Abstract
A dozen genes/regions have been confirmed as genetic risk factors for oral clefts in human association and linkage studies, and animal models argue even more genes may be involved. Genomic sequencing studies should identify specific causal variants and may reveal additional genes as influencing risk to oral clefts, which have a complex and heterogeneous etiology. We conducted a whole exome sequencing (WES) study to search for potentially causal variants using affected relatives drawn from multiplex cleft families. Two or three affected second, third, and higher degree relatives from 55 multiplex families were sequenced. We examined rare single nucleotide variants (SNVs) shared by affected relatives in 348 recognized candidate genes. Exact probabilities that affected relatives would share these rare variants were calculated, given pedigree structures, and corrected for the number of variants tested. Five novel and potentially damaging SNVs shared by affected distant relatives were found and confirmed by Sanger sequencing. One damaging SNV in CDH1, shared by three affected second cousins from a single family, attained statistical significance (P = 0.02 after correcting for multiple tests). Family-based designs such as the one used in this WES study offer important advantages for identifying genes likely to be causing complex and heterogeneous disorders.
Keywords: whole exome sequencing, oral clefts, pedigree studies, rare variants
NONSYNDROMIC oral clefts [including cleft lip (CL), cleft palate (CP), and cleft lip and palate (CLP)] are common craniofacial malformations with a complex and heterogeneous etiology, reflecting both genetic and environmental risk factors (Dixon et al. 2011). Both genome-wide linkage and association studies have shown multiple genes influence risk to oral clefts (Mangold et al. 2011; Marazita 2012) and recently at least a dozen different genes have been identified as genetic risk factors in genome-wide association studies (GWASs) (Ludwig et al. 2012; Beaty et al. 2013). Few of these genes, however, have causal variants identified. Association studies using case-control or case-parent trio designs have little power to detect rare variants (RVs) that may be causal in a fraction of cases (or their families). Linkage studies have better power to detect regions of the genome harboring RVs exerting a large effect on risk within a family, but genome-wide linkage studies of oral clefts have revealed a high degree of “locus heterogeneity,” where different families show evidence of linkage to different genes, and the statistical signals generated from linkage analysis span large chromosomal regions (Marazita et al. 2004; 2009). In either linkage or association analysis, the specific markers yielding statistical evidence are rarely directly causal themselves; rather they tag unobserved causal variants, either through linkage disequilibrium (LD) or cosegregation within families (measured as a low estimated recombination fraction or excess allele sharing between affected relatives).
Our goal was to identify rare potentially causal variants among a large list of candidate genes for oral clefts [334 biologically plausible, autosomal candidate genes for oral clefts assembled by Jugessur et al. (2009) supplemented with confirmed GWAS “hits” (Ludwig et al. 2012; Beaty et al. 2013)] from whole exome sequencing (WES) data on affected individuals drawn from multiplex families originally ascertained for linkage studies. Our inferences assume damaging RVs shared between such distant affected relatives may be causal. Some of these multiplex families had been genotyped in previous genome-wide linkage screens (Wyszynski et al. 2003; Mangold et al. 2009), but marker panels varied. Other families were not genotyped previously.
Materials and Methods
Ethics statement
Multiplex families were recruited by separate research groups under protocols reviewed and approved by their own institutional review board (IRB). Collaborations between US and foreign investigators were subject to review and approval by both the appropriate local IRB and the corresponding IRB of the US investigator. Each participant was advised of the purpose of the research project and provided informed consent for themselves and, when appropriate, for their minor children.
Genotyping
Exome sequencing and genotyping was done at the Center for Inherited Disease Research (CIDR). Genomic DNA was isolated by the original research team, and DNA aliquots were sent to CIDR for sequencing. All affected subjects included in the sequencing study were genotyped using Illumina’s Human OmniExpress SNP array as a quality control step. Genotypes were called using Illumina’s software package GenomeStudio (version 2010.2, Genotyping Module version 1.7.4, and GenTrain version 1.0). Six subjects were genotyped in duplicate (four family members and two HapMap controls). Single nucleotide polymorphic (SNP) markers with call rate <98%, cluster separation value <0.2, or with discrepant genotypes in more than one duplicate pair were dropped.
Library preparation and exome sequence capture
DNA fragmentation was performed on 200 ng of genomic DNA using a Covaris E210 system, which shears DNA into fragments 150–200 bp in length with 3′ or 5′ overhangs. End repair was performed where 3′ to 5′ exonuclease activity of enzymes removes 3′ overhangs, and the polymerase activity fills in the 5′ overhangs. An “A” base is then added to the 3′ end of the blunt phosphorylated DNA fragments to prepare fragments for ligation to the sequencing adapters, which have a single “T” base overhang at their 3′ end. Ligated fragments are subsequently size selected through purification using SPRI beads and undergo PCR amplification techniques to prepare “libraries.” The Caliper LabChip GX was used for quality control (QC) of libraries to ensure adequate concentration and appropriate fragment size.
Exon capture was done using the Agilent SureSelect Human All Exon Target Enrichment system (kit S0297201), which results in ∼51 Mb of targeted sequence capture per sample. Under standard procedures, biotinylated RNA oligonucleotides were hybridized with 500 ng of the library. Magnetic bead selection was used to capture the resulting RNA–DNA hybrids. RNA was digested and remaining DNA capture PCR amplified. Sample indexing was introduced at this step. The Agilent Bioanalyzer (HiSensitivity) was used for QC of adequate fragment sizing and quantity of DNA capture.
DNA sequencing
DNA sequencing was performed on an Illumina HiSequation 2500 instrument using standard protocols for a 100-bp paired-end run. Six samples were run per flowcell, guaranteeing >90–95% completeness at a minimum of 20× coverage.
Variant calling
Illumina HiSeq reads were processed through Illumina’s Real-Time Analysis (RTA) software generating base calls and corresponding quality scores. Resulting data were aligned to a reference genome with the Burrows–Wheeler Alignment (BWA) tool creating a sequence alignment map/binary alignment map file. Postprocessing of the aligned data includes local realignment around indels, base call quality score recalibration performed by the Genome Analysis Toolkit (GATK) and flagging of molecular/optical duplicates using software from the Picard program suite. Multisample variant calling was performed using GATK 2.0’s Unified Genotyper. Variant quality score recalibration (VQSR) was done in GATK 2.0 and only variants passing this step were included. CIDR required a minimum mean of 8× coverage before calling any single nucleotide variant (SNV), but the overall coverage averaged 84× over all exons.
Analyzing called variants
In this work, we focused on SNVs in 334 autosomal candidate genes for oral clefts (Jugessur et al. 2009) plus 14 recently confirmed genes/regions yielding genome-wide significance in a meta-analysis (Ludwig et al. 2012) and a replication study (Beaty et al. 2013). Thus, 348 candidate genes were considered. To minimize the multiple comparisons burden and to focus on potentially causal variants with high penetrance (i.e., variants rare in the population), our analysis was restricted to SNVs not found in build 137 of the SNP Database (dbSNP), and predicted to be damaging based on a sorting intolerant from tolerant (SIFT) score <0.05 (Ng and Henikoff 2003). Variants not seen in either the sequence of 5379 subjects in the Exome Sequencing Project (ESP; esp.gs.washington.edu/drupal/) database or the 1000 Genomes data (www.1000genomes.org, April 2012 release) were retained if they also had a minor allele frequency (MAF) <0.1 in an internal database of variants in all exomes previously sequenced at CIDR, to help filter out variant calls resulting solely from technical artifacts.
Assessing evidence for potentially causal SNVs
Evidence that a rare SNV could cause oral clefts was based on two or more affected distant relatives sharing that particular variant. More precisely, we quantified this evidence by computing the exact probability a RV would be shared by all sequenced relatives in a family, given it occurred in any one of them, under the null hypothesis of a complete absence of linkage and association. For variants seen in only one family, this probability can be interpreted directly as a P-value from a Bernoulli trial. For variants seen in M families and shared by affected relatives in m of them, the appropriate P-value was obtained as the sum of the probability of events as or more extreme than the observed sharing in m out of M families (Bureau et al. 2014).
RV sharing probabilities based on known pedigree structure
Assuming the known pedigree structure accurately describes the relationships between sequenced affected individuals (implying all founders are unrelated), copies of any RV in two or more relatives are almost certainly identical by descent (IBD). Letting Ci be the number of copies of a RV received by sequenced subject i out of n sequenced subjects, and Fj be the event a founder j introduced one copy of this RV into the pedigree, then the probability of interest can be expressed as
| (1) |
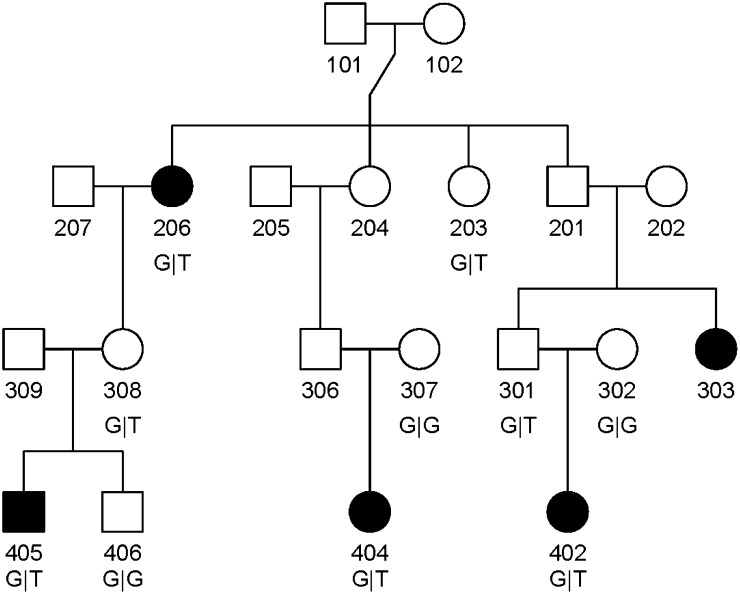
where the expression on the second line results from assuming a single copy of the RV existed among all alleles in the nf founders. The probabilities P(Fj) cancel from the numerator and the denominator of Equation 1. Mathematical expressions have been derived for the other terms, namely the probabilities that all sequenced subjects and at least one sequenced subject received the RV, given it was introduced into the pedigree by founder j (Bureau et al. 2014). As an example, for three sequenced second cousins shown in Figure 1 (individuals 402, 404, and 405), the probability P(C1 = C2 = C3 = 1|Fj) = ((1/2)3)3 = 1/512 when j is one of the two great-grandparents (individuals 101 or 102 in Figure 1), i.e., a probability of (1/2)3 of transmitting the variant through three meioses to each great-grandchild, raised to the power 3 because this event had to happen for all three of the second cousins. Other founders are ancestors of only one of the sequenced subjects, so the probability they transmitted this RV to all three subjects becomes zero. The probability P(C1 + C2 + C3 > 1|Fj) = 1 − P(C1 = C2 = C3 = 0|Fj), the event that no sequenced subject received this RV, is (1 − (1/2)3)3 = 343/512 for each of the two great-grandparents who must not have transmitted the RV to any of their great-grandchildren, 1 − (1/2)2 = 1/4 for each of the three grandparents of one of the sequenced subjects (individuals 202, 205, and 207 in Figure 1) and 1/2 for each of the three parents of one of the three sequenced subjects (individuals 302, 307, and 309 in Figure 1). Putting all this together gives
Equation 1 is a generalization of this sharing probability for two subjects: 1/(2(D+1) − 1), where D is the degree of relationship between the two subjects (Feng et al. 2011) (for example, 1/15 for a pair of first cousins). It is important to stress this RV sharing event considered here has a lower probability under the Ho: complete independence between RV sharing and affected status compared to the predicted IBD sharing under the null hypothesis of no linkage only, which is also the P-value of an allele-sharing linkage test in one family where IBD sharing between affected relatives is observed. For the sake of comparison, the null probability that three second cousins would share one allele IBD is 3/512 and that for two first cousins is 1/4 (note that the chance of one allele being shared IBD between two first cousins is 1/16, but since the IBD sharing events of the four grandparental alleles are mutually exclusive, the probability of any one allele being shared IBD becomes 1/4).
Figure 1.
Structure of pedigree where three affected second cousins shared a rare variant in CDH1. Affected subjects are represented by filled symbols. Individuals 402, 404, and 405 were sequenced.
Defining the set of RV tested
The lowest possible P-value for a RV being found in only one or very few families always depends on family structure. Sharing probabilities between sequenced subjects in small or highly inbred families may be high, and so is the potential P-value for a RV being seen only in one such family (for instance, it is 1/7 for a grandparent–grandchild pair). We therefore decided to test the null hypothesis only for those RVs achieving a sufficiently low P-value if shared by all affected subjects in the family (or families) where they were seen. These potential P-values are independent of the actual sharing pattern among affected relatives and therefore of the subsequent testing of RV sharing. We obtain this subset of RVs by ordering the potential P-values of all RVs in decreasing order and stopping at the last potential P-value lower than the type I error level 0.05 divided by the rank t of that P-value. The P-value critical threshold is then 0.05/t.
Confirmation with Sanger sequencing
For each family identified as sharing a damaging RV between distant affected relatives, Sanger sequencing was used to confirm the existence of the RV using all available family members. Primers were designed to amplify a 400- to 1000-bp region flanking each variant of interest using Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi). PCR products were sent for sequencing using an ABI 3730XL (Functional Biosciences, Madison, WI). Chromatograms were transferred to a UNIX workstation, base-called with PHRED (v. 0.961028), assembled with PHRAP (v. 0.960731), scanned by POLYPHRED (v. 0.970312), and viewed with the CONSED program (v. 4.0).
Results
Multiplex cleft families
Fifty-six multiplex oral cleft families from diverse populations [Germany, the Philippines, India, the Syrian Arab Republic, plus two of Chinese origin (one each from Taiwan and Shanghai) and one European American family] were selected because they included affected second or third degree relatives (note that second degree relatives included half-sibs, avuncular, or grandparental pairs; third degree relatives included first cousins and great-avuncular pairs). Some more distant relatives such as second cousins and first cousins once removed were also included. One member of an affected relative pair failed, so 114 affected members from 55 families were available for analysis. Fifty-one families provided 2 affected individuals and 4 families provided 3 affected individuals each (Table 1).
Table 1. Ethnic origin of multiplex families used in whole exome sequencing and total number of subjects sequenced.
| Ethnicity | Families | Subjects | CL/P | CP | Unknown |
|---|---|---|---|---|---|
| Indian | 12 | 26 | 26 | 0 | 0 |
| Filipino | 11 | 22 | 19 | 2 | 1 |
| German | 19 | 38 | 31 | 7 | 0 |
| Syrian | 10 | 22 | 22 | 0 | 0 |
| European-American | 1 | 2 | 1 | 1 | 0 |
| Chinese | 2 | 4 | 2 | 2 | 0 |
Novel SNVs predicted to be damaging
A total of 183 novel variants were predicted by SIFT score to damage the final gene product in these 348 candidate genes (Supporting Information, Table S1), but only 5 were shared by the affected distant relatives sequenced in this study. Table 2 lists five novel SNVs predicted to be damaging where two or three affected family members had the same genotype. Each shared SNV listed in Table 2 was checked using the Integrative Genomics Viewer, and all showed good alignment patterns (see Figure S1). All SNVs listed in Table 2 occurred in heterozygotes, except the SNV in FTCD where genotype call could not be made with full confidence due to reduced coverage, but all reads contained the variant G allele. Each of these variants were predicted to be “possibly damaging” using Polyphen2 (>0.15) also (Abxhubei et al. 2013).
Table 2. Novel and damaging SNVs where the genotype was shared by affected distant relatives in multiplex cleft families.
| Gene | Chr | Position (HG19) | Ref | Alt | Quality | Amino acid change (no. transcripts) | SIFT score | Polyphen2 | Type of affected relatives | Ethnic origin of family |
|---|---|---|---|---|---|---|---|---|---|---|
| CDH1 | 16q22.1 | 68,857,508 | G | T | 1659 | G→stop (3) | 0.01 | 0.735 | Three second cousins | Indian |
| FGF8 | 10q24 | 103,531,236 | C | A | 1522 | G→V (4) | 0.00 | 0.888 | Grandparent–grandchild | German |
| FGFR4 | 5q35.1 | 176,524,621 | G | C | 513 | D→H (6) | 0.00 | 0.676 | First cousins | Indian |
| TRPS1 | 8q24.12 | 116,616,313 | T | C | 1073 | D→H (4) | 0.00 | 0.984 | Great-avuncular | Filipino |
| FTCD | 21q22.3 | 47,572,892 | A | G | 147 | V→A (4) | 0.01 | 0.899 | First cousins (inbred) | Syrian |
Ref: Reference allele; Alt: Alternate allele.
Probabilities of rare variant sharing
Sharing probabilities based on the reported pedigree structure were computed for all 183 novel variants predicted to be damaging. P-values of a test of the null hypothesis of a complete absence of linkage and association were derived from sharing probabilities in one or more families. Sixteen of these RVs had a potential P-value below the Bonferroni-adjusted significance threshold of 0.05/16, making them eligible for further statistical testing. Among the 55 families in this study, 22 had a sufficiently low RV sharing probability among sequenced members to achieve this significance threshold on their own. Only one of these 16 RVs was actually shared, the CDH1 variant listed in Table 2. The null probability that a RV would be shared by three second cousins is 1/745 = 0.0013 following the computation shown above, giving a Bonferroni-adjusted P-value of 0.0013 × 16 = 0.0208.
Confirmation by Sanger sequencing
Exonic regions containing the five variants in Table 2 were sequenced in all family members with available DNA using Sanger sequencing to confirm genotypes from WES. Each individual used for WES was confirmed as heterozygous for their respective RVs, and some additional unaffected relatives were also carriers. For example, the affected grandparent–grandchild pair sharing a RV in FGF8 was confirmed, and the intervening unaffected parent also carried this RV. In the Indian family segregating for the damaging RV in CDH1, the presence of a T allele was confirmed in the three affected second cousins used in WES, and two of their parents, 301 and 308, who are unaffected first cousins (see Figure 1). Parent 306, who is a first cousin of 301 and 308, did not have DNA available, but his spouse 307 was GG. In total, Sanger sequencing revealed three unaffected GT carriers (the two unaffected parents and one unaffected great-aunt), one additional affected relative who was GT (individual 206), and three unaffected relatives with the wild-type GG genotype (subject 406 sibling of 405, and subjects 302 and 307, two married-in mothers of 402 and 404, respectively).
Discussion
Whole exome sequencing data on distantly related affected individuals from multiplex families revealed five novel SNVs predicted to be damaging and shared by two or three distant affected relatives from the same family. Evidence that a RV could be causal was based on the probability that such a RV would be shared by the two or three affected relatives conditional on its presence in the family and given the pedigree structure. Focusing on 348 established candidate genes maximized the a priori chance that any novel, damaging variant would actually be causal and lowered the threshold for statistical significance. Indeed, by restricting statistical testing to the 16 SNVs showing some potential to achieve a sufficiently low P-value, a novel SNV in gene CDH1 yielded significant evidence of cosegregation with cleft status in one family from India (see Figure 1). We also examined all rare and low frequency SNVs in exons and splice junctions with a MAF < 0.01 from all annotated genes (Bureau et al. 2014). That exome-wide analysis required a much steeper correction for multiple testing, and only SNVs that could potentially achieve the significance level of 2.2 × 10−5 were included in that analysis. This excluded the novel SNV in CDH1 and all SNVs seen in single families. Highlights of these results were reported elsewhere (Bureau et al. 2014) to illustrate our analytical approach based on RV sharing probabilities. We list in Table S2 the 80 SNVs yielding a P < 0.05.
Sanger sequencing revealed these likely damaging RVs in a number of unaffected relatives (as well as confirming the results of WES), which suggests considerable incomplete penetrance. Cooper et al. (2013) recently reviewed incomplete penetrance for many recognized Mendelian disorders and pointed out multiple biological mechanisms may be responsible. For disorders (such as oral clefts) that have a complex and heterogeneous etiology, incomplete penetrance should be expected. Based on our pedigree structures alone, where most or all distant affected relatives used in WES had unaffected parents, some incomplete penetrance must exist, so we did not extend our calculation of sharing probabilities to include these unaffected relatives.
Frebourg et al. (2006) first reported two different splicing site mutations in CDH1 in two families where some relatives had CLP and diffuse gastric cancer, while other relatives had only gastric cancer. Studies of polymorphic SNPs in and near CDH1 have shown equivocal evidence of association in case-control studies from various populations (Letra et al. 2008; Rafighdoost et al. 2013). Recently, Vogelaar et al. (2012) sequenced 81 cleft cases and found four distinct missense mutations and four intronic variants in CDH1 among 13 cases, all distinct from the RV reported here.
We must caution, however, unobserved relationships between founders could lead to false positive findings under this strategy because the probability of sharing a RV among family members would thus be higher than calculated based on pedigree structure alone. There is also the possibility that two families recruited from the same population could be related to one another in some unrecognized fashion. We investigated the extent to which sequenced subjects were related to each other (beyond their reported familial relationships) by estimating kinship coefficients between affected subjects from genome-wide markers using an estimator robust to population stratification as implemented in the King package (Manichaikul et al. 2010). The family segregating for the novel CDH1 variant listed in Table 2 was Bengali. Estimates of kinship between sequenced relatives in all families from this population showed little deviation from expected values based on reported pedigree structures (see Figure S2). Additionally, no evidence of unexpected relatedness between the 12 Indian families was detected (results not shown). Still, we cannot exclude the possibility the T allele at this novel SNV could be identical by state (IBS) but not IBD in all three sequenced subjects, rendering the reported sharing probability too optimistic. We carried out a sensitivity analysis by calculating IBS sharing probabilities as a function of population frequency of the T allele. As long as the true allele frequency in the Indian Bengali population is <2.0%, our finding retains statistical significance after multiple comparison correction (Figure S3). The absence of evidence of any unobserved relationships among these Indian multiplex cleft families and the tolerance of the sharing probability to low allele frequencies (unlikely to be exceeded by a protein-truncating variant) corroborates the statistical significance of our finding. Examining the empirical distribution of P-values from the exome-wide analysis revealed a good agreement with the uniform distribution, evidence of the general accuracy of these RV sharing probabilities (Bureau et al. 2014, Figure S3).
The number of novel SNVs predicted to be damaging (183 damaging SNVs in 348 candidate genes) was too small to undertake any analysis combining all SNVs in any one gene. In addition to the shared SNVs listed in Table 2, the genes CDH1, FGFR4, TRPS1, and FTCD each contained one novel SNV predicted to be damaging and not shared by all sequenced affected relatives within the family.
In summary, among five novel and likely damaging SNVs shared by affected distant relatives found by sequencing 348 recognized candidate genes for oral clefts, one SNV in CDH1 was very unlikely to have occurred by chance alone. This finding adds to the mounting evidence that mutations in CDH1 may cause oral clefts, but finding truly causal genes for complex and heterogeneous disorders (such as oral clefts) remains a daunting challenge (Rao 2008). This study illustrates how families originally recruited for linkage studies can be used to search for causal variants using whole exome sequencing.
Supplementary Material
Acknowledgments
We thank the members of the families who participated in this study and the field and laboratory staff who made this analysis possible. This work was supported by the National Institutes of Health (NIH) (R01-DE-014581 and U01-DE-018993 to T.H.B., R01-DE-016148 to M.L.M., and P50-DE-016215 to J.C.M.), with additional support from X01 HG006177 to T.H.B., M.L.M., and J.C.M. for whole exome sequencing at the Center for Inherited Disease Research, which is funded through a federal contract from the NIH to Johns Hopkins University (contract no. HHSN268200782096C). A.B. is supported by a research fellowship from the Fonds de Recherche du Québec - Santé. I.R. was further supported by NIH grant R01 GM083084.
Footnotes
Communicating editor: L. Jorde
Literature Cited
- Abxhubei I., Jordan D. M., Sunyaev S. R., 2013. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 7:Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty T. H., Taub M. A., Scott A. F., Murray J. C., Marazita M. L., et al. , 2013. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum. Genet. 132: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau A., Younkin S., Parker M. M., Bailey-Wilson J. E., Marazita M. L., et al. , 2014. Inferring rare disease risk variants based on exact probabilities of sharing by multiple affected relatives. Bioinformatics DOI: 10.1093/bioinformatics/btu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. N., Krawczak M., Polychronakos C., Smith C. T., Kehrer-Sawatzki H., 2013. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet. 132: 1077–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M. J., Marazita M. L., Beaty T. H., Murray J. C., 2011. Cleft lip and palate: understanding genetic and environmental influences. Nat. Rev. Genet. 12: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B. J., Tavtigian S. V., Southey M. C., Goldgar D. E., 2011. Design considerations for massively parallel sequencing studies of complex human disease. PLoS ONE 6: e23221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frebourg T., Oliveira C., Hochain P., Karam R., Manouvrier S., et al. , 2006. Cleft lip/palate and CDH1/E-cadherin mutations in families with hereditary diffuse gastric cancer. J. Med. Genet. 43: 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A., Shi M., Gjessing H. K., Lie R. T., Wilcox A. J., et al. , 2009. Genetic determinants of facial clefting: analysis of 357 candidate genes using two national cleft studies from Scandinavia. PLoS ONE 4: e5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letra A., Menezes R., Cranjeiro J. M., Vieira A. R., 2008. AXIN2 and CDH1 polymorphisms, tooth agenesis and oral clefts. Birth Defects Res. A Clin. Mol. Teratol. 85: 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig K. U., Mangold E., Hermes S., Nowak S., Ruetter H., et al. , 2012. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet. 44: 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E., Reutter H., Birnbaum S., Walier M., Mattheisen M., et al. , 2009. Genome-wide linkage scan of nonsyndromic orofacial clefting in 91 families of central European origin. Am. J. Med. Genet. A. 149A: 2680–2694. [DOI] [PubMed] [Google Scholar]
- Mangold E., Ludwig K. U., Noethen M., 2011. Breakthroughs in the genetics of orofacial clefting. Trends Mol. Med. 17: 1–9. [DOI] [PubMed] [Google Scholar]
- Manichaikul A., Mychaleckyj J. C., Rich S. S., Daly K., Sale M., et al. , 2010. Robust relationship inference in genome-wide association studies. Bioinformatics 26: 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita M. L., 2012. The evolution of human genetic studies of cleft lip and cleft palate. Annu. Rev. Genomics Hum. Genet. 13: 263–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita M. L., Murray J. C., Lidral A. C., Arcos-Bargos M., Cooper M. E., et al. , 2004. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q and 2q. Am. J. Hum. Genet. 75: 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita M. L., Lidral A. C., Murray J. C., Field L. L., Maher B. S., et al. , 2009. Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals phenotype specific differences in linkage and association results. Hum. Hered. 68: 151–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. C., Henikoff S., 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafighdoost H., Hashemi M., Narouei A., Eshanadri-Nasab E., Dashti-Khadivake G., et al. , 2013. Association between CDH1 and MSX1 gene polymorphisms and the risk of monsyndromic cleft lip and/or cleft palate in a southeast Iranian population. Cleft Palate Craniofac. J. 50: e98–e104. [DOI] [PubMed] [Google Scholar]
- Rao D. C., 2008. An overview of the genetic dissection of complex traits. Adv. Genet. 60: 3–34. [DOI] [PubMed] [Google Scholar]
- Vogelaar I. P., Figueiredo J., van Rooij I. A., Simões-Correia J., van der Post R. S., et al. , 2012. Identification of germline mutations in the cancer predisposing gene CDH1 in patients with orofacial clefts. Hum. Mol. Genet. 22: 919–926. [DOI] [PubMed] [Google Scholar]
- Wyszynski D. F., Albacha-Hejazi H., Aldirani M., Hammod M., Shkair H., et al. , 2003. A genome-wide scan for loci predisposing to non-syndromic cleft lip with or without cleft palate in two large Syrian families. Am. J. Med. Genet. A. 123A: 140–147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.