Abstract
To identify novel genomic regions that regulate sex determination, we utilized the powerful C57BL/6J-YPOS (B6-YPOS) model of XY sex reversal where mice with autosomes from the B6 strain and a Y chromosome from a wild-derived strain, Mus domesticus poschiavinus (YPOS), show complete sex reversal. In B6-YPOS, the presence of a 55-Mb congenic region on chromosome 11 protects from sex reversal in a dose-dependent manner. Using mouse genetic backcross designs and high-density SNP arrays, we narrowed the congenic region to a 1.62-Mb genomic region on chromosome 11 that confers 80% protection from B6-YPOS sex reversal when one copy is present and complete protection when two copies are present. It was previously believed that the protective congenic region originated from the 129S1/SviMJ (129) strain. However, genomic analysis revealed that this region is not derived from 129 and most likely is derived from the semi-inbred strain POSA. We show that the small 1.62-Mb congenic region that protects against B6-YPOS sex reversal is located within the Sox9 promoter and promotes the expression of Sox9, thereby driving testis development within the B6-YPOS background. Through 30 years of backcrossing, this congenic region was maintained, as it promoted male sex determination and fertility despite the female-promoting B6-YPOS genetic background. Our findings demonstrate that long-range enhancer regions are critical to developmental processes and can be used to identify the complex interplay between genome variants, epigenetics, and developmental gene regulation.
Keywords: sex determination, disorders of sex development, gonad, testis, congenic, sex reversal
THE formation of a testis or an ovary from an undifferentiated bipotential gonad relies on the expression of a single Y-chromosome gene, SRY, to trigger male sex determination (Sinclair et al. 1990; Koopman et al. 1991). In mammals, SRY expression is required for development of the testis, secondary sex characteristics, and, ultimately, fertility. In both humans and mice, mutations and deletions of SRY result in XY sex reversal (Berta et al. 1990; Sinclair et al. 1990), while translocations of SRY onto the X chromosome or an autosome result in XX males (Berkovitz et al. 1992). Similarly, overexpression of various SOX (Sry-related HMG box) gene family members within the bipotential gonad results in XX males (Bishop et al. 2000; O’Bryan et al. 2008; Polanco et al. 2010), indicating that increased SOX gene expression during critical periods in fetal sex determination activates the male sex determination pathway. Disorders of sex development (DSD) are among the most common genetic disorders, occuring in ∼1/100 births when patients with hypospadias are included (Lee et al. 2006).
One of the most powerful models in mammalian sex determination is a mouse strain in which XY males are sensitized to XY sex reversal. The strain’s autosomes and X chromosome are from the Mus m. musculus C57BL/6J (B6) strain while the Y chromosome is from one of the Mus m. domesticus (DOM) substrains. B6 males with their native Y chromosome, YB6, develop as normal fertile males. In contrast, B6-YDOM strains show a range of sex determination phenotypes from full sex reversal (female phenotype) to delayed testicular development and incompletely masculinized males. Placing the M. m. domesticus poschiavinus Y chromosome (YPOS), named after the wild-derived strain from the Poschiavo valley in Switzerland, on the B6 background results in a nearly 100% adult XY female phenotype. At the morphologic and molecular levels, all of the B6-YPOS animals have abnormal early testis development and a significant delay in Sry expression (Bullejos and Koopman 2005).
The interaction between genetic background and Sry expression is critical to initiation of testis development in utero. Yet, on other genetic backgrounds, such as 129S1/SvIMJ, DBA/2J, C58/J, or BALB/cBy, YPOS does not result in XY sex reversal (Eicher 1988). The altered testis phenotype detected during early gonadal development in YPOS is specific to the B6 background.
Genetic differences in the Sry gene from musculus vs. domesticus strains do not explain the B6-YDOM sex reversal (Albrecht and Eicher 1997). For example, the M. musculus isoform is nearly twice as large (395 aa) as the M. domesticus Sry (230 aa) due to a premature termination codon located in the glutamine repeat region of the M. domesticus Sry. Additionally, the size of Sry glutamine repeat alone does not correlate with propensity toward B6-YDOM sex reversal.
Full testis development requires both spatiotemporally appropriate Sry levels and a receptive microenvironment within the somatic cells on which Sry acts to converge in a short 6-hr window (Hiramatsu et al. 2009). Sry expression is required within a specific proportion of somatic cells of the bipotential gonad and at a certain level to initiate and propagate male sex determination (Bullejos and Koopman 2005; Kashimada and Koopman 2010). In humans, genetic background modulates the phenotypic expression of SRY gene mutations and results in variable gonadal and genital phenotypes among family members who share the same SRY mutation (Maier et al. 2003). In mouse models, the interaction between the B6 background and the YPOS chromosome results in a 4-hr delay in peak Sry expression in B6-YPOS compared to B6-YB6 embryos (Bullejos and Koopman 2005). While the genetic underpinnings of the spatiotemporal regulation of SRY remain to be fully elucidated, recent work has shown that upregulation of Sry is coordinated by MAP kinase signaling (Bogani et al. 2009; Gierl et al. 2012; Warr et al. 2012), as well as chromatin-modifying factors such as CBX2 (Katoh-Fukui et al. 1998; Katoh-Fukui et al. 2012) and Jmjd1a (Kuroki et al. 2013). The multiple layers of genetic and epigenetic regulation underscore the tight regulation of developmental gene expression.
In addition to the intricate regulation of SRY expression, the receptiveness of the downstream genetic environment is equally important, yet poorly understood. Of the downstream genes, the best studied is the regulatory region of SOX9, which spans >2.5 Mb upstream and downstream of the SOX9 ORF (Wagner et al. 1994; Pfeifer et al. 1999). Autosomal dominant mutations in SOX9 result in campomelic dysplasia with XY sex reversal (Online Mendelian Inheritance in Man #114290) due to the importance of SOX9 in both cartilage and testis determination (Wright et al. 1995). Point mutations within the human syntenic region of the testis-specific enhancer located 5′ to the Sox9 open reading frame, TESCO, have not been found (Sekido and Lovell-Badge 2008; Georg et al. 2010). However, duplications and deletions limited to the SOX9 regulatory regions have recently been identified in several familial and isolated cases of 46,XY DSD or 46,XX DSD (Benko et al. 2011; Cox et al. 2011; White et al. 2011). These deletions and duplications cause dysregulation of SOX9 expression with either loss of testis-specific SOX9 expression or abnormal ectopic expression in an XX individual resulting in either 46,XY gonadal dysgenesis or 46,XX testicular DSD, respectively. Furthermore, these copy-number changes indicate that there are testis-specific regulatory regions of human SOX9. In addition to mutations, transgenic insertions in mice disrupting regulation of Sox9 have been shown to result in aberrant activation of Sox9 and XX males (Bishop et al. 2000).
In the congenic B6-YPOS mouse model, the incompatibility between the gonadal microenvironment created by the B6 genetic background and the low levels of SryPOS prevents timely activation of male sex determination (Bullejos and Koopman 2005). Early genetic linkage studies using B6 and a non-sex-reversing strain, DBA2/J, to identify genetic regions contributing to the formation of XYPOS hermaphrodites, identified three autosomal testis-determining loci on mouse chromosomes 2, 4, and 5, respectively, and demonstrated that homozygosity for the B6 allele is one of the major contributing factors in B6-YPOS sex reversal (Eicher et al. 1996).
More recently, comparative studies of gene expression within B6 and another non-sex-reversing strain, 129S1, in developing testis showed that, compared to 129-Y129 gonads, B6-YB6 gonads had a higher average expression of “female”-related genes and a lower expression of “male” genes (Munger et al. 2009, 2013). On the B6 genetic background, Sox9 expression was elevated by nearly twofold, which may be compensation for the B6 strain’s “pro-female” gene expression profile. Therefore, to develop as a phenotypic male, the strength of the interaction between the Y chromosome and autosomal loci must overcome the “pro-female” balance intrinsic to the B6 genetic background. These studies highlight the complex networks that control gonadal development and indicate that there exists a normal flux of gene expression between various genetic backgrounds. The maintenance of testis and ovary simply requires that the equilibrium is driven toward the correct developmental program and is evolutionarily maintained by fertility. Genetic mutations within the majority of genes implicated in either male or female mouse sex determination do not result in complete sex reversal, but instead in variable degrees of sex reversal, the severity of which can be modified by genetic background (Eicher 1988; Eicher et al. 1996; Bogani et al. 2009). Therefore, an independent model appears to be flawed as the interacting networks of genes with some prominent driver genes have large effects on gonadal developmental programs.
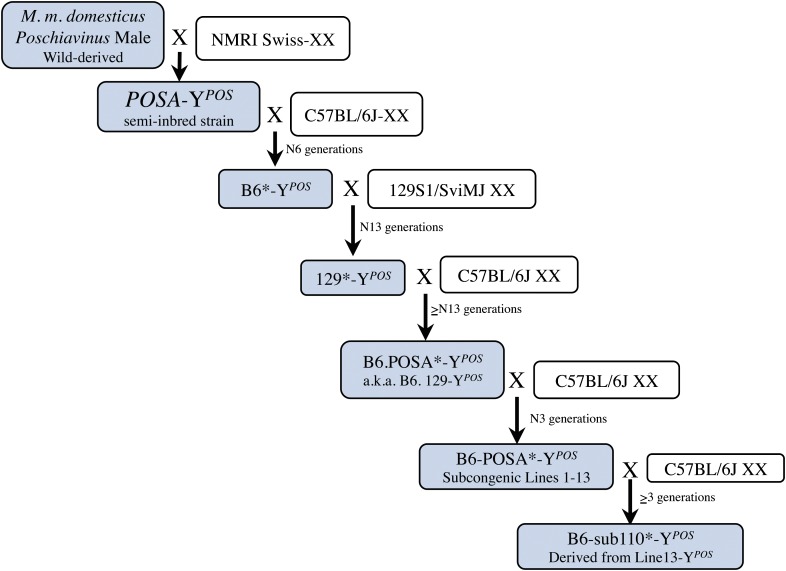
In this study, we utilized a congenic strain, previously developed by multiple backcross generations, in which a homozygous non-B6 region on chromosome 11 was identified that conferred 100% protection from B6-YPOS sex reversal (Whitney et al. 2000; Nikolova et al. 2008). To fully understand the genetic origin of the protection from B6-YPOS sex reversal, we retraced the 30-year history of this congenic strain (Figure 1). The initial discovery of B6-YPOS sex reversal occurred after backcrossing semi-inbred POSA males (cross between XY wild-derived M. m. domesticus poschiavinus and Naval Medical Research Institute Swiss XX mice) that carried the Hbath-J trait on chromosome 11 for α-thalassemia to the C57BL6/J background (Eicher et al. 1982). After 5 generations, the male-to-female sex ratio was highly skewed in favor of females, and half of the phenotypic females had an XY karyotype. Subsequent studies showed that the Hbath-J trait was not responsible for the sex reversal and that the sex reversal trait was Y-linked (Eicher et al. 1982). In the breeding of the protected congenic strain, fertile B6-YPOS males were backcrossed onto 129S1 for 13 generations, resulting in a normal sex ratio in the consomic strain 129-YPOS. At this point, it was hypothesized that the 129 genetic background protected from B6-YPOS sex reversal, and therefore 129-YPOS was backcrossed to B6 to initiate the current congenic strain. At each generation, only the most masculinized males were mated to B6 females, with the goal of selecting for autosomal regions that protect from B6-YPOS sex reversal (Whitney et al. 2000). In a previous publication, we used SNP markers polymorphic between B6 and 129 and identified large regions on chromosome 11 that resulted in dose-dependent protection from B6-YPOS sex reversal (Nikolova et al. 2008). While previous publications called this strain B6.129-YPOS, we show in this article that the origins of the congenic region are unlikely to be from the 129 background and, based on our analysis, most likely are derived from POSA. Therefore, we will refer to the B6.129-YPOS strain more accurately as B6.POSA-YPOS (see Figure 1 and Figure 2).
Figure 1.
History of the protected B6-POSA-YPOS strains. The original POSA strain was a semi-inbred strain created when Naval Medical Research Institute (NMRI) Swiss females were intercrossed to M. musculus domesticus poschiavinus wild-derived males. A POSA male was backcrossed to B6 over 6 generations, and one of the B6-YPOS males that was not sex-reversed was backcrossed onto the 129 background for 13 generations. Finally, a 129-YPOS was backcrossed over >13 generations to B6, and at each generation only the most masculinized males were selected for breeding, creating the B6.POSA-YPOS congenic line (previous studies referred to this line as B6.129), which was then further crossed to B6 females to create subcongenic chromosomes 1–13. In each backcrossing experiment, a region of the POSA genome remained, undetected, conferring protection from sex reversal (asterisk) on all breeding males. The shaded boxes follow the transmission of the YPOS chromosome throughout the generations.
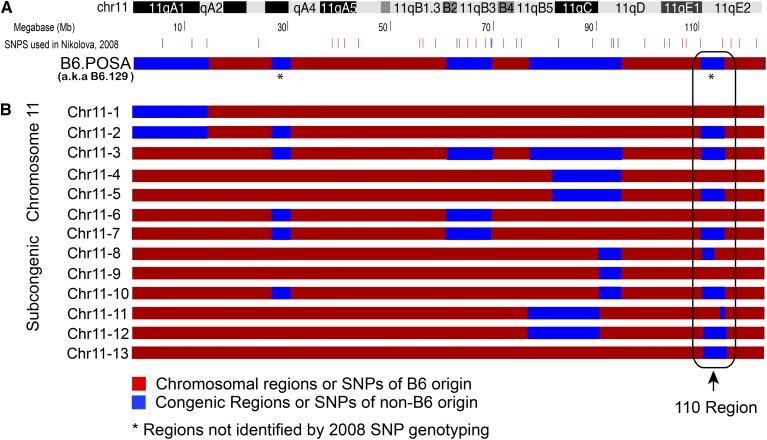
Figure 2.
One congenic and 13 subcongenic chromosomes generated through breeding. (A) Location of SNPs polymorphic between the B6 (red) and non-B6 (blue) strains used to identify the original congenic region are shown next to a representation of mouse chromosome 11. The non-B6 markers are polymorphic between B6 and 129 and chosen because of the previous hypothesis that the protection was derived from the 129 strain. Two regions, one located at 30 Mb and the other located at 110 Mb, were not identified by the 2008 SNP scan (asterisk). (B) Thirteen different subcongenic configurations of chromosome 11 (Chr11-1 through Chr11-13), with varying degrees of non-B6 DNA, were created by backcrossing the B6.POSA- YPOS with B6-XX females. Region 110 is circled.
To narrow down the genomic element important in protection from B6-YPOS sex reversal, we backcrossed heterozygous B6.POSA-YPOS males to B6 XX females and, at each generation, performed SNP genotyping to identify recombinants within the congenic region. Animals with recombination events within the congenic region were used to found 12 subcongenic lines. In this study, we narrowed the large 60-Mb region originally identified in Nikolova et al. (2008) to a small 1.62-Mb noncoding region that is responsible for protection from B6-YPOS sex reversal. These results correct some longstanding assumptions about B6-YPOS and the origins of previously reported protection from sex reversal phenotype. Additionally, they reveal the importance of the promoter of Sox9 in the protection from B6-YPOS sex reversal.
Materials and Methods
Mouse strains
Animals were housed according to the guidelines of the University of California at Los Angeles (UCLA) Division of Laboratory Animal Medicine. All experiments were approved by the Institutional Animal Care and Use Committees of UCLA. C57BL/6J females and males used for breeding and subcongenic line generation were obtained from the Jackson Laboratory. The Jackson Laboratory is fully accredited by the American Association for Accreditation of Laboratory Animal Care.
Construction of the C57BL/6J.POSA-YPOS subcongenic strain
The original C57BL/6J.POSA-YPOS (a.k.a. C57BL/6J.129-YPOS in previous publication) strain was produced as described previously (Whitney et al. 2000; Nikolova et al. 2008). To generate the subcongenic strains, fertile heterozygous C57BL/6J.POSA-YPOS males were backcrossed to B6-XX females from the Jackson Laboratory (Figure 1). At each generation, offspring were genotyped with SNP markers polymorphic between B6 and 129 on chromosome 11 identified in the SNP scan published in Nikolova et al. (2008) (Supporting Information, Table S1). A region surrounding each SNP was PCR-amplified from genomic DNA and then sequenced to determine genotype at each informative SNP. Animals in which a meiotic recombination occurred within the congenic region were used to generate new subcongenic chromosomes (Figure 2). Subcongenic founder YPOS males were bred to B6-XX females, and all offspring were genotyped to determine the presence of the subcongenic region. Heterozygous brothers and sisters were mated together to generate animals that were homozygous for each subcongenic region.
Phenotyping of adults
A mouse was classified (1) as a female, if female external genitalia, yellow mammary-associated hair pigmentation, bilateral uterine horns, and normal-appearing ovaries were present; (2) as a hermaphrodite, if ambiguous genitalia, some yellow mammary-associated hair pigmentation, and an ovary and a contralateral ovotestis or testis were present; or (3) as a male, if normal-appearing male genitalia, no yellow mammary-associated hair pigmentation, and two testes were present.
Classification of fetuses
Midday of the day of a vaginal plug was considered embryonic day 0.5 (E0.5), and the fetal stage of development was verified by limb morphology (Theiler 1989). Individual gonads from E14.5–E16 fetuses were classified as an ovary, an ovotestis, or a testis (Eicher et al. 1982). Finally, the approximate amount of testicular tissue in each ovotestis was estimated at the time of dissection and confirmed using a captured image at 25× under a dissection microscope (Nikolova et al. 2008). If 50% or greater testicular tissue was present, the ovotestis was designated as “oT.” If <50% testicular tissue was present, the ovotestis was designated as “Ot.”
Sex chromosome genotyping
Chromosomal sex was determined with a PCR-based assay using a single primer pair, which detects the X-linked Smc-x gene (330 bp) and the Y-linked Smc-y gene (301 bp) (Bullejos and Koopman 2005). Immomix Red 2× (Bioline, London) was used for the PCR amplification as per the manufacturer’s guidelines with an annealing temperature of 57°. Samples were resolved by electrophoresis on a 2% agarose gel.
Statistical analysis
All data analyses were conducted using Stata (version 11, StataCorp). Categorical and ordinal data analyses for the genotype–phenotype correlations were assessed using Fisher’s exact tests or likelihood-ratio tests for the ordinal treatment of the data. The dose-effect hypothesis was tested using polytomous logistic regression (categorical outcomes) or ordered logistic regression (ordinal outcomes) and allowed for inclusion of covariates.
Mouse diversity genotyping array (Affymetrix)
Genomic DNA from 10 founder homozygous males from the subcongenic lines was extracted using the Gentra Puregene Mouse Tail Kit (Qiagen) and analyzed at JAX Mouse Diversity Genotyping Array Service (Bar Harbor, ME) according to the manufacturer’s protocol. SNP and copy number variation (CNV) analyses were performed by the Jackson Laboratory Bioinformatics Core (Yang et al. 2009) using the R MouseDivGeno software package (http://cgd.jax.org/tools/MouseDivGeno/). The MouseDivGeno package assumes three possible genotypes—homozygous for allele A, homozygous for allele B, or heterozygous for alleles AB—and can also declare missing (N) calls and genomic location information using SNPs annotated on Build 37 mm9 mouse genome. In total, this array contains 623,000 SNPs and 916,000 CNV probes. On chromosome 11, there are a total of 28,000 SNPs with a mean distance between SNPs of 24 kb. The additions to chromosome 11 in the mouse build 39 (mm10) released in 2011 did not directly affect the 110 region or our interpretation of the SNP array results. For CNV analysis, the SimpleCNV function within the R MouseDivGeno package was used and normalized against a subset of 249 high-quality control arrays generated with the Diversity Array by the Jackson Laboratory and CEL files generated from this experiment.
Immunofluorescence
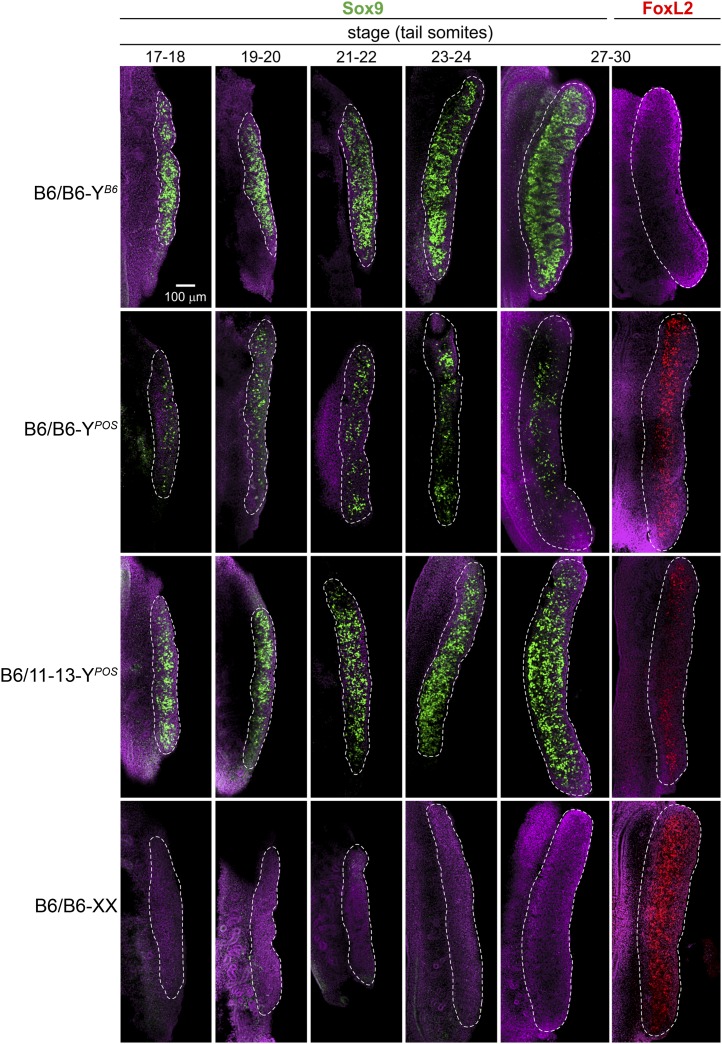
The time course of Sox9 expression was analyzed by whole-mount immunofluorescence (WM-IF) following the experimental design of Wilhelm et al. (2009) and using standard WM-IF protocols previously established in our laboratory (described in Fleming et al. 2012). The number of tail somites was used to assess the precise developmental stage of each embryo (Hacker et al. 1995; Bullejos and Koopman 2001). We examined B6-YB6, B6-YPOS, LineB6/11-13-YPOS, and B6-XX gonads at 17–18, 19–20, 21–22, 23–24, and 27–30 tail somites, roughly corresponding to E11.5–E12.5. Sox9 (rabbit polyclonal, SC-20095, Santa Cruz Lot#G0709) and FoxL2 (rabbit polyclonal, supplied by Dagmar Wilhelm, Monash University, Clayton, VIC, Australia) antibodies were used. ToproRed (Invitrogen) was used to counterstain nuclei. All images were taken on a Zeiss LSM 510 Meta confocal microscope. All genotypes and time points had n = 3–4, and the gonad shown is representative of the set.
Database use
The National Center for Biotechnology Information (http:/www.ncbi.nlm.nih.gov/), University of California at Santa Cruz Genome Bioinformatics (http:/genome.ucsc.edu/), and Mouse Genome Informatics (http:/www.informatics.jax.org/) websites were used to retrieve gene, marker, and sequence information. MatInspector Version 8.0.4 was used to identify predicted transcription-factor-binding sites within the promoter of Sox9. The genomic location information is based on Mouse Build 37 (July 2007).
Results
Generation of subcongenic lines
The original congenic strain, B6.POSA-YPOS, generated as described in Materials and Methods (Figure 1), contained three major congenic regions on chromosome 11 located between the following base pairs: 1–14,267,131, 58,783,266–71,981,592, and 76,380,940–94,191,519, respectively (Figure 2A). Identification of these three large regions relied on 149 microsatellite markers and 404 SNPs polymorphic between the B6 and 129 strains, which is equivalent to approximately one marker per 2 Mb (Nikolova et al. 2008). Over three generations, we generated subcongenic lines through backcrossing to B6-XX females. Animals with recombination within the congenic region were identified by genotyping of SNP markers from Nikolova et al. (2008) (Table S1A). Subcongenic lines were bred to homozygosity over two generations.
Our previous study had concluded that protection from B6-YPOS sex reversal was 100% in animals that were homozygous for the chromosome 11 congenic regions (Figure 2A). However, since that study we have noted that, compared to our published data, the same congenic B6.POSA-YPOS line began to show decreased rates, approaching 50%, of protection from sex reversal after 1.5 years of sibling matings. This loss of protection from B6-YPOS sex reversal was present only in offspring from specific fathers. All breeding males were confirmed to have SryPOS and regenotyped with markers from the original article, which confirmed the presence of a homozygous congenic region (Figure 2A and Table S1A). We hypothesized that there was another non-B6 genomic region affecting protection that had not been identified due to either the choice of markers being specifically polymorphic between B6 and 129, the low density of SNP markers, or an undetected de novo event in one of our founder males. To address these hypotheses, we performed whole-genome SNP array analysis in the founder males to identify all of the non-B6 regions and their genetic origin.
Genomic analysis of the subcongenic lines
The Affymetrix Mouse SNP Diversity Array developed at the Jackson Laboratory (Yang et al. 2009) was performed on genomic DNA from homozygous, breeding, subcongenic males and two of our phenotypically male B6-YPOS animals. This array was designed to identify regions of non-B6 origin and to determine from which inbred strain the region was derived. Call rates were >97% for all arrays, and the congenic regions on chromosome 11 had a large concentration of non-B6-SNPs (>100 SNPs/Mb) compared with all other chromosomes. All subcongenic regions identified through SNP genotyping were present, and no CNV regions were identified in more than one of the founder males (Table S2 and Table S3).
Our analysis confirmed the presence of the three previously identified regions and identified two new congenic regions on chromosome 11 that had a high density of non-B6 SNPs: the first spanning base pairs 20,652,664–29,502,320 and the second spanning base pairs 110,023,548–114,635,519 (Figure 2B and Figure 3). The region spanning base pairs 20,652,664–29,502,320 was present as heterozygous or homozygous in only half of the genotyped males, and thus its presence was not required for protection from B6-YPOS sex reversal.
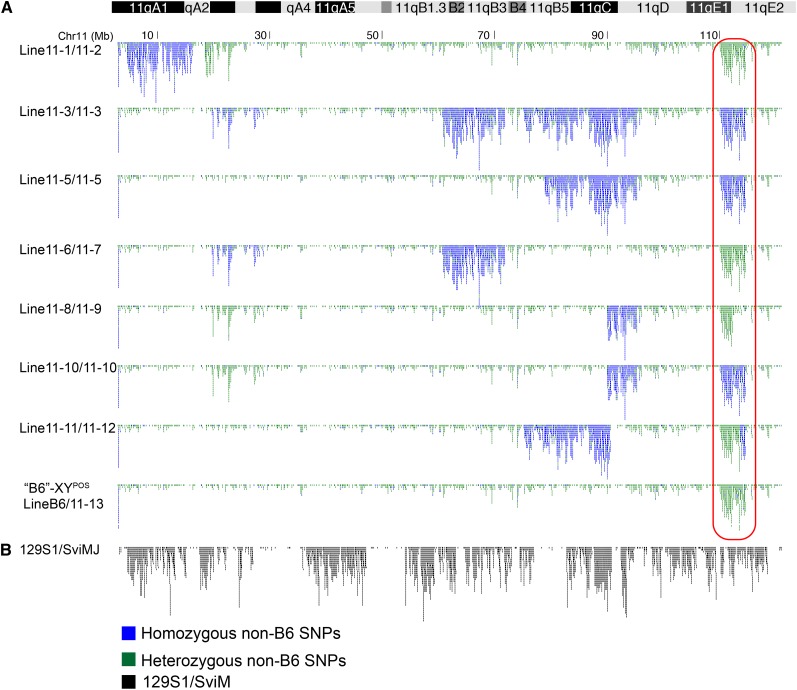
Figure 3.
All subcongenic founder, non-sex-reversed B6-YPOS male mice had a non-B6 region encompassing Sox9 and its promoter that is not of B6 or 129 origin. (A) Each schematic represents a YPOS male used for breeding and generation of subcongenic lines and is labeled for each version of chromosome 11 that the animal has (e.g., Line11-1/11-2 is an animal with two different copies of chromosome 11, Chr11-1 and Chr11-2 (see Figure 2B for notations). All of the subcongenic XYPOS breeding males were either heterozygous (green SNPs) or homozygous (blue SNPs) for a large 4.5-Mb region, which encompasses Sox9 and its promoter (red box). (Top) Ideogram of mouse chromosome 11. (B) Diversity array analysis of the 129S1 inbred strain has a SNP profile (black) that has a different pattern from the SNPs in the subcongenic protected males (compare to A). This shows that the congenic regions were not derived from the 129S1 strain, as previously described.
The second region from megabases 110 to 114.5, which we will refer to as the “110 region,” was present as either heterozygous or homozygous in all founder XYPOS males that were protected from sex reversal (Figure 3A). Even the animals that we originally believed to be “B6-YPOS” males were heterozygous for this 4.5-Mb region and were renamed B6/Line11-13-YPOS (Figure 3A). We hypothesized that this newly identified 110 region resulted in the originally observed dose-dependent protection from sex reversal. The 110 region was present in every male protected from B6-YPOS sex reversal and used for breeding. Furthermore, the 110 region contains a well-described sex determination gene, Sox9, as well as the entirety of its promoter region. The presence of the 110 region in all of our phenotypically male and breeding mice and the presence of Sox9 within the 110 region strongly suggested that the 110 region was associated with protection from B6-YPOS sex reversal.
Chr11-13, which has only the 110-region of non-B6 origin, protects from B6-YPOS sex reversal
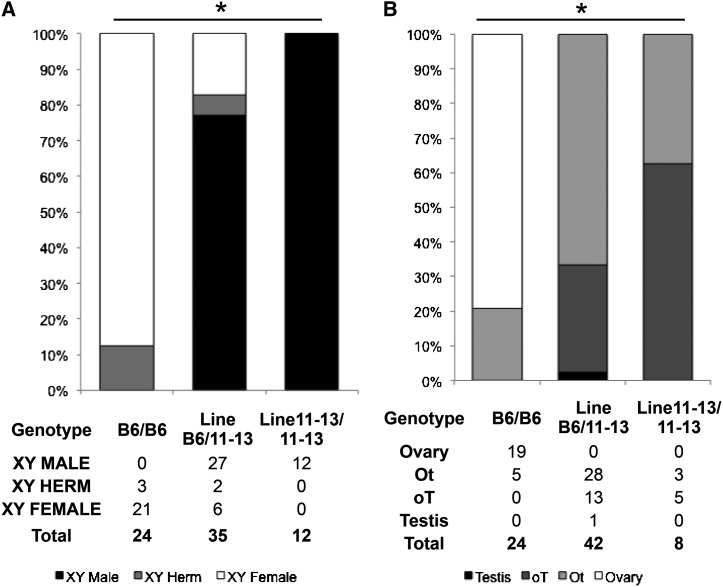
Based on our new genotypic data, we reanalyzed our subcongenic lines at adult and embryonic stages to determine the role of the 110 region in protection from B6-YPOS sex reversal. Adult phenotypic analysis of animals that were heterozygous (LineB6/11-13-YPOS) or homozygous (Line11-13/11-13-YPOS) for Chr11-13 (Figure 2B), whose only non-B6 region is the 110 region, showed significant (Fisher exact test, P-value < 0.001) protection from B6-YPOS sex reversal (Figure 4A). Adult animals homozygous for the 110 region were 100% protected from sex reversal. All embryonic gonads heterozygous or homozygous for the 110 region had some degree of testicular tissue (Figure 4B, Fisher exact test, P-value < 0.001). Thus, heterozygosity for the 110 region was sufficient to prevent the formation of complete embryonic B6-YPOS ovaries. These data show that the 110 region alone is both necessary and sufficient to direct testis determination above the spontaneous rate in the B6-YPOS strain.
Figure 4.
Genotype–phenotype correlation show that heterozygous and homozygous presence of Chr11-13, which has only the 110 region of non-B6 origin, provides significant protection from B6-YPOS sex reversal in adult and embryonic gonads. (A) B6-YPOS adult animals, none of the animals are YPOS males with testes. For adult animals that are heterozygous or homozygous for Chr11-13, there is significantly increased protection from B6-YPOS sex reversal. (B) In B6-YPOS animals, the majority of gonads in XYPOS animals are Ot (>50% ovary) or complete ovary. Embryonic gonad analysis showed that all LineB6/11-13 and Line11-13/Line11-13 gonads have some degree of testicular cord development and that none are XYPOS ovaries. *Fisher exact P-value < 0.001 for the three-way comparison.
To confirm that the 110 region was the only region responsible for the protection from B6-YPOS sex reversal, we also performed analyses for each of the subcongenic lines in both embryos and adults. Our data showed that the subcongenic lines that did not contain the 110 region (Chr11-1, 4, 6, 8, 9, and 11) did not confer significant protection from YPOS sex reversal compared to B6-YPOS animals (Figure S1). Lines heterozygous or homozygous for the 110 region were associated with a significant protection from B6-YPOS sex reversal (P < 0.001). When all subcongenic lines in which the 110 region is present (Chr11-2, 3, 5, 7, 10, 12, and 13) were grouped together, heterozygosity for the 110 region resulted in 80% (102/127) developing as adult phenotypic YPOS males, and homozygosity for the 110 region resulted in 98.9% (109/111) YPOS males. In the absence of the 110 region, only 6.9% (5/72) animals developed as XYPOS males. Embryonic gonad analysis corroborated our adult data analysis, with significantly more testis formation conferred by heterozygosity (∼60–70% of gonads were oT or T) for the 110 region compared to B6-YPOS gonads (0% were oT or T) (P-value < 0.001). However, heterozygosity for the 110 region does not result in fully normal embryonic testis, with the majority of gonads with some degree of ovarian formation (either oT or Ot) (Figure S2). Our data prove that the 110 region is required to drive testis formation in the majority of the gonad in the presence of the B6-YPOS genetic background.
Sox9 protein expression in B6-YPOS and B6-YB6
The 110 region spans 4.5 Mb and includes a well-studied sex determination gene, Sox9, and the entirety of its >2-Mb regulatory region. Sox9 belongs to the SOX gene family and, along with Sry, plays a major role in male sex determination. Sox9 is part of the initial genetic cascade that pushes the bipotential gonad toward the male pathway. A loss of either Sry or Sox9 results in XY sex reversal in humans and mice (Berta et al. 1990; Foster et al. 1994). The importance of Sox9 in sex determination led us to further explore the protein expression levels of Sox9 in B6-YB6, B6-YPOS, LineB6/11-13-YPOS, and B6-XX embryonic gonads early in sex differentiation: at 17–18, 19–20, 21–22, 23–24, and 27–30 tail somites (ts) (E11.5–E12.5). Whole-mount immunofluorescence was performed with antibodies to Sox9 to assess if protein expression was delayed or decreased in the B6-YPOS gonads and to determine whether Sox9 expression was rescued in animals that were heterozygous for Chr11-13.
Compared to the wild-type B6-YB6 gonads, B6-YPOS gonads showed decreased levels and fewer somatic cells expressing Sox9 at all time points (Figure 5). By E12.5, or 27–30 ts, when Sox9 is normally highly expressed and seminiferous tubules have begun to form in B6-YB6 gonads, expression in B6-YPOS is limited to a few scattered cells within the gonad. In contrast, a rescue of the testis molecular phenotype was observed in heterozygous LineB6/11-13-YPOS animals with a significantly greater number of cells actively expressing high levels of Sox9 throughout the length of the gonad. However, heterozygous LineB6/11-13-YPOS animals (Figure 5, row 3) were delayed in the tubule formation as compared to the wild-type B6-YB6 gonads.
Figure 5.
Timing and expression levels of the male marker Sox9 is rescued in the B6/Line11-13-YPOS across critical stages of early gonadogenesis, but female marker FoxL2 is not fully extinguished. Using fluorescence immunohistochemistry, we stained embryonic gonads from B6-YB6, B6-YPOS, LineB6/11-13-YPOS, and B6-XX animals during sex determination (E11.5–E12.5). B6-YPOS gonads showed diminished expression of Sox9 protein (green) compared to B6-YB6 at all time points assayed and had significant expression of FoxL2 (red). Both timing and qualitative expression levels of Sox9 were rescued in the heterozygous LineB6/11-13-YPOS embryonic gonads. FoxL2 expression in LineB6/11-13-YPOS gonads was strongly diminished to much lower levels than in the B6-YPOS gonads.
FoxL2 is a female somatic cell marker that is upregulated in female gonads at 27–30 ts. Studies of ovotestes have demonstrated that expression of Sox9 and FoxL2 is mutually exclusive at the cellular level and that FoxL2 expression suppresses Sox9 (Wilhelm et al. 2009). At 27–30 ts, B6-YPOS gonads had a female-similar phenotype with more FoxL2 and decreased Sox9 expression compared to both wild-type B6-YB6 and protected LineB6/11-13-YPOS animals. Probably because of the remaining minimal Sox9 expression, the number of cells expressing FoxL2 in the B6-YPOS group was still lower than in the B6-XX female control (Figure 5; compare right-most panels in first and third rows). In the protected LineB6/11-13-YPOS, female-specific FoxL2 expression is not completely suppressed, as it is in the wild-type B6-YB6. Therefore, the LineB6/11-13-YPOS gonads are incompletely masculinized. Although the temporal and masculinized expression of Sox9 matches that of wild-type B6-YB6 gonads, the female-specific marker FoxL2 was not extinguished.
These molecular phenotypes are concordant with the phenotypic findings above in which B6/11-13-YPOS gonads at later stages in development have more testicular tissue at E15.5 and a male phenotype in adulthood (Figure 4).
Narrowing down the 110 region
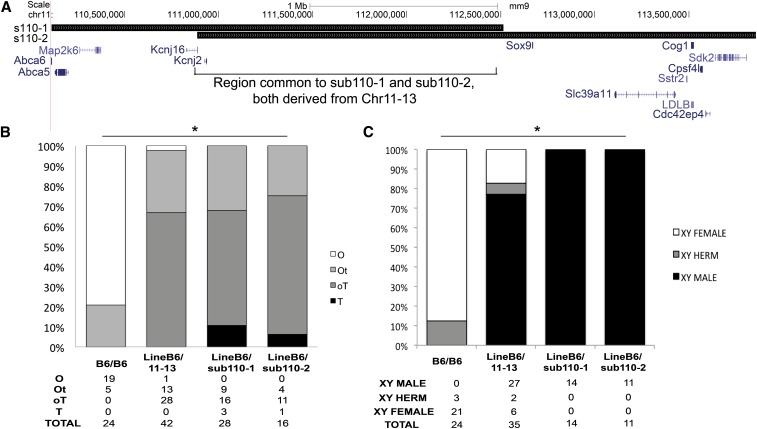
To confirm that Sox9 was solely responsible for protection from B6-YPOS sex reversal, we aimed to narrow the Chr11-13 region through backcrossing to B6-XX females. We identified two mice with recombination events within the 110 region, creating two smaller subcongenic regions. The first animal had sub110-1, which spanned base pairs 110,111,305–112,514,446 with a boundary 120 kb 5′ to the Sox9 open reading frame. The second mouse had a subcongenic region, sub110-2, which spanned base pairs 110,887,739–113,856,611 including Sox9 and several telomeric genes (Figure 6A). Sub110-1 and sub110-2 had an overlap of 1.62 Mb, which is localized entirely within the upstream Sox9 regulatory region. This overlapping region has no known open reading frames and does not include the only described testis-specific enhancer of Sox9, TESCO (Sekido and Lovell-Badge 2008). We performed embryonic analysis on both sub110-1 and sub110-2 to determine whether one or both lines was sufficient to confer protection from YPOS sex reversal. Phenotype analysis showed that both sub110-1 and sub110-2 regions independently protect from B6-YPOS sex reversal, producing both testicular tissue in embryonic gonads (Figure 6B, Fisher exact P-value < 0.001 for all comparisons to B6/B6) and fertile YPOS adult males. These data further narrow the smallest congenic region required for protection from B6-YPOS sex reversal to a minimal 1.62-Mb region encompassing the 5′ Sox9 regulatory region. Our results contribute to the evidence of additional testis enhancers regulating Sox9 expression in male sex determination.
Figure 6.
The 5′ regulatory region of Sox9 is responsible for protection from B6-YPOS sex reversal. (A) The region displayed here shows the entirety of the non-B6 region spanning megabases 110–114 on chromosome 11, and all known genes in this region are shown below in blue. Two smaller subcongenic lines, sub110-1 and sub110-2, were generated through breeding of LineB6/11-13 animals. Sub110-1 and sub110-2 overlap only over the 5′ regulatory region of Sox9 (black box). Within the overlap region, there are no known genes. (B) Embryonic gonad analysis of the heterozygous animal lines showed significant protection compared to B6/B6 animals. The levels of protection between heterozygous animals LineB6/11-13, LineB6/sub110-1, and LineB6/sub110-2 were not significantly different. *Fisher exact P-value < 0.001 for the three way comparison. (C) Adult analysis of the heterozygous subcongenic lines showed significant protection from XYPOS sex reversal compared to B6/B6-YPOS animals. The levels of protection between heterozygous animals LineB6/11-13, LineB6/sub110-1, and LineB6/sub110-2 were not significantly different. *Fisher exact P-value < 0.001 for the three-way comparison.
The protective subcongenic region is not of 129S1 origin
Inspection of the SNPs in the congenic strains as compared to the 129S1 genotyping data (Figure 3B) showed marked differences in the SNP distribution. Thus, we suspected that the origin of the protective congenic regions was not from 129S1, as originally hypothesized, but instead from the original wild-type strain from which the YPOS chromosome was derived >30 years ago (Figure 1). Further analysis comparing our strain to diversity array genotyping of 60 wild-derived strains (Yang et al. 2009, 2011) demonstrates that the protective region is most closely related to M. musculus domesticus poschiavinus (Figure S3). Therefore, based on comparison of genotyping data from our congenic strains with that of the 129 inbred strain as well as wild-derived strains, we conclude that the entire congenic region is not of 129 origin, as originally hypothesized, and that the origin of the protective genomic elements is from the wild-derived M. musculus domesticus poschiavinus (Figure 1).
Discussion
The congenic approaches that we used to identify genetic loci that protect from B6-YPOS sex reversal have highlighted the effect of interaction between the poschiavinus Y chromosome and a small, 1.62-Mb, noncoding regulatory region upstream of Sox9 on male sex determination. This study redefined and narrowed the minimal protective region from >60 to 1.62 Mb, a nearly 37-fold decrease in size. The homozygous presence of the 1.62-Mb protective region rescues both the embryonic and the adult B6-YPOS sex reversal phenotype. At the molecular level, embryonic gonads that are heterozygous for the protected region show increased protein expression of the male marker Sox9 and decreased expression of female marker FoxL2, compared to B6-YPOS gonads.
Our study also reexamines the history of this congenic strain. Previously, it was hypothesized that the protection from B6-YPOS sex reversal was conferred by genetic elements of the 129S1/SviM (129) strain. However, analysis of several of our breeding males with the SNP Diversity Array proved that the protective congenic region on chromosome 11 was not of 129 origin but rather closely related to wild-derived strains of M. m. domesticus poschiavinus genotyped by the Jackson Laboratory. Therefore, we conclude that the genetic origins of the protection likely came from the semi-inbred strain POSA (Figure 1).
Revised history of the B6-YPOS strain
In 1988, prior to the discovery of SRY as the testis-determining gene, mouse geneticist Eva Eicher postulated that, if there existed a single autosomal locus in the heterozygous state from the original POSA strain that allowed for some testicular development and fertility in B6-YPOS, it would be “kept in a forced heterozygous state to perpetuate the B6-YPOS strain” (Eicher 1988). This is exactly what we believe happened in our protected B6-YPOS strain. Twenty-five years ago, Eicher hypothesized the scenario by which the wild-derived strain was able to maintain fertility and breeding through the presence of a heterozygous protective region.
The B6-YPOS strain examined in this article diverged from existing colonies of B6-YPOS animals in the mid-1980s, shortly after the initial publication of B6-YPOS sex reversal (Eicher et al. 1982). Unlike our animals, most other colonies of B6-YPOS are maintained in the presence of a transgenic Sry (Bullejos and Koopman 2005; Correa et al. 2012). It is highly unlikely that the protective genomic element identified in our animals continues to exist in other B6-YPOS colonies because, in the presence of transgenic Sry, the selective pressure to maintain POSA-protective genomic elements would have been lost.
This minimal 1.62-Mb region located 5′ to Sox9, which we show protects from B6-YPOS sex reversal, initially came as a surprise to us as this region was not identified by our SNP scan published in Nikolova et al. (2008) This region remained unidentified because we were limited by our then-incomplete knowledge about the genetic diversity among mouse strains. By testing only SNPs polymorphic between B6 and 129, we could not reliably pick up regions that were present in other strains, such as M. m. domesticus poschiavinus, unless the SNP was, by chance, also polymorphic between B6 and M. m. domesticus poschiavinus. Similarly, previous linkage studies focused on identifying genomic regions important for promoting sex reversal were limited by genotyping of only SNPs polymorphic between B6 and DBA2 (Eicher et al. 1996). In both studies, strain-specific SNPs were limiting as they did not account for the above hypothesis that a surreptitious heterozygous autosomal genomic element derived from the original POSA strain may be responsible for protection from the YPOS sex-reversal phenotype.
Improved genetic technologies have allowed for deeper genomic analyses of the ancestral diversity of current laboratory strains and paved the way for our greater genetic understanding of the congenic B6.POSA-YPOS strain. Throughout the 30-year breeding history of these congenic mice, the assumption that one had achieved a pure consomic B6-YPOS was based on statistical evidence, which holds true unless, such as in our case, there is a strong selective pressure to maintain specific genomic elements for fertility and breeding (Silver 1995). Reanalysis of our animals and close examination of the background history of the B6-YPOS strain identified POSA-specific genomic elements required for B6-YPOS male fertility. This region likely was important for propagation of the B6-YPOS strain prior to the introduction of transgenic Sry into other existing colonies of B6-YPOS. This region confers significant protection from B6-YPOS sex reversal, but heterozygosity for the protective region is necessary but not sufficient to confer full protection from B6-YPOS sex reversal. We do observe a dose-dependent protection, and homozygosity is sufficient for protection from B6-YPOS sex reversal.
The protective effect of the POSA Sox9 promoter on B6-YPOS sex reversal
The mechanisms underlying the protective effect of the POSA Sox9 promoter region remain unclear. The entire mouse Sox9 regulatory region spans nearly 2.4 Mb, both centromeric and telomeric of the Sox9 open reading frame. Sox9 is involved in development not only of the gonad (Sekido and Lovell-Badge 2008), but also of the neuroectoderm, brain, gut (Bagheri-Fam et al. 2006), skeletal (Bi et al. 1999), and craniofacial tissues (Benko et al. 2011). As such, Sox9 has many tissue- and temporal-specific promoters that regulate its expression. Point mutations, deletions, and duplications limited to the Sox9 regulatory regions can result in abnormal expression of Sox9, resulting in a variety of phenotypes, ranging from XX males (Bishop et al. 2000; Cox et al. 2011) to isolated craniofacial dysmorphism with Pierre Robin Sequence (Benko et al. 2009) to skeletal anomalies (Foster et al. 1994).
Developmentally important genes, such as SHH, FOXL2, and SOX9, are known to have long-range enhancers that function from >1 Mb away and from introns of neighboring genes (Beysen et al. 2005; Gurnett et al. 2007; Benko et al. 2009). Further studies will be required to precisely identify enhancers of Sox9 and how the small region that we have identified here results in protection from B6-YPOS sex reversal. TESCO, the only testis-specific enhancer identified in mouse (Sekido and Lovell-Badge 2008), does not cause any known cases of human 46,XY gonadal dysgenesis. Therefore, other testis-specific enhancers of Sox9 may work in synergy with the TESCO enhancer to promote continued Sox9 expression within the developing testis. Attempts to narrow down the location for a second testis-specific SOX9 enhancer from human data with small deletions in the SOX9 regulatory region have identified a minimal overlap of a 67-kb noncoding region 584 kb upstream of SOX9 (Benko et al. 2011; Xiao et al. 2013). Both the 67-kb region and the entirety of the SOX9 regulatory region are active lines of investigation within the field of sex determination. Our narrowed minimal protective region encompasses the mouse region syntenic to the smallest 67-kb region identified in human studies, but not TESCO.
Using a predictive transcription-factor-binding algorithm (Cartharius et al. 2005), we have identified putative binding sites for Nr5a1/Sf-1, Sry, and Sox9 transcription factors within the 67-kb minimal region and its flanking genomic region (Figure S4). Potential Sry- and Sf1-binding sites appear to be well represented within this region (approximately one binding site per 2 kb), while Sox9 transcription-factor-binding sites are more sparse (approximately one binding site per 7.5 kb). Although several sites fall into regions of the genome that are under selective constraint (Davydov et al. 2010), further interpretation of these data would require evidence to show that any of these transcription-factor-binding sites play a role in human sex development or are causative in our B6-YPOS protective model as many mechanisms outside of transcription factor binding work within the noncoding genome to regulate transcription, including long noncoding RNAs, methylation, and chromatin conformation, to name a few.
Our SNP array studies show that the congenic strain’s 110 region is very similar, but not identical, to M. m. domesicus poschiavinus. Therefore, further studies to characterize the base-pair sequence, CNV, presence of epigenetic marks, and noncoding RNA transcripts may shed light on the complex regulation of this developmentally important region.
Following Sry up-regulation, Sox9 up-regulation in the somatic cells of the bipotential gonad is one of the initial and critical steps in testis sex determination. Previous studies have demonstrated that, within B6-YPOS gonads, Sry messenger RNA expression is delayed beyond the critical window to initiate testis sex determination (Bullejos and Koopman 2005; Hiramatsu et al. 2009). While that might be the primary cause of B6-YPOS sex reversal, the protection conferred by the 110 region rescues the sex reversal phenotype farther downstream, through up-regulating Sox9 expression despite decreased Sry expression. High levels of Sox9 have been shown in both humans and mice to be both necessary and sufficient for testis formation in both XX and XY individuals (Huang et al. 1999; Vidal et al. 2001).
What is remarkable is the strength of the interaction between YPOS and the concomitant POSA regulatory region of Sox9. Only co-inheritance of the SryPOS and the POSA regulatory region of Sox9 allowed for male sex determination, fertility, and breeding. Inheritance of YPOS but not the POSA-protective elements results in line extinction through sex reversal and infertility of the male mice. Fertility in XX mice is unaffected by this genomic element. Only B6-YPOS mice possessing this region were capable of reproducing, and thus we hypothesize that YPOS and promoter regions co-evolved with YPOS depending on or requiring the POSA promoter of Sox9 to perpetuate the testis-specific gene network within the spermatic cords of an XYPOS mouse.
Our findings highlight the complex network of genes that can work to promote the key pathways within male sex determination. Mice harboring YPOS require the POSA region to maintain the male-specific network equilibrium and ensure the up-regulation of Sox9, testis development, and fertility. YPOS in the presence of the B6 genetic background shifts the balance toward female sex determination, and our findings indicate that this is due to decreased activation of Sox9. The co-evolution of the POSA Sox9 promoter occurred because the presence of both these elements (YPOS and POSA promoter) maintained male fertility; hence, it is likely that SryPOS, in addition to other factors, is specifically geared to activate the POSA promoter of Sox9. While Sry from other mouse strains may be able to activate POSA, if the genetic background is not inherently “pro-female” as the B6 strain is, the presence of the POSA promoter may simply be a redundant mechanism pushing the network equilibrium further toward testis development and providing a buffer for minor genetic alleles that promote ovary formation. Unraveling the nature of these interactions will require evaluation of the entire Sox9 promoter region to identify critical regions and to dissect how SNPs, DNAse hypersensitivity sites, and transcription-factor-binding sites specifically interact with SryPOS and the entire gene network within the developing gonad.
Finally, these types of studies may provide a glimpse into the genetic heterogeneity that we may expect to find in human cases of disorders of sex development. To date, it is estimated that only 30–40% of all cases of 46,XY gonadal dysgenesis can be explained by single gene mutations, deletions, or duplications. The remaining 60% may involve more complex inheritance of multiple alleles required to promote sex determination. Thus, in the absence of one gene, the entire sex determination program may be delayed or, if the effect of the gene is large, may result in sex reversal. Early studies in humans have began to unravel these coexisting mutations and modifiers that influence early developmental processes (Sykiotis et al. 2010) and the intricate web in which cell-intrinsic processes as well as the external environment must come together in a delicate balance to promote proper sex determination.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants HD044513 (to E.V.) and F31HD068136 and by University of California at Los Angeles institutional funds (to V.A.).
Footnotes
Communicating editor: D. Charlesworth
Literature Cited
- Albrecht K. H., Eicher E. M., 1997. DNA sequence analysis of Sry alleles (subgenus Mus) implicates misregulation as the cause of C57BL/6J-Y(POS) sex reversal and defines the SRY functional unit. Genetics 147: 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri-Fam S., Barrionuevo F., Dohrmann U., Gunther T., Schule R., et al. , 2006. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev. Biol. 291: 382–397. [DOI] [PubMed] [Google Scholar]
- Benko S., Fantes J. A., Amiel J., Kleinjan D. J., Thomas S., et al. , 2009. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat. Genet. 41: 359–364. [DOI] [PubMed] [Google Scholar]
- Benko S., Gordon C. T., Mallet D., Sreenivasan R., Thauvin-Robinet C., et al. , 2011. Disruption of a long distance regulatory region upstream of SOX9 in isolated disorders of sex development. J. Med. Genet. 48: 825–830. [DOI] [PubMed] [Google Scholar]
- Berkovitz G. D., Fechner P. Y., Marcantonio S. M., Bland G., Stetten G., et al. , 1992. The role of the sex-determining region of the Y chromosome (SRY) in the etiology of 46,XX true hermaphroditism. Hum. Genet. 88: 411–416. [DOI] [PubMed] [Google Scholar]
- Berta P., Hawkins J. R., Sinclair A. H., Taylor A., Griffiths B. L., et al. , 1990. Genetic evidence equating SRY and the testis-determining factor. Nature 348: 448–450. [DOI] [PubMed] [Google Scholar]
- Beysen D., Raes J., Leroy B. P., Lucassen A., Yates J. R., et al. , 2005. Deletions involving long-range conserved nongenic sequences upstream and downstream of FOXL2 as a novel disease-causing mechanism in blepharophimosis syndrome. Am. J. Hum. Genet. 77: 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W., Deng J. M., Zhang Z., Behringer R. R., de Crombrugghe B., 1999. Sox9 is required for cartilage formation. Nat. Genet. 22: 85–89. [DOI] [PubMed] [Google Scholar]
- Bishop C. E., Whitworth D. J., Qin Y., Agoulnik A. I., Agoulnik I. U., et al. , 2000. A transgenic insertion upstream of sox9 is associated with dominant XX sex reversal in the mouse. Nat. Genet. 26: 490–494. [DOI] [PubMed] [Google Scholar]
- Bogani D., Siggers P., Brixey R., Warr N., Beddow S., et al. , 2009. Loss of mitogen-activated protein kinase kinase kinase 4 (MAP3K4) reveals a requirement for MAPK signalling in mouse sex determination. PLoS Biol. 7: e1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullejos M., Koopman P., 2001. Spatially dynamic expression of Sry in mouse genital ridges. Dev. Dyn. 221: 201–205. [DOI] [PubMed] [Google Scholar]
- Bullejos M., Koopman P., 2005. Delayed Sry and Sox9 expression in developing mouse gonads underlies B6-Y(DOM) sex reversal. Dev. Biol. 278: 473–481. [DOI] [PubMed] [Google Scholar]
- Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., et al. , 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942. [DOI] [PubMed] [Google Scholar]
- Correa S. M., Washburn L. L., Kahlon R. S., Musson M. C., Bouma G. J., et al. , 2012. Sex reversal in C57BL/6J XY mice caused by increased expression of ovarian genes and insufficient activation of the testis determining pathway. PLoS Genet. 8: e1002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. J., Willatt L., Homfray T., Woods C. G., 2011. A SOX9 duplication and familial 46,XX developmental testicular disorder. N. Engl. J. Med. 364: 91–93. [DOI] [PubMed] [Google Scholar]
- Davydov E. V., Goode D. L., Sirota M., Cooper G. M., Sidow A., et al. , 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLOS Comput. Biol. 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher E. M., 1988. Autosomal genes involved in mammalian primary sex determination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 322: 109–118. [DOI] [PubMed] [Google Scholar]
- Eicher E. M., Washburn L. L., Whitney J. B., III, Morrow K. E., 1982. Mus poschiavinus Y chromosome in the C57BL/6J murine genome causes sex reversal. Science 217: 535–537. [DOI] [PubMed] [Google Scholar]
- Eicher E. M., Washburn L. L., Schork N. J., Lee B. K., Shown E. P., et al. , 1996. Sex-determining genes on mouse autosomes identified by linkage analysis of C57BL/6J-YPOS sex reversal. Nat. Genet. 14: 206–209. [DOI] [PubMed] [Google Scholar]
- Fleming A., Ghahramani N., Zhu M. X., Delot E. C., Vilain E., 2012. Membrane beta-catenin and adherens junctions in early gonadal patterning. Dev. Dyn. 241: 1782–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W., Dominguez-Steglich M. A., Guioli S., Kwok C., Weller P. A., et al. , 1994. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372: 525–530. [DOI] [PubMed] [Google Scholar]
- Georg I., Bagheri-Fam S., Knower K. C., Wieacker P., Scherer G., et al. , 2010. Mutations of the SRY-responsive enhancer of SOX9 are uncommon in XY gonadal dysgenesis. Sex. Dev. 4: 321–325. [DOI] [PubMed] [Google Scholar]
- Gierl M. S., Gruhn W. H., von Seggern A., Maltry N., Niehrs C., 2012. GADD45G functions in male sex determination by promoting p38 signaling and Sry expression. Dev. Cell 23: 1032–1042. [DOI] [PubMed] [Google Scholar]
- Gurnett C. A., Bowcock A. M., Dietz F. R., Morcuende J. A., Murray J. C., et al. , 2007. Two novel point mutations in the long-range SHH enhancer in three families with triphalangeal thumb and preaxial polydactyly. Am. J. Med. Genet. A 143: 27–32. [DOI] [PubMed] [Google Scholar]
- Hacker A., Capel B., Goodfellow P., Lovell-Badge R., 1995. Expression of Sry, the mouse sex determining gene. Development 121: 1603–1614. [DOI] [PubMed] [Google Scholar]
- Hiramatsu R., Matoba S., Kanai-Azuma M., Tsunekawa N., Katoh-Fukui Y., et al. , 2009. A critical time window of Sry action in gonadal sex determination in mice. Development 136: 129–138. [DOI] [PubMed] [Google Scholar]
- Huang B., Wang S., Ning Y., Lamb A. N., Bartley J., 1999. Autosomal XX sex reversal caused by duplication of SOX9. Am. J. Med. Genet. 87: 349–353. [DOI] [PubMed] [Google Scholar]
- Kashimada K., Koopman P., 2010. Sry: the master switch in mammalian sex determination. Development 137: 3921–3930. [DOI] [PubMed] [Google Scholar]
- Katoh-Fukui Y., Tsuchiya R., Shiroishi T., Nakahara Y., Hashimoto N., et al. , 1998. Male-to-female sex reversal in M33 mutant mice. Nature 393: 688–692. [DOI] [PubMed] [Google Scholar]
- Katoh-Fukui Y., Miyabayashi K., Komatsu T., Owaki A., Baba T., et al. , 2012. Cbx2, a polycomb group gene, is required for Sry gene expression in mice. Endocrinology 153: 913–924. [DOI] [PubMed] [Google Scholar]
- Koopman P., Gubbay J., Vivian N., Goodfellow P., Lovell-Badge R., 1991. Male development of chromosomally female mice transgenic for Sry. Nature 351: 117–121. [DOI] [PubMed] [Google Scholar]
- Kuroki S., Matoba S., Akiyoshi M., Matsumura Y., Miyachi H., et al. , 2013. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science 341: 1106–1109. [DOI] [PubMed] [Google Scholar]
- Lee P. A., Houk C. P., Ahmed S. F., Hughes I. A., 2006. Consensus statement on management of intersex disorders. Pediatrics 118: e488–e500. [DOI] [PubMed] [Google Scholar]
- Maier E. M., Leitner C., Lohrs U., Kuhnle U., 2003. True hermaphroditism in an XY individual due to a familial point mutation of the SRY gene. J. Pediatr. Endocrinol. Metab. 16: 575–580. [DOI] [PubMed] [Google Scholar]
- Munger S. C., Aylor D. L., Syed H. A., Magwene P. M., Threadgill D. W., et al. , 2009. Elucidation of the transcription network governing mammalian sex determination by exploiting strain-specific susceptibility to sex reversal. Genes Dev. 23: 2521–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger S. C., Natarajan A., Looger L. L., Ohler U., Capel B., 2013. Fine time course expression analysis identifies cascades of activation and repression and maps a putative regulator of mammalian sex determination. PLoS Genet. 9: e1003630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova G., Sinsheimer J. S., Eicher E. M., Vilain E., 2008. The chromosome 11 region from strain 129 provides protection from sex reversal in XYPOS mice. Genetics 179: 419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bryan M. K., Takada S., Kennedy C. L., Scott G., Harada S., et al. , 2008. Sox8 is a critical regulator of adult Sertoli cell function and male fertility. Dev. Biol. 316: 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer D., Kist R., Dewar K., Devon K., Lander E. S., et al. , 1999. Campomelic dysplasia translocation breakpoints are scattered over 1 Mb proximal to SOX9: evidence for an extended control region. Am. J. Hum. Genet. 65: 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanco J. C., Wilhelm D., Davidson T. L., Knight D., Koopman P., 2010. Sox10 gain-of-function causes XX sex reversal in mice: implications for human 22q-linked disorders of sex development. Hum. Mol. Genet. 19: 506–516. [DOI] [PubMed] [Google Scholar]
- Sekido R., Lovell-Badge R., 2008. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453: 930–934. [DOI] [PubMed] [Google Scholar]
- Silver, L. M., 1995 Mouse Genetics: Concepts and Applications. Oxford University Press, Oxford. [Google Scholar]
- Sinclair A. H., Berta P., Palmer M. S., Hawkins J. R., Griffiths B. L., et al. , 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346: 240–244. [DOI] [PubMed] [Google Scholar]
- Sutton E., Hughes J., White S., Sekido R., Tan J., et al. , 2011. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J Clin Invest. 121: 328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis G. P., Plummer L., Hughes V. A., Au M., Durrani S., et al. , 2010. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc. Natl. Acad. Sci. USA 107: 15140–15144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiler K., 1989. The House Mouse: Atlas of Embryonic Development. Springer, New York. [Google Scholar]
- Vidal V. P., Chaboissier M. C., de Rooij D. G., Schedl A., 2001. Sox9 induces testis development in XX transgenic mice. Nat. Genet. 28: 216–217. [DOI] [PubMed] [Google Scholar]
- Wagner T., Wirth J., Meyer J., Zabel B., Held M., et al. , 1994. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79: 1111–1120. [DOI] [PubMed] [Google Scholar]
- Warr N., Carre G. A., Siggers P., Faleato J. V., Brixey R., et al. , 2012. Gadd45gamma and Map3k4 interactions regulate mouse testis determination via p38 MAPK-mediated control of Sry expression. Dev. Cell 23: 1020–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S., Ohnesorg T., Notini A., Roeszler K., Hewitt J., et al. , 2011. Copy number variation in patients with disorders of sex development due to 46,XY gonadal dysgenesis. PLoS ONE 6: e17793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney J. B., Mills T. M., Lewis R. W., Wartell R., Abney T. O., 2000. A single genetic determinant that prevents sex reversal in C57BL-YPOS congenic mice. Biochem. Genet. 38: 119–137. [DOI] [PubMed] [Google Scholar]
- Wilhelm D., Washburn L. L., Truong V., Fellous M., Eicher E. M., et al. , 2009. Antagonism of the testis- and ovary-determining pathways during ovotestis development in mice. Mech. Dev. 126: 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E., Hargrave M. R., Christiansen J., Cooper L., Kun J., et al. , 1995. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat. Genet. 9: 15–20. [DOI] [PubMed] [Google Scholar]
- Xiao B., Ji X., Xing Y., Chen Y. W., Tao J., 2013. A rare case of 46, XX SRY-negative male with approximately 74-kb duplication in a region upstream of SOX9. Eur. J. Med. Genet. 56: 695–698. [DOI] [PubMed] [Google Scholar]
- Yang H., Ding Y., Hutchins L. N., Szatkiewicz J., Bell T. A., et al. , 2009. A customized and versatile high-density genotyping array for the mouse. Nat. Methods 6: 663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A., et al. , 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.