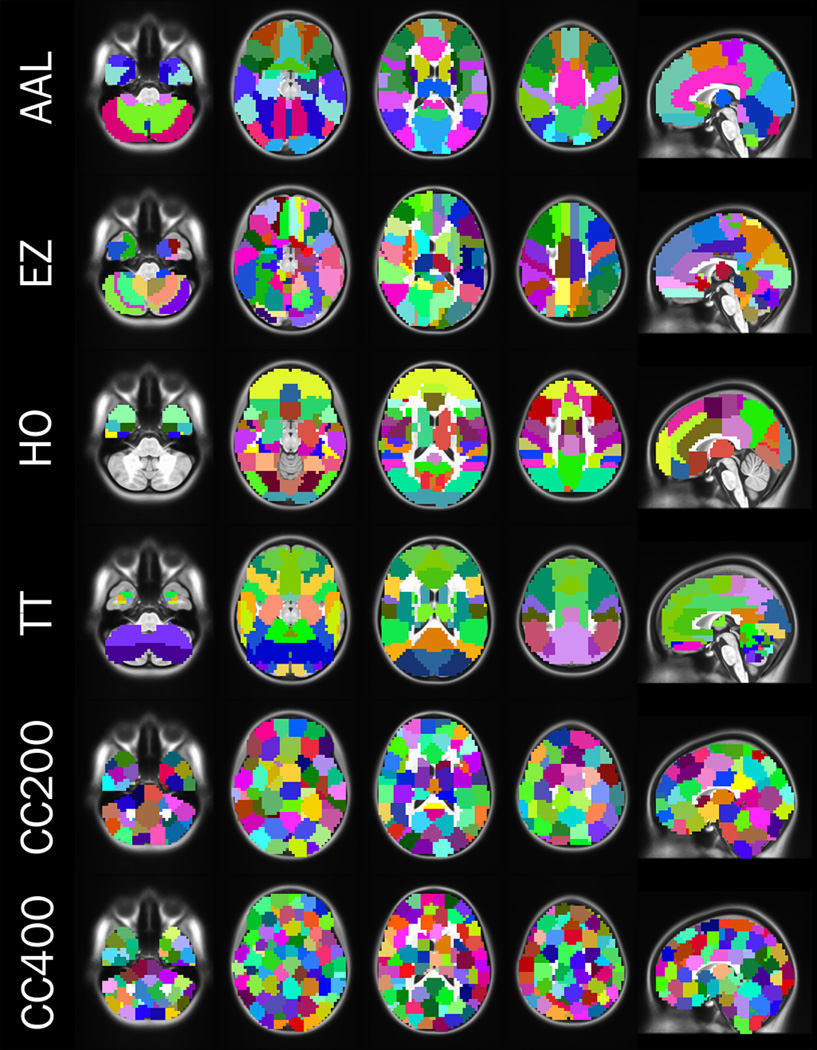
Defining the nodes of a macroscale connectome is a complex task as we lack agreement on how best to define the constituent brain units. Depending on the scope of the investigation, the specific brain subunits represented by nodes can range from the small patches of cortex contained in individual MRI voxels to larger brain areas (e.g., dorsolateral prefrontal cortex). Early parcellation efforts utilized post-mortem architectonic measurements (e.g. cell morphology) of single individuals. Although these resulting atlases are central to neuroscience, no functional or structural connectivity-based information was used in their construction, thus limiting their ability to accurately represent connectomes. For example, despite being represented as a single area in anatomical atlases6, the anterior cingulate region contains a number of subregions, each of which are characterized by dramatically different functional7and structural8connectivity patterns. As demonstrated in figure 1, while the large-scale pattern captured by different parcellation strategies may bear a gross similarity to one another, the specific details conveyed vary substantially (Fig. 1).
Figure 1. Different Parcellations.
We depict several different atlases of brain areas generated using anatomical (rows 1 – 4) and functional (rows 5 – 6) parcellation schemes. AAL (automated anatomical labeling)109and Harvard Oxford (HO)110are derived from anatomical landmarks (sulci and gyral). The EZ (Eickhoff - Zilles)111and TT (Talariach Daemon)112atlases are derived from post-mortem cyto- and myelo-architectonic segmentations. The CC200 and CC400 atlases are derived from 200 and 400 unit functional parcellations11.
Ideally, information from brain function and structural connectivity should be exploited to delineate brain areas. Meta-analytic approaches can define nodes based on task-based fMRI (T-fMRI) studies9. Alternatively, data-driven clustering techniques can subdivide the brain into areas based on homogeneity of functional time series10, 11, or functional or structural connectivity profiles8, 11, 12. Blind source separation techniques can also define network nodes using spatial independence13. These methods commonly pool information across individuals, and most can enforce specific properties on resulting brain areas11. One drawback is the need for pre-specification of the number of areas to be generated, which can be estimated based upon homogeneity, accuracy, reproducibility, or stability of the brain areas10, 11. Optimal comparison of connectomes is likely to require parcellation strategies that incorporate information across individuals and modalities, potentially at the cost of quality of fit for single subjects and modalities.
Estimating Structural Connectivity
Structural connectivity encompasses the collection of axonal and dendritic connections among neurons1. Despite definitional simplicity, structural connectivity is difficult to measure with non-invasive, in vivo imaging approaches. Prior to dMRI, our knowledge was primarily derived from lesions and blunt dissection in humans, or invasive tracing in non-humans. These methodologies remain the gold standard for establishing connectivity, but the non-invasive nature of dMRI makes it the de facto standard for human structural connectivity.
Acquisition
The basic principle underlying the inference of structural connectivity from dMRI is that water diffusion in white matter is hindered, occurring primarily along the path of axons. In contrast, water diffusion in grey matter and cerebral spinal fluid occurs (almost) equally in all directions. By following the motion of water, it is possible to map the orientation(s) of fibers passing through each white matter voxel. In dMRI, a series of images are acquired, each sensitive to diffusion along a specific direction. The number of unique directions acquired varies from six into the hundreds. When combined, these images contain the information necessary to estimate the orientation(s) of fibers passing through each voxel; this information is used to reconstruct large-scale white matter tracts (tractography). See Box 1 for further information about dMRI acquisition.
Box 1. Optimizing fMRI and dMRI image acquisition.
Echo planar imaging (EPI) is the most common method for acquiring functional (fMRI) and diffusion magnetic resonance imaging (dMRI) data. An EPI volume is a set of slices, each acquired after a single excitation. EPI volumes are collected in sequence, representing time-points in fMRI and diffusion directions in dMRI. Gradient echo EPI in fMRI only involves excitation and readout. However, in dMRI an additional 180° rephasing (spin echo) radio frequency pulse, and bipolar diffusion encoding gradients are applied.
| (1) |
Signal-to-noise ratio (SNR), spatial distortions and artifacts, and image contrast determine image quality. Image SNR can be represented in terms of parameters such as available signal (I0), voxel dimensions (Δx, Δy, Δz), number of image acquisitions (Nacq), number of samples in each dimension (Nx, Ny, Nz), time between samples (Δt) and parallel imaging factor (Pf) (Eq 1)117. These parameters are described in Table 3. Equation 1 demonstrates how different parameter settings affect SNR; for example, acquiring and averaging 2 volumes (N↓acq = 2) improves SNR by SNR for modern imaging technology is greater than required for most imaging applications, and can be a traded-off to optimize other imaging aspects (i.e., to reduce spatial distortions). Importantly, beyond spatial image considerations, parameter optimizations must be evaluated in terms of impact on the temporal (fMRI) or diffusion (dMRI) signal.
General Considerations for EPI
Spatial distortion
The EPI readout scheme leads to extended readout times and asymmetric bandwidth between areas, in turn increasing T2 decay and in-plane smoothing. Bandwidth corresponds to the number of frequencies allocated to a voxel; any shift in frequency due to magnet field imperfections spatially shifts the signal in proportion to the inverse of the bandwidth118. The bandwidth in the phase encoding direction is a fraction of the bandwidth in the readout direction, making it particularly susceptible to spatial distortions. Increasing bandwidth can minimize spatial distortions, though costing SNR. Parallel imaging techniques increase effective bandwidth and image acquisition rate119. These techniques require calibration scans for proper reconstruction, and any misalignment (e.g., head motion) between these scans and image data produce reconstruction errors119. Alternatively, spatial distortions can be mathematically corrected using maps of the spatial variation in the magnetic field15.
Image resolution
Image resolution can be increased by reducing the field of view (FOV) or increasing the number of samples along a dimension. Decreasing the FOV produces a proportional decrease in SNR; the FOV must remain larger than the head to avoid excessive ghosting. Increasing the number of samples has less impact on SNR (reduced by square root of the increase117). However, increasing the number of samples in the phase encoding direction substantially increases read out time, which increases the amount of T2 decay experienced during slice acquisition, and leads to increased in-plane smoothness118; parallel imaging mitigates these effects119.
Considerations for fMRI
BOLD contrast
T2* contrast is the most sensitive to the BOLD effect and is optimized using an echo time equal to the T2* of grey matter (TE~30ms at 3T). BOLD can also be measured with spin echo sequences, by using a TE near the T2 of grey matter, though BOLD contrast is low120.
Minimizing dropout
Tilting, and positioning imaging slices to prevent slices from intersecting air-tissue interfaces can minimize signal dropout due to susceptibility121. Slice positioning should focus on areas most vital to the experiment. Reducing TE, which decreases BOLD contrast, and increasing resolution, which decreases SNR, should be avoided. Specialty sequences (e.g., spiral-in/out122and z-shim techniques123) acquire multiple echoes with different dephasing characteristics to reduce susceptibility effects. Alternatively, spin-echo EPI avoids susceptibility artifacts but is much less sensitive to BOLD120.
Motion sensitivity
Beyond image misalignment, head motion induces fMRI signal fluctuations due to partial voluming and spin-history effects39. Lower flip angles124, longer TR125, and interleaved slices minimize spin-history effects by allowing the signal in a slice to fully relax before acquiring its neighboring slice. Spiral sequences are less sensitive to motion during slice acquisition126. 3D imaging techniques minimize the impact of within-volume motion, but between-volume motions remain problematic unless a long TR is utilized; long readouts increase the interaction between magnetic field inhomogeneity and motion127. Parallel imaging can improve acquisition time, resolution, and bandwidth but requires calibration scans119. Misalignments between the acquired data and calibration scans induce reconstruction errors and motion correlated imaging artifacts119.
Temporal resolution
Number of slices acquired determines temporal resolution. TE fundamentally limits slice acquisition time. Increasing bandwidth, partial Fourier imaging, and parallel imaging can offer marginal speed gains118.
Considerations for dMRI
Diffusion Contrast
Minimizing echo time, and therefore T2 decay, is essential to dMRI. T2 reduces overall MR signal and image SNR, and increases in-plane image blurring. Increasing bandwidth, partial Fourier imaging, parallel imaging, and using stronger diffusion gradients (b-values; high b-values require longer diffusion gradient pulses) can minimize TE118. Higher strength gradient systems enable high b-values (desirable for fiber tracking) in shorter time, though requiring hardware upgrade for most systems. Echo shifting also reduces TE, by relaxing the spin echo condition and shifting the echo train earlier in time118.
Scan Time
TE and number of slices determine dMRI volume TR. The number of diffusion directions, b-values, and averages acquired determine total scan time.
Emerging Acquisition sequences
Multiband and multi-echo imaging are emerging acquisition strategies. In multiband, multiple slices are acquired simultaneously, substantially improving temporal resolution, or increases in spatial resolution per unit time128, though at the cost of SNR and spatial smoothness. This improvement enables higher fMRI sampling rates, and more diffusion directions per b-value acquisitions in dMRI129– substantially improving estimation of macroscale connectomes130. In multi-echo fMRI, two echoes of a slice are acquired at each acquisition (one with low TE, one with a TE optimized for BOLD131). Systematic noise (heartbeat, respiration, head motion, scanner instability) are measured in the first echo, and removed from the second, enabling signal denoising with practically no SNR cost.
Preprocessing
Following acquisition, dMRI data must be preconditioned before directional information can be extracted, and tractography performed. Little debate exists regarding dMRI data preprocessing (see14for an exception), though this may reflect the difficulties of preprocessing and its complex impact rather than field consensus.
Correcting image distortions
In vivo dMRI data are plagued with spatial distortions, which are a central focus of preprocessing. In particular, magnetic field inhomogeneities are a major contributor. Areas where materials that differ with respect to magnetic susceptibility (i.e., degree of magnetization achieved in the MRI) interface with one another (e.g. air-tissue interfaces) are particularly prone to such inhomogeneities. These local variations in the magnetic field generate spatial distortions, which can be reduced through parallel imaging techniques or mathematically corrected using estimates of the field variations15. Another cause of spatial distortion is the interaction between the static magnetic field and currents induced by the rapid switching of gradients with the magnetic field, known as eddy-currents. These artifacts can be reduced using bipolar gradients16. Image coregistration, i.e. spatial alignment of brain scans, is commonly employed in preprocessing to correct for eddy current distortions and subject head motion17. However, this method fails for images acquired using very strong diffusion gradients. Model-based approaches that explicitly account for the effects of eddy-currents during image acquisition are emerging as the preferred option18.
Overlooked Issues
The above preprocessing steps, combined with visual inspection, constitute standard preprocessing. A few important details are often overlooked (see14for more examples). First, modern scanners are often equipped with antennas that acquire data in parallel through multiple-channels that are subsequently combined. Such parallel imaging techniques can elevate noise levels if the data are submitted to standard reconstruction19. This can artifactually lower diffusivity estimates along the axons, which increases the tendency of tractography approaches to overfit and generate false positive connections. An alternative reconstruction method has recently been proposed to address this important issue19. Second, any distortion correction must account for voxel-wise compression or expansion during image coregistration. This is particularly important for dMRI, where distortions vary in direction and magnitude between different gradient orientations, but is largely ignored by current software packages. Finally, image coregistrations contain a rotation component that is applied to each volume, and must therefore be applied to the concurrent gradient orientation20.
Estimating fiber orientation
Prior to delineating tracts and bundles via tractography, fiber orientation(s) must be inferred for white matter voxels individually. The dMRI signal’s sensitivity to water diffusion properties (i.e., rate, direction) enables the estimation of fiber orientation at each voxel. Importantly, each voxel contains thousands of axonal fibers, not just one. Thus, the goal of dMRI analysis is to infer a probability function for each voxel, which captures the different fiber orientations present, and their relative proportions21. Referred to as the fiber orientation density function (fODF), estimation of this function at each voxel is the first step in estimating structural connectivity.
The diffusion tensor is a simplistic but viable model for the diffusion profile that provides a simple approximation to the fODF22. Diffusion tensor imaging (DTI) uses a 3×3 matrix to provide an abstract ellipsoid representation of the water diffusion profile for a given voxel. Mathematical decomposition of this matrix yields information regarding the directions (x, y, z) of maximum and minimum water motion (eigenvectors), as well as the amount of diffusion that occurs along each direction (eigenvalues). The direction of maximal diffusion, referred to as the principal diffusion direction, is taken as the best estimate of fiber orientation within a voxel. Formally speaking, in the case of DTI, the fODF is approximated using a delta function or peak aligned with the principal diffusion direction.
The diffusion tensor provides a good estimate of fiber orientations when axons are homogeneously aligned within a voxel. However, this is not always the case. Fibers are known to disperse (i.e., fan), cross, merge, and kiss (i.e., temporarily run adjacent to one another) – all of which can happen within a single voxel and lead to heterogeneity not accounted for by a simple delta function. More complex fODF approximations can account for such heterogeneity within a voxel, though require higher angular coverage23, 24(i.e., more directions) and models that either explicitly or implicitly account for interactions between fiber orientation and the diffusion signal (See21for example methods). Complex fODF models better estimate fiber trajectories, particularly when several white matter tracts intersect, and allow recovery of non-dominant pathways invisible to DTI25.
Estimating Edges
Following estimation of voxel-wise fiber orientations, tractography approaches are used to establish structural connectivity between connectome nodes. Three-dimension trajectories, referred to as “streamlines”, are used to trace putative white matter paths. Local fiber orientation information guides the construction of streamlines along the fODF, allowing us to trace major white matter bundles26. Results are typically visualized as 3D renderings of thin curves grouped into bundles (Fig. 2), reminiscent of post-mortem dissection photographs. Importantly, the individual streamlines do not represent actual axons; they depict estimates of the average trajectories of axon bundles, given our assumption that diffusion is least hindered along axons.
Figure 2. Diffusion Imaging of structural connectivity maps.
(A) dMRI-based map of principal tensor orientations of the human brain viewed from below; blue: Superior-inferior, green: anterior-posterior and red: medial-lateral. (B) DTI fiber orientation estimates (red lines) from a region of corpus callosum superimposed on a fractional anisotropy image. Sample streamline depicted in yellow. (C) Pyramidal tract streamlines based on deterministic (left) and probabilistic (right) approaches are depicted (results superimposed on fractional anisotropy image). Image courtesy: Stam Sotiropoulos, http://etheses.nottingham.ac.uk/1164/.
The specific process by which streamlines are developed varies depending on the complexity of the fODF approximations available. With diffusion tensor modeling, the principal diffusion direction at each voxel guides the formation of the streamline; specifically, it provides a candidate for the tangent to the streamline at each voxel. For more complex fODF models, streamlining follows the same principle, though with multiple peak orientations available at each voxel, rather than a single principal diffusion direction. This allows streamlines with differing orientations to pass through the same voxel – which is crucial when heterogeneous fibers are present. While traditional tractography approaches are deterministic, probabilistic approaches account for uncertainty in estimates of local fiber orientations, allowing for estimation of probabilities for any given streamline.
Using streamlining methodologies, it is theoretically possible to measure all connections between grey matter areas. We can estimate both the trajectories and the end points of anatomical pathways. In practice, however, inference of point-to-point connectivity using streamlining is imprecise and error-prone27; improvements in both data quality and modeling are needed to yield more accurate structural connectomes.
Ideally, we should not only be able to infer the existence or absence of connections between nodes, but estimate connection (edge) strengths as well. Anatomical connections are made up of axons; features of these axons, such as density, size, length, and myelination, have important consequences on the propagation of action potentials, and hence information transfer. Measures of microstructural features based on more complex diffusion MRI experiments are emerging28, and may become an important constituent of connectomics. Related measures, such as fractional anisotropy and diffusivity, serve as common proxies for these microstructural complexities, but are extremely sensitive to confounding factors such as partial volume (e.g. voxels containing a mixture of white matter and grey matter) and axonal dispersion27, 29. Accordingly, these measures should be used cautiously to quantify connection strength. Other anatomical factors such as dendrite and spine densities, number of synapses at axon terminals, and synaptic efficacy are much harder to determine noninvasively, though potentially more relevant.
Unfortunately, tractography does not quantify any of the above properties. Often, probabilistic tractography, which estimates the uncertainty of the streamline trajectories, is used to quantify connection strength. Strong connections are expected to have a more significant trace in the diffusion data, and therefore lower uncertainty in their trajectories. This approximation, however, can easily break. For instance, locally non-dominant pathways (e.g., those that cross larger bundles), have higher uncertainty. Uncertainty is also affected by non-relevant factors such as signal-to-noise and partial volume effects. Another issue specific to streamlining is that uncertainty in the streamline’s path increases with the length of tract. Because streamlining operates by propagating uncertainty spatially, connection probabilities inevitably decrease with distance. As a result, tractography-based structural connections are difficult to quantify, threshold, compare between groups, and use for other types of statistical analyses27, 29.
Interpretation and considerations
Tractography pitfalls can be divided into two categories: accuracy (correctness) and precision (reproducibility)14. Accuracy refers to our ability to infer axonal organization from measurement of water diffusion. In the ideal case, there is no instrumental or physiological noise, yet we can still make erroneous inferences regarding microstructure due to inaccurate modeling. A white matter voxel contains hundreds of thousands of axons, which do not necessarily align30. Fiber ODF models, which account for multiple directions within a voxel (crossing fibers) are replacing tensor models for tractography. However, the sub-voxel organization of axons can be more complex than a simple crossing, and may not always be easily recovered from the diffusion profile. For instance, a collection of axons that bend within a voxel will create a diffusion pattern that may not be easily distinguishable from that of fiber dispersion. One can easily imagine even more complex situations where all these configurations (e.g., bending, dispersion, crossing) happen within the same voxel. Diffusion data from a single voxel cannot unambiguously resolve these complexities. Future approaches may benefit from semi-global models that aggregate diffusion data across multiple adjacent voxels to infer sub-voxel features.
Regarding precision, measurement noise (e.g., instrumental or physiological) and inadequate water diffusion modeling can compromise tensor and fODF estimation, inducing spurious variations in the generated streamlines. Additionally, use of a fixed step size in the generation of streamlines (despite local variations in anatomy), and the discrete nature of voxels (when tracts are continuous) increase measurement error. Probabilistic tractography algorithms try to quantify these errors by estimating the uncertainty in the entire process. Uncertainty in voxel wise fiber orientation can be quantified31and propagated into uncertainties regarding the location of streamlines. This process turns 3D point estimates of streamline trajectories into spatial histograms of their locations (Fig. 2c).
Beyond concerns regarding accuracy and precision, identification of fibers in their entirety requires knowledge of where tracts terminate throughout cortex. This remains a challenge for tractography algorithms, which are very good at estimating the location of bundles in deep white matter, but not so good (yet) at identifying where they project into grey matter27. These difficulties result from a lack of detail in white matter architecture modeled through diffusion and biases in cortical projections.
Estimating Functional Connectivity
While the concept of structural connectivity is relatively intuitive given the presence of physical connections between brain areas, its functional counterpart can be more challenging to define. The macroconnectomics field has adopted a neurophysiological perspective of functional connectivity, defining it as the synchronization of neurophysiological events between spatially remote brain areas32. First quantified in early electroencephalography (EEG) and multiunit recordings studies, functional connectivity analyses were adopted by positron emission tomography (PET) and fMRI in 199332. Although functional connectivity can be measured using a variety of neuroimaging modalities (e.g., PET, fMRI, magnetoencephalography) and different indices related to physiological function (e.g., blood oxygenation level dependent [BOLD], cerebral blood flow, glucose metabolism), BOLD-based fMRI is the most popular technique for inferring functional connectivity.
Functional connectivity studies may be dichotomized based upon the presence or absence of a task (i.e., task-based fMRI [T-fMRI] vs. task-free or “resting state” [R-fMRI]). Task-based approaches focus on the detection of synchronous responses to extrinsic stimulation or tasks, referred to herein as evoked functional connectivity (eFC) or coactivation33. Evoked functional connectivity can be quantified across the entire period of task performance, or in response to specific event types. Approaches using R-fMRI focus on the detection of synchronized spontaneous activity occurring in the absence of experimenter controlled tasks or stimuli, referred to as intrinsic functional connectivity (iFC)34. Although iFC and eFC patterns can be strikingly similar, especially when eFC is assessed using meta-analytic techniques35or broad comparisons (e.g., task vs. rest), these analyses probe different aspects of the functional architecture33. Importantly, eFC patterns obtained using one task will not necessarily generalize to another, and aspects of iFC obtained during one state (e.g., wakefulness) may not necessarily generalize to another (e.g., sleep) (see Box 2 for a discussion of states other than wakeful rest).
Box 2 – iFC and Conscious States.
An emerging literature has demonstrated the impact of cognitive97, physiological132and pathological states133on functional connectivity. Consciousness has received particular attention, with numerous studies examining physiological states (e.g., sleep), induced states (e.g., anesthesia, hypnosis) and pathological states (e.g., coma, vegetative syndrome, and minimally conscious state133). iFC patterns detected in these states are grossly similar to those observed during wakefulness, though direct comparison reveals state-related iFC changes. For example, the default and lateral networks each exhibit decreased functional connectivity among network components (i.e., within network connectivity) during sleep, suggesting decreased integration of information. Complementary findings of decreased negative iFC between these networks and others may suggest decreased segregation as well,134. These results suggest that while sleep-based studies may be useful in populations not amenable to examination in wakeful states (e.g., toddlers), comparison of findings across states may be problematic. Of note, changes in thalamocortical connectivity are reported during anesthesia, non-rapid eye movement (non-REM) sleep and vegetative states135, but the specific role of thalamocortical circuitry in consciousness remains underexplored. These studies also suggest potential clinical applications of iFC, such as improving recognition of consciousness following recovery from coma.
Acquisition
BOLD is the predominant fMRI technique in functional connectivity (see36for alternative cerebral blood flow-based technique). BOLD is measured using ultra-fast imaging sequences that are sensitive to relative concentrations of deoxyhemoglobin, which is paramagnetic (i.e., dephases the MR signal) in contrast to oxyhemoglobin, which is diamagnetic37; the resulting measurement is an indirect measure of neural activation. Datasets for functional connectivity analysis using fMRI are typically obtained in 5–30 minutes, as a participant either performs an experimental task (T-fMRI), or rests quietly in the scanner (typically while awake; R-fMRI). See Box 1 for a discussion of determinants of BOLD imaging acquisition quality.
Preprocessing
Preprocessing aims to remove confounding variation from data and facilitate comparison across subjects. Structured nuisance signals and anatomical variation can obscure functional connectivity measurements if left unaccounted for. Despite considerable effort, we lack consensus regarding the optimal set of preprocessing steps, their ordering and their implementation. Most preprocessing steps originated with task activation approaches. However, their usage and implications are greater for functional connectivity approaches due to the increased risk of spurious findings given that the independent and dependent variables can be contaminated by the same noise signals (e.g., motion, respiration). Comprehensive comparison of preprocessing strategies and their implications for eFC and iFC analyses remain elusive, in part due to lack of objective benchmarks. The preprocessing steps described below are post-hoc corrections. However, optimization of acquisition strategies to minimize the impact of noise sources is preferred (see Box 1).
Slice Timing Correction
The slices of an fMRI volume are acquired at different times, creating effective shifts in time-series obtained at different slices. Although some question its necessity38, correction by temporal-interpolation is recommended to avoid the potential for deleterious impact of these lags on signal denoising and time-series extraction from brain areas.
Motion Correction
Head motion results in a misalignment of brain areas between volumes typically accounted for using 3D image registration techniques. Additionally, head motion induces artifactual fMRI signal fluctuations due to changes in slice tissue composition (partial voluming), and residual magnetization from prior slice excitations (spin history effects)39. These motion artifacts are typically modeled and removed in a regression framework, containing predictors calculated from motion parameters estimated during coregistration40. Although effective, modeling based approaches do not completely remove motion-related fluctuations in the fMRI signal41–43. To address this issue, the “scrubbing” of offending volumes via removal41or spike regression44has been proposed. Importantly, excluding time-points alters the temporal structure of the data, thereby compromising analyses that rely on this structure (e.g., temporal dynamics, spectral analysis, estimating temporal autocorrelation). Regardless of the motion correction scheme employed, it is necessary to account for motion in group level analyses42, 45.
Physiological Noise Correction
Cardiac pulsation and respiration can induce fMRI signal fluctuations, which led to early criticisms attributing iFC to these physiological signals rather than neural signals46. The cardiac cycle generates pulsatile motion throughout the brain47. Respiratory movement of the chest and abdomen induce changes in the magnetic field, producing intensity fluctuations in fMRI images47. Additionally, changes in cardiac rhythm as well as rate and depth of breathing create longer-term effects. Respiration and pulse can be recorded to model and subsequently remove their impact47. Although accepted as ideal, this is not commonly performed. Instead, signals present in white matter and cerebrospinal fluid are taken as surrogates for respiration and cardiac effects, and regressed from the fMRI time-series. Incorporating spatial variation in the noise captured by the white matter signal provides superior denoising (e.g., ANATICOR48). Blind source separation techniques provide another means of physiological correction (e.g., Corsica49).
Global Signal Regression (GSR)
The mean time-series across the whole brain is commonly regressed from the data. In this model, the global signal is considered a non-specific measure of noise whose removal improves the specificity of iFC50, decreases motion-effects44, and removes inter-session and inter-site effects. However, awareness that GSR centers the correlation distribution at zero, and thus introduces negative connections51and can alter inter-individual differences52, has made its use controversial. In this regard, it is important to note that functional correlation coefficients obtained after GSR are relative values, not absolute. Additionally, electrophysiological demonstrations of globally synchronous neural signals in grey matter53call into question the interpretation of the global signal as simply noise.
Temporal Filtering
Bandpass filtering (0.001<f<0.08) is usually included. This frequency range targets removal of low frequency scanner drift and frequencies above those traditionally associated with functional connectivity34, 54. However, complete removal of physiological noise is unlikely, due to artifacts induced by the low temporal resolution of fMRI (e.g. aliasing)46. Concerns about temporal filtering include reductions in the degrees of freedom for the time-series and recent demonstrations of functional connectivity at frequencies higher than 0.1 Hz for several brain areas, suggesting that low-pass filtering is removing valuable signal55. Thus, despite historical precedent, inclusion of low-pass filtering merits further consideration.
Spatial Normalization and Smoothing
Another aspect of preprocessing is conditioning the data for comparison across subjects. Spatial normalization addresses morphological variation across individuals by transforming them to a common stereotactic space; population- and study-specific templates are increasingly used to optimize correspondence. Spatial smoothing further improves the correspondence of brain areas across individuals, and increases the signal-to-noise ratio56.
Estimating Edges
Several mathematical modeling techniques can be used to define functional relationships, differing primarily in the stringency with which they define functional connectivity. Functional connectivity simply implies a statistical dependency between activities observed in brain areas, and is an umbrella term for a wide range of dependency measures, each providing a different perspective32. For example, mutual information measures statistical dependency from the joint probability distribution function and is sensitive to linear and non-linear relationships, whereas Pearson’s correlation is primarily sensitive to linear relationships57. Effective connectivity, on the other hand, requires a mathematically precise (directional) description of the interactions between brain areas58. Importantly, this leads to a plurality of graphs that can be derived from functional connectivity, each characterizing functional interactions from a different perspective.
Estimating iFC from R-fMRI typically begins with extraction of the mean time-series across voxels in each brain node (i.e., parcellation-defined brain area). Intrinsic functional connectivity is commonly estimated from bivariate tests for statistical dependency (e.g., Pearson’s correlation, mutual information, spectral coherence) between every possible pairing of time-series59. Although these approaches perform well in simple simulations59, the limited number of observations results in noisy estimates of statistical relationships, which can be reduced using regularization (shrinkage) methods60. A limitation of bivariate approaches is that they do not take into account multiple brain areas simultaneously. Hence they cannot distinguish direct from indirect interactions (mediated by shared relationships with other areas). Partial correlation (related to the inverse covariance matrix) estimates the conditional linear dependency between two brain areas, after accounting for interactions with every other area61. Although considered preferential to bivariate approaches, the number of brain areas commonly exceeds the degrees of freedom, preventing unique specification of partial correlations. In these cases, regularization techniques (e.g., graphical lasso, elastic net) can assist in finding a solution59. Additionally, information can be pooled across individuals to optimize estimation parameters62. Note that some, but not all, of these approaches enforce symmetry; in other words, one can obtain dependency in one direction but not the other (see Box 3).
Box 3 - Directionality in functional connectomics.
Commonly referred to as effective connectivity, the mapping of directional relationships is essential for characterizing information flow in functional connectivity studies136. Identification of neural drivers can facilitate our understanding of control systems, as well as ectopic foci leading to pathological conditions (e.g., epilepsy137). While invasive tracing and stimulation techniques are powerful tools for mapping directional relationships in non-human populations138, they are not generally applicable in humans. Here, we provide an overview of non-invasive approaches to establishing directionality.
Statistical Techniques
Before reviewing statistical effective connectivity approaches, we note that despite nomenclature, their findings should not be interpreted as indicating causality, but rather the directionality of information flow. Structural Equation Modeling (SEM) and Dynamic Causal Modeling (DCM) approaches evaluate the fit of hypothesized models of directional interactions among nodes with measured fMRI data. SEM is a covariance-based approach that represents each node as an exogenous variable (i.e., predicting the activity of another node), endogenous variable (i.e., activity is predicted by another node), or both139; effective connectivity is modeled at the hemodynamic (BOLD response) level. In contrast, DCM is a generative approach, modeling effective connectivity at the neuronal level based upon a given biophysical model and employs a forward model to produce the downstream fMRI activity. Models can be evaluated individually; however, common-practice is to compare models representing competing hypotheses regarding causality to identify the “best fit” model. Concerns exist regarding the feasibility of successfully identifying a “best fit” model from a large population of putative models140. SEM and DCM approaches are limited in the number of nodes and interactions they can model efficiently. Primarily intended for confirmatory analysis, exploratory implementations of SEM141and DCM136are emerging for use with R-fMRI.
Granger causality analysis (GCA) is a model-free, data-driven effective connectivity approach142that investigates whether past values of the time-series for one node could improve the prediction of the current value in another143. In contrast to SEM and DCM, GCA can assess numerous nodes simultaneously. Application of GCA to R-fMRI144is particularly controversial due to: 1) limitations imposed by the sluggish and variable hemodynamic response function (HRF), 2) slow sampling rate (e.g., TR ≥ 2s)59, and 3) assumptions that HRF characteristics are constant across nodes59. Attempts to rehabilitate GCA for R-fMRI include: HRF deconvolution prior to GCA137, accounting for regional variation in the HRF using breath-hold scans, and faster sampling rates145. Nonetheless, using GCA with R-fMRI remains problematic.
Lesion Studies
Lesion studies directly perturb the system and therefore can be compelling in establishing causality when carried out in a controlled manner. From this perspective, changes in the activity of a node following disruption of inputs from another are taken to infer causal influences. In the ideal paradigm, scans are obtained pre- and post-occurrence of a given lesion. For example, Johnston et al.146scanned a 6-year-old child pre- and post-callosotomy, revealing disruptions specific to interhemispheric connectivity. Lesion studies are, however, commonly limited to post-lesion scans, taken after naturally-occurring lesions (e.g., stroke, congenital abnormality, neoplasm, seizure focus). Although useful for testing predictions or generating novel hypotheses, findings of such studies cannot be considered definitive.
Brain Stimulation
The gold standard for establishing directional influences is the demonstration that direct stimulation of node A impacts node B, but not the converse. In this regard, intraoperative studies of corticocortical evoked potentials (CCEP) –which involve the tracking of electrical signals from stimulation sites to other locations– are powerful tools147, though with limited applicability (i.e., neurosurgical patients). Non-invasive stimulation techniques with a broader range of applications are slowly evolving. Transcranial magnetic stimulation (TMS) is capable of producing temporary, and reversible neural excitation or suppression in targeted cortical areas. Concurrent TMS–fMRI studies have revealed causal relationships within motor circuits148and the visual system149. Transcranial direct current stimulation (tDCS) and transcranial alternating current stimulation (tACS) approaches deliver weak currents to targeted brain areas, and are emerging as less expensive and technically demanding alternatives to TMS, though delivering focal stimulation is challenging150.
A number of approaches exist for estimating eFC. Several authors have borrowed approaches from iFC, assuming the time-series spans the entire task63or from concatenated blocks of specific task conditions64. Psychophysiological interaction (PPI)39analyses directly model interactions between patterns of functional connectivity and the experimental stimulus design, potentially offering greater specificity of findings. Others have measured eFC from “coactivation” using a series of fitted regression coefficients65or binarized (by applying a threshold to regression coefficients) time-series66generated from a first level task analysis. Regression coefficient-series are then compared using correlation or partial correlation33, 65. Binarized time-series can be compared from the joint distribution of the two values using measures akin to mutual information66. Finally, meta-analytical approaches provide a means of measuring eFC across studies and often tasks, detecting patterns of coactivation across statistical maps generated from the literature35, 67.
A variety of data-driven techniques are also used for identifying iFC and eFC patterns. Examples include self-organizing maps68, principal component analysis, normalized cut clustering69, and independent component analysis70. These methods are more appropriate for identifying nodes of connectome graphs (see node section) than edges, although exceptions exist59.
Once functional connectivity is estimated, some applications require it to be thresholded or binarized (i.e., a connection is present or not). Threshold selection is not straightforward, but can be accomplished by applying a test of statistical significance to each edge. When using parametric statistics, care must be taken to adjust the degrees of freedom for temporal autocorrelation. Alternatively, this can be addressed using non-parametric tests of significance such as wavestrapping71or circular block bootstrap72. Sparse covariance estimation methods can also be employed.
Interpretations and Considerations
A common pursuit of T-fMRI and R-fMRI studies is to fractionate the connectome into a set of spatially and functionally distinct networks that can each be annotated in terms of the specific functional (e.g. cognitive, affective, or visceral) domain they subserve. Impressively, they have converged on similar definitions of 8–20 spatially and functionally distinct networks - though numerous studies have suggested the actual number of networks is substantially greater13. The concordance of T-fMRI and R-fMRI findings suggests the brain's intrinsic functional architecture provides a framework for moment-to-moment responses to the external world. As summarized by Smith et al.67, it appears that “the full repertoire of functional networks utilized by the brain in action is continuously and dynamically ‘active’ even when at ‘rest’.”
When considering the visualization of functional connectivity (Fig. 3), an important question is: “what are we missing?” While functional connectivity is often represented with static graphs, neurophysiological models have long asserted the transient nature of many functional interactions. Specifically, distributed neural assemblies appear to change their patterns of interaction with one another from one cognitive act or state to another. Consistent with this notion, eFC studies have noted significant task-dependency in their findings, even when looking at the same regions33, 39. Perhaps most exciting, recent iFC studies have observed dynamic changes in iFC patterns over a 5–minute scan73. These findings suggest that commonly employed metrics of iFC are incomplete, only capturing the “mean” connectivity over time. If true, the implications would be multifold: 1) findings of hypo- or hyper-connectivity in population studies would need to be reassessed, as they may reflect a different distribution of time spent in the various iFC configurations between populations, 2) the detection of changing eFC patterns over the course of task performance may prove to be a means of explaining observed behavioral variability. Of note, an increasing number of studies are highlighting the potential value of examining transition zones between functional areas in the brain5. Examination of temporal dynamics may inform such efforts, by mapping changes in the boundary over time and providing greater clarity for findings.
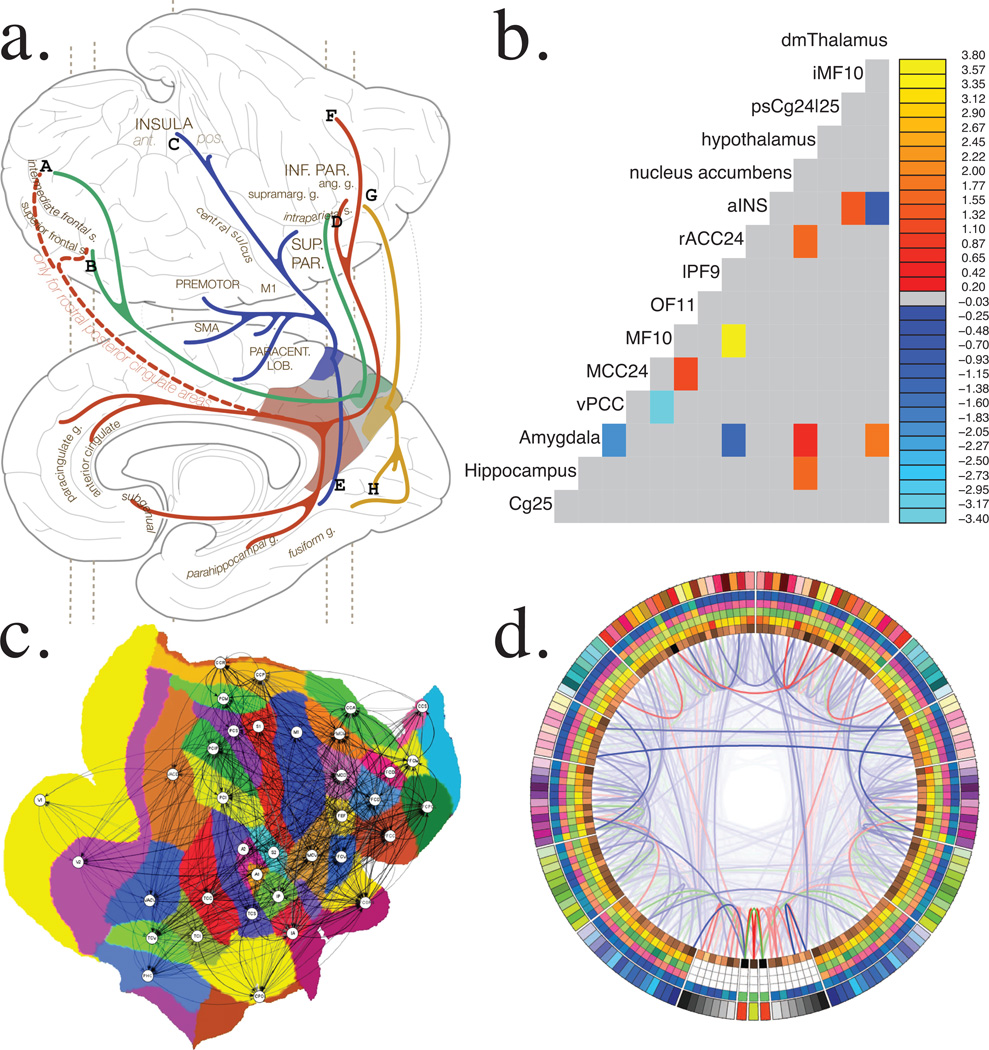
Figure 3. Visualizing the Connectome.
(A) Classical anatomical-tracing style depiction of iFC for posteromedial cortex subdivisions (reproduced with permission from Margulies et al., 2009113). (B) Matrix representation of functional brain connections predictive of diagnostic status in depression (reproduced with permission from Craddock et al., 200990). (C) Flatmap-based representation of the CoCoMac atlas of the Macaque connectome (reproduced with permission from Knock et al., 2009114). (D) Connectogram depicts brain areas (nodes) as columns in the circular band, differing connectivity metrics in separate layers and connections with lines, lobes are differentiated by color and left/right halves corresponds to hemispheres (reproduced with permission from van Horn et al., 2012115).
In addition to naturally occurring variations in iFC over time, studies have suggested that iFC can be systematically impacted by cognitive demands prior to R-fMRI data acquisition. By comparing iFC during R-fMRI scans collected before and after task performance74, studies have shown that iFC strength within and between networks is altered in a task-dependent manner. For example, R-fMRI based functional connectivity between the inferior frontal gyrus and visual areas was found to vary depending on the category of stimuli viewed prior to the R-fMRI scan75. Further, across participants, the degree to which iFC was modulated correlated with subsequent memory for the stimuli. In addition to exhibiting plasticity related to tasks performed close in time to the measurements, iFC is modulated by direct stimulation protocols including median nerve stimulation76, heat pain77, transcranial magnetic stimulation (TMS)78and transcranial direct current stimulation (tDCS)79. This suggests that R-fMRI may have utility in identifying targets for stimulation protocols as well as assessing their efficacy (e.g., in the context of the treatment of depression80).
This iFC-based evidence of experience-induced plasticity provide strong support for the hypothesis that iFC reflects a history of coactivation among areas. However, they also suggest a corollary - correlated intrinsic activity plays a role in learning and memory consolidation81. The demonstration of brain-behavior correlations between task-related modulations of iFC and subsequent behavior (e.g., recall) supports this hypothesis. If short-term iFC alterations reflect experience-induced plasticity, then enduring changes would be expected following extended practice or training. Several studies suggest this is the case82. Studies of long-term training-induced plasticity have the potential to inform our understanding of mechanisms involved in remediation-based recovery of function, or even to index the efficacy of treatment interventions. For example, in a preliminary retrospective study, differences in iFC were observed between children with dyslexia who were successfully remediated by reading interventions versus children who had received no treatment83.
Statistical analysis of the connectome
Once connectomes graphs are estimated, the next goal is to annotate them in terms of their relevance to higher order cognitive processes, neuropsychiatric diagnoses, or other phenotypic variables5. These associations are most often inferred by performing a categorical or dimensional statistical analysis that compares connectivity across a population of individuals, or within an individual across time or treatments84. Many of the same statistical approaches are appropriate for the analysis of connectome graphs regardless of whether they were constructed with functional or structural datasets. However, the interpretation of the results must always consider the idiosyncratic differences between structural and functional connectivity. For example, functional connections are typically weighted, and can be positive or negative. Structural connectivity graphs tend to be unweighted and are strictly non-negative27. Additionally, structural connections can be thought of as pathways along which information can flow, but functional connections cannot be interpreted in the same manner85.
A Bag of Edges
The simplest approach to comparing graphs is to treat them as a bag, or collection of edges, and perform statistical analyses at each edge one at a time, without taking into account interactions or relationships between them (i.e., edgewise statistics)84. Such univariate approaches (e.g., T-tests, F-tests, or regression) allow researchers to identify easily interpretable relationships between categorical or dimensional variables and edge weights. However, this approach results in a large number of statistical tests, which require correction for multiple comparisons to adequately control for the number of false positives. Standard correction techniques such as false discovery rate86that do not model the dependencies between edges may result in overly liberal or conservative corrections87. Alternate correction techniques such as the network-based statistic88or group Benjamini-Hochberg89leverage information about the group structure of connectome graphs to increase statistical power, while maintaining control of false positives.
Alternatively, multivariate regression and classification techniques evaluate the relationship between the entire connectome graphs and their associated phenotypic variables with a single statistical test90, 91. Although powerful for connectome-phenotype relationships, they obscure information about the involvement of individual edges. Extracting this information, if desired, requires a return to edge-specific tests, and the need for multiple comparison correction90. Of note, while these multivariate techniques tend to be applied to bag-of-edges representations, which ignore graph structure, they can also be performed using graph distance measures that preserve topological information when comparing graphs92.
Node and graph-level statistics: Invariants
Graphical representations of connectomes contain a wealth of information about brain architecture beyond the presence and strength of bivariate connections, which can be described using a variety of node-level and graph-level statistics. These measures are called invariants in graph theory parlance, or topological measures in network theory, because they are not unique to particular representations of the graph. The most commonly employed node invariants are centrality measures that indicate a node’s relative influence in a graph. Several different centrality metrics are available that measure a node’s importance based on the number and strength of direct connections (degree centrality93), the importance of neighboring nodes (eigenvector94or Page Rank95centrality), and their role in connecting other pairs of nodes (betweenness85). The various measures provide different perspectives on a node’s role in the graph, and when combined can provide a more holistic understanding of connectome-phenotype relationships95.
Similarly, a range of graph-level invariants is employed for studying structural and functional connectivity. In particular, graphs are commonly assessed in terms of their local and global efficiency. Local efficiency assesses the degree to which neighbors are densely interconnected, while global efficiency captures the number of connections that must be traversed to connect any two nodes93. The relationships of these two measures to what would be obtained from random graphs with similar properties can be combined to assess the small-worldness of a graph93. Small-world graphs balance integration and segregation to obtain fast and cost-efficient propagation of information through the graph, as well as robustness to single-node failures96. The cost-efficiency of a graph can be inferred from the difference between global efficiency and the number of edges in the graph93. An additional invariant is modularity, which quantifies the degree to which a graph can be segregated into densely intraconnected but sparsely interconnected modules and allows direct comparison of module membership between graphs85.
Each of the previously described node and graph invariants can be statistically evaluated to identify relationships with categorical and dimensional phenotypes. Although invariants can increase statistical power by decreasing the number of multiple comparisons, the resulting relationships can be more difficult to interpret. When comparing invariants between graphs it is important to consider the impact of potential differences in graph properties (e.g. number of edges) that can systematically differ between individuals or groups and confound interpretation of findings93. Additionally, since the distributional properties of most invariants are poorly characterized, non-parametric statistical tests are preferred93.
Finally, researchers frequently aim to identify connectivity patterns predictive of a phenotypic variable (e.g., diagnosis90, age91or brain state97). Predictive modeling directly assesses the ability of a connectivity pattern to predict the phenotype of an individual, in contrast to inferential statistics, which evaluate improbability of a set of relationships arising by chance98. Predictive modeling is typically supervised, with the training set consisting of connectivity graphs and their associated phenotypes99. One can assess the predictive accuracy via cross-validation99or other model selection techniques. Predictive modeling has primarily focused on invariants and bags-of-edges90, 100style approaches.
Translational Connectomics
MRI-based approaches to connectomics research are rapidly transforming neuroscience in animal models as well, by removing barriers to longitudinal examinations associated with invasive techniques (e.g., animal sacrifice, injection of toxic chemicals). The recent Mouse BIRN initiative (http://www.loni.ucla.edu/BIRN/Projects/Mouse/) provides an initial demonstration of the potential to complement cross-sectional atlases of the developing brain generated using histological approaches with longitudinal atlases obtained using dMRI. Simultaneously, R-fMRI is emerging as a powerful tool for comparative functional neuroanatomy studies. Initial work has demonstrated impressive correspondence between the iFC observed in humans and macaques for homologous functional networks supporting an array of functions, including those that are putatively “human” (e.g., language, self-referential, cognition)101. Evidence of homologies with patterns of iFC in lower mammals, such as rats, further underscores this translational potential102. Armed with increasingly powerful imaging-based tools, macroscale connectomic studies in animal models are poised to provide a mechanistic understanding of brain function through the combination of non-invasive imaging with direct structural, pharmacological, molecular, and genetic manipulations that are impossible in humans.
Despite the rich promise of translational connectomics, methodological issues must also be addressed. For example, iFC can be examined in awake rats that have been habituated to restraint in the loud MRI environment103. However, most studies are conducted under anesthesia – in particular, using the general anesthetic isoflurane104which can confound findings due to its effects on neural excitability. The sedating alpha-2 adrenergic agonist medetomidine may be preferable, as it avoids such confounds105. Dose-response studies are few104and are essential. Initial translational studies in monkeys, rats and mice have relied on preprocessing and analytical approaches identical to those developed in humans103. While their success is encouraging, differences in physiological (e.g., cardiac, respiration) and imaging parameters must be explored to arrive at optimal strategies. Finally, we note that the many questions raised regarding the interpretation of dMRI and R-fMRI techniques in humans also apply to animal studies.
Towards Neurophenotypes and Clinical Applications
An overarching goal of the connectomics era is the derivation of “neurophenotypes”106, a concept that remains poorly specified despite increasing investigator enthusiasm. An individual’s macroscale connectome and its subgraphs contribute to the specification of their neurophenotype. A central goal of connectomics is to catalog neurophenotypes and relate them to phenotypic profiles4. This can be accomplished through data-driven approaches focused on the detection of commonalities and distinctions in connectomes, or that can be reverse-engineered from phenotypic profiles. The breadth of phenotyping can vary depending on the application, though it typically consists of some combination of cognitive, affective, behavioral, neurological or psychiatric variables. When cataloging neurophenotypes based upon macroscale connectomes, the specificity of findings will depend on their nature, granularity of node definitions and quality of neuroimaging data employed. Similarly, when sorting neurophenotypes based on phenotypic profiles, specificity will be determined by the precision and comprehensiveness (i.e., number and breadth of independent features) of the phenotyping available to statistical analysis. Importantly, future work will need to find a balance between categorical and dimensional perspectives of neurophenotypes.
Beyond the derivation of a fundamental understanding of brain architecture and its implications for behavior and cognition, a major reason for the excitement surrounding connectomics is the promise of clinical utility because of the ability to obtain individually-relevant reliable brain indices (see Table 1 for initiatives that are accelerating the pace of macroconnectomics research). Recent years have witnessed an explosion in the number of neurological and psychiatric disorders studied with dMRI and R-fMRI (see Table 2). Hopes of attaining clinically useful diagnostic tools are increasingly espoused in the literature. However, leaders in the field have recently suggested that the attainment of tools capable of stratifying individuals based upon disease risk, prognosis and treatment response may prove to be a more fruitful goal than focusing on diagnosis107. Regardless, a key requirement remains: attaining large-scale datasets representative of the human population. In this regard, the macroconnectomics community has supported several large-scale data-sharing initiatives dedicated to rapidly aggregating the necessary data108.
Table 1.
Large-scale initiatives macroscale connectomics inititatives (See116 for additional information regarding these initiatives and others).
| INITIATIVES PROMISING TO ACCELERATE THE PACE OF MACROSCALE CONNECTOMICS RESEARCH |
|---|
| Brain Genomics Superstruct (US). Aims to collect a large-scale imaging dataset to explore brain-behavior relationships and their genetic influences. The initiative has collected R-fMRI, dMRI, and saliva samples from over 3,000 adults, along with comprehensive phenotyping (cognitive, personality, lifestyle), and the resultant repository containing 1500 completed, quality pass datasets is expected to be publicly available within the next year. |
| Brainnetome (http://www.brainnetome.org; China). Attempts to characterize brain networks with multimodal neuroimaging techniques, from the microscale (microtechnique, ultramicrotomy, staining and visualization techniques) to the most macroscale (EEG, fMRI, dMRI). R-fMRI and diffusion imaging datasets, along with behavioral and blood data from more than 1000 schizophrenia patients, 300 Alzheimer’s Disease / mild cognitive impairment (MCI) patients, 120 stroke patients, 50 glioma patients and 2000 healthy controls collected from eleven hospitals and imaging centers. |
| Consortium of Neuroimagers for the Non-invasive Exploration of Brain Connectivity and Tracts (http://www.brain-connect.eu; EU). Consortium focused on studying the brain's microstructure, tracts and connectivity using dMRI. Target deliverables include optimized acquisition protocols, analytic tools and a connectivity atlas. |
| Developing Human Connectome Project (EU). Recently awarded initiative that will comprehensively map and model the human connectome for 1000 babies, including in utero and in vivo imaging (from 20 to 44 weeks post-conceptional age). |
| NIH Human Connectome Project: WU-Minn consortium (http://humanconnectome.org; US). State-of-the-art multimodal imaging initiative (MEG, EEG, R-fMRI, T-fMRI, dMRI), that makes use of a twin-design (1200 healthy aduts, including twin pairs and their siblings from 300 families) to provide insights into the genetic bases of the human connectome. The initiative is making use of “low TR” multiband imaging sequences for R-fMRI and dMRI, which it has refined and is currently distributing to interested centers. All data and tools developed through the initiative will be openly shared. |
| NIH Human Connectome Project: MGH/Harvard-UCLA consortium (http://humanconnectome.org; US). Initiative focusing on the unraveling the full connectivity map using the first “Connectome Scanner”, which is designed to carry out diffusion using ultrahigh gradient strength (4–8 times the strength of conventional systems). Efforts to optimize dMRI technology will focus on increasing the spatial resolution, quality, and speed of acquisition. |
| 1000 Functional Connectomes Project (FCP; http://fcon_1000.projects.nitrc.org; global). Grass-roots data-sharing initiative that brought together over 1200 previously collected R-fMRI datasets from 33 independent sites |
Table 2.
Neuropsychiatric disorders most commonly studied using macroscale functional and structural connectivity. Determined by the number (count) of R-fMRI and dMRI papers dedicated to the neuropsychiatric diagnosis. Calculated from the Child Mind Institute Librarian (Resting State and DTI libraries; http://www.mendeley.com/profiles/cmi-librarian/).
| Neuropsychiatric Diagnoses | R-fMRI Count |
dMRI Count |
|---|---|---|
| Schizophrenia | 100 | 283 |
| Alzheimer's Disease | 89 | 201 |
| Depression | 81 | 101 |
| Epilepsy and Seizures | 62 | 179 |
| Mild Cognitive Impairment | 58 | 115 |
| Other Neurological Disorders | 45 | 50 |
| Substance Dependence | 40 | 94 |
| Attention Deficit Hyperactivity Disorder | 32 | 37 |
| Autism Spectrum Disorders | 26 | 86 |
| Multiple Sclerosis | 22 | 211 |
| Parkinson's Disease | 19 | 73 |
| Traumatic Brain Injury | 18 | 259 |
| Sleep Disorders | 17 | 7 |
| Stroke | 15 | 182 |
| Anxiety Disorders | 15 | 16 |
| Coma and Vegetative State | 15 | 9 |
| Obsessive Compulsive Disorder | 12 | 27 |
| Amyotrophic Lateral Sclerosis | 10 | 84 |
| Bipolar Disorder | 10 | 61 |
Conclusion
The connectomics era is the culmination of more than a century of conceptual and methodological innovation. MRI-based approaches to mapping and annotating the connectome at the macroscale are transforming basic, translational and clinical neuroscience research by overcoming barriers to progress faced by more traditional invasive methodologies. This review has broadly surveyed the many challenges that remain at hand in the acquisition, preprocessing and analysis of brain-imaging data. Failure to consider the many complexities could jeopardize this burgeoning field through the introduction of spurious, irreproducible findings associated with suboptimal methodologies. Conversely, increased attention to the acquisition of high quality data, combined with optimized preprocessing and analytic methodologies can serve to accelerate the pace at which connectomes can be meaningfully annotated, and their variations catalogued.
Table 3.
Magnetic Resonance Imaging Parameters
| Parameter | Definition | Impact |
|---|---|---|
| Echo Time (TE) | The time between slice excitation and acquiring the center of k-space. | Determines impact of T2 and T2* on the images. |
| Repetition Time (TR) | The time between acquisitions of adjacent fMRI volumes (sampling period). | Impacts the signal available for imaging (I0), impacted by the number of slices. Longer TRs reduce motion sensitivity. |
| Bandwidth (BW; 1/Δt) | The range of frequencies mapped to a voxel. | Lower bandwidth settings can increase artifacts due to inadequate shimming or susceptibility, and distortions in the phase-encoding direction (e.g. ‘scalloping’) |
| Flip Angle (α) | The amount of rotation applied to proton spins by the excitation pulse. | Impacts the signal available for imaging (I0). Flip angles larger or smaller that the Ernst angle for a given TR will reduce I0. Low flip angles may reduce motion sensitivity, and in-flow effects and improve T1 contrast of images. |
| Spatial Resolution (Δx, Δy, Δz, Nx, Ny) | The volume of tissue sampled in a given voxel. Determined by field of view (FOV) and the number of points sampled in a slice. | SNR is significantly impacted by voxel volume; higher spatial resolution have lower SNR, e.g. a 2mm iso voxel has only ~30% of the SNR of a 3mm iso voxel, holding all other factors constant. |
| Parallel Imaging (Pf) | Methods such as GRAPPA and SENSE can decrease TR, and reduce spatial distortions by sampling k-space lines in parallel. Decreases in TR are rate-limited by TE required for BOLD contrast. | Can allow faster image acquisition, but at a reduction of SNR by 1/parallelization factor, holding all other factors constant. Will increase temporal noise resulting from head motion, respiration, or pulsatile effects. |
| Nacq | The number of acquisitions that are acquired and subsequently averaged. | Improves SNR, does not make sense for fMRI, but commonly used for dMRI. |
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institute of Mental Health (BRAINS R01MH094639 to M.P.M. and K23MH087770 to A.D.M.), the Stavros Niarchos Foundation (M. P. M), the Brain and Behavior Research Foundation (R.C.C.), and the Leon Levy Foundation (C.K. and A.D.M).. J.T.V. receives funding from the London Institute for Mathematical Sciences HDTRA1-11-1-0048 and NIH R01ES017436. Additional support provided by a gift from Joseph P. Healey to the Child Mind Institute (M.P.M.). Thanks to Daniel Lurie for his assistance in the preparation of the manuscript and references, as well as to Zarrar Shehzad, Zhen Yang and Sebastian Urchs for their helpful comments. We would additionally like to acknowledge our colleagues who have allowed us to re-use their figures, and the helpful suggestions of the reviewers.
Footnotes
COMPETING FINANCIAL INTERESTS
Keith Heberlein, Ph.D. is a full time employee of Siemens Medical Solutions USA, and owns shares in Siemens, AG.
References
- 1.Sporns O, Tononi G, Kötter R. The human connectome: A structural description of the human brain. PLoS Comput. Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varela F, Lachaux JP, Rodriguez E, Martinerie J. The brainweb: phase synchronization and large-scale integration. Nat. Rev. Neurosci. 2001;2:229–239. doi: 10.1038/35067550. [DOI] [PubMed] [Google Scholar]
- 3.Biswal BB, et al. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens TEJ, Sporns O. Human connectomics. Curr. Opin. Neurobiol. 2011:1–10. doi: 10.1016/j.conb.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly AMC, Biswal BB, Craddock RC, Castellanos XF, Milham MP. Characterizing variation in the functional connectome: promise and pitfalls. Trends Cogn. Sci. 2012;16:1–8. doi: 10.1016/j.tics.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. New York: Georg Thieme, Stuttgart; 1988. [Google Scholar]
- 7.Margulies DS, et al. Mapping the functional connectivity of anterior cingulate cortex. Neuroimage. 2007;37:579–588. doi: 10.1016/j.neuroimage.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 8.Beckmann M, Johansen-Berg H, Rushworth MFS. Connectivity-based parcellation of human cingulate cortex and its relation to functional specialization. J. Neurosci. 2009;29:1175–1190. doi: 10.1523/JNEUROSCI.3328-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dosenbach NU, et al. A core system for the implementation of task sets. Neuron. 2006;50:799–812. doi: 10.1016/j.neuron.2006.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellec P, et al. Identification of large-scale networks in the brain using fMRI. Neuroimage. 2006;29:1231–1243. doi: 10.1016/j.neuroimage.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 11.Craddock RC, James GA, Holtzheimer PE, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum. Brain Mapp. 2012;33:1914–1928. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen AL, et al. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage. 2008;41:45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiviniemi V, et al. Functional segmentation of the brain cortex using high model order group PICA. Hum. Brain Mapp. 2009;30:3865–3886. doi: 10.1002/hbm.20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones DK. Challenges and limitations of quantifying brain connectivity in vivo with diffusion MRI. Imaging. 2010;2:341–355. [Google Scholar]
- 15.Andersson JLR, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. 2003;20:870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- 16.Alexander AL, Tsuruda JS, Parker DL. Elimination of eddy current artifacts in diffusion-weighted echo-planar images: the use of bipolar gradients. Magn. Reson. Med. 1997;38:1016–1021. doi: 10.1002/mrm.1910380623. [DOI] [PubMed] [Google Scholar]
- 17.Haselgrove JC, Moore JR. Correction for distortion of echo-planar images used to calculate the apparent diffusion coefficient. Magn. Reson. Med. 1996;36:960–964. doi: 10.1002/mrm.1910360620. [DOI] [PubMed] [Google Scholar]
- 18.Anderson J, et al. International Society for Magnetic Resonance in Medicine (ISMRM) Melbourne, Australia: 2012. [Google Scholar]
- 19.Sotiropoulos SN, et al. Effects of image reconstruction on fibre orientation mapping from multichannel diffusion MRI: Reducing the noise floor using SENSE. Magn. Reson. Med. 2013 doi: 10.1002/mrm.24623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leemans A, Jones DK. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn. Reson. Med. 2009;61:1336–1349. doi: 10.1002/mrm.21890. [DOI] [PubMed] [Google Scholar]
- 21.Seunarine K, Alexander D. Diffusion MRI. San Diego: Academic Press; 2009. pp. 55–72. [Google Scholar]
- 22.Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J. Magn. Reson. B. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 23.Tuch DS, et al. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn. Reson. Med. 2002;48:577–582. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- 24.Wedeen VJ, Hagmann P, Tseng WY, Reese TG, Weisskoff RM. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn. Reson. Med. 2005;54:1377–1386. doi: 10.1002/mrm.20642. [DOI] [PubMed] [Google Scholar]
- 25.Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage. 2007;34:144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catani M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. Neuroimage. 2002;17:77–94. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- 27.Jbabdi S, Johansen-Berg H. Tractography: where do we go from here? Brain Connect. 2011;1:169–183. doi: 10.1089/brain.2011.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Assaf Y, Blumenfeld-Katzir T, Yovel Y, Basser PJ. AxCaliber: a method for measuring axon diameter distribution from diffusion MRI. Magn. Reson. Med. 2008;59:1347–1354. doi: 10.1002/mrm.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones DK, Knosche TR, Turner R. White matter integrity, fiber count, and other fallacies: The do's and don'ts of diffusion MRI. Neuroimage. 2013;73:239–254. doi: 10.1016/j.neuroimage.2012.06.081. [DOI] [PubMed] [Google Scholar]
- 30.Mikula S, Binding J, Denk W. Staining and embedding the whole mouse brain for electron microscopy. Nat. Methods. 2012;9:1198–1201. doi: 10.1038/nmeth.2213. [DOI] [PubMed] [Google Scholar]
- 31.Behrens T, Jbabdi S. Diffusion MRI. San Diego: Academic Press; 2009. pp. 333–351. [Google Scholar]
- 32.Friston KJ, Frith CD, Liddle PF, Frackowiak RS. Functional connectivity: the principal-component analysis of large (PET) data sets. J. Cereb. Blood Flow Metab. 1993;13:5–14. doi: 10.1038/jcbfm.1993.4. [DOI] [PubMed] [Google Scholar]
- 33.Mennes M, Kelly C, Colcombe S, Castellanos FX, Milham MP. The extrinsic and intrinsic functional architectures of the human brain are not equivalent. Cereb. Cortex. 2013;23:223–229. doi: 10.1093/cercor/bhs010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 35.Toro R, Fox PT, Paus T. Functional coactivation map of the human brain. Cereb. Cortex. 2008;18:2553–2559. doi: 10.1093/cercor/bhn014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chuang K-H, et al. Mapping resting-state functional connectivity using perfusion MRI. Neuroimage. 2008;40:1595–1605. doi: 10.1016/j.neuroimage.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa S, et al. Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging. Proc. Natl. Acad. Sci. U. S. A. 1992;89:5951–5955. doi: 10.1073/pnas.89.13.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu CW, et al. Empirical evaluations of slice-timing, smoothing, and normalization effects in seed-based, resting-state functional magnetic resonance imaging analyses. Brain Connect. 2011;1:401–410. doi: 10.1089/brain.2011.0018. [DOI] [PubMed] [Google Scholar]
- 39.Friston KJ, et al. Psychophysiological and modulatory interactions in neuroimaging. Neuroimage. 1997;6:218–229. doi: 10.1006/nimg.1997.0291. [DOI] [PubMed] [Google Scholar]
- 40.Friston KJ, Williams S, Howard R, Frackowiak RS, Turner R. Movement-related effects in fMRI time-series. Magn. Reson. Med. 1996;35:346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- 41.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Dijk KRA, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satterthwaite TD, et al. Impact of in-scanner head motion on multiple measures of functional connectivity: Relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satterthwaite TD, et al. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan CG, et al. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage. 2013;76C:183–201. doi: 10.1016/j.neuroimage.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lund TE. fcMRI--mapping functional connectivity or correlating cardiac-induced noise? Magn. Reson. Med. 2001;46:628–629. doi: 10.1002/mrm.1238. [DOI] [PubMed] [Google Scholar]
- 47.Birn RM. The role of physiological noise in resting-state functional connectivity. Neuroimage. 2012;62:864–870. doi: 10.1016/j.neuroimage.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 48.Jo HJ, Saad ZS, Simmons WK, Milbury LA, Cox RW. Mapping sources of correlation in resting state FMRI, with artifact detection and removal. Neuroimage. 2010;52:571–582. doi: 10.1016/j.neuroimage.2010.04.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perlbarg V, et al. CORSICA: correction of structured noise in fMRI by automatic identification of ICA components. Magn. Reson. Imaging. 2007;25:35–46. doi: 10.1016/j.mri.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 50.Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. J. Neurophysiol. 2009;101:3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy K, Birn RM, Handwerker Da, Jones TB, Bandettini Pa. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 2009;44:893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saad Z, et al. Trouble at Rest: How Correlation Patterns and Group Differences Become Distorted After Global Signal Regression. Brain Connect. 2012;2:25–32. doi: 10.1089/brain.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schölvinck ML, Maier A, Ye FQ, Duyn JH, Leopold DA. Neural basis of global resting-state fMRI activity. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10238–10243. doi: 10.1073/pnas.0913110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cordes D, et al. Frequencies contributing to functional connectivity in the cerebral cortex in "resting-state" data. AJNR. Am. J. Neuroradiol. 2001;22:1326–1333. [PMC free article] [PubMed] [Google Scholar]
- 55.Niazy RK, Xie J, Miller K, Beckmann CF, Smith SM. Spectral characteristics of resting state networks. Prog. Brain Res. 2011;193:259–276. doi: 10.1016/B978-0-444-53839-0.00017-X. [DOI] [PubMed] [Google Scholar]
- 56.Worsley KJ, Marrett S, Neelin P, Evans AC. Searching scale space for activation in PET images. Hum. Brain Mapp. 1996;4:74–90. doi: 10.1002/(SICI)1097-0193(1996)4:1<74::AID-HBM5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 57.Marrelec G, et al. Regions, systems, and the brain: hierarchical measures of functional integration in fMRI. Med. Image Anal. 2008;12:484–496. doi: 10.1016/j.media.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 58.Friston KJ, Frith CD. Time dependent changes in effective connectivity measured with PET. Hum. Brain Mapp. 1993;1:69–79. [Google Scholar]
- 59.Smith SM, et al. Network modelling methods for FMRI. Neuroimage. 2011;54:875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- 60.Varoquaux G, Gramfort A, Poline JB. Advances in Neural Information Processing Systems. Vancouver, Canada: 2010. [Google Scholar]
- 61.Marrelec G, et al. Partial correlation for functional brain interactivity investigation in functional MRI. Neuroimage. 2006;32:228–237. doi: 10.1016/j.neuroimage.2005.12.057. [DOI] [PubMed] [Google Scholar]
- 62.Varoquaux G, et al. A group model for stable multi-subject ICA on fMRI datasets. Neuroimage. 2010;51:288–299. doi: 10.1016/j.neuroimage.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 63.Lowe MJ, Dzemidzic M, Lurito JT, Mathews VP, Phillips MD. Correlations in low-frequency BOLD fluctuations reflect cortico-cortical connections. Neuroimage. 2000;12:582–587. doi: 10.1006/nimg.2000.0654. [DOI] [PubMed] [Google Scholar]
- 64.Sun FT, Miller LM, D'Esposito M. Measuring interregional functional connectivity using coherence and partial coherence analyses of fMRI data. Neuroimage. 2004;21:647–658. doi: 10.1016/j.neuroimage.2003.09.056. [DOI] [PubMed] [Google Scholar]
- 65.Rissman J, Gazzaley A, D'Esposito M. Measuring functional connectivity during distinct stages of a cognitive task. Neuroimage. 2004;23:752–763. doi: 10.1016/j.neuroimage.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 66.Patel RS, Bowman FD, Rilling JK. A Bayesian approach to determining connectivity of the human brain. Hum. Brain Mapp. 2006;27:267–276. doi: 10.1002/hbm.20182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith SM, et al. Correspondence of the brain's functional architecture during activation and rest. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peltier SJ, Polk TA, Noll DC. Detecting low-frequency functional connectivity in fMRI using a self-organizing map (SOM) algorithm. Hum. Brain Mapp. 2003;20:220–226. doi: 10.1002/hbm.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van den Heuvel M, Mandl R, Hulshoff Pol H. Normalized cut group clustering of resting-state FMRI data. PLoS ONE. 2008;3:e2001. doi: 10.1371/journal.pone.0002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beckmann CF. Modelling with independent components. Neuroimage. 2012;62:891–901. doi: 10.1016/j.neuroimage.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 71.Breakspear M, Brammer MJ, Bullmore ET, Das P, Williams LM. Spatiotemporal wavelet resampling for functional neuroimaging data. Hum. Brain Mapp. 2004;23:1–25. doi: 10.1002/hbm.20045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bellec P, Marrelec G, Benali H. A bootstrap test to investigate changes in brain connectivity for functional MRI. Stat. Sin. 2008;18:1253–1268. [Google Scholar]
- 73.Smith SM, et al. Temporally-independent functional modes of spontaneous brain activity. Proc. Natl. Acad. Sci. U. S. A. 2012;109:3131–3136. doi: 10.1073/pnas.1121329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waites AB, Stanislavsky A, Abbott DF, Jackson GD. Effect of prior cognitive state on resting state networks measured with functional connectivity. Hum. Brain Mapp. 2005;24:59–68. doi: 10.1002/hbm.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stevens WD, Buckner RL, Schacter DL. Correlated low-frequency BOLD fluctuations in the resting human brain are modulated by recent experience in category-preferential visual regions. Cereb. Cortex. 2010;20:1997–2006. doi: 10.1093/cercor/bhp270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klingner CM, Hasler C, Brodoehl S, Axer H, Witte OW. Perceptual plasticity is mediated by connectivity changes of the medial thalamic nucleus. Hum. Brain Mapp. 2012 doi: 10.1002/hbm.22074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Riedl V, et al. Repeated pain induces adaptations of intrinsic brain activity to reflect past and predict future pain. Neuroimage. 2011;57:206–213. doi: 10.1016/j.neuroimage.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 78.Vercammen A, Knegtering H, Liemburg EJ, den Boer JA, Aleman A. Functional connectivity of the temporo-parietal region in schizophrenia: effects of rTMS treatment of auditory hallucinations. J. Psychiatr. Res. 2010;44:725–731. doi: 10.1016/j.jpsychires.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 79.Keeser D, et al. Prefrontal transcranial direct current stimulation changes connectivity of resting-state networks during fMRI. J. Neurosci. 2011;31:15284–15293. doi: 10.1523/JNEUROSCI.0542-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fox MD, Buckner RL, White MP, Greicius MD, Pascual-Leone A. Efficacy of Transcranial Magnetic Stimulation Targets for Depression Is Related to Intrinsic Functional Connectivity with the Subgenual Cingulate. Biol. Psychiatry. 2012 doi: 10.1016/j.biopsych.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tambini A, Ketz N, Davachi L. Enhanced brain correlations during rest are related to memory for recent experiences. Neuron. 2010;65:280–290. doi: 10.1016/j.neuron.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lewis CM, Baldassarre A, Committeri G, Romani GL, Corbetta M. Learning sculpts the spontaneous activity of the resting human brain. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17558–17563. doi: 10.1073/pnas.0902455106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koyama MS, et al. Cortical signatures of dyslexia and remediation: an intrinsic functional connectivity approach. PLoS ONE. 2013;8:e55454. doi: 10.1371/journal.pone.0055454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Varoquaux G, Craddock RC. Learning and comparing functional connectomes across subjects. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 85.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 86.Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 87.Efron B. When should hypothesis testing problems be combined? Ann. Appl. Stat. 2008;2:197–223. [Google Scholar]
- 88.Zalesky A, Fornito A, Bullmore ET. Network-based statistic: identifying differences in brain networks. Neuroimage. 2010;53:1197–1207. doi: 10.1016/j.neuroimage.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 89.Hu JX, Zhao H, Zhou HH. False Discovery Rate Control With Groups. J. Am. Stat. Assoc. 2010;105:1215–1227. doi: 10.1198/jasa.2010.tm09329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Craddock RC, Holtzheimer PE, Hu XP, Mayberg HS. Disease state prediction from resting state functional connectivity. Magn. Reson. Med. 2009;62:1619–1628. doi: 10.1002/mrm.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dosenbach NUF, et al. Prediction of individual brain maturity using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bunke H. A graph distance metric based on the maximal common subgraph. Pattern Recognition Letters. 1998;19:255–259. [Google Scholar]
- 93.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 94.Bullmore ET, Bassett DS. Brain graphs: graphical models of the human brain connectome. Annu. Rev. Clin. Psychol. 2011;7:113–140. doi: 10.1146/annurev-clinpsy-040510-143934. [DOI] [PubMed] [Google Scholar]
- 95.Zuo XN, et al. Network centrality in the human functional connectome. Cereb. Cortex. 2012;22:1862–1875. doi: 10.1093/cercor/bhr269. [DOI] [PubMed] [Google Scholar]
- 96.Bassett DS, Meyer-Lindenberg A, Achard S, Duke T, Bullmore E. Adaptive reconfiguration of fractal small-world human brain functional networks. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19518–19523. doi: 10.1073/pnas.0606005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Richiardi J, Eryilmaz H, Schwartz S, Vuilleumier P, Van De Ville D. Decoding brain states from fMRI connectivity graphs. Neuroimage. 2011;56:616–626. doi: 10.1016/j.neuroimage.2010.05.081. [DOI] [PubMed] [Google Scholar]
- 98.Hansen LK. Multivariate strategies in functional magnetic resonance imaging. Brain Lang. 2007;102:186–191. doi: 10.1016/j.bandl.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 99.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. Springer; 2001. [Google Scholar]
- 100.Zhu CZ, et al. Discriminative analysis of brain function at resting-state for attention-deficit/hyperactivity disorder. Medical Image Computing and Computer Assisted Interventions. 2005;8:468–475. doi: 10.1007/11566489_58. [DOI] [PubMed] [Google Scholar]
- 101.Vincent JL, et al. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- 102.Pawela CP, et al. Resting-state functional connectivity of the rat brain. Magn. Reson. Med. 2008;59:1021–1029. doi: 10.1002/mrm.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Becerra L, Pendse G, Chang P-C, Bishop J, Borsook D. Robust reproducible resting state networks in the awake rodent brain. PLoS ONE. 2011;6:e25701. doi: 10.1371/journal.pone.0025701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang K, et al. Temporal scaling properties and spatial synchronization of spontaneous blood oxygenation level-dependent (BOLD) signal fluctuations in rat sensorimotor network at different levels of isoflurane anesthesia. NMR Biomed. 2011;24:61–67. doi: 10.1002/nbm.1556. [DOI] [PubMed] [Google Scholar]
- 105.Pawela CP, et al. A protocol for use of medetomidine anesthesia in rats for extended studies using task-induced BOLD contrast and resting-state functional connectivity. Neuroimage. 2009;46:1137–1147. doi: 10.1016/j.neuroimage.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sörös P, Stanton SG. On Variability and Genes: Inter-individual Differences in Auditory Brain Function. Front. Hum. Neurosci. 2012;6:150. doi: 10.3389/fnhum.2012.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kapur S, Phillips AG, Insel TR. Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Mol. Psychiatry. 2012;17:1174–1179. doi: 10.1038/mp.2012.105. [DOI] [PubMed] [Google Scholar]
- 108.Mennes M, Biswal BB, Castellanos FX, Milham MP. Making data sharing work: The FCP/INDI experience. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tzourio-Mazoyer N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 110.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 111.Eickhoff SB, et al. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. Neuroimage. 2005;25:1325–1335. doi: 10.1016/j.neuroimage.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 112.Lancaster JL, et al. Automated Talairach atlas labels for functional brain mapping. Hum. Brain Mapp. 2000;10:120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Margulies DS, et al. Precuneus shares intrinsic functional architecture in humans and monkeys. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20069–20074. doi: 10.1073/pnas.0905314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Knock SA, et al. The effects of physiologically plausible connectivity structure on local and global dynamics in large scale brain models. J. Neurosci. Methods. 2009;183:86–94. doi: 10.1016/j.jneumeth.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 115.Van Horn JD, et al. Mapping connectivity damage in the case of Phineas Gage. PLoS ONE. 2012;7:e37454. doi: 10.1371/journal.pone.0037454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jiang T. Brainnetome: A new -ome to understand the brain and its disorders. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 117.Haacke EM, Bornw RW, Thompson MR, Venkatesan R. Magnetic Resonance Imaging: Physical Principles and Sequence Design. Wiley-Liss; 1999. [Google Scholar]
- 118.Bernstein MA, King KF, Zhou XJ. Handbook of MRI pulse sequences. Academic Press; 2004. [Google Scholar]
- 119.Blaimer M, et al. SMASH, SENSE, PILS, GRAPPA: how to choose the optimal method. Top. Magn. Reson. Imaging. 2004;15:223–236. doi: 10.1097/01.rmr.0000136558.09801.dd. [DOI] [PubMed] [Google Scholar]
- 120.Yacoub E, et al. Spin-echo fMRI in humans using high spatial resolutions and high magnetic fields. Magn. Reson. Med. 2003;49:655–664. doi: 10.1002/mrm.10433. [DOI] [PubMed] [Google Scholar]
- 121.Weiskopf N, Hutton C, Josephs O, Deichmann R. Optimal EPI parameters for reduction of susceptibility-induced BOLD sensitivity losses: a whole-brain analysis at 3 T and 1.5 T. Neuroimage. 2006;33:493–504. doi: 10.1016/j.neuroimage.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 122.Glover GH, Law CS. Spiral-in/out BOLD fMRI for increased SNR and reduced susceptibility artifacts. Magn. Reson. Med. 2001;46:515–522. doi: 10.1002/mrm.1222. [DOI] [PubMed] [Google Scholar]
- 123.Heberlein KA, Hu X. Simultaneous acquisition of gradient-echo and asymmetric spin-echo for single-shot z-shim: Z-SAGA. Magn. Reson. Med. 2004;51:212–216. doi: 10.1002/mrm.10680. [DOI] [PubMed] [Google Scholar]
- 124.Gonzalez-Castillo J, Roopchansingh V, Bandettini Pa, Bodurka J. Physiological noise effects on the flip angle selection in BOLD fMRI. Neuroimage. 2011;54:2764–2778. doi: 10.1016/j.neuroimage.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Grootoonk S, et al. Characterization and correction of interpolation effects in the realignment of fMRI time series. Neuroimage. 2000;11:49–57. doi: 10.1006/nimg.1999.0515. [DOI] [PubMed] [Google Scholar]
- 126.Noll DC, Cohen JD, Meyer CH, Schneider W. Spiral K-space MR imaging of cortical activation. J. Magn. Reson. Imaging. 1995;5:49–56. doi: 10.1002/jmri.1880050112. [DOI] [PubMed] [Google Scholar]
- 127.Rabrait C, et al. High temporal resolution functional MRI using parallel echo volumar imaging. J. Magn. Reson. Imaging. 2008;27:744–753. doi: 10.1002/jmri.21329. [DOI] [PubMed] [Google Scholar]
- 128.Moeller S, et al. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn. Reson. Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Setsompop K, et al. Improving diffusion MRI using simultaneous multi-slice echo planar imaging. Neuroimage. 2012;63:569–580. doi: 10.1016/j.neuroimage.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Feinberg DA, et al. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS ONE. 2010;5:e15710. doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bright MG, Murphy K. Removing motion and physiological artifacts from intrinsic BOLD fluctuations using short echo data. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heine L, et al. Resting state networks and consciousness: alterations of multiple resting state network connectivity in physiological, pharmacological, and pathological consciousness States. Front. Psychol. 2012;3:295. doi: 10.3389/fpsyg.2012.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Boly M, et al. Brain connectivity in disorders of consciousness. Brain Connect. 2012;2:1–10. doi: 10.1089/brain.2011.0049. [DOI] [PubMed] [Google Scholar]
- 134.Horovitz SG, et al. Decoupling of the brain's default mode network during deep sleep. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11376–11381. doi: 10.1073/pnas.0901435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Spoormaker VI, et al. Development of a large-scale functional brain network during human non-rapid eye movement sleep. J. Neurosci. 2010;30:11379–11387. doi: 10.1523/JNEUROSCI.2015-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Friston K. Dynamic causal modeling and Granger causality Comments on: the identification of interacting networks in the brain using fMRI: model selection, causality and deconvolution. Neuroimage. 2011;58:303–305. doi: 10.1016/j.neuroimage.2009.09.031. author reply 310–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.David O, et al. Identifying neural drivers with functional MRI: an electrophysiological validation. PLoS Biol. 2008;6:2683–2697. doi: 10.1371/journal.pbio.0060315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Scannell JW, Burns GA, Hilgetag CC, O'Neil MA, Young MP. The connectional organization of the cortico-thalamic system of the cat. Cereb. Cortex. 1999;9:277–299. doi: 10.1093/cercor/9.3.277. [DOI] [PubMed] [Google Scholar]
- 139.Mclntosh AR, Gonzalez-Lima F. Structural equation modeling and its application to network analysis in functional brain imaging. Hum. Brain Mapp. 1994;2:2–22. [Google Scholar]
- 140.Lohmann G, Erfurth K, Müller K, Turner R. Critical comments on dynamic causal modelling. Neuroimage. 2012;59:2322–2329. doi: 10.1016/j.neuroimage.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 141.Zhuang J, LaConte S, Peltier S, Zhang K, Hu X. Connectivity exploration with structural equation modeling: an fMRI study of bimanual motor coordination. Neuroimage. 2005;25:462–470. doi: 10.1016/j.neuroimage.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 142.Goebel R. Investigating directed cortical interactions in time-resolved fMRI data using vector autoregressive modeling and Granger causality mapping. Magn. Reson. Imaging. 2003;21:1251–1261. doi: 10.1016/j.mri.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 143.Granger CW. Investigating causal relations by econometric models and cross-spectral methods. Econometrica. 1969:424–438. [Google Scholar]
- 144.Sridharan D, Levitin DJ, Menon V. A critical role for the right fronto-insular cortex in switching between central-executive and default-mode networks. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12569–12574. doi: 10.1073/pnas.0800005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Deshpande G, Hu X. Investigating Effective Brain Connectivity from fMRI Data: Past Findings and Current Issues with Reference to Granger Causality Analysis. Brain Connect. 2012 doi: 10.1089/brain.2012.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Johnston JM, et al. Loss of resting interhemispheric functional connectivity after complete section of the corpus callosum. J. Neurosci. 2008;28:6453–6458. doi: 10.1523/JNEUROSCI.0573-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Matsumoto R, et al. Functional connectivity in the human language system: a corticocortical evoked potential study. Brain. 2004;127:2316–2330. doi: 10.1093/brain/awh246. [DOI] [PubMed] [Google Scholar]
- 148.Bohning DE, et al. Echoplanar BOLD fMRI of brain activation induced by concurrent transcranial magnetic stimulation. Invest. Radiol. 1998;33:336–340. doi: 10.1097/00004424-199806000-00004. [DOI] [PubMed] [Google Scholar]
- 149.Ruff CC, et al. Distinct causal influences of parietal versus frontal areas on human visual cortex: evidence from concurrent TMS-fMRI. Cereb. Cortex. 2008;18:817–827. doi: 10.1093/cercor/bhm128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Datta A, et al. Gyri-precise head model of transcranial direct current stimulation: improved spatial focality using a ring electrode versus conventional rectangular pad. Brain Stimulat. 2009;2:201–207. doi: 10.1016/j.brs.2009.03.005. 207.e201. [DOI] [PMC free article] [PubMed] [Google Scholar]