Abstract
A large number of collagen-like proteins have been identified in bacteria during the past ten years, principally from analysis of genome databases. These bacterial collagens share the distinctive Gly-Xaa-Yaa repeating amino acid sequence of animal collagens which underlies their unique triple-helical structure. A number of the bacterial collagens have been expressed in E. coli, and they all adopt a triple-helix conformation. Unlike animal collagens, these bacterial proteins do not contain the post-translationally modified amino acid, hydroxyproline, which is known to stabilize the triple-helix structure and may promote self-assembly. Despite the absence of collagen hydroxylation, the triple-helix structures of the bacterial collagens studied exhibit a high thermal stability of 35–39 °C, close to that seen for mammalian collagens. These bacterial collagens are readily produced in large quantities by recombinant methods, either in the original amino acid sequence or in genetically manipulated sequences. This new family of recombinant, easy to modify collagens could provide a novel system for investigating structural and functional motifs in animal collagens and could also form the basis of new biomedical materials with designed structural properties and functions.
Keywords: collagen, triple helix, recombinant expression, thermal stability, prokaryote, biomedical material
1. Discovery of bacterial collagens
Collagen is the most abundant protein in mammals, and plays a critical role in extracellular matrix structural properties and cell signaling. The defining feature of a collagen is its molecular structure, which is the unique supercoiled triple-helix. This conformation is made up of three left-handed polyproline-like chains twisted together into a right-handed triple-helix (Brodsky and Ramshaw, 1997). The tight packing of the triple helix requires that every third residue in the primary sequence be Gly, because there is no space for any larger amino acid in the interior axis of the triple-helix. This leads to the repetitive sequence pattern (Gly-Xaa-Yaa)n, which is a distinguishing feature of collagens. Another characteristic of animal collagens is the presence of a high content of Pro and, notably, a high content (>10% of residues) of the post-translationally formed hydroxyproline (Hyp) (Myllyharju, 2003). The enzyme prolyl hydroxylase hydroxylates all Pro residues in the Yaa position of the Gly-Xaa-Yaa repeat in collagens. Hyp residues make a critical contribution to the stability of the triple helix through stereoelectronic effects (Bretscher et al. 2001) and/or hydration (Bella et al. 1994), and also appear essential for collagen self-association (Perret et al. 2001) and for some receptor interactions. Although collagens were originally thought to be found only in multicellular animals and to require the Hyp residue, it has recently been demonstrated that there are collagen-like proteins in bacteria and that they can form triple-helix structures even though they lack Hyp (Table 1).
Table 1.
Comparison of bacterial collagens which have been characterized with mammalian collagens
| Mammalian collagens | Bacterial collagens | ||
|---|---|---|---|
| Similarities | Gly-Xaa-Yaa repeats | Yes | Yes |
| Triple-helix | Yes | Yes | |
| Trypsin resistance | Yes | Yes | |
| Thermal stability | Tm = ~37 °C | Tm = ~35-39 °C | |
| Calorimetric enthalpy | Very high | Relatively high, but lower than mammalian collagen | |
| Trimerization domain | At either end | At either end | |
| Differences | Length | ~350 triplets for fibrillar collagens | ~35-95 triplets (characterized so far) |
| Hydroxyproline | Yes | No | |
| Heterotrimer | Some of them | Not found yet | |
| Sequence and stabilization | Pro/Hyp rich, important for stabilization | Strategies include electrostatic interactions; glycosylation of Thr residues; very high Pro | |
| Types | 28 different types in human | Species dependent – can be many species variants | |
| Interruptions | In some of them | None characterized | |
| Fibril formation | In vivo | Probably limited | |
In 2000, two proteins in the gram negative bacterium Streptococcus pyogenes were found to contain a substantial length of (Gly-Xaa-Yaa)n amino acid sequence and it was postulated that these form collagenous structures (Lukomski et al. 2000; Rasmussen et al. 2000). As increasing numbers of genomic sequences were being reported, an analysis was carried out on 136 eubacterial genomes (Rasmussen et al. 2003), searching for sequences with homology to (Gly-Pro-Pro)n. Hits were found for 56 proteins in 25 bacterial genomes, with none seen in any of the 15 archeobacterial genomes. The number of Gly-Xaa-Yaa tripeptides varied from 7 to 745, with an average length of 76 triplets, and these collagen-like sequences are always flanked by non-collagenous domains. The collagen-like sequences from different bacteria all had a relatively high Pro content, and Rasmussen et al. (2003) found distinctive amino acid compositions for different potential proteins which could be categorized as Thr-rich, Pro-rich, or rich in charged residues. Pro was preferentially found in the X position in bacterial proteins in contrast to mammalian collagens where there are typically half or more of the Pro residues in the Yaa-position, which are subsequently hydroxylated. Conversely, in the bacterial collagens Thr and Gln are much more frequent in the Yaa-position than observed for mammalian collagens (Rasmussen et al. 2003). Several bacterial genomes contained multiple collagen-like sequences, up to 9 in some cases, so it is possible that heterotrimers, with two or three different chains, are formed in these cases. It has been suggested that bacterial collagen sequences arise from horizontal gene transfer from eukaryotes to bacteria (Rasmussen et al. 2003).
Clearly, in the 10 years since this initial study was reported, the extent of the genomic information has increased many-fold and a large number of additional genomes are available for interrogation. Further studies on several of these bacterial proteins have confirmed that they have the characteristic triple-helix structure of collagen and suggest they may play important roles in pathogenesis. These proteins, which are being recognized in increasing numbers, are no longer unexpected curiosities, but represent an opportunity for approaching basic science problems in collagen and for biomaterial applications.
2. Biological role of bacterial collagen-like proteins
While there are many bacterial species that contain collagen-like sequences in the genome (Rasmussen et al. 2003), there is evidence for their natural expression for only a few cases (Karlstrom et al. 2004; 2006). A few pathogenic bacterial systems have been well characterized and these cases suggest the collagen protein may interact with the host to assist invasion or help a pathogen evade the host immune system. The two S. pyogenes bacterial collagens, Scl1 and Scl2, have sequences indicating they are anchored on the cell surface and have been shown to bind to a variety of host proteins. Depending on the specific serotype, the non-collagenous V-domain of Scl1 may bind to high-density lipoprotein (HDL) (Gao et al. 2010), low-density lipoprotein (LDL) (Han et al. 2006a), factor H (Caswell et al. 2008a), complement factor H-related protein 1 (CFHR1) (Reuter et al. 2010), or the extra cellular matrix (ECM) proteins fibronectin and laminin (Caswell et al. 2009). Binding to these components helps S. pyogenes escape from complement-regulated phagocytosis and enhances its adherence to the macrophages and ECM. Both Scl1 and Scl2 bind to thrombin-activatable fibrinolysis inhibitor (TAFI, procarboxypeptidase) and recruit it to S. pyogenes cell surface, counteracting the host response through regulating the proteolysis by activated TAFI (Pahlman et al. 2007) and redirecting inflammation from a transient state to a chronic state (Seron et al. 2011). The collagenous domain of Scl1 (denoted CL) mimics mammalian collagens by interacting with collagen receptor integrins α2β1 and α11β1 through a GLPGER binding site (Caswell et al. 2008b). This interaction facilitates S. pyogenes adherence to host cells and activates intracellular signaling (Humtsoe et al. 2005). It also enhances the internalization of S. pyogenes by host cells and reemergence from host cells into extracellular environment (Caswell et al. 2007). More recently, it was found that Scl1 protein plays an important role in biofilm formation by targeting EDA-containing cellular fibronectin (Oliver-Kozup HA et al. 2011; 2013).
A very different role appears for the two collagen like proteins, BclA and BclB, found in the pathogenic bacteria Bacillus anthracis (Sylvestre et al. 2002; Waller et al. 2005). These glycosylated proteins are structural components of the Bacillus exosporium and have been shown to be present in thin hair-like surface filaments. Similar to Scl1 and Scl2, the central part of BclA and BclB is the collagenous region with a (Gly-Xaa-Yaa)n sequence (Boydston et al. 2005). The length of the central collagenous domain is highly polymorphic, with 17–91 Gly-Xaa-Yaa tri-peptides, and the variation of exosporium filament hair length is dependent on the length of BclA collagenous domain (Sylvestre et al. 2003). A globular C-terminal domain is located at the distal end of the filaments and forms a rugged permeability barrier or shield around the spore (Boydston et al. 2005).
Even systems which have only been partly characterized hint at the complexities of quaternary structure, interactions and function that may be involved with bacterial collagen-like proteins. For example, collagen-like sequences have been found as part of the spore appendages of Clostridium taeniosporum (Walker et al. 2007). Two of the 4 appendage proteins have collagen-like sequences: GP85 has 53 Gly-Xaa-Yaa repeats, while CL2 has 43 Gly-Xaa-Yaa repeats (Walker et al. 2007). In other species, such as B. anthracis (Steichen et al. 2003), an external nap has been associated with triple helical collagen, so this may also prove to be the case for C. taeniosporum, but the formation of triple helical structure has not yet been shown. Another partly characterized system is the collagen like domains reported in Pasteuria ramosa (Mouton et al. 2009; McElroy et al. 2011), where a triple-helical structures has been inferred by comparison to the Bacillus structure (Mouton et al. 2009; McElroy et al. 2011). Recent studies (McElroy et al. 2011), using analysis of an incomplete genome analysis for P. ramosa, have suggested huge complexity for the collagens in this species.
The bacterial collagens are frequently associated with the outer membrane of the organisms. In mammalian systems there are also certain collagens, for example types XIII, XVII, XXIII and XXV that are transmembrane collagens (Franzke et al. 2005; Ricard-Blum, 2011). The ectodomains of mammalian transmembrane collagens and certain bacterial collagens both show cell adhesive properties. The mammalian collagens are all type II transmembrane proteins, with a short cytosolic N-terminal and a longer C-terminal ectodomain including multiple triple-helical domains. The orientation is more variable among bacterial collagens. For example, the B. anthracis collagen-like proteins having a C-terminal triple-helix domain and globular domain extending out as hair-like filaments, and the S. pyogenes collagen-like proteins have an N-terminal globular domain and triple-helix extending outside of the cell wall.
3. Bacterial collagens that are known to form a triple helix structure
Despite the large number of putative collagen structures in bacteria, only eight have been confirmed as having a triple helical structure. The eight proteins all consist of a single uninterrupted collagen triple helical domain, flanked by both N-terminal and C-terminal non-collagenous domains. In most cases, the triple-helix has been confirmed for a single bacterial species, but this is likely to be representative of multiple polymorphisms of these genes in different strains of these species. Although collagen-like proteins from pathogenic organisms were the earliest characterized (S. pyogenes and B. anthracis), the collagen triple-helix structure has also been confirmed in nonpathogenic organisms. In the study by Xu and colleagues (Xu et al. 2010) a group of potential collagens was selected by database searches, using a selection parameter that the number of triplet repeats should be greater than 35. This size was selected from knowledge of the stability characteristics of other collagen domains and peptides. A final selection was made after the potential stability of sequences was assessed using a collagen stability predictor algorithm (Persikov et al. 2005). This approach allowed the selection and study of collagen structure from three soil bacteria that are not pathogens, S. usitatus, R. palustris and Methylobacterium sp 4–46. There were no characteristics that set the non-pathogen structures apart from pathogenic ones.
The structures confirmed to date (Table 2) are from the following species:
Streptococcus pyogenes, SclA/Scl1 gene. This is a pathogenic organism that is responsible for a variety of diseases such as superficial skin and throat infections, but also can lead to more serious invasive conditions such as acute rheumatic fever. Both SclA and SclB (see below) contain the cell wall anchoring motif LAPTGE.
Streptococcus pyogenes, SclB/Scl2 gene. This second gene from S. pyogenes is distinct from and larger than SclA.
Bacillus anthracis, BclA gene. This pathogenic bacterium is the causative agent of anthrax. BclA is a glycosylated protein that is one of two collagen-like structural components of the bacillus exosporium filaments.
Legionella pneumophila, Lcl gene. This species is a gram negative, facultative intracellular pathogen that is the agent involved in Legionnaires’ Disease.
Clostridium perfringens. This gram positive pathogen is the causative agent for gas gangrene.
Solibacter usitatus. This non-pathogenic, gram negative bacterium is abundant in soils.
Rhodopseudomonas palustris. This non-pathogenic, gram negative bacterium is a phototrophic species that is found in both marine and soil environments.
Methylobacterium sp 4–46. This non-pathogenic bacterium is found in soils, where it can utilize methanol derived from plants and can stimulate plant development.
Table 2.
Bacterial collagens known to form a stable triple-helix.
| Bacterium | Gene | N-terminal domain* |
Collagen domain* |
C- terminal domain* |
Calculated pI |
Tm (°C) (CL domain) |
Triple –helix validation |
Reference(s) |
|---|---|---|---|---|---|---|---|---|
| Streptococcus pyogenes | SclA/Scl1 | 68 | 150 | 93 | 5.1 | 36.4 | CD and trypsin | Xu et al. 2002 |
| Streptococcus pyogenes | SclB/Scl2 | 74 | 237 | 100 | 5.4 | 37.6 | CD and trypsin | Xu et al. 2002 |
| Bacillus anthracis | wt BclA | 19 | 228 | 134 | 3.1 | 37.0 | CD and trypsin | Boydson et al. 2005 |
| Legionella pneumophila | Lcl | n.d. | 105 | n.d. | 5.3 | n.d. | Trypsin | Vandersmissen et al. 2010 |
| Clostridium perfringens | 53 | 189 | 162 | 4.7 | 38.8 | CD and trypsin | Xu C, et al. 2010 | |
| Solibacter usitatus | 42 | 246 | 147 | 5.6 | 38.5 | CD and trypsin | Xu C, et al. 2010 | |
| Rhodopseudomonas palustris | 9 | 117 | 86 | 9.3 | 37.0 | CD and trypsin | Xu C, et al. 2010 | |
| Methylobacterium sp 4-46 | 102 | 147 | 74 | 8.6 | 35.0 | CD and trypsin | Xu C, et al. 2010 |
n.d. Not determined.
The numbers of residues given are for a specific reported isolate, but extensive polymorphisms are present for many, if not all species.
The triple-helix structures characterized show a wide variety in the size of the triple helical domain, ranging from 105 to 285 amino acid residues (Table 2). Similarly, the N- and C-terminal non-collagenous ends of these proteins also show wide variations in size, ranging from 9 to 102 residues (N-terminal domain after signal cleavage) and 74 to 162 residues (C-terminal domain) (Table 2).
The bacterial collagens that have been characterized are diverse in amino acid composition characteristics, with very different amino acids in the Xaa and Yaa positions of the constant (Gly-Xaa-Yaa)n pattern. A wide range of (calculated) isoelectric points are present, ranging from acid pI values, such as seen for both S. pyogenes and S. usitatus proteins, to basic pI values, such as seen for collagen-like proteins from R. palustris and Methylobacterium sp 4–46 (Table 2). One striking feature of bacterial collagens, which is never seen in animal collagens, is the frequent presence of repeating amino acid sequence patterns, such as GKDGKDGQNGKDGLP in S. pyogenes Scl2, (GPKGEP)n in M. sp4–46, and the repeating (GPT)5GDTGTT sequence in B. anthracis BclA. The number of repeats is seen to vary in different strains of the bacteria. Considering only the non-Gly residues in the protein, all bacterial collagen domains have a significant Pro content of more than 20% (Figure 1). Examination of the distributions shows that some proteins have a very high content of charged residues, while others are highly enriched in polar residues, compared to animal collagens. The lack of hydroxyproline suggests that bacterial collagens use a different approach than mammalian collagens for achieving a similar triple helical stabilization, using combinations of other stabilizing amino acid sequences (Persikov et al. 2005) (see below).
Fig. 1.
A pie chart representation of the non-Gly amino acid composition of bacterial collagen-like domains. Only the amino acids at Xaa and Yaa positions were taken into consideration. The groups include: hydrophobic (Val, Ile, Leu, Met, Phe), polar (Ser, Thr, Asn, Gln), charged (Asp, Glu, Lys, Arg, His), small (Gly and Ala) and Pro. The amino acid composition of the α1 chain of human4type I collagens is shown for comparison.
Finally, an interesting group of collagens has been reported that form triple-helical structures associated with E. coli (Ghosh et al. 2012). These collagens often, but not always, comprise around 111 amino acids, and are particularly stable, with an example showing a Tm of 42 °C. However, these collagens are not found in the common laboratory strains, such as K-12. Rather they are found in various pathogenic strains, such as O157:H7. It has been suggested that these collagens have a probable role as a trimeric phage side-tail protein that participates in the attachment of phage particles to target E. coli cells (Ghosh et al. 2012). Many pathogenic strains have genomes that are 0.9 Mb larger than non-pathogenic strains, with the additional genetic material, including collagen-like sequences and also virulence factors, arising from horizontal gene transfer (Ghosh et al. 2012). However, since these collagen sequences are included in prophages embedded in the bacterial genome and may be considered as bacteriophage sequences rather than bacterial ones, they will not, along with other collagen-like proteins described in fungi and viruses (Rasmussen et al. 2003; Wang and St Leger, 2006), be considered further in this review. Rather this review will focus on the small number of the proteins found to have Gly-Xaa-Yaa repeating sequences in bacteria which have been expressed and shown to form triple helical structures.
4. Structural Studies of recombinant bacterial collagens which form a collagen-triple helix
4.1 Triple-helix structure and stability
Thus far, no direct studies have been carried out on any collagen-like proteins extracted from their natural bacteria. However, a number of the genes have been expressed in E. coli as recombinant proteins and their properties studied. A triple-helical region is identified by two major criteria. Native triple-helical structures are resistant to digestion by trypsin, chymotrypsin, pepsin and other common proteases. Therefore, enzyme digestion followed by SDS-PAGE is a routine assay which can be done on a small amount of purified material. In addition, the triple-helix has a characteristic CD spectrum, with a maximum near 220 nm and a minimum near 198 nm. When this typical CD spectrum is seen, the mean residue ellipticity at 220 nm can be followed with increasing temperature to measure thermal stability. Enzyme digestion and/or CD studies have been done for the various proteins described above, in Section 3, and all bacterial proteins with (Gly-Xaa-Yaa)n reading frames which have been expressed in E. coli in a soluble form have turned out to form stable triple-helical structures (Table 2). In addition, the protein from L. pneumophila, as well as the B. anthracis BclA protein and the S. pyogenes Scl1 and Scl2 proteins, were all shown to be susceptible to bacterial (C. histolyticum) collagenase digestion (Boydsen et al. 2005; Vandersmissen et al. 2010).
In general, bacteria appear to lack the prolyl hydroxylase enzyme necessary for the formation of hydroxyproline, although a prolyl hydroxylase has been reported in B. anthracis (Culpepper et al. 2010). The bacterial collagens expressed in E. coli do not contain Hyp, and presumably Hyp is not present in the original bacterial protein either. Despite the absence of Hyp, these bacterial collagens formed typical triple-helices that were highly stable (Table 2). Even with the varying amino acid compositions described in Figure 1, the melting temperatures of all of the bacterial collagen-like proteins fell into the range of ~35–39 °C, similar to Tm~39 °C for human collagens. The relatively high content of Pro residues in all of these proteins is an important stabilizing factor for the triple-helix structure, but different bacterial collagens appear to maintain thermal stabilities via different additional strategies. Some bacterial collagens, e.g. S. pyogenes, are rich in charged residues and stabilized by electrostatic interactions (Mohs et al. 2007), while polar residues may contribute to the stability of other proteins (Xu et al. 2010). Threonine residues in the Yaa-position, some of which are glycosylated, appear to stabilize the triple-helix in the BclA protein of B. anthracis (Boydston et al. 2005), as well as contributing to the adhesion of the spores to target cells (Daubenspeck et al. 2004; Lequette et al. 2011). The positive effect for stabilization is probably because the hydroxyl group of Thr or the many hydroxyl groups of the Thr-bound oligosaccharides could be involved in water bridges that lead to triple helix stabilization (Mann et al. 1996; Bann and Bachinger, 2000; Sylvestre et al. 2002).
It is tempting to suggest that bacterial collagens evolved to maintain thermal stabilities close to 38 °C to promote bacterial attachment to host tissues and other pathological processes, but three of these collagen-like proteins come from non-pathogenic soil bacteria (M. sp 4–46, S. usitatus and R. palustris). Although there are no Hyp residues, a high calorimetric enthalpy was still observed for S. pyogenes Scl2 bacterial collagen (Yoshizumi et al. 2009), suggesting a high degree of hydrogen bonding mediated by hydration. Thus, bacterial collagens might have retained some of the striking hydration of animal collagens (Bella et al. 1994). In general, the presence of non-triple-helical N-terminal or C-terminal domains had little effect on the stability of the triple-helix (Xu et al. 2010; Yu et al. 2010). However, it was striking that the presence of the C-terminal trimerization domain in B. anthracis BclA raised the melting temperature from 37 °C to more than 80 °C (Boydston et al. 2005).
4.2 Formation of higher order structures
In animals, self-assembly of collagen molecules to form fibrils or networks is a key aspect of collagen biology (Ricard-Blum, 2011). For bacterial collagens, many appear to be membrane components and there is no natural higher order structure observed so far. This may be due, in part, to the absence of Hyp, which is implicated in the self-association of triple-helical molecules and collagen (Kramer at al., 2000; Perret et al. 2001). Since the bacterial collagens expressed successfully in E. coli to date contain no interruptions in the (Gly-Xaa-Yaa)n sequence, it may be possible to induce them to form higher order structures in vitro (Yoshizumi et al. 2009). Under some conditions, S. pyogenes Scl2 bacterial collagen triple-helical domains CL (with a length ¼ of human fibrillar collagens) and CL-CL (duplicate of CL, with a length ½ of human fibrillar collagens) can self-assemble to form twisted and staggered fibrillar structures at neutral pH. For CL-CL fibrils, the length of aggregated units is around 140 nm (the length of CL-CL molecule) and the diameter is 4–5 nm indicating the alignment of more than one molecule (with a diameter of around 1.5 nm) in parallel or anti-parallel register (Yoshizumi et al. 2009). These fibrillar structures are small and do not show the periodic banding pattern typical for animal collagen fibrils, but it is possible that wider and larger bacterial collagen fibrils with periodic banding could be obtained through manipulation of the sequence, such as selectively distributing charged residues and further increasing the length of the collagenous domain.
5. Manipulation of triple-helix in recombinant bacterial collagens: a tool for understanding animal collagen structure and function
Since human collagens cannot be expressed easily in a recombinant system, it has been difficult to modify sequences and lengths, and to produce large quantities of modified recombinant materials. Laboratory scale production of various constructs of different types of human collagens has been achieved and has allowed the definition of functions and properties of various sequence elements (Ito et al. 2006). Examples include constructs of human collagen types I and II with either deletions or tandem repeats of D period segments (Zafarullah et al. 1997; Arnold et al. 1998; Steplewski et al. 2004); single amino acid substitutions to mimic disease causing mutations inhuman collagens (Brittingham et al. 2005; Adachi et al. 1999); and single amino acid replacements near the MMP cleavage site in type III collagen (Williams and Olsen, 2010). Alternatively, homologous sets of collagen model peptides can be used to probe structure and function, but may be limited by the length. On the other hand, the recombinant bacterial collagen system brings the potential to easily alter the triple-helix sequence and vary the triple-helix length, as well as the ability to insert biologically active sequences, in a system where large yields of protein are practical. This facilitates the investigation of features found in normal and pathological human collagens, and enables amino acid sequence/structure correlations as well as sequence/function relationships to be elucidated.
5.1 Effect of triple-helix length on structural properties
The triple-helix is a linear polymer type structure, and its structural properties will depend on its length as well as its amino acid sequence. Studies on collagen-like peptides show there must be a minimum length of (Gly-Xaa-Yaa)n in order to form a triple-helix and then stability levels off with increasing length, fitting a single exponential curve (Persikov et al. 2005). The triple-helix length of bacterial collagens varies in different strains, and it has also been possible to manipulate the length of the triple-helix. Han et al. (2006) studied S. pyogenes collagen-like proteins of different lengths, and found that the Tm values of most of them were close to 37.5–39 °C, suggesting a pressure for stability near body temperature. The shortest protein (n=20) showed a Tm ~ 5 °C lower than the longer constructs, indicating again that some minimum length is required to form a stable triple-helix. However, the stability was unchanged for lengths n=60–129, showing that, as seen for peptides, there is an exponential approach to a maximum stability value, near 39oC in this case. The triple-helix stability of all longer constructs is similar to that of hydroxylated mammalian collagens even though Hyp is absent. The Scl2.28 based protein with a duplication of the collagen domain V-CL-CL (n=158) had a Tm value near that of the original V-CL (n=79) construct (36.5 °C), suggesting both proteins have a length sufficient to reach the maximal stability (Yoshizumi et al. 2009).
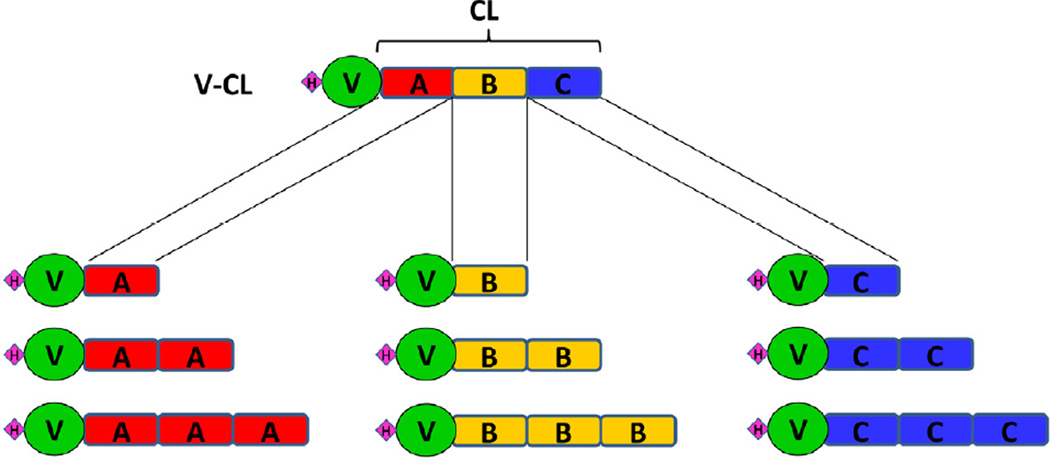
To investigate more closely how length and amino acid sequence influenced stability, segments equal to about 1/3 length of the original CL were expressed and studied (Yu et al. 2011) (Figure 2). The CL domain of Scl2 protein can be considered as being composed of three approximately equal segments with distinctive amino acid features: N-terminal A (lowest charge), middle B (highest Pro content) and C-terminal C (very high charge concentration). Each domain was expressed alone or adjacent to a trimerization domain, and also as homodimers (AA, BB, CC) and homotrimers (AAA, BBB, CCC), although V-CC and V-CCC were insoluble and not purified (Yu et al. 2011). The stabilities of these constructs were observed to depend upon their amino acid sequences and increased as the triple helix got longer. The B module was more stable than A and C, and the BBB construct had the same stability as the original CL domain. The V trimerization domain promoted refolding, but the folding rate of each construct again depended upon the sequence and became reduced for longer constructs. The folding rates of all the other constructs were lower than that of the natural V-ABC protein (=V-CL) (Yu et al. 2011). The ability to express fragments of a collagen, as well as create new tandem repeats presents a way to dissect out the contributions to triple-helix stability and folding.
Fig. 2.
A diagram of the constructs including A, B, or C fragments of the S. pyogenes Scl2 CL domain, showing the rod-like CL domain, the globular V domain, and a small diamond to represent the His6-tag at the N terminus. Fragments A, B and C were expressed individually in E. coli, or fused with the V domain to promote trimerization. Each fragment was also tandemly fused to construct a dimer (AA, BB, or CC) or a trimer (AAA, BBB, or CCC) appended to the V domain. Although CCC was successfully expressed it was insoluble (Yu et al. 2011
5.2. Effect of Gly missense mutations and interruptions on triple-helix properties
A number of hereditary connective tissue disorders, including Osteogenesis Imperfecta, Ehlers Danlos Syndrome type IV, and some chondrodysplasias, are due to mutations in collagen, and the most frequent mutations are single base substitutions that replace one Gly residue in the Gly-Xaa-Yaa repeat (Marini et al. 2007). The precise sequence of events that leads from a Gly missense mutation in collagen to the clinical phenotype has not been easy to unravel, and it is not understood why a Gly→Ser missense mutation at one site in the triple-helix may lead to a severe clinical phenotype while a nearby Gly→Ser mutation may lead to milder symptoms. The following factors might be important for symptom severity: the identity of the residue replacing Gly, the immediate sequence environment, and the location of mutation with respect to initiation point. Peptides have been used as models to study the impact of Gly substitutions (Beck et al. 2000) and have provided important information about the conformational perturbation and stability changes due to replacement of Gly by different residues (Hyde et al. 2006; Bryan et al. 2011), but peptides are not good models for animal collagen folding, which requires nucleation followed by linear propagation of the triple-helix.
The recombinant bacterial collagen system has been applied to characterize the effects of a Gly mutation, since a mutation can be introduced at any location within the triple-helix while controlling the sequence surrounding it (Cheng et al. 2011). Site-directed mutagenesis was used to introduce a Gly→Arg or a Gly→Ser mutation at a site near the middle or near the N-terminus of the triple-helix adjacent to the trimerization domain. All mutations led to small decreases in stability ~2oC, but the Gly→Arg mutation very close to the N-terminus introduced a trypsin sensitive site within the triple-helix, highlighting the presence of a locally destabilized region with limited effect on the overall Tm value. The bacterial collagen-like protein represents a good folding model for mammalian collagens, since it contains an N-terminal globular trimerization domain which is essential for the folding of the adjacent collagen domain and hence allows study of collagen folding in presence of the mutations. A Gly→Arg mutation near the center of the triple-helix led to a significant folding delay, (t1/2 = 10 min to 55 min), while the Gly→Arg mutation very close to the N-terminal trimerization domain led to a dramatic decrease in the folding rate (t½> 1000 min) and the extent of refolding, suggesting disruption of the triple helix nucleation process.
The recombinant bacterial collagen system was also used to investigate the effect of interruptions in the Gly-Xaa-Yaa repeating sequence on triple-helix conformation, stability and folding (Hwang and Brodsky, 2012). Although human fibril forming collagens all have a perfect (Gly-Xaa-Yaa)n amino acid sequence pattern in their triple-helix region, non-fibrillar collagens (such as basement membrane type IV collagen) contain sites where this repeat is broken. These interruptions in the perfect repeating sequence may be functional, and have been suggested to play a role in molecular flexibility, formation of network structures, and sites of enzymatic degradation. A single interruption was inserted between the two S. pyogenes collagen domains, incorporating either a 4-residue (GAAVM) or 15-residue (GQISEQKRPIDVEFQK) interruption sequence from the a5 chain of type IV collagen. It was observed that the type IV collagen interruptions were successfully incorporated into the bacterial triple-helix protein with little impact on overall structure or stability. The resistance of the construct with the 4 residue interruption to trypsin and thermolysin supports the retention of a tightly wound triple-helix, while enzyme susceptibility within the 15 residue interruption suggests a loosened or alternative conformation which transitions from a susceptible state to an enzyme resistant triple-helix state as the susceptibility boundary between interruption and (Gly-Xaa-Yaa)n sequence is approached. Triple-helix folding was significantly slower in the presence of an interruption, and the longer 15 residue interruption showed a greater delay than the 4-residue interruption. A mutation introduced near an interruption sequence found within type IV collagen also led to significant delay in folding (Hwang and Brodsky, 2012).
5.3. Introduction of biological functional sequences
Animal collagen is the major structural protein providing physical support of tissues, and also has various biological functions by interacting with cell surface receptors and other extracellular matrix molecules. In the sequence of fibril-forming collagens, there are known binding sites for more than 50 biologically functional molecules (Sweeney et al. 2008), including cell receptors such as integrins, DDR and GPVI, (Leitinger and Hohenester, 2007) and other important ECM molecules, such as fibronectin, laminin and proteoglycans. In addition, collagens interact with matrix metalloproteinases (MMPs), which degrade the matrix and play an important role in physiological and pathological processes, such as wound healing, tissue repair, angiogenesis, arthritis and metastasis (Visse and Nagase, 2003).
Since bacteria are single-cell organisms without an extracellular matrix, bacterial collagens do not have similar functions as animal collagens. However, it has been discovered that several type of bacterial collagens can interact with mammalian proteins and may play important roles in bacterial living processes. For example, in some S. pyogenes strains, the CL domain of Scl1 protein can interact with integrins α2β1 and α11β1 through the sequence GLPGER (Caswell et al. 2008b). In contrast, the bacterial collagen Scl2.28 from S. pyogenes does not contain any known ligand binding sites and has been suggested as a collagen “blank slate”. Using recombinant DNA technology, the biologically inert Scl2 collagen-like protein has been modified to incorporate human collagen derived sequences (Seo et al. 2010; Yu et al. 2012). These engineered bacteria-human collagen chimeras were expressed recombinantly in E. coli and have been shown to successfully confer corresponding biological activities of the inserted human collagen sequences. Figure 3 shows the recombinant Scl2.28 variants and sites for the integrated human collagen motifs. Current research on bacteria-human collagen chimeras based on Scl2.28 is twofold. On one hand, it has been used as a collagen model system to study human collagen fragments in terms of regional folding behaviors or biological functions. On the other hand, building various recombinant collagen chimeras with multiple human collagen functions may establish a new collagen source for a variety of medical applications.
Fig. 3.
A diagram illustrating various substitutions and insertions of functional domains from animal collagens that have been made in S. pyogenes Scl2 constructs, including integrin (Seo et al. 2010; An et al. 2013; Peng et al. 2013); heparin (Peng et al. 2013); fibronectin (An et al. 2013), and MMP-1 (Yu et al. 2012).
As a collagen model system, the bacterial collagen sequence provides a triple helical platform for the incorporated human collagen sequences to form stable secondary structure in the absence of hydroxyproline. Enzymes and receptors that require collagen triple helix structure for their interactions with collagen have been shown to have similar behavior to the recombinant bacterial collagen chimeras. Integrin binding sequences (GLPGER, GFPGER, GFPGEN, GERGFP, GERGVE) have been introduced in Scl2.28 through site-direct mutagenesis by various groups (Seo et al. 2010; An et al 2013, Peng et al 2013). The affinity to integrins has been demonstrated in those modified bacterial collagens by both solid state binding assays and cell culture experiments. Similarly, a four triplet GRPGKPGKQGQK sequence corresponding to the heparin binding site of collagen has also been introduced by serial mutations to the Scl2.28 and has subsequently showed binding to the fluorescently labeled heparin (Peng et al. 2013). Moreover, a putative fibronectin binding region from human collagen type II has been inserted into the bacterial collagen within a CL domain or between the two tandem CL domain repeats (An et al., 2013; An et al. 2014). A solid-state binding assay indicated that by incorporating a minimum of a 6 triplet GLAGQRGIVGLPGQRGER sequence, this recombinant bacterial collagen could bind fibronectin specifically at its gelatin binding domain. Cell culture based assays also showed a remarkable improvement of this fibronectin binding-bacterial collagen in supporting human mesenchymal stem cell (hMSC) spreading and proliferation as well as megakaryocytes attachment and differentiation. A specific enzyme cleavage site could also be incorporated into the bacterial collagen system. Various lengths of collagen type III sequence around the MMP cleavage triplet have been inserted in between two CL domains. It was revealed that in order for MMP-1 to cleave at the inserted sequence, a minimum of 1 triplet preceding and 3 triplets succeeding the cleavage triplet were required. The MMP and trypsin susceptibility of the inserted sequence in this bacterial collagen context was found to mimic that of human collagen type III (Yu et al. 2012). It is noteworthy that the lack of post-translational proline hydroxylation in bacterial collagen likely indicates that hydroxyproline is not essential for the above mentioned collagen functional sites. However, its absence may account, in part, for the differences in affinity compared to mammalian collagen.
A chimeric structure where a silk tag (GAGAGS)n was added to the bacterial collagen C-terminus enabled specific non-covalent binding to fabricated silk porous scaffolds. This enabled stable structures to be formed without introduced chemical crosslinking. The excellent mechanical properties of silk in addition to the various functional domains of the engineered bacterial collagens made the first step towards developing a multifunctional artificial extracellular matrix for various biomedical needs (An et al. 2013).
6. Characterization and manipulation of trimerization domains adjacent to triple-helices
The characteristic (Gly-Xaa-Yaa)n sequence has difficulty folding into a triple-helix efficiently unless it is flanked by a non-collagenous trimerization or registration domain. The trimerization domains of most types of mammalian collagens are located C-terminus to the triple-helix domain. For example, in type I collagen folding, three C-propeptides trimerize, determining the chain selection of two α1 chains and one α2 chain; the register is then set for the adjacent triple-helix (Khoshnoodi et al. 2006), followed by triple-helix zippering from C- to N- terminus. In addition, the non-collagenous domains of most collagen types have been implicated in a wide range of biological functions, such as inhibiting angiogenesis and promoting cell proliferation (Ortega and Werb, 2002). All (Gly-Xaa-Yaa)n triple-helix domains of bacterial collagens are flanked by variable lengths of sequence that could represent independent trimerization domains and/or have distinct structural and functional roles. In S. pyogenes, the N-terminal globular domains (V domains) of the Scl1 and Scl2 proteins are of variable lengths and amino acid sequences in different strains, although all V domains share a high content of α-helical secondary structure (Han et al. 2006b; Yu et al. 2010). Recently, the crystal structure of Scl2.3 globular domain has been reported as a compact trimeric six-helix bundle (Squeglia et al. 2014) which is unique among any known trimerization domains of collagen. The V domains of S. pyogenes have been shown to promote the refolding of the triple-helix domain. Interestingly, the triple-helix domain of S. pyogenes can fold by itself when initially expressed in E. coli but cannot refold in vitro unless it is adjacent to the V domain. As discussed in Section 2, the V domains were also found to bind to extracellular matrix proteins and to a number of plasma components, with interactions likely to be important in the pathogenesis of this bacterium. In B. anthracis, the highly stable beta-sheet-containing C-terminal globular domain is likely to be important for folding and stability of the BclA triple-helix, whereas its N-terminal non-collagenous domain is essential for basal layer attachment (Boydston et al. 2005; Rety et al. 2005; Tan and Turnbough, 2009).
It has been shown that the trimerization domains of bacterial collagen-like proteins act as modular units which can be exchanged or manipulated at either end of collagen-like domains. Movement of the V domain of Streptococcal Scl2 protein from the N-terminus to the C-terminus resulted in molecules with similar conformation and stability as the original V-CL protein, but the ability of in vitro refolding was compromised. By fusion to the N-terminus, Scl2-V domain could also facilitate correct folding of the collagen-like domain from Clostridium perfringens, which could not fold in its original context. The ability of the V domain to fold a collagen-like molecule from a different bacteria species supports its modular nature (Yu et al. 2010). In a more recent study, Scl2-V was replaced with a hyperstable three-stranded coiled-coil, either at the N-terminus or the C-terminus of the triple-helix. The chimeric proteins retain their distinctive melting temperatures, but the rate of refolding was faster when the coiled-coil was at C-terminus (Yoshizumi et al. 2011).
7. Products and Applications
7.1 Biological properties related to biomaterials of recombinant collagens
To be suitable as a biomedical material, bacterial collagen must meet certain key safety criteria. For example, they must be non-cytotoxic. This has been demonstrated for the collagen domain of S. pyogenes Scl2 protein using a Live/Dead Cytotoxicity/Viability assay and Neutral Red assay on three different mammalian fibroblast cell lines (Peng et al. 2010b). Also collagen used as biomaterial should be non-immunogenic. Medical grade bovine collagen, which is not or only slightly cross-linked, does show a limited immunological response in humans, with about 3% showing some level of response (Werkmeister and Ramshaw, 2000). The immunological response of the purified collagenlike domain of S.pyogenes has been examined in two different mouse strains (both outbred and inbred) (Peng et al. 2010b). In the absence of adjuvant, Scl2 CL domain was non-immunogenic; in the presence of adjuvant, there was a negligible response observed (Peng et al. 2010), but this immunogenicity of bacterial collagen Scl2 was certainly less than that had been observed for both medical grade bovine and avian collagens (Peng et al. 2010a; Peng et al. 2010b) in the same experimental approach, suggesting that bacterial collagen Scl2, is a particularly poor immunogen. For mammalian collagens, the non-collagenous telopeptide domains appear to be more immunogenic than the triple helical domain (Furthmayr et al. 1971). Based on this observation it is probably better to remove any non-collagenous domains, as was done above, prior to using bacterial collagens for biomedical applications. On the other hand, while there is little, if any, immunological response to the purified collagen domain from S. pyogenes (Peng et al. 2010b), observation of positive immune responses to the collagen domain in vivo has been observed, in response to infection by S. pyogenes (Hoe et al. 2007), S. equi, which causes strangles in horses (Karlstrom et al. 2006), and B. anthracis (Steichen et al. 2003), perhaps due to an adjuvant-like effect from the other adjacent bacterial proteins.
7.2 Production of recombinant collagens
Recombinant bacterial collagen would potentially have a very high value for biomedical and regenerative medicine applications (Werkmeister and Ramshaw, 2012). To date, most collagen products used for biomaterials or biomedical devices are extracted from animal sources (Ramshaw et al. 1996). Application of animal collagens always has the risk of pathogen or prion contamination and the possibility of causing allergy. Other problems include the lack of standardization for animal collagen extraction processes and the inability to modify collagen sequences to achieve different biological purposes. Compared with collagens extracted from animal tissues, recombinant collagens are highly pure, disease free, consistent among batches, and amendable to sequence modifications and large scale production (Werkmeister and Ramshaw, 2012).
Production of recombinant mammalian collagens in high yield has not been easy, in part because full replication of mammalian collagen synthesis requires a variety of specific post-translational modifications and proteolytic cleavage processes before forming insoluble higher-order structures. Various systems have been examined including mammalian/insect cell culture, transgenic animals where endogenous prolyl hydroxylation is present and various yeast and transgenic plant systems where prolyl hydroxylase activity needs to be introduced (Werkmeister and Ramshaw, 2012). Of these, the most successful has been a Pichia expression system (Nokelainen et al. 2001) although opportunities for plant-based production are increasing (Brodsky and Kaplan, 2013). It has proved difficult to produce active prolyl hydroxylase in prokaryotes, so reports of mammalian collagen expression in E.coli are limited. It has been suggested that modification of the media to include hydroxyproline can lead to protein including this amino acid (Buechter et al. 2002) but selectivity for the Y position would not be possible. In a more recent study, human prolyl hydroxylase was introduced into E.coli to allow proline hydroxylase during expression of a collagen peptide (Pinkas et al. 2011). Other developments that may prove useful include the characterization of active prolyl hydroxylases from a range of microbial sources, including a novel enzyme from B. anthracis (Culpepper et al. 2010).
If a bacterial collagen or a modified bacterial collagen sequence is to be used as a biomedical or specialty material, it is important that they can be produced in commercially feasible quantities at a competitive cost. If the bacterial collagen were to be used as a coating for a medical device or as a composite with another material, then the production requirement would be less than that if the bacterial collagen were to be used as a single major component, such as in a material for wound management.
Studies on the production of the V-CL Scl2 protein from S. pyogenes have been reported (Peng et al. 2102). These studies used a pColdIII (Takara Bio Inc.) vector for expression in E. coli. The pCold vectors have been proved to be very useful for expression of bacterial collagens from various species (Xu et al. 2010). However, it is possible that other vectors could give better commercial yields than observed using the pCold system.
Production in shake flask cultures gives low yields of recombinant product, < 1 g/L. Increased yields, of around 1 g/L, can be obtained when the shake flask process is transferred to a stirred tank bioreactor. However, more relevant yields around 10 g/L are reported through use of a high cell density fed-batch process and the use of a suitably formulated fully defined media (Table 3) (Peng et al. 2012). This approach was applicable to different constructs of the S. pyogenes Scl2 product (Peng et al. 2012). The best yields, of up to 19 g/L, were reported using this high cell density strategy and extended 24 h production time (Table 3). These yields compare favorably with the average yields reported for other bacterial expression studies of 14 g/L (Adrio and Demain, 2010), although there may be higher commercial yields that remain commercial-in-confidence. This capacity for good fermentation yield, however, still needs to be matched with an equivalent downstream purification protocol. While the use of a His6-tag protocol is efficient for laboratory purifications (Yoshizumi et al. 2009; Peng et al. 2010b), along with other methods such as gel permeation chromatography, it is not suitable for large scale commercial preparations.
Table 3.
Fermentation conditions and protein yields for V-CL in a pCold vector using defined medium.
| Process | Pre-induction phase |
Induction temperature |
Induction phase |
Final OD (600 nm) |
Wet ce ll paste (g/L) |
Volumetric productivity (g/L) |
|---|---|---|---|---|---|---|
| Flask | 24 h at 37°C | 25 °C | 25°C for 10 h 15°C for 14 h |
5.6 | 8.3 | 0.2 |
| Batch | 5 h at 37°C | 25 °C | 25°C for 7 h | 5.5 | 5.3 | 0.7 |
| Fed batch | 24 h at 37°C | 25 °C | 25°C for 10 h | 88 | 113 | 9.5 |
| Fed batch | 24 h at 37°C | 25 °C | 25°C for 10 h 20°C for 5 h 15°C for 9 h |
96 to 144 | 137 to 148 | 13.0 to 19.3 (av. (3) 17.1) |
From Peng et al. (2012)
7.3 Application as a biomedical material
Animal collagens in different forms are used widely as biomaterials in medical products and have been shown to be safe and effective in various clinical applications (Ramshaw et al. 1996). They have also been proposed as materials in the emerging area of tissue engineering (Mafi et al. 2012). There are opportunities for producing new collagen based products using bacterial collagens, especially if an animal-free system is preferred, but so far there is no commercially available product made from bacterial collagens. However, the scientific data presented to date suggest that it has significant potential to emerge as a clinically effective biomedical material. Thus, as noted above (Section 7.1), the collagen domain of the bacterial collagen Scl2 from S. pyogenes is neither cytotoxic nor immunogenic (Peng et al. 2010b). It can also be produced, including the V-domain, by fermentation in E. coli in good yields, of up to ~19 g/L (Peng et al. 2012), equivalent to a yield of around 14 g/L for the collagen CL domain.
To date, there have been limited reports of fabrication of bacterial collagens into formats suitable for use in medical applications. For bulk materials, a collagen scaffold produced by freeze drying will almost certainly need cross-linking. This will increase its thermal stability as (Ramshaw et al. 1996) well as extending its turnover time. Thus, lyophilized Scl2 collagen cross-linked by glutaraldehyde vapour formed sponge-like material, which had increased stability and supported cell attachment and proliferation (Peng et al. 2010b).
Bacterial collagens can be readily modified to introduce a variety of new biological functions (Section 5.4). In a recent study, a composite material comprising a polyurethane network integrated with polyethylene glycol (PEG) hydrogel containing modified bacterial collagen has been reported (Cosgriff-Hernandez et al. 2010; Browning et al. 2012). The collagen contained a substitution to include an integrin binding domain that supported endothelial attachment but was resistant to platelet adhesion and aggregation (Browning et al. 2012). The material was based on reaction of the collagen with acrylate-PEG-N-hydroxysuccinimide and its subsequent incorporation by photo-polymerisation into a 3-D poly(ethylene glycol) diacrylate (PEGDA) hydrogel (Browning et al. 2012). However, for any ‘off the shelf’ product, sterilization and storage conditions are important. Recent studies have shown that dry storage of these modified materials is better than wet storage (Luong et al. 2013), as under wet conditions, ester hydrolysis of the protein linker has been attributed to the slow loss of the bioactive collagen component (Luong et al. 2013, Browning et al. 2013). However, this problem can be potentially resolved through use of an alternative modification reagent, acrylamide-PEG-isosyanate (Browning et al. 2013).
7. Conclusions
Hundreds of protein sequences containing (Gly-Xaa-Yaa)n domains have been found in bacterial genomic databases, and eight of these proteins, coming from both pathogenic and non-pathogenic bacteria, have been expressed as recombinant proteins in E. coli and characterized in detail. For these expressed bacterial collagens, it has been shown that all the predicted collagen-like structures do form stable triple-helices with protease resistance and melting temperatures similar to animal collagens. This suggests that most, if not all, of the (Gly-Xaa-Yaa)n regions of sufficient length in bacterial proteins are likely to be triple-helical, and surprisingly, that they may all have a thermal stability in the ~35–38°C range. Unlike animal collagens, bacterial collagens have no stabilizing Hyp residues, so, depending on individual amino acid composition, their high thermal stability is due in part to contributions from electrostatic interactions or a high content of glycosylated Thr or a very high polar residue content. For bacterial collagens, no natural, higher order structure has been observed so far, but some of them are able to form aggregated structures in vitro. The recombinant bacterial collagens represent an opportunity for exploring basic questions about collagen structure and function, and also provide potential material for biomedical applications. Recombinant protein production in E. coli is already a mature industrial process, free from pathogen contamination. Purified Scl2 collagen is neither immunogenic in mice nor cytotoxic to various human cell lines. The ease of production, and production of structural variants, suggests that it may be useful as a new biomaterial as an alternative to mammalian collagen. With proper fabrication methods, a large library of recombinant bacterial collagens with tunable bioactive motifs may open up the potential to build multifunctional artificial extracellular matrix for many biomedical applications.
Acknowledgments
This work was supported through NIH grant #EB011620.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Zhuoxin Yu, Email: zhuoxin_yu@yahoo.com.
Bo An, Email: Bo.An@tufts.edu.
John A.M. Ramshaw, Email: John.Ramshaw@csiro.au.
Barbara Brodsky, Email: Barbara.Brodsky@tufts.edu.
References
- Adachi E, Katsumato O, Yamashina S, Prockop DJ, Fertala A. Collagen II containing a Cys substitution for Arg-alpha1-519. Analysis by atomic force microscopy demonstrates that mutated monomers alter the topography of the surface of collagen II fibrils. Matrix Biol. 1999;18:189–196. doi: 10.1016/s0945-053x(99)00011-6. [DOI] [PubMed] [Google Scholar]
- An B, DesRochers TM, Qin G, Xia X, Thiagarajan G, Brodsky B, Kaplan DL. The influence of specific binding of collagen-silk chimeras to silk biomaterials on hMSC behavior. Biomaterials. 2013;34:402–412. doi: 10.1016/j.biomaterials.2012.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B, Abbonante V, Yigit S, Balduini A, Kaplan DL, Brodsky B. Definition of the native and denatured type II collagen binding site for fibronectin using a rcombinant collagen system. J. Biol. Chem. 2014 doi: 10.1074/jbc.M113.530808. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrio JL, Demain AL. Recombinant organisms for production of industrial products. Bioeng. Bugs. 2010;1:116–131. doi: 10.4161/bbug.1.2.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold WV, Fertala A, Sieron AL, Hattori H, Mechling D, Bächinger HP, Prockop DJ. Recombinant procollagen II: Deletion of D period segments identifies sequences that are required for helix stabilization and generates a temperature-sensitive N-proteinase cleavage site. J. Biol. Chem. 1998;273:31822–31828. doi: 10.1074/jbc.273.48.31822. [DOI] [PubMed] [Google Scholar]
- Bann JG, Bachinger HP. Glycosylation/Hydroxylation-induced stabilization of the collagen triple helix. 4-trans-hydroxyproline in the Xaa position can stabilize the triple helix. J. Biol.Chem. 2000;275:24466–24469. doi: 10.1074/jbc.M003336200. [DOI] [PubMed] [Google Scholar]
- Beck K, Chan VC, Shenoy N, Kirkpatrick A, Ramshaw JAM, Brodsky B. Destabilization of osteogenesis imperfecta collagen-like model peptides correlates with the identity of the residue replacing glycine. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4273–4278. doi: 10.1073/pnas.070050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 Å resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- Boydston JA, Chen P, Steichen CT, Turnbough CL. Orientation within the exosporium and structural stability of the collagen-like glycoprotein BclA of Bacillus anthracis . J. Bacteriol. 2005;187:5310–5317. doi: 10.1128/JB.187.15.5310-5317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher LE, Jenkins CL, Taylor KM, DeRider ML, Raines RT. Conformational stability of collagen relies on a stereoelectronic effect. J. Am. Chem. Soc. 2001;123:777–778. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]
- Brittingham R, Colombo M, Ito H, Steplewski A, Birk DE, Uitto J, Fertala A. Single amino acid substitutions in procollagen VII affect early stages of assembly of anchoring fibrils. J. Biol.Chem. 2005;280:191–198. doi: 10.1074/jbc.M406210200. [DOI] [PubMed] [Google Scholar]
- Brodsky B, Kaplan DL. Shining light on collagen: expressing collagen in plants. Tissue Eng Part A. 2013;19:1499–1501. doi: 10.1089/ten.tea.2013.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky B, Ramshaw JAM. The collagen triple-helix structure. Matrix Biol. 1997;15:545–554. doi: 10.1016/s0945-053x(97)90030-5. [DOI] [PubMed] [Google Scholar]
- Browning MB, Dempsey D, Guiza V, Becerra S, Rivera J, Russell B, Höök M, Clubb F, Miller M, Fossum T, Dong JF, Bergeron AL, Hahn M, Cosgriff-Hernandez E. Multilayer vascular grafts based on collagen-mimetic proteins. Acta Biomater. 2012;8:1010–1021. doi: 10.1016/j.actbio.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Browning MB, Russell B, Rivera J, Höök M, Cosgriff-Hernandez EM. Bioactive hydrogels with enhanced initial and sustained cell interactions. Biomacromolecules. 2013;14:2225–2233. doi: 10.1021/bm400634j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan MA, Cheng H, Brodsky B. Sequence environment of mutation affects stability and folding in collagen model peptides of osteogenesis imperfecta. Biopolymers. 2011;96:4–13. doi: 10.1002/bip.21432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechter DD, Paolella DN, Leslie BS, Brown MS, Mehos KA, Gruskin EA. Co-translational incorporation of trans-4-hydroxyproline into recombinant proteins in bacteria. J. Biol. Chem. 278:645–650. doi: 10.1074/jbc.M209364200. [DOI] [PubMed] [Google Scholar]
- Caswell CC, Lukomska E, Seo NS, Höök M, Lukomski S. Scl1-dependent internalization of group A Streptococcus via direct interactions with the α2β1 integrin enhances pathogen survival and re-emergence. Mol. Microbiol. 2007;64:1319–1331. doi: 10.1111/j.1365-2958.2007.05741.x. [DOI] [PubMed] [Google Scholar]
- Caswell CC, Han R, Hovis KM, Ciborowski P, Keene DR, Marconi RT, Lukomski S. The Scl1 protein of M6-type group A Streptococcus binds the human complement regulatory protein, factor H, and inhibits the alternative pathway of complement. Mol. Microbiol. 2008a;67:584–596. doi: 10.1111/j.1365-2958.2007.06067.x. [DOI] [PubMed] [Google Scholar]
- Caswell CC, Barczyk M, Keene DR, Lukomska E, Gullberg DE, Lukomski S. Identification of the first prokaryotic collagen sequence motif that mediates binding to human collagen receptors, integrins α2β1 and α11β1. J. Biol.Chem. 2008b;283:36168–36175. doi: 10.1074/jbc.M806865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell CC, Oliver-Kozup H, Han R, Lukomska E, Lukomski S. Scl1, the multifunctional adhesin of group A Streptococcus, selectively binds cellular fibronectin and laminin, and mediates pathogen internalization by human cells. FEMS Microbiol. Lett. 2009;303:61–68. doi: 10.1111/j.1574-6968.2009.01864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Rashid S, Yu Z, Yoshizumi A, Hwang E, Brodsky B. Location of glycine mutations within a bacterial collagen protein affects degree of disruption of triple-helix folding and conformation. J. Biol.Chem. 2011;286:2041–2046. doi: 10.1074/jbc.M110.153965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culpepper MA, Scott EE, Limburg J. Crystal structure of prolyl 4-hydroxylase from Bacillus anthracis . Biochemistry. 2010;2010:124–133. doi: 10.1021/bi901771z. 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgriff-Hernandez E, Hahn MS, Russell B, Wilems T, Munoz-Pinto D, Browning MB, Rivera J, Höök M. Bioactive hydrogels based on designer collagens. Acta Biomater. 2010;6:3969–3977. doi: 10.1016/j.actbio.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Daubenspeck JM, Zeng H, Chen P, Dong S, Steichen CT, Krishna NR, Pritchard DG, Turnbough CL. Novel oligosaccharide side chains of the collagen-like region of BclA, the major glycoprotein of the Bacillus anthracis exosporium. J. Biol.Chem. 2004;279:30945–30953. doi: 10.1074/jbc.M401613200. [DOI] [PubMed] [Google Scholar]
- Franzke CW, Bruckner P, Bruckner-Tuderman L. Collagenous transmembrane proteins: recent insights into biology and pathology. J. Biol.Chem. 2005;280:4005–4008. doi: 10.1074/jbc.R400034200. [DOI] [PubMed] [Google Scholar]
- Furthmayr H, Beil W, Timpl R. Different antigenic determinants in the polypeptide chains of human collagen. FEBS Lett. 1971;12:341–344. doi: 10.1016/0014-5793(71)80010-8. [DOI] [PubMed] [Google Scholar]
- Gao Y, Liang C, Zhao R, Lukomski S, Han R. The Scl1 of M41-type group A Streptococcus binds the high-density lipoprotein. FEMS Microbiol. Lett. 2010;309:55–61. doi: 10.1111/j.1574-6968.2010.02013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh N, McKillop TJ, Jowitt TA, Howard M, Davies H, Holmes DF, Roberts IS, Bella J. Collagen-like proteins in pathogenic E. coli strains. PLoS One. 2012;7:e37872. doi: 10.1371/journal.pone.0037872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R, Caswell CC, Lukomska E, Keene DR, Pawlowski M, Bujnicki JM, Kim JK, Lukomski S. Binding of the low-density lipoprotein by streptococcal collagen-like protein Scl1 of Streptococcus pyogenes . Mol. Microbiol. 2006a;61:351–367. doi: 10.1111/j.1365-2958.2006.05237.x. [DOI] [PubMed] [Google Scholar]
- Han R, Zwiefka A, Caswell CC, Xu Y, Keene DR, Lukomska E, Zhao Z, Höök M, Lukomski S. Assessment of prokaryotic collagen-like sequences derived from streptococcal Scl1 and Scl2 proteins as a source of recombinant GXY polymers. Appl. Microbiol. Biotechnol. 2006b;72:109–115. doi: 10.1007/s00253-006-0387-5. [DOI] [PubMed] [Google Scholar]
- Hoe NP, Lukomska E, Musser JM, Lukomski S. Characterization of the immune response to collagen-like proteins Scl1 and Scl2 of serotype M1 and M28 group A Streptococcus. FEMS Microbiol Lett. 2007;277:142–149. doi: 10.1111/j.1574-6968.2007.00955.x. [DOI] [PubMed] [Google Scholar]
- Humtsoe JO, Kim JK, Xu Y, Keene DR, Höök M, Lukomski S, Wary KK. A streptococcal collagen-like protein interacts with the alpha2beta1 integrin and induces intracellular signaling. J. Biol.Chem. 2005;280:13848–13857. doi: 10.1074/jbc.M410605200. [DOI] [PubMed] [Google Scholar]
- Hwang ES, Brodsky B. Folding delay and structural perturbations caused by type IV collagen natural interruptions and nearby Gly missense mutations. J. Biol.Chem. 2012;287:4368–4375. doi: 10.1074/jbc.M111.269084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde TJ, Bryan MA, Brodsky B, Baum J. Sequence dependence of renucleation after a Gly mutation in model collagen peptides. J. Biol.Chem. 2006;281:36937–36943. doi: 10.1074/jbc.M605135200. [DOI] [PubMed] [Google Scholar]
- Ito H, Steplewski A, Alabyeva T, Fertala A. Testing the utility of rationally engineered recombinant collagen-like proteins for applications in tissue engineering. J. Biomed. Mater. Res. A. 2006;76:551–560. doi: 10.1002/jbm.a.30551. [DOI] [PubMed] [Google Scholar]
- Karlstrom A, Jacobsson K, Flock M, Flock JI, Guss B. Identification of a novel collagen-like protein, SclC, in Streptococcus equi using signal sequence phage display. Vet. Microbiol. 2004;104:179–188. doi: 10.1016/j.vetmic.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Karlstrom A, Jacobsson K, Guss B. SclC is a member of a novel family of collagen-like proteins in Streptococcus equi subspecies equi that are recognised by antibodies against SclC. Vet. Microbiol. 2006;114:72–81. doi: 10.1016/j.vetmic.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Khoshnoodi J, Cartailler JP, Alvares K, Veis A, Hudson BG. Molecular recognition inthe assembly of collagens: terminal noncollagenous domains are key recognition modules in the formation of triple helical protomers. J. Biol.Chem. 2006;281:38117–38121. doi: 10.1074/jbc.R600025200. [DOI] [PubMed] [Google Scholar]
- Leitinger B, Hohenester E. Mammalian collagen receptors. Matrix Biol. 2007;26:146–155. doi: 10.1016/j.matbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Lequette Y, Garénaux E, Combrouse T, Dias Tdel L, Ronse A, Slomianny C, Trivelli X, Guerardel Y, Faille C. Domains of BclA, the major surface glycoprotein of the B. cereus exosporium: glycosylation patterns and role in spore surface properties. Biofouling. 2011;27:751–761. doi: 10.1080/08927014.2011.599842. [DOI] [PubMed] [Google Scholar]
- Lukomski S, Nakashima K, Abdi I, Cipriano VJ, Ireland RM, Reid SD, Adams GG, Musser JM. Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect. Immun. 2000;68:6542–6553. doi: 10.1128/iai.68.12.6542-6553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong PT, Browning MB, Bixler RS, Cosgriff-Hernandez E. Drying and storage effects on poly(ethylene glycol) hydrogel mechanical properties and bioactivity. J. Biomed. Mater. Res. A. 2013 doi: 10.1002/jbm.a.34977. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafi P, Hindocha S, Mafi R, Khan WS. Evaluation of biological protein-based collagen scaffolds in cartilage and musculoskeletal tissue engineering--a systematic review of the literature. Curr. Stem Cell Res. Ther. 2012;7:302–309. doi: 10.2174/157488812800793045. [DOI] [PubMed] [Google Scholar]
- Mann K, Mechling DE, Bachinger HP, Eckerskorn C, Gaill F, Timpl R. Glycosylated threonine but not 4-hydroxyproline dominates the triple helix stabilizing positions in the sequence of a hydrothermal vent worm cuticle collagen. J. Mol. Biol. 1996;261:255–266. doi: 10.1006/jmbi.1996.0457. [DOI] [PubMed] [Google Scholar]
- Marini JC, Forlino A, Cabral WA, Barnes AM, San Antonio JD, et al. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum. Mutat. 2007;28:209–221. doi: 10.1002/humu.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy K, Mouton L, Du Pasquier L, Qi W, Ebert D. Characterisation of a large family of polymorphic collagen-like proteins in the endospore-forming bacterium Pasteuria ramosa . Res. Microbiol. 2011;162:701–714. doi: 10.1016/j.resmic.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Mohs A, Silva T, Yoshida T, Amin R, Lukomski S, Inouye M, Brodsky B. Mechanism of stabilization of a bacterial collagen triple helix in the absence of hydroxyproline. J. Biol.Chem. 2007;282:29757–29765. doi: 10.1074/jbc.M703991200. [DOI] [PubMed] [Google Scholar]
- Mouton L, Traunecker E, McElroy K, Du Pasquier L, Ebert D. Identification of a polymorphic collagen-like protein in the crustacean bacteria Pasteuria ramosa . Res. Microbiol. 2009;160:792–799. doi: 10.1016/j.resmic.2009.08.016. [DOI] [PubMed] [Google Scholar]
- Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003;22:15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- Nokelainen M, Tu H, Vuorela A, Notbohm H, Kivirikko KI, Myllyharju J. High-level production of human type I collagen in the yeast Pichia pastoris . Yeast. 2001;18:797–806. doi: 10.1002/yea.730. [DOI] [PubMed] [Google Scholar]
- Oliver-Kozup HA, Elliott M, Bachert BA, Martin KH, Reid SD, Schwegler-Berry DE, Green BJ, Lukomski S. The streptococcal collagen-like protein-1 (Scl1) is a significant determinant for biofilm formation by group A Streptococcus. BMC Microbiol. 2011;11:262. doi: 10.1186/1471-2180-11-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Kozup H, Martin KH, Schwegler-Berry D, Green BJ, Betts C, Shinde AV, Van De Water L, Lukomski S. The group A streptococcal collagen-like protein-1, Scl1, mediates biofilm formation by targeting the extra domain A-containing variant of cellular fibronectin expressed in wounded tissue. Mol Microbiol. 2013;87:672–689. doi: 10.1111/mmi.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega N, Werb Z. New functional roles for non-collagenous domains of basement membrane collagens. J. Cell Sci. 2002;115:4201–4214. doi: 10.1242/jcs.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlman LI, Marx PF, Morgelin M, Lukomski S, Meijers JC, Herwald H. Thrombin-activatable fibrinolysis inhibitor binds to Streptococcus pyogenes by interacting with collagenlike proteins A and B. J. Biol.Chem. 2007;282:24873–24881. doi: 10.1074/jbc.M610015200. [DOI] [PubMed] [Google Scholar]
- Peng YY, Glattauer V, Ramshaw JAM, Werkmeister JA. Evaluation of the immunogenicity and cell compatibility of avian collagen for biomedical applications. J. Biomed. Mater. Res. A. 2010a;93:1235–1244. doi: 10.1002/jbm.a.32616. [DOI] [PubMed] [Google Scholar]
- Peng YY, Yoshizumi A, Danon SJ, Glattauer V, Prokopenko O, Mirochnitchenko O, Yu Z, Inouye M, Werkmeister JA, Brodsky B, Ramshaw JAM. A Streptococcus pyogenes derived collagen-like protein as a non-cytotoxic and non-immunogenic cross-linkable biomaterial. Biomaterials. 2010b;31:2755–2761. doi: 10.1016/j.biomaterials.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YY, Howell L, Stoichevska V, Werkmeister JA, Dumsday GJ, Ramshaw JAM. Towards scalable production of a collagen-like protein from Streptococcus pyogenes for biomedical applications. Microb. Cell Fact. 2012;11:146. doi: 10.1186/1475-2859-11-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YY, Stoichevska V, Schacht K, Werkmeister JA, Ramshaw JAM. Engineering multiple biological functional motifs into a blank collagen-like protein template from Streptococcus pyogenes . J. Biomed. Mater. Res. 2013 doi: 10.1002/jbm.a.34898. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Perret S, Merle C, Bernocco S, Berland P, Garrone R, Hulmes DJ, Theisen M, Ruggiero F. Unhydroxylated triple helical collagen I produced in transgenic plants provides new clues on the role of hydroxyproline in collagen folding and fibril formation. J. Biol.Chem. 2001;276:43693–43698. doi: 10.1074/jbc.M105507200. [DOI] [PubMed] [Google Scholar]
- Persikov AV, Ramshaw JAM, Brodsky B. Prediction of collagen stability from amino acid sequence. J. Biol.Chem. 2005;280:19343–19349. doi: 10.1074/jbc.M501657200. [DOI] [PubMed] [Google Scholar]
- Pinkas DM, Ding S, Raines RT, Barron AE. Tunable, post-translational hydroxylation of collagen domains in Escherichia coli . ACS Chem Biol. 2011;6:320–324. doi: 10.1021/cb100298r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramshaw JAM, Werkmeister JA, Glattauer V. Collagen-based biomaterials. Biotechnol. Genet. Eng. Rev. 1996;13:335–382. doi: 10.1080/02648725.1996.10647934. [DOI] [PubMed] [Google Scholar]
- Rasmussen M, Eden A, Bjorck L. SclA, a novel collagen-like surface protein of Streptococcus pyogenes . Infect. Immun. 2000;68:6370–6377. doi: 10.1128/iai.68.11.6370-6377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Jacobsson M, Bjorck L. Genome-based identification and analysis of collagen-related structural motifs in bacterial and viral proteins. J. Biol.Chem. 2003;278:32313–32316. doi: 10.1074/jbc.M304709200. [DOI] [PubMed] [Google Scholar]
- Reuter M, Caswell CC, Lukomski SZipfel PF. Binding of the human complement regulators CFHR1 and factor H by streptococcal collagen-like protein 1 (Scl1) via their conserved C termini allows control of the complement cascade at multiple levels. J. Biol.Chem. 2010;285:38473–38485. doi: 10.1074/jbc.M110.143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rety S, Salamitou S, Garcia-Verdugo I, Hulmes DJ, Le Hegarat F, Chaby R, Lewit-Bentley A. The crystal structure of the Bacillus anthracis spore surface protein BclA shows remarkable similarity to mammalian proteins. J. Biol.Chem. 2005;280:43073–43078. doi: 10.1074/jbc.M510087200. [DOI] [PubMed] [Google Scholar]
- Ricard-Blum S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011;3:a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo N, Russell BH, Rivera JJ, Liang X, Xu X, Afshar-Kharghan V, Höök M. An engineered alpha1 integrin-binding collagenous sequence. J. Biol.Chem. 2010;285:31046–31054. doi: 10.1074/jbc.M110.151357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seron MV, Plug T, Marquart JA, Marx PF, Herwald H, de Groot PG, Meijers JC. Binding characteristics of thrombin-activatable fibrinolysis inhibitor to streptococcal surface collagen-like proteins A and B. Thromb. Haemost. 2011;106:609–616. doi: 10.1160/TH11-03-0204. [DOI] [PubMed] [Google Scholar]
- Squeglia F, Bachert B, Simone AD, Lukomski S, Berisio R. The crystal structure of the streptococcal collagen-like protein 2 globular domain from invasive M3-type group A Streptococcus shows significant similarity to immunomodulatory HIV protein gp41. J. Biol. Chem. 2014 doi: 10.1074/jbc.M113.523597. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen C, Chen P, Kearney JF, Turnbough CL. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 2003;185:1903–1910. doi: 10.1128/JB.185.6.1903-1910.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steplewski A, Majsterek I, McAdams E, Rucker E, Brittingham RJ, Ito H, Hirai K, Adachi E, Jimenez SA, Fertala A. Thermostability gradient in the collagen triple helix reveals its multi-domain structure. J. Mol. Biol. 2004;338:989–998. doi: 10.1016/j.jmb.2004.03.037. [DOI] [PubMed] [Google Scholar]
- Sweeney SM, Orgel JP, Fertala A, McAuliffe JD, Turner KR, Di Lullo GA, Chen S, Antipova O, Perumal S, Ala-Kokko L, Forlino A, Cabral WA, Barnes AM, Marini JC, San Antonio JD. Candidate cell and matrix interaction domains on the collagen fibril, the predominant protein of vertebrates. J. Biol.Chem. 2008;283:21187–21197. doi: 10.1074/jbc.M709319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre P, Couture-Tosi E, Mock M. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 2002;45:169–178. doi: 10.1046/j.1365-2958.2000.03000.x. [DOI] [PubMed] [Google Scholar]
- Sylvestre P, Couture-Tosi E, Mock M. Polymorphism in the collagen-like region of the Bacillus anthracis BclA protein leads to variation in exosporium filament length. J. Bacteriol. 2003;185:1555–1563. doi: 10.1128/JB.185.5.1555-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Turnbough CL. Sequence motifs and proteolytic cleavage of the collagen-like glycoprotein BclA required for its attachment to the exosporium of Bacillus anthracis . J. Bacteriol. 2010;192:1259–1268. doi: 10.1128/JB.01003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandersmissen L, De Buck E, Saels V, Coil DA, Anné J. A Legionella pneumophila collagen-like protein encoded by a gene with a variable number of tandem repeats is involved in the adherence and invasion of host cells. FEMS Microbiol. Lett. 2010;306:168–176. doi: 10.1111/j.1574-6968.2010.01951.x. [DOI] [PubMed] [Google Scholar]
- Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ. Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- Waller LN, Stump MJ, Fox KF, Harley WM, Fox A, Stewart GC, Shahgholi M. Identification of a second collagen-like glycoprotein produced by Bacillus anthracis and demonstration of associated spore-specific sugars. J. Bacteriol. 2005;187:4592–4597. doi: 10.1128/JB.187.13.4592-4597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JR, Gnanam AJ, Blinkova AL, Hermandson MJ, Karymov MA, Lyubchenko YL, Graves PR, Haystead TA, Linse KD. Clostridium taeniosporum spore ribbon-like appendage structure, composition and genes. Mol. Microbiol. 2007;63:629–643. doi: 10.1111/j.1365-2958.2006.05494.x. [DOI] [PubMed] [Google Scholar]
- Wang C, St Leger RJ. A collagenous protective coat enables Metarhizium anisopliae to evade insect immune responses. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6647–6652. doi: 10.1073/pnas.0601951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werkmeister JA, Ramshaw JAM. Immunology of collagen-based biomaterials. In: Wise DL, editor. Handbook of biomaterials engineering. Marcel Dekker Inc; New York: 2000. pp. 739–759. [Google Scholar]
- Werkmeister JA, Ramshaw JAM. Recombinant protein scaffolds for tissue engineering. Biomed. Mater. 2012;7:012002. doi: 10.1088/1748-6041/7/1/012002. [DOI] [PubMed] [Google Scholar]
- Williams KE, Olsen DR. Matrix metalloproteinase-1 cleavage site recognition and binding in full-length human type III collagen. Matrix Biol. 2009;28:373–379. doi: 10.1016/j.matbio.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Xu Y, Keene DR, Bujnicki JM, Höök M, Lukomski S. Streptococcal Scl1 and Scl2 proteins form collagen-like triple helices. J. Biol.Chem. 2002;277:27312–27318. doi: 10.1074/jbc.M201163200. [DOI] [PubMed] [Google Scholar]
- Xu C, Yu Z, Inouye M, Brodsky B, Mirochnitchenko O. Expanding the family of collagen proteins: recombinant bacterial collagens of varying composition form triple-helices of similar stability. Biomacromolecules. 2010;11:348–356. doi: 10.1021/bm900894b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi A, Yu Z, Silva T, Thiagarajan G, Ramshaw JAM, Inouye M, Brodsky B. Self-association of Streptococcus pyogenes collagen-like constructs into higher order structures. Protein Sci. 2009;18:1241–1251. doi: 10.1002/pro.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi A, Fletcher JM, Yu Z, Persikov AV, Bartlett GJ, Boyle AL, Vincent TL, Woolfson DN, Brodsky B. Designed coiled coils promote folding of a recombinant bacterial collagen. J. Biol.Chem. 2011;286:17512–17520. doi: 10.1074/jbc.M110.217364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Mirochnitchenko O, Xu C, Yoshizumi A, Brodsky B, Inouye M. Noncollagenous region of the streptococcal collagen-like protein is a trimerization domain that supports refolding of adjacent homologous and heterologous collagenous domains. Protein Sci. 2010;19:775–785. doi: 10.1002/pro.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Brodsky B, Inouye M. Dissecting a bacterial collagen domain from Streptococcus pyogenes: sequence and length-dependent variations in triple helix stability and folding. J. Biol.Chem. 2011;286:18960–18968. doi: 10.1074/jbc.M110.217422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Visse R, Inouye M, Nagase H, Brodsky B. Defining requirements for collagenase cleavage in collagen type III using a bacterial collagen system. J. Biol.Chem. 2012;287:22988–22997. doi: 10.1074/jbc.M112.348979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafarullah K, Sieron AL, Fertala A, Tromp G, Kuivaniemi H, Prockop DJ. A recombinant homotrimer of type I procollagen that lacks the central two D-periods. The thermal stability of the triple helix is decreased by 2 to 4 degrees C. Matrix Biol. 1997;16:245–253. doi: 10.1016/s0945-053x(97)90013-5. [DOI] [PubMed] [Google Scholar]