Summary
A successful cellular response to virus infection is essential for evolutionary survival. In plants, arthropods, and nematodes, cellular antiviral defenses rely on RNA interference (RNAi). Interestingly, the mammalian response to virus is predominantly orchestrated through interferon (IFN)-mediated induction of antiviral proteins. Despite the potency of the IFN system, it remains unclear whether mammals also have the capacity to employ antiviral RNAi. Here we investigate this by disabling either IFN, small RNA function or both activities in the context of virus infection. We find that loss of small RNAs in the context of an in vivo RNA virus infection lowers titers due to reduced transcriptional repression of the host antiviral response. In contrast, enabling a virus with the capacity to inhibit the IFN system results in increased titers. Taken together, we conclude that small RNA silencing is not a physiological contributor to the IFN-mediated cellular response to virus infection.
Introduction
Productive replication of a virus demands access to the raw materials of the host cell for successful generation of progeny virions. In response, the host cell must rapidly recognize the presence of the virus and employ a defense aimed at halting the infection. This arms race has resulted in an evolutionary track record of countless measures and countermeasures employed by both entities. In plants, arthropods, and nematodes, cells recognize the formation of double stranded RNA (dsRNA) as a foreign structure indicative of virus infection (Kemp and Imler, 2009). This pathogen associated molecular pattern (PAMP) is then processed in a variety of means to generate virus–specific small interfering RNAs (vsiRNAs) through an RNAse III family of nucleases such as Dicer (Ding and Voinnet, 2007; Hutvagner and Zamore, 2002; Sabin et al., 2013). vsiRNAs are then loaded into an RNA induced silencing complex (RISC) and subsequently used to guide it to complementary RNA (Ding and Voinnet, 2007). This system, generally referred to as RNA interference (RNAi), is capable of cleaving viral mRNA in an enzymatic fashion and successfully inhibiting replication (Ratcliff et al., 1999; Zamore et al., 2000). In response to this effective defense, plant and arthropod viruses have evolved antagonists to many aspects of the vsiRNA biogenesis pathway (Chao et al., 2005; Li et al., 2002; Nayak et al., 2010; Qi et al., 2012; van Rij et al., 2006).
Interestingly, the cellular response to virus in mammals is also initiated by the detection of dsRNA or other foreign nucleic-acid based structures (Bowie and Unterholzner, 2008). However, unlike in plants and arthropods, detection of dsRNA results in the culmination of a cytokine-mediated response. That is, PAMP detection in mammalian cells results in the activation of host kinases and transcription factors that result in the induction of Type I and III interferons (IFN-I and IFN-III) (Rauch et al., 2013). IFN induction can act in both an autocrine and paracrine manner to promote the upregulation of hundreds of IFN stimulated genes (ISGs) (Schoggins and Rice, 2011). These genes work in concert to slow virus replication and provide the necessary time for the adaptive response to clear the infection. Included in the list of ISGs are host products that inhibit transcription, translation, cellular transport as well as genes involved in cell death and the release of chemokines to recruit immune cells to the site of infection (Schoggins and Rice, 2011). As is the case for arthropod and plant pathogens, viruses that infect mammals also have evolved proteins to antagonize many aspects of the IFN-I and –III responses (Weber et al., 2003).
Despite extensive research aimed at defining the mammalian response to virus infection, it still remains controversial whether RNAi is also a component in mammalian cells (Umbach and Cullen, 2009). Evidence for RNAi in mammals includes the evolutionary conservation and utilization of small RNAs in the form of microRNAs (miRNAS) (Bartel, 2004). These 19–21nt duplex RNAs, like vsiRNAs, are generated by RNAse III nucleases, load into RISC, and mediate posttranscriptional silencing (Bartel, 2004). Given the conservation of this pathway and the required nucleases, it remains tempting to speculate small RNAs, or the machinery itself, could function in an antiviral fashion. This concept is further supported by the fact that many dsRNA binding proteins that antagonize virus detection in mammals, also disrupt RNAi (Cullen et al., 2013; Fabozzi et al., 2011; Garcia-Sastre et al., 1998; Haasnoot et al., 2007; Li et al., 2004; Prins et al., 2010). While this data may reflect the fact that both systems are dependent on dsRNA detection and processing, a recent paper from our own group identified a bona fide inhibitor of small RNAs from poxviruses (Backes et al., 2012) leading many to speculate that aspects of RNAi are indeed conserved in mammals.
While limited evidence has supported a claim for RNAi in mammals, other experiments have strongly suggested that small RNAs are not a component of the vertebrate response to infection. For example, engineering influenza A virus (IAV) lacking its dsRNA antagonist regains full virulence when administered to mice lacking IFN signaling capacity (Garcia-Sastre et al., 1998). Should RNAi contribute to the mammalian response to virus infection, one would suppose that in an IFN-independent model system, lack of a dsRNA antagonist should still demonstrate some level of attenuation. Furthermore, incorporation of miRNA target sites into the genomes of countless viruses can induce attenuation suggesting these viruses have not needed to evolve countermeasures for small RNA silencing (tenOever, 2013). The debate about RNAi in mammals however has been further intensified with the advent of deep sequencing. Profiling of small RNAs from virus infection has demonstrated detectable pools of virus-derived small RNAs that could theoretically serve a role in RNAi, although these small RNA pools remain unchanged in the absence of Dicer and may actually be the by-product of ISGs such as RNaseL (Girardi et al., 2013; Parameswaran et al., 2010). Despite any clear consensus for how these small RNAs are generated and how they might function, the idea of mammalian RNAi has recently gained significant traction in the scientific community following the characterization of a mutant Nodavirus in both stem cells and immortalized fibroblasts (Li et al., 2013; Maillard et al., 2013).
In an effort to formally address whether small RNAs significantly contribute to the mammalian response to virus infection, we generated a recombinant RNA virus with the capacity to disrupt either RISC-associated silencing or the IFN-mediated response. We demonstrate that we can confer onto Vesicular Stomatitis Virus (VSV) the capacity to eliminate RISC-associated small RNAs through the expression of Vaccinia virus (VACV) VP55 (VSV-VP55) or block detection of IFN induction with the addition of IAV NS1 (VSV-NS1). Interestingly, we find that while VSV-NS1 replicates to higher titers in vivo, VSV-VP55 is attenuated. Furthermore, we find attenuation is the result of an enhancement of the IFN response in agreement with a recent report that found miRNAs to suppress basal levels of antiviral transcripts (Seo et al., 2013). Profiling the transcriptome of these infections also provided us with a comprehensive list of endogenous miRNA targets in the context of infection. To determine if small RNAs contributed in the absence of the IFN response, VSV-VP55 was also administered to IFN-I and -III receptor knockout mice. In this model system, virus attenuation of VP55 was lost but the degradation of small RNAs still failed to increase overall virus titers. Taken together, we conclude that the cellular response to virus infection is independent of small RNA silencing and is mediated exclusively by IFNs.
Results
Small RNA profiling of virus infected cells
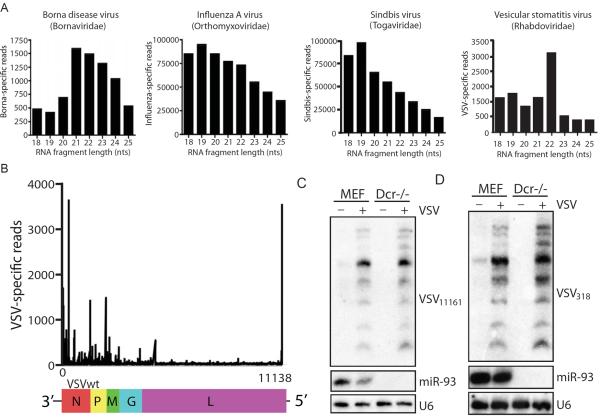
Given the recent claims of RNAi in mammals, we sought to define the profile of small RNAs in the context of a spectrum of RNA virus infections. To this end, we cloned and deep sequenced total nucleotide pools in the range of 18–25nts from cells infected with viruses from diverse families including: Orthomyxoviridae (Influenza A virus (IAV)), Togaviridae (Sindbis virus (SV)), Bornaviridae (Borna disease virus, (BDV)), and Rhabdoviridae (Vesicular Stomatitis Virus (VSV)) (Table S1). Small RNA sequencing reads were then consolidated and mapped to the respective virus genomes. For each virus infection, small RNA reads could be assembled into contigs with complete genomic coverage. In agreement with published literature, captured small RNA reads where readily detectable in all infections: 0.04%, 18.4%, 24.1% and 0.05% of total small RNA reads mapped to the BDV, IAV, SV and VSV genomes, respectively (Figure 1A) (Donaszi-Ivanov et al.; Girardi et al., 2013; Parameswaran et al., 2010; Sabin et al., 2013). With the exception of VSV, virus-derived small RNAs failed to show a size preference indicative of RNAi activity (Figure 1A). Interestingly, when the size distribution of the VSV reads were plotted, the dominant size was 22nts and there was an enrichment for sequence reads that mapped to the ends of the genome, reminiscent of the RNAi-like activity recently described in mammalian cells (Figures 1A and B) (Li et al., 2013). However, it should be noted that these two most abundant VSV-derived small RNAs, which could be detected by small RNA Northern blot, were Dicer-independent (Figure 1C and D). Taken together, these data suggest VSV may be a good model to ascertain additional evidence for or against a small RNA mediated antiviral defense in mammals.
Figure 1. Small RNA profiling of virus-infected cells.
(A) 18–25 nucleotides (nts) RNA sequencing reads obtained from glial cells persistently infected with Borna disease virus, or fibroblasts infected with Influenza A virus, Sindbis virus and Vesicular stomatitis virus (VSV) (MOI of 1 for 12hrs) were analyzed for size distribution. (B) VSV-derived reads from (A) were aligned for their distribution across the genome. (C) Northern blot analysis of wildtype (MEF) and dicer-deficient cells (Dcr−/−) infected with VSV (MOI of 1 for 9 hrs) for VSV-derived small RNA VSV318, miR-93 and U6. (D) Northern blot analysis of MEF and Dcr−/− infected with VSV (MOI of 1 for 9 hrs) for VSV-derived small RNA VSV11161, miR-93 and U6.
VSV does not encode an inhibitor of small RNA silencing
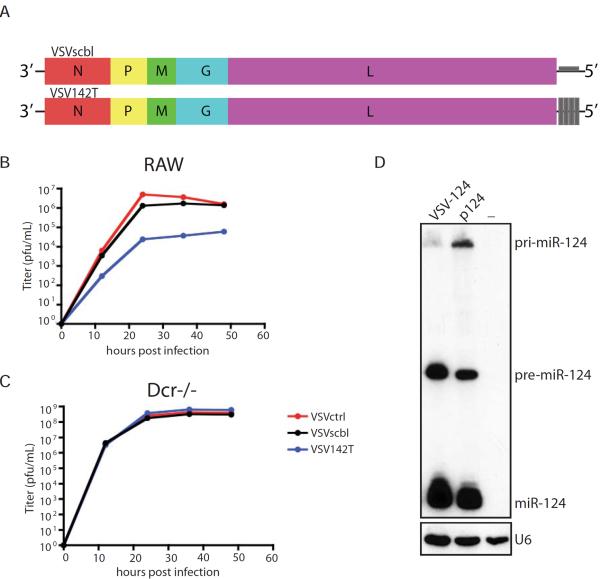
To probe whether RNAi activity contributed to the cellular response to VSV infection, we next determined whether VSV encoded a suppressor of RNAi. While VSV has been found to be sensitive to the RNAi system of arthropods and nematodes, we wanted to ensure small RNAs could additionally target VSV in the context of our mammalian systems (Mueller et al., 2010; Wilkins et al., 2005). To this end, we cloned four perfect target sites for the host hematopoietic cell-specific miR-142 as a 3'UTR of the Large (L) polymerase mRNA as previously described (Figure 2A) (Kelly et al., 2010). In agreement with published literature, we found that endogenous miR-142 processing and silencing potential was unperturbed in miR-142 expressing macrophage-derived RAW cells in response to VSV infection as titers were reduced by ~2 logs when the miR-142 target sites were present (Figure 2B) (Kelly et al., 2008; Kelly et al., 2010). In contrast, in the absence of miR-142, all viruses replicated to comparable levels (Figure 2C and Figure S1). Furthermore, insertion of a primary miRNA (miR-124) into VSV demonstrates that, in the context of infection, the capacity to process hairpins is not compromised (Figure 2D). These results are in agreement with numerous published studies demonstrating the successful engineering of mammalian viruses to be silenced by, or produce, small RNAs in the mammalian host (Barnes et al., 2008; Cawood et al., 2009; Chua et al., 2013; Edge et al., 2008; Kelly et al., 2008; Langlois et al., 2013; Langlois et al., 2012a; Langlois et al., 2012b; Perez et al., 2009; Pham et al., 2012; Shapiro et al., 2012; Shapiro et al., 2010; Varble et al., 2013; Varble et al., 2010). Taken together, these data suggest that RNA virus infection in mammalian cells results in large pools of small RNAs and that RISC function is not impaired during the early stages of infection.
Figure 2. VSV does not encode an inhibitor of RNAi.
(A) Schematic of VSV genome engineered to encode 4 perfectly complementary miR-142 target (VSV142T) or 4 scrambled sites (VSVscbl) in the 3' UTR of the mRNA encoding for the L protein. (B) and (C) Multicycle growth curve of VSVctrl, VSV142T and VSVscbl on (B) RAW cells or (C) dicer-deficient cells (Dcr−/−). Supernatants were analyzed at the indicated time points by plaque assay. (D) Northern blot analysis of BHK transfected with miR-124 expressing plasmid (p124) or infected with VSV expressing miR-124 (VSV-124, MOI of 1 for 16 hr) for miR-124 and U6.
Engineering an RNA virus with the capacity to disrupt small RNA function
We recently characterized a single Vaccinia virus (VACV) protein, termed VP55, that was both necessary and sufficient for the tailing of RISC-associated host miRNAs (Backes et al., 2012). We found that VP55 indiscriminately added non-templated adenosines specifically to RNAs that are associated with RISC, resulting in their rapid degradation (Backes et al., 2012). While we postulated that this activity stemmed from the evolutionary ancestry of entomopox viruses, which needed to block vsiRNAs, it remained possible that virus-derived small RNAs themselves provided an inherent protection to virus in mammals given the recent reports on such activity (Li et al., 2013; Maillard et al., 2013).
To assess the contribution of small RNAs in the mammalian response to virus, we sought to insert VP55 into VSV to enable the virus the capacity to block small RNA-mediated silencing. To this end, we utilized a VP55 variant that was identified following mutagenesis studies and was found to have increased protein stability and activity (Figure S2). VP55 T109A (herein referred to as simply VP55) was introduced between the Glycoprotein (G) and L RNA dependent RNA polymerase (RdRp) genes using the canonical initiation and termination sequence of the Nucleoprotein (NP) gene (Figure 3A). Incorporation of VP55 into VSV (VSV-VP55), as well as a matched control virus with a comparable RNA insert but lacking an open reading frame (VSVctrl) were generated and used to infect fibroblasts (Figure 3B). Approximately 12hrs post infection, VSV-VP55 demonstrated robust expression of the VACV protein. Furthermore, this expression correlated with the capacity to tail and degrade exogenously expressed miR-124 while not impacting VSV leader RNA or the splicing RNA component U6 (Figure 3C).
Figure 3. Engineering VSV to antagonize small RNAs.
(A) Schematic of Vaccinia virus (VACV) VP55 insertion into the VSV genome. Each independent transcript is shown as it would be generated. “An”, polyadenylated tail.
(B) Western blot analysis of VACV, VSVctrl or VSV-VP55-infected BHK for VACV VP55 and E3 and for VSV G expression. (C) Northern blot analysis of BHK co-infected with VSV expressing miR-124 (VSV-124) and VACV, VSVctrl or VSV-VP55 (MOI of 1 for 12hrs) for miR-124, VSV-leader RNA and U6. Relative density depicts mature miR-124/U6. (D) Analysis of murine fibroblasts transfected with control (scbl) or VSV-N-specific (VSV-N) siRNAs. Six hours posttransfection, cells were infected with VSVctrl or VSV-VP55 for 10 hrs (MOI of 1). Western blot was probed for VSV proteins and actin. (E) Schematic of premiR-146b-5p processing and corresponding miRNA duplex (middle) and mature miRNAs (bottom). The mature miRNA sequence-specific reads were determined by sequencing of the 20–25 nt fraction of VSV-VP55-infected cells. Adenosines (A) in black depict nontemplated bases and percent representation reflects the portion of the corresponding sequence in the total miRNA-specific tailed fraction.
To determine whether VSV-based expression of VP55 was sufficient to confer onto the virus the capacity to block RNAi, we next transfected exogenous vsiRNAs directed against NP. To permit VP55-mediated tailing of the vsiRNA, we synthesized duplex RNA lacking 2'O-methylation as this chemistry renders small RNAs resistant to VP55 tailing (Backes et al., 2012). Transfection of unmodified 21nt RNA duplexes composed of a scrambled sequence (scbl) or a complement of NP was followed by virus infection at a high multiplicity of infection (MOI). Ten hours post infection, whole cell lysate was analyzed by western blot using a polyclonal antibody that recognizes the viral glycoprotein (G), the viral matrix protein (M), and NP (Figure 3D). These analyses demonstrated that VSVctrl was highly susceptible to NP vsiRNA silencing in contrast to VSV-VP55, which showed no evidence of NP targeting. Furthermore, sequencing of miR-146b, a miRNA shown to be induced in response to virus infection (Taganov et al., 2006), demonstrated in vivo evidence of VP55-mediated polyadenylation (Figure 3E).
Determining the interplay between miRNAs, RNAi and RNA virus infection
In an effort to characterize the biology of VSV-VP55, independently of miRNA activity, we first compared multi-cycle growth curves to VSVctrl in cells lacking Dicer (Figures 4A and B). Administering the virus at a low MOI of 0.01 for a 48hr period resulted in no significant change in viral protein levels (Figure 4A) nor viral load as both VSVctrl and VSV-VP55 replicated to comparable titers of 1×108 plaque forming units (pfu)/ml (Figure 4B). These data demonstrate that, in the absence of Dicer-generated small RNAs, VSV-VP55 and VSVctrl replicate at comparable levels. As a result, we subsequently used these two viruses to compare virus replication in Dicer expressing fibroblasts (Figures 4C and D). Surprisingly, despite publications implicating host miRNAs in the direct silencing of VSV and RNAi activity in fibroblasts, we found VSV-VP55 replication was indistinguishable from VSVctrl replication, again reaching approximately equal viral protein levels (Figure 4C) and titers (Figure 4D) (Li et al., 2013; Otsuka et al., 2007). This was also true at earlier time points post infection (Figure S3). Taken together, these results suggest that small RNAs do not significantly impact the mammalian cellular response to virus infection in vitro.
Figure 4. Small RNAs do not impact VSV replication.
(A) and (B) Multicycle growth curve of VSVctrl and VSV-VP55 (MOI of 0.01) on dicer-deficient cells (Dcr−/−). (A) Cells were analyzed at the indicated time points for expression of VSV proteins. (B) Supernatants were analyzed at the indicated time points by plaque assay. (C) and (D) Multicycle growth curve of VSVctrl and VSV-VP55 (MOI of 0.01) on wildtype fibroblasts (MEF). (C) Cells were analyzed at the indicated time points for expression of VSV proteins. (D) Supernatants were analyzed at the indicated time points by plaque assay.
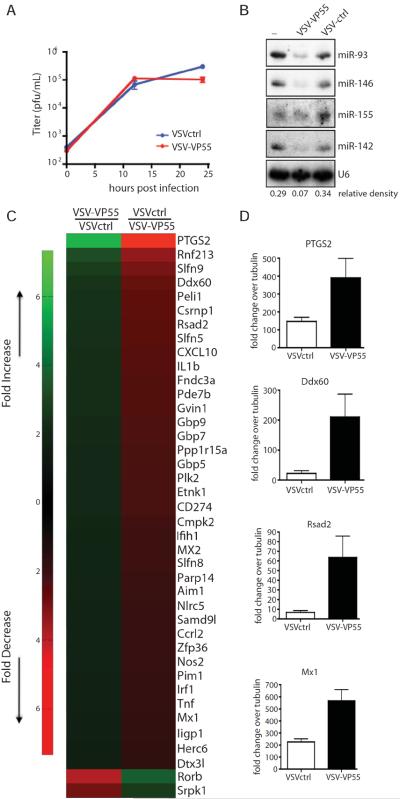
Next, we chose to study a homogenous population of physiologically relevant primary cells. As macrophages are an important cell type for in vivo replication of VSV, we decided to evaluate VSVctrl and VSV-VP55 in the context of a monocyte-derived primary macrophage cell population (Junt et al., 2007). Ex vivo infection of bone-marrow derived macrophages (BMMs) demonstrated comparable levels of infection as measured by plaque assay (Figure 5A). Not surprisingly, small RNA analysis of infected cohorts demonstrated a dramatic loss of host miRNAs including miR-142, miR-146, miR-155, and miR-93, without impacting U6 RNA (Figure 5B). To ascertain how loss of small RNAs in primary macrophages impacted the cellular response to virus infection, we performed mRNAseq and compared the transcriptomes of VSV infection in the presence and absence of small RNAs (Figure 5C and Table S2). In agreement with recent reports concerning the indirect role of miRNAs in enhancing expression of host antiviral genes, we found the majority of transcripts that were impacted the greatest by the expression of VP55 were canonical ISGs (Seo et al., 2013). Interestingly, this virus-induced miRNA `targetome' included numerous guanylate-binding proteins (Mx1, Mx2, Gbp5, Gbp7, and Gbp9), cytokines (TNFα, Cxcl10 and Ccrl2), pyrogens (Il1b) and known components of the antiviral sensing and signaling machinery (Rsad2, Ifih1, Aim1, Peli1, Csrnp1, Nlrc5, IRF1, Herc6 and Ddx60). Furthermore, the differential host expression of these genes in the absence of small RNAs could be independently corroborated by qPCR (Figure 5D). Interestingly, this list includes transcripts like PTGS2, IRF1, Pim1, Nos2, Fndc3a and Peli1 which have already been independently implicated as miRNA targets (Guo et al., 2012; Liu et al., 2013a; Liu et al., 2013b; Marquez et al., 2010; Shan et al., 2009; Thomas et al., 2012). It is also noteworthy that we identified only two genes that were expressed at elevated levels in VSVctrl (Table S2) - in agreement with the idea that miRNAs are negative regulators of transcription (Bartel, 2004).
Figure 5. Host miRNAs target interferon-stimulated genes during VSV-infection.
(A) to (D) Bone-marrow derived macrophages (BMMs) were infected with VSVctrl or VSV-VP55 (MOI of 5). (A) Viral replication was analyzed by plaque assay at the indicated time points. (B) 24 hrs post infection, BMMs were analyzed by Northern blot for presence of cellular miRNAs (miR-93, miR-146, miR-155, miR142) and U6. Relative density depicts miR-146/U6. (C) 10 hours post infection, BMMs were analyzed by mRNA sequencing. Top 40 of protein-coding transcripts differentially regulated by VSV-VP55 infection are shown. (D) Quantitative reverse transcriptase PCR depicting levels of PTGS2, Ddx60, Rsad2 and Mx1 induction over endogenous tubulin levels from BMMs infected for 10 hrs with VSVctrl or VSV-VP55.
Defining the contribution of small RNAs in vivo in the host response to virus infection
Given the lack of an in vitro phenotype following disruption of small RNA silencing (Figures 4 and 5), we next sought to determine whether RNAi contributes to the physiological response to virus infection in vivo. To formally evaluate this possibility we first examined whether in vivo infection of VSV resulted in the appearance of virus-specific small RNAs. To this end, we infected mice intranasally with VSVctrl or VSV-VP55 and deep sequenced small RNAs between 18–25nts. These data demonstrated the appearance of comparable small RNAs as those observed in fibroblasts, albeit at vanishing rare levels (Figure 6A and Table S3). Next we investigated whether miRNA tailing and degradation was observed in vivo. For this, we chose to again ascertain the levels of miR-146. 24hrs post infection, total lung tissue from three independent animals demonstrated a significant loss of miR-146 demonstrating that, as previously reported, VP55 is sufficient for targeting of small RNAs (Figure 6B) (Backes et al., 2012).
Figure 6. Functional IFN-response impacts VSV replication.
(A) 18–25 nucleotides (nts) RNA sequencing reads obtained from the lungs of mice infected intranasally with VSVctrl or VSV-VP55 (1×107 PFU for 24hrs) aligned for their distribution across the VSV-VP55 genome. “goi”, gene of interst. (B) Wildtype mice were infected intranasally with VSVctrl or VSV-VP55 (1×107 PFU; 3 mice per virus). 24hrs post infection lungs were analyzed for miR-146 and U6 expression by Northern blot. “M1, M2, M3”, mouse 1, mouse 2, mouse 3. Relative density depicts mature miR-146/U6. (C) Quantitative reverse transcriptase PCR depicting levels of IFN-β induction over endogenous tubulin levels from wildtype fibroblasts (MEF) infected with VSVctrl, VSV-VP55 or VSVNS1 (MOI of 1 for 16hrs). (D) Wildtype mice were infected intranasally with VSVctrl, VSV-VP55 or VSV-NS1 (1×107 PFU). 24hrs post infection lungs and spleens were analyzed by plaque assay. (E) Interferon I and III receptor knockout mice (Ifnar1−/−/Il28r−/−) were infected intranasally with VSVctrl, VSV-VP55 or VSV-NS1 (1×104 PFU). 48hrs post infection lungs and spleens were analyzed by plaque assay.
In an effort to directly compare the RNAi response to IFN signaling, we generated a VSV strain to encode the NS1 protein of IAV, a potent inhibitor of RIG-I and the subsequent induction of IFN (Mibayashi et al., 2007; Pichlmair et al., 2006). VSV-NS1 was generated in an identical manner to that of VSV-VP55 (Figure 3A). In vitro infections of VSV-NS1 demonstrated that it produced robust levels of NS1 and replicated to comparable levels compared to VSVctrl and VSV-VP55 in immortalized fibroblasts where IFN signaling is impaired (Figures S4A and B) (Stojdl et al., 2003). A similar phenotype was observed in dicer-deficient cells (Dcr−/−) (Figure S4C). However, while infections of these fibroblasts with either VSVctrl or VSV-VP55 resulted in robust induction of IFN-beta (IFNβ), VSV-NS1 demonstrated a dramatic loss of cytokine production suggesting NS1 was successfully targeting RIG-I (Figure 6C).
The induction of IFNβ in vitro also inversely correlated to virus titers in both the lung and spleen (Figure 6D). That is, VSV-NS1, in the absence of IFNβ induction, replicated to levels greater than one log when compared to VSVctrl virus. Furthermore, VSV-VP55, which demonstrated a modest increase in IFNβ induction when compared to VSVctrl, was reduced by a log in vivo (Figure 6D). These results clearly illustrate the importance of IFN in the mammalian response to virus.
In an effort to address whether redundancy in the antiviral response systems failed to provide a fitness advantage to VSV-VP55 in vivo, we next repeated our studies in mice lacking both type I and III IFN systems (Figure 6E). To this end, intranasal infection of IFN-I and -III receptor knockout mice (Ifnar1−/−/Il28r−/−) were performed with either VSVctrl, -VP55, or –NS1 but demonstrated no significant differences in viral titers in the lung or spleens of infected mice (Figure 6E). These data allow us to conclude that the enhanced titers of VSV-NS1 in wild type mice was the result of muting the IFN response and, conversely, that the attenuation of VSV-VP55 was a result of an enhanced IFN response. Furthermore, the inability of VSV-VP55 to replicate to titers that exceeded that of VSVctrl, even in the absence of IFN-I and –III signaling, strongly suggests that a functional antiviral RNAi system is not a physiological contributor to the mammalian antiviral defenses.
Discussion
The discovery that small RNAs have the capacity to influence protein levels post transcriptionally ushered in a new era for biology (Fire et al., 1998). Small RNA silencing can be observed through diverse biological processes but predominantly involves pathogen defense, achieving `transcriptional robustness', or is involved in maintaining distinct cellular lineages (Bartel and Chen, 2004; Ebert and Sharp 2012; Nayak et al. 2013).
The silencing potential of small RNAs is largely determined by its extent of target complementarity. Therefore, genome-encoded miRNAs, which demonstrate only partial complementary to target genes, have significant less capacity to silence a transcript as compared to a vsiRNA, which would be perfectly complementary (tenOever, 2013). For this reason, the concept of miRNAs contributing to the antiviral defenses of the cell are unlikely because the modest repression coupled to the average half-life of cellular proteins would yield limited overall impact during the context of acute virus infection. This is not the case for viruses that can persist (Gottwein and Cullen, 2008). However, while miRNAs may not have the capacity to directly silence viral message, the cellular capacity to process and utilize small RNAs has led to extensive speculation as to whether mammals were capable of eliciting an RNAi response. The idea of RNAi in mammals has been further supported by reports that viral dsRNA binding proteins such as NS1, E3L and VP35 can inhibit RNAi activity in both plants and arthropods (Bucher et al., 2004; Chou et al., 2007; Delgadillo et al., 2004; Li et al., 2004). Furthermore, with the advent of deep sequencing, small RNA profiling demonstrated that many virus infections result in the accumulation of genome-derived small RNAs, although in the case of mammals, these RNAs were found to be independent of Dicer (Parameswaran et al., 2010). It is also noteworthy that Drosha has been reported to translocate from the nucleus to the cytoplasm in response to virus infection but this activity did not correlate with the appearance of vsRNAs (Shapiro et al., 2014).
Most recently, two reports were published with evidence for RNAse III generated vsiRNAs against EMCV and a mutant Nodavirus (Li et al., 2013; Maillard et al., 2013). Interestingly, one of these studies suggested RNAi was only a component of undifferentiated stem cells and that this activity diminished as the cells become responsive to IFN (Maillard et al., 2013). In contrast, the second paper found similar evidence for small RNA processing in terminally differentiated hamster fibroblasts (Li et al., 2013). Together, these papers put forth some provocative data hinting at the existence of an RNAi pathway in mammals, but neither paper demonstrated that the small RNAs detected mediated viral cleavage in an antiviral fashion (Li et al., 2013). Furthermore, both papers utilize multifunctional virus antagonists (B2 of Nodavirus and VP35 of Ebola), which have been shown to profoundly impact the mammalian host response to virus independently of small RNAs (Petrillo et al., 2013; Prins et al., 2010). Given the importance of this question, we sought to formally evaluate the mammalian response to virus to determine whether RNAi was a physiological contributor to this process. To this end, we generated two virus model systems using a vaccine strain of VSV that is exquisitely sensitive to both IFN and RNAi (Mueller et al., 2010; Vogel and Fertsch, 1987; Wilkins et al., 2005). For this, we inserted the only known mammalian virus protein capable of degrading small RNAs loaded into RISC (VACV VP55) or a RIG-I antagonist of IAV (IAV NS1). Insertion of VP55 or NS1 into VSV provided us a unique tool to ascertain the physiological role of small RNAs or IFN, respectively. Interestingly, a VSV strain capable of eliminating host small RNAs was found to replicate at a diminished capacity as compared to control virus in contrast to VSV-NS1 which replicated to levels that exceeded one log over control. While these results argue against a role for RNAi in mammals, it is in agreement with a recent publication concerning virus-induced polyribosylation and shut down of RISC, which also would argue against RNAi functioning in mammals during times of stress (Leung et al., 2011; Seo et al., 2013). Transcriptome profiling of VSV-VP55 and VSVctrl infections support the idea that loss of RISC function results in a significant increase in a broad range of transcripts including a subset of ISGs. It should be noted that this data does not suggest the loss of miRNAs leads to a specific upregulation in this class of genes, but rather that in the context of infection, changes in ISG levels would be most prominent. Lastly, to determine whether RNAi perhaps contributed a secondary response to IFN that would be masked in the context of wild type mice, we also tested our engineered viruses in mice lacking both Type I and III IFN systems. These results also demonstrated all viruses replicated to comparable levels suggesting that the small virus-derived RNAs that can be detected in vivo, are unlikely to contribute to the antiviral response under physiological conditions.
While proving the absence of a biological activity is difficult, we feel our results put forth a provocative argument against a physiological RNAi antiviral response in mammals. In addition to the results presented here, there are other studies that suggest a lack of small RNA silencing in our antiviral defenses. Perhaps the greatest support for this idea comes from the fact that viruses can be targeted through the exploitation of host miRNAs (tenOever, 2013). This technique, which has been employed by countless groups and on a wide range of viruses, demands that RISC function be intact during infection, at least for the initial hours proceeding viral transcription. Should RNAi in mammals be detrimental to virus infection, one would postulate that this aspect of RISC function would be antagonized by viruses as is the case for IFN. In addition to this anecdotal support, there are other aspects of mammalian biology that suggest RNAi is unlikely to contribute to the antiviral response. First, it has been demonstrated that expression of an RdRp in mammalian cells is sufficient for the induction of IFN-I in the absence of infection (Yu et al., 2012). RdRp function in plants and worms is used to generate and amplify vsiRNAs (Carthew and Sontheimer, 2009; Schwach et al., 2005). While flies maintain an RNAi response in the absence of vsiRNA amplification or an RdRp, they compensate for this by modifying their small RNAs via 2' O-methylation to extend half-life and use these in a systemic manner (Ameres et al., 2010; Saleh et al., 2009). However, in contrast to flies and plants, mammals only utilize this chemistry on germline encoded small RNAs (Aravin and Hannon, 2008). Taken together, these studies all suggest that the evolution of chordates involved a dramatic modification of how our cells respond to virus infection.
It is noteworthy however that while RNAi does not perform a physiological role in the antiviral response, this lack of interplay provides us with some valuable resources. One example of such a resource is the VSV-VP55 virus used in these studies. Transcriptome profiling of infected cells allowed us to accurately map the miRNA targetome in infected cells from an in vivo infection. The addition of VP55 to more inert vectors, like a lentivirus or adenovirus, may also be useful for similar studies. Furthermore, the lack of interplay between small RNAs and mammalian viruses allows for the exploitation of the small RNA pathway to control tropism, produce vaccines, or create layers of biocontainment (Lauring et al., 2010; tenOever, 2013). In addition, the lack of interplay between mammalian viruses and the small RNA machinery also makes these vectors ideal for the delivery of small RNAs (Schmid et al., 2014). The delivery of siRNAs is a critical need to the medical field and the recent discovery that RNA viruses can also be engineered to encode siRNAs and utilize the small RNA host machinery makes this a provocative option for future therapeutics.
Experimental Procedures
Cells, Viruses and Infections
Vero, BHK, RAW, murine wildtype fibroblasts (MEF) and murine dicer-deficient cells (DCR−/−) were grown at 37°C in DMEM media supplemented with 10% Fetal Bovine Serum and 1% penicillin-streptomycin. Dicer-deficient cells were a kind gift from Dr. P. Sharp, Massachusetts Institute of Technology, USA (Calabrese et al., 2007). For the generation of Bone Marrow Macrophages, femurs were removed from naive mice and bone marrow cells were cultured for a minimum of 10 days in RPMI containing 20% (vol/vol) FBS, L-glutamine, and 30% (vol/vol) conditioned media from L929 cells. VSV142T and VSVscbl were generated as previously described (Kelly et al., 2008). VSV expressing Vaccinia virus (VACV) VP55 (VSV-VP55), Influenza A virus NS1 (VSV-NS1) and a same size RNA insert not encoding for an open reading frame (VSVctrl) were generated and rescued as previously described (Stojdl et al., 2003). Generation of VSV expressing miR-124 has been described elsewhere (Langlois et al., 2012a). Infections with VSV (strain Indiana), VACV (strain Western Reserve), Sindbis virus and Influenza A virus (A/PR/8/34) were performed at the indicated multiplicity of infection (MOI) at 37°C.
In vivo Infections
C57BL/6 mice were purchased from Taconic. Ifnar1−/−/Il28r−/− mice have been described elsewhere (Mordstein et al., 2008). Mice were anesthetized with isofluorane and infected intranasally with 1×104 (Ifnar1−/−/Il28r−/−) or 1×107 (C57BL/6) plaque-forming units (pfu) of VSV. Lungs and spleens were removed on the indicated days post infection and homogenized in 500μl PBS for analysis by plaque assay or in 1mL Trizol for analysis by Northern blot and small RNA deep sequencing. All experiments involving animals were done in accordance with Mount Sinai School of Medicine Institutional Animal Care and Use Committee.
Western Blot and small RNA Northern Blot
Western blots were generated from total protein separated on a 12% SDS-PAGE gel. Resolved protein was transferred to nitrocellulose (Bio-Rad), blocked for 1 h with 5% skim milk at 25°C, and then incubated with the indicated antibody overnight at 4°C. The polyclonal VSV antibody (a ki nd gift from Dr. J. Bell, University of Ottawa, Canada) and Vaccinia virus (VACV) VP55 antibody (kind gift from Dr. R. Condit, University of Florida, Gainesville) were used at a 1:5000 dilution. The VACV E3L antibody (BEI Resources, NR-4547), the Influenza A virus NS1 antibody (MSSM Hybridoma Center:#1A7) and anti pan-actin antibody (Neomarkers) were used at a 1:2000 dilution. Secondary mouse and rabbit antibodies (GE Healthcare) were used at a 1:5000 dilution for 1 h at 25°C. All Antibodies were diluted in 5% skim milk. Immobilon Western Chemoluminescent HRP substrate (Millipore) was used as directed.
Small RNA northern blots and probe labeling were performed as described previously (Pall and Hamilton, 2008). Probes used include anti-miR-124: 5'-TGGCATTCACCGCGTGCCTT AA-3', anti-U6: 5'-GCCATGCTAATCTTCTCTGTATC-3', anti-miR-146: 5'-AACCCATGGAATTCAGTTCTCA-3', anti-miR-155: 5'-ACCCCTATCACAATTAGCATTAA-3', anti-miR-142: 5'-TCCATAAAGTAGGAAACACTACA -3', anti-miR-93: 5'-CTACCTGCACGAACAGCACTT TG-3', anti-miR-21: 5'-TCAACATCAGTCTGATAAGCTA-3', anti-miR-24: 5'-CTGTTCCTGCTGAACTGAGCCA-3', anti-VSV leader RNA: 5'-GTTTCTCCTGAGCCTTTTAATGATAATAATGGTTTGTTTGTCTTCGT -3', anti-VSV318: 5'- TCCGAAACTTGACCAATCTTTA-3', anti-VSV11161: 5'-TATCTGGTTTTGTGGTCTTCGT-3'.
VSV Plaque Assay
VSV was inoculated into indicated cell lines containing serum-free DMEM for 1 h. Inoculum was then aspirated off and replaced with complete medium for the indicated times. Briefly, cells were infected with VSV (MOI as indicated) and 0.25 mL of supernatant was removed at the indicated times. Supernatant was plaqued in Vero cells in serial dilutions in triplicate in 1% methylcellulose. Plaques were counted after 3 d post-infection. P-values were calculated based on a two-tailed t-test. Error bars represent the standard deviation (n=3).
Quantitative Reverse Transcriptase-PCR
Quantitative reverse transcriptase PCR for VSV M (primers 5'-GCGGTATTGGCAGATCAAGGT-3' and 5'- CCCCATCCTATGTGGCAAAT-3'), IFN-β (primers 5'- AGATGTCCTCAACTGCTCTC-3' and 5'-AGATTCACTACCAGTCCCAG-3'), PTGS2 (primers 5'-GTCAGGACTCTGCTCACGAA-3' and 5'-AGGATTTGCTGCCTGGCTGA-3', Ddx60 (primers 5'- GTCTCCTGTGGTCGACTGTG-3' and 5'-AATGTCGTATCGGGAAGCCC-3'), Rsad2 (primers 5'-TGGCCGTGGTCAAGGAAAAA-3' and 5'- GGAAAACCTTCCAGCGCAC-3') and MX1 (primers 5'-ACCTCCCACATCTGTAAATCACT-3' and 5'-GTATGTCTGCACCGTACTTCTG-3') of complementary DNA samples was performed using KAPA SYBR FAST qPRC Master Mix (KAPA Biosystems, Boston, MA). PCRs were performed on a Mastercycler ep realplex (Eppendorf). Murine tubulin (primers 5'-TGCCTTTGTGCACTGGTATG-3 and 5'-CTGGAGCAGTTTGACGACAC-3') was used as the endogenous housekeeping gene and Delta delta cycle threshold (ΔΔCT) values were calculated with replicates over tubulin. Values represent the fold change over mock-infected samples.
RNA Interference and Transfections
Chemically unmodified anti VSV-NP siRNA (IDT; 5'-TTTCCCGATGTTTATTCC-3') was transfected into BHK cells in suspension using RNAiMAX (Invitrogen). Cells were infected with VSV 6 hrs posttransfection and harvested 10 hrs postinfection. Plasmids were transfected into cells in suspension using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Small RNA Deep Sequencing and mRNA-seq analysis
Deep sequencing was performed on wild-type murine fibroblasts (18–25 nt fraction) infected with VSV, Sindbis virus or Influenza A virus at an MOI of 1 for 12hours, on C6 glial cells persistently infected with Borna disease virus (He/80) (18–25 nt fraction) or on mouse lungs infected with VSVctrl or VSV-VP55 (18–25 nt fraction). Isolation, purification, and amplification of small RNA species were performed as previously described (Langlois et al., 2012b). Small RNA libraries were generated as previously described (Pfeffer et al., 2004). Briefly, total RNA from indicated samples was isolated using TRIzol (Invitrogen) and spiked with radiolabeled size markers prior to size fractionation on a 12% denaturing Trisurea gel (SequaGel, National Diagnostics). RNA was separated by electrophoresis on a 15% TBE-urea gel, and RNA molecules •17–26 nt were excised and eluted from the gel fragments. Following ethanol precipitation, smRNA-seq libraries were produced using the Small RNA Sample Prep v1.5 kit (Illumina, San Diego, CA) as per manufacturer's instructions. RNA sequencing was performed on Bone-marrow derived macrophages infected with VSV at an MOI of 5 for 10hrs. RNA extracts were prepared using standard mRNA-seq protocols (Illumina, Cat. No1502062). Briefly, mRNA was isolated from one microgram of RNA using sera oligo-dT beads. This was then used for cDNA synthesis with SuperScript II reverse transcriptase (Life Technologies, Grand Island, NY). This is followed by second strand synthesis, end-repair, A-tailing, ligation and PCR using the Illumina Truseq kit. Amplification of the cDNA library is checked using the Bioanalyzer DNA 1000 Assay. mRNA-seq libraries were clustered with cBOT (Illumina) and then run on HiSeq (Illumina) for 100 base single read sequencing.
Statistical analysis
Statistical analysis was performed on indicated samples using a two-tailed, unpaired Students-t test. Data are considered significant if P value is <0.05.
Supplementary Material
Highlights.
Direct comparison of IFN and small RNAs in the mammalian response to virus infection.
Engineering of an RNA virus to selectively degrade RISC-bound small RNAs
Comprehensive analysis of the virus-induced, RISC-mediated, targetome
Characterization of the antiviral response in the absence of IFN and/or RISC function
Acknowledgements
This material is based upon work supported in part by the US Army Research Laboratory and the US Army Research Office under grant numbers W911NF-12-R-0012 and W911NF-07-R-0003. SB is supported by Deutsche Forschungsgemeinschaft DFG BA 4878/1-1. RAL is supported by the Research Training Program in Molecular and Cellular Hematology (T32-HL094283). BRtO is supported in part through a Burroughs Wellcome Fund for Investigators in the Pathogenesis of Infectious Disease. We also acknowledge the Mount Sinai Genomics Core Facility for deep sequencing analyses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283–290. doi: 10.1101/sqb.2008.73.058. [DOI] [PubMed] [Google Scholar]
- Backes S, Shapiro JS, Sabin LR, Pham AM, Reyes I, Moss B, Cherry S, tenOever BR. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe. 2012;12:200–210. doi: 10.1016/j.chom.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 2008;4:239–248. doi: 10.1016/j.chom.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol. 2004;85:983–991. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawood R, Chen HH, Carroll F, Bazan-Peregrino M, van Rooijen N, Seymour LW. Use of tissue-specific microRNA to control pathology of wild-type adenovirus without attenuation of its ability to kill cancer cells. PLoS Pathog. 2009;5:e1000440. doi: 10.1371/journal.ppat.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Chou YT, Tam B, Linay F, Lai EC. Transgenic inhibitors of RNA interference in Drosophila. Fly (Austin) 2007;1:311–316. doi: 10.4161/fly.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua MA, Schmid S, Perez JT, Langlois RA, Tenoever BR. Influenza A virus utilizes suboptimal splicing to coordinate the timing of infection. Cell Rep. 2013;3:23–29. doi: 10.1016/j.celrep.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR, Cherry S, tenOever BR. Is RNA interference a physiologically relevant innate antiviral immune response in mammals? Cell Host Microbe. 2013;14:374–378. doi: 10.1016/j.chom.2013.09.011. [DOI] [PubMed] [Google Scholar]
- Delgadillo MO, Saenz P, Salvador B, Garcia JA, Simon-Mateo C. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J Gen Virol. 2004;85:993–999. doi: 10.1099/vir.0.19735-0. [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaszi-Ivanov A, Mohorianu I, Dalmay T, Powell PP. Small RNA analysis in Sindbis virus infected human HEK293 cells. PLoS One. 2013;8:e84070. doi: 10.1371/journal.pone.0084070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge RE, Falls TJ, Brown CW, Lichty BD, Atkins H, Bell JC. A let-7 MicroRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication. Mol Ther. 2008;16:1437–1443. doi: 10.1038/mt.2008.130. [DOI] [PubMed] [Google Scholar]
- Fabozzi G, Nabel CS, Dolan MA, Sullivan NJ. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J Virol. 2011;85:2512–2523. doi: 10.1128/JVI.01160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Girardi E, Chane-Woon-Ming B, Messmer M, Kaukinen P, Pfeffer S. Identification of RNase L-dependent, 3'-end-modified, viral small RNAs in Sindbis virus-infected mammalian cells. MBio. 2013;4:e00698–00613. doi: 10.1128/mBio.00698-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Shao L, Zheng L, Du Q, Li P, John B, Geller DA. miRNA-939 regulates human inducible nitric oxide synthase posttranscriptional gene expression in human hepatocytes. Proc Natl Acad Sci U S A. 2012;109:5826–5831. doi: 10.1073/pnas.1118118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. RNAi: nature abhors a double-strand. Curr Opin Genet Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Junt T, Moseman EA, Iannacone M, Massberg S, Lang PA, Boes M, Fink K, Henrickson SE, Shayakhmetov DM, Di Paolo NC, et al. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 2007;450:110–114. doi: 10.1038/nature06287. [DOI] [PubMed] [Google Scholar]
- Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- Kelly EJ, Nace R, Barber GN, Russell SJ. Attenuation of vesicular stomatitis virus encephalitis through microRNA targeting. J Virol. 2010;84:1550–1562. doi: 10.1128/JVI.01788-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C, Imler JL. Antiviral immunity in drosophila. Curr Opin Immunol. 2009;21:3–9. doi: 10.1016/j.coi.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RA, Albrecht RA, Kimble B, Sutton T, Shapiro JS, Finch C, Angel M, Chua MA, Gonzalez-Reiche AS, Xu K, et al. MicroRNA-based strategy to mitigate the risk of gain-of-function influenza studies. Nat Biotechnol. 2013;31:844–847. doi: 10.1038/nbt.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RA, Shapiro JS, Pham AM, tenOever BR. In vivo delivery of cytoplasmic RNA virus-derived miRNAs. Mol Ther. 2012a;20:367–375. doi: 10.1038/mt.2011.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RA, Varble A, Chua MA, Garcia-Sastre A, tenOever BR. Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proc Natl Acad Sci U S A. 2012b;109:12117–12122. doi: 10.1073/pnas.1206039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring AS, Jones JO, Andino R. Rationalizing the development of live attenuated virus vaccines. Nat Biotechnol. 2010;28:573–579. doi: 10.1038/nbt.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ru J, Zhang J, Zhu LH, Liu M, Li X, Tang H. miR-23 a targets interferon regulatory factor 1 and modulates cellular proliferation and paclitaxel-induced apoptosis in gastric adenocarcinoma cells. PLoS One. 2013a;8:e64707. doi: 10.1371/journal.pone.0064707. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu Z, Wang D, Hu Y, Zhou G, Zhu C, Yu Q, Chi Y, Cao Y, Jia C, Zou Q. MicroRNA-146a negatively regulates PTGS2 expression induced by Helicobacter pylori in human gastric epithelial cells. J Gastroenterol. 2013b;48:86–92. doi: 10.1007/s00535-012-0609-9. [DOI] [PubMed] [Google Scholar]
- Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez RT, Wendlandt E, Galle CS, Keck K, McCaffrey AP. MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets Pellino-1, and inhibits NF-kappaB signaling. Am J Physiol Gastrointest Liver Physiol. 2010;298:G535–541. doi: 10.1152/ajpgi.00338.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr., Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol. 2008;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Gausson V, Vodovar N, Deddouche S, Troxler L, Perot J, Pfeffer S, Hoffmann JA, Saleh MC, Imler JL. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc Natl Acad Sci U S A. 2010;107:19390–19395. doi: 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A, Berry B, Tassetto M, Kunitomi M, Acevedo A, Deng C, Krutchinsky A, Gross J, Antoniewski C, Andino R. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol. 2010;17:547–554. doi: 10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A, Tassetto M, Kunitomi M, Andino R. RNA interference-mediated intrinsic antiviral immunity in invertebrates. Curr Top Microbiol Immunol. 2013;371:183–200. doi: 10.1007/978-3-642-37765-5_7. [DOI] [PubMed] [Google Scholar]
- Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, et al. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010;6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JT, Pham AM, Lorini MH, Chua MA, Steel J, tenOever BR. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat Biotechnol. 2009;27:572–576. doi: 10.1038/nbt.1542. [DOI] [PubMed] [Google Scholar]
- Petrillo JE, Venter PA, Short JR, Gopal R, Deddouche S, Lamiable O, Imler JL, Schneemann A. Cytoplasmic granule formation and translational inhibition of nodaviral RNAs in the absence of the double-stranded RNA binding protein B2. J Virol. 2013;87:13409–13421. doi: 10.1128/JVI.02362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Pham AM, Langlois RA, TenOever BR. Replication in cells of hematopoietic origin is necessary for Dengue virus dissemination. PLoS Pathog. 2012;8:e1002465. doi: 10.1371/journal.ppat.1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Prins KC, Delpeut S, Leung DW, Reynard O, Volchkova VA, Reid SP, Ramanan P, Cardenas WB, Amarasinghe GK, Volchkov VE, et al. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J Virol. 2010;84:3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi N, Zhang L, Qiu Y, Wang Z, Si J, Liu Y, Xiang X, Xie J, Qin CF, Zhou X, et al. Targeting of dicer-2 and RNA by a viral RNA silencing suppressor in Drosophila cells. J Virol. 2012;86:5763–5773. doi: 10.1128/JVI.07229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff FG, MacFarlane SA, Baulcombe DC. Gene silencing without DNA. rna-mediated cross-protection between viruses. Plant Cell. 1999;11:1207–1216. doi: 10.1105/tpc.11.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch I, Muller M, Decker T. The regulation of inflammation by interferons and their STATs. JAKSTAT. 2013;2:e23820. doi: 10.4161/jkst.23820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin LR, Zheng Q, Thekkat P, Yang J, Hannon GJ, Gregory BD, Tudor M, Cherry S. Dicer-2 processes diverse viral RNA species. PLoS One. 2013;8:e55458. doi: 10.1371/journal.pone.0055458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S, Zony LC, tenOever BR. A versatile RNA vector for delivery of coding and noncoding RNAs. J Virol. 2014;88:2333–2336. doi: 10.1128/JVI.03267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, Hsiang TY, Krug RM, Sullivan CS. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe. 2013;14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee SP, Siragam V, et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;11:1031–1038. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- Shapiro JS, Schmid S, Aguado LC, Sabin LR, Yasunaga A, Shim JV, Sachs D, Cherry S, Tenoever BR. Drosha as an interderon-independent antiviral factor. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1319635111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JS, Langlois RA, Pham AM, Tenoever BR. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA. 2012;18:1338–1346. doi: 10.1261/rna.032268.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JS, Varble A, Pham AM, Tenoever BR. Noncanonical cytoplasmic processing of viral microRNAs. RNA. 2010;16:2068–2074. doi: 10.1261/rna.2303610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever BR. RNA viruses and the host microRNA machinery. Nat Rev Microbiol. 2013;11:169–180. doi: 10.1038/nrmicro2971. [DOI] [PubMed] [Google Scholar]
- Thomas M, Lange-Grunweller K, Weirauch U, Gutsch D, Aigner A, Grunweller A, Hartmann RK. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene. 2012;31:918–928. doi: 10.1038/onc.2011.278. [DOI] [PubMed] [Google Scholar]
- Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varble A, Benitez AA, Schmid S, Sachs D, Shim JV, Rodriguez-Barrueco R, Panis M, Crumiller M, Silva JM, Sachidanandam R, et al. An in vivo RNAi screening approach to identify host determinants of virus replication. Cell Host Microbe. 2013;14:346–356. doi: 10.1016/j.chom.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Varble A, Chua MA, Perez JT, Manicassamy B, Garcia-Sastre A, tenOever BR. Engineered RNA viral synthesis of microRNAs. Proc Natl Acad Sci U S A. 2010;107:11519–11524. doi: 10.1073/pnas.1003115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel SN, Fertsch D. Macrophages from endotoxin hyporesponsive (Lpsd) C3H/HeJ mice are permissive for vesicular stomatitis virus because of reduced levels of endogenous interferon: possible mechanism for natural resistance to virus infection. J Virol. 1987;61:812–818. doi: 10.1128/jvi.61.3.812-818.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Kochs G, Haller O, Staeheli P. Viral evasion of the interferon system: old viruses, new tricks. J Interferon Cytokine Res. 2003;23:209–213. doi: 10.1089/107999003765027410. [DOI] [PubMed] [Google Scholar]
- Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- Yu GY, He G, Li CY, Tang M, Grivennikov S, Tsai WT, Wu MS, Hsu CW, Tsai Y, Wang LH, et al. Hepatic expression of HCV RNA-dependent RNA polymerase triggers innate immune signaling and cytokine production. Mol Cell. 2012;48:313–321. doi: 10.1016/j.molcel.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.