Abstract
Grb7 is an adaptor molecule mediating signal transduction from multiple cell surface receptors to diverse downstream pathways. Grb7, along with Grb10 and Grb14, make up the Grb7 protein family. This protein family has been shown to be overexpressed in certain cancers and cancer cell lines. Grb7 and a receptor tyrosine kinase, ErbB2, are overexpressed in 20–30% of breast cancers. Grb7 overexpression has been linked to enhanced cell migration and metastasis, although the participants in these pathways have not been fully determined. In this study, we report the Grb7 protein interacts with Filamin-a, an actin-crosslinking component of the cell cytoskeleton. Additionally, we have demonstrated the interaction between Grb7 and Flna is specific to the RA-PH domains of Grb7, and the immunoglobulin-like repeat 16–19 domains of Flna. We demonstrate that full-length Grb7 and Flna interact in the mammalian cellular environment, as well as in vitro. Immunofluorescent microscopy shows potential co-localization of Grb7 and Flna in membrane ruffles upon epidermal growth factor stimulation. These studies are amongst the first to establish a clear connection between Grb7 signaling and cytoskeletal remodeling.
Keywords: Grb7, ErbB2, HER2, neu, cell migration, cytoskeleton, actin binding, Filamin-a
INTRODUCTION
Grb7, Grb10, and Grb14 comprise a family of SH2 domain-containing Grb proteins discovered through cloning of receptor targets screening (Daly et al., 1996; Ooi et al., 1995). A homologous Caenorhabditis elegans protein, Mig-10, is involved in neuronal migration during development (Manser et al., 1997). The members of this family all contain an N-terminal Pro-rich region, an RA-like domain (Wojcik et al., 1999), a PH domain, a BPS domain, and a C-terminal SH2 domain. The RA, PH, and BPS domains are collectively known as the Grbs and Mig domain. Mig-10 does not contain a C-terminal SH2 domain but instead replaces this with a Pro-rich sequence at the C-terminus (Manser et al., 1997).
Grb7 has been shown to bind to ErbB2 (avian erythroblastosis oncogene B 2) and FAK through its SH2 domain. ErbB2 and Grb7 are both overexpressed in a subset (20–30%) of breast cancer patients who share a poor long-term survival rate and a statistically high incidence of metastases (Seshadri et al., 1993; Slamon, 1987). The FAK/Grb7 association results in FAK phosphorylation of Grb7 at tyrosine residues and enhanced cell migration effects (Han et al., 2000). Chu et al. (2009) have also shown the tyrosine phosphorylation state of Grb7 is important in Grb7-mediated cell migration. Overexpression of Grb7 can lead to increased cell migration and development of metastases in cancer, as seen in the overexpression of Grb7 in lymph node metastases (Tanaka et al., 1998). In addition, a Grb7 protein variant lacking its C-terminal region and termed Grb7V has been linked to invasive esophageal carcinoma (Tanaka et al., 1998) and high-grade ovarian cancer (Wang et al., 2010). Grb7 expression upregulation in breast cancer cell lines has also been observed as a result of treatment with the ErbB2 inhibitor lapatinib (Nencioni et al., 2010).
The tyrosine phosphorylation state of Grb7 has been shown important in Grb7's ability to interact with other signaling molecules. We have shown the Grb7 protein binds to both four and a half LIM domains 2 and Hax-1, and the tyrosine phosphorylation state of Grb7 is an important determinant in binding to both proteins (Siamakpour-Reihani et al., 2009; Siamakpour-Reihani et al., 2011). Tsai et al. (2007) showed Grb7 acts as a translational repressor by binding to the mRNA 5’UTR of the kappa opioid receptor (KOR), and Grb7 binding to KOR mRNA is controlled by its tyrosine phosphorylation state.
The Filamins (a, b, c with Filamin-a also known as filamin-α or filamin-1) are a family of homologous proteins whose common function serves to crosslink actin filaments during cytoskeletal remodeling, but who also function as scaffolding and binding partners for a myriad of other proteins, including receptors, signaling molecules and adaptors, channels, and transcription factors (for a review see Nakamura et al., 2011). Genetic mutations in Filamin-a are associated with human diseases, such as periventricular heterotopia (Fox et al., 1998; Sheen et al., 2001), Ehlers–Danlos syndrome (Sheen et al., 2005), otopalatodigital syndrome types 1 and 2 (Robertson, 2007), frontometaphyseal dysplasia (Robertson et al., 2006), and Melnik–Needles syndrome (Robertson, 2007). Filamin-a mutations are common in cardiac valvular dystrophy (Kyndt et al., 2007), and a secreted Filamin-a variant has been associated with breast and brain cancers (Alper et al., 2009).
The studies presented here describe an initial characterization of the Grb7/Filamin-a interaction and offer insight into a direct effect of Grb7 on cytoskeletal remodeling.
MATERIALS AND METHODS
Materials
Yeast two-hybrid media, reagents, and strains were purchased from Clontech Laboratories, Inc. (Mountain View, CA). All other chemicals and reagents were from Sigma-Aldrich (St. Louis, MO). The following antibodies were obtained from Santa Cruz Biotechnology Inc, (Santa Cruz, CA): mouse monoclonal Filamin 1 (3 F180), mouse monoclonal Filamin 1 (PM6/317), rabbit polyclonal Grb7 (H-70), rabbit polyclonal Grb7 (C-20), mouse monoclonal GST (B-14), mouse monoclonal c-Myc (9E10), mouse monoclonal Actin (C-2), mouse monoclonal p-Tyr (PY20), goat anti-mouse IgG-HRP, and non-specific rabbit or mouse control IgG. Goat anti-rabbit IgG-HRP (Product #31460) was obtained from Thermo Fisher Scientific (aka Pierce, Rockford, IL). Fluorophore conjugated secondary antibodies Alexa Fluor 488, Alexa Fluor 546, Phalloidin 647, and Hoechst nuclear staining reagent were purchased from Invitrogen Life Technologies (Grand Island, NY). Coimmunoprecipitation kit and chemiluminescence substrates for Western blot development were obtained from Thermo Fisher Scientific (aka Pierce, Rockford, IL).
Yeast two-hybrid library screening and assays
Yeast strains used in the studies were Saccharomyces cerevisiae Y187, Y2HGold, and AH109. A pretransformed mouse embryonic stem cell cDNA library (normalized) in yeast strain Y187 was obtained from Clontech Laboratories. The clones in the stem cell cDNA library were fused within the prey GAL4 AD vector, PGADT7-RecAB. The Grb7-RA, Grb7-PH, and Grb7-RAPH domain genes were ligated into the bait GAL4 BD vector, pGBKT7-BD (Siamakpour-Reihani et al., 2009; Siamakpour-Reihani et al., 2011). The pGBKT7-BD bait vector containing the Grb7- PH domain gene was transformed into yeast strain Y2HGold following the Yeastmaker Yeast Transformation System 2 protocol. Yeast two-hybrid library screening was carried out by yeast mate and plate technique following standard protocols (Yeast two-hybrid Gold system, Clontech Laboratories). Briefly, 1 ml of normalized cDNA library strain yeast (provided as is by Clontech Laboratories) was combined with 4–5 ml of bait strain yeast (produced by resuspending the cell pellet from a 50 ml SD/-Trp culture grown to OD600 = 0.8) in a sterile 2 L flask and grown in 2X yeast peptone dextrose (YPD) containing kanamycin at 30 °C for 24 hr. Cells were harvested by centrifugation, and the cell pellet suspended in a 0.5X YPD containing kanamycin. The culture was plated on selective quadruple synthetic defined drop-out media (SD/-Leu/-Trp and SD/-Ade/-His/-Leu/-Trp) containing aureobasidin antibiotic and incubated at 30 °C until colonies were visually apparent (approximately 3 days).
Library plasmid isolation and rescue from yeast
Positive interaction yeast prey plasmids were isolated using a Zymoprep Yeast Plasmid Miniprep II kit (Zymo Research Corp., Irvine, CA) and transformed into Escherichia coli. for plasmid amplification. Purified DNA inserts were sequenced and analyzed using the National Center for Bioinformatics, Blast nucleotide query program. Positive interactions were again confirmed by transforming DNA-BD/bait and positive AD/library plasmids into the Y2HGold yeast strain and demonstration of growth on SD/-Ade/-His/-Leu/-Trp media.
β-galactosidase liquid assay
For positive yeast 2-hybrid interactions, the presence of soluble β-galactosidase (β-gal) was assayed using O -nitrophenyl —D- galactoside (ONPG) as the substrate following standard protocols for liquid assay (Clontech Laboratories). Briefly, three isolated colonies were grown on 5 ml of SD/-Leu/-Trp media overnight at 30 °C until reaching an OD600 = 0.6. Each reaction was carried out in triplicate. Cells were centrifuged, suspended in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgSO4), and lysed by sequential freeze-thawing cycles. The ONPG substrate was added to the reaction, and the mixture incubated at 30 °C until a persistent yellow color developed. At this point, the reaction was terminated by adding 0.5 ml of a 1 M Na2CO3 solution. The signal at OD420 was measured spectrophotometrically and quantified as unitless relative β-gal units.
Protein expression and purification
Full-length Filamin-a encoding plasmid vector pREP4-Filamin-a was a gift from Dr. Fumihiko Nakamura (Brigham and Women's Hospital, Department of Medicine, Harvard, Boston, MA). The Filamin IgG repeats 16-19 protein expression construct (IgFlna-16-19) was prepared by PCR amplification using the pREP4-Filamin-a plasmid as a template, followed by ligation into the pGEX-2 T vector. The resulting expressed GST-IgFlna-16-19 fusion protein has a molecular weight of 65 kDa. The GST-IgFlna-16-19 fusion protein was expressed from 100 ml of E. coli Rosetta 2 cells (EMD Millipore, Billerica, MA). Cells were pelleted and solubilized in lysis buffer (5 ml 1X PBS containing 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 5 mM DTT) followed by sonication. One per cent (v/v) Triton X-100 was added to the sonicated solution and incubated for 30 min at 4 °C with shaking. After centrifugation at 15 000 g for 15 min, the supernatant was added to 1.0 ml of a 50% (w/v) slurry of glutathione-sepharose 4B beads (GE Healthcare, Piscataway, NJ). The mixture was then incubated for 2 h at 4 °C shaker with gentle mixing, centrifuged at 600 g for 5 min, and washed extensively with wash buffer (1 X PBS, 10% v/ v Glycerol, 5 mM DTT, 1 mM PMSF, 1% v/v Triton X-100, 1 μg/ml leupeptin, and 1 μg/ml pepstatin). The GST-IgFlna-16-19 fusion protein was then eluted with elution buffer (20 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0, 5% v/v glycerol, and 3 mM DTT).
The Grb7RAPH-6His protein (Siamakpour-Reihani et al., 2009; Siamakpour-Reihani et al., 2011) was expressed in Rosetta cells and purified by affinity using Ni-NTA agarose beads (Qiagen, Valencia, CA) according to the manufacturer's instructions and our previous published protocol (Siamakpour-Reihani et al., 2009; Siamakpour-Reihani et al., 2011).
In vitro GST tagged protein binding assay
The Grb7-RAPH-6His protein was incubated with glutathione sepharose 4B beads prebound to the GST-IgFlna16-19 or GST only (negative control) overnight at 4 °C. Protein complexes were eluted with GST Elution buffer (20 mM reduced glutathione, 50 mM Tris-HCl, 5% v/v glycerol, 1 mM DTT, and pH 8.0), separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and stained using Coomassie brilliant blue.
In vitro pull down assay
The myc-tagged Grb7-RAPH domains were expressed in vitro using a TNT-T7 Quick Coupled Transcription/Translation system (Promega Corporation, Madison, WI), using the pGBKT7-Grb7RAPH vector as a template. Five hundred nanogram of GST (negative control) or GST-IgFlna16-19 protein bound to 10 μl of a 50% (w/v) slurry of glutathione sepharose beads was combined with 25 μl in vitro translated Grb7-RAPH protein TNTT7 reaction brought to a total volume of 200 μl with wash buffer (4.2 mM Na2HPO4 , 2 mM KH2PO4, 140 mM NaCl, and 10 mM KCl). The mixture was incubated at 4 °C with gentle shaking for 1.5 h. After washing, the protein complexes were eluted with 20 μl of 20 mM reduced glutathione solution. Eluted samples were analyzed by SDS-PAGE followed by staining with Coomassie brilliant blue.
Cell culture and transfection
SKBR3 and HeLa cells were cultured on DMEM/F12 containing 10% v/v FBS. For investigation of the effect of Grb7 tyrosine mutants, the pCMV constructs encoding different Grb7 full-length mutants (see Site directed mutagenesis in the Materials and Methods) were transfected into HeLa cells using Lipofectamine LTX according to the manufacturer's instructions (Invitrogen Life Technologies). For EGF stimulation studies, serum-starved SKBR3 cell cultures were treated with 0.1 μg/ml EGF for 30 min. In both cases, cells were lysed with cell lysis buffer (25 mM Tris, 100 mM NaCl, 1% v/v Triton X-100, 1 mM EDTA, 1 mM DTT, 1 mM NaVO4, 10% v/v glycerol and a 1:100 dilution of Halt Protease inhibitor cocktail (Thermo Fisher Scientific product #78410), or fixed with 3.7% v/v formaldehyde, depending upon the intended subsequent experiment.
Antibody immobilization
Twenty microgams each of mouse monoclonal Filamin-a antibody (3 F180), rabbit polyclonal Grb7 antibody (H-70), and non-specific IgG were used for immobilization. Before immobilization, primary amines (e.g., tris and glycine) in the antibody solution were removed using Thermo Scientific Zeba Spin Desalting Columns while gelatin and carrier proteins were removed using a Thermo Scientific Pierce Antibody Clean-up Kit. Respective antibodies were covalently coupled onto amine-reactive agarose resins using the Pierce coimmunoprecipitation method according to the manufacturer's protocol (Thermo Fisher Scientific).
Site directed mutagenesis
Selected tyrosine residues were mutated to phenylalanine residues by polymerase chain reaction following the guidelines given in the Quick Change II site directed mutagenesis kit (Agilent technologies, Everett, WA) and confirmed by DNA sequencing. The following Grb7 full-length mutants were prepared from the wild-type pCMV-Grb7 plasmid: Grb7 (Y188F), Grb7 (Y338F), and Grb7 (Y188F/Y338F). The tyrosine residues chosen for mutation were selected based upon their demonstrated importance in FAK-mediated Grb7 signaling (Chu et al., 2009).
Reciprocal coimmunoprecipitation and Western blot analysis
The SKBR3 breast cancer cell line endogenously expresses Grb7 and Filamin-a proteins. SKBR3 cells were lysed with cell lysis buffer and precleared with control agarose resins. 400 μl of cell lysate (approximately 400 μg total protein) was diluted further with an additional 100 μl of lysis buffer, and the mixture was added to either the Grb7(H-70), Filamin-a(3 F180), or control IgG antibody-immobilized resins and incubated at 4 °C with gentle shaking. After washing the antibody-immobilized beads extensively, proteins were eluted with elution buffer.
Immunoblot analysis
For all Western blot analyses, proteins were resolved by SDS-PAGE and transferred onto PVDF membranes. After the complete transfer of proteins, membranes were rinsed once with 1X Tris-buffered saline with Tween-20 (TBST) (50 mM Tris; 150 mM NaCl; 0.05% Tween 20) and blocked with 5% (w/v) non-fat dry milk for 1 h at room temperature. Membranes were washed once with 1X TBST for 5 min and incubated with the indicated primary antibodies diluted in 1X TBST buffer. Thereafter, membranes were washed three times, 5 min/wash in 1X TBST buffer and incubated with corresponding horseradish-peroxidase-conjugated secondary antibodies. Membranes were then washed four times, 7 min/wash in 1X TBST buffer followed by the addition of Supersignal West Dura chemiluminescent reagent substrate (Thermo Fisher Scientific). Visualization and quantification of Western blot signals was achieved using a Chemidoc XRS system loaded with Quantity One software (Bio-Rad, Hercules, CA).
Immunofluorescence
Cells were grown to 80–90% confluence on glass cover slips, fixed with 3.7% formaldehyde/PBS (15 min), permeabilized in 0.5% v/v Triton X-100 (20 min), and blocked in 3% w/v BSA/ PBS (1 h). Simultaneous double immunofluorescence was carried out. Cover slips were incubated with mouse anti-Filamin-a (3 F180) and rabbit anti-Grb7 (H-70) antibodies at 1:150 and 1:250 dilutions, respectively. These primary antibodies were detected with Alexa Fluor-labeled secondary antibodies: anti-mouse Alexa Fluor 488 for Filamin-a and anti-rabbit Alexa Fluor 546 for Grb7 were used in 1:500 dilution in 1% w/v BSA/PBS. Alexa Fluor 647 Phalloidin was used for F-actin filaments staining in 1:35 dilution. Hoechst 33342 was used for nuclear staining at the concentration of 1 μg/ml. After washing cells extensively with 1X PBS, cover slips were mounted in 90% glycerol/PBS and imaged using a Zeiss Axiovert 200 M fluorescence microscope (Carl Zeiss, Thornwood, NY) equipped for standard epifluorescence and Apotome structured illumination capabilities. Images were acquired using a 63X-Plan-achromat 1.4 NA oil objective and a 12-bit AxioCam MrM CCD camera driven by Axiovision 4.5 software (Zeiss) Figures were prepared from exported 8-bit images using Adobe Photoshop CS2 software.
shRNA knockdown and wound healing scratch assay
The Grb7shRNA vector was a gift from Dr. Shen (National Taiwan University, Taipei, Taiwan; (Chu et al., 2009)). SKBR3 cells were cultured in 12-well plates to 80% confluence and subsequently transfected with Grb7shRNA or scramble shRNA plasmids using Lipofectamine LTX (Invitrogen Life Technologies) according to the manufacturer's instructions. After 6 h of growth, the media were removed, and fresh growth media were added to the cell culture plate. After 24 h, the cell monolayer was gently scratched with a 10 μl pipette tip across the center of the well. Growth media were removed, cells were washed once with 1X PBS and fresh growth media were added. Cells were incubated at 37 °C in a 5% CO2 environment, and cell scratch images were taken at different time points as indicated. Cells were then lysed, and Western blot analysis was performed to assess the Grb7 knockdown effect.
RESULTS
The RA and PH domains of Grb7 interact with Filamin-a by yeast 2-hybrid assay
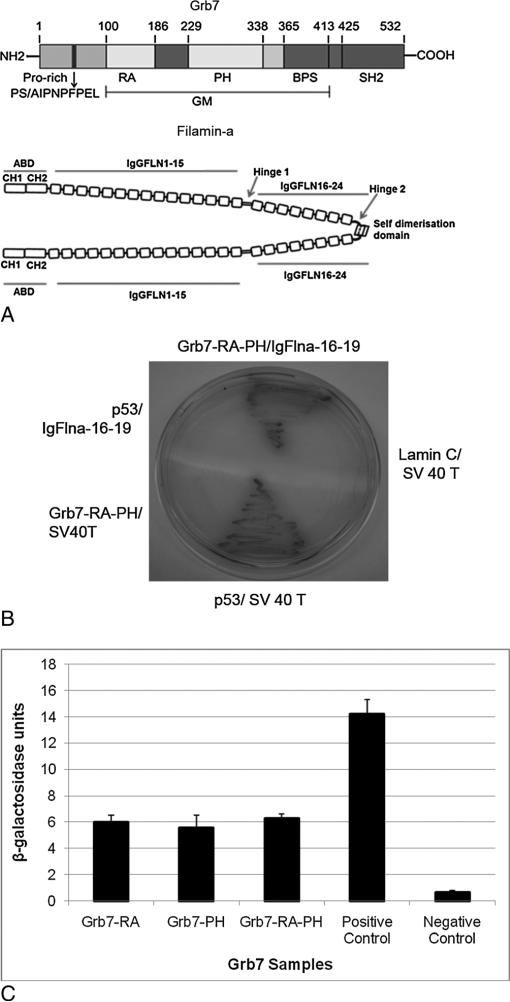
The Grb7 PH domain (Figure 1A) was used as a bait construct to screen a mouse embryonic stem cell cDNA library by yeast 2-hybrid analysis, which revealed positive interaction with a protein potentially involved in Grb7-mediated cellular signaling: Filamin-a (Flna). Specifically, three Fln-a clones were identified as binding positively to the Grb7-PH and Grb7- RA-PH region (out of thirty positive clones, the remaining positive clones will be reported elsewhere). Yeast 2-hybrid assays were performed using the Grb7-RA, and PH domains independently indicated the Grb7-RA, or the Grb7-PH domains alone were sufficient for binding to Flna, with almost equal β-gal activity in comparison with the Grb7-RA-PH two-domain construct (Figure 1C).
Figure 1.
A-C. A) Top: Domain topology of the Grb7 protein. The approximate locations of the Grb7 protein domains described in the text are identified by residue numbers located above the Grb7 schematic. The GM (Grbs and Mig) domain corresponds roughly to the homologous regions between the Grb7 and Mig 10 proteins. Bottom: Domain topology of the Filamin-a protein (dimer). The Filamin-a figure was adapted from Nakamura et al., 2011. B-C: β-galactosidase assays. B) The prey vector containing the IgFlna-16-19-AD (activation domain) was cotransformed in yeast with bait vectors containing the Grb7-RA-DBD (DNA binding domain), Grb7-PH-DBD, or Grb7-RA-PH-DBD, respectively (Figure shows the Grb7-RAPH-DBD experiment only). The p53/SV-40-T interaction serves as a positive control. C) β-galactosidase assay activity was determined by liquid assays using ONPG as substrate. The data represent averages with standard deviations from three independent experiments each done in triplicate.
Grb7 and Filamin-a interact both in vitro and in vivo
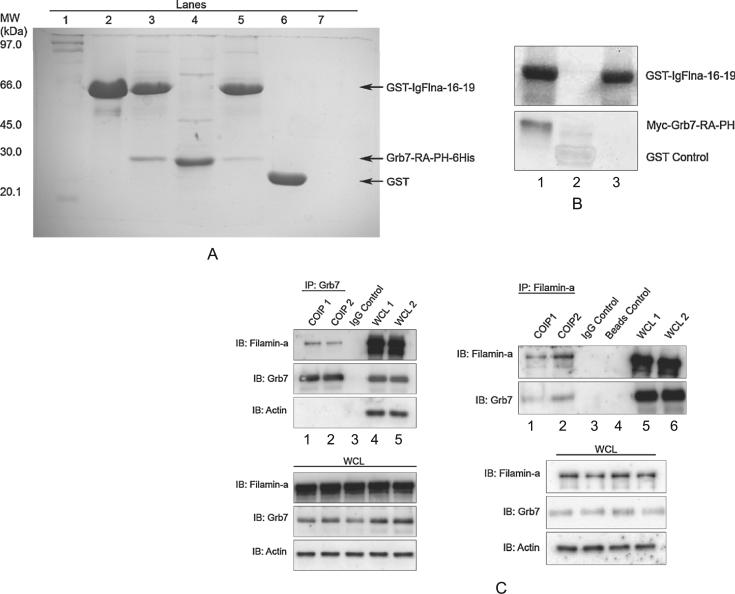
in vitro Grb7/Flna interaction. Immunoprecipitation of Grb7 and Flna using purified protein constructs verify that the two proteins interact directly (Figure 2, A–B) and not through possible additional binding partners pulled down in complex from cell lysates (Figure 2C). Figure 2, A–B shows pull down of Grb7-RA-PH by GST-tagged Flna domains, followed by either staining with Coomasie brilliant blue (Figure 2A), or Western blot analysis using GST and myc antibodies (Figure 2B). Figure 2A–B also provides partial localization of the Grb7 and Flna domains responsible for the interaction. Pull down using the Hexa-His-tagged Grb7-RA-PH domains demonstrates immunoglobulin-like repeat domains 16-19 (IgFlna-16-19) of Flna are sufficient for binding between the two proteins. The result provides evidence the Grb7-RA-PH and IgFlna-16-19 domains contain the regions responsible for protein-protein binding.
Figure 2.
A) GST binding assay: the IgFlna-16-19 domains bind with the Grb7-RA-PH domains. GST-IgFlna-16-19 and Grb7-RA-PH-6His were bacterially expressed and purified using glutathione (Glu-) sepharose beads and Ni-NTA agarose beads, respectively. Grb7-RA-PH-6His was incubated with GST-IgFlna-16-19 coupled glutathione beads and the eluted fraction was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained using Coomassie brilliant blue. Lanes: 1: protein standards, 2: purified GST-IgFlna-16-19, 3: Glu-sepharose beads elution fraction of GST-IgFlna-16-19 and Grb7-RA-PH-6His mixture, 4: purified Grb7-RA-PH-6His, 5: Glu-sepharose beads elution fraction of GST-IgFlna-16-19 and Grb7-RA-PH-6His mixture, 6: Glu-sepharose beads elution fraction of GST and Grb7-RA-PH-6His mixture, 7: Glu-sepharose beads elution fraction of Grb7-RAPH-6His alone. B) in vitro pull down of myc-Grb7-RA-PH by IgFlna-16-19. in vitro translated myc-tagged Grb7- RA-PH was incubated with purified glutathione coupled GST-IgFlna-16-19 (Lane 1) or GST alone (Lane 2). Lane 3 represents the elution fraction from glutathione coupled GST-IgFlna-16-19 only. Elution fractions were resolved by SDS-PAGE, and proteins were identified by Western blot analysis using mouse myc monoclonal antibody for Grb7-RA-PH detection and mouse GST monoclonal antibody for IgFlna-16-19 detection. C) In vivo coimmunoprecipitation of endogenous Grb7 and Filamin-a proteins. Left panels: coimmunoprecipitation with resin-coupled Grb7 antibody was performed on SKBR3 cell extracts endogenously expressing Grb7 and Filamin-a (Lanes 1–2, left upper panel). The whole cell lysate (WCL, lower left panel) and the resulting immunocomplexes (IP: Grb7) were immunoblotted with anti Grb7 (H-70), anti-Filamin-a (3 F-180), and anti-Actin (C-2) antibodies. A non-specific IgG control antibody immunoprecipitation (Lane 3, upper left panel) was also carried out on the extracts to show the interaction was specific. These experiments were also performed in MCF7 and Hela cells (Grb7 transfected), and similar results were observed (unpublished data). Right panels: the reverse process, that is, coimmunoprecipitation with resin-coupled Flna antibody was also performed (Lanes 1–2, upper right panel). The whole cell lysate (WCL, lower right panel), and the resulting immunocomplexes (IP: Filamin-a) were immunoblotted with Filamin-a and Grb7 antibodies. Non-specific IgG control and “non-antibody coupled beads” control coimmunoprecipitations (Lanes 3–4, upper right panel) show the interaction were specific. (COIP1: coimmunoprecipitation reaction1, COIP2: coimmunoprecipitation reaction 2, WCL1: whole cell lysate sample1, WCL2: whole cell lysate sample 2).
In vivo Grb7/Flna interaction. SKBR3 cells endogenously express both Grb7 and Flna. Cell lysates from SKBR3 cells were passed over immobilized Grb7 antibody and sequentially probed with Flna and Grb7 antibodies (Figure 2C). The result demonstrates full-length Grb7 and Flna endogenously expressed in a mammalian cell environment bind to each other.
Mutation of Grb7 tyrosine residues affects binding to Flna
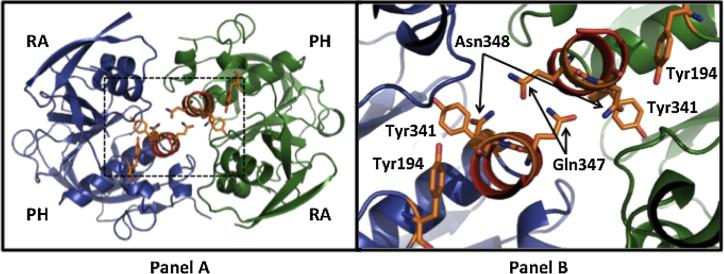
Two Grb7 tyrosine phosphorylation sites (Y188 and Y338) previously shown to affect Grb7 function (Chu et al., 2009) were mutated to phenylalanine residues. Specifically, Chu et al. (2009) demonstrated Y188 and Y338 are involved (in a phosphorylation-dependent manner) in FAK-mediated regulation of cell migration, proliferation, and tumorigenesis. Y188 is located at the end of the Grb7-RA domain while Y338 is at the end of the Grb7-PH domain (Figure 1A). The putative spatial location of these residues in the Grb7-RA-PH domains can be seen in Figure 6, based upon homology to the sister protein Grb10. The corresponding Grb10 residues are Y194 (Y188 in Grb7) and Y341 (Y338 in Grb7).
Figure 6.
The Grb10 RA-PH domain structure (Depetris et al., 2009, PDB# 3HK0). The left panel depicts the dimerized structure of the Grb10-RA-PH domains, with each RA and PH domains annotated. The right panel is an enlargement of the region outlined by dotted lines in the left panel. The Grb10 residues Y194 and Y341 (corresponding to Grb7 residues Y188 and Y338, respectively) are labeled for each RA-PH structure. The Grb10-RA-PH domain residues Q347 and N348, when mutated to alanine diminish dimerization, are marked by arrows.
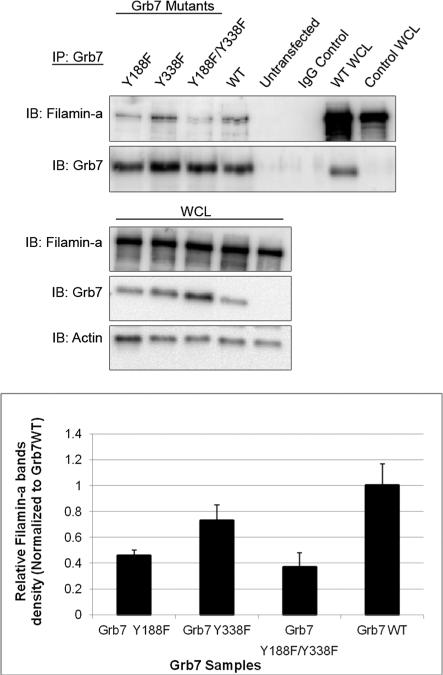
Wild-type Grb7 as well as single or double tyrosine phosphorylation mutants were transfected into HeLa cells, and the same Flna immunoprecipitation studies were performed. HeLa cells endogenously express Flna but do not express Grb7. The ability of both Grb7 (Y188F) and Grb7 (Y338F) to immunoprecipitate Flna is somewhat diminished, as can be seen by the quantified Western blot signals of Flna in Figure 3. The double mutant Grb7 (Y188/338 F) shows the largest decrease in the Flna signal at approximately 40% of the Flna immunoprecipitated by wild type Grb7. In addition, probing with an antiphosphotyrosine antibody demonstrates wild type Grb7 and all three Grb7 mutants [Grb7 (Y188F), Grb7 (Y338F), and Grb7 (Y188/338 F)] remain tyrosine phosphorylated (supplemental data). This result shows removal of potential tyrosine phosphorylation sites within Grb7, affects Grb7's ability to immunoprecipitate Flna, but loss of phosphorylation at the specific residues Y188 and/or Y338 does not completely abolish the ability of Grb7 to interact with Flna in HeLa cells.
Figure 3.
The Filamin-a interaction with Grb7 tyrosine mutants is decreased. Upper panels: Western blot analysis. HeLa Cells were transfected with pCMV-FLGrb7- Y188F (full-length Grb7-Y188F mutant), pCMV-FLGrb7-Y338F, and pCMV-FLGrb7- Y188F/Y338F (double mutant) constructs. Cells were lysed and immunoprecipitated by resin-coupled rabbit polyclonal anti-Grb7 (H-70), followed by Western blot analysis with Filamin-a antibody (3 F-180) and Grb7 antibody (H-70). Whole cell lysates were also probed with the same antibodies and anti-Actin(c-2) antibody. The figure shown is a representative blot of three independent experiments, each displaying similar results. Lower panel: the results of three different immunoprecipitation experiments were analyzed by densitometry. Filamin-a band densities were divided by corresponding Grb7 mutant band densities and were shown to be normalized to Grb7 WT. Standard deviations were calculated for each sample data set and plotted in bar-graph format.
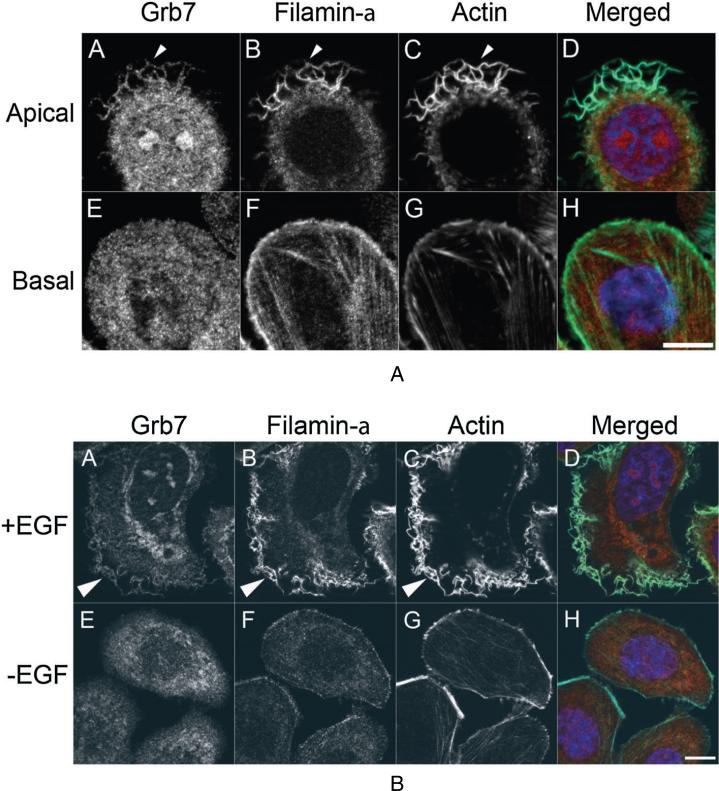
Grb7 and Flna potentially colocalize to membrane ruffles in SKBR3 cells upon EGF stimulation
Flna and Grb7 interact in vitro and in vivo (Figures 2 and 3), providing a possible direct link between extracellular signaling and the actin cytoskeleton. To further explore this notion, localization studies were performed in SKBR3 cells grown either in the presence of 10% serum (Figure 4A) or shifted into low-serum media and stimulated with EGF (Figure 4B). Imaging of the apical and basal surfaces revealed that while little colocalization could be detected on the basal surface, Grb7 and Flna could be detected in membrane ruffles (Figure 4A). Because membrane ruffling occurs in response to extracellular stimuli, cells were shifted down to low serum media for 24 h, and stimulated for 30 min with EGF. Whereas unstimulated cells in low serum expressed few ruffles and displayed no colocalization between Grb7 and Flna (Figure 4B, panels E–H), EGF-stimulated SKBR3 cells elaborated extensive membrane ruffles containing both Grb7 and Flna (Figure 4B, Panels A–D). It is acknowledged preferential Grb7 staining in the membrane ruffles versus the cell cytoplasm of EGF-stimulated SKBR3 cells is not grossly obvious. This may be a function of the endogenously over-expressed Grb7/ErbB2 protein levels in this cell line.
Figure 4.
Grb7 and Filamin-a colocalize within SKBR3 cell membrane ruffles. A) SKBR3 Cells grown on 18 mm cover slips were fixed and probed for Grb7 (red), Filamin-a (green), filamentous actin (cyan) and DNA (blue). Colocalization was observed on the membrane ruffles at the apical domain of cells (Panels A–D, arrowheads), whereas no colocalization was observed on the ventral or basal surface (Panels E–H). Representative fluorescent images of the cells are shown, and example colocalization of Grb7 and Filamin-a is indicated by an arrowhead. Bar 10 μm. B) EGF stimulation results in Grb7/ Filamin-a colocalization to cell membrane ruffles. Serum starved SKBR3 cells treated for 30 min with 100 ng/ml EGF resulted in profuse cell membrane ruffling (Panel A–D), whereas serum starved, unstimulated cells lacked any detectable colocalization. Bar 10 μm.
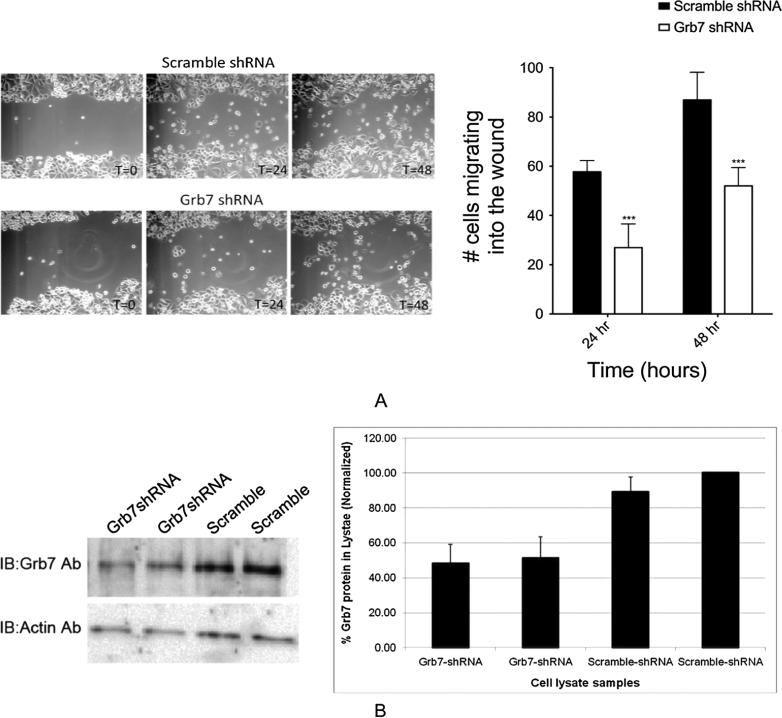
Knockdown of Grb7 expression diminishes wound healing rate in scratch tests
Scratch tests using SKBR3 cells transfected with either Grb7 specific shRNA or scrambled shRNA were performed, and the number of cells invading the wound area at different time points was quantified (Figure 5). An appreciable increase in the time required for cells to invade the wound area is observed for the Grb7 specific shRNA in comparison with the scrambled shRNA. Quantification of the knockdown of Grb7 protein expression in the shRNA transfected SKBR3 cells was also measured and showed an approximate decrease of 60% (Figure 5). Thus, while the effect of Grb7 expression knockdown on wound healing in SKBR3 cells is presumably related to decreased cell migration rates, we have not completely ruled out the possibility that the decrease in Grb7 expression is affecting the SKBR3 cell proliferation rate, and not cell migration.
Figure 5.
A) Grb7 knockdown significantly decreases wound closure rate in a wound scratch assay. SKBR3 cells were transfected with Grb7shRNA or Scramble shRNA. A wound was made with a 10 μl pipette tip across each well of a 12-well plate, and images were captured at different time points (in hours) as indicated. The invasion of wound-space over time was assessed by counting the number of cells present within the original wound boundaries at respective time points. The effect of Grb7shRNA on wound closure was compared with that of scramble shRNA treated cells. B) Quantification of the comparative decrease in Grb7 expression in SKBR3 cells upon either transfection with Grb7-specific shRNA or scrambled shRNA is shown in both Western Blot and densitometric bar-graph format.
DISCUSSION
For these studies, an embryonic stem cell cDNA library was selected for yeast 2-hybrid screening using Grb7 as bait, with the thought Grb7 could be involved in cell migration functions related to normal development and/or development of stem cell-based cancers. As our results show, Grb7 and Filamin-a directly bind each other both in vitro and in vivo, and their colocalization in SKBR3 cells is affected by EGF stimulation. Establishing a clear connection between Grb7 and the cytoskeleton has until now proven elusive. The study presented here has provided definitive evidence that Grb7-mediated cell signaling through ErbB2 and FAK could result in cytoskeletal remodeling. Filamin-a plays a direct role in the regulation of cytoskeletal restructuring by connecting actin filaments to the signaling molecules responding to stimuli, for example, integrins, receptor tyrosine kinases, etc.
Grb7 binds to ErbB2, a receptor tyrosine kinase whose overexpression is considered a marker for aggressive breast cancer (Seshadri et al., 1993; Slamon, 1987). Grb7 also binds to FAK; and FAK, through its interactions with integrins, is involved in the regulation of focal adhesions and cellular adhesion in general (Chu et al., 2009). With the knowledge that Grb7 also binds to Flna, the “middle man” between Filamin-a and either ErbB2 or FAK, may be Grb7. Because ErbB2 does not have a known extracellular ligand, the response of Grb7/Flna colocalization to EGF is likely through ErbB heterodimers (e.g., EGFR/ErbB2). It is noted that we have not ruled out the possibility that EGF may be acting to affect Grb7/Flna colocalization through some other (less direct) signaling route.
Mutations in Filamin-a are seen in genetic-based diseases exhibiting cell migration defects. Truncation of Flna, and thus loss of the IgG-Flna domains (Figure 1A) that bind to Grb7, is associated with periventricular heterotopia and Ehlers–Danlos syndrome (Fox et al., 1998; Sheen et al., 2005). Periventricular heterotopia is characterized by improper migration of neurons along the brain ventricles, and can result in central nervous system effects (epilepsy, mild to severe mental retardation), blood vessel abnormalities, and recurrent infections. Ehlers–Danlos syndrome is characterized by overly flexible joints and skin and fragile blood vessels.
Additionally, Flna, ErbB2, and FAK all have links to normal and carcinoma cell development. For example, the reinduction of ErbB2 in astrocytes promotes radial glial progenitor identity in the cerebral cortex (Ghashghaei et al., 2007). These progenitor cells are capable of development into new neurons and support neuronal migration. ErbB2 is necessary for induction of carcinoma cell invasion by ErbB family receptor tyrosine kinases (Spencer et al., 2000). Filamin-a regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion, with this disassembly regulation occurring at the leading edge of motile cells (Xu et al., 2010).
Hummolgaard et al. (Hommelgaard et al., 2004) showed ErbB2 is present at lamellipodial protrusions and is resistant to clathrin-mediated endocytosis and degradation. A link between Grb7 and non-clathrin-mediated endocytosis has been suggested previously, in that c-Src/Cav-1/Grb7 form a signaling cassette (Lee et al., 2000). The association between ErbB2, Grb7, caveolin-1, and now Flna may offer a potential route for ErbB2 escape from clathrin-mediated lysosomal degradation. Alternatively, the same association could potentially provide a mechanism for ErbB2 recycling at membrane ruffles.
We, and others, have shown the tyrosine phosphorylation state of Grb7 affects it ability to interact with other signaling partners (Chu et al., 2009; Siamakpour-Reihani et al., 2009; Siamakpour-Reihani et al., 2011; Tsai et al., 2007). For example, by mutating specific tyrosine residues to phenylalanine, we showed previously Grb7 is non-phosphorylated on tyrosines and is incapable of immunoprecipitating either four and a half LIM domains 2 or Hax-1 from transfected HeLa cells. With this knowledge in mind, we undertook a similar approach with the Grb7/Flna interaction. Figure 3 shows mutation of Tyr188 and Tyr338 affects the ability of Grb7 to immunoprecipitate Flna from Grb7-transfected HeLa cells. However, mutation of either tyrosine, or both, does not completely negate Grb7 binding to Flna. As well, the Grb7 (Y188F), Grb7 (Y388F), and Grb7 (Y188/388 F) mutants indicate other Grb7 tyrosine residues remain phosphorylated (supplemental data).
Although Y188 and Y338 were selected for mutation based upon their ability to alter Grb7 function (Chu et al., 2009), it is not immediately obvious why the phosphorylation states of these two tyrosines in particular can affect binding to Flna. Tyrosine residues 188 and 338 are also conserved in the Grb7 sister protein Grb10 (Y194 and Y341, respectively, numbering in accordance with PDB structure 3HK0). Depetris et al. (2009) solved the structure of the Grb10-RA-PH domains (PDB# 3HK0) and found some evidence to suggest dimerization may occur between the RA-PH domains, as well as the SH2 domain, in Grb10. The authors also demonstrated mutation of two residues (Q347 and N348, Figure 6 right panel) located in the C-terminal α-helix of the PH domain diminish RA-PH domain dimerization. Although the significance is unknown at this point, it is noted that Y194 and Y341 (corresponding to Y188 and Y338 in Grb7) are located close in space to Q347 and N348, the residues affecting RA-PH dimerization. In particular, Y194 is 4.2 Å distant and Y341 is 6.5 Å distant from N348. We have proposed previously that dimerization (possibly controlled by phosphorylation state) may play an important role in Grb7 regulation and function (Peterson et al., 2012).
Finally, we sought to demonstrate whether a forced decrease in Grb7 protein expression in SKBR3 cells could affect migration rates in this cell line. Transient transfection of SKBR3 cells with Grb7-specific shRNA demonstrates a statistically significant decrease in the wound closure rate in scratch tests compared with transfection with scrambled shRNA (Figure 5). Although not definitive, the result implies Grb7-mediated signaling pathways directly affect cell migration in the cell line used for our studies.
In summary, our studies have provided a direct link between ErbB2, Grb7, and cytoskeletal remodeling mediated through growth factor stimulation. The implications of the newly discovered Grb7/Flna interaction could be far reaching, with potential mechanistic clarifications in neuronal maturation and migration, ErbB2 membrane localization and cycling, and growth factor-stimulated cancer cell migration.
Supplementary Material
Acknowledgements
The financial support of NIH grant numbers 5U54 CA132383 (BAL) and 1SC1 HD063917 (CBS) is gratefully acknowledged.
Abbreviations and Symbols
- abd
actin binding domain
- AD
activation domain
- BD
DNA binding domain
- BSA
bovine serum albumin
- ErbB2
erythroblastic leukemia viral (v-erb-b) oncogene homolog 2, aka HER2
- FAK
focal adhesion kinase
- DMEM/F12
Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12
- DTT
dithiothreitol
- EGF
epidermal growth factor
- FBS
fetal bovine serum
- Flna
Filamin-a
- GAL4
yeast transcription factor regulating galactose inducible genes
- Grb7,10,14
growth factor receptor bound 7, 10, and 14 protein, respectively
- GST
glutathione S-transferase
- HeLa
cervical cancer cell line derived from the patient Henrietta Lacks
- IgG
immunoglobulin G
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- PVDF
polyvinylidene difluoride
- PH
pleckstrin homology
- PMSF
phenylmethylsulfonyl fluoride
- pY
phosphorylated tyrosine
- RA
Ras-Associating
- SEC
size exclusion chromatography, aka gel filtration
- SH2
Src homology 2
- SKBR3
an epithelial breast cancer cell line endogenously expressing ErbB2 and Grb7
- Y2H
yeast 2-hybrid
Footnotes
Supporting information may be found in the online version of this paper.
REFERENCES
- Alper O, Stetler-Stevenson WG, Harris LN, Leitner WW, Ozdemirli M, Hartmann D, Raffeld M, Abu-Asab M, Byers S, Zhuang Z, Oldfield EH, Tong Y, Bergmann-Leitner E, Criss WE, Nagasaki K, Mok SC, Cramer DW, Karaveli FS, Goldbach-Mansky R, Leo P, Stromberg K, Weil RJ. Novel anti-filamin-A antibody detects a secreted variant of filamin-A in plasma from patients with breast carcinoma and high-grade astrocytoma. Canc Sci. 2009;100:1748–1756. doi: 10.1111/j.1349-7006.2009.01244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu PY, Huang LY, Hsu CH, Liang CC, Guan JL, Hung TH, Shen TL. Tyrosine phosphorylation of Grb7 by FAK in the regulation of cell migration, proliferation, and tumorigenesis. J Biol Chem. 2009;284:20215–20226. doi: 10.1074/jbc.M109.018259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly RJ, Sanderson GM, Janes PW, Sutherland RL. Cloning and characterization of GRB14, a novel member of the GRB7 gene family. J Biol Chem. 1996;271:12502–12510. doi: 10.1074/jbc.271.21.12502. [DOI] [PubMed] [Google Scholar]
- Depetris RS, Wu J, Hubbard SR. Structural and functional studies of the ras-associating and pleckstrin-homology domains of grb10 and grb14. Nat. Struct. Mol. Biol. 2009;16:833–839. doi: 10.1038/nsmb.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JW, Lamperti ED, Ekşioğlu YZ, Hong SE, Feng Y, Graham DA, Scheffer IE, Dobyns WB, Hirsch BA, Radtke RA, Berkovic SF, Huttenlocher PR, Walsh CA. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- Ghashghaei HT, Weimer JM, Schmid RS, Yokota Y, McCarthy KD, Popko B, Anton ES. Reinduction of ErbB2 in astrocytes promotes radial glial progenitor identity in adult cerebral cortex. Genes Dev. 2007;21:3258–3271. doi: 10.1101/gad.1580407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DC, Shen TL, Guan JL. Role of Grb7 targeting to focal contacts and its phosphorylation by focal adhesion kinase in regulation of cell migration. J Biol Chem. 2000;275:28911–28917. doi: 10.1074/jbc.M001997200. [DOI] [PubMed] [Google Scholar]
- Hommelgaard AM, Lerdrup M, van Deurs B. Association with membrane protrusions makes ErbB2 an internalization-resistant receptor. Molecular biology of the cell. 2004;15:1557–1567. doi: 10.1091/mbc.E03-08-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyndt F, Gueffet JP, Probst V, Jaafar P, Legendre A, Le Bouffant F, Toquet C, Roy E, McGregor L, Lynch SA, Newbury-Ecob R, Tran V, Young I, Trochu JN, Le Marec H, Schott JJ. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 2007;115:40–49. doi: 10.1161/CIRCULATIONAHA.106.622621. [DOI] [PubMed] [Google Scholar]
- Lee H, Volonte D, Galbiati F, Iyengar P, Lublin DM, Bregman DB, Wilson MT, Campos-Gonzalez R, Bouzahzah B, Pestell RG, et al. Constitutive and growth factor-regulated phosphorylation of caveolin-1 occurs at the same site (Tyr-14) in vivo: identification of a c-Src/Cav-1/Grb7 signaling cassette. Mol Endocrinol. 2000;14:1750–1775. doi: 10.1210/mend.14.11.0553. [DOI] [PubMed] [Google Scholar]
- Manser J, Roonprapunt C, Margolis B. C. elegans cell migration gene mig-10 shares similarities with a family of SH2 domain proteins and acts cell nonautonomously in excretory canal development. Dev Biol. 1997;184:150–164. doi: 10.1006/dbio.1997.8516. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adhes Migrat. 2011;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nencioni A, Cea M, Garuti A, Passalacqua M, Raffaghello L, Soncini D, MOran E, Zoppoli G, Pistoia V, Patrone F, Ballestrero A. Grb7 upregulation is a molecular adaptation to HER2 signaling inhibition due to removal of Akt-mediated gene repression. PLoSONE. 2010;5(2):e9024. doi: 10.1371/journal.pone.0009024. doi: 10.1371/journal.pone.0009024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi J, Yajnik V, Immanuel D, Gordon M, Moskow JJ, Buchberg AM, Margolis B. The cloning of Grb10 reveals a new family of SH2 domain proteins. Oncogene. 1995;10:1621–1630. [PubMed] [Google Scholar]
- Peterson T, Benallie R, Bradford A, Pias S, Yazzie J, Lor S, Haulsee Z, Park C, Johnson D, Rohrschneider L, Spuches A, Lyons B. Dimerization in the Grb7 protein. J. Mol. Recognit. 2012;25:427–34. doi: 10.1002/jmr.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SP. Otopalatodigital syndrome spectrum disorders: otopalatodigital syndrome types 1 and 2, frontometaphyseal dysplasia and Melnick–Needles syndrome. Eur J Hum Genet. 2007;15:3–9. doi: 10.1038/sj.ejhg.5201654. [DOI] [PubMed] [Google Scholar]
- Robertson SP, Jenkins ZA, Morgan T, Ades L, Aftimos S, Boute O, Fiskerstrand T, Garcia-Minaur S, Grix A, Green A, et al. Frontometaphyseal dysplasia: mutations in FLNA and phenotypic diversity. Am J Med Genet A. 2006;140:1726–1736. doi: 10.1002/ajmg.a.31322. [DOI] [PubMed] [Google Scholar]
- Seshadri R, Firgaira FA, Horsfall DJ, McCaul K, Setlur V, Kitchen P. Clinical significance of HER-2/neu oncogene amplification in primary breast cancer. The South Australian Breast Cancer Study Group. J. Clin. Oncol. 1993;11:1936–1942. doi: 10.1200/JCO.1993.11.10.1936. [DOI] [PubMed] [Google Scholar]
- Sheen VL, Dixon PH, Fox JW, Hong SE, Kinton L, Sisodiya SM, Duncan JS, Dubeau F, Scheffer IE, Schachter SC, Wilner A, Henchy R, Crino P, Kamuro K, DiMario F, Berg M, Kuzniecky R, Cole AJ, Bromfield E, Biber M, Schomer D, Wheless J, Silver K, Mochida GH, Berkovic SF, Andermann F, Andermann E, Dobyns WB, Wood NW, Walsh CA. Mutations in the X-linked filamin 1 gene cause periventricular nodular heterotopia in males as well as in females. Hum Mol Genet. 2001;10:1775–1783. doi: 10.1093/hmg/10.17.1775. [DOI] [PubMed] [Google Scholar]
- Sheen VL, Jansen A, Chen MH, Parrini E, Morgan T, Ravenscroft R, Ganesh V, Underwood T, Wiley J, Leventer R, Vaid RR, Ruiz DE, Hutchins GM, Menasha J, Willner J, Geng Y, Gripp KW, Nicholson L, Berry-Kravis E, Bodell A, Apse K, Hill RS, Dubeau F, Andermann F, Barkovich J, Andermann E, Shugart YY, Thomas P, Viri M, Veggiotti P, Robertson S, Guerrini R, Walsh CA. Filamin A mutations cause periventricular heterotopia with Ehlers–Danlos syndrome. Neurology. 2005;64:254–262. doi: 10.1212/01.WNL.0000149512.79621.DF. [DOI] [PubMed] [Google Scholar]
- Siamakpour-Reihani S, Argiros HJ, Wilmeth LJ, Haas LL, Peterson TA, Johnson DL, Shuster CB, Lyons BA. The cell migration protein Grb7 associates with transcriptional regulator FHL2 in a Grb7 phosphorylation-dependent manner. J. Mol. Recognit. 2009;22:9–17. doi: 10.1002/jmr.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siamakpour-Reihani S, Peterson TA, Bradford AM, Argiros HJ, Haas LL, Lor SN, Haulsee ZM, Spuches AM, Johnson DL, Rohrschneider LR, Shuster CB, Lyons BA. Grb7 binds to Hax-1 and undergoes an intramolecular domain association that offers a model for Grb7 regulation. J. Mol. Recognit. 2011;24:314–321. doi: 10.1002/jmr.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DJ. Proto-oncogenes and human cancers. New Engl J Med. 1987;317:955–957. doi: 10.1056/NEJM198710083171509. [DOI] [PubMed] [Google Scholar]
- Spencer KS, Graus-Porta D, Leng J, Hynes NE, Klemke RL. ErbB2 is necessary for induction of carcinoma cell invasion by ErbB family receptor tyrosine kinases. J Cell Biol. 2000;148:385–397. doi: 10.1083/jcb.148.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Mori M, Akiyoshi T, Tanaka Y, Mafune K, Wands JR, Sugimachi K. A novel variant of human Grb7 is associated with invasive esophageal carcinoma. J Clin Investig. 1998;102:821–827. doi: 10.1172/JCI2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai NP, Bi J, Wei LN. The adaptor Grb7 links netrin-1 signaling to regulation of mRNA translation. EMBO J. 2007;26:1522–1531. doi: 10.1038/sj.emboj.7601598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chan DW, Liu VW, Chiu P, Ngan HY. Differential functions of growth factor receptor-bound protein 7 (GRB7) and its variant GRB7v in ovarian carcinogenesis. Clin. Cancer Res. 2010;16:2529–2539. doi: 10.1158/1078-0432.CCR-10-0018. [DOI] [PubMed] [Google Scholar]
- Wojcik J, Girault JA, Labesse G, Chomilier J, Mornon JP, Callebaut I. Sequence analysis identifies a ras-associating (RA)-like domain in the N-termini of band 4.1/JEF domains and in the Grb7/10/14 adapter family. Biochem Biophys Res Comm. 1999;259:113–120. doi: 10.1006/bbrc.1999.0727. [DOI] [PubMed] [Google Scholar]
- Xu Y, Bismar TA, Su J, Xu B, Kristiansen G, Varga Z, Teng L, Ingber DE, Mammoto A, Kumar R, Alaoui-Jamali MA. Filamin A regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J Exp Med. 2010;207:2421–2437. doi: 10.1084/jem.20100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.