Abstract
Knowledge of the nature and extent of karyotypic differences between species provides insight into the evolutionary history of the genomes in question and, in the case of closely related species, the potential for genetic exchange between taxa. We constructed high-density genetic maps of the silverleaf sunflower (Helianthus argophyllus) and Algodones Dune sunflower (H. niveus ssp. tephrodes) genomes and compared them to a consensus map of cultivated sunflower (H. annuus) to identify chromosomal rearrangements between species. The genetic maps of H. argophyllus and H. niveus ssp. tephrodes included 17 linkage groups each and spanned 1337 and 1478 cM, respectively. Comparative analyses revealed greater divergence between H. annuus and H. niveus ssp. tephrodes (13 inverted segments, 18 translocated segments) than between H. annuus and H. argophyllus (10 inverted segments, 8 translocated segments), consistent with their known phylogenetic relationships. Marker order was conserved across much of the genome, with 83 and 64% of the H. argophyllus and H. niveus ssp. tephrodes genomes, respectively, being syntenic with H. annuus. Population genomic analyses between H. annuus and H. argophyllus, which are sympatric across a portion of the natural range of H. annuus, revealed significantly elevated genetic structure in rearranged portions of the genome, indicating that such rearrangements are associated with restricted gene flow between these two species.
Keywords: genome rearrangement, karyotypic evolution, gene flow, reproductive isolation, comparative mapping
CHROMOSOMAL rearrangements are of considerable interest because they are often associated with barriers to gene flow between related species, either due to their direct effects on the fitness of heterozygotes or through the indirect effects of genic barriers embedded within them (White 1978; Barton and Bengtsson 1986; Rieseberg et al. 1995b, 1999; Rieseberg 2001; Navarro and Barton 2003; Kirkpatrick and Barton 2006; reviewed in Faria and Navarro 2010; Gimenez et al. 2012). As such, detailed information on karyotypic differences between species provides insight into the nature of reproductive isolation and, more pragmatically, informs attempts to introgress beneficial alleles from wild species into crop gene pools (Chetelat and Meglic 2000; Foulongne et al. 2003; Dirlewanger et al. 2004). An improved understanding of synteny across species can also facilitate the identification and localization of functionally important genes in a taxon of interest through the extrapolation of gene order from model species (e.g., Choi et al. 2004; Dilbirligi et al. 2006).
The genus Helianthus, which is composed of 49 species native to the Americas (Timme et al. 2007) and includes cultivated sunflower (Helianthus annuus L.; 2n = 2x = 34; hereafter referred to as ANN), has emerged as a model for genetic studies of adaptation, hybridization, and speciation (Rieseberg et al. 1995a,b; Lai et al. 2005; Reagon and Snow 2006; Massinga et al. 2009; Gutierrez et al. 2010; Vekemans 2010; Roumet et al. 2013). Insight into the nature and extent of reproductive barriers within Helianthus will provide valuable understanding of how these species arose and will also aid in the development of strategies for the introgression of beneficial alleles from related wild species (e.g., silverleaf sunflower and the Algodones Dune sunflower) into the cultivated sunflower gene pool.
Silverleaf sunflower (H. argophyllus Torrey and Gray; 2n = 2x = 34; hereafter referred to as ARG) is the sister species to ANN. ARG is native to the sandy soils of coastal Texas where it overlaps (Supporting Information, Figure S1) with the southern portion of the native range of wild ANN, which is the progenitor of cultivated sunflower. In cultivated sunflower breeding programs, ARG has been used widely as a donor of advantageous alleles for disease resistance (Heiser 1951; Rogers et al. 1982; Gulya and Miller 1991; Slabaugh et al. 2003; Dussle et al. 2004; Radwan et al. 2004; Seiler et al. 2007; Wieckhorst et al. 2010), fertility restoration of the PET1 cytoplasm (Chepurnaya et al. 2003), and cytoplasmic male sterility (Horn et al. 2002). ARG has also been identified as a possible source of favorable alleles for salt and drought tolerance (Richards 1992) and insect resistance (Rogers and Thompson 1980; Rogers et al. 1982, 1987; Sujatha and Lakshminarayana 2007). Crosses between ANN and ARG produce vigorous offspring with reduced pollen viability (F1 = 5–50% viable, BC1 = 24–97% viable) and chromosomal abnormalities (Heiser 1951; Chandler et al. 1986; Quillet et al. 1995), resulting in restricted introgression in experimental crossing programs. Cytological studies identified meiotic abnormalities (e.g., univalents, rod bivalents, and tetravalents) in the interspecific hybrids, indicating that ANN and ARG differ by at least two reciprocal translocations (Heiser 1951; Chandler et al. 1986; Quillet et al. 1995). A recent comparative mapping study identified five nonreciprocal translocations and two inversions between ANN and ARG (Heesacker et al. 2009); however, this analysis was based on an ARG map with just 299 markers (i.e., SSRs, indels, and SSCPs), of which only 131 were orthologous to loci that had been mapped in ANN.
The Algodones Dune sunflower [H. niveus (Benth.) Brandegee ssp. tephrodes (A. Gray) Heiser; 2n = 2x = 34; hereafter referred to as NIV] is a mostly perennial, sometimes annual, xerophytic species that inhabits sandy dunes in Arizona, Baja California, and Sonora, Mexico (Figure S1) (Rogers et al. 1982; Bowers 1996). NIV is sister to H. petiolaris, with which ANN is known to hybridize in the wild. Interspecific hybrids between NIV and both ARG and ANN exhibit low pollen viability (<10%) and mispairing (i.e., univalents) during meiosis (Chandler et al. 1986). Thus far, NIV has not been used as a source of advantageous alleles for improving cultivated sunflower, though it demonstrates resistance to aphid nymphs and adults (Masonaphis masoni) (Rogers et al. 1982) and is a potential source of alleles for traits related to drought resistance (e.g., leaf pubescence, germination/establishment in a desert environment, etc.). Prior to this study, no genetic maps of this species had been developed.
Previous comparative mapping studies in sunflower have resulted in the identification of numerous rearrangements across species, though these studies have been limited by relatively low marker density (Burke et al. 2004; Lai et al. 2005; Heesacker et al. 2009). In this article, we describe the construction of the first high-density linkage maps of H. argophyllus and H. niveus ssp. tephrodes using single nucleotide polymorphisms (SNPs) derived from expressed sequence tags (ESTs). We then compare these maps to the 10,000+ locus consensus SNP map of cultivated sunflower (Bowers et al. 2012) to produce a detailed picture of synteny among ANN, ARG, and NIV. Finally, we describe the results of a population genomic analysis of ANN and ARG to determine the extent to which observed chromosomal rearrangements are associated with restricted interspecific gene flow between these species.
Materials and Methods
Mapping populations
Intraspecific F1 hybrids of ARG and NIV were produced by crossing individuals from two different accessions of each species with each other. A single individual of ARG1820 (PI 494580) was crossed with a single individual of ARG1834 (PI 494582) and a single individual of NIV58 (PI 613758) was crossed with a single individual of NIV20 (PI 650020). The goal was to produce a highly heterozygous individual of each species that could be used in a pseudo-testcross mapping design (Grattapaglia and Sederoff 1994; Burke et al. 2004). Pollen from a randomly selected intraspecific F1 from each species was then used to pollinate a nuclear male-sterile ANN inbred line (NMS373; PI 597362) to produce ARG × ANN and NIV × ANN interspecific mapping populations. This allowed the segregation of alleles from the ARG or NIV intraspecific hybrids to be tracked against a mostly homozygous ANN background.
DNA extraction and genotyping
DNA was isolated from leaves of 94 F1 seedlings of each interspecific mapping population and genotyped using a custom array designed to target ∼10,000 cultivated sunflower SNPs following established methods (Bachlava et al. 2012). This array was previously used to produce a high-density consensus map of the ANN genome based on multiple crosses (Bowers et al. 2012).
Genetic mapping
The genetic maps were initially constructed manually using spreadsheet software to sequentially sort genotypes and cluster linked loci to minimize the number of apparent recombination events (Bowers et al. 2012). Map order was then verified using MapDisto v. 1.75 (Lorieux 2007, 2012) using the group, order, and ripple commands. When combined with the biallelic nature of all markers, the use of a pseudo-testcross mapping design meant that each heterozygous locus in ARG or NIV randomly shared either its maternal (ARG1834 or NIV58) or paternal (ARG1820 or NIV20) allele with the ANN mapping parent. Thus, prior to map construction, all loci in the dataset were duplicated and recoded (i.e., SNPs scored as heterozygous were recoded homozygous and vice versa). The resulting genetic maps consisted of 34 linkage groups (LGs) with 17 pairs of “mirror image” linkage groups that contained identical sets of loci separated by the same map distances, but with reversed genotype scores. One linkage group from each of these pairs was retained for inclusion in the final ARG and NIV maps, each containing 17 linkage groups. The numbering and orientation of the ARG and NIV linkage groups followed the standard nomenclature developed for ANN (Tang et al. 2002; Bowers et al. 2012). ARG and NIV LGs with translocated segments relative to ANN were labeled according to the ANN LGs that were involved (e.g., ARG6/15 consisted of portions of LGs 6 and 15 from the ANN map; NIV17/16/12 consisted of portions of LGs 17, 16, and 12 from the ANN map; see below for details).
Synteny assessment
Homologous linkage groups were plotted in MapChart v. 2.2 (Voorrips 2002) and synteny was assessed using two datasets: an overall subset of 295 SNPs mapped in all three species and species-pair subsets consisting of all homologous markers mapped in ANN and ARG (n = 1455 markers), ANN and NIV (n = 1058 markers), and ARG and NIV (n = 318 markers) (Figure S2). Map coverage using the overall subset of 295 SNPs mapped in all three species was estimated for ARG and NIV after subtracting gaps of >20 cM, as well as gaps at the end of LGs (Wu et al. 2010). For the sake of comparison, the ANN genome was used as a standard reference and structural rearrangements were identified vs. this reference. This should not, however, be taken to imply that the ANN orders are necessarily ancestral. A conserved block of synteny was defined as two or more independent markers (ignoring those without a known homologous position in the ANN genome) present as uninterrupted strings of collinear loci. Structural rearrangements (i.e., inverted and/or translocated segments) were defined using a modified version of the guidelines used by Wu et al. (2009a,b, 2010). Inverted segments were identified as two or more independent markers that exhibited reversed ordering between species; inverted segments at the terminal ends of LGs required the misordering of at least one terminal marker and two or more internal markers. Because establishing the correct map position of tightly linked markers in high-density linkage maps can be difficult due to duplicated loci, genotyping errors, segregation distortion, and chiasma interference (Hackett and Broadfoot 2003; Ferreira et al. 2006; Cheema and Dicks 2009; Collard et al. 2009), minor ordering errors can arise. As such, a 2-cM threshold was applied for declaring noncollinearity (Hudson et al. 2011). Thus, markers were only considered noncollinear when a shift in marker order and position exceeded 2 cM in both maps. Translocated segments were identified as regions with two or more independent markers assigned to the “wrong” LG relative to the ANN consensus map; single, terminal, nonsyntenic markers were not considered to be translocated segments. Synteny was assumed in regions of conflicting data and markers violating this assumption were identified and locus names were printed in bold and underlined. The lengths of inverted and/or translocated segments were defined as the distance between the first and last loci within the block based on the ARG or NIV map, respectively. The percentages of the ARG and NIV genomes that were syntenic, inverted, or translocated relative to ANN were calculated by summing all of the individual segments that were assigned to each of these categories and dividing these values by the total length of the ARG or NIV map, as appropriate.
Population genomic analyses
Genome-wide estimates of population genetic divergence between ANN and ARG were based on previously generated transcriptome resequencing data derived from 40 and 28 individuals of these species, respectively (see Renaut et al. 2013 for details). Briefly, the raw transcriptome data (produced using either the Roche 454 FLX or Illumina GAII platforms) were aligned against a reference transcriptome of 51,468 contigs using the Burrows–Wheeler aligner (BWA) (Li and Durbin 2009). SAMtools (Li et al. 2009) was then used to call SNPs. Genotypes with Phred-scaled genotype likelihoods <30, which correspond to a minimum genotyping accuracy of 99.9%, were considered as missing. Questionable SNPs were removed due to poor sequence quality, low coverage, potential sequencing errors, and paralogy. The data were also filtered to remove SNPs with low expected heterozygosity (i.e., He < 0.20) because, due to the sample sizes employed here, they may represent sequencing errors. Likewise, SNPs with very high observed heterozygosity (i.e., Ho > 0.60) were removed because they likely result from paralogous sequence variants. From this curated dataset, FST (Weir and Cockerham 1984) and Jost’s (2008) D were both calculated (Noor and Bennett 2009; Meirmans and Hendrick 2011) for each marker, using the package HIERFSTAT (FST, Goudet 2005) and diveRsity (D, Keenan et al. 2013) in the programming language R (R Development Core Team 2012). BLASTN was then used to assign nearly half of the transcriptome contigs (24,406 of 51,468 total) to 3047 unique genomic map locations on a sequence-based genetic map of the H. annuus genome (Renaut et al. 2013), and FST and D for each map position were calculated by averaging the values for all markers within ±0.5 cM of the position of interest. In cases where diversity and differentiation are high, conclusions regarding population differentiation may be wrong when using measures of differentiation such as FST, which are based on additive between-group heterozygosity (Jost 2008). While FST is proportional to the variance of allele frequency among populations, Jost (2008) introduced another measure of differentiation, D, which indicates the proportion of allelic diversity that lies among populations. Therefore, D is a genetic distance measure more related to the distance between populations than to the variance in allele frequencies (Whitlock 2011).
The effect of chromosomal rearrangements on interspecific gene flow was investigated by comparing the extent of population structure (i.e., the magnitude of FST and D) for the rearranged vs. nonrearranged portions of the genome. For this analysis, we identified the 12 most well-supported rearrangements from the ANN vs. ARG comparison (i.e., those that were supported by three or more markers), including seven inverted segments and five translocated segments, and compared their average FST and D values against the balance of the genome. FST and D values were also calculated for the 5-cM regions adjacent to the breakpoints separating the rearranged and nonrearranged segments.
Results
Genetic linkage maps
The ARG genetic map (Figure 1 and Figure S3) consisted of 1626 EST-SNP markers and 17 LGs covering 1337 cM (Table S1). This represents an increase of >1300 loci relative to the previous ARG map constructed by Heesacker et al. (2009) and a decrease from 21 to 17 LGs (i.e., the haploid chromosome number of ARG). The average distance between markers (excluding colocalizing markers) was 2.4 cM with a maximum gap of 45.7 cM (ARG10). The ARG map consisted of 567 unique marker locations with 285 (50%) of these positions having two or more colocalized markers per position [average 4.8, maximum (max) 98] (Table S2). The NIV genetic map (Figure 1 and Figure S3) consisted of 1194 markers, 17 LGs, and spanned 1478 cM (Table S1). The average intermarker distance was 2.7 cM with a maximum gap of 22.7 cM on NIV9. The NIV map showed similar levels of marker colocalization with 562 unique marker locations with 249 (44%) of these positions having two or more colocalized markers per position (average 3.6, max 26) (Table S2).
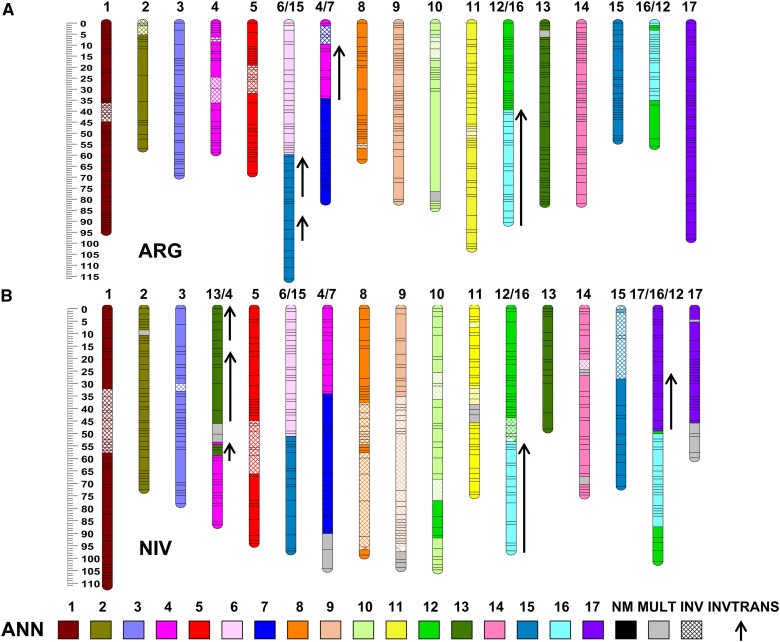
Figure 1.
Genetic linkage maps of Helianthus argophyllus (ARG) and H. niveus ssp. tephrodes (NIV). ARG and NIV linkage groups (LGs) are labeled and color coded based on macrosynteny with H. annuus (ANN) chromosomes ANN1–17 (Bowers et al. 2012). Gray segments contain markers that are mapped to multiple ANN LGs not including that particular ARG or NIV LG; cross-hatching indicates a region that is inverted relative to ANN; black arrows indicate translocated segments that are also inverted relative to ANN. The scale on the left is in centimorgans (cM). See Figure S2 for more detail.
Synteny estimates
Synteny between ANN, ARG, and NIV for both the species-pair sets of markers (ANN/ARG, ANN/NIV, and ARG/NIV) and the overall set of 295 homologous markers mapped in all three species is presented in Figure 2, Figure S4, and Figure S5. Segments that were noncollinear between species were classified as inverted and segments that were nonsyntenic were classified as translocated (Table S3). Note that these designations were made with reference to the ANN consensus map for consistency with existing chromosomal nomenclature and should not be interpreted as indicating ancestral vs. derived states. The ability to identify rearranged regions is dictated by marker resolution, which is defined by both the number and distribution of shared markers. The smallest inverted and/or translocated segment that was detected was 1.1 cM and 2.1 cM, respectively, for the ARG and NIV maps. The average size was 11.1 cM and 16.7 cM (Table S4). Approximately 70% of both maps had adequate marker resolution to detect rearranged segments of >6 cM and ∼20% of the maps had marker resolution to detect rearrangements of <2 cM (Figure S6 and Figure S7).
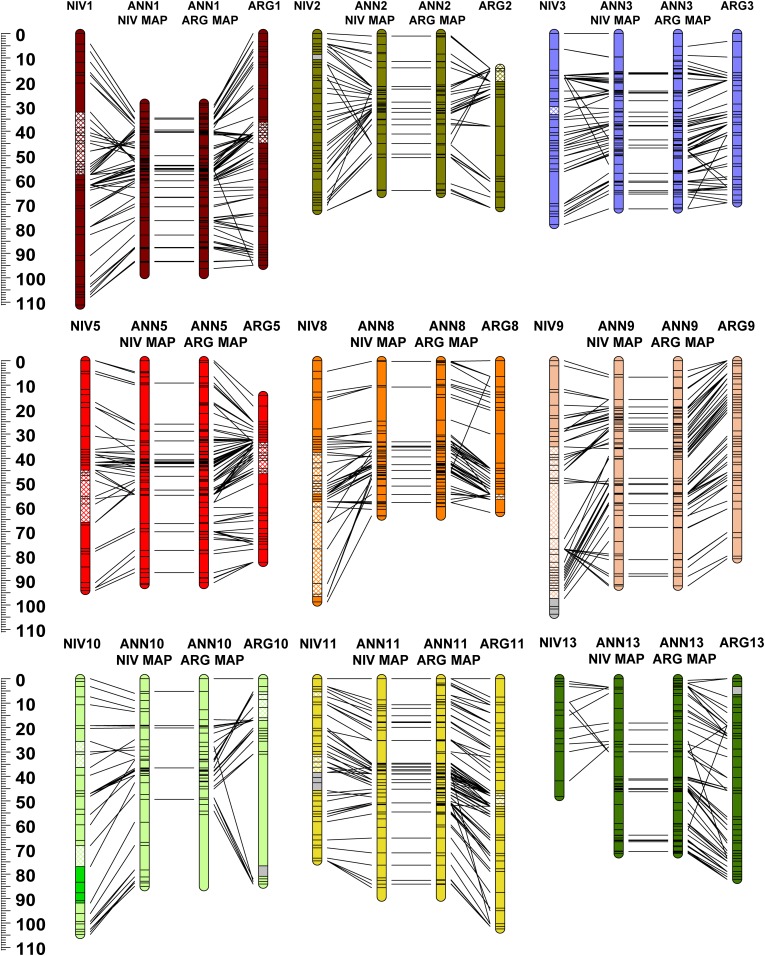
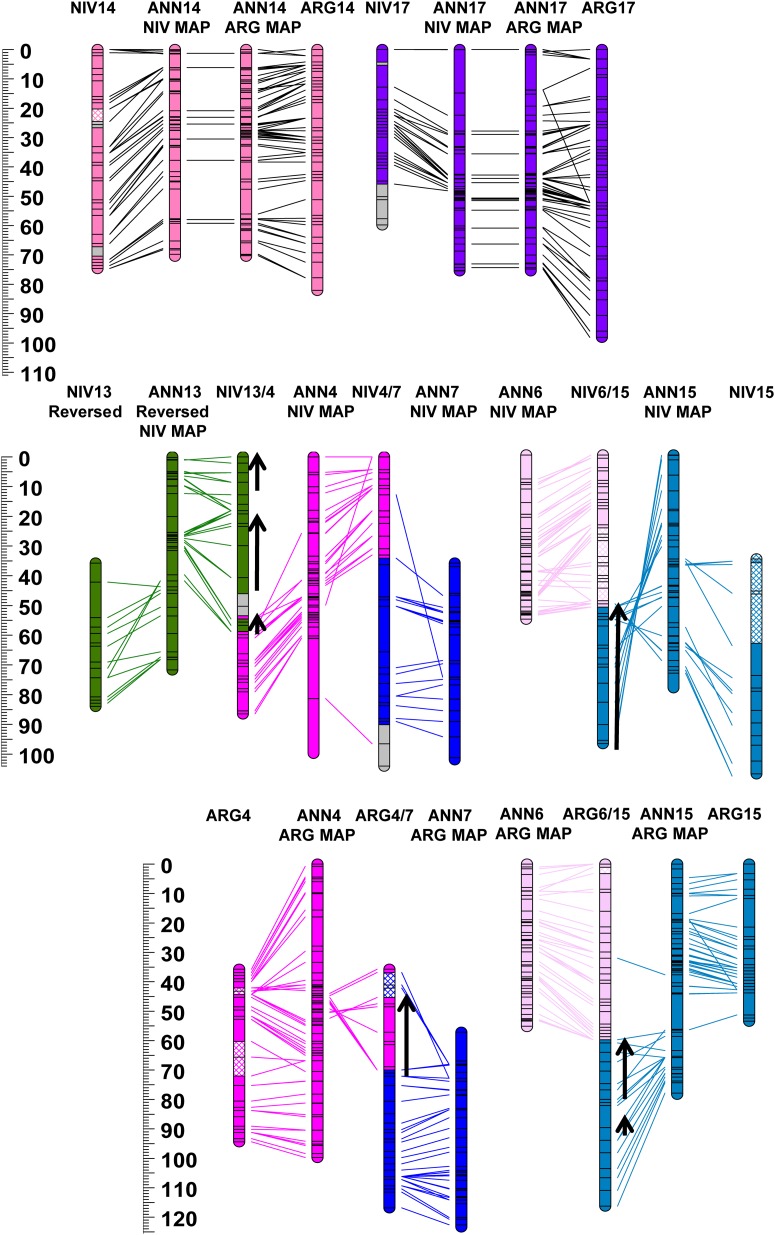
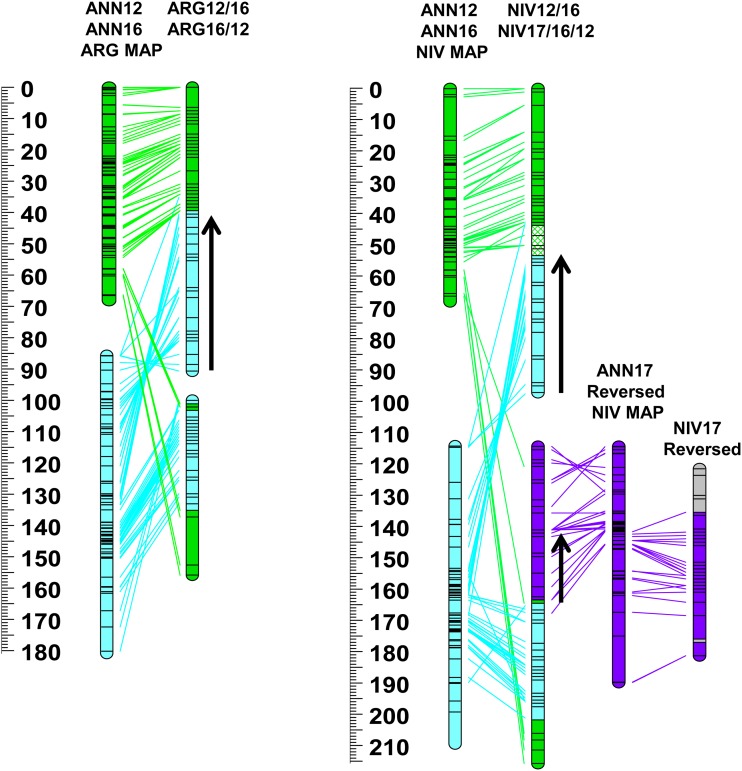
Figure 2.
Genetic maps of Helianthus argophyllus (ARG) and H. niveus ssp. tephrodes (NIV) compared to a consensus map of H. annuus (ANN) from Bowers et al. 2012. Color coding and chromosome nomenclature follow Figure 1. Homologous markers are connected by lines. Only ANN markers mapped in ARG or NIV are included. See Figure S3 and Figure S4 for more detail.
ANN vs. ARG:
The ANN/ARG species-pair markers (n = 1455) revealed the presence of 12 largely syntenic LGs (1–3, 5, 8–11, 13–15, and 17), 10 inverted segments, and eight translocated segments (Table S1 and Table S3). On a genome-wide basis (Figure S8), 83% of the ARG map was syntenic with the ANN consensus map, 12% was translocated, and 5% inverted. Four major translocated segments were identified: two nonreciprocal translocated segments involving LGs ARG4/7 and ARG6/15 and one reciprocal translocated segment involving LGs ARG12/16 and ARG16/12. Linkage group ARG4/7 was composed of a segment of the proximal portion of ANN4 inserted as two pieces into the proximal region of ANN7. The translocated segment of LG4 spanned 6 cM in ANN and 21 cM in ARG. Linkage group ARG6/15 consisted of ANN6 and the distal end of ANN15. Linkage groups ARG12/16 and ARG16/12 formed a reciprocally translocated LG of ANN12 and ANN16. Inverted segments were identified on several LGs (1, 2, 4, 5, and 8–11).
The ANN/ARG synteny estimate based on the subset of 295 markers mapped in all three species was slightly higher (89 vs. 83%) with only three inverted segments and three translocated segments (Figure S5, Figure S8, and Figure S9). The lower number of rearranged segments estimated using this marker subset was likely due to the reduced marker density (1626 markers vs. 295 markers) and map coverage (only 62%), with limited coverage (<25%) on LGs 4, 4/7, 10, and 15 (Table S5).
ANN vs. NIV:
The full set of homologous EST-SNP markers (n = 1058) mapped in NIV and ANN showed the presence of 10 largely syntenic LGs (1, 2, 5, 8–11, and 13–15), 13 inverted segments, and 18 translocated segments (Table S1). On a genome-wide basis (Figure S8), 64% of the NIV map was syntenic with the ANN consensus map (vs. 83% for ARG), 19% was translocated, and 17% inverted. The same four major translocated LGs identified in ARG (i.e., ARG4/7, ARG6/15, ARG12/16, and ARG16/12) were also identified in NIV (i.e., NIV4/7, NIV6/15, NIV12/16, and NIV17/16/12), in addition to a nonreciprocal translocated segment of the distal end of ANN13 to the proximal end of ANN4 forming LG NIV13/4. Similar to ARG4/7, NIV4/7 also contained a translocated segment of the proximal portion of ANN4 inserted as a single piece (vs. two pieces in ARG) into the proximal region of ANN7. Interestingly, NIV6/15 was composed of ANN6 and the inverted proximal end of ANN15, whereas, in ARG, this translocated segment involved the opposite (distal) end of ANN15 (Figure 2, Figure S4, and Figure S5). NIV12/16 and NIV17/16/12 formed a reciprocally translocated LG of ANN12 and ANN16 much like in ARG; however, in NIV the distal end of ANN17 was also translocated to the proximal end of one of the NIV reciprocal LGs forming NIV17/16/12. Additionally, a small translocated segment of ANN16 inserted into the distal end of NIV10 was also identified, as well as a number of other small, translocated regions containing markers from multiple ANN LGs on NIV LGs 2, 13/4, 4/7, 9, 11, 14, and 17. Inverted segments relative to ANN were identified on NIV LGs 1, 3, 13/4, 5, 8–11, and 14 with large inverted segments on NIV LGs 8 and 9 covering >50 cM.
The ANN/NIV synteny estimate based on the subset of 295 markers mapped in all three species was 69%, with five inverted segments and 10 translocated segments (Figure S5, Figure S8, and Figure S9). Map coverage in NIV using this marker subset was better (74 vs. 62%) than in ARG, with poor coverage limited only to NIV10 (7%) (Table S5).
ARG vs. NIV:
ARG and NIV were mostly syntenic (71–75%) to each other with minor inverted or translocated segments on LGs 1–3, 5, 8, 11, and 13, and a major inverted segment on LG 9 (Figure S10). As mentioned previously, ARG and NIV appear to share a number of translocated LGs relative to ANN, and within these rearranged segments synteny was mostly conserved (e.g., proximal portion of 6/15, 12/16 and distal portions of ARG16/12 and NIV17/16/12). In general, marker coverage was adequate to evaluate the synteny between ARG and NIV except for LG 10, which only shared two homologous markers with the remaining markers (four in ARG and three in NIV) mapping to other LGs (5, 8, and 12/16 in ARG and 2, 5, and 6/15 in NIV).
Population genomic divergence
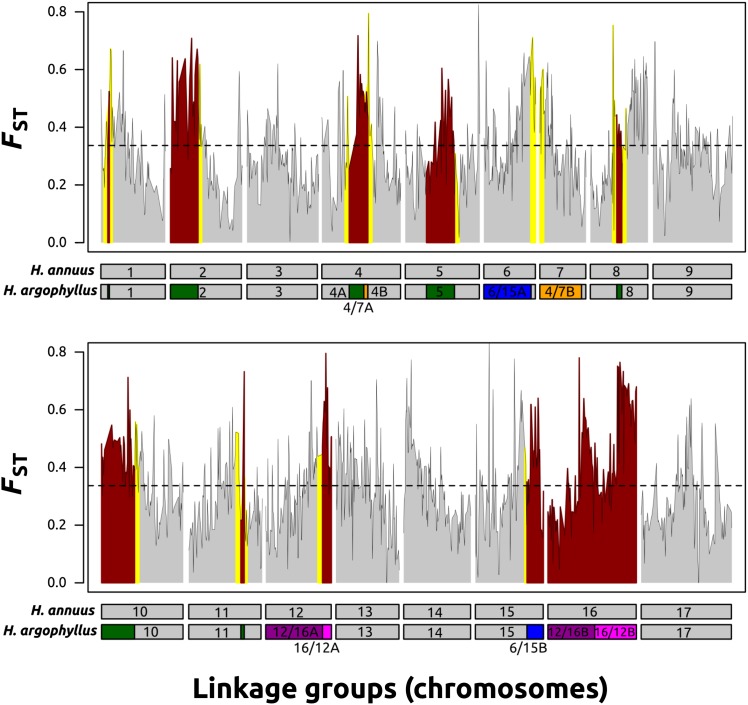
We identified 205,372 SNPs between ANN and ARG based on our strict quality controls. From this curated dataset, the genome-wide average FST between ANN and ARG was 0.34 ± 0.14 (mean ± SD), with an average of 0.43 ± 0.14 across the most well-supported rearrangements (0.45 ± 0.12 for the inverted segments, 0.42 ± 0.15 for the translocated segments) vs. 0.31 ± 0.13 for the balance of the genome (see Figure 3 for a visual summary of these results). The distribution and values of D were well correlated with the observed FST values (Figure S11 and Figure S12). The genome-wide average D between ANN and ARG was 0.26 ± 0.13 (mean ± SD) with an average of 0.36 ± 0.14 across the most well-supported rearrangements (0.36 ± 0.12 for the inverted segments and 0.35 ± 0.14 for the translocated segments) vs. 0.24 ± 0.12 for the balance of the genome (see Figure S11 for a visual summary of these results). Based on a randomization test where FST (or D) statistics were randomly assigned genomic map positions, and the average values for the rearranged vs. nonrearranged portions of the genome were recalculated (this was done 1,000,000 times for both FST and D), the observed differences were highly significant (P < 0.0001). Similar values (P < 0.0001) were obtained using a nonparametric Wilcoxon rank sum test comparing rearranged vs. nonrearranged portions of the genome for FST (or D) statistics. FST and D for the regions within 5 cM of the breakpoints between the rearranged and nonrearranged segments (0.39 ± 0.15 and 0.31 ± 0.14, respectively) were slightly less than the values observed for the whole rearranged regions, yet they were still significantly (P < 0.0001; Figure 3 and Figure S11) elevated relative to the nonrearranged regions.
Figure 3.
Genome-wide divergence (FST) between Helianthus annuus (ANN) and H. argophyllus (ARG) in syntenic and rearranged regions of the genome. Syntenic portions of the genome are colored in gray, rearranged portions are in red, and the 5-cM regions neighboring the rearrangements are in yellow. The horizontal dashed line represents mean genome-wide FST. Linkage groups in ANN and their corresponding location in ARG are pictured below the x-axis. Inverted chromosomal segments are pictured in green, while translocated segments in ARG are in blue, orange, light purple, or dark purple.
Discussion
The relative levels of synteny observed in this study accord well with the known phylogenetic relationships among these species, with ARG being sister to ANN, and NIV being somewhat more distantly related (Timme et al. 2007). In several cases, ARG and NIV possessed similar rearrangements (e.g., inverted segments on LGs 1 and 5 and the 12/16 reciprocally translocated LGs) relative to ANN, indicating that the ANN configuration, upon which the standard linkage group nomenclature is based (Tang et al. 2002; Yu et al. 2003; Bowers et al. 2012) may be a derived condition. Interestingly, ARG and NIV both exhibited translocated segments involving LGs 6 and 15. However, these translocated segments involved opposite ends of LG 15 (in opposite orientations). This result suggests that the central portion of LG 15 may be predisposed (or favored by adaptation to similar environmental conditions) to translocation onto LG 6.
Given that ANN and ARG differ by as many as 10 inverted segments and 8 translocated segments and diverged from one another ∼1.5 MYA [90% HPD (highest posterior density) range = 1.2–2.0 MYA; J. L. Strasburg and L. H. Rieseberg, unpublished data], the estimated rate of karyotypic evolution (K) between these species ranges from 4.5 to 7.5 chromosomal rearrangements/MY. Similarly, ANN and NIV differ by as many as 13 inverted segments and 18 translocated segments and are diverged from one another ∼1.8 MYA (90% HPD range = 1.5–2.1 MYA based on the fact that NIV is sister to H. petiolaris (Sambatti et al. 2012). As such, the estimated rate of karyotypic evolution (K) between these species ranges from 7.4 to 10.3 rearrangements/MY. The fact that these values are considerably higher than previous estimates for ANN vs. ARG (7 rearrangements total, including 5 translocation and two inversions; Heesacker et al. 2009) and for ANN vs. H. petiolaris (11 rearrangements total, including 8 translocations and three inversions; Burke et al. 2004), is likely due to the much higher marker density in the present study, which improved our ability to detect rearrangements. We recognize that even with the marker resolution achieved in this study, we are likely missing smaller rearrangements, which will only be resolved once the full genome (or physical maps) become available. Regardless, the overall rate of occurrence of large-scale chromosomal rearrangements in Helianthus appears to be quite high.
In terms of the impact of these rearrangements on genetic exchange, FST and D values were clearly and significantly elevated within the rearrangements vs. elsewhere in the genome. This result stands in contrast to an earlier study, based on many fewer loci, that found evidence of increased divergence between ANN and H. petiolaris in the vicinity of chromosomal breakpoints, but not within rearranged regions overall (Strasburg et al. 2009). In fact, the effects documented in the present study extended into regions bordering the rearrangements, with collinear regions within 5 cM of chromosomal breakpoints also exhibiting significantly elevated FST and D values. Our results are thus consistent with the view that rearrangements effectively suppress genetic exchange between chromosomally differentiated taxa, though the root cause of this effect remains unclear. It has long been argued that chromosomal rearrangements act to limit gene flow through their underdominant effects on fitness (White 1978; Barton and Bengtsson 1986; King 1995; Levin 2002), though this view faces significant theoretical challenges (Rieseberg 2001; reviewed in Faria and Navarro 2010). Most notably, rearrangements must be strongly underdominant to effectively reduce gene flow, but strongly underdominant rearrangements are difficult to fix through drift except in small, inbred populations (Walsh 1982; Lande 1985). In contrast, weakly underdominant rearrangements can go to fixation more easily via drift, but are expected to have minimal effects on gene flow. In this case, however, the drift-based establishment of even weakly underdominant rearrangements is improbable due to the occurrence of sporophytic self-incompatibility (resulting in obligate outcrossing) and large effective population sizes in wild Helianthus species (Strasburg et al. 2011).
An alternative view that has gained prominence in recent years is that the primary effect of rearrangements is to limit recombination, thereby allowing adaptive differences to accrue within rearrangements, which then extends the effects of genic barriers to gene flow across larger genomic regions (Rieseberg 2001; reviewed in Jackson 2011). In fact, Kirkpatrick and Barton (2006) provided theoretical evidence that adaptive differentiation can drive the fixation of chromosomal differences by suppressing recombination between loci carrying locally favorable alleles. In this context, it is worth noting that ARG and ANN exhibit major adaptive differences. ARG flowers in late summer and exhibits a strong preference for the sandy, coastal soils in its native range in south Texas, as well as Florida, where it is thought to be a naturalized weed (Heiser 1951). This species also has densely pubescent leaves covered with long, silky hairs, exhibits a woody growth habit under certain environmental conditions, and is tolerant of drought conditions and saline soils (Richards 1992; Baldini and Vannozzi 1999). In contrast, wild ANN flowers much earlier in the summer, has less pubescent leaves that are green in appearance, is rarely found in sandy soils, and is typically salt sensitive (Welch and Rieseberg 2002).
Unfortunately, we know relatively little about the history of contact between these species. Moreover, the genetic architecture of the aforementioned trait differences remains largely unexplored in wild Helianthus species. As such, a more complete understanding of the causes and effects of the rearrangements identified herein thus awaits further study.
Supplementary Material
Acknowledgments
This project was supported in part by the National Research Initiative of the US Department of Agriculture’s National Institute of Food and Agriculture (2008-35300-04579 and 2008-35300-19263) and the National Science Foundation’s Plant Genome Research Program (DBI-0820451).
Footnotes
Communicating editor: N. H. Barton
Literature Cited
- Bachlava E., Taylor C. A., Tang S., Bowers J. E., Mandel J. R., et al. , 2012. SNP discovery and development of a high-density genotyping array for sunflower. PLoS ONE 7: e29814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini M., Vannozzi G. P., 1999. Yield relationships under drought in sunflower genotypes obtained from a wild population and cultivated sunflowers in rain-out shelter in large pots and field experiments. Helia 22: 81–96. [Google Scholar]
- Barton N., Bengtsson B. O., 1986. The barrier to genetic exchange between hybridising populations. Heredity 56: 357–376. [DOI] [PubMed] [Google Scholar]
- Bowers J. E., 1996. Seedling emergence on Sonoran Desert dunes. J. Arid Environ. 33: 63–72. [Google Scholar]
- Bowers J. E., Bachlava E., Brunick R. L., Rieseberg L. H., Knapp S. J., et al. , 2012. Development of a 10,000 locus genetic map of the sunflower genome based on multiple crosses. G3 (Bethesda) 2: 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Lai Z., Salmaso M., Nakazato T., Tang S., et al. , 2004. Comparative mapping and rapid karyotypic evolution in the genus Helianthus. Genetics 167: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J., Jan C., Beard B., 1986. Chromosomal differentiation among the annual Helianthus species. Syst. Bot. 11: 354–371. [Google Scholar]
- Cheema J., Dicks J., 2009. Computational approaches and software tools for genetic linkage map estimation in plants. Briefings Bioinf. 10: 595–608. [DOI] [PubMed] [Google Scholar]
- Chepurnaya A. L., Sherstyuk S. V., Tikhomirov V. T., 2003. CMS-Rf system for sunflower breeding. Helia 26: 59–65. [Google Scholar]
- Chetelat R., Meglic V., 2000. Molecular mapping of chromosome segments introgressed from Solanum lycopersicoides into cultivated tomato (Lycopersicon esculentum). Theor. Appl. Genet. 100: 232–241. [Google Scholar]
- Choi H. K., Mun J. H., Kim D. J., Zhu H., Baek J. M., et al. , 2004. Estimating genome conservation between crop and model legume species. Proc. Natl. Acad. Sci. USA 101: 15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard B., Mace E., McPhail M., Wenzl P., Cakir M., et al. , 2009. How accurate are the marker orders in crop linkage maps generated from large marker datasets? Crop Pasture Sci. 60: 362–372. [Google Scholar]
- Dilbirligi M., Erayman M., Campbell B. T., Randhawa H. S., Baenziger P. S., et al. , 2006. High-density mapping and comparative analysis of agronomically important traits on wheat chromosome 3A. Genomics 88: 74–87. [DOI] [PubMed] [Google Scholar]
- Dirlewanger E., Graziano E., Joobeur T., Garriga-Calderé F., Cosson P., et al. , 2004. Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc. Natl. Acad. Sci. USA 101: 9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussle C., Hahn V., Knapp S., Bauer E., 2004. Pl Arg from Helianthus argophyllus is unlinked to other known downy mildew resistance genes in sunflower. Theor. Appl. Genet. 109: 1083–1086. [DOI] [PubMed] [Google Scholar]
- Faria R., Navarro A., 2010. Chromosomal speciation revisted: rearranging theory with pieces of evidence. Trends Ecol. Evol. 25: 660–669. [DOI] [PubMed] [Google Scholar]
- Ferreira A., da Silva M. F., Silva L. C., Cruz C. D., 2006. Estimating the effects of population size and type on the accuracy of genetic maps. Genet. Mol. Biol. 29: 187–192. [Google Scholar]
- Foulongne M., Pascal T., Arús P., Kervella J., 2003. The potential of Prunus davidiana for introgression into peach [Prunus persica (L.) Batsch] assessed by comparative mapping. Theor. Appl. Genet. 107: 227–238. [DOI] [PubMed] [Google Scholar]
- Gimenez M. D., White T. A., Hauffe H. C., Panithanarak T., Searle J. B., 2012. Understanding the basis of diminished gene flow between hybridizing chromosome races of the house mouse. Evolution 67: 1446–1462. [DOI] [PubMed] [Google Scholar]
- Goudet J., 2005. HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5: 184–186. [Google Scholar]
- Grattapaglia D., Sederoff R., 1994. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137: 1121–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulya T., Miller J., 1991. Inheritance of resistance to race 4 of downy mildew derived from interspecific crosses in sunflower. Crop Sci. 31: 40–43. [Google Scholar]
- Gutierrez A., Carrera A., Basualdo J., Rodriguez R., Cantamutto M., et al. , 2010. Gene flow between cultivated sunflower and Helianthus petiolaris (Asteraceae). Euphytica 172: 67–76. [Google Scholar]
- Hackett C., Broadfoot L., 2003. Effects of genotyping errors, missing values and segregation distortion in molecular marker data on the construction of linkage maps. Heredity 90: 33–38. [DOI] [PubMed] [Google Scholar]
- Heesacker A. F., Bachlava E., Burke J. M., Brunick R. L., Rieseberg L. H., et al. , 2009. Karyotypic evolution of the common and silverleaf sunflower genomes. Plant Genome 2: 233–246. [Google Scholar]
- Heiser C. B., Jr, 1951. Hybridization in the annual sunflowers: Helianthus annuus x H. argophyllus. Am. Nat. 85: 65–72. [Google Scholar]
- Horn R., Kusterer B., Lazarescu E., Prüfe M., Özdemir N., et al. , 2002. Molecular diversity of CMS sources and fertility restoration in the genus: Helianthus. Helia 25: 29–40. [Google Scholar]
- Hudson C. J., Kullan A. R. K., Freeman J. S., Faria D. A., Grattapaglia D., et al. , 2011. High synteny and colinearity among Eucalyptus genomes revealed by high-density comparative genetic mapping. Tree Genet. Genomes 8: 339–352. [Google Scholar]
- Jackson B. C., 2011. Recombination-supression: How many mechanisms for chromosomal speciation? Genetica 139: 393–402. [DOI] [PubMed] [Google Scholar]
- Jost L., 2008. GST and its relatives do not measure differentiation. Mol. Ecol. 17: 4015–4026. [DOI] [PubMed] [Google Scholar]
- Keenan K., McGinnity P., Cross T. F., Crozier W. W., Prodöhl P. A., 2013. diveRsity: an R package for the estimation of population genetics parameters and their associated errors. Methods Ecol. Evol. 10.1111/2041-210X.12067. [DOI] [Google Scholar]
- King M., 1995. Species Evolution: The Role of Chromosome Change, Cambridge University Press, Boston. [Google Scholar]
- Kirkpatrick M., Barton N., 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Nakazato T., Salmaso M., Burke J. M., Tang S., et al. , 2005. Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics 171: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., 1985. The fixation of chromosomal rearrangements in a subdivided population with local extinction and colonization. Heredity 54: 323–332. [DOI] [PubMed] [Google Scholar]
- Levin D. A., 2002. The Role of Chromosomal Change in Plant Evolution, Oxford University Press, New York. [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux, M., 2007 MapDisto, a free user-friendly program for computing genetic maps, computer demonstration. Plant and Animal Genome XV Conference. San Diego. [Google Scholar]
- Lorieux M., 2012. MapDisto: fast and efficient computation of genetic linkage maps. Mol. Breed. 30: 1231–1235. [Google Scholar]
- Massinga R. A., Al-Khatib K., Amand P. S., Miller J. F., 2009. Gene flow from imidazolinone-resistant domesticated sunflower to wild relatives. Weed Sci. 51: 854–862. [Google Scholar]
- Meirmans P. G., Hendrick P. W., 2011. Assessing population structure: FST and related measures. Mol. Ecol. Resour. 11: 5–18. [DOI] [PubMed] [Google Scholar]
- Navarro A., Barton N. H., 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57: 447–459. [DOI] [PubMed] [Google Scholar]
- Noor M. A. F., Bennett S. M., 2009. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity 103: 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillet M., Madjidian N., Griveau Y., Serieys H., Tersac M., et al. , 1995. Mapping genetic factors controlling pollen viability in an interspecific cross in Helianthus sect. Helianthus. Theor. Appl. Genet. 91: 1195–1202. [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2012 R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna. Open access available at: http://cran.r-project. org.
- Radwan O., Bouzidi M., Nicolas P., Mouzeyar S., 2004. Development of PCR markers for the Pl5/Pl8 locus for resistance to Plasmopara halstedii in sunflower, Helianthus annuus L. from complete CC-NBS-LRR sequences. Theor. Appl. Genet. 109: 176–185. [DOI] [PubMed] [Google Scholar]
- Reagon M., Snow A. A., 2006. Cultivated Helianthus annuus (Asteraceae) volunteers as a genetic “bridge” to weedy sunflower populations in North America. Am. J. Bot. 93: 127–133. [Google Scholar]
- Renaut S., Grassa C., Yeaman S., Moyers B., Lai Z., et al. , 2013. Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nature Commun. 4: 1827. [DOI] [PubMed] [Google Scholar]
- Richards R., 1992. Increasing salinity tolerance of grain crops: Is it worthwhile? Plant Soil 146: 89–98. [Google Scholar]
- Rieseberg L. H., 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16: 351–358. [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H., Desrochers A. M., Youn S. J., 1995a Interspecific pollen competition as a reproductive barrier between sympatric species of Helianthus (Asteraceae). Am. J. Bot. 82: 515–519. [Google Scholar]
- Rieseberg L. H., Linder C. R., Seiler G. J., 1995b Chromosomal and genic barriers to introgression in Helianthus. Genetics 141: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Whitton J., Gardner K., 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152: 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C., Gershenzon J., Ohno N., Mabry T., Stipanovic R., et al. , 1987. Terpenes of wild sunflowers (Helianthus): an effective mechanism against seed predation by larvae of the sunflower moth, Homoeosoma electellum (Lepidoptera: Pyralidae). Environ. Entomol. 16: 586–592. [Google Scholar]
- Rogers C. E., Thompson T., 1980. Helianthus resistance to the sunflower beetle (Coleoptera: Chrysomelidae). J. Kans. Entomol. Soc. 53: 727–730. [Google Scholar]
- Rogers C. E., Thompson T. E., Seiler G. J., 1982. Sunflower Species of the United States. National Sunflower Association, Fargo, ND. [Google Scholar]
- Roumet M., Noilhan C., Latreille M., David J., Muller M. H., 2013. How to escape from crop-to-weed gene flow: phenological variation and isolation-by-time within weedy sunflower populations. New Phytol. 197: 642–654. [DOI] [PubMed] [Google Scholar]
- Sambatti J., Strasburg J. L., Ortiz-Barrientos D., Baack E. J., Rieseberg L. H., 2012. Reconciling extremely strong barriers with high levels of gene exchange in annual sunflowers. Evolution 66: 1459–1473. [DOI] [PubMed] [Google Scholar]
- Seiler, G., T. Gulya, and L. Marek, 2007 Re-collection of Helianthus argophyllus, source of the P1Arg gene for downy mildew resistance, surviving for 25 years on Daytona Beach, Florida. 29th Sunflower Research Workshop, Fargo, ND. [Google Scholar]
- Slabaugh M. B., Yu J. K., Tang S., Heesacker A., Hu X., et al. , 2003. Haplotyping and mapping a large cluster of downy mildew resistance gene candidates in sunflower using multilocus intron fragment length polymorphisms. Plant Biotechnol. J. 1: 167–185. [DOI] [PubMed] [Google Scholar]
- Strasburg J. L., Scotti-Saintagne C., Scotti I., Lai Z., Rieseberg L. H., 2009. Genomic patterns of adaptive divergence between chromosomally differentiated sunflower species. Mol. Biol. Evol. 26: 1341–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasburg J. L., Kane N. C., Raduski A. R., Bonin A., Michelmore R., et al. , 2011. Effective population size is positively correlated with levels of adaptive divergence among annual sunflowers. Mol. Biol. Evol. 28: 1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujatha M., Lakshminarayana M., 2007. Resistance to Spodoptera litura (Fabr.) in Helianthus species and backcross derived inbred lines from crosses involving diploid species. Euphytica 155: 205–213. [Google Scholar]
- Tang S., Yu J.-K., Slabaugh M., Shintani D., Knapp S., 2002. Simple sequence repeat map of the sunflower genome. Theor. Appl. Genet. 105: 1124–1136. [DOI] [PubMed] [Google Scholar]
- Timme R. E., Simpson B. B., Linder C. R., 2007. High-resolution phylogeny for Helianthus (Asteraceae) using the 18S–26S ribosomal DNA external transcribed spacer. Am. J. Bot. 94: 1837–1852. [DOI] [PubMed] [Google Scholar]
- Vekemans X., 2010. What’s good for you may be good for me: evidence for adaptive introgression of multiple traits in wild sunflower. New Phytol. 187: 6–9. [DOI] [PubMed] [Google Scholar]
- Voorrips R., 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Walsh J. B., 1982. Rate of accumulation of reproductive isolation by chromosome rearrangements. Am. Nat. 120: 510–532. [Google Scholar]
- Weir B. S., Cockerham C. C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Welch M. E., Rieseberg L. H., 2002. Habitat divergence between a homoploid hybrid sunflower species, Helianthus paradoxus (Asteraceae), and its progenitors. Am. J. Bot. 89: 472–478. [DOI] [PubMed] [Google Scholar]
- White M. J. D., 1978. Modes of Speciation. W. H. Freeman, San Francisco. [Google Scholar]
- Whitlock M. C., 2011. G′ST and D do not replace FST. Mol. Ecol. 20: 1083–1091. [DOI] [PubMed] [Google Scholar]
- Wieckhorst S., Bachlava E., Dußle C., Tang S., Gao W., et al. , 2010. Fine mapping of the sunflower resistance locus Pl ARG introduced from the wild species Helianthus argophyllus. Theor. Appl. Genet. 121: 1633–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Eannetta N. T., Xu Y., Durrett R., Mazourek M., et al. , 2009a A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor. Appl. Genet. 118: 1279–1293. [DOI] [PubMed] [Google Scholar]
- Wu F., Eannetta N. T., Xu Y., Tanksley S. D., 2009b A detailed synteny map of the eggplant genome based on conserved ortholog set II (COSII) markers. Theor. Appl. Genet. 118: 927–935. [DOI] [PubMed] [Google Scholar]
- Wu F., Eannetta N. T., Xu Y., Plieske J., Ganal M., et al. , 2010. COSII genetic maps of two diploid Nicotiana species provide a detailed picture of synteny with tomato and insights into chromosome evolution in tetraploid N. tabacum. Theor. Appl. Genet. 120: 809–827. [DOI] [PubMed] [Google Scholar]
- Yu J.-K., Tang S., Slabaugh M. B., Heesacker A., Cole G., et al. , 2003. Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci. 43: 367–387. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.