Abstract
Purpose
To investigate if delays in resting-state spontaneous fluctuations of the BOLD (sfBOLD) signal can be used to create maps similar to time-to-maximum of the residue function (Tmax) in Moyamoya patients and to determine whether sfBOLD delays affect the results of brain connectivity mapping.
Methods
Ten patients were scanned at 3T using a gradient-echo EPI sequence for sfBOLD imaging. Cross correlation analysis was performed between each brain voxel signal and a reference signal comprised of either the superior sagittal sinus (SSS) or whole brain (WB) average time course. sfBOLD delay maps were created based on the time shift necessary to maximize the correlation coefficient, and compared to dynamic susceptibility contrast Tmax maps. Standard and time-shifted resting-state BOLD connectivity analyses of the default mode network were compared.
Results
Good linear correlations were found between sfBOLD delays and Tmax using the SSS as reference (r2=0.8, slope=1.4, intercept=-4.6) or WB (r2=0.7, slope=0.8, intercept=-3.2). New nodes of connectivity were found in delayed regions when accounting for delays in the analysis.
Conclusions
Resting-state sfBOLD imaging can create delay maps similar to Tmax maps without the use of contrast agents in Moyamoya patients. Accounting for these delays may affect the results of functional connectivity maps.
Keywords: BOLD contrast, Perfusion, Moyamoya disease, MRI, resting-state fMRI, functional connectivity
Introduction
Studies focused on the spontaneous fluctuations of the MR BOLD signal (sfBOLD), also known as resting-state functional MRI (rs-fMRI), are increasingly being used to explore the brain’s functional organization (1,2). Synchronicity between low-frequency time evolutions of the MR signal in spatially distinct regions have been used to detect resting state functional networks (such as default mode, sensorimotor, or visual networks) in normal subjects (3).
While sfBOLD is typical used for brain functional studies that evaluate connectivity, they could also have an impact in the field of perfusion measurements. In a typical rs-fMRI experiment, high frequencies of the signal originating from respiration and cardiac are considered as noise and removed either by spectral filtering or retrospective corrections using data from external sensors (4). However, each heartbeat or inhalation can be viewed as an internal challenge to the vasculature, in some way equivalent to a tiny bolus of oxygenated blood. As such, BOLD fluctuations contain information about brain perfusion, vascular reactivity, and oxygenation. Temporal relationships between BOLD-based fluctuations in different parts of the brain may also allow the identification of clinically relevant arterial arrival delay information (5). Typically, such regions are detected using gadolinium-based dynamic susceptibility contrast (DSC) methods (6). Since not all patients can receive contrast, a non-contrast alternative to measure delay may prove valuable for managing patients with cerebrovascular disease, for example, possibly helping identify patients who would benefit from thrombolysis (7).
In this study, we aimed to compare non contrast sfBOLD delay maps (obtained using 2 different approaches) to time to maximum of the residue function (Tmax) maps (6) obtained with DSC in patients with Moyamoya disease. This cerebrovascular disease is associated with terminal internal carotid artery (ICA) stenosis and occlusion with collateral formation (8). As such, these patients have Tmax prolongation, primarily in the affected anterior circulation (9), but do not usually present with acute infarcts. Furthermore, we investigated if accounting for delays in sfBOLD affects the results of functional connectivity mapping.
Materials and Methods
Patient Population
This prospective study was approved by the institutional review board and was Health Insurance Portability and Accountability Act (HIPAA) compliant. After giving informed written consent, 10 patients with newly diagnosed Moyamoya disease (5 men, 5 women; mean age 42 years; range 26–71 years) were included as part of their preoperative assessment for possible superficial temporal artery to middle cerebral artery bypass. All patients were symptomatic, with the most common symptoms including transient ischemic attacks, chronic small deep white matter infarcts, seizures, and headache.
Imaging Protocol
Magnetic resonance imaging was performed using a whole-body 3.0T scanner (MR750, GE Healthcare Systems, Milwaukee, WI) and an 8-channel head coil. The protocol included a 3D T1-weighted fast spoiled gradient echo sequence (TR=9.2ms, TE=3.7ms, FOV=22x22 cm2, slice thickness ST=1.2mm, acquisition matrix =256x256, 130 slices) used to acquire structural information of the whole brain. Diffusion-weighted imaging (DWI) was performed with a b-value of 1000 s/mm2; TR/TE 5.000/83.3 ms; FOV 24 cm; matrix 128 × 128; slice thickness 5 mm; and gradients in three tetrahedrally-encoded directions to create isotropic DWI and apparent diffusion coefficient maps. A gradient echo EPI sequence (TE=40ms, TR=1800ms, 20 slices, FOV=20x20 cm, ST=5mm, 128x128) with 120 repetitions was used for resting state BOLD acquisitions (acquisition time: 3 min 36 sec). The same sequence was repeated to acquire DSC maps during injection of 10 ml (body-weight independent) of gadobenate dimeglumine (Gd-BOPTA, Bracco, Milan, Italy) delivered using a power injector at 4 mL/sec and followed by a 20 ml saline chaser. Data from the scanner were imported into Matlab (MathWorks Inc., Natick, MA, USA) and SPM8 (Wellcome Department of Imaging Neuroscience, UCL, London, England) was used for co-registration.
Perfusion Maps
Hemodynamic maps (CBF, CBV, MTT, and Tmax) were created using automatic arterial input function and venous output function detection. Delay-insensitive deconvolution with block-circulant singular value decomposition was used with a regularization threshold of 15% of the maximum singular value. The entire process was done automatically using ‘RAPID’, a software developed for automatic perfusion/diffusion mismatch detection (10).
sfBOLD Delay Maps
The resting-state BOLD images were corrected for head motion (least squares approach, 6 parameters spatial transformation) using SPM8. The first ten time points were discarded to avoid transient signal changes before magnetization reached a steady state. After linear signal drift correction, the data were spatially smoothed with a Gaussian kernel (3mm FWHM) and low-pass filtered (<0.1 Hz). Note that given the relatively long TR (1.8s) of the MR sequence, most of the cardiac or respiration contributions are aliased into the low frequency parts of the spectrum.
A region of interest (ROI) was manually placed within the superior sagittal sinus (SSS) in one slice of the anatomic T1w-weighted image in each patient (approximately 4 voxels). ROI placement was guided using the map of temporal standard deviation of the BOLD signal to look for regions of high spontaneous fluctuations. The time evolution averaged over the SSS ROI was recorded as a reference or “seed” time course. Another reference time course was determined from the average signal of the 10 central slices as an approximation of whole brain signal time course. Cross correlation analysis was then performed between the reference time course and the time courses for all other voxels in the brain. The analysis was performed shifting the time course signal for each voxel between −5TR to +5TR (−9 to +9 s) to determine the offset associated with the highest correlation coefficient with the reference signal. The lag range was selected to improve analysis speed. It was empirically based on pilot studies showing that the maximum correlation occurred in this window of delays for the Moyamoya patients. The voxel value for the sfBOLD delay maps was assigned to the offset time that yielded the highest correlation coefficient, provided that the latter was higher than a confidence bound (>95 percent confidence interval) (Fig2a). The color scale was inverted to facilitate visual comparison with the delay maps. This is because Tmax values, which represent an estimate of time delay for blood delivery between a main feeding artery and tissue at a given spatial location, are positive. The sfBOLD delay maps are referenced to the venous system, and are mostly negative in the affected regions.
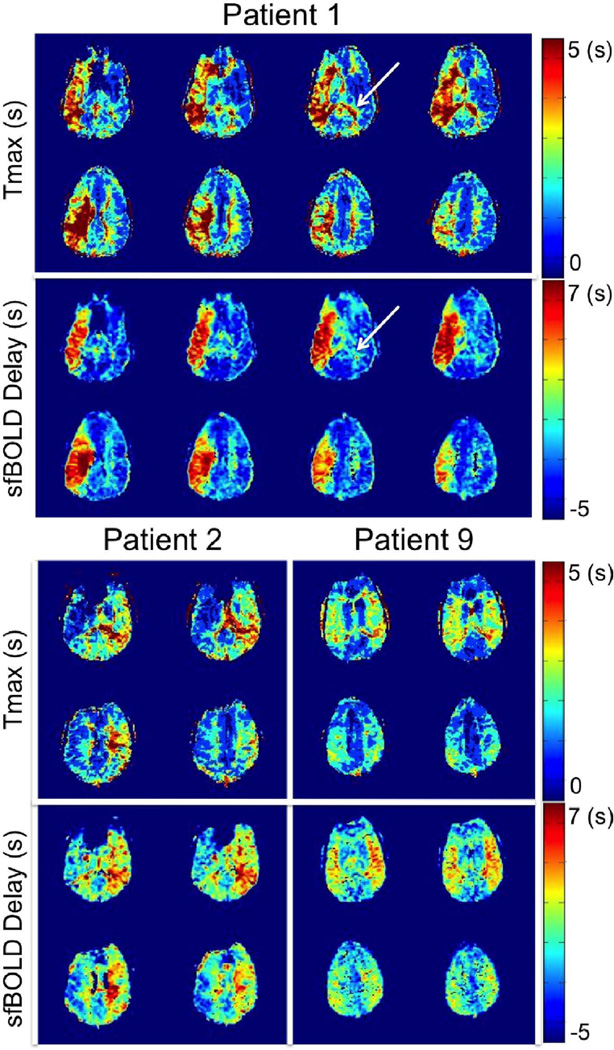
Figure 2.
Multislice comparison between Tmax maps and resting-state sfBOLD delay maps in 3 Moyamoya patients with unilateral (patient 1 and patient 2) or mild bilateral (patient 9) delays. While good agreement was found between both approaches, white arrows indicate regions showing high Tmax values but normal sfBOLD delays.
Resting-state connectivity analysis
Pre-processed data were normalized to the Montreal Neurological Institute (MNI) template space using FSL (http://www.fmrib.ox.ac.uk/fsl). 6 rigid-body movement parameter time-courses (estimated in the motion correction step) along with the whole brain average time series were removed from the data by linear regression.
Default Mode Network (DMN) was probed using a standard seed-based functional connectivity analysis (11). The ROI of reference was defined as a ~10 mm-radius sphere centered at (0, −56, 28) mm in the precuneus/PCC in the MNI space (12). The reference time course was generated by extracting the mean time series within the DMN ROI and was correlated against all brain voxels to obtain the ‘In phase’ functional connectivity map. Delayed reference time courses were also obtained by shifting the reference time course by values ranging from −5 TR to +5 TR. For each of these delayed reference time courses, we derived the corresponding ‘delayed’ functional connectivity maps.
Image Analysis
The DWI maps were scrutinized in each patient to identify any possible regions of acute infarct. The regions in which the assumptions of the DSC model are not met (i.e., where the gadolinium tracer does not remain intravascular) were also visually observed. To analyze the lag maps, in each patient, the 10 central slices were divided into left and right hemispheres to define 20 large ROI’s. Scatterplots of the sfBOLD delay values (averaged in the ROI’s) against corresponding Tmax values were created for each patient. Linear regression analysis was performed and the Pearson correlation coefficient, slope, and intercept of the linear regression lines were recorded. Unpaired Student t-tests were used to compare results obtained using either the SSS or the whole brain signal time courses as the reference time course.
Results
There were no regions of reduced diffusion in any of the patients that might correspond to acute infarcts. An overview of the sfBOLD delay analysis in one representative patient is presented Figure 1.
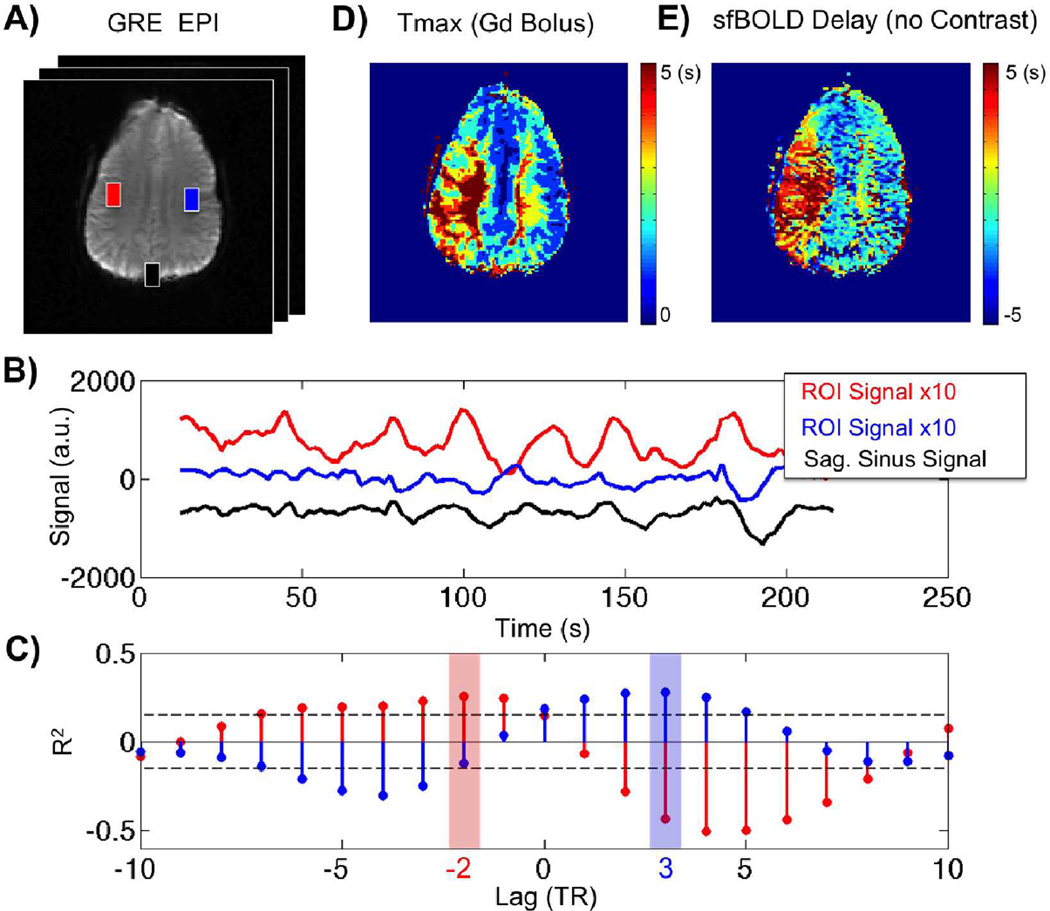
Figure 1.
Temporal correlation of resting-state BOLD spontaneous fluctuations in one patient with Moyamoya disease. a) Gradient-echo echo-planar image (EPI) image at a single time point, Tmax map obtained with gadolinium-based dynamic susceptibility contrast, and corresponding sfBOLD delay map obtained without contrast agent injection. b) sfBOLD signal time courses corresponding to the ROIs defined in (a): red: affected hemisphere, blue: unaffected hemisphere, black: SSS seed region. Note that different scales are used for easier visualization. c) Cross correlation analysis between ROI signal in tissue ROI’s (red and blue) with the SSS seed region (black). Maximum correlation is found at −2TR for the affected tissue (red) while +3TR is found for healthy tissue (blue). Dashed lines denote the confidence interval. Both maxima have a correlation coefficient (R2=0.25, R2=0.30 for blue and red ROI respectively) higher than the 95% confidence bound (R2=0.18). The (d) Tmax map (obtained with DSC) and (e) sfBOLD delay maps (obtained with the SSS as the seed region) from the same patient display similar characteristics, with delays present in the left hemisphere.
Figure 2 presents a comparison between Tmax maps and sfBOLD delay maps (obtained with the SSS reference approach) in 3 representative patients with significant unilateral hemisphere delays (patients 1 and 2) or mild bilateral delays (patient 9). Good agreement was found between both approaches across multiple slices. In all patients, sfBOLD delay detected prolonged Tmax values in the affected hemispheres. Among the entire cohort, the maximum delay found was 6 sec, while the average value in normal tissue was −2 sec. While overall, there was good concordance between the two approaches, a few differences may be observed. For example, the choroid plexus (as indicated by the white arrows in patient 1) typically shows a prolonged Tmax (in both affected and unaffected hemispheres), but the sfBOLD delay maps are normal in this region. As expected, the sagittal sinus vein shows a Tmax delay but not an sfBOLD delay as a result of the difference in the reference time point between the methods (artery versus vein).
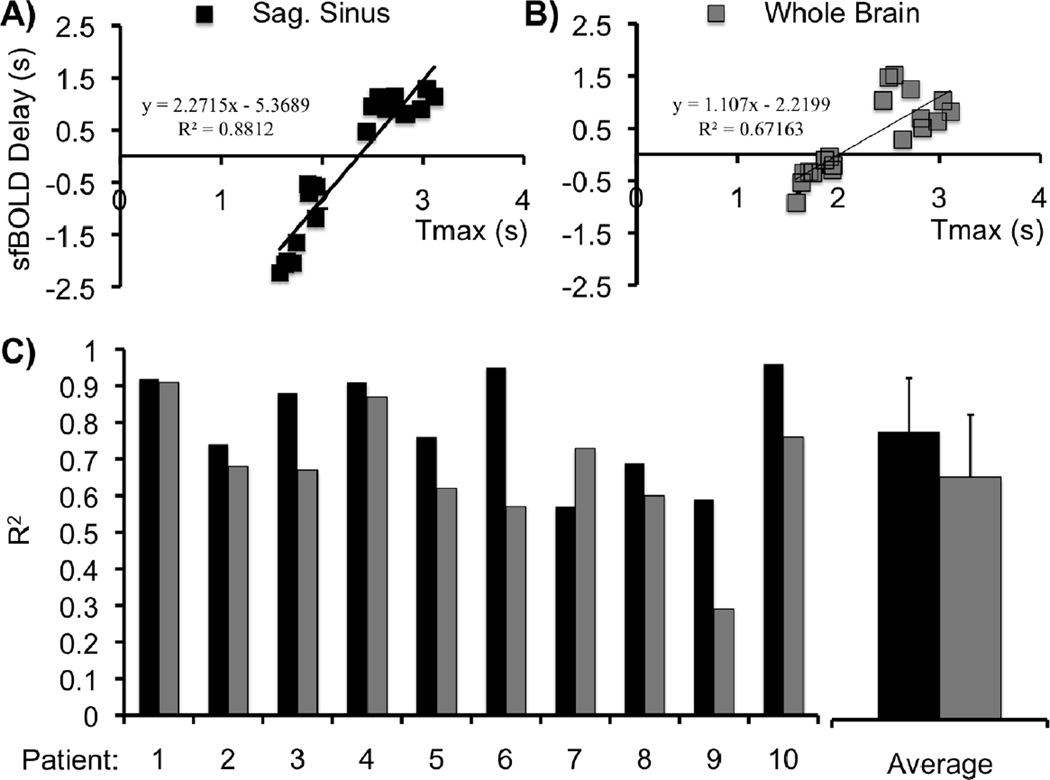
Figure 3 shows scatter plots between Tmax and sfBOLD delay in one patient created by defining the seed region as (a) the SSS or (b) the whole brain average. The correlation coefficients obtained for all patients are summarized in Figure 3c. Except for patient 7, the sfBOLD delay values are more strongly correlated to Tmax values when using the SSS as the reference. All the subjects showed a correlation coefficient higher than 0.55 for SSS-based maps and higher than 0.25 for whole brain-based maps. On average R2=0.80 ± 0.15 for the SSS and R2=0.67±0.17 for whole brain reference approaches, respectively. Slope and intercept of the linear fit are also showed in Table 1. It can be seen that the values changed from patient-to-patient and were also different between the two approaches. The SSS approach tended to overestimate Tmax while whole brain tended to underestimate it. The average value of the slope across patients is however close to 1 (1.4±0.8 vs 0.8±0.6 for SSS and WB, respectively; p=0.06). The intercept was negative for all patients; larger values were found with the SSS methodology, though the difference was not statistically significant (−4.6±2.1 vs −3.2±2.6, p=0.09).
Figure 3.
Correlation between Tmax and sfBOLD delay maps in one patient using different reference seed regions: (a) SSS, (b) whole brain average. Points correspond to hemispheric ROI’s obtained in 10 central slices. Corresponding correlation coefficients and linear fit equations are also indicated. c) Correlation coefficients for similar plots obtained for all patients. Except for patient 7, sfBOLD delay maps using the SSS seed approach had higher correlation to Tmax delay maps than those created with the whole brain average time series used as the reference region.
Table 1.
Results of cross correlation analysis between Tmaxand sfBOLD delay maps in 10 moyamoya patients.
| Patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Av. | Std. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sag.Sinus | R2 | 0.92 | 0.74 | 0.88 | 0.91 | 0.76 | 0.95 | 0.57 | 0.69 | 0.59 | 0.96 | 0.80 | 0.15 |
| Slope | 2.1 | 0.9 | 2.3 | 2.6 | 1.1 | 0.7 | 0.9 | 0.5 | 0.5 | 1.9 | 1.4 | 0.8 | |
| Intercept | −6.7 | −3.5 | −5.4 | −5.9 | −2.6 | −3.2 | −6.5 | −3.4 | −1.1 | −7.9 | −4.6 | 2.1 | |
| WholeBrain | R2 | 0.91 | 0.68 | 0.67 | 0.87 | 0.62 | 0.57 | 0.73 | 0.60 | 0.29 | 0.76 | 0.67 | 0.17 |
| Slope | 1.7 | 0.9 | 1.1 | 1.9 | 0.6 | 0.4 | 0.8 | 0.5 | 0.1 | 0.3 | 0.8 | 0.6 | |
| Intercept | −5.4 | −2.5 | −2.2 | −4.4 | −1.8 | −1.3 | −9.4 | −1.6 | −1.2 | −1.7 | −3.2 | 2.6 |
Discussion
In this study, we showed that resting-state BOLD signal fluctuations acquired without contrast agents contain temporal information that can be used to create delay maps that are strikingly similar to Tmax maps obtained using contrast-based DSC in patients with Moyamoya disease. This confirms similar observations made by Lv and colleagues in stroke patients (5), where it was shown that the maximum of correlation between the global signal averaged over the whole brain and the signal from tissues was delayed (up to 9 s) in regions with concurrent prolonged mean transit time (MTT).
sfBOLD delay maps obtained with the SSS seed show a global agreement with Tmax maps obtained using a bolus of contrast agent. In every patient, sfBOLD delays were able to detect regions with high bolus delays. A good correlation was found between the two approaches in large regions of interests with a good coefficient of correlation across patients, a slope close to unity, and intercept corresponding to the typical delay observed between veins and arteries. Yet, some discrepancies were also found. First some spatial differences were observed in healthy regions. While this might be due to a low sensitivity of BOLD contrast in white matter (13), it could also be due to the failure of the assumptions of the DSC model in the choroid plexus, which is highly permeable to gadolinium contrast (14). Different amount of smoothing between the two methods, as well as co-registrations errors, might also explain some differences in the locations of lesions with high delays. Second, the relation between the Tmax and sfBOLD delay values varies between individuals. This could be explained by variations in the definition of the arterial signal of reference for the DSC studies between individuals. A more detailed analysis needs to be performed before applying a threshold approach for the delineation of the ischemic penumbra. A better comparison between Tmax and sfBOLD delay could be performed if the same seed signal was chosen for both approaches, yet this is complicated due to low fluctuations of oxygenation in the arterial side of the vasculature.
The imaging time required to produce the sfBOLD delay maps in our study was significantly shorter than the study of Lv et al. (3 min 36 sec vs. 5 min 50 sec), which makes the approach suitable for an inpatient stroke workup. We evaluated the delay maps created using two different seed signals: the average signal from the whole brain (as in the prior report) and a new method: using a seed region located in the SSS. While the results from both methods compared well with Tmax, we found that the SSS approach was more strongly correlated with the DSC results. This could be explained by the large amplitudes of fluctuations found in venous part of the vasculature. A priori, the whole brain average seed is easier to define and does not necessitate an operator intervention, though this can be problematic in the presence of a large lesion. If the affected region is very large, a new ROI that focused on normal tissue must be chosen, as was sometimes necessary in the case in the previous stroke study (5). While not shown in this report, it is possible to automatically identify the SSS using both anatomical cues and search patterns that identify the maximum variations of sfBOLD fluctuations.
Several potential ways exist to improve the method. First, a long TR (1.8s) was used in our study. This allows the acquisition of multiple slices and was convenient to compare directly to the results to the perfusion parameters. Reducing the TR will improve the precision of the delay maps, at the expense of reduced SNR in the individual images and less craniocaudal slice coverage. Also, a shorter TR (< around 400ms) should enable a true frequency analysis of the resting-state BOLD power spectrum (e.g., due to spontaneous neural activity, respiration, cardiac) which are aliased using the current TR=1.8 sec approach. This could be used to understand the origin of the delays and permit a better filtering of the data by removing the unwanted components. The cost of a limited spatial coverage could be mitigated using multiband acquisitions created using multi-channel coils that are already available in the clinical environment (15). External physiologic sensors such as photoplethysmograph and pneumatic belt (which are standard equipment of clinical scanners) might also help for deciphering the origins of the fluctuations. Instead of removing the contributions as is usually done in rs-fMRI analysis, one could also use the information to determine a cardiac (16) or respiration response function (17) for each voxel. In fact, small delays in the BOLD signal have already been observed in volunteers participating in resting state connectivity using this approach (18). Large head motion during the acquisition can also degrade the quality of the maps and lead to unusable dataset (e.g., 6 patients were excluded in Lv study). These effects could be mitigated using multi-echo sequences and short echo time (19) images as regressors for motion. Multiple echoes can also be combined for better SNR and sensitivity (20).
An attractive feature of resting-state BOLD imaging is that the same data can be used for multiple purposes. In theory, amplitude of fluctuations could be used to study cerebral blood volume and/or blood oxygenation. A few experimental studies have attempted to use resting-state signals in context of perfusion in experimental tumors (21) or stroke in animals (22), and demonstrated variations of amplitude of fluctuations in the lesions. A retrospective analysis was conducted in humans (23), and while limited by the MR sequence used for the analysis (17 time points), it was shown that the magnitude of fluctuations appeared to be lower in ischemic regions compared to normal tissue.
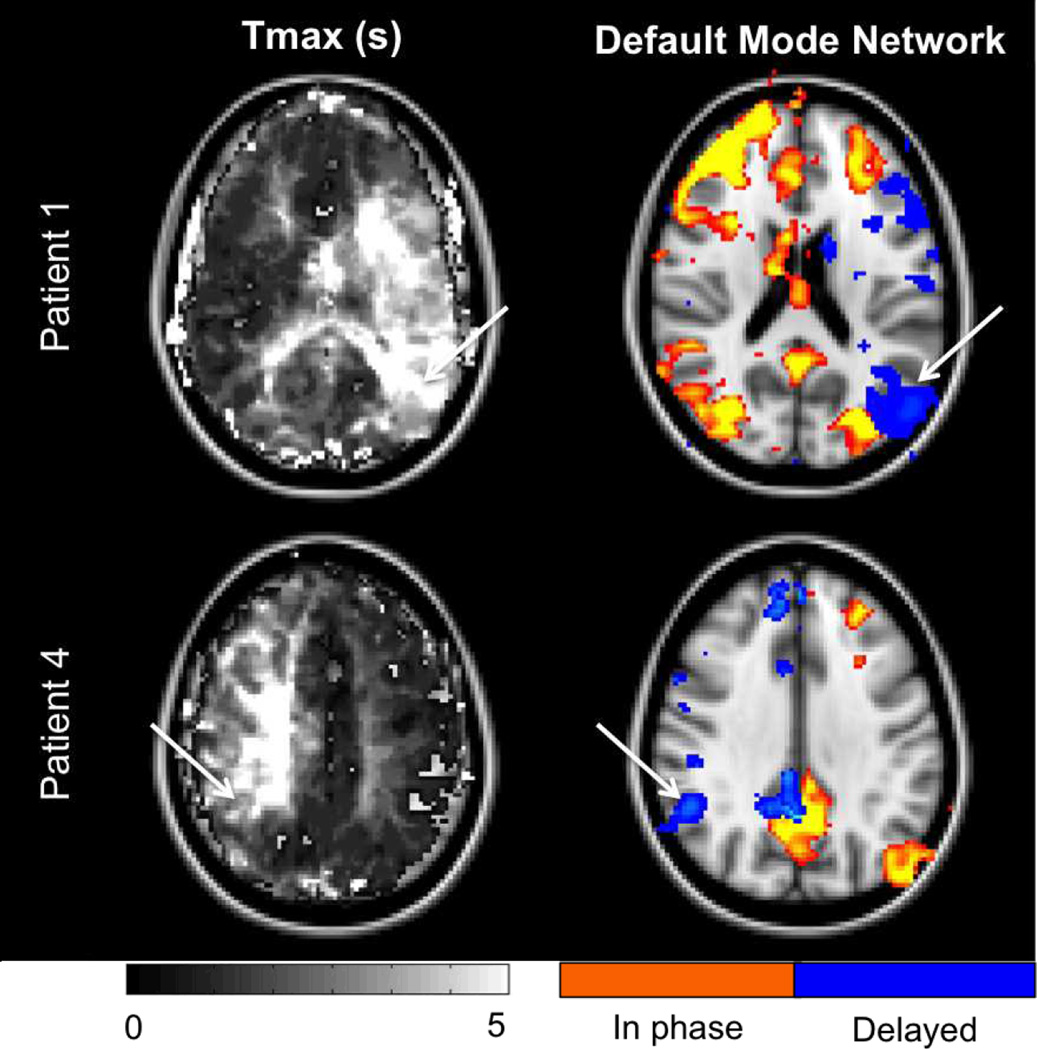
Finally, perhaps the most exciting use of the resting-state sfBOLD images is the potential to study both perfusion and brain connectivity at the same time. Connectivity analyses could provide insight into network reorganization in acute and chronic phases after stroke and may help design treatment strategies to promote recovery. Currently, rs-fMRI approaches to mapping connectivity require whole brain coverage and longer acquisition times (usually >8 min at 3T). It is however worth to notice that we were able to identify major networks such as the default mode using our data. Similar to the perfusion analysis, we used a seed based approach with ROI placed in the precuneus and looked for regions presenting high in-phase correlation with this reference signal. We also observed that by introducing delays in the connectivity analysis (seed signal shifted from +-5TR), some regions emerged as new ‘nodes’ in the affected hemisphere (white arrows in Fig 4 corresponding to high Tmax values). While definite conclusions cannot be drawn from this limited data acquisition, further investigations are warranted to see if apparent disruption of networks in patients could be due to spatial differences in arterial arrival delay and if accounting for these delays in the analysis could help to recover them. This may have wide applicability as many older subjects, even in the absence of identifiable pathology, have such spatially distinct regions of long arrival delay, particularly in the borderzone regions (24–26).
Figure 4.
Resting-state connectivity analysis in two patients. Default mode network was probed using a seed based approach with the reference signal chosen in the precuneus. While our acquisition was suboptimal for functional analysis (<4min), most of the network can be seen (red colorbar, in phase). Interestingly, accounting for the delays in the reference signal leads to the identification of new nodes (blue colorbar, delayed) in the affected hemisphere (see white arrows in connectivity maps and corresponding Tmax maps). This suggests that functionally connected networks may not be identified in their entirety in such patients if processing does not account for the intrinsic delays present.
The present study suffers from several limitations in addition to those mentioned above. First, the repeatability of results was not explored. The problem of data acquisition might explain the variability of slopes found between rsBOLD delay and Tmax. Second, the origin of the delay in resting state signal was not investigated. This temporal delay may come from tissue perfusion and/or neural impairment. It is however interesting to note that Moyamoya patients, unlike stroke patients, have presumably normal cognition/neural activity in the delayed regions. This would suggest that the major contribution is from the perfusion changes, particularly delay. Finally, there is clearly further room for improvement in performance of the approach by optimizing imaging parameters and post-processing techniques.
In conclusion, the findings of this study demonstrate that resting-state sfBOLD imaging can create delay maps similar to Tmax maps without the need for contrast agents in Moyamoya patients. In addition, our results suggest that accounting for this delay affects the results of functional connectivity maps.
Footnotes
Disclosure/Conflict of Interest: None
References
- 1.Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007 Sep;8(9):700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- 2.Van den Heuvel MP, Hulshoff Pol HE. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol J Eur Coll Neuropsychopharmacol. 2010 Aug;20(8):519–534. doi: 10.1016/j.euroneuro.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 3.De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM. fMRI resting state networks define distinct modes of long-distance interactions in the human brain. NeuroImage. 2006 Feb 15;29(4):1359–1367. doi: 10.1016/j.neuroimage.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 4.Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med Off J Soc Magn Reson Med Soc Magn Reson Med. 2000 Jul;44(1):162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 5.Lv Y, Margulies DS, Cameron Craddock R, Long X, Winter B, Gierhake D, et al. Identifying the perfusion deficit in acute stroke with resting-state functional magnetic resonance imaging. Ann Neurol. 2013 Jan;73(1):136–140. doi: 10.1002/ana.23763. [DOI] [PubMed] [Google Scholar]
- 6.Calamante F, Christensen S, Desmond PM, Østergaard L, Davis SM, Connelly A. The Physiological Significance of the Time-to-Maximum (Tmax) Parameter in Perfusion MRI. Stroke. 2010 Jun 1;41(6):1169–1174. doi: 10.1161/STROKEAHA.110.580670. [DOI] [PubMed] [Google Scholar]
- 7.Lansberg MG, Straka M, Kemp S, Mlynash M, Wechsler LR, Jovin TG, et al. MRI profile and response to endovascular reperfusion after stroke (DEFUSE 2): a prospective cohort study. Lancet Neurol. 2012 Oct;11(10):860–867. doi: 10.1016/S1474-4422(12)70203-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott RM, Smith ER. Moyamoya Disease and Moyamoya Syndrome. N Engl J Med. 2009;360(12):1226–1237. doi: 10.1056/NEJMra0804622. [DOI] [PubMed] [Google Scholar]
- 9.Lee M, Zaharchuk G, Guzman R, Achrol A, Bell-Stephens T, Steinberg GK. Quantitative hemodynamic studies in moyamoya disease: a review. Neurosurg Focus. 2009 Apr;26(4):E5. doi: 10.3171/2009.1.FOCUS08300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Straka M, Albers GW, Bammer R. Real-time diffusion-perfusion mismatch analysis in acute stroke. J Magn Reson Imaging JMRI. 2010 Nov;32(5):1024–1037. doi: 10.1002/jmri.22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biswal B, Zerrin Yetkin F, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn Reson Med. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 12.Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD. Decoding Subject-Driven Cognitive States with Whole-Brain Connectivity Patterns. Cereb Cortex. 2012 Jan 1;22(1):158–165. doi: 10.1093/cercor/bhr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logothetis NK, Wandell BA. Interpreting the BOLD Signal. Annu Rev Physiol. 2004;66(1):735–769. doi: 10.1146/annurev.physiol.66.082602.092845. [DOI] [PubMed] [Google Scholar]
- 14.Bouzerar R, Chaarani B, Gondry-Jouet C, Zmudka J, Balédent O. Measurement of choroid plexus perfusion using dynamic susceptibility MR imaging: capillary permeability and age-related changes. Neuroradiology. 2013 Dec;55(12):1447–1454. doi: 10.1007/s00234-013-1290-2. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Glasser MF, et al. Multiplexed Echo Planar Imaging for Sub-Second Whole Brain FMRI and Fast Diffusion Imaging. PLoS ONE. 2010 Dec 20;5(12):e15710. doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang C, Cunningham JP, Glover GH. Influence of heart rate on the BOLD signal: the cardiac response function. NeuroImage. 2009 Feb 1;44(3):857–869. doi: 10.1016/j.neuroimage.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birn RM, Smith MA, Jones TB, Bandettini PA. The respiration response function: the temporal dynamics of fMRI signal fluctuations related to changes in respiration. NeuroImage. 2008 Apr 1;40(2):644–654. doi: 10.1016/j.neuroimage.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang C, Thomason ME, Glover GH. Mapping and correction of vascular hemodynamic latency in the BOLD signal. NeuroImage. 2008 Oct 15;43(1):90–102. doi: 10.1016/j.neuroimage.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bright MG, Murphy K. Removing motion and physiological artifacts from intrinsic BOLD fluctuations using short echo data. NeuroImage. 2013 Jan 1;64:526–537. doi: 10.1016/j.neuroimage.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poser BA, Versluis MJ, Hoogduin JM, Norris DG. BOLD contrast sensitivity enhancement and artifact reduction with multiecho EPI: parallel-acquired inhomogeneity-desensitized fMRI. Magn Reson Med Off J Soc Magn Reson Med Soc Magn Reson Med. 2006 Jun;55(6):1227–1235. doi: 10.1002/mrm.20900. [DOI] [PubMed] [Google Scholar]
- 21.Baudelet C, Ansiaux R, Jordan BF, Havaux X, Macq B, Gallez B. Physiological noise in murine solid tumours using T2*-weighted gradient-echo imaging: a marker of tumour acute hypoxia? Phys Med Biol. 2004 Aug 7;49(15):3389–3411. doi: 10.1088/0031-9155/49/15/006. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, D’Arceuil H, He J, Duggan M, Gonzalez G, Pryor J, et al. MRI of spontaneous fluctuations after acute cerebral ischemia in nonhuman primates. J Magn Reson Imaging JMRI. 2007 Oct;26(4):1112–1116. doi: 10.1002/jmri.21131. [DOI] [PubMed] [Google Scholar]
- 23.Wang HH, Menezes NM, Zhu MW, Ay H, Koroshetz WJ, Aronen HJ, et al. Physiological noise in MR images: an indicator of the tissue response to ischemia? J Magn Reson Imaging JMRI. 2008 Apr;27(4):866–871. doi: 10.1002/jmri.21007. [DOI] [PubMed] [Google Scholar]
- 24.MacIntosh BJ, Lindsay AC, Kylintireas I, Kuker W, Günther M, Robson MD, et al. Multiple inflow pulsed arterial spin-labeling reveals delays in the arterial arrival time in minor stroke and transient ischemic attack. AJNR Am J Neuroradiol. 2010 Nov;31(10):1892–1894. doi: 10.3174/ajnr.A2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaharchuk G, Bammer R, Straka M, Shankaranarayan A, Alsop DC, Fischbein NJ, et al. Arterial spin-label imaging in patients with normal bolus perfusion-weighted MR 17 imaging findings: pilot identification of the borderzone sign. Radiology. 2009 Sep;252(3):797–807. doi: 10.1148/radiol.2523082018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell AM, Beaulieu C. Pulsed arterial spin labeling parameter optimization for an elderly population. J Magn Reson Imaging JMRI. 2006 Mar;23(3):398–403. doi: 10.1002/jmri.20503. [DOI] [PubMed] [Google Scholar]