Abstract
Our understanding of the evolutionary consequences of mutation relies heavily on estimates of the rate and fitness effect of spontaneous mutations generated by mutation accumulation (MA) experiments. We performed a classic MA experiment in which frequent sampling of MA lines was combined with whole genome resequencing to develop a high-resolution picture of the effect of spontaneous mutations in a hypermutator (ΔmutS) strain of the bacterium Pseudomonas aeruginosa. After ∼644 generations of mutation accumulation, MA lines had accumulated an average of 118 mutations, and we found that average fitness across all lines decayed linearly over time. Detailed analyses of the dynamics of fitness change in individual lines revealed that a large fraction of the total decay in fitness (42.3%) was attributable to the fixation of rare, highly deleterious mutations (comprising only 0.5% of fixed mutations). Furthermore, we found that at least 0.64% of mutations were beneficial and probably fixed due to positive selection. The majority of mutations that fixed (82.4%) were base substitutions and we failed to find any signatures of selection on nonsynonymous or intergenic mutations. Short indels made up a much smaller fraction of the mutations that were fixed (17.4%), but we found evidence of strong selection against indels that caused frameshift mutations in coding regions. These results help to quantify the amount of natural selection present in microbial MA experiments and demonstrate that changes in fitness are strongly influenced by rare mutations of large effect.
Keywords: Pseudomonas aeruginosa, hypermutator, whole genome resequencing, spontaneous mutation, experimental evolution
MUTATIONS are the ultimate source of genetic variation that natural selection acts upon. Understanding the rate at which mutations arise and the distribution of fitness effects of spontaneous mutations is therefore of central importance to the study of evolutionary biology (Haldane 1937; Kondrashov 1988; Partridge and Barton 1993; Charlesworth and Hughes 1996, 2000; Hughes 2010; Bank et al. 2014). One of the most widely used methods for determining the rate and fitness effect of spontaneous mutations is the MA experiment. Following the pioneering work Bateman (1959) and Mukai (1964), MA experiments involve propagating many replicate lines at very small effective population sizes so that the effect of natural selection is swamped out by that of genetic drift, allowing weakly selected mutations to accumulate randomly. The decline in mean fitness and increase in among-line variance in fitness are then used to indirectly infer mutation rate and effect estimates (Bateman 1959; Mukai 1964; Keightley 1994; García-Dorado 1997; Shaw et al. 2002).
Recently, whole genome resequencing of MA lines has been used to directly measure the mutation rate in microorganisms (Lynch et al. 2008; Lee et al. 2012; Ness et al. 2012; Sung et al. 2012a,b; Long et al. 2013). In line with classic mutation rate estimates from reporter gene assays, the emerging consensus is that the genomic mutation rate is remarkably constant across DNA-based microbes, ∼3 × 10−3 mutations/genome/generation (Drake 1991; Lynch 2010). Accurate estimates of the fitness effects of spontaneous mutation, however, have remained elusive (Eyre-Walker and Keightley 2007; Halligan and Keightley 2009).
Because MA experiments rely on making comparisons among lines, they have traditionally focused on studying how fitness changes across as many lines as possible. An alternative approach is to combine whole genome resequencing in a smaller number of MA lines of a hypermutator strain to allow a greater number of mutations to accumulate, thus increasing our ability to detect and quantify the amount of natural selection that occurs during microbial mutation accumulation experiments. Furthermore, whole genome resequencing directly determines the average number of mutations that accumulate between fitness measurements, allowing for improved estimates of the distribution of fitness effects of spontaneous mutations.
Natural selection must occur to some extent during microbial mutation accumulation experiments because colonies must grow big enough to become visible, resulting in an effective population size (Ne) >1. Beneficial and deleterious mutations should be subject to effective selection when Nes > 1, where s is the absolute value of the fitness effect of the mutation, and the fluctuating population size of microbial MA experiments may further increase the efficacy of selection (Otto and Whitlock 1997). This may explain why many microbial MA experiments have reported results that are consistent with the fixation of some beneficial mutations as a result of positive selection (Shaw et al. 2000; Joseph and Hall 2004; Perfeito et al. 2007; Dickinson 2008; Trindade et al. 2010; Stevens and Sebert 2011). Studies have begun to combine both MA and whole genome resequencing in microorganisms (Lynch et al. 2008; Lee et al. 2012; Ness et al. 2012; Sung et al. 2012a,b; Long et al. 2013), but none have detected a genomic signature of natural selection.
Using detailed fitness measurements and whole genome resequencing, we studied the evolutionary dynamics of eight replicate mutation accumulation lines of a hypermutator strain of the pathogenic bacterium Pseudomonas aeruginosa. MA lines were passaged through 28 single-cell bottlenecks followed by rapid population growth over a period of ∼644 generations. Under this regime, we estimate that the effective population size of MA lines had a lower limit of ∼16, which should be sufficient to prevent natural selection on the vast majority of spontaneous mutations. We determined the evolutionary dynamics of our lines with a high degree of precision by (1) directly measuring competitive fitness instead of a component of fitness such as growth rate, and (2) measuring fitness at every second bottleneck to capture a small number of mutations between each time point. In line with recent work, we used deep whole genome sequencing to determine the genetic consequences of population bottlenecking, infer the molecular basis of altered fitness, and test for genomic signatures of natural selection during the MA procedure.
Consistent with previous MA experiments, we found that mean fitness decayed linearly over time. Detailed trajectories of fitness in individual lines coupled to whole genome sequencing revealed that rare, strongly deleterious mutations account for nearly half of the total loss of fitness. Furthermore, we found that positive selection resulted in the fixation of beneficial mutations, and that purifying selection was able to remove the majority of frameshift mutations.
Materials and Methods
Strains
The eight replicate clones used in this study were founded from the P. aeruginosa hypermutator strain PAO1ΔmutS, which was created by replacing mutS—part of the methyl-directed mismatch repair pathway—with the antibiotic resistance marker aac1 using the Cre-lox system for gene deletion and antibiotic resistance marker recycling following the methods of Mandsberg et al. (2011). Deleting mutS increases the mutation rate by ∼70-fold in P. aeruginosa (Torres-Barcelo et al. 2013), primarily by increasing the rate of transitions (Miller 1996). The reference strain used to assess competitive fitness was PAO1-GFP. This strain was generated by integrating a constitutively expressed GFP marker at the chromosomal tn7 insertion site in P. aeruginosa PAO1 using the methods of Choi and Schweizer (2006).
Mutation accumulation
Eight replicate mutation accumulation lines were generated by streaking randomly selected colonies of PAO1ΔmutS onto individual M9KB agar plates (glycerol, 10 g/liter; peptone, 20 g/liter; M9 salts, 10.5 g/liter; agar, 12 g/liter; and MgSO4, 2 mL/liter). Plates were incubated at 37° for 18 hr before repeating the process of picking a random colony and streaking it on a fresh plate. This process was repeated daily for 30 days. Each day, colonies would form from a single cell, which had doubled ∼23 times, resulting in an Ne of ∼16. Every second day, a portion of the randomly selected colony was suspended in a 50% v/v solution of glycerol and frozen at −80° to be stored for competition assays. To ensure random selection of colonies, the last colony of the streak, which was not touching another colony, was selected. It is unlikely that random colony selection suffered a detection bias due to missing extremely small colonies; we sampled 14 regions between the visible colonies of our streaked plates and restreaked them, but did not detect a single instance of colony growth after 10 days.
Competitive fitness assay
Fitness of each line at each time point was determined relative to the PAO1-GFP strain. Strains were precultured in M9KB medium from frozen 50% glycerol stocks. Overnight cultures of each strain were mixed in M9KB broth at a ratio of ∼80% mutant to 20% PAO1-GFP. The exact initial proportions were confirmed via flow cytometry. Mixtures were competed for 18 hr at 37°, with agitation at 200 rpm, and the final proportion was again measured by flow cytometry. We define the relative fitness of the mutant as the number of doublings that the mutant strain undergoes during the 18-hr competition divided by the number of doublings of the wild-type strain, given by the formula
where wmutant is the fitness of the mutant relative to the wild-type and Nij is the number of either the mutant or the wild-type cells at either the beginning or the end of the competition. Each competition assay was performed in two experimental blocks with three replicate competitions per block. In some mutation accumulation lines, fitness became too low to accurately measure (final mutant proportion <10%) and thus these data have been excluded from all analyses except those pertaining to Figure 2 and the decay in average fitness over time. The inclusion of these inaccurate points does not change the statistical significance of any of the results presented.
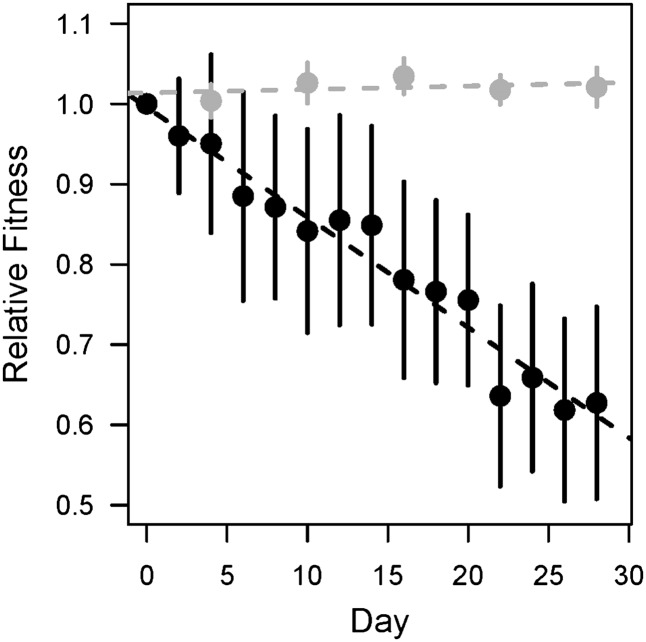
Figure 2.
Average fitness decays in mutation accumulation lines. Plotted points show the mean fitness (± SE) of hypermutator lines (solid symbols, n = 8) and control lines (shaded symbols, n = 4) that were passaged through 28 daily bottlenecks, which correspond to ∼644 generations of mutation accumulation. The fitness of hypermutator lines rapidly declined, but the fitness of control lines did not change over the course of the experiment (ANOVA: F1,3 = 0.436, P = 0.556). Note that in some MA lines, fitness decayed to the point where it was not possible to measure fitness reliably, but these data are included to prevent bias.
Flow cytometry
Flow cytometry was used to determine the relative proportions of mutant and wild-type strains at the beginning and end of the competitive fitness assays. Bacterial cultures, diluted 200-fold in sterile filtered M9 salts, were prepared using deionized water to minimize background signal in the flow cytometer. Diluted mixtures were run on an Accuri C6 Flow Cytometer Instrument (BD Accuri, San Jose, CA) until 10,000 cells had been assayed. Events with a forward scatter value <10,000 or a side scatter value <8000 were excluded to prevent the false detection of small particles in the medium and electrical noise. To discriminate between GFP-tagged and untagged cells, cells were excited at a wavelength of 488 nm and fluorescence emissions between 518 and 548 nm were measured. There was a small overlap in the fluorescence profiles of tagged and untagged cells (i.e., the most fluorescent untagged cells were slightly more fluorescent than the least fluorescent GFP-tagged cells), so pure cultures of PAO1 and PAO1-GFP were used as controls to correct for such spillover.
Whole genome sequencing
Illumina whole genome sequencing was performed on the first and last time point of each line, as well as on the five pairs of adjacent time points that showed the largest decrease in fitness. Raw sequencing data were analyzed using an in-house pipeline. Briefly, raw reads were filtered using the NGS QC Toolkit (Patel and Jain 2012) and aligned against the reference genome using BWA (Li and Durbin 2009). Two approaches were used to call variants, GATK’s Unified Genotyper (Depristo et al. 2011) and SAMtools’s Mpileup (Li et al. 2009). Identified variants were annotated with SnpEff (Cingolani et al. 2012). To detect structural variants, we combined two algorithms, Breakdancer (Chen et al. 2009) and Pindel (Ye et al. 2009). Finally, copy number variants (CNVs) were detected using Control-FREEC (Boeva et al. 2012).
All differences between the P. aeruginosa PAO1 reference genome and the first time point of each bacterial line were excluded, leaving only mutations that accumulated throughout the experiment. Sequences from intermediate time points were treated as sequences from end points. All mutations found in intermediate time points were found at the end points except for one that fell in a mutation hotspot.
Testing for selection on base substitutions
To test for selection on base substitutions in protein coding genes, we estimated the expected number of protein altering mutations, under the assumption that synonymous mutations are effectively neutral. Specifically, since almost all base substitutions in our experiment were transitions (99.5%), we calculated the neutral mutation rate of each of the four bases to its partner (A→G, G→A, C→T, and T→C) using the observed synonymous mutations in our experiment. Given these mutation rates, we used the nucleotide composition and codon usage of P. aeruginosa proteins to estimate the rates of nonsynonymous and synonymous mutations (dN/dS ratio), as well as the rates of stop-gain, stop-loss, and intergenic mutations. To test for a deviation from the neutral expectation, we tested the null hypothesis that the proportion of mutations in a given class (nonsynonymous, truncation, or intergenic) relative to the number of observed synonymous mutations is equal to the predicted ratio calculated using the synonymous mutation rate. This hypothesis was tested using the normal approximation of the binomial distribution (Zar 2010).
Repetitive regions
The RepeatMasker program (Smit et al. 1996–2010) was used to screen the PAO1 genome for simple repeats, interspersed repeats, and low-complexity DNA sequences. Homopolymeric tracts of single nucleotide repeats ranging from 4 to 20 bases were identified using the dreg program, implemented in the EMBOSS package (Rice et al. 2000).
Magnitude of selection against indels in coding regions
The percentage of indels in repetitive coding regions removed by natural selection was calculated under the assumption that indels in the repetitive noncoding genome are neutral. The expected number of indels in repetitive coding regions before natural selection was calculated by dividing the observed number of “neutral” mutations in repetitive noncoding regions by the fraction of repetitive elements that are in noncoding regions (21.8%) and multiplied this value by the fraction of repetitive elements that are in coding regions (78.2%). The percentage of indels removed due to natural selection is then 1 − observed/expected. If mutations in noncoding repetitive regions are not neutral, then this method will generate a lower limit estimate.
Core genes
Precomputed pairwise reciprocal best BLAST hits for 36 Pseudomonas species were downloaded from the Pseudomonas Genome Database (Winsor et al. 2011). The core genome for P. aeruginosa PAO1 was defined as the set of PAO1 genes that had pairwise reciprocal best BLAST hits in the 35 remaining Pseudomonas species. We found a total of 1435 core genes.
Clusters of Orthologous Groups analysis
A list of P. aeruginosa PAO1 genes with annotated Clusters of Orthologous Groups (COGs) categories (Tatusov et al. 2000) was downloaded from the National Center for Biotechnology Information. This list was intersected with the list of genes that had experienced at least one mutation during our experiment. Genes with annotated mutations and COG categories were compared to the rest of the genes in the PAO1 genome that were unmutated, but had been assigned a COG category. P-values were computed using Fisher’s exact test and corrected for multiple testing using the false discovery rate method (Benjamini and Hochberg 1995).
Statistical analysis and simulations
All statistical analyses were conducted in R (version 2.15.0) (R Development Core Team 2012). All statistical tests are reported as a P-value and the value for the test statistic with a subscript indicating the degrees of freedom. All tests use α = 0.05 and, where applicable, are two tailed.
Simulations were used to generate the expected distribution of the number of mutations per gene, given the substantial variation in gene length in the P. aeruginosa genome (mean: 830 bp, 95% confidence interval: 247–2786 bp). The lengths of all genes in the P. aeruginosa genome were obtained from the Pseudomonas Genome Database (Winsor et al. 2011). In each simulation, mutations (either synonymous or nonsynonymous) were randomly distributed across a simulated genome, using the same number of mutations as was detected in our experiment. The number of mutations per gene was recorded and results were averaged across 100 simulations.
Results
Here we present the results from a ∼644-generation-long mutation accumulation experiment in eight replicate MA lines. We measured the fitness of each MA line every 2 days, providing a high-resolution picture of the evolutionary dynamics of heavily bottlenecked bacterial populations. We performed whole genome resequencing on multiple time points of each line to determine the molecular nature of mutations fixed under conditions of relaxed natural selection.
Whole genome resequencing identified 944 mutations in the eight mutation accumulation lines. Sanger sequencing of a random sample of these mutations confirmed 35/35 mutations (Supporting Information, Table S1), indicating a very low false positive rate. As expected, mutations were Poisson distributed across MA lines (one-sample Kolmogorov–Smirnoff test: P = 0.521, D = 0.270) with an average of 118 mutations fixed per line and an average of 8.4 mutations fixed between each adjacent time point. This equates to a per base pair mutation rate of 2.95 (± 0.21 SE) × 10−8 mutations/site/generation and a genomic mutation rate of 0.18 (± 0.01 SE) mutations/genome/generation. Given that the hypermutator strain used in this study increases the mutation rate by ∼70-fold (Torres-Barcelo et al. 2013), this estimated genomic mutation rate is in line with the consensus bacterial genomic mutation rate of ∼3 × 10−3 mutations/genome/generation (Drake 1991; Lynch 2010).
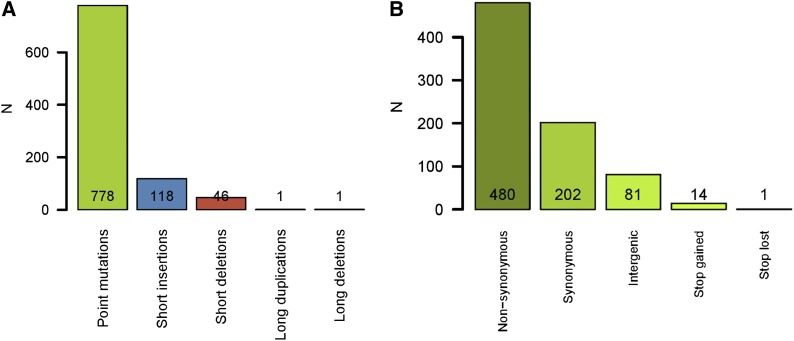
Of the 944 mutations, 778 (82.4%) were base substitutions, 164 (17.4%) were short indels (<10 bp), and 2 (0.2%) were large structural variations, consisting of a partial gene duplication event (pvdD) and a 1880-bp intergenic deletion. Insertions were ∼2.5-fold more common than deletions (118 insertions vs. 46 deletions) (Figure 1). As is typical for a ΔmutS hypermutator strain, almost all base substitutions were transitions (774/778 = 99.5%), and G:C→A:T transitions (478) were ∼60% more common than A:T→G:C transitions (298).
Figure 1.
Types of mutations accumulated. (A) The distribution of accumulated mutations according to type of mutation. Indels <10 base pairs long were considered to be “short.” (B) Further information on the effects of point mutations.
As expected, the average fitness of the hypermutator populations decreased significantly over time (Figure 2; ANOVA: P = 1.68 × 10−6, F1,13 = 67.409), indicating that the average effect of spontaneous mutations was deleterious and that recurrent population bottlenecks inhibited the action of natural selection (mean mutational fitness effect = −0.16%). In fact, in some lines, fitness became so low that it was no longer possible to reliably measure (Figure 3). These data are included in Figure 2 to prevent bias, but excluded from subsequent analyses. The average fitness of bottlenecked nonhypermutator control lines did not change significantly over the course of the experiment (ANOVA: P = 0.712, F1,118 = 0.137), indicating that the loss of fitness in hypermutator lines was due to mutation accumulation.
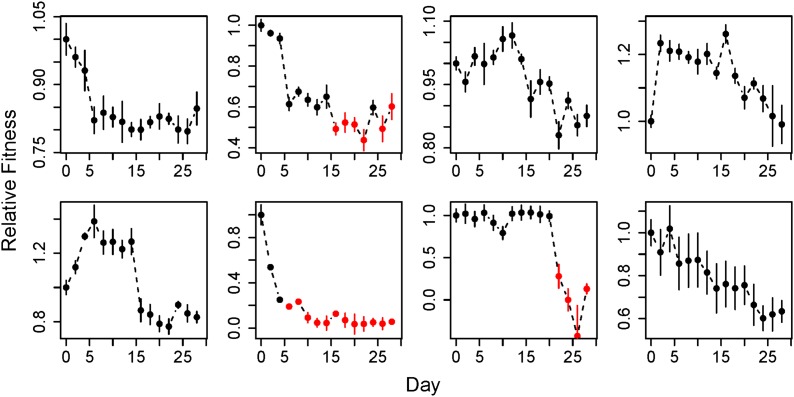
Figure 3.
Fitness trajectories for individual mutation accumulation lines. The mean (± SE; n = 6) fitness of individual hypermutator lines through time. Red data points indicate that fitness is too low to measure accurately. The mean fitness (± SE; n = 6) of individual hypermutator lines through time. Red data points indicate that fitness is too low to measure accurately. The y-axis of each plot is scaled differently to maximize the resolution of evolutionary dynamics within a single line.
Fitness data
Unlike the linear decrease observed for average fitness, the evolutionary trajectory of individual lines was much more complex (Figure 3). The net change in the fitness of MA lines ranged from −1 to −27% (mean: −13% ± 9 SD). A large portion of the net decrease in fitness of each line was due to a single drop between adjacent time points (we hereafter refer to a pair of adjacent time points as a “step”). Specifically, on average, 42.2% (± 12.9% SD) of the total decrease in fitness between the first and last time point in an individual MA line (excluding any beneficial steps) was due to the largest deleterious step in that line. Furthermore, the four most deleterious steps across all lines accounted for 42.3% of the total fitness decrease throughout the entire experiment. To determine whether these large drops in fitness were caused by (1) the accumulation of a greater number of mutations than other steps or (2) the accumulation of mutations of larger effect, we performed whole genome sequencing on the four largest deleterious steps across all MA lines, as well as on an exceptionally large deleterious step, which caused the fitness of its MA line to drop to an undetectable level. These steps did not contain a significantly greater number of mutations than the remaining steps (mean of five largest steps: 9.0 mutations, mean of remainder: 7.9 mutations, paired t-test: P = 0.285, t4 = 1.235). However, these large deleterious steps showed a significantly higher frequency of mutations in highly conserved core genes than other steps (χ2 goodness-of-fit test: P = 0.049, χ21 = 3.882; Table S2). Therefore, large drops in fitness are due to mutations in more important genes rather than due to a greater number of mutations.
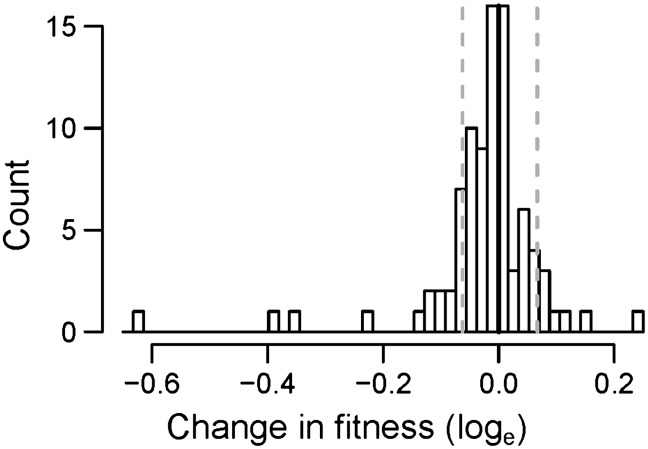
Although the average fitness effect of a step was deleterious, there were numerous steps in which fitness increased (Figure 4). To confirm the presence of steps containing beneficial mutations, we repeated the competitive fitness assays for the 11 steps with the largest increases in fitness. Even after false discovery rate correction (Benjamini and Hochberg 1995), fitness increased significantly (P < 0.05) in 6/89 (6.7%) of the measurable steps. Because steps where fitness increased were rare, it is likely that each of these steps only contained a single beneficial mutation. This implies that at least six beneficial mutations were fixed during the mutation accumulation experiment, which corresponds to 0.64% of all mutations that were fixed during the experiment.
Figure 4.
Changes in fitness for individual “steps.” The distribution of fitness changes for each step in the mutation accumulation experiment across all eight hypermutator lines. Each step represents the difference in fitness between successive assays for an MA line (∼8.4 mutations accumulated/step). The solid line depicts no change in fitness and the area between the dashed shaded lines is the area in which Nes < 1, where Ne is the harmonic mean of population size over time (although this may be an underestimate) (Otto and Whitlock 1997).
Signatures of natural selection
Selection on base substitutions in protein coding genes:
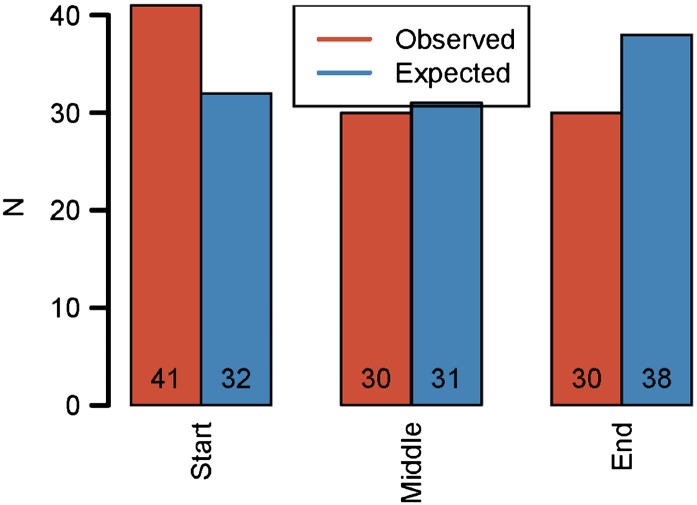
The vast majority of protein-altering base substitutions were nonsynonymous mutations, but the ratio of the rate of nonsynonymous mutations to silent mutations (dN/dS = 1.08) did not differ significantly from the neutral expectation of 1 (Table 1; Z-test: Z = 0.92, P = 0.26). We observed only a single loss-of-stop mutation, but this was similar to our predicted number of 1.4. Truncation mutations that introduce a premature stop codon were much more frequent (n = 14), but this was not significantly different from the neutral expectation of nine truncation mutations (Z-test: Z = 1.63, P = 0.10).
Table 1. Testing for selection on single base pair substitutions.
| Protein effect | Observed | Expected |
|---|---|---|
| Nonsynonymous | 480 | 444.38 |
| Intergenic | 80 | 84.33 |
| Stop-gain | 14 | 8.94 |
| Stop-loss | 1 | 1.41 |
The number of observed single base pair substitutions relative to the neutral expectation, as determined from the synonymous mutation rate and genome composition of P. aeruginosa. The observed number of mutations does not differ from the neutral expectation for any functional category of mutation.
Selection on coding and noncoding regions:
Protein coding sequence accounts for 89.4% of the P. aeruginosa genome and so we expected that if no natural selection has occurred during the MA experiment then ∼89.4% of mutations will have occurred in protein coding sequences. We found that the percentage of mutations (short indels and base substitutions) that occurred in coding regions was 85.4% (804/942), which was significantly different from the neutral expectation of 89.4% (χ2 goodness-of-fit test: P < 0.001, χ21 = 15.888). Interesting patterns arose when we analyzed base substitutions and short indels separately.
We found that the percentage of base substitutions in coding regions (89.6%, 697/778) and intergenic regions (10.41%, 81/778) was not significantly different from the neutral expectation (χ2 goodness-of-fit test: P = 0.833, χ21 = 0.045). This result may be confounded because intergenic regions contain a larger proportion of repetitive DNA than coding regions (intergenic: 7.2%, coding: 3.1%), but when we restricted our analysis to repetitive regions we still observed that the percentage of base substitutions that fell in coding (4.3%) and intergenic (16.5%) repetitive regions did not differ from the neutral expectation (χ2 goodness-of-fit test: P = 0.181, χ21 = 1.791).
Selection on indels:
In contrast to base substitutions, we found significantly fewer indels in coding regions than expected (observed: 107/164 = 65.2%; expected: 89.4%; χ2 goodness-of-fit test: P < 0.0001, χ21 = 100.236). Again, this difference could be confounded because intergenic regions contain a larger proportion of indel-prone repetitive DNA, but we also found significantly fewer indels in repetitive coding regions (observed: 103/160 = 64.4%; expected: 78.2%; χ2 goodness-of-fit test: P < 0.0001, χ21 = 17.920) than expected in the absence of selection. This indicates strong purifying selection against frameshift mutations. In fact, these data suggest that at least 49.6% of frameshift mutations are sufficiently deleterious to be removed by natural selection, even under a regime of intense bottlenecking. Despite selection against frameshift mutations, we still found 106 frameshifts in our experiment. Almost all of them (101/106 = 95.3%) overlapped with homopolymeric tracts of C (ranging from 4C to 8C) or G (ranging from 5G to 8G). There were significantly more frameshifts located near the N terminus of the protein than expected, given the distribution of homopolymeric tracts in the P. aeruginosa genes (Figure 5; one-sided exact binomial test: P = 0.037). We found no significant difference for frameshifts near the middle (one-sided exact binomial test: P = 0.453) or near the C terminus of the protein (one-sided exact binomial test: P = 0.063).
Figure 5.
The distribution of indel mutations in proteins. Comparison between the observed and expected position of frameshifts in coding regions. Proteins were divided into three equal pieces and we counted the number of frameshifts (overlapping with homopolymeric tracts) that fell in each section. Expected frequencies were computed by counting the number of homopolymeric tracts in the P. aeruginosa PAO1 proteome that fall in each section. The differences between observed and expected values were statistically significant for the N-terminal third of proteins (one-sided exact binomial test: P = 0.037).
Tests for parallel evolution:
Previous work has shown that exposing replicate microbial populations to a similar selective pressure results in parallel adaptation at a molecular level in both lab experiments (Wichman et al. 2000; Segrè et al. 2006; Barrick et al. 2009) and clinical populations (Huse et al. 2010; Lieberman et al. 2011). To test for parallel evolution at the level of individual genes, we compared the distribution of the number of mutations fixed per gene in the eight MA lines, with the distribution expected based on the lengths of the genes in the P. aeruginosa genome (Figure S1; see Materials and Methods for details on calculating the expected distribution). We found no deviation from the expected distribution for synonymous mutations (χ2 goodness-of-fit test: P = 0.643, χ22 = 0.883). On the other hand, we found significantly fewer parallel nonsynonymous mutations than expected (χ2 goodness-of-fit test: P < 0.0001, χ22 = 19.302), which does not support the hypothesis that natural selection was capable of causing parallel evolution on the genomic scale in these MA lines. Rather, longer genes simply had more mutations than smaller genes (Figure S2): genes with one or more mutations were significantly longer than genes without mutations (Kolmogorov–Smirnov test, P < 0.001).
It is also possible that parallel evolution could act on levels higher than the gene. We analyzed our mutation data for evidence of over- or underenrichment of mutations in COGs—genes that share a common function. After false discovery rate correction (Benjamini and Hochberg 1995), we found a significant underrepresentation of mutated genes involved in transcription (Table S3; Fisher’s exact test: P = 0.023, Fisher’s odds ratio1 = 0.530), suggesting that mutations in these genes tend to have highly deleterious effects.
Core genes:
We observed that large drops in fitness during the MA experiment were associated with the accumulation of mutations in core genes (Figure 2), and so we sought to determine whether natural selection was effective against mutations in these genes. Surprisingly, there was no significant underrepresentation of mutations in core genes (Fisher’s exact test: P = 0.611, Fisher’s odds ratio1 = 1.051) despite their potentially large deleterious effects on fitness.
Discussion
Mutations are rare events that often lead to small changes in fitness, and these properties of mutations make it intrinsically difficult to directly study the evolutionary consequences of mutation. Our experiment, which combined a classic mutation accumulation experiment with powerful whole genome resequencing technology, found that 42.3% of the decrease in fitness in our lines was driven by 4.5% of the steps with highly deleterious effects on fitness. Given the rarity of large drops in fitness, the most parsimonious explanation is that each one of these drops was driven by a single highly deleterious mutation. Under this assumption, the 42.3% of the decrease in fitness in our experiment was driven by 0.5% of the mutations fixed, which is consistent with previous work in Caenorhabditis elegans (Davies et al. 1999). The mean mutational effect, s = −1.6 × 10−3, is similar to previous work in Saccharomyces cerevisiae (s = −6 × 10−3), in which whole genome resequencing and MA were combined (Lynch et al. 2008) and, as expected, is approximately one to two orders of magnitude smaller than previous microbial MA studies that did not use whole genome resequencing and were therefore unable to detect neutral mutations (Halligan and Keightley 2009; Trindade et al. 2010). We also found evidence of both positive and negative selection in our MA experiment, demonstrating that the results of our experiment cannot be interpreted as a proxy for the effects of spontaneous mutation alone.
Beneficial mutations
Previous studies in Arabidopsis thaliana (Shaw et al. 2000), Escherichia coli (Perfeito et al. 2007; Trindade et al. 2010), Streptococcus pneumoniae (Stevens and Sebert 2011), and S. cerevisiae (Joseph and Hall 2004; Dickinson 2008) have also found evidence that beneficial mutations are fixed during mutation accumulation experiments. Our experimental approach allowed us to experimentally demonstrate that it is highly likely that at least 0.64% of the mutations that fixed during our MA experiment were beneficial. For these mutations to have been fixed by drift, the beneficial mutation rate in a nonhypermutator population with a genomic mutation rate of 3 × 10−3 mutations/genome/generation would have to have been ∼5 × 10−6 mutations/genome/generation, which is two to three orders of magnitude higher than existing estimates (Gerrish and Lenski 1998; Miralles et al. 1999; Imhof and Schlotterer 2001; Rozen et al. 2002; Barrett et al. 2006; but for exceptions, see Perfeito et al. 2007). Instead, we argue that positive selection was able to drive the fixation of beneficial mutations in our experiment. Consistent with this idea, five of the six significantly beneficial mutations that fixed were sufficiently beneficial that Nes was >1.
Tests for selection at a molecular level
In agreement with recent microbial mutation accumulation experiments that have used whole genome resequencing, we found no evidence of selection on base substitutions, including nonsynonymous mutations (Lynch et al. 2008; Lee et al. 2012; Ness et al. 2012; Sung et al. 2012a,b; Long et al. 2013). Additionally, we found no evidence of positive selection on the same genes in different MA lines. Surprisingly, we found that nonsynonymous mutations in highly conserved core genes can have strong deleterious effects on fitness (Figure 2), and yet we found no evidence that these mutations were removed by natural selection. The most striking evidence of selection at a genetic level comes from the lack of short indel mutations in coding regions. We estimate that negative selection prevented the fixation of at least 50% of indels in coding regions. In contrast, we did not find any evidence of an underrepresentation of base substitutions that generated a premature stop codon, implying that the absence of indels in coding regions is due to selection against frameshifts, and not selection against gene loss.
Despite strong selection, we still found that frameshifts comprise 13.2% of all mutations in coding regions. This high incidence of frameshifting could be because 95.3% of frameshifts overlapped with homopolymeric tracts. Homopolymeric tracts are hypermutable: they are highly prone to gaining or losing repeats through slippage, thereby producing indels. Consistent with recent work (Orsi et al. 2010; Lin and Kussell 2012), we found a significant overrepresentation of frameshifts at the 5′ end of genes and underrepresentation at the 3′ end (given the distribution of homopolymeric tracts in the PAO1 genome). Although the reasons for the enrichment of 5′ frameshifts is unclear, possible explanations include: (1) 5′ frameshifts tend to create shorter proteins and thus may be less prone to forming toxic aggregations; (2) intergenic regions in P. aeruginosa are very short and 3′ indels may knock out downstream genes; and/or (3) 5′ indels are more likely to destroy gene function, which may be beneficial in some circumstances. For example, Moxon et al. (2006) have proposed that simple sequence repeats (such as homopolymeric tracts) are localized hypermutation targets and a mechanism for adaptation. Moreover, standing genetic variation in homopolymeric tracts has been shown to drive the adaptation of Campylobacter jejuni to a novel host (Jerome et al. 2011).
Implications for mutation accumulation experiments
It is important to emphasize that our experiment differed from most previous MA experiments because we used a hypermutator strain. To what extent is this likely to have biased our results? Hypermutators produce an altered spectrum of spontaneous mutations (e.g., bias toward transitions), which can have important evolutionary implications when strong selection acts on a small number of sites in the genome (Couce et al. 2013) (e.g., some cases of high-level antibiotic resistance). In our system, frameshifts experienced much stronger selection than any other class of mutation, and it is possible that using a ΔmutS hypermutator altered the rate of appearance of indel mutations relative to base substitutions (Marvig et al. 2013). However, by using a hypermutator we were able to detect a sufficiently large number of mutations to analyze the effects of relatively rare types of mutation, such as indels, which have traditionally been overlooked in MA studies.
Conclusion
In conclusion, we find that fitness decays in recurrently bottlenecked populations of hypermutator P. aeruginosa because of the fixation of many weakly deleterious mutations and a few highly deleterious mutations. We argue that this pattern of punctuated decay of fitness arises for two reasons. First, most mutations carry little, if any, fitness cost in a laboratory environment, but a substantial fraction of mutations are highly deleterious. Our results suggest that weakly deleterious mutations tend to be intergenic and nonsynonymous mutations, while highly deleterious mutations tend to be indels and mutations in core genes. Second, we find that recurrent bottlenecking does not completely compromise the efficacy of natural selection in microbial mutation accumulation experiments, although large deleterious mutations are unlikely to play a substantial role in the evolution of natural populations. We hope that this study will pave the way for future work aimed at understanding: (1) why frameshift mutations are subject to such strong selection, (2) how bacteria adapt to the deleterious effects of spontaneous mutations, and (3) how the molecular basis of spontaneous mutation is linked to the fitness effects of mutations in natural populations.
Supplementary Material
Acknowledgments
We thank Antonio Oliver for providing us with the PAO1ΔmutS strain. We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant 090532/Z/09/Z and Medical Research Council hub grant G0900747 91070) for the generation of the sequencing data.
Footnotes
Communicating editor: J. Bull
Literature Cited
- Bank C., Hietpas R. T., Wong A., Bolon D. N., Jensen J. D., 2014. A Bayesian MCMC approach to assess the complete distribution of fitness effects of new mutations: uncovering the potential for adaptive walks in challenging environments. Genetics 196: 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett R. D., MacLean R. C., Bell G., 2006. Mutations of intermediate effect are responsible for adaptation in evolving Pseudomonas fluorescens populations. Biol. Lett. 2: 236–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E., Yu D. S., Yoon S. H., Jeong H., Oh T. K., et al. , 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461: 1243–1247. [DOI] [PubMed] [Google Scholar]
- Bateman A., 1959. The viability of near-normal irradiated chromosomes. Int. J. Radiat. Biol. 1: 170–180. [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple Testing. J Roy Stat Soc B Met 57: 289–300. [Google Scholar]
- Boeva V., Popova T., Bleakley K., Chiche P., Cappo J., et al. , 2012. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28: 423–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Hughes K. A., 1996. Age-specific inbreeding depression and components of genetic variance in relation to the evolution of senescence. Proc. Natl. Acad. Sci. USA 93: 6140–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., and K. A. Hughes, 2000 The maintenance of genetic variation in life-history traits, pp. 369–392 in Evolutionary Genetics: From Molecules to Morphology, edited by R. S. Singh and C. Krimbas. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Chen K., Wallis J. W., McLellan M. D., Larson D. E., Kalicki J. M., et al. , 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6: 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. H., Schweizer H. P., 2006. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1: 153–161. [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couce A., Guelfo J. R., Blazquez J., 2013. Mutational spectrum drives the rise of mutator bacteria. PLoS Genet. 9: e1003167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies E. K., Peters A. D., Keightley P. D., 1999. High frequency of cryptic deleterious mutations in Caenorhabditis elegans. Science 285: 1748–1751. [DOI] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson W. J., 2008. Synergistic fitness interactions and a high frequency of beneficial changes among. Genetics 178: 1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88: 7160–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. [DOI] [PubMed] [Google Scholar]
- García-Dorado A., 1997. The rate and effects distribution of viability mutation in Drosophila: minimum distance estimation. Evolution 51: 1130–1139. [DOI] [PubMed] [Google Scholar]
- Gerrish P. J., Lenski R. E., 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102–103: 127–144. [PubMed] [Google Scholar]
- Haldane J., 1937. The effect of variation on fitness. Am. Nat. 71: 337–349. [Google Scholar]
- Halligan D. L., Keightley P. D., 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 40: 151–172. [Google Scholar]
- Hughes K. A., 2010. Mutation and the evolution of ageing: from biometrics to system genetics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1273–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse H. K., Kwon T., Zlosnik J. E., Speert D. P., Marcotte E. M., et al. , 2010. Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. MBio 1(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof M., Schlotterer C., 2001. Fitness effects of advantageous mutations in evolving Escherichia coli. Proc. Natl. Acad. Sci. USA 98: 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome J. P., Bell J. A., Plovanich-Jones A. E., Barrick J. E., Brown C. T., et al. , 2011. Standing genetic variation in contingency loci drives the rapid adaptation of Campylobacter jejuni to a novel host. PLoS ONE 6: e16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. B., Hall D. W., 2004. Spontaneous mutations in diploid Saccharomyces cerevisiae: more beneficial than expected. Genetics 168: 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., 1994. The distribution of mutation effects on viability in Drosophila melanogaster. Genetics 138: 1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A. S., 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336: 435–440. [DOI] [PubMed] [Google Scholar]
- Lee H., Popodi E., Tang H., Foster P. L., 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 109: E2774–E2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman T. D., Michel J. B., Aingaran M., Potter-Bynoe G., Roux D., et al. , 2011. Parallel bacterial evolution within multiple patients identifies candidate. Nat. Genet. 43: 1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. H., Kussell E., 2012. Evolutionary pressures on simple sequence repeats in prokaryotic coding regions. Nucleic Acids Res. 40: 2399–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H. A., Paixão T., Azevedo R. B., Zufall R. A., 2013. Accumulation of spontaneous mutations in the ciliate Tetrahymena thermophila. Genetics 195: 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2010. Evolution of the mutation rate. Trends Genet. 26: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandsberg L. F., Macia M. D., Bergmann K. R., Christiansen L. E., Alhede M., et al. , 2011. Development of antibiotic resistance and up-regulation of the antimutator gene. FEMS Microbiol. Lett. 324: 28–37. [DOI] [PubMed] [Google Scholar]
- Marvig R. L., Johansen H. K., Molin S., Jelsbak L., 2013. Genome analysis of a transmissible lineage of Pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet. 9: e1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., 1996. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu. Rev. Microbiol. 50: 625–643. [DOI] [PubMed] [Google Scholar]
- Miralles R., Gerrish P. J., Moya A., Elena S. F., 1999. Clonal interference and the evolution of RNA viruses. Science 285: 1745–1747. [DOI] [PubMed] [Google Scholar]
- Moxon R., Bayliss C., Hood D., 2006. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial. Annu. Rev. Genet. 40: 307–333. [DOI] [PubMed] [Google Scholar]
- Mukai T., 1964. The genetic structure of natural populations of Drosophila melanogaster. I. Spontaneous mutation rate of polygenes controlling viability. Genetics 50: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness R. W., Morgan A. D., Colegrave N., Keightley P. D., 2012. Estimate of the spontaneous mutation rate in Chlamydomonas reinhardtii. Genetics 192: 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi R. H., Bowen B. M., Wiedmann M., 2010. Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genomics 11: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P., Whitlock M. C., 1997. The probability of fixation in populations of changing size. Genetics 146: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Barton N. H., 1993. Optimality, mutation and the evolution of ageing. Nature 362: 305–311. [DOI] [PubMed] [Google Scholar]
- Patel R. K., Jain M., 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7: e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfeito L., Fernandes L., Mota C., Gordo I., 2007. Adaptive mutations in bacteria: high rate and small effects. Science 317: 813–815. [DOI] [PubMed] [Google Scholar]
- R Development Core Team , 2012. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Rice P., Longden I., Bleasby A., 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Rozen D. E., de Visser J. A., Gerrish P. J., 2002. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12: 1040–1045. [DOI] [PubMed] [Google Scholar]
- Segrè A. V., Murray A. W., Leu J. Y., 2006. High-resolution mutation mapping reveals parallel experimental evolution in yeast. PLoS Biol. 4: e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw F. H., Geyer C. J., Shaw R. G., 2002. A comprehensive model of mutations affecting fitness and inferences for Arabidopsis thaliana. Evolution 56: 453–463. [DOI] [PubMed] [Google Scholar]
- Shaw R. G., Byers D. L., Darmo E., 2000. Spontaneous mutational effects on reproductive traits of Arabidopsis thaliana. Genetics 155: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A., R. Hubley and P. Green, 1996–2010 RepeatMasker Open-3.0. http://www.repeatmasker.org.
- Stevens K. E., Sebert M. E., 2011. Frequent beneficial mutations during single-colony serial transfer of Streptococcus pneumoniae. PLoS Genet. 7: e1002232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Miller S. F., Doak T. G., Lynch M., 2012a Drift-barrier hypothesis and mutation-rate evolution. Proc. Natl. Acad. Sci. USA 109: 18488–18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Tucker A. E., Doak T. G., Choi E., Thomas W. K., et al. , 2012b Extraordinary genome stability in the ciliate Paramecium tetraurelia. Proc. Natl. Acad. Sci. USA 109: 19339–19344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V., 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28: 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Barcelo C., Cabot G., Oliver A., Buckling A., Maclean R. C., 2013. A trade-off between oxidative stress resistance and DNA repair plays a role in the evolution of elevated mutation rates in bacteria. Proc. Biol. Sci. 280: 20130007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade S., Perfeito L., Gordo I., 2010. Rate and effects of spontaneous mutations that affect fitness in mutator Escherichia coli. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman H. A., Scott L. A., Yarber C. D., Bull J. J., 2000. Experimental evolution recapitulates natural evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1677–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor G. L., Lam D. K., Fleming L., Lo R., Whiteside M. D., et al. , 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39: D596–D600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K., Schulz M. H., Long Q., Apweiler R., Ning Z., 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar J. H., 2010. Biostatistical Analysis, Pearson Prentice-Hall, Upper Saddle River, NJ. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.