Abstract
PCR detection of intestinal protozoa is often restrained by a poor DNA recovery or by inhibitors present in feces. The need for an extraction protocol that can overcome these obstacles is therefore clear. QIAamp® DNA Stool Mini Kit (Qiagen) was evaluated for its ability to recover DNA from oocysts/cysts directly from feces. Twenty-five Giardia-positive, 15 Cryptosporidium-positive, 15 Entamoeba histolytica-positive, and 45 protozoa-free samples were processed as control by microscopy and immunoassay tests. DNA extracts were amplified using 3 sets of published primers. Following the manufacturer's protocol, the kit showed sensitivity and specificity of 100% towards Giardia and Entamoeba. However, for Cryptosporidium, the sensitivity and specificity were 60% (9/15) and 100%, respectively. A series of optimization experiments involving various steps of the kit's protocol were conducted using Cryptosporidium-positive samples. The best DNA recoveries were gained by raising the lysis temperature to the boiling point for 10 min and the incubation time of the InhibitEX tablet to 5 min. Also, using a pre-cooled ethanol for nucleic acid precipitation and small elution volume (50-100 µl) were valuable. The sensitivity of the amended protocol to Cryptosporidium was raised to 100%. Cryptosporidium DNA was successfully amplified by either the first or the second primer set. When applied on parasite-free feces spiked with variable oocysts/cysts counts, ≈ 2 oocysts/cysts were theoretically enough for detection by PCR. To conclude, the Qiagen kit with the amended protocol was proved to be suitable for protozoan DNA extraction directly from feces and support PCR diagnosis.
Keywords: Cryptosporidium, Giardia, Entamoeba histolytica, DNA extraction, feces, diagnostic PCR
INTRODUCTION
PCR has gained an increasing importance in diagnostic laboratories for diagnosing bacterial and viral infections [1]. However, for protozoan pathogens, such as Cryptosporidium, Giardia, and Entamoeba histolytica, the 3 common intestinal protozoa infecting humans, development of a diagnostic PCR assay is challenged by a number of factors: First, feces, where the diagnostic stages of these protozoa are present, is a very complex specimen [2]. Second, the genetic material of these protozoa, to be isolated, is enclosed mainly in oocysts/cysts which possess very robust cell walls [3]. Last, some fecal constituents, such as heme, bilirubins, bile salts, and carbohydrates inhibit PCR [4]. These constituents of feces impair oocysts/cysts lysis, degrade the nucleic acid, and/or inhibit polymerase activity if co-extracted with the target pathogen DNA [5].
As a result, processing procedures to feces have been frequently adopted, in many studies, prior to protozoan oocysts/cysts DNA extraction [6,7,8,9]. For examples, salt flotation and formol-ether concentration techniques have been approached to purify oocysts/cysts from the complex fecal matrix [6]. In other studies, fecal samples, have been exposed to variable number of freeze-thaw cycles or bursts of Fast Prep® instrument or ultrasound liquid processor 'sonicator', to facilitate oocyst/cyst wall disruption and nucleic acid isolation [6,8,9]. Sometimes, more than 1 preparatory step has been used before the extraction method [8,9]. The purification step of oocysts/cysts present in feces is proved to be useful in reducing the carry-over of material that impairs their nucleic acid extraction. The physical and/or mechanical agitations of oocysts/cysts in feces were proved helpful for oocysts/cysts wall disruption. However, these preparatory steps add significantly more cost, labor, and time to the extraction method [8,9]. In addition, purification and concentration steps cause some loss of oocysts/cysts in the original fecal specimen.
In-house DNA extraction methods, such as the phenol-chloroform extraction method [9], and the guanidinium thiocyanate-silica method [8], has been adopted for protozoan DNA extraction from oocysts/cysts in feces. Also, few commercially-available DNA extraction kits have been used for the same purpose. Although the majority of these kits was originally designed for nucleic acid extraction from pathogens other than enteric protozoa, these kits were tried for protozoan DNA extraction from feces [10,16].
The QIAamp stool Mini Kit (Qiagen, Hilden, Germany), one of these commercial kits, was originally assembled for DNA isolation from metabolically active cells found in feces. Its buffer system permits direct cell lysis and allows optimal binding of nucleic acids to a silica gel membrane. Inclusion of an initial heating step, InhibitEX tablets and 2 successive wash steps are employed in the manufacturer's instructions to remove contaminants that are commonly found in feces. Recent studies have investigated the utility of the kit as a DNA extraction tool for a range of entero-pathogenic bacteria directly from human stool. Both spore-forming and non-spore forming bacteria were subjected to DNA extraction and subsequent PCR amplifications [16,17]. The kit has also been employed for DNA extraction from purified protozoan oocysts/cysts suspensions [18,19]. Only a few studies have reported the use of the kit for E. histolytica and Giardia lamblia DNA extraction directly from whole stool specimens [20,21]. For Cryptosporidium DNA extraction directly from fecal specimens, the standard kit protocol is usually preceded by several preparatory steps.
In this study, the Qiagen kit was initially evaluated for its ability to purify DNA of Cryptosporidium oocysts, G. lamblia, and E. histolytica cysts present in feces. Then, an effort was made to maximize its DNA recovery and purity by introducing modifications over the manufacturer's protocol. Finally, the kit with the amended protocol was evaluated more through its application on whole feces and on feces subjected to oocysts/cysts purification step or to a few freeze/thaw cycles. Further validation of the extraction procedure was carried out through its application on random stool samples from Al-Taif, Saudi Arabia.
MATERIALS AND METHODS
Collection of clinical samples and storage
Two-hudred test samples were randomly collected between January and August 2013 for evaluation of the fully optimized extraction protocol. Fecal samples were collected from those submitted to various governmental hospitals in Al-Taif, Saudi Arabia for laboratory diagnosis. Fresh feces, without preservatives, were properly labeled and sent to the medical laboratory at College of Applied Medical sciences, Al-Taif University within 2-3 hr of collection. On arrival, in the laboratory, feces were stored at 4℃ for microscopic and immunoassay testing. An aliquot of each specimen was stored at -20℃ for PCR testing.
Preparation of control samples
One-hundred protozoan-positive and negative samples were collected for use as controls; 25 Giardia-positive, 15 Cryptosporidium-positive, 15 E. histolytica-positive, and 45 protozoa-free samples were prepared using a combined gold standard test comprising of microscopy and immunoassay tests. Wet mount smears stained with iodine were subjected to microscopic diagnosis for Giardia and E. histolytica/dispar cysts as done earlier [22]. Detection of Cryptosporidium oocysts was carried out using the modified Ziehl-Neelsen (ZN) stain as formerly prescribed [23]. All fecal samples were subjected to protozoan coproantigen detection by RIDA® Quick Giardia (R-Biopharm, Darmstadt, Germany), E. histolytica II ELISA (TechLab, Blacksburg, Virginia, USA), and RIDA® Quick Cryptosporidium (R-Biopharm) kits for detection of Giardia, E. histolytica, and Cryptosporidium, respectively. Immunoassays were performed following the manufacturers' directions. The 2 rapid test results were interpreted visually by the naked eye while the E. histolytica II ELISA (TechLab) test results were analyzed in a multi-well scanning spectrophotometer (ELISA reader) with the cutoff of ≥0.150 for the positive sample at an optical density of 450 nm.
Purification of oocysts/cysts from feces
A purified preparation of ≈ 8×105 Cryptosporidium parvum oocysts with PBS in volume of 1 ml was purchased from Moredun Animal Health, Scotland, UK. In contrary, Giardia and E. histolytica cyst suspensions were prepared in the study. Briefly, highly positive stool specimens were pooled, concentrated, and purified, at first with modified formol-ether concentration technique [22,23] and then by the sucrose density-gradient centrifugation technique [24]. Cysts were counted under the microscope using a modified Fuchs-Rosenthal counting chamber. Preparations, 1 ml of PBS each, containing ≈ 4×105 of Giardia cysts and ≈ 3×104 of E. histolytica cysts were formed. These oocysts/cysts suspensions were used for seeding experiments and as sources of protozoan genomic DNA (gDNA) samples.
Spiking (seeding) experiments
For estimation of the lower detection limit for the extraction protocol together with the corresponding PCR test, seeding experiments were performed. Aliquots of protozoa-free feces, 200 µl each, containing approximately 1,700, 1,500, 1,000, 500, 100, 50, and 10 of the Cryptosporidium oocysts, Giardia cysts, or E. histolytica cysts were prepared. Each set of spiked samples was subjected to DNA extraction by the amended extraction protocol, and subsequently amplified by the target-matching PCR.
DNA extraction and optimization experiments
Early DNA extraction experiments were done using the Qiagen kit following the manufacturer's protocol. DNA extracts were subjected to amplification by the matching PCRs. DNA recovery was measured based on the intensity of ethidium bromide-stained DNA bands on agarose gels and compared with controls of known molecular weight. Three experiments were done to rule in or rule out the amplification failure of DNA extracted from known oocysts/cysts positive fecal samples as follows: First, DNA samples were diluted (1:10 and 1:100) with nanopure water prior PCR retesting. Second, DNA extracts were subjected to PCR amplification using 16SrDNA broad range universal primers [25,26]. Last, gDNA samples were spiked into the PCR reaction tube with the DNA extract. After ruling out PCR inhibition as a cause of amplification failure, a series of optimization experiments were performed in an attempt to increase the DNA recovery. Different lysis temperatures, lysis duration, centrifugation time, incubation time, and elution volumes were individually assessed using multiple aliquots of a single positive stool sample. All next DNA extractions were accomplished by the QIAamp® kit with the amended protocol for its evaluation.
PCR amplification and product analysis
As seen in Table 1, several primer pairs were adopted in PCR amplification reactions in the study. Protozoan DNA extracts were amplified using target-matching PCR assays as previously described [27,28,29]. Samples with discordant results were subjected to reamplification with a target matching reference PCR with high reported sensitivities [30,31,32]. A broad-range bacterial universal primers were adopted in the study to amplify bacterial DNA found in the DNA extracts, as done earlier [25,26]. Primers were synthesized by the VHBio (Gateshead, UK), dissolved in dH2O for stock preparation (100 pmol/µl) and stored at -20℃ until use. PCR reactions were carried out in Techne™ TC-4000 thermal cycler.
Table 1.
Primers used in this study
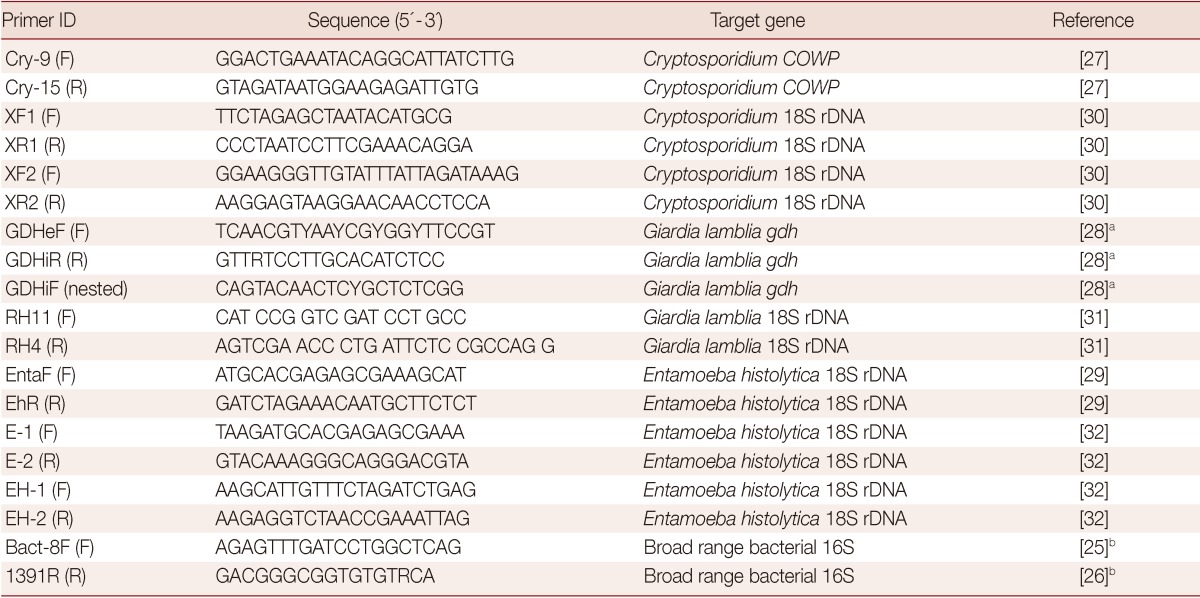
(F) stands for forward, (R) stands for reverse, (COWP) for Cryptosporidium oocyst wall protein gene, and (gdh) Giardia lamblia glutamate dehydrogenase gene.
aPrimers with degenerate bases; 'Y' indicates a 50:50 mix of 'C' and 'T', while 'R' is an equivalent mix of 'A' and 'G' in the degenerate primer mixes produced.
bPrimer pairs was used early in the study to rule in or rule out PCR failure duo to the presence of impurities in DNA extracted from feces.
Application of the extraction protocol on processed samples
Aliquots of oocysts/cysts suspension, 200 µl each, were subjected to vigorous agitation with FastPrep® Instrument (Qbiogene, Irvine, California, USA) prior to DNA extraction as done elsewhere [8,33]. Aliquots of the same fecal samples, 200 µl each, were subjected to 6 rounds of freeze/thaw cycles as carried out previously [8,33]. Each cycle required exposing samples to dry ice-ethanol bath for 1 min and to heating at 97℃ for another 1 min. The remaining of each sample and the aforementioned aliquots were subjected to DNA extraction, amplification by the corresponding PCR, and the results were then compared.
Validation of the extraction protocol by random clinical samples from Al-Taif
Fecal aliquots of 200 µl volume were subjected to DNA extraction using the modified protocol. PCR amplifications and subsequent analysis were performed blindly to minimize bias from immunoassay kits' results. Samples with discordant results between PCR and the target matching immunoassay kit were retested with the corresponding reference PCR assay.
RESULTS
Preliminary DNA extraction using the manufacturer's protocol
All control samples (n=100) were subjected to DNA extraction using the standard kit's protocol prior to PCR amplification. Giardia DNA was detected in all the Giardia-positive samples. Similarly, Entamoeba DNA was detected in all Entamoeba-positive samples. However, of all Cryptosporidium-positive samples, 6 showed no amplification products on gel electrophoresis. Importantly, no amplification products were shown on agarose gel for all the negative control samples. In principle, the kit's protocol with the target matching PCR showed specificity of 100% (45/45) towards the 3 protozoa. However, the diagnostic sensitivities were 100% (25/25), 100% (15/15), and 60% (9/15) for Giardia, Entamoeba, and Cryptosporidium, respectively. Aliquots of the Cryptosporidium-positive clinical samples with false negative results were subjected to a series of experiments to rule out PCR inhibition and to increase the extraction efficiency of the kit. The Cryptosporidium gDNA samples, spiked into the PCR reaction tube with the crude DNA extracts, were successfully amplified. Also, the bacterial DNA found in the crude DNA extracts was successfully amplified using the broad range bacterial primers. Last, decimal dilutions of the DNA extracts prior PCR amplification did not affect the false negative results.
Modifications of the manufacturer's protocol
Four changes over the kit's instructions were carried out as follows: First, a lysis temperature of 100℃ for 10 min was used instead of 97℃ for 7 min (Fig. 1, lane 4). Second, the incubation time for the InhibitEX tablet in the DNA lysate solution was raised to 3-5 min instead of 1 min. Third, pre-cooling ethanol before its use was adopted in the protocol. Last, eluting the purified DNA sample was carried out with 50-100 µl of elution buffer instead of 200 µl as specified after a 3-min incubation time at room temperature. To check out the efficiency of the boiling step on oocysts/cysts disruption, Cryptosporidium/Giardia-positive stool samples were subjected to DNA extraction following the modified kit's protocol. After initial heating of the stool homogenate and subsequent centrifugation, 20 µl of each cell lysate was mounted on a microscopic slide and examined by bright field microscope. Few oocysts/cysts with intact cell membranes (0-3) were seen. Similarly, 4 smears were prepared, but this time, from the fecal pellet of each sample and examined by the bright field microscope. Few oocysts/cysts with intact cell walls (0-2) were also identified. To assess the efficiency of using 50-100 µl elution buffer for recovering all DNA from the spin column, a second elution step with another 50 µl elution buffer was adopted for a batch of samples and subjected to PCR amplification. No amplification of the target DNA was shown in the DNA samples recovered by the second elution step.
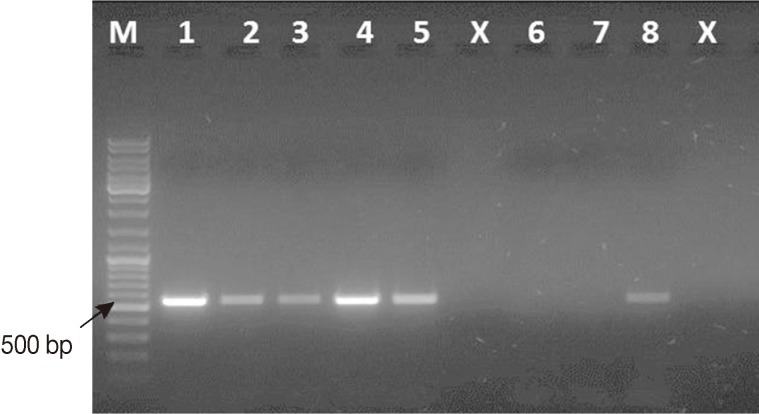
Fig. 1.
Representative ethidium bromide-stained 1% agarose gel picture showing PCR amplification products of Cryptosporidium-positive feces subjected to different lysis temperatures and durations. Amplicons of cowp gene sequence (≈ 550 bp) were generated using Cry-9/Cry-15 primers. M, GeneRuler™ 100 bp DNA marker; Lane 1, PCR product of Cryptosporidium gDNA (Just for comparison); Lane 2, PCR product of DNA sample recovered from fecal aliquot subjected to lysis at 97℃ for 15 min; Lane 3, PCR product of DNA sample recovered from fecal aliquot subjected to lysis at 97℃ for 20 min; Lane 4, PCR product of DNA sample recovered from fecal aliquot subjected to lysis at 100℃ for 10 min; Lane 5, PCR product of DNA sample recovered from fecal aliquot subjected to lysis at 100℃ for 15 min; Lane x, Empty; Lane 6, Cryptosporidium-negative stool sample (extraction negative control); Lane 7, no-template master mix sample (PCR negative control); Lane 8, PCR product of DNA sample recovered from fecal aliquot subjected to lysis at 97℃ for 5 min as originally mentioned in the kit's protocol.
Impact of prior sample processing on the amended kit's protocol
The amended kit's protocol was tried on the purified, mechanically agitated, oocysts suspension, and the results were comparable to those obtained from PCR amplification of DNA recovered directly from the whole stool samples. On the contrary, DNA recovered directly from the whole stool samples exposed to 6 rounds of freeze-thaw cycles showed more amplification products on agarose gel (Fig. 2). Both sample processing procedures increased the assay time by ≥20 min and led to a greater complexity of the extraction protocol. Neither the freeze/thaw nor the oocysts/cysts purification procedure was included as a part of the amended DNA extraction protocol.
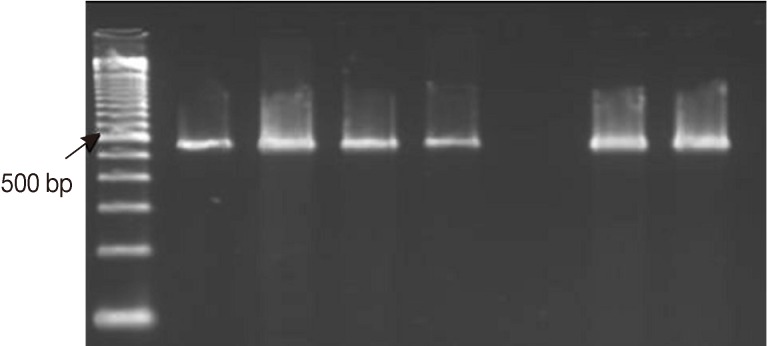
Fig. 2.
Representative ethidium bromide-stained 2% agarose gel picture showing PCR amplification products of Cryptosporidium COWP gene sequence (≈ 550 bp) with primers Cry-9/Cry-15. Extracts of DNA were retrieved by the amended kit's protocol from 1 Cryptosporidium-positive fecal sample using 3 different approaches. M: GeneRuler™ 100 bp DNA marker; Lanes 1, 2, PCR products of 2 fecal aliquots subjected to direct DNA extraction; Lanes 3, 4, PCR products of 2 aliquots subjected to oocysts purification step prior to DNA extraction; Lane 5, A Cryptosporidium negative stool sample (extraction negative control). Lanes 6, 7, PCR products of two aliquots subjected to 6 freeze/thaw cycles prior to DNA extraction.
Diagnostic performance of the extraction protocol
The amended kit's protocol was retried on all Cryptosporidium-positive fecal specimens (n=15), and the retrieved DNA were successfully amplified by either the first or the second target matching PCR primer set. By principle, the sensitivity of the amended extraction protocol with the corresponding PCR was raised to 100% (15/15), similar to that was reported, earlier in the study, for Giardia and Entamoeba. Also, the target protozoan DNA sequence was successfully extracted and subsequently amplified from fecal samples seeded with oocysts/cysts down to≈100 oocysts/cysts, corresponding to ≈ 500 oocysts per gram of stool and to ≈ 2 oocysts/cysts by PCR reaction (Fig. 3).
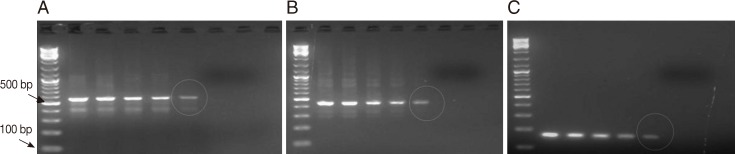
Fig. 3.
Representative ethidium bromide-stained agarose gel pictures showing PCR amplification products of protozoan DNA extracts recovered from feces seeded with various counts of oocysts/cysts by the amended kit's protocol. Encircled amplicons were the lowest number of oocysts/cysts present per extract (i.e., 200 µl) and could be detected by PCR (i.e., the lower detection limits) (A) PCR amplification products of Cryptosporidium COWP gene sequence (≈ 550 bp) using primers Cry-9/Cry-15. (B) PCR amplification products of G. lamblia gdh gene sequence (≈ 450 bp) with primers GDHeF/GDHiR. (C) PCR amplification products of E. histolytica 18S rDNA gene sequence (≈ 170 bp) with primers EntaF/EhR. Lane 1, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 1,700 oocysts/cysts; Lane 2, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 1,500 oocysts/cysts; Lane 3, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 1,000 oocysts/cysts; Lane 4, with ≈ 500 oocysts/cysts; Lane 5, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 100 oocysts/cysts; Lane 6, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 50 oocysts/cysts; Lane 7, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 10 oocysts/cysts; M, GeneRuler™ 100 bp DNA marker.
Validation of the extraction protocol with clinical samples:
Of all randomly collected clinical samples (n=200), protozoan DNA was found in 51 samples (25.5%) by the target matching first PCR assay. Cryptosporidium, Giardia, and Entamoeba DNA were detected in 31 (15.5%), 17 (8.5%), and 3 (1.5%) samples, respectively. In comparison, the protozoan coproantigens were detected in 46 (23%) fecal samples. Coproantigens of Cryptosporidium, Giardia, and Entamoeba were detected in 30 (15%), 14 (7%), and 2 (1%) samples, respectively. Samples with discordant results (n=5) were subjected to PCR amplification by the corresponding second confirmatory primer set, and the results were constant with that of the first matching primer set.
DISCUSSION
In this study, an extraction protocol based on QIAamp® DNA Stool Mini Kit was developed for protozoan DNA extraction directly from the diarrheic stool specimens. The protocol was proved to be simple and economical as it did not require hazardous reagents, such as phenol, or additional preparatory steps, such as concentration techniques or application of mechanical force using instruments, such as FastPrep disruptor or Mini Beadbeater for oocysts/cysts disruption. The small volume of sample subjected to extraction (200 µl) allowed the extraction procedure to be carried out at 1-2 ml scale, hence permitting the use of inexpensive table-top microcentrifuge and heating block.
Following the kit manufacturer's protocol, DNA extracts of all Giardia/Entamoeba-positive control fecal samples gave satisfactory results with the corresponding PCR test. The extraction efficiency of the kit towards Giardia and Entamoeba, revealed in this study, was consistent with previous studies [35,36]. However, this was not the case for Cryptosporidium. The causes of amplification failure with the 6 initial Cryptosporidium-positive samples subjected to the standard manufacturer's DNA extraction protocol remains speculative. It could be due to different reasons; Reasons related to the PCR protocol, such as the low sensitivity of the PCR assay or inhibition of the reaction by impurities present in the stool samples and co-purified with the target DNA, were considered. Reasons related to the extraction procedure, such as inefficient nucleic acid isolation or purification, was also reviewed. Bacterial DNA present in these extracts was successfully amplified using the broad range bacterial primers. However, interpretation of these data is problematic as fecal samples contain massive numbers of bacteria, potentially yielding high loads of bacterial 16SrDNA that are likely to exceed by many orders the quantities of protozoan DNA present in the samples. Also, gDNA samples were successfully amplified using Cryptosporidium primers in the presence of the crude DNA extract. Similarly, 10-fold dilutions of the DNA extracts before PCR amplification did not change the results. Dilution of the nucleic acid sample can be useful in decreasing the load of potential inhibitory substances, if present, on the Taq polymerase.
At that stage, an effort was made through a series of optimization experiments to increase the Cryptosporidium DNA yield of the extraction kit. First, to ease to isolate genetic material enclosed inside the robust cell walls like oocysts, the lysis temperature was raised to the boiling point for 10 min. First, raising the boiling step of stool homogenate to the boiling point for 10 min helped to isolate genetic material enclosed inside the very robust cell walls like oocysts. Boiling samples in the presence of the lysis buffer proved to be effective on oocyst disruption, as few oocysts with intact cell membranes were seen in the cell lysate or in the sediment mounted on the microscopic slides. Second, increasing the incubation time of the InhibitEX tablet step to 5 min was purposed to allow for improved adsorption of the DNA damaging substances and PCR inhibitors present in feces. Also, the use of pre-cooled ethanol for nucleic acid precipitation appeared to improve yields, but no obvious explanation was clear. Finally, the use of the smaller elution volume without any obvious loss of elution efficiency allowed for concentrating the final DNA sample by 2-4 folds.
The kit's protocol, with the changes introduced, was retried on 6 Cryptosporidium-positive fecal specimens, which showed amplification failure, and the DNA extract of all these samples were successfully extracted and amplified by either the first or the second Cryptosporidium PCR primer set. By principle, the sensitivity of the amended extraction protocol with both primer sets was raised to 100%. Elwin and her colleagues [6] have used the spin columns to purify DNA extract from semi-purified Cryptosporidium oocysts suspension after boiling step and reported higher performance, but on expense of cost, time, and simplicity of the extraction method [6]. In comparison, the extraction protocol, developed in the study, together with the target matching PCR displayed comparable results with no sample processing step and without use of the FastPrep cell disruptor.
As a trial to know the effects of 2 widely used sample-processing steps on the efficiency of the extraction procedure, the amended kit's protocol was tried on purified oocyst suspension and on whole stool samples exposed to 6 rounds of freeze-thaw cycles as previously reported [8,9]. Based on the intensities of the ethidium bromide stained DNA bands on the agarose gel, the results were comparable to those gained from PCR amplification of DNA recovered directly from the whole stool samples. Use of the freeze/thaw procedure appeared to increase DNA recoveries slightly, but the diagnostic significance of apparent refinement remains to be explored. Importantly, the 2 extrapreparatory steps added more time and costs to the extraction procedure without clear evidence of big gains. These results were in line with those obtained from other previous studies [7,8,9].
Accordingly, neither the freeze/thaw nor the oocysts/cysts purification procedure was included as a part of the amended DNA extraction protocol. The fecal-derived DNA samples showed amplification of the specific target gene sequence in all of the protozoan-positive control samples demonstrating that the QIAamp® kit effectively removed fecal impurities that can inhibit amplification or degrade DNA. These results were consistent with previously published studies [8,9,37,38]. The target DNA sequence of each PCR assay was successfully extracted and afterwards amplified from feces seeded with oocysts/cysts down to ≈ 100 oocysts/cysts. Assuming that the oocysts/cysts count in all spiked stool aliquots were precise, and the DNA extraction was carried out from all the seeded oocysts/cysts with equal efficiency, which was ruled out earlier for few Cryptosporidium-positive stool samples, the lower detection limit of the PCR assays was ≤100 oocysts/cysts per 200 µl stool extract which corresponds to ≈ 500 oocysts/cysts per gram of stool. Because the purified DNA was eluted in 50 µl of the elution buffer and only 1 µl of the fecally derived DNA was subjected to PCR amplification, the lower detection limit of each PCR with the extraction protocol was ≈ 2 oocysts/cysts per reaction.
Finally, the developed extraction protocol with the subsequent PCR was validated using many clinical stool samples and proved to be a simple and an economic. Importantly, the amended kit's protocol together with the matching PCR tests picked more positive samples than immunoassay tests. Taking the more sensitive confirmatory PCR tests results as a gold standard, it was clear that the amended DNA extraction protocol with the PCR test were more sensitive than the target matching coproantigen detection kit, agreeing with previous reports [12,19,23]. Equally important, none of the figures given in this study reflect the actual prevalence rate of each parasite in the studied populations because of the short duration of the study.
In conclusion, based on the QIAamp stool Mini kit (Qiagen), an extraction protocol was developed in this study. The protocol was proved useful in extracting DNA from the 3 predominant pathogenic enteric protozoa found in human infections. The protozoan DNA, recovered directly from oocysts/cysts found in feces, was sufficiently purified and proved to be compatible with diagnostic PCR.
ACKNOWLEDGMENTS
The author would like to thank Dr. Ghaffar M. Abdel, Faculty of Medicine, Menoufia University, Egypt and Dr. M. Younes, King Faisal Hospital, Saudi Arabia, for providing me with clinical specimens. He is also grateful to the 2 anonymous reviewers for their valuable comments and suggestions to improve the quality of this paper.
Footnotes
The author has no conflict of interest related to this study.
References
- 1.Mackay IM. Real-time PCR in the microbiology laboratory. Clin Microbiol Infect. 2004;10:190–212. doi: 10.1111/j.1198-743x.2004.00722.x. [DOI] [PubMed] [Google Scholar]
- 2.Stephen AM, Cummings JH. The microbial contribution to human fecal mass. J Med Microbiol. 1980;13:45–56. doi: 10.1099/00222615-13-1-45. [DOI] [PubMed] [Google Scholar]
- 3.Surl CG, Jung BD, Park BK, Kim HC. Resistance of Cryptosporidium parvum oocysts following commercial bleach treatment. Korean J Vet Res. 2011;51:101–105. [Google Scholar]
- 4.Oikarinen S, Tauriainen S, Viskari H, Simell O, Knip M. PCR inhibition in stool samples in relation to age of infants. J Clin Virol. 2009;44:211–221. doi: 10.1016/j.jcv.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 5.Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors - occurrence, properties and removal. J Appl Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 6.Elwin K, Robinson G, Hadfield SJ, Fairclough HV, Gomara MI, Chalmers RM. A comparison of two approaches to extracting Cryptosporidium DNA from human stools as measured by a real-time PCR assay. J Microbiol Methods. 2012;89:38–40. doi: 10.1016/j.mimet.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Halstead FD, Lee AV, Couto-Parada X, Polley SD, Ling C, Jenkins C, Chalmers RM, Elwin K, Gray JJ, Iturriza-Gómara M, Wain J, Clark DA, Bolton FJ, Manuel RJ the Olympics GI Group. Universal extraction method for gastrointestinal pathogens. J Med Microbiol. 2013;62:1535–1539. doi: 10.1099/jmm.0.058743-0. [DOI] [PubMed] [Google Scholar]
- 8.Elwin K, Fairclough HV, Hadfield SJ, Chalmers RM. Giardia duodenalis typing from stools: a comparison of three approaches to extracting DNA, and validation of a probe-based real-time PCR typing assay. J Med Microbiol. 2014;63:38–44. doi: 10.1099/jmm.0.066050-0. [DOI] [PubMed] [Google Scholar]
- 9.Babaei Z, Oormazdi H, Rezaie S, Rezaeian M, Razmjou E. Giardia intestinalis: DNA extraction approaches to improve PCR results. Exp Parasitol. 2011;128:159–162. doi: 10.1016/j.exppara.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, Das P, Lal AA, Xiao L. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. 2003;9:1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooshyar H, Rezaian M, Kazemi B, Jeddi-Tehrani M, Solaymani-Mohammadi S. The distribution of Entamoeba histolytica and Entamoeba dispar in northern, central, and southern Iran. Parasitol Res. 2004;94:96–100. doi: 10.1007/s00436-004-1175-8. [DOI] [PubMed] [Google Scholar]
- 12.Santos HL, Peralta RHS, de Macedo HW, Barreto MGM, Peralta JM. Comparison of multiplex-PCR and antigen detection for differential diagnosis of Entamoeba histolytica. Braz J Infect Dis. 2007;11:365–370. doi: 10.1590/s1413-86702007000300013. [DOI] [PubMed] [Google Scholar]
- 13.Pedraza-Diaz S, Amar C, Iversen AM, Stanley PJ, McLauchlin J. Unusual Cryptosporidium species recovered from human feces: first description of Cryptosporidium felis and Cryptosporidium 'dog type' from patients in England. J Med Microbiol. 2001;50:293–296. doi: 10.1099/0022-1317-50-3-293. [DOI] [PubMed] [Google Scholar]
- 14.Amar CF, Dear PH, McLauchlin J. Detection and genotyping by real-time PCR/RFLP analyses of Giardia duodenalis from human feces 2003. J Med Microbiol. 2003;52:681–683. doi: 10.1099/jmm.0.05193-0. [DOI] [PubMed] [Google Scholar]
- 15.Limor JR, Lal AA, Xiao L. Detection and differentiation of Cryptosporidium parasites that are pathogenic for humans by real-time PCR. J Clin Microbiol. 2002;40:2335–2338. doi: 10.1128/JCM.40.7.2335-2338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subrungruang I, Mungthin M, Petmitr PC, Rangsin R, Naaglor T, Leelayoova S. Evaluation of DNA extraction and PCR methods for detection of Enterocytozoon bienuesi in stool specimens. J Clin Microbiol. 2004;42:3490–3494. doi: 10.1128/JCM.42.8.3490-3494.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaGier MJ, Joseph LA, Passaretti TV, Musser KA, Cirino NM. A real-time multiplexed PCR assay for rapid detection and differentiation of Campylobacter jejuni and Campylobacter coli. Mol Cell Probes. 2004;18:275–282. doi: 10.1016/j.mcp.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Stroup SE, Roy S, Mchele J, Maro V, Ntabaguzi S, Siddique A, Guerrant RL, Kirkpatrick BD, Fayer R, Herbein J, Ward H, Haque R, Houpt ER. Real-time PCR detection and speciation of Cryptosporidium infection using Scorpion probes. J Med Microbiol. 2006;55:1217–1222. doi: 10.1099/jmm.0.46678-0. [DOI] [PubMed] [Google Scholar]
- 19.Furrows SJ, Moody AH, Chiodini PL. Comparison of PCR and antigen detection methods for diagnosis of Entamoeba histolytica infection. J Clin Pathol. 2004;57:1264–1266. doi: 10.1136/jcp.2004.017822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. PCR detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii in stool samples from Sydney, Australia. J Clin Microbiol. 2007;45:1035–1037. doi: 10.1128/JCM.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berrilli F, Di Cave D, D'Orazi C, Orecchia P, Xhelilaj L, Bejko D, Çaça P, Bebeci D, Cenko F, Donia D, Divizia M. Prevalence and genotyping of human isolates of Giardia duodenalis from Albania. Parasitol Int. 2006;55:295–297. doi: 10.1016/j.parint.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Garcia LS. Practical Guide to Diagnostic Parasitology. 2nd ed. Washington DC, USA: ASM Press; 2009. [Google Scholar]
- 23.McHardy IH, Wu M, Cohen RS, Couturier MR, Humphries RM. Clinical laboratory diagnosis of intestinal protozoa. J Clin Microbiol. 2014;52:712–720. doi: 10.1128/JCM.02877-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heyman MB, Shigekuni LK, Ammann AJ. Separation of Cryptosporidium oocysts from fecal debris by density gradient centrifugation and glass bead columns. J Clin Microbiol. 1986;23:789–791. doi: 10.1128/jcm.23.4.789-791.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucl Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Nat Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spano F, Putignani L, McLauchlin J, Casemore DP, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 28.Read CM, Monis PT, Thompson RC. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect Genet Evol. 2004;4:125–130. doi: 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Hamzah Z, Petmitr S, Mungthin M, Leelayoova S, Petmitr PC. Differential detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii by a single-round PCR assay. J Clin Microbiol. 2006;44:3196–3200. doi: 10.1128/JCM.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao L, Bern C, Limor J, Sulaiman I, Roberts J, Checkley W, Cabrera L, Gilman RH, Lal AA. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J Infect Dis. 2001;183:492–497. doi: 10.1086/318090. [DOI] [PubMed] [Google Scholar]
- 31.Hopkins RM, Meloni BP, Groth DM, Wetherall JD, Reynoldson JA, Thompson RC. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J Parasitol. 1997;83:44–51. [PubMed] [Google Scholar]
- 32.Khairnar K, Parija SC. A novel nested multiplex polymerase chain reaction (PCR) assay for differential detection of Entamoeba histolytica, E. moshkovskii and E. dispar DNA in stool samples. BMC Microbiol. 2007;7:47. doi: 10.1186/1471-2180-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adamska M, Duniec AL, Maciejewska A, Sawczuk M, Skotarczak B. Comparison of efficiency of various DNA extraction methods from cysts of Giardia intestinalis measured by PCR and TaqMan real time PCR. Parasite. 2010;17:299–305. doi: 10.1051/parasite/2010174299. [DOI] [PubMed] [Google Scholar]
- 34.Gardner AL, Roche JK, Weikel CS, Guerrant RL. Intestinal cryptosporidiosis: pathophysiologic alterations and specific cellular and humoral immune responses in rnu/+ and rnu/rnu (athymic) rats. Am J Trop Med Hyg. 1991;44:49–62. doi: 10.4269/ajtmh.1991.44.49. [DOI] [PubMed] [Google Scholar]
- 35.da Silva AJ, Bornay-Llinares FJ, Moura IN, Slemenda SB, Tuttle JL, Pieniazek NJ. Fast and reliable extraction of protozoan parasite DNA from fecal specimens. Mol Diagn. 1999;4:57–64. doi: 10.1016/s1084-8592(99)80050-2. [DOI] [PubMed] [Google Scholar]
- 36.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. PCR detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii in stool samples from Sydney, Australia. J Clin Microbiol. 2007;45:1035–1037. doi: 10.1128/JCM.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonin P, Trudel L. Detection and differentiation of Entamoeba histolytica and Entamoeba dispar isolates in clinical samples by PCR and enzyme-linked immunosorbent assay. J Clin Microbiol. 2003;41:237–241. doi: 10.1128/JCM.41.1.237-241.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaki M, Verweij JJ, Clark CG. Entamoeba histolytica: direct PCR-based typing of strains using fecal DNA. Exp Parasitol. 2003;104:77–80. doi: 10.1016/s0014-4894(03)00121-8. [DOI] [PubMed] [Google Scholar]