Abstract
Phylogenetic analysis groups mammalian odorant receptors into two broad classes and numerous subfamilies. These subfamilies are proposed to reflect functional organization. Testing this idea requires an assay allowing detailed functional characterization of odorant receptors. Here we show that a variety of Class I and Class II mouse odorant receptors can be functionally expressed in Xenopus laevis oocytes. Receptor constructs included the N-terminal 20 residues of human rhodopsin and were coexpressed with Gαolf and the cystic fibrosis transmembrane regulator to allow electrophysiological measurement of receptor responses. For most mouse odorant receptors tested, these conditions were sufficient for functional expression. Co-expression of accessory proteins was required to allow functional surface expression of some mouse odorant receptors. We used this assay to examine the receptive ranges of all members of the MOR42 subfamily. MOR42-1 responded to dicarboxylic acids, preferring a 10–12 carbon chain length. MOR42-2 responded to monocarboxylic acids (7–10 carbons). MOR42-3 responded to dicarboxylic acids (8–10 carbons) and monocarboxylic acids (10–12 carbons). Thus, the receptive range of each receptor was unique. However, overlap between the individual receptive ranges suggests that the members of this subfamily form one contiguous subfamily receptive range, suggesting that odorant receptor subfamilies do constitute functional units.
Keywords: olfactory receptors, Xenopus oocytes, electrophysiology
Introduction
The mammalian olfactory system can detect and distinguish thousands of diverse chemical structures. This challenging ligand recognition task is accomplished by a vast family of odorant receptors (ORs) (Buck and Axel, 1991). The ORs, rhodopsin-like G-protein coupled receptors (GPCRs) residing on the cilia of olfactory sensory neurons, act through a G-protein (Gαolf) to activate an adenylate cyclase (ACIII). The increase in cAMP activates a cyclic nucleotide gated channel and the resulting Ca++ influx then activates a Ca++-activated Cl− channel (Mombaerts, 2004; Reed, 2004). ORs are identified by several characteristic sequence motifs and constitute the largest gene family in the mammalian genome (Young et al., 2002; Zhang and Firestein, 2002; Godfrey et al., 2004). Sequence analysis groups these receptors into two broad classes, which are further divided into many subfamilies (Zhang and Firestein, 2002; Godfrey et al., 2004). The ORs show remarkable sequence diversity, with the greatest variability in the transmembrane domains, likely accounting for the high diversity in ligand specificity (Buck and Axel, 1991).
While the OR family is large, the number of detectable odorants is far larger. The olfactory system is thought to surmount this problem by using ORs combinatorially, with each odorant being recognized by an array of ORs and each OR recognizing an array of odorants (Malnic et al., 1999). This combinatorial coding is reflected at the glomerular level (Rubin and Katz, 1999). The subfamily organization of ORs suggests functional organization (Godfrey et al., 2004) and several studies show that receptors in the same subfamily can recognize similar chemical structures. Several receptors in the MOR31 and MOR32 subfamilies respond to aliphatic monocarboxylic acids, two members of the MOR42 subfamily respond to nonanedioic acid, two receptors in the MOR171 subfamily (M71 and M72) respond to acetophenone and two receptors in the MOR174 subfamily respond to similar cyclic structures (Bozza et al., 2002; Feinstein et al., 2004; Malnic et al., 1999; Kajiya et al., 2001; Saito et al., 2004). To improve our understanding of the functional organization of ORs, it is important to determine the ligand specificity of more ORs. In particular, it is important to analyze the function of all members of OR subfamilies. However, progress in this area has been slow due to difficulties in functionally expressing ORs in heterologous systems (McClintock and Sammeta, 2000; Lu et al., 2004). Recently, several accessory proteins have been identified that promote functional expression of mammalian ORs in heterologous cells, allowing large-scale screening of odorants and ORs to be contemplated (Saito et al., 2004).
The Xenopus oocyte expression system is particularly useful for detailed analysis of the ligand specificity of receptors and there have been several reports showing that a few ORs from various species can be expressed in oocytes (Speca et al., 1999; Wetzel et al., 1999; Wetzel et al., 2001; Katada et al., 2003). Here we demonstrate that Xenopus oocytes are a generally useful expression system for functional characterization of mammalian ORs. We show that a wide variety of mouse ORs (MORs) can be functionally expressed. Using this assay system, we examine the receptive ranges of all members of an OR subfamily.
Experimental Procedures
Materials
Xenopus laevis frogs were purchased from Nasco (Fort Atkinson, WI). The care and use of X. laevis frogs in this study was approved by the University of Miami Animal Research Committee and meet the guidelines of the National Institutes of Health. RNA transcription kits were from Ambion (Austin, TX). Collagenase B was from Boehringer-Mannheim (Indianapolis, IN). All other compounds and all odorants were from Sigma–Aldrich (St. Louis, MO).
Expression Constructs
We refer to MORs using the nomenclature of Zhang and Firestein (2002). Here we also provide the GI (GenInfo Identifier), the Olfactory Receptor Database designation (Crasto et al., 2002) and common names (if any) for each MOR used in this study. These receptors are: MOR23-1 (18480025; ORL1500), MOR31-4 (18479311; ORL1527), MOR32-11 (18480767; ORL1574), MOR42-1 (18479803; ORL469; S50), MOR42-2 (18481334; ORL1668), MOR42-3 (18481356; ORL463; S6), MOR174-9 (18480203; ORL828; mOR-EG), MOR203-1 (18479747; ORL1138) and MOR258-5 (18480853; ORL432; olfr62, H12). Constructs containing the MORs 23-1, 31-4, 32-11, 42-1, 42-3, 174-9, 203-1 and 258-5, and the mouse accessory proteins RTP1, RTP2 and REEP1, each in the pCI expression vector (Promega), were generated as previously described (Saito et al., 2004). The coding region of MOR42-2 was amplified by PCR from mouse genomic DNA (BD Biosciences/Clontech), subcloned into the pCI vector and confirmed by sequencing. Constructs containing human Gαolf, Gα15, Gβ1, Gγ3 subunits and β2AR each in the pcDNA3.1 vector were purchased from the UMR cDNA Resource Center. The human CFTR clone was kindly provided by Dr. Ian Dickerson (University of Rochester). The rat GIRK subunit clones Kir3.1 and Kir3.4 were kindly provided by Dr. Lily Jan (University of California, San Francisco) and Dr. John Adelman (Vollum Institute), respectively. Unless otherwise noted, all OR constructs contain an N-terminal extension consisting of the N-terminal 20 amino acid residues of human rhodopsin. In preliminary experiments, we tested constructs containing various N-terminal extensions of 5-HT3 serotonin receptor sequence, similar to the extension employed by Wetzel et al. (1999). However, no functional expression was observed.
Preparation of oocytes and cRNA injection
Oocytes were surgically removed from mature Xenopus laevis frogs (Nasco, Fort Atkinson, WI). Follicle cells were removed by treatment with Collagenase B (Boehringer Mannhem, Indianapolis, IN) for 2 hours at room temperature. Stage V oocytes were injected with cRNA in 25 nl of water. Capped cRNA encoding each protein was generated using mMessage mMachine kits (Ambion, Austin, TX). cRNA quantities injected per oocyte: ORs, 25 ng; Gαolf; 10 ng, CFTR, 1 ng; RTP1, 10 ng; RTP2, 10 ng; REEP1, 10 ng. Optimal quantities of each cRNA were determined empirically. For each set of oocytes, the desired set of cRNAs were combined and injected together. Oocytes were incubated at 18°C in Barth’s saline (in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.3 CaNO3, 0.41 CaCl2, 0.82 MgSO4, 15 HEPES, pH 7.6 and 12 µg/ml tetracycline) for 2–4 days prior to electrophysiological recording. IBMX-induced CFTR current amplitudes, which first became apparent 1 day after cRNA injection, increased until reaching a plateau approximately 2 days after cRNA injection (data not shown).
Electrophysiology and Data Analysis
Odorant induced Cl− currents, resulting from cAMP induced activation of co-expressed CFTR, were measured 2–4 days after cRNA injection using two-electrode voltage clamp in an automated parallel electrophysiology system (OpusExpress 6000A, Molecular Devices). Micropipettes were filled with 3M KCl and had resistances of 0.2–2.0 MΩ. The holding potential was −70 mV. Current responses, filtered (4-pole, Bessel, low pass) at 20 Hz (-3db) and sampled at 100 Hz, were captured and stored using OpusXpress 1.1 software (Molecular Devices). Initial analysis was done using Clampfit 9.1 software (Molecular Devices).
Oocytes were perfused with ND96 (in mM: 96 NaCl, 2 KCl, 1 CaCl2, 1 MgCl2, 5 HEPES, pH 7.5). Odorants were stored under argon and high concentration stock solutions (usually 1M) of each odorant were freshly prepared in ethanol or DMSO on the day of each experiment. Each odorant, diluted in ND96, was applied for 15 sec (Uezono et al., 1993). In preliminary experiments, application durations ranging from 15 sec to 10 min were tested and found to yield current responses of similar amplitude, but with the longer applications yielding prolonged responses (data not shown). Thus, all experiments in this study we done with 15 sec odorant applications. IBMX (1 mM) was used to activate the CFTR in a receptor independent manner. This occurs both through inhibition of phosphodiesterase and the consequent increase in cAMP concentration, and through a direct action on the CFTR (Schultz et al., 1999). When IBMX was applied to uninjected oocytes, no current responses were observed (data not shown). Application of forskolin (10 µM) could also activate CFTR through direct stimulation of adenylate cyclase (data not shown). CFTR can be directly activated by a wide variety of structures (Ma et al., 2002). Thus, all compounds (at all concentrations) used in our studies were tested with oocytes expressing Gαolf and CFTR, but no odorant receptors, to guard against false positives. Only eugenol (100 µM) yielded current responses in oocytes expressing Gαolf and CFTR, but no odorant receptors. These responses were small, having current amplitudes that were less that 10% of the response amplitudes of oocytes expressing MOR174-9 (Figure 4B).
Figure 4. Functional expression of a variety of MORs in Xenopus oocytes.
A) Current recordings from oocytes injected with cRNA encoding Gαolf, CFTR and one of a variety of different MORs (or no receptor). Nonanedioic acid (NDA), nonanoic acid (NA) and 2-coumaranone (2-C) are applied at 100 µM. Scale: 0.5 µA, 200 sec. B) Current recordings from oocytes injected with cRNA encoding Gαolf and CFTR, and either MOR174-9 (mOR-EG) or no receptor. Eugenol (EG) and methyl isoeugenol (MIEG) are applied at 100 µM. Scale: 0.25 µA, 200 sec. All applications were 15 sec in duration.
In preliminary experiments, MOR42-3 was found to be able to activate CFTR in the absence of Gαolf, presumably through endogenous Gαs (Wetzel et al., 1999; Katada et al., 2003). However, coexpression of Gαolf significantly increased current amplitudes in response to MOR42-3 activation (3.5-fold, p<0.01). Addition of Gβ1 and Gγ3 did not result in any further changes in current amplitude (data not shown). Thus, we included Gαolf, but not Gβ1 or Gγ3, in all experiments.
To achieve functional expression of some of the MORs (MOR23-1 and MOR258-5), co-expression of one or more accessory proteins (RTP1, RTP2 and REEP1) was required. While in Figure 5 we co-express all three accessory proteins with MOR23-1, and RTP1 and RTP2 with MOR258-5, we have found that co-expression of RTP1 is sufficient to allow functional expression of MOR23-1 (A. Nichols and C.W. Luetje, unpublished experiments).
Figure 5. Accessory proteins are required for functional expression of some ORs.
A) A cryosection of an oocyte injected with cRNA encoding MOR23-1, Gαolf, CFTR, RTP1, RTP2 and REEP1, and labeled with the 4D2 anti-rhodopsin antibody and a Cy3-conjugated secondary antibody, viewed in transmitted light. B) The same section as in panel A, viewed at 570 nm. C) A cryosection of an oocyte injected with cRNA encoding MOR23-1, Gαolf and CFTR, and labeled with the 4D2 anti-rhodopsin antibody and a Cy3-conjugated secondary antibody, viewed in transmitted light. D) The same section as in panel C, viewed at 570 nm. E) A cryosection of an oocyte injected with cRNA encoding Gαolf, CFTR, RTP1, RTP2 and REEP1 (but no OR cRNA), and labeled with the 4D2 anti-rhodopsin antibody and a Cy3-conjugated secondary antibody, viewed in transmitted light. F) The same section as in panel E, viewed at 570 nm. In panels A–F, the cytoplasm is indicated by “c” and the scale bar is 5 µm. G) Upper traces, an oocyte injected with cRNA encoding MOR23-1, Gαolf and CFTR fails to respond to 100µM Octanoic acid (OA). Lower traces, functional expression is achieved by coexpressing the accessory proteins RTP1, RTP2 and REEP1 (RTPs). Scale: 0.5 µA, 200 sec. H) Responses of MOR23-1 injected oocytes to 100µM OA, in the presence and absence of RTPs, are plotted as the ratio of OR response to IBMX response (mean ± SEM, n=7, p<0.001). I) Top traces, an oocyte injected with cRNA encoding MOR258-5, Gαolf and CFTR fails to respond to 2-Coumaranone (2-C). Lower traces, functional expression is achieved by coexpressing the accessory proteins RTP1 and RTP2. Scale: 0.5 µA, 200 sec. J) Responses of MOR258-5 injected oocytes to 100µM 2-C, in the presence and absence of RTPs, are plotted as the ratio of OR response to IBMX response (mean ± SEM, n=5, p<0.05). In panels G and I, all applications were 15 sec in duration.
Dose-response analysis of decanedioic acid activation of MOR42-1 and nonanedioic acid activation of MOR42-3 was performed as follows. For each receptor, data from individual oocytes were fit to a Hill equation (see below), normalized to the fit maximum and then averaged to produce the final curve. The responses of MOR42-1 and MOR42-3 to other dicarboxylic acids were normalized to the maximal response to decanedioic acid or nonanedioic acid, respectively. For each oocyte, the dicarboxylic acid responses were preceded and followed by the normalizing applications of either 100µM decanedioic acid (EC90 for MOR42-1) or 100 µM nonanedioic acid (EC90 for MOR42-3). During the course of these experiments, we found that the order of application did not affect the response profiles observed in Figure 6.
Figure 6. Ligand specificities of the MOR42 subfamily.
A) Oocytes expressing Gαolf and CFTR, and either MOR42-1 (left), MOR42-2 (middle), or MOR42-3 (right) are challenged with 30µM of dicarboxylic acids of varying carbon length. The MOR42-2 expressing oocyte responded to 30 µM octanoic acid (not shown). Scale: 0.25 µA, 300 sec. B) Oocytes expressing Gαolf and CFTR, and either MOR42-1 (left), MOR42-2 (middle), or MOR42-3 (right) are challenged with 30µM of monocarboxylic acids of varying carbon length. The MOR42-1 expressing oocyte responded to 30 µM decanedioic acid and the MOR42-3 expressing oocyte responded to 30 µM nonanedioic acid (not shown). Scale: 0.25 µA, 300 sec. C) Responses of MOR42-1, MOR42-2 and MOR42-3 to dicarboxylic acids, monocarboxylic acids, aldehydes and alcohols of varying carbon chain length. Responses were normalized to the response of the same oocyte to 30µM decanedioic acid (MOR42-1), octanoic acid (MOR42-2) or nonanedioic acid (MOR42-3) and are presented as the mean of 6–8 separate oocytes. Standard errors are provided in Table 1. Unfilled squares on the floor of each graph represent compounds that were tested but yielded no response. These non-functional compounds were tested with oocytes that were expressing MOR42-1, MOR42-2 or MOR42-3, as confirmed by robust responses to 30 µM decanedioic acid, octanoic acid or nonanedioic acid, respectively. In panels A and B, all applications were 15 sec in duration.
Dose-response and statistical analyses were done using Prism 4 (Graphpad, San Diego, CA). Odorant dose-response curves were fit according to the equation: I = Imax/(1+(EC50/X)n) where I represents the current response at a given concentration of odorant, X; Imax is the maximal response; EC50 is the concentration of odorant yielding a half maximal response; n is the apparent Hill coefficient. Statistical significance was assessed using a two-tailed unpaired t-test, or a one-way ANOVA followed by the Dunnett’s post-test, as appropriate.
In addition to the compounds presented in Table 1, the following compounds were tested and found to be non-functional. Compounds (tested at 30µM) that failed to activate MORs 42-1, 42-2 and 42-3 were pentanol, hexanol, heptanol, octanol, nonanol, decanol, pentanal, hexanal, heptanal, octanal, nonanal, decanal, undecanal, pentanoic acid, hexanoic acid, butanedioic acid, pentanedioic acid, hexanedioic acid and heptanedioic acid. Compounds (tested at 100µM) found to be non-functional at MORs 42-1 and 42-3 were heptanoic acid, nonanoic acid, suberoyl chloride, azelaoyl chloride, 1,2 phenylene diacetic acid, 1,3 phenylenediacetic acid, 2,2’ biquinoline-4,4’-dicarboxylic acid and 2,2’ bipyridine-5,5’-dicarboxylic acid.
Table 1.
Agonists for MOR42-1, MOR42-2 and MOR42-3.
| MOR42-1 | MOR42-2 | MOR42-3 | ||
|---|---|---|---|---|
| Octanedioic acid | 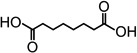 |
0 | 0 | 13 ± 3 |
| Nonanedioic acid | 13 ± 3 | 0 | 100 | |
| Decanedioic acid |  |
100 | 0 | 23 ± 5 |
| Undecanedioic acid | 90 ± 11 | 0 | 1.3 ± 0.9 | |
| Dodecanedioic acid |  |
55 ± 10 | 0 | 0 |
| Heptanoic acid | 0 | 98 ± 5 | 0 | |
| Octanoic acid | 0 | 100 | 0 | |
| Nonanoic acid | 0 | 22 ± 3 | 0 | |
| Decanoic acid | 0 | 8 ± 2 | 7 ± 2 | |
| Undecanoic acid | 0 | 0 | 13 ± 3 | |
| Dodecanoic acid | 0 | 0 | 8 ± 2 | |
| 5-oxononanedioic acid |
0 | nt | 13 ± 4 | |
| 1,4 Phenylene dipropionic acid |
 |
97 ± 26 | nt | 51 ± 5 |
Each compound was tested at 30µM, and responses are expressed as a percent of the response of the same oocyte to 30 µM decanedioic acid (MOR42-1), octanoic acid (MOR42-2) or nonanedioic acid (MOR42-3). 5-oxononanedioic and 1,4 phenylenedipropionic acids were tested at 100µM, and responses are expressed as a percent of the response of the same oocyte to 100µM decanedioic acid (MOR42-1) nonanedioic acid (MOR42-3). Data are the mean ± SEM (n= 6–8). 0, no detectable response; nt, not tested.
Immunocytochemistry
Vitelline membranes were manually removed from oocytes 2–4 days after cRNA injection. The peeled oocytes were incubated in blocking solution (Barth’s saline + 1mg/ml BSA) for 15 min at room temperature. After rinsing, the oocytes were incubated with a 1:30 dilution of hybridoma supernatant containing the anti-rhodopsin antibody 4D2 (Hicks and Molday, 1986) for 15 min. Oocytes were washed and transferred into cryo-embedding medium and frozen overnight. 15 µm sections were cut on a Leica microtome. The sections were thawed and incubated with Cy3-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories) at a 1:400 dilution for 15 min. Sections were rinsed and mounted in 40% glycerol. The sections were visualized on a Leica DMIRB Epifluorescent inverted microscope equipped with a Q-Imaging color CCD camera and imaging software. The 4D2 antibody was kindly provided by Dr. Robert Molday (University of British Columbia).
Results
Functional surface expression of a mammalian odorant receptor in Xenopus oocytes
Xenopus oocytes have been used to express a wide variety of receptors and channels and are highly amenable to electrophysiological analysis. However, functional assay of the G-protein coupled ORs in this system requires installation of a signal transduction pathway that provides an electrophysiological output in response to OR activation. Use of three such pathways has been reported for characterization of ORs from a variety of species. Expression of Gα15 can provide linkage to release of Ca2+ from internal stores and activation of an endogenous Ca2+-activated Cl− channel (Wetzel et al., 2001; Katada et al., 2003). Expression of Gαolf can provide linkage to a co-expressed G-protein regulated inward rectifying K+ (GIRK) channel (Speca et al., 1999). Expression of Gαolf can also provide linkage to a coexpressed Cl− channel, the cystic fibrosis transmembrane regulator (CFTR) (Wetzel et al., 1999; Katada et al., 2003). We used the MOR42-3 receptor (commonly known as S6), which responds to nonanedioic acid (Malnic et al., 1999; Saito et al., 2004; Shirokova et al., 2005), to examine each of these approaches. Our MOR42-3 construct was extended at the N-terminus with the N-terminal 20 amino acid residues of human rhodopsin (Krautwurst et al., 1998; Saito et al., 2004), which serves as both an epitope tag and as an essential determinant of functional expression in oocytes (see below). We obtained functional responses for MOR42-3 using all three transduction systems. However, the Gα15 and Gαolf/GIRK pathways yielded relatively small responses (data not shown) and concerns have been raised regarding the use of Gα15 when characterizing the ligand specificities of ORs (Shirokova et al., 2005). The Gαolf/CFTR pathway yielded robust current responses, leading us to use this transduction system in all subsequent experiments.
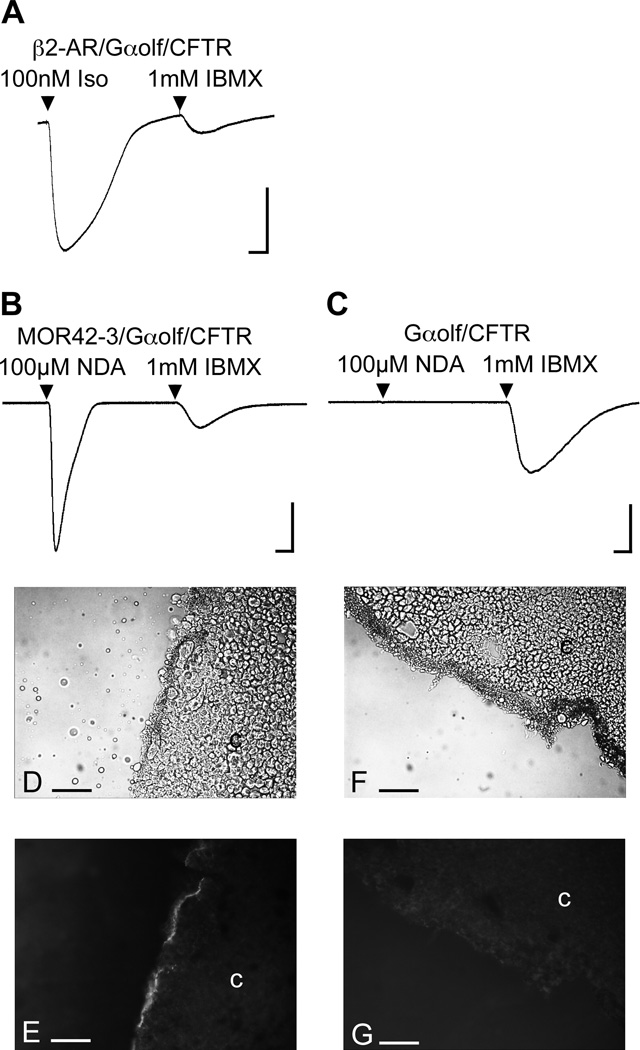
When a GPCR is coexpressed with a GαS–type G-protein and the CFTR in Xenopus oocytes, receptor activation operates through the G-protein to stimulate endogenous adenylate cyclase and the increase in the cAMP activates endogenous protein kinase A. Phosphorylation and ATP binding then activates CFTR, yielding slow Cl− currents lasting several minutes (Uezono et al., 1993). We used the human β2-adrenergic receptor (β2-AR), which can couple to the GαS-like Gαolf, to test this approach (Jones et al., 1990; Uezono et al., 1993). When the β2-AR is coexpressed with human Gαolf and human CFTR, 15 sec application of 100nM isoproterenol yields a large current response (Fig. 1A). Similarly, when the MOR42-3 receptor is coexpressed with Gαolf and CFTR, 15 sec nonanedioic acid (see Table 1 for structure) application yields a large current response (Fig. 1B). In the absence of receptor (no OR cRNA was injected), nonanedioic acid fails to yield a response (Fig. 1C). In each case, the phosphodiesterase inhibitor, IBMX could be used to activate CFTR in a receptor independent manner.
Figure 1. Functional surface expression of MOR42-3 in Xenopus oocytes.
A) A TEVC recording of an oocyte injected with cRNA encoding β2-AR, Gαolf and CFTR responding to 100 nM isoproterenol (Iso) and to 1 mM IBMX. Scale: 0.5 µA, 100 sec. B) A TEVC recording of an oocyte injected with cRNA encoding MOR42-3, Gαolf and CFTR responding to 100µM nonanedioic acid (NDA) and to 1 mM IBMX. Scale: 0.5 µA, 100 sec. C) A TEVC recording of an oocyte injected with cRNA encoding Gαolf and CFTR (but no OR cRNA) failing to respond to 100µM nonanedioic acid, but responding to 1 mM IBMX. Scale: 0.5 µA, 100 sec. D) A cryosection of an oocyte injected with cRNA encoding MOR42-3, Gαolf and CFTR, and labeled with the 4D2 anti-rhodopsin antibody and a Cy3-conjugated secondary antibody, viewed in transmitted light. E) The same section as in panel D, viewed at 570 nm. F) A cryosection of an oocyte injected with cRNA encoding Gαolf and CFTR (but no OR cRNA), and labeled with the 4D2 anti-rhodopsin antibody and a Cy3-conjugated secondary antibody, viewed in transmitted light. G) The same section as in panel E, viewed at 570 nm. In panels A – C, all applications were 15 sec in duration. In panels D – G, the cytoplasm is indicated by “c” and the scale bar is 5 µm.
The rhodopsin-tag allows cell surface localization of ORs to be assessed (Krautwurst et al., 1998; Saito et al., 2004). We used the 4D2 anti-rhodopsin monoclonal antibody (Hicks and Molday, 1986) to demonstrate cell surface localization of MOR42-3. After removal of the vitelline membrane, whole live oocytes were incubated with the 4D2 antibody. Cryosections were then incubated with a Cy-3 conjugated secondary antibody and examined with fluorescent microscopy. Oocytes injected with MOR42-3 cRNA show surface staining (Figure 1E), while oocytes not injected with receptor cRNA do not (Figure 1G). When the 4D2 antibody was excluded from the protocol, oocytes injected with receptor cRNA did not show surface staining (data not shown).
In addition to serving as an epitope tag, the rhodopsin-tag is essential for functional expression of MOR42-3 in oocytes. When the tag is removed from the MOR42-3 construct, functional expression is eliminated (Figure 2), possibly due to a lack of surface expression. The presence of the rhodopsin-tag was also found to be essential for functional expression of MORs 42-1 and 23-1 (data not shown). Thus, all subsequent work was conducted in the presence of the rhodopsin tag.
Figure 2. The rhodopsin tag is essential for functional expression of MOR42-3.
A) TEVC recordings from an oocyte injected with cRNA encoding untagged MOR42-3, Gαolf and CFTR failing to respond to 100µM nonanedioic acid, but responding to 1 mM IBMX. Scale: 0.5 µA, 100 sec. B) TEVC recordings from an oocyte injected with cRNA encoding tagged MOR42-3, Gαolf and CFTR responding to 100µM nonanedioic acid and to 1 mM IBMX. Scale: 0.5 µA, 100 sec. C) Functional responses from multiple oocytes are shown as the ratio of NDA responses to IBMX responses (mean ± SEM, n = 6–11). The NDA responses of oocytes injected with cRNA encoding tagged MOR42-3 are significantly greater than those of oocytes injected with cRNA encoding untagged MOR42-3 or oocytes not injected with receptor cRNA (p<0.05). In panels A and B, all applications were 15 sec in duration.
In Figure 3 we examine the responsiveness of MOR42-3 to nonanedioic acid in more detail. Repeated application of a relatively high concentration of ligand (30 µM) does not result in desensitization (Figure 3A), with the fifth response being 107 ± 3 % of the first response (n = 3). This simplifies the generation of dose-response data. Oocytes expressing MOR42-3 were challenged with a series of nonanedioic acid concentrations ranging from 100nM to 1mM (Figure 3B, C). Fitting the data to a Hill equation (see Experimental Procedures) yielded an EC50 of 5.9 ± 0.9 µM and an apparent Hill coefficient of 0.9 ± 0.2 (n = 8).
Figure 3. Functional responses of MOR42-3 to nonanedioic acid.
A) An oocyte expressing MOR42-3, Gαolf and CFTR is challenged with repeated applications of 30µM nonanedioic acid. Scale: 0.2 µA, 5 min. B) An oocyte expressing MOR42-3, Gαolf and CFTR is challenged with a range of nonanedioic acid concentrations. Scale: 0.2 µA, 10 min. C) Dose-response curve for MOR42-3 responding to nonanedioic acid (mean ± SEM, n = 8). Data is fit to a Hill equation (see Experimental Procedures). In panels A and B, all applications were 15 sec in duration.
Many different odorant receptors can be functionally expressed in Xenopus oocytes
We tested the general utility of the Xenopus oocyte system by attempting to express additional MORs, belonging to both Class I (23-1, 31-4, 32-11) and Class II (174-9, 203-1, 258-5). Each MOR was coexpressed with Gαolf and CFTR, and was challenged with 100 µM of the appropriate ligand. MORs 31-4, 32-11 and 203-1, all previously shown to respond to nonanoic acid (Saito et al., 2004), were each functionally expressed (Figure 4A). However, MOR23-1, previously shown to respond to monocarboxylic acids such as heptanoic, octanoic and nonanoic acid (Saito et al., 2004), failed to functionally express (but see below). Similarly, MOR258-5 (olfr62), previously shown to respond to 2-coumaranone (Saito et al., 2004), also failed to functionally express (but see below).
MOR174-9 (mOR-EG) is activated by eugenol and antagonized by methyl isoeugenol (Kajiya et al., 2001; Oka et al., 2004). These ligand recognition properties of this receptor have been demonstrated in both transfected mammalian cells and mouse olfactory neurons (Oka et al., 2004), making MOR174-9 a useful test of the accuracy of the Xenopus oocyte expression system. In Figure 4B, we find that 100µM eugenol activates this receptor. Methyl isoeugenol (100 µM), while not able to activate the receptor, antagonizes the response to eugenol. The eugenol response in the presence of methyl isoeugenol was 55.7 ± 9.0% of the response to eugenol alone (n=5), a significant reduction (p<0.05) when compared to a second eugenol response in the absence of methyl isoeugenol (105 ± 20% of the first eugenol response, n=3, not shown). Our results are consistent with the reported IC50s for methyl isoeugenol antagonism of the responses to 100µM eugenol of MOR174-9 expressed in transfected cells (66µM) or responses to 300µM eugenol of MOR174-9 expressing mouse olfactory neurons (119µM) (Oka et la., 2004). Unlike the other odorants tested in this study, application of eugenol to oocytes expressing Gαolf and CFTR (but not receptor) did yield small current responses (Figure 4B), perhaps through a direct effect on the CFTR. However, these responses were intermittent and small, with amplitudes less than 10% of the eugenol responses in oocytes expressing receptor.
Accessory proteins are required for functional expression of some odorant receptors in Xenopus oocytes
The failure of MORs 23-1 and 258-5 to functionally express in Xenopus oocytes was reminiscent of the difficulties encountered with ORs in a variety of systems. The failure of many ORs to function in exogenous expression systems appears to be due to a failure to exit the endoplasmic reticulum and thus a failure to appear on the cell surface (McClintock and Sammeta, 2003; Lu, 2004). Recently, Receptor Transporting Proteins 1 and 2 (RTP1 and RTP2) and Receptor Expression Enhancing Protein 1 (REEP1) were identified as accessory proteins that promote functional surface expression of ORs (Saito et al., 2004). In Figure 5, we show that in the absence of the accessory proteins, rhodopsin-tagged MOR23-1 does not appear on the oocyte surface (Figure 5D). However, when RTP1, RTP2 and REEP1 are coexpressed with MOR23-1, cell surface expression of receptor is observed (Figure 5B).
The ability of the accessory proteins to promote cell surface expression of MOR23-1 prompted us to reevaluate the functional expression of MORs 23-1 and 258-5. In Figure 5 (panels G and H), oocytes injected with cRNA encoding MOR23-1, Gαolf and CFTR (but no accessory proteins) fail to respond to octanoic acid. However, when RTP1, RTP2 and REEP1 are coexpressed, large responses to octanoic acid are observed. Similarly, 2-coumaranone activation of MOR258-5 is observed in the presence of RTP1 and RTP2 (Figure 5, I and J). In the absence of the accessory proteins, no responses are observed.
While some ORs can be expressed in oocytes without the accessory proteins (Figure 4) and other ORs require the presence of accessory proteins (Figure 5), the presence of the rhodopsin tag appears to be required in all cases for functional expression in Xenopus oocytes. The accessory proteins were not able to “rescue” functional expression of either MOR 23-1 or 42-3 when the rhodopsin tag was removed (data not shown).
Functional analysis of the MOR42 subfamily
We have demonstrated that the Xenopus oocyte expression system provides a generally applicable method for functional characterization of mammalian odorant receptors (Figures 1–5). With this assay, we analyzed the ligand specificities of the members of an OR subfamily, MOR42. In addition to MOR42-3, the MOR42 subfamily also contains MOR42-1 and MOR42-2 (Zhang and Firestein, 2002; Godfrey et al., 2004). While, MOR42-1 and MOR42-3 are closely related (89% amino acid identity), MOR42-2 shows only 55% and 57% identity with MOR42-1 and MOR42-3, respectively. Similar to MOR42-3, MOR42-1 (also known as S50) has been shown to respond to nonanedioic acid (Malnic et al., 1999; Saito et al., 2004), while MOR42-2 has not been characterized. To investigate the ligand specificities of these receptors in more detail (and to “de-orphanize” MOR42-2), we screened with a series of aliphatic compounds (dicarboxylic acids, monocarboxylic acids, aldehydes and alcohols) of varying carbon chain length, each at 30 µM. Similar to our results with MOR42-3, we found that MOR42-1 and MOR42-2 could be functionally expressed without the accessory proteins. While MOR42-1 did respond to nonanedioic acid, much larger responses could be obtained with longer carbon chain lengths (Figure 6A, C). MOR42-1 was most responsive to decanedioic and undecanedioic acids. MOR42-3 was most responsive to nonanedioic acid and also responded to octanedioic and decanedioic acids, but showed only modest responses to undecanedioic acid (Figure 6A, C). While MOR42-1 did not respond to any of the monocarboxylic acids, MOR42-3 showed small responses to decanoic, undecanoic and dodecanoic acids (Figure 6B, C). MOR42-1 and MOR42-3 did not respond to any of the aldehydes or alcohols (Figure 6C).
In contrast to the robust responses observed for MOR42-1 and MOR42-3, MOR42-2 did not respond to aliphatic dicarboxylic acids with carbon chain lengths ranging from 4 to 12 (each at 30 µM) (Figure 6A, C). However, MOR42-2 did respond to monocarboxylic acids (Figure 6B, C). This receptor was most responsive to heptanoic acid and octanoic acid. Smaller responses were also observed with nonanoic acid and decanoic acid. This receptor did not respond to any of the aldehydes or alcohols (Figure 6C).
Similar to many of the MORs that we have tested (Figure 4), the members of the MOR42 subfamily did not require the accessory proteins (RTP1, RTP2, REEP1) for functional expression in Xenopus oocytes. However, it is possible that while the accessory proteins are not required for expression they might still associate with and alter the function of these receptors. We examined this possibility in detail for MOR42-2. When MOR42-2 expressing oocytes were exposed to monocarboxylic acids with carbon lengths varying from 4 to 10, the ratio of responses did not vary whether cRNAs encoding the accessory proteins were coinjected or not. When oocytes were not coinjected with accessory protein cRNA, the heptanoic, nonanoic and decanoic acid responses were 97.5±5.1%, 21.6±3.1% and 7.9±1.5%, respectively, of the octanoic acid response (mean ± SEM, n=6). When oocytes were coinjected with accessory protein cRNA, the heptanoic, nonanoic and decanoic acid responses were 104.0±5.3%, 25.5±5.7% and 10.8±2.2%, respectively, of the octanoic acid response (mean ± SEM, n=6). Furthermore, MOR42-2 did not respond to 100 µM applications of aliphatic dicarboxylic acids with carbon chain lengths ranging from 2 to 12 whether cRNA for the accessory proteins were coinjected (n = 11, data not shown) or not (n = 14, data not shown). We have also examined the responsiveness of MOR42-1 and MOR42-3 to dicarboxylic acids when cRNA encoding the accessory proteins are coinjected and find no differences from receptor responses in oocytes that have not had the accessory protein cRNA coinjected (data not shown). We conclude that the accessory proteins are either not associating with these receptors, or they are associating but are not affecting ligand specificity in any obvious way.
We continued our analysis by screening the closely related MOR42-1 and MOR42-3 with a variety of additional compounds. 5-oxononanedioic acid, with an additional oxygen at the 5 position was capable of modest activation of MOR42-3 (Table 1). This suggested that these receptors might be able to accommodate additional structure in the center of the ligand. For this reason, we also examined a series of phenylene dicarboxylic acids. While no phenylene diacetic acid configurations were able to activate either receptor, 1,4 phenylene dipropionic acid proved to be good agonist for both MOR42-1 and MOR42-3 (Table 1). In contrast, 1,4 phenylenediacrylic acid was unable to activate either receptor. Additional compounds that failed to activate these receptors are listed in Experimental Procedures.
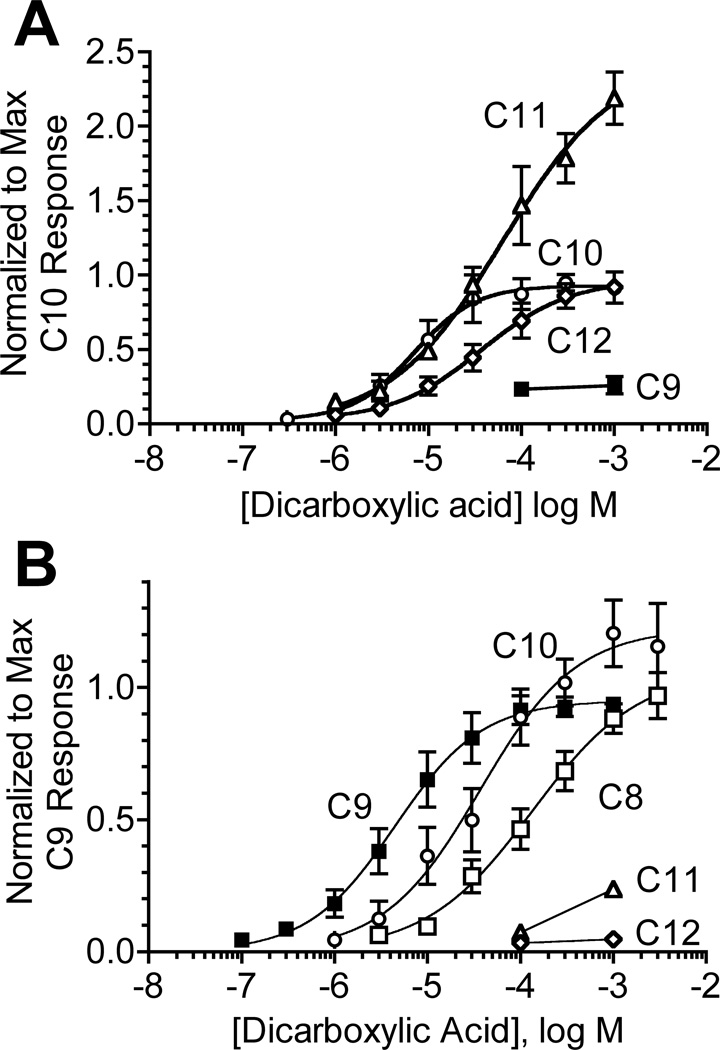
The variation in the responses of the MOR42 receptors to various compounds could be due to differences in efficacy or functional potency. For this reason, we examined the responsiveness of MOR42-1 and 42-3 to a range of dicarboxylic acid concentrations (Figure 7 and Table 2). By normalizing each current response to the maximal response to decanedioic acid (in the case of MOR42-1) or nonanedioic acid (in the case of MOR42-3), we can compare both the functional potencies and relative efficacies of these compounds.
Figure 7. Dose-response analysis of MOR42-1 and MOR42-3.
A) Responses of MOR42-1 to a range of dicarboxylic acid concentrations. The dose-response curve for decanedioic acid (C10) was generated as described in Experimental Procedures. Responses to nonanedioic acid (C9), undecanedioic acid (C11) and dodecanedioic acid (C12) were normalized to the maximal response to decanedioic acid (see Experimental Procedures). Data are the mean ± SEM (n = 3–11). B) Responses of MOR42-3 to a range of dicarboxylic acid concentrations. The dose-response curve for nonanedioic acid (C9) is from Figure 3C. Responses to octanedioic acid (C8), decanedioic acid (C10), undecanedioic acid (C11) and dodecanedioic acid (C12) are normalized to the maximal response to nonanedioic acid (see Experimental Procedures). Data are the mean ± SEM (n = 3–12).
Table 2.
Functional potencies and relative efficacies for activation of MOR42-1 and MOR42-3.
| Receptor | Compound | EC50 (µM) | nH | Relative Efficacy |
|---|---|---|---|---|
| Decanedioic acid | 6.5 ± 1.7 | 1.2 ± 0.3 | 0.93 ± 0.06 | |
| MOR42-1 | Undecanedioic acid | 63 ± 31††† | 0.7 ± 0.2 | 2.82 ± 0.36††† |
| Dodecanedioic acid | 36 ± 16††† | 0.9 ± 0.3 | 1.13 ± 0.13 | |
| Octanedioic acid | 146 ± 28*** | 0.8 ± 0.2 | 1.07 ± 0.11 | |
| MOR42-3 | Nonanedioic acid | 5.9 ± 0.9 | 0.9 ± 0.2 | 0.95 ± 0.05 |
| Decanedioic acid | 47 ± 11*** | 0.8 ± 0.2 | 1.22 ± 0.10** | |
EC50 and Hill coefficient (nH) values were derived by fitting the data in Figure 8 to a Hill equation (see Experimental Procedures). Relative efficacies are the maximum values derived from fitting the data and are expressed as a ratio to the maximal responses to decanedioic acid (for MOR42-1) or nonanedioic acid (for MOR42-3). Values are the mean ± SEM derived from dose-response data from 3–7 separate oocytes. Significant differences from nonanedioic acid:
p<0.001,
p<0.01. Significant differences from decanedioic acid:
p<0.001.
For MOR42-1, decanedioic acid is the most potent agonist, with an EC50 of 6.5 ± 1.7 µM. Undecanedioic acid and dodecanedioic acid were 10-fold and 6-fold less potent, respectively. Dodecanedioic acid displayed an efficacy similar to that of decanedioic acid. In contrast, the relative efficacy of undecanedioic acid was nearly 3-fold greater than that of decanedioic acid. MOR42-1 was only weakly responsive to nonanedioic acid.
For MOR42-3, nonanedioic acid was the most potent agonist, with an EC50 of 5.9 ± 0.9 µM. Octanedioic acid and decanedioic acid were less potent (25-fold and 8-fold, respectively), but displayed similar efficacies relative to nonanedioic acid. MOR42-3 was only weakly responsive to undecanedioic acid and was unresponsive to dodecanedioic acid.
As might be expected, the ability of these receptors to distinguish among odorants varies with odorant concentration. However, the way in which concentration affects the ability to distinguish among the odorants differs between the two receptors (Figure 7). At low concentrations (<10µM), MOR42-1 cannot distinguish between decanedioic and undecanedioic acids, but can distinguish these from nonanedioic and dodecanedioic acids. At high concentrations (>100µM), decanedioic and dodecandioic acids are indistinguishable, while higher efficacy makes undecanedioic acid easily distinguishable from the others. At low concentrations, MOR42-3 can readily distinguish among octanedioic, nonanedioic and decanedioic acids. However, at high concentrations these three odorants are indistinguishable to MOR42-3. This variation in the way that concentration affects the ability of ORs to distinguish among odorants adds to the complexity of odorant coding.
Discussion
To understand how the phylogenetic classification of mammalian ORs relates to functional organization, it is necessary to express these receptors in a system that allows detailed functional analysis. Here we show that Xenopus oocytes provide a generally useful system for expression and functional characterization of mammalian ORs. A variety of Class I and Class II ORs can be expressed and this assay allows detailed analysis of the receptor specificity, functional potency and relative efficacy of odorant ligands acting on mammalian ORs.
Functional expression of mammalian ORs in Xenopus oocytes was first demonstrated by injecting mRNA isolated from rat olfactory epithelium (Dahmen, 1992), but the resulting receptor heterogeneity precluded detailed analysis of receptor specificity. Human OR17–40 and rat I7, expressed in oocytes with the CFTR, were shown to respond to helional and octanal, respectively (Wetzel et al., 1999). However, the receptor responses were small and could only be observed in the presence of IBMX, complicating the analysis due to the ability of IBMX to activate CFTR both directly and indirectly. Eugenol activation of MOR174-9 (mOR-EG), tagged with a Flag/rhodopsin N-terminal extension and co-expressed with CFTR in oocytes has been reported (Katada et al., 2003) and receptor responses were robust, but this group opted to use a HEK293 cell based assay in subsequent studies. The lack of additional reports using oocytes to express mammalian odorant receptors may be due to a failure to use appropriate combinations of components (such as N-terminal extensions, G-proteins, accessory proteins, etc.) to achieve functional expression. Nevertheless, the general utility of Xenopus oocytes for functional expression of a wide variety of receptors prompted our interest in using this system.
The N-terminal extension of rhodopsin sequence (Kaushal et al., 1994; Krautwurst et al., 1998) is an essential factor in achieving functional expression of MORs in Xenopus oocytes, raising the concern that such modification might alter the ligand recognition properties of ORs. While we can’t address this concern directly in oocytes, this issue has been addressed in other studies. N-terminal modification with a variety of sequences (including a bovine rhodopsin sequence) did not alter the specificity of MOR174-9 (mOR-EG) expressed in HEK293T cells (Katada et al., 2004). Also, an N-terminal Flag-tag modified rat I7 receptor, expressed in rat olfactory neurons through adenoviral infection, displayed appropriate ligand sensitivity (Ivic et al., 2002).
Some MORs require co-expression of accessory proteins for functional expression in Xenopus oocytes, while others do not. It is unclear why MORs display this differential dependence on the accessory proteins. A comparison of receptor sequences did not reveal any obvious motifs. It is possible that oocytes express a protein that can substitute for the accessory proteins. It is also unclear whether MORs that do not require exogenous accessory proteins for expression might still be associating with them. This raises the concern that association with an accessory protein might alter the ligand specificity of an OR, leading to results that vary depending on the presence or absence of the various accessory proteins. However, we have not found any differences in the ligand specificity of MORs 42-1, 42-2 and 42-3 in the presence or absence of these proteins (see Results), in agreement with previous work showing that the accessory proteins do not alter the ligand specificity of other ORs (Saito et al., 2004). Thus, the relative functional potencies and relative efficacies that we have obtained using the oocyte expression system should be generally useful, regardless of whether or not the accessory proteins are present.
A common concern for heterologous expression systems is one of accuracy. Specifically, is the ligand specificity of a receptor expressed in a heterologous system an accurate representation of the properties that the receptor displays in vivo? We find that the ligand specificity of MOR174-9 (mOR-EG) expressed in oocytes (activation by eugenol and antagonism by methyl isoeugenol) recapitulates the ligand specificity of this receptor when expressed in olfactory neurons (Oka et al., 2004). We also find that the ligand specificities of MOR42-1 (S50) and MOR42-3 (S6) expressed in oocytes agree well with the properties of these receptors expressed in olfactory neurons (Malnic et al., 1999). A related concern that is particularly important for olfaction is the observation that functionally characterized ORs seem much less sensitive than might be expected, given the extraordinary sensitivity of mammalian olfaction (Mombaerts, 2004). In our work, the most potent ligands for MOR42-1 and MOR42-3 activate these receptors with EC50s in the low micromolar range, and ligand sensitivities in the low to mid micromolar range are common for other ORs, whether expressed in heterologous cells (Kajiya et al., 2001; Saito et al., 2004) or isolated olfactory neurons (Touhara et al., 1999; Bozza et al., 2002; Oka et al., 2004). Thus, this relatively low sensitivity does not appear to be due to the particular cell type in which an OR is assayed. One possible explanation is that while ligands have been identified for some ORs, additional higher potency ligands for these receptors await discovery, while the known ligands may act with higher potency on as yet uncharacterized ORs (Bozza et al., 2002). Also, isolated olfactory neurons may be less sensitive than olfactory neurons residing in intact olfactory epithelium due to an impaired ability to accumulate intracellular Cl− (Kaneko et al., 2004), which would reduce the main signal amplification mechanism within these cells (Lowe and Gold, 1993; Reisert et al., 2005). In addition, components in the mucus environment surrounding olfactory cilia, such as odorant binding proteins, may play a role in increasing sensitivity (Pelosi, 1998). Finally, the sensitivity of mammalian olfaction may be higher than the sensitivity of any individual OR due to the convergence, amplification and noise reduction that results from the circuit properties of the olfactory system (Bozza et al., 2002; Bhandawat et al., 2005). Thus, the mammalian olfactory system may achieve high sensitivity while using relatively low sensitivity odorant receptors.
Using the Xenopus oocyte system, we have examined the receptive range of all members of one MOR subfamily. The closely related MOR42-1 and MOR42-3 have overlapping ligand specificities, but can distinguish among odorants based on small structural features. The general requirements for agonists of these receptors are similar. Two carboxylic acid moieties are preferred and a 10-carbon chain length activates both receptors. 1,4-phenylenedipropionic acid, which proved to be a strong agonist for both MOR42-1 and 42-3, provides a carbon chain length similar to that of decanedioic acid, but with the introduction of a large, rigid central structure. In contrast, 1,4-phenylenediacrylic acid was unable to activate either receptor, suggesting that while a rigid central structure can be accommodated, some flexibility is required near the ends of the ligand. Despite the similarities, these receptors do differ in their ligand specificities, with MOR42-1 preferring carbon chain lengths ≥10 and MOR42-3 preferring carbon chain lengths ≤10.
The MOR42 subfamily also contains MOR42-2, a receptor with no previously known ligands. Because MOR42-2 is in the MOR42 subfamily, we reasoned that a narrowly focused screen of closely related compounds would be the most efficient way to de-orphanize this receptor. MOR42-2 did not respond to a wide range of dicarboxylic acids, but was instead activated by monocarboxylic acids (Figure 6). The receptive range of MOR42-2 overlapped only slightly with the receptive range of MOR42-3 (both responded to decanoic acid). This divergent ligand specificity is consistent with the divergence seen at the amino acid level. While MORs 42-1 and 42-3 display 89% amino acid identity, MOR42-2 shows only 55% and 57% identity with MOR42-1 and MOR42-3, respectively. The ligand specificities of two members of the MOR171 subfamily and two members of the MOR174 subfamily have also been examined in detail. MOR171-2 (M71) and MOR171-3 (M72) are 96% identical at the amino acid level and both respond to acetophenone (Bozza et al., 2002; Feinstein et al., 2004). MOR174-9 (mOR-EG) and MOR174-4 (mOR-EV) are 78% identical and respond to similar cyclic structures Kajiya et al., 2001). While both MORs 174-9 and 174-4 respond to vanillin and ethylvanillin, MOR174-9 also responds to a variety of additional compounds that did not activate MOR174-4. This overlap in ligand specificity appears to be less extensive than what we have observed between MORs 42-1 and 42-3, but is greater than what we have observed when comparing MOR42-2 to other subfamily members. Thus, OR receptive range divergence may correlate with sequence divergence. Much of the sequence divergence among ORs lies within the transmembrane domains where the ligand binding site is likely to be located (Buck and Axel, 1991; Zhang and Firestein, 2002)
The patterns of dicarboxylic acid specificity for MOR42-1 and 42-3 are primarily due to differences in functional potency, a reflection of binding affinity. For example, while 42-1 responds well to undecanedioic and dodecanedioic acids, 42-3 responds poorly to these compounds (Figure 7). Modest responses can be seen when a high concentration of undecanedioic acid (1mM) is applied to 42-3, suggesting weak functional potency. The inability of dodecanedioic acid, at any tested concentration, to either activate or antagonize 42-3 indicates that functional potency is very weak or absent for this compound. These results emphasize that even small structural changes in the ligand can yield large changes in functional potency. In addition, small variations in ligand structure can also affect the relative efficacy of receptor activation. While decanedioic acid and dodecanedioic acid activate MOR42-1 with similar relative efficacies, undecanedioic acid activates this receptor with roughly 3-fold greater relative efficacy (Figure 7A). Thus, while undecanedioic acid may be a full agonist for MOR42-1, decanedioic and dodecandioic acids are partial agonists. For any particular odorant, both functional potency and relative efficacy may contribute to the overall responsiveness of an OR, affecting the contribution of that receptor to odorant perception.
Our results provide insight into the participation of an OR subfamily in the combinatorial coding of odorant recognition. While each receptor in this subfamily recognizes a unique range odorants, these receptive ranges overlap, with some odorants being recognized by two receptors. Thus, the individual receptors appear to be contributing to one contiguous subfamily receptive range. These results support the proposal that OR subfamilies constitute functional units (Zhang and Firestein, 2002; Godfrey et al., 2004). However, it is clear that analysis of additional subfamilies will be required to come to a definitive conclusion on this point. Use of the Xenopus oocyte expression system will allow such characterization and will expand our understanding of the functional relationships among members of the mammalian OR gene family.
Acknowledgements
This work was supported by National Institutes of Health grants MH66038 and DA08102 (to C.W.L.) and DC005782 (to H.M.). We would like to thank Ana Mederos and Floyd Maddox for excellent technical assistance, Dr. Gerhard Dahl for help with immunohistochemical techniques, Sarah Repicky and Dr. Stephen Roper for helpful comments on the manuscript, Dr. Ian Dickerson for the CFTR construct, Dr. Lily Jan for the Kir3.1 construct, Dr. John Adelman for the Kir3.4 construct and Dr. Robert Molday for the 4D2 anti-rhodopsin antibody.
Abbreviations
- CFTR
cystic fibrosis transmembrane regulator
- GIRK
G-protein regulated inward rectifying K+ channel
- GPCR
G-protein coupled receptor
- MOR
mouse odorant receptor
- OR
odorant receptor
- RTP
receptor transporting protein
- REEP
receptor expression enhancing protein
References
- Bhandawat J, Reisert J, Yau KW. Elementary response of olfactory receptor neurons to odorants. Science. 2005;308:1931–1934. doi: 10.1126/science.1109886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J. Neurosci. 2002;22:3033–3043. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Crasto C, Marenco L, Miller PL, Shepherd GS. Olfactory Receptor Database: a metadata-driven automated population from sources of gene and protein sequences. Nucleic Acids Res. 2002;1:354–360. doi: 10.1093/nar/30.1.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmen N, Wang HL, Margolis FL. Expression of olfactory receptors in Xenopus oocytes. J. Neurochem. 1992;58:1176–1179. doi: 10.1111/j.1471-4159.1992.tb09379.x. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Bozz T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the β2 adrenergic receptor. Cell. 2004;117:833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. USA. 2004;101:2156–2161. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks D, Molday RS. Differential immunogold-dextran labeling of bovine and frog rod and cone cells using monoclonal antibodies against bovine rhodopsin. Exp. Eye Res. 1986;42:55–71. doi: 10.1016/0014-4835(86)90017-5. [DOI] [PubMed] [Google Scholar]
- Ivic L, Zhang C, Zhang X, Yoon SO, Firestein S. Intracellular trafficking of a tagged and functional mammalian olfactory receptor. J. Neurobiol. 2002;50:56–68. doi: 10.1002/neu.10016. [DOI] [PubMed] [Google Scholar]
- Jones DT, Masters SB, Bourne HR, Reed RR. Biochemical characterization of three stimulatory GTP-binding proteins. The large and small forms of Gs and the olfactory-specific G-protein, Golf. J. Biol. Chem. 1990;265:2671–2676. [PubMed] [Google Scholar]
- Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K. Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. J. Neurosci. 2001;21:6018–6025. doi: 10.1523/JNEUROSCI.21-16-06018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, Putzier I, Frings S, Kaupp UB, Gensch T. Chloride accumulation in mammalian olfactory sensory neurons. J. Neurosci. 2004;24:7931–7938. doi: 10.1523/JNEUROSCI.2115-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Tanaka M, Touhara K. Structural determinants for membrane trafficking and G protein selectivity of a mouse olfactory receptor. J. Neurochem. 2004;90:1453–1463. doi: 10.1111/j.1471-4159.2004.02619.x. [DOI] [PubMed] [Google Scholar]
- Katada S, Nakagawa T, Kataoka H, Touhara K. Odorant response assays for a heterologously expressed olfactory receptor. Biochem. Biophys. Res. Commun. 2003;305:964–969. doi: 10.1016/s0006-291x(03)00863-5. [DOI] [PubMed] [Google Scholar]
- Kaushal S, Ridge KD, Khorana HG. Structure and function in rhodopsin: The role of asparagine-linked glycosylation. Proc. Natl. Acad. Sci. USA. 1994;91:4024–4028. doi: 10.1073/pnas.91.9.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautwurst D, Yau KW, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- Lowe G, Gold GH. Nonlinear amplification by calcium-dependent chloride channels in olfactory receptor cells. Nature. 1993;366:283–286. doi: 10.1038/366283a0. [DOI] [PubMed] [Google Scholar]
- Lu M, Staszewski L, Echeverri F, Xu H, Moyer BD. Endoplasmic reticulum degradation impedes olfactory G-protein coupled receptor functional expression. BMC Cell Biol. 2004;5:34. doi: 10.1186/1471-2121-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Vetrivel L, Yang H, Pedemonte N, Zegarra-Moran O, Galietta LJ, Verkman AS. High-affinity activators of cystic fibrosis transmembrane conductance regulator (CFTR) chloride conductance identified by high-throughput screening. J. Biol. Chem. 2002;277:37235–37241. doi: 10.1074/jbc.M205932200. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- McClintock TS, Sammeta N. Trafficking prerogatives of olfactory receptors. Neuroreport. 2003;14:1547–1552. doi: 10.1097/00001756-200308260-00001. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Oka Y, Omura M, Kataoka H, Touhara K. Olfactory receptor antagonism between odorants. EMBO J. 2004;23:120–126. doi: 10.1038/sj.emboj.7600032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P. Odorant-binding proteins: structural aspects. Ann. N.Y. Acad. Sci. 1998;855:281–293. doi: 10.1111/j.1749-6632.1998.tb10584.x. [DOI] [PubMed] [Google Scholar]
- Reed RR. After the holy grail: establishing a molecular basis for Mammalian olfaction. Cell. 2004;116:329–336. doi: 10.1016/s0092-8674(04)00047-9. [DOI] [PubMed] [Google Scholar]
- Reisert J, Lai J, Yau K-Y, Bradley J. Mechanism of the excitatory Cl response in mouse olfactory receptor neurons. Neuron. 2005;45:553–561. doi: 10.1016/j.neuron.2005.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BD, Katz LC. Optical imaging of odorant representations in the mammalian olfactory bulb. Neuron. 1999;23:499–511. doi: 10.1016/s0896-6273(00)80803-x. [DOI] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Schultz BD, Singh AK, Devor DC, Bridges RJ. Pharmacology of CFTR chloride channel activity. Physiol. Rev. 1999;79:S109–S144. doi: 10.1152/physrev.1999.79.1.S109. [DOI] [PubMed] [Google Scholar]
- Shirokova E, Schmiedeberg K, Bedner P, Niessen H, Willecke K, Raguse JD, Meyerhof W, Krautwurst D. Identification of specific ligands for orphan olfactory receptors. G protein-dependent agonism and antagonism of odorants. J. Biol. Chem. 2005;280:11807–11815. doi: 10.1074/jbc.M411508200. [DOI] [PubMed] [Google Scholar]
- Speca DJ, Lin DM, Sorensen PW, Isacoff EY, Ngai J, Dittman AH. Functional identification of a goldfish odorant receptor. Neuron. 1999;23:487–498. doi: 10.1016/s0896-6273(00)80802-8. [DOI] [PubMed] [Google Scholar]
- Touhara K, Sengoku S, Inaki K, Tsuboi A, Hirono J, Sato T, Sakano H, Haga T. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proc Natl Acad Sci USA. 1999;96:4040–4045. doi: 10.1073/pnas.96.7.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezono Y, Bradley J, Min C, McCarty NA, Quick M, Riordan JR, Chavkin C, Zinn K, Lester HA, Davidson N. Receptors that couple to 2 classes of G proteins increase cAMP and activate CFTR expressed in Xenopus oocytes. Receptors Channels. 1993;1:233–241. [PubMed] [Google Scholar]
- Wetzel CH, Oles M, Wellerdieck C, Kuczkowiak M, Gisselmann G, Hatt H. Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and Xenopus Laevis oocytes. J. Neurosci. 1999;19:7426–7433. doi: 10.1523/JNEUROSCI.19-17-07426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel CH, Behrendt HJ, Gisselmann G, Stortkuhl KF, Hovemann B, Hatt H. Functional expression and characterization of a Drosophila odorant receptor in a heterologous cell system. Proc. Natl. Acad. Sci. USA. 2001;98:9377–9380. doi: 10.1073/pnas.151103998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Friedman C, Williams EM, Ross JA, Tonnes-Priddy L, Trask BJ. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum Mol Genet. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]