Abstract
Glycogen synthase kinase 3 beta (GSK3β) is highly inactivated in epithelial cancers and is known to inhibit tumor migration and invasion. The zinc-finger-containing transcriptional repressor, Slug, represses E-cadherin transcription and enhances epithelial-mesenchymal transition (EMT). In this study, we find that the GSK3β-pSer9 level is associated with the expression of Slug in non-small cell lung cancer (NSCLC). GSK3β-mediated phosphorylation of Slug facilitates Slug protein turnover. Proteomic analysis reveals that the C-terminus of Hsc70-interacting protein (CHIP) interacts with wild-type Slug (wtSlug). Knockdown of CHIP stabilizes the wtSlug protein and reduces Slug ubiquitylation and degradation. In contrast, nonphosphorylatable Slug-4SA is not degraded by CHIP. The accumulation of nondegradable Slug may further lead to the repression of E-cadherin expression and promote cancer cell migration, invasion, and metastasis. Our findings provide evidence of a de novo GSK3β-CHIP-Slug pathway that may be involved in the progression of metastasis in lung cancer.
Keywords: GSK3β, Slug, CHIP, post-translational modification
Introduction
Lung cancer is associated with a high metastatic rate, making it the leading cause of cancer deaths worldwide.1 Metastasis complicates therapeutic treatment as it transforms a locally growing tumor into a systemic disease that is resistant to existing therapeutic agents.2–4 In the first step of metastasis, which is termed invasion, tumor cells dissociate and migrate from the primary tumors.5 Such a process is controlled by multiple cellular and molecular signaling pathways that together change cell adhesion and migration. This change is known as the epithelial-mesenchymal transition (EMT).5, 6
E-cadherin-mediated intercellular junctions tightly knit epithelial cells together and prevent them from dissociating. Therefore, suppression of E-cadherin is critical for EMT and is associated with the development of malignant epithelial cancers.7–10
A fundamental mechanism that downregulates the expression of E-cadherin during tumor progression is transcriptional repression.5 Slug (also known as SNAI2), a zinc-finger transcriptional repressor of the Snail family, has been shown to repress E-cadherin expression and trigger EMT.11, 12 Recently, we have demonstrated the importance of the Slug-E-cadherin axis in non-small-cell lung cancers (NSCLCs) where aberrant upregulation of Slug promotes cancer invasion and malignant transformation.13–15 However, less is known about the regulation of Slug by post-translational modification in lung cancer.16 Hence, it is of critical interest to investigate the scenario in which Slug is post-translationally regulated. To date, phosphorylation and ubiquitylation have been shown to modify numerous transcriptional repressors, such as Snail, NF-κB, and cAMP response element binding protein (CREB), etc.17–19 We postulated that Slug, as a labile protein, may undergo phosphorylation-dependent degradation as well.
CHIP (the carboxyl terminus of Hsc70-interacting protein) is a U-box-type ubiquitin ligase that was originally identified to be associated with Hsp70 and Hsp90 chaperones, and to target misfolded proteins for degradation.20–23 Accumulating evidence has suggested that CHIP also regulates oncogenic pathways as it induces ubiquitylation and degradation of several oncogenic proteins, such as cystic fibrosis transmembrane conductance regulator (CFTR), estrogen receptor (ER), and ErbB2.24–27 Some of the substrates require phosphorylation before degradation.28 Phosphorylated tau, for example, is mainly ubiquitylated by CHIP and overexpression of CHIP can rescue phosphorylated tau-induced cell death.29, 30
The serine/threonine kinase, GSK3β, is involved in regulating the stability of various oncogenic transcriptional factors by phosphorylation.31–36 Signaling pathways that inactivate GSK3β, such as PI3K/Akt and MAPK, may promote the cell cycle, anti-apoptosis, and invasion, thus facilitating tumor progression.37–41 Indeed, inactivation of GSK3β has been observed in various tumors.42–45 Here by proteomic study, we show that Slug proteins can be post-translationally modified by GSK3β and later ubiquitylated by the E3 ligase, CHIP, for proteasomal degradation in lung cancer. Dysregulation of the GSK3β-CHIP-Slug pathway may lead to the accumulation of nondegradable Slug and display higher invasive capabilities, which may promote cancer metastasis.
Results
The activity of GSK3β inversely correlates with Slug protein levels
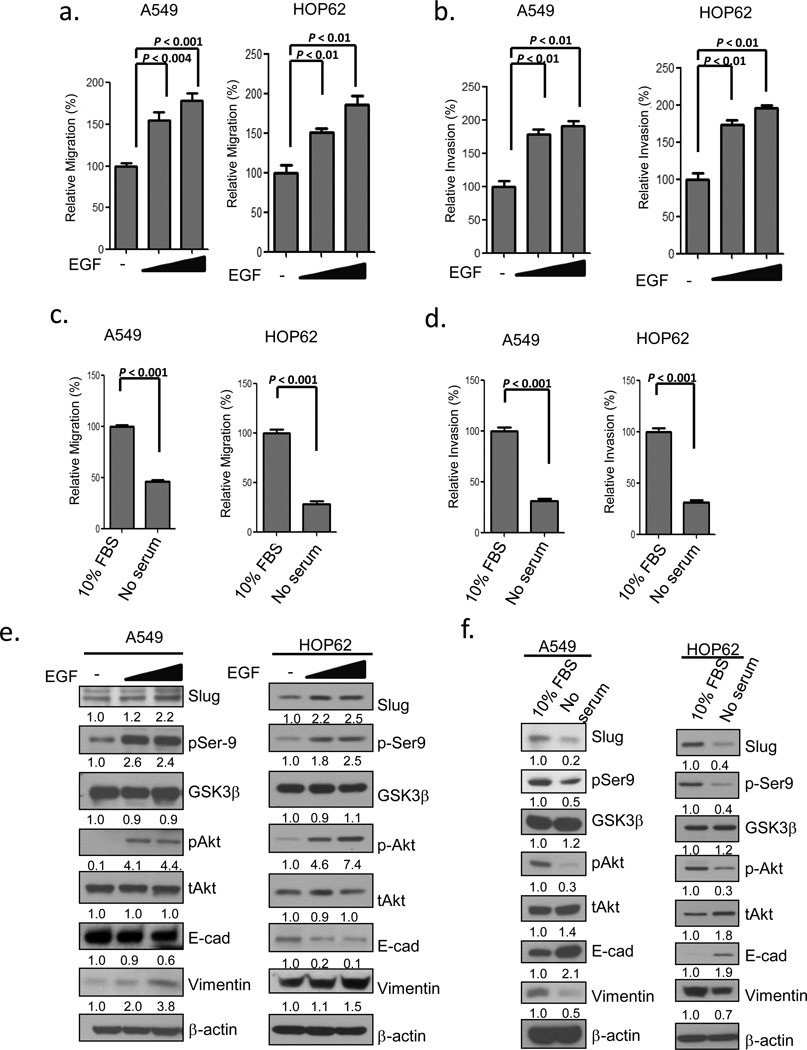
To elucidate whether the activity of GSK3β correlates with Slug expression, we treated A549 cells and HOP62 cells with recombinant epidermal growth factor (EGF) or serum-free medium and analyzed Slug levels by immunoblotting. It has been indicated that Ser9 phosphorylation renders GKS3β an inactive kinase and unphosphorylation, an active one.46 We found that the treatment of elevated EGF caused the suppression of GSK3β (through phosphorylation of Ser9) by the activation of Akt and a marked increase in Slug protein levels (Figure 1e). The reduction in E-cadherin and upregulation of vimentin were accompanied by a spindle-like morphological change (Figure 1e; Supplementary Figure 1a, upper panel), increased migration and invasion (Figure 1a-b). Blockage of the EGF-activated signaling pathways with an Akt inhibitor, LY294002, reduced Ser9 phosphorylation on GSK3β, leading to the elevated activity of GSK3β the decreased Slug protein levels (Supplementary Figure 1c, lane 3), and the retraction of filopodia (Supplementary Figure 1b). To investigate whether the GSK3β activity is required for the EGF-mediated effects on cell morphology and Slug destabilization, we added GSK3β inhibitor VIII in EGF-treated cells and found that the Slug protein levels were accumulated (Supplementary Figure 1c, lane 4) while cells morphologically maintained an elongated shape (Supplementary Figure 1b). Conversely, serum withdrawal induced the activation of GSK3β and a conspicuous decrease in Slug protein levels (Figure 1f). Serum-starved cells aggregated locally (Supplementary Figure 1a, lower panel) and manifested reduced migration and invasion (Figures 1c-d). The inverse correlation between the GSK3β activity and Slug protein levels was also observed in another lung adenocarcinoma cell line (CL1-5) (Supplementary Figures 1d-f). These data indicate that mitogen-stimulated pathways can regulate the activity of GSK3β and influence cell migration and invasion. We inferred that the inverse correlation between the activity of GSK3β and the expression of Slug suggests that GSK3β may in turn regulate the expression of Slug.
Figure 1.
The activity of GSK3β inversely correlates with Slug protein levels. (a-b) EGF induces cell migration and invasion. A549 and HOP62 cells were cultured in serum-free medium for 12 h with or without EGF treatment for 6 h. Cell migration (a) and invasion (b) were determined by the Transwell assay at 6 h or 12 h after treatment. (c-d) Serum starvation reduces cell migration and invasion. A549 and HOP62 cells were cultured in DMEM or RPMI medium supplemented with 10% FBS or in serum-free medium for 6 h. Cell migration (c) and invasion (d) were determined by the Transwell assay at 6 h or 12 h after treatment. Data are shown as mean ± s.e.m. in three different experiments (n=3). (e-f) Correlation between the Slug protein level and the activity of GSK3β. Cell lysates of the experiments in (a-d) were analyzed by Western blotting.
GSK3β-mediated Slug phosphorylation affects Slug protein turnover
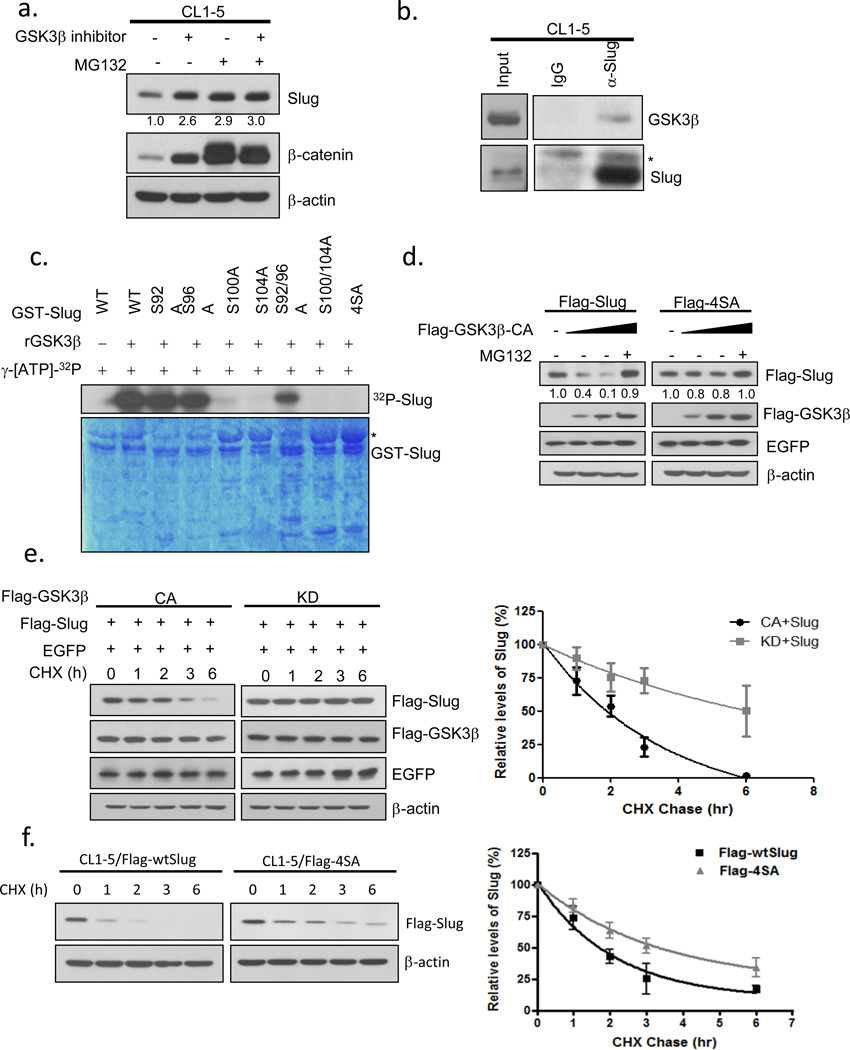
To further investigate whether GSK3β regulates the turnover of Slug, CL1-5 cells were treated with GSK3β inhibitor VIII with or without MG132, an inhibitor of the 26S proteasome. Both GSK3β and proteasome inhibition resulted in increased Slug steady-state protein levels (Figure 2a). Ectopically-produced Flag-tagged Slug also accumulated in cells treated with the GSK3β inhibitor (Supplementary Figure 2a), suggesting that Slug may be regulated by GSK3β in a transcription-independent manner. In confirmation of this observation, GSK3β-specific shRNAs were transduced into cells and the knockdown of GSK3β stabilized endogenous Slug proteins without a significant change in Slug mRNA levels (Supplementary Figures 2b-e). Although GSK3α and GSK3β are structurally similar, we did not detect any effect of GSK3αon Slug using GSK3α-specific shRNAs (Supplementary Figures 2g-h). These data suggest that Slug may be regulated by GSK3β-dependent post-translational modification and later targeted for proteasome degradation.
Figure 2.
GSK3β phosphorylates Slug and modulates its protein stability. (a) Inhibition of GSK3β and proteasome pathways stabilizes Slug. CL1-5 cells were treated with or without 10 μM MG132 or 25 μM GSK3β inhibitor VIII, or both, for 6 h and analyzed by Western blotting. (b) Interaction of endogenous GSK3β and Slug. CL1-5 cells incubated with MG132 for 6 h and crosslinked were lysed and either analyzed directly by Western blotting or subjected to immunoprecipitation with control and anti-Slug antibodies. The asterisk indicates a non-specific band. (c) GSK3β phosphorylates Slug at serine-92, -96, -100, and -104. Recombinant GSK3β (50U) was incubated with glutathione S-transferase (GST)-Slug fusion proteins (GST-WT, -92A, -96A, -100A, -104A, -92/96A, -100/104A, -4SA) in the presence of γ-[32P]-ATP. The protein kinase reaction products were resolved by SDS-PAGE, and phosphorylation was detected by autoradiography. The asterisk indicates a non-specific band. (d) Lysates were prepared from CL1-5 cells transfected with plasmids expressing Flag-wtSlug or Flag-4SA in the presence or absence of increasing amounts of Flag-tagged constitutively-active GSK3β for 24 h. A plasmid encoding EGFP was used as a negative control for transfection efficiency. Proteins were analyzed by Western blotting. (e) GSK3β modulates Slug protein stability. H1299 cells were transfected with the Flag-Slug construct together with the indicated Flag-GSK3β-expressing plasmids. A plasmid encoding EGFP was used as a negative control for transfection efficiency. Twenty-four hours after transfection, the cells were treated with 300 μgml−1 cycloheximide (CHX). At the indicated time points, lysates were prepared and Western blotting analysis was performed with antibodies specific for the indicated proteins (left panel). The Slug band intensity was normalized to EGFP and then normalized to the t=0 controls (right panel). (f) CL1-5 cells were stably transduced with viruses expressing Flag-Slug-WT or Flag-Slug-4SA for 24 h. Cycloheximide treatment and Western blotting (left panel) were conducted as in (e). The Slug band intensity was normalized to actin and then normalized to the t=0 controls (right panel). Data are shown as mean ± s.e.m. in three different experiments (n=3).
Next, we determined whether GSK3β and Slug could form a complex in vivo. We overexpressed GSK3β and Slug proteins in H1299 cells. Interactions between ectopically-expressed GSK3β and Slug were detected in vivo (Supplementary Figure 2i). Complex formation between endogenous Slug and GSK3β was demonstrated by co-immunoprecipitation (Figure 2b). To clarify whether GSK3β mediates Slug phosphorylation directly, we performed the in vitro kinase assay using recombinant GSK3β and GST-tagged Slug purified from Escherichia coli (Supplementary Figure 3a). GSK3β dose-dependently phosphorylated Slug and the phosphorylation sites were further determined by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Supplementary Figure 3b-c). Interestingly, two serine sites were identified that matched the consensus GSK3β phosphorylation motif whereas in silico analysis predicted two more within this region. To confirm the phosphorylation sites, we constructed and purified bacterially- or cellularly-expressed Slug mutants as the substrates in the in vitro kinase assay. We found that phosphorylation on GST-S104A and GST-S100/104A mutants was equivalently reduced while that on GST-S92/96A was obviously retained, indicating that Slug was sequentially phosphorylated by GSK3β (Fig. 2c, Supplementary Discussion). Kinase-refractile substitutions of alanine for serine at positions 92, 96, 100, and 104 (4SA) displayed markedly reduced levels of phosphorylation in vitro, whereas single or double mutations (except for GST-S104A and GST-S100/104A) partially reduced phosphorylation (Figure 2c; Supplementary Figure 3d). We therefore reasoned that the 4SA mutant may better represent the nonphosphorylated status of Slug. Noticeably, phosphorylation of wild-type Slug also occurred in vivo and was enhanced by the co-expression of constitutively-active GSK3β (GSK3β-CA) as determined by the in vivo kinase assay (Supplementary Figure 3e). Slug-4SA was marginally increased in the presence of GSK3β-CA and we speculated that the non-specific phosphorylation in Slug-4SA may have been caused by the robust activity of GSK3β-CA. These data suggest that Slug interacts with GSK3β and is phosphorylated by GSK3β at four sequential sites.
To examine whether GSK3β affects Slug stability, we expressed elevated levels of constitutively-active GSK3β with Slug-WT or Slug-4SA mutant. Expression of wild-type Slug was dose-dependently depleted whereas the 4SA mutant was more resistant to such depletion (Figure 2d). In further confirmation of the negative regulation of Slug by GSK3β, we blocked de novo protein synthesis with cycloheximide in cells overexpressing Flag-tagged Slug with either constitutively-active or kinase-inactive GSK3β and analyzed the amount of Slug protein by immunoblotting. Active GSK3β markedly facilitated Slug protein turnover; however inactive GSK3β did not, as the amount of Slug protein was still abundant at 6 h after cycloheximide treatment (Figure 2e). In support of this, knockdown of GSK3β resulted in an extension of the half-life of endogenous Slug (Supplementary Figure 2f). To determine whether GSK3β enhances Slug protein turnover through phosphorylation, we compared the half-life of wild-type Slug to that of the nonphosphorylatable Slug-4SA. As chased by cycloheximide, the half-life of the 4SA mutant was approximately two times longer than that of wild-type Slug (Figure 2f). Consistent with its longer half-life, Slug-4SA was less ubiquitylated than Slug-WT (Supplementary Figure 2j). These results suggest that the activity of GSK3β limits the intracellular concentration of Slug and modulates Slug protein turnover by direct phosphorylation.
Phosphorylation of Slug by GSK3β promotes CHIP binding and ubiquitin-mediated proteolysis
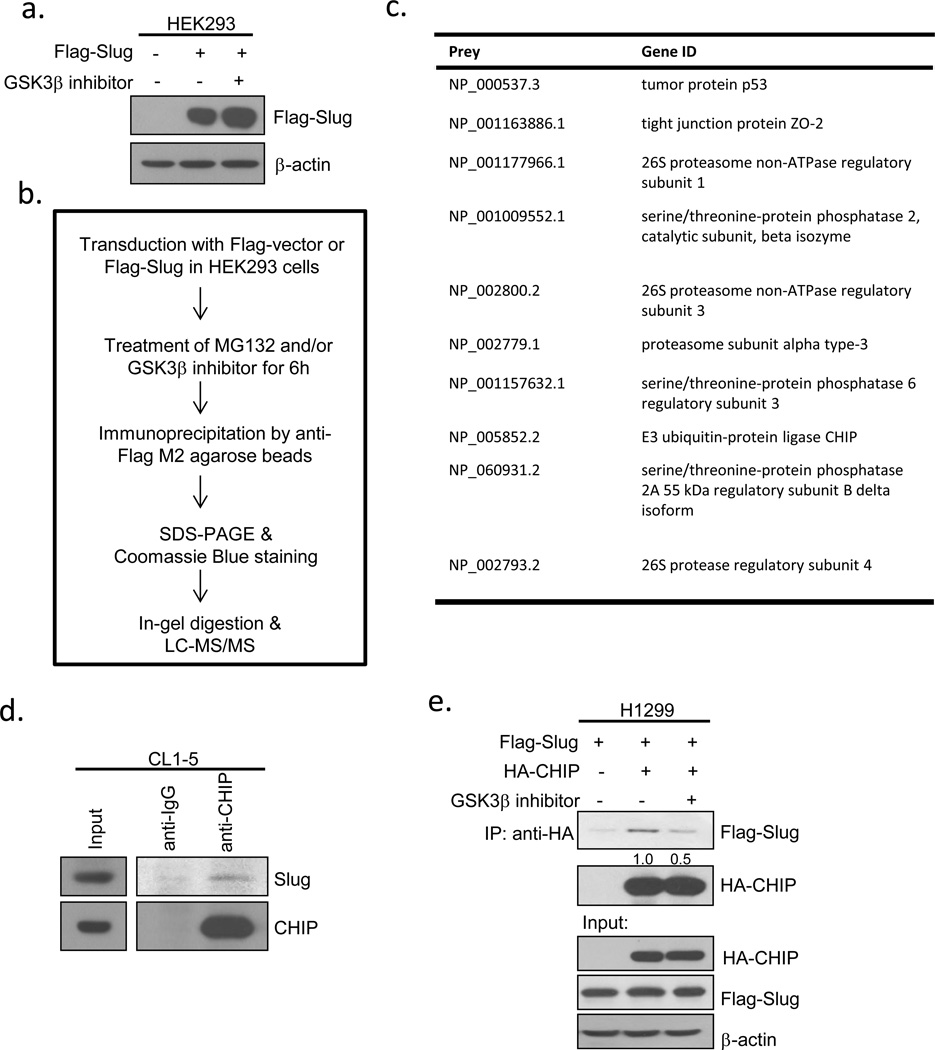
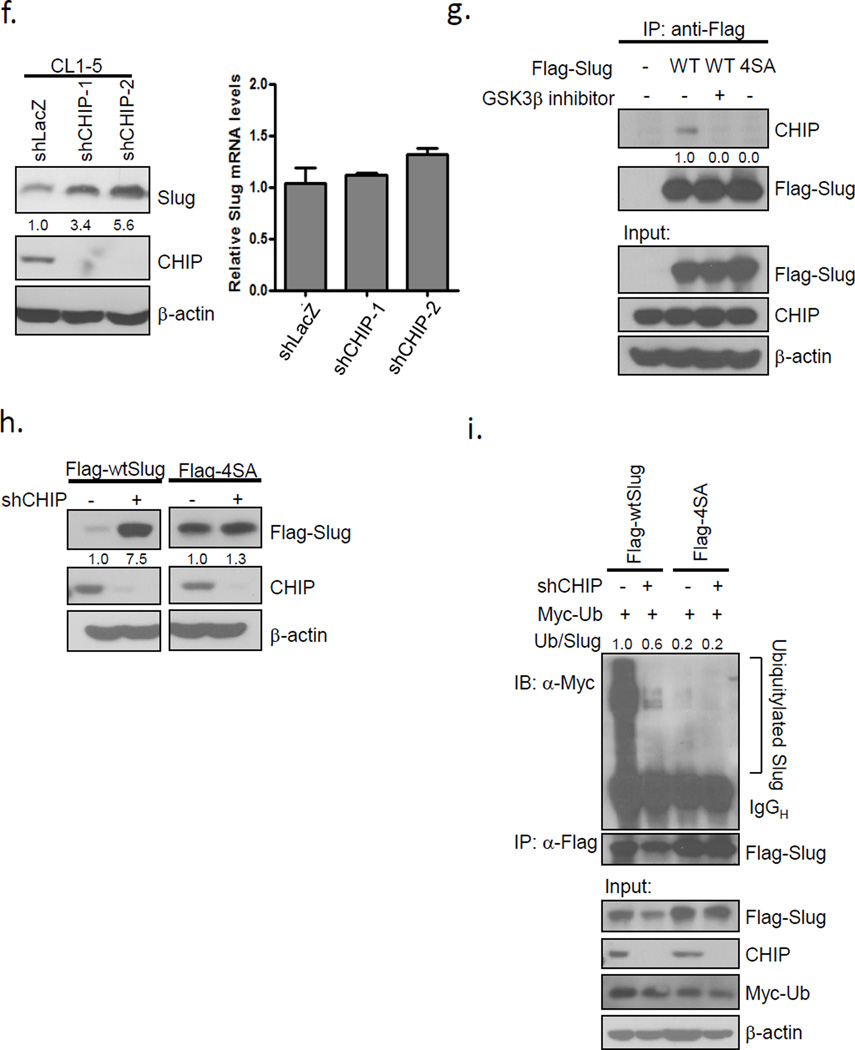
Since Slug is unstable when phosphorylated, we hypothesized that an unknown ubiquitin ligase may regulate Slug turnover. To identify the ligase, we performed co-immunoprecipitation and mass spectrometry using Flag-tagged wild-type Slug in the presence or absence of a GSK3β inhibitor (Figures 3a-b). We used the SAINT method to compute the interaction probabilities among each group47 and the interactors that had a higher association probability in the Slug alone group than that in the Slug+inhibitor group are shown in Figure 3c. We found that Slug without GSK3β phosphorylation inhibition was associated with more subunits of proteasomes after MG132 treatment, indicating that the Slug protein is truly labile by proteasomal degradation (Figure 3c). Among several Slug-interacting proteins, CHIP, a U box-containing E3 ubiquitin ligase implicated in cancer progression and metastasis24, was identified (Figure 3c). To determine whether CHIP and Slug form a complex in vivo, we examined the interaction between endogenous CHIP and Slug in lung cancer cells., The protein complexes involving CHIP and Slug could be detected in CL1-5 cells pre-treated with MG132 (Fig. 3d). Furthermore, the interaction between ectopically-expressed Flag-tagged Slug and HA-tagged CHIP was weakened by GSK3β inhibition (Figure 3e; Supplementary Figure 4a), suggesting that GSK3β-mediated Slug phosphorylation enhances CHIP-Slug interaction. To investigate whether CHIP participates in Slug degradation, we knocked down CHIP by its specific shRNAs in CL1-5 cells transduced by lentiviruses. Depletion of CHIP caused a conspicuous elevation of Slug protein levels without a significant change in Slug mRNA levels (Figure 3f), suggesting that Slug is not transcriptionally regulated by CHIP. To assess whether the inactivation of GSK3β-mediated phosphorylation sites is less associated with CHIP, we expressed Flag-tagged Slug-WT or Slug-4SA in HEK293 cells. The Flag-tagged 4SA mutant abolished the interaction with endogenous CHIP, as did Flag-Slug-WT under the inhibition of GSK3β activity (Figure 3g). Moreover, knockdown of CHIP led to the accumulation of the protein level of ectopically-expressed Flag-Slug-WT, but not that of Flag-Slug-4SA, indicating that CHIP may participate in the GSK3β-mediated turnover of Slug (Figure 3h). A well-known F-box protein, β-trcp, has been reported to play a role in many GSK3β-degraded substrates48. However, we did not observe any significant elevation of endogenous or ectopically-expressed Slug protein levels in cells transduced with β-trcp shRNAs (Supplementary Figures 4b-c). Finally, to analyze whether CHIP is involved in Slug ubiquitylation, we knocked down CHIP by lentiviral infection in cells overexpressing Flag-tagged Slug-WT or Slug-4SA. Loss of CHIP expression led to reduced ubiquitylated Slug-WT compared to the control, whereas Slug-4SA was not affected (Figure 3i), indicating that CHIP-mediated degradation of Slug may depend on phosphorylation. These results demonstrate that CHIP is involved in Slug protein degradation in a GSK3β-mediated phosphorylation-dependent manner.
Figure 3.
The E3 ligase CHIP is involved in GSK3β-mediated phosphorylation-dependent Slug degradation. (a) HEK293 cells were infected with control or Flag-Slug-expressing viruses in the presence or absence of GSK3β inhibitor for 8 h. The Slug expression was assessed by immunoblotting. (b) Schematic representation of the experimental procedure of immunoprecipitation. (c) List of the associated genes of Flag-Slug. The probability of an interaction partner of the wild-type Slug immunoprecipitate was computed and compared against that of the vector control and GSK3β inhibitor using the SAINT method. (d) Slug interacts with CHIP. CL1-5 cells were treated with 10 μM MG132 for 6 h before cell collection. The lysates were immunoprecipitated with anti-CHIP antibodies and resolved by SDS-PAGE. (e) H1299 cells were transfected with the plasmids expressing the indicated proteins, HA-CHIP and Flag-Slug (or the control). Twenty-four hours after transfection, the cells were treated with 10 μM MG132 with or without 25 μM GSK3β inhibitor VIII for 8 h before cell collection. The lysates were immunoprecipitated with anti-HA antibodies and resolved by SDS-PAGE. (f) CHIP knockdown leads to the accumulation of Slug protein levels. CL1-5 cells were infected with control shLacZ or two different shCHIP viruses. The Slug expression was assessed by real-time PCR (right) and immunoblotting (left). GAPDH and β-actin were used as the internal controls, respectively. Data are shown as mean±s.e.m. in three different experiments (n=3). (g) HEK293 cells were transfected with the vectors expressing Flag-Slug-WT or -4SA. Twenty-four hours after transfection, the cells were treated with 10 μM MG132 and 25 μM GSK3β inhibitor VIII (where indicated) for 8 h before cell collection. The lysates were subjected to immunoprecipitation using anti-Flag antibodies. Western blot analysis was conducted with the indicated antibodies to determine the protein interactions. (h) A549 cells were infected with control shLacZ or shCHIP viruses. Seventy-two hours after infection, the cells were re-seeded for the transduction with Flag-Slug-WT or -4SA lentiviruses. Forty-eight hours after transduction, the lysates were prepared and Western analysis was performed. (i) A549 cells infected in (h) were transfected with the plasmid expressing Myc-ubiquitin. Twenty-four hours after transfection, the cells were treated with 10 μM MG132 for 6 h before cell collection. The lysates were subjected to immunoprecipitation using anti-Flag antibodies. Western blot analysis was conducted with the indicated antibodies to determine the protein interactions. Ub, ubiquitin.
Stabilized Slug promotes cell migration and invasion
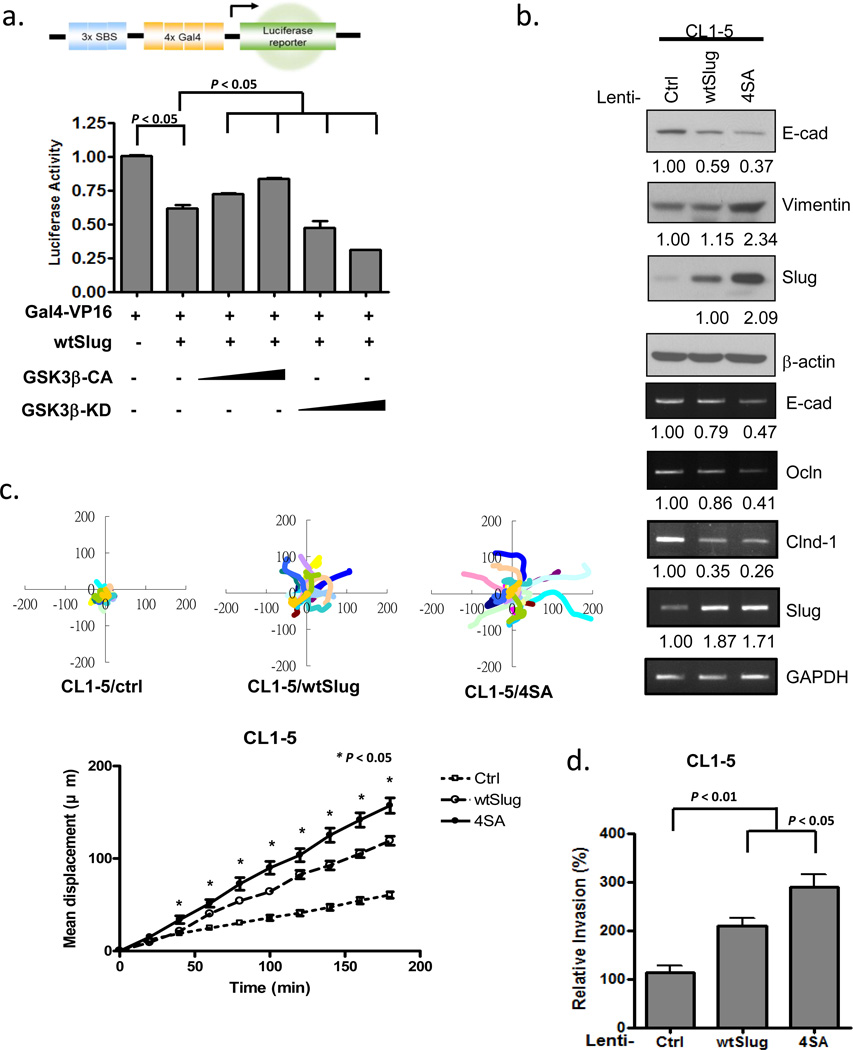
We next explored whether GSK3β regulates the transcription activity of Slug by the reporter assay. A Slug-responsive luciferase reporter was repressed in the presence of a Slug expression construct. This repression was alleviated in the presence of constitutively-active GSK3β (GSK3β-CA), but coexpression of kinase-inactive GSK3β (GSK3β-KD) enhanced the repression activity of Slug (Figure 4a), supporting the notion that GSK3β-KD acts as a dominant negative form of the normal enzyme.44, 49, 50 Since Slug transcriptionally represses E-cadherin, we then established stable cells expressing Slug-WT or Slug-4SA in CL1-5 or HEK293 cells by lentiviral transduction. Expressions of E-cadherin, along with two other tight junction molecules regulated by Slug, occludin and claudin-1, were downregulated in cells overexpressing wild-type Slug and were further decreased in those overexpressing nonphosphorylatable Slug-4SA (Figure 4b; Supplementary Figure 5a). Loss of epithelial markers and induction of the fibroblast marker, vimentin, indicated that these cells underwent EMT (Figure 4b; Supplementary Figure 5a). The Slug-4SA mutant stable cells were more motile than Slug-WT cells, as reflected by the wound-healing assay and the total displacement in the single-cell tracking assay (Figure 4c; Supplementary Figure 4b-e). Cell invasiveness was increased in cells overexpressing wild-type Slug and further enhanced in those overexpressing the nonphosphorylatable Slug-4SA mutant (Figure 4d). These data indicate that stabilized Slug-4SA, which abolishes GSK3β-mediated phosphorylation, can enhance cell motility and invasiveness.
Figure 4.
Non-degradable Slug promotes cell migration and invasion. (a) CL1-5 cells transfected with the luciferase reporter plasmid 3xSBS-Luc were co-transfected with plasmids expressing the indicated proteins. The luciferase activity was assayed 24 h later and normalized to the β-galactosidase activity, with pSV-β-gal used as an internal control plasmid. Each data point represents the mean ± s.d. The experiments were performed twice, in triplicate. (b) CL1-5 cells were stably transduced with the control, Slug-WT, or -4SA lentiviruses. The indicated RNA and protein expressions were assessed by RT-PCR and immunoblotting. GAPDH and β-actin were used as the internal controls, respectively. (c) Single-cell tracking migration analysis of control, wtSlug, and 4SA-overexpressing CL1-5 cells by time-lapse video microscopy. The movement of individual cells was followed using cell-tracking software and representative trajectories are shown (upper panel). Total displacement of the indicated cell lines was quantified from the track plots (mean ± s.e.m. of 20 cells analyzed in two independent experiments.) (d) Cell invasion was determined by the Transwell assay. Data are shown as mean ± s.e.m. in three different experiments (n=3).
Nonphosphorylatable Slug mutant (4SA) increases cancer metastasis in vivo
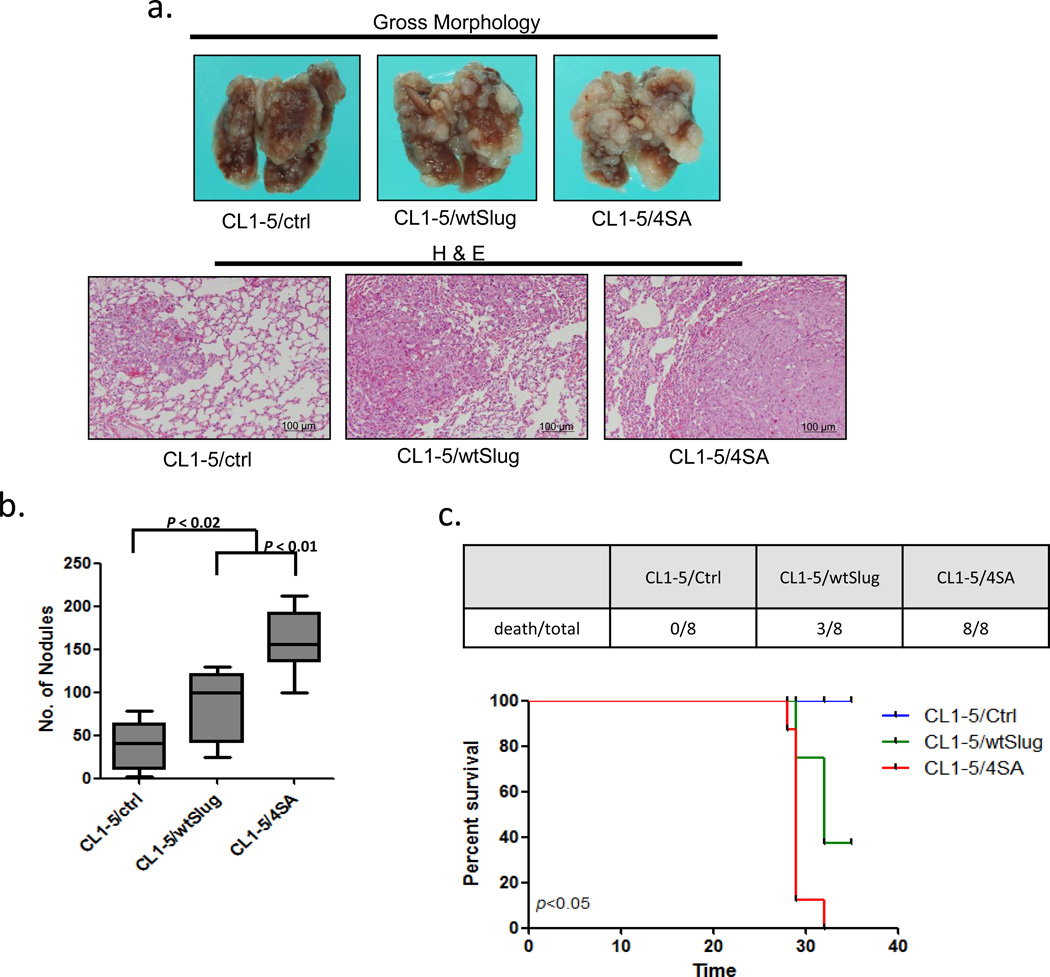
To investigate whether phosphorylation of Slug affects cancer metastasis in vivo, we injected wild-type or mutant (4SA) Slug-overexpressing CL1-5 cells intravenously into non-obese diabetic-severe combined immunodeficiency (NOD-SCID) mice. As shown in Figure 5a-b, mice injected intravenously with CL1-5/wtSlug and CL1-5/4SA cells developed more pulmonary nodules than those injected with control cells. We also found that mice with CL1-5/4SA cells formed more metastatic lung tumor nodules than those with CL1-5/wtSlug cells. Likewise, all mice with CL1-5/4SA were dead at 35 days post tumor injection (Figure 5c, upper panel). Survival analysis demonstrated that mice with CL1-5/4SA cells had the worst survival (Figure 5c, lower panel). These results suggest that Slug-4SA may increase the metastasis of lung adenocarcinoma cells in vivo.
Figure 5.
Non-degradable Slug increases in vivo cancer metastasis of lung adenocarcinoma cells. (a) Effects of Slug overexpression on metastasis in vivo. Upper panel: representative lungs of mice i.v. injected with CL1-5/control, CL1-5/wtSlug, or CL1-5/4SA cells. Lower panel: Histological examination of the lungs by hematoxylin-eosin staining. (b) Quantitative evaluation of lung metastatic nodules 4 weeks after tail-vein injection. Data are expressed as the mean±s.e.m. (n=8 per group). (c) The survival curve (lower panel) of the mice i.v. injected with CL1-5/control (n=8), CL1-5/wtSlug (n=8), or CL1-5/4SA cells (n=8). The number of death/total in each group at 35 days post i.v. injection is presented in the upper panel. Data are expressed as the mean±s.d. *P < 0.05 by the log-rank (Mantel-Cox) test.
The GSK3β-pSer9 level is associated with the expression of Slug in NSCLC tumor specimens
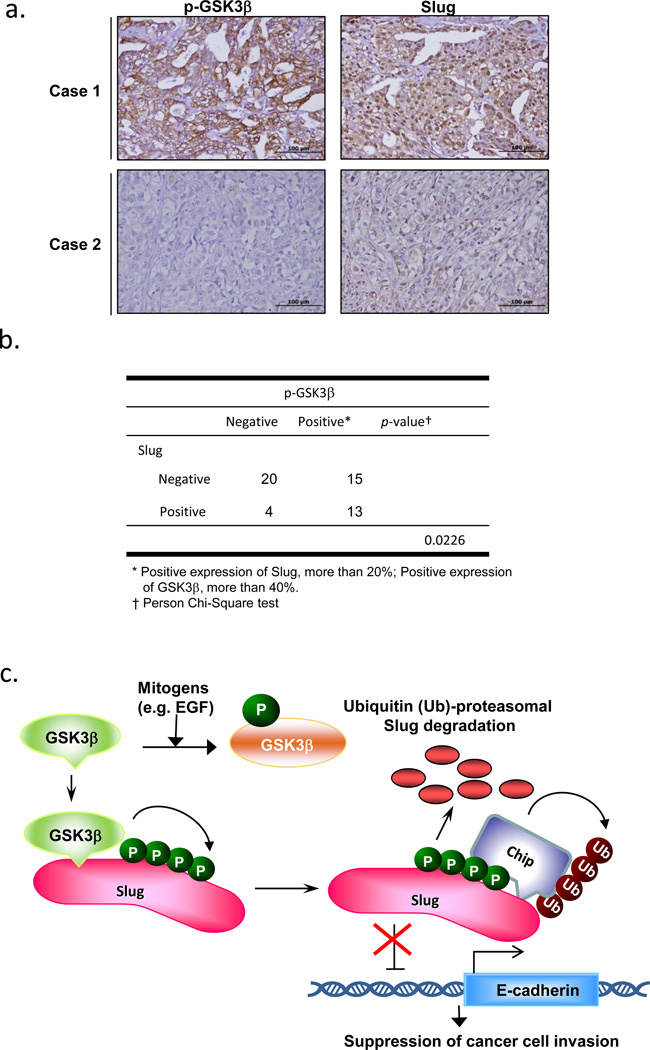
To assess whether the correlation between GSK3β and Slug also exists in lung cancer progression, we collected a cohort of 52 patients with NSCLC and determined the protein expressions of GSK3β-pSer9 and Slug by immunohistochemistry (Figure 6a-b). The clinical characteristics of the patients with NSCLC are shown in Supplementary Information, Table S1. Among the 52 patients with NSCLC, 28 (53.8%) samples stained positive for GSK3β-pSer9, in agreement with the reports that GSK3β is highly inactivated in various tumors.42 Slug was highly expressed in 13 out of 28 samples with GSK3β inactivation (46.4%). On the other hand, of the samples with low GSK3β-pSer9 staining, most (83.3%) displayed a low expression of Slug in patients with NSCLC (P=0.0226; Figure 6b). Interestingly, among these 20 patients with a low Slug protein level and a low expression of GSK3β-pSer9, CHIP was highly expressed in 12 (60%) of the samples (Supplementary Figure 6a; Table S2), implicating that high CHIP expression may facilitate GSK3β-mediated Slug degradation in vivo.
Figure 6.
The GSK3β-pSer9 level is associated with the expression of Slug in NSCLC tumor specimens. (a-b) Immunohistochemistry of GSK3β-pSer9 and Slug in serial sections of NSCLC tumor specimens. Representative specimens of p-GSK3β-Ser9 and Slug staining of the same case are shown in (a). Scale bar represents 100 μm. (c) Schematic representation depicting that Slug is phosphorylated by GSK3β at four consecutive serine sites and is bound by CHIP for ubiquitylation. GSK3β suppression by mitogens, i.e. EGF, releases Slug from this negative regulation, thus enhancing Slug protein stability and resulting in EMT of cancer cells, their migration/invasion, and metastasis.
Discussion
In this study, we found a novel post-translational mechanism that controls Slug protein turnover in lung cancer. By proteomic analysis, we successfully identified GSK3β and CHIP as the components of Slug degradation machinery (Figure 6c). As revealed by recent studies, GSK3β-mediated phosphorylation of Slug affects its protein stability, localization, and functions in breast cancer.51,52 We found that GSK3β interacts with Slug in vivo in the CL1-5 lung cancer cell line and reduces Slug protein stability by CHIP-mediated degradation. Accumulation of nondegradable Slug, on the other hand, manifests increased migratory and invasive capabilities in lung cancer cells. These enhanced EMT functions further promote tumor metastasis, as demonstrated by the in vivo cancer metastasis mouse model, suggesting that Slug needs to be tightly controlled in tumor progression and further development of cancer metastasis.
Increasing evidence has indicated that the tumor microenvironment may provide a variety of signals to stimulate EMT, including numerous growth factors, pro-oxidant species, and cytokines. During cancer progression, these signals help reorganize the structure and composition of the connective tissue and in turn influence cancer cell functions, usually accelerating tumor growth and invasion and aggravating therapy.53 It has been demonstrated that the PI3K/Akt and Erk1/2 signaling pathways are activated by EGF and p38 mitogen-activated protein kinase (MAPK) by enhanced oxidative stress, all of which induce migration and invasion mediated by the up-regulation of Snail, Slug, ZEB1, and β-catenin in association with GSK3β inhibition in various cancers.41, 54–58 In this study, we showed that the activity of GKS3β inversely correlates with the Slug protein level in EGF treatment or serum starvation. Treatment of an Akt inhibitor reverses such a correlation and cell morphology. Further treatment of a GSK3β inhibitor led to the accumulation of the protein level of Slug and kept the cells in their spindle-like shapes, suggesting that GSK3β may be involved in Slug degradation and the process of EMT.
Interestingly, John et al. has recently reported that GSK3β inhibition is associated with the decreased expressions of Slug and N-cadherin in melanoma.59 Although GSK3β is often considered inactivated in most cancers and deemed a tumor suppressor, this report may raise the possibility that GSK3β acts as a tumor promoter instead of a tumor suppressor in certain tumors, depending on the cell type.60, 61
We found that in our cohort examined, active GSK3β highly correlates with Slug downregulation (83.3%). Moreover, up to 60% of patients with a low Slug level and a low GSK3β-pSer9 level have a high expression of CHIP, implying that CHIP may participate in the GSK3β-mediated Slug degradation in NSCLC. On the other hand, inactive GSK3β is partially associated with Slug overexpression (46.4%). We inferred that besides GSK3β, other mechanisms may regulate the turnover of Slug in NSCLC, such as the p53-MDM2-Slug pathway identified earlier by our group.14 GSK3β inactivation and upregulation of Slug, however, can partially account for the clinically observed cancer progression and metastasis in patients with NSCLC.
Although E3 ligases, such as FBW7 and β-trcp, often degrade the substrates of GSK3β, their well-characterized degradation sequences (degrons) are not found within Slug, Moreover, our preliminary data showed that knock-down of neither FBW7 (data not shown) nor β-trcp affected the Slug protein level in lung cancer cell lines. We thus speculated that other E3 ligases might be involved in GSK3β-mediated Slug degradation. CHIP newly identified by the proteomic assay has been implicated in the degradation of a variety of oncogenic proteins, many of which play an important role in tumorigenesis, migration, and invasion.62–64 We found that CHIP interacts with Slug in vivo and that the interaction weakens in the presence of GSK3β inhibitor or nonphosphorylatable Slug. Knockdown of CHIP accumulates both endogenous and ectopically-expressed wtSlug (but not 4SA). In addition, knockdown of CHIP reduces the ubiquitylation of wild-type Slug, but not that of nonphosphorylatable Slug, suggesting that CHIP degrades Slug in a phosphorylation-dependent manner. Wu et al. has indicated that β-trcp participates in the GSK3β-mediated degradation of Slug in breast cancer.52 However, we did not detect an increase in the Slug protein level in β-trcp-knockdown lung cancer cell lines. We inferred that different cancers or environments may display preferential regulations by their natures. For example, Mcl-1, a Bcl-2-like antiapoptotic protein, is bound and degraded by β-trcp after stress-induced phosphorylation by GSK3.65, 66 A recent report reveals that loss of FBW7, but not other kinds of F-box proteins, accounts for the accumulation of Mcl-1 in T-cell acute lymphoblastic leukemia (T-ALL) and FBW7 promotes Mcl-1 destruction in a GSK3-mediated phosphorylation-dependent manner.67 Therefore, cells may indeed orchestrate these destruction components in the context required.
Slug has long been held as an invasion-promoter, but the post-translational regulation of Slug has been less discussed. The inactivation of GSK3β in epithelial cancers stabilizes several oncogenic transcriptional repressors, i.e. Slug, in our finding. Nonphosphorylatable Slug enhances migration and invasion; moreover, it promotes metastasis and aggravates the survival rate of the experimental models. Conacci-Sorrell et al. has indicated that Slug can be transcriptionally regulated by β-catenin in a cell-density manner.68 Post-translational modification and protein degradation may be equally significant in modulating the amount of Slug in cells that require migration and invasion—in this case, lung cancers. In summary, the finding that GSK3β phosphorylates Slug through the CHIP pathway to mediate Slug degradation may offer new therapeutic targets to prevent cancer metastasis.
Material and Methods
Plasmids and transfection
The Slug expression plasmids, p3xFlag-CMV-7.1-2-Slug, -S100A, -S104A, -2SA, and -4SA, were constructed by cloning full-length human Slug cDNA into the p3xFlag-CMV-7.1-2 vector and its mutant expression plasmids were generated by site-directed mutagenesis with a QuikChange kit (Stratagene), using the primers: for S100A, forward 5’-CTCCTCCAAGGACCACGCTGGCTCAGAAAGCCCC-3’ and reverse 5’-GGGGCTTTCTGAGCCAGCGTGGTCCTTGGAGGAG-3’; for S104A, forward 5’-CCACAGTGGCTCAGAAGCCCCCATTAGTGATGAAG-3’ and reverse 5’-CTTCATCACTAATGGGGGCTTCTGAGCCACTGTGG-3’; for Slug-2SA, forward5’- GACCACGCTGGCTCAGAAGCCCCCATTAGTGATGAAGAG-3’ and reverse 5’- CCTCTTCATCACTAATGGGGGCTTCTGAGCCAGCGTGGTC-3’; for Slug-4SA, forward 5’- CCCCCTCCTCCAGCTGACACCTCCGCCAAGGACCACGCT-3’and reverse5’- AGCGTGGTCCTTGGCGGAGGTGTCAGCTGGAGGAGGGGG-3’. Flag-tagged GSK3β (WT, CA, and KD) expression plasmids were subcloned from pCMV-5A-GSK3β (WT, CA, and KD plasmids, a gift from Dr. M.-C. Hung) into pFlag-CMV5a-vector (Sigma). All transfection experiments were performed with Lipofectamine™ 2000 reagents (Invitrogen, Eugene, OR) in accordance with the manufacturer’s protocols.
Viruses and transduction
Slug-WT and Slug-4SA were subcloned into pAS2.neo lentiviral vectors. LacZ- and GSK3α-, GSK3β-, β-trcp-, CHIP-shRNA-containing lentiviral vectors were obtained from the National RNAi Core Facility (Academia Sinica, Taiwan) and prepared in accordance with standard protocols. In brief, HEK293T cells were cotransfected with pLKO.1-shRNA, pCMVΔR8.91, and pMD.G. Virus-containing medium was collected at 24, 48, and 72 h post-transfection. To generate the stably-transduced clones, cells were infected with lentivirus in medium containing polybrene (8 μg ml−1). At 24 h after infection, cells were treated with 0.75 μg ml−1 puromycin or 400 μg ml−1 G418 to allow for the selection of a pool of antibiotic-resistant clones.
Cell culture
HEK293, HEK293T, and H1299, A549 cells were cultured in DMEM containing 10% FBS. HOP62 and CL1-5 cells were cultured in RPMI medium containing 10% FBS and L-glutamine. Stably transduced, pooled clones were maintained in the medium supplemented with 0.75 μg ml−1 puromycin or 400 μg ml−1 G418, as appropriate for the corresponding selection markers. For serum starvation, cells were maintained in RPMI medium without 10% FBS. For EGF treatment, fresh medium supplemented with the indicated concentrations of EGF was added after 24 h starvation.
Cell lysate preparation, immunoprecipitation and immunoblotting
Cell lysates for immunoblotting or immunoprecipitation were prepared in IP lysis buffer (20 mM Tris pH7.5, 100 mM NaCl, 1% IGEPAL CA-630, 100 μM Na3VO4, 50 mM NaF, 30 mM sodium pyrophosphate) containing 1×complete protease inhibitor cocktail with or without EDTA (Roche). For co-immunoprecipitation, H1299 cells were transfected with plasmids expressing GSK3β and Slug with the indicated tags for 24 h, and then treated with MG132 (10 µM) for 5 h. The supernatant was incubated with anti-Flag M2 antibodies overnight at 4°C and with protein A agarose for 1 h at 4°C. For endogenous co-immunoprecipitation, cells were cross-linked with 5 mM Dimethyl 3,3’-dithiobisproprionamidate-2-HCl (DTBP, Pierce) at 37°C for 30 min and Slug was immunoprecipitated by anti-Slug antibodies as described previously.14, 69 Protein samples were separated by SDS-PAGE, transferred to a poly(vinylidene difluoride) membrane and probed with the indicated antibodies. Proteins were detected by chemiluminescence.
Antibodies
The primary antibodies used for immunoblotting were as follows: monoclonal anti-HA (HA11; Covance), anti-GSK3α/β (Santa Cruz), anti-GSK3β (BD Biosciences), anti-CHIP (Epitomics), anti-E-cadherin (BD Biosciences), anti-Flag (M2; Sigma), anti-GFP (BD Biosciences), anti-Vimentin (BD Biosciences); poly-clonal anti-Slug (Santa Cruz), anti-β-Trcp (Cell Signaling), anti-Myc (9E11; Millipore), and anti-β-actin (Sigma).
Cycloheximide protein synthesis inhibition assay
The turnover of proteins was determined using cycloheximide inhibition of protein synthesis. Cells were treated with 300 µg ml−1 cycloheximide (Sigma) for the indicated time points.
Reverse-transcriptase polymerase chain reaction (RT-PCR) and real-time quantitative PCR
Total RNAs were extracted by TRIzol (Invitrogen) and cDNAs were prepared and underwent PCR as described previously.14 The amount of PCR products was determined by agarose gel electrophoresis in the presence of ethidium bromide. For real-time quantitative PCR, the primer sets for Slug (Hs00161904_m1) and the internal control, GAPDH (Hs99999905_m1), were purchased from Applied Biosystems (Foster City, CA). Data were acquired using an ABI PRISM 7500 system (Applied Biosystems). The expression level of Slug relative to that of GAPDH was defined as −ΔCT=−[CTSlug-CTGAPDH]. The Slug cDNA/GAPDH cDNA ratio was calculated as 2−ΔCT×K, in which K is a constant. Experiments were performed in triplicates.
Time-lapse microscopy
Time-lapse images of migrating cells in serum-containing medium at 37°C/5% CO2 were taken on an inverted Zeiss Axiovert 200M microscope (Carl Zeiss, Jena, Germany) over the course of 20 hours. Images were obtained with a CoolSNAP HQ CCD camera (Roper Scientific, NJ) at 15-minute intervals and analyzed using MetaMorph 5.0 software (Universal Imaging, Downingtown, PA).
Luciferase activity assays
CL1-5 cells (8x105 cells per well) were seeded into 6-well plates. Plasmids were transfected into cells using Lipofectamine™ 2000 reagents (Invitrogen). The luciferase reporter 3xSBS-Luciferase was cotransfected with a β-galactosidase construct, pGal4-VP16 at a DNA ratio of 10:1. The luciferase activity and β-galactosidase activity were measured by a Dual-Luciferase® Reporter Assay System (Promega, WI) at 24 h post-transfection. Transfection efficiency was normalized with β-galactosidase activity. Data were expressed as relative luciferase activity (firefly luciferase activity divided by β-galactosidase activity.)
In vitro kinase assay
For the in vitro kinase assay, Flag-tagged wtSlug and its various mutant proteins were obtained from HEK293T cells transfected with each expression plasmid for 24 h. Cells were treated with MG132 (10 µM) for 5 h before they were lysed with RIPA buffer containing protease inhibitor (Roche). Lysates were sonicated and centrifuged at 12,000 rpm for 20 min at 4°C. The supernatants were incubated with 1 µg anti-Flag antibodies overnight at 4°C followed by the addition of protein A-Sepharose for 1 h at 4 ° C. The immunocomplexes were collected by centrifugation at 3,000 rpm and finally washed three times with the RIPA buffer and once with the indicated 1x kinase buffer. GST-fused Slug proteins were obtained by 200 µM IPTG induction in the E. coli strain, BL21, transformed with pGEX-5x-1-CPO expression plasmids. Cell pellets were resuspended in 100 ml Buffer A (20 mM Tris pH7.4, 1M NaCl, 10 mM β-mercaptoethanol, 0.2 mM EDTA pH8.0, and 1 mM PMSF), sonicated for 10 min, and centrifuged at 12,000 rpm for 20 min at 4°C. Supernatants were incubated with 0.5 ml Glutathione agarose for 30 min at 4° C, washed with Buffer A three times, followed by Buffer D (20 mM HEPES pH8.0, 20% glycerol, 100 mM KCl, 0.2 mM EDTA pH8.0, 0.5 mM PMSF, and 0.5 mM DTT) three times. Immunoprecipitated cellularly-expressed Flag-tagged wtSlug and mutants or bacterially-expressed GST-fused Slug proteins were incubated with recombinant GSK3β (New England Biolabs, MA) at 30°C for 30 min in the indicated kinase buffer containing 0.2 mM unlabeled ATP and 10 µCi of γ-[32P]-ATP (Perkin Elmer). Reactions were stopped with sample buffer at 100°C for 10 min and analyzed by SDS-PAGE. The amount of 32P-labeled-Slug was detected by autoradiography and input by Coomassie blue staining or immunoblotting.
Experimental metastasis in vivo
For the in vivo tail vein metastasis assay, a single-cell suspension containing 106 cells in 0.1 ml of PBS was injected into the lateral tail veins of ten 8-week-old NOD-SCID mice (supplied by the Laboratory Animal Facilities in the Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan). After four weeks (CL1-5/wtSlug and 4SA mutant overexpression groups; at least eight mice per group), the mice were sacrificed and the lungs were examined for metastasis. The lungs were removed and fixed in 10% formalin, and the number of lung tumor colonies were counted under a dissecting microscope.
Statistical analysis
Data are presented as mean ± s.e.m. Comparisons of data between two groups were made with the Student’s t test. Statistical analyses were performed with SPSS software (version 10.0; SPSS, Inc., Chicago, IL). All statistical tests were two-sided, and P values of <0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank M.C. Hung (Department of Molecular and Cellular Oncology, University of Texas M.D. Anderson Cancer Center, Houston, USA) for providing the plasmids for GSK3β, and Wen-Lung Wang, Yi-Ying Wu, Chi-Yuan Chen for technical assistance. This work was supported by grants from the National Science Council, Taiwan (NSC99-2628-B-006-031-MY3 NSC101-2325-B-006-018, NSC100-2321-B-002-071, and NSC101-2321-B002-068), National Taiwan University, Taiwan (10R71601-2), and National Institute of Health, USA (R01-GM-094231, to AIN). S.P. Wang is supported by a Human Frontier Science Program long-term fellowship. shRNA constructs were obtained from the National RNAi Core Facility at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, Taipei, Taiwan.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no competing financial interests.
References
- 1.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12(8):895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 4.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 6.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68(10):3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 8.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28(1-2):151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 9.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 10.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 11.Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62(6):1613–1618. [PubMed] [Google Scholar]
- 12.Ye Y, Xiao Y, Wang W, Yearsley K, Gao JX, Shetuni B, et al. ERalpha signaling through slug regulates E-cadherin and EMT. Oncogene. 2010;29(10):1451–1462. doi: 10.1038/onc.2009.433. [DOI] [PubMed] [Google Scholar]
- 13.Shih JY, Tsai MF, Chang TH, Chang YL, Yuan A, Yu CJ, et al. Transcription repressor slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin Cancer Res. 2005;11(22):8070–8078. doi: 10.1158/1078-0432.CCR-05-0687. [DOI] [PubMed] [Google Scholar]
- 14.Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11(6):694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- 15.Shih JY, Yang PC. The EMT regulator slug and lung carcinogenesis. Carcinogenesis. 32(9):1299–1304. doi: 10.1093/carcin/bgr110. [DOI] [PubMed] [Google Scholar]
- 16.Shih JY, Yang PC. The EMT regulator slug and lung carcinogenesis. Carcinogenesis. 2011;32(9):1299–1304. doi: 10.1093/carcin/bgr110. [DOI] [PubMed] [Google Scholar]
- 17.Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6(10):931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 18.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 19.Taylor CT, Furuta GT, Synnestvedt K, Colgan SP. Phosphorylation-dependent targeting of cAMP response element binding protein to the ubiquitin/proteasome pathway in hypoxia. Proc Natl Acad Sci U S A. 2000;97(22):12091–12096. doi: 10.1073/pnas.220211797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esser C, Alberti S, Hohfeld J. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim Biophys Acta. 2004;1695(1-3):171–188. doi: 10.1016/j.bbamcr.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 21.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3(1):93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 23.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8(4):303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kajiro M, Hirota R, Nakajima Y, Kawanowa K, So-ma K, Ito I, et al. The ubiquitin ligase CHIP acts as an upstream regulator of oncogenic pathways. Nat Cell Biol. 2009;11(3):312–319. doi: 10.1038/ncb1839. [DOI] [PubMed] [Google Scholar]
- 25.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3(1):100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 26.Tateishi Y, Kawabe Y, Chiba T, Murata S, Ichikawa K, Murayama A, et al. Ligand-dependent switching of ubiquitin-proteasome pathways for estrogen receptor. EMBO J. 2004;23(24):4813–4823. doi: 10.1038/sj.emboj.7600472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci U S A. 2002;99(20):12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rees I, Lee S, Kim H, Tsai FT. The E3 ubiquitin ligase CHIP binds the androgen receptor in a phosphorylation-dependent manner. Biochim Biophys Acta. 2006;1764(6):1073–109. doi: 10.1016/j.bbapap.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Shimura H, Schwartz D, Gygi SP, Kosik KS. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J Biol Chem. 2004;279(6):4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 30.Dickey CA, Yue M, Lin WL, Dickson DW, Dunmore JH, Lee WC, et al. Deletion of the ubiquitin ligase CHIP leads to the accumulation, but not the aggregation, of both endogenous phospho- and caspase-3-cleaved tau species. J Neurosci. 2006;26(26):6985–6996. doi: 10.1523/JNEUROSCI.0746-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Groot RP, Auwerx J, Bourouis M, Sassone-Corsi P. Negative regulation of Jun/AP-1: conserved function of glycogen synthase kinase 3 and the Drosophila kinase shaggy. Oncogene. 1993;8(4):841–847. [PubMed] [Google Scholar]
- 32.Buss H, Dorrie A, Schmitz ML, Frank R, Livingstone M, Resch K, et al. Phosphorylation of serine 468 by GSK-3beta negatively regulates basal p65 NF-kappaB activity. J Biol Chem. 2004;279(48):49571–49574. doi: 10.1074/jbc.C400442200. [DOI] [PubMed] [Google Scholar]
- 33.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23(10):2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciani L, Salinas PC. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat Rev Neurosci. 2005;6(5):351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- 35.Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8(12):1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 36.M GA, Uddin S, Mahmud D, Damacela I, Lavelle D, Ahmed M, et al. Regulation of myeloma cell growth through Akt/Gsk3/forkhead signaling pathway. Biochem Biophys Res Commun. 2002;297(4):760–764. doi: 10.1016/s0006-291x(02)02278-7. [DOI] [PubMed] [Google Scholar]
- 37.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378(6559):785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 38.Thornton TM, Pedraza-Alva G, Deng B, Wood CD, Aronshtam A, Clements JL, et al. Phosphorylation by p38 MAPK as an alternative pathway for GSK3beta inactivation. Science. 2008;320(5876):667–670. doi: 10.1126/science.1156037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HY, Oh SH, Suh YA, Baek JH, Papadimitrakopoulou V, Huang S, et al. Response of non-small cell lung cancer cells to the inhibitors of phosphatidylinositol 3-kinase/Akt- and MAPK kinase 4/c-Jun NH2-terminal kinase pathways: an effective therapeutic strategy for lung cancer. Clin Cancer Res. 2005;11(16):6065–6074. doi: 10.1158/1078-0432.CCR-05-0009. [DOI] [PubMed] [Google Scholar]
- 40.Ellerbroek SM, Halbleib JM, Benavidez M, Warmka JK, Wattenberg EV, Stack MS, et al. Phosphatidylinositol 3-kinase activity in epidermal growth factor-stimulated matrix metalloproteinase-9 production and cell surface association. Cancer Res. 2001;61(5):1855–1861. [PubMed] [Google Scholar]
- 41.Al-Mulla F, Bitar MS, Al-Maghrebi M, Behbehani AI, Al-Ali W, Rath O, et al. Raf kinase inhibitor protein RKIP enhances signaling by glycogen synthase kinase-3beta. Cancer Res. 2011;71(4):1334–1343. doi: 10.1158/0008-5472.CAN-10-3102. [DOI] [PubMed] [Google Scholar]
- 42.Leis H, Segrelles C, Ruiz S, Santos M, Paramio JM. Expression, localization, and activity of glycogen synthase kinase 3beta during mouse skin tumorigenesis. Mol Carcinog. 2002;35(4):180–185. doi: 10.1002/mc.10087. [DOI] [PubMed] [Google Scholar]
- 43.Kang T, Wei Y, Honaker Y, Yamaguchi H, Appella E, Hung MC, et al. GSK-3 beta targets Cdc25A for ubiquitin-mediated proteolysis, and GSK-3 beta inactivation correlates with Cdc25A overproduction in human cancers. Cancer Cell. 2008;13(1):36–47. doi: 10.1016/j.ccr.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farago M, Dominguez I, Landesman-Bollag E, Xu X, Rosner A, Cardiff RD, et al. Kinase-inactive glycogen synthase kinase 3beta promotes Wnt signaling and mammary tumorigenesis. Cancer Res. 2005;65(13):5792–5801. doi: 10.1158/0008-5472.CAN-05-1021. [DOI] [PubMed] [Google Scholar]
- 45.Zheng H, Saito H, Masuda S, Yang X, Takano Y. Phosphorylated GSK3beta-ser9 and EGFR are good prognostic factors for lung carcinomas. Anticancer Res. 2007;27(5B):3561–3569. [PubMed] [Google Scholar]
- 46.Karrasch T, Spaeth T, Allard B, Jobin C. PI3K-dependent GSK3ss(Ser9)-phosphorylation is implicated in the intestinal epithelial cell wound-healing response. PLoS One. 2011;6(10):e26340. doi: 10.1371/journal.pone.0026340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nature methods. 2011;8(1):70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu C, Kim NG, Gumbiner BM. Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle. 2009;8(24):4032–4039. doi: 10.4161/cc.8.24.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He X, Saint-Jeannet JP, Woodgett JR, Varmus HE, Dawid IB. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature. 1995;374(6523):617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- 50.Patel R, Gao M, Ahmad I, Fleming J, Singh LB, Rai TS, et al. Sprouty2, PTEN, and PP2A interact to regulate prostate cancer progression. J Clin Invest. 2013;123(3):1157–1175. doi: 10.1172/JCI63672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JY, Kim YM, Yang CH, Cho SK, Lee JW, Cho M. Functional regulation of Slug/Snail2 is dependent on GSK-3beta-mediated phosphorylation. FEBS J. 2012;279(16):2929–2939. doi: 10.1111/j.1742-4658.2012.08674.x. [DOI] [PubMed] [Google Scholar]
- 52.Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci U S A. 2012;109(41):16654–16659. doi: 10.1073/pnas.1205822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 54.Cheng JC, Auersperg N, Leung PC. EGF-induced EMT and invasiveness in serous borderline ovarian tumor cells: a possible step in the transition to low-grade serous carcinoma cells? PLoS One. 7(3):e34071. doi: 10.1371/journal.pone.0034071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Come C, Arnoux V, Bibeau F, Savagner P. Roles of the transcription factors snail and slug during mammary morphogenesis and breast carcinoma progression. J Mammary Gland Biol Neoplasia. 2004;9(2):183–193. doi: 10.1023/B:JOMG.0000037161.91969.de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang L, Amann JM, Kikuchi T, Porta R, Guix M, Gonzalez A, et al. Inhibition of epidermal growth factor receptor signaling elevates 15-hydroxyprostaglandin dehydrogenase in non-small-cell lung cancer. Cancer Res. 2007;67(12):5587–5593. doi: 10.1158/0008-5472.CAN-06-2287. [DOI] [PubMed] [Google Scholar]
- 57.Byles V, Zhu L, Lovaas JD, Chmilewski LK, Wang J, Faller DV, et al. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene. 2012;31(43):4619–4629. doi: 10.1038/onc.2011.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Current molecular medicine. 2009;9(7):873–886. doi: 10.2174/156652409789105570. [DOI] [PubMed] [Google Scholar]
- 59.John JK, Paraiso KH, Rebecca VW, Cantini LP, Abel EV, Pagano N, et al. GSK3beta inhibition blocks melanoma cell/host interactions by downregulating N-cadherin expression and decreasing FAK phosphorylation. The Journal of investigative dermatology. 2012;132(12):2818–2827. doi: 10.1038/jid.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo J. Glycogen synthase kinase 3beta (GSK3beta) in tumorigenesis and cancer chemotherapy. Cancer letters. 2009;273(2):194–200. doi: 10.1016/j.canlet.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rajakishore M. Glycogen synthase kinase 3 beta: can it be a target for oral cancer. Molecular Cancer. 2010;9(144):1–15. doi: 10.1186/1476-4598-9-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmed SF, Deb S, Paul I, Chatterjee A, Mandal T, Chatterjee U, et al. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J Biol Chem. 2012;287(19):15996–16006. doi: 10.1074/jbc.M111.321083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esser C, Scheffner M, Hohfeld J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J Biol Chem. 2005;280(29):27443–27448. doi: 10.1074/jbc.M501574200. [DOI] [PubMed] [Google Scholar]
- 64.Wang S, Wu X, Zhang J, Chen Y, Xu J, Xia X, et al. CHIP functions as a novel suppressor of tumour angiogenesis with prognostic significance in human gastric cancer. Gut. 2012 doi: 10.1136/gutjnl-2011-301522. [DOI] [PubMed] [Google Scholar]
- 65.Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27(11):4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Molecular cell. 2006;21(6):749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 67.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471(7336):104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze'ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163(4):847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winter M, Sombroek D, Dauth I, Moehlenbrink J, Scheuermann K, Crone J, et al. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol. 2008;10(7):812–824. doi: 10.1038/ncb1743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.