Abstract
Single-stranded DNA binding proteins (SSBs) selectively bind single-stranded DNA (ssDNA) and facilitate recruitment of additional proteins and enzymes to their sites of action on DNA. SSB can also locally diffuse on ssDNA, which allows it to quickly reposition itself while remaining bound to ssDNA. In this work, we used a hybrid instrument that combines single-molecule fluorescence and force spectroscopy to directly visualize the movement of Escherichia coli SSB on long polymeric ssDNA. Long ssDNA was synthesized without secondary structure which can hinder quantitative analysis of SSB movement. The apparent diffusion coefficient of E. coli SSB thus determined ranged from 70,000 to 170,000 nt2/s, which is at least six hundred times higher than that determined from SSB diffusion on short ssDNA oligomers, and is within the range of values reported for protein diffusion on double stranded DNA. Our work suggests that SSB can also migrate via a long range intersegment transfer on long ssDNA. The force dependence of SSB movement on ssDNA further supports this interpretation.
Introduction
A variety of proteins associate with single-stranded DNA (ssDNA) and play crucial roles in DNA replication, recombination, replication restart, and repair.1; 2; 3 Single-stranded DNA binding proteins (SSBs) form a class of such proteins. SSB binds selectively to ssDNA in a sequence-independent manner4; 5 and protects transiently formed ssDNA from degradation. SSB is also likely to coordinate a multitude of proteins competing for access to ssDNA during their functions.2; 6; 7; 8; 9; 10 Escherichia coli SSB also has the ability to diffuse locally along ssDNA, at least up to ~ 60 nt.11; 12 It has also been inferred from indirect evidence that the phage T4 SSB protein (gene 32) can diffuse along ssDNA.13; 14 Diffusion of SSB might facilitate SSB’s recruitment of other proteins to their sites of action.2
E. coli SSB is a representative homotetrameric SSB consisting of 177 amino acids.15 It forms a stable homotetramer16 which binds and wraps ssDNA around its subunits,17; 18 and is essential for cell viability due to its multiple roles in genome maintenance.2; 5; 17 The N-terminal domain of E. coli SSB, comprised of 112 amino acids, forms an OB-fold that contains the ssDNA-binding sites,4; 15; 18 while the C-terminus consists of an unstructured linker region, the last eight amino acids of which selectively bind and recruit its partner proteins to ssDNA.2; 10; 19 Thus, the tetramer has four ssDNA binding domains enabling it to bind ssDNA in a variety of modes depending on the salt concentration.17; 20 The (SSB)35 binding mode, which is favored in low salt concentrations (<10 mM Na+) and high protein binding density, uses an average of only two subunits for ssDNA binding, occludes ~35 nucleotides21 and binds ssDNA cooperatively,22; 23 whereas the (SSB)65 binding mode, favored in moderately high salt concentrations (≥ 2 mM Mg2+ or ≥ 200 mM Na+), uses all four ssDNA binding sites, occludes ~65 nucleotides and binds ssDNA with little cooperativity.17; 18 The two binding modes and real-time interconversion between them have also been examined using single molecule fluorescence resonance energy transfer (FRET).24 The diffusional migration along ssDNA of E. coli SSB in its (SSB)65 binding mode was observed and it was found that SSB diffusion stimulates the elongation of RecA filaments on DNA that can form secondary structures by transiently melting DNA hairpin structures and that SSB migrates on DNA via reptation.11; 12
Using a hybrid instrument that combines single molecule fluorescence and optical trapping,25 we have now visualized the dynamics of E. coli SSB on long ssDNA molecules that are void of secondary structure. We found that the apparent diffusion of the SSB tetramer in its (SSB)65 binding mode follows a 1D random walk, but with a diffusion coefficient that is at least six hundred times larger than was estimated on short poly(dT) ssDNA,13 suggesting that on long ssDNA, SSB can also reposition itself via a long range intersegment transfer mechanism.26; 27 The force dependence of the apparent diffusion coefficient further supports this interpretation.
Results
Preparation of secondary-structure-free ssDNA constructs
To quantitatively study the movement of E. coli SSB on long ssDNA, we reasoned that secondary structures formed within the ssDNA should be avoided because melting and rezipping of these structures could add undesirable noise to the position trajectory of SSB. Unzipping secondary structures by applying high forces (>10 pN) is not a viable option because the dissociation rate of SSB tetramers in the (SSB)65 binding mode is force-dependent, and SSB will dissociate at approximately 10 pN of force applied to the ends of ssDNA.12
In this study, we used rolling circle replication (RCR) to synthesize long ssDNA (10,000–20,000 nt) with only deoxythymidines and deoxycytidines, thus preventing intramolecular base pairing. In RCR, a user-defined template is first hybridized to a short oligonucleotide primer in order to circularize the template strand. The template strand is subsequently ligated to form a closed circular ssDNA template. Next, a highly processive DNA polymerase capable of strand-displacement synthesis is added. During the reaction, DNA polymerase binds to the 3′-terminus of the primer and replicates around the circular template for thousands of cycles, thereby producing long polymeric ssDNA.28; 29; 30 We generated ssDNA constructs with only dT and dC by creating a circular template whose sequence consists of only dA (20 out of 28 nucleotides) and dG (8 out of 28 nucleotides) (Materials and Methods).
Visualization of E. coli SSB motion on ssDNA
Total internal reflection fluorescence (TIRF) microscopy combined with a single-beam optical trap was employed to visualize the dynamic movement of E. coli SSB tetramers along ssDNA25 (Figure 1). The biotinylated 5′-end of a ssDNA construct is immobilized on the surface via a biotin-neutravidin bond and is stretched by a laser-trapped bead (anti-digoxigenin coated) attached to the digoxigenin attached to the 3′-end of the DNA. A stable homotetramer of E. coli SSB possessing a single cysteine in its intrinsically disordered C-terminal tail (A122C) was labeled with ~1 Alexa555 fluorophore per subunit11 and was imaged while bound to the ssDNA construct.
Figure 1. Schematic of the experimental setup.
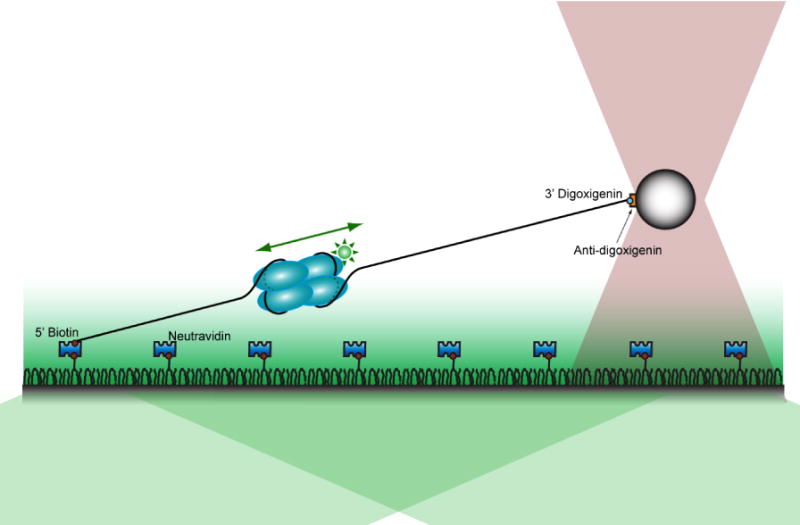
One end of a ssDNA construct is immobilized on a PEG-coated surface, and the other end is attached to a polystyrene bead confined in the optical trap (pink gray). A green excitation laser selectively illuminates molecules within a few hundred nanometers from the coverslip surface by total internal reflection (TIR).
We analyzed the motion of only the first SSB tetramer that binds DNA because if there are multiple SSB tetramers bound on the same strand, it is possible that they might perturb the motion of each other via collision. In order to prevent simultaneous binding of multiple SSB tetramers, a very low concentration (< 20 pM) of SSB tetramers was used. The salt concentration in the experiment was 500 mM NaCl: in this condition at equilibrium, the SSB tetramers bind ssDNA exclusively in the (SSB)65 binding mode, allowing us to make comparisons to our previous studies performed under the same condition.11; 12 In addition, the (SSB)65 binding mode is favored at moderately high salt (≥ 2 mM Mg2+ or ≥ 200 mM Na+)21; 24, and because both mono- and di-valent cations exist in the cell, it is likely that the data presented below for the (SSB)65 mode is physiologically relevant. Prior to each measurement, all SSB tetramers already bound to a ssDNA construct were removed by applying high tension (over 25 pN) for 1 min, taking advantage of the force-dependent dissociation of SSB tetramers in the (SSB)65 binding mode (full SSB dissociation occurs at forces < 13 pN).12 This process ensured that there were no SSB tetramers bound to the ssDNA constructs before the force is reduced and data acquisition started. At the force values used in this experiment (2–5 pN), the wrapping of ssDNA around an SSB tetramer is partially disrupted due to the tension such that 10~30 nt is unraveled31 as reflected in Figure 1.
Figure 2 shows position trajectories, in the form of kymograms, of individual SSB tetramers along ssDNA at various forces. The movement along ssDNA is bidirectional and appears to resemble a random walk. It should be noted that the range of movement is very large, such that an SSB tetramer can travel over 2,000 nm during a 74 s observation (Figure 2A). This is surprising since 2,000 nm is a much longer distance than the expected ~100 nm based on the previously estimated diffusion coefficient of 270 nt2/s for the E. coli SSB tetramer at 37 °C.11 The fluorescence intensity of the SSB, in general, increases (decreases) as it approaches (moves away from) the surface, due to the exponentially decaying excitation profile of TIRF microscopy25 (although sometimes, this is not the case, for example, ~30 s in Figure 2A). One possible explanation is that initially there were two fluorophores on that SSB tetramer quenching each other until one became photobleached32). For further analysis, we fitted the fluorescence images to 2D Gaussian functions and obtained the position trajectory of each SSB tetramer. The position trajectories (in nanometers) were then converted to the number of nucleotides using the force-extension curve of the ssDNA construct. To achieve this, we obtained the average force-extension curve of multiple ssDNA constructs from all experiments (total of 62 molecules) (Figure S2A). Although the length of each ssDNA construct varies because the construct was made via the RCR method, we still could average them because normalized (fractional) force-extension curves superimpose. The averaged force-extension curves were fit to the worm-like chain model to determine the extension per nucleotide and this was used to calculate the total extension of ssDNA in terms of nucleotides (Figure S2B).
Figure 2. Visualization of SSB movement along ssDNA.
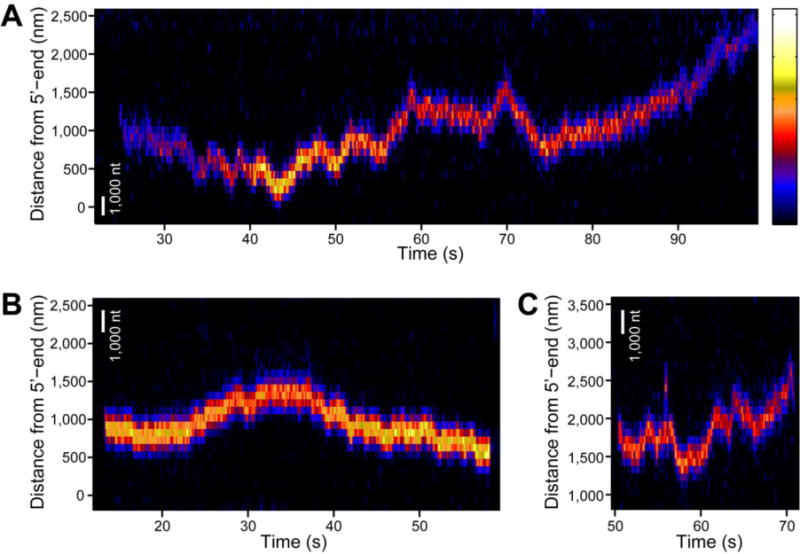
(A) A kymogram of a single SSB homotetramer moving along a long ssDNA molecule stretched at 3 pN tension. This SSB travels across large distance over 2,000 nm. A scale bar representing the length of 1,000 nt is shown in white. Fluorescence intensity is color-coded, and the color scale is shown on the right (minimum intensity: black (bottom); maximum intensity: white (top)). (B) A kymogram of a single SSB homotetramer recorded at 4 pN tension. A scale bar in white corresponds to the length of 1,000 nt at this tension. (C) A kymogram of a single SSB homotetramer observed at 5 pN. A scale bar at this tension is shown in white. The Figure 2B and 2C share the same color scale for fluorescence intensity with Figure 2A.
Ultrafast migration of E. coli SSB and force-dependence of diffusion
To check if the movement we observe for SSB follows the expected behavior for 1D diffusion, and to calculate an apparent diffusion coefficient to compare to the previously reported value,11 we collected many position trajectories of SSB. Figure 3 shows a subset of the position trajectories collected at the applied force, 4.1 pN. Only a small number of the 82 position trajectories examined are shown in the figure so that individual trajectories can be discerned. The mean of 82 trajectories is shown as a black dashed curve. The mean displacement does not change over time and is very small compared to the displacements observed for individual trajectories, which suggests that the motion along the ssDNA is random and not biased in either direction. Although we collected 82 trajectories, the fluorescence signal from ~50% of them was lost after a few seconds (likely due to photobleaching of the Alexa 555 fluorophore on the SSB). As a result, the number of trajectories contributing to the average decreased with time, which makes the mean displacement appear to be more rugged. We therefore used only data within the first few seconds for the following analysis.
Figure 3. Single-molecule position trajectories of several SSBs at 4.1 pN tension.
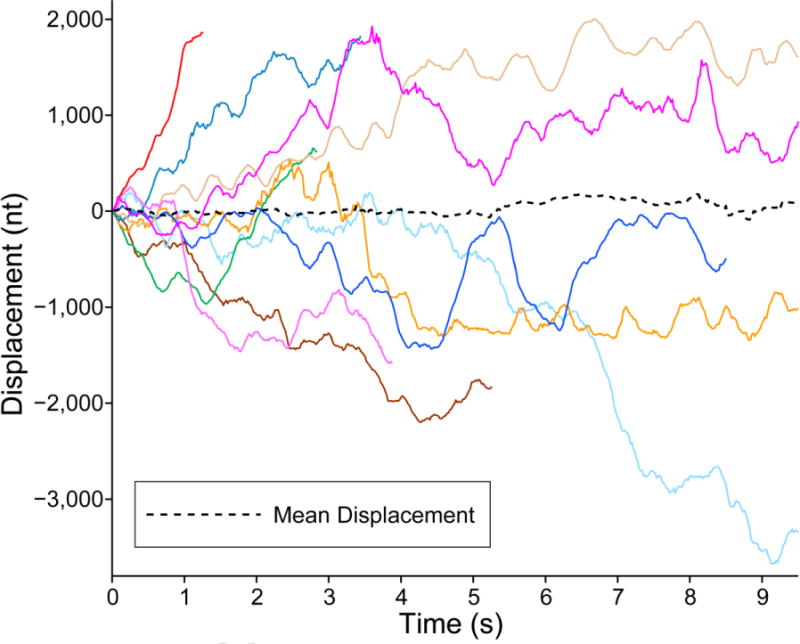
The position trajectories of different SSBs are drawn in solid lines of different colors. A black dashed curve represents the mean displacement of all trajectories. The displacement was measured in nanometers, and then converted into number of nucleotides by using the extension per nucleotide at the corresponding force (Figure S2B).
In Figure 4A, the mean square displacements (MSD) obtained from at least 50 molecules at various forces are plotted against the elapsed time. For each MSD curve, the number of trajectories used in calculating the MSD at a certain time point is shown in Figure S3A). The MSD data at four applied forces (2.1 pN, 3.1 pN, 4.1 pN and 5.1 pN) are shown. For all four forces, the MSD initially increases linearly with respect to the elapsed time, suggesting that the motion of SSB along ssDNA resembles 1D diffusion for the observed time and length scales. For each applied force, a straight line was fit to this initial section and an apparent diffusion coefficient Dapp was determined and plotted in Figure 4B. Dapp = 171,000 nt2/s at 2.1 pN, six-hundred times higher than the previously reported value of 270 nt2/s at 37 °C on short (<95 nt) poly(dT) at the same NaCl concentration but at zero force.11 Considering that local diffusion as measured by single molecule FRET is faster at higher temperature11 and lower forces12 and that our measurement was performed at 25 °C with an applied force of at least 2 pN, the true difference is likely to be greater.
Figure 4. Force-dependence of SSB diffusion.
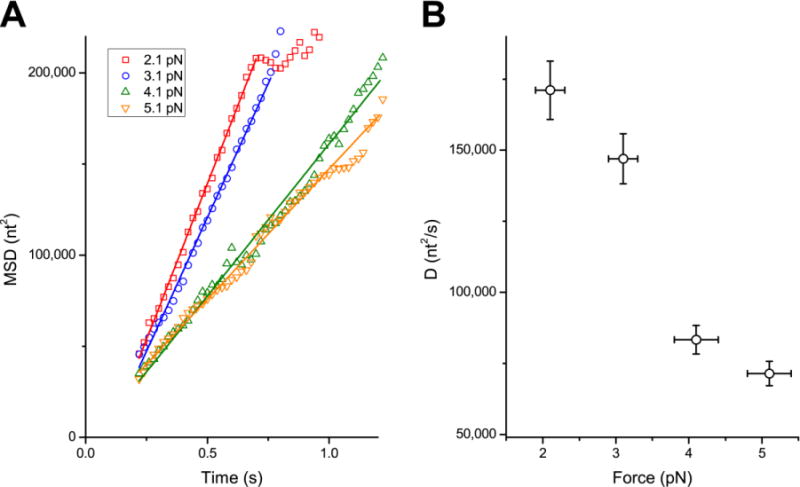
(A) The time courses of mean squared displacement (MSD) at various forces. The MSDs are the mean of 80 to 110 trajectories. In the scatter plot, the MSD at 2.1 pN, 3.1 pN, 4.1 pN and 5.1 pN are plotted against elapsed time in red, blue, green and orange, respectively. All the MSDs are linear in the beginning. The linear section is fit to a straight line, and the fits are drawn in respective colors. (B) The diffusion coefficients at various forces. The error bars in X and Y represent the standard deviation of applied force among trajectories and the uncertainty in diffusion coefficients propagated from the uncertainty in the extension per nucleotide (Figure S2B), respectively. The diffusion coefficient increases as the applied force is decreased.
Discussion
We visualized the dynamics of E. coli SSB tetramer movement along ssDNA while bound in its (SSB)65 binding mode, and SSB exhibited 1D random walk behavior. We also calculated an apparent diffusion coefficient to compare to the previously reported 1D diffusion coefficients of other proteins11; 33; 34; 35 (Figure 5). Although the comparison is limited to proteins that diffuse along double-stranded DNA (dsDNA) since method to probe long-range protein dynamics on ssDNA have not been available until recently25, our estimate is within the range of those values. However, the apparent diffusion of SSB is ultrafast compared to the previously reported 1D diffusion coefficient of SSB on short ssDNA: the coefficients determined here are at least six hundred times larger than the previously reported value of 270 nt2/s at 37 °C, determined from SSB diffusion on oligo(dT) with lengths only slightly longer than the occluded site size of 65 nucleotides.11 This large difference in apparent diffusion coefficients led us to hypothesize that SSB can migrate along long ssDNA through a mechanism that cannot occur during local diffusion on short ssDNA. It has been shown that E. coli SSB tetramers can undergo a direct transfer between two ssDNA molecules without an intermediate step of complete dissociation of the protein from one ssDNA molecule.26; 27 Since direct transfer is possible between two separate ssDNA molecules, SSB might also be able to transfer between two segments within the same ssDNA molecule when the ssDNA is very long. This process is termed intersegment transfer36 and has been observed for several proteins.26; 27; 37; 38; 39 A schematic representation of intersegment transfer of an SSB tetramer is depicted in Figure 6. An SSB tetramer bound to one segment of a ssDNA via one DNA binding site is transferred to another segment of the same DNA, when the other segment interacts with the bound SSB at a second DNA binding site on the SSB. Such a long range repositioning of the SSB tetramer would conceivably result in the appearance of an ultrafast diffusion process.
Figure 5. Comparison to 1D Diffusion coefficients of other proteins.
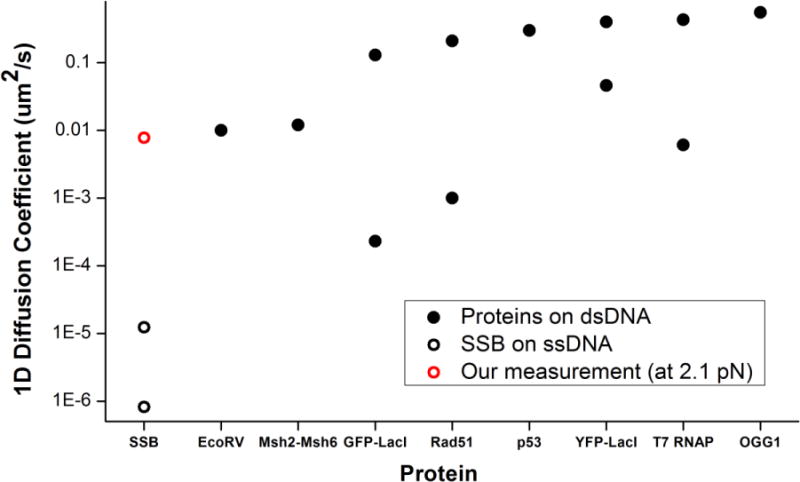
Comparison of the 1D diffusion coefficients reported for various proteins11; 33; 34; 35 (black disc) on dsDNA and our measurement of the apparent diffusion coefficient of SSB on ssDNA (red circle). Previously reported diffusion coefficients of SSB11 (black circle) are the smallest of all. (These were converted from units of nt2s−1 to μm2s−1, for comparison using the conversion factor at 2.1 pN. The higher of the two was an extrapolation to 37 °C, while the lower was measured at 4 °C.11)
Figure 6. Schematic depicting the concept of intersegment transfer.
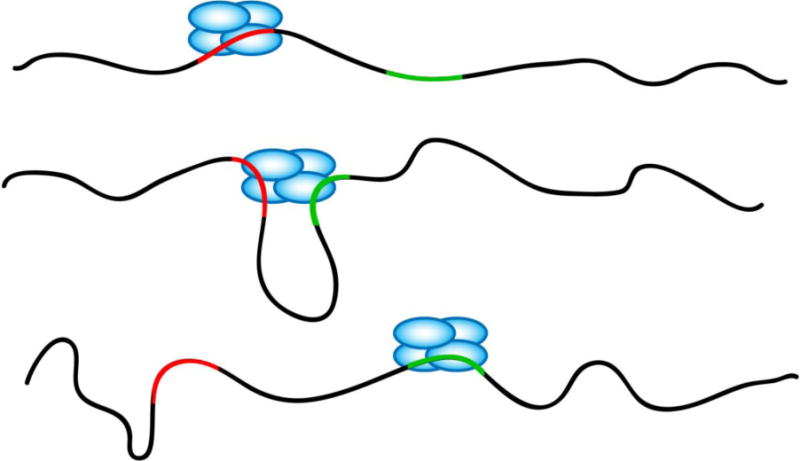
In this model, SSB migrates from one location to the other via a long range intersegment transfer. A SSB is bound at a location (red) on a long ssDNA. When the other segment (green) of the same strand on which the SSB is bound makes contact to the SSB via random fluctuations, the SSB then transfers to the new location.
We also observed that the apparent diffusion coefficient of the SSB tetramer is dependent on the force applied to the ends of the ssDNA. The apparent diffusion becomes faster as the applied tension to the ssDNA is reduced. To test if this force-dependence can be explained by the intersegment transfer model, we calculated the apparent diffusion coefficient by analyzing simulated diffusion trajectories for an SSB tetramer that can undergo intersegment transfer at various forces. In these simulations, an SSB bound to one segment of ssDNA was allowed to be transferred to another ssDNA segment whenever the second segment approached the SSB (to be more precise, the first segment to which the SSB is bound) within a certain threshold radius, which was chosen to be 5 nm because the width and the height of E. coli SSB tetramer is about 4.4 nm and 7.1 nm in the crystal structure.40 We used the probability distribution of the end-to-end distance of a free ssDNA molecule41; 42 after modifying it to include the influence of applied tension. The time scale of the simulation (the conversion factor which translates one step in the simulation into seconds) was the only parameter which was adjusted to fit the simulated diffusion coefficients to the experimentally determined values. A comparison of the force-dependence measured experimentally (black circle) to the force-dependence from the simulation (red dots) is shown in Figure 7. The force-dependence of the diffusion coefficients predicted by the intersegment transfer mechanism is reasonably close to the experimental result. These simulations suggest that the decrease in apparent diffusion coefficient with increasing force is due to the increased tension and thus lower flexibility of the ssDNA causing a decrease in the probability of a second segment of ssDNA folding back and interacting with a bound SSB.
Figure 7. Comparison of diffusion coefficients at various forces fromexperiments and intersegment transfer simulations.
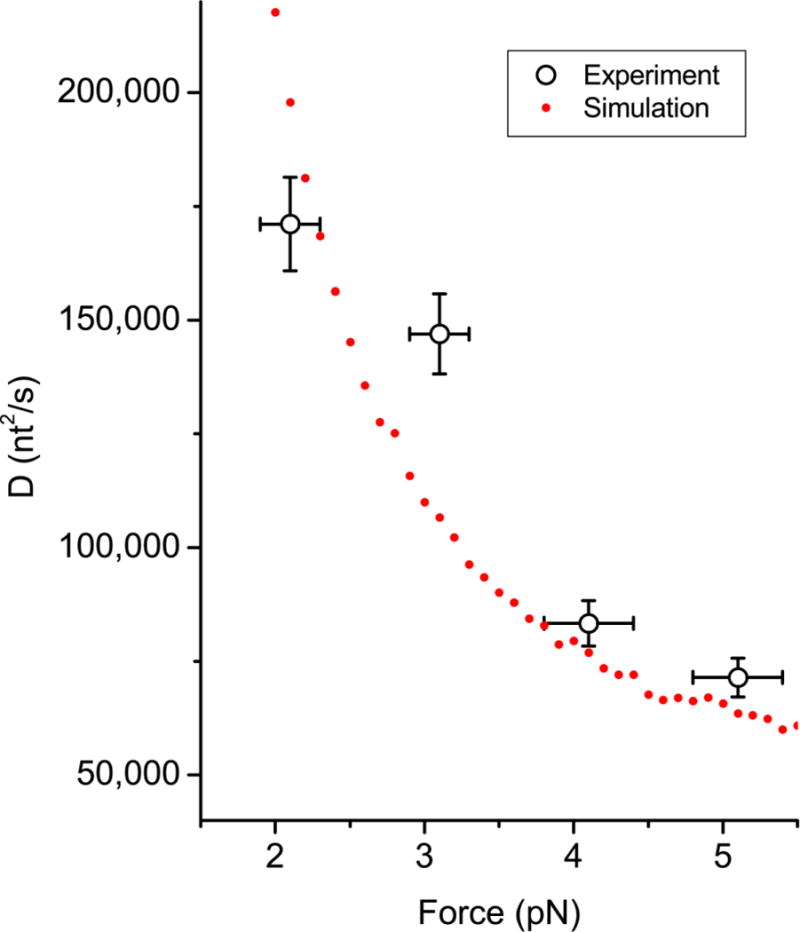
Black circles with X and Y errorbars are the experimental data, whereas the red disks are the result of the simulations. In the simulations, it was assumed that a SSB migrates to other segments of ssDNA, when the segment approaches the SSB within a threshold radius. The simulation results were obtained using a threshold radius of 5 nm and WLC model with a persistence length of 1.5 nm for ssDNA. The unknown conversion factor between one step in the simulation and one second was determined as the value which yields the least square error between the experiments and the simulations.
A critical requirement for any protein to undergo an intersegment transfer process is that it must possess at least two distinct sites for binding DNA since the transfer occurs through a doubly DNA ligated protein intermediate.27 This is clearly met by SSB since it contains four ssDNA binding sites (subunits) due to its homo-tetrameric nature. Kozlov and Lohman27 showed that direct transfer of an SSB tetramer between two separate ssDNA molecules is much faster when the SSB is bound in its (SSB)35 binding mode such that only 2 of the 4 subunits are initially bound to ssDNA, allowing the other 2 free subunits to be available to bind a second ssDNA molecule. When bound in the (SSB)65 binding mode, where ssDNA occupies all 4 subunits, the observed rate of direct transfer is ~103 – 104-fold slower since transfer can happen only when the ssDNA becomes partially unwrapped from one of the subunits, allowing the second ssDNA molecule to bind transiently.27 This aspect of the intersegment transfer of SSB was not included in the model used to generate the simulated trajectories analyzed above. This is the case when the applied force on the ssDNA is zero, however, in a crowded cellular environment in which many other proteins bind on the same strand of ssDNA to which SSB is bound, the force on the ssDNA may be high enough to partially unravel the DNA.43 Increasing forces should increase the probability of partial unwrapping of ssDNA and exposing free SSB subunits that can interact with another ssDNA segment. This aspect of the intersegment transfer reaction would be expected to show the opposite effect of force, i.e., an increase in intersegment transfer at higher forces. Based on the data in Figure 6, it appears the decrease in ssDNA flexibility with increasing force dominates the effect of force on SSB intersegment transfer. More detailed studies as a function of force will be needed to resolve the balance between these two effects.
Through the intersegment transfer mechanism, SSB should be able to be repositioned on long ssDNA much more efficiently than if it had to diffuse entirely along the contour length of the DNA. This mechanism may facilitate the important role of SSB protein, which is to recruit other partner proteins to their sites of action and reposition itself for the function of the partner proteins. As proposed previously, this mechanism may also allow SSB to be rapidly transferred and thus recycled between old and new Okazaki fragments during replication.27 Of course, this fast mode of repositioning should occur along with the relatively slower, local mode of diffusion11; 12 and would complement each other: the local diffusion could be used to move SSB locally on ssDNA that is tightly packed with protein to protect the DNA from degradation and move SSB to allow access of the partner proteins to the ssDNA. In contrast, intersegment transfer should be more advantageous when SSB needs to be redistributed over long distances. This type of long range redistribution might also be useful in order for a translocating enzyme (e.g., a helicase) to bypass SSB.44; 45 SSB could be transferred to a DNA segment behind the helicase via intersegment transfer, effectively removing the SSB as a barrier. This mechanism is reminiscent of the proposed mechanism of histone core transfer from a location ahead of an RNA polymerase to a location behind it.46 Recent technical advances in imaging individual DNA binding proteins on densely covered DNA47 are anticipated to shed light on detailed mechanisms for how DNA binding proteins move on crowded DNA during replication and repair.
For long-range movement of SSB via intersegmental transfer to occur in vivo, three requirements need to be satisfied. First, the DNA binding surface of the SSB tetramer should be partially exposed to capture the destination DNA segment. Second, the destination DNA segment should be unoccupied to receive the incoming SSB tetramer. Third, the original and destination DNA segment should be brought close enough to be bridged by an SSB tetramer. We suggest that all three requirements are likely satisfied in vivo because our previous studies have shown that the multivalent interaction between DNA and SSB is highly dynamic, breaking off and reforming little by little, and because ssDNA and SSB/ssDNA complexes are likely to be more dynamic and compact compared to dsDNA of same length.
Although there have been many studies of protein-ssDNA interactions at a nanometer level, direct observation of single protein dynamics on on longer lengths of ssDNA has been limited. As shown here in the case of SSB, observations made on exactly the same biological system but using much longer ssDNA molecules can reveal a fundamentally new activity. Our approach enables direct visualization of protein dynamics on long ssDNA and should provide access to biological processes that have not been observable on shorter DNA molecules.
Materials and Methods
Instrument design details
The optical layout of the experimental setup is outlined in Figure S1. The setup is based on an inverted microscope (Olympus IX71), with slight modification of a condenser and a mount for a dichroic mirror (D3). A sample is placed on top of a XYZ piezo-nanostage (Mad City Labs) mounted on a manual stage (Semprex) with two micrometers in the XY axis, which is fixed on top of the microscope. An oil immersion objective lens (100X/1.40, Olympus) is used for objective-type TIRF microscopy and optical trapping. A 532-nm diode-pumped solid state laser (Spectra Physics) and a 1,064-nm Nd-YAG laser (Spectra physics) are used for fluorescence excitation and optical trapping, respectively. Fluorescence emission is imaged onto an EMCCD (electron-multiplying charge-coupled device) camera (Andor iXon) and is recorded at the frame rate of 50 Hz. Antidigoxigenin coated polystyrene beads (Spherotech) are used for optical trapping and scattered laser light from a trapped bead is collected by condenser lens and imaged onto a quadrant photodiode detector (Pacific Sensors).
Preparation of secondary-structure-free ssDNA constructs
Long ssDNA molecules without secondary structures were synthesized following the method demonstrated by Brockman et al.30 The following template and primer oligonucleotide sequences were used: 5′- AGG AGA AAA AGA AAA AAA GAA AAG AAG G -3′ and 5′-Biotin – TCT CCT CCT TCT -3′. The 5′-end of the primer oligonucleotide is labeled with a biotin moiety, for surface immobilization. The product ssDNA polymers have only pyrimidine nucleobases (thymine and cytosine), which prevents intramolecular base-pairing interactions. To initiate the reaction, template oligonucleotide (0.2 μM) is annealed to a primer oligonucleotide (0.2 μM) to form a circular partial duplex DNA with a nick. During the hybridization reaction, the solution is heat treated to 70 °C for 2.5 minutes and then slowly cooled to room temperature. Next, 600 units of T4 DNA ligase (NEB) is used to ligate the nick and to make a covalently closed circular ssDNA template (5 hrs @ 16 °C). Replication is initiated by adding 50 nM closed circular ssDNA template, 25 uM each of dTTP and dCTP, 20 μg bovine serum albumin and 5 units phi29 DNA polymerase (NEB) in 1X phi29 DNA polymerase buffer (NEB). After incubating the reaction for 12 min, digoxigenin-11-ddUTP (Roche) is added to quench the reaction and label the 3′-end of the ssDNA product with digoxigenin to attach anti-digoxigenin coated beads. Finally, 200 mM EDTA was then added to quench phi29 activity. We determined that these reaction conditions typically yielded ssDNA products with an average size of ~10,000 nt, as determined by agarose gel electrophoresis. Phi29 DNA polymerase exhibits superb processivity and strand displacement activity,48; 49 and is able to generate a very long ssDNA with lengths upwards of 65,000 nt.28
Experimental solution conditions
All experiments were performed in 20 mM Tris-HCl (pH 8.3), 500 mM NaCl, and 0.5 mM EDTA at 24 °C. Preparation and characterization of Alexa555-labeled SSB was described previously.11 The concentration of E. coli SSB tetramers was varied between 10 pM and 20 pM. To enhance the stability and longevity of Alexa555 fluorophore on the SSB, we used 3 mM Trolox (Sigma-Aldrich), 0.8 % dextrose with glucose oxidase (Sigma-Aldrich) and catalase (Calbiochem).50; 51
Supplementary Material
Highlights.
We directly visualize the movement of Escherichia coli SSB on long polymeric single-stranded DNA (ssDNA).
We observed ultrafast redistribution of SSB along ssDNA, which is at least 600 times faster than previously reported local diffusion.
Our work suggests that SSB can also migrate via a long range intersegment transfer on long ssDNA.
Acknowledgments
This work was supported by the US National Institutes of Health (GM065367 and RR025341 to T.H. and GM030498 to T.M.L.) and by the National Science Foundation (0822613 and 0646550 to T.H.). T.H. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kowalczykowski SC, Bear DG, Von Hippel PH. 21 Single-Stranded DNA Binding Proteins. In: Paul DB, editor. The Enzymes. Vol. 14. Academic Press; 1981. pp. 373–444. [Google Scholar]
- 2.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ha T, Kozlov AG, Lohman TM. Single-Molecule Views of Protein Movement on Single-Stranded DNA. Annual Review of Biophysics. 2012;41 doi: 10.1146/annurev-biophys-042910-155351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams KR, Spicer EK, LoPresti MB, Guggenheimer RA, Chase JW. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. Journal of Biological Chemistry. 1983;258:3346–3355. [PubMed] [Google Scholar]
- 5.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–80. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hobbs MD, Sakai A, Cox MM. SSB protein limits RecOR binding onto single-stranded DNA. J Biol Chem. 2007;282:11058–67. doi: 10.1074/jbc.M611007200. [DOI] [PubMed] [Google Scholar]
- 7.Baitin DM, Gruenig MC, Cox MM. SSB antagonizes RecX-RecA interaction. J Biol Chem. 2008;283:14198–204. doi: 10.1074/jbc.M801511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu D, Myers AR, George NP, Keck JL. Mechanism of Exonuclease I stimulation by the single-stranded DNA-binding protein. Nucleic Acids Research. 2011;39:6536–45. doi: 10.1093/nar/gkr315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marceau AH, Bahng S, Massoni SC, George NP, Sandler SJ, Marians KJ, Keck JL. Structure of the SSB-DNA polymerase III interface and its role in DNA replication. EMBO J. 2011;30:4236–47. doi: 10.1038/emboj.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antony E, Weiland E, Yuan Q, Manhart CM, Nguyen B, Kozlov AG, McHenry CS, Lohman TM. Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J Mol Biol. 2013;425:4802–19. doi: 10.1016/j.jmb.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–7. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou R, Kozlov AG, Roy R, Zhang J, Korolev S, Lohman TM, Ha T. SSB functions as a sliding platform that migrates on DNA via reptation. Cell. 2011;146:222–32. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohman TM, Kowalczykowski SC. Kinetics and mechanism of the association of the bacteriophage T4 gene 32 (helix destabilizing) protein with single-stranded nucleic acids. Evidence for protein translocation. J Mol Biol. 1981;152:67–109. doi: 10.1016/0022-2836(81)90096-6. [DOI] [PubMed] [Google Scholar]
- 14.Lohman TM. Kinetics and mechanism of dissociation of cooperatively bound T4 gene 32 protein-single-stranded nucleic acid complexes. 2. Changes in mechanism as a function of sodium chloride concentration and other solution variables. Biochemistry. 1984;23:4665–75. doi: 10.1021/bi00315a023. [DOI] [PubMed] [Google Scholar]
- 15.Sancar A, Williams KR, Chase JW, Rupp WD. Sequences of the ssb gene and protein. Proceedings of the National Academy of Sciences. 1981;78:4274–4278. doi: 10.1073/pnas.78.7.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiner JH, Bertsch LL, Kornberg A. The deoxyribonucleic acid unwinding protein of Escherichia coli. Properties and functions in replication. Journal of Biological Chemistry. 1975;250:1972–1980. [PubMed] [Google Scholar]
- 17.Ferrari ME, Lohman TM. Apparent heat capacity change accompanying a nonspecific protein-DNA interaction. Escherichia coli SSB tetramer binding to oligodeoxyadenylates. Biochemistry. 1994;33:12896–910. doi: 10.1021/bi00209a022. [DOI] [PubMed] [Google Scholar]
- 18.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7:648–52. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 19.Kozlov AG, Cox MM, Lohman TM. Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. The Journal of biological chemistry. 2010;285:17246–52. doi: 10.1074/jbc.M110.118273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohman TM, Bujalowski W. Effects of base composition on the negative cooperativity and binding mode transitions of Escherichia coli SSB-single-stranded DNA complexes. Biochemistry. 1994;33:6167–76. doi: 10.1021/bi00186a016. [DOI] [PubMed] [Google Scholar]
- 21.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–603. [PubMed] [Google Scholar]
- 22.Lohman TM, Overman LB, Datta S. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J Mol Biol. 1986;187:603–15. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- 23.Ferrari ME, Bujalowski W, Lohman TM. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J Mol Biol. 1994;236:106–23. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 24.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic Structural Rearrangements Between DNA Binding Modes of E. coli SSB Protein. Journal of Molecular Biology. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee KS, Balci H, Jia H, Lohman TM, Ha T. Direct imaging of single UvrD helicase dynamics on long single-stranded DNA. Nat Commun. 2013;4:1878. doi: 10.1038/ncomms2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider RJ, Wetmur JG. Kinetics of transfer of Escherichia coli single strand deoxyribonucleic acid binding protein between single-stranded deoxyribonucleic acid molecules. Biochemistry. 1982;21:608–615. doi: 10.1021/bi00533a002. [DOI] [PubMed] [Google Scholar]
- 27.Kozlov AG, Lohman TM. Kinetic mechanism of direct transfer of Escherichia coli SSB tetramers between single-stranded DNA molecules. Biochemistry. 2002;41:11611–27. doi: 10.1021/bi020361m. [DOI] [PubMed] [Google Scholar]
- 28.Zhao W, Ali MM, Brook MA, Li Y. Rolling circle amplification: applications in nanotechnology and biodetection with functional nucleic acids. Angewandte Chemie. 2008;47:6330–7. doi: 10.1002/anie.200705982. [DOI] [PubMed] [Google Scholar]
- 29.Reiss E, Holzel R, Bier FF. Synthesis and stretching of rolling circle amplification products in a flow-through system. Small. 2009;5:2316–22. doi: 10.1002/smll.200900319. [DOI] [PubMed] [Google Scholar]
- 30.Brockman C, Kim SJ, Schroeder CM. Direct observation of single flexible polymers using single stranded DNA. Soft Matter. 2011;7:8005–8012. doi: 10.1039/C1SM05297G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khafizov R. Single molecule force spectroscopy of single stranded DNA binding protein and Rep helicas 2012 [Google Scholar]
- 32.Zhou R, Kunzelmann S, Webb MR, Ha T. Detecting intramolecular conformational dynamics of single molecules in short distance range with subnanometer sensitivity. Nano Lett. 2011;11:5482–8. doi: 10.1021/nl2032876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonnet I, Biebricher A, Porté PL, Loverdo C, Bénichou O, Voituriez R, Escudé C, Wende W, Pingoud A, Desbiolles P. Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA. Nucleic Acids Research. 2008;36:4118–4127. doi: 10.1093/nar/gkn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 35.Tafvizi A, Huang F, Leith JS, Fersht AR, Mirny LA, van Oijen AM. Tumor Suppressor p53 Slides on DNA with Low Friction and High Stability. Biophysical Journal. 2008;95:L01–L03. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berg OG, von Hippel PH. Diffusion-Controlled Macromolecular Interactions. Annual Review of Biophysics and Biophysical Chemistry. 1985;14:131–158. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 37.Menetski JP, Kowalczykowski SC. Transfer of recA protein from one polynucleotide to another. Effect of ATP and determination of the processivity of ATP hydrolysis during transfer. Journal of Biological Chemistry. 1987;262:2093–2100. [PubMed] [Google Scholar]
- 38.Aragay AM, Diaz P, Daban JR. Association of nucleosome core particle DNA with different histone oligomers: Transfer of histones between DNA-(H2A,H2B) and DNA-(H3,H4) complexes. Journal of Molecular Biology. 1988;204:141–154. doi: 10.1016/0022-2836(88)90605-5. [DOI] [PubMed] [Google Scholar]
- 39.Ruusala T, Crothers DM. Sliding and intermolecular transfer of the lac repressor: kinetic perturbation of a reaction intermediate by a distant DNA sequence. Proceedings of the National Academy of Sciences. 1992;89:4903–4907. doi: 10.1073/pnas.89.11.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-Å resolution. Proceedings of the National Academy of Sciences. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thirumalai D, Ha BY. Statistical mechanics of semiflexible chains. In: Grosberg AIU, editor. Theoretical and Mathematical Models in Polymer Research: Modern Methods in Polymer Research and Technology. Academic Press; 1998. [Google Scholar]
- 42.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophysical journal. 2004;86:2530–7. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brenner MD, Zhou R, Ha T. Forcing a connection: impacts of single-molecule force spectroscopy on in vivo tension sensing. Biopolymers. 2011;95:332–44. doi: 10.1002/bip.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honda M, Park J, Pugh RA, Ha T, Spies M. Single-molecule analysis reveals differential effect of ssDNA-binding proteins on DNA translocation by XPD helicase. Mol Cell. 2009;35:694–703. doi: 10.1016/j.molcel.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spies M, Ha T. Inching over hurdles: how DNA helicases move on crowded lattices. Cell Cycle. 2010;9:1742–9. doi: 10.4161/cc.9.9.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M, Bustamante C. The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nat Struct Mol Biol. 2011;18:1394–9. doi: 10.1038/nsmb.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heller I, Sitters G, Broekmans OD, Farge G, Menges C, Wende W, Hell SW, Peterman EJ, Wuite GJ. STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nat Methods. 2013;10:910–6. doi: 10.1038/nmeth.2599. [DOI] [PubMed] [Google Scholar]
- 48.Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. The Journal of biological chemistry. 1989;264:8935–40. [PubMed] [Google Scholar]
- 49.Baner J, Nilsson M, Mendel-Hartvig M, Landegren U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Research. 1998;26:5073–8. doi: 10.1093/nar/26.22.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ha T. Single-Molecule Fluorescence Resonance Energy Transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 51.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Meth. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


