Abstract
The assumption that pleiotropic mutations are more deleterious than mutations with more restricted phenotypic effects is an important premise in models of evolution. However, empirical evidence supporting this assumption is limited. Here, we estimated the strength of stabilizing selection on mutations affecting gene expression in male Drosophila serrata. We estimated the mutational variance (VM) and the standing genetic variance (VG) from two well-matched panels of inbred lines: a panel of mutation accumulation (MA) lines derived from a single inbred ancestral line and a panel of inbred lines derived from an outbred population. For 855 gene-expression traits, we estimated the strength of stabilizing selection as s = VM/VG. Selection was observed to be relatively strong, with 17% of traits having s > 0.02, a magnitude typically associated with life-history traits. Randomly assigning expression traits to five-trait sets, we used factor analytic mixed modeling in the MA data set to identify covarying traits that shared pleiotropic mutations. By assigning traits to the same trait sets in the outbred line data set, we then estimated s for the combination of traits affected by pleiotropic mutation. For these pleiotropic combinations, the median s was three times greater than s acting on the individual component traits, and 46% of the pleiotropic trait combinations had s > 0.02. Although our analytical approach was biased toward detecting mutations with relatively large effects, likely overestimating the average strength of selection, our results provide widespread support for the prediction that stronger selection can act against mutations with pleiotropic effects.
THE extent to which new mutations have pleiotropic effects on multiple traits, and ultimately on fitness is central to our understanding of the maintenance of genetic variation and the process of adaptation (Kondrashov and Turelli 1992; Otto 2004; Johnson and Barton 2005; Zhang and Hill 2005). Analyses of Fisher’s (1930) geometric model of adaptation have shown that a mutation with effects on many traits will have a reduced probability of contributing to adaptive evolution (Orr 2000; Welch and Waxman 2003; see also Haygood 2006). For a population close to its optimum under mutation–selection balance, a direct corollary of this is that selection must act more strongly against mutations with wider pleiotropic effects (Zhang 2012).
Evidence for the strength of selection increasing with the number of traits that are pleiotropically affected by a mutation is limited. At a phenotypic level, nonlinear (stabilizing) selection is much stronger on combinations of metric traits than on each individual trait contributing to the combination (Blows and Brooks 2003; Walsh and Blows 2009). Given that genetic correlations among such traits are expected to be a consequence of pleiotropic alleles (Lande 1980), stronger selection on trait combinations is consistent with stronger selection on pleiotropic mutations that are likely to underlie the genetic covariance among such traits. There is some evidence that per-trait allelic effects might be greater for alleles with more widespread pleiotropic effects (Wagner et al. 2008; Wang et al. 2010); as mutations with larger phenotypic effects might be more effectively targeted by selection, this also suggests stronger selection against more pleiotropic mutation.
Mutation accumulation (MA) breeding designs, in which the opportunity for selection is reduced, allowing new mutations to drift to fixation, provide an opportunity to characterize the strength of selection acting directly against new mutations. Rice and Townsend (2012) proposed an approach for determining the strength of selection acting against mutations at individual loci, combining information from QTL mapping and MA studies. This approach could conceivably be extended to associate the strength of selection with the number of traits a QTL affects. More typically, estimates of selection from MA designs are focused on traits, rather than alleles. Under the assumption that most mutations are deleterious, an assumption supported by MA studies (Halligan and Keightley 2009), the strength of selection acting on mutations affecting quantitative traits can be measured as the ratio of the mutational to the standing genetic variance, s = VM/VG, where s is the selection coefficient of the mutation in heterozygous form (Barton 1990; Houle et al. 1996). While estimating s in this way provides a framework for estimating selection on pleiotropic combinations of traits, we are not aware of any studies adopting this approach to directly estimate the strength of selection acting on mutations affecting multiple traits.
Within an MA framework, Estes and Phillips (2006) manipulated the opportunity for selection, providing rare direct evidence of stronger selection against mutations with pleiotropic effects. In a DNA repair-deficient strain of Caenorhabditis elegans, Estes and Phillips (2006) observed lower mutational covariance among life-history components when selection was allowed (larger populations) than when the opportunity for selection was limited (small populations). Similarly, McGuigan et al. (2011) compared Drosophila serrata MA lines accumulating mutations in the presence or absence of sexual selection on males, reporting reduced covariance between two fitness components in the selection treatment. These studies reveal that selection can eliminate nonlethal alleles with pleiotropic effects, but whether traits other than life-history components exhibit similar evidence of selection against pleiotropic alleles remains unknown.
In parallel to the quantitative genetic predictions that pleiotropic alleles will be under stronger selection, molecular genetic theory predicts that the rate of gene evolution will be negatively correlated with pleiotropy (Pal et al. 2006; Salathe et al. 2006). More highly pleiotropic genes, as identified through the extent of connectivity (the number of interactions) in protein–protein interaction networks (Jeong et al. 2001), or the number of gene ontology (GO) terms (Jovelin and Phillips 2009) are more likely to be essential (i.e., knockout mutations result in lethality), suggesting that selection is stronger against large-effect (knockout) mutations in more highly pleiotropic genes. However, the selection acting against small-effect, nonlethal mutations in pleiotropic genes is less clear (Pal et al. 2006). Several studies have found an association between gene pleiotropy indices, such GO annotation of the number of biological processes or tissue specificity of expression, and the rate of sequence evolution (e.g., Pal et al. 2001; Salathe et al. 2006; Jovelin and Phillips 2009; Su et al. 2010). These pleiotropy indices typically explain little of the variation in sequence evolutionary rates, and it remains unclear whether more highly pleiotropic mutations are typically under stronger selection (Pal et al. 2006; Salathe et al. 2006).
Here, we estimate the selection coefficients acting against naturally occurring mutations affecting gene-expression traits in male D. serrata to quantitatively test if selection is stronger on mutations that affect multiple traits. Gene-expression phenotypes are uniquely positioned to enable detailed investigations of pleiotropy: there are many of them, they represent a broad coverage of biological function, they can be analyzed to quantify developmental pleiotropy in the same way as traits traditionally considered in quantitative genetics, and GO information can be used to index molecular genetic pleiotropy. We use multivariate mixed-model analyses of expression traits in a set of inbred lines from a mutation accumulation experiment to estimate the mutational variance in individual expression traits, and the pleiotropic mutational covariance among random sets of five expression traits. Using a second panel of inbred lines, derived from a natural, outbred, population, we estimate the standing genetic variance in the same individual traits and five-trait combinations. From these estimates of mutational and standing genetic variance, we calculate s for each of the individual traits and trait combinations to determine whether selection has typically been stronger on mutations with pleiotropic effects than on other mutations affecting each trait. We complement this quantitative genetic analysis of developmental pleiotropy with an analysis of molecular genetic pleiotropy (Paaby and Rockman 2013), determining whether the strength of selection acting on individual expression traits can be predicted from the number of biological functions that the gene annotates to in the GO database or to the range of tissues in which the gene is expressed.
Materials and Methods
Populations and data collection
Here, we estimate the selection coefficient of new mutations using two populations of D. serrata, which were established in a similar manner, maintained under the same culture conditions, and assayed in a similar way. The first, referred to as the mutation (M) population, consisted of a set of 45 inbred lines that were derived from a single, ancestral line that was inbred using full-sib mating for 13 generations and maintained through full-sib inbreeding for a further 27 generations in a MA experiment (McGuigan et al. 2011). The second, genetic (G) population, consisted of a set of 42 inbred lines, which were derived from females collected from a natural outbred population, with each iso-female line subjected to 15 generations of full-sib mating (Allen et al. 2013).
RNA sample collection was similar between the two sets of lines. In the generation prior to sample collection, replicate vials were established per inbred line to ensure that random microenvironmental variation was not confounded with variation among lines. Within each data set, all replicate breeding vials of all lines were established on the same day, and subsequent collection of virgins and RNA extractions were also performed on the same day for all lines within a data set. From the replicate rearing vials, a total of 40 (M-lines) or 60 (G-lines) virgin males were collected and held in groups of 5 until 3 (G) or 4 (M) days posteclosion. At that time, two random subsamples of 20 (M) or 30 (G) males were snap frozen on liquid nitrogen, and total RNA was extracted using Trizol (Invitrogen), and then purified using RNeasy kits (Qiagen), all according to manufacturer’s instructions. Pooling of RNA from multiple individuals is a standard approach for small organisms, such as Drosophila (Rifkin et al. 2005; Ayroles et al. 2009; Yang et al. 2011), particularly when individuals within a line are genetically nearly identical after many (here, at least 15) generations of full-sib inbreeding.
A microarray was designed for D. serrata as described in Allen et al. (2013) and manufactured by NimbleGen (Roche). Briefly, each array had 20K random probes, plus 11,631 features from ESTs, targeted by five 60-mer oligonucleotide probes; this set of 5 probes per 11,631 genes was fully replicated an each array, with each probe appearing twice. A single sample, with a single color (Cy-3), was hybridized to each array. Twelve arrays appeared on each slide, and samples within a data set were randomly assigned to a slide, with the stipulation that replicate extractions from a single line did not appear on the same slide. cDNA synthesis, labeling, hybridization, and microarray scanning were performed by the Centre for Genomics and Bioinformatics, Bloomington, Indiana. We determined the quality of the array data using the BioConductor oligo package probe-level models (Gentleman et al. 2004; Carvalho and Irizarry 2010; Draghici 2012) and using the experimental metrics report provided by NimbleGen (Roche Nimblegen 2011). Due to poor quality data (e.g., high mean empty signal or strong local stains) in at least one replicate of a line, we discarded several lines, resulting in 41 M-lines and 30 G-lines retained for analysis. Due to lack of probe replication, 27 features on the microarrays were not included in analyses, resulting in 11,604 traits for further analysis. The expression data are available through the NCBI Gene Expression Omnibus (Edgar et al. 2002; Barrett et al. 2011) (G-lines, GSE45801; M-lines, GSE49815).
Analytical approach and preliminary analyses
We used a series of linear mixed models to allow us to characterize the mutational and standing genetic variation and covariation among traits. As described in detail in McGuigan et al. (2014), we took the mean of the log10 expression of the two replicates of each probe on an array, and variance standardized (mean = 0; SD = 1) these data prior to analyses to facilitate the multivariate analyses, where large differences in the scale of each trait within a single model can inhibit convergence. We did not perform any other preprocessing steps, preferring to allow the mixed model to partition variation (including technical variation) in signal intensity.
We describe the modeling approach taken in detail below, but here give an overview of preliminary results to explain how we defined a subset of expression traits for which we are able to contrast selection against pleiotropic mutation vs. the total selection on traits. McGuigan et al. (2014) presented a detailed description of the mutational variance in the M-lines. Of the 11,604 expression traits analyzed in that study, only 3385 (29%) exhibited among-M-line variance, with statistical support for mutational variance in 1035 traits, 533 of which remained significant after false discovery rate (FDR) correction (McGuigan et al. 2014). For the remaining 71% of traits, the restricted maximum likelihood (REML) estimates of among-mutation line variance were zero; negative estimates of variance were not permitted. There could be several causes for the large number of zero REML estimates of mutational variance. First, these expression traits might not have a genetic basis. McGuigan et al. (2014) also analyzed the G-lines to demonstrate that the vast majority of these same traits were heritable in that population of outbred lines, with only 489 (4.6%) traits displaying zero REML estimates of among-G-line variance components. Therefore we can discount this possibility. Second, these traits might have been affected by mutations, but with effect sizes too small to be detected by the analyses. Third, none of the 41 MA lines accumulated any mutations affecting these traits over the 27 generations that the experiment ran, during which an average of 34 mutations per MA line are expected to have accumulated (McGuigan et al. 2014). We cannot distinguish between a large class of mutations of very small effect, vs. a large class of traits with a relatively low mutation rate. Irrespective of which of these two explanations is responsible for the zero variance components, estimates of s for these traits (with a zero numerator) would be zero, but it is not known if they are under negligible selection or if we simply failed to sample any alleles in our study.
For the 3385 traits with nonzero variance among the M-lines, McGuigan et al. (2014) identified pleiotropic covariance among traits through multivariate mixed-model analyses (detailed below). This was done by assigning each of the 3385 variable traits to one of 677 five-trait sets; each trait appeared in only one five-trait set. Traits were assigned randomly, with no consideration of their biological function. Since the probability of detecting covariance among random sets of five traits will depend on the true number of traits that a mutation affects, this approach is biased toward detecting mutations with relatively widespread pleiotropic effects. The size (five) of each trait set was selected as a consequence of the mixed-model convergence behavior; models with more traits sometimes did not converge or had convergence times that were prohibitively long.
Although for individual traits we can estimate s, irrespective of whether the mutational variance surpasses any statistical threshold, identifying pleiotropic covariance does depend on statistical testing. If we want to understand how much of the selection a trait might experience as a consequence of pleiotropic mutations, we need to ensure that we are looking at mutations that demonstrably affected more than one trait. To detect mutational pleiotropic covariance among traits (details below) McGuigan et al. (2014) fit a model with a covariance structure that partitioned variation that is shared among traits in the five-trait set (covariance) from variation that is unique to each trait. Statistical support for the presence of a shared component of the variation reveals mutational covariance among traits. McGuigan et al. (2014) detected significant pleiotropic covariance in 245 of the 677 five-trait sets, 145 of which remained significant after FDR correction. These 245 five-trait sets therefore provide a subset of traits in which we can clearly characterize the effect of selection through pleiotropic effects.
As noted previously, 489 expression traits were invariant among the genetic lines; 303 of these invariant traits were not coincident with invariant traits in the mutation data set. Since selection coefficients are undefined when the standing genetic variance (denominator) is zero, we excluded any of the 245 five-trait sets that included traits that were not variable among G-lines. These matrices were excluded without reference to the magnitude of estimated mutational pleiotropy. An alternative analytical strategy would have been to reanalyze the M-lines for a new set of random five-trait matrices constructed from traits that were variable in both data sets, but we discounted this approach for two reasons. First, retaining the same five-trait sets retained comparability to the published results (McGuigan et al. 2014) and further analyses to be conducted on these data in future work. The assignment of traits to matrices can be found in supporting information, File S1. Second, the computational demands of repeating the very large number of multivariate mixed-model analyses of the two sets of lines (to construct the LRT; see below) for the 616 matrices in the M-data set was prohibitive.
The above criteria resulted in 855 (520 at FDR), individual expression traits for which we could estimate the strength of selection in a univariate fashion and for which we could then contrast the strength of selection acting on combinations of those traits within the 171 (104 at FDR) five-trait matrices. We therefore base our estimates of selection coefficients on the traits for which we have statistical evidence of pleiotropic effects, with the caveat that there might be a class of smaller-effect pleiotropic mutations in the matrices that were excluded by the statistical significance testing on mutational pleiotropic variance. We further consider in the Discussion how various factors, including our data subsetting approach, has resulted in our analyses describing only part of the total distribution of selection acting on mutations.
Estimating selection coefficients of individual gene-expression traits
To estimate the mutational and standing genetic variation in each trait, we ran separate mixed models for each trait in each data set. Analyses were implemented on the variance-standardized data within a restricted maximum-likelihood framework in SAS (v. 9.3), using the univariate model,
| (1) |
where, for each gene, the variance in standardized log10 mean expression, Y, of the kth replicate probe in the jth replicate extraction and hybridization of the ith line was partitioned to among-line variance (Line), among replicate extractions within lines (Rep), and the residual (ε) variance among the five replicate probes of each gene. To account for substantial differences in mean signal intensity among the five replicate probes of some genes in the G-data set, we fit replicate probe as a fixed effect. We observed a segregating factor in the M-lines that must have been present in the ancestor and was therefore not a product of mutation during the experiment (see McGuigan et al. 2014, Supporting Information). To prevent this factor from contributing to estimates of mutational (among-line) variance, a fixed effect was fit to remove the mean difference in trait expression between the two groups of lines with the two alternative forms of this segregating factor.
Selection coefficients (s) for each gene were estimated as the mutational variance divided by the genetic variance (VM/VG), where VG was the among-line variance from model 1, and VM was defined as the among-M-line variance from model 1 divided by twice the number of generations of mutation accumulation (here, 27 generations) to give the per-generation rate of input of mutational variance to this diploid population (Lynch and Walsh 1998). To return the estimates of s to the original scale (log10 signal intensity), we multiplied the estimates of s by the corresponding ratio (M/G) of the total phenotypic variance, estimated on the unstandardized data (i.e., on the log10 scale) using model 1. We note that our estimates of VG are biased upward compared to estimates of the additive genetic variance (VA) in an outbred population as a consequence of the inclusion of nonadditive sources of variance. It is therefore likely that our estimates of s are biased downward by this component of the experimental design when considering the strength of selection under more natural conditions.
Estimating selection coefficients of pleiotropic trait combinations
To allow us to directly compare mutational and standing genetic variances for the same multivariate combination traits, expression traits in the genetic data set were assigned to the same five-trait matrices as those previously used in the mutation data set. These random sets of five traits were analyzed separately in each data set using the multivariate form of model (1),
| (2) |
where X is a design matrix for the fixed effects (replicate probe per gene in the G-data set and the groups segregating the two different ancestral variants in the M-data set), ε is a diagonal matrix containing the residual (among probe mean) variances for each trait, Zl and Zr are design matrices for the line and replicate within line random effects respectively, and δl and δr are the covariance matrices for these effects. We fit two different types of covariance structures in these analyses. The among-line variance (δl) in the M-data set and the within-line variance (δr) in both data sets were modeled using a reduced rank factor-analytic (FA) structure,
| (3) |
where Λ was a lower triangular matrix of factor loadings for a single factor capturing the covariance shared among the five traits, and ψ was a diagonal matrix containing specific variances for each trait (Meyer 2009; McGuigan and Blows 2010). The FA structure has two important benefits over other covariance structures for our analyses. First, the relatively low variation in the M-data set caused problems with model convergence, and fitting a reduced rank model at the within-line level improved behavior of the models. To minimize differences in the way that the two data sets were handled, we also fitted a reduced rank model to the within-line level of the G-data set. In the G-data set, we confirmed that this resulted in almost identical estimates of the variance component used to estimate s as fitting an unstructured covariance matrix at the within-line level.
Second, and most importantly, fitting the FA model structure at the among-mutation-line level allowed us to partition the variation shared among traits (the pleiotropic mutational variance) from the mutational variance unique to each trait. The single factor contained in Λ can then be used to estimate a reduced rank mutational covariance matrix with a single dimension, M (M = ΛΛT, where T indicates transpose). The single dimension of mutational covariance captured by this model (the eigenvector of M, m1) is the combination of the five randomly combined traits in a set that exhibits the greatest mutational covariance. Note that the trait-specific variances, which make up the diagonal of ψ, might still contain variation that is due to mutational pleiotropy; we do not test for the presence of more than one pleiotropically affected trait combination per matrix (i.e., we do not test for improved goodness of fit from FA[2] model over an FA[1] model). Our goal in this analysis was to identify some trait combinations corresponding to mutational pleiotropy for further investigation, but not to exhaustively identify all pleiotropic trait combinations within each five-trait set (an enormous task given the multivariate models involved).
As outlined above in preliminary analyses, McGuigan et al. (2014) analyzed the M-data set using model 2. Statistical support for pleiotropic covariance between at least two traits in a set was determined using log-likelihood ratio tests (LRT), comparing a model in which the common factor (i.e., pleiotropy) was fit vs. a model in which only trait-specific variances were estimated. This LRT has 5 d.fr. and is considered a conservative test because the precise mixture of χ2 distributions is unknown (Self and Liang 1987; Littell et al. 2006). McGuigan et al. (2014) applied a 5% false-discovery rate (FDR) (Benjamini and Hochberg 2000; Storey and Tibshirani 2003) correction based on these P- values, using the MULTTEST procedure in SAS.
In the G-data set, the among-line (i.e., genetic) variance was modeled using an unstructured covariance structure, resulting in a 5 × 5 unstructured covariance matrix (G). This analytical difference between the two data sets was required to allow us to directly compare the level of genetic variance found in the major axis of mutational variance, m1, with the standing genetic variance for the same trait combination in the outbred population. This required a full-rank covariance to be estimated in the G-data set. We then estimated the standing genetic variance in m1 by projecting the normalized (m1T m1 = 1) vector through the total genetic space to find the genetic variance in this trait combination, , using
| (4) |
We calculated the strength of stabilizing selection acting on the multivariate combination of traits as , where λm1 was the among-M-line variance (the eigenvalue of the common variance component from model 2), divided by twice the number of generations (here, 27 generations) to give the per-generation input of variance (Lynch and Walsh 1998), and came from Equation 4. To return s to the original log10 scale, we multiplied it by the ratio (M/G) of phenotypic variances in the m1 trait combination, on the original scale. To do this, we first calculated values of the index trait m1 by applying the equation yijkTm1i, where m1i was the normalized vector from the ith five-trait set, and yijk is the ith trait set five-element vector of log10 (unstandardized) expression trait replicate probe means for the kth replicate of the jth MA or G line. We subjected these univariate trait scores for the m1 index to analysis through model 1 to estimate the phenotypic variance associated with each m1 trait combination in the M- and G-data sets.
Exploring the estimation bias in m1
There are several potential sources of bias that could inflate the estimate of s associated with the pleiotropic trait combinations over the univariate trait estimates. First, we determined whether s might be inflated simply by including more traits (and more variance) in the estimate. To address this possibility, we estimated s from the sum of the mutational and genetic trait variances for the five traits in each set.
Second, known biases are associated with estimation of eigenvalues (Hill and Thompson 1978; Meyer and Kirkpatrick 2008), and we took two separate approaches to explore the effect of these potential biases on our estimates. First, different statistical approaches were used to estimate the mutational and standing multivariate variance in m1; the mutational variance in m1 was estimated as an eigenvalue from fitting a reduced-rank covariance matrix, but the standing genetic variance in m1 was estimated using projection through a full-rank estimate of G. If the eigenanalysis overestimated the mutational variance in the trait combination m1, as might commonly occur in eigenanalyses, then s would be upwardly biased. To test whether the eigenvalue of m1 was inflated, we reestimated the mutational variance in m1 by creating a univariate trait representing m1 (as described above for estimating the phenotypic variance in the m1 traits on the log10 scale). These univariate trait values corresponding to m1 were then subjected to the same univariate mixed model as the original individual expression traits (model 1), and we compared the model 1 and model 2 estimates of mutational variance.
Finally, overestimation of the mutational variance in m1 in the M-data set could have occurred as a consequence of greater sampling error in this direction being misconstrued as mutational variance by the analysis. To directly estimate the amount of sampling error that could inflate our estimates of mutational variance in m1, we employed a randomization approach. Observations for each trait were shuffled among MA lines, while keeping replicate measures within each line together. This shuffling ensured that estimates of variance of each trait remained the same at the among-line, within-line, and residual levels, but covariances among traits were disrupted. McGuigan et al. (2014) used this approach to demonstrate that the significant pleiotropic mutational variance detected by model 2 is lost when covariances among traits were disrupted. Here, the shuffled data set, subdivided into the same five-trait sets, was then subjected to analysis using model 2, fitting an unconstrained covariance matrix at the among-line level. We then projected the m1 vector estimated for that trait set in the analysis of the observed (not shuffled) M-data through the shuffled matrix using Equation 4. This analysis provided a direct estimate of variation in m1 due to random associations generated by sampling error in the data set and consequently allowed a quantitative correction to be applied to our estimates of mutational variance in m1.
Molecular genetic pleiotropy and selection
Information on the products of a gene, such as obtained from annotations in the GO project, can be used to determine pleiotropy. Importantly, the inference of pleiotropy from GO corresponds to Paaby and Rockman’s (2013) “molecular gene pleiotropy,” as opposed to the “developmental pleiotropy” that we characterize through the mixed-model analyses of variance in expression intensity. Therefore, in this study we define pleiotropy in two independent ways and determine the relationship between the strength of selection and pleiotropy from both gene and allele (mutation)-centric views.
Stand-alone BLASTX 2.2.27+ (Altschul et al. 1990) was used locally to search the nonredundant protein database (downloaded April 17, 2013) (Pruitt et al. 2012) for matching sequences; we did not restrict the search to specific taxa, but searched the entire nonredundant protein database. Gene ontology terms (Ashburner et al. 2000) (release September 2013) were then annotated to the genes via Blast2GO 2.7.0 (Conesa et al. 2005) using default settings. A total of 1813 genes were successfully annotated to at least one GO biological process (BP) term. Because here we consider each individual gene, and define pleiotropy through the molecular properties of that gene, we consider all genes for which a univariate selection coefficient could also be estimated (i.e., among-line variance components in both the M- and G-data sets were greater than zero in model 1). This resulted in a subset of 1436 genes for which we could estimate both s and which annotated to at least one BP GO term. Although theoretically both BP and molecular function (MF) terms could indicate gene pleiotropy, we restrict consideration to BP terms. Pleiotropy in yeast has been correlated with the number of biological processes a gene participates in, but not with the number of molecular functions of the gene (He and Zhang 2006). From the perspective of trait covariances, when a gene is involved in multiple functions, pleiotropy might be caused by different (linked) sites within the gene; for genes with a single function (although potentially involved in multiple processes), pleiotropy might be generated by a single, unlinked polymorphism (Chen and Lubberstedt 2010). Therefore, defining pleiotropy by the number of BP is expected to be most consistent with the pleiotropy defined through the mixed modeling of among mutation line variance.
We also considered the range of tissues in which a gene might affect biological processes as a measure of pleiotropy. Extent of expression is correlated with the rate of nonsynonymous substitutions (Hastings 1996; Duret and Mouchiroud 2000). τ is a metric describing the tissue specificity of gene expression, ranging from 0, for so-called housekeeping genes that are expressed broadly across many tissue types (i.e., highly pleiotropic), to 1 for genes with expression restricted to one tissue type (Yanai et al. 2005). As described in Allen et al. (2013), we calculated τ for the same subset of 1436 D. serrata genes based on measures of gene expression in nine different tissue types (male- and female-only dissections of head, thorax, and abdomen plus ovaries, testes, and accessory gland); the tissue-specific expression data can be found in the Gene Expression Omnibus G-line accession no. GSE45801.
Because of the nonnormal distribution of s and of the two molecular metrics of pleiotropy, we used Spearman’s rank coefficient to determine whether the strength of selection increased with the number of annotated processes or decreased with increasing tissue specificity of the genes. We used a permutation test, implemented in PopTools (Hood 2009) to determine whether the strength of selection was significantly correlated with either molecular measure of pleiotropy. Gene rank was shuffled 10,000 times for each metric (s, τ, and the number of annotated terms), and Spearman’s rank coefficient was calculated for each replicate. Significance was assessed through one-tailed tests at α = 0.05; estimated correlations were considered significantly different if they were greater/less than the 500th highest/lowest random correlation.
Results
Selection coefficients of individual gene-expression traits
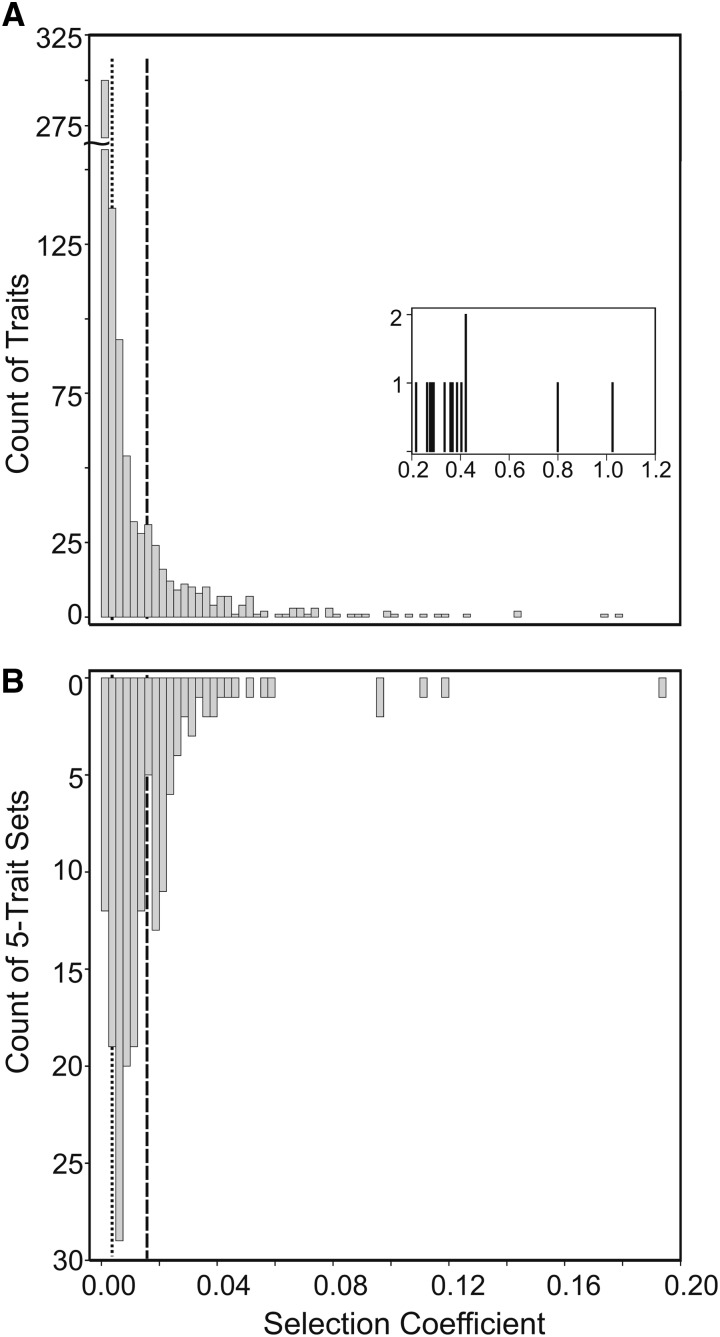
The distribution of individual selection coefficients was L shaped, with most traits under relatively weak stabilizing selection and relatively few traits under strong selection (Figure 1A). The median strength of selection on each of the 855 expression traits was 0.0047. A similar distribution of the strength of selection was observed for mutations affecting expression of 3696 genes in C. elegans, where the median s was ∼0.005 (Denver et al. 2005, Supplementary Figure 3). The strength of selection acting against mutations affecting expression traits appears comparable to the strength of selection acting on mutations affecting morphological traits, where the median s estimated from published data on a range of traits and taxa is 0.0087 (Houle et al. 1996). The median strength of selection acting against viability mutations has been estimated as 0.02 (Houle et al. 1996); 17% of the estimates of s for individual expression traits in male D. serrata exceeded this value (Figure 1A).
Figure 1.
The distribution of selection coefficients (on the original log10 expression intensity scale) for (A) univariate traits and (B) m1. Sixteen individual expression traits in A had s >0.2; with the exception of one extreme, unplotted, univariate estimate (s = 16.2), these have been plotted in the inset to enhance visualization of the lower part of the distribution. The dotted and dashed lines respectively indicate the median s for individual traits (dotted line, 0.0047) and the three-times higher median s for m1 (dashed line, 0.0159).
More than 75% of the estimates of s (Figure 1A) were larger than the neutral expectation (s = 1/2Ne), assuming an effective population size (Ne) of 500 for population of D. serrata sampled for the outbred lines. This is a conservative assumption because 500 would be considered a small population size for most Drosophila species (Barker 2011), and assuming a larger Ne decreases the strength of selection expected under neutrality, resulting in a higher proportion of the estimates of s being greater than the neutral expectation. It should be remembered, however, that we have only included in this analysis traits that contributed to significant mutational covariance; individual traits that were randomly assigned to sets with four other traits with which they did not share statistically significant mutational covariance are missing from this distribution. For these other 2223 traits for which s could be estimated (i.e., nonzero genetic and mutational variance), but were not included in the multivariate analyses, the median s was slightly lower (0.0040). However, 81% of these traits had estimates of s larger than the neutral expectation, and 15% of these estimates of s were >0.02. This indicates that conclusions about selection on individual traits were not markedly biased by our pleiotropy selection criterion.
Selection coefficients of pleiotropic trait combinations
The median selection coefficient for the five-trait vectors was 0.0359, suggesting substantially stronger selection on the pleiotropic trait combination than on the individual constituent traits (for which the median s was 0.0047). We explored three potential biases that might have inflated the estimated strength of selection against the m1 trait combination. First, and most simplistically, the observation of stronger selection on m1 than on individual traits was not a consequence of considering more traits (and variance) in these trait combinations. Selection acting on the sum of the five individual trait variances was of a similar magnitude to the univariate traits themselves, with a median s of 0.0032.
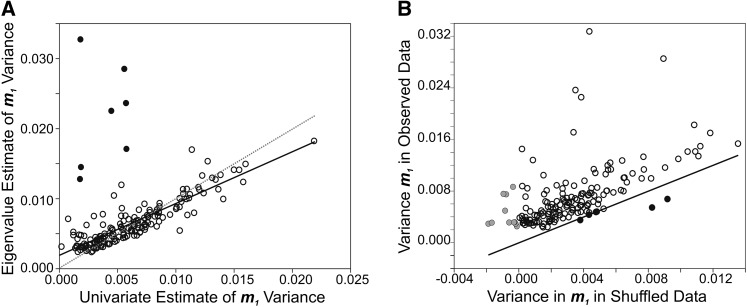
Second, to determine whether the multivariate analysis overestimated the variance in the trait combination m1, we compared the FA(1) model eigenvalue estimate of variance in m1 with the variance estimated through the univariate model analysis of m1 index trait scores. There were seven m1 trait combinations for which the eigenvalue estimate of variance was >2 SD greater than the univariate model estimate (Figure 2A). Excluding these seven, the average relationship between eigenanalysis and univariate index trait estimates for the remaining 164 m1 was close to the 1:1 expected in the absence of any bias (Figure 2A) (paired t-test: mean difference [eigenanalysis m1 – univariate model m1] = 0.00025, t163 = 1.859, P = 0.065). On average, the eigenanalysis m1 variance was 9.6% higher than the univariate estimate of variance in m1; applying this average bias correction reduced the m1 median s from 0.0359 to 0.0324, which was still 6.9 times greater than the univariate median estimate (0.0047). Therefore, estimating the mutational variance in m1 as the eigenvalue of the FA(1) M and the genetic variance in m1 by projection through an unconstrained G is unlikely to have strongly biased the estimates of s.
Figure 2.
Examining the bias in the eigenvalue estimates of pleiotropic covariance. (A) We estimated the per-generation mutational variance in the trait combination defined by the eigenvector m1 both as the eigenvalue of m1 (from multivariate analysis using model 2; y-axis) and from the univariate analysis of index traits defined by m1 (the univariate mutational variance from model 1; x-axis). Estimates for all 171 m1 are plotted, with points shaded in black falling more than two standard deviations from the mean difference between the two estimates per trait. The dotted line indicates the 1:1 relationship between estimates, and the solid line indicates the linear regression through the observed points (excluding the seven, shaded, outliers). (B) The per-generation mutational variance in m1 in the observed data (eigenvalue of m1, y-axis) and in shuffled data (after projection, x-axis). The solid line indicates equality (1:1) of variance in shuffled and observed data; six m1 fall below this line and are shown in solid black. The 11 m1 for which there was no variance in the shuffled data (variance less than zero) are shaded.
Finally, we explored the bias that could exist in our data if the direction of m1 was biased by sampling variation in the data. The magnitude of variance in the direction of m1 in the shuffled data set was substantial, corresponding to ∼50% of the observed variance in m1 (Figure 2B). Nonetheless, the variance in the shuffled data was less than in the observed data in all but six cases (Figure 2B). Because we estimated the among-shuffled-line covariance matrices using an unconstrained structure, they could be negative definite; for 11 trait sets, the vector m1 fell within the null space, and the estimated variance in the shuffled data was negative (Figure 2B), indicating that there was no sampling variation in that direction.
We used the estimates of variance in the shuffled data to rescale our estimates of per-generation mutational variance in m1, using the difference between the two estimates (observed – shuffled). To do this, we set negative estimates of shuffled variance to zero (a variance cannot truly be negative, and allowing a negative estimate would have inflated the corrected estimate), and we set negative estimates of the difference to zero (i.e., when there was more variance in the direction of m1 in the shuffled than observed data, we assumed that there was no mutational variance).
Following this correction, the median selection coefficient acting against mutational variance in the five-trait combination m1 vectors was 0.0159 (Figure 1B). That is, selection acting on the pleiotropic mutations causing variance in the trait combination described by m1 was 3.4 times stronger than the median selection acting on the individual traits comprising the five-trait sets. To determine if selection was significantly stronger on trait combinations that shared mutational covariance than individual traits, we used a nonparametric approach based on sign tests as the distribution of the s values is nonnormal (Figure 1), and the univariate and multivariate values for each matrix are paired. For each five-trait matrix, we applied a sign test on the comparison of the five individual univariate trait values of s to the multivariate s value. The P-values from the 171 sign tests were then combined using Fisher’s combined probability test (χ2 = 482, d.f. = 342, P = 1.3 × 10−6), indicating that selection on the multivariate combination was consistently stronger than on univariate traits across the total set of traits.
Molecular genetic pleiotropy and selection
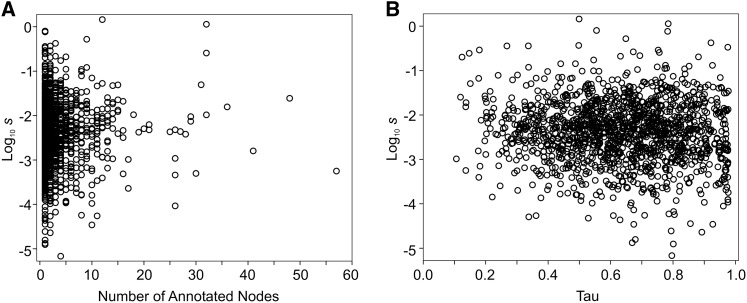
Finally, we complimented our quantitative genetic analyses of developmental pleiotropy with an assessment of the relationship between the strength of selection and molecular gene pleiotropy. The 1436 genes under consideration annotated to between 1 and 57 biological process GO terms, with a median of 2 (Figure 3A). That is, most of the genes considered were inferred by this metric to have relatively low levels of pleiotropy. Consistent with this, tissue specificity was also typically relatively high, with a median of 0.6 (Figure 3B). The relationship between the strength of selection and the number of GO terms to which a gene was annotated was in the predicted direction, but was very weak and nonsignificant (Spearman’s ρ = 0.035, P = 0.0862). Similarly, as the tissue specificity of the gene’s expression decreased, potentially allowing effects on more processes, the strength of selection increased, but the relationship was again weak (Spearman’s ρ = −0.047, P = 0.0369). Nonneutral sequence divergence between D. serrata and organisms from which gene ontology data are available, such as D. melanogaster, might have biased these analyses if genes with sufficient sequence similarity to be annotated were those that experienced stronger stabilizing selection. The median s for the 1436 genes included in the GO analyses was 0.0045, very similar to that observed for the remaining traits that we could estimate s for, suggesting little bias from this source.
Figure 3.
The relationship between the strength of selection estimated from mutational and standing genetic variance and two metrics of molecular gene pleiotropy. The strength of selection is plotted on a log scale for illustration, but was not transformed in this way for analyses. (A) s plotted against the number of biological process GO terms to which the gene annotated (Spearman’s rank coefficient 0.035, P = 0.0862). (B) s plotted against the tissue specificity metric, τ, which ranges from 0 (broadly expressed and more pleiotropic) to 1 (tissue specific with low pleiotropy) (Spearman’s rank coefficient = −0.047, P = 0.0369).
Discussion
Since Fisher’s (1930) development of the geometric model of the process of adaptation, it has been assumed that a mutation that affects many parts of a complex organism is less likely to be beneficial than a mutation of more restricted effect. Direct evidence supporting the generality of this supposition has been difficult to obtain, in part because of the challenge of studying high-dimensional phenotypes (Houle 2010) and because of the lack of information on the relationship between effects of pleiotropic alleles on phenotypes vs. on fitness (Paaby and Rockman 2013). By using multivariate statistical modeling of the genetic variance generated by new mutations, and of the standing genetic variance in a natural population, we have shown that selection is consistently stronger on pleiotropic mutations, supporting Fisher’s model of adaptation for populations in the vicinity of an adaptive optimum (Zhang 2012).
In general, we inferred strong selection acting against new mutations affecting the adult male D. serrata expression of genes assayed in this study. Many individual traits appeared to be under stronger selection than expected under neutral evolution, and 17% had selection coefficients >0.02, a magnitude of selection commonly associated with life-history traits, and with viability in D. melanogaster (Houle et al. 1996). The proportion of traits with estimated selection coefficients comparable to those of fitness traits increased to 46% (after bias correction) for the pleiotropic trait combinations. Analyses of divergence in gene-expression profiles among taxa have also suggested a prevailing pattern of relatively strong stabilizing selection within taxa (Denver et al. 2005; Rifkin et al. 2005; Bedford and Hartl 2009; Warnefors and Eyre-Walker 2012), although per-trait stabilizing selection might be very weak (nearly neutral) (Bedford and Hartl 2009). It is possible that purifying selection acts directly against mutations affecting gene expression through energy costs of transcription (Wagner 2005). Alternatively, the fitness consequences of gene expression might be indirect, mediated through the effect of expression on other traits, including morphology, behavior, and physiology.
Evidence for widespread stabilizing selection acting on phenotypes within populations has been surprisingly difficult to detect (Kingsolver et al. 2012), despite the overriding importance of stabilizing selection for explaining patterns of divergence over long time periods (Estes and Arnold 2007). Nonlinear (stabilizing) selection has previously been observed to be much stronger on combinations of standard metric traits than on each individual trait contributing to the combination (Blows and Brooks 2003; Walsh and Blows 2009). Our results suggest that pleiotropic mutations are likely to be under stabilizing selection stronger than that of mutations with more limited phenotypic effects and that the strength of selection acting against such pleiotropic alleles cannot be inferred from the analysis of individual traits in isolation.
We found that many expression traits are under strong stabilizing selection and that selection acts more strongly against mutations affecting multiple traits than would be expected on the basis of selection acting on each of the traits individually. This conclusion is based on a subset of all the traits that we measured, and there are a number of aspects of the experimental design and analysis that must be considered when placing these results into a broader context. First, the classical mutation accumulation design that we employ allows us to infer mutation from the distribution of line means, and the relatively few generations (27) and inferred mutations per line (34) suggest that it is reasonable to assume that each line segregates only a single mutation affecting any particular trait. However, our analyses are focused on the traits, not the mutations themselves, and consequently, we cannot exclude the possibility that lines are segregating mutations with antagonistic pleiotropic effects, canceling one another out and resulting in no observed covariance among traits. We cannot infer the selection acting on this class of pleiotropic mutations.
Second, we defined a subset of traits for analysis based on several criteria. First, we excluded all traits for which we could detect no variation among M-lines. These traits might have been segregating mutations of very small effect (or multiple antagonistic mutations, canceling each other’s effects). Considering all 11,604 expression traits that we measured, the median s is zero, with no evidence of selection. The median selection acting on individual expression traits is therefore likely to be lower than the value of 0.0047 we have reported. Nonetheless, many expression traits were observed to be under strong selection.
Third, our approach to defining a subset of data will have excluded many mutations with pleiotropic effects, but where the covariance generated was small and not statistically detected. Because we are observing variation in phenotypes, not the mutations themselves, we require statistically significant covariance among traits to infer that a pleiotropic mutation has occurred. Excluding other mutations with smaller pleiotropic effects across traits might have resulted in an overestimation of the strength of selection acting on pleiotropic mutations. Importantly, we measure both the total selection acting on each of the 855 included traits and the selection that is acting through a known pleiotropic mutation affecting those traits. The univariate estimate of s for each trait includes mutational variance that is due just to mutations affecting that trait and any mutations that might also have pleiotropic effects on other traits. The key result of this article is that, for the same set of traits, selection acting on pleiotropic combinations of traits is, on average, three times stronger than the selection acting on each trait in isolation.
A final aspect of mutation accumulation experiments that should be considered is the selection acting within the population of lines. Mutations causing a >10% decline in fitness are unlikely to accumulate in MA experiments employing brother–sister mating (Lynch et al. 1999), and selection against these mutations can act either through line extinction or against further accumulation within a line (Schaack et al. 2013). We previously demonstrated that lines that went extinct during the experiment were phenotypically distinct from extant lines (McGuigan and Blows 2013). Mutations eliminated by viability selection (by definition, under strong purifying selection) might have differed in their pleiotropic effects from mutations fixed (and assayed here).
The number of annotated gene ontology terms for each gene, a pleiotropy metric that has previously been associated with gene knockout effects across multiple environments (He and Zhang 2006), and with the rate of sequence evolution (Salathe et al. 2006; Jovelin and Phillips 2009) was weakly associated with our estimate of the strength of selection, as was the tissue specificity of gene expression. Although previous studies have been suggestive of stronger selection on mutations in more pleiotropic genes, these relationships have typically been weak, with the pleiotropy metric explaining very little of the variance in evolutionary rates (Pal et al. 2006; Salathe et al. 2006). It remains to be determined whether this weak relationship reflects the incomplete information available to determine gene pleiotropy or whether the true relationship is between the pleiotropic effects of a locus and the selection acting on mutations affecting the expression of that locus. Other metrics derived from GO might have been utilized in this study. For instance, the strength of selection on gene expression varies with the functional categories a gene annotates to in nematodes (Denver et al. 2005); however, no independent information on pleiotropy is used to define these categorical divisions, and such comparisons are difficult to interpret if genes annotate to multiple categories (Rhee et al. 2008). Finally, it is not clear how metrics describing connections among genes based on analyses of GO-directed acyclic graphs (Dameron et al. 2013) relate to real interactions among genes, given the parent–child nature of these graphs (Rhee et al. 2008). There has been considerable debate in the literature about the effect of pleiotropy (inferred in many ways) on the rate of protein sequence evolution (Pal et al. 2006). Useful characterization of molecular interactions underlying pleiotropy might typically require more nuanced detailed focus, rather than relying on broad-scale patterns (Kopp and Mcintyre 2012).
In conclusion, selection on pleiotropic combinations of traits was found to be over three times stronger than that on the individual expression traits. An important consequence of strong selection on pleiotropic mutations is that the pattern of standing genetic variance is substantially different from how new genetic variance is generated by mutation. That selection appears to shape the standing genetic variance to such an extent implies that a mutation–selection balance (Johnson and Barton 2005; Zhang and Hill 2005) may be a key process in the maintenance of genetic variance in natural populations.
Supplementary Material
Acknowledgments
We thank J. Thompson, C. Oesch-Lawson, D. Petfield, F. Frentiu, O. Sissa-Zubiate, and T. Gosden for line maintenance and data collection. Comments from Nick Barton and two anonymous reviewers greatly improved the clarity of the article, and we are grateful to one reviewer who encouraged us to fully explore the sources of bias in the estimation of the multivariate mutational variance. This work was funded by the Australian Research Council.
Footnotes
Communicating editor: N. H Barton
Literature Cited
- Allen S. L., Bonduriansky R., Chenoweth S. F., 2013. The genomic distribution of sex-biased genes in Drosophila serrata: X-chromosome demasculanistaion, feminisation, and hyper-expression in both sexes. Genome Biol. Evol. 5: 1986–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., et al. , 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J. S. F., 2011. Effective population size of natural populations of Drosophila buzzatii, with a comparative evaluation of nine methods of estimation. Mol. Ecol. 20: 4452–4471. [DOI] [PubMed] [Google Scholar]
- Barrett T., Troup D. B., Wilhite S. E., Ledoux P., Evangelista C., et al. , 2011. NCBI GEO: archive for functional genomics data sets-10 years on. Nucleic Acids Res. 39: D1005–D1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 1990. Pleiotropic models of quantitative variation. Genetics 124: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T., Hartl D. L., 2009. Optimization of gene expression by natural selection. Proc. Natl. Acad. Sci. USA 106: 1133–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 2000. On the adaptive control of the false discovery fate in multiple testing with independent statistics. J. Educ. Behav. Stat. 25: 60–83. [Google Scholar]
- Blows M. W., Brooks R., 2003. Measuring nonlinear selection. Am. Nat. 162: 815–820. [DOI] [PubMed] [Google Scholar]
- Carvalho B. S., Irizarry R. A., 2010. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26: 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. S., Lubberstedt T., 2010. Molecular basis of trait correlations. Trends Plant Sci. 15: 454–461. [DOI] [PubMed] [Google Scholar]
- Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., et al. , 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Dameron O., Bettembourg C., andLe Meur N., 2013. Measuring the evolution of ontology complexity: the gene ontology case study. PLoS ONE 8.: e75993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver D. R., Morris K., Streelman J. T., Kim S. K., Lynch M., et al. , 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat. Genet. 37: 544–548. [DOI] [PubMed] [Google Scholar]
- Draghici S., 2012. Statistics and Data Analysis for Microarrays Using R and Bioconductor. Chapman & Hall, Boca Raton, FL. [Google Scholar]
- Duret L., Mouchiroud D., 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17: 68–74. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E., 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes S., Phillips P. C., 2006. Variation in pleiotropy and the mutational underpinnings of the G-matrix. Evolution 60: 2655–2660. [PubMed] [Google Scholar]
- Estes S., Arnold S. J., 2007. Resolving the paradox of stasis: models with stabilizing selection explain evolutionary divergence on all timescales. Am. Nat. 169: 227–244. [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford. [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., et al. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan D. L., Keightley P. D., 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 40: 151–172. [Google Scholar]
- Hastings K. E. M., 1996. Strong evolutionary conservation of broadly expressed protein isoforms in the troponin I gene family and other vertebrate gene families. J. Mol. Evol. 42: 631–640. [DOI] [PubMed] [Google Scholar]
- Haygood R., 2006. Mutation rate and the cost of complexity. Mol. Biol. Evol. 23: 957–963. [DOI] [PubMed] [Google Scholar]
- He X. L., Zhang J. Z., 2006. Toward a molecular understanding of pleiotropy. Genetics 173: 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Thompson R., 1978. Probabilities of non-positive definite between-group or genetic covariance matrices. Biometrics 34: 429–439. [Google Scholar]
- Hood, G. M., 2009 PopTools, CSIRO. http://www.cse.csiro.au/poptools.
- Houle D., 2010. Numbering the hairs on our heads: the shared challenge and promise of phenomics. Proc. Natl. Acad. Sci. USA 107: 1793–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D., Morikawa B., Lynch M., 1996. Comparing mutational variabilities. Genetics 143: 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Mason S. P., Barabasi A. L., Oltvai Z. N., 2001. Lethality and centrality in protein networks. Nature 411: 41–42. [DOI] [PubMed] [Google Scholar]
- Johnson T., Barton N., 2005. Theoretical models of selection and mutation on quantitative traits. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 1411–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R., Phillips P. C., 2009. Evolutionary rates and centrality in the yeast gene regulatory network. Genome Biol. 10.: R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsolver J. G., Diamond S. E., Siepielski A. M., Carlson S. M., 2012. Synthetic analyses of phenotypic selection in natural populations: lessons, limitations and future directions. Evol. Ecol. 26: 1101–1118. [Google Scholar]
- Kondrashov A. S., Turelli M., 1992. Deleterious mutations, apparent stabilizing selection and the maintenance of quantitative variation. Genetics 132: 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A., McIntyre L. M., 2012. Transcriptional network structure has little effect on the rate of regulatory evolution in yeast. Mol. Biol. Evol. 29: 1899–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., 1980. The genetic covariances between characters maintained by pleiotropic mutations. Genetics 94: 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littell R. C., Milliken G. A., Stroup W. W., Wolfinger R. D., Schabenberger O., 2006. SAS System for Mixed Models. SAS Institute, Cary, NC. [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer, Sunderland, MA. [Google Scholar]
- Lynch M., Blanchard J., Houle D., Kibota T., Schultz S., et al. , 1999. Spontaneous deleterious mutation. Evolution 53: 645–663. [DOI] [PubMed] [Google Scholar]
- McGuigan K., Blows M. W., 2010. Evolvability of individual traits in a multivariate context: partitioning the additive genetic variance into common and specific components. Evolution 64: 1899–1911. [DOI] [PubMed] [Google Scholar]
- McGuigan K., Blows M. W., 2013. Joint allelic effects on fitness and metric traits. Evolution 67: 1131–1142. [DOI] [PubMed] [Google Scholar]
- McGuigan K., Petfield D., Blows M. W., 2011. Reducing mutation load through sexual selection on males. Evolution 65: 2816–2829. [DOI] [PubMed] [Google Scholar]
- McGuigan K., Collet J., McGraw E. A., Ye Y. H., Allen S. L., et al. , 2014. The nature and extent of mutational pleiotropy in gene expression of male Drosophila serrata. Genetics 196: 911–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., 2009. Factor-analytic models for genotype x environment type problems and structured covariance matrices. Genet. Sel. Evol. 41: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Kirkpatrick M., 2008. Perils of parsimony: properties of reduced rank estimates of genetic covariance matrices. Genetics 180: 1153–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 2000. Adaptation and the cost of complexity. Evolution 54: 13–20. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., 2004 Two steps forward, one step back: the pleiotropic effects of favoured alleles. Proc. R. Soc. B Biol. Sci. 271: 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., Rockman M. V., 2013. The many faces of pleiotropy. Trends Genet. 29: 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C., Papp B., Hurst L. D., 2001. Highly expressed genes in yeast evolve slowly. Genetics 158: 927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C., Papp B., Lercher M. J., 2006. An integrated view of protein evolution. Nat. Rev. Genet. 7: 337–348. [DOI] [PubMed] [Google Scholar]
- Pruitt K. D., Tatusova T., Brown G. R., Maglott D. R., 2012. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40: D130–D135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S. Y., Wood V., Dolinski K., Draghici S., 2008. Use and misuse of the gene ontology annotations. Nat. Rev. Genet. 9: 509–515. [DOI] [PubMed] [Google Scholar]
- Rice D. P., Townsend J. P., 2012. A test for selection employing quantitative trait locus and mutation accumulation data. Genetics 190: 1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin S. A., Houle D., Kim J., White K. P., 2005. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature 438: 220–223. [DOI] [PubMed] [Google Scholar]
- Roche NimbleGen, 2011 NimbleGen Software: Guide to Interpreting the Experimental Metrics Report. Roche, Madison, WI. [Google Scholar]
- Salathe M., Ackermann M., Bonhoeffer S., 2006. The effect of multifunctionality on the rate of evolution in yeast. Mol. Biol. Evol. 23: 721–722. [DOI] [PubMed] [Google Scholar]
- Schaack S., Allen D. E., Latta L. C., Morgan K. K., Lynch M., 2013. The effect of spontaneous mutations on competitive ability. J. Evol. Biol. 26: 451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self S. G., Liang K.-Y., 1987. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J. Am. Stat. Assoc. 82: 605–610. [Google Scholar]
- Storey J. D., Tibshirani R., 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z. X., Zeng Y. W., Gu X., 2010. A preliminary analysis of gene pleiotropy estimated from protein sequences. J. Exp. Zool. B 314B: 115–122. [DOI] [PubMed] [Google Scholar]
- Wagner A., 2005. Energy constraints on the evolution of gene expression. Mol. Biol. Evol. 22: 1365–1374. [DOI] [PubMed] [Google Scholar]
- Wagner G. P., Kenney-Hunt J. P., Pavlicev M., Peck J. R., Waxman D., et al. , 2008. Pleiotropic scaling of gene effects and the ’cost of complexity’. Nature 452: 470–473. [DOI] [PubMed] [Google Scholar]
- Walsh B., Blows M. W., 2009. Abundant genetic variation + strong selection = multivariate genetic constraints: a geometric view of adaptation. Annu. Rev. Ecol. Evol. Syst. 40: 41–59. [Google Scholar]
- Wang Z., Liao B. Y., Zhang J. Z., 2010. Genomic patterns of pleiotropy and the evolution of complexity. Proc. Natl. Acad. Sci. USA 107: 18034–18039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnefors M., Eyre-Walker A., 2012. A selection index for gene expression evolution and its application to the divergence between humans and chimpanzees. PLoS ONE 7.: e34935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J. J., Waxman D., 2003. Modularity and the cost of complexity. Evolution 57: 1723–1734. [DOI] [PubMed] [Google Scholar]
- Yanai I., Benjamin H., Shmoish M., Chalifa-Caspi V., Shklar M., et al. , 2005. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21: 650–659. [DOI] [PubMed] [Google Scholar]
- Yang Y. J., Graze R. M., Walts B. M., Lopez C. M., Baker H. V., et al. , 2011. Partitioning transcript variation in Drosophila: abundance, isoforms, and alleles. Genes Genom. Genet. 1: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. S., 2012. Fisher’s geometrical model of fitness landscape and variance in fitness within a changing environment. Evolution 66: 2350–2368. [DOI] [PubMed] [Google Scholar]
- Zhang X. S., Hill W. G., 2005. Genetic variability under mutation selection balance. Trends Ecol. Evol. 20: 468–470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.