1. Introduction
All life forms have an absolute requirement for metals, as metals play critical roles in fundamental processes, including osmotic regulation, catalysis, metabolism, biomineralization, and signaling. Group I and II metals (alkali and alkaline earth metals such as sodium, potassium, calcium, and magnesium) are highly abundant in most biological organisms. Gradients of group I and II metals across membranes represent a classical way to store potential energy, and these ions play roles in osmotic regulation, generation of action potentials, and signaling. Transition metals that are generally recognized as playing critical roles in biology include iron, zinc, copper, manganese, cobalt, nickel, molybdenum, tungsten, chromium, and vanadium.1 These elements are often referred to as trace elements because they are present at much lower levels than the group I and II metals, although it is important to note that iron and zinc are often found in substantial amounts and hence their characterization as trace elements is sometimes misleading. Transition metal abundance and usage differs notably across different superkingdoms. For example, eukaryotes devote a higher proportion of their proteome to binding zinc than bacteria or archaea, but the reverse is true for iron, manganese, and cobalt.2 A growing number of comparative genomics studies suggest that iron and zinc are widely used in biology, whereas other metals such as copper, molybdenum, tungsten, nickel, and cobalt are used more sporadically across groups of organisms.3 To add an additional level of complexity, a recent proteomics study suggested the microbial metallome, that is, the full distribution of metals used by an organism, is largely uncharacterized, and there may be additional uses of transition metals, such as cadmium, uranium, arsenic, and lead not commonly recognized as being beneficial biometals.4
One of the first steps in defining the usage of metals by different organisms is to establish a metal inventory by quantifying the metal content of cells and tissues. Biological metals may exist in different forms, including as hydrated ions, tightly bound forms such as metal-bound cofactors and protein- or nucleic-acid bound species, or loosely bound forms in association with a diverse heterogeneous buffer, which can consist of low molecular weight species such as amino acids, glutathione, or citric acid, and labile species. The total metal content consists of the sum of all of these diverse forms. Historically elemental analysis was carried out by either flame or graphite furnace atomic absorption spectroscopy (AAS), a technique that enables quantification of the average total metal content from a digested sample at parts per billion (μg/L) sensitivity, one metal at a time.5 Since its introduction in the 1980s, inductively coupled plasma mass spectrometry (ICP-MS) has largely surpassed AAS as the analytical method of choice for quantification of metals in a bulk sample due to its ability to measure multiple metals at once, increased sensitivity (0.1–10 parts per trillion, i.e., ng/L, for most transition metals), and increased dynamic range.6 While these techniques are instrumental in defining metal abundance in a bulk sample, they do not permit single cell analysis or subcellular analysis of the location of metals within a sample. Yet to fundamentally gain insight into the mechanisms by which cells and organisms regulate and use metals, it is essential to go beyond quantification of total metal content in a bulk sample, and to define the speciation, distribution, and accessibility of metals in individual cells, tissues, and whole organisms.
Elemental mapping of metals involves measurement of the distribution of metals in a biological sample in a spatially resolved manner. One method for accomplishing this is to adapt mass spectrometry techniques to permit spatial resolution of total metal content in fixed biological specimens at the cellular and subcellular levels.5,7 Some of the more widely used techniques include secondary ion mass spectrometry (SIMS), nano-SIMS,8 and laser ablation coupled with ICP-MS (LA-ICP-MS).9 Additional analytical techniques that permit mapping of total metal content with high sensitivity and spatial resolution involve synchrotron or focused ion-beam microprobes.10 Many of these techniques have recently been comprehensively reviewed elsewhere and will not be the focus of this Review.5,7−11
As a complement to the above techniques, it is important to define the chemical form or speciation of metal ions in biological samples and the distribution between free hydrated ions, loosely bound ions, and a tightly bound, largely inaccessible, pool. Currently, there is no single technique available that permits measurement of all of these different species within the same specimen. Yet there are some techniques that permit measurement of different subsets of these pools, for example, the use of fluorescent sensors as detailed below. Thus, combinations of complementary methods will be required for a comprehensive view of cellular metal regulation. Another important factor is the measurement of metal ions in live samples. Life is by definition dynamic, and this dynamism is key to understanding the mechanisms between cause and effect for biological processes. Analytical methods that permit examination of accessible metal pools in live samples would enable identification of metal ion fluxes, dynamics, and movements in response to environmental perturbations, a critical step in defining how metals are regulated and used in cells. An analogy that has often been used to emphasize the importance of visualization of living specimens is that reconstructing the basic rules and their consequences of a sports game such as football from a series of still images taken at different times from different games would be exceedingly challenging, if not impossible.12 This is because events are not simply a factor of time, but are also a consequence of factors that happened earlier within the same game.
Light microscopy is an indispensible tool for cell and molecular biology and is compatible with visualization of living specimens. The human eye can only resolve objects on the order of 0.1 mm, but cells are orders of magnitude smaller, often ranging from 5 to 30 μm. Moreover, bacteria (1 μm), viruses (10–100 nm), and subcellular structures such as the nucleus (10 μm), mitochondrion (2–5 μm), or microvilli (1 μm) are smaller still.13 Because a traditional light microscope can resolve objects on the order of 250 nm, it has been an instrumental tool for studying the microscopic world. Recent advances in super-resolution microscopy have extended the resolution limit, permitting visualization and analysis of nanoscale structures.14 The biggest challenge with microscopy is differentiating the interesting (i.e., a specific object, structure, molecule, or metal) from the uninteresting (i.e., the background).
Metals have long been identified and classified by colorimetric methods due to their light absorption properties, which lead to rich and highly characteristic optical transitions.1 Yet in the complex environment of a cell, where multiple metals and other absorbing species are present in differing quantities, additional approaches are required to visualize the metal of interest. One strategy for accomplishing this is to use a chromogenic stain or dye for the metal of interest to isolate the metal and enhance contrast between the signal (i.e., presence of the metal) and background. Since the introduction of Perls’ Prussian blue in 1867 as a stain for nonheme iron,15 chromogenic dyes have been widely used histology tools for visualizing the presence of metals in fixed cells.10a Yet dyes that rely on absorption of light have limited sensitivity as compared to fluorescence, thus driving the development of fluorescent sensors for metals to be used in conjunction with fluorescence microscopy to map metals in cells.
This Review focuses on fluorescent sensors for transition metals commonly found in biological organisms. Generally speaking, such sensors are designed to measure the accessible or labile pool of metals (free hydrated and loosely bound, buffered ions), and thus access a subset of the total metal content of a cell. For sensors to be minimally perturbing, they should not engage in competitive exchange with tightly bound endogenous metal complexes, a property that depends on the affinity of the sensor, its concentration within the cell, and the nature of the diverse bound-metal pool. A deeper discussion of this point and strategies for critically evaluating whether sensors perturb metal speciation will be discussed in section 3.2. We start this Review by giving a basic overview of fluorescence imaging and sensor design, followed by a critical analysis of parameters and properties to consider when using sensors in biological systems. We then present a historical perspective of how the field has evolved. While this Review focuses on transition metals, we discuss some of the key advances/milestones achieved in the development of fluorescent Ca2+ indicators as these helped lay the groundwork for much of the subsequent work developing sensors for transition metals. Finally, we highlight progress in sensor development for biological metals, emphasizing recent advances, while including a discussion of the most widely used sensors. To demonstrate what kind of measurements can be made and what kind of information can be learned from using fluorescent sensors, we review several applications of sensors for defining metal homeostasis and dynamics in cells or organisms. We would also like to call readers’ attention to several excellent prior16 reviews that focus on different aspects of sensor development.17 Additionally, important practical considerations for using probes and experimental protocols have been reported elsewhere.18
One of the most exciting and powerful possibilities of fluorescence microscopy is that it can provide a window into the intracellular metabolism of metals in live intact systems. Fluorescence microscopy permits visualization of an object of interest in unicellular organisms, individual cells from multicellular organisms, cells encapsulated in 3D matrices, organotypic cultures, ex vivo models, and, with the right instrumentation, whole organisms (bacteria, yeast, plants, flies, worms, fish, and mice).19 The application of fluorescent sensors and fluorescence microscopy, in combination with other analytical techniques for mapping total metal content, offers researchers the opportunity to address fundamental questions about cellular metal homeostasis. Some of these basic unanswered questions include: What is the amount and speciation of metals in cells? Where are metals located? How do metal ion concentrations change in response to cellular events, environmental changes, or onset of disease? Finally, how do cells regulate metal dynamics, and how do metal dynamics impact cellular function?
2. General Features of Fluorescent Sensors for Metal Ions
Fluorescence involves the emission of photons that occurs nanoseconds after an absorption event. A fluorescence microscope takes advantage of the shift in wavelength between the absorbed and emitted light by filtering out light due to the excitation source without blocking the emitted light.20 Fluorescent sensors for metals contain two essential features: a metal chelating or binding moiety and at least one fluorophore capable of absorbing and emitting light. To function as a sensor, metal binding must alter either the electronic structure or the molecular structure of the sensor. Changes in the electronic structure can lead to a change in the intensity or wavelength of light absorption or emission, while changes in the molecular structure can alter the distance or orientation between a pair of fluorophores that serve as a donor–acceptor pair. A fluorescence microscope permits visualization of changes in fluorescence, and hence the target of a particular sensor, which in this case is a specific metal ion of interest, in a spatially resolved manner.
2.1. Photophysical Properties of Fluorophores
Arguably the most important property of a fluorescent sensor is its ability to be detected within the complex environment of a cell or organism. The sensitivity and signal-to-noise ratio of a sensor are highly dependent on the brightness and stability of the sensor’s fluorophore(s), as well as the characteristics of the instrumentation.21 Brighter fluorophores require less excitation light, thus causing less photodamage to the living specimen. Additionally, brighter sensors can be used at lower concentrations, thus minimizing perturbation of metal ion homeostasis. The stability of a fluorophore is particularly important for time-lapse imaging. While in principle all fluorophores can cycle between the ground and excited state many times, repeated exposure to light inevitably leads to photobleaching, where bleaching is a generic term for all of the myriad processes that cause permanent decay in fluorescence intensity. Photobleaching not only limits the length of time a process can be monitored, it can contribute to phototoxicity as well.
The theoretical brightness of a fluorophore is defined as the product of the extinction coefficient and the quantum yield.22 The extinction coefficient is the efficiency with which a chromophore absorbs light, while the quantum yield represents the efficiency with which a fluorophore emits light after absorption. In this Review, we calculate the theoretical brightness of sensors by multiplying the quantum yield and extinction coefficient reported in the literature. Because many metal sensors involve a change in brightness upon metal binding, we report brightness in the metal-free and metal-bound state. However, there may be differences in the photophysical properties of a sensor in vitro (i.e., in a cuvette) versus in situ (i.e., inside a cell) due to differences in viscosity, pH, solvent, accessibility to oxygen, or other factors associated with the cellular environment.23 Moreover, the theoretical brightness of the fluorophore is not the only factor to consider when defining the detection sensitivity.
An additional important factor that impacts detection sensitivity in cells is the wavelength of excitation and emission. Many biomolecules absorb light in the UV and visible spectrum. Because excited molecules can react with molecular oxygen to produce free radicals, exposure to electromagnetic radiation can produce reactive oxygen species, which are damaging to biological samples.24 Generally speaking, higher energy, lower wavelength light causes greater photodamage than lower energy, longer wavelength light.25 In addition, because many biomolecules emit light in the UV and visible range, the background signal from the cellular milieu is higher at higher energy.26 Light is also scattered when it encounters matter, and this scattering depends on the nature of the tissue and wavelength of light.27 Scattering limits the depths to which light can penetrate a biological specimen; for example, a photon is scattered once for every 47 μm that it transits through an adult rat brain, limiting the effective imaging depth to ∼50 μm using confocal laser scanning microscopy.19 As a general rule, fluorescent sensors that absorb and emit at longer wavelengths give rise to less phototoxicity, decreased background autofluorescence, and are subject to decreased scattering.
Of course it is important to note that the detection sensitivity, often referred to as the contrast between signal and background, will depend not just on the inherent properties of the sensor and the biological specimen, but also on the instrumentation available. Excitation source (intensity and nature of the source – for example, how well a laser line overlaps with the excitation of the fluorophore), filter sets (both the bandwidth and the transmission), camera sensitivity, and the objective are all factors that influence the intensity of a measured fluorescence signal.20
2.2. Mechanisms of Altering a Fluorescence Signal
As stated above, metal binding must alter the electronic and/or molecular structure of the sensor to induce changes in fluorescence properties that can be detected by a fluorescence microscope. Two common mechanisms by which a metal can modulate the electronic structure and hence fluorescence are energy transfer or electron transfer between the metal and photoexcited fluorophore. Both processes can give rise to either a “turn-off” or a “turn-on” fluorescence response, due to fluorescence quenching or enhancement, respectively. A variety of clever approaches have been used to manipulate these properties to design platforms for optical detection of metal ions. There is an extensive body of literature on chemosensors whose optical properties are altered by analyte binding, and that make use of small-molecule fluorophores, polymers, solids and gels, material surfaces (quantum dots, glass or gold surfaces, carbon nanotubes), and mesoporous materials.28 Such probes exploit a variety of different mechanisms for chemical or environmental detection of metal ions. In some cases, such probes have been used for biological detection of transition metals. This Review focuses on fluorescent sensors for metals that have been applied to biology, and so the discussion below focuses on the mechanisms that are prevalent in the subset of probes that have been applied for biological detection of transition metals.
Energy transfer can occur between transition metals with partially filled d-orbitals of appropriate energy and a photoexcited fluorophore by a double electron exchange process (Figure 1A). This type of energy transfer, first postulated by Dexter, is also referred to as short-range or collisional.29 It is a form of quenching whereby an excited electron from one molecule (the donor) is transferred to another molecule (the acceptor). Figure 1A displays a schematic of Dexter energy transfer. The process is active only at very short distances, typically less than 10 Å, because it requires wave function overlap. This electron exchange is one of the primary mechanisms by which the emission of organic fluorophores can be quenched by metal ions.30 While this quenching property means that most metal ions are capable of directly modulating fluorescence emission, it also poses a challenge in distinguishing between different metals if multiple metals capable of quenching are present in a complex sample. It also complicates the design of “turn-on” sensors in which a fluorescence signal is increased in response to metal ions.
Figure 1.
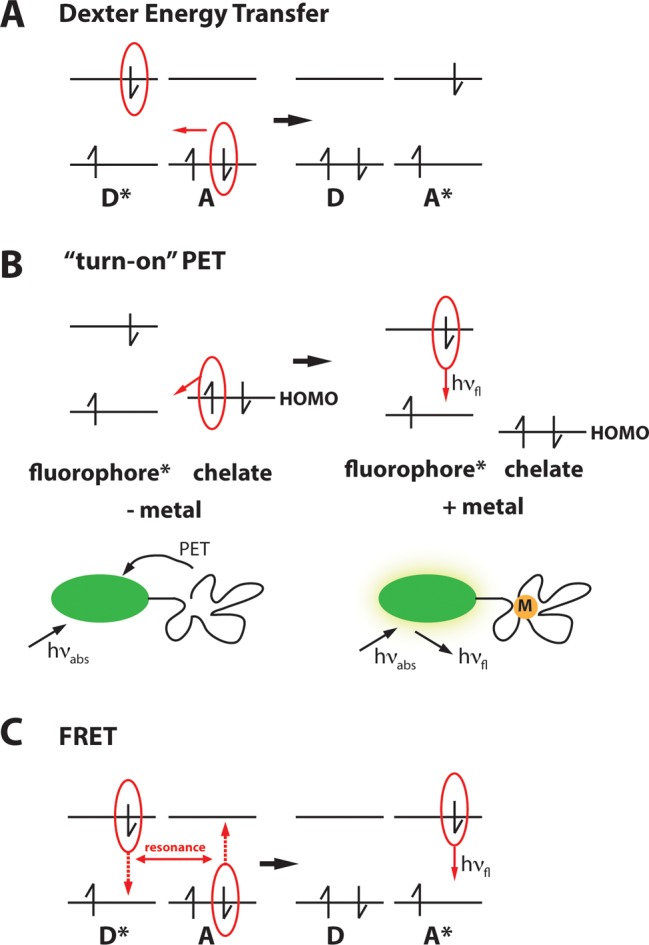
Schematic of Dexter energy transfer (A), “turn-on” PET (B), and FRET (C).
In addition to energy transfer, fluorescence properties can also be modulated by electron transfer between the metal and the fluorophore or modulation of electron transfer within a self-contained fluorophore–chelate unit upon metal binding (Figure 1B).30a,30b This process requires separation of charge and therefore excitation of the donor by light; hence it is typically referred to as photoinduced electron transfer (PET). As with energy transfer, electron transfer can lead to either quenching or enhancement of fluorescence. Direct electron transfer between a photoexcited fluorophore and a metal ion with low energy empty or partially filled d-orbitals typically leads to quenching.
Fluorescence quenching by metal ions does not have to be deleterious, and the right sensor design can turn it into a benefit. As one example, Kool and co-workers created polyfluorophore sensors on a DNA backbone that take advantage of quenching properties.28d The molecular design of these sensors incorporates fluorophores and metal binding ligands into DNA-like oligomers. A variety of fluorescence responses were observed including fluorescence enhancement and red- and blue-shifts. A panel of sensors was then used to differentiate eight metal ions that are typically implicated in fluorescence quenching, including Hg2+, Cu2+, Co2+, Ni2+, Pb2+, Ag+, Cr3+, and Fe3+. While this approach was only employed for chemical detection of metals in solution, recent efforts by the same research group have demonstrated that polyfluorophores can be fused to a protein of interest in a mammalian cell using the HaloTag technology, opening the possibility that this sensor platform could be adapted for cellular detection of metal ions.31
PET can also give rise to fluorescence enhancement (Figure 1B). This phenomenon is most commonly observed in small-molecule sensors comprised of a fluorophore, linker domain, and an electron-rich metal chelate. In the absence of a metal, excitation leads to separation of charges, and PET between the fluorophore and the chelate competes with fluorescence emission. Thus, PET gives rise to an efficient relaxation pathway, decreasing the quantum yield of the fluorophore. Modulation of PET can occur when binding of a metal ion to an electron-rich chelating moiety shifts the charge density, effectively quenching the PET decay pathway and increasing the quantum yield.28a,28c,32 The development of fluorescent Ca2+ sensors in 1980 by Roger Tsien was one of the first examples of how tuning of this photophysical mechanism can lead to robust fluorescent sensors, in this case for Ca2+, demonstrating the potential of this design for biological fluorescence imaging.33 The modularity of the PET platform has been exploited for the development of sensors with enhanced properties. This platform consists of three components: a fluorophore, linker, and chelator that can all be individually modified to alter PET within the probe. In particular, tuning of electron density by incorporation of electron-withdrawing groups, altering the nature of the PET “switch”, and changing the linker between the chelate and fluorophore can tune the PET efficiency, thus influencing the relative brightness of the sensor in the unbound and bound state.17b,28c
A slightly modified sensor platform characterized by an integrated fluorophore–chelate system without a clear spacer can also be exploited for metal sensing.32 Although this design sacrifices some of the modularity and tunability of the classical three-component system, internal charge transfer (ICT) can lead to a shift in wavelength of excitation or emission, which if large enough can result in a ratiometric sensor. For ratiometric sensors, fluorescence images are collected at two different wavelengths, typically the wavelength maxima in the metal-free and bound state, enabling the free and bound states of the indicator to be monitored simultaneously. Such sensors permit normalization for perturbations of fluorescence that are not related to changes in metal ions such as changes in the path length, sample thickness, dye concentration, or movement of the sample. Such sensors also allow researchers to quantify the concentration of dye within cells, which is an important control when assessing whether the sensor perturbs metal homeostasis.
Another mechanism that has been exploited for the development of metal sensors is Förster resonance energy transfer (FRET). This phenomenon was described by Theodor Förster in 1948 and involves dipole–dipole coupling between a photoexcited donor and an acceptor.34 This is a radiationless process in which energy is transmitted by coupling of the two oscillating dipoles (Figure 1C). The probability of energy transfer is described by the FRET efficiency, which is highly dependent on the distance between the two chromophores (inversely proportional to the sixth power of the distance between the donor and acceptor), overlap between the donor emission and acceptor absorption, and the relative orientation of the transition dipoles of the donor and acceptor (maximal transfer for collinear dipoles, zero transfer for perpendicular dipoles). The acceptor can either be a chromophore, simply capable of absorbing energy, or a fluorophore in which case the excited molecule emits a photon upon relaxation to the ground state due to sensitized emission. FRET causes a decrease in donor emission and a decrease in the donor lifetime, and hence can be monitored at the donor wavelength only. However, all FRET-based sensors for metal ions employ two fluorophores so that measurement of both donor emission and sensitized emission from the acceptor yields a ratiometric sensor. For such sensors, the FRET ratio is the ratio of the sensitized acceptor emission and the donor emission and can either reported as either acceptor/donor or donor/acceptor. A typical sensor design employs two fluorophores (a donor and acceptor) and a metal chelating unit. Metal binding induces a change in the molecular structure that alters either distance, orientation, or both so as to either promote or disrupt FRET.
A final mechanism that is increasingly employed is to exploit the unique chemical reactivity of different metal ions to generate probes in which a metal-catalyzed reaction leads to a change in fluorescence. Specificity in such probes is encoded by the fact that only a certain metal (or small subset of metals) is capable of mediating the reaction. Two classic examples are the chelation-induced spirolactam ring-opening employed in sensors for Cu2+, Fe3+, and Hg2+.17e,35 Another chelation-enhanced fluorescence was used early on to develop a probe for the toxic metal Pb2+.36
2.3. Classes of Sensors for Live-Cell Imaging
2.3.1. Molecular Probes
Molecular probes are compromised of small-molecule fluorophores coupled to a metal chelating unit. They may be entirely chemical in nature or comprised of peptide or nucleic acid components. The distinguishing feature of these probes is that they cannot be synthesized within a living cell or organism and hence must be delivered in some way. Some molecular sensors are naturally membrane permeable, and hence delivery simply involves adding the sensor to cells and waiting an appropriate length of time for the sensor to diffuse into the cell. However, many metal chelates contain charged carboxylate moieties, which prevent cell entry. In 1981, Roger Tsien introduced a clever trick of masking the four carboxylates in a Ca2+ sensor by esterifying them with an acetoxymethyl (AM) ester, thus rendering the sensor cell permeable.37 Upon entry into cells, exposure to cellular esterases led to hydrolysis of the AM ester, thus trapping the charged indicator in cells and rendering it Ca2+ sensitive once again. This approach has subsequently been used to facilitate cell permeability of some fluorescent sensors for transition metals, as detailed in sections 5–9 of this Review. In an exciting recent development, Tian et al. tested a series of synthetic branched esters against a panel of esterases and identified selective enzyme–substrate pairs.38 Expression of different esterases in different cell types then permits cell-specific delivery of small-molecule fluorophores. Such an approach could be used to trap metal sensors to permit monitoring of metal homeostasis in specific subsets of cells in intact multicellular organisms.
Another method of delivery is attachment of a molecular sensor to a cell penetrating peptide. An array of naturally occurring and synthetic peptides have been shown to be spontaneously transported into mammalian cells and are capable of carrying along cargo as large as a 120 kDa protein.39 While the mechanisms of entry remain controversial, and the ultimate destination of cargo is complicated and in some cases unpredictable, nevertheless there have been many successes of using cell penetrating peptides as an efficient delivery method.39 For example, this approach was employed in early generations of Zn2+ sensors based on carbonic anhydrase covalently linked to a small-molecule fluorophore, AlexaFluor.40
Finally, molecular sensors can be microinjected into individual cells.41 This method allows delivery of a well-defined concentration of sensor into single cells as long as the cells are robust enough to withstand penetration by a micropipet. This approach is not widely used because the invasive nature may lead to sustained damage of the plasma membrane, dyes must be loaded one cell at a time, and this approach does not permit delivery into multiple cells in whole tissues or organisms.42
A convenient feature of molecular sensors is their modular construction and the opportunity to exploit a large repertoire of well-characterized fluorophores. Small-molecule fluorophores tend to have excellent photophysical properties (brightness and photostability), where the best organic dyes emit 10–100-times more photons before bleaching when compared to fluorescent proteins.43 Although such photophysical properties may not be maintained within a sensor, there are a large number of excellent dyes from which to choose as the basic building blocks for sensor construction. The majority of molecular platforms rely on coumarin, fluorescein, boron-dipyrromethene (BODIPY), and rhodamine.
2.3.2. Genetically Encoded Probes
Genetically encoded probes are fluorescent sensors that are encoded by a nucleic acid sequence and are synthesized entirely by a cell. The largest category of genetically encoded sensors is comprised of protein-based probes that utilize one or more fluorescent proteins (FP) as the fluorophore. The sensors also contain a peptide or protein moiety that serves as a metal binding domain. For single FP-based sensors, metal binding induces a change in the chemical or electronic environment around the chromophore, causing either a change in intensity or a shift in the excitation or emission spectrum.44 Sensors containing two FPs typically exploit the principle of FRET, where metal binding induces a conformational change, thereby either promoting or disrupting FRET between the two FPs.
An under-explored platform for creating genetically encoded sensors is the use of nucleic acids. The recent discovery of naturally occurring metal-sensing RNAs, called riboswitches, that sense Mg2+ levels and regulate the expression of metal transporters, demonstrates that nucleic acids can function as robust metal-dependent switches in cells.45 Structure–function studies on the Mg2+-sensing riboswitches, so-called M-box riboswitches, revealed that in vitro these riboswitches bind different metal ions with varying affinity, but similar cooperativity.46 While the naturally occurring riboswitches control Mg2+ regulatory genes, one can imagine engineering a riboswitch to drive the expression of a fluorescent reporter, thus generating a genetically encoded nucleic acid-based Mg2+ sensor. Furthermore, tuning of the ligand binding site might enable the development of sensors specific for different transition metals.
2.3.3. Hybrid Probes
Probes that involve a combination of genetically encoded and small molecular elements are referred to as hybrid probes. Such probes involve introduction of the genetically encoded component by transfection, viral transduction, or some other transgenic technology and introduction of the small molecular component by the means described above. This approach typically makes use of protein or peptide tags, although nucleic acid-based targeting could be an area of future development. A number of peptide/protein tags have been developed that are capable of binding small-molecule agents in cells, including the FlAsH/ReAsH system,47 SNAP-tag,48 HaloTag, and peptides selected for sensor binding.49 There are a handful of fluorescent probes for different analytes that fall in this category, although only the SNAP-tag technology has been used to genetically target metal-based sensors. One of the first examples was the use of SNAP-tag technology to target the small-molecule ZP1 sensor to mitochondria and Golgi.50 To be compatible with SNAP-tag technology, the ZP1 probe was modified to incorporate a benzylguanine moiety that could serve as a substrate for O6-alkylguanine-DNA alkyltransferase (AGT). AGT acts on benzylguanine-tethered sensors through an active site cysteine, which attacks the O6-benzylguanine, leading to covalent attachment of the sensor to AGT and release of guanine.48 Transfection of cells with AGT that is genetically targeted to a specific compartment (such as mitochondria or Golgi) provides the opportunity to localize a small-molecule sensor in a particular location. This approach has been used to target Ca2+ sensors,51 Zn2+ sensors,50 and H2O2 sensors52 to specific cellular compartments, and in principle is generalizable to any sensor platform that can be modified with an O6-BG moiety.
Another example of a hybrid probe platform is that of the carbonic anhydrase (CA) family of Zn2+ probes. The CA-probes were recently re-engineered to replace the covalently attached small-molecule fluorophore with a red FP.53 The CA-FP fusion protein has been expressed in both bacterial and mammalian cells. Addition of dapoxyl sulfonamide, a cell permeable probe that binds to an open coordination position on Zn2+ when it is bound to CA, leads to FRET between the dapoxyl sulfonamide and red FP. Because the CA-FP fusion is synthesized by the cell, signal peptides can be used to target the sensor to intracellular organelles, and this sensor was successfully targeted to mitochondria of mammalian cells.53a
3. Important Considerations for Introduction of Sensors
In addition to the photophysical properties of sensors (brightness, photostability, wavelength range) and biochemical properties (affinity and specificity for the target metal), there are a number of factors that influence the use of fluorescent sensors for mapping accessible pools of metal ions in cells. For such applications, factors such as the intracellular concentration of the sensor, where it is located within cells, and the extent to which metal ions are buffered in the cellular milieu will strongly influence the resulting measurements. For example, if the sensor concentration greatly exceeds the metal ion concentration, the sensor can sequester the entire metal ion pool and perturb the system. However, this effect can be mitigated if there is a large reservoir of buffered metal ion and the sensor concentration is substantially less than this reservoir. Likewise, if the sensor affinity is high and the concentration is substantial, the sensor may engage in competitive exchange with endogenous bound metal complexes. A discussion of these factors is presented below.
3.1. Factors Affecting the Intracellular Concentration of Sensors
The intracellular concentration of a sensor is governed by a combination of how much of the probe is incorporated or expressed in cells, and how well the sensor is retained. Molecular probes are applied to cells or tissues, and either diffuse passively into cells if they are sufficiently hydrophobic, or are aided by the processes described above. It is important to recognize that the amount of dye applied to cells, tissues, or organisms may differ substantially from the intracellular concentration, and the only way to truly define how much dye is present is to measure the concentration inside cells, although this is challenging unless the probe is ratiometric. There are some mechanisms by which probes become trapped in cells, leading to accumulation and concentration in the cellular milieu. Such mechanisms may also affect the localization of probes within cells, as detailed in section 3.3.1. Cleavable esters, which when hydrolyzed by cellular esterases yield a charged probe that does not freely diffuse out of cells, are often used to promote accumulation of probes within cells.37,38 In fact, AM-ester-based probes are often concentrated at least 100-fold inside cells, yielding intracellular concentrations in the hundreds of micromolar up to millimolar.54 Intracellular accumulation can also be facilitated by pH for dyes that are generally lipophilic and hence membrane permeable, but that are also weak acids or bases.55 Such probes tend to concentrate in either basic or acidic compartments, respectively, and are further discussed in section 3.3.1. The unfortunate reality is that cell loading remains poorly understood and still poses a challenge for many otherwise promising molecular sensors.
The retention of probes is also an important consideration, as over time all molecular probes will be expelled from cells, either by active extrusion or by passive leakage. Probes with poly carboxylates (such as the free acid form of AM-ester based probes) can be extruded by nonspecific anion transporters by a mechanism that is similar to organic anions.56 This process can be minimized by probenecid and sulfinpyrazone, which inhibit uric acid transport, and increase the retention of probes within cells.57 However, it is not only the free acid form that is expelled from cells as one multidrug resistance protein (MDR1) has been shown to extrude the AM-ester, but not the hydrolyzed free acid form of sensors, suggesting multiple mechanisms for expulsion of dyes.58 There are also many examples of leakage of fluorescein-based probes from cells, where the rate of leakage is often dependent on the charge of the molecule with more highly charged probes leaking more slowly.55,59
Genetically encoded sensors are most commonly incorporated into cells as plasmid DNA. Transient transfection of cells with plasmid DNA results in expression of genetically encoded sensors anywhere from 1 to 5 days, whereas viral transduction can result in the stable expression of a sensor due to genomic incorporation. The amount of sensor present in cells depends on the method of incorporation (transfection versus viral transduction) and the strength of the promoter that drives sensor expression.
3.2. Buffering
Defining the concentration of sensor in cells is an important consideration when evaluating the extent to which the sensor perturbs what you are trying to measure, the free, labile, or accessible metal pool. If the concentration of sensor is too high, this could lead to buffering of the metal, perturbation of cellular metal pools, and an inner filter effect. One method to determine whether the sensor perturbs the free ion pool is to measure the metal concentration as a function of sensor concentration. Such an analysis has been carried out for the small-molecule Zn2+ sensor FluoZin-3 AM in two different cell types54b,60 and the genetically encoded Zn2+ sensor (ZapCY platform) targeted to a variety of locations.60,61 For these two probes, it was revealed that treatment of cells with increasing concentrations of FluoZin-3 AM led to depletion of the Zn2+ pool, perhaps because high levels of accumulation of the dye led to intracellular concentrations that rivaled the buffered Zn2+ pool (i.e., hundreds of micromolar). On the other hand, the ZapCY sensor, which was present at concentrations in the low micromolar range, did not lead to measurable perturbations of the Zn2+ pool. While little is known about the buffering capacity of different kinds of cells for different metal ions, a reasonable guideline is to minimize the sensor concentration. Moreover, for quantitative measurements, that is, determination of metal ion concentrations within cells, it is essential to perform measurements at a range of concentrations to define whether the resulting measurements are influenced by the sensor concentration. Finally, the inner filter effect arises if the concentration of dye molecules is sufficiently high that the excitation light is not constant over the illumination spot.62 Again, inner filter effects can be minimized by minimizing dye concentrations.
3.3. Localization
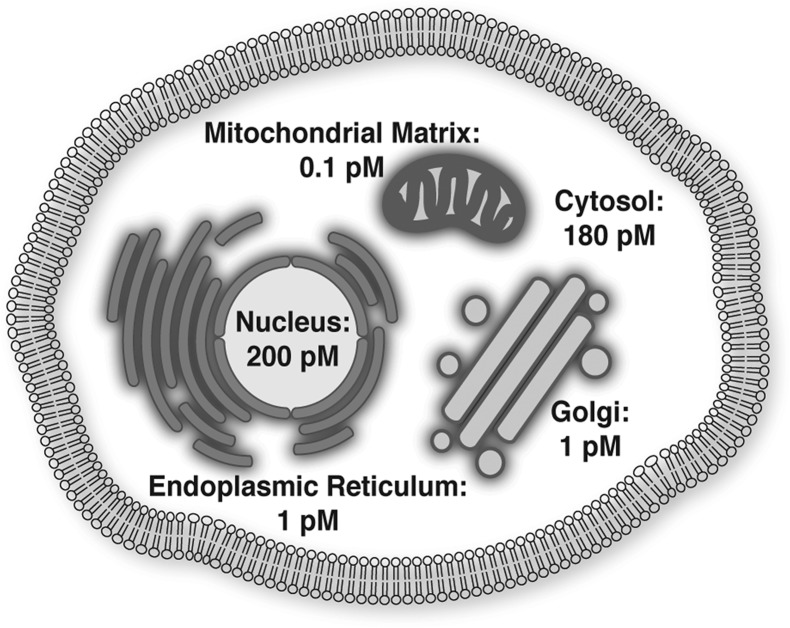
One of the primary applications of fluorescent sensors is that they permit measurement of metal ions in a spatially defined manner. Eukaryotic cells are by definition compartmentalized, containing a nucleus that is separated from the cytoplasm as well as membrane enclosed organelles. Even bacteria display compartmentalization with the cytoplasm separated from the periplasm. Compartmentalization leads to different chemical environments, with changes in pH, reduction potential, and of course biochemistry. It is well established that different metalloproteins and metalloenzymes localize to different cellular compartments, for example, zinc-dependent polymerases in the nucleus, iron–sulfur cluster biogenesis machinery in mitochondria, and manganese-dependent photosystem II in the thylakoid membrane of chloroplasts. Just as different cells and organisms have different metal requirements,2 so too will compartments within cells. In fact, even in cells with minimal compartmentalization such as bacteria, differences between metal availability in the cytosol and periplasm may play a critical role in ensuring proper metalation of proteins. In a proof of principle study, Robinson and co-workers demonstrated that the compartment in which a protein folds can determine which metal is bound to the protein, suggesting that one important feature of compartmentalization is to segregate metals to ensure that the right proteins have access to the right metals.63 One of the exciting applications of fluorescent metal sensors is the potential to visualize and quantify the accessible metal pool in the cytoplasm as well as in distinct compartments.
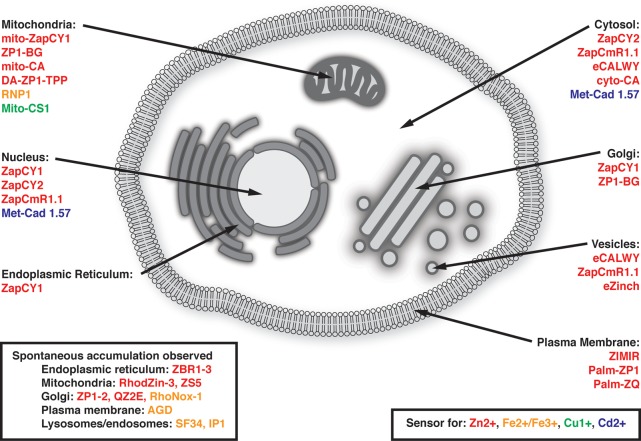
Given the compartmentalized nature of cells and the growing evidence that metal distribution is heterogeneous, it is essential to define the precise localization of fluorescent probes, and to assess whether the probe reports on multiple compartments. The location of a fluorescent sensor can result from either direct targeting or serendipitous localization. Localization is typically defined by comparing the colocalization of the probe with a well-established organelle marker and quantifying the overlap using some sort of correlation coefficient, such as Pearson’s correlation coefficient. While colocalization is a standard practice in light microscopy, it is important to note that not all cellular organelles have clearly defined and unique markers and likewise not all markers are restricted to single cellular compartment, or even have homogeneous distribution within a single compartment. A notorious example relates to defining vesicle populations, where RabGTPases generally mark vesicular populations, but these proteins are rarely restricted to a single type of vesicle.64 The discussion below will be divided into molecular probes, whose localization is governed by chemical nature of the probes, and genetically targeted sensors (genetically encoded and hybrid probes), whose localization is directed by signal peptides or fusion to other proteins. A summary of sensors that have been targeted to specific subcellular locations is presented in Figure 2.
Figure 2.
Diagram of sensors that have been targeted to specific organelles for subcellular metal ion imaging, either with peptide signaling motifs or with chemical groups known to associate with a particular subcellular location. Additionally, probes for which spontaneous accumulation in an organelle has been verified by colocalization studies are shown. More detailed descriptions of particular targeting strategies are discussed in later sections.
3.3.1. Factors Governing Localization of Molecular Probes
Even after many years of study on fluorescent indicators, particularly those for Ca2+ and pH, we still lack a comprehensive understanding of the principles that govern the intracellular distribution of fluorescent probes.55,59,65 Molecular probes must be sufficiently lipophilic to pass through the plasma membrane, but not so lipophilic that they accumulate within membranes. Plasma membrane permeability often means the probes will cross intracellular membranes as well, which, given the altered chemical environment of intracellular compartments, may lead to trapping of sensors in intracellular organelles. For some dyes, particularly those that are weak acids or bases, accumulation and hence cellular distribution depends on pH.55 The neutral form of the probe may readily diffuse through membranes; however, the charged form does not diffuse through membranes as readily, and instead accumulates in subcellular compartments. For example, weak bases that become protonated cations in acid compartments may be trapped in compartments such as endosomes, lysosomes, Golgi, and secretory vesicles, whereas weak acids that become anions in more basic compartments may accumulate in mitochondria.55
Many AM-ester based probes also exhibit complex and heterogeneous localization. In addition to passing through the plasma membrane, AM-ester probes can often penetrate intracellular membranes, and it has been shown that enzymatic hydrolysis of AM esters can occur within subcellular compartments.66 Moreover, de-esterification is often not complete, influencing both localization and dye retention.23b,54a,55,65 These probes have been detected in an array of intracellular compartments including endosomes/lysosomes, vesicles, Golgi, ER, mitochondria, and plasma membrane.23b,54a,60,65 Moreover, it is common for a probe to exhibit different spontaneous localization in different cell types.60,65 In an effort to predict properties of dye uptake and intracellular localization, Thompson et al. examined the molecular charge and lipophilicity/hydrophobicity by the logarithm of the octanol–water partition coefficient (logP) for a series of fluorescent probes, and found that both of these parameters play a role.65 In addition, some cell types can endocytose sensors, which may or may not be able to escape from the endosome. For example, AM-ester-based probes can be endocytosed and then hydrolyzed in the lumen of the vesicle, thus trapping the sensor in the endocytic pathway.66
Finally, tweaks in molecular design of small-molecule probes often result in changes in localization. A commonly employed technique for promoting accumulation of a probe into mitochondria is the incorporation of lipophilic delocalized cations such as phosphonium ions or use of positively charged rhodamine derivatives, whose uptake into mitochondria is enhanced by the negative mitochondrial membrane potential.67 However, as shown by Chyan et al., a cation such as triphenylphosphonium alone is not sufficient for mitochondrial targeting, as probes require a minimum level of lipophilicity to prevent endo/lysosomal accumulation.68 Two clever approaches for targeting the plasma membrane involved addition of dodecyl alkyl chains or a peptide-targeting motif to a Zn2+ sensor that facilitated the anchoring of the sensor on the extracellular side of the plasma membrane, facilitating measurement of Zn2+ release from cells.69 In another example that resulted in a serendipitous change in localization, recently developed benzoresorufrin-based probes accumulate in the ER, whereas similar fluorescein-based probes do not.70
Another potentially complicating factor that could influence intracellular properties and localization is solubility. Fahrni et al. recently demonstrated that a number of small-molecule Cu+ probes formed colloidal aggregates in aqueous buffer.71 While it remains to be seen whether this affects the cellular properties of these probes, it is an important reminder that all probes are prone to potential artifacts, and careful controls must be conducted to minimize artifacts.
Finally, spontaneous localization may change between different types of cells. This is well documented for calcium probes,65 but occurs for metal sensors as well. Qin et al. demonstrated that while the small-molecule sensor FluoZin-3 AM shows the strongest colocalization with the Golgi in HeLa cells, it shows the strongest colocalization with VAMP2 (a marker of vesicles) in cortical neurons.60 However, in both cell types, there was also FluoZin-3 present in the cytosol. Moreover, the bright signal of FluoZin-3 in the Golgi was unresponsive to perturbations of cellular Zn2+, revealing that the high fluorescence intensity resulted from a high dye concentration, rather than a high Zn2+ concentration.
While the uncertainty in dye localization can give rise to numerous artifacts, it is possible to empirically change experimental conditions (concentration of the probe, loading time and temperature, cell type) to minimize intracellular compartmentalization. Moreover, compartmentalization can be an advantage for measuring metal ions within compartments.
3.3.2. Genetic Targeting of Probes
The localization of genetically encoded probes and hybrid probes is defined by genetic targeting, such as attachment of the sensor to a signal peptide, or fusion to a protein of interest to direct the sensor to a particular location. Common targeting motifs are presented in Table 1. Such strategies can be used to localize a probe with high fidelity. Localization should always be confirmed by visual comparison with well-established organelle markers and quantification of colocalization, as sometimes genetic targeting fails to properly localize the probe.
Table 1. Signal Peptides and Fusions Commonly Used for Genetic Targetinga.
| targeted location | signal peptide (source: sequence) | refs |
|---|---|---|
| nucleus | NLS: PKKKRKVEDA (at C-terminus) | (73) |
| ER lumen | calreticulin ss: MLLSVPLLLGLLGLAAAD (at N-terminus) | (61a,73a,74) |
| bovine prolactin ss + 10 aa of mature domain: MDSKGSSQKAGSRLLLLLVVSNLLLCQGVVS-TPVCPNGPGN | ||
| KDEL (at C-terminus) | ||
| mitochondrial matrix | CytCOx ss: MSVLTPLLLRGLTGSARRLPVPRAKIHSLGDP (N-term) | (61b,75) |
| DAKAP1a ss: MAIQLRSLFPLALPGMLALLGWWWFFSRKK (N-term) | (73b) | |
| mitochondrial membrane | Tom20 ss: MVGRNSAIAAGVCGALFIGYCIYFDRKRRSDPN (N-term) | (73c,76) |
| Golgi lumen | fusion to GalT (at N-terminus) | (61a,77) |
| Golgi membrane | eNOS ss: MGNLSKSVAQEPGPPCGLGLGLGLGLCGKQCPA (N-term) | (73c,76) |
| plasma membrane, intracellular surface | MGCIKSKRKDNLNDDGVDMKT (at N-term, MyrPalm) | (73c,75,78) |
| KKKKKSKTKCVIM (at C-terminus, polybasic + Farn) | (73b) | |
| KLNPPDESGPGCMSCKCVLS (at C-terminus) | (79) | |
| MLCCMRRTKQVEKNDEDQKI (at N-terminus, PalmPalm) | (79) | |
| plasma membrane | secretion singal: MGTDTLLLWVLLLWVPGSTGD (N-terminus) | (80) |
| extracellular surface | transmembrane anchoring domain of PDGFR | |
| vesicles | VAMP2 (at N-terminus) fusion | (81) |
| synaptophysin fusion | (82) | |
| VGLUT-1 fusion | (82) | |
| endosomes | VAMP3 fusion | (81a) |
Abbreviations used in the table are as follows: NLS, nuclear localization signal; CytCOx, cytochrome c oxidase; Tom20, mitochondrial import receptor subunit Tom20; eNOS, endothelial nitric oxide synthase; GalT, human galactosyltransferase type II; MyrPalm, myristoylation and palmitoylation; Farn, farnesylation; PalmPalm palmitoylation and palmitoylation; PDGFR, platelet-derived growth factor receptor; VAMP2, vesicle associated membrane protein 2 (also known as synaptobrevin 2); VGLUT-1, vesicular glutamate transporter 1; VAMP3, vesicle associated membrane protein 3 (also known as synaptobrevin 3).
4. A Brief History of Visualizing Cellular Metal Ion Distribution with Probes
To place the current efforts in the development of metal sensors in perspective, it is instructive to look at how the field of mapping metals in biological organisms has evolved to where it is today. Figure 3 presents a historical timeline that highlights some of the key landmarks in the past 150 years, and we elaborate on these discoveries below. Transition metals were first shown to be necessary for life when Raulin demonstrated that zinc was essential for growth of the common bread mold Aspergillus niger.72 This discovery catalyzed active research into the concentration of different metal ions and their distribution throughout cells and tissues. The history of visualizing metal ions in cells begins with histological staining. Histology typically involves sectioning and staining cells or tissues before examination under a light or electron microscope. Specific structures can be visualized by staining with certain dyes, among which hematoxylin (nuclei) and eosin (cytoplasm) are some of the most widely used. One of the earliest histological stains for a biological trace metal is the Prussian blue method pioneered by Perls, who first described staining tissues for nonheme iron in 1867.15 Perls treated tissue samples first with potassium ferrocyanide followed by HCl. The acid released iron from the tissue, which then could react with the ferrocyanide ion to generate the insoluble Prussian blue precipitate. The resulting tissues samples were stained a vivid blue-green in the presence of iron. Around the same time, Quincke used ammonium sulfide to visualize iron in tissues as black iron sulfide.83 Another alternative, the Turnbull method, uses acid-ferricyanide instead of the acid-ferrocyanide reagent of Perls.84 The ferricyanide ion reacts with Fe2+ to produce the insoluble Turnbull blue precipitate. These basic methods are still employed today for the detection of nonheme iron but have been subjected to various optimizations and improvements, the details of which can be found elsewhere.85
Figure 3.
Timeline of historical developments in visualizing metal ions in cells.
At the end of the 19th century, one of first reports of copper distribution was enabled by the cytoplasmic dye hematoxylin to stain copper in diseased oysters.86 This dye was also used by Mendel and Bradley to visualize the distribution of metals in hepatic tissues of the sea snail Sycotypus canaliculatus.87 This study also employed sodium nitroprusside in what was the first demonstration of labile Zn2+ in these tissues. Although this technique suffered from low sensitivity and therefore attracted little attention at the time, the reaction was later shown to be specific for Zn2+ detection.88 These studies represent some of the earliest attempts to visualize the distribution of transition metals throughout tissues using exogenous probes, and they set the stage for imaging these ions in living cells with fluorescent sensors.
Subsequent improvements in histological staining methods allowed for new discoveries concerning the distribution of trace metals throughout tissues. Okamoto developed the use of rubenic acid-based methods for the detection of copper in the late 1930s.89 Although it can form colored complexes with other metal ions, notably Ni2+, Ag+, and Co2+, these complexes have different colors and solubility in acetate and ethanol than the dark green precipitate that forms upon reaction with Cu2+ ions. With a detection limit in the low micromolar range, this technique is not suitable for examining healthy concentrations of Cu2+ in tissues; however, it has been used to visualize excess Cu2+ accumulation in tissues from Wilson’s disease patients.90 In later years, rhodanine was established for the selective staining of Cu+ over divalent ions.91 Other methods for histochemical staining of copper include diethyldithiocarbamate, dithizone, and orcein, but all of these methods are only able to detect abnormally high concentrations of copper in tissues and often produced conflicting results.92
In the early 1940s, Okamoto applied dithizone for the histochemical visualization of Zn2+ in islets of Langerhans of the pancreas.93 For many years, this was one of the few histochemical methods available for Zn2+ visualization and was used to identify labile Zn2+ pools in numerous tissues. For example, the presence of labile Zn2+ in the brain was first demonstrated in 1955 by Maske and co-workers with intraperitonial dithizone injections.94
In addition to chromogenic dyes, autometallographic methods have been used for visualizing metals in tissues. Briefly, autometallography involves the silver-amplified detection of selenide or sulfide nanocrystals formed with endogenous or toxic metal ions. The large silver nanocrystals can be visualized via light or electron microscopy. This technique for transition metal detection was originally proposed by Timm in 1958,95 and has subsequently been optimized for visualization of labile Zn2+ pools, most notably by Danscher and co-workers.96 While such techniques have been used mostly for Zn2+ detection, they have been applied to other metals as well.97
The last 50 years have given rise to a gradual evolution in the development of fluorescent indicators for imaging metal ions in cells, tissues, and organisms. Such probes offer greater optical sensitivity than the chromogenic stains discussed above and the potential for imaging metal ions in live specimens. The use of fluorescent indicators for metal ions dates back to 1968, when Mahanand and Houck used 8-hydoxyquinoline as a fluorescent stain for Zn2+ in human plasma.98 In an attempt to find a stain that combined the sensitivity and resolution of silver-amplification methods with the specificity of dithizone, Frederickson and co-workers screened several quinoline-based compounds.99 In vitro experiments revealed that 6-methoxy-8-p-toluenesulfonamido-quinoline (TSQ) had the most intense fluorescence when complexed with Zn2+ as compared to related molecules. Building on previous work with Zn2+-containing neurons, this study highlighted the use of TSQ for selectively labeling Zn2+-rich regions of the central nervous system for both quantitative estimates of labile Zn2+ pools and qualitative assessments of localization. While TSQ improved upon earlier histological stains, it was never successfully used in live-cell experiments.
Live-cell imaging of metal ions began not with transition metals, but with the development of the Ca2+ sensor Quin2 by Roger Tsien in the early 1980s. At the time, a regulatory role for cytosolic Ca2+ had been proposed, but measurement of the free Ca2+ concentration in live cells was a challenging analytical problem. Tsien and co-workers developed a fluorescent probe with high affinity for Ca2+ over other ions such as Mg2+ and H+ that had a large increase in fluorescence intensity in response to Ca2+ binding,33 and a way to trap the probe in cells with nonpolar ester groups that were cleaved by intracellular esterases to reveal membrane-impermeable carboxylate anions.37 This new tool allowed for real-time, noninvasive measurements of cytoplasmic free Ca2+ in intact lymphocytes.100 Further optimization of the sensor platform revealed the possibility of systematically modifying the modular chelate-linker-fluorophore platform and resulted in the first ratiometric fluorescent sensors for Ca2+.101 Over 10 years after Quin2 was introduced, a similar tool became available for Zn2+. Building on the histofluorescence studies by Frederickson and the probe-trapping technique pioneered by Tsien, Zalewski and co-workers developed Zinquin by adding an ethyl ester to the 6-methoxy group of TSQ, improving its solubility and cellular retention.102 This new probe, the first fluorescent transition metal sensor used in live cells, was used to study the correlation between apoptosis and intracellular Zn2+ levels. To address some of the shortcomings of Zinquin, in particular phototoxicity caused by the UV-wavelength excitation light, many groups have worked on making the plethora of improved small-molecule sensors for Zn2+ that will be discussed in section 5. Another major development in the field was the creation of the first genetically encoded sensor for a transition metal (also for Zn2+) by the Eide laboratory in 2006.103 The design platform was based on previously developed Ca2+ sensors,73a and resulted in a probe that could be introduced into cells, tissues, or organisms as a DNA fragment that is subsequently transcribed and translated by cellular machinery into a fully functional sensor. Shortly thereafter, it was demonstrated that genetically encoded sensors could be targeted to subcellular compartments,80 offering the exciting possibility of constructing a complete map of labile metal ion distribution throughout the cell.
The historical timeline in Figure 3 illustrates how studies on Ca2+ set the framework for cellular imaging of metal ions. Tools for imaging labile Zn2+ have expanded substantially in the last 10 years. These probes possess a range of chemical and photophysical properties, and it is now possible to define the concentration of accessible Zn2+ in the cytosol, nucleus, ER, Golgi, and mitochondria, and visualize Zn2+ fluxes. Likewise, the arsenal of fluorescent probes for Cu+ continues to grow, and these probes are sufficiently sophisticated to permit imaging Cu+ in vivo. It is also apparent that probes for other metal ions have lagged substantially behind those for Zn2+ and Cu+. Still developments in the last two decades are promising. The lessons learned from the tuning of small-molecule sensors, as well as the development of genetically encoded sensors, should prove fruitful for expanding the repertoire of fluorescent sensors for other transition metals.
5. Probes for Zinc
5.1. Zinc Homeostasis
Zinc (Zn2+) is ubiquitous in all forms of life and is the second most abundant transition metal in the human body after iron. Zn2+ is not redox active in the cellular environment and is present in the +2 oxidation state.104 Mammalian cells sequester high levels of Zn2+ from the extracellular environment: an average cellular concentration of total Zn2+ in a mammalian cell is in the hundreds of micromolar range, while the concentration of Zn2+ in serum or plasma is approximately 1–10 μM.105 The vast majority of the cellular Zn2+ pool is bound to proteins, enzymes, metabolites, and other low molecular weight ligands such that the labile or accessible pool of Zn2+ in the cell is in the picomolar range.54b,61a,81b,106 This pool represents biological Zn2+ available for newly synthesized proteins or potential signaling functions. Bioinformatics work by Andreini et al. has suggested that up to 10% of the proteins encoded by the human genome contain a putative Zn2+ binding motif,107 underscoring the importance of Zn2+ in biological systems. Given the importance of Zn2+ in biology and the growing evidence that Zn2+ levels are both heterogeneous and dynamic, it is perhaps not surprising that Zn2+ sensors constitute the largest family of fluorescent indicators for transition metals. In the large arsenal of fluorescent Zn2+ sensors, there are probes in all three main classes: small-molecule probes, genetically encoded sensors, and hybrid probes. Small-molecule sensors constitute the largest class by far, and this group can be further subdivided into two categories: intensity-based probes, where Zn2+ binding induces an increase in fluorescence intensity, or ratiometric probes, where Zn2+ binding shifts the excitation and/or emission wavelength. There are multiple families of genetically encoded Zn2+ sensors based on FRET between two fluorescent proteins, and many of these have been targeted to different cellular locations. Finally, there are a handful of hybrid probes, which as the name suggests are a combination of the aforementioned classes and have both genetically encoded and exogenous elements. These hybrid probes include small molecules with targeting groups that interact with specific enzymes and genetically encoded proteins whose signal output is modulated by binding of a small-molecule fluorophore. This Review will focus on recent advances in these areas, but we encourage readers to refer to several recent reviews for further information regarding the development of Zn2+ probes.16,17,108
5.2. Small-Molecule Probes for Zn2+
5.2.1. Intensity-Based Probes
The majority of small-molecule probes for Zn2+ undergo a change in fluorescence intensity upon Zn2+ binding. Most of these sensors operate on the principle of PET between the fluorophore and the metal-binding group. In the absence of Zn2+, the fluorophore is quenched by PET from the electron-rich chelating group. Upon binding Zn2+, PET between the chelating moiety and the fluorophore is disrupted, leading to an increase in fluorescence emission. Manipulation of the fluorophore and binding motif platform can tune the efficiency of PET, affecting the brightness in both the bound and the unbound states of the sensor and therefore the magnitude of fluorescence change with Zn2+ binding. This can be accomplished by the incorporation of electron-withdrawing groups, alteration of the linker between the chelator and fluorophore, and by changing the nature of the PET “switch” itself. Table 2 presents a comprehensive list of the photophysical and biochemical parameters of many current small-molecule intensity-based Zn2+ sensors.
Table 2. Intensity-Based, Small-Molecule Fluorescent Sensors for Zn2+.
| excitation |
emission |
brightnessb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sensor | λfree (nm) | εfreea | λbound (nm) | εbounda | λfree (nm) | φfree | λbound (nm) | φbound | free | bound | DRc | KD (M) | ref |
| TSQ | 380 | ND | 380 | ND | 495 | ND | 495 | 0.1 | ND | ND | 100 | ND | (99) |
| Zinquin | 370 | ND | 370 | ND | 490 | ND | 490 | ND | ND | ND | ND | 2.0 × 10–10 | (102) |
| 3-Zn | 343 | 7.6 | 343 | 6.8 | 450 | 0.038 | 450 | 0.88 | 0.2888 | 5.984 | 23 | 5.0 × 10–7 | (109) |
| ZP1 | 515 | 67 | 507 | 78 | 531 | 0.38 | 527 | 0.87 | 25.46 | 67.86 | 3.1 | 7.0 × 10–10 | (110) |
| ZP2 | 498 | 44 | 490 | 53 | 522 | 0.25 | 517 | 0.92 | 11 | 48.76 | 6 | 5.0 × 10–10 | (111) |
| ZP3 | 502 | 75 | 492 | 85 | 521 | 0.15 | 516 | 0.92 | 11.25 | 78.2 | 6 | 7.0 × 10–10 | (112) |
| ZP4 | 506 | 61 | 495 | 67 | 521 | 0.06 | 515 | 0.34 | 3.66 | 22.78 | 5 | 6.5 × 10–10 | (113) |
| ZP5 | 504 | 83 | 495 | 91 | 520 | 0.29 | 517 | 0.48 | 24.07 | 43.68 | 1.6 | 5.0 × 10–10 | (114) |
| ZP6 | 506 | 89 | 495 | 98 | 519 | 0.1 | 517 | 0.34 | 8.9 | 33.32 | 2.7 | 5.0 × 10–10 | (114) |
| ZP7 | 505 | 68 | 495 | 77 | 521 | 0.04 | 517 | 0.05 | 2.72 | 3.85 | 0.4 | 5.0 × 10–10 | (114) |
| ZP8 | 500 | 81 | 489 | 78 | 516 | 0.03 | 510 | 0.35 | 2.43 | 27.3 | 11 | 6.0 × 10–10 | (115) |
| ZP9 | 505 | 51 | 494 | 44 | 526 | 0.02 | 521 | 0.41 | 1.02 | 18.04 | 12 | 6.9 × 10–7 | (116) |
| ZP10 | 506 | 55 | 497 | 45 | 523 | 0.08 | 516 | 0.33 | 4.4 | 14.85 | 10 | 1.9 × 10–6 | (116) |
| ZPF1 | 533 | 99 | 525 | 120 | 547 | 0.11 | 544 | 0.55 | 10.89 | 66 | 5 | 9.0 × 10–10 | (112) |
| ZPCl1 | 534 | 97 | 527 | 120 | 550 | 0.22 | 549 | 0.5 | 21.34 | 60 | 1.8 | 1.1 × 10–9 | (112) |
| ZPBr1 | 534 | 45 | 528 | 86 | 549 | 0.25 | 547 | 0.36 | 11.25 | 30.96 | 1.7 | 9.0 × 10–10 | (112) |
| ZPF3 | 520 | 87 | 510 | 93 | 537 | 0.14 | 533 | 0.6 | 12.18 | 55.8 | 3.6 | 8.0 × 10–10 | (112) |
| Me2ZP1 | 515 | 74 | 505 | 80.6 | 528 | 0.18 | 524 | 0.61 | 13.32 | 49.166 | 4 | 3.3 × 10–9 | (117) |
| Me4ZP1 | 516 | 56 | 506 | 47.4 | 529 | 0.17 | 525 | 0.35 | 9.52 | 16.59 | 2 | 6.0 × 10–7 | (117) |
| ZPP1 | 505 | ND | 500 | ND | 532 | 0.052 | 523 | 0.7 | ND | ND | 13 | 5.1 × 10–9 | (118) |
| DA-ZP1-TPP | 510 | ND | 510 | ND | 529 | 0.001 | 529 | 0.75 | ND | ND | 12 | 6.0 × 10–10 | (68) |
| ZS1 | 510 | 83.9 | 501 | 75.2 | 531 | 0.5 | 526 | 0.7 | 41.95 | 52.64 | 1.4 | ND | (119) |
| ZS2 | 499 | 66.9 | 489 | 67.6 | 523 | 0.39 | 516 | 0.69 | 26.091 | 46.644 | 2 | ND | (119) |
| ZS3 | 500 | 86.9 | ND | ND | 525 | 0.71 | 525 | NA | 61.699 | ND | 1 | ND | (119) |
| ZS4 | 507 | 81.1 | 495 | ND | 522 | 0.12 | 520 | 0.5 | 9.732 | ND | 4.5 | ND | (119) |
| ZS5 | 497 | 33 | 490 | 42 | 522 | 0.36 | 517 | 0.8 | 11.88 | 33.6 | 4.6 | 1.5 × 10–6 | (120) |
| ZS6 | 515 | ND | 505 | ND | 533 | 0.44 | 527 | 0.64 | ND | ND | 3.3 | ND | (120) |
| ZS7 | 500 | 62 | 490 | 66 | 524 | 0.25 | 518 | 0.79 | 15.5 | 52.14 | 2.7 | 3.7 × 10–6 | (120) |
| ZSF6 | 532 | 63 | 522 | 70 | 549 | 0.19 | 545 | 0.63 | 11.97 | 44.1 | 2 | 4.6 × 10–6 | (120) |
| ZSF7 | 521 | 62 | 511 | 70 | 535 | 0.24 | 527 | 0.68 | 14.88 | 47.6 | 2.5 | 5.0 × 10–6 | (120) |
| QZ1 | 505 | 68.9 | 498 | 69.8 | 524 | 0.024 | 524 | 0.78 | 1.6536 | 54.444 | 42 | 3.3 × 10–5 | (121) |
| QZ2 | 499 | 37.2 | 489 | 33.6 | 520 | 0.005 | 518 | 0.7 | 0.186 | 23.52 | 150 | 4.1 × 10–5 | (121) |
| QZ2E | 499 | 27.2 | 496 | 16 | 519 | 0.004 | 514 | 0.73 | 0.1088 | 11.68 | 120 | 1.8 × 10–3 | (122) |
| QZ2A | 498 | 64.1 | 492 | 40 | 515 | 0.012 | 515 | 0.51 | 0.7692 | 20.4 | 30 | 1.3 × 10–4 | (122) |
| FluoZin-1 | 496 | ND | 496 | ND | 515 | ND | 515 | ND | ND | ND | 200 | 7.8 × 10–6 | (123) |
| FluoZin-2 | 495 | ND | 495 | ND | 525 | ND | 525 | ND | ND | ND | 12 | 2.1 × 10–6 | (123) |
| FluoZin-3 | 495 | ND | 495 | ND | 516 | ND | 516 | ND | ND | ND | 200 | 1.5 × 10–8 | (123) |
| ZnAF-12 | 492 | 74 | 492 | 63 | 514 | 0.02 | 514 | 0.23 | 1.48 | 14.49 | 17 | 7.8 × 10–10 | (124) |
| ZnAF-23 | 492 | 78 | 492 | 76 | 514 | 0.02 | 514 | 0.32 | 1.56 | 24.32 | 51 | 2.7 × 10–9 | (124) |
| ZNAF-1F | 489 | 77 | 492 | 70 | 515 | 0.004 | 515 | 0.17 | 0.308 | 11.9 | 69 | 2.2 × 10–9 | (125) |
| ZnAF-2F | 490 | 74 | 492 | 73 | 515 | 0.006 | 515 | 0.24 | 0.444 | 17.52 | 60 | 5.5 × 10–9 | (125) |
| ZnAF-2M | 490 | 53 | 492 | 52 | 515 | 0.03 | 515 | 0.27 | 1.59 | 14.04 | 6.8 | 3.8 × 10–8 | (126) |
| ZnAF-2MM | 490 | 111 | 493 | 88 | 515 | 0.01 | 515 | 0.1 | 1.11 | 8.8 | 12.3 | 3.9 × 10–6 | (126) |
| ZnAF-3 | 490 | 71 | 493 | 62 | 515 | 0.03 | 515 | 0.38 | 2.13 | 23.56 | 10.4 | 7.9 × 10–7 | (126) |
| ZnAF-4 | 490 | 68 | 492 | 64 | 515 | 0.01 | 515 | 0.22 | 0.68 | 14.08 | 16.3 | 2.5 × 10–5 | (126) |
| ZnAF-5 | 490 | 64 | 492 | 43 | 515 | 0.004 | 515 | 0.21 | 0.256 | 9.03 | 34.3 | 6.0 × 10–4 | (126) |
| Newport Green DCF | 505 | ND | 505 | ND | 535 | ND | 535 | ND | ND | ND | 5 | 4.0 × 10–5 | (127) |
| Newport Green PDX | 495 | ND | 495 | ND | 520 | ND | 520 | ND | ND | ND | ND | 3.0 × 10–4 | (123) |
| ZIMIR | 493 | 73 | 493 | 73 | 515 | 0.0032 | 515 | 0.225 | 0.2336 | 16.425 | 70 | 4.5 × 10–6 | (69a) |
| ZnAB | 499 | ND | 499 | ND | 509 | 0.003 | 509 | 0.058 | ND | ND | ND | ND | (128) |
| BDA | 491 | 19.5 | 491 | 18 | 509 | 0.077 | 509 | 0.857 | 1.5015 | 15.426 | 10.5 | 1.0 × 10–9 | (129) |
| WZS | 499 | 17.1 | 499 | 17.1 | 550 | 0.03 | 550 | 0.19 | 0.513 | 3.249 | 6 | 6.2 × 10–10 | (130) |
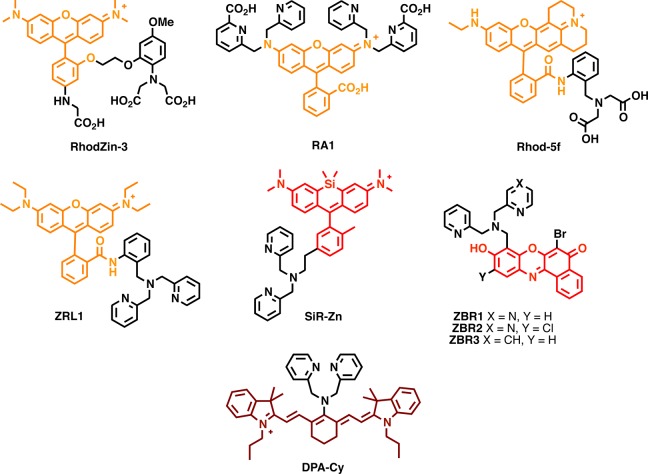
| RhodZin-3 | 550 | ND | 550 | ND | 575 | ND | 575 | ND | ND | ND | 75 | 6.5 × 10–5 | (67a) |
| RA1 | 535 | 45 | 535 | 1.3 | 561 | 0.7 | 561 | 0.78 | 31.5 | 1.014 | ND | 1.3 × 10–6 | (131) |
| ZRL1 | 569 | ND | 569 | 20.8 | 595 | <0.001 | 595 | 0.22 | ND | 4.576 | 220 | 7.3 × 10–5 | (132) |
| Rhod-5f | 571 | ND | 571 | ND | 594 | 0.28 | 594 | 0.13 | ND | ND | 11.6 | 4.1 × 10–6 | (133) |
| ZBR1 | 514 | 19.3 | 525 | 26.4 | 625 | 0.067 | 628 | 0.41 | 1.2931 | 10.824 | 8.4 | 6.9 × 10–10 | (70) |
| ZBR2 | 550 | 16.9 | 530 | 25.6 | 630 | 0.069 | 630 | 0.22 | 1.1661 | 5.632 | ND | 7.0 × 10–10 | (70) |
| ZBR3 | 530 | 13.3 | 535 | 19.3 | 623 | 0.342 | 628 | 0.6 | 4.5486 | 11.58 | ND | 1.0 × 10–12 | (70) |
| DPA-CY | 606 | 150 | 606 | 190 | 800 | 0.02 | 800 | 0.41 | 3 | 77.9 | 20 | 6.3 × 10–8 | (70) |
| SiR-Zn | 650 | 98 | 651 | 110 | 665 | 0.009 | 665 | 0.12 | 0.882 | 13.2 | 15 | 1.4 × 10–9 | (134) |
Molar extinction coefficients given as ε/103 M–1 cm–1.
Brightness is defined as the product of the molar extinction coefficient and the quantum yield (ε × φ).
There is no systematic way to present dynamic range (DR), so we encourage readers to refer to the original publications for more details about this value. For intensity-based probes, this number is generally the maximum fold change in fluorescence intensity upon Zn2+ binding. ND, not determined.
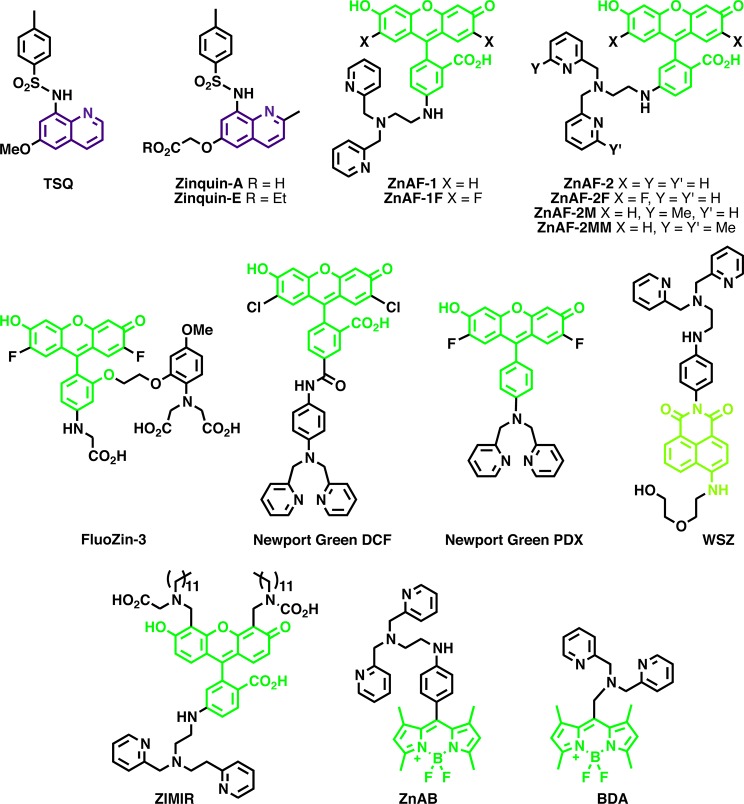
Early imaging studies of cellular Zn2+ were carried out with probes based on the UV-excitable quinoline fluorophore and a sulfonamide Zn2+ chelating group (Figure 4). The use of this class of probes began with histochemical studies in fixed tissues. In 1987, Frederickson and co-workers used N-(6-methoxy-8-quinolyl)-p-toluenesulfonamide to identify a pool of histochemically reactive Zn2+ in the vesicles of axon boutons.99 TSQ staining was found to not only correspond very well with previous studies of Zn2+ visualization in the brain, but improved on earlier histochemical methods by combining the sensitivity and resolution of silver-amplification methods (i.e., Timm’s stain) with the specificity of dithizone. Building on the work with TSQ, the related probe Zinquin was developed by Zalewski and co-workers as a probe of labile Zn2+ in living cells.102 This study found that decreased labile Zn2+ levels lead to apoptotic events in mammalian cells. Conversely, it appeared that increasing cellular Zn2+ levels could prevent DNA fragmentation upon pharmacological induction of apoptosis. While these probes permitted visualization of cellular pools of labile Zn2+ in living cells, they were hampered by their UV-range excitation wavelength, which leads to photodamage and high background fluorescence in the cell. Recent work from the Petering lab has demonstrated that these probes form ternary complexes with Zn2+-containing proteins.135 Thus, it appears that instead of imaging the free or labile pool of Zn2+ within cells, these probes actually image part of the Zn2+ proteome. While this indicates such probes do not report on the accessible Zn2+ pool, it suggests they may have an unintended use in examining the proteome. It is becoming increasing clear that within the complex environment of the cell, probes can be involved in interactions that are difficult to predict based on the conditions used for in vitro biochemical characterization. Similar studies on other small-molecule probes have not been reported, but it is possible that other probes interact with cellular components.135b
Figure 4.
Quinoline-, fluorescein-, 4-aminonapthalimide-, and BODIPY-based Zn2+ sensors.
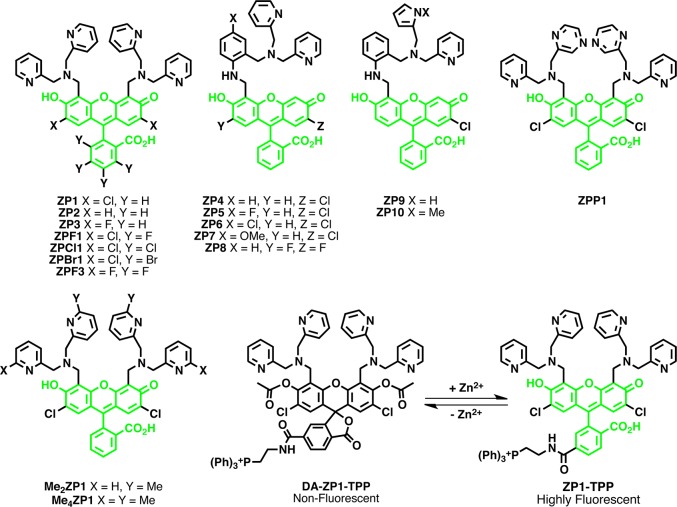
To overcome the limitations of these quinoline-based probes, there has been a surge in development of sensors based on other fluorophore platforms. Fluorescein, which has a high quantum yield and lower energy excitation and emission profiles more amenable for live-cell imaging, has been used to design numerous Zn2+ sensors. One of the largest families of fluorescein-based sensors is the Zinpyr or ZP family (Figure 5). In the past decade, this family of sensors has undergone extensive tuning of photophysical, chemical, and thermodynamic properties. The first iteration of this probe ZP1 featured a di-2-picolylamine (DPA) Zn2+ chelator and a dichlorofluorescein (DCF) fluorophore.136 This probe was more suited to live-cell imaging than previous quinoline-based probes as it featured excitation and emission wavelengths above 490 nm and could be passively incorporated into cells. ZP2 was created shortly thereafter in an attempt to lay out a more general strategy for the construction of fluorescein-based sensors.111 While ZP2 improved upon the dynamic range of its predecessor (6-fold versus 3.1-fold for ZP1), both probes still had relatively small changes in fluorescence intensity upon Zn2+ binding and were pH sensitive. In an effort to control the pH sensitivity of these sensors, Chang et al. explored the effect of electronegative substitution on the fluorescein backbone and generated ZP3, a new probe with a lower pKa (6.8) as compared to previous ZP sensors.112 ZP3 has a dynamic range similar to that of ZP2, but can be prepared in a single synthetic step instead of the many steps required for the construction of ZP2.112 By incorporating a modified Zn2+ binding moiety onto an unsymmetrically functionalized fluorescein scaffold, Burdette et al. generated ZP4, a sensor with lower background fluorescence that formed only mononuclear Zn2+ complexes.113 It was initially thought that this probe was unable to cross cell membranes, a property that was exploited for detailed imaging of damaged neurons: tissue sample preparation allowed the dye to enter brain slices, but only neurons damaged by Zn2+ release during seizures showed fluorescent staining. Visualization of such tissue damage would have been much more difficult with TSQ or even ZP1, which would stain healthy and damaged neurons indiscriminately. Follow-up work with ZP4 indicated that it may in fact be able to enter cells, albeit less efficiently than some other probes.114 Such studies highlight the need for rigorous experimentation before assigning localization of any small-molecule dye. Further work on this asymmetric scaffold led to the development of ZP5–7 and ZP8, which demonstrated how electron-withdrawing groups on the Zn2+ binding moiety and fluorophore could alter pH sensitivity and dynamic range.114,137 The binding affinity for Zn2+ could be manipulated by incorporating a pyrrole into the Zn2+-chelating group of the asymmetric probes (ZP9 and 10) or methylating four pyridyl groups in the symmetric ZP1 scaffold (Me4ZP1).116,117 ZPP1, created by replacing one pyridine at each DPA group of ZP1 with a pyrazine, featured lower background fluorescence, increased dynamic range (13-fold), and decreased affinity for Zn2+ than its predecessor.118
Figure 5.
The ZP family of Zn2+ sensors.
ZP1 has recently been delivered to the mitochondria by a Zn2+-depended ester cleavage reaction and tryphenylphosphonium (TPP) targeting.68 Addition of a TPP group is a widely used method of targeting a molecule to the mitochondria, but this strategy is dependent on mitochondrial membrane potential, and therefore the respiratory state of the cells can affect probe localization.67b,67c Conjugation of a TPP motif to the 6-position of the benzoic acid group of diacetylated ZP1 (DA-ZP1-TPP) allowed for successful delivery of the probe to mitochondria. DA-ZP1-TPP is nonfluorescent and resistant to intracellular esterases over a 2 h period, but Zn2+-mediated hydrolysis of the acetyl groups reveals ZP1-TPP, which localizes to mitochondria and has a 12-fold increase in fluorescence intensity in response to Zn2+. Using this probe, Chyan et al. were able to observe decreased mitochondrial Zn2+ uptake in cancerous prostate cells lines as compared to healthy cells.
Concurrent with the development of the ZP family, the Nagano laboratory developed another fluorescein-based sensor platform (Figure 4). One issue with early ZP sensors was high background fluorescence in the absence of a bound Zn2+ ion, due to incomplete PET quenching of the fluorophore in the apo-state. In an effort to reduce this background, the DPA chelating group was attached to various positions of the benzoic acid moiety.124 Amino-substituted fluoresceins have a very low quantum yield, so in the absence of Zn2+ this probe exhibits very low background fluorescence. The first generation of ZnAF probes, ZnAF-1 and ZnAF-2, featured low quantum yields in the absence of Zn2+ (0.022 and 0.023, respectively) and high turn-on responses to Zn2+ (17-fold and 51-fold increases in fluorescence intensity, respectively) and a pKa value of 6.2 for the phenolic hydroxyl group of the fluorophore. Although the original probes were not able to cross the cell membrane, diacetyl derivatives masked the negative charge on the probes, allowing them to be used to stain intracellular Zn2+. In an effort to lower the pKa and further decrease background fluorescence, ZnAF-1F and ZnAF-2F were created by substitution of fluorine at the ortho-position of the phenolic hydroxyl group.125 The probes had extremely low quantum yields in the absence of Zn2+: 0.004 for ZnAF-1F and 0.006 for ZnAF-2F. Additionally, Zn2+ binding increased the fluorescence by 69-fold for ZnAF-1F and 60-fold for ZnAF-2F, yielding some of the highest turn-on responses available. However, the quantum yields in the Zn2+-bound state of ZnAF-1F and ZnAF-2F are still fairly low, rendering the probes somewhat dim for intensity-based sensors. Addition of the fluorine atoms decreased the pKa of these probes to 4.9, affording stable fluorescence under neutral and slightly acidic conditions. Later, a suite of ZnAF probes with a range of affinities for Zn2+ from 10–10 to 10–3 M were developed by modifying the Zn2+ chelating group.126 To our knowledge, this family of probes still features some of the lowest levels of background fluorescence and largest magnitude turn-on responses upon Zn2+ binding of available probes.
Many more intensity-based Zn2+ probes have been developed in the past decade using fluorescein and other fluorophores (Figure 4). Several Zn2+ probes have been based on existing sensors for Ca2+, including FluoZin-3.123 FluoZin-3 in particular was generated by removing one of the acetate groups on the well-characterized Ca2+ chelator bis(o-aminophenoxy)-ethane-N,N,N′,N′-tetraacetic acid (BAPTA) to reduce affinity for Ca2+. FluoZin-3 has high affinity for Zn2+ (KD 15 nM), shows minimial Ca2+, and approximately a 200-fold fluorescence increase upon Zn2+ binding. Furthermore, the FluoZin-3 AM variant is membrane-permeable and trapped in cells upon cleavage of the AM groups by esterases. FluoZin-3 is a very widely used small-molecule Zn2+ sensor and has been used in hundreds of publications. The fluorescein-based dyes Newport Green DCF127 and Newport Green PDX123 have been developed with lower affinities for Zn2+ (KD = 1 μM for DCF and KD = 30–40 μM for PDX). In addition to the ZP family, the Lippard lab has also developed the Zinalkylpyr (ZAP),138 Zinspy (ZS),119,120 and QZ families of sensors (Figure 6).121,122 The ZAP analogues of ZP1 have an alkyl group in place of one of the Zn2+-binding picolyl moieties.138 This change increased the quantum yield of the probes in the Zn2+-free state such that they no longer exhibit a fluorescent response to Zn2+ binding. Although these analogues are not useful as live-cell probes, they helped demonstrate how the picolyl groups were involved in pH-dependent quenching in ZP sensors. The DPA ligands of the early ZP sensors possess high affinity for other first row transition metals such as Fe2+ and Cu2+; to improve selectivity, the ZS family was constructed with pyridyl-amine-thioether ligands for Zn2+.119 Early iterations of this sensor platform (ZS1–4) suffered from high background fluorescence and narrow dynamic range. To address these issues, the thioether ligands were replaced with thiophene moieties. These less basic groups are unable to coordinate Zn2+ and confer both lower background fluorescence (and thus higher dynamic range) and decreased affinity for Zn2+ to the new probes while maintaining the improved Zn2+ selectivity of the first ZS sensors. Other modifications of the Zn2+-chelating group led to the generation of QZ1 and QZ2, which used 8-aminoquinoline moieties to bind Zn2+.121 These sensors have exceptionally large dynamic ranges (42 and 150, respectively) and micromolar affinities for Zn2+, rendering them useful for exploring larger pools of labile Zn2+. Derivatives of these sensors were later generated that were cell trappable (QZ2E) and cell-impermeable (QZ2A).122
Figure 6.
The ZS, QZ, and ZAP families of Zn2+ sensors.
While the majority of Zn2+ sensors have used a fluorescein-based scaffold, many sensors have employed other fluorophore platforms, including coumarin,109,139 boron dipyrromethene (BODIPY) derivatives,128,129 4-aminonapthalimide,130 rhodamine,131−134 and tricarbocyanine.140 The use of coumarins for Zn2+ probes in live-cell applications has been largely underdeveloped, but a handful of these probes have been generated and tested in vitro. In particular, the DPA-coumarin probe 3-Zn2+ has a respectable dynamic range (23-fold) and good selectivity toward Zn2+, but was never used in cells.139 BODIPY-based sensors are generally less sensitive to pH than those based on fluorescein: the probe BDA has a very low pKa (2.1), high quantum yield in the Zn2+-bound state (0.857), nanomolar affinity for Zn2+, and was used to visualize Zn2+ in mammalian cells.129 In the past few years, several rhodamine-based Zn2+ probes have been developed. Rhodamine-based platforms feature longer-wavelength excitation and emission profiles than fluoresceins, generally good quantum yields, and can be highly photostable. The probes RA1 and Rhod-5f were well-characterized in vitro, but never used in live cells.131,133 On the other hand, ZRL1 is cell-permeable and was used to detect Zn2+ in HeLa cells.132 One particularly interesting rhodamine probe contains silicon at the 10 position of the xanthene chromophore (SiR-Zn).134 Placement of the group 14 element in the rhodamine background resulted in a probe that was active in the near-infrared region (near-IR) while retaining the high quantum yield and water solubility of original rhodamines. Near-IR probes are attractive because their low-energy excitation and emission profiles are well-suited for imaging deeper in tissues and have reduced interference from cellular autofluoresence. SiR-Zn features nanomolar affinity for Zn2+, a 15-fold increase in fluorescence upon Zn2+ binding, and is functional in cells. Another near-IR probe uses DPA as a Zn2+-binding group and tricarbocyanine as a fluorophore and was also shown to be functional in cells.140 These sensors based on longer wavelength fluorophores are shown in Figure 7. Expanding the repertoire of probes for Zn2+ by exploring new fluorophores opens the possibility of simultaneously monitoring different metal pools, whether for different metals or different concentrations of the same metal.
Figure 7.
Rhodamine-, resorufin-, and cyanine-based small-molecule Zn2+ sensors.
One complication with small-molecule sensors is their unpredictable localization within a cell. For example, FluoZin-3 AM has been shown to localize to the cytosol,141 Golgi,60 lysosomes,142 and vesicles,143 with different locations in different cell types.60 While this has led to the use of FluoZin-3 for monitoring multiple Zn2+ pools, it can lead to complications because the probe may report on Zn2+ within multiple cellular locations. Several strategies have been used to address this. One of the first available targeted sensors for Zn2+ was RhodZin-3, which accumulates in active mitochondria as confirmed by colocalization with MitoTracker dye.67a,123 This probe was generated by replacing the fluorescein group of FluoZin-3 with the positively charged rhodamine fluorophore, which accumulates in mitochondria due to the negative membrane potential. However, this probe requires proper mitochondrial membrane potential for localization, making it dependent on the metabolic state of the cell.80
In an effort to design probes with longer-wavelength excitation and emission profiles more suitable for prolonged live-cell imaging experiments, Lin and co-workers developed the benzoresorufin-based probes ZBR1–3 (Figure 7).70 Intriguingly, colocalization experiments with a number of established dyes in several cell lines revealed that these probes localized spontaneously to the ER. These probes were used to visualize labile Zn2+ released from the ER in response to peroxynitrite-induced stress in neural stem cells. Recently, Li and co-workers developed ZIMIR, a sensor displayed on the extracellular side of the membrane (Figure 4).69a The probe consists of fluorescein attached to a DPA Zn2+-binding moiety and two dodecyl alkyl chains that anchor it in the plasma membrane. Because it is not membrane permeable, the probe remains anchored to the extracellular side of the cell membrane and was used to detect Zn2+ release from insulin-secreting cells. An alternative method of localizing sensors to the plasma membrane was used to construct Palm-ZP1 and Palm-ZQ.69b These probes feature a peptide with an N-terminal palmitoyl group, a polyproline helix, and two Asp residues covalently attached to Zinpyr-1 (Palm-ZP1) or Zinquin (Palm-ZQ) with a C-terminal Lys residue. These sensors were used to visualize Zn2+ addition to the extracellular milieu of HeLa and prostate cells. The modularity of this approach led to the proposal that this system could, in theory, be used to attach other peptide targeting motifs to other small-molecule Zn2+ sensors.
The intensity-based small-molecule probes described above have been instrumental in revealing exciting new processes in Zn2+ biology. To emphasize the kind of information that can be learned using small-molecule Zn2+ sensors and highlight the complexity of the studies that can be carried out, we profile four examples of how small-molecule sensors have transformed our understanding of zinc homeostasis and signaling. In the first example, ZP1 was used in Arabadopsis to study the effects of Zn2+-deficiency mutations.144 Although Zinquin was previously used to study Zn2+ excretion from the roots of tobacco,145 the work by Sinclair et al. was the first reported use of a small-molecule sensor to visualize Zn2+ within plants. They studied how Zn2+ localization in plants was affected by mutations in two transporters that lead to Zn2+ accumulation in root tissue and a Zn2+-deficient growth phenotype due to lack of Zn2+ translocation throughout the plant. Treatment of wild-type seedlings with ZP1 revealed Zn2+ localization in the xylem, whereas mutant plants showed high fluorescence in pericycle cells adjacent to the phloem. Furthermore, treatment with TPEN or exogenous ZnCl2 led to expected changes in ZP1 fluorescence, indicating that the probe was functional in Arabadopsis. This work demonstrated the utility of small-molecule Zn2+ sensor as tools to investigate Zn2+ homeostasis in plants.
In the next example, the Kornfeld laboratory used FluoZin-3 to visualize Zn2+ in the gut granules of C. elegans,146 demonstrating the feasibility of using this small-molecule Zn2+ probe and imaging Zn2+ in an optically transparent living organism. By simply incubating C. elegans on plates supplemented with FluoZin-3 AM, distinct fluorescent puncta could be seen. Treatment with additional Zn2+ or the Zn2+ chelator N,N,N′,N′-tetrakis-(2-pyridylmethyl)-ethylenediamine (TPEN) increased or decreased FluoZin-3 fluorescence, respectively, indicating that the dye was monitoring accessible Zn2+ pools and not aggregating in the subcellular compartments. The goal of the study was to define the relationship between cellular storage of excess Zn2+ and the response to Zn2+ deficiency at the organismal level. By using the Zn2+ probe in conjunction with LysoTracker and fluorescent protein fusions of lysosomal proteins, Kornfeld and co-workers were able to see a remarkable bilobal morphology of the gut granules when C. elegans were fed a high Zn2+ diet. This led to the hypothesis that these granules may be storing excess Zn2+ for later utilization during times of Zn2+ deficiency. To this end, they investigated the genetic pathway for the formation of these bilobal granules and showed Zn2+ could be indeed be mobilized from these cellular stores in response to Zn2+ deficiency. This result suggested that Zn2+ may be sequestered into these granules when Zn2+ is abundant (high Zn2+ diet) and can be mobilized when the worms are switched to a low Zn2+ diet.
In the third example, the O’Halloran laboratory used FluoZin-3 to study Zn2+ dynamics during oocyte fertilization, demonstrating the power of time lapse imaging and revealing exquisite Zn2+ transients.147 Using a cell-impermeable version of the dye, they were able to observe a release of Zn2+ into the extracellular environment upon fertilization or chemical activation. These bursts of Zn2+ were dubbed sparks, by analogy to Ca2+ sparks produced upon release of Ca2+ from the ER. These sparks were also associated with an intracellular Ca2+ signal, which was monitored by using a cell-permeable Ca2+ probe in conjunction with the impermeable FluoZin-3. Furthermore, the Zn2+ sparks did not occur without the Ca2+ transient, suggesting some level of coordination between the signaling and control of these two metal ions. Using a combination of X-ray fluorescence microscopy and live-cell imaging with intracellular FluoZin-3 and Zinquin, the distribution of Zn2+ was mapped to cortically polarized puncta within the cell. When cellular Zn2+ levels were elevated by treatment with Zn2+-pyrithione after activation, the eggs re-established metaphase arrest. Conversely, chelation of intracellular Zn2+ allowed for cell cycle resumption. Visualization of the extracellular Zn2+ sparks, as well as pharmacological manipulation of Zn2+ levels, gave way to a model where a decrease in Zn2+ availability for the oocyte is necessary for proper cell cycle resumption after fertilization.
In a final example, a recent clinical application of a fluorescent Zn2+ probe came from the Lippard lab. Ghosh et al. used a new sensor from the Zinpyr family (ZPP1) to look at Zn2+ levels in prostate cancer.148 In a cell culture model, they found significantly decreased ZPP1 fluorescence in a cancerous versus healthy prostate cell line when Zn2+ was added. Furthermore, they found ZPP1 fluorescence accumulated in the prostate of mice injected with ZPP1 by both epifluorescence whole-body imaging and intravital microscopy of dissected glands. Co-injection with ZPP1 and Zn2+ chelator revealed that the sensor responded specifically to Zn2+ in the animals. The group found substantially decreased ZPP1 fluorescence in a mouse model of prostate cancer, leading them to suggest that Zn2+ levels could potentially be used as an imaging biomarker for detection and progression of prostate cancer.
5.2.2. Ratiometric Probes for Zn2+
For ratiometric Zn2+ probes, Zn2+ binding alters the excitation wavelength, emission wavelength, or both. With such probes, fluorescent images are typically collected at the wavelength maxima for both the free and the bound states, and fluorescence changes are reported as a ratio of the fluorescence intensity at the two wavelengths. While this approach minimizes artifacts from cellular movements, sample thickness, and sensor concentration, these probes typically have smaller changes in signal upon Zn2+ binding than intensity-based probes. Furthermore, acquisition of images at two different wavelengths requires more sophisticated microscopy instrumentation. However, a major advantage of this class of probes is that they are more suitable for accurate quantification of Zn2+ levels.
There is currently a much more limited repertoire of ratiometric small-molecule probes than intensity-based ones, likely because of the challenge in engineering probes that undergo a shift in wavelength upon Zn2+ binding. As such, this class of probes has undergone far less optimization in terms of tuning photophysical and Zn2+-binding properties. For many of these probes, Zn2+ binding alters the electronic structure of the molecule, thus modifying internal charge transfer or excited state proton transfer, which in turn affects the excitation and/or emission profiles.17b The Ca2+ sensors fura-2 and indo-1 have been modified for use as ratiometric Zn2+ sensors. In the presence of increasing Zn2+ levels, FuraZin exhibits an excitation wavelength shift from 378 to 330 nm, while IndoZin shifts emission wavelength from 480 to 395 nm.123 The O’Halloran laboratory developed Zinbo-5, a probe built around a fluorescent benzoxazole core.149 This probe has nanomolar affinity for Zn2+, is functional in cells, and can be used in two-photon experiments. Upon Zn2+ binding, the emission wavelength undergoes a red shift from 407 to 443 nm. This probe was used to image exogenously added Zn2+ in fibroblast cells, as well as Zn2+ fluxes in Plasmodium falciparum parasites.150 The parameters of these and a handful of other ratiometric sensors are shown in Table 3.131,139,151
Table 3. Ratiometric Small-Molecule Zn2+ Sensors.
| excitation |
emission |
brightnessb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| name | λfree (nm) | εfreea | λbound (nm) | εbounda | λfree (nm) | φfree | λbound (nm) | φbound | free | bound | KD (M) | DR (Rmax/Rmin) | ref |
| FuraZin | 378 | ND | 330 | ND | 510 | ND | 510 | ND | ND | ND | 2.1 × 10–6 | 9 | (123) |
| IndoZin | 350 | ND | 350 | ND | 480 | ND | 395 | ND | ND | ND | 3.0 × 10–6 | ND | (123) |
| ZnAF-R1 | 359 | ND | 329 | ND | 532 | 0.088 | 528 | 0.031 | ND | 10.199 | 7.9 × 10–10 | ND | (151a) |
| ZnAF-R2 | 365 | ND | 335 | ND | 495 | 0.17 | 495 | 0.1 | ND | 33.5 | 2.8 × 10–9 | 7 | (151a) |
| Zinbo-5 | 337 | ND | 376 | ND | 407 | 0.02 | 443 | 0.1 | ND | 37.6 | 2.2 × 10–9 | 33 | (150) |
| ZNP1 | 503/539 | 7.2/6.7 | 547 | ND | 528/604 | 0.02 | 624 | 0.05 | ND | ND | 5.5 × 10–10 | 17.8 | (151b) |
| RF3 | 514 | 9.5 | 495 | 4.4 | 540 | 0.62 | 523 | 0.52 | 5.89 | 257.4 | 2.2 × 10–5 | 2.4 | (131) |
| DIPCY | 627 | 70 | 671 | 85 | 758 | 0.02 | 765 | 0.02 | 1.4 | 13.42 | 9.8 × 10–8 | 1.5 | (151c) |
| 4-Zn | 400 | 16.9 | 431 | ND | 484 | 0.64 | 505 | ND | 10816 | ND | 5.0 × 10–7 | 17.8 | (139) |
| CZ1 | 445 | 37.2 | 445 | 41 | 488 | 0.01 | 488 | 0.01 | 0.372 | 4.45 | 2.5 × 10–10 | 8 | (152a) |
| 505 | 38.6 | 505 | 38.1 | 534 | 0.02 | 534 | 0.04 | 0.772 | 20.2 | ||||
| CZ2 | 450 | 26 | 448 | 26.6 | 490 | 0.01 | 490 | 0.02 | 0.26 | 8.96 | 2.5 × 10–10 | 4 | (152b) |
| 526 | 22.4 | 521 | 24.3 | 535 | 0.01 | 535 | 0.04 | 0.224 | 20.84 | ||||
Molar extinction coefficients given as ε/103 M–1 cm–1.
Brightness is defined as the product of the molar extinction coefficient and the quantum yield (ε × φ). ND, not determined.
The Coumazin sensors use a different mechanism: the dyes Zinpyr (ZP) and coumarin are joined together by an ester linker that can be cleaved by intracellular esterases upon internalization of the probe.152 The esterases split the original molecule into ZP and coumarin fragments. The ZP fluorescence intensity changes in response to Zn2+ levels, but the coumarin intensity stays constant and thus acts as an internal standard. One can take the ratio of the correct optical channel for each fluorophore, but careful controls and analysis must be taken to ensure that the dyes are not differentially localized or extruded from the cell. Representative ratiometric probes for Zn2+ are shown in Figure 8.
Figure 8.
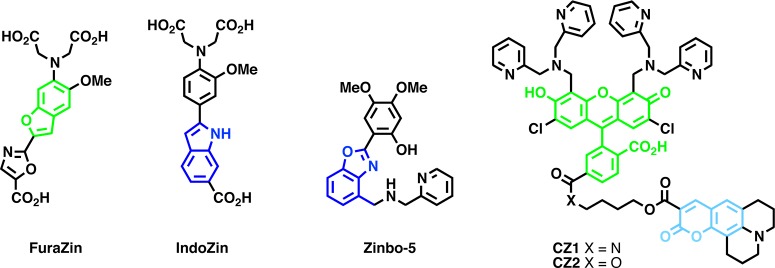
Ratiometric small-molecule Zn2+ sensors.
5.3. Genetically Encoded Sensors for Zn2+
Recently, significant work has led to the generation of Zn2+ sensors based entirely on protein or peptide motifs. Such constructs can be introduced into cells, tissues, or whole organisms as DNA by transient transfection or viral transduction. The sensors are then transcribed and translated by the machinery of the cell and do not require the addition of any exogenous cofactors for functionality. Currently, all genetically encoded Zn2+ sensors operate by Förster resonance energy transfer (FRET) between donor and acceptor fluorescent proteins (FPs). As a general design, the donor and acceptor FPs are joined by a domain that binds Zn2+ and changes conformation in such a way that the FRET efficiency is altered (Figure 9A,B). Thus, changes in Zn2+ levels can be monitored by changes in FRET efficiency. Experimentally, researchers excite the donor fluorophore and measure the resulting emission from the acceptor fluorophore, and then take the ratio (R) of FRET emission intensity to donor emission intensity. The ratiometric nature of these sensors means they can allow for more accurate quantification of labile Zn2+ levels than intensity-based sensors. The overall sensitivity and dynamic range are defined by the ΔR and Rmax/Rmin parameters. Current genetically encoded Zn2+ sensors and their biophysical parameters are summarized in Table 4.
Figure 9.
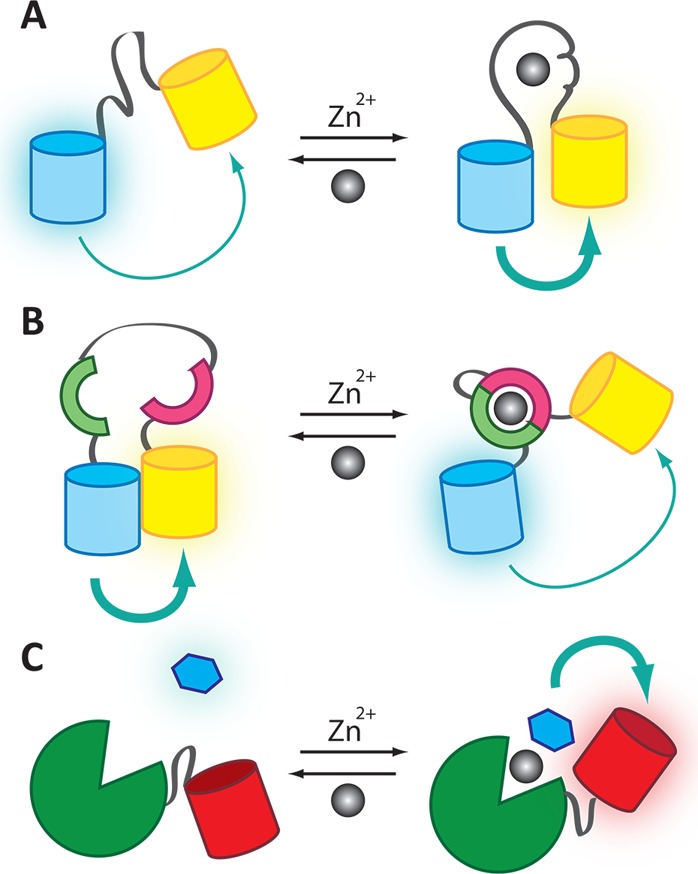
Mechanisms of metal ion sensing by genetically encoded and hybrid probes for Zn2+. (A) The Zap and Zif families consist of one or two Zn2+-finger domains between two FPs. Zn2+ binding induces a conformational change in the Zn2+-finger that leads to a change in FRET ratio. (B) The eCALWY family uses Zn2+ binding domains from Atox1 and WD4. The two FPs associate in the absence of Zn2+, but Zn2+ binding causes association of the binding domains and reduces the FRET efficiency. (C) The hybrid probe CA-FP has an FP linked to CA. When Zn2+ binds to CA, an exogenously added dapoxyl sulfonamide (blue hexagon) can bind to an open site on the Zn2+ ion, leading to a FRET response between the small-molecule fluorophore and the FP.
Table 4. Genetically Encoded Zn2+ Sensors.
| FP |
excitation |
emission |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| name | donor | acceptor | λdonor | εdonora | λacceptor | εacceptora | λdonor | .φdonor | λacceptor | φacceptor | KD (M) | DR (Rmax/Rmin) | ref |
| ZifCY1 | ECFP | mCitrine | 439 | 32.5 | 516 | 77.0 | 476 | 0.40 | 529 | 0.57 | 1.0 × 10–6 | 1.4 | (80) |
| ZifCY2 | ECFP | mCitrine | 439 | 32.5 | 516 | 77.0 | 476 | 0.40 | 529 | 0.57 | 1.0 × 10–4 | 4.0 | (80) |
| ZapCY1 | ECFP | mCitrine | 439 | 32.5 | 516 | 77.0 | 476 | 0.40 | 529 | 0.57 | 2.5 × 10–12 | 3.0 | (61a) |
| ZapCY2 | ECFP | mCitrine | 439 | 32.5 | 516 | 77.0 | 476 | 0.40 | 529 | 0.57 | 8.1 × 10–10 | 1.5 | (61a) |
| eCALWY-1 | Cerulean | Citrine | 433 | 43.0 | 516 | 77.0 | 475 | 0.62 | 529 | 0.57 | 2.0 × 10–12 | 2.0 | (81b) |
| eCALWY-2 | Cerulean | Citrine | 433 | 43.0 | 516 | 77.0 | 475 | 0.62 | 529 | 0.57 | 9.0 × 10–12 | 2.0 | (81b) |
| eCALWY-3 | Cerulean | Citrine | 433 | 43.0 | 516 | 77.0 | 475 | 0.62 | 529 | 0.57 | 4.5 × 10–11 | 1.7 | (81b) |
| eCALWY-4 | Cerulean | Citrine | 433 | 43.0 | 516 | 77.0 | 475 | 0.62 | 529 | 0.57 | 6.3 × 10–10 | 2.0 | (81b) |
| eCALWY-5 | Cerulean | Citrine | 433 | 43.0 | 516 | 77.0 | 475 | 0.62 | 529 | 0.57 | 1.8 × 10–9 | 1.8 | (81b) |
| eCALWY-6 | Cerulean | Citrine | 433 | 43.0 | 516 | 77.0 | 475 | 0.62 | 529 | 0.57 | 2.9 × 10–9 | 1.8 | (81b) |
| eZinCH | Cerulean | Citrine | 433 | 43.0 | 516 | 77.0 | 475 | 0.62 | 529 | 0.57 | 8.0 × 10–6 | 4.0 | (81b) |
| ZapSM2 | tSapphire | mKO | 399 | 44.0 | 548 | 51.6 | 511 | 0.60 | 559 | 0.60 | ND | 1.1 | (153) |
| ZapSR2 | tSapphire | tagRFP | 399 | 44.0 | 555 | 98.0 | 511 | 0.60 | 584 | 0.41 | ND | 1.2 | (153) |
| ZapOC2 | mOrange2 | mCherry | 549 | 58.0 | 587 | 72.0 | 565 | 0.60 | 610 | 0.022 | ND | 1.1 | (153) |
| ZapOK2 | mOrange2 | mKate | 549 | 58.0 | 588 | 31.5 | 565 | 0.60 | 635 | 0.28 | ND | 1.1 | (153) |
| ZapCmR1.1 | Clover | mRuby2 | 505 | 111 | 559 | 113 | 515 | 0.77 | 600 | 0.38 | ND | 1.5 | (153) |
Molar extinction coefficients given as ε/103 M–1 cm–1. ND, not determined.
The first genetically encoded sensors to monitor Zn2+ in cells were developed by the Eide laboratory and consisted of pairs of Zn2+ fingers from the yeast transcription factor Zap1 between CFP and YFP.103 These sensors were expressed in yeast and demonstrated that manipulation of Zn2+ levels could induce a change in FRET signal, thus demonstrating the feasibility of such a sensor platform. Merkx and co-workers introduced an alternative design strategy in their CALWY family of sensors.154 Instead of a Zn2+ finger motif that folds into a compact three-dimensional structure in the presence of Zn2+, these sensors rely on Zn2+-induced association of metal-binding domains from the copper ATPase ATP7B (fourth domain referred to as WD4) and the copper chaperone protein Atox1. The name of these sensors derives from the molecular components: CFP-Atox1-Linker-WD4-YFP. Through engineering of the metal binding domains and linker region, the Merkx group was able to generate a panel of sensors that was specific for Zn2+ and had a wide range of affinities. While these first generation sensors were never tested in cells, they showed the functionality of this platform. By enhancing the dynamic range, the Merkx laboratory created the eCALWY family and used these improved sensors to measure cytosolic Zn2+ levels in a variety of mammalian cell types.81b The Palmer lab has continued work on the Zn2+ finger-based sensor platform. Current sensors include the ZifCY and ZapCY family that feature single or double Zn2+ fingers derived from the transcription factors Zif268 or Zap1, respectively, and a cyan-yellow FRET pair (hence the designation “CY”) comprised of a truncated CFP and citrine variant of YFP or circularly permuted Venus FP.61,80,153 By mutating the metal ion-coordinating residues, the lab has generated sensors with affinities for Zn2+ that range from a Kd of 2.5 pM to hundreds of micromolar, and used the sensors to measure Zn2+ in a variety of cell types. Both the Palmer and the Merkx laboratories have enhanced the dynamic range and other properties of their sensors by optimizing the linker between the FPs and Zn2+ binding domains, manipulating the dimerization tendency of the FPs, and exploring alternate FP FRET pairs.
While the majority of genetically encoded Zn2+ sensors utilize CFP and YFP variants as the FRET pair, there have been recent attempts to develop alternatively colored sensors based on green-red or orange-red platforms. Two advantages of this system over the CFP-YFP platform are an increase in theoretical brightness and ability to excite the donor with a common 488 nm laser line. Additionally, an expanded color palette of sensors can allow for simultaneous imaging of Zn2+ in different subcellular compartments. The Palmer lab generated sensors that used mOrange2/mCherry as well as Clover/mRuby2, a newly designed FRET pair.153,155 These sensors, ZapOC and ZapCmR, were used in conjunction with ZapCY sensors to monitor Zn2+ uptake into the nucleus and other organelles simultaneously. Additionally, the Merkx laboratory developed redCALWY-1 and redCALWY-4 by introducing an R125I mutation in both fluorescent proteins that promotes self-association of the two FPs.156 Dimerization of the two FPs and Zn2+-induced association of the Atox1 and WD4 domains are mutually exclusive, so Zn2+ binding decreases the FRET ratio of this sensor platform.
A major advantage of genetically encoded sensors is the ability for relatively easy and precise targeting to specific subcellular compartments. By incorporating localization sequences such as those listed in Table 1 into the constructs, the Palmer lab has been able to target sensors to the cytoplasm, nucleus, endoplasmic reticulum (ER), Golgi apparatus, mitochondria, and extracellular side of the plasma membrane in various cell types.61,80,153 Meanwhile, the Merkx lab targeted their eCALWY sensors to vesicles by fusing the sensor to the single-pass transmembrane protein synaptobrevin 2, also known as vesicle-associated membrane protein 2 (VAMP2).81b As emphasized previously, when using a targeted sensor, it is critical to verify proper localization by comparing sensor expression with established organelle markers. Additionally, it is necessary to carefully inspect images to ensure expression of the sensor does not disrupt proper organelle structure. For example, Snapp and co-workers have observed altered ER morphology in cells that express weakly dimerizing FPs in the secretory pathway.157
Because of their ratiometic nature and the fact that they do not appear to perturb accessible Zn2+ pools, ZapCY sensors targeted to different organelles within mammalian cells have revealed substantial heterogeneity in the spatial distribution of accessible Zn2+ pools. As shown in Figure 10, the level of buffered or accessible Zn2+ in the ER and Golgi of HeLa cells is approximately 100-fold lower than in the cytosol, and the accessible pool is lower still in the matrix of mitochondria. An important point to consider is that fluorescent sensors only access a subset of the total Zn2+ pool, the so-called accessible/buffered/labile pool, and a full accounting of the Zn2+ status of these organelles would require measurement of the total Zn2+ and/or the tightly bound pool as a complement to the above studies. Intriguingly, measurements on different types of cells have revealed different levels of buffered Zn2+ in different cell types, for example, a decrease in cytosolic Zn2+ in RWPE1 normal prostate cells as compared to an increase in a cancerous prostate cell line LNCaP,60 and an increase in Zn2+ within the mitochondrial matrix of insulin secreting Min6 cells and neurons as compared to other cell types measured.61b Furthermore, ER-targeted ZapCY1 helped reveal a degree of coregulation and crosstalk between Ca2+ and Zn2+ in the ER.61a Influx of Ca2+ into the cytosol led to a concomitant loss of Zn2+ from the ER. Similarly, exogenously added Zn2+ led to Ca2+ release from the ER. While the precise mechanisms behind these processes are still unclear, this study provides some direct evidence that fluctuations in Ca2+ or Zn2+ levels can affect the homeostasis of other metal ions within the cell.
Figure 10.
Heterogeneous distribution of Zn2+ throughout the mammalian cell. Genetically encoded sensors can be targeted to specific compartments with a signaling sequence to selectively monitor the Zn2+ pool of that organelle.
5.4. Hybrid Probes for Zn2+
The last class of currently available Zn2+ sensors is comprised of hybrid probes, which have genetically encoded components and an exogenous cofactor. Two basic designs have been utilized for Zn2+ probes: the SNAP-tag system and the carbonic anhydrase platform. The general features of the SNAP-tag system have been described elsewhere in detail,48b,48c,158 and are introduced in section 2.3.3. The power of this technique is that it allows small-molecule Zn2+ probes to be targeted to specific cellular locations. While the Zn2+ sensing and photophysical properties of the particular probe are unchanged, this strategy does overcome the issues with ambiguous localization inherent to small-molecule sensors. The Lippard lab used a BG-conjugated version of ZP1 and targeted it to the mitochondria and Golgi with the SNAP-tag system, demonstrating the feasibility of this approach for targeting small-molecule sensors.50 One potential challenge of this technique is that modification of a fluorescent probe with the BG moiety can impact cell permeability, as demonstrated by the Chang lab for fluorescent probes designed to sense hydrogen peroxide.52
The second hybrid system is based on a variant of carbonic anhydrase (CA), an enzyme that binds Zn2+ with a KD of approximately 4 pM. In elegant molecular engineering work, the Fierke laboratory was able to tune the specificity and affinity of CA to create a series of probes that could respond to physiological concentrations of Zn2+.159 Early generations of these probes featured a small-molecule fluorophore covalently linked to carbonic anhydrase. When Zn2+ binds carbonic anhydrase, a second cofactor (the fluorophore dapoxyl sulfonamide) binds to an open site on the Zn2+ ion, allowing energy transfer from the dapoxyl moiety to the fluorophore on the enzyme. Recently, Zeng et al. reported a long wavelength, emission ratiometric modification of the carbonic anhydrase Zn2+ sensor that could be amenable to imaging in tissues.160 This new system uses Alexa Fluor 594 as a FRET donor and Chesapeake Blue sulfonamide as the acceptor fluorophore. One limitation of these versions of the sensor is the requirement for microinjection or the attachment of cell-penetrating peptides to introduce it into cells, because covalent attachment of the fluorophore prevented the probe from being genetically encodable.40 However, recent iterations have replaced the small-molecule fluorophore with a FP, thus allowing part of the probe (CA-FP) to be genetically encoded and transfected into cells. The membrane-permeable dapoxyl sulfonamide can be added directly to cells to complete the hybrid system (Figure 9C).
So far, this platform has been used to image Zn2+ in both E. coli and in the cytosol and mitochondria of mammalian cells.53,161 A CA-RFP variant was used to measure the median labile Zn2+ concentration in E. coli at 20 pM,53b and to monitor transient spikes in Zn2+ concentration upon sudden exposure to toxic levels of Zn2+.161 After exposure to Zn2+ shock, intracellular free Zn2+ increased to nanomolar levels for approximately 1 h. The activity of the Zn2+-responsive transcription factor ZntA was also measured in response to Zn2+ shock. In vitro studies of this transcription factor had previously shown that it senses femtomolar concentrations of free Zn2+ ions, but in live bacteria ZntR appeared to be activated by nanomolar Zn2+ spikes. This difference may be due to competition with and regulation by other factors within the cell. This study highlights how measurements of labile Zn2+ levels in live cells can be coupled with other cellular experiments to more precisely define protein functions.
6. Probes for Copper
Copper is a trace metal nutrient essential for most forms of life and is the third most abundant transition metal in humans.162 Copper serves as a structural and catalytic cofactor for many proteins and enzymes including important metabolic factors such as cytochrome c oxidase and copper–zinc superoxide dismutase.162b,162c,163 Copper occurs in two oxidation states within biological systems, either oxidized (Cu2+) or reduced (Cu+). Cu+ is thought to be the dominant oxidation state of labile copper in cells, where this speciation is largely ascribed to the function of membrane reductases that reduce extracellular Cu2+ prior to import as well as the reducing environment maintained within the cytosol.162,164 The redox activity of copper is critical for several key physiological processes; however, unregulated levels of copper can induce oxidative stress and toxicity in cells. Like zinc, dysregulation of copper homeostasis is associated with disease, including the following neurodegenerative disorders: Alzheimer’s disease, amyotrophic lateral sclerosis, Menkes disease, Parkinson’s disease, and Wilson’s disease.165 Cells must maintain optimal concentrations and speciation of copper by tightly regulating the uptake, distribution, storage, mobility, and efflux of this ion. Much of the total cellular copper is associated with high affinity binding proteins, and what is considered labile copper is effectively buffered by a plethora of cellular ligands that minimize free copper ions.162,164
Live-cell fluorescence microscopy using copper selective sensors provides a valuable method to better understand the complex handling of copper in cells. However, there are added challenges posed by targeting copper ions over Zn2+ due to the need for selectivity between different oxidation states, the fluorescence quenching activity of Cu2+, and the fact that sensors must have high enough affinities to compete for copper within its biological window (10–21–10–17 M).166 As a result, only a handful of copper sensors have been generated for biologically accessible copper. Most of the probes designed for biological systems target Cu+.17b It is noteworthy that a substantial body of work has been devoted toward production of small molecule, nucleic acid, and protein-based fluorescent sensors for both mono- and divalent copper; however, this Review will focus only on the sensors applied to imaging Cu+ in biological systems. The biophysical parameters of these Cu+ probes are displayed in Table 5.
Table 5. Small-Molecule Sensors for Cu+.
| excitation |
emission |
brightnessb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sensor | λfree (nm) | εfreea | λbound (nm) | εbounda | λfree (nm) | φfree | λbound (nm) | φbound | free | bound | DRc | KD (M) | ref |
| CTAP-1 | 365 | ND | 365 | 480 | 0.003 | 480 | 0.14 | ND | ND | 4.6 | 4.00 × 10–8 | (167) | |
| CS1 | 540 | 30.0 | 540 | 40.0 | 566 | 0.016 | 561 | 0.13 | 0.48 | 5.20 | 10 | 3.60 × 10–12 | (168) |
| CS3 | 550 | 31.0 | 540 | 46.0 | 560 | 0.007 | 548 | 0.4 | 0.22 | 18.40 | 75 | 8.90 × 10–14 | (169) |
| RCS1 | 480 | 43.0 | 548 | 40.0 | 505, 570 | 0.002, 0.003 | 556 | 0.002, 0.05 | 0.13 | 2.00 | 20 | 4.00 × 10–11 | (170) |
| Mito-CS1 | 555 | 28.0 | 550 | 26.0 | 569 | 0.009 | 558 | 0.05 | 0.25 | 1.30 | 10 | 7.20 × 10–12 | (67c) |
| ACu1 | 359 | ND | 363 (750)* | ND | 492 | 0.028 | 482 | 0.13 | ND | ND | 4 | 2.00 × 10–11 | (171) |
| CS790 | 760 | ND | 760 | ND | 790 | 0.0042 | 790 | 0.072 | ND | ND | 15 | 3.00 × 10–11 | (172) |
| CTAP-2 | ND | ND | 396 | 29.0 | ND | ND | 508 | 0.083 | ND | 2.41 | 65 | 4.00 × 10–12 | (71) |
| Cao Cu-3 | 696 | ND | 750 | ND | 792 | ND | ND | ND | ND | ND | 9.6 | 6.10 × 10–12 | (173) |
Molar extinction coefficients given as ε/103 M–1 cm–1.
Brightness is defined as the product of the molar extinction coefficient and the quantum yield (ε × φ).
There is no systematic way to present dynamic range (DR), so we encourage readers to refer to the original publications for more details about this value. For intensity-based probes, this number is generally the maximum fold change in fluorescence intensity upon Cu+ binding. ND, not determined.
The first small-molecule sensor for detecting labile copper in cells, CTAP-1, was developed by the Fahrni group in 2005.167 CTAP-1 uses an azatetrathiacrown Cu+ binding motif linked to a pyrazoline-based dye that excites in the UV region. This sensor is selective for Cu+ over Cu2+ or other cellular ions and displays up to a 4.6-fold increase in fluorescence intensity upon Cu+ binding. The fluorescence enhancement upon Cu+ binding is consistent with a mechanism that involves modulation of PET between the chelate and fluorophore. Experiments using fixed NIH 3T3 fibroblast cells stained with CTAP-1 and complemented with organellar costains and X-ray fluorescence microscopy mapped cellular copper for the first time and showed that labile copper is localized largely to mitochondria and the Golgi apparatus in these cell types under acute copper overload.10b,167
Soon after this study, the Chang laboratory presented a live-cell Cu+ responsive fluorescent probe called Coppersensor-1 (CS1).168,174 CS1 is composed of a boron-dipyrromethene (BODIPY) dye platform and a thioether-rich receptor chemically similar to the azatetratiocrown used by CTAP-1. CS1 is excited and emits in the visible region, offering decreased phototoxicity as compared to CTAP-1, and it undergoes a greater increase in fluorescence intensity in the presence of Cu+ (10-fold vs 4.6-fold). Using CS1, dynamic changes in copper pools could be visualized in real time during Cu+ uptake of HEK 293T cells under acute copper overload.168 However, some challenges associated with the use of this sensor in neuronal and glial cell lines treated with CuCl2 or Cu(II)(gtsm) were identified in a recent study by Price et al., including pH sensitivity and lysosomal uptake.175
Tuning the BODIPY scaffold of CS1 to improve optical brightness and turn-on enhancement produced the second-generation sensor CS3.169 Exchanging the fluoro substituents with methoxy groups increased electron density over the fluorophore, yielding a higher dynamic range and a brighter Cu+–dye complex. CS3 undergoes a 75-fold increase in fluorescence intensity upon binding Cu+.169 These improvements over the first generation sensors, CTAP-1 and CS1, provided the ability to detect Cu+ under both basal and depleted levels, whereas the previous sensors only detected Cu+ overload. In conjunction with synchrotron-based X-ray fluorescence microscopy, CS3 was successfully used to reveal that hippocampal neurons redistribute large pools of copper from somatic regions to peripheral processes upon depolarization. This study established a link between copper mobilization and calcium release, suggesting that some aspects of copper regulation might be correlated with major cell signaling pathways.
Mitochondria require Cu+ to function due to its role as an essential cofactor in aerobic respiration.162a Cells tightly control Cu+ uptake and transport to avoid accumulation of reactive free ions. Cu+ is highly buffered in the cytoplasm and shuttled to mitochondria by chaperone proteins where it has been shown to collect in the matrix.176 To monitor the accessible or labile Cu+ pool in the mitochondrial matrix, the Chang group developed a mitochondrial-localized sensor, Mito-CS1, using a modified BODIPY platform, similar to the other CS sensors. By incorporating a TPP moiety into the CS1 platform, Chang and co-workers were able to monitor mitochondrial Cu+ in live HEK 293T and human fibroblast cells.67c The results of this study suggest that cells maintain mitochondrial copper homeostasis in a narrower range relative to other areas of the cell as mitochondrial Cu+ levels were only moderately altered as compared to total Cu+ levels between states of Cu+ deficiency, mitochondrial metallochaperone malfunction, and healthy cells.
To further the characterization of copper homeostasis and obtain a more cohesive picture of copper regulation, sensors can be used to image Cu+ in cells over longer durations as well as in more complex samples, such as tissues or intact multicellular organisms. However, as discussed earlier, sensor excitation with short wavelength light limits the penetration depth, increases cellular autofluorescence, and inevitably induces photodamage to cells and photobleaching of the probe. These phenomena limit the usefulness of the above sensors for long-term imaging or imaging of tissues and organisms. One way to circumvent these limitations is to use two-photon excitation microscopy.177 Most of the examples discussed thus far involve standard microscopy, where two-photon excitation microscopy differs as a nonlinear optical technique that uses low energy infrared photons. The first Cu+ selective probe designed specifically for two-photon excitation, ACu1, uses a naphthalene-based reporter that can be excited by two near-infrared photons.171 This is an intensity-based probe that gives rise to a 4-fold change in fluorescence intensity upon Cu+ binding. ACu1 was successfully used to visualize Cu+ distribution in rat hippocampal slices at depths up to 200 μm.
Another way to achieve increased sample penetration, reduced phototoxicity, and reduced autofluorescence is by single-photon excitation in the near-infrared. Using different functionalized tricarbocyanine derivatives, two near-infrared “turn-on” sensors for Cu+ have recently been developed. One of these, which we refer to as Cao Cu-3, after the author and sensor number, uses the thio-rich bis(2-((2-(ethylthio)ethyl)-thio)ethyl)amine (BETA) moiety as a high affinity receptor for Cu+. Cao Cu-3 undergoes a 9.6-fold increase in fluorescence upon Cu+ binding and was used to image labile Cu+ mobilized by ascorbic acid treatment in MG63 cells.173 By adjusting the fluorophore of the CS series to a cyanine dye (Cy7)-based platform, Chang and co-workers produced another far red Cu+ sensor CS790, which as its name suggests has an emission maximum at a wavelength of 790 nm.172 CS790 is available in an aqueous compatible and membrane permeable acetoxymethyl ester form (CS790AM) that was used to visualize dynamic copper fluctuations at endogenous levels in living mice. CS790AM showed promise for monitoring labile copper in a single mouse over time, tracking different stages of health and disease. Used in a Wilson disease mouse model (Atp7b–/–), CS790AM showed atypical copper accumulation over time, consistent with progression of this disease. As CS790AM permitted visualization of fluctuations in bioavailable copper in living animals, it enables the potential for tracking copper accumulation and distribution throughout disease development.
As with all metal sensors, there is a continual push to optimize the photophysical properties of copper sensors as well as features such as metal-sensor stoichiometry. One recently recognized issue along these lines concerns many of the current small-molecule copper sensors: the BODIPY-based CS sensors as well as CTAP-1 can form spontaneous dimers or colloidal aggregates at low micromolar concentrations in aqueous solutions.71 These sensor–sensor interactions affect the sensitivity and photophysical properties of the probes. BODIPY dimers show blue-shifted absorption and emission spectra, and aggregates are completely nonfluorescent. In response to these findings, the Fahrni group produced a series of new Cu+ selective sensors, including CTAP-2, which remains monomeric up to 10 μM in aqueous environments.71 CTAP-2 contains a modified thiocrown receptor that incorporates four hydroxymethyl groups and is combined with a triarylpyrazoline fluorophore functionalized with a solubilizing sulfonate group. CTAP-2 undergoes a 65-fold fluorescence enhancement upon binding Cu+, although the quantum yield is not improved over other pyrazoline-based sensors. A great comprehensive review of small-molecule Cu+ sensors discusses in detail how sensor design corresponds to these and other observed photophysical properties.164
Although the above intensity-based probes have been useful for visualizing the localization and redistribution of Cu+ in cells, variations in probe concentrations between cells, heterogeneous distribution within cells, and issues associated with cell thickness and movement complicate their use in detecting quantifiable changes in copper concentrations. These issues can be addressed by ratiometric imaging. The Chang group developed a small-molecule ratiometric reporter for Cu+, based on an asymmetric BODIPY platform and referred to as ratiometric CS1 (RCS1).170 Upon binding Cu+, RCS1 undergoes an impressive 20-fold fluorescence ratio change with excitation and emission in the visible regime. Treatment of HEK 293T cells with RCS1 enabled monitoring transient increases in cytosolic Cu+ that originated from intracellular stores after stimulation with ascorbate.170 The molecular sensors for Cu+ are shown in Figure 11.
Figure 11.
Molecular Cu+ sensors.
Continuing efforts to expand the palette of ratiometric Cu+ sensors have led to the generation of a few genetically encodable Cu+ FRET sensors (Figure 12). FRET-based sensors can overcome some of the challenges small-molecule fluorophores face due to water insolubility and cytotoxicity. In addition, genetically encodable FRET sensors provide the opportunity to target specific subcellular organelles. The He laboratory has developed three FRET-based sensors for visualizing cellular Cu+: AMT1-FRET, Ace1-FRET, and Mac1-FRET.166b,178 These sensors were constructed with a cysteine-rich Cu+ binding domain placed between a CFP and YFP FRET pair (Figure 12A). All three sensors bind up to four equivalents of Cu+, where the eight cysteines contained in each copper binding domain form a tetracopper(I) cluster. The different binding domains that give these sensors their names derive from different yeast-based Cu+-dependent transcriptional regulators. Amt1 activates genes for detoxification and efflux in the presence of excess Cu+. Ace1 is a homologue of Amt1. In contrast, Mac1 activates the expression of Cu+ uptake factors during copper depletion.178 These three genetically encoded Cu+ sensors offer a range of affinities, which has enabled quantification of the window of biologically available copper in yeast (10–21–10–17 M).166b
Figure 12.
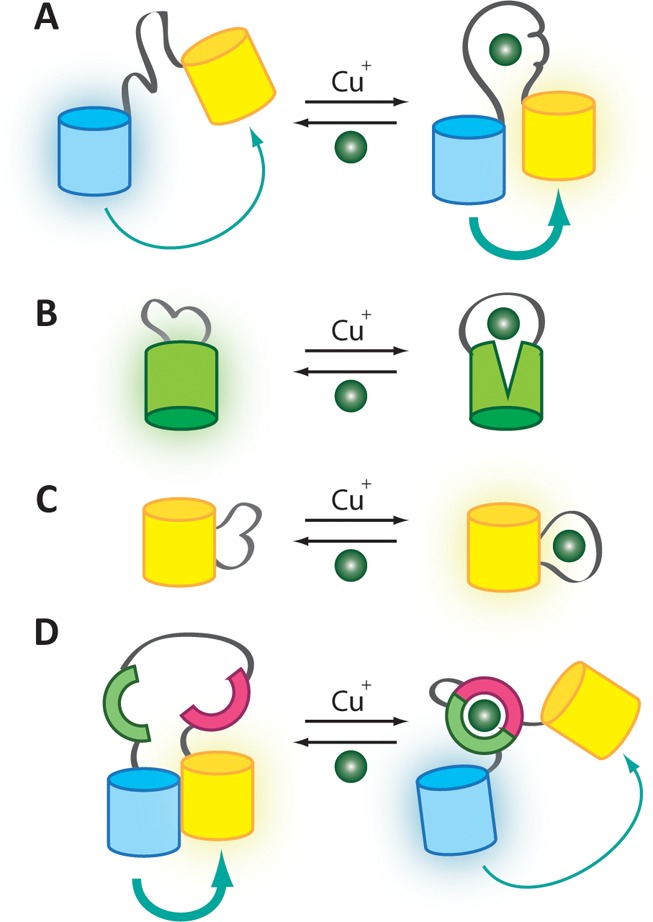
Mechanism of metal ion sensing by genetically encoded probes for Cu+. (A) AMT1-FRET, Ace1-FRET, and Mac1-FRET have a cysteine-rich Cu+-binding domain between a CFP/YFP FRET pair such that metal binding results in an increased FRET signal. (B) Cu+ binding to EGFP-Amt1 distorts the β-barrel of EGFP and decreases fluorescence. (C) YFP-Ace1 and related sensors have the Cu+-binding domain of Ace1 inserted between two strands of EYFP. Cu+ binding alters the local environment of the chromophore and leads to an increased fluorescent signal. (D) The eCALWY Zn2+ sensor platform can be tuned for improved selectivity toward Cu+. In the absence of Cu+, association between two FPs produces a FRET signal. CU+-induced association between the metal binding domains of Atox1 and WD4 changes the structure of the sensor and results in a decreased FRET signal.
More recently, a unique approach has been used to design a single FP sensor, EGFP-145Amt1.179 In this sensor, the Amt1 Cu+ binding domain was inserted between residues 145 and 146 of EGFP. Cu+ binding induces a structural distortion of the EGFP β-barrel structure, decreasing the fluorescence intensity of this sensor by about 50% (Figure 12B).180 Recently, another single fluorescent protein Cu+ reporter, YFP-Ace1, was created using a similar approach.181 YFP-Ace1 consists of the Cu+ binding domain of Ace1 inserted between the residues 145 and 146 of EYFP. The inserted Cu+ binding domain is positioned close enough to the fluorophore of EYFP to cause a change in the local environment and detectably alter its fluorescent properties when Cu+ binds (Figure 12C). In this case, Cu+ binding results in up to a 40% increase in fluorescence intensity. YFP-Ace1 was then used to generate a family of sensors called YAGn, where n denotes the linker length used to connect the Cu+ binding domain. The YAGn series offers a variety of Cu+ binding affinities ranging from 8.29 × 10–21 to 8.61 × 10–16 M, and was successfully used to visualize Cu+ in HeLa cells.
Because of the unique coordination chemistry of transition metals, binding sites can be manipulated to design sensors selective for one metal over another. The Merkx lab recently took advantage of the differential coordination chemistry of Cu+, Cu2+, and Zn2+ to reversibly tweak the affinity of a Zn2+ sensor toward Cu+ (Figure 12D).182 Cu+ preferentially binds to soft ligands such as the sulfur donors cysteine or methionine and forms either a 2-coordinate linear or a 3-coordinate trigonal geometry.162a,182 Cu2+ and Zn2+ will accommodate harder ligands such as nitrogen donors like histidine or oxygen donors like aspartate or glutamate. Furthermore, Zn2+ prefers tetrahedral four-coordinate binding sites.162a,182 With this in mind, Merkx and co-workers designed a class of sensors called eCALWYs that combine two Cu+ binding motifs (ATOX1 and the fourth domain of ATP7B (WD4)) oriented to form a tetra cysteine Zn2+ binding site. The small Cu+ binding motifs are separated by a flexible linker and bridge the FRET pair Cerulean (donor) and Citrine (acceptor). This unique design was recently shown to be tunable for selecting either Cu+ or Zn2+. Systematically replacing the binding site cysteines with methionines produced conformational variants that regained affinity for Cu+ and lost the ability to form stable tetrahedral Zn2+ complexes. So far, the affinities of the eCALWY mutants remain outside the biological window for Cu+(∼10–15 M); however, they may be suited for monitoring cells under conditions of extreme Cu+ stress. Table 6 lists some of the important features of the genetically encoded sensors discussed above.
Table 6. Genetically Encoded Cu+ Sensors.
| FP |
excitation |
emission |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| name | donor | acceptor | λdonor (nm) | εdonora | λacceptor (nm) | εacceptora | λdonor (nm) | φdonor | λacceptor (nm) | φacceptor | DR (Rmax/Rmin) | KD (M) | ref |
| Amt1-FRET | ECFP | EYFP | 439 | 32.5 | 514 | 83.4 | 476 | 0.60 | 527 | 0.61 | ND | 2.50 × 10–18 | (178) |
| Ace1-FRET | ECFP | EYFP | 439 | 32.5 | 514 | 83.4 | 476 | 0.60 | 527 | 0.61 | ND | 4.70 × 10–18 | (166b) |
| Mac1-FRET | ECFP | EYFP | 439 | 32.5 | 514 | 83.4 | 476 | 0.60 | 527 | 0.61 | ND | 9.70 × 10–20 | (166b) |
| eCALWY-C2M/C3M | Cerulean | Citrine | 433 | 43.0 | 516 | 77.0 | 475 | 0.62 | 529 | 0.76 | ND | <1 × 10–17 | (182) |
| YAG(n), n = 0 | EYFP | ND | 514 | 83.4 | ND | ND | 527 | 0.61 | ND | ND | ND | 8.20 × 10–18 | (181) |
| YAG(n), n = 1 | EYFP | ND | 514 | 83.4 | ND | ND | 527 | 0.61 | ND | ND | ND | 2.00 × 10–18 | (181) |
| YAG(n), n = 2 | EYFP | ND | 514 | 83.4 | ND | ND | 527 | 0.61 | ND | ND | ND | 1.20 × 10–18 | (181) |
| YAG(n), n = 3 | EYFP | ND | 514 | 83.4 | ND | ND | 527 | 0.61 | ND | ND | ND | 4.60 × 10–19 | (181) |
| YAG(n), n = 4 | EYFP | ND | 514 | 83.4 | ND | ND | 527 | 0.61 | ND | ND | ND | 3.30 × 10–19 | (181) |
| EGFP-145Amt1 | EGFP | ND | 484 | 32.5 | ND | ND | 507 | 0.60 | ND | ND | ND | 4.60 × 10–19 | (179) |
Molar extinction coefficients given as ε/103 M–1 cm–1. ND, not determined.
7. Probes for Iron
7.1. Iron Homeostasis
Iron is the most abundant transition metal in the human body: the average adult human contains approximately 3–5 g of this trace element, and the total cellular concentration is approximately 50–100 μM.183 Iron is involved in numerous cellular processes from metabolism, electron transport, and DNA synthesis.184 In myoglobin and hemoglobin, Fe2+ bound to a heme cofactor is critical for oxygen transport throughout the body. Iron is also found in iron–sulfur cluster proteins (e.g., aconitase), heme enzymes (e.g., cytochrome P450), and nonheme iron enzymes (e.g., ribonucleotide reductase). Biological iron almost exclusively exists in the ferrous (Fe2+) or ferric (Fe3+) state, although other oxidation states are possible during catalytic cycles. Cells must carefully control iron levels, distribution, and speciation. Disruption of iron regulation has been linked to disorders such as anemia, hemochromatosis, and Alzheimer’s.185 The reduction potential in the cytosol favors Fe2+ over Fe3+, and, furthermore,186 Fe3+ is poorly soluble at neutral pH in aqueous media. On the other hand, free Fe2+ ions are capable of participating in Fenton chemistry, leading to the generation of harmful free radicals; therefore, the amount of free Fe2+ in cells must be kept to a minimum.187
The idea of a labile iron pool was first suggested by Greenberg and Winthrop in 1946,188 and later by Jacobs in 1977,189 and there has been substantial interest in defining it further since then.190 However, in the absence of good methods to monitor this iron pool, its biological function is not completely clear. Fluorescent probes are attractive tools to visualize the distribution and speciation of labile iron. Ideally, such probes should be able to selectively respond to either Fe2+ or Fe3+ or be able to detect iron species such as heme–iron or iron–sulfur clusters. One major challenge with detection of iron using fluorescent sensors is the paramagnetic quenching nature of both ions; as such, many early probes for iron exhibit “turn off” fluorescence response to iron binding. Moreover, probes must be able to distinguish the two oxidation states of iron. In the last several years, new probes have been developed for Fe2+ and Fe3+ that may allow new discoveries regarding cellular iron homeostasis. Although most of these probes have only been used for modest applications in living cells so far, further developments on these tools will undoubtedly yield new insights into iron biology. A summary of the photophysical properites of a number of iron probes is presented in Table 7.
Table 7. Fluorescent Probes for Fe2+ and Fe3+.
| excitation |
emission |
brightnessb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| name | λfree (nm) | εfreea | λbound (nm) | εbounda | λfree (nm) | φfree | λbound (nm) | φbound | free | bound | KD (M) | DRc | ref |
| calcein | 486 | ND | 486 | ND | 517 | ND | 517 | ND | ND | ND | 2.2 × 10–7 | 43% | (191) |
| Phen Green SK | 507 | ND | 507 | ND | 532 | ND | 532 | ND | ND | ND | ND | 96% | (192) |
| RhoNox-1 | 492 | 24 | 555 | ND | 575 | 0.01 | 575 | 0.3 | ND | ND | ND | 30 | (193) |
| BDP-Cy-Tpy | 485 | ND | 485 | ND | 507 | ND | 507 | ND | ND | ND | 2.5 × 10–6 | ND | (194) |
| 569 | ND | 596 | ND | 635 | ND | 635 | ND | ND | ND | ||||
| AGD | 430 | 5.2 | 430 | ND | 480 | 0.103 | 480 | 0.0057 | ND | ND | 2.4 × 10–5 | 33% | (195) |
| IP1 | 470 | 0.4 | 470 | 0.4 | 508 | ND | 508 | ND | ND | ND | ND | ND | (196) |
| OuYang Fe-1 | 520 | ND | 520 | ND | 561 | ND | 583 | 0.13 | ND | ND | ND | 1000 | (197) |
| NBD-DFO | 475 | 7 | 475 | ND | 548 | 0.79 | 548 | ND | ND | ND | ND | ND | (198) |
| SF34 | 491 | 49.6 | 491 | ND | 514 | ND | 514 | ND | ND | ND | ND | 77% | (199) |
| Pyochelin-1 | 470 | ND | 470 | ND | 545 | 6 | 545 | 0.5 | ND | ND | 1.6 × 10–11 | 290 | (200) |
| Pyochelin-2 | 470 | ND | 470 | ND | 545 | ND | 545 | ND | ND | ND | 3.8 × 10–20 | 320 | (200) |
| RNP1 | 371 | ND | 371 | ND | 431 | 0.004 | 594 | 0.14 | ND | ND | 5 × 10–5 | ND | (201) |
Molar extinction coefficients given as ε/103 M–1 cm–1.
Brightness is defined as the product of the molar extinction coefficient and the quantum yield (ε × φ).
There is no systematic way to present dynamic range, so we encourage readers to refer to the original publications for more details about this value. For turn-on probes, this number is generally the maximum fold change in fluorescence intensity upon Fe2+/Fe3+ binding. For turn-off probes, this is the % quenching of maximal signal upon Fe2+/Fe3+ binding. ND, not determined.
7.2. Early Probes for Labile Iron
Among the first reports of visualizing the labile iron pool was the use of calcein, a fluorescein derivative, by Breuer et al. in 1995, and it is still one of the most common protocols for measuring labile iron.191 Cells are loaded with the nonfluorescent, membrane-permeable acetoxymethyl derivative of calcein. Upon entering the cell, the ester groups are removed by intracellular enzymes to reveal the fluorescent calcein probe (Figure 13A). Metal ion binding quenches fluorescence, and the emission from the probe can be restored by subsequent addition of a membrane-permeable iron chelator such as salicylaldehyde isonicotinoylhydrazone (SIH). In this way, the labile pool of iron can be quantified. This method suffers from a number of drawbacks: the probe can form redox-active and potentially toxic complexes with iron,202 addition of a chelator is required for quantification, and the probe cannot adequately distinguish between oxidation states. As an alternative, Petrat et al. used the fluorescein-based probe Phen Green SK (Figure 13B), which includes a 1,10-phenanthroline chelating group to study the labile iron pool in cultured hepatocytes.192,202 This probe has a greater turn-off response to iron than calcein (93% versus 46% quenching, respectively). Using a novel ex situ confocal microscopy technique, Petrat et al. measured the concentration of labile iron in hepatocytes to be approximately 2.5–9.8 μM, which accounts for roughly 1% of the total iron content. However, both of these probes are not specific for either Fe2+ or Fe3+ and can also interact with other metal ions, leading to an interfering signal.
Figure 13.
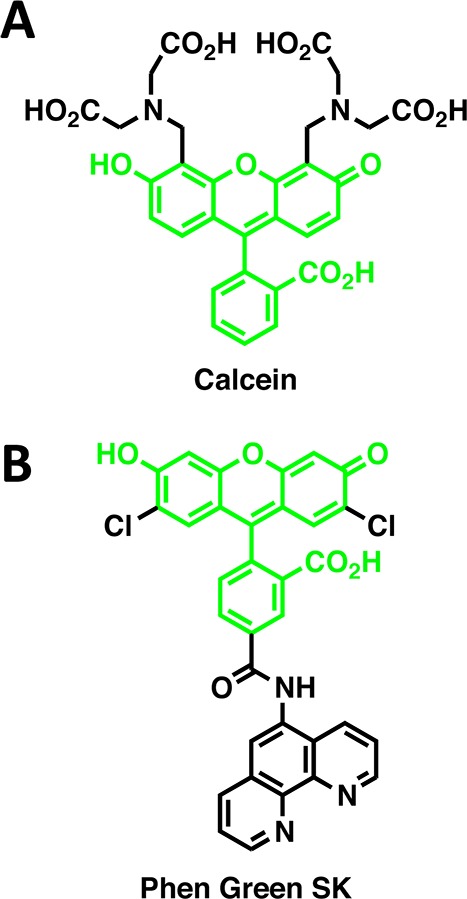
Calcein (A) and Phen Green SK (B) represent early fluorescent tools for visualizing cellular iron homeostasis.
7.3. Probes for Fe2+
Because of the propensity for Fe2+ to be oxidized to Fe3+ in aerobic aqueous environments, it has been difficult to design probes specific for ferrous ions. Two early Fe2+ probes, pyrene-TEMPO and DansSQ, exhibit turn-on responses to Fe2+ by different mechanisms. When linked to pyrene, the organic radical TEMPO quenches fluorescence, but Fe2+ is able to reduce TEMPO in aqueous solution and restore fluorescence to pyrene.203 While selective for Fe2+, this probe has limited application for intact biological systems for two reasons: the reaction must be carried out in acidic solution, and it can be triggered by other radicals. DansSQ consists of a dansyl group linked to styrylquinoline.204 Binding of Fe2+ disrupts internal charge transfer between the two fragments and results in a 15-fold increase in fluorescence at 460 nm. However, the probe is not entirely selective for Fe2+ and is only soluble in acetonitrile and 10% H2O, making biological application of DansSQ challenging.
The past few years have seen a growing number of new Fe2+ probes suitable for live-cell imaging experiments (Figure 14). The first turn-on, Fe2+-selective probe that was used in live cells was developed by Hirayama et al. in 2013.193 RhoNox-1 is a rhodamine-based probe that makes use of the chemical reactivity of Fe2+: the metal ion reduces an N-oxide group on the probe to reveal a tertiary amine. This reaction does not occur in the presence of Fe3+. Cells treated with a chelator displayed significantly lower levels of fluorescence than untreated cells, indicating that RhoNox-1 was able to detect endogenous levels of Fe2+. Co-localization experiments revealed that this probe localized to the Golgi apparatus. Despite the attractiveness of a Fe2+-selective sensor for live-cell imaging, the mechanism of sensing was not shown to be reversible, limiting the ability of RhoNox-1 to monitor fluxes in labile Fe2+ levels. Another Fe2+ sensor has been developed by the Chang laboratory that utilizes a biomimetic oxidative dealkylation to reveal a fluorescent fluorescein derivative.196 Iron probe 1 (IP1) is selective for Fe2+ over other metal ions at their biological concentrations and can detect endogenous levels of labile Fe2+. Furthermore, IRP1 was used to demonstrate elevated intracellular labile Fe2+ levels in a liver cell line caused by treatment with hepcidin or ascorbic acid. However, like RhoNox-1, this reaction-based probe was not shown have a reversible mode of detection. AGD is a coumarin-based probe with 2-amino-2-(hydroxylmethyl)propane-1,3-diol as a ferrous binding domain that has a fluorescence quenching response to Fe2+ binding.195 While selective for Fe2+ over other transition metal ions, the probe still exhibited some quenching by excess Fe3+. This probe appeared to be concentrated to the plasma membrane, an idea that was supported by molecular dynamics simulations. This probe was used in cells to detect exogenously added ferric nitriloacetic acid complexes. Fluorescence was restored by treatment with bipyridyl and quenched by subsequent addition of ferrous ammonium sulfate, demonstrating the reversible mode of action of this sensor.
Figure 14.

Small-molecule Fe2+ sensors.
Li and co-workers developed a new ratiometric sensor for Fe2+, BDP-Cy-Tpy.194,205 As described in previous sections, dual-emission ratiometric probes have the advantages of minimizing artifacts from cellular movement, variable sample thickness, and sensor concentration. This probe links the near-IR fluorophore cyanine (Cy) to the Fe2+-binding group 4′-(aminomethylphenyl)-2,2′,6′,2″-terpyridine (Tpy) such that Fe2+ binding leads to PET-induced quenching of Cy fluorescence. The probe also features a BODIPY fluorophore that is unaffected by the presence of metal ions. This sensor was used in mammalian cells to monitor an ascorbic acid-induced increase in labile Fe2+ levels.
7.4. Probes for Fe3+
In contrast to the relatively small number of Fe2+ probes available, there are many probes selective for Fe3+. A recent review by Sahoo et al. exhaustively profiles the development of molecular and supramolecular probes for Fe3+; however, the vast majority of these have not been used for biological investigations and will not be included in this Review.206 A representative sample of Fe3+ sensors is shown in Figure 15. Some microorganisms, especially bacteria and some fungi, produce siderophores to scavenge Fe3+ from their environments. Uptake of this essential nutrient is hampered by the very low solubility of the Fe3+ ion in oxidizing environments such as the sea or soil. Secreted siderophores form soluble complexes with Fe3+ that can then be taken up by the organism by active transport mechanisms. These compounds are among the tightest known binders of Fe3+, and have been exploited to develop several fluorescent probes for this ion. One of the first reported fluorescent sensors for iron was modeled on the siderophore desferrioxamine B (DFO) linked to the fluorophore 7-nitrobenz-2-oxa-1,3-diazole (NBD).198 This probe, appropriately designated NBD-DFO, was demonstrated to extract metals from Fe3+-loaded siderophores and ferriproteins in vitro. Addition of the hydrophobic fluorophore improved the membrane permeability of the probe over DFO alone, and thus NBD-DFO could be used to monitor Fe3+ extraction in cultured hepatoma cells. Although the probe was not used in imaging experiments, Fe3+ extraction could be quantified by measuring NBD-DFO fluorescence in a cuvette in a fluorometer. This probe was also used to monitor Fe3+ uptake in cotton (Gossypium spp.) and maize (Zea mays L.) plants.207 DFO has also been conjugated to other fluorophores: for example, fluorescein-DFO has been used to assess the presence of serum Fe3+ in patients undergoing chelation therapy.208
Figure 15.
Small-molecule Fe3+ sensors.
Two new siderophore-based probes have been developed on the basis of pyochelin, a molecule produced by the Gram-negative bacteria Pseudomonas aeruginosa and Burkholderia cepacia.200 These probes feature a 4-nitro-benzo[1,2,5]oxadiazole (NBD) fluorophore and exhibit a fluorescence increase upon Fe3+ binding. On the basis of quantum yield measurements, it was speculated that pyochelin quenches the fluorescence of the NBD group, but in the presence of Fe3+, the intrinsic fluorescence of the siderophore is completely quenched. The end result of Fe3+ binding is an increase in NBD fluorescence. Wild-type bacteria were able to uptake the probes, but a strain lacking the appropriate outer membrane receptor for the siderophore were not labeled. It should be noted that the physiological role of pyochelin is not completely clear, and recent studies have re-evaluated the affinity of this molecule for Fe3+, Cu2+, and Zn2+.209 These or similar tools could nevertheless be useful for exploring siderophore-dependent uptake pathways. Fakih et al. introduced a fluorescein-based probe, SF34, with a 3-hydroxy-4-one Fe3+ chelating unit.199 The fluorescence of SF34 is quenched by 77.4% upon Fe3+ binding and is selective for ferric ions over other biological metals including Zn2+, Ca2+, and Mg2+. While the probe could be introduced into mammalian cells, the charged nature of the molecule led to the proposal that it was engulfed by pinocytosis or macropinocytosis and unable to diffuse across the plasma membrane. Further analysis revealed that the probe was concentrated in an endosomal or lysosomal compartment. Probes for this compartment, which may be involved in trafficking and storage of labile iron,210 have not been previously available. Additionally, SF34 was used to monitor the compartment-specific effects of different chelating agents on iron homeostasis.
Several “turn-on” fluorescent probes for Fe3+ have been developed on the basis of the equilibrium between a nonfluorescent spirolactam and fluorescent ring-opened amide forms of rhodamine. The basic premise of such probes is that Fe3+ can shift the equilibrium toward the ring-opened form, leading to an increase in fluorescence intensity. In the past few years, several groups have developed variants of these rhodamine-based probes by exploring the use of different chelating groups.211 Although these studies have generated a new series of probes and demonstrated some degree of modularity for the rhodamine-based platform, the biological applications of these probes have been mostly limited to proof-of-principle detection of exogenously added Fe3+ in cells. Recently, OuYang et al. developed a novel optical-electrochemical multichannel sensor for Fe3+.197 Some rhodamine-based sensors are subject to interference by Cr3+, but the multichannel nature of this probe allowed OuYang et al. to use differential pulse voltammetry to understand the different electrochemical responses to these two ions. A ratiometric sensor, RNP1, has also been developed featuring a naphthalene chromophore attached to the rhodamine backbone.212 In the presence of Fe3+, emission from naphthalene can excite rhodamine via FRET, giving rise to ratiometric signals. Furthermore, the probe localizes to mitochondria because of its TPP group. The probe was functional in cells and could detect exogenously loaded Fe3+ in mitochondria. Although iron probes have not been as thoroughly investigated and optimized as probes for zinc and copper, these recent developments will undoubtedly set the foundation for future imaging studies of the labile iron pool in living cells, tissues, and eventually whole organisms.
8. Available Fluorescent Probes for Other Biological Metals
8.1. Manganese (Mn2+)
Manganese is one of the most abundant elements in the earth’s crust,213 and it is an essential micronutrient to all three kingdoms of life.214 Manganese is a necessary cofactor for a variety of critical biological processes, such as oxygen evolution in chloroplasts, maintenance of redox balance in mitochondria, formation of deoxyribonucleotides from ribonucleotides, thus generating the essential building blocks for DNA synthesis, and many other enzymatic transformations. In addition, there is growing evidence that manganese plays a role in cellular adaptation to oxidative stress,215215 vesicle trafficking in mammalian cells,216 and neuron function.217217 While manganese can exist in a variety of oxidation states, the most abundant form in cells is presumed to be Mn2+. As with most metals, excess Mn2+ is toxic. One of the primary sites of toxicity in humans is the brain, where it has been known for over 150 years that Mn2+ is a neurotoxic agent, and manganese overload gives rise to a neurological disorder that resembles Parkinson’s disease.217 The drive to better understand the cellular distribution of manganese and follow the flux of manganese in cells and intact organisms has led to progress in recent years in the development of selective and sensitive fluorescent probes for detection and quantification of Mn2+ (Figure 16).
Figure 16.
Molecular sensors for the biological metals Mn2+, Ni2+, and Co2+.
Historically, techniques for free Mn2+ detection have been available using atomic absorption,218 ion chromatography,219 or capillary zone electrophoresis.220 A few colorimetric probes have also been used to detect Mn2+ in aqueous samples. These are based on photochemical oxidation reactions catalyzed by the presence of Mn2+.219,221 However, only recently have fluorescent tools become available for detecting Mn2+ (Table 8). The development of selective “turn-on” sensors for Mn2+ has been limited due to the fact that paramagnetic Mn2+ quenches fluorescence. In addition, Mn2+ is 3d5 and can be difficult to distinguish from other metal ions, and can compete with Mg2+ and Ca2+, which are far more abundant in cells, posing a direct challenge for the selectivity of all Mn2+ sensors.
Table 8. Fluorescent Sensors for Ni2+, Mg2+, and Co2+.
| excitation |
emission |
brightnessb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sensor | λfree (nm) | εfreea | λbound (nm) | εbounda | λfree (nm) | φfree | λbound (nm) | φbound | free | bound | DRc | KD (M) | ref |
| NS1 | 495 | 5.8 | 495 | 5.5 | 507 | 0.002 | 507 | 0.055 | 0.01 | 0.30 | 25 | 1.90 × 10–8 | (222) |
| Liang Mn-1 | 256 | ND | 256 | ND | 286 | ND | 286 | ND | ND | ND | ND | 2.40 × 10–9 | (223) |
| Liang Mn-2 | 493 | ND | 493 | ND | 519 | 0.1 | 519 | 0.37 | ND | ND | ND | 9.70 × 10–8 | (223) |
| Liang Mn-3 | 505 | ND | 505 | ND | 530 | 0.13 | 530 | 0.49 | ND | ND | 2.4 | 1.00 × 10–8 | (223) |
| Lin Co-1 | 340/429 | ND | 340/429 | ND | 430/606 | ND | 430/606 | ND | ND | ND | 85 | 7.9 × 10–12 | (224) |
Molar extinction coefficients given as ε/103 M–1 cm–1.
Brightness is defined as the product of the molar extinction coefficient and the quantum yield (ε × φ).
There is no systematic way to present dynamic range (DR), so we encourage readers to refer to the original publications for more details about this value. For intensity-based probes, this number is generally the maximum fold change in fluorescence intensity upon metal ion binding. ND, not determined.
One method of fluorescent detection of free Mn2+ described by the Canary laboratory in 2009 involves an ion displacement reaction. This multicomponent sensory system uses Mn2+ to displace Zn2+ from a chelating agent for a turn-on fluorescence response.225 The chelator EGTA is used as a masking agent that prevents Zn2+ binding to a small-molecule fluorescent Zn2+ reporter such as PAR. Under favorable conditions, Mn2+ displaces Zn2+ from the EGTA chelator, and the liberated Zn2+ then reacts with PAR to form a fluorescent complex. In an indirect manor, this multicomponent system effectively detects Mn2+ using fluorescence response. Although this specific system is not applicable for use in cells due to lack of selectivity over Ca2+, later an analogous technique was employed for detecting Mn2+ in HEK and DMT-1 cells treated with exogenous Mn2+.180 Additionally, the Canary group presented a more direct sensor design for Mn2+ based on the Ca2+ indicator calcium green.223 Calcium green includes a BAPTA (1,2-bis(o-aminophenoxy)ethane-N,N,N,N-tetraacetic acid) chelating group that was tuned for Mn2+ selectivity by substitution of carboxylate groups with pyridines. The resulting family of Mn2+ sensors (Liang Mn-1–Liang Mn-3) was shown to be 2.3–4.8 times more selective for Mn2+ over Ca2+ and was used to detect endogenous levels of Mn2+ in HeLa cells as well as HeLa cells treated with micromolar MnCl2.
Another group offered a pH-dependent recognition technique that uses quantum dot (QD) fluorescence quenching to discriminate between Mn2+ and Cu2+ in vitro.226 Mn2+ diffusion and adsorption onto the surface of the QDs was found to be highly pH sensitive. Additionally, the energy transfer from the QDs to Mn2+ is pH-dependent. Because the mechanism of QD fluorescence sensing for Mn2+ is strongly pH dependent, where the sensing of Cu2+ is less pH sensitive, a recognizable detection of the two ions is possible by adjusting solution pH. For example, selective detection of Cu2+ by QD fluorescence can be achieved by using pH conditions that are outside of the Mn2+ responsive window. Measuring the sample at both Mn2+-sensitive and Mn2+-insensitive pH and deducting the previously measured influence of Cu2+ then reveals the fluorescence response of Mn2+. Although this platform was only utilized in vitro, it is one of the few metal sensor platforms to make use of quantum dots and may help inform the design of sensors that are ultimately biocompatible.
Recently, a unique bipyrene-functionalized graphene sensor for Mn2+ detection in cells was presented. The pyrene derivative 1,2-bis-(2-pyren-1-ylmethylamino-ethoxy) ethane (NPEY) was synthesized and associated with the surface of graphene nanosheets (GNs) through π–π stacking interactions, which provided a turn-on fluorescent sensor for Mn2+ (NPEY-GNs).227 In the absence of Mn2+, the interaction between NPEY and the GNs renders the NPEY dye nonfluorescent. Mn2+ binding disrupts the direct π–π stacking interactions and produces a fluorescence response. NPEY-GNs was applied both as a turn-on fluorescence sensor for Mn2+ in vitro as well as in living HeLa cells.
8.2. Nickel (Ni2+)
Nickel is an essential cofactor for a variety of enzymes that play important roles in microorganisms (bacteria, archea, fungi, algea)228 and plants, particularly in energy and nitrogen metabolism.229 As compared to other biological metals, nickel is used at low levels; its use is relatively widespread in microorganisms but more limited in eukaryotes.230 Yet, there are very few Ni2+ selective indicators for live-cell imaging (Table 8 and Figure 16). The Chang group presented one of the first and only fluorescent sensors for cellular Ni2+, Nickelsensor-1 (NS1).222 NS1 is composed of a BODIPY-derived fluorophore conjugated to a mixed N/O/S receptor. This probe displays a 25-fold increase in fluorescence upon binding Ni2+, although it binds with relatively low affinity (Kd = 193 μM). NS1 staining of human lung carcinoma A549 cells incubated in extracellular NiCl2 (1 mM) was able to display an increase in the intracellular Ni2+, suggesting the sensor can detect increases over basal Ni2+ levels in mammalian cells. NS1 remains a platform from which more Ni2+ sensors can be modeled to achieve Ni2+ detection in live cells with higher dynamic range and altered affinities, for application in microorganisms.
8.3. Cobalt (Co2+)
Co2+ is an essential trace element for all organisms; however, it occurs less frequently in nature than other transition metals.213,248 Biological forms of Co2+ are dominantly found in corrinoid metal complexes.249 Corrinoid rings are porphyrin-like prosthetic groups essential for the function of biologically significant metalloenzymes found throughout bacteria and eukarya. Co2+ associated corrinoid metalloenzymes catalyze events like electron transfer, transmethylation, and rearrangement reactions. Most notably among these is cobalamin (vitamin B12). Cobalamin is an essential coenzyme for a wide range of metabolic processes as well as DNA synthesis.250 In animals it is required for red blood cell formation, growth and development, and maintenance of the nervous system.249 However, consistent with all redox-active metal ions, unregulated Co2+ is toxic to cells. Co2+ toxicity can arise from the generation of reactive oxygen species (ROS), but more commonly it is due to competition with other biologically essential metal ions.249,251 In humans, this toxicity is associated with various diseases and conditions including allergic asthma, contact dermatitis, pneumonia, and even cancer.249,251
The underlying mechanisms of how Co2+ is regulated by cells in both healthy and disease states remain poorly understood. Very few chemical probes for fluorescence or colorimetric cobalt detection in cells have been reported to date (Figure 16), in part due to the quenching nature of this paramagnetic ion. In an effort to circumvent this problem, Cobalt probe 1 (CP1)252 was designed using a reaction-based strategy to exploit the redox activity of Co2+. The CP1 probe features a fluorescein dye linked to a polypyridine-based ligand and is only weakly fluorescent in the absence of Co2+. C–O bond cleavage mediated by Co2+ affords an 18-fold increase in fluorescence by releasing the ligand and reconstituting the fully conjugated fluorescein fluorophore. CP1 was used to selectively detect aqueous Co2+ as well as endogenous cellular and Co2+ treated A549 human carcinoma cells. The parameters of this probe are listed in Table 8. Another type of Co2+ sensor is Lin Co-1, a small-molecule FRET sensor derived from coumarin-conjugated porphyrins.224 A series of these sensors were developed so that in the absence of Co2+, the donor excitation energy from coumarin is efficiently transferred to the porphyrin acceptor. Upon Co2+ coordination to the porphyrin component, there is a detectable decrease in FRET efficiency. In addition to this ratiometric fluorescence response, these sensors can be used for colorometric detection of Co2+. The free probe displays a red color; however, upon Co2+ binding, the color dramatically shifts to blue, the emission color of the coumarin component.
9. Probes for Toxic Metals
9.1. Lead (Pb2+)
Pb2+ poses a serious health hazard because it is directly sequestered by cells from the environment and even at very low concentrations (15 ppb) interferes with numerous processes that ultimately result in toxicity.253 The increasing concern about the neurological, cardiovascular, reproductive, and developmental effects of Pb2+ in humans has prompted investigations into the primary molecular targets and mechanisms responsible for Pb2+ toxicity. It has been suggested that Pb2+ is able to enter cells by a variety of mechanisms, including the use of uptake and transport pathways of essential metal ions.253 Pb2+ then interacts with a host of proteins by either mimicking or antagonizing the physiological effects of divalent metals, predominantly Ca2+ and Zn2+. Some of the factors susceptible to Pb2+ include voltage-gated ion channels, ligand-gated ion channels, calmodulin, protein kinase C, calcineurin, Ca2+/calmodulin-dependent protein kinase II, and synaptotagmin.254 However, unraveling more mechanistic detail on the effects of Pb2+ toxicity has been hindered by the lack of reliable tools for monitoring intracellular tracking and quantification.
The probes for selective fluorescence detection of cellular Pb2+ are few (Figure 17). However, a number of fluorescent sensors for aqueous Pb2+ have been developed over recent years. Some of these contain unique platforms consisting of fluorophore and peptide,255 protein and DNA duplex,256 DNAzyme,257 and nanoparticles.258 A review of the sensors used for environmental detection or other non-cell-based applications has been published previously.259
Figure 17.
Molecular sensors for the toxic metals Pb2+, Cd2+, and Hg2+.
One of the early small-molecule-based sensors applied to imaging Pb2+ in live cells, Leadfluor-1 (LF1), combines a Pb2+-selective dicarboxylate pseudocrown ether and a fluorescein-like xanthenone reporter that undergo an 18-fold fluorescence enhancement upon binding.231,260 LF1 forms a 1:1 complex with Pb2+ with micromolar affinity. Even though LF1 is not sensitive enough to track toxicologically relevant concentrations (15 ppb per EPA standards), studies using confocal microscopy revealed that it is capable of monitoring changes in cytosolic Pb2+ within living cells.231,260 Additionally, the commercially available probe Leadmium is available in a cell-permeant form that becomes fluorescent in the presence of nanomolar levels of Pb2+ and micromolar levels of Cd2+. Photophysical properites of molecular Pb2+ probes are given in Table 9.
Table 9. Small-Molecule Toxic Metal Probes.
| excitation |
emission |
brightnessb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sensor | λfree (nm) | εfreea | λbound (nm) | εbounda | λfree (nm) | φfree | λbound (nm) | φbound | free | bound | DRc | KD (M) | ref |
| Lead (Pb2+) | |||||||||||||
| LF1 | 490 | 25.0 | 490 | 28.0 | 514 | 0.001 | 514 | 0.013 | 0.03 | 0.36 | 18 | 2.3 × 10–7 | (231) |
| Leamium | 490 | 520 | |||||||||||
| Cadmium (Cd2+) | |||||||||||||
| Liu Cd-1 | 503 | 80.0 | 503 | 80.0 | 518 | 0.006 | 518 | ND | 0.48 | ND | 2 | 8.1 × 10–6 | (232) |
| Peng Cd-1 | 600 | ND | 571 | ND | 656 | 0.12 | 597 | 0.59 | ND | ND | ND | 5–7 × 10–5 | (233) |
| Cheng Cd-1 | 550 | ND | 550 | ND | 578 | 0.003 | 578 | 0.3 | ND | ND | 195 | 6.0 × 10–7 | (234) |
| Mameli Cd-L | 332 | 37.0 | 332 | ND | 510 | 0.0001 | 510 | ND | ND | ND | 13 | 1.0 × 10–6 | (235) |
| DBITA | 362 | ND | 362 | ND | 493/534 | 0.18 | 493/587 | 0.42 | ND | ND | 3 | 2.5 × 10–13 | (236) |
| DQCd1 | 405 | ND | 405 | ND | 558 | 0.15 | 495 | 0.11 | ND | ND | 15 | 4.1 × 10–13 | (237) |
| CYP-1 | 766 | 158.0 | 766 | 156.0 | 791 | 0.0012 | 791 | 0.035 | 0.19 | 5.46 | ND | 4.4 × 10–7 | (238) |
| CYP-2 | 771 | 152.0 | 771 | 147.0 | 793 | 0.0059 | 793 | 0.015 | 0.90 | 2.21 | ND | 5.0 × 10–6 | (238) |
| Mercury (Hg2+) | |||||||||||||
| 8H-BDP | 470 | ND | 470 | ND | 509 | 0.002 | 509 | ND | ND | ND | 27 | 6.3 × 10–19 | (239) |
| MF1 | 485 | 20.0 | 495 | 49.0 | 514 | 0.001 | 517 | 0.16 | 0.02 | 7.84 | 170 | 7.0 × 10–11 | (240) |
| Hatai Hg-1 | 340 | ND | 340 | ND | 590 | 0.022 | 540 | 0.4 | ND | ND | 19 | 8.0 × 10–7 | (241) |
| Zhao Hg-1 | 530 | ND | 530 | ND | 586 | ND | 586 | ND | ND | ND | 1200 | 4.6 × 10–7 | (242) |
| RG1 | 500 | ND | 500 | ND | 550 | ND | 550 | 0.48 | ND | ND | ND | 2.0 × 10–6 | (243) |
| Lin Hg-1 | 500 | ND | 500 | ND | 561 | ND | 561 | ND | ND | ND | 1000 | 2.5 × 10–5 | (244) |
| RS1 | 500 | ND | 500 | ND | 564 | ND | 564 | ND | ND | ND | 120 | 1.9 × 10–6 | (245) |
| reaction-based Hg2+ probes | λ-unreacted (nm) | ε-unreacted | λ-reacted (nm) | ε-reacted | λ-unreacted (nm) | Φ-unreacted | λ-reacted (nm) | Φ-reacted | unreacted | reacted | DRc | detection limit (M) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RBPH | 530 | ND | 530 | ND | 630 | ND | 580 | 0.61 | ND | ND | ND | 1.9 × 10–7 | (246) |
| RBC1 | 365 | ND | 365 | ND | 468/590 | 0.31/ND | 590 | ND/0.13 | ND | ND | 240 | 2.1 × 10–9 | (247) |
Molar extinction coefficients given as ε/103 M–1 cm–1.
Brightness is defined as the product of the molar extinction coefficient and the quantum yield (ε × φ).
There is no systematic way to present dynamic range (DR), so we encourage readers to refer to the original publications for more details about this value. For intensity-based probes, this number is generally the maximum fold change in fluorescence intensity upon metal ion binding. ND, not determined.
More recently, a genetically encoded FRET-based Pb2+ sensor, Met-lead 1.59, was developed for cellular detection (Table 10).211c Met-lead 1.59 consists of an ECFP with the last 11 amino acids deleted (ECFPΔC11) and cp173Venus FRET pair connected by a Pb2+ binding domain derived from the bacterial metalloregulatory protein PbrR. Live-cell imaging of HEK293 cells transfected with Met-lead 1.59 and exposed to high concentrations (10–50 μM) of Pb2+ showed that this sensor was able to detect cellular increase after a delay time of a few hours.211c The development of new and more sensitive genetically encoded FRET sensors is a promising direction for the cellular detection of Pb2+.
Table 10. Genetically Encoded Toxic Metal Sensors.
| FP |
excitation |
emission |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| name | donor | acceptor | λdonor(nm) | εdonora | λacceptor (nm) | εacceptora | λdonor (nm) | φdonor | λacceptor (nm) | φacceptor | DR (Rmax/Rmin) | KD (M) | ref |
| Met-lead 1.59 | ECFP(ΔC11) | cp173Venus | 439 | 32.5 | ND | ND | 476 | 0.40 | 535 | ND | 1.49 | 69 × 10–9/22 × 10–6 | (211c) |
| Met-lead 2.58 | ECFP(ΔC11) | cp173Venus | 439 | 32.5 | ND | ND | 476 | 0.40 | 535 | ND | 1.46 | 18 × 10–9/20 × 10–6 | (211c) |
| Met-lead 3.62 | ECFP(ΔC11) | cp173Venus | 439 | 32.5 | ND | ND | 476 | 0.40 | 535 | ND | 1.18 | 15 × 10–10/74 × 10–6 | (211c) |
| Cd-FRET-1 | Cerulean | Citrine | 433 | 43.0 | 516 | 0.77 | 475 | 0.62 | 529 | 0.76 | 2 | 1.5 × 10–10 | (261) |
| Cd-FRET-2 | Cerulean | Citrine | 433 | 43.0 | 516 | 0.77 | 475 | 0.62 | 529 | 0.76 | 1.3 | 1.0 × 10–8 | (261) |
| Cd-FRET-3 | Cerulean | Citrine | 433 | 43.0 | 516 | 0.77 | 475 | 0.62 | 529 | 0.76 | 1.2 | 2.4 × 10–9 | (261) |
| Met-cad1.57 | ECFP(ΔC11) | cp173Venus | 439 | 32.5 | ND | ND | 476 | 0.40 | ND | ND | ND | 2.5 × 10–10 | (262) |
| Met-cad1.77 | ECFP(ΔC11) | cp173Venus | 439 | 32.5 | ND | ND | 476 | 0.40 | ND | ND | ND | 2.2 × 10–10 | (262) |
Molar extinction coefficients given as ε/103 M–1 cm–1. ND, not determined.
9.2. Cadmium (Cd2+)
Like Pb2+, Cd2+ is toxic to cells due to interference with Ca2+- and Zn2+-dependent processes. Fluorescent tools for studying Cd2+ toxicity are similarly rare (Figure 17). One of the main challenges for developing Cd2+ probes comes from the very similar binding properties of Cd2+ and Zn2+.263 It remains difficult to develop sensors selective for Cd2+ that do not show interference from physiological Zn2+ levels.264
The first intracellular fluorescent sensor responsive to Cd2+, Liu Cd-1, was a small-molecule dye composed of fluorescein and a thiosemicarbazide. This sensor was applied to HK-2 cells in the presence of micromolar concentrations of Cd2+.232 Around the same time, the BODIPY-based Cd2+ sensor Peng Cd-1 was generated.233 This sensor was the first fluorophore used for imaging Cd2+ in living systems with a selective response to Cd2+ over Zn2+ based on internal charge transfer and shifted emission. It was used to monitor Cd2+ uptake into PC12 and DC cells using both intensity and ratiometric image collection parameters in the presence of micromolar Cd2+.233 Shortly thereafter, an alternative BODIPY-based sensor, Cheng Cd-1, with a higher affinity for Cd2+ was produced and used in HeLa cells.234 Another cell permeable compound based on 8-hydroxyquinoline was applied to imaging Cd2+ in cultured cells, but this sensor (Mameli Cd-1) showed a low dynamic range.235
A ratiometric sensor, DBITA, showed high selectivity and sensitivity (picomolar) to Cd2+.236 Another sensor DQCd1, based on the fluorophore 4-isobutoxy-6-(dimethylamino)-8-ethoxyquinaldine, showed both a high dynamic ratio (15-fold) and high sensitivity (picomolar) to Cd2+.237 Two near-infrared fluorescent sensors (CYP-1 and CYP-2) for Cd2+ detection based on a tricabocyanine platform have also been generated. CYP-1 is functionalized with sulfate groups to increase water solubility and decrease aggregation where CYP-2 is much more cell permeable and was used for detecting Cd2+ in HeLa cells.238 Small-molecule Cd2+ sensors are profiled in Table 9.
The first genetically encoded Cd2+ sensor, Cd-FRET, was actually produced by modifying a Zn2+-FRET sensor. By introducing four cysteine residues on the dimerization interface of the eCFP–eYFP FRET pair of ZinCh-9, the resulting Cd-FRET sensor exhibited high selectivity for Cd2+ over Zn2+.261 More recently, the FRET-based Cd2+ selective indicator Met-cad1.57 was presented for use in imaging human kidney HEK cells.262 The same group that generated the FRET-based Pb2+ sensor Met-lead 1.59 produced Met-cad 1.57, which uses a similar ECFP (ΔC11) and cpVenus FRET pair platform bridged by a metal binding domain, specifically the bacterial Cd2+ binding protein CadR. Met-cad 1.57 and organelle-targeted variants were successfully used to monitor cytosolic entry and subcellular accumulation of Cd2+ into the nucleus of HEK cells treated with micromolar concentrations of Cd2+.262 Looking forward, these types of sensors could be applied to investigate the cellular toxicology of Cd2+. The properties of these genetically encoded Cd2+ sensors are presented in Table 10.
9.3. Mercury (Hg2+)
Hg2+ detection is of great significance because this metal ion is highly toxic, widely distributed in the environment, and bioaccumulates through the food chain. Seminal work in this field by the Czarnik and Lippard laboratories led to the development of numerous turn-off, and later turn-on fluorescent sensors for Hg2+. For a comprehensive review on the development of colorimetric and fluorescent sensors for detecting free environmental Hg2+ that lead to fluorescent detection of Hg2+ in cells, we refer readers to Kim et al.265 More recent work has led to the development of fluorescent Hg2+ indicators based on rhodamine spirolactams (Figure 18). Rhodamine spirolactam sensors are nonfluorescent and colorless in the absence of Hg2+, where Hg2+ exposure and subsequent coordination induces a turn-on response involving spirolactam ring-opening that results in strong fluorescence emission and a pink color. These rhodamine-based chemosensors are appropriate for live-cell applications due to large molar extinction coefficients, high emission quantum yields, and low energy absorption and emission wavelengths (Table 9); consequently, some of these sensors have been applied for imaging Hg2+ in living cells. For example, the rhodamine-based chemosensor for Hg2+ (Zhao Hg-1)242 and RG1, a water-soluble sugar-conjugated rhodamine spirolactam construct, were used to image Hg2+ in HeLa cells.243 In addition, several sensors based on modified versions of rhodamine spirolactams have been used for bioimaging. One type of modification was designed on the basis of the thiophilic nature of Hg2+ and uses a thioamide–alkyne scaffold that substitutes the lactam carbonyl with a thiocarbonyl to generate a thiospirolactam sensor derivative. The rhodamine thiospirolactam sensor “Lin Hg-1” was used to monitor Hg2+ in HeLa cells,244 and a similar sensor, RS1, was applied to rat Schwann cells.245 Another type of modified rhodamine spirolactam sensor (Chen Hg-1) incorporates a thiolactone moiety, and this was the first sensor used to visualize Hg2+ in C. elegans.35c
Figure 18.
Hg2+ sensor built on a spirolactam ring-opening platform.
More recently, a FRET-based fluorescent Hg2+ sensor called RBC1 was presented for use in human melanoma A375 cells.247 RBC1 uses a 7-diethylcoumarin and rhodamine FRET pair. In the absence of Hg2+, the rhodamine acceptor fluorophore is in the same closed nonfluorescent spirolactam form as described above. This closed ring form is supported by the presence of a thiosemicarbazide-protecting group. Interaction with Hg2+ promotes turn-on fluorescence of the rhodamine FRET acceptor and shifts the FRET ratio. This fluorescence response reaction is not through a Hg2+ coordination as described above but rather via Hg2+-mediated conversion of the thiosemicarbazide protection group to oxadiazole, which in turn induces the opening of the rhodamine spirolactam producing the fluorescent form of rhodamine. Using a similar reaction-based technique, the sensor RBPH was used as a chemodosimeter to detect Hg2+ in MCF-7 cells.246
Other types of fluorescent Hg2+ sensors include the BODIPY-derived detector 8H-BDP,239 Mercuryfluor-1 (MF1),240 which combines a fluorescein reporter with a thioether-rich crown receptor, and a histidine-based detector “Hatai Hg-1” that was used to image HeLa cells as well as zebra fish.241 Hatai Hg-1 includes histidine residues appended to a bipodal thiocarbamate scaffold that provide sulfur atoms as donors to increase selectivity for Hg2+. In summary, there are currently sensitive and selective fluorescent detectors for Hg2+ available, most of which can also be used for naked eye detection of water samples, and the field continues to push for improved sensors to combat growing environmental pollution concerns.
10. Outlook
We have come a long way since 1993 when Zinquin was used to measure the first transition metal ion (Zn2+) in living cells. In the past 20 years, we have witnessed an explosion in the number of probes available, an expansion in the range of metals that can be detected, increasing sophistication in the types of measurements that can be performed, and remarkably creative design platforms to ensure specificity (reaction-based probes) and overcome traditional challenges associated with detecting paramagnetic ions. While many existing tools could benefit from improved brightness, higher dynamic range, and increased specificity, the current toolbox has nevertheless provided an unprecedented view of accessible metal pools in live cells and organisms. These tools have been used to quantify accessible metal ion pools, map their location, and monitor dynamics and fluxes of metal ions. Such studies have revealed that metal ion pools are more widespread and dynamic than previously imagined, that such pools can be systematically perturbed in disease states, and that metal ions are intimately connected to canonical signaling pathways, suggesting a rich connection between transition metals and cell physiology.
It is an exciting time to imagine where the field might go from here. Looking forward, it is clear that the ability to perform simultaneous multianalyte imaging to elucidate interactions between different metal pools will be instrumental for dissecting the mechanism of crosstalk between different ions. Such measurements would require an expanded repertoire of probes of different colors, increasing rigor in metal specificity, a greater number of localized probes, and careful attention to ensure no perturbation of the cellular pools. An additional frontier is to expand the toolkit for imaging metal ions in whole organisms, in which the three-dimensional tissue architecture and multiple complex interacting systems is preserved, would enable us to generate a more comprehensive understanding of metal homeostasis at the organismal level. Finally, while there is much to do in the development of probes themselves, dissecting detailed mechanistic questions would benefit immensely from an expanded toolbox for manipulating metal ion fluxes in cells, such as a repertoire of specific metal chelators and photoactive caged metal compounds.
Biographies

Kyle P. Carter was born in Spokane, WA in 1989. He attended Western Washington University where he worked in the laboratory of Professor Greg O’Neil for two years studying masked alkene reactions and their use in natural product synthesis. After graduating with his B.S. in 2011, he entered the biochemistry Ph.D. program at the University of Colorado, Boulder, where he subsequently joined the laboratory of Professor Amy Palmer. His research is focused on using and developing fluorescent sensors to dissect zinc homeostasis and dysregulation in breast cancer.

Alexandra M. Young received her undergraduate education and a Masters degree in chemistry with an organic focus at Illinois State University performing research under the guidance of Professor Timothy D. Lash. She entered the biochemistry Ph.D. program at the University of Colorado, Boulder, in 2010 and joined the laboratory of Professor Amy E. Palmer. Her graduate studies are focused on fluorescent techniques to study Salmonella pathogenesis.

Amy E. Palmer received her B.A. cum laude in 1994 from Dartmouth College where she worked with Karen E. Wetterhahn on the toxicity of chromium compounds. She received her Ph.D. in 2001 in Chemistry from Stanford University, working under the direction of Edward I. Solomon characterizing the structure and function of multicopper oxidases. From 2001–2005, Dr. Palmer was an NIH-postdoctoral fellow in the lab of Roger Y. Tsien at the University of California San Diego. During this time, she developed a family of genetically encoded fluorescent calcium sensors and used them to examine localized calcium signaling. In 2005 she began her independent career in the Department Chemistry and Biochemistry and BioFrontiers Institute at the University of Colorado–Boulder, where she is currently an Associate Professor. Her research focuses on developing fluorescent tools to visualize, measure, and quantify biochemical processes in living cells, including sensors for metal ions to probe metal distribution and dynamics, host–pathogen interactions, and optically integrated microfluidics as tools for discovery in biology.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Biological Inorganic Chemistry, 1st ed.; Bertini I., Gray H. B., Stiefel E. I., Valentine J. S., Eds.; University Science Books: Sausalito, CA, 2007. [Google Scholar]
- Dupont C. L.; Yang S.; Palenik B.; Bourne P. E. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 17822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev V. N.; Zhang Y. Met. Ions Life Sci. 2013, 12, 529. [DOI] [PubMed] [Google Scholar]
- a Cvetkovic A.; Menon A. L.; Thorgersen M. P.; Scott J. W.; Poole F. L. II; Jenney F. E. Jr.; Lancaster W. A.; Praissman J. L.; Shanmukh S.; Vaccaro B. J.; Trauger S. A.; Kalisiak E.; Apon J. V.; Siuzdak G.; Yannone S. M.; Tainer J. A.; Adams M. W. Nature 2010, 466, 779. [DOI] [PubMed] [Google Scholar]; b Yannone S. M.; Hartung S.; Menon A. L.; Adams M. W.; Tainer J. A. Curr. Opin. Biotechnol. 2012, 23, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchiaro G.; Manieri T. M.; Bertuchi F. R.. Metallomics 2013. [DOI] [PubMed]
- a Ammann A. A. J. Mass Spectrom. 2007, 42, 419. [DOI] [PubMed] [Google Scholar]; b Profrock D.; Prange A. Appl. Spectrosc. 2012, 66, 843. [DOI] [PubMed] [Google Scholar]
- Qin Z.; Caruso J. A.; Lai B.; Matusch A.; Becker J. S. Metallomics 2011, 3, 28. [DOI] [PubMed] [Google Scholar]
- Moore K. L.; Lombi E.; Zhao F. J.; Grovenor C. R. Anal. Bioanal. Chem. 2012, 402, 3263. [DOI] [PubMed] [Google Scholar]
- Becker J. S.; Zoriy M.; Matusch A.; Wu B.; Salber D.; Palm C.; Becker J. S. Mass Spectrom. Rev. 2010, 29, 156. [DOI] [PubMed] [Google Scholar]
- a McRae R.; Bagchi P.; Sumalekshmy S.; Fahrni C. J. Chem. Rev. 2009, 109, 4780. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Fahrni C. J. Curr. Opin. Chem. Biol. 2007, 11, 121. [DOI] [PubMed] [Google Scholar]; c Ralle M.; Lutsenko S. BioMetals 2009, 22, 197. [DOI] [PubMed] [Google Scholar]
- Penner-Hahn J. E. Met. Ions Life Sci. 2013, 12, 15. [DOI] [PubMed] [Google Scholar]
- Lichtman J. W.; Fraser S. E. Nat. Neurosci. 2001, 4, 1215. [DOI] [PubMed] [Google Scholar]
- Masters B. R.History of the Optical Microscope in Cell Biology and Medicine; John Wiley & Sons, Ltd.: Chichester, 2008. [Google Scholar]
- a Hell S. W. Science 2007, 316, 1153. [DOI] [PubMed] [Google Scholar]; b Huang B.; Bates M.; Zhuang X. Annu. Rev. Biochem. 2009, 78, 993. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Sengupta P.; Van Engelenburg S.; Lippincott-Schwartz J. Dev. Cell 2012, 23, 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perls M. Virchows Arch. Pathol. Anat. 1867, 39, 42. [Google Scholar]
- Pluth M. D.; Tomat E.; Lippard S. J. Annu. Rev. Biochem. 2011, 80, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Domaille D. W.; Que E. L.; Chang C. J. Nat. Chem. Biol. 2008, 4, 168. [DOI] [PubMed] [Google Scholar]; b Que E. L.; Domaille D. W.; Chang C. J. Chem. Rev. 2008, 108, 1517. [DOI] [PubMed] [Google Scholar]; c Vinkenborg J. L.; Koay M. S.; Merkx M. Curr. Opin. Chem. Biol. 2010, 14, 231. [DOI] [PubMed] [Google Scholar]; d Dean K. M.; Qin Y.; Palmer A. E.. Biochim. Biophys. Acta 2012. [DOI] [PMC free article] [PubMed]; e Hyman L. M.; Franz K. J. Coord. Chem. Rev. 2012, 256, 2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kao J. P. Y. In Methods in Cell Biology: A Practical Guide to the Study of Calcium in Living Cells; Nuccitelli R., Ed.; Academic Press: San Diego, CA, 1994; Vol. 40. [Google Scholar]; b Palmer A. E.; Tsien R. Y. Nat. Protoc. 2006, 1, 1057. [DOI] [PubMed] [Google Scholar]; c Park J. G.; Palmer A. E. Methods Mol. Biol. 2014, 1071, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson P.; McGhee E. J.; Anderson K. I. J. Cell Sci. 2011, 124, 2877. [DOI] [PubMed] [Google Scholar]
- Lichtman J. W.; Conchello J. A. Nat. Methods 2005, 2, 910. [DOI] [PubMed] [Google Scholar]
- a Lakowicz J. R.Principles of Fluorescence Spectroscopy, 2nd ed.; Kluwer Academic Press: New York, 1999. [Google Scholar]; b Murray J. M. J. Microsc. 1998, 191, 128. [DOI] [PubMed] [Google Scholar]; c Stelzer E. H. K. J. Microsc. 1998, 189, 15. [Google Scholar]
- Shaner N. C.; Steinbach P. A.; Tsien R. Y. Nat. Methods 2005, 2, 905. [DOI] [PubMed] [Google Scholar]
- a Tsien R. Y. Annu. Rev. Neurosci. 1989, 12, 227. [DOI] [PubMed] [Google Scholar]; b Roe M. W.; Lemasters J. J.; Herman B. Cell Calcium 1990, 11, 63. [DOI] [PubMed] [Google Scholar]; c Poenie M. Cell Calcium 1990, 11, 85. [DOI] [PubMed] [Google Scholar]
- a Grzelak A.; Rychlik B.; Bartosz G. Free Radical Biol. Med. 2001, 30, 1418. [DOI] [PubMed] [Google Scholar]; b Dixit R.; Cyr R. Plant J. 2003, 36, 280. [DOI] [PubMed] [Google Scholar]; c Hoebe R. A.; Van Oven C. H.; Gadella T. W. Jr.; Dhonukshe P. B.; Van Noorden C. J.; Manders E. M. Nat. Biotechnol. 2007, 25, 249. [DOI] [PubMed] [Google Scholar]
- a Godley B. F.; Shamsi F. A.; Liang F. Q.; Jarrett S. G.; Davies S.; Boulton M. J. Biol. Chem. 2005, 280, 21061. [DOI] [PubMed] [Google Scholar]; b Pattison D. I.; Davies M. J. EXS 2006, 131. [DOI] [PubMed] [Google Scholar]
- Aubin J. E. J. Histochem. Cytochem. 1979, 27, 36. [DOI] [PubMed] [Google Scholar]
- Cheong W.-F.; Prahl S. A.; Welch A. J. IEEE J. Quantum Electron. 1990, 26, 2166. [Google Scholar]
- a Fluorescent Chemosensors for Ion and Molecule Recognition; American Chemical Society: Washington, DC, 1993. [Google Scholar]; b Basabe-Desmonts L.; Reinhoudt D. N.; Crego-Calama M. Chem. Soc. Rev. 2007, 36, 993. [DOI] [PubMed] [Google Scholar]; c de Silva A. P.; Moody T. S.; Wright G. D. Analyst 2009, 134, 2385. [DOI] [PubMed] [Google Scholar]; d Tan S. S.; Kim S. J.; Kool E. T. J. Am. Chem. Soc. 2011, 133, 2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter D. L. J. Chem. Phys. 1953, 21, 836. [Google Scholar]
- a Kemlo J. A.; Shepherd T. M. Chem. Phys. Lett. 1977, 47, 158. [Google Scholar]; b Fabbrizzi L.; Licchelli M.; Pallavicini P.; Sacchi D.; Taglietti A. Analyst 1996, 121, 1763. [Google Scholar]; c Brox D.; Kiel A.; Worner S. J.; Pernpointer M.; Comba P.; Martin B.; Herten D.-P. PLoS One 2013, 8, e58049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V.; Wang S.; Kool E. T. J. Am. Chem. Soc. 2013, 135, 6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva A. P.; Fox D. B.; Moody T. S.; Weir S. M. Trends Biotechnol. 2001, 19, 29. [DOI] [PubMed] [Google Scholar]
- Tsien R. Y. Biochemistry 1980, 19, 2396. [DOI] [PubMed] [Google Scholar]
- Forster T. Ann. Phys. 1948, 437, 55. [Google Scholar]
- a Ko S. K.; Yang Y. K.; Tae J.; Shin I. J. Am. Chem. Soc. 2006, 128, 14150. [DOI] [PubMed] [Google Scholar]; b Yang H.; Zhou Z.; Huang K.; Yu M.; Li F.; Yi T.; Huang C. Org. Lett. 2007, 9, 4729. [DOI] [PubMed] [Google Scholar]; c Chen X.; Nam S.-W.; Jou M. J.; Kim Y.; Kim S.-J.; Park S.; Yoon J. Org. Lett. 2008, 10, 5235. [DOI] [PubMed] [Google Scholar]; d Kumar K. S.; Ramakrishnappa T.; Balakrishna R. G.; Pandurangappa M.. J. Fluoresc. 2013. [DOI] [PubMed]
- Chae M. Y.; Yoon J.; Czarnik A. W. J. Mol. Recognit. 1996, 9, 297. [DOI] [PubMed] [Google Scholar]
- Tsien R. Y. Nature 1981, 290, 527. [DOI] [PubMed] [Google Scholar]
- Tian L.; Yang Y.; Wysocki L. M.; Arnold A. C.; Hu A.; Ravichandran B.; Sternson S. M.; Looger L. L.; Lavis L. D. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara C.; Sagan S. FEBS Lett. 2013, 587, 1693. [DOI] [PubMed] [Google Scholar]
- Bozym R. A.; Thompson R. B.; Stoddard A. K.; Fierke C. A. ACS Chem. Biol. 2006, 1, 103. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Yu L. C. BioEssays 2008, 30, 606. [DOI] [PubMed] [Google Scholar]
- Tsien R. Y. In Calcium as a Cellular Regulator; Carafoli E., Klee C., Eds.; Oxford University Press: New York, 1999. [Google Scholar]
- Harms G. S.; Cognet L.; Lommerse P. H.; Blab G. A.; Schmidt T. Biophys. J. 2001, 80, 2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Baird G. S.; Zacharias D. A.; Tsien R. Y. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 11241. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Nagai T.; Sawano A.; Park E. S.; Miyawaki A. Proc. Natl. Acad. Sci. U.S.a. 2001, 98, 3197. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Tian L.; Hires S. A.; Mao T.; Huber D.; Chiappe M. E.; Chalasani S. H.; Petreanu L.; Akerboom J.; McKinney S. A.; Schreiter E. R.; Bargmann C. I.; Jayaraman V.; Svoboda K.; Looger L. L. Nat. Methods 2009, 6, 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Cromie M. J.; Shi Y.; Latifi T.; Groisman E. A. Cell 2006, 125, 71. [DOI] [PubMed] [Google Scholar]; b Dann C. E. III; Wakeman C. A.; Sieling C. L.; Baker S. C.; Irnov I.; Winkler W. C. Cell 2007, 130, 878. [DOI] [PubMed] [Google Scholar]
- Ramesh A.; Wakeman C. A.; Winkler W. C. J. Mol. Biol. 2011, 407, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tour O.; Adams S. R.; Kerr R. A.; Meijer R. M.; Sejnowski T. J.; Tsien R. W.; Tsien R. Y. Nat. Chem. Biol. 2007, 3, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Keppler A.; Gendreizig S.; Gronemeyer T.; Pick H.; Vogel H.; Johnsson K. Nat. Biotechnol. 2003, 21, 86. [DOI] [PubMed] [Google Scholar]; b Keppler A.; Kindermann M.; Gendreizig S.; Pick H.; Vogel H.; Johnsson K. Methods 2004, 32, 437. [DOI] [PubMed] [Google Scholar]; c Keppler A.; Pick H.; Arrivoli C.; Vogel H.; Johnsson K. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks K. M.; Rosinov M.; Nolan G. P. Chem. Biol. 2004, 11, 347. [DOI] [PubMed] [Google Scholar]
- Tomat E.; Nolan E. M.; Jaworski J.; Lippard S. J. J. Am. Chem. Soc. 2008, 130, 15776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Bannwarth M.; I. R. Correa J.; Sztretye M.; Pouvreau S.; Fellay C.; Aebischer A.; Royer L.; Rios E.; Johnsson K. ACS Chem. Biol. 2009, 4, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ruggiu A. A.; Bannwarth M.; Johnsson K. Org. Biomol. Chem. 2010, 8, 3398. [DOI] [PubMed] [Google Scholar]; c Kamiya M.; Johnsson K. Anal. Chem. 2010, 82, 6472. [DOI] [PubMed] [Google Scholar]
- Srikun D.; Albers A. E.; Nam C. I.; Iavarone A. T.; Chang C. J. J. Am. Chem. Soc. 2010, 132, 4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a McCranor B. J.; Bozym R. A.; Vitolo M. I.; Fierke C. A.; Bambrick L.; Polster B. M.; Fiskum G.; Thompson R. B. J. Bioenerg. Biomembr. 2012, 44, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang D.; Hurst T. K.; Thompson R. B.; Fierke C. A. J. Biomed. Opt. 2011, 16, 087011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Moore E. D.; Becker P. L.; Fogarty K. E.; Williams D. A.; Fay F. S. Cell Calcium 1990, 11, 157. [DOI] [PubMed] [Google Scholar]; b Krezel A.; Maret W. J. Biol. Inorg. Chem. 2006, 11, 1049. [DOI] [PubMed] [Google Scholar]; c Dineley K. E.; Malaiyandi L. M.; Reynolds I. J. Mol. Pharmacol. 2002, 62, 618. [DOI] [PubMed] [Google Scholar]
- Tsien R. Y. Methods Cell Biol. 1989, 30, 127. [DOI] [PubMed] [Google Scholar]
- Di Virgilio F.; Steinberg T. H.; Swanson J. A.; Silverstein S. C. J. Immunol. 1988, 140, 915. [PubMed] [Google Scholar]
- Di Virgilio F.; Steinberg T. H.; Silverstein S. C. Cell Calcium 1990, 11, 57. [DOI] [PubMed] [Google Scholar]
- Homolya L.; Hollo Z.; Germann U. A.; Pastan I.; Gottesman M. M.; Sarkadi B. J. Biol. Chem. 1993, 268, 21493. [PubMed] [Google Scholar]
- Han J.; Burgess K. Chem. Rev. 2010, 110, 2709. [DOI] [PubMed] [Google Scholar]
- Qin Y.; Miranda J. G.; Stoddard C. I.; Dean K. M.; Galati D. F.; Palmer A. E. ACS Chem. Biol. 2013, 8, 2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Qin Y.; Dittmer P. J.; Park J. G.; Jansen K. B.; Palmer A. E. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 7351. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Park J. G.; Qin Y.; Galati D. F.; Palmer A. E. ACS Chem. Biol. 2012, 7, 1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S.; Asanov A. N.; Oldham P. B. Instrum. Sci. Technol. 1998, 26, 375. [Google Scholar]
- Tottey S.; Waldron K. J.; Firbank S. J.; Reale B.; Bessant C.; Sato K.; Cheek T. R.; Gray J.; Banfield M. J.; Dennison C.; Robinson N. J. Nature 2008, 455, 1138. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Nat. Rev. Mol. Cell Biol. 2009, 10, 513. [DOI] [PubMed] [Google Scholar]
- Thompson K.; Dockery P.; Horobin R. W. Biotech. Histochem. 2012, 87, 468. [DOI] [PubMed] [Google Scholar]
- Malgaroli A.; Milani D.; Meldolesi J.; Pozzan T. J. Cell Biol. 1987, 105, 2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Sensi S. L.; Ton-That D.; Weiss J. H.; Rothe A.; Gee K. R. Cell Calcium 2003, 34, 281. [DOI] [PubMed] [Google Scholar]; b Smith R. A.; Hartley R. C.; Murphy M. P. Antioxid. Redox Signaling 2011, 15, 3021. [DOI] [PubMed] [Google Scholar]; c Dodani S. C.; Leary S. C.; Cobine P. A.; Winge D. R.; Chang C. J. J. Am. Chem. Soc. 2011, 133, 8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyan W.; Zhang D. Y.; Lippard S. J.; Radford R. J. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Li D.; Chen S.; Bellomo E. A.; Tarasov A. I.; Kaut C.; Rutter G. A.; Li W. H. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 21063. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Radford R. J.; Chyan W.; Lippard S. J. Chem. Sci. 2013, 4, 3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.; Buccella D.; Lippard S. J. J. Am. Chem. Soc. 2013, 135, 13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. T.; Bagchi P.; Fahrni C. J. J. Am. Chem. Soc. 2011, 133, 15906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulin J. Ann. Sci. Nat., Bot. 1869, 11. [Google Scholar]
- a Miyawaki A.; Llopis J.; Heim R.; McCaffery J. M.; Adams J. A.; Ikura M.; Tsien R. Y. Nature 1997, 388, 882. [DOI] [PubMed] [Google Scholar]; b DiPilato L. M.; Cheng X.; Zhang J. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 16513. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Gallegos L. L.; Kunkel M. T.; Newton A. C. J. Biol. Chem. 2006, 281, 30947. [DOI] [PubMed] [Google Scholar]
- a Palmer A. E.; Jin C.; Reed J. C.; Tsien R. Y. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 17404. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Levine C. G.; Mitra D.; Sharma A.; Smith C. L.; Hegde R. S. Mol. Biol. Cell 2005, 16, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A. E.; Giacomello M.; Kortemme T.; Hires S. A.; Lev-Ram V.; Baker D.; Tsien R. Y. Chem. Biol. 2006, 13, 521. [DOI] [PubMed] [Google Scholar]
- Sasaki K.; Sato M.; Umezawa Y. J. Biol. Chem. 2003, 278, 30945. [DOI] [PubMed] [Google Scholar]
- Griesbeck O.; Baird G. S.; Campbell R. E.; Zacharias D. A.; Tsien R. Y. J. Biol. Chem. 2001, 276, 29188. [DOI] [PubMed] [Google Scholar]
- Kunkel M. T.; Ni Q.; Tsien R. Y.; Zhang J.; Newton A. C. J. Biol. Chem. 2005, 280, 5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim N.; Griesbeck O. J. Biol. Chem. 2004, 279, 14280. [DOI] [PubMed] [Google Scholar]
- Dittmer P. J.; Miranda J. G.; Gorski J. A.; Palmer A. E. J. Biol. Chem. 2009, 284, 16289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Miesenbock G.; Angelis D. A. D.; Rothman J. E. Naure 1998, 394, 192. [DOI] [PubMed] [Google Scholar]; b Vinkenborg J. L.; Nicolson T. J.; Bellomo E. A.; Koay M. S.; Rutter G. A.; Merkx M. Nat. Methods 2009, 6, 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Foss S. M.; Dobryy Y. L.; Park C. K.; Hires S. A.; Shaner N. C.; Tsien R. Y.; Osborne L. C.; Voglmaier S. M. Front. Mol. Neurosci. 2011, 4, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quincke H. Arch. Exp. Pathol. Pharmakol. 1896, 37, 183. [Google Scholar]
- Schmeltzer W. Z. Wiss. Mikrosk. 1933, 50. [Google Scholar]
- Meguro R.; Asano Y.; Odagiri S.; Li C.; Iwatsuki H.; Shoumura K. Arch. Histol. Cytol. 2007, 70, 1. [DOI] [PubMed] [Google Scholar]
- Boyce R.; Herdman W. Philos. Trans. London 1897, 62. [Google Scholar]
- Mendel L. B.; Bradley H. C. Am. J. Physiol. 1905, 14, 313. [Google Scholar]
- Sebruyns M. C. R. Soc. Biol. Paris 1946, 140. [Google Scholar]
- Okamoto K.; Utamura M. Trans. Soc. Pathol. Jpn. 1938, 20. [Google Scholar]
- a Green C. L. Am. J. Pathol. 1955, 31, 545. [PMC free article] [PubMed] [Google Scholar]; b Uzman L. L. Lab. Invest. 1956, 5, 299. [PubMed] [Google Scholar]; c Butt E. M.; Nusbaum R. E.; Gilmour T. C.; Didio S. L. Am. J. Clin. Pathol. 1958, 30, 479. [DOI] [PubMed] [Google Scholar]
- Irons R. D.; Schenk E. A.; Lee J. C. Arch. Pathol. Lab. Med. 1977, 101, 298. [PubMed] [Google Scholar]
- Jain S.; Scheuer P. J.; Archer B.; Newman S. P.; Sherlock S. J. Clin. Pathol. 1978, 31, 784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Okamoto K. Trans. Soc. Pathol. Jpn. 1943, 33. [Google Scholar]; b Okamoto K. Trans. Soc. Pathol. Jpn. 1942, 32. [Google Scholar]
- Maske H. Naturwissenshaften 1955, 42, 424. [Google Scholar]
- a Timm F. Z. Zellforsch. Mikrosk. Anat. Histochem. 1960, 2, 150. [DOI] [PubMed] [Google Scholar]; b Timm F. Dtsch. Z. Gesamte Gerichtl. Med. 1958, 46, 706. [PubMed] [Google Scholar]
- a Danscher G.; Zimmer J. Histochemistry 1978, 55, 27. [DOI] [PubMed] [Google Scholar]; b Danscher G. Histochemistry 1981, 71, 1. [DOI] [PubMed] [Google Scholar]
- Danscher G. Prog. Histochem. Cytochem. 2006, 41. [DOI] [PubMed] [Google Scholar]
- Mahanand D.; Houck J. C. Clin. Chem. 1968, 14, 6. [Google Scholar]
- Frederickson C. J.; Kasarskis E. J.; Ringo D.; Frederickson R. E. J. Neurosci. Methods 1987, 20, 91. [DOI] [PubMed] [Google Scholar]
- Tsien R. Y.; Pozzan T.; Rink T. J. J. Cell Biol. 1982, 94, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grynkiewicz G.; Poenie M.; Tsien R. Y. J. Biol. Chem. 1985, 260, 3440. [PubMed] [Google Scholar]
- Zalewski P. D.; Forbes I. J.; Betts W. H. Biochem. J. 1993, 296, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao W.; Mooney M.; Bird A. J.; Winge D. R.; Eide D. J. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Holm R. H.; Kennepohl P.; Solomon E. I. Chem. Rev. 1996, 96, 2239. [DOI] [PubMed] [Google Scholar]; b Auld D. S. BioMetals 2001, 14, 271. [DOI] [PubMed] [Google Scholar]
- Maret W.; Li Y. Chem. Rev. 2009, 109, 4682. [DOI] [PubMed] [Google Scholar]
- a Outten C. E.; O’Halloran T. V. Science 2001, 292, 2488. [DOI] [PubMed] [Google Scholar]; b Changela A.; Chen K.; Xue Y.; Holschen J.; Outten C. E.; O’Halloran T. V.; Mondragon A. Science 2003, 301, 1383. [DOI] [PubMed] [Google Scholar]; c Thompson R. B. Curr. Opin. Chem. Biol. 2005, 9, 526. [DOI] [PubMed] [Google Scholar]
- Andreini C.; Banci L.; Bertini I.; Rosato A. J. Proteome Res. 2006, 5, 196. [DOI] [PubMed] [Google Scholar]
- a Tomat E.; Lippard S. J. Curr. Opin. Chem. Biol. 2010, 14, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mbatia H. W.; Burdette S. C. Biochemistry 2012, 51, 7212. [DOI] [PubMed] [Google Scholar]
- Lim N. C.; Yao L.; Freake H. C.; Bruckner C. Bioorg. Med. Chem. Lett. 2003, 13, 2251. [DOI] [PubMed] [Google Scholar]
- Walkup G. K.; Burdette S. C.; Lippard S. J.; Tsien R. Y. J. Am. Chem. Soc. 2000, 1222, 5644. [DOI] [PubMed] [Google Scholar]
- Burdette S. C.; Walkup G. K.; Spingler B.; Tsien R. Y.; Lippard S. J. J. Am. Chem. Soc. 2001, 123, 7831. [DOI] [PubMed] [Google Scholar]
- Chang C. J.; Nolan E. M.; Jaworski J.; Burdette S. C.; Sheng M.; Lippard S. J. Chem. Biol. 2004, 11, 203. [DOI] [PubMed] [Google Scholar]
- Burdette S. C.; Frederickson C. J.; Bu W.; Lippard S. J. J. Am. Chem. Soc. 2003, 125, 1778. [DOI] [PubMed] [Google Scholar]
- Nolan E. M.; Burdette S. C.; Harvey J. H.; Hilderbrand S. A.; Lippard S. J. Inorg. Chem. 2004, 43, 2624. [DOI] [PubMed] [Google Scholar]
- Bassik M. C.; Scorrano L.; Oakes S. A.; Pozzan T.; Korsmeyer S. J. EMBO J. 2004, 23, 1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan E. M.; Jaworski J.; Racine M. E.; Sheng M.; Lippard S. J. Inorg. Chem. 2006, 45, 9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith C. R.; Lippard S. J. Inorg. Chem. 2006, 45, 555. [DOI] [PubMed] [Google Scholar]
- Zhang X. A.; Hayes D.; Smith S. J.; Friedle S.; Lippard S. J. J. Am. Chem. Soc. 2008, 130, 15788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan E. M.; Lippard S. J. Inorg. Chem. 2004, 43, 8310. [DOI] [PubMed] [Google Scholar]
- Nolan E. M.; Ryu J. W.; Jaworski J.; Feazell R. P.; Sheng M.; Lippard S. J. J. Am. Chem. Soc. 2006, 128, 15517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan E. M.; Jaworski J.; Okamoto K.; Hayashi Y.; Sheng M.; Lippard S. J. J. Am. Chem. Soc. 2005, 127, 16812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade L. E.; Lippard S. J. Inorg. Chem. 2010, 49, 9535. [DOI] [PubMed] [Google Scholar]
- Gee K. R.; Zhou Z. L.; Ton-That D.; Sensi S. L.; Weiss J. H. Cell Calcium 2002, 31, 245. [DOI] [PubMed] [Google Scholar]
- Hirano T.; Kikuchi K.; Urano Y.; Higuchi T.; Nagano T. J. Am. Chem. Soc. 2000, 122, 12399. [Google Scholar]
- Hirano T.; Kikuchi K.; Urano Y.; Nagano T. J. Am. Chem. Soc. 2002, 124, 6555. [DOI] [PubMed] [Google Scholar]
- Komatsu K.; Kikuchi K.; Kojima H.; Urano Y.; Nagano T. J. Am. Chem. Soc. 2005, 127, 10197. [DOI] [PubMed] [Google Scholar]
- Haugland R. P.Handbook of Fluorescent Probes and Research Products, 9th ed.; Molecular Probes, Inc.: Eugene, OR, 2002. [Google Scholar]
- Koutaka H.; Kosuge J.; Fukasaku N.; Hirano T.; Kikuchi K.; Urano Y.; Kojima H.; Nagano T. Chem. Pharm. Bull. (Tokyo) 2004, 52, 700. [DOI] [PubMed] [Google Scholar]
- Wu Y.; Peng X.; Guo B.; Fan J.; Zhang Z.; Wang J.; Cui A.; Gao Y. Org. Biomol. Chem. 2005, 3, 1387. [DOI] [PubMed] [Google Scholar]
- Wang J.; Xiao Y.; Zhang Z.; Qian X.; Yang Y.; Xu Q. J. Mater. Chem. 2005, 15, 2836. [Google Scholar]
- Tomat E.; Lippard S. J. Inorg. Chem. 2010, 49, 9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P.; Lippard S. J. Inorg. Chem. 2010, 49, 10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H.; Hanaoka K.; Urano Y.; Terai T.; Nagano T. Bioorg. Med. Chem. 2011, 19, 1072. [DOI] [PubMed] [Google Scholar]
- Koide Y.; Urano Y.; Hanaoka K.; Terai T.; Nagano T. ACS Chem. Biol. 2011, 6, 600. [DOI] [PubMed] [Google Scholar]
- a Meeusen J. W.; Tomasiewicz H.; Nowakowski A.; Petering D. H. Inorg. Chem. 2011, 50, 7563. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Nowakowski A. B.; Petering D. H. Inorg. Chem. 2011, 50, 10124. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Meeusen J. W.; Nowakowski A.; Petering D. H. Inorg. Chem. 2012, 51, 3625. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Nowakowski A.; Petering D. Metallomics 2012, 4, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkup G. K.; Burdette S. C.; Lippard S. J.; Tsien R. Y. J. Am. Chem. Soc. 2000, 122, 5644. [DOI] [PubMed] [Google Scholar]
- Chang C. J.; Nolan E. M.; Jaworski J.; Okamoto K.; Hayashi Y.; Sheng M.; Lippard S. J. Inorg. Chem. 2004, 43, 6774. [DOI] [PubMed] [Google Scholar]
- Goldsmith C. R.; Lippard S. J. Inorg. Chem. 2006, 45, 6474. [DOI] [PubMed] [Google Scholar]
- Lim N. C.; Bruckner C. Chem. Commun. (Cambridge, U. K.) 2004, 1094. [DOI] [PubMed] [Google Scholar]
- Tang B.; Huang H.; Xu K.; Tong L.; Yang G.; Liu X.; An L. Chem. Commun. (Cambridge, U. K.) 2006, 3609. [DOI] [PubMed] [Google Scholar]
- a Nicolson T. J.; Bellomo E. A.; Wijesekara N.; Loder M. K.; Baldwin J. M.; Gyulkhandanyan A. V.; Koshkin V.; Tarasov A. I.; Carzaniga R.; Kronenberger K.; Taneja T. K.; da Silva Xavier G.; Libert S.; Froguel P.; Scharfmann R.; Stetsyuk V.; Ravassard P.; Parker H.; Gribble F. M.; Reimann F.; Sladek R.; Hughes S. J.; Johnson P. R.; Masseboeuf M.; Burcelin R.; Baldwin S. A.; Liu M.; Lara-Lemus R.; Arvan P.; Schuit F. C.; Wheeler M. B.; Chimienti F.; Rutter G. A. Diabetes 2009, 58, 2070. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mato S.; Sanchez-Gomez M. V.; Bernal-Chico A.; Matute C. Glia 2013, 61, 750. [DOI] [PubMed] [Google Scholar]; c Kiedrowski L. J. Neurochem. 2012, 121, 438. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Kiedrowski L. J. Neurochem. 2011, 117, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. J.; Lee S. J.; Kim T. Y.; Cho J. H.; Koh J. Y. J. Neurosci. 2008, 28, 3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Qian J.; Noebels J. L. J. Physiol. 2005, 566, 747. [DOI] [PMC free article] [PubMed] [Google Scholar]; b McCormick N.; Velasquez V.; Finney L.; Vogt S.; Kelleher S. L. PLoS One 2010, 5, e11078. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Jeong J.; Walker J. M.; Wang F.; Park J. G.; Palmer A. E.; Giunta C.; Rohrbach M.; Steinmann B.; Eide D. J. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, E3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair S. A.; Sherson S. M.; Jarvis R.; Camakaris J.; Cobbett C. S. New Phytol. 2007, 174, 39. [DOI] [PubMed] [Google Scholar]
- Sarret G.; Harada E.; Choi Y. E.; Isaure M. P.; Geoffroy N.; Fakra S.; Marcus M. A.; Birschwilks M.; Clemens S.; Manceau A. Plant Physiol. 2006, 141, 1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh H. C.; Collier S.; Guthrie J.; Robertson J. D.; Kornfeld K. Cell Metab. 2012, 15, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. M.; Bernhardt M. L.; Kong B. Y.; Ahn R. W.; Vogt S.; Woodruff T. K.; O’Halloran T. V. ACS Chem. Biol. 2011, 6, 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S. K.; Kim P.; Zhang X. A.; Yun S. H.; Moore A.; Lippard S. J.; Medarova Z. Cancer Res. 2010, 70, 6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki M.; Wolford J. L.; O’Halloran T. V. J. Am. Chem. Soc. 2004, 126, 712. [DOI] [PubMed] [Google Scholar]
- Marvin R. G.; Wolford J. L.; Kidd M. J.; Murphy S.; Ward J.; Que E. L.; Mayer M. L.; Penner-Hahn J. E.; Haldar K.; O’Halloran T. V. Chem. Biol. 2012, 19, 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Maruyama S.; Kikuchi K.; Hirano T.; Urano Y.; Nagano T. J. Am. Chem. Soc. 2002, 124, 10650. [DOI] [PubMed] [Google Scholar]; b Chang C. J.; Jaworski J.; Nolan E. M.; Sheng M.; Lippard S. J. Proc. Natl. Acad. Sci. U.S.a. 2004, 101, 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Kiyose K.; Kojima H.; Urano Y.; Nagano T. J. Am. Chem. Soc. 2006, 128, 6548. [DOI] [PubMed] [Google Scholar]
- a Woodroofe C. C.; Lippard S. J. J. Am. Chem. Soc. 2003, 125, 11458. [DOI] [PubMed] [Google Scholar]; b Woodroofe C. C.; Won A. C.; Lippard S. J. Inorg. Chem. 2005, 44, 3112. [DOI] [PubMed] [Google Scholar]
- Miranda J. G.; Weaver A. L.; Qin Y.; Park J. G.; Stoddard C. I.; Lin M. Z.; Palmer A. E. PLoS One 2012, 7, e49371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a van Dongen E. M.; Dekkers L. M.; Spijker K.; Meijer E. W.; Klomp L. W.; Merkx M. J. Am. Chem. Soc. 2006, 128, 10754. [DOI] [PubMed] [Google Scholar]; b van Dongen E. M.; Evers T. H.; Dekkers L. M.; Meijer E. W.; Klomp L. W.; Merkx M. J. Am. Chem. Soc. 2007, 129, 3494. [DOI] [PubMed] [Google Scholar]
- Lam A. J.; St-Pierre F.; Gong Y.; Marshall J. D.; Cranfill P. J.; Baird M. A.; McKeown M. R.; Wiedenmann J.; Davidson M. W.; Schnitzer M. J.; Tsien R. Y.; Lin M. Z. Nat. Methods 2012, 9, 1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenburg L. H.; Hessels A. M.; Ebberink E. H.; Arts R.; Merkx M.. ACS Chem. Biol. 2013. [DOI] [PMC free article] [PubMed]
- Snapp E. L.; Hegde R. S.; Francolini M.; Lombardo F.; Colombo S.; Pedrazzini E.; Borgese N.; Lippincott-Schwartz J. J. Cell Biol. 2003, 163, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juillerat A.; Gronemeyer T.; Keppler A.; Gendreizig S.; Pick H.; Vogel H.; Johnsson K. Chem. Biol. 2003, 10, 313. [DOI] [PubMed] [Google Scholar]
- Hurst T. K.; Wang D.; Thompson R. B.; Fierke C. A. Biochim. Biophys. Acta 2010, 1804, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H. H.; Matveeva E. G.; Stoddard A. K.; Fierke C. A.; Thompson R. B. J. Fluoresc. 2013, 23, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Hosteen O.; Fierke C. A. J. Inorg. Biochem. 2012, 111, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kim B.-E.; Nevitt T.; Thiele D. J. Nat. Chem. Biol. 2008, 4, 176. [DOI] [PubMed] [Google Scholar]; b Lutsenko S. Curr. Opin. Chem. Biol. 2010, 14, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Bertini I.; Cavallaro G.; McGreevy K. S. Coord. Chem. Rev. 2010, 254, 506. [Google Scholar]
- a Arnesano F.; Banci L.; Bertini I.; Ciofi-Baffoni S. Eur. J. Inorg. Chem. 2004, 2004, 1583. [Google Scholar]; b Puig S.; Thiele D. J. Curr. Opin. Chem. Biol. 2002, 6, 171. [DOI] [PubMed] [Google Scholar]
- Fahrni C. J. Curr. Opin. Chem. Biol. 2013, 17, 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Gaggelli E.; Kozlowski H.; Valensin D.; Valensin G. Chem. Rev. 2006, 106, 1995. [DOI] [PubMed] [Google Scholar]; b Mercer J. F. Trends Mol. Med. 2001, 7, 64. [DOI] [PubMed] [Google Scholar]; c Donnelly P. S.; Xiao Z.; Wedd A. G. Curr. Opin. Chem. Biol. 2007, 11, 128. [DOI] [PubMed] [Google Scholar]; d Madsen E.; Gitlin J. D. Annu. Rev. Neurosci. 2007, 30, 317. [DOI] [PubMed] [Google Scholar]
- a Rae T. D.; Schmidt P. J.; Pufahl R. A.; Culotta V. C.; O’Halloran T. V. Science 1999, 284, 805. [DOI] [PubMed] [Google Scholar]; b Wegner S. V.; Sun F.; Hernandez N.; He C. Chem. Commun. 2011, 47, 2571. [DOI] [PubMed] [Google Scholar]
- Yang L.; McRae R.; Henary M. M.; Patel R.; Lai B.; Vogt S.; Fahrni C. J. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.; Miller E. W.; Pralle A.; Isacoff E. Y.; Chang C. J. J. Am. Chem. Soc. 2006, 128, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodani S. C.; Domaille D. W.; Nam C. I.; Miller E. W.; Finney L. A.; Vogt S.; Chang C. J. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domaille D. W.; Zeng L.; Chang C. J. J. Am. Chem. Soc. 2010, 132, 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. S.; Han J. H.; Kim C. W.; Kang M. Y.; Kang D. W.; Cho B. R. Chem. Commun. 2011, 47, 7146. [DOI] [PubMed] [Google Scholar]
- Hirayama T.; Van de Bittner G. C.; Gray L. W.; Lutsenko S.; Chang C. J. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X.; Lin W.; Wan W. Chem. Commun. (Cambridge, U. K.) 2012, 48, 6247. [DOI] [PubMed] [Google Scholar]
- Miller E. W.; Zeng L.; Domaille D. W.; Chang C. J. Nat. Protoc. 2006, 1, 824. [DOI] [PubMed] [Google Scholar]
- Price K. A.; Hickey J. L.; Xiao Z.; Wedd A. G.; James S. A.; Liddell J. R.; Crouch P. J.; White A. R.; Donnelly P. S. Chem. Sci. 2012, 3, 2748. [Google Scholar]
- Cobine P. A. J. Biol. Chem. 2004, 279, 14447. [DOI] [PubMed] [Google Scholar]
- a Zipfel W. R.; Williams R. M.; Webb W. W. Nat. Biotechnol. 2003, 21, 1369. [DOI] [PubMed] [Google Scholar]; b Hoover E. E.; Squier J. A. Nat. Photon. 2013, 7, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner S. V.; Arslan H.; Sunbul M.; Yin J.; He C. J. Am. Chem. Soc. 2010, 132, 2567. [DOI] [PubMed] [Google Scholar]
- Liang J.; Qin M.; Xu R.; Gao X.; Shen Y.; Xu Q.; Cao Y.; Wang W. Chem. Commun. 2012, 48, 3890. [DOI] [PubMed] [Google Scholar]
- Gruppi F.; Liang J.; Bartelle B. B.; Royzen M.; Turnbull D. H.; Canary J. W. Chem. Commun. 2012, 48, 10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Karpus J.; Wegner S. V.; Chen P. R.; He C. J. Am. Chem. Soc. 2013, 135, 3144. [DOI] [PubMed] [Google Scholar]
- Koay M. S.; Janssen B. M. G.; Merkx M. Dalton Trans. 2013, 42, 3230. [DOI] [PubMed] [Google Scholar]
- Epsztejn S.; Glickstein H.; Picard V.; Slotki I. N.; Breuer W.; Beaumont C.; Cabantchik Z. I. Blood 1999, 94, 3593. [PubMed] [Google Scholar]
- Hider R. C.; Kong X. Dalton Trans. 2013, 42, 3220. [DOI] [PubMed] [Google Scholar]
- a Barnham K. J.; Masters C. L.; Bush A. I. Nat. Rev. Drug Discovery 2004, 3, 205. [DOI] [PubMed] [Google Scholar]; b Kell D. B. BMC Med. Genomics 2009, 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. J. FEBS Lett. 1982, 140, 3. [DOI] [PubMed] [Google Scholar]
- Koppenol W. H. Free Radical Biol. Med. 1993, 15, 645. [DOI] [PubMed] [Google Scholar]
- Greenberg G. R.; Wintrobe M. M. J. Biol. Chem. 1946, 165, 397. [PubMed] [Google Scholar]
- Jacobs A. Blood 1977, 50, 433. [PubMed] [Google Scholar]
- Kakhlon O.; Cabantchik Z. I. Free Radical Biol. Med. 2002, 33, 1037. [DOI] [PubMed] [Google Scholar]
- Breuer W.; Epsztejn S.; Millgram P.; Cabantchik I. Z. Am. J. Physiol. 1995, 268, C1354. [DOI] [PubMed] [Google Scholar]
- Petrat F.; Rauen U.; de Groot H. Hepatology 1999, 29, 1171. [DOI] [PubMed] [Google Scholar]
- Hirayama T.; Okuda K.; Nagasawa H. Chem. Sci. 2013, 4, 1250. [Google Scholar]
- Li P.; Fang L.; Zhou H.; Zhang W.; Wang X.; Li N.; Zhong H.; Tang B. Chemistry 2011, 17, 10520. [DOI] [PubMed] [Google Scholar]
- Garcia-Beltran O.; Mena N.; Yanez O.; Caballero J.; Vargas V.; Nunez M. T.; Cassels B. K. Eur. J. Med. Chem. 2013, 67, 60. [DOI] [PubMed] [Google Scholar]
- Au-Yeung H. Y.; Chan J.; Chantarojsiri T.; Chang C. J.. J. Am. Chem. Soc. 2013. [DOI] [PMC free article] [PubMed]
- OuYang H.; Gao Y.; Yuan Y. Tetrahedron Lett. 2013, 54. [Google Scholar]
- Lytton S. D.; Mester B.; Libman J.; Shanzer A.; Cabantchik Z. I. Anal. Biochem. 1992, 205, 326. [DOI] [PubMed] [Google Scholar]
- Fakih S.; Podinovskaia M.; Kong X.; Schaible U. E.; Collins H. L.; Hider R. C. J. Pharm. Sci. 2008, 98, 2212. [DOI] [PubMed] [Google Scholar]
- Noel S.; Guillon L.; Schalk I. J.; Mislin G. L. Org. Lett. 2011, 13, 844. [DOI] [PubMed] [Google Scholar]
- Chen I.; Ting A. Y. Curr. Opin. Biotechnol. 2005, 16, 35. [DOI] [PubMed] [Google Scholar]
- Petrat F.; de Groot H.; Rauen U. Arch. Biochem. Biophys. 2000, 376, 74. [DOI] [PubMed] [Google Scholar]
- Chen J.-L.; Zhuo S.-J.; Wu Y.-Q.; Fang F.; Li L.; Zhu C.-Q. Spectrochim. Acta, Part A 2006, 63, 438. [DOI] [PubMed] [Google Scholar]
- Praveen L.; Reddy M. L. P.; Varma R. L. Tetrahedron Lett. 2010, 51, 6626. [Google Scholar]
- Andreu N.; Zelmer A.; Wiles S. FEMS Microbiol. Rev. 2011, 35, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo S. K.; Sharma D.; Bera R. K.; Crisponi G.; Callan J. F. Chem. Soc. Rev. 2012, 41, 7195. [DOI] [PubMed] [Google Scholar]
- Bar-Ness E.; Hadar Y.; Chen Y.; Shanzer A.; Libman J. Plant Physiol. 1992, 99, 1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer W.; Ermers M. J.; Pootrakul P.; Abramov A.; Hershko C.; Cabantchik Z. I. Blood 2001, 97, 792. [DOI] [PubMed] [Google Scholar]
- Brandel J.; Humbert N.; Elhabiri M.; Schalk I. J.; Mislin G. L.; Albrecht-Gary A. M. Dalton Trans. 2012, 41, 2820. [DOI] [PubMed] [Google Scholar]
- Kurz T.; Terman A.; Gustafsson B.; Brunk U. T. Histochem. Cell Biol. 2008, 129, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Chereddy N. R.; Suman K.; Korrapati P. S.; Thennarasu S.; Mandal A. B. Dyes Pigm. 2012, 95, 606. [Google Scholar]; b Botella H.; Stadthagen G.; Lugo-Villarino G.; de Chastellier C.; Neyrolles O. Trends Microbiol. 2012, 20, 106. [DOI] [PubMed] [Google Scholar]; c Chiu T. Y.; Yang D. M. Toxicol. Sci. 2012, 126, 436. [DOI] [PubMed] [Google Scholar]; d Yang Z.; She M.; Yin B.; Cui J.; Zhang Y.; Sun W.; Li J.; Shi Z. J. Org. Chem. 2012, 77, 1143. [DOI] [PubMed] [Google Scholar]
- Chen W.; Gong W.; Ye Z.; Lin Y.; Ning G. Dalton Trans. 2013, 42, 10093. [DOI] [PubMed] [Google Scholar]
- Taylor S. R. Geochim. Cosmochim. Acta 2002, 28, 1273. [Google Scholar]
- Kehl-Fie T. E.; Skaar E. P. Curr. Opin. Chem. Biol. 2010, 14, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre J. D.; Culotta V. C. J. Biol. Chem. 2012, 287, 13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S.; Bachert C.; Smith D. R.; Linstedt A. D. Mol. Biol. Cell 2010, 21, 1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A. Brain Res. Rev. 2003, 41, 79. [DOI] [PubMed] [Google Scholar]
- Viñas P.; Pardo-Martínez M.; Hernández-Córdoba M. J. Agric. Food Chem. 2000, 48, 5789. [DOI] [PubMed] [Google Scholar]
- Schnell S.; Ratering S.; Jansen K.-H. Environ. Sci. Technol. 1998, 32, 1530. [Google Scholar]
- Motomizu S.; Oshima M.; Kuwabara M.; Obata Y. Analyst 1994, 119, 1787. [Google Scholar]
- a Beyer J.; Wayne F.; Fridovich I. Anal. Biochem. 1988, 170, 512. [DOI] [PubMed] [Google Scholar]; b Serrat F. B. Microchim. Acta 1998, 129, 77. [Google Scholar]
- Dodani S. C.; He Q.; Chang C. J. J. Am. Chem. Soc. 2009, 131, 18020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J.; Canary J. W. Angew. Chem., Int. Ed. 2010, 49, 7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.; Yuan L.; Long L.; Guo C.; Feng J. Adv. Funct. Mater. 2008, 18, 2366. [Google Scholar]
- Dai Z.; Khosla N.; Canary J. W. Supramol. Chem. 2009, 21, 296. [Google Scholar]
- Xu S.; Wang C.; Zhang H.; Sun Q.; Wang Z.; Cui Y. J. Mater. Chem. 2012, 22, 9216. [Google Scholar]
- Mao X.; Su H.; Tian D.; Li H.; Yang R. ACS Appl. Mater. Interfaces 2013, 5, 592. [DOI] [PubMed] [Google Scholar]
- Mulrooney S. B.; Hausinger R. P. FEMS Microbiol. Rev. 2003, 27, 239. [DOI] [PubMed] [Google Scholar]
- a Ragsdale S. W. J. Biol. Chem. 2009, 284, 18571. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Yruela I. Metallomics 2013, 5, 1090. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Rodionov D. A.; Gelfand M. S.; Gladyshev V. N. BMC Genomics 2009, 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q.; Miller E. W.; Wong A. P.; Chang C. J. J. Am. Chem. Soc. 2006, 128, 9316. [DOI] [PubMed] [Google Scholar]
- Liu W.; Xu L.; Sheng R.; Wang P.; Li H.; Wu S. Org. Lett. 2007, 9, 3829. [DOI] [PubMed] [Google Scholar]
- Peng X.; Du J.; Fan J.; Wang J.; Wu Y.; Zhao J.; Sun S.; Xu T. J. Am. Chem. Soc. 2007, 129, 1500. [DOI] [PubMed] [Google Scholar]
- Cheng T.; Xu Y.; Zhang S.; Zhu W.; Qian X.; Duan L. J. Am. Chem. Soc. 2008, 130, 16160. [DOI] [PubMed] [Google Scholar]
- Mameli M.; Aragoni M. C.; Arca M.; Caltagirone C.; Demartin F.; Farruggia G.; De Filippo G.; Devillanova F. A.; Garau A.; Isaia F.; Lippolis V.; Murgia S.; Prodi L.; Pintus A.; Zaccheroni N. Chem.—Eur. J. 2009, 16, 919. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Zhang C.; He W.; Yang Z.; Gao X.; Guo Z. Chem. Commun. 2010, 46, 6138. [DOI] [PubMed] [Google Scholar]
- Xue L.; Li G.; Liu Q.; Wang H.; Liu C.; Ding X.; He S.; Jiang H. Inorg. Chem. 2011, 50, 3680. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Cheng T.; Zhu W.; Xu Y.; Qian X. Org. Lett. 2011, 13, 264. [DOI] [PubMed] [Google Scholar]
- Lu H.; Xiong L.; Liu H.; Yu M.; Shen Z.; Li F.; You X. Org. Biomol. Chem. 2009, 7, 2554. [DOI] [PubMed] [Google Scholar]
- Yoon S.; Albers A. E.; Wong A. P.; Chang C. J. J. Am. Chem. Soc. 2005, 127, 16030. [DOI] [PubMed] [Google Scholar]
- Hatai J.; Pal S.; Jose G. P.; Bandyopadhyay S. Inorg. Chem. 2012, 51, 10129. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Sun Y.; Lv X.; Liu Y.; Chen M.; Guo W. Org. Biomol. Chem. 2010, 8, 4143. [DOI] [PubMed] [Google Scholar]
- Huang W.; Zhou P.; Yan W.; He C.; Xiong L.; Li F.; Duan C. J. Environ. Monit. 2009, 11, 330. [DOI] [PubMed] [Google Scholar]
- a Lin W.; Cao X.; Ding Y.; Yuan L.; Yu Q. Org. Biomol. Chem. 2010, 8, 3618. [DOI] [PubMed] [Google Scholar]; b Lin W.; Cao X.; Ding Y.; Yuan L.; Long L. Chem. Commun. 2010, 46, 3529. [DOI] [PubMed] [Google Scholar]
- Wang H.; Li Y.; Xu S.; Li Y.; Zhou C.; Fei X.; Sun L.; Zhang C.; Li Y.; Yang Q.; Xu X. Org. Biomol. Chem. 2011, 9, 2850. [DOI] [PubMed] [Google Scholar]
- Kumar K. S.; Ramakrishnappa T.; Balakrishna R. G.; Pandurangappa M.. J. Fluoresc. 2013, [Epub ahead of print] [DOI] [PubMed]
- Zhou Y.; Chu K.; Zhen H.; Fang Y.; Yao C. Spectrochim. Acta, Part A 2013, 106, 197. [DOI] [PubMed] [Google Scholar]
- Andreini C.; Bertini I.; Cavallaro G.; Holliday G. L.; Thornton J. M. J. Biol. Inorg. Chem. 2008, 13, 1205. [DOI] [PubMed] [Google Scholar]
- Okamoto S.; Eltis L. D. Metallomics 2011, 3, 963. [DOI] [PubMed] [Google Scholar]
- Raux E.; Schubert H. L.; Warren M. J. Cell. Mol. Life Sci. 2000, 57, 1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M.; Shimizu S. Eur. J. Biochem. 1999, 261, 1. [DOI] [PubMed] [Google Scholar]
- Au-Yeung H. Y.; New E. J.; Chang C. J. Chem. Commun. 2012, 48, 5268. [DOI] [PubMed] [Google Scholar]
- Martinez-Finley E. J.; Chakraborty S.; Fretham S. J. B.; Aschner M. Metallomics 2012, 4, 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Neal A. P.; Guilarte T. R. Mol. Neurobiol. 2010, 42, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Vijverberg H. P.; Oortgiesen M.; Leinders T.; van Kleef R. G. Environ. Health Perspect. 1994, 102, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo S.; Godwin H. A. J. Am. Chem. Soc. 2000, 122, 174. [Google Scholar]
- Chen P.; Greenberg B.; Taghavi S.; Romano C.; van der Lelie D.; He C. Angew. Chem., Int. Ed. 2005, 44, 2715. [DOI] [PubMed] [Google Scholar]
- Liu J.; Lu Y. J. Am. Chem. Soc. 2004, 126, 12298. [DOI] [PubMed] [Google Scholar]
- Wang L.; Jin Y.; Deng J.; Chen G. Analyst 2011, 136, 5169. [DOI] [PubMed] [Google Scholar]
- a Vijverberg H. P. M.; Westerink R. H. S. Toxicol. Sci. 2012, 130, 1. [DOI] [PubMed] [Google Scholar]; b Deibler K.; Basu P. Eur. J. Inorg. Chem. 2012, 2013, 1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E. W.; He Q.; Chang C. J. Nat. Protoc. 2008, 3, 777. [DOI] [PubMed] [Google Scholar]
- Vinkenborg J. L.; van Duijnhoven S. M. J.; Merkx M. Chem. Commun. 2011, 47, 11879. [DOI] [PubMed] [Google Scholar]
- Chiu T.-Y.; Chen P.-H.; Chang C.-L.; Yang D.-M. PLoS One 2013, 8, e65853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Bridges C. C.; Zalups R. K. Toxicol. Appl. Pharmacol. 2005, 204, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Thévenod F. Toxicol. Appl. Pharmacol. 2009, 238, 221. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Li P.; Shi Z.; Tang X.; Chen C.; Liu W. Inorg. Chem. 2012, 51, 9226. [DOI] [PubMed] [Google Scholar]
- Kim H. N.; Ren W. X.; Kim J. S.; Yoon J. Chem. Soc. Rev. 2012, 41, 3210. [DOI] [PubMed] [Google Scholar]