Abstract
Studies of X chromosome evolution in various organisms have indicated that sex-biased genes are nonrandomly distributed between the X and autosomes. Here, to extend these studies to nematodes, we annotated and analyzed X chromosome gene content in four Caenorhabditis species and in Pristionchus pacificus. Our gene expression analyses comparing young adult male and female mRNA-seq data indicate that, in general, nematode X chromosomes are enriched for genes with high female-biased expression and depleted of genes with high male-biased expression. Genes with low sex-biased expression do not show the same trend of X chromosome enrichment and depletion. Combined with the observation that highly sex-biased genes are primarily expressed in the gonad, differential distribution of sex-biased genes reflects differences in evolutionary pressures linked to tissue-specific regulation of X chromosome transcription. Our data also indicate that X dosage imbalance between males (XO) and females (XX) is influential in shaping both expression and gene content of the X chromosome. Predicted upregulation of the single male X to match autosomal transcription (Ohno’s hypothesis) is supported by our observation that overall transcript levels from the X and autosomes are similar for highly expressed genes. However, comparison of differentially located one-to-one orthologs between C. elegans and P. pacificus indicates lower expression of X-linked orthologs, arguing against X upregulation. These contradicting observations may be reconciled if X upregulation is not a global mechanism but instead acts locally on a subset of tissues and X-linked genes that are dosage sensitive.
Keywords: X chromosome evolution, dosage compensation, genomics, nematode, sex-biased gene expression
IN an XY sex-determination system, male and female genomes are identical with the exception of the male-specific Y chromosome, which bears few genes (Charlesworth et al. 2005). This is particularly true when the Y chromosome is thought to be completely lost, as is the case for C. elegans and many other nematodes (Walton 1940). Because gene content is the same, phenotypic differences between males and females, termed “sexual dimorphisms,” must be caused by differential gene expression between the two sexes (Connallon and Knowles 2005; Ellegren and Parsch 2007). Throughout the article, such differentially expressed genes are referred to as “sex biased.”
As males and females have different fitness optima, a trait that is beneficial to one sex can be harmful to the other (termed sexual antagonism) (Rice and Chippindale 2001; Arnqvist 2004; Connallon and Knowles 2005; Ellegren and Parsch 2007; Mank et al. 2008a; Rice 1984). The evolution of sex-biased gene expression is thought to mediate the effects of sexual antagonism and allow for achievement of sex-specific fitness. Previous studies have indicated that anywhere between 30 and 60% of metazoan genes may be sex biased (Ranz et al. 2003; Parisi et al. 2004; Reinke et al. 2004; Yang et al. 2006; Reinius et al. 2008; Small et al. 2009; Innocenti and Morrow 2010; Assis et al. 2012; Reinius et al. 2012; Thomas et al. 2012). Genes with sex-biased expression contribute to both somatic and gonadal sexual dimorphisms (Ranz et al. 2003; Parisi et al. 2004; Yang et al. 2006; Mank et al. 2008a; Reinius et al. 2008).
As the number of X chromosomes differs between males and females, the X plays a large role in the evolution of sexual dimorphisms (Rice 1984; Mank et al. 2008a). Due to both sex-specific natural selection and the unique life cycle of the X chromosome, it is predicted that sex-biased genes should accumulate on the X (Rice 1984). Because males are monosomic for the X (they bear only one copy of the X chromosome to two copies of each autosome), recessive alleles that emerge on the X chromosome are immediately visible to male-specific selection. Recessive male-benefitting alleles are predicted to increase in frequency on the X, even if the fitness cost to females is much greater (Rice 1984). Additionally, because the X spends two-thirds of its evolutionary history in females, dominant female-benefitting alleles are predicted to accumulate on the X chromosome.
Indeed, in many organisms, sex-biased genes are nonrandomly distributed between the X chromosome and the autosomes (Saifi and Chandra 1999; Wang et al. 2001; Lercher et al. 2003; Reinke et al. 2004; Parisi et al. 2003; Ranz et al. 2003; Khil et al. 2004; Divina et al. 2005; Yang et al. 2006; Sturgill et al. 2007; Reinius et al. 2012; Meisel et al. 2012a; Allen et al. 2013). However, it is not always the case that both male and female-biased genes are enriched on the X. Enrichment of female-biased genes on the X chromosome (“feminization of the X”) is observed in mammals, Drosophila, and Caenorhabditis elegans (Parisi et al. 2003; Ranz et al. 2003; Khil et al. 2004; Reinke et al. 2004; Reinius et al. 2012; Allen et al. 2013). The same studies also provide evidence for “demasculinization of the X chromosome,” where male-biased genes are largely excluded from the X. The degree to which the Drosophila X chromosome is depleted of male-biased genes is still being studied (Vibranovski et al. 2009a,b; Meiklejohn and Presgraves 2012; Meisel et al. 2012a). Older male-biased genes are underrepresented on the Drosophila X chromosome, while younger male-biased genes [those emerged after the melanogaster split ∼3–6 million years ago (Russo et al. 1995)] are enriched (Zhang et al. 2010). This suggests that X demasculinization is an evolutionary process (Gao et al. 2014). While enrichment of female-biased genes on the X is predicted, demasculinization of the X opposes the expectation that male-benefitting genes should accumulate on the X chromosome (Rice 1984).
One often-cited explanation for the depletion of male-biased genes from the X chromosome is meiotic sex chromosome inactivation (MSCI). In many species, the single unpaired X chromosome is transcriptionally silenced during male meiosis (Fong et al. 2002; Kelly et al. 2002; Turner 2007; Maine 2010). MSCI begins in early meiosis and persists throughout varying stages of spermatogenesis in different species (Turner 2007). Thus, genes necessary for spermatogenesis should move off of the X chromosome. Studies in mice and humans have found that male-biased genes expressed in the testes are enriched on the X chromosome (Wang et al. 2001; Lercher et al. 2003). It has been subsequently suggested that this enrichment may be true only for genes expressed during the earliest stages of spermatogenesis before the onset of MSCI (Khil et al. 2004). In C. elegans, sperm-enriched genes are depleted from the X chromosome (Reinke et al. 2000, 2004). Immunofluorescence analyses of various histone modifications in conjunction with microarray expression analyses provide strong evidence for the presence of MSCI in C. elegans (Kelly et al. 2002; Reuben and Lin 2002; Bean et al. 2004; Bessler et al. 2010). Throughout C. elegans spermatogenesis, histone modifications associated with active transcription are depleted from the X chromosome while those associated with transcriptional repression are enriched (Kelly et al. 2002; Reuben and Lin 2002; Bean et al. 2004; Bessler et al. 2010).
The C. elegans X chromosome is also repressed in the hermaphrodite germline (Reinke et al. 2000; Kelly et al. 2002; Reinke et al. 2004; Bender et al. 2006; Maine 2010). Germline X repression is restricted to early meiotic cells where the X is largely depleted of active chromatin marks, including H3K4me2 (Kelly et al. 2002). Early germline X silencing appears to be conserved in nematodes, since H3K4me2 is depleted from the X chromosome in each of the Pristionchus pacificus, C. briggsae, and C. remanei germlines (Kelly et al. 2002). Transcriptional repression of the X in both male and female germlines explains the observation that germline-expressed genes are underrepresented on the X chromosome in C. elegans (Reinke et al. 2000, 2004; Wang et al. 2009; Tabuchi et al. 2011; Gaydos et al. 2012).
Constraints on X chromosome dosage compensation have been cited as another potential explanation for the depletion of male-biased genes from the X chromosome (Bachtrog et al. 2010). Dosage compensation equalizes X chromosome transcript levels between XX females and single X males. Although strategies differ, dosage-compensation mechanisms have been found in mammals, Drosophila, and C. elegans (Meyer 2010; Conrad and Akhtar 2012; Disteche 2012; Ferrari et al. 2014). Mammalian females inactivate most of the genes on one of their two X chromosomes to match transcriptional output from the single male X (Pollex and Heard 2012; Dupont and Gribnau 2013). In C. elegans, the dosage-compensation complex (DCC) binds to both of the hermaphrodite X chromosomes and represses transcription of each by one-half (Csankovszki 2009; Ercan and Lieb 2009; Meyer 2010). The Drosophila dosage compensation machinery binds to the single X chromosome in males and increases transcription twofold (Gelbart and Kuroda 2009). In Drosophila, it has been hypothesized that functional or mechanistic constraints on the X could limit the upregulation of already transcriptionally upregulated genes in males, thus preventing them from becoming male biased (Bachtrog et al. 2010). However, the mechanism of such a constraint remains unclear.
Susumu Ohno hypothesized that, during evolution of sex chromosomes, transcription from the X was upregulated to compensate for the degeneration of the Y (Ohno 1967; Adler et al. 1997; Gupta et al. 2006; Nguyen and Disteche 2006; Lin et al. 2007; Deng et al. 2011; Gribnau and Grootegoed 2012). Ohno further hypothesized that X upregulation was not specific to males but also occurred in XX females, resulting in overexpression of the female X above autosomal levels. He predicted that dosage compensation in XX females (i.e., the downregulation of X transcription in mammals) evolved to counter X upregulation. Supporting Ohno’s hypothesis, microarray and mRNA-seq data from mouse, Drosophila, and C. elegans have indicated that overall transcript levels are similar between the X chromosome and the autosomes in both males (X:AA) and females (XX:AA). Further, hermaphrodite X chromosome transcription increases to above autosomal levels when the DCC is mutated in C. elegans embryos (Kruesi et al. 2013).
To test Ohno’s hypothesis more directly, attempts have been made to define those genes present on the autosomal progenitor of the X chromosome using species from major mammalian and bird lineages (Julien et al. 2012; Lin et al. 2012). If the X chromosome is upregulated, transcriptional output from the present-day X and its autosomal progenitor should be similar. These studies found transcription from the current eutherian (the mammalian clade that includes humans and other placental mammals) X chromosome to be significantly lower compared to its autosomal progenitor (Julien et al. 2012; Lin et al. 2012). However, X and autosomal progenitor expression levels were similar when analyses were limited to highly expressed genes and those encoding for proteins involved in large (more than seven subunits) complexes (Julien et al. 2012; Lin et al. 2012). This result suggests that X upregulation is restricted to dosage-sensitive genes.
Much of the comparative work regarding evolution of sex-biased gene expression and the X chromosome has been performed in Drosophila and mammals (Wang et al. 2001; Lercher et al. 2003; Parisi et al. 2003; Khil et al. 2004; Parisi et al. 2004; Yang et al. 2006; Sturgill et al. 2007; Vicoso and Charlesworth 2009; Ellegren 2011; Meisel et al. 2012a; Allen et al. 2013; Parsch and Ellegren 2013). Here, to expand our understanding of X evolution, we compared X-linked gene expression in five nematode species that include both gonochoristic (outcrossing) and hermaphroditic mating systems (Figure 1A). Hermaphroditism has arisen independently in several nematode species (Kiontke et al. 2011), a process that strongly influences evolution of the genome. The gonochoristic Caenorhabditis species that we analyzed have larger genomes and greater magnitude of sex-biased expression compared to the hermaphroditic species (Thomas et al. 2012).
Figure 1.
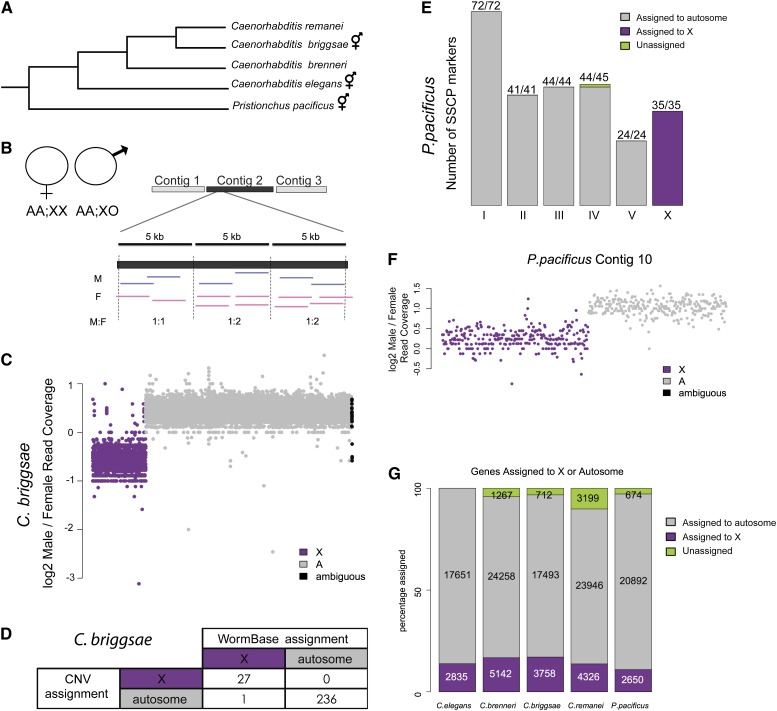
Copy-number analysis allows for assignment of sequencing contigs to either the X or autosomes. (A) Phylogeny showing the evolutionary relationship of the five nematode species (Kiontke et al. 2004, 2011). Pristionchus pacificus is estimated to have split from Caenorhabditis ∼300 million years ago (Dieterich et al. 2006). C. briggsae, C. elegans, and P. pacificus are androgynous. C. brenneri and C. remanei are gonochoristic. Estimates of C. elegans and C. briggsae divergence times range between 40 and 80 million years. (B) Overview of the copy-number approach. DNA-seq data were generated from males (AA;XO) and females (AA;XX). Contigs were broken into 5-kb windows and male-to-female coverage ratio was evaluated for each window. (C) Plotting male-to-female coverage ratio shows clear difference between X and autosomal contigs. The log2 male-to-female coverage ratio is plotted for C. briggsae. Each point represents one 5-kb window. X chromosome (purple) and autosomal (gray) contigs are plotted along the x-axis. Contigs that could not be assigned unambiguously are plotted in black. (D) Overlap of C. briggsae copy-number assignment with WormBase assignment indicates high accuracy. (E) Overlap of P. pacificus copy-number assignment with previously reported SSCP markers (Srinivasan et al. 2002; Dieterich et al. 2006) indicates high accuracy. Chromosomal assignments by SSCP are indicated below each bar. Bars are colored to indicate copy-number assignment. Numbers above each bar indicate agreement. (F) Copy-number analysis allows for identification of a split contig in P. pacificus. The male-to-female coverage ratio of the left and right half of P. pacificus contig 10 are assigned to the X and autosome, respectively. (G) Ratio of X to autosomal gene content is roughly unchanged across species. The number of assigned genes in each category (X, purple; autosomal, gray; unassigned, green) is indicated. For each of the five species, ∼10–14% of coding genes are assigned to the X chromosome.
For those species without a genetic map, we first assigned genes to the X chromosome using high-throughput sequencing of DNA isolated from XO males and XX females/hermaphrodites. Subsequently, we used mRNA-seq to analyze sex-biased gene expression in young adult males and females/hermaphrodites. Overall, we found that the genomic distribution of the sex-biased genes is conserved at varying degrees between C. elegans and the other four nematode species. Generally, those genes with high male-biased expression are underrepresented on the X chromosome. Genes with high female-biased expression are enriched on the X. To test Ohno’s hypothesis, we compared expression of orthologous genes that are differentially located between C. elegans and P. pacificus. Transcription is higher from the autosomal-linked orthologs, suggesting that there is no upregulation of these genes on the X chromosome. However, in all nematode species analyzed here, the level of transcription from the single X chromosome is similar compared to the autosomes, supporting X upregulation. These contradicting observations can be reconciled if the mechanism of X upregulation does not act chromosome-wide but specifically regulates dosage-sensitive genes.
Materials and Methods
Worm strains and growth
C. elegans (N2), C. brenneri (PB2801), C. briggsae (AF16), C. remanei (PB4641), and P. pacificus (PS312) strains were maintained at 20° on NGM agar plates using standard C. elegans growth methods.
DNAseq
One female and two or more male replicates were collected per species (summarized in Supporting Information, Table S1). At least 50 young adult worms were hand picked per replicate. Worms were washed by settling three to five times with 1 ml of M9, starved overnight to eliminate gut bacterial contamination, and resuspended in 100 μl TE. A total of 400 μl of lysis buffer (0.1 M Tris–HCl; 0.1 M NaCl; 50 mM EDTA; 1.25% SDS) was added and worms were sonicated for 30 min using the Bioruptor at high setting, 30 sec on/off. Sonicated DNA was isolated and cleaned up using Qiagen MinElute kit. Illumina DNA sequencing libraries between 250 and 500 bp were prepared from the purified DNA using Illumina TruSeq DNA kit with the following modifications. Briefly, after end repair and A tailing, adapters were ligated and the resulting DNA was purified using AmpureXP beads. Ligated DNA was amplified by PCR and DNA library between 300 and 500 bp was gel purified. Fifty base pair paired-end or single-end sequencing (see Table S1) was performed using Illumina HiSeq-2000. The raw data can be found at Gene Expression Omnibus (GEO) under series number GSE53144. For paired-end data, quality scores of the reverse reads were much lower than those of the forward reads. As such, only forward reads were used for analysis.
Copy-number approach: X and autosomal gene assignments
For each species, forward reads were aligned to WS228 with Bowtie version 0.12.7 (Langmead et al. 2009). We supplied the parameter (m = 4) to suppress all alignments with more than four hits in the genome. The resulting alignment files (in BAM format, a binary file type containing sequence alignment data) were converted to SAM format (tab-delimited text version of a BAM file) using SAMtools v. 0.1.18 (Li et al. 2009). The SAM files were sorted and used to generate bedgraph files (BEDTools v. 2.15.0) (Quinlan and Hall 2010). For each species, the contigs were split into 5-kb windows and the bedgraph file was used to calculate the sequencing coverage for each window. The male-to-female coverage ratio was computed for each window by taking the log2 of male coverage divided by female coverage. Baseline was set as the mean male-to-female coverage ratio. Windows whose log2 ratio fell one standard deviation below the mean were initially assigned to the X chromosome. If the majority of 5-kb windows contained within a contig were assigned to the X, then the contig was assigned to the X. If there was no majority, or if the contig was <15 kb, we could not assign the contig. Assignments were given confidence scores ranging from 0 to 2 based on both length of sequencing contig and agreement of assignment between replicates. One point was given to those contigs with lengths >50 kb. A second point was given if the final contig assignment obtained by combining all replicates matched the assignment obtained when the replicates were analyzed separately. The gene assignments and confidence scores are given in Table S2.
mRNA-seq
C. elegans (N2) and P. pacificus (PS312) worms were synchronized by bleaching gravid adults and allowing worms to hatch overnight. Larval worms were plated and grown at 20°. At least 750 young adult worms were hand picked for each of three biological replicates. Worms were washed three to five times in M9. Ten volumes of Trizol (Invitrogen) was added. Samples were freeze-cracked five times and RNA purification was performed according to the manufacturer protocol. Isolated RNA was cleaned up using the Qiagen RNeasy kit. mRNA was purified using Sera-Mag oligo(dT) beads (ThermoScientific) from at least 1 μg of total RNA. Stranded mRNA-seq libraries were prepared based on incorporation of dUTPs during cDNA synthesis using a previously described protocol (Parkhomchuk et al. 2009). Single-end 50-bp sequencing was performed using the Illumina HiSeq-2000. Data for C. elegans (fog-2), C. brenneri, C. briggsae (she-1), and C. remanei were downloaded from GEO (accession no. GSE41367). Reads were aligned to genome version WS228 with tophat v. 2.0.0 (Trapnell et al. 2012). Default parameters allow up to 20 hits for each read. Read numbers and mapping percentages (which refer to the percentage of unique reads with at least one alignment) can be found in Table S1. Gene expression was estimated using Cufflinks v. 2.0.2 with default parameters and supplying gene annotations for WS228. Average male and female expression (FPKM, fragments per kilobase per million mapped reads) was determined (Table S6). The raw read data and the average Cufflinks FPKM data can be obtained from GEO accession no. GSE53144. Differential expression analysis between males and females was performed using the R package DESeq (Anders and Huber 2010; R Development Core Team 2012). These results are available in Table S6.
Defining sex-biased gene expression
To define sex bias, we first identified those genes that were differentially expressed between the two sexes (DESeq qvalue < 0.05). Genes with FPKM >1 in at least one sex were considered “expressed” and used for subsequent analyses. We categorized “sex-biased” genes as those having FPKM >1 in one sex and FPKM >0 in the other. For each sex-biased gene, the magnitude of sex bias was calculated as the log2 ratio of FPKM values between the two sexes. Those genes that have FPKM >1 in one sex and FPKM = 0 in the other were categorized as “sex-specific” genes. We categorized “nonbiased” genes as those genes not called significant by DESeq (qvalue > 0.05) with FPKM >1 in both sexes and with less than twofold expression difference between the sexes. We categorized genes with high and low sex-biased expression based on a log2 sex-expression ratio cutoff of 3 (Table S3). The cutoff was selected based on a breakpoint in the distribution of sex-biased expression ratios driven largely by the gonadal expression of highly sex-biased genes (discussed further in Results and Discussion).
Results and Discussion
Copy-number approach for gene assignment to the X chromosome
To compare X chromosome gene content and sex-biased gene expression, we focused on five nematode species (Figure 1A). Four species are from the Caenorhabditis genus: C. elegans, C. brenneri, C. briggsae, and C. remanei. Determination of nematode divergence times is difficult owing to a lack of fossil records. Previous calculations based on the assumption of a universal molecular clock estimated that C. elegans and C. briggsae diverged 80–110 million years ago (Coghlan and Wolfe 2002; Stein et al. 2003). More recently, estimates of neutral mutation rates were used to approximate the divergence time between C. elegans and C. briggsae to ∼40 million years (Cutter 2008). The fifth species, P. pacificus, is estimated to have diverged ∼300 million years ago (Dieterich et al. 2006, 2008).
Three of the species, C. elegans, C. briggsae, and P. pacificus, are androdioecious and consist of self-fertile hermaphrodites and outcrossing males. The other two species, C. remanei and C. brenneri, are gonochoristic and have true outcrossing males and females. For simplicity, we refer to hermaphrodites as females except where the distinction is necessary. Although all five genomes have been previously sequenced, only the C. elegans and C. briggsae chromosomes are fully assembled. The other genomes remain on sequencing contigs.
To compare X and autosomal expression, we needed first to assign genes to either the X chromosome or the autosomes. To accomplish this task, we used a copy-number approach taking advantage of the fact that the X chromosome is present in one copy in males and two copies in females. The same principle was used recently for assigning genes to the Z chromosome of snakes and trematodes (Vicoso and Bachtrog 2011; Vicoso et al. 2013). We performed Illumina sequencing of DNA isolated from hand-picked male and female adults from C. remanei, C. brenneri, C. briggsae, and P. pacificus. Table S1 lists the number of reads and genome coverage for each replicate. We split each contig into 5-kb windows and calculated the log2 ratio of male-to-female read coverage for each window (Figure 1B). Figure 1C shows the log2 ratio of male-to-female read coverage for C. briggsae. A contig was assigned to the X chromosome if the majority of windows had a male-to-female coverage ratio one standard deviation below the mean.
When we compared our C. briggsae gene assignments to the current WormBase assignment [based on a high-density recombination map (Hillier et al. 2007; Koboldt et al. 2010; Ross et al. 2011)] we found that we correctly assigned 236 of 236 contigs to autosomes and 27 of 28 contigs to the X chromosome (Figure 1D). The inconsistent contig, cb25.fpc2310b, is assigned to an autosome by our method and to the X chromosome by WormBase. It contains no genes and was previously assigned to chromosome V as part of a larger sequencing contig, cb25.fpc2310. This contig was split into three parts for the current release, two of which are still assigned to chromosome V.
We also compared our P. pacificus contig assignments to a limited linkage map that was generated using a single-strand conformational polymorphism technique (SSCP; Orita et al. 1989). Recombinant inbred lines from two P. pacificus strains (wild-type P. pacificus var. California and the polymorphic Washington strain) were used to generate the linkage map (Srinivasan et al. 2002; Dieterich et al. 2006). Our assignment matched all of the X chromosome SSCP markers and all but one autosomal marker (Figure 1E). This last marker was unassigned by our copy-number approach because its contig is less than the minimum 15 kb required by our method. Of note, our method also allowed for identification of contigs that may be incorrectly assembled. By our method, the left half of PPA contig10 (WS228) should be assigned to the X chromosome while the right half should be assigned to an autosome (Figure 1F).
If we assigned a contig to the X chromosome, then all of its annotated genes were also assigned to the X. We correctly assigned 3758 genes to the C. briggsae X chromosome and 17493 genes to the autosomes (Figure 1G). A total of 712 genes were left unassigned because their sequencing contigs were too short to be reliably called by our method. With this copy-number approach, we were able to assign between 90 and 97% of genes to either the X or the autosomes in C. brenneri, C. briggsae, C. remanei, and P. pacificus. A list of our gene assignments can be found in Table S2.
Gonochoristic nematode species (C. brenneri and C. remanei) have larger genomes than those of hermaphroditic species (C. elegans, C. briggsae, and P. pacificus) in terms of both sequence and gene content (Stein et al. 2003; Barriere et al. 2009; Thomas et al. 2012). Despite difference in size, all four Caenorhabditis genomes have roughly the same distribution of X and autosomal genes. P. pacificus stands out as having fewer X-linked genes than the other species (Figure 1G). This result is consistent with the observation that one arm of P. pacificus chromosome I contains genes orthologous to those located on the C. elegans X, indicative of a major translocation event that occurred after the Pristionchus–Caenorhabditis split (Srinivasan et al. 2002).
This type of translocation event is rare within Rhabditina (the phylogenetic clade that contains both Pristionchus and Caenorhabditis). Among the estimated 4000 rearrangement events that have occurred since C. elegans and C. briggsae split, few were interchromosomal (Coghlan and Wolfe 2002; Stein et al. 2003). Similarly, an 11-gene region on P. pacificus chromosome III has 10/11 orthologs on the C. elegans chromosome III (Lee et al. 2003). Further, members of Rhabditina have a stable chromosome number of six (Mitreva et al. 2005). These observations suggest that for the species analyzed here, the X chromosome descended from a common ancestor. Additionally, there is no apparent pseudoautosomal region on the nematode X chromosome. As the Y chromosome is absent in all five nematode species examined here (Walton 1940), all X chromosome genes are sex linked.
Determining genes with sex-biased gene expression
To investigate the relationship between sex-biased gene expression and the nematode X chromosome, we analyzed previously published mRNA-seq data from young adult males and females in each of the four Caenorhabditis species (Thomas et al. 2012; GEO accession no. GSE41367) and produced a similar data set for P. pacificus. The published data used C. elegans (fog-2) and C. briggsae (she-1) mutants, which lack XX spermatogenesis but produce otherwise normal developing female worms. Because a similar feminizing mutation was not available for P. pacificus, we used J4/young adult males and hermaphrodites of the wild-type California strain (PS312) and produced a comparable data set for wild-type C. elegans (N2) L4/young adult males and hermaphrodites. Expression values (FPKM) for each gene were calculated using Cufflinks (Trapnell et al. 2012) and differential expression was determined by DESeq (Anders and Huber 2010).
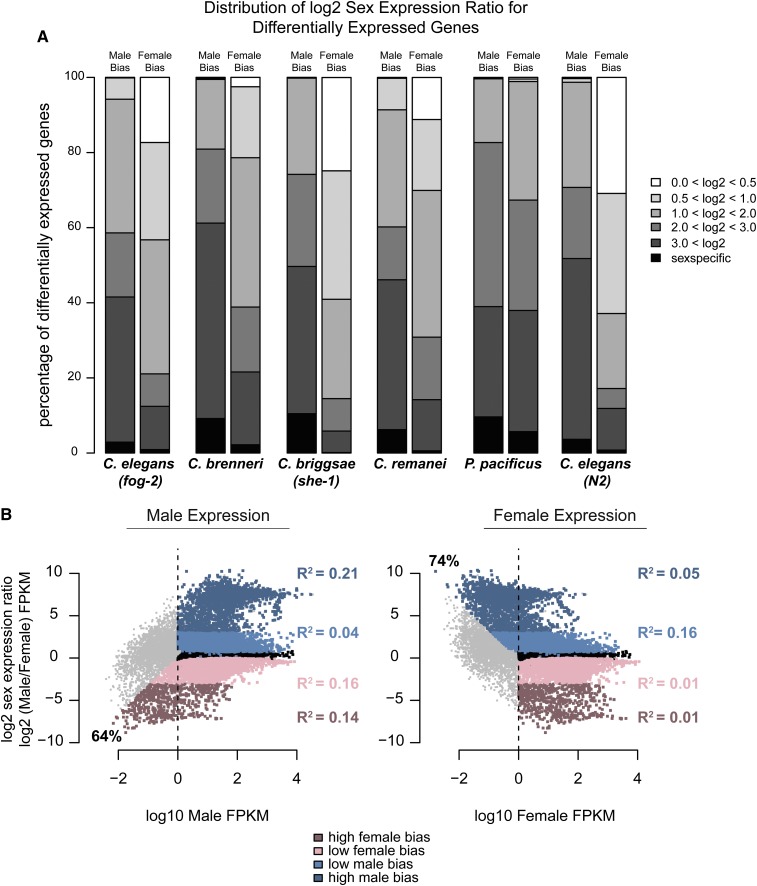
For differentially expressed genes, we calculated the magnitude of sex bias as the log2 ratio of expression between the two sexes (see Materials and Methods). In all five species, male-biased genes have a higher magnitude of bias compared to female-biased genes. Between 38 and 60% of male-biased genes are expressed at least eight times more in males than in females (Figure 2A). Conversely, the majority of female-biased genes have a less than twofold difference in expression between the sexes.
Figure 2.
Distribution of the magnitude of sex bias (log2 sex expression ratio) is different for male- and female-biased genes. (A) Differential expression was determined using DESeq, q-value <0.05. Only genes with FPKM >1 in at least one sex were considered. Male and female expression was determined using cufflinks and reported as FPKM. Magnitude of sex bias was determined by taking the log2 of the sex expression ratio. For each category, male or female, genes were binned by magnitude of bias. Percentage of differentially expressed genes contained within each bin is plotted on the y-axis. (B) For C. elegans (fog-2) the magnitude of sex-biased expression is plotted against the level of expression in males (left) and females (right). Dashed line indicates FPKM = 1. Percentages of high-sex-biased genes with low expression (FPKM <1) in the opposite sex are indicated. Nonbiased genes are plotted in black. Male-biased genes are plotted in blue; female-biased genes are plotted in pink. Darker colors indicate high magnitude of bias. R2 values between magnitude of bias and expression levels are indicated for each category.
We categorized genes with high and low sex-biased expression using a log2 sex expression ratio cutoff of 3 (see Materials and Methods). For each species and sex, we plotted the magnitude of sex bias against the overall expression levels (Figure S1). Figure 2B shows the male/female expression ratio plotted against male expression (left) and female expression (right) for C. elegans (fog-2). As expected, genes that are highly sex biased have higher expression in the corresponding sex. Further, a large proportion of the high sex-biased genes have very low expression (FPKM <1) in the opposite sex (Figure 2B). However, regression analysis revealed low correlation between magnitude of sex-biased expression and overall expression levels (for all species, R-squared values ranged between 0.01 and 0.38). This relationship between sex bias and overall expression levels is similar for the X chromosome and autosomes (Figure S2A, Pearson correlation values between 0.6 and 0.9).
Overall tendency toward male-biased expression
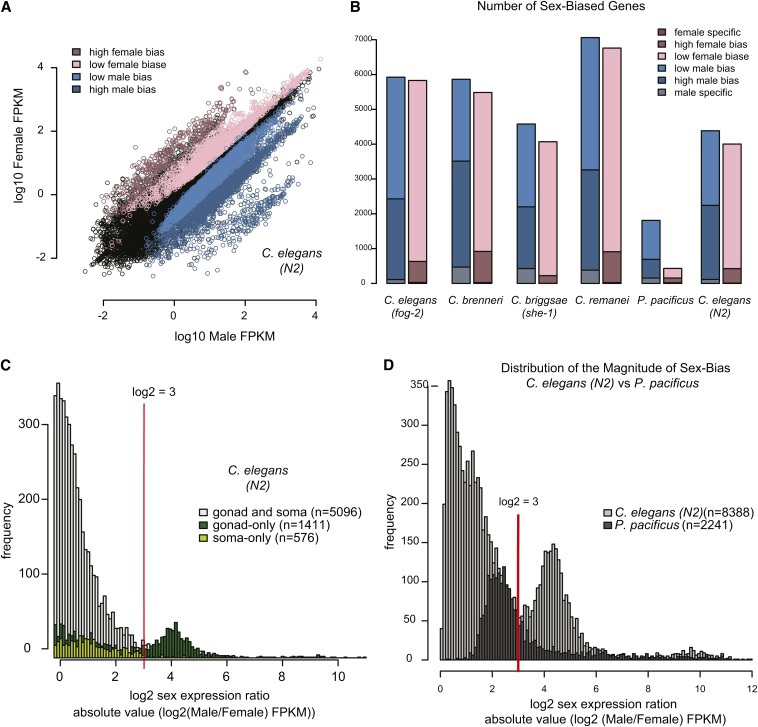
Previous work on Caenorhabditis species noted a higher prevalence of genes with male-biased expression (Thomas et al. 2012) (Figure 3A). Our analyses replicated these findings and indicated the same trend in Pristionchus worms. In all five species, the number of male-specific and male-biased genes is greater than the number of female-specific and female-biased genes (Figure 3B). Overall, sex-biased expression is significantly more likely to be male-biased (P-value = 0.03 by paired t-test).
Figure 3.
Majority of sex-biased genes are male biased. High magnitude of sex bias is linked to gonadal expression. (A) For C. elegans (N2), male (x-axis) and female (y-axis) expression values (log10 FPKM) are plotted. (B) There is a greater tendency toward male bias in all five species. Number of genes with male (blue) and female (pink) biased expression is plotted for each species. (C) Genes expressed in the gonad show higher magnitude of sex-biased expression. Lists of genes with gonadal and somatic expression were taken from Spencer et al. (2011). Histogram shows the frequency of the magnitude of sex bias for gonad-only (dark green), soma-only (light green), and gonad and soma-expressed (gray) genes. Number of genes in each category is indicated. Red line indicates log2 expression ratio of 3. (D) Distribution of magnitude of sex bias for P. pacificus and C. elegans YA worms. Absolute value of the log2 sex expression ratio was calculated for all sex-biased genes. Histogram shows the frequency of the magnitude of sex bias for C. elegans (light gray) and P. pacificus (dark gray).
Compared to Caenorhabditis species, we identified fewer sex-biased genes in P. pacificus. (Figure 3B). We noted that P. pacificus mRNA-seq data were collected from male and hermaphrodite populations while Caenorhabditis data came from male and female populations. We reasoned that the use of hermaphrodite worms, which produce both sperm and eggs, might have reduced our ability to call sex bias in P. pacificus. To test this, we compared sex-biased gene expression in C. elegans using data from wild-type (N2) hermaphrodites or fog-2 mutant females. More than 92% of the sex-biased genes identified in N2 were also identified as sex biased in the fog-2 mutant analysis (Figure S3, A and B). Overall, we identified 8388 genes as differentially expressed between C. elegans males and hermaphrodites. This was fewer than the number of sex-biased genes identified using fog-2 females (11,753) but substantially more than the number of biased genes identified in our P. pacificus analysis (2241). We concluded that the reduced number of sex-biased genes identified in the P. pacificus genome was not entirely due to the use of hermaphrodite worms.
We also noted that our P. pacificus data sets contained fewer mRNA-seq reads compared to C. elegans (fog-2). We randomly resampled the C. elegans (fog-2) reads to match P. pacificus and reanalyzed the distribution of biased genes. After resampling, we identified 11,484 sex-biased genes, slightly less than 11,753 genes identified before resampling (Figure S3C). The overlap of sex-biased genes was high: 5733 (97%) male-biased genes and 5555 (95%) female-biased genes were reidentified upon resampling (Figure S3, D and E). Thus, the reduced number of P. pacificus sex-biased genes was not a consequence of having fewer sequencing reads.
Gonad-specific genes show higher sex-biased expression
Previous work in Drosophila indicated that gonadal tissues account for most of the sex-biased expression, both in number of genes that are differentially expressed between the sexes and in magnitude of bias (Parisi et al. 2004). Using previously defined gene sets with somatic and gonadal expression in C. elegans (Spencer et al. 2011), we found that sex-biased genes expressed in the soma have a lower magnitude of bias compared to sex-biased genes expressed in the gonad (Figure 3C). Genes with low sex-biased expression are composed of both gonad and soma-expressed genes. Highly sex-biased genes are largely gonad specific (Figure S4A).
In contrast to C. elegans, we observed that the majority of P. pacificus sex-biased genes had a log2 sex expression ratio <3 (Figure 3D). We can think of two possible explanations for this. First, it could be that Pristionchus gonads are less sexually dimorphic than those of Caenorhabditis. Alternatively, a delay in the timing of gonad development in Pristionchus young adults (compared to the Caenorhabditis species) could mean that our P. pacificus collections contained young adults without full gonadal expression. In C. elegans, sperm production begins just prior to the L4/adult molt and hermaphrodites switch to oocyte production within the first 2 hr of young adulthood (Kimble 1981; Kimble and White 1981). In contrast, the first P. pacificus sperm are produced after the J4/adult molt. Hermaphrodite oogenesis does not switch on until 4 to 6 hr after this final molt (Rudel et al. 2005); thus young adults might not yet have full gonadal expression. Further experiments using older hermaphrodites are required to distinguish these two possibilities.
Demasculinization and feminization of the X chromosome
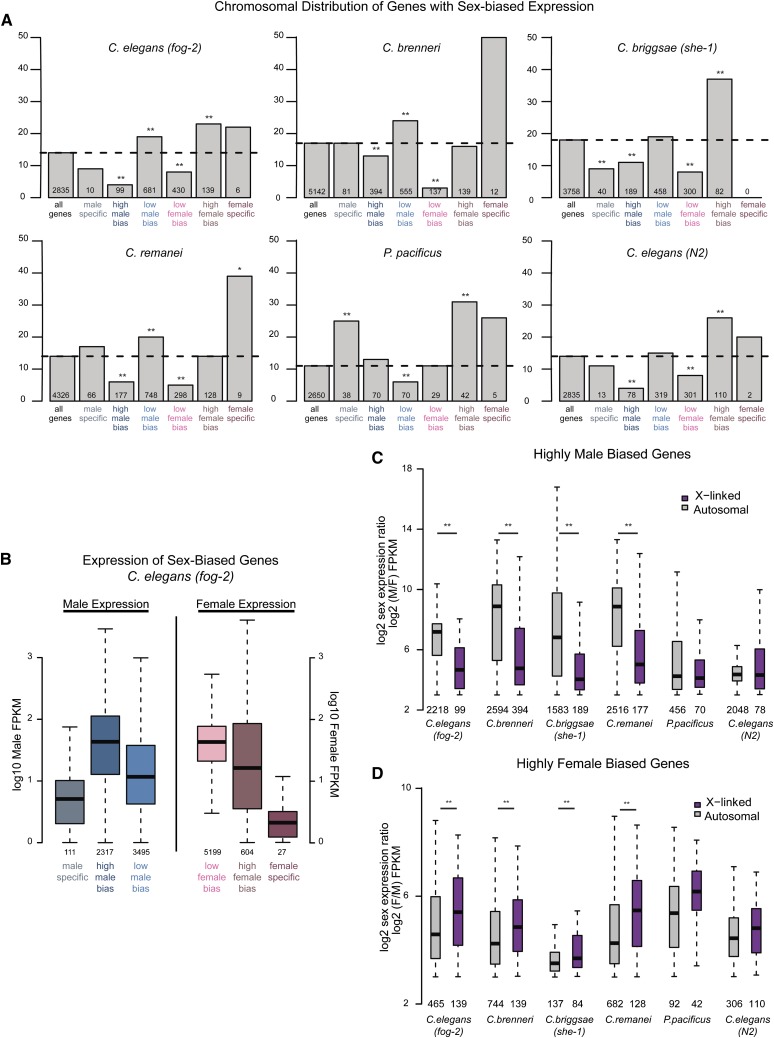
Genes with sex-biased expression tend to have nonrandom chromosomal distribution (Saifi and Chandra 1999; Wang et al. 2001; Khil et al. 2004; Lercher et al. 2003; Parisi et al. 2003; Ranz et al. 2003; Parisi et al. 2004; Reinke et al. 2004; Divina et al. 2005; Yang et al. 2006; Sturgill et al. 2007; Meisel et al. 2012a; Allen et al. 2013). Microarray analysis of gene expression in C. elegans has shown the X chromosome to be both feminized (enriched for genes with female-biased expression) and demasculinized (depleted of genes with male-biased expression) (Reinke et al. 2004). Consistent with this previous work, we found that only 4% (99/2317) of high male-biased genes are on the X chromosome in C. elegans (fog-2). This is significantly less than expected by chance (14% of all C. elegans genes are on the X chromosome, P-value < 1e–40 by Fisher test). Conversely, 23% (139/604) of the high female-biased genes are X linked, significantly more than expected (P-value <3.2 e–7 by Fisher test).
Figure 4A plots the percentage of genes on the X chromosome that show sex-biased expression for each of the five nematode species. In the Caenorhabditis species, highly sex-biased genes reveal a pattern of X chromosome demasculinization and feminization. Because those genes with high sex-biased expression are mainly expressed from the gonad, depletion of high male-biased genes from the X chromosome may be due to MSCI in the male germline.
Figure 4.
Sex-biased genes are nonrandomly distributed between the X and autosomes. (A) Genes for each species were placed into one of seven categories: all genes, male-specific, high male bias, low male bias, low female bias, high female bias, or female specific (see Materials and Methods). For each category, percentage of X-linked genes is plotted. Number of X-linked genes is indicated below each bar. Significance of enrichment or depletion was calculated using Fisher test: (*)P-value <0.05; (**)P-value <0.001. (B) For C. elegans (fog-2), expression levels of sex-biased genes in each category are plotted. Male expression is plotted for male-biased genes (left three boxes) and female expression is plotted for female-biased genes (right three boxes). (C) Magnitude of male-biased expression (log2 male over female expression) was calculated for each high-male-biased gene. X-linked genes are plotted in purple; autosomal genes are plotted in gray. Number of genes analyzed is indicated below each box. High-male-biased genes located on autosomes showed significantly greater magnitude of sex bias compared to those located on the X chromosome as calculated by t-test: (*)P-value <0.01. (D) As in C, magnitude of female-biased expression (log2 female over male expression) was calculated for each high female-biased gene.
Genes with low male-biased expression are enriched on the Caenorhabditis X chromosome. For C. elegans (fog-2), 19% (681/3495) of genes with low male-biased expression are on the X (significant enrichment over the 14% expected, P-value <4.28 e–13 by Fisher test,), consistent with the predicted X accumulation of sex-biased genes (Rice 1984). However, only 8% (430/5199) of genes with low female-biased expression are on the X, indicating a surprising depletion (P-value <6.9 e–24). One explanation for this observation is that the female benefit for these low-biased genes is not large enough to drive X accumulation. Female-biased genes are predicted to accumulate on the X chromosome only if the fitness cost to males is minimal. It could be that the fitness cost of having a single copy of these genes in males is greater than the fitness benefit to females.
Unlike high-male-biased genes, male-specific genes are not particularly depleted from the X chromosome (Figure 4A), nor is their expressions specific to the gonad (Figure 4A). In addition, expression levels of sex-specific genes are, on average, lower than those of sex-biased genes (Figure 4B) This suggests that these genes may be expressed only in a small number of sex-specific somatic cells. For example, among the set of identified C. elegans male-specific genes is ceh-7 (FPKM = 1.6), which encodes for a homeodomain transcription factor that is transcribed in cells around the rectum of the male tail (Kagoshima et al. 1999). The observation that male-specific genes with low somatic expression are not depleted from the X chromosome suggests that X demasculinization is largely due to highly expressed male-biased genes avoiding MSCI.
We observed the X chromosome of P. pacificus to be significantly enriched for both male and female highly biased genes. In C. elegans, gonad-specific genes tend to be autosomal whereas somatic genes tend to be X linked (Figure S4B) (Reinke et al. 2000; Spencer et al. 2011). The enrichment of both male and female-biased genes on the Pristionchus X might be due either to somatic bias in the data or to differences in regulation of X transcription between Pristionchus and Caenorhabditis.
Magnitude of sex bias differs between X and autosomes and between the sexes
We found that the magnitude of sex bias depends both on chromosomal location as well as on sex. For high-male-biased genes, the magnitude of sex bias is greater when the gene is autosomal (Figure 4C). Notably, in the two male/hermaphrodite data sets (P. pacificus and C. elegans, N2), the magnitude of male bias is similar for X and autosomal genes, suggesting that sperm production in hermaphrodites reduces the observable magnitude of autosomal male-biased expression. Overall, our analyses indicate that Caenorhabditis male-biased genes on the X chromosome tend to be more weakly biased than those located on the autosomes.
The opposite trend is true for high-female-biased genes (Figure 4D). Magnitude of female bias is greatest for those genes that are X linked. The overall trends were the same when we included all biased genes in the analysis (Figure S4, C and D). In summary, we found the nematode X chromosome to be generally demasculinized and feminized with respect to both gene content and magnitude of bias.
Hermaphroditism and the preferential loss of autosomal male-biased genes
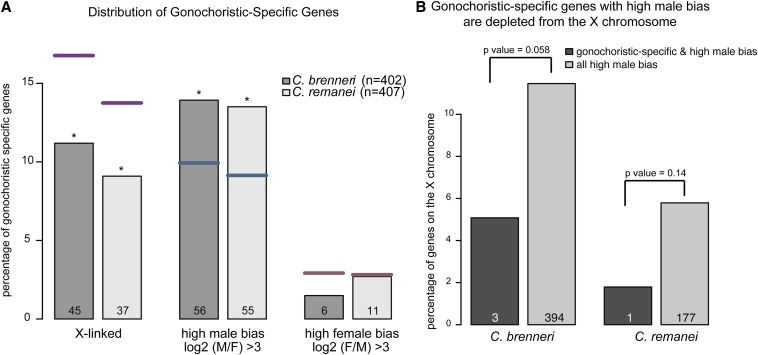
Gonochoristic species C. remanei and C. brenneri have substantially larger genomes than the hermaphroditic C. elegans and C. briggsae (Barriere et al. 2009). Hermaphrodite genome shrinking may be linked to a phenomenon whereby autosomes carrying large deletions preferentially segregate with the X chromosome during male meiosis (Wang et al. 2010). Previous work has also indicated that genes with high-sex-biased expression are preferentially lost in hermaphrodite lineages (Thomas et al. 2012). We wanted to see if there was a difference between X and autosomes with respect to this gene loss. To do so, we generated a list of gonochoristic-specific orthologs composed of genes that have a one-to-one ortholog in C. brenneri, C. remanei, and a third gonochorite, C. japonica, but lack an ortholog in both C. elegans and C. briggsae (Table S4). Similar to the previous report (Thomas et al. 2012), we found the set of gonochoristic-specific orthologs to be significantly enriched for genes with high male bias (Figure 5A). We also observed the set to be significantly depleted of X-linked genes, indicating that genes are preferentially lost from autosomes in the self-fertile species.
Figure 5.
Evolution of hermaphroditism is linked to the preferential loss of autosomal-linked highly male-biased genes. (A) Gonochoristic-specific genes are enriched for genes with high male bias. Gonochoristic-specific genes were identified by taking the overlap of genes that had an ortholog in each of three gonochoristic species (C. brenneri, C. remanei, and C. japonica), but lacked an ortholog in two hermaphroditic species (C. elegans and C. briggsae). For C. brenneri (dark gray) and C. remanei (light gray), the percentage of gonochoristic-specific genes that are X linked, show high male bias, or show high female bias is plotted. The number of genes identified in each category is indicated in each bar. Significance of enrichment or depletion was calculated using Fisher test: (*)P-value <0.05; (**)P-value <0.001. From left to right: purple, blue, and pink lines demark the genome-wide percentage of X-linked, high-male-biased, and high-female-biased genes for C. brenneri and C. remanei respectively. (B) Gonochoristic-specific genes with high male bias are underrepresented on the X chromosome. Plotted is the percentage of high male-bias genes that are X linked. Gonochoristic-specific genes are plotted in dark gray. Numbers below each bar indicates the number of genes that are in each category. Significance of depletion was calculated using Fisher test.
Because high-male-biased genes are significantly underrepresented on the X chromosome, we tested whether the tendency toward autosomal gene loss was simply a consequence of losing male-biased genes or if it reflected a true evolutionary difference between the X and autosomes. Among the high-male-biased genes, those that are gonochoristic specific are underrepresented on the X chromosome (Figure 5B). For example, in C. remanei, 6% of high-male-biased genes are on the X (177/2878), but only 2% (1 of 55) of male-biased gonochoristic-specific genes are on the X chromosome (Figure 5B). This result indicates that high-male-biased genes located on the X chromosome are better conserved among Caenorhabditis species, as further discussed below.
Conservation of chromosomal location and sex bias of one-to-one orthologs
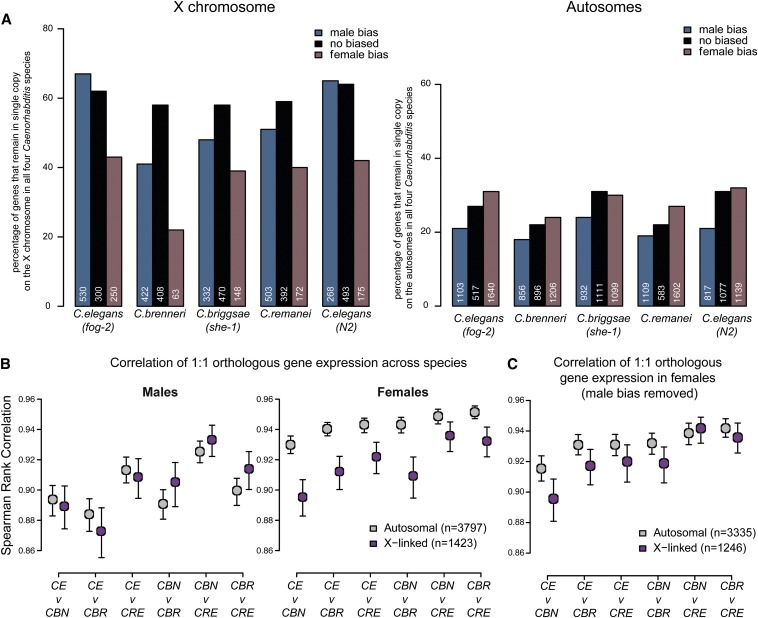
Previous work comparing the C. elegans and C. briggsae genomes indicated substantial conservation of macrosynteny between the two organisms (Hillier et al. 2007). Overall, 95% of one-to-one orthologs are located on the same autosome and 97% on the X chromosome in both species. To understand how evolutionary pressures that maintain macrosynteny of sex-biased genes differ between the X and autosomes, we compared the conservation of chromosomal location for genes with male, female, and nonbiased expression. From orthologous gene data downloaded from WormBase, we extracted a set of 5383 1:1:1:1 orthologs between the four Caenorhabditis species (Table S5). We limited analysis to Caenorhabditis species because inclusion of evolutionarily distant P. pacificus to generate a 1:1:1:1:1 ortholog list greatly reduced the number of comparable orthologs. For male, female, and nonbiased genes, we determined the percentage of genes that had both a 1:1:1:1 ortholog and that also retained chromosomal location (X or autosome) across all four species (Figure 6A). For C. elegans and C. briggsae, we could compare each chromosome separately (Figure S4E). A high percentage indicates that most genes in the category are in single copy and remained on the X or autosome in all four Caenorhabditis species.
Figure 6.
Conservation of orthologous gene expression across species. (A) On the X, male-biased genes are more likely to be conserved than female-biased genes. For each species, genes were divided into three categories: male-biased (blue), female-biased (purple), and nonbiased (black). Within each category, we identified 1:1:1:1 Caenorhabditis orthologs that are located on the same chromosome (X, left) or autosome (right) in all four species. X-linked genes have a higher tendency than autosomal genes to remain on the same chromosome and in single copy. Similarly, X-linked male-biased genes show greater tendency to remain in single copy compared to X-linked female-biased genes. (B) Correlation of 1:1 orthologous gene expression between Caenorhabditis indicates that X-linked expression is evolving faster in females. Between any two species, Spearman rank correlation of 1:1 orthologous gene expression was plotted for males (left) and females (right). As determined by bootstrapping, 95% confidence intervals are indicated. (CE, C. elegans; CBN, C. brenneri; CBR, C. briggsae; CRE, C. remanei). (C) High male-biased genes are removed from correlation analysis shown in B, right.
This analysis led to two main observations. First, genes on the X chromosome are better conserved compared to autosomal genes (Figure 6A, compare left to right). In C. elegans (N2), there are 2835 X-linked genes. Among these, 1423 (50.2%) have an X-linked 1:1:1:1 ortholog in all four Caenorhabditis species. Conversely, among the 17642 autosomal genes, only 3797 (21.5%) have an autosomal 1:1:1:1 ortholog in the other species. Because one-to-one orthologs usually have the same functional role in both species (Altenhoff and Dessimoz 2009; Verster et al. 2014), the observed high percentage of genes retaining X-linked single copy across the four Caenorhabditis species highlights the importance of conservation of X-linked genes. Second, genes with male-biased expression have higher incidence of conservation on the X chromosome than genes with high female bias. This observation agrees with a previous observation that male-related genes on the X have significantly lower rates of protein evolution compared to those on the autosomes (Cutter and Ward 2005). Lower conservation of female-biased genes on the X chromosome (with respect to both chromosomal location and copy number) may be due to greater functional pleiotropy of female-biased genes compared to male-biased genes (Zhang et al. 2007; Mank et al. 2008b; Mank and Ellegren 2009; Meiklejohn and Presgraves 2012). If an X-linked female-biased gene has a non-sex-biased role in many different tissues, there may be pressure to undergo gene duplication to allow one copy to carry out the sex-biased function and the other the pleiotropic functions (Maciejowski et al. 2005).
Better conservation of male-biased gene expression on the nematode X chromosomes
We next compared conservation of sex-biased gene expression between C. elegans (fog-2) and each of the other four nematode species. Among the 742 high male-biased genes in C. elegans (fog-2) that have a one-to-one ortholog in C. briggsae, 474 (64%) also show high male bias in C. briggsae (Figure S5A). Conversely, only 32 of 164 (19.5%) C. elegans (fog-2) high female-biased orthologs also show high female-biased expression in C. briggsae. For orthologs with low-sex-biased expression, conservation is higher for female-biased genes.
Sex-biased genes often show greater amino acid sequence divergence than nonbiased genes (reviewed in Ellegren and Parsch 2007). This relationship between sex bias and the rate of molecular evolution has been observed in Drosophila, C. elegans, and mammals (Torgerson et al. 2002; Cutter and Ward 2005; Khaitovich et al. 2005; Richards et al. 2005). Although most of the previous studies focused on molecular evolution of sex-biased genes, recent studies in Drosophila have also indicated higher divergence of expression for male-biased genes, particularly for those located on the X chromosome (Meiklejohn et al. 2003; Llopart 2012).
To analyze expression divergence of sex-biased genes, we calculated the coefficient of variation (σ/μ) between species for each Caenorhabditis 1:1:1:1 ortholog. Unlike in the Drosophila studies, we did not observe higher divergence of male-biased gene expression (Figure S5B). Instead, we found that male-biased genes tend to have higher expression divergence in females, and female-biased genes have higher expression divergence in males. This suggests that sex-biased genes experience high selective pressure in the opposite sex (Jiang and Machado 2009).
Evaluation of the faster-X hypothesis based on gene expression
As it is present in only one copy in males, the X chromosome has a smaller effective population size compared to the autosomes, leaving it more susceptible to genetic drift (Avery 1984). This, in combination with the unique selective pressures acting on the X in males and females, led to the prediction that adaptive evolution may proceed faster on the X chromosome [the faster X hypothesis (Charlesworth et al. 1987; Wu and Davis 1993; Orr 1997; Turelli 1998)]. This hypothesis is supported by observations of higher rates of protein evolution for X-linked genes in both mammals and Drosophila (Thornton and Long 2002; Counterman et al. 2004; Lu and Wu 2005; Mikkelsen et al. 2005; Baines et al. 2008; Hvilsom et al. 2012; Kousathanas et al. 2013; Hu et al. 2013). Notably, no such faster X evolution was observed when comparing nonsynonymous substitution rates between C. elegans and C. briggsae (Artieri et al. 2008). However, adaptive changes in the genome frequently occur in cis-regulatory regions (Wittkopp et al. 2004; Schaefke et al. 2013) resulting in interspecies expression differences. Recent studies of gene expression evolution in Drosophila and mammals have found evidence of faster-X evolution in the form of greater divergence of X-linked expression across species (Brawand et al. 2011; Kayserili et al. 2012; Meisel et al. 2012b).
To evaluate faster-X evolution of gene expression in nematodes, we compared expression correlation of 1:1:1:1 orthologs between the four Caenorhabditis species. Between any two Caenorhabditis species, we calculated the Spearman rank correlation coefficient (as in Brawand et al. 2011 and Meisel et al. 2012b). In males, expression levels of orthologs on the X and autosomes are similarly correlated across species (Figure 6B, left). In females, X-linked ortholog expression is less correlated than autosomal ortholog expression (Figure 6B right, P-value = 0.00046, by paired t-test: Figure S5B). This result suggests that female gene expression diverges faster on the X compared to the autosomes.
Lower correlation of X expression in females may be due to differences in dosage compensation across species. Although orthologs of C. elegans dosage compensation complex subunits have been identified in the three other Caenorhabditis species, female X expression divergence may be due to differences in DCC binding or downstream effectors. It is worth noting that male-biased genes located on the X chromosome are largely responsible for the observed female expression divergence. When we removed high-male-biased genes from analysis, female X expression correlation between species resembled autosomal correlation (Figure 6C), consistent with the observation that male-biased genes show high levels of expression divergence in females (Figure S5B).
Effect of X localization on gene expression
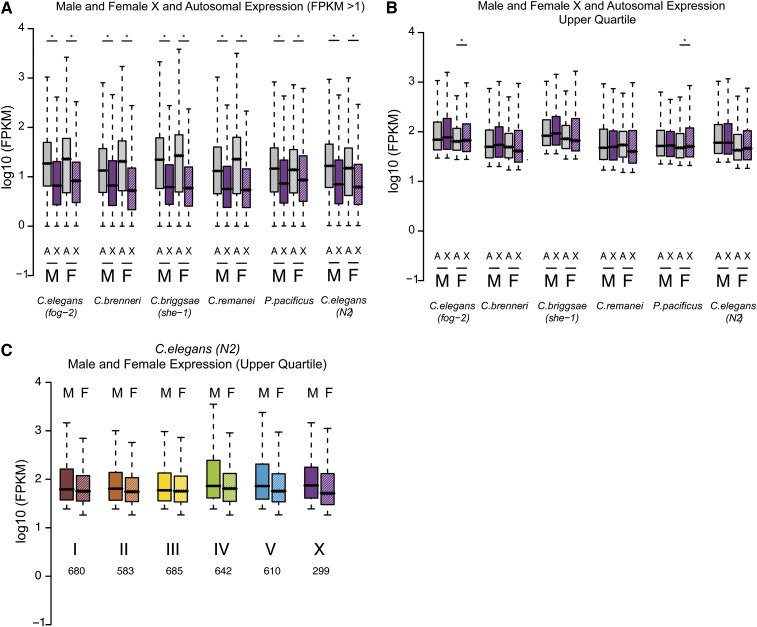
Ohno’s hypothesis and the extent of X upregulation are currently being debated in the field. The unique gene content of the X chromosome makes it difficult to compare X and autosomal transcription (Deng et al. 2011). In C. elegans, analysis is further complicated by germline repression of the X chromosome in young adult worms, which contain almost twice as many germline cells than somatic cells. As our data come from young adult worms, X chromosome repression in the germline explains the observation that overall X expression is lower than autosomal expression (Figure 7A). In support of this explanation, X and autosomal expression levels are similar in C. elegans worms lacking a germline (Deng et al. 2011). To remove those genes whose expression may be repressed in the germline, we limited analysis to only highly expressed genes (upper quartile of expression values). In agreement with previous observations (Deng et al. 2011), overall transcript levels of highly expressed genes are similar between X and autosomes in males and females (Figure 7B) and for each chromosome in C. elegans (Figure 7C).
Figure 7.
Analysis of overall expression levels from the X chromosome and autosomes. (A) Expression level of genes with FPKM >1 is plotted. X-linked genes are shown in purple. Autosomal genes are in gray. Male expression data are plotted in solid bars. Female data are in dashed bars. As measured in young adults, X expression is significantly lower than autosomal expression. (*)P-value <0.01 by t-test. (B) Genes whose expression is in the upper quartile in both males and females are plotted (at least 225 X-linked and 2890 autosomal-linked genes). X and autosomal expression is similar except in C. brenneri and C. remanei, where female X expression is significantly lower than autosomal expression. (*)P-value <0.01 by t-test. (C) C. elegans (fog-2) expression values plotted for each chromosome separately. Analysis is limited to the upper quartile genes. Number of genes analyzed is indicated below each bar.
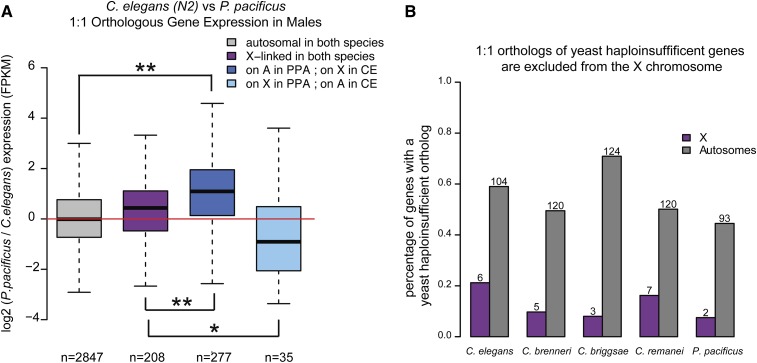
To more directly test if the X chromosome is upregulated in males during the course of X evolution, we compared expression of one-to-one orthologs located on the autosome in one species and the X chromosome in another. Unfortunately, as reported above, macrosynteny among the Caenorhabditis species is high. Between C. elegans and each of the other three Caenorhabditis species, <50 genes have changed chromosomal location between X and autosomes (Figure S4F). However, owing to a large chromosomal translocation event that occurred after their split, 466 genes have moved between X and autosomes in C. elegans and P. pacificus. Of these orthologs, 317 are expressed (FPKM >1) in the males of both species, which allowed us to analyze the effect of X and autosomal localization on gene expression. If the single male X chromosome is upregulated, there should be no difference in gene expression when the ortholog is differentially located on X and autosomes. To reduce any effects of sex bias on analysis of dosage compensation, we removed all genes with high sex-biased expression in either species. One-to-one orthologs that are similarly located (on either the X chromosome or on an autosome) are similarly expressed in both species (Figure 8A). For orthologs that are differentially located, expression is higher in the species where the ortholog is on an autosome. This result suggests that expression of the X-linked orthologs is not upregulated to match autosomal expression.
Figure 8.
Monosomy of the male X results in lower expression of X-linked orthologs in males. (A) Comparison of gene expression between C. elegans and P. pacificus indicates higher expression of the autosomal ortholog. Log2 male expression ratio (P. pacificus/C. elegans) is plotted for four groups of genes: autosomal in both species (gray), X-linked in both species (purple), autosomal in P. pacificus and X in C. elegans (dark blue), X in P. pacificus and autosomal in C. elegans (light blue). Number of genes in each category is indicated below each bar. (B) Haplo-insufficient genes are excluded from the X chromosome. Orthologs of yeast haplo-insifficient genes were identified in each of the five species (Table S7). The percentage of orthologs that are on X (purple) and autosomes (gray) is plotted. Numbers above each bar indicate the number of haplo-insufficient 1:1 orthologs identified.
Orthologs of yeast haplo-insufficient genes are excluded from the X chromosome
If X upregulation does not compensate for the single X in males, then selective pressure should move dosage-sensitive genes off of the X chromosome. Haplo-insufficient genes are dosage sensitive. A reduction in their gene copy number from two to one results in a significant reduction of overall fitness. Haplo-insufficient genes have been well characterized in Saccharomyces cerevisae and can be used to predict haplo-insufficiency in other metazoan species (de Clare et al. 2011). Orthologs of yeast haplo-insufficient genes are excluded from the X chromosome in humans and C. elegans (de Clare et al. 2011). Here, we found that this trend holds for the four other nematode species (Figure 8B).
Previous studies have noted two additional differences in the organization of gene content between the X chromosome and the autosomes in C. elegans. First, genes essential for early embryonic development are underrepresented on the X chromosome (Piano et al. 2000). Second, ∼15% of all C. elegans genes are expressed within operons, which are underrepresented on the X chromosome (Blumenthal et al. 2002). Underrepresentation of operons on the X is also observed in C. briggsae (Uyar et al. 2012).
Our analysis of X chromosome gene content with respect to sex-biased gene expression supports a general trend whereby the nematode X chromosome is enriched for female-biased genes and depleted of male-biased genes. This observation holds for highly sex-biased genes, which are mainly expressed in the gonad, but not for genes with low-sex-biased expression, which are expressed in both the soma and the gonad. In the male germline the X chromosome is silenced, potentially driving movement of high-male-biased genes from the X and resulting in the observed demasculinization. In the soma, X chromosome transcription is more subtly regulated by dosage compensation mechanisms that do not restrain the accumulation of male-biased genes on the X. Since the distribution of sex-biased genes is generally conserved between the five nematode species, regulation of X chromosome transcription in the germline and soma may also be conserved. However, we noted differences in sex-biased gene expression and X chromosome content between C. elegans and P. pacificus, as well as a lower correlation of X chromosome gene expression in females between the four Caenorhabditis species. These observations suggest the existence of species-specific differences in the mechanisms that regulate X chromosome transcription. Our analyses indicate that monosomy of the X chromosome is an important player in shaping X chromosome gene content. While similarity of overall transcript levels from the X and autosomes support X chromosome upregulation, a shortage of haplo-insufficient genes and lower expression of differentially located X-linked orthologs suggests that potential mechanisms of X chromosome upregulation do not act on every X-linked gene.
Supplementary Material
Acknowledgments
We thank Karin Kiontke for an initial discussion on this project and Mark Siegal for comments on the manuscript. We thank New York University—Center for Genomics and Systems Biology High Throughput Sequencing Facility for sequencing and raw data processing. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). S.E. and S.E.A. conceived and designed the experiments. S.E.A., P.R., and M.K. performed the experiments. S.E.A., A.K., and C.D. analyzed the data. S.E. and S.E.A. wrote the manuscript.
Footnotes
Communicating editor: D. Charlesworth
Literature Cited
- Adler D. A., Rugarli E. I., Lingenfelter P. A., Tsuchiya K., Poslinski D., et al. , 1997. Evidence of evolutionary up-regulation of the single active X chromosome in mammals based on Clc4 expression levels in Mus spretus and Mus musculus. Proc. Natl. Acad. Sci. USA 94: 9244–9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S. L., Bonduriansky R., Chenoweth S. F., 2013. The genomic distribution of sex-biased genes in Drosophila serrata: X chromosome demasculinization, feminization, and hyperexpression in both sexes. Genome Biol. Evol. 5: 1986–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoff A. M., and Dessimoz C., 2009. Phylogenetic and functional assessment of orthologs inference projects and methods. PLOS Comput. Biol. 5: e1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., and Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist G., 2004. Sexual conflict and sexual selection: lost in the chase. Evolution 58: 1383–1393. [DOI] [PubMed] [Google Scholar]
- Artieri C. G., Haerty W., Gupta B. P., Singh R. S., 2008. Sexual selection and maintenance of sex: evidence from comparisons of rates of genomic accumulation of mutations and divergence of sex-related genes in sexual and hermaphroditic species of Caenorhabditis. Mol. Biol. Evol. 25: 972–979. [DOI] [PubMed] [Google Scholar]
- Assis R., Zhou Q., Bachtrog D., 2012. Sex-biased transcriptome evolution in Drosophila. Genome Biol. Evol. 4: 1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery P. J., 1984. The population-genetics of haplo-diploids and X-linked genes. Genet. Res. 44: 321–341. [Google Scholar]
- Bachtrog D., Toda N. R., Lockton S., 2010. Dosage compensation and demasculinization of X chromosomes in Drosophila. Curr. Biol. 20: 1476–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines J. F., Sawyer S. A., Hartl D. L., Parsch J., 2008. Effects of X-linkage and sex-biased gene expression on the rate of adaptive protein evolution in Drosophila. Mol. Biol. Evol. 25: 1639–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriere A., Yang S. P., Pekarek E., Thomas C. G., Haag E. S., et al. , 2009. Detecting heterozygosity in shotgun genome assemblies: lessons from obligately outcrossing nematodes. Genome Res. 19: 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean C. J., Schaner C. E., Kelly W. G., 2004. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet. 36: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender L. B., Suh J., Carroll C. R., Fong Y., Fingerman I. M., et al. , 2006. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 133: 3907–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessler J. B., Andersen E. C., Villeneuve A. M., 2010. Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS Genet. 6: e1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T., Evans D., Link C. D., Guffanti A., Lawson D., et al. , 2002. A global analysis of Caenorhabditis elegans operons. Nature 417: 851–854. [DOI] [PubMed] [Google Scholar]
- Brawand D., Soumillon M., Necsulea A., Julien P., Csardi G., et al. , 2011. The evolution of gene expression levels in mammalian organs. Nature 478: 343–348. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Coyne J. A., Barton N. H., 1987. The relative rates of evolution of sex-chromosomes and autosomes. Am. Nat. 130: 113–146. [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G., 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. [DOI] [PubMed] [Google Scholar]
- Coghlan A., Wolfe K. H., 2002. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 12: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T., Knowles L. L., 2005. Intergenomic conflict revealed by patterns of sex-biased gene expression. Trends Genet. 21: 495–499. [DOI] [PubMed] [Google Scholar]
- Conrad T., Akhtar A., 2012. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat. Rev. Genet. 13: 123–134. [DOI] [PubMed] [Google Scholar]
- Counterman B. A., Ortiz-Barrientos D., Noor M. A. F., 2004. Using comparative genomic data to test for fast-x evolution. Evolution 58: 656–660. [PubMed] [Google Scholar]
- Csankovszki G., 2009. Condensin function in dosage compensation. Epigenetics 4: 212–215. [DOI] [PubMed] [Google Scholar]
- Cutter A. D., 2008. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 25: 778–786. [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Ward S., 2005. Sexual and temporal dynamics of molecular evolution in C. elegans development. Mol. Biol. Evol. 22: 178–188. [DOI] [PubMed] [Google Scholar]
- de Clare M., Pir P., and Oliver S. G., 2011. Haploinsufficiency and the sex chromosomes from yeasts to humans. BMC Biol. 9: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. X., Hiatt J. B., Nguyen D. K., Ercan S., Sturgill D., et al. , 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 43: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C., Clifton S. W., Schuster L. N., Chinwalla A., Delehaunty K., et al. , 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 40: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C., Roeseler W., and Srinivasan J., 2006. Pristionchus pacificus genomics: from genetics to genome sequence. WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.7.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche C. M., 2012. Dosage compensation of the sex chromosomes. Annu. Rev. Genet. 46(46): 537–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divina P., Vlcek C., Strnad P., Paces V., Forejt J., 2005. Global transcriptome analysis of the C57BL/6J mouse testis by SAGE: evidence for nonrandom gene order. BMC Genomics 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C., Gribnau J., 2013. Different flavors of X-chromosome inactivation in mammals. Curr. Opin. Cell Biol. 25: 314–321. [DOI] [PubMed] [Google Scholar]
- Ellegren H., 2011. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat. Rev. Genet. 12: 157–166. [DOI] [PubMed] [Google Scholar]
- Ellegren H., Parsch J., 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 8: 689–698. [DOI] [PubMed] [Google Scholar]
- Ercan S., Lieb J. D., 2009. C. elegans dosage compensation: a window into mechanisms of domain-scale gene regulation. Chromosome Res. 17: 215–227. [DOI] [PubMed] [Google Scholar]
- Ferrari F., Alekseyenko A. A., Park P. J., Kuroda M. I., 2014. Transcriptional control of a whole chromosome: emerging models for dosage compensation. Nat. Struct. Mol. Biol. 21: 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y., Bender L., Wang W., Strome S., 2002. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science 296: 2235–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Vibranovski M. D., Zhang L., Li Z., Liu M., et al. , 2014. A long-term demasculinization of X-linked intergenic noncoding RNAs in Drosophila melanogaster. Genome Res. 4: 629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos L. J., Rechtsteiner A., Egelhofer T. A., Carroll C. R., Strome S., 2012. Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep 2: 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart M. E., Kuroda M. I., 2009. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136: 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J., Grootegoed J. A., 2012. Origin and evolution of X chromosome inactivation. Curr. Opin. Cell Biol. 24: 397–404. [DOI] [PubMed] [Google Scholar]
- Gupta V., Parisi M., Sturgill D., Nuttall R., Doctolero M., et al. , 2006. Global analysis of X-chromosome dosage compensation. J. Biol. 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L. W., Miller R. D., Baird S. E., Chinwalla A., Fulton L. A., et al. , 2007. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 5: 1603–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T. T., Eisen M. B., Thornton K. R., Andolfatto P., 2013. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 23: 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvilsom C., Qian Y., Bataillon T., Li Y. R., Mailund T., et al. , 2012. Extensive X-linked adaptive evolution in central chimpanzees. Proc. Natl. Acad. Sci. USA 109: 2054–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti P., Morrow E. H., 2010. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 8: e1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z. F., Machado C. A., 2009. Evolution of sex-dependent gene expression in three recently diverged species of Drosophila. Genetics 183: 1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P., Brawand D., Soumillon M., Necsulea A., Liechti A., et al. , 2012. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 10: e1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoshima H., Cassata G., Burglin T. R., 1999. A Caenorhabditis elegans homeobox gene expressed in the male tail, a link between pattern formation and sexual dimorphism? Dev. Genes Evol. 209: 59–62. [DOI] [PubMed] [Google Scholar]
- Kayserili M. A., Gerrard D. T., Tomancak P., and Kalinka A. T., 2012. An excess of gene expression divergence on the X chromosome in Drosophila embryos: implications for the faster-x hypothesis. PLoS Genet. 8: e1003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Schaner C. E., Dernburg A. F., Lee M. H., Kim S. K., et al. , 2002. X-chromosome silencing in the germline of C. elegans. Development 129: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P., Hellmann I., Enard W., Nowick K., Leinweber M., et al. , 2005. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309: 1850–1854. [DOI] [PubMed] [Google Scholar]
- Khil P. P., Smirnova N. A., Romanienko P. J., Camerini-Otero R. D., 2004. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 36: 642–646. [DOI] [PubMed] [Google Scholar]
- Kimble J., 1981. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev. Biol. 87: 286–300. [DOI] [PubMed] [Google Scholar]
- Kimble J. E., White J. G., 1981. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81: 208–219. [DOI] [PubMed] [Google Scholar]
- Kiontke K., Gavin N. P., Raynes Y., Roehrig C., Piano F., et al. , 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101: 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K. C., Felix M. A., Ailion M., Rockman M. V., Braendle C., et al. , 2011. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 11: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D. C., Staisch J., Thillainathan B., Haines K., Baird S. E., et al. , 2010. A toolkit for rapid gene mapping in the nematode Caenorhabditis briggsae. BMC Genomics 11: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A., Halligan D. L., Keightley P. D., 2013. Faster-x adaptive protein evolution in house mice. Genetics 196: 1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi W. S., Core L. J., Waters C. T., Lis J. T., Meyer B. J., 2013. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife 2: e00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Z., Eizinger A., Nandakumar R., Schuster S. C., Sommer R. J., 2003. Limited microsynteny between the genomes of Pristionchus pacificus and Caenorhabditis elegans. Nucleic Acids Res. 31: 2553–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher M. J., Urrutia A. O., Hurst L. D., 2003. Evidence that the human X chromosome is enriched for male-specific but not female-specific genes. Mol. Biol. Evol. 20: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Xing K., Zhang J., He X., 2012. Expression reduction in mammalian X chromosome evolution refutes Ohno’s hypothesis of dosage compensation. Proc. Natl. Acad. Sci. USA 109: 11752–11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Gupta V., VerMilyea M. D., Falciani F., Lee J. T., et al. , 2007. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 5: 2809–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart A., 2012. The rapid evolution of X-linked male-biased gene expression and the large-X effect in Drosophila yakuba, D. santomea, and their hybrids. Mol. Biol. Evol. 29: 3873–3886. [DOI] [PubMed] [Google Scholar]
- Lu J., Wu C. I., 2005. Weak selection revealed by the whole-genome comparison of the X chromosome and autosomes of human and chimpanzee. Proc. Natl. Acad. Sci. USA 102: 4063–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J., Ahn J. H., Cipriani P. G., Killian D. J., Chaudhary A. L., et al. , 2005. Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics 169: 1997–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine E. M., 2010. Meiotic silencing in Caenorhabditis elegans. Int. Rev. Cell Mol. Biol. 282(282): 91–134. [DOI] [PubMed] [Google Scholar]
- Mank J. E., Ellegren H., 2009. Are sex-biased genes more dispensable? Biol. Lett. 5: 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank J. E., Hultin-Rosenberg L., Webster M. T., Ellegren H., 2008a The unique genomic properties of sex-biased genes: insights from avian microarray data. BMC Genomics 9: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank J. E., Hultin-Rosenberg L., Zwahlen M., Ellegren H., 2008b Pleiotropic constraint hampers the resolution of sexual antagonism in vertebrate gene expression. Am. Nat. 171: 35–43. [DOI] [PubMed] [Google Scholar]
- Meiklejohn C. D., Presgraves D. C., 2012. Little evidence for demasculinization of the drosophila x chromosome among genes expressed in the male germline. Genome Biol. Evol. 4: 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Parsch J., Ranz J. M., Hartl D. L., 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 100: 9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel R. P., Malone J. H., Clark A. G., 2012a Disentangling the relationship between sex-biased gene expression and X-linkage. Genome Res. 22: 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel R. P., Malone J. H., Clark A. G., 2012b Faster-X evolution of gene expression in Drosophila. PLoS Genet. 8: e1003013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. J., 2010. Targeting X chromosomes for repression. Curr. Opin. Genet. Dev. 20: 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T. S., Hillier L. W., Eichler E. E., Zody M. C., Jaffe D. B., et al. , 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87. [DOI] [PubMed] [Google Scholar]
- Mitreva M., Blaxter M. L., Bird D. M., McCarter J. P., 2005. Comparative genomics of nematodes. Trends Genet. 21: 573–581. [DOI] [PubMed] [Google Scholar]
- Nguyen D. K., Disteche C. M., 2006. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 38: 47–53. [DOI] [PubMed] [Google Scholar]
- Ohno S., 1967. Sex Chromosomes and Sex-Linked Genes. Springer-Verlag, New York. [Google Scholar]
- Orita M., Iwahana H., Kanazawa H., Hayashi K., Sekiya T., 1989. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc. Natl. Acad. Sci. USA 86: 2766–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 1997. Haldane’s rule. Annu. Rev. Ecol. Syst. 28: 195–218. [Google Scholar]
- Parisi M., Nuttall R., Edwards P., Minor J., Naiman D., et al. , 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M., Nuttall R., Naiman D., Bouffard G., Malley J., et al. , 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299: 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., et al. , 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J., Ellegren H., 2013. The evolutionary causes and consequences of sex-biased gene expression. Nat. Rev. Genet. 14: 83–87. [DOI] [PubMed] [Google Scholar]
- Piano F., Schetter A. J., Mangone M., Stein L., Kemphues K. J., 2000. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10: 1619–1622. [DOI] [PubMed] [Google Scholar]
- Pollex T., Heard E., 2012. Recent advances in X-chromosome inactivation research. Curr. Opin. Cell Biol. 24: 825–832. [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2012. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Ranz J. M., Castillo-Davis C. I., Meiklejohn C. D., Hartl D. L., 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300: 1742–1745. [DOI] [PubMed] [Google Scholar]
- Reinius B., Saetre P., Leonard J. A., Blekhman R., Merino-Martinez R., et al. , 2008. An evolutionarily conserved sexual signature in the primate brain. PLoS Genet. 4: e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius B., Johansson M. M., Radomska K. J., Morrow E. H., Pandey G. K., et al. , 2012. Abundance of female-biased and paucity of male-biased somatically expressed genes on the mouse X-chromosome. BMC Genomics 13: 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V., Smith H. E., Nance J., Wang J., Van Doren C., et al. , 2000. A global profile of germline gene expression in C. elegans. Mol. Cell 6: 605–616. [DOI] [PubMed] [Google Scholar]
- Reinke V., Gil I. S., Ward S., Kazmer K., 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323. [DOI] [PubMed] [Google Scholar]
- Reuben M., Lin R., 2002. Germline X chromosomes exhibit contrasting patterns of histone H3 methylation in Caenorhabditis elegans. Dev. Biol. 245: 71–82. [DOI] [PubMed] [Google Scholar]
- Rice W. R., 1984. Sex-chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742. [DOI] [PubMed] [Google Scholar]
- Rice W. R., Chippindale A. K., 2001. Intersexual ontogenetic conflict. J. Evol. Biol. 14: 685–693. [Google Scholar]
- Richards S., Liu Y., Bettencourt B. R., Hradecky P., Letovsky S., et al. , 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 15: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. A., Koboldt D. C., Staisch J. E., Chamberlin H. M., Gupta B. P., et al. , 2011. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genet. 7: e1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel D., Riebesell M., Sommer R. J., 2005. Gonadogenesis in Pristionchus pacificus and organ evolution: development, adult morphology and cell–cell interactions in the hermaphrodite gonad. Dev. Biol. 277: 200–221. [DOI] [PubMed] [Google Scholar]
- Russo C. A., Takezaki N., Nei M., 1995. Molecular phylogeny and divergence times of Drosophilid species. Mol. Biol. Evol. 12: 391–404. [DOI] [PubMed] [Google Scholar]
- Saifi G. M., Chandra H. S., 1999. An apparent excess of sex- and reproduction-related genes on the human X chromosome. Proc. Biol. Sci. 266: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefke B., Emerson J. J., Wang T. Y., Lu M. Y., Hsieh L. C., et al. , 2013. Inheritance of gene expression level and selective constraints on trans- and cis-regulatory changes in yeast. Mol. Biol. Evol. 30: 2121–2133. [DOI] [PubMed] [Google Scholar]
- Small C. M., Carney G. E., Mo Q., Vannucci M., Jones A. G., 2009. A microarray analysis of sex- and gonad-biased gene expression in the zebrafish: evidence for masculinization of the transcriptome. BMC Genomics 10: 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer W. C., Zeller G., Watson J. D., Henz S. R., Watkins K. L., et al. , 2011. A spatial and temporal map of C. elegans gene expression. Genome Res. 21: 325–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J., Sinz W., Lanz C., Brand A., Nandakumar R., et al. , 2002. A bacterial artificial chromosome-based genetic linkage map of the nematode Pristionchus pacificus. Genetics 162: 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L. D., Bao Z. R., Blasiar D., Blumenthal T., Brent M. R., et al. , 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 1: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill D., Zhang Y., Parisi M., and Oliver B., 2007. Demasculinization of X chromosomes in the Drosophila genus. Nature 450: 238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi T. M., Deplancke B., Osato N., Zhu L. J., Barrasa M. I., et al. , 2011. Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 7: e1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. G., Li R. H., Smith H. E., Woodruff G. C., Oliver B., et al. , 2012. Simplification and desexualization of gene expression in self-fertile nematodes. Curr. Biol. 22: 2167–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K., Long M., 2002. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol. Biol. Evol. 19: 918–925. [DOI] [PubMed] [Google Scholar]
- Torgerson D. G., Kulathinal R. J., Singh R. S., 2002. Mammalian sperm proteins are rapidly evolving: evidence of positive selection in functionally diverse genes. Mol. Biol. Evol. 19: 1973–1980. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., 1998. Evolutionary genetics: the causes of Haldane’s rule. Science 282: 889–891. [DOI] [PubMed] [Google Scholar]
- Turner J. M., 2007. Meiotic sex chromosome inactivation. Development 134: 1823–1831. [DOI] [PubMed] [Google Scholar]
- Uyar B., Chu J. S., Vergara I. A., Chua S. Y., Jones M. R., et al. , 2012. RNA-seq analysis of the C. briggsae transcriptome. Genome Res. 22: 1567–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verster A. J., Ramani A. K., McKay S. J., Fraser A. G., 2014. Comparative RNAi screens in C. elegans and C. briggsae reveal the impact of developmental system drift on gene function. PLoS Genet. 10: e1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski M. D., Lopes H. F., Karr T. L., Long M., 2009a Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 5: e1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski M. D., Zhang Y., Long M., 2009b General gene movement off the X chromosome in the Drosophila genus. Genome Res. 19: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Bachtrog D., 2011. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol. Evol. 3: 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Charlesworth B., 2009. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: A consequence of dosage compensation? J. Mol. Evol. 68: 576–583. [DOI] [PubMed] [Google Scholar]
- Vicoso B., Emerson J. J., Zektser Y., Mahajan S., Bachtrog D., 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11: e1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton A. C., 1940. Gametogenesis, pp. 205–215 in An Introduction to Nematology, edited by Chitwood G. B., Chitwood M. B. Babylon Press, New York. [Google Scholar]
- Wang J., Chen P. J., Wang G. J., Keller L., 2010. Chromosome size differences may affect meiosis and genome size. Science 329: 293. [DOI] [PubMed] [Google Scholar]
- Wang P. J., McCarrey J. R., Yang F., Page D. C., 2001. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27: 422–426. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhao Y., Wong K., Ehlers P., Kohara Y., et al. , 2009. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics 10: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. [DOI] [PubMed] [Google Scholar]
- Wu C. I., Davis A. W., 1993. Evolution of postmating reproductive isolation: the composite nature of Haldane’s rule and its genetic bases. Am. Nat. 142: 187–212. [DOI] [PubMed] [Google Scholar]
- Yang X., Schadt E. E., Wang S., Wang H., Arnold A. P., et al. , 2006. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 16: 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sturgill D., Parisi M., Kumar S., and Oliver B., 2007. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature 450: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. E., Vibranovski M. D., Krinsky B. H., Long M., 2010. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 20: 1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.