Abstract
Down syndrome (DS) is due to increased copy number of human chromosome 21. The contribution of different genetic regions has been tested using mouse models. As shown previously, the Abcg1-U2af1 genetic region contributes to cognitive defects in working and short-term recognition memory in Down syndrome mouse models. Here we analyzed the impact of monosomy of the same genetic interval, using a new mouse model, named Ms2Yah. We used several cognitive paradigms and did not detect defects in the object recognition or the Morris water maze tests. However, surprisingly, Ms2Yah mice displayed increased associative memory in a pure contextual fear-conditioning test and decreased social novelty interaction along with a larger long-term potentiation recorded in the CA1 area following stimulation of Schaffer collaterals. Whole-genome expression studies carried out on hippocampus showed that the transcription of only a small number of genes is affected, mainly from the genetic interval (Cbs, Rsph1, Wdr4), with a few additional ones, including the postsynaptic Gabrr2, Gabbr1, Grid2p, Park2, and Dlg1 and the components of the Ubiquitin-mediated proteolysis (Anapc1, Rnf7, Huwe1, Park2). The Abcg1–U2af1 region is undeniably encompassing dosage-sensitive genes or elements whose change in copy number directly affects learning and memory, synaptic function, and autistic related behavior.
Keywords: aneuploidy, genetic dosage, mouse model, intellectual disabilities, trisomy 21
ANEUPLOIDY, defined as an abnormal number of copies of a genomic region, is a common cause of human genetic disorders associated with intellectual disabilities and autism (Mefford et al. 2012). Down syndrome (DS) is a chromosomal aneuploidy characterized by the presence of a supernumerary copy of the whole or part of human chromosome 21 (Hsa21) that gives rise to a syndromic intellectual disability. Although classically research has focused on the search of candidate genes, exciting new findings demonstrate that in addition to direct and indirect alteration of expression of Hsa21 and non-Hsa21 genes, alteration in copy number of functional, nontraditional genomic elements may affect the variability of the DS phenotype (Letourneau et al. 2014).
Several “dosage sensitive” regions, including genes and noncoding conserved elements, have been mapped across the length of Hsa21 and shown to be sufficient for induction of the complete phenotype of DS (Korbel et al. 2009; Lyle et al. 2009). The results of such studies provide initial evidence supporting that some of the phenotypes of DS are not due to specific genes, but rather to the over/underexpression of a whole chromosomal domain or even to other long noncoding RNA (Umlauf et al. 2008). Until recently, the mouse models available have not explored the contribution of the gene-rich telomeric part of Hsa21 that maps to the mouse chromosomes Mmu17, Mmu16, and Mmu10 and do not completely explain the complexity and variability of some phenotypes observed in patients with DS (Reeves et al. 1995; Yu et al. 2010b; Duchon et al. 2011). We previously demonstrated the contribution of trisomy of the Abcg1–U2af1 genetic interval from the telomeric part of Hsa21 to DS, in a mouse model named Ts1Yah (Lopes Pereira et al. 2009). Ts1Yah mice showed defects in novel object recognition, open-field, and Y-maze tests, similar to other DS models, but the trisomy induced an improvement of the hippocampal-dependent spatial memory in the Morris water maze along with enhanced and longer-lasting long-term potentiation in vivo in the hippocampus. Although DS induces several deleterious effects, Hsa21 also carries protective genes or regions, which may explain some “advantageous” phenotypes (Sussan et al. 2008).
To further analyze the dosage effect of this particular region, we generated the corresponding monosomy (Ms2Yah) model to the Ts1Yah model and we characterized various functional domains, using behavioral and electrophysiological studies (Lopes Pereira et al. 2009). We used several cognitive paradigms, non-hippocampal-dependent [object recognition, conditioned stimulus (CS) freezing] and hippocampal-dependent (water maze, context freezing) tests, with locomoter (rotarod) and social tests to evaluate defects observed in people with DS. We expected to find an opposite phenotype to that observed in the Ts1Yah model, but we could not detect alterations in the performance in either the object recognition or the Morris water maze test. However, surprisingly, monosomy of the Abcg1–U2af1 genetic interval led to increased associative memory in a pure contextual fear-conditioning test and altered social interaction along with a significant increase in the long-term potentiation in vivo. To understand the possible underlying mechanisms in the hippocampus, we used a transcriptome approach and we observed a very small effect with the decreased expression of genes associated with the Abcg1–U2af1 monosomy that highlights the contribution of genes from the interval to the learning and memory phenotypes. These changes were detected in the absence of abnormalities in the size or density of pyramidal neurons.
Materials and Methods
Mouse line and ethical statement
The Ms2Yah mouse line carried a deletion of the region located downstream of Abcg1 and upstream of U2af1 and was bred with C57BL/6J for >10 generations (Lopes Pereira et al. 2009). Mice were handled with the agreement of the local ethical committee and in accordance with the European Council Guidelines for the Care and Use of Laboratory animals (accreditation 7320). They were housed under a 12-hr/12-hr light/dark cycle in the Institut Clinique de la Souris mouse facility in Illkirch (certificate C45-234-6) and shipped by authorized carriers to Barcelona for the behavioral assessment or to Seville for the in vivo LTP experiments. Mice were housed in standard cages with access to food and water ad libitum, under a controlled environment (temperature = 20° ± 1°; humidity = 60%), and with a light/dark cycle of 12 hr (8:00 am to 8:00 pm). All experimental protocols were performed under authorization [Ethical Committee of Animal Experimentation (CEEA) of the Parc de Recerca Biomédica de Barcelona (PRBB); MDS-08-1060P1 and JMC-07-1001P1-MDS] and met the guidelines of the local (law 32/2007) and European regulations [European Union (EU) directive 86/609, EU decree 2001-486] and Statement of Compliance with Standards for Use of Laboratory Animals by Foreign Institutions no. A5388-01, National Institutes of Health. All investigators involved in animal experimentations have the appropriate training (Department de Medi Ambient i Habitatge, 214/1997). The Centre for Genomic Regulation is authorized to work with genetically modified organisms (A/ES/05/I-13 and A/ES/05/14). All the behavioral testing was conducted in an isolated room by the same experimenter at the same time of the day (between 8:30 am and 3:30 pm). The experimenter was blinded to the genetic status of the animals. Yann Herault, the principal investigator in this study, was granted the accreditations 45-31 and 67-369 to perform the reported experiments with collaborators Mara Dierssen and José Maria Delgado-Garcia.
Mouse genotyping
Genomic DNA was isolated from tail biopsies and the Ms2Yah allele was identified by a 3′ PCR, using one common forward (Fwd) primer (5′-CCAGCTGAAGATGGGTGTGTCTGC-3′), one reverse (Rev) primer (5′-AGCCTTCCCTGGGGACCTGAAA-3′) specific for the wild-type (WT) allele, and a Ms2Yah Rev (5′-AACGACCGAGCGCAGCGA-3′) primer specific for the mutant. PCR reactions gave WT and Ms2Yah products of length 468 and 272 bp, respectively.
Behavioral analysis
All the experimental procedures for motor and cognitive assessments have been previously described (Besson et al. 2007; Lopes Pereira et al. 2009; Duchon et al. 2011). Fourteen WT and 13 Ms2Yah male littermates were obtained from a monosomic male crossed with wild-type females. They were 3 months old when they entered the test battery with the following order: 1, rotarod; 2, treadmill; 3, novel object recognition; 4, Morris water maze; and 5, fear conditioning; with 1 week between tests.
Briefly, motor performance was tested on the rotarod and treadmill tests, to evaluate motor coordination and learning. The experimental design consisted of two training trials on a rotarod (LE8500; Panlab-Harvard Apparatus, Barcelona, Spain) at 4 rpm followed by consecutive 1-min trials at increasing fixed rotating speed (4, 10, 14, 19, 24, and 34 rpm; intertrial period 5 min) and one accelerating rod trial (4–40 rpm over 40 sec). For each trial, the elapsed time until the mouse fell off the rod was recorded. The treadmill (LE8708; Panlab-Harvard Apparatus) consisted of a belt (50 cm long and 20 cm wide) with adjustable speed (5–150 cm/sec) and slope (0°–45°) and an electrified grid delivering a foot shock (0.6 mA) whenever the mice fell off the belt. Mice received eight 1-min trials on a single-day session: two training trials (5 cm/sec at 0°) and six test trials (5, 10, 20, 30, 40, and 50 cm/sec at 20°). Number and duration of shocks were quantified.
In the novel object recognition task, a 10-min habituation session was performed in an open field (OF) (70 × 70 × 40 cm) (TBS, Barcelona, Spain) with two multicolored interlocking plastic bricks (Tente; EXIN-LINES BROS, Barcelona, Spain) with the same preference levels. The familiar one was a simple colored column and the unfamiliar one was a colored abstract shape construction. Twenty-four hours later, a familiarization trial where mice were presented with a pair of identical objects for a maximum of 10 min was performed. Any investigative behavior (head orientation or sniffing) or deliberate contact with each object in a distance ≤2 cm is considered as exploration of the objects. Animals not exploring the objects for at least 20 sec were discarded. After 1 hr, one of the familiar objects was changed to a new one, and the animals explored the OF for 5 min. Long-term memory was operationally defined by the discrimination index (DI): time spent investigating the novel object minus the familiar one [DI = (novel object exploration time/total exploration time) – (familiar object exploration time/total exploration time) × 100].
Animals were also tested in a visuo-spatial learning paradigm in the Morris water maze (MWM) test over 9 days (four trials per session, 10-min intertrial intervals). The water maze consisted of a circular pool (diameter, 1.50 m; height, 0.35 m; TBS) filled with tepid water (24°) opacified with nontoxic white ink. A white escape platform (8 cm diameter, height 24 cm) was located 1 cm below the water surface in a fixed position (northeast quadrant, 25 cm away from the wall). The maze was surrounded by white curtains with three black patterns affixed to provide spatial cues. In a pretraining session the platform was visible in the center of the pool (day 1). In the five acquisition sessions (days 2–6), mice were introduced in the pool through different locations (north, south, east, and west) and were allowed to swim until they located the platform. Mice failing to find the platform within 60 sec were placed on it for 20 sec. A probe trial (day 7) in which the platform was removed was followed by a cued session (day 8) where the platform position was visible. Escape latencies, length of the swimming paths, and swimming speed for each animal and trial were monitored and computed by a software tracking system, SMART (Panlab-Harvard Apparatus), connected to a video camera placed above the pool, and Jtrack, an in-house designed software (Arqué et al. 2008).
Finally, in the pure contextual fear-conditioning paradigm, each mouse was placed in a test chamber (67 × 53 × 55 cm) inside a sound-attenuated compartment (Panlab-Harvard Apparatus) with a weight transducer, two audio generators, an electrified grid, and an incandescent light of 40 W. In the conditioning session mice were allowed to explore the chamber freely for 2 min. Thereafter, an 8-kHz 100-dB pure tone, which served as the CS, was presented for 30 sec, followed by a mild foot shock (2 sec, 0.2 mA), which served as the unconditioned stimulus (US) (training phase). Fear testing was conducted 24 hr after conditioning during 2 min of free exploration and 3 min of exposure to the CS (test phase). Freezing time (seconds) was recorded and analyzed. The extinction was further evaluated by scoring freezing time in 2-min sessions over a 6-min period.
Social preference and discrimination were evaluated using a specific test with three successive and identical chambers (Stoelting, Dublin). The protocol is similar to the one described by Moy et al. (2004, 2008) and was described previously (Duchon et al. 2011) In the habituation, tested mice were allowed to explore the three chambers freely for 10 min starting from the intermediate compartment and with the two side and opposite chambers having an empty wire cage. In the second phase, the test mouse was placed in the central box, while an unfamiliar mouse (stranger 1) was now put in one of the wire cages in a random and balanced manner. The doors were reopened and the test mouse was allowed to explore the three chambers for 10 min. Time spent in each of the chambers, the number of entries into each chamber, and the time spent sniffing each wire cage were recorded for social preference. In a third phase, the social discrimination was evaluated with a new stranger mouse (stranger 2) placed into the empty wire cage and the test mouse was allowed to explore again the entire arena for 6 min, having the choice between the first, already-investigated mouse (stranger 1) and the novel unfamiliar mouse (stranger 2). The same measures were taken as for the social preference. The olfactory tests were described previously (Duchon et al. 2011).
Electrophysiology
Another experiment in which we found interesting results in Ts1Yah was the activity of hippocampal synapses; change was obviously expected in Ms2Yah correlatively with Abcg1–U2af1 dosage diminution. Six-month-old control and mutant males (n = 10 per group) were anesthetized with 0.8–3% halothane at a flow rate of 1 liter/min oxygen. Animals were implanted with bipolar stimulating electrodes aimed at the right Schaffer collateral–commissural pathway of the dorsal hippocampus [2 mm lateral and 1.5 mm posterior to bregma; depth from brain surface, 1.0–1.5 mm (Paxinos and Franklin 2001)] and with two recording electrodes aimed at the CA1 area (1.2 mm lateral and 2.2 mm posterior to bregma; depth from brain surface, 1.0–1.5 mm). The final position of hippocampal electrodes was determined using the field potential depth profile evoked by paired (40-msec interval) pulses presented at the Schaffer collateral pathway as a guide. A bare silver wire (0.1 mm) was affixed to the skull as a ground. The four wires were connected to a four-pin socket and the socket was fixed to the skull with the help of two small screws and dental cement (see Gruart et al. 2006 for details of this chronic preparation).
Recording and stimulation procedures
Field excitatory postsynaptic potentials (fEPSPs) were recorded with Grass P511 differential amplifiers. Electrical stimulus presented to Schaffer collaterals consisted of 100-µsec, square, biphasic pulses presented alone, paired, or in trains. Stimulus intensities ranged from 0.02 to 500 µA for the construction of the input/output curves. For paired pulse facilitation, the stimulus intensity was set well below the threshold for evoking a population spike, usually 35% of the intensity necessary for evoking a maximum fEPSP response (Gureviciene et al. 2004). Paired pulses were presented at six (10, 20, 40, 100, 200, and 500) different pulse intervals. The stimulus intensity was also set at 35% of its asymptotic value for long-term potentiation (LTP) induction. For LTP induction, each animal was presented with a high-frequency stimulation (HFS) protocol consisting of five trains (200 Hz, 100 msec) of pulses at a rate of 1/sec. This protocol was presented six times in total, at intervals of 1 min. The 100-µsec, square, biphasic pulses used to evoke LTP were applied at the same intensity used for the single pulse presented following CS presentation. Further details of this chronic preparation can be found in previous work (Gruart et al. 2006).
Histological procedures and cresyl violet/Luxol blue staining
Animals were anesthetized with pentobarbital (25 mg/kg) and intracardially perfused with sterile PBS (0.01 M) followed by 4% paraformaldehyde (PFA). Then brains were dissected and kept in 4% PFA solution at 4° (overnight). Brains were dehydrated and embedded in paraffin and 5- to 7-µm coronal sections were processed using microtome (Leica). Sections were selected between bregma + 2.00 and + 2.50 mm. Deparaffinized sections were colored using the classical procedure for Luxol blue and cresyl violet. Slides were dried overnight at room temperature and then mounted with pertex. The sections were imaged in brightfield, using the NanoZoomer Digital Pathology-2.0 HT (C9600; Hamamatsu Photonics, Hamamatsu, Japan). The CA1, CA2–3, and dentate gyrus areas were estimated using the NDP.view software. On the same sections, relative neuronal density was estimated by counting the cresyl violet-positive cells in three randomized fields at magnification ×40 for each hippocampal area. For each individual, five sections were used and the mean was calculated with five animals per genotype (WT and Ms2Yah).
Expression profiling analysis
Hippocampi were isolated from five male mice at 3 months of age and flash frozen. Total RNA was extracted using RNeasy mini kit extraction (QIAGEN, Valencia, CA) according to the manufacturer’s instructions. The quality of all RNA samples was controlled using an Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA was converted to complementary DNA (cDNA), using Superscript II (Invitrogen, Carlsbad, CA). Hybridization was carried out on Affymetrix Mouse Gene 1.0 ST. Raw data of fluorescence were then normalized with the robust multiarray average (RMA) algorithm developed by Irizarry et al. (2003) and we obtained 35,557 probe RMA values. We fixed the background with RMA signals >4.36 and selected 24,697 probes. Values were log transformed and finally we calculated the fold change between the two groups for analysis. Hierarchical clustering was carried out with Cluster 3.0 software, using Euclidian distances to calculate the distances between the genes and between the samples. Functional annotation was performed using the mouse genome database (Blake et al. 2014) The microarray data were submitted to the Array Express Home under accession no. E-MTAB-988. We applied selection filters on the raw data provided by the RMA algorithm to extract relevant differences between our two groups. The final data file retained recapitulates all the genes that had a statistical difference (t-test, P < 0.05) between WT and Ms2Yah, without unknown transcript (no mRNA accession number) and multiple probes for a single gene. We estimated the false positive rate, with n1 = α × N(total genes selected) and n2 = number of values < α; α is the threshold P-value used in Student’s t-test. We applied this to the first batch of probes after the background threshold. For α = 0.05, we had n1 = 80 and n2 = 171, so the allowed error is n1/n2 = 0.46.
For the quantitative RT-PCR experiments we considered 14 genes mapped on Mmu17, between the Abcg1 and U2af1 genes (mm9 database, ENSEMBL); assays for the Umodl1, Abcg1, U2af1, and Cryaa genes were also designed to check the expression of genes located at the borders of the interval. In addition, we included assays for 17 genes located outside the region of interest. Among these, 3 genes for each tissue were selected for normalization and the remaining genes were used as additional controls. Selection of normalization genes was performed with GeNorm software (Vandesompele et al. 2002). To assess the differences in gene expression values between monosomic and WT samples, we performed Student’s t-test.
Results
General behavior and motor learning
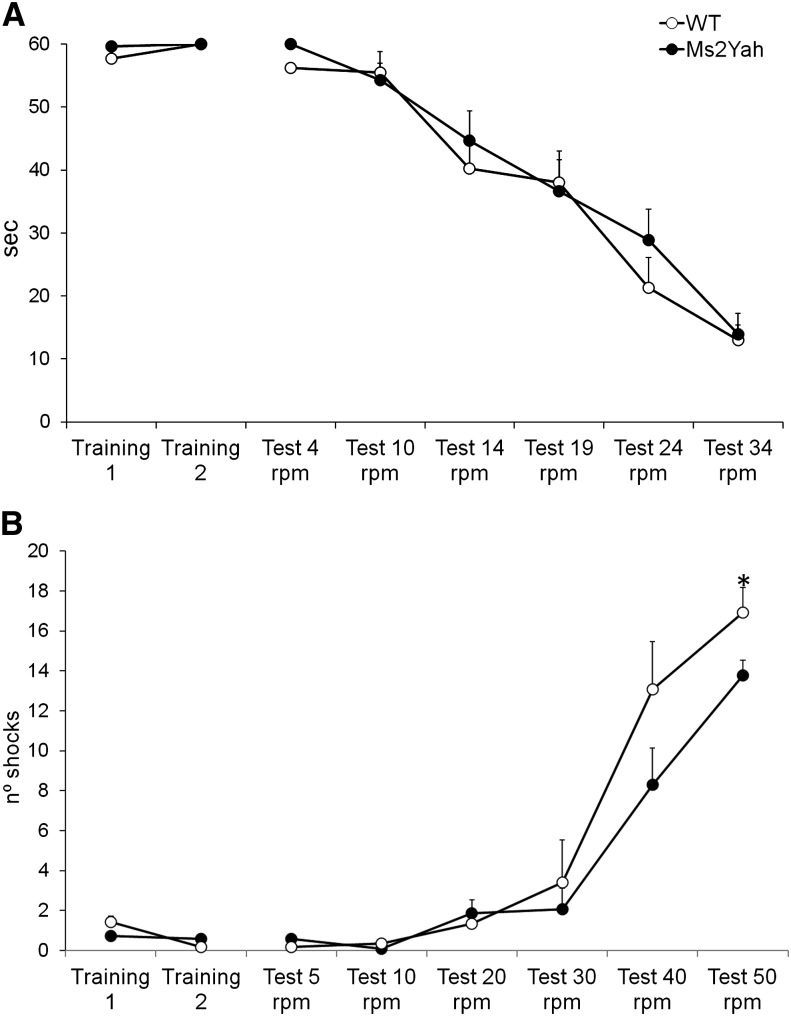
The Ms2Yah mice were generated like the Ts1Yah mice (Lopes Pereira et al. 2009) after chromosomal engineering in embryonic stem cells. Ms2Yah individuals are heterozygous for the deletion of the Abcg1–U2af1 region encompassing 16 genes. The carrier mice were obtained and appeared normal without any gross abnormal phenotypes or stereotypic behavior in our standard battery of tests. However, to assess the motor learning and behavior we compared the Ms2Yah mice with controls, using the rotarod and treadmill paradigms. No significant differences between genotypes were detected in the rotarod test (Figure 1A). The latency to fall off the rod was similar both at constant speeds (4, 10, 14, 19, 24, and 34 rpm) and in the acceleration test (4–40 rpm in 40 sec). However, in the treadmill test, we observed a worse performance in the last session [ANOVA, F(1,26) = 4.980, P = 0.035; Figure 1B], in which Ms2Yah mice receive a higher number of shocks than wild-type mice, suggesting a poorer performance on high task demands. Locomotion is not affected in mutant mice, but it appears that Ms2Yah have difficulties maintaining their effort for a long period. No coordination and motor learning impairments were found but muscle activity might be affected.
Figure 1.
Locomotor and behavioral assessment of the Ms2Yah monosomic model for the Abcg1–U2af1 region. (A) Rotarod test. No differences were observed between genotypes in the two training sessions prior to the test. In addition, no differences were observed between both genotypes in the latency of falling from the rotating rod. All mice acquired the criteria to perform the task at the same time. (B) Treadmill test. In the treadmill test, all mice showed a normal pattern of locomotion, without apparent alterations. But in the last trial of the test, at the highest-demanding speed of 50 rpm, significant difference between genotypes was shown [ANOVA, F(1,26) = 4.980, P = 0.035]. Ms2Yah mice received significantly more shocks than control wild-type mice.
Cognitive tests
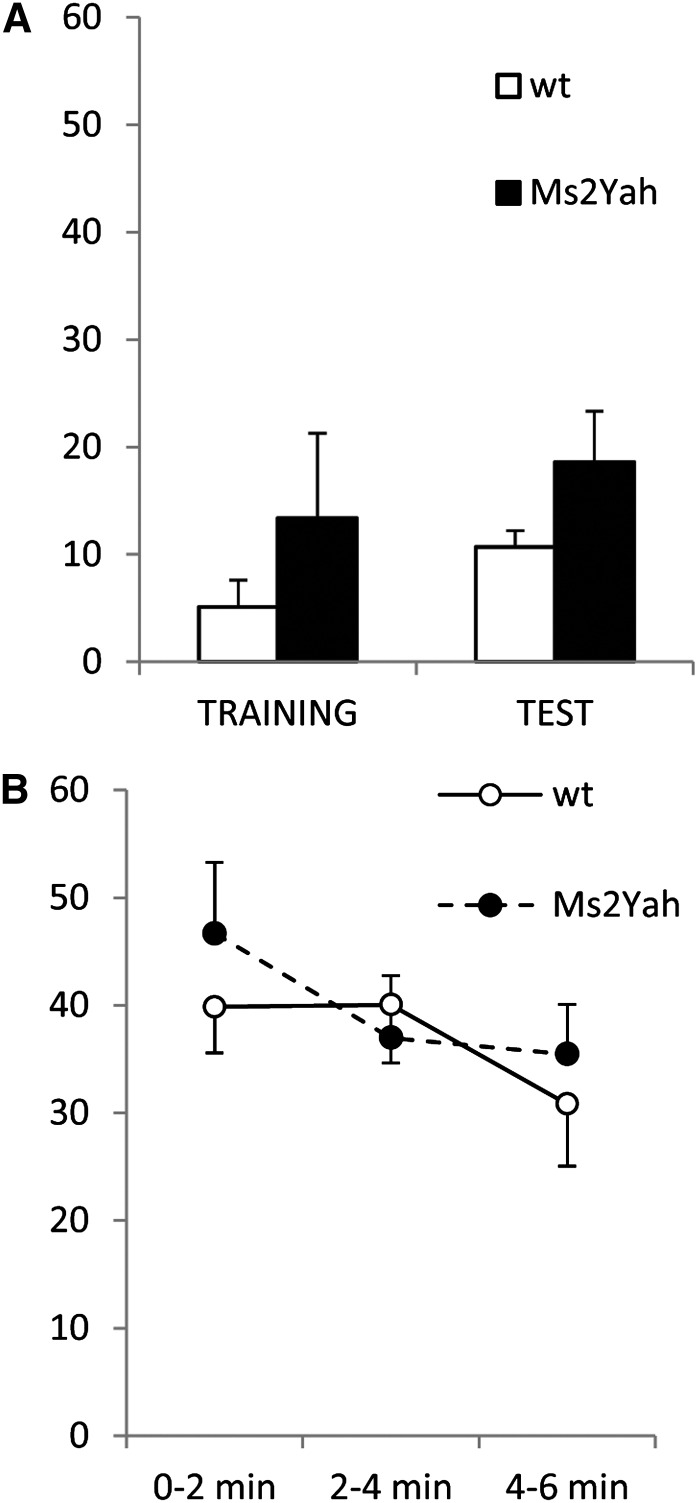
We then investigated whether the decrease in copy number of the Abcg1–U2af1 region has an impact on cognitive functions as observed for the Ts1Yah mice (Lopes Pereira et al. 2009). No differences were observed between genotypes in the object recognition test [ANOVA, F(1,26) = 0.015, P = 0.903] or in the Morris water maze, during the acquisition and cued sessions [repeated measures ANOVA, F(1,26) = 0.070, P = 0.794]. Mice of both genotypes reached the platform with similar latencies [repeated measures ANOVA, F(1,26) = 0.010, P = 0.923] and no differences in swimming speed were detected. In the removal session, no differences were observed in the preference for the trained quadrant [ANOVA, F(1,26) = 0.013, P = 0.909]. In the reversal session no differences in learning the new platform position were observed between genotypes [ANOVA, F(1,26) = 0.040 P = 0.842]. However, in the pure contextual fear-conditioning test a tendency to increase the freezing time in test session in Ms2Yah mice was found [ANOVA, F(1,26) = 2.963, P < 0.05]. This increase is significantly higher in Ms2Yah than in wild-type mice [repeated measures F(1,26) = 5.680, P = 0.025] with respect to the freezing in training sessions (Figure 2).
Figure 2.
Altered pure contextual fear conditioning in the Ms2Yah mice. (A) When we analyzed the total freezing time during the test phase, no differences between both genotypes were observed, beyond a nonsignificant trend to increase the percentage of freezing in Ms2Yah mice. However, when we analyzed the freezing during the training phase compared to the test phase, a significant difference between both genotypes was observed [repeated measures F(1,26) = 5.680, P = 0.025], being much higher in Ms2Yah mice compared to wild-type mice. (B) We then further analyzed the extinction of contextual fear memory by scoring freezing time in 2-min sessions over 6 min but did not detect any significant changes.
Social interest and social discrimination
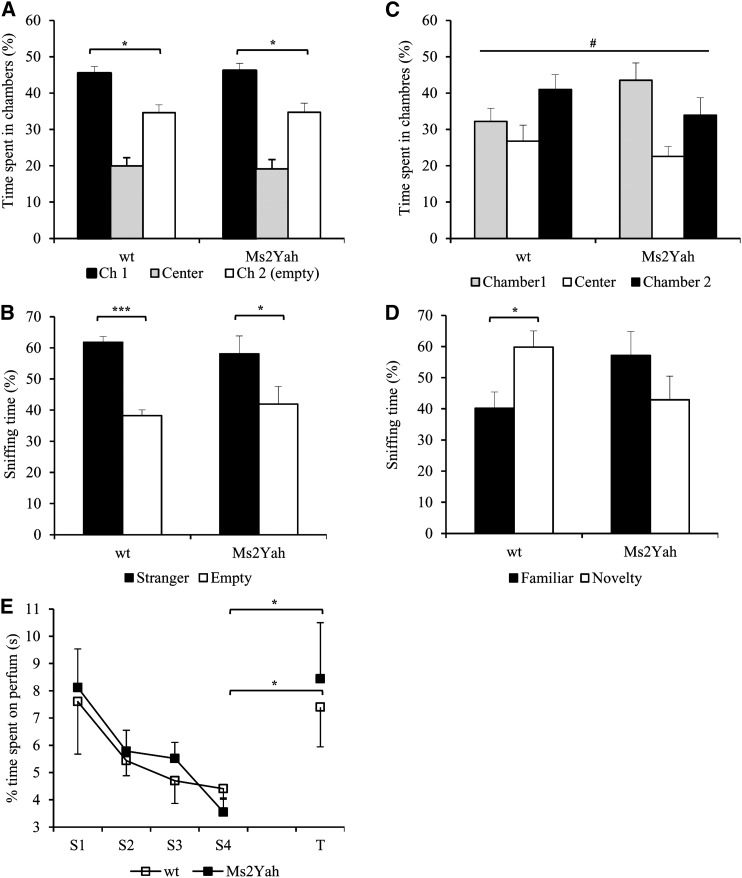
We evaluated the social behavior of Ms2Yah mice, using the three-compartment test (Moy et al. 2004, 2008; Duchon et al. 2011). The wild-type and monosomic animals explored the three chambers similarly [ANOVA “genotype × chambers,” F(2,36) = 2.588, P = 0.089], excluding a place preference in the arena. During the social interest session, control and Ms2Yah mice displayed strong preference for a congener mouse and spent more time exploring the chamber with the congener compared to the empty cage chamber [two-way ANOVA “congener (chamber 1) vs. empty cage (chamber 2),” F(1,24) = 14.17, P < 0.001; posthoc Tukey “congener (chamber 1) vs. empty cage (chamber 2),” WT, P = 0.016; Ms2Yah, P = 0.012; Figure 3A]. Likewise, the percentage of time spent by the control littermate sniffing at the wire cage containing the mouse is increased vs. sniffing the empty one [ANOVA, “chamber 1 vs. chamber 2,” F(1,24) = 21.78, P < 0.001; posthoc Tukey “stranger 1 vs. empty cage,” WT, P < 0.001; Ms2Yah, P = 0.013; Figure 3B]. We conclude that there was no interference for social interaction due to the monosomic condition.
Figure 3.
Social novelty performance is altered in the Ms2Yah mice. (A–D) Percentage of time spent in the different chambers (A and C) and of time spent sniffing the cages (B and D). In A and B, chamber 1 contained a mouse (familiar) and chamber 2 is empty. The WT mice showed a strong preference for the chambers and the cage containing an unfamiliar mouse while monosomic mice showed no significant interaction with the congener. In C and D, a new mouse (novel) was placed in chamber 2. Here the WT mice had a preference for the chambers or spent more time sniffing the cage with the novel individual whereas the Ms2Yah mice failed to interact with the new mouse and spent more time in the chamber with the familiar individual. In E, odor learning and discrimination were controlled to exclude the olfactory defect in the Ms2Yah mice. Data shown are mean + SEM for each strain. Statistical analyses are based on Student’s t-test (***P < 0.001; **P > 0.01; *P > 0.05); # n.s. Chamber1 versus Chamber2.
During social discrimination there was no difference in the time spent in the chamber with the newly introduced congener (chamber 2) for both groups [ANOVA, “familiar (chamber 1) vs. novel congener (chamber 2),” F(1,24) = 0.001, P = 0.923; posthoc Tukey “familiar vs. novel,” WT, P = 0.167; Ms2Yah, P = 0.131; Figure 3C). The tendency for social discrimination was certainly due to a slight difference between the control and the Ms2Yah group in exploring chamber 2 [ANOVA, “genotype × chamber,” F(1,24) = 4.467, P = 0.045]; the WT and Ms2Yah displayed different exploratory patterns, confounding the altered pattern of the monosomics. Similarly, the percentage of time spent sniffing the new congener in chamber 2 was affected [ANOVA, “stranger 1 chamber vs. stranger 2 chamber,” F(1,24) = 6.690, P = 0.016] (Figure 3C) but the WT spent more time exploring the new congener than the Ms2Yah mice (posthoc Tukey “stranger 1 cage vs. stranger 2 cage,” WT, P = 0.045; Ms2Yah, P = 0.08; time of sniffing the new congener, WT, 1.91 ± 0.27; Ms2Yah, 1.44 ± 0.31). Then we checked that the Ms2Yah have no alteration in odor recognition (Student’s t-test, “trial 4 vs. test session,” WT, P = 0.045; Ms2Yah, P = 0.024; Figure 3E). This experiment demonstrated that Ms2Yah animals had a lower interaction with new congeners.
Input/output curves and paired-pulse facilitation
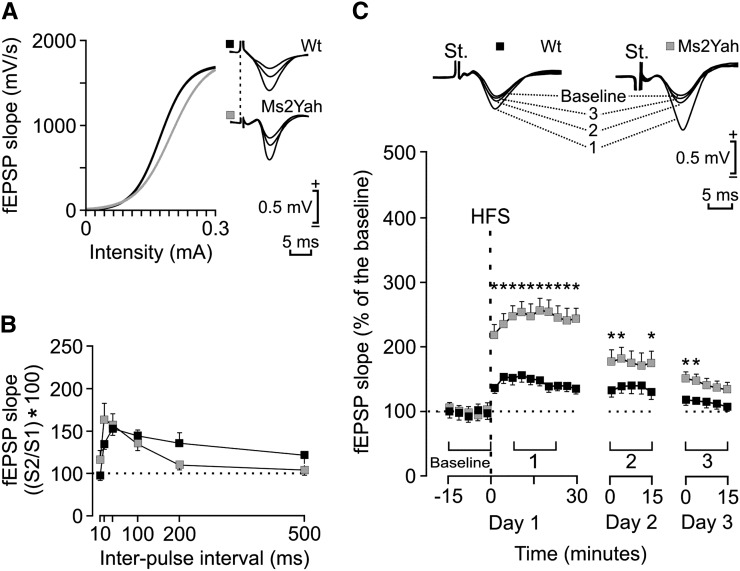
Since overexpression of genes located in the Abcg1–U2af1 region led to increased LTP in the hippocampal CA3–CA1 synapse (Lopes Pereira et al. 2009), we explored the effect of monosomy on this phenotype. In our current experiment, both WT and transgenic (Ms2Yah) mice presented similar increases in the slope of fEPSP evoked at the CA1 area following the presentation of singles pulses of increasing intensity at the ipsilateral Schaffer collaterals (Figure 4). In all cases, these relationships were fitted by sigmoid curves (r > 0.9, P < 0.0001), thus suggesting normal functioning of the CA3–CA1 synapse in the two genotypes.
Figure 4.
Changes in long-term potentiation in Ms2Yah mice compared to wild-type littermates. (A) Input/output curves for the CA3–CA1 synapse. A single (100 μsec, biphasic) pulse was presented to Schaffer collaterals at increasing intensities (in milliamperes) while recording the evoked fEPSP at the CA1 area for wild-type (wt, solid squares) and transgenic (Ms2Yah, shaded squares) Ms2 mice. Some fEPSPs collected from the two types of mouse are illustrated at the right. Ms2Yah mice presented input/output curves similar to those presented by Ms2Wt animals. (B) Paired-pulse facilitation. The data shown are mean ± SEM slopes of the second fEPSP expressed as a percentage of the first for six (10, 20, 40, 100, 200, 500) interstimulus intervals. The two groups of mice presented similar paired-pulse facilitation at intervals of 20–100 msec. (C) Top, examples of fEPSPs collected from selected Ms2Wt (solid square) and Ms2Yah (shaded square) animals before (baseline) and after (days 1–3) high-frequency stimulation (HFS) of Schaffer collaterals. Bottom, time course of LTP evoked in the CA1 area (fEPSP mean ± SEM) following HFS for Ms2Wt and Ms2Yah mice. The HFS was presented after 15 min of baseline recordings, at the time marked by the dashed line. The fEPSP is given as a percentage of the baseline (100%) slope. Although the two groups presented a significant increase (ANOVA, two-tailed) in fEPSP slope following HFS when compared with baseline records, values collected from the MS2Yah group were significantly [*P < 0.01; F(24,216) = 10.785] larger than those collected from Ms2Wt mice at the indicated times.
As illustrated in Figure 4B, both wild-type and monosomic animals presented a significant [F(12,135) = 56.126, P < 0.001] increase of the response to the second pulse at short (20–100 msec) time intervals. No significant differences between groups were observed at any of the selected (10, 20, 40, 100, 200, and 500 msec) intervals [F(12,135) = 0.735, P = 0.540], thus suggesting normal short-term hippocampal plasticity.
Comparison of LTP evoked in alert behaving mice
For the LTP study, and to obtain a baseline, animals were stimulated every 20 sec for ≥15 min at Schaffer collaterals (Figure 4C). After a stable baseline was obtained, the experimental animal was presented with the HFS protocol (see Materials and Methods). After HFS, the same single stimulus used to generate baseline records was presented at the initial rate (3/min) for another 30 min. Recording sessions were repeated 24 and 48 hr later for 15 min each (Figure 4C). Using this HFS and recording protocols, both experimental groups presented a significant LTP for the first 24 hr [P < 0.001; F(24,216) = 10.785], but with some interesting differences between them. Thus, the LTP response presented by Ms2Yah mice was significantly larger than that presented by the Ms2Wt group for most of the recording period (P < 0.01).
Morphological analysis of hippocampus
We measured hippocampal areas (whole and subregions): the mean surface of the entire hippocampus for wild type was 2.01 and 1.92 mm2 for the mutant and the selected region, CA1 stretched to 0.88 ± 0.09 mm2 (WT) and 0.83 ± 0.07 mm2 (Ms2Yah), CA2–3 was 0.54 ± 0.07 mm2 (WT) and 0.51 ± 0.06 mm2 (Ms2Yah), and the dentate gyrus was 0.59 ± 0.02 mm2 (WT) and 0.58 ± 0.03 mm2 (Ms2Yah). The cell count was performed to evaluate the neuronal density in hippocampal subregions, by counting cresyl violet-stained cells: in CA1, we calculated 1140 cells/mm2 in the WT, similar to 1199 cells/mm2 in Ms2Yah; in the CA2–3 region we quantified 1225 cells/mm2 in wild type compared to 1245 in the mutant group. Finally, in the dentate gyrus area, the density was evaluated to be 1469 cells/mm2 in the WT and to 1565 cells/mm2 in Ms2Yah. Student’s t-test was performed and we did not observe any size difference in the hippocampus or in neuronal density. There are neither signs of volume changes nor layer disorganization. We concluded that Ms2Yah did not show abnormal morphology of hippocampus.
Whole-genome expression analysis
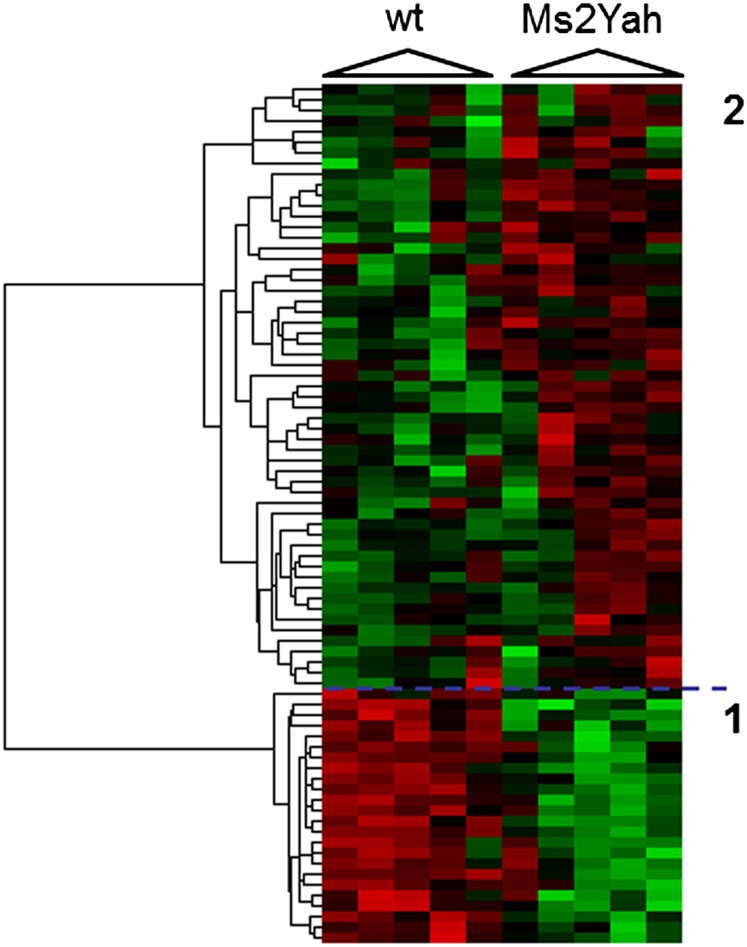
To better characterize the mechanisms underlying the behavioral and LTP phenotype observed in the Abcg1–U2af1 monosomy, we carried out whole-genome expression analysis on adult WT and Ms2Yah hippocampi, using Affymetrix Gene Chip technology (Raveau et al. 2012). A total of 24,697 transcripts were found expressed in the hippocampi over our threshold background of which 12,009 were known with an mRNA accession number. After normalization, we found only a subset of 80 genes that were statistically misexpressed with a fold change >1.20 or <0.80 (Table 1). We also performed a cluster analysis showing a clear discrimination between WT and Ms2Yah samples with two groups of genes (Figure 5) and the first group corresponds to 33 downregulated genes, with as expected Cbs [fold change (FC) = 0.69 with P = 0.02] and Rsph1/Tsga2 (FC = 0.80 with P = 0.02) from the monosomic interval. The second group encompassed a series of 139 upregulated genes. Most of the genes were located outside the aneuploid regions, indicating a small genome-wide effect on gene expression in the Ms2Yah adult hippocampi. We analyzed the enrichment of functional annotation, using DAVID (Huang et al. 2009), and found an enrichment for postsynaptic cell membrane proteins encoded by Gabrr2, Gabbr1, Grid2p, Park2, and Dlg1 [fold enrichment = 13.75; P-value = 7.46E-04; false discovery rate (FDR) = 0.87] and with members of the Ubiquitin-mediated proteolysis (Anapc1, Rnf7, Huwe1, and Park2; fold enrichment = 7.67; P-value = 0.012; FDR = 11.13).
Table 1. Genes deregulated in the Ms2Yah mice hippocampi.
| Group | Gene name | Chr | WT: mean ± SEM | Ms2Yah: mean ± SEM | FC | Test: posthoc |
|---|---|---|---|---|---|---|
| 1 | Cldn5 | 16 | 0.77 ± 0.15 | 0.52 ± 0.09 | 0.68 | * |
| 1 | Cbs | 17 | 0.9 ± 0.12 | 0.62 ± 0.05 | 0.69 | * |
| 1 | Tmem234 | 4 | 0.84 ± 0.1 | 0.63 ± 0.07 | 0.75 | * |
| 1 | Rps2 | 17 | 0.98 ± 0.08 | 0.74 ± 0.07 | 0.76 | * |
| 1 | Fam40b/Strip2 | 6 | 0.95 ± 0.06 | 0.72 ± 0.08 | 0.76 | * |
| 1 | Tek | 4 | 0.85 ± 0.1 | 0.66 ± 0.14 | 0.77 | ** |
| 1 | Ggcx | 6 | 0.99 ± 0.04 | 0.76 ± 0.09 | 0.77 | * |
| 1 | Sox2 | 3 | 0.86 ± 0.11 | 0.67 ± 0.15 | 0.78 | * |
| 1 | Hist1h1c | 13 | 1.05 ± 0.15 | 0.82 ± 0.08 | 0.78 | * |
| 1 | Hyou1 | 9 | 0.89 ± 0.08 | 0.69 ± 0.09 | 0.78 | * |
| 1 | Copg | 6 | 0.9 ± 0.07 | 0.7 ± 0.08 | 0.78 | ** |
| 1 | Slco1c1 | 6 | 0.95 ± 0.1 | 0.74 ± 0.12 | 0.79 | * |
| 1 | Gm12696 | 4 | 0.71 ± 0.13 | 0.56 ± 0.12 | 0.79 | * |
| 1 | Elk1 | X | 0.9 ± 0.08 | 0.71 ± 0.06 | 0.79 | * |
| 1 | Gm11599 | 1 | 1.02 ± 0.07 | 0.8 ± 0.06 | 0.79 | * |
| 1 | Spsb3 | 17 | 0.93 ± 0.04 | 0.73 ± 0.09 | 0.79 | * |
| 1 | Slc39a1 | 3 | 1.04 ± 0.09 | 0.82 ± 0.15 | 0.79 | * |
| 1 | 2810006K23Rik | 5 | 0.92 ± 0.06 | 0.73 ± 0.1 | 0.79 | * |
| 1 | Tbl3 | 17 | 1.07 ± 0.12 | 0.85 ± 0.07 | 0.79 | * |
| 1 | Msl3l2 | 10 | 0.96 ± 0.04 | 0.76 ± 0.12 | 0.8 | * |
| 1 | Slc37a3 | 6 | 0.88 ± 0.05 | 0.7 ± 0.12 | 0.8 | * |
| 1 | Smyd5 | 6 | 0.93 ± 0.09 | 0.74 ± 0.09 | 0.8 | * |
| 1 | Hmcn1 | 1 | 0.87 ± 0.08 | 0.7 ± 0.09 | 0.8 | *** |
| 1 | Anapc1 | 2 | 0.97 ± 0.05 | 0.77 ± 0.17 | 0.8 | * |
| 2 | Dlg1 | 16 | 0.87 ± 0.08 | 1.04 ± 0.1 | 1.2 | * |
| 2 | Kcnn3 | 3 | 1.04 ± 0.05 | 1.25 ± 0.13 | 1.2 | * |
| 2 | B430305J03Rik | 3 | 1.08 ± 0.07 | 1.29 ± 0.1 | 1.2 | * |
| 2 | Trp53inp1 | 4 | 1.12 ± 0.13 | 1.35 ± 0.15 | 1.2 | * |
| 2 | Gdpd2 | X | 0.95 ± 0.04 | 1.14 ± 0.07 | 1.21 | * |
| 2 | Gabrr2 | 4 | 1.28 ± 0.13 | 1.55 ± 0.09 | 1.21 | * |
| 2 | Gm7820 | X | 1.09 ± 0.04 | 1.31 ± 0.12 | 1.21 | * |
| 2 | Kitl | 10 | 0.88 ± 0.07 | 1.06 ± 0.08 | 1.21 | * |
| 2 | AI836003 | 15 | 1.05 ± 0.03 | 1.26 ± 0.1 | 1.21 | * |
| 2 | Cav3 | 6 | 0.97 ± 0.04 | 1.17 ± 0.1 | 1.21 | * |
| 2 | S100a10 | 3 | 0.91 ± 0.07 | 1.1 ± 0.1 | 1.21 | * |
| 2 | Vma21 | X | 1.08 ± 0.15 | 1.31 ± 0.03 | 1.21 | * |
| 2 | Figf | X | 0.93 ± 0.15 | 1.13 ± 0.12 | 1.22 | * |
| 2 | 2210404J11Rik | 17 | 0.98 ± 0.07 | 1.2 ± 0.13 | 1.23 | * |
| 2 | Trhr | 15 | 0.66 ± 0.14 | 0.81 ± 0.09 | 1.23 | * |
| 2 | Tsc22d3 | X | 1.23 ± 0.26 | 1.52 ± 0.19 | 1.23 | * |
| 2 | Gm4876 | 6 | 0.91 ± 0.09 | 1.12 ± 0.11 | 1.24 | * |
| 2 | Arrdc2 | 8 | 1.19 ± 0.11 | 1.47 ± 0.21 | 1.24 | * |
| 2 | Cpsf7 | 19 | 0.85 ± 0.11 | 1.06 ± 0.07 | 1.24 | * |
| 2 | Cntn6 | 6 | 0.92 ± 0.08 | 1.14 ± 0.13 | 1.24 | * |
| 2 | Dclre1b | 3 | 1.05 ± 0.07 | 1.31 ± 0.16 | 1.25 | ** |
| 2 | Cacna1g | 11 | 0.92 ± 0.06 | 1.16 ± 0.14 | 1.26 | * |
| 2 | Huwe1 | X | 1.1 ± 0.11 | 1.39 ± 0.13 | 1.26 | * |
| 2 | Fam70a | X | 0.92 ± 0.09 | 1.16 ± 0.12 | 1.26 | * |
| 2 | Gm129 | 3 | 1.12 ± 0.26 | 1.41 ± 0.24 | 1.26 | * |
| 2 | Gabrr1 | 4 | 1.03 ± 0.05 | 1.31 ± 0.05 | 1.27 | ** |
| 2 | Gm9801 | 7 | 0.95 ± 0.08 | 1.21 ± 0.14 | 1.27 | * |
| 2 | Anapc1 | 2 | 0.95 ± 0.15 | 1.2 ± 0.08 | 1.27 | * |
| 2 | Thpo | 16 | 1.03 ± 0.1 | 1.31 ± 0.1 | 1.27 | * |
| 2 | Park2 | 17 | 1.21 ± 0.14 | 1.55 ± 0.08 | 1.28 | ** |
| 2 | Gm5148 | 3 | 0.96 ± 0.08 | 1.22 ± 0.13 | 1.28 | * |
| 2 | Rnf7 | 9 | 0.96 ± 0.04 | 1.23 ± 0.13 | 1.28 | * |
| 2 | Bmp2 | 2 | 0.99 ± 0.06 | 1.26 ± 0.17 | 1.28 | * |
| 2 | Cit | 5 | 0.92 ± 0.06 | 1.18 ± 0.09 | 1.28 | * |
| 2 | Uevld | 7 | 0.86 ± 0.07 | 1.11 ± 0.14 | 1.28 | * |
| 2 | Usp54 | 14 | 1.12 ± 0.09 | 1.44 ± 0.25 | 1.29 | * |
| 2 | Rnu1b1 | 3 | 1.01 ± 0.02 | 1.31 ± 0.09 | 1.29 | ** |
| 2 | 1700048O20Rik | 9 | 1.07 ± 0.03 | 1.39 ± 0.13 | 1.29 | * |
| 2 | Brunol5 | 4 | 1 ± 0.11 | 1.3 ± 0.15 | 1.3 | * |
| 2 | Eltd1 | 3 | 1.08 ± 0.12 | 1.42 ± 0.31 | 1.32 | * |
| 2 | Carhsp1 | 16 | 1.06 ± 0.09 | 1.4 ± 0.17 | 1.32 | * |
| 2 | Dhrs1 | 14 | 0.98 ± 0.12 | 1.31 ± 0.14 | 1.33 | * |
| 2 | Mybl1 | 1 | 1.07 ± 0.11 | 1.44 ± 0.22 | 1.35 | * |
| 2 | Dnahc3 | 7 | 0.98 ± 0.07 | 1.33 ± 0.25 | 1.36 | * |
| 2 | Mir692-1 | 17 | 0.8 ± 0.14 | 1.09 ± 0.11 | 1.36 | * |
| 2 | Gpr4 | 7 | 0.89 ± 0.09 | 1.22 ± 0.14 | 1.37 | * |
| 2 | mIRNAAL589870 | UN | 1.08 ± 0.13 | 1.49 ± 0.16 | 1.37 | ** |
| 2 | Rin3 | 12 | 1.06 ± 0.04 | 1.48 ± 0.25 | 1.39 | * |
| 2 | Camk2d | 3 | 0.97 ± 0.06 | 1.36 ± 0.2 | 1.4 | * |
| 2 | Erdr1 | UN | 1.13 ± 0.14 | 1.59 ± 0.07 | 1.41 | * |
| 2 | Grid2ip | 5 | 0.84 ± 0.08 | 1.19 ± 0.1 | 1.41 | * |
| 2 | Taf1d | 9 | 1.09 ± 0.17 | 1.56 ± 0.1 | 1.43 | ** |
| 2 | Ccl28 | UN | 0.81 ± 0.19 | 1.16 ± 0.1 | 1.44 | * |
| 2 | Plekhf1 | 7 | 1.39 ± 0.25 | 2.05 ± 0.32 | 1.47 | ** |
| 2 | Meg3 | 12 | 0.85 ± 0.17 | 1.25 ± 0.15 | 1.48 | * |
| 2 | 3110052M02Rik | 17 | 1.05 ± 0.16 | 1.59 ± 0.04 | 1.52 | ** |
Groups of genes downregulated (1) or upregulated (2) for which the statistical test is <0.05 are listed. Chr, chromosome; FC, fold change. The posthoc statistical test used is a bilateral t-test. ***P < 0.001; **P < 0.01; *P < 0.05.
Figure 5.
Clustering derived from statistically deregulated genes in Ms2Yah hippocampi. Whole-genome microarray expression analysis was performed on RNA isolated from whole adult mouse hippocampi and statistically deregulated genes were assessed using GeneSpring software. Underexpression and overexpression are represented in green and red, respectively, and expression levels were calculated by comparison to mean expression level of all arrays for each gene. The genes were selected with a statistical significant level (P-value <0.05) and with an up/down cutoff on the fold change (0.8 < x for group 1 and x > 1.2 for group 2).
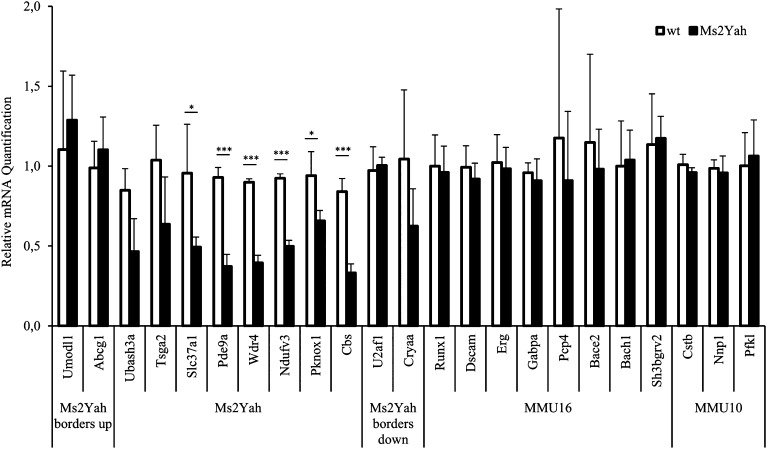
To confirm the expression pattern detected in the whole-genome approach, we performed quantitative RT-PCR on RNA isolated from monosomic hippocampi and their respective controls. All methods from RNA extraction to cDNA quantification have been already described in previous work (Lopes Pereira et al. 2009; Raveau et al. 2012). We focused on the Abcg1–U2af1 genes plus genes located at the borders and a few genes among Mmu10 and -16 homologous regions. Genes expressed from the monosomic interval were downregulated as anticipated, with a strong and significant decrease for Ubash3a, Pde9a, Wdr4, Ndufv3, Pknox1, and Cbs (Figure 6), whereas the expression of a subset of genes, located at the bordering regions (Umodl1, Abcg1, U2af1, and Cryaa) or outside in the genome in regions homologous to human chromosomes or elsewhere, was not affected. We conclude that a limited change in the genetic expression was induced by the monosomy of the Abcg1–U2af1 interval.
Figure 6.
Quantitative PCR expression study focusing on genes located in the Abcg1–U2af1 regions and on genes homologous to Hsa21 located in additional genetic intervals. We tested Ms2Yah region genes, borders, and relevant genes on mouse chromosomes 10 and 16. These results confirm the transcriptome analysis showing underexpressed genes from the Abcg1–U2af1 region compared to all the other genes tested.
Discussion
We evaluated here the cognitive and electrophysiological phenotypes of a mouse model monosomic for the Abcg1–U2af1 genetic interval. This model helps us to evaluate the contribution of not only the 16 encompassed coding genes but also other functional elements within this chromosomal region toward cognition. Previous studies of trisomic mice for the same region (Ts1Yah) showed a complex cognitive phenotype that combined alteration in specific domains that are affected in well-characterized DS models such as nonassociative learning and short-term spatial and recognition memory (Lopes Pereira et al. 2009). Similar results were obtained for a model encompassing the same region (Yu et al. 2010b). However, trisomy of the interval improved the performance in spatial learning, along with larger and long-lasting LTP, suggesting improved hippocampal function. Thus, we concluded that, whatever the copy number, the Abcg1–U2af1 region is necessary to maintain normal LTP activation in hippocampus.
The Df(17)1Yey/+ monosomic mouse model, encompassing the Abcg1–Rpr1b interval, displayed reduced swimming speed in the Morris water maze and decreased freezing time in the contextual fear-conditioning paradigm (Yu et al. 2010a). In the present report, the Ms2Yah, mice showed slightly different phenotypes: a mild motor defect found in the treadmill, no spatial or recognition memory impairment with no changes in the swimming activity, improvement in pure contextual fear associative memory along with increased LTP, and a light sociability discrimination defect. The phenotypic differences observed in the two models certainly result from the size of the deficiency: shorter in the Ms2Yah with four more disomic genes, namely Cryaa, Sik1, Hsf2bp, and Rrp1b, compared with the Df(17)1Yey. According to the Allen brain atlas, Cryaa, Sik1, and Rrp1b are expressed in the adult mouse brain, including the hippocampus (all three), the cortex (Sik1 and Rrp1b), and the cerebellum (Rrp1b). Cryaa is more likely a molecular chaperone, stabilizing other partners, but with yet unknown brain function (Fan et al. 2014). Sik1, a HDAC kinase contributing to the CREB pathway, with a wide impact on brain function (Berdeaux et al. 2007; Takemori et al. 2007), and Rrp1b, a chromatin-associated factor that is a substrate of MAPK14 (Varjosalo et al. 2013), are more interesting candidates.
Interestingly, Ms2Yah does not mirror the Ts1Yah phenotypes with no changes in the MWM and the novel object recognition (Lopes Pereira et al. 2009). Short-term plastic processes are not affected in Ms2Yah animals, since the synaptic facilitation evoked by the presentation of a pair of pulses, a typical presynaptic short-term plastic property of the hippocampal CA3–CA1 synapse, which has been related to the process of neurotransmitter release, was not affected in monosomic mice. However, similar to what was previously described in the homologous trisomy, Ms2Yah mice present larger and longer-lasting LTPs than their respective littermate wild types, thus suggesting a dissociation of hippocampal function between spatial memory, novelty discrimination, and fear memory in the Ms2Yah model. Comparing those two functions affected, we could hypothesize that spatial and working memory is related to short-term plasticity because it is associated only with the trisomic mouse model, but associative memory, evaluated by fear conditioning, is implicated more in neuronal circuits than ones structuring around the hippocampus. The high LTP levels recorded are present only in mouse models restricted to the syntenic region between Mmu17 and Hsa21. This means that the role of those few common genes could have an underestimated impact on Down syndrome cognition disabilities.
We further explored the molecular origins of the monosomic phenotypes. The transcriptomic study performed comparing the expression profile in wild-type and mutant hippocampi points to the extreme sensitivity of the modifications generated by the deletion. On enhancing the stringency and the power of the statistical tests, the analysis highlighted 80 misregulated coding genes (Table 1). Some of them are involved in cognitive function, such as Gabrr1, Gabbr2, Grid2p, Park2, and Dlg1 encoding postsynaptic cell membrane proteins. Grid2ip is an interacting protein of the δ-subunit of glutamate receptor expressed in the cerebellum and in the hippocampus (Miyagi et al. 2002; Matsuda et al. 2006). Inactivation of Grid2ip facilitates long term depression (LTD) in the cerebellum with more sensitivity to calcium response compared to WT (Takeuchi et al. 2008). Interestingly, Grid2ip is a partner of GluRd2, which is involved in motor learning and motor coordination (Kashiwabuchi et al. 1995). Grid2ip could be a good candidate to explore change in the excitability of the neurons and the motor function defects observed in the Ms2Yah mice. We detected also variations of Park2, a regulator of dopamine production in the striatum, whose loss decreases synaptic excitability (Goldberg et al. 2003). Here, Park2 was overexpressed in Ms2Yah and may contribute to the enhanced LTP. Dlg1, the discs large homolog 1 multidomain scaffolding protein, is involved in synaptogenesis and postsynaptic signaling and potentially involved in brain pathologies (de Bartolomeis and Fiore 2004; Nithianantharajah et al. 2013). Interestingly, Dlg1 and Pak2 were recently found deleted in autism (Quintero-Rivera et al. 2010). However, both Gabrr1 and Gabrr2 or both Park2 and Dlg1 are located at proximity, respectively, on mouse chromosome 4 or 16 and are coregulated in our Ms2Yah hippocampi. In addition Sox2, a well-known transcription factor controlling neural stem cells, is downregulated in the Ms2Yah brain (Wegner and Stolt 2005; Shi et al. 2008).
From the Abcg1–U2af1 deleted genetic interval, six of eight genes expressed in the hippocampus, namely Ubash3a, Pde9a, Wdr4, Ndufv3, Pknox1, and Cbs, were found downregulated compared to the control. Little is known about the functions of Ubash3a, Wdr4, and Pknox1 in the brain, whereas Pde9a encodes a phosphodiesterase specifically expressed in hippocampus, whose inhibition increases early LTP after stimulation (Kroker et al. 2012). Thus Pde9a, with Grid2ip, is a candidate for the enhanced LTP observed in vivo. The Ndufv3 dehydrogenase is a member of a family that contributes to bipolar disorders or depression (Ben-Shachar and Karry 2008) but the role of this particular protein is actually unknown. Cbs is mutated in the intellectual disability homocystinuria. Cbs could affect neuronal function through various mechanisms including the synthesis of H2S, a new gaseous neurotransmitter (Kimura 2002; Chen et al. 2004).
During the social interest and discrimination test, we found an unexpected phenotype with the Ms2Yah mice showing reduced interaction with a new congener. This behavior is not due to a deficiency in odor system but suggests memory impairment. Interestingly the Gabrr1 and Gabrr2 genes are associated with several cognitive defects, including schizophrenia (Green et al. 2010) and autism (Ma et al. 2005). Their altered expression observed in the Ms2Yah hippocampi could be linked to the abnormal phenotypes observed in fear conditioning and social interaction. With this lack of interest for a novel individual, the Ms2Yah mice develop a specific autistic phenotype. Several autistic spectrum disorders (ASD) are linked with copy number variants (Moreno-De-Luca et al. 2010; Sanders et al. 2011; Luo et al. 2012). In the autism database (Basu et al. 2009), we found a few duplications and one deletion in patients that included segments from the Abcg1–U2af1 region (Jacquemont et al. 2006; Marshall et al. 2008; Pinto et al. 2010; Levy et al. 2011; Sanders et al. 2011). Thus the Ms2Yah mice might represent a model for autism with several functions affected in the brain without any notable cellular defects, but with changes in the expression of genes involved in the postsynaptic functions without any stereotypies detected so far.
Partial deletions of 21q are rare and these patients display a highly variable phenotype, depending on the size and position of the deletion. A review of the literature identified 38 cases with pure 21q deletions. Partial monosomy 21 was recently segregated into three regions associated with variable clinical severity. Terminal deletions (21q22.2-ter and 21q22.3-ter) or interstitial hemizygous loss of 16.4 Mb (21q21.1–q22.11) gave rise to relatively mild phenotypes while other 21q deletions produce a greater number of dysmorphic features and some major malformations related to genes in the interval (Lindstrand et al. 2010; Roberson et al. 2011). The combined information of the 38 available cases suggests that the ITSN1 gene is involved in severe mental retardation in patients with 21q deletion. In addition, a critical region of 0.56 Mb containing four genes, KCNE1, DSCR1, CLIC6, and RUNX1, is associated with severe congenital heart defects, and deletions of the most proximal 15–17 Mb of 21q are associated with mild or no cognitive impairment, but may lead to problems with balance and motor function. Several cases of chromosomal aberration encompassing the human chromosome 21q22.3 up to the qter regions have been reported (Lyle et al. 2009; Hannachi et al. 2011; Melis et al. 2011; Roberson et al. 2011) with mild phenotypes. In the ECARUCA database (www.ecaruca.net), 8 of 16 cases with complex aberration encompassing the 22q3 region of Hsa21 displayed mental retardation (Ferrante et al. 1983; McGinniss et al. 1992; Feenstra et al. 2006). In our previous analysis of the monosomy of the most telomeric part of human chromosome 21, we found no phenotypes attached to the deletion of the Cstb–Prmt2 genetic interval (Duchon et al. 2011), similar to the mild effect of the 21qter monosomy in humans. With this new report, the Abcg1–U2af1 region should be considered an important genetic interval, containing key dosage-sensitive elements controlling neurological phenotypes. The Ms2Yah mice should be further explored as a model of Hsa21 terminal deletion for which little consequence has been described so far (Lyle et al. 2009; Lindstrand et al. 2010; Roberson et al. 2011). On the other hand, an increase in the Abcg1–U2af1 region dosage may be a determinant for the social behavior observed in Down syndrome.
Acknowledgments
We thank members of the research group, of the Institut de Génétique de Biologie Moléculaire et Cellulaire (IGBM), of the Institut Clinique de la Souris (ICS), and of the AnEUploidy consortium for their helpful comments (www.aneuploidy.org). We are grateful to the animal caretakers of the Centre National de la Recherche Scientifique (CNRS) UPS44 TAAM unit and of the ICS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This project was supported by the French CNRS; the French Institut National de la Santé et de la Recherche Médicale; the University of Strasbourg and the “Centre Europeen de Recherche en Biomedecine”; the “Fondation Jerome Lejeune”; Koplowitz, Fragile X Research Foundation (FRAXA), and Association Française contre les Myopathies (AFM) Foundations; the Spanish Ministry of Economy (SAF2010-16427, SAF2007-31093-E, and FIS-PI 082038); Marató TV3; and the European Commission (AnEUploidy project LSHG-CT-2006-037627 to Y.H. and M.D.) The laboratory of M.D. is supported by DURSI (Grups consolidats 09 2009SGR1313).
Footnotes
Communicating editor: T. R. Magnuson
Literature Cited
- Arqué G., Fotaki V., Fernández D., Martínez de Lagrán M., Arbonés M., et al. , 2008. Impaired spatial learning strategies and novel object recognition in mice haploinsufficient for the dual specificity tyrosine-regulated kinase-1A (Dyrk1A). PLoS ONE 3: e2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S. N., Kollu R., Banerjee-Basu S., 2009. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 37: D832–D836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar D., Karry R., 2008. Neuroanatomical pattern of mitochondrial complex I pathology varies between schizophrenia, bipolar disorder and major depression. PLoS ONE 3: e3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdeaux R., Goebel N., Banaszynski L., Takemori H., Wandless T., et al. , 2007. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat. Med. 13: 597–603. [DOI] [PubMed] [Google Scholar]
- Besson V., Brault V., Duchon A., Togbe D., Bizot J.-C., et al. , 2007. Modeling the monosomy for the telomeric part of human chromosome 21 reveals haploinsufficient genes modulating the inflammatory and airway responses. Hum. Mol. Genet. 16: 2040–2052. [DOI] [PubMed] [Google Scholar]
- Blake J. A., Bult C. J., Eppig J. T., Kadin J. A., Richardson J. E., Genome Database Group The Mouse, 2014. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 42: D810–D817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. L., Jhee K. H., Kruger W. D., 2004. Production of the neuromodulator H2S by cystathionine beta-synthase via the condensation of cysteine and homocysteine. J. Biol. Chem. 279: 52082–52086. [DOI] [PubMed] [Google Scholar]
- de Bartolomeis A., Fiore G., 2004. Postsynaptic density scaffolding proteins at excitatory synapse and disorders of synaptic plasticity: implications for human behavior pathologies. Int. Rev. Neurobiol. 59: 221–254. [DOI] [PubMed] [Google Scholar]
- Duchon A., Pothion S., Brault V., Sharp A. J., Tybulewicz V. L. J., et al. , 2011. The telomeric part of the human chromosome 21 from Cstb to Prmt2 is not necessary for the locomotor and short-term memory deficits observed in the Tc1 mouse model of Down syndrome. Behav. Brain Res. 217: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q., Huang L. Z., Zhu X. J., Zhang K. K., Ye H. F., et al. , 2014. Identification of proteins that interact with alpha A-crystallin using a human proteome microarray. Mol. Vis. 20: 117–124. [PMC free article] [PubMed] [Google Scholar]
- Feenstra I., Fang J., Koolen D. A., Siezen A., Evans C., et al. , 2006. European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations (ECARUCA); an online database for rare chromosome abnormalities. Eur. J. Med. Genet. 49: 279–291. [DOI] [PubMed] [Google Scholar]
- Ferrante E., Vignetti P., Antonelli M., Bruni L., Bertasi S., et al. , 1983. Partial monosomy for a 21 chromosome. Report of a new case of r(21) and review of the literature. Helv. Paediatr. Acta 38: 73–80. [PubMed] [Google Scholar]
- Goldberg M. S., Fleming S. M., Palacino J. J., Cepeda C., Lam H. A., et al. , 2003. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 278: 43628–43635. [DOI] [PubMed] [Google Scholar]
- Green E. K., Grozeva D., Moskvina V., Hamshere M. L., Jones I. R., et al. , 2010. Variation at the GABA(A) receptor gene, Rho 1 (GABRR1) associated with susceptibility to bipolar schizoaffective disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 153B: 1347–1349. [DOI] [PubMed] [Google Scholar]
- Gruart A., Munoz M. D., Delgado-Garcia J. M., 2006. Involvement of the CA3–CA1 synapse in the acquisition of associative learning in behaving mice. J. Neurosci. 26: 1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gureviciene I., Ikonen S., Gurevicius K., Sarkaki A., van Groen T., et al. , 2004. Normal induction but accelerated decay of LTP in APP+PS1 transgenic mice. Neurobiol. Dis. 15: 188–195. [DOI] [PubMed] [Google Scholar]
- Hannachi H., Mougou-Zerelli S., BenAbdallah I., Mama N., Hamdi I., et al. , 2011. Clinical and molecular characterization of a combined 17p13.3 microdeletion with partial monosomy 21q21.3 in a 26-year-old man. Cytogenet. Genome Res. 135: 102–110. [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., et al. , 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Jacquemont M. L., Sanlaville D., Redon R., Raoul O., Cormier-Daire V., et al. , 2006. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J. Med. Genet. 43: 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwabuchi N., Ikeda K., Araki K., Hirano T., Shibuki K., et al. , 1995. Impairment of motor coordination, Purkinje cell synapse formation, and cerebellar long-term depression in GluR delta 2 mutant mice. Cell 81: 245–252. [DOI] [PubMed] [Google Scholar]
- Kimura H., 2002. Hydrogen sulfide as a neuromodulator. Mol. Neurobiol. 26: 13–19. [DOI] [PubMed] [Google Scholar]
- Korbel J. O., Tirosh-Wagner T., Urban A. E., Chen X. N., Kasowski M., et al. , 2009. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc. Natl. Acad. Sci. USA 106: 12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroker K. S., Rast G., Giovannini R., Marti A., Dorner-Ciossek C., et al. , 2012. Inhibition of acetylcholinesterase and phosphodiesterase-9A has differential effects on hippocampal early and late LTP. Neuropharmacology 62: 1964–1974. [DOI] [PubMed] [Google Scholar]
- Letourneau A., Santoni F. A., Bonilla X., Sailani M. R., Gonzalez D., et al. , 2014. Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature 508: 345–350. [DOI] [PubMed] [Google Scholar]
- Levy D., Ronemus M., Yamrom B., Lee Y. H., Leotta A., et al. , 2011. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 70: 886–897. [DOI] [PubMed] [Google Scholar]
- Lindstrand A., Malmgren H., Sahlen S., Schoumans J., Nordgren A., et al. , 2010. Detailed molecular and clinical characterization of three patients with 21q deletions. Clin. Genet. 77: 145–154. [DOI] [PubMed] [Google Scholar]
- Lopes Pereira P., Magnol L., Sahún I., Brault V., Duchon A., et al. , 2009. A new mouse model for the trisomy of the Abcg1–U2af1 region reveals the complexity of the combinatorial genetic code of Down syndrome. Hum. Mol. Genet. 18: 4756–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Sanders S. J., Tian Y., Voineagu I., Huang N., et al. , 2012. Genome-wide transcriptome profiling reveals the functional impact of rare de novo and recurrent CNVs in autism spectrum disorders. Am. J. Hum. Genet. 91: 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyle R., Bena F., Gagos S., Gehrig C., Lopez G., et al. , 2009. Genotype-phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur. J. Hum. Genet. 17: 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D. Q., Whitehead P. L., Menold M. M., Martin E. R., Ashley-Koch A. E., et al. , 2005. Identification of significant association and gene-gene interaction of GABA receptor subunit genes in autism. Am. J. Hum. Genet. 77: 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C. R., Noor A., Vincent J. B., Lionel A. C., Feuk L., et al. , 2008. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 82: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K., Matsuda S., Gladding C. M., Yuzaki M., 2006. Characterization of the delta 2 glutamate receptor-binding protein delphilin - splicing variants with differential palmitoylation and an additional PDZ domain. J. Biol. Chem. 281: 25577–25587. [DOI] [PubMed] [Google Scholar]
- McGinniss M. J., Kazazian H. H., Stetten G., Petersen M. B., Boman H., et al. , 1992. Mechanisms of ring chromosome formation in 11 cases of human ring chromosome 21. Am. J. Hum. Genet. 50: 15–28. [PMC free article] [PubMed] [Google Scholar]
- Mefford H. C., Batshaw M. L., Hoffman E. P., 2012. Genomics, intellectual disability, and autism. N. Engl. J. Med. 366: 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis D., Genesio R., Cappuccio G., V. MariaGinocchio, R. D. Casa et al, 2011. Mental retardation, congenital heart malformation, and myelodysplasia in a patient with a complex chromosomal rearrangement involving the critical region 21q22. Am. J. Med. Genet. A. 155A: 1697–1705. [DOI] [PubMed] [Google Scholar]
- Miyagi Y., Yamashita T., Fukaya M., Sonoda T., Okuno T., et al. , 2002. Delphilin: a novel PDZ and formin homology domain-containing protein that synaptically colocalizes and interacts with glutamate receptor delta 2 subunit. J. Neurosci. 22: 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca D., Mulle J. G., Kaminsky E. B., Sanders S. J., Myers S. M., et al. , 2010. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am. J. Hum. Genet. 87: 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy S. S., Nadler J. J., Perez A., Barbaro R. P., Johns J. M., et al. , 2004. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 3: 287–302. [DOI] [PubMed] [Google Scholar]
- Moy S. S., Nadler J. J., Young N. B., Nonneman R. J., Segall S. K., et al. , 2008. Social approach and repetitive behavior in eleven inbred mouse strains. Behav. Brain Res. 191: 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithianantharajah J., Komiyama N. H., McKechanie A., Johnstone M., Blackwood D. H., et al. , 2013. Synaptic scaffold evolution generated components of vertebrate cognitive complexity. Nat. Neurosci. 16: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G., Franklin K. B. J., 2001. The mouse brain in stereotaxic coordinates, Ed. 2, Academic Press, San Diego. [Google Scholar]
- Pinto D., Pagnamenta A. T., Klei L., Anney R., Merico D., et al. , 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466: 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero-Rivera F., Sharifi-Hannauer P., Martinez-Agosto J. A., 2010. Autistic and psychiatric findings associated with the 3q29 microdeletion syndrome: case report and review. Am. J. Med. Genet. A. 152A: 2459–2467. [DOI] [PubMed] [Google Scholar]
- Raveau M., Lignon J. M., Nalesso V., Duchon A., Groner Y., et al. , 2012. The App-Runx1 region is critical for birth defects and electrocardiographic dysfunctions observed in a Down syndrome mouse model. PLoS Genet. 8: e1002724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. H., Irving N. G., Moran T. H., Wohn A., Kitt C., et al. , 1995. A mouse model for down-syndrome exhibits learning and behavior deficits. Nat. Genet. 11: 177–184. [DOI] [PubMed] [Google Scholar]
- Roberson E. D., Wohler E. S., Hoover-Fong J. E., Lisi E., Stevens E. L., et al. , 2011. Genomic analysis of partial 21q monosomies with variable phenotypes. Eur. J. Hum. Genet. 19: 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S. J., Ercan-Sencicek A. G., Hus V., Luo R., Murtha M. T., et al. , 2011. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70: 863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Sun G., Zhao C., Stewart R., 2008. Neural stem cell self-renewal. Crit. Rev. Oncol. Hematol. 65: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussan T. E., Yang A. N., Li F., Ostrowski M. C., Reeves R. H., 2008. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down’s syndrome. Nature 451: 73–75. [DOI] [PubMed] [Google Scholar]
- Takemori H., Kajimura J., Okamoto M., 2007. TORC-SIK cascade regulates CREB activity through the basic leucine zipper domain. FEBS J. 274: 3202–3209. [DOI] [PubMed] [Google Scholar]
- Takeuchi T., Ohtsuki G., Yoshida T., Fukaya M., Wainai T., et al. , 2008. Enhancement of both long-term depression induction and optokinetic response adaptation in mice lacking delphilin. PLoS ONE 3: e2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D., Fraser P., Nagano T., 2008. The role of long non-coding RNAs in chromatin structure and gene regulation: variations on a theme. Biol. Chem. 389: 323–331. [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., et al. , 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M., Keskitalo S., Van Drogen A., Nurkkala H., Vichalkovski A., et al. , 2013. The protein interaction landscape of the human CMGC kinase group. Cell Rep. 3: 1306–1320. [DOI] [PubMed] [Google Scholar]
- Wegner M., Stolt C. C., 2005. From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 28: 583–588. [DOI] [PubMed] [Google Scholar]
- Yu T., Clapcote S., Li Z., Liu C., Pao A., et al. , 2010a Deficiencies in the region syntenic to human 21q22.3 cause cognitive deficits in mice. Mamm. Genome 21: 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Li Z. Y., Jia Z. P., Clapcote S. J., Liu C. H., et al. , 2010b A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum. Mol. Genet. 19: 2780–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]