Abstract
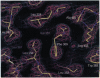
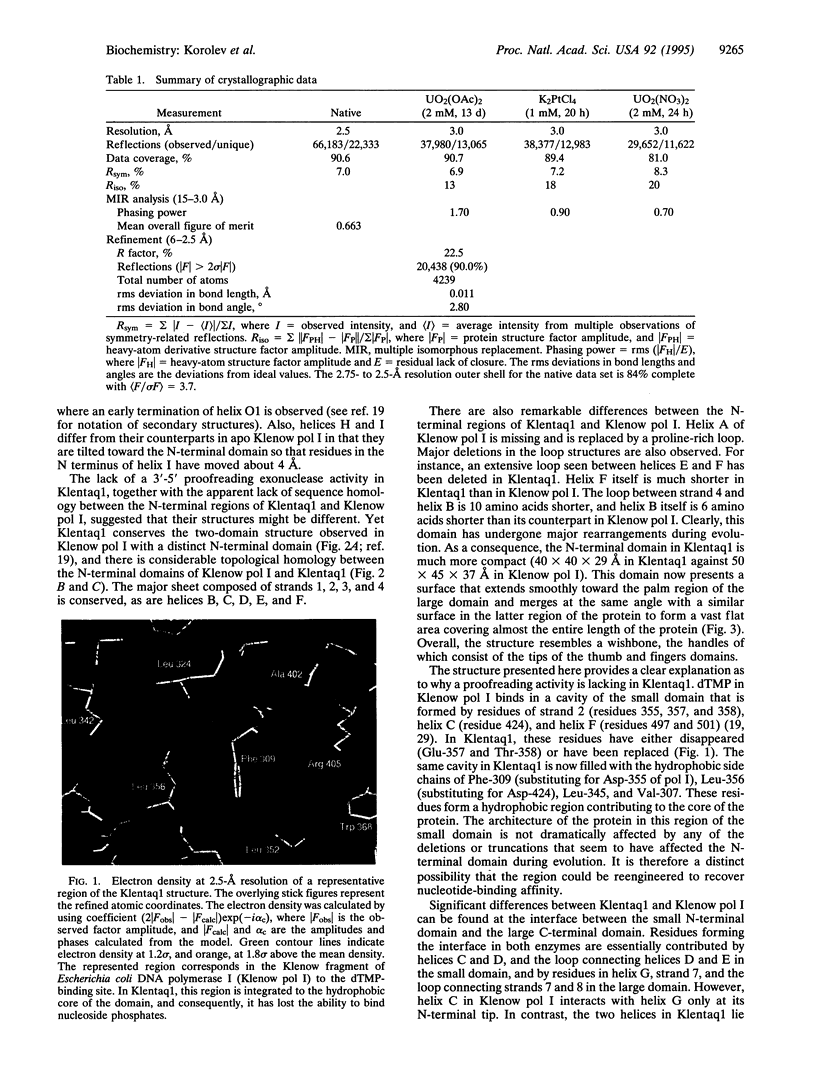
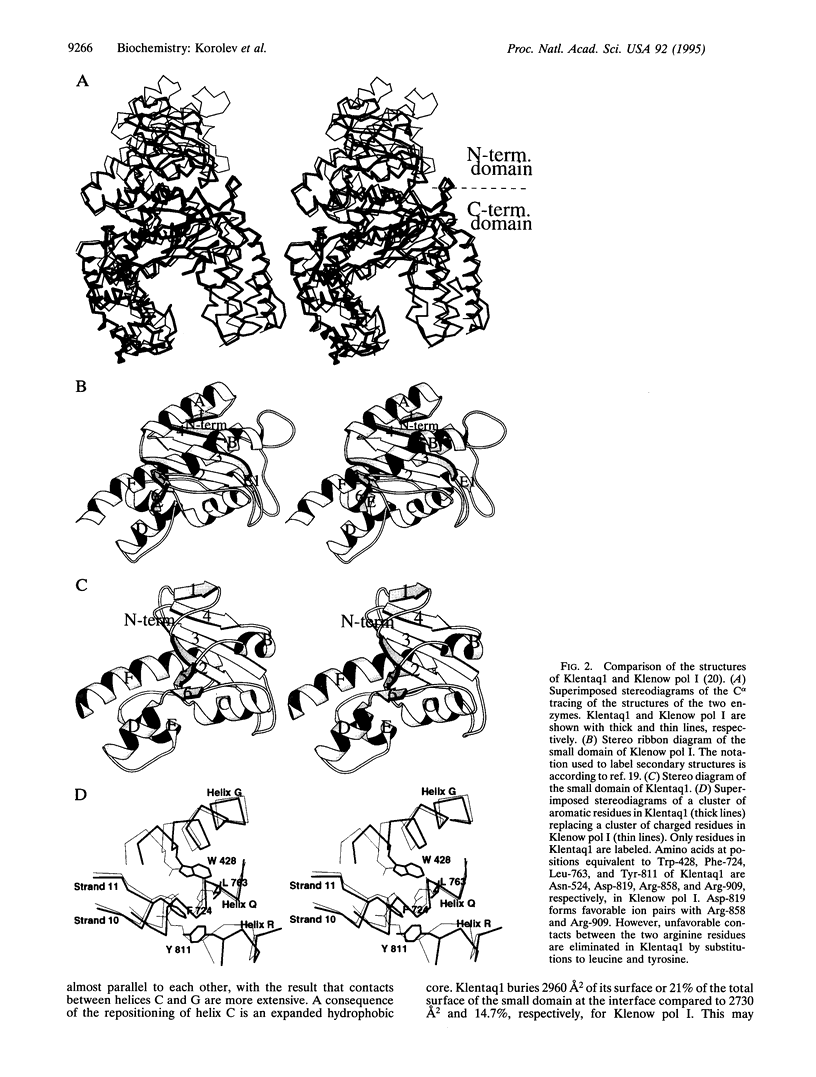
The crystal structure of the large fragment of the Thermus aquaticus DNA polymerase (Klentaq1), determined at 2.5-A resolution, demonstrates a compact two-domain architecture. The C-terminal domain is identical in fold to the equivalent region of the Klenow fragment of Escherichia coli DNA polymerase I (Klenow pol I). Although the N-terminal domain of Klentaq1 differs greatly in sequence from its counterpart in Klenow pol I, it has clearly evolved from a common ancestor. The structure of Klentaq1 reveals the strategy utilized by this protein to maintain activity at high temperatures and provides the structural basis for future improvements of the enzyme.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T. Mutational effects on protein stability. Annu Rev Biochem. 1989;58:765–798. doi: 10.1146/annurev.bi.58.070189.004001. [DOI] [PubMed] [Google Scholar]
- Argos P., Rossman M. G., Grau U. M., Zuber H., Frank G., Tratschin J. D. Thermal stability and protein structure. Biochemistry. 1979 Dec 11;18(25):5698–5703. doi: 10.1021/bi00592a028. [DOI] [PubMed] [Google Scholar]
- Barnes W. M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes W. M. The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene. 1992 Mar 1;112(1):29–35. doi: 10.1016/0378-1119(92)90299-5. [DOI] [PubMed] [Google Scholar]
- Beese L. S., Derbyshire V., Steitz T. A. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993 Apr 16;260(5106):352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3' leads to 5' exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972 Jan 10;247(1):241–248. [PubMed] [Google Scholar]
- Burley S. K., Petsko G. A. Amino-aromatic interactions in proteins. FEBS Lett. 1986 Jul 28;203(2):139–143. doi: 10.1016/0014-5793(86)80730-x. [DOI] [PubMed] [Google Scholar]
- Eckert K. A., Kunkel T. A. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990 Jul 11;18(13):3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson M. K., Honig B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins. 1988;4(1):7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- Herning T., Yutani K., Inaka K., Kuroki R., Matsushima M., Kikuchi M. Role of proline residues in human lysozyme stability: a scanning calorimetric study combined with X-ray structure analysis of proline mutants. Biochemistry. 1992 Aug 11;31(31):7077–7085. doi: 10.1021/bi00146a008. [DOI] [PubMed] [Google Scholar]
- Innis M. A., Myambo K. B., Gelfand D. H., Brow M. A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr, Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Lawyer F. C., Stoffel S., Saiki R. K., Chang S. Y., Landre P. A., Abramson R. D., Gelfand D. H. High-level expression, purification, and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5' to 3' exonuclease activity. PCR Methods Appl. 1993 May;2(4):275–287. doi: 10.1101/gr.2.4.275. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Hodel A., Sauer R. T., Richards F. M. The crystal structure of a mutant protein with altered but improved hydrophobic core packing. Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):423–427. doi: 10.1073/pnas.91.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W. A., Sauer R. T. Alternative packing arrangements in the hydrophobic core of lambda repressor. Nature. 1989 May 4;339(6219):31–36. doi: 10.1038/339031a0. [DOI] [PubMed] [Google Scholar]
- Lundberg K. S., Shoemaker D. D., Adams M. W., Short J. M., Sorge J. A., Mathur E. J. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene. 1991 Dec 1;108(1):1–6. doi: 10.1016/0378-1119(91)90480-y. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Becktel W. J., Matthews B. W. Hydrophobic stabilization in T4 lysozyme determined directly by multiple substitutions of Ile 3. Nature. 1988 Aug 4;334(6181):406–410. doi: 10.1038/334406a0. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Wozniak J. A., Sun D. P., Matthews B. W. Structural studies of mutants of T4 lysozyme that alter hydrophobic stabilization. J Biol Chem. 1989 Sep 25;264(27):16059–16066. [PubMed] [Google Scholar]
- Matthews B. W., Nicholson H., Becktel W. J. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6663–6667. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson A. Current approaches to macromolecular crystallization. Eur J Biochem. 1990 Apr 20;189(1):1–23. doi: 10.1111/j.1432-1033.1990.tb15454.x. [DOI] [PubMed] [Google Scholar]
- Menéndez-Arias L., Argos P. Engineering protein thermal stability. Sequence statistics point to residue substitutions in alpha-helices. J Mol Biol. 1989 Mar 20;206(2):397–406. doi: 10.1016/0022-2836(89)90488-9. [DOI] [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994 Jun 24;264(5167):1891–1903. [PubMed] [Google Scholar]
- Perutz M. F. Electrostatic effects in proteins. Science. 1978 Sep 29;201(4362):1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- Perutz M. F., Raidt H. Stereochemical basis of heat stability in bacterial ferredoxins and in haemoglobin A2. Nature. 1975 May 15;255(5505):256–259. doi: 10.1038/255256a0. [DOI] [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sawaya M. R., Pelletier H., Kumar A., Wilson S. H., Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994 Jun 24;264(5167):1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- Sharp K., Fine R., Honig B. Computer simulations of the diffusion of a substrate to an active site of an enzyme. Science. 1987 Jun 12;236(4807):1460–1463. doi: 10.1126/science.3589666. [DOI] [PubMed] [Google Scholar]
- Sousa R., Chung Y. J., Rose J. P., Wang B. C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution. Nature. 1993 Aug 12;364(6438):593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]