Abstract
Fibrocytes are a unique population of circulating cells reported to exhibit characteristics of both hematopoietic and mesenchymal cells, and play an important role in wound healing. However putative fibrocytes have been found to lose expression of hematopoietic surface markers such as CD45 during differentiation, making it difficult to track these cells in vivo with conventional methodologies. In this study, to distinguish hematopoietic and non-hematopoietic cells without surface markers, we took advantage of the gene vav 1, which is expressed solely on hematopoietic cells but not on other cell types, and established a novel transgenic mouse, in which hematopoietic cells are irreversibly labeled with green fluorescent protein (GFP) and non-hematopoietic cells with red fluorescent protein (RFP). Use of single-cell transcriptional analysis in this mouse model revealed two discrete types of collagen I (Col I) expressing cells of hematopoietic lineage recruited into excisional skin wounds. We confirmed this finding on a protein level, with one subset of these Col I synthesizing cells being CD45+ and CD11b+, consistent with the traditional definition of a fibrocyte, while another was CD45− and Cd11b−, representing a previously unidentified population. Both cell types were found to initially peak, then reduce post-healing, consistent with a disappearance from the wound site and not a loss of identifying surface marker expression. Taken together we have unambiguously identified two cells of hematopoietic origin that are recruited to the wound site and deposit collagen, definitively confirming the existence and natural time-course of fibrocytes in cutaneous healing.
Keywords: Fibrocytes, wound healing, hematopoietic cells, surface markers, collagen
INTRODUCTION
A multitude of cell types are involved in the process of wound healing, including resident cells from the injured tissue as well as others recruited via the circulation. While inflammatory cells clearly comprise the majority of circulating cells that are recruited to the wound, recent studies have described the involvement of a cell type referred to as a fibrocyte in these processes. First reported in 1994 [1], it has been postulated that fibrocytes possess characteristics of both hematopoietic and mesenchymal cells, are recruited via the blood to sites of injury, and play a role in physiologic wound healing as well as pathologic fibroses [2–8]. Specifically, it has been suggested that fibrocytes are hematopoietic and derived from monocyte/macrophage lineage cells because they express the monocyte surface markers CD11b, CD13, and the leukocyte marker CD45 [9,10]. However, in both in vitro and in vivo studies, fibrocytes have also been shown to possess characteristics typical of mesenchymal cells, such as spindle-shaped morphology and expression of collagen. Unfortunately, research efforts to more clearly elucidate the origin and in vivo role of fibrocytes in wound healing are greatly hindered by both this complex cell surface signature, as well as the transient expression of the identifying cell surface molecules. In fact, fibrocytes lose the expression of hematopoietic surface markers during differentiation [4,6,11,12], and can become indistinguishable from resident tissue fibroblasts using traditional tracing methodologies. This has made their long-term fate in tissues an important area of controversy [13].
To better track recruited cells in vivo, several studies have employed bone marrow transplantation and/or parabiosis to a sex-mismatched or green fluorescent protein (GFP) labeled mouse prior to wounding [4,14–16]. Although these reports largely support the involvement of bone marrow-derived collagen producing cells in wound healing, the true lineage of these cells could not be distinguished, as both mesenchymal and hematopoietic bone marrow derived cells are tracked similarly in these models. Moreover, the long-term role of the fibrocyte in wounds remained unclear, as surface marker independent tracking of these cells throughout healing requires the simultaneous demonstration of hematopoietic origin and collagen production, a level of granularity not achievable using standard bone-marrow tracing approaches.
In order to unambiguously determine the existence and long-term fate of hematopoietic lineage cells capable of producing collagen (i.e. fibrocytes) during wound healing, the ability to clearly and permanently distinguish hematopoietic cells and non-hematopoietic cells without surface markers is required. Unfortunately, previously available transgenic mouse models for hematopoietic lineage tracking only target selected lineages or particular differentiation stages [17]. For example, CD45, which is currently regarded as the most specific marker for hematopoietic cells, is expressed only during later stages of hematopoietic progenitor cell development and therefore incompletely identifies these cells [18].
In this study, we therefore bred a cre-activated dual fluorescence mouse strain with cre- expressing mice under the vav 1 gene promoter. Vav 1 is a pan-hematopoietic marker expressed almost exclusively in the hematopoietic system, but not in other tissues [19,20]. Its activity can be detected during early embryologic stages of hematopoiesis and in more ancestral uncommitted progenitors, and when used in this system, facilitates a ubiquitous and robust differentiation between hematopoietic and non-hematopoietic cells in vivo.
Utilizing this novel double transgenic mouse model, in which hematopoietic lineage cells are irreversibly labeled with green fluorescent protein and non-hematopoietic cells are labeled with red fluorescent protein (RFP), we examined the recruitment of circulatory cells in wound healing, and identified the presence of two transient, hematopoietic-derived cell types with distinct characteristics of a fibrocyte, definitively confirming their existence and natural time-course in wound healing.
METHODS
Mice
Animal care was provided in accordance with institutional guidelines and all animal protocols were approved by the Administrative Panel on Laboratory Animal Care at the Stanford University School of Medicine. A mouse strain containing a CMV beta-actin enhancer-promoter, driving a loxP-flanked red fluorescent protein (RFP) preceding GFP [21], was obtained from Jackson Laboratories (B6.129(Cg) -Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J) or (mTmG). This mouse is designed to express cell membrane localized RFP (membrane tomato/mT) ubiquitously prior to Cre recombinase exposure, and cell membrane-localized GFP (membrane green/mG) in Cre recombinase expressing cells. Prior to cross-breeding this mouse, all cells in the mTmG were RFP labeled. A second mouse containing Cre recombinase driven by a global hematopoietic gene vav 1 (B6.Cg-Tg(Vav1-cre)A2Kio/J) (Vav-Cre) [22] was obtained from Jackson Laboratories (Bar Harbor, ME). By crossbreeding Vav-Cre and mTmG strains, a double transgenic strain (Vav-Cre/mTmG; termed Vav-recombinant, or VavR) was created, in which all hematopoietic cells driven by vav were labeled with GFP after a Cre-mediated excision of the floxed RFP, and non-hematopoietic cells with no Cre activity remained RFP labeled (Figure 1A). Importantly, any cells that ever transcribed vav 1 at any point during development were permanently labeled with GFP, eliminating the problem of transient expression of hematopoietic lineage markers. The Vav- Cre strain was maintained hemizygously in order to reduce Cre-associated toxicity, while the mTmG strain was maintained homozygously. Transgenic progeny were identified with common genotyping techniques as outlined in the corresponding Jackson Laboratories protocols. C57BL/6 wild-type mice (Jackson Laboratories) were used for comparison with the double transgenic mice. Experiments were performed with female mice at the age of 8 to 12 weeks.
Figure 1.
VavR double transgenic mice express GFP in hematopoietic cells and RFP in nonhematopoietic cells. (A): On cross-breeding the mTmG mouse with the Vav-Cre, all cells expressing vav 1 (hematopoietic) in the progeny exclude RFP due to cleavage at loxP sites and instead express GFP. Cells not expressing vav 1 (nonhematopoietic) remain RFP+. (B): Flow cytometry of peripheral blood cells is nonfluorescent in wild-type mice and GFP+ in VavR mice. Circulating RFP+ cells are not detected in the peripheral blood of VavR mice. (C): Histological images of spleen, liver, heart, and skin showing distribution of RFP and GFP cells. Nuclei are stained with 4′,6-diami-dino-2-phenylindole. Scale bar =75 μm. (D): Flow cytometry of cells isolated from the heart, spleen, liver, and skin of VavR mice indicates relative proportions of GFP+ and RFP+ cells in each of these organs. (E, F): Immunohistochemistry for CD45 in the spleen (E) and cytokeratin in the skin (F) shows CD45 (purple) colocalizing with GFP and cytokeratin (purple) colocalizing with RFP. White arrows highlight areas of colocalization. Gray arrows illustrate a small population of GFP+/C45− cells in the spleen, indicating that CD45 is not a ubiquitous marker of hematopoietic lineage cells. Gray scale bar =75 μm. White scale bar =150 μm. Abbreviations: GFP, green fluorescent protein; RFP, red fluorescent protein.
Cell culture
Bone marrow was flushed from the tibias and femurs of mice and passed through a 70-μm filter. Cells were plated in a 10-cm dish at 1 × 106 cells per cm2 in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS). Cells were passaged at 80% confluence. To isolate CD11b+ cells from bone marrow, MACS Separation Columns and CD11b MicroBeads (both from Miltenyi Biotec, Auburn, CA) were used.
Excisional wound healing model
The excisional wound healing model was utilized as previously described [23]. Briefly, the dorsal surface of the mouse was shaved and depilated. Two 6-mm full thickness wounds were created on either side of the midline. A donut-shaped silicon splint was placed with 6-0 nylon sutures to prevent wound contraction. Wounds were followed until closure and tissue samples were harvested at 7, 14 and 28 days after surgery for histologic analyses. Normal skin was also collected as a control.
Histology
Tissues were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) overnight. The tissues were then cryoprotected with 30% sucrose for 24 hours, and embedded in OCT Compound (Sakura Finetek USA, Torrance, CA). Cryosections (10-μm thickness) were prepared and used for immunohistochemistry. Briefly, antigen retrieval was performed in 1% SDS in PBS, followed by blocking in 1% goat serum in PBS. The following primary antibodies were used at a 1:100 dilution and incubated overnight at 4°C: rabbit anti-CD31 (ab28364 Abcam, Cambridge MA), rabbit anti-αSMA (ab5694 Abcam), rat anti-F4/80 (ab16911 Abcam), rabbit anti-collagen I (ab34710 Abcam) and rabbit anti-cytokeratin (ab97764 Abcam). Alexa Fluor 350-congugated goat antibodies for rat or rabbit IgG (Molecular Probes, Eugene, OR) at a 1:250 dilution for 1hr at room temperature were used as secondary antibodies. Fluorescent conjugated antibodies for CD11b (15-0112-83 Pe-Cy5 conjugated, eBioscience, San Diego, CA), F4/80 (48-4801-82 eFluor 450 conjugated, eBioscience) and CD45 (559864 APC conjugated, BD Pharmigen, Franklin Lakes, NJ) were also used at a 1:50 dilution. Nuclei were stained with 4′,6-diamidino- 2-phenylindole (DAPI). Sections were finally mounted with ProLong Gold antifade reagent (Molecular Probes) and photomicrophs were taken on a Leica DM5000 B upright or Leica SP2 AOBS confocal microscope (Leica Microsystems, Buffalo Grove, IL). For quantitative analysis of histology, three randomly selected fields (at 400x magnification) per section were used.
Flow cytometry
For skin samples, tissues were cut into small pieces and digested with 0.5 mg/ml Liberase TL (Roche Diagnostics, Indianapolis, IN) for 60 minutes at 37°C. Cells were then filtered through a 70-μm mesh. For bone marrow cells, bone marrow was flushed from the tibias and femurs of mice and passed through a 70-μm filter. Mature hematopoietic cells were depleted when indicated using a Lineage Cell Depletion Kit in MACS Separation Columns (both from Miltenyi Biotec). Mononuclear cells from blood were extracted from buffy coat by Ficol-Paque density separation [24]. Isolated cells were stained with the following fluorochrome-conjugated antibodies at a 1:20 dilution on ice for 30 minutes: rat anti-CD11b-PE-Cy7 (15-0112-83, eBioscience), rat anti-CD45-Pacific Blue (103125, BioLegend, San Diego, CA) or rat anti-CD45-PE-Cy7 (25-0451-81, eBioscience), rat anti-F4/80-eFluor 450 (48-4801-82, eBioscience) and rat anti-Sca-1-PE-Cy7 (122514, BioLegend) antibodies. When co-staining with collagen I, cells were first stained with CD45 and CD11b on ice for 30 minutes. Cells were then fixed in 4% PFA for 15 minutes at room temperature, permeabilized with 0.1% TritonX in PBS on ice for 5 minutes, blocked in FACS buffer, incubated with 1:25 rabbit anti-mouse collagen I antibody (ab34710, Abcam) for 1 hour at room temperature and finally followed by counter-incubation with 1:100 goat anti- rabbit Alexa Fluor 350 secondary antibody (Molecular Probes). Samples were analyzed on a BD LSR II flow cytometry machine having a UV laser.
Single cell transcriptional analysis
Single cell transcriptional analysis was performed as previously described [25,26]. Briefly, single cells were sorted into each well of a 96-well plate preloaded with 10 μl of a master mix containing Tris-EDTA buffer (pH 7.0), Cells Direct reaction mix (Invitrogen), and SUPERase-In RNAse inhibitor (Applied Biosystems, Foster City, CA). Cells were lysed and reverse transcription was performed (20 minutes at 50°C, 2 minutes at 95°C) using Superscript III reverse transcriptase enzyme (Invitrogen, Carlsbad, CA) and target gene-specific TaqMan assay probes (Applied Biosystems, Foster City, CA), followed by a 22-cycle pre-amplification (denature at 95°C for 15 seconds and anneal at 60°C for 4 minutes) using Platinum Taq Polymerase (Invitrogen). Resultant single cell cDNA was mixed with sample loading agent (Fluidigm, South San Francisco, CA) and Universal PCR Master Mix (Applied Biosystems) and loaded into 48.48 Dynamic Array chips (Fluidigm) along with target gene-specific TaqMan assays and assay loading agent (Fluidigm). Quantitative polymerase chain reaction (PCR) and analysis were performed on the BioMark reader system (Fluidigm). A list of genes and primers is shown in Supplementary Table 1.
Immunoblot
To evaluate whether hematopoietic cells in the wound deposit collagen I, VavR mice were wounded and the tissue was harvested as above after 7 days. 250,000 GFP+ or RFP+ cells were isolated by flouresence-activated cell sorting (FACS) using a BD Aria II flow cytometry machine, with 250,000 RFP+ and GFP+ cells from unwounded VavR skin were isolated simultaneously as controls. The cells were lysed in RIPA buffer containing protease inhibitor. Cell lysates containing equal cell numbers were loaded with NuPAGE LDS Sample Buffer (Life Technologies, Grand Island, NY) on 12% polyacrylamide gels (Life Technologies) in SDS-Laemmli buffers (Life Technologies), and transferred to Nitrocellulose membrane (Life Technologies). After the blots were blocked with 5% milk in PBS, target proteins were probed with primary antibodies at 1:1000 for collagen I (ab21286 Abcam) or 1:2000 for α-tubulin (ab52866 Abcam). The blots were probed with horseradish peroxidase-conjugated secondary antibodies, and the bound peroxidase was visualized by enhanced chemiluminescence (GE Life Sciences, Piscataway, NJ).
Chemotaxis assay
CD11b+ cells isolated from cultured bone marrow cells were suspended in serum free medium and 1 × 105 cells in 200 μl were seeded into the upper chamber of a Transwell insert (8.0-μm pore size, Corning Life Sciences, Lowell, MA). Medium alone or medium containing stromal cell-derived factor-1α (SDF-1α) (100 ng/ml), monocyte chemoattractant protein-1 (MCP-1) (100 ng/ml), and transforming growth factor-β1 (TGF-β1) (10 ng/ml) (all from R&D Systems, Minneapolis, MN) was added to the lower chamber (600 μl). The cells were then allowed to migrate for 4 hours at 37°C, after which they were fixed in 100% methanol for 15 minutes and stained with hematoxylin for 10 minutes. Cells retained on the top surface of the membrane were eliminated using a cotton swab. The number of migrated cells was counted using a light microscope (200x magnification) and plotted as numbers per field.
Quantitative real-time PCR
CD11b+ cells isolated from cultured bone marrow cells were serum-starved overnight, and cultured with SDF-1α (100 ng/ml), MCP-1 (100 ng/ml), and TGF- β1 (10 ng/ml) for 24 hours. One microgram of total RNA was isolated from the cells using an RNeasy Mini Kit (Quiagen, Hilden, Germany), followed by reverse transcription. We amplified cDNA for 40 cycles with the ABI PRISM 7900HT sequence detection system and a TaqMan PCR Master Mix (both from Applied Biosystems). Expression levels were calculated by the comparative CT method using GAPDH as an endogenous reference gene.
Statistical analysis
Data are expressed as mean ± standard error of the mean. Comparisons of multiple groups were done by analysis of variance with Bonferroni’s multiple t-test. A value of * p < 0.05 was considered significant. For single cell transcriptional data, we employed a partitional clustering algorithm to separate distinct functional subtypes.
RESULTS
The VavR double transgenic reporter mouse model can distinguish hematopoietic and non-hematopoietic lineage cells
In the VavR mouse model, all cells that express the enzyme Cre recombinase under the control of the vav 1 gene promoter have RFP excised and the membrane bound green fluorescent protein (GFP) activated (Figure 1A). The VavR mice were viable, fertile and appeared phenotypically normal, yet possessed a distinctive labeling of GFP in the entire hematopoietic cell lineage (Figure 1B–D). Accordingly, the peripheral blood of the VavR mice showed universal expression of GFP. Moreover, a histological and flow cytometric analysis of solid organs revealed that the spleen was composed mainly of GFP+ hematopoietic cells (94.9% GFP+). Conversely, the liver and heart were mainly positive for RFP (57.3 and 78.8% RFP+), consistent with the non-hematopoietic origin of the hepatocytes and cardiomyocytes largely composing these organs. The majority of skin cells were also RFP+ (89.7%), including epithelial cells and fibroblasts, with GFP+ hematopoietic cells sparsely distributed in the dermis, likely representing transient or resident inflammatory cells.
Performing immunohistochemistry to further confirm this model, staining for the hematopoietic cell marker CD45 in the spleen demonstrated specificity to GFP+ cells (Figure 1E), while staining for non-hematopoietic cell markers, including CD31+ endothelial cells in the heart and cytokeratin+ epithelial cells in the skin, demonstrated specificity to RFP+ cells (Supplemental Figure 1A and Figure 1F). Interestingly, not all GFP+ cells co-localized with CD45 in the spleen, indicating that CD45 is not a ubiquitous marker of all hematopoietic cells.
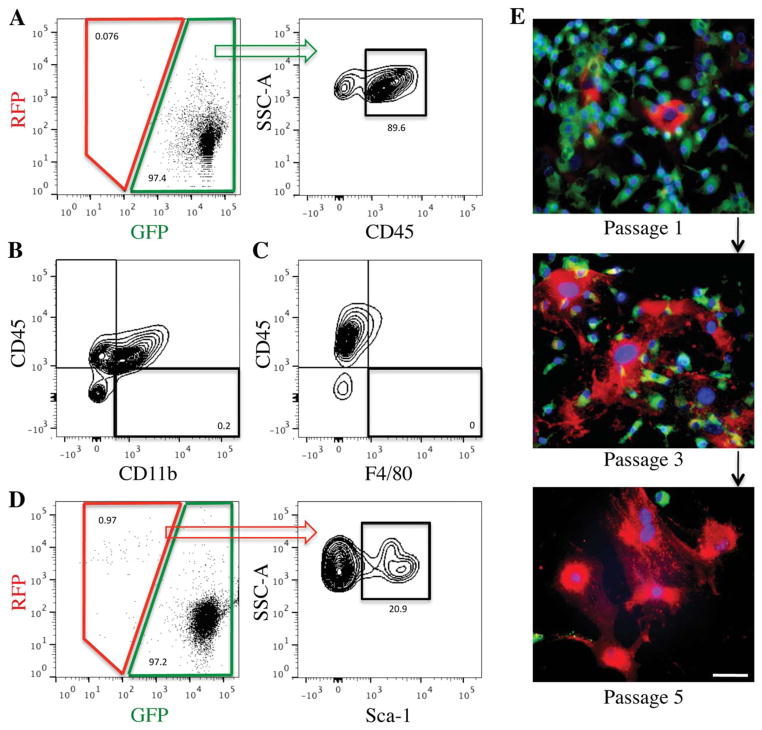
Within the bone marrow, flow cytometry confirmed that the vast majority of cells (~90%) were both GFP+ and CD45+ (Figure 2A). Similar to the spleen, a small GFP+/CD45− population (<10% of total cells) was observed, yet these GFP+/CD45− cells were negative for the monocyte and macrophage markers CD11b and F4/80 (Figure 2B–C, Supplemental Figure 1B), excluding them from being traditionally defined fibrocytes. Additionally, we found that early passages of cultured bone marrow cells showed a high correlation between the expression of the monocyte/fibrocyte marker CD11b and GFP positivity (Supplemental Figure 1C), allowing for a CD11b-based enrichment of hematopoietic-lineage fibrocytes from cultured bone marrow cells for subsequent in vitro analyses.
Figure 2.
Bone marrow cells in the VavR double transgenic mouse are primarily GFP+ but display a population of RFP+ bone marrow mesenchymal stem/progenitor cells (BMSCs). (A): Flow cytometry analysis of the whole VavR bone marrow without Lineage depletion reveals negligible RFP+ cells. Most GFP+ cells are CD45+ (percentage relative to total GFP+ cells). (B, C): Flow cytometry analysis of GFP+ bone marrow cells demonstrates that GFP+/CD45− cells do not display markers for either monocytes, CD11b+, or macrophages, F4/80+ (percentage relative to total GFP+ cells). (D): Lineage depletion of the bone marrow cells allows for detection of RFP+ cells, a portion of which are positive for the BMSC marker Sca-1 (percentage relative to total RFP+ cells). (E): Culture of bone marrow cells in vitro increases prevalence of RFP+ cells over passages. Scale bar =50 μm. Abbreviations: GFP, green fluorescent protein; RFP, red fluorescent protein.
Also of note, the bone marrow was found to contain a small population of RFP+ stromal cells (approximately 1% of lineage-negative cells) (Figure 2D). Some of these RFP+ cells (approximately 20%) expressed Sca-1, consistent with traditional definitions of bone marrow mesenchymal stem/progenitor cells (BMSCs). Moreover, culture of bone marrow cells based on plastic adherence demonstrated that RFP+ cells expanded in number and overgrew GFP+ cells following serial passage (Figure 2E), which is compatible with previous reports on culture expansion of BMSCs [27,28].
Hematopoietic and non-hematopoietic cells in VavR wound healing
Following these confirmatory studies in normal tissues, an excisional wound model was utilized on VavR double transgenic mice to confirm cell dynamics following injury. Compatible with our previous report in wild-type mice [23], wound closure was observed by day 14, and hematoxylin-eosin staining showed primarily cellular infiltration with inflammatory cells and granulation tissue formation around day 7 (data not shown). Moreover, fluorescence microscopy and flow cytometry demonstrated that the number of GFP+ hematopoietic cells infiltrating into the wound peaked on day 7, and returned to near baseline levels by day 28 (Figure 3A–C), consistent with the recruitment of inflammatory cells in the early stages of wound healing. As expected, the majority of these infiltrating GFP+ cells were also CD45+ (Figure 3C, Supplemental Figure 1D), although the GFP+/45- cell fraction was found to peak at day 7 and return to baseline levels by day 28, indicative of a dynamic recruitment and remodeling process. Interestingly, the total number of RFP+ non-hematopoietic cells, as well as the number of RFP+/CD31+ endothelial cells, showed a gradual increase in number until day 14, indicating an active process of granulation tissue formation, neovascularization and remodeling throughout the wound healing process (Figure 3A–E, Supplemental Figure 1E).
Figure 3.
Hematopoietic and nonhematopoietic cells are well distinguished during wound healing in VavR double transgenic mice. (A, B): Histological images and quantification during wound healing show GFP+ hematopoietic cells peaking at day 7, while RFP+ nonhematopoietic cells peak at day 14. Scale bar =50 μm. Quantification includes number of RFP+ and GFP+ cells per high power field (hpf). (C): Flow cytometry demonstrates that majority of the GFP+ cells in the wound were CD45+ (numbers relative to total GFP+ cells). (C–E): RFP+/CD31+ endothelial cells are seen to peak in the wound at day 14 by flow cytometry (C) (percentage relative to total RFP+ cells), and quantification of immunostaining (D, E). Blue: CD31. White arrows highlight areas of RFP/CD31 colocalization. Scale bar =100 μm. Of note, RFP and GFP percentages in (C) are relative numeric values (i.e., RFP counts vs. RFP +GFP counts), and do not reflect absolute cell number changes. Abbreviations: GFP, green fluorescent protein; RFP, red fluorescent protein.
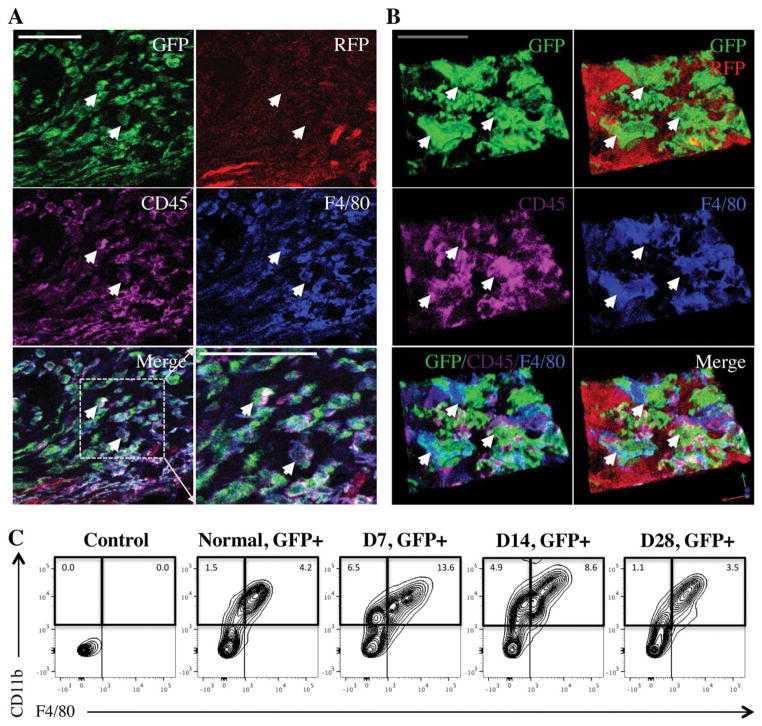
Looking more closely at the GFP+ cells, immunohistochemistry and flow cytometry confirmed that the majority of the GFP+ hematopoietic cells in the wound were monocyte/macrophage lineage cells, demonstrating positive expression of the markers CD45, CD11b and F4/80 (Figure 4A–C, Supplemental Figure 2A–D), a finding which is consistent with the normal wound healing progression. Additionally, these recruited GFP+ hematopoietic cells were often found to localize around the vasculature, as demonstrated via immunohistochemical staining with the pericyte marker Desmin (Supplemental Figure 3A), consistent with prior reports suggesting a role for hematopoietic derived cells in the support of neovascularization [29].
Figure 4.
Monocyte/macrophage lineage cells are the major population recruited to the wound. (A, B): Standard and three-dimensional immunohistochemical imaging of F4/80 (blue) on day 7 demonstrating that F4/80+ cells in the wound colocalize with GFP+ and CD45+ (purple) cells. White scale bar =40 μm; gray scale bar =20 μm. White arrows highlight areas of GFP+/CD45+/F4/80+ colocalization. (C): Flow cytometry of wound lysate demonstrating the GFP+/CD11b+/F4/80+ cell fraction (percentage relative to total cells) peaks at day 7 and retracts to baseline levels by day 28. Abbreviations: GFP, green fluorescent protein; RFP, red fluorescent protein.
Single cell transcriptional analysis of GFP+ hematopoietic cells in the wound identifies the putative fibrocyte
To further analyze the function of the multitude of GFP+ hematopoietic cells recruited to the wound, a single cell transcriptional analysis of this population was performed as previously described [25,26]. This process allowed for the transcriptional characterization of neutrophils, monocytes and other hematopoietic lineage cells in the wound, such as fibrocytes, without the need for a priori assumptions based upon cell surface marker expression. Employing a partitional clustering of the single cell data, GFP+ cells were subdivided into three populations (Figure 5A). The first population represented putative monocyte/macrophage lineage cells and was characterized by high expression of the surface markers CD11b (Itgam) and F4/80 (Emr1). The second population consisted of putative T cells and was characterized by high expression levels of the T-helper cell surface antigen CD4. The third population consisted of other hematopoietic lineage cells recruited to the wound, primarily granulocytes.
Figure 5.
Single-cell transcriptional analysis reveals importance of monocyte/macrophage lineage cells and their expression of collagen in wound healing. (A): Partitional clustering of green fluorescent protein+ cells in the wound. Columns represent individual cells and rows correspond to genes. Yellow indicates increased expression and blue decreased expression. Monocyte/macrophage lineage cells are characterized by high expression of CD11b (Itgam) and F4/80 (Emr1), and express type I and type III collagen. (B): The relative percentage of collagen I expressing monocyte/macrophage cells in the wound increases dramatically at day 7 postwounding and slowly regresses toward baseline levels at subsequent time points (percentage relative to total Col I+ monocytes/macrophages). (C): Scatter plots representing correlation between collagen I expression and surface markers. There is heterogeneity in the expression of CD45 and CD11b among collagen I expressing hematopoietic cells.
Interestingly, a number of the monocyte/macrophage lineage cells expressed type I collagen (relative frequency: normal skin (8%), day 7 (43%), day 14 (29%) and day 28 (20%); Figure 5B), in addition to type III collagen and other fibrotic genes. Since these cells were by definition hematopoietic lineage, and collagen I and III expression is believed to be a defining feature of fibrocytes that is commonly absent from other hematopoietic cells [1], the presence of high expression levels of these collagen genes supported their existence as fibrocytes. Consistent with the reported secretory profile of fibrocytes [4], many of these monocyte/macrophage lineage cells also showed high expression levels of growth factors, such as TGF-β1, MCP-1 (Ccl2), and vascular endothelial growth factor (VEGF). Moreover, these cells also expressed receptors for growth factors, such as receptors for SDF-1α (Cxcr4), MCP-1 (Ccr2), and TGF-β1 (Tgfr1), similar to traditionally defined fibrocytes [30,31].
Upon further examining the correlation between expression of collagen I and specific cell surface markers within these hematopoietic lineage cells, approximately 5% of collagen I expressing cells across all time points were low in CD45, while 60% of collagen I expressing cells were low for CD11b (Figure 5C), indicating that hematopoietic lineage collagen expressing cells (fibrocytes) are incompletely defined by traditional surface marker profiling.
Protein confirmation of the hematopoietic lineage fibrocyte and identification of a novel population of blood-derived collagen producing cells
An immunoblot, as well as immunohistochemical and flow cytometric analyses were next performed to confirm these transcriptional findings on a protein level. Consistent with the presence of a dynamically recruited collagen producing hematopoietic cell, immunoblot of hematopoietic lineage (GFP+) cells isolated from normal skin and day 7 wounds demonstrated the presence of collagen I producing cells at day 7 post wounding, with an absence of this cell type in normal skin (Figure 6B, Supplemental Figure 3B). As a control for this assay, stromal cell (RFP+) collagen I production was also found to increases following wounding. Looking more closely at recruited GFP+ cells, the existence of the traditionally defined fibrocyte was corroborated by immunohistochemical co-localization of hematopoietic lineage GFP expression with collagen I and the monocyte/macrophage marker F4/80 (Figure 6A). Moreover, flow cytometry demonstrated a dynamic population of GFP+/CD45+/Cd11b+/Col I+ cells within the wound, with these cells peaking in frequency at day 7, and returning to near-baseline levels by day 28 (Figure 6C).
Figure 6.
Two hematopoietic-derived fibrocyte populations are identified: GFP+/CD45+/CD11b+/Col I+ and GFP+/CD45−/CD11b−/Col I+. (A): Immunohistochemistry confirms the presence of a monocyte/macrophage subpopulation that deposits type I collagen. Purple: Collagen I, Blue: F4/80. Scale bar =20 μm. (B): Immunoblot demonstrating the recruitment of hematopoietic lineage (GFP+) Col I producing cells at day 7 postwounding, with an absence of this cell type in normal skin. Stromal cell (RFP+) Col I production also increases following wounding. (C): Flow cytometric analysis demonstrating that GFP+/CD45+/CD11b+/Col I+ cells are recruited to the wound, peak at day 7, and reduce in numbers by day 28 (percentage based on total GFP+ cells). (D): A second, non-monocyte hematopoietic cell subpopulation (GFP+/CD45−/CD11b−/Col I+) depositing type I collagen and demonstrating a similar recruitment pattern is also identified via flow cytometry (percentage based on total GFP+ cells). Abbreviations: GFP, green fluorescent protein; RFP, red fluorescent protein.
Leveraging the surface marker independent hematopoietic lineage tracing capacities of the VavR model, we also looked at the GFP+/CD45− fraction of cells recruited to the wound site, to determine if hematopoietic lineage cells other than those defined by traditional surface marker definitions possessed the ability to deposit collagen. Consistent with our single-cell transcriptional data, a rare population of GFP+/CD45−/CD11b−/Col I+ cells was identified via flow cytometry, and displayed a similar recruitment profile as their GFP+/CD45+/Cd11b+/Col I+ counterparts (Figure 6D). This previously unidentified cell population demonstrates the heterogeneity of hematopoietic cells contributing to the wound environment, and highlights the difficulty of utilizing surface markers for cell identification in this setting.
Fibrocytes can be isolated from bone marrow-derived CD11b+ precursors, migrate towards an SDF1α signal and differentiate following in vitro and in vivo stimuli
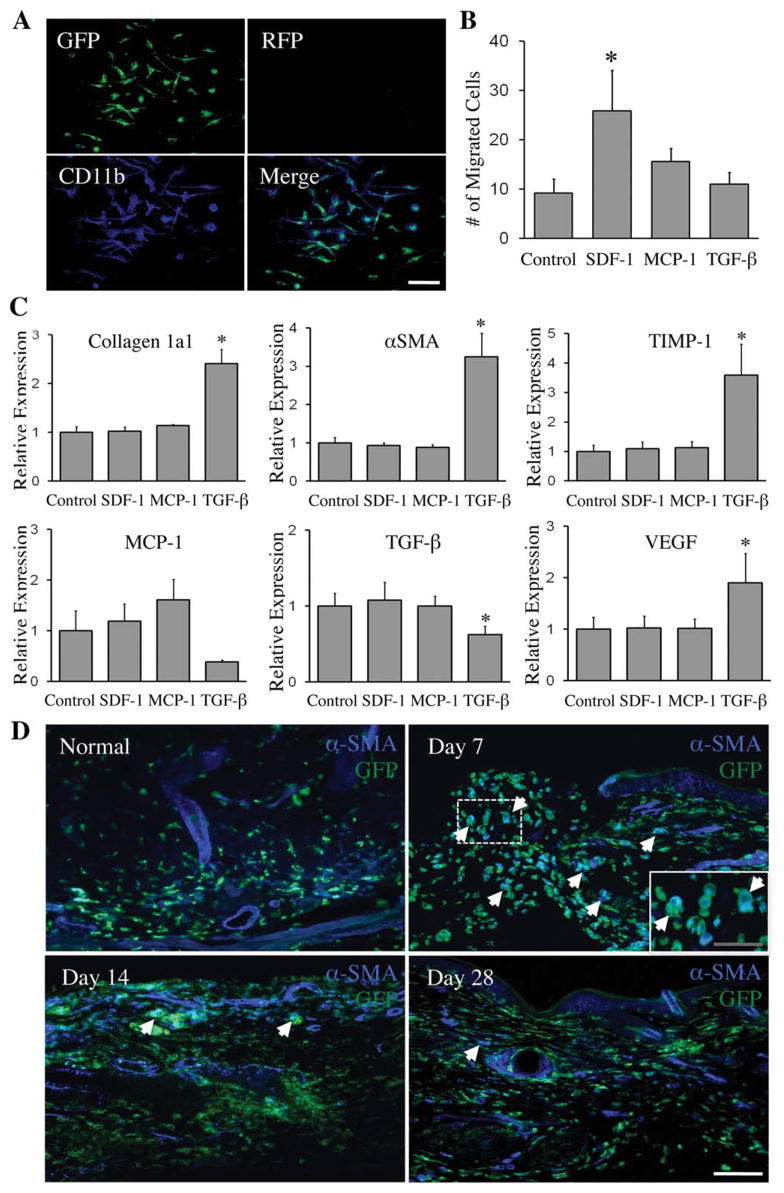
Following confirmation of the existence and dynamic recruitment of hematopoietic-lineage collagen producing cells to the wound, we focused next on the mobilization mechanism and differentiation capacity of these cells. Building upon reports that traditionally defined fibrocytes can be derived from CD11b+ leukocyte/monocyte fractions found in the circulation and secondary lymphatic organs [32], we focused our studies on CD11b+ cells isolated from primary cultured bone marrow, first confirming their hematopoietic lineage based on GFP co-expression (Figure 7A). Because monocyte/macrophage lineage cells in the wound expressed receptors for SDF-1α, MCP-1, and TGF-β1 in the single-cell transcriptional analysis, which are known to be responsible for directing fibrocyte recruitment, we next examined the effects of these growth factors on the GFP+/CD11b+ bone marrow cells in vitro. Chemotaxis assays showed that SDF-1α induced migration of the bone marrow-derived GFP+/CD11b+ cell population (Figure 7B), providing a mechanism for recruitment of fibrocytes to the wound site, where SDF-1 is know to be upregulated [33].
Figure 7.
SDF-1α causes bone marrow-derived CD11b+ cell migration, TGF-β1 regulates gene expression in CD11b+ cells and αSMA, the myofibroblast marker is confirmed in a subpopulation of hematopoietic cells infiltrating the wound in vivo. (A): Primary cultures of VavR bone marrow were MACS sorted for CD11b and recultured. Immunofluorescence confirmed plated cells as GFP+/CD11b+. Blue: CD11b. Scale bar =50 μm. (B): Chemotaxis assay with SDF-1α, MCP-1, and TGF-β1 shows migration of CD11b+ cells in response to SDF-1α, with *, p <.05 compared to control (n =4). (C): SDF-1α, MCP-1, and TGF-β1 treatment of CD11b+ cells shows TGF-β1 upregulating fibrosis-related genes (type I collagen, αSMA, and TIMP-1) and VEGF by qPCR. *, p <.05 compared to control (n =4). (D): αSMA is expressed in a subpopulation of infiltrating hematopoietic cells at earlier time points in the wound, but regresses with the healing of wounds. White arrows highlight areas of αSMA/GFP colocalization. White scale bar =75 μm; gray scale bar =40 μm. Abbreviations: αSMA, smooth muscle actin; GFP, green fluorescent protein; RFP, red fluorescent protein; TIMP-1, tissue inhibitor of metalloproteinase-1; VEGF, vascular endothelial growth factor.
Providing insight into fibrocyte differentiation, TGF-β1 was found to up-regulate the expression of type I collagen, α-smooth muscle actin (αSMA), and tissue inhibitor of metalloproteinase-1 (TIMP-1) in these GFP+/CD11b+ cells, all markers of mature fibrocytes (Figure 7C). Furthermore, TGF-β1 regulated the expression of growth factors, decreasing MCP-1 and TGB-β1, and increasing VEGF. These findings demonstrate that functional fibrocytes can be derived from a C11b+ subpopulation of hematopoietic bone marrow cells.
Finally, to assess whether hematopoietic lineage cells differentiate into myofibroblasts after recruitment into the wound in vivo, expression of the myofibroblast marker αSMA was evaluated using immunohistochemistry. Interestingly, GFP+ hematopoietic cells showed co-localization with αSMA across all post-injury time points, peaking at day 7 (Figure 7D and Supplemental Figures 4–5). These data indicate that hematopoietic lineage cells can not only contribute to the extracellular matrix via production of collagens I and III, but also play a role in wound contraction.
DISCUSSION
In this paper, we have established a new double transgenic reporter mouse model (VavR) that uses the vav 1 gene as a specific marker for hematopoietic lineage cells. Utilizing this system, we unambiguously confirmed the existence and natural time-course of two hematopoietic- derived, collagen producing cell types that are recruited to cutaneous wounds.
One of the ongoing challenges in the field of fibrocyte biology is the complex definition of these cells. Defined as being both hematopoietic and collagen producing, fibrocytes are traditionally identified in part by co-expression of the putative hematopoietic cell marker CD45 and collagen I. However, reliance on CD45 for identification of hematopoietic lineage cells is not ideal, as this surface marker is mostly expressed during later stages of hematopoietic progenitor cell development, potentially missing specific cell populations in vivo [21]. Moreover, CD45 and CD11b, another common fibrocyte marker, have been shown to be down-regulated in vitro as these cells transition to a more collagen-producing phenotype [11]. Similarly, because it has been shown that CD45 expression is lost by many cells recruited from the bone marrow throughout the course of normal wound healing [15], it is unclear whether the previously described transience of fibrocytes in skin wounds is the result of a loss of CD45 expression [5], or the actual disappearance of these cells from the healed wound.
Because the vav 1 gene is ubiquitously expressed by all hematopoietic lineage cells [20,22,34,35], including during embryologic hematopoiesis [19], utilization of this gene to irreversibly label all cells within the hematopoietic lineage enabled us to definitively confirm the existence, as well as characterize the natural time course of blood-derived collagen producing cells in the wound. The GFP+/CD45+/CD11b+/Col I+ cells first identified in this work are consistent with traditional definitions of the fibrocyte [9,11], and were found to be actively recruited to the wound site. Importantly, the long-term decrease in numbers of these cells was not found to be the result of a down-regulation of CD45 or CD11b, as the GFP+/CD45+/CD11b−/Col I+ and GFP+/CD45−/CD11b−/Col I+ populations did not display a long-term increase in numbers, as would be required with the loss of these surface markers from the originally recruited GFP+/CD45+/CD11b+Col I+ cells. As such, the loss of this specific hematopoietic collagen producing population over time likely results from either an apoptotic or migratory event, with the alterative explanation of a concomitant downregulation in collagen production not consistent with the overall low numbers of GFP+ hematopoietic lineage cells found in the healed wound.
In this study, we also identify a novel population of CD45−/CD11b−/Col I+ hematopoietic lineage (GFP+) cells that display a similar recruitment profile to cutaneous wounds as the CD45+/CD11b+/Col I+ fibrocyte, but would have been missed by conventional surface marker-based definitions. The existence of a recruited, bone-marrow derived CD45− cell in cutaneous wounds has been previously demonstrated through the utilization of bone marrow transplantation of GFP+ cells [15]. However, a definitive determination of collagen expression in these cells was not performed in this work, and they were deemed to be distinct from fibrocytes as they lacked the defining surface marker CD45. Moreover, experiments from this same study demonstrated the ability of multiple bone marrow compartments to home to site of cutaneous injury [15], further illustrating how a definitive lineage tracing of the CD45−/CD11b−/Col I+ cells identified herein would be impossible without the use of the VavR model. Interestingly, this GFP+/CD45−/CD11b−/Col I+ population displayed a comparable transience in the long-term as the traditionally defined fibrocyte, suggestive of a similar functional role for these cells within the healing wound.
Given the early post-wounding recruitment profiles of both the traditionally and non-traditionally defined hematopoietic lineage collagen producing cells identified in this study, their contributions are likely greatest in the early phase of wound healing. Granulation tissue formation and neovascularization are critical elements of the wound healing process during this time, and it has been suggested that fibrocytes can contribute to angiogenesis through secretion of several pro-angiogenic factors such as VEGF [11,36]. Focusing on the CD11b positive fraction of bone marrow derived GFP+ hematopoietic cells, we were able to confirm this pro-vasculogenic phenotype in our in vitro analysis. Furthermore, our data indicating the presence of GFP+ hematopoietic cells in the perivascular space further support a paracrine contribution of these cells to neovascular processes within the wound.
Aside from their obvious role in extracellular matrix deposition, another potential role of fibrocytes within the wound is differentiation into myofibroblasts. While some groups report that fibrocytes can make significant contributions to the myofibroblast population, especially during the early phases of wound healing [4,14], others suggest that bone marrow derived fibrocytes do not play a major role in the accumulation of these cells [16]. This discrepancy may be due in part to differences in the cell identification methods used in these studies, as fibrocytes defined via negative selection demonstrated the highest expression of the myofibroblast marker αSMA [4], while fibrocytes defined by via bone marrow tracking of GFP or Col Ia I/GFP labeled cells demonstrated a lower frequency or absence of αSMA production [14,16]. In this work, we observed significant levels of αSMA expression in hematopoietic lineage GFP+ recruited cells, particularly during the early phases of wound healing. These data confirm that the hematopoietic compartment can contribute to the wound myofibroblast population. However, given the rapid global decrease in GFP+ cells from the remodeling wound, these contributions may be greatest in the acute post-injury phase.
The cytokine regulation and recruitment mechanism of fibrocytes was also examined in this work. Fibrocytes are believed to be regulated by several cytokines [37–39], and we specifically examined the effects of SDF-1α, MCP-1, and TGF-β1 because these are felt to be critical mediators of circulatory cell recruitment and mesenchymal differentiation [40]. In these in vitro studies, SDF-1α induced the migration of bone marrow-derived CD11b+ cells, suggesting an involvement of this signaling pathway in the mobilization and recruitment of fibrocytes from the bone marrow. Moreover, TGF-β1 was found to up-regulate the expression of type I collagen and αSMA, indicative of a mesenchymal differentiation that is consistent with previous reports [38,39]. Not previously described, TGF-β1 was also found to up-regulate TIMP-1 and VEGF. Given this data, we believe that several cytokines including SDF-1α recruit monocyte/macrophage lineage cells from the bone marrow to the wound, and once the cells reach the wound site, other cytokines such as TGF-β1 induce a pro-fibrotic and pro-angiogenic phenotype. Because monocyte/macrophage lineage cells including fibrocytes can express TGF-β1 by themselves, as indicated by us and other researchers [3,41], autocrine mechanisms may be involved in this phenotypic change.
While the focus of our study was to examine the fate of hematopoietic lineage cells in the wound healing process, recent evidence suggests that non-hematopoietic mesenchymal cells derived from the bone marrow (BMSCs) can circulate in the blood and contribute to wound healing [42–44]. However, their fate in the wound healing process has been difficult to quantify due to the low prevalence of this population in the bone marrow and circulation, and the inability to specifically label these cells. Here, too, the VavR double transgenic model may overcome these limitations and enable their specific detection in tissues. Our data showed that BMSCs existed in bone marrow as RFP+ cells and were clearly discernable from GFP+ hematopoietic cells. However, in the wound RFP+ cells were indistinguishable from local non-circulatory cells and therefore a different model would be required to examine their involvement in wound healing.
In summary, we established a new double transgenic reporter mouse model to distinguish hematopoietic and non-hematopoietic cells without surface markers, using the vav 1 gene to specifically and permanently label hematopoietic cells. This new methodology allowed us to track hematopoietic cells in the wound and clearly demonstrate the existence and natural time-course of two types of collagen expressing hematopoietic lineage cells. We believe a similar approach will be useful in identifying the existence and lineage of other circulating cell populations with transitory surface marker expression.
Supplementary Material
(A) Immunohostochemistry for CD31 (blue) in the heart shows endothelial cells co-localizing with RFP. Scale bar = 150μm. (B) Flow cytometry control for CD45 vs CD11b GFP+ bone marrow derived cells shown in figure 2B. (C) Flow cytometry after primary culture of bone marrow cells shows the major percentage of GFP+ cells displaying the macrophage marker CD11b (% in CD45 vs CD11b plot relative to total GFP+ cells). (D) Flow cytometry control for CD45 GFP+ cells shown in figure 3C. (E) Flow cytometry control for CD31 RFP+ cells shown in figure 3C.
(A–B) Macrophages in our model are defined as GFP+, CD11b+ (purple) and F4/80+ (blue), and can be visualized via high (A) and low (B) power immunohistochemistry. White arrows highlight areas of GFP/F4/80/CD11b co-localization. White scale bar = 40μm; Gray Scale bar = 75μm. (C) Flow cytometry of GFP+ cells recruited to the wound site demonstrates that GFP+/F4/80+ cells are CD45+, while GFP+/45- cells are negative for the macrophage marker F4/80 (day 14). (D) Low power immunohistochemical imaging of F4/80 (blue) on day 7 demonstrating that F4/80+ cells in the wound co-localize with GFP+ and CD45+ (purple) cells. Gray scale bar = 75μm.
(A) GFP+ hematopoietic cells localize to the perivascular space (indicated by white arrows), visualized by overlays of photomicrographs of GFP+ hematopoietic cells, RFP+ blood vessels and desmin (purple), a pericyte marker. Scale bar = 50μm. (B) Entire immunoblot from Figure 6B. Molecular weight (MW) of type I collagen precursor: 140–210 kilodaltons (kDa). MW of mature type I collagen: 70–90 kDa. Arrows indicate protein bands in the expected ranges for precursor and mature type I collagen.
(A–B) Immunofluorescence for αSMA (blue) at days 7, 14 and 28 post-wounding reveals distinct co-localization with GFP+ hematopoietic cells, peaking at day 7. Normal skin and day 7 visualized in this figure. Scale Bar = 75 μm.
(A–B) Immunofluorescence for αSMA (blue) at days 7, 14 and 28 post-wounding reveals distinct co-localization with GFP+ hematopoietic cells, peaking at day 7. Day 14 and 28 visualized in this figure. Scale Bar = 75 μm.
List of tested genes and their primers for single-cell transcriptional analysis.
Acknowledgments
This study was supported by the U.S. National Institutes of Health grant RO1 DK074095-07 to G.C.G. H.S. was supported by the Uehara Memorial Foundation, M.R was supported partially by the Wound Healing Society 3M grant and J.P.G. was supported by the U.S. National Institute of Health NRSA grant F32DK088448-01. We acknowledge the supports of the Oak Foundation and the Hagey Laboratory for Pediatric Regenerative Medicine to G.C.G. and M.T.L. Photomicrographs were taken at the Stanford Cell Sciences Imaging Facility, and flow cytometry was conducted at the Stanford Shared FACS Facility. The authors would like to thank Ms. Yujin Park and Ms. Kitty Li for their technical assistance.
Footnotes
Disclaimer
The authors indicate no potential conflicts of interest.
Author contributions
H.S.: Collection and assembly of data, data analysis and interpretation, and manuscript writing; R.C.R.: Collection and assembly of data, data analysis and interpretation and manuscript writing; M.R.: Collection and assembly of data, data analysis and interpretation and manuscript writing; M.S.: Collection and assembly of data, data analysis and interpretation, and manuscript writing; J.P.G.: Collection and assembly of data, data analysis and interpretation; M.J.: data analysis and interpretation; T.F Collection and assembly of data; M.T.L.: Conception and design, final approval of manuscript; G.C.G.: Conception and design, financial support, manuscript writing, and final approval of manuscript.
References
- 1.Bucala R, Spiegel LA, Chesney J, et al. Circulating fibrocytes define a new leukocyte subpopulation that mediates tissue repair. Molecular medicine. 1994;1(1):71–81. [PMC free article] [PubMed] [Google Scholar]
- 2.Yang L, Scott PG, Dodd C, et al. Identification of fibrocytes in postburn hypertrophic scar. Wound repair and regeneration. 2005;13(4):398–404. doi: 10.1111/j.1067-1927.2005.130407.x. [DOI] [PubMed] [Google Scholar]
- 3.Yang L, Scott PG, Giuffre J, et al. Peripheral blood fibrocytes from burn patients: identification and quantification of fibrocytes in adherent cells cultured from peripheral blood mononuclear cells. Laboratory investigation. 2002;82(9):1183–1192. doi: 10.1097/01.lab.0000027841.50269.61. [DOI] [PubMed] [Google Scholar]
- 4.Mori L, Bellini A, Stacey MA, et al. Fibrocytes contribute to the myofibroblast population in wounded skin and originate from the bone marrow. Experimental cell research. 2005;304(1):81–90. doi: 10.1016/j.yexcr.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Ishida Y, Kimura A, Takayasu T, et al. Detection of fibrocytes in human skin wounds and its application for wound age determination. International journal of legal medicine. 2009;123(4):299–304. doi: 10.1007/s00414-009-0320-4. [DOI] [PubMed] [Google Scholar]
- 6.Phillips RJ, Burdick MD, Hong K, et al. Circulating fibrocytes traffic to the lungs in response to CXCL12 and mediate fibrosis. The Journal of clinical investigation. 2004;114(3):438–446. doi: 10.1172/JCI20997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto N, Jin H, Liu T, et al. Bone marrow-derived progenitor cells in pulmonary fibrosis. The Journal of clinical investigation. 2004;113(2):243–252. doi: 10.1172/JCI18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kisseleva T, Uchinami H, Feirt N, et al. Bone marrow-derived fibrocytes participate in pathogenesis of liver fibrosis. Journal of hepatology. 2006;45(3):429–438. doi: 10.1016/j.jhep.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Reilkoff RA, Bucala R, Herzog EL. Fibrocytes: emerging effector cells in chronic inflammation. Nature reviews Immunology. 2011;11(6):427–435. doi: 10.1038/nri2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellini A, Mattoli S. The role of the fibrocyte, a bone marrow-derived mesenchymal progenitor, in reactive and reparative fibroses. Laboratory investigation. 2007;87(9):858–870. doi: 10.1038/labinvest.3700654. [DOI] [PubMed] [Google Scholar]
- 11.Kao HK, Chen B, Murphy GF, et al. Peripheral blood fibrocytes: enhancement of wound healing by cell proliferation, re-epithelialization, contraction, and angiogenesis. Annals of surgery. 2011;254(6):1066–1074. doi: 10.1097/SLA.0b013e3182251559. [DOI] [PubMed] [Google Scholar]
- 12.Pilling D, Fan T, Huang D, et al. Identification of markers that distinguish monocyte-derived fibrocytes from monocytes, macrophages, and fibroblasts. PloS one. 2009;4(10):e7475. doi: 10.1371/journal.pone.0007475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-de-Alba C, Selman M, Pardo A. Hematopoietic Derived Fibrocytes: Emerging Effector Cells in Fibrotic Disorders. In: Pelayo R, editor. Advances in Hematopoietic Stem Cell Research. Intech; 2012. [Google Scholar]
- 14.Ishii G, Sangai T, Sugiyama K, et al. In vivo characterization of bone marrow-derived fibroblasts recruited into fibrotic lesions. Stem cells. 2005;23(5):699–706. doi: 10.1634/stemcells.2004-0183. [DOI] [PubMed] [Google Scholar]
- 15.Fathke C, Wilson L, Hutter J, et al. Contribution of bone marrow-derived cells to skin: collagen deposition and wound repair. Stem cells. 2004;22(5):812–822. doi: 10.1634/stemcells.22-5-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barisic-Dujmovic T, Boban I, Clark SH. Fibroblasts/myofibroblasts that participate in cutaneous wound healing are not derived from circulating progenitor cells. Journal of cellular physiology. 2010;222(3):703–712. doi: 10.1002/jcp.21997. [DOI] [PubMed] [Google Scholar]
- 17.Adams JM, Harris AW, Strasser A, et al. Transgenic models of lymphoid neoplasia and development of a pan-hematopoietic vector. Oncogene. 1999;18(38):5268–5277. doi: 10.1038/sj.onc.1202997. [DOI] [PubMed] [Google Scholar]
- 18.Zambidis ET, Peault B, Park TS, et al. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. Blood. 2005;106(3):860–870. doi: 10.1182/blood-2004-11-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bustelo XR, Rubin SD, Suen KL, et al. Developmental expression of the vav protooncogene. Cell growth & differentiation. 1993;4(4):297–308. [PubMed] [Google Scholar]
- 20.Moores SL, Selfors LM, Fredericks J, et al. Vav family proteins couple to diverse cell surface receptors. Molecular and cellular biology. 2000;20(17):6364–6373. doi: 10.1128/mcb.20.17.6364-6373.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muzumdar MD, Tasic B, Miyamichi K, et al. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 22.de Boer J, Williams A, Skavdis G, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. European journal of immunology. 2003;33(2):314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 23.Galiano RD, Michaels Jt, Dobryansky M, et al. Quantitative and reproducible murine model of excisional wound healing. Wound repair and regeneration. 2004;12(4):485–492. doi: 10.1111/j.1067-1927.2004.12404.x. [DOI] [PubMed] [Google Scholar]
- 24.Jaatinen T, Laine J. Isolation of mononuclear cells from human cord blood by Ficoll-Paque density gradient. Current protocols in stem cell biology. 2007;Chapter 21(Unit 2A):1. doi: 10.1002/9780470151808.sc02a01s1. [DOI] [PubMed] [Google Scholar]
- 25.Glotzbach JP, Januszyk M, Vial IN, et al. An information theoretic, microfluidic-based single cell analysis permits identification of subpopulations among putatively homogeneous stem cells. PloS one. 2011;6(6):e21211. doi: 10.1371/journal.pone.0021211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levi B, Wan DC, Glotzbach JP, et al. CD105 protein depletion enhances human adipose-derived stromal cell osteogenesis through reduction of transforming growth factor beta1 (TGF-beta1) signaling. The Journal of biological chemistry. 2011;286(45):39497–39509. doi: 10.1074/jbc.M111.256529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gnecchi M, Melo LG. Bone marrow-derived mesenchymal stem cells: isolation, expansion, characterization, viral transduction, and production of conditioned medium. Methods in molecular biology. 2009;482:281–294. doi: 10.1007/978-1-59745-060-7_18. [DOI] [PubMed] [Google Scholar]
- 28.Mosna F, Sensebe L, Krampera M. Human bone marrow and adipose tissue mesenchymal stem cells: a user’s guide. Stem cells and development. 2010;19(10):1449–1470. doi: 10.1089/scd.2010.0140. [DOI] [PubMed] [Google Scholar]
- 29.Okuno Y, Nakamura-Ishizu A, Kishi K, et al. Bone marrow-derived cells serve as proangiogenic macrophages but not endothelial cells in wound healing. Blood. 2011;117(19):5264–5272. doi: 10.1182/blood-2011-01-330720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris DA, Zhao Y, LaPar DJ, et al. Inhibiting CXCL12 blocks fibrocyte migration and differentiation and attenuates bronchiolitis obliterans in a murine heterotopic tracheal transplant model. The Journal of thoracic and cardiovascular surgery. 2013;145(3):854–861. doi: 10.1016/j.jtcvs.2012.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ekert JE, Murray LA, Das AM, et al. Chemokine (C-C motif) ligand 2 mediates direct and indirect fibrotic responses in human and murine cultured fibrocytes. Fibrogenesis & tissue repair. 2011;4(1):23. doi: 10.1186/1755-1536-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niedermeier M, Reich B, Rodriguez Gomez M, et al. CD4+ T cells control the differentiation of Gr1+ monocytes into fibrocytes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(42):17892–17897. doi: 10.1073/pnas.0906070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh SA, Chang EI, Galvez MG, et al. SDF-1 alpha expression during wound healing in the aged is HIF dependent. Plastic and reconstructive surgery. 2009;123(2 Suppl):65S–75S. doi: 10.1097/PRS.0b013e318191bdf4. [DOI] [PubMed] [Google Scholar]
- 34.Ogilvy S, Metcalf D, Gibson L, et al. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. 1999;94(6):1855–1863. [PubMed] [Google Scholar]
- 35.Chamson-Reig A, Arany EJ, Hill DJ. Lineage tracing and resulting phenotype of haemopoietic-derived cells in the pancreas during beta cell regeneration. Diabetologia. 2010;53(10):2188–2197. doi: 10.1007/s00125-010-1835-4. [DOI] [PubMed] [Google Scholar]
- 36.Hartlapp I, Abe R, Saeed RW, et al. Fibrocytes induce an angiogenic phenotype in cultured endothelial cells and promote angiogenesis in vivo. FASEB. 2001;15(12):2215–2224. doi: 10.1096/fj.01-0049com. [DOI] [PubMed] [Google Scholar]
- 37.Chesney J, Metz C, Stavitsky AB, et al. Regulated production of type I collagen and inflammatory cytokines by peripheral blood fibrocytes. Journal of immunology. 1998;160(1):419–425. [PubMed] [Google Scholar]
- 38.Abe R, Donnelly SC, Peng T, et al. Peripheral blood fibrocytes: differentiation pathway and migration to wound sites. Journal of immunology. 2001;166(12):7556–7562. doi: 10.4049/jimmunol.166.12.7556. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt M, Sun G, Stacey MA, et al. Identification of circulating fibrocytes as precursors of bronchial myofibroblasts in asthma. Journal of immunology. 2003;171(1):380–389. doi: 10.4049/jimmunol.171.1.380. [DOI] [PubMed] [Google Scholar]
- 40.Ceradini DJ, Kulkarni AR, Callaghan MJ, et al. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nature medicine. 2004;10(8):858–864. doi: 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- 41.Aung H, Sherman J, Tary-Lehman M, et al. Analysis of transforming growth factor-beta 1 (TGF-beta1) expression in human monocytes infected with Mycobacterium avium at a single cell level by ELISPOT assay. Journal of immunological methods. 2002;259(1–2):25–32. doi: 10.1016/s0022-1759(01)00485-9. [DOI] [PubMed] [Google Scholar]
- 42.He Q, Wan C, Li G. Concise review: multipotent mesenchymal stromal cells in blood. Stem cells. 2007;25(1):69–77. doi: 10.1634/stemcells.2006-0335. [DOI] [PubMed] [Google Scholar]
- 43.Wu Y, Zhao RC, Tredget EE. Concise review: bone marrow-derived stem/progenitor cells in cutaneous repair and regeneration. Stem cells. 2010;28(5):905–915. doi: 10.1002/stem.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Y, Chen L, Scott PG, et al. Mesenchymal stem cells enhance wound healing through differentiation and angiogenesis. Stem cells. 2007;25(10):2648–2659. doi: 10.1634/stemcells.2007-0226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Immunohostochemistry for CD31 (blue) in the heart shows endothelial cells co-localizing with RFP. Scale bar = 150μm. (B) Flow cytometry control for CD45 vs CD11b GFP+ bone marrow derived cells shown in figure 2B. (C) Flow cytometry after primary culture of bone marrow cells shows the major percentage of GFP+ cells displaying the macrophage marker CD11b (% in CD45 vs CD11b plot relative to total GFP+ cells). (D) Flow cytometry control for CD45 GFP+ cells shown in figure 3C. (E) Flow cytometry control for CD31 RFP+ cells shown in figure 3C.
(A–B) Macrophages in our model are defined as GFP+, CD11b+ (purple) and F4/80+ (blue), and can be visualized via high (A) and low (B) power immunohistochemistry. White arrows highlight areas of GFP/F4/80/CD11b co-localization. White scale bar = 40μm; Gray Scale bar = 75μm. (C) Flow cytometry of GFP+ cells recruited to the wound site demonstrates that GFP+/F4/80+ cells are CD45+, while GFP+/45- cells are negative for the macrophage marker F4/80 (day 14). (D) Low power immunohistochemical imaging of F4/80 (blue) on day 7 demonstrating that F4/80+ cells in the wound co-localize with GFP+ and CD45+ (purple) cells. Gray scale bar = 75μm.
(A) GFP+ hematopoietic cells localize to the perivascular space (indicated by white arrows), visualized by overlays of photomicrographs of GFP+ hematopoietic cells, RFP+ blood vessels and desmin (purple), a pericyte marker. Scale bar = 50μm. (B) Entire immunoblot from Figure 6B. Molecular weight (MW) of type I collagen precursor: 140–210 kilodaltons (kDa). MW of mature type I collagen: 70–90 kDa. Arrows indicate protein bands in the expected ranges for precursor and mature type I collagen.
(A–B) Immunofluorescence for αSMA (blue) at days 7, 14 and 28 post-wounding reveals distinct co-localization with GFP+ hematopoietic cells, peaking at day 7. Normal skin and day 7 visualized in this figure. Scale Bar = 75 μm.
(A–B) Immunofluorescence for αSMA (blue) at days 7, 14 and 28 post-wounding reveals distinct co-localization with GFP+ hematopoietic cells, peaking at day 7. Day 14 and 28 visualized in this figure. Scale Bar = 75 μm.
List of tested genes and their primers for single-cell transcriptional analysis.